Abstract
In this investigation, we report identification and characterization of a 95 kDa postsynaptic density protein (PSD-95)/discs-large/ZO-1 (PDZ) domain-containing protein termed tamalin, also recently named GRP1-associated scaffold protein (GRASP), that interacts with group 1 metabotropic glutamate receptors (mGluRs). The yeast two-hybrid system and in vitro pull-down assays indicated that the PDZ domain-containing, amino-terminal half of tamalin directly binds to the class I PDZ-binding motif of group 1 mGluRs. The C-terminal half of tamalin also bound to cytohesins, the members of guanine nucleotide exchange factors (GEFs) specific for the ADP-ribosylation factor (ARF) family of small GTP-binding proteins. Tamalin mRNA is expressed predominantly in the telencephalic region and highly overlaps with the expression of group 1 mGluR mRNAs. Both tamalin and cytohesin-2 were enriched and codistributed with mGluR1a in postsynaptic membrane fractions. Importantly, recombinant and native mGluR1a/tamalin/cytohesin-2 complexes were coimmunoprecipitated from transfected COS-7 cells and rat brain tissue, respectively. Transfection of tamalin and mutant tamalin lacking a cytohesin-binding domain caused an increase and decrease in cell-surface expression of mGluR1a in COS-7 cells, respectively. Furthermore, adenovirus-mediated expression of tamalin and dominant-negative tamalin facilitated and reduced the neuritic distribution of endogenous mGluR5 in cultured hippocampal neurons, respectively. The results indicate that tamalin plays a key role in the association of group 1 mGluRs with the ARF-specific GEF proteins and contributes to intracellular trafficking and the macromolecular organization of group 1 mGluRs at synapses.
Keywords: yeast two-hybrid system, protein interaction, PDZ domain, metabotropic glutamate receptor, tamalin, cytohesin, guanine nucleotide exchange factor (GEF), ADP-ribosylation factor (ARF), cell-surface expression, receptor trafficking
Glutamate is a major excitatory neurotransmitter in the mammalian CNS and plays a key role in neuronal plasticity, neural cell development, and neurotoxicity (Choi and Rothman, 1990; Bliss and Collingridge, 1993; Nakanishi and Masu, 1994). Glutamate neurotransmitter exerts physiological actions through two distinct types of receptors, namely, ionotropic and metabotropic glutamate receptors (mGluRs) (Nakanishi, 1992). The ionotropic receptors are glutamate-gated ion channels, whereas the mGluRs belong to G-protein-coupled receptors and exert their effects on intracellular second messengers and ion channels through G-proteins. Eight different subtypes of mGluRs have been identified so far that can be subdivided into three groups according to sequence similarities, intracellular signaling mechanisms, and pharmacological profiles (Nakanishi, 1992;Pin and Duvoisin, 1995). Group 1 receptors (mGluR1 and mGluR5) activate phospholipase C and couple to inositol 1,4,5-triphosphate/Ca2+ signal transduction (Abe et al., 1992; Aramori and Nakanishi, 1992). Group 2 (mGluR2 and mGluR3) and group 3 (mGluR4, mGluR6, mGluR7, and mGluR8) receptors show different pharmacological profiles but are both linked to the inhibitory cAMP cascade (Tanabe et al., 1992; Pin and Duvoisin, 1995).
Recent studies have indicated that the 95 kDa postsynaptic density protein (PSD-95)/discs-large/ZO-1 (PDZ) domain-containing proteins interact with the C-terminal motifs of glutamate receptors and play an important role in receptor trafficking and clustering as well as coupling of receptors to signaling proteins (Craven and Bredt, 1998;Garner et al., 2000; Sheng and Sala, 2001). For example, PSD-95 interacts with NR2 subunits of NMDA receptors (Kornau et al., 1995;Niethammer et al., 1996), whereas glutamate receptor-interacting protein (GRIP), AMPA receptor binding protein (ABP), and the protein interacting with C-kinase (PICK1) bind to AMPA receptor subunits (Dong et al., 1997; Srivastava et al., 1998; Dev et al., 1999; Xia et al., 1999). In addition, PICK1 has been shown to interact with the PDZ-binding motif of mGluR7a (Boudin et al., 2000; Dev et al., 2000;El Far et al., 2000). Group 1 and group 2 mGluRs possess a typical PDZ-binding motif (STL or SSL) at their C-terminal sequences. Individual receptor subtypes of the mGluR family are localized distinctly at axonal terminals, dendritic processes, or somatodendritic regions (Shigemoto and Mizuno, 2000). However, targeting proteins involved in mGluR subtypes still remain to be clarified.
This investigation concerns identification of a PDZ domain-containing protein that interacts with the PDZ-binding motif of the mGluR family. We here report that a PDZ domain-containing protein, termed tamalin, interacts with group 1 mGluRs and forms a protein complex comprising cytohesins, the members of guanine nucleotide exchange factors (GEFs) for the ADP-ribosylation factor (ARF) family of small GTP-binding proteins. Furthermore, we show that tamalin causes an increase in not only cell-surface expression of mGluR1a in transfected cells but also in neuritic distribution of endogenous mGluR5 in cultured hippocampal neurons.
MATERIALS AND METHODS
Yeast two-hybrid system. The C-terminal regions of mGluRs (ct-mGluRs) and the GABAB2 subunit of GABA receptors were amplified by PCR from plasmids encoding the full-length rat receptors or by reverse transcriptase (RT)-mediated PCR from rat brain total RNA. The amplified fragments were subcloned in frame into a bait plasmid, pAS2–1 (Clontech, Palo Alto, CA). The appropriate integrity of inserts was verified by DNA sequencing. In yeast two-hybrid screening, an adult rat brain cDNA library fused to the GAL4 activation domain (MATCHMAKER GAL4 cDNA library; Clontech) was cotransformed into yeast Y190 (Clontech) with the bait plasmid containing the ct-mGluR2 sequence. Colony selection and β-galactosidase reporter gene assay were performed as described previously (Dev et al., 1999). Yeast two-hybrid screening of tamalin-binding proteins from a rat brain cDNA library was conducted with use of the bait plasmid containing the full-length rat tamalin sequence.
cDNA cloning and sequence analysis. Four independent clones encoding the full-length tamalin were isolated from 5 × 105 phage clones of a rat forebrain cDNA library by hybridization with the PstI fragment of the tamalin cDNA. Two mouse expressed sequence tag (EST) clones encoding mouse tamalin, MNCb-4428 (accession number AB041603) and MNCb-4442 (accession number AU079540), were obtained from the Japanese National Institute of Infectious Disease (Tokyo, Japan). A mouse cDNA clone in GenBank database (accession number AF192525) encoded cytohesin-binding protein (CBP) but lacked the amino-terminal portion of this protein. Appropriate primers for RT-PCR were designed from the GenBank EST database and used for cloning of the deleted amino-terminal portion of CBP from mouse brain RNA. Both strands of the cDNA clones were determined by the chain-termination method with BigDye Terminator Cycle Sequencing (Applied Biosystems, Foster City, CA). The full-length rat tamalin nucleotide sequence and the supplemented 5′-nucleotide sequence of mouse CBP were deposited in the GenBank database as accession numbers AF374272 and AF374273, respectively. Sequence database analysis was performed with the BLAST program (National Center for Biotechnology Information, National Institutes of Health, Bethesda, MD) and the ISREC ProfileScan server (http://www.isrec.isb-sib.ch/profile). Sequence alignment was conducted with the ClustalW algorithm (http://www.clustalw.genome.ad.jp/).
Northern blotting. Multiple Tissue Northern blots (2 μg poly(A+) RNA per lane; Clontech) were probed with 32P-labeled cDNA fragments under high-stringency conditions using ExpressHyb Hybridization Solution (Clontech); the cDNA fragments used corresponded to amino acid residues 10–200 of mouse tamalin and residues 36–236 of mouse CBP. The blots were exposed on x-ray films at −80°C with intensifying screens overnight.
In situ hybridization. In situ hybridization was performed as described previously (Watanabe et al., 1998). Briefly,35S-labeled antisense and sense riboprobes corresponding to amino acid residues 10–200 of mouse tamalin were hybridized with cryostat sections (10 μm) of adult mouse brain. After washing and RNase A treatment, sections were exposed to Hyperfilm βmax (Amersham Biosciences, Buckinghamshire, UK) for 1 week, dipped into photographic emulsion NTB-3 (Eastman Kodak, Rochester, NY), and exposed for 10 d.
Glutathione S-transferase pull-down assay. Fusion constructs were generated by inserting appropriate restriction fragments isolated from cloned cDNAs or PCR products in frame into pGEX-4T-1, pGEX-4T-2 (Amersham Biosciences), pMALc2X (New England Biolabs, Beverly, MA), or pET thioredoxin (Trx) fusion system vector 32a(+) (Novagen, Darmstadt, Germany). GlutathioneS-transferase (GST) fusion proteins, maltose-binding protein (MAL) fusion proteins, and hexahistidine-tagged Trx (His-Trx) fusion proteins were expressed in Escherichia coli and purified by glutathione-Sepharose 4B beads (Amersham Biosciences), Amylose Resin (New England Biolabs), and Ni-NTA Agarose (Qiagen, Hilden, Germany), respectively. The purified proteins were dialyzed against PBS (137 mm NaCl, 2.68 mm KCl, 8.10 mmNa2HPO4, 1.47 mmKH2PO4) at 4°C. GST fusion proteins were immobilized on glutathione-Sepharose 4B beads and incubated with either MAL fusion proteins or His-Trx fusion proteins in the solution (500 μl) containing PBS and 1% Triton X-100 at 4°C for 2 hr; 1 μg of each fusion protein was used. The beads were washed four times with the incubation buffer. Bound proteins were eluted with 2× SDS-PAGE loading buffer, separated by SDS-PAGE (12% gel), and immunoblotted.
Immunoprecipitation. Mammalian expression vectors for the full-length and deletion mutants of rat mGluR1a were constructed in pEF-BOS (Kawabata et al., 1996). Other expression vectors were generated by inserting appropriate restriction fragments isolated from either cDNAs or PCR fragments into pcDNA3.1 (Invitrogen, San Diego, CA), pCMV-Tag3B, pCMV-Tag3C (Stratagene, La Jolla, CA), or pCI-neo (Promega, Madison, WI). Expression vectors were transfected into either human embryonic kidney (HEK) 293 cells or COS-7 cells on a six-well plate by lipofectamine or lipofectamine 2000 (Invitrogen, Grand Island, NY). Cells were lysed 36–48 hr after transfection with the following lysis buffer: 50 mm Tris-Cl, pH 7.4, 150 mm NaCl, 2 mm EDTA, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, and protease inhibitor mixture Complete (Roche Diagnostics, Mannheim, Germany). For measurement of cell-surface expression of mGluR1a in transfected cells, cell-surface proteins were biotinylated with membrane-impermeable biotinamidocaproate N-hydroxysuccinamide ester for 30 min at 4°C as described by the vendor (Amersham Biosciences). Cell lysates were centrifuged at 10,000 × g for 10 min, and supernatants were precleared with protein A Sepharose. They were incubated with 2 μl antiserum or 1 μg antibody and attached to protein A Sepharose. Precipitates were washed three times with 500 μl lysis buffer and dissolved into 2× SDS-PAGE loading buffer. Immunoblots were reacted with horse radish peroxidase (HRP)-conjugated NeutrAvidin (Pierce, Rockford, IL) and detected with HRP reaction. Band intensities were quantified by GS-710 densitometer (Bio-Rad, Hercules, CA). Approximately 6% of mGluR1a was recovered at cell-surface membranes in cells transfected with mGluR1a cDNA alone. Adult rat whole brains were homogenized and fractionated into cytosols and P1 and P2 membrane fractions (Dunah et al., 1998). The P2 fraction was solubilized in 50 mm Tris-Cl, pH 7.4, containing 1% sodium deoxycholate at 36°C for 30 min and dialyzed against 50 mm Tris-Cl, pH 7.4, and 0.1% Triton X-100 overnight. The solubilized fraction was immunoprecipitated as described (Naisbitt et al., 1999). In immunoblot analysis of brain extracts, tamalin comigrated with the heavy chain of the antibody used for immunoprecipitation. To solve this problem, rabbit anti-tamalin antibody was biotinylated with biotinamidocaproateN-hydroxysuccinamide ester, and immunoprecipitates were blotted with the biotin-conjugated anti-tamalin antibody. Postsynaptic membrane fractions were extracted from synaptosomes with Triton X-100 once (PSD I) or twice (PSD II) or with Triton X-100 followed by sarkosyl (PSD III) as described (Cho et al., 1992).
Immunocytochemistry. Transfected COS-7 cells were plated on a coverslip 24 hr after transfection and grown for 1 d. Cells were washed with PBS and immunoreacted with human antibody directed against the extracellular portion of mGluR1 (Sillevis Smitt et al., 2000) in PBS containing 5% normal goat serum for 1 hr at room temperature. They were washed, fixed with 4% paraformaldehyde in PBS for 10 min at room temperature, and permeabilized with 0.1% Triton X-100 in PBS containing 5% normal goat serum overnight at 4°C. They were immunoreacted with anti-tamalin antibody and then secondary antibodies and visualized as described (Dev et al., 1999), using an Olympusconfocal microscopy (Tokyo, Japan).
Hippocampal cell culture and adenoviral infection. CA1/CA3 hippocampal neurons were cultured according to the procedures described by Bito et al. (1996) with minor modifications. Briefly, dissociated cells from the CA1/CA3 regions of hippocampi of 1-d-old ICR mice were maintained in culture for 7 d on a coverslip coated with Matrigel substrate (Becton Dickinson, Franklin Lakes, NJ). Proliferation of non-neuronal cells was prevented by the addition of 4 μm cytosine arabinoside (Sigma, St. Louis, MO) from the second day in culture. To promote selective expression of recombinant proteins in neurons, the tetracycline (tet) gene expression system was used (Gossen and Bujard, 1992) by generating two types of recombinant adenoviruses. One carried tet transactivator (tTA) with nuclear localization signal (Yoshida and Hamada, 1997) immediately downstream of the neuron-specific enolase promoter (NSE-tTA), whereas the other contained myc-tagged full-length rat tamalin (myc-tamalin), myc-N-tam, and enhanced green fluorescent protein (EGFP) downstream of the tet response element (TRE; Clontech). The NSE promoter was a kind gift from H. Bito (Department of Pharmacology, Kyoto University Faculty of Medicine) (Sakimura et al., 1987). Recombinant adenoviral plasmids were generated by homologous recombination in E. coli according to the procedures described by He et al. (1998); pShuttle vector, pAdEasy vector, and E. coli BJ5183 were obtained from B. Vogelstein (The Johns Hopkins Oncology Center) (He et al., 1998). The NSE-tTA, TRE-myc-tamalin, TRE-myc-N-tam, and TRE-EGFP genes were inserted into pShuttle vector, linearized, and subjected to homologous recombination with an adenoviral backbone plasmid pAdEasy in E. coli BJ5183 (He et al., 1998). The adenoviral plasmids were linearized and transfected into HEK 293 cells by lipofectamine. Ten days after transfection, cell lysate was obtained from HEK 293 cells, and recombinant adenoviruses were amplified by repeating adenoviral infection twice. Hippocampal neurons were coinfected at 7 d in culture with two types of adenoviruses, one carrying NSE-tTA and the other bearing TRE-myc-tamalin, TRE-myc-N-tam, or TRE-EGFP. Forty-eight to sixty hours after infection, neurons were washed with PBS, fixed with 4% paraformaldehyde in PBS for 10 min, and immunostained with primary and then secondary antibodies as described above. Immunoreactive soma and neurites were visualized with a confocal microscopy LSM510 (Carl Zeiss, Göttingen, Germany).
Antibodies. Rabbit tamalin antisera were raised against a bovine serum albumin (BSA)-conjugated synthetic peptide corresponding to the amino-terminal sequence (residues 1–20) of rat tamalin; this sequence significantly diverges from the corresponding sequence of CBP. Rabbit cytohesin-2 antisera were raised against a BSA-conjugated synthetic peptide of the amino-terminal sequence (residues 1–26) of rat cytohesin-2. Anti-cytohesin-2 antibody preferentially reacted with cytohesin-2 but slightly (<5%) cross-reacted with cytohesin-1 and cytohesin-3. Anti-tamalin and anti-cytohesin-2 antisera were used for immunoprecipitation and immunocytochemistry of transfected cells. For brain extracts, these antisera were further affinity purified on HiTrap columns (Amersham Biosciences) coupled to GST fused to the amino-terminal half of tamalin or His-Trx fused to cytohesin-2. Other primary antibodies used were polyclonal rabbit anti-mGluR5 and anti-mGluR1a antibodies (Shigemoto et al., 1993, 1997), polyclonal rabbit anti-MAL antibody (New England Biolabs), monoclonal mouse anti-mGluR1a antibody (BD PharMingen, San Diego, CA), monoclonal mouse anti-myc antibody (Clontech), polyclonal rabbit anti-EGFP antibody (Clontech), monoclonal mouse anti-PSD-95 antibody (BD Transduction Laboratories, Lexington, KY), and monoclonal mouse anti-synaptophysin antibody (Sigma). Immunoblotting bands and immunostaining patterns were detected with HRP-conjugated anti-rabbit or anti-mouse IgG antibody (Jackson ImmunoResearch, West Grove, PA), followed by HRP reaction, biotinylated anti-human IgG antibody, followed by FITC-conjugated Avidin D (Vector Laboratories, Burlingame, CA), Texas Red-conjugated anti-rabbit IgG antibody (Molecular Probes, Eugene, OR), Cy3-conjugated anti-rabbit IgG antibody (Jackson ImmunoResearch), and FITC-conjugated anti-mouse IgG antibody (Vector Laboratories).
RESULTS
Isolation of an mGluR-interacting protein using the yeast two-hybrid system
Group 1 and group 2 mGluRs contain a common ST/SL motif that belongs to the class I PDZ-binding motif (Songyang et al., 1997; Sheng and Sala, 2001). In contrast, the C-terminal sequences of group 3 mGluRs resemble the class II PDZ-binding motif or diverge from these motifs (Songyang et al., 1997; Sheng and Sala, 2001). To identify the PDZ domain-containing proteins that interact with the C-terminal sequences of group 1 and group 2 mGluRs, the last 53 amino acid C-terminal sequence (residues 820–872) of mGluR2 (ct-mGluR2) was used as a bait. We screened 3.8 × 105 clones of a rat brain cDNA library fused to the GAL4 activation domain. Of 30 β-galactosidase-positive clones, 24 clones shared an identical sequence that encoded a PDZ domain-containing protein termed tamalin (tagging metabotropic glutamate receptors and ARF-GEF), and one representative tamalin clone was further analyzed. To determine the specificity of interaction, the tamalin clone was transformed into yeast harboring plasmids containing ct-mGluRs and subjected to filter β-galactosidase assays. Tamalin interacted with the ct-mGluRs possessing the class I PDZ-binding motif (ct-mGluR1a, 2, 3, and 5), whereas no such interaction was observed for other ct-mGluRs lacking the class I PDZ-binding motif (ct-mGluR4, 6, 7a, and 8) (Fig. 1). Tamalin also interacted with the C terminus of GABAB2, the last 3-amino acid sequence (SGL) of which is similar to that of group 1 and group 2 mGluRs.
Fig. 1.
Interaction of tamalin with the C-terminal regions of mGluRs and GABAB2 in the yeast two-hybrid system. Interaction of the complete C-terminal regions of mGluR2, 3, 4, 6, 7a, and 8 was tested by filter β-galactosidase assay in the yeast two-hybrid system. For mGluR1a and 5, the last seven and five amino acid C-terminal sequences were used, respectively, because the complete C-terminal sequences of both receptors displayed false positive data without addition of a prey plasmid. ct-GABAB2 contained the last 80 amino acid sequence of the C terminus of GABAB2. Positive interaction is indicated as +, and negative is indicated as −.
Sequence analysis of tamalin
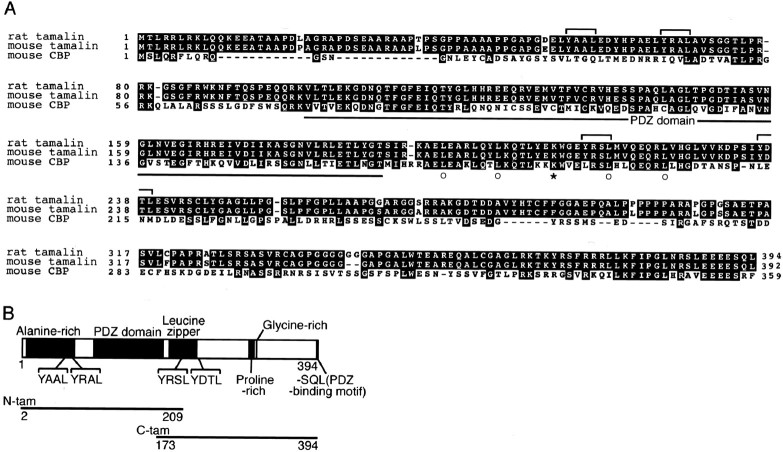
Clones encoding the full-length tamalin were isolated from an adult rat forebrain cDNA library and subjected to DNA sequencing. The tamalin cDNA insert encoded the protein sequence composed of 394 amino acid residues (Fig.2A). BLAST search analysis showed that several mouse EST clones shared a highly homologous nucleotide sequence with the rat tamalin cDNA. Two clones (accession numbers AB041603 and AU079540) were obtained, and they showed an identical protein sequence of 392 residues and were highly homologous to the tamalin rat counterpart with a sequence identity of 97% positions (Fig. 2A). The nucleotide sequences surrounding the first AUG codon in both rat and mouse tamalin mRNAs agreed well with the Kozak consensus sequence (Kozak, 1987), and the mouse mRNA possessed an in-frame stop codon in the putative 5′-untranslated region.
Fig. 2.
Structure of tamalin. A, Amino acid sequence alignment of rat tamalin, mouse tamalin, and mouse CBP is indicated. Identical amino acids are shown by black boxes, and conserved amino acids are shaded by gray boxes. The PDZ domain is underlined, and YXXL motifs are indicated by brackets. Four leucine residues and one lysine residue present every seven amino acids in the leucine zipper sequence are displayed by open circles and anasterisk, respectively. B, A schematic structure indicating the characteristic structural domains and sequence motifs of rat tamalin. Two separate regions used for delineating protein–protein interacting domains of tamalin are shown below the schematic structure.
Further BLAST search analysis identified a closely related protein sequence, termed CBP, from both human and mouse. CBP was initially isolated as a gene product specific to human natural killer cells by subtraction cloning techniques (Dixon et al., 1993). Tamalin and CBP are highly homologous in several characteristic domains of this protein family (56% match in the common structural domains) (Fig.2A). Recently, Nevrivy et al. (2000) reported molecular cloning of a retinoic acid-induced gene, termed GRASP, from P19 embryonal carcinoma cells. Mouse tamalin and GRASP showed a single amino acid substitution (L/P) at position 299, indicating that they represent the same gene product. The term of tamalin has been kept in this study to avoid confusion with a distinct GRASP that was identified as a GRIP-associated protein (Ye et al., 2000).
Tamalin possesses a PDZ domain and an alanine-rich, proline-rich, and glycine-rich region (Fig. 2B). Additionally, tamalin has a leucine zipper sequence, four YXXL motifs involved in clathrin-mediated endocytosis (Ohno et al., 1995; Mellman, 1996;Heilker et al., 1999), and a PDZ-binding motif (SQL) at the end of the C terminus. Interestingly, CBP possesses a PDZ domain and a leucine zipper domain but lacks all other protein motifs characteristic of tamalin, suggesting that both tamalin and CBP serve as scaffold proteins but may have distinct functions.
Distribution of tamalin and CBP mRNAs
Tissue distribution of mRNAs for mouse tamalin and CBP was examined by Northern blot analysis (Fig.3A). Tamalin mRNA was ∼2 kilonucleotides long and expressed at high levels in the brain and at moderate or low levels in the heart, lung, and kidney. This tissue distribution of mouse tamalin mRNA was consistent with the result of RT-PCR analysis reported by Nevrivy et al. (2000). In contrast, mouse CBP mRNA was undetectable in the brain and distributed in other tissues such as the spleen, lung, kidney, testis, heart, and liver.
Fig. 3.
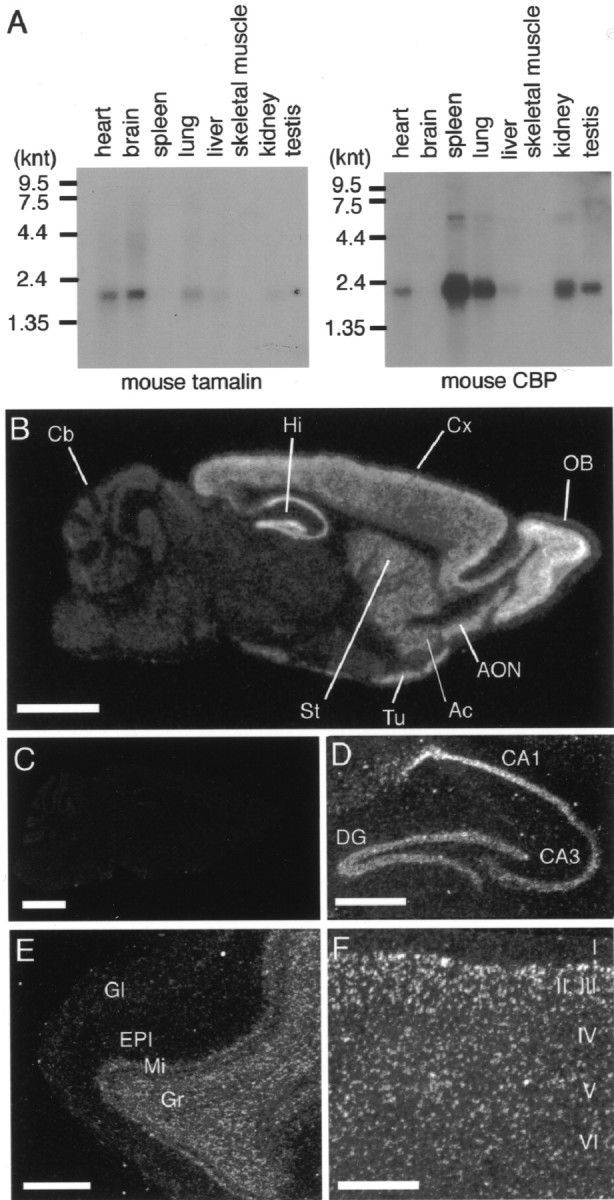
Expression patterns of mRNAs for mouse tamalin and CBP. A, Northern blot analysis of mouse tamalin and CBP mRNAs. A mouse Multiple Tissue Northern blot was probed with the mouse tamalin (left) and CBP (right) cDNAs. Positions of RNA molecular size markers are shown in kilonucleotides (knt). B–F, In situ hybridization analysis of tamalin mRNA in the adult mouse brain. Negative film images of in situ hybridization of parasagittal sections analyzed with the tamalin antisense riboprobe (B) and sense riboprobe (C) are shown.OB, Main olfactory bulb; Cx, cerebral cortex; Hi, hippocampus; Cb, cerebellum;AON, anterior olfactory nucleus; Ac, nucleus accumbens; St, striatum; Tu, olfactory tubercle. Dark-field photomicrographs show tamalin mRNA in the hippocampus (D), olfactory bulb (E), and cerebral cortex (F). DG, Dentate gyrus;Gl, glomerular layer; EPl, external plexiform layer; Mi, mitral cell layer;Gr, internal granular layer. Scale bars:B, C, 2 mm; D–F, 500 μm.
In situ hybridization analysis with an antisense riboprobe revealed a wide distribution of tamalin mRNA in the mouse brain (Fig.3B). No hybridization signals were observed in parallel experiments using the sense riboprobe (Fig. 3C). Prominent expression of tamalin mRNA was detected mainly in the telencephalic regions, including the olfactory bulb, cerebral cortex, hippocampus, anterior olfactory nucleus, olfactory tubercle, striatum, and nucleus accumbens (Fig. 3B). No appreciable expression was noted in the cerebellum, thalamus, or hypothalamus. In the hippocampus, intense signals were seen in pyramidal cells throughout CA1–CA3 regions as well as in granule cells of the dentate gyrus (Fig. 3D). In the olfactory bulb, prominent expression was observed in mitral cells and internal granular cells (Fig. 3E). In the cerebral cortex, signals were distributed in all but layer I, with the highest intensity at layers II–III (Fig. 3F). Interestingly, this cellular distribution of tamalin mRNA correlates well with that of mGluR5 mRNA and partially overlaps that of mGluR1a mRNA (Shigemoto and Mizuno, 2000).
GST pull-down assay
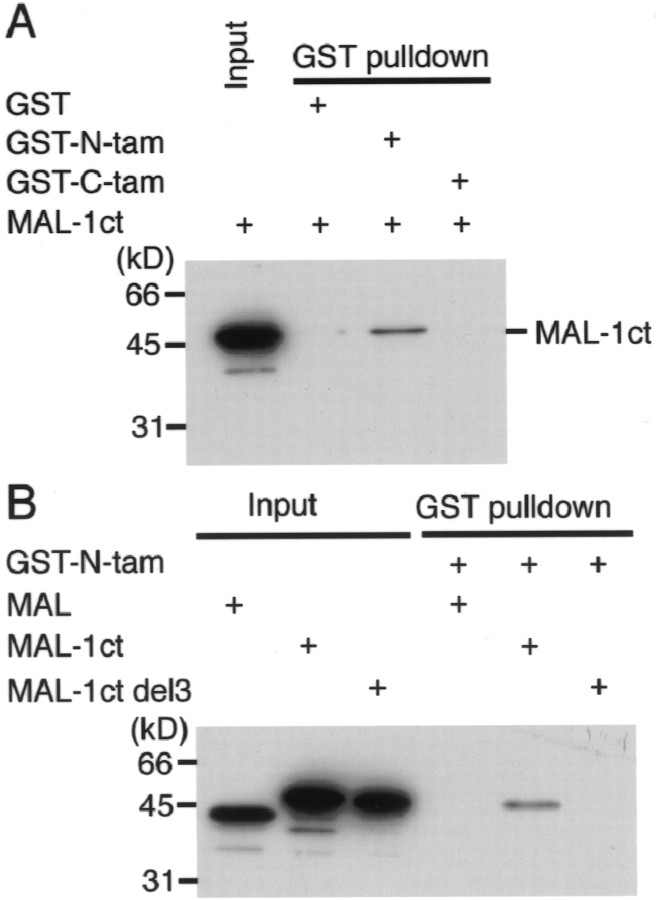
The specific interaction between mGluR subtypes and tamalin was biochemically examined by GST pull-down assays using the GST-tamalin fusion proteins. GST fused to the full-length tamalin resided in insoluble fractions in E. coli, and only a small amount of GST-tamalin was recovered by solubilization with detergents. Although the solubilized GST-tamalin was found to bind to the last 59 amino acid sequence of ct-mGluR1a fused to MAL (MAL-1ct), it was difficult to quantify the interaction between tamalin and ct-mGluR1a. We therefore separated rat tamalin into two fragments covering the PDZ-containing and PDZ-lacking domains and fused each domain to GST (GST-N-tam and GST-C-tam, respectively) (Fig. 2B). The two domains became soluble in E. coli extracts. GST pull-down assays showed that MAL-1ct effectively bound to GST-N-tam but not to either GST-C-tam or GST alone (Fig.4A). An effective interaction of GST-N-tam was also observed for ct-mGluR2, ct-mGluR3, and ct-mGluR5, but not for ct-mGluR4, ct-mGluR7a, and ct-mGluR8 (all ct-mGluRs used were the MAL fusion proteins) (data not shown). This selectivity in tamalin interaction agreed well with that determined by yeast two-hybrid analysis. Furthermore, deletion of the last three amino acid residues of ct-mGluR1a (MAL-1ct del3) abolished the ability of MAL-1ct to bind to GST-N-tam (Fig. 4B). The results indicated that the PDZ domain of tamalin recognizes the class I PDZ-binding motif of ct-mGluRs and allows a specific complex formation between tamalin and group 1 and group 2 mGluRs.
Fig. 4.
In vitro binding of tamalin with ct-mGluR1a. A, GST fusion proteins containing the amino-terminal sequence (residues 2–209, GST-N-tam) and the C-terminal sequence (residues 173–394, GST-C-tam) of rat tamalin as well as GST alone were immobilized on glutathione-Sepharose 4B beads and tested for their ability to bind to the MAL fusion protein containing the last 59 amino acid residues of ct-mGluR1a (MAL-1ct). Bound proteins were detected by immunoblotting with anti-MAL antibody.B, A mutant lacking the last three amino acid residues of ct-mGluR1a (MAL-1ct del3), MAL-1ct, and MAL were tested as in A. Inputs show equivalent amounts of MAL and MAL fusion proteins used for pull-down assays. The sizes (in kilodaltons) of molecular markers are indicated on theleft.
Coimmunoprecipitation of group 1 mGluRs and tamalin
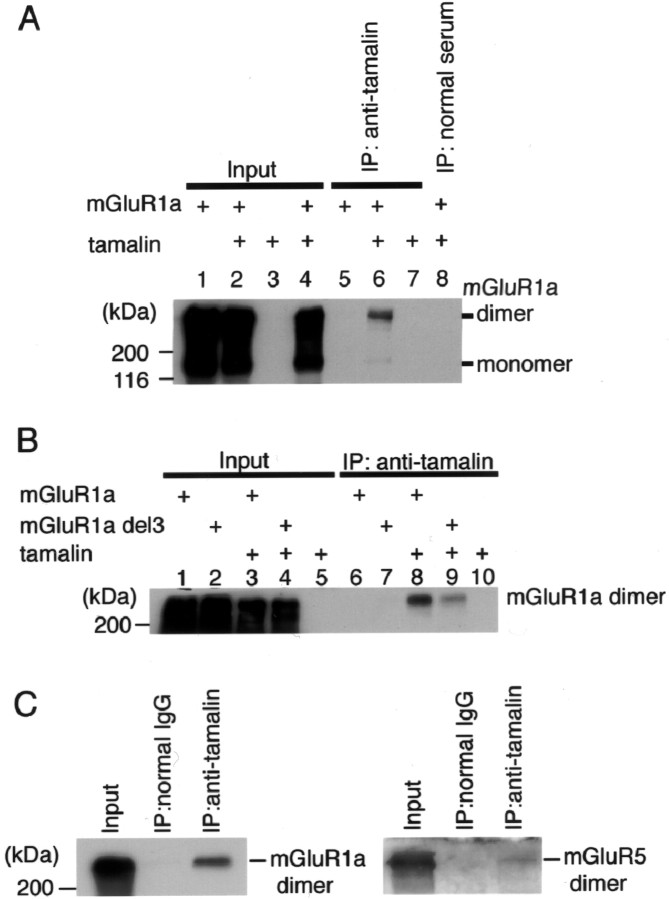
To investigate the interaction between tamalin and mGluR1a in mammalian cells, cell lysates of HEK293 cells transiently coexpressing tamalin and mGluR1a were immunoprecipitated with anti-tamalin antibody, followed by immunoblotting with anti-mGluR1a antibody (Fig.5A). Anti-tamalin antibody coimmunoprecipitated a dimeric form of mGluR1a, whereas no such mGluR1a immunoprecipitate was detected in cells transfected with either mGluR1a or tamalin alone. In addition, nonimmunized serum did not precipitate mGluR1a from cells coexpressing mGluR1a and tamalin. We then tested the interaction between tamalin and mGluR1a lacking either the last 3 amino acid residues (mGluR1a del3) (Fig. 5B) or the last 10 residues (mGluR1a del10) (data not shown). Both deletions showed a reduced but small amount of coimmunoprecipitates of mutant mGluR1a (Fig. 5B). The coimmunoprecipitation of tamalin and mGluR1a del3 or mGluR1a del10 may result from an interaction with an additional site other than the last 10 amino acid residues of mGluR1a either directly or through as yet unidentified scaffold proteins. More importantly, the reduction of mGluR1a coimmunoprecipitates was significant in both deletions, indicating that the binding between the PDZ domain and the PDZ-binding motif plays a major role in interaction between tamalin and mGluR1a.
Fig. 5.
Interaction between mGluR1a and tamalin in transfected HEK293 cells. A, HEK293 cells were transfected with full-length mGluR1a, mouse tamalin, or both. Cell lysates were immunoprecipitated with either anti-tamalin antibody (lanes 5–7) or nonimmunized serum (lane 8) and immunoblotted with anti-mGluR1a antibody. Inputs (lanes 1–4) show of cell lysates used for immunoprecipitation. A dimeric form of mGluR1a was detected by immunoprecipitation with anti-tamalin antibody. B, HEK293 cells were transfected with either mGluR1a or mGluR1a del3 with or without full-length tamalin. Immunoprecipitation and immunoblotting were performed as in A. C, Solubilized rat brain P2 membrane fraction was immunoprecipitated with either anti-tamalin antibody or normal IgG and immunoblotted with anti-mGluR1a or anti-mGluR5 antibody. Inputs show equivalent amounts of extracts used for immunoprecipitation.
We next addressed whether tamalin and mGluRs form a protein complex in the rat brain. Solubilized rat brain membrane fractions were immunoprecipitated with an affinity-purified anti-tamalin antibody and analyzed with anti-mGluR antibodies. Anti-tamalin antibody coimmunoprecipitated dimeric forms of both mGluR1a and mGluR5 in brain extracts, whereas these receptors were not precipitated with normal rabbit IgG (Fig. 5C). Also noteworthy is the finding that no appreciable coimmunoprecipitates were detected by immunoblotting with anti-mGluR2/3 antibody (data not shown). This was consistent with an overlapping expression between tamalin and group 1 mGluR mRNAs in various brain regions (Fig. 3B–F). The results indicated that tamalin mainly forms a complex with group 1 mGluRs in the brain, although the possible assembly between tamalin and group 2 mGluRs in limited neuronal cell subpopulations cannot be excluded.
A complex formation of mGluR1a, tamalin, and cytohesin-2
Because tamalin possesses many protein-interacting motifs, we searched for tamalin-binding proteins, using the full-length rat tamalin as a bait for yeast two-hybrid screening of an adult rat brain cDNA library. Four of 69 positive clones encoded the sequence of cytohesin-2/sec7B/ARNO (ARF nucleotide-binding-site opener) that is a GEF member for the ARF family. Recently, Nevrivy et al. (2000) reported a protein interaction between the C-terminal half of mouse tamalin (GRASP) and the coiled-coil (CC) domain of cytohesins, using GST pull-down assays as well as heterologously transfected HEK293 cells. We also reached the same conclusion with similar experiments (data not shown): in pull-down assays, the His-Trx fusion protein containing the full-length as well as the CC domain of rat cytohesin-2 specifically bound to GST-C-tam, but not to GST-N-tam; anti-myc antibody immunoprecipitated EGFP-cytohesin-2 from COS-7 cell lysates transfected with EGFP-cytohesin-2 and myc-tagged rat tamalin.
Because these results indicated that tamalin interacts with group 1 mGluRs and cytohesin-2 via its amino-terminal and C-terminal domains, respectively, we investigated the existence of a ternary complex of mGluR1a/tamalin/cytohesin-2 by several different approaches. mGluR1a is mostly localized at the postsynaptic membrane in neuronal cells (Martin et al., 1992). We examined the postsynaptic membrane localization of tamalin and cytohesin-2 by immunoblotting of serially extracted synaptosomal membrane proteins (Fig.6A). Both mGluR1a and PSD-95 were mostly concentrated at the PSD III fraction, whereas the presynaptic marker protein, synaptophysin, was deprived from synaptosome by detergent extraction. Both tamalin and cytohesin-2 were enriched up to the PSD II fraction but slightly and completely deprived from the PSD III fraction, respectively. The results indicated that both tamalin and cytohesin-2 are localized at postsynaptic membrane fractions and are relatively tightly and loosely associated with postsynaptic membranes, respectively.
Fig. 6.
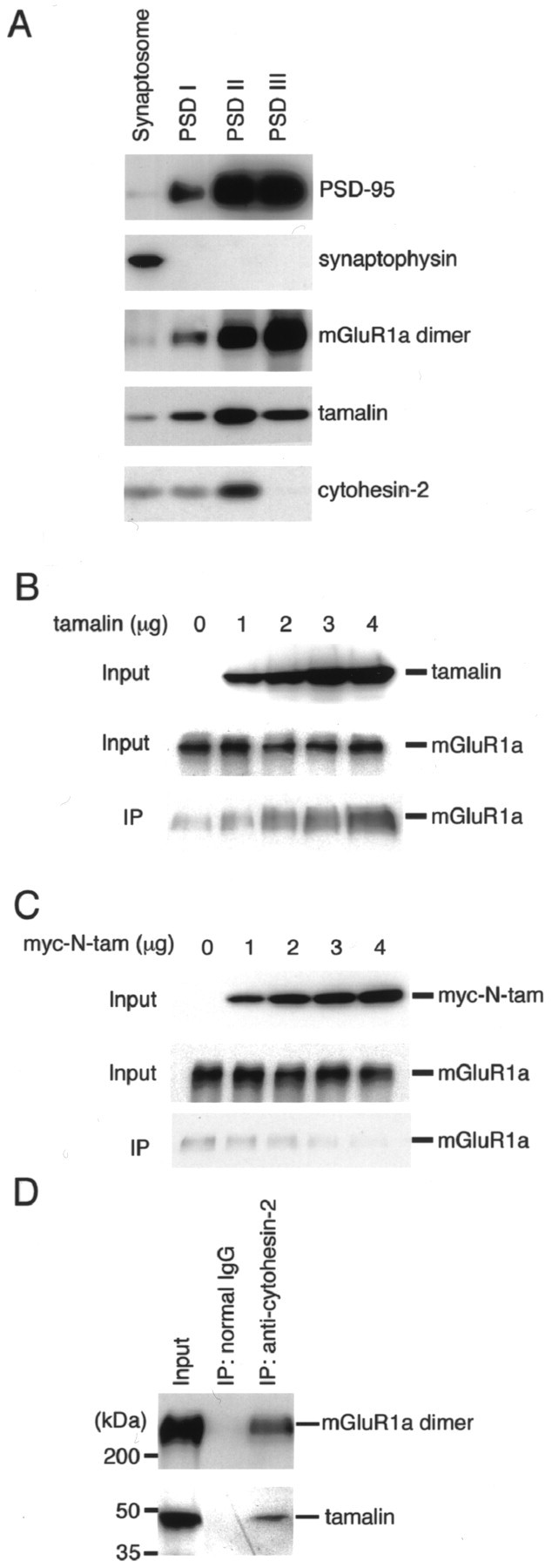
A complex formation of mGluR1a, tamalin and cytohesin-2. A, Synaptosomal and PSD I, II, and III fractions (1 μg protein each) were immunoblotted with the indicated antibodies. B, COS-7 cells were transfected with mGluR1a, myc-cytohesin-2, and tamalin. Amounts of transfected cDNAs for mGluR1a and myc-cytohesin-2 were fixed (0.5 μg each), whereas those of tamalin cDNA were increased as indicated. Cell lysates were solubilized and immunoprecipitated with anti-cytohesin-2 antibody. Immunoprecipitates and cell lysates were blotted with anti-tamalin and anti-mGluR1a antibodies. Inputs show of cell lysates used for immunoprecipitation. C, COS-7 cells were transfected with mGluR1a, myc-cytohesin-2, and myc-N-tam. Amounts of transfected cDNAs for mGluR1a and myc-cytohesin-2 were fixed (0.5 μg each), whereas those of myc-N-tam cDNA were increased as indicated. Cell lysates were immunoprecipitated with anti-cytohesin-2 antibody. Immunoprecipitates and cell lysates were blotted with anti-tamalin and anti-mGluR1a antibodies as in B. D, Solubilized brain P2 membrane fraction was immunoprecipitated with anti-cytohesin-2 antibody or normal IgG. Immunoprecipitated mGluR1a was blotted with anti-mGluR1a antibody, followed by immunoreaction with HRP-conjugated anti-mouse IgG. Immunoprecipitated tamalin was blotted with biotinylated anti-tamalin antibody, followed by reaction with avidin-HRP. mGluR1a and tamalin were then detected by HRP reaction. Inputs show (mGluR1a) and an equivalent amount (tamalin) of membrane fraction used for immunoprecipitation.
Next, we triply transfected mGluR1a, tamalin, and myc-cytohesin-2 into COS-7 cells and analyzed coimmunoprecipitation of mGluR1a by anti-cytohesin-2 antibody. Anti-cytohesin-2 antibody coimmunoprecipitated mGluR1a, but a small amount of coimmunoprecipitates was also observed without tamalin transfection (Fig. 6B). Importantly, coimmunoprecipitation of mGluR1a was increased by increasing concentrations of tamalin transfected into COS-7 cells (Fig. 6B). Therefore, formation of an mGluR1a/cytohesin-2 complex without tamalin transfection seemed to be mediated by endogenous tamalin-related proteins. To confirm this possibility, we triply transfected myc-N-tam, mGluR1a, and myc-cytohesin-2 into COS-7 cells and examined whether this mutant lacking a cytohesin-binding domain competes with endogenous tamalin-like proteins for complex formation of mGluR1a and cytohesin-2. Transfection of myc-N-tam significantly reduced coimmunoprecipitation of mGluR1a and cytohesin-2 in a concentration-dependent manner (Fig.6C). The results strongly indicated that tamalin links a complex formation between mGluR1a and cytohesin-2.
The interaction between cytohesin-2 and mGluR1a was further addressed in solubilized rat brain membrane fractions (Fig.6D). Immunoblotting showed that anti-cytohesin-2 antibody coimmunoprecipitated not only tamalin but also a dimeric form of mGluR1a in brain membrane fractions, whereas no proteins were precipitated with normal IgG (Fig. 6D). The results indicated that a protein complex of mGluR1a/tamalin/cytohesin-2 exists in the rat brain.
Effects of tamalin on intracellular trafficking of group 1 mGluRs
To study the effects of tamalin on cell-surface expression of mGluR1a, we transfected fixed amounts of mGluR1a and myc-cytohesin-2 in combination with different amounts of tamalin. We then biotinylated cell-surface proteins of transfected cells with a membrane-impermeable biotin ester. No cytoplasmic myc-cytohesin-2 was biotinylated under the experimental condition (data not shown), indicating that the biotin ester did not penetrate through the cell membrane. We immunoprecipitated mGluR1a from cell lysates and quantified biotinylated cell-surface mGluR1a by avidin-HRP reaction. Amounts of cell-surface mGluR1a increased in a concentration-dependent manner by more than threefold when tamalin was cotransfected with mGluR1a (Fig.7A,B). Furthermore, when myc-N-tam was coexpressed with mGluR1a and myc-cytohesin-2, cell-surface expression of mGluR1a was conversely reduced in a concentration-dependent manner of transfected myc-N-tam (Fig. 7A,B). In both cases, the amounts of total mGluR1a that immunoprecipitated with anti-mGluR1a antibody remained unchanged (Fig. 7A). This result indicated that the interaction between mGluR1a and tamalin is critical for cell-surface expression of mGluR1a.
Fig. 7.
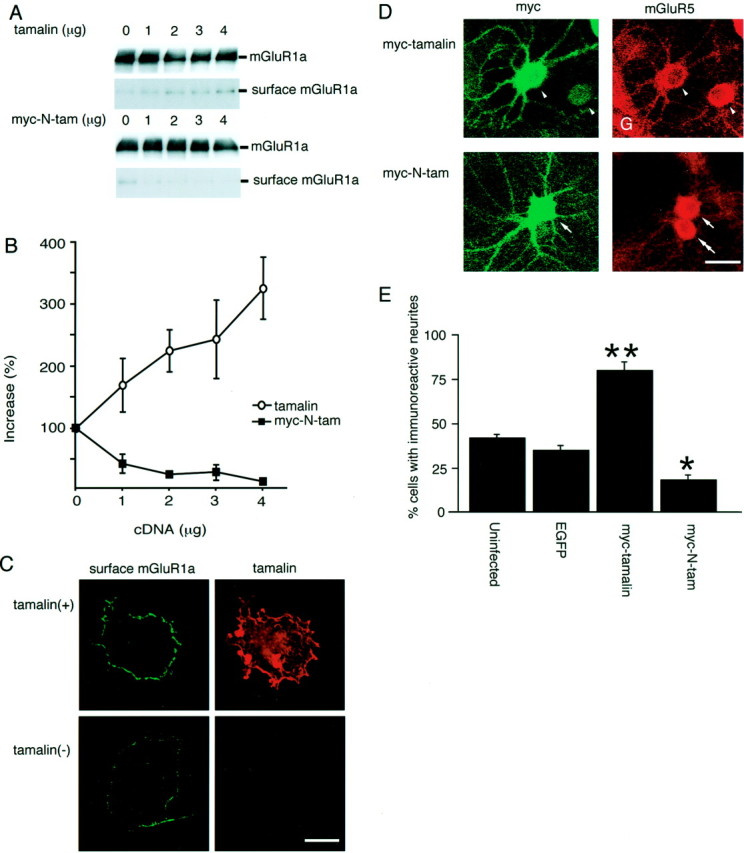
Tamalin-mediated increase in cell-surface mGluR1a expression in COS-7 cells and neuritic mGluR5 targeting in hippocampal neurons. A, COS-7 cells were transfected with fixed amounts of cDNAs for mGluR1a and myc-cytohesin-2 (0.5 μg each) in combination with indicated amounts of cDNA for either tamalin or myc-N-tam. Cell-surface proteins were biotinylated with a membrane-impermeable biotin ester. Cell lysates were immunoprecipitated with anti-mGluR1a antibody. An aliquot of immunoprecipitates was immunoblotted with anti-mGluR1a antibody, followed by immunoreaction with HRP-conjugated anti-mouse IgG. An equivalent amount of immunoprecipitates was run on a separate gel and reacted with HRP-conjugated avidin. Both total mGluR1a and biotinylated cell-surface mGluR1a were detected with HRP reaction. B, Relative amounts of cell-surface mGluR1a were quantified and plotted against concentrations of tamalin cDNA transfected into COS-7 cells. Data represent mean ± SD (n = 3).C, COS-7 cells were transfected with mGluR1a and myc-cytohesin-2 with and without tamalin. Cell-surface mGluR1a was immunostained on nonpermeabilized cells with antibody against the extracellular domain of mGluR1, whereas tamalin was immunostained on permeabilized cells with anti-tamalin antibody. Scale bar, 20 μm. D, Cultured hippocampal neurons were infected with adenovirus containing the TRE-myc-tamalin or TRE-myc-N-tam together with the NSE-tTA adenovirus. Two days after infection, neurons were immunostained with anti-myc antibody (left panel) and anti-mGluR5 antibody (right panel). Intense mGluR5 immunoreactivity was seen in neurites of most of the myc-tamalin-expressing neurons (arrowheads). Little mGluR5 immunoreactivity was observed in neurites of either myc-N-tam-expressing neurons (arrow) or adenovirus-uninfected neurons (double arrows).G, Glial cells. Scale bar, 20 μm. E, Neurons displaying positive mGluR5 immunostaining in neurites were counted. Data indicate percentages of neurons exhibiting neuritic mGluR5 immunostaining for each virus construct (mean ± SD;n = 2 with 30 neurons per experiment).Asterisks indicate statistical significance as compared with both uninfected neurons and EGFP-transfected neurons (**p < 0.01, *p < 0.05; unpaired Student's t test).
The cell-surface expression of mGluR1a was further assessed by immunofluorescence staining of nonpermeabilized COS-7 cells, using anti-mGluR1 antibody directed against the extracellular portion of mGluR1. In this experiment, anti-mGluR1 antibody was first reacted with the cell-surface mGluR1a on nonpermealized cells. Anti-tamalin antibody was then reacted with intracellular tamalin in permeabilized cells. No tamalin immunoreactivity was detected without tamalin transfection, indicating that tamalin per se is not expressed in COS-7 cells (Fig. 7C). When tamalin was cotransfected with mGluR1a, tamalin immunoreactivity was distributed in the cytoplasm and plasma membrane as well as perinuclear compartments (Fig.7C). In these cells, a significant increase in cell-surface distribution of mGluR1a was detected as compared with cells transfected with mGluR1a alone (Fig. 7C).
We next examined the role of tamalin in intracellular trafficking of endogenous mGluR5 in cultured hippocampal neurons, using adenovirus-mediated gene expression of recombinant proteins. In this experiment, the neuronal expression of recombinant proteins was enforced by combining the tet gene expression system. tTA was expressed under the control of the neuron-specific enolase promoter and drove neuron-selective expression of recombinant proteins through an interaction of tTA with the tet response element located upstream of each recombinant construct (TRE-myc-tamalin, TRE-myc-N-tam, or TRE-EGFP). The combination of the adenoviral gene delivery with the tet expression system specified an expression of recombinant proteins in neurons, giving rise to 20–25% expression in a neuronal cell population without any appreciable expression in glial cells (data not shown). In adenovirus-uninfected culture, mGluR5 immunoreactivity was confined to soma in a large population of cultured neurons with little or only weak mGluR5 immunoreactivity in their neuritic processes (Fig.7D, double arrow, E). In contrast, coinfection of adenoviruses carrying NSE-tTA and TRE-myc-tamalin exhibited extensive distribution of mGluR5 immnoreactivity not only in soma but also throughout neuritic processes of most of the adenovirus-infected neurons (Fig. 7D, arrowheads,E). In cultured hippocampal neurons, moderate expression of endogenous tamalin was detected by immunoblotting analysis (data not shown). Consistent with this observation, coinfection of adenoviruses bearing NSE-tTA and TRE-myc-N-tam significantly reduced a cell population displaying neuritic localization of mGluR5 immunoreactivity (Fig. 7D, arrow, E). In control, the adenoviral infection per se, as detected by EGFP expression, had no effect on mGluR5 distribution or morphological characteristics of cultured neurons (Fig. 7E). The results indicated that tamalin plays an important role in intracellular trafficking of group 1 mGluRs in both heterologously transfected cells and cultured neurons.
DISCUSSION
A number of PDZ domain-containing proteins have been reported to interact with target proteins via the recognition of a PDZ-binding motif located at their C-terminal regions. This investigation concerns identification and characterization of the PDZ domain-containing tamalin that interacts with group 1 mGluRs. Both yeast two-hybrid screening and in vitro GST pull-down assay showed that tamalin interacts directly with the C-terminal sequences of group 1 and group 2 mGluRs but not with those of group 3 mGluRs. Structural analysis of tamalin and ct-mGluR1a by truncation and deletion mutations further indicated that the PDZ domain of tamalin is crucial for interaction with the PDZ-binding motif at the C-terminal sequence of mGluR1a. The expression patterns of mRNAs for tamalin and mGluR5 correlate well with a preferential distribution of both mRNAs at the telencephalic region. Additionally, tamalin is enriched and codistributed with group 1 mGluRs in postsynaptic membrane fractions. More decisively, a complex formation of native tamalin and mGluR1a/mGluR5 in the rat brain was verified by coimmunoprecipitation of these proteins. Our data indicate that tamalin forms a protein complex with group 1 mGluRs at the postsynaptic site of specific neuronal cells.
Cotransfection of the deletional mGluR1a (mGluR1a del3 or del10) and tamalin, however, suggested the involvement of an additional sequence of mGluR1a in interaction with tamalin. It has recently been shown that Shank and munc18-interacting protein 1 (Mint 1), in addition to their direct binding to the PDZ-binding motifs of group 1 mGluRs and calcium channels, respectively, interact indirectly with these target proteins via Homer and CASK, respectively (Maximov et al., 1999;Tu et al., 1999). Our immunoprecipitation analysis showed that anti-tamalin antibody coimmunoprecipitated several scaffold proteins from brain membrane fractions, such as CASK, guanylate kinase-associated protein, and Mint2 (data not shown). Although an interaction between mGluR1a and these proteins was not yet determined, it is possible that some of these scaffold proteins may contribute to a stable complex formation between tamalin and mGluRs.
Recently, Nevrivy et al. (2000) reported that tamalin (GRASP) interacts with cytohesins. This assembly occurs via the C-terminal domain of tamalin and the CC domains of cytohesins. Therefore, tamalin interacts with mGluR1a and cytohesins via two separate structural domains of this scaffold protein. Importantly, a complex formation of mGluR1a/tamalin/cytohesin-2 was verified in not only transfected cells but also in rat brain tissue. Furthermore, N-tam that lacks a cytohesin-binding domain competed with endogenous tamalin-like proteins and blocked assembly between mGluR1a and cytohesin-2 in transfected cells. Therefore, tamalin serves as a key scaffold protein that links a complex formation between mGluR1a and cytohesins. Furthermore, tamalin forms intermolecular self-associations via its PDZ domain and PDZ-binding motif (data not shown). It is thus conceivable that tamalin contributes to a macromolecular complex formation at synapses.
Tamalin and N-tam caused an increase and a decrease in not only cell-surface expression of mGluR1a in transfected cells but also in neuritic distribution of endogenous mGluR5 in cultured hippocampal neurons, respectively. These changes in intracellular trafficking of group 1 mGluRs are in good agreement with the potency of tamalin and N-tam to increase and decrease a complex formation of mGluR1a and cytohesin-2 in transfected cells. Cytohesins are GEF proteins for the ARF family of small GTP-binding proteins (Jackson and Casanova, 2000;Jackson et al., 2000). These GEFs activate ARFs by facilitating a switch from an inactive GDP-bound to an active GTP-bound state of ARFs. The ARFs fall into three classes and control both nonclathrin [coatomers for coat protomer complex (COP) vesicles] and clathrin (clathrin-associated protein adaptor complexes) coat recruitments at distinct steps of intracellular membrane transport (Moss and Vaughan, 1998; Chavrier and Goud, 1999; Roth, 1999). For example, ARF1, the best-characterized ARF, has been shown to regulate the assembly/disassembly of COPI coatomer onto Golgi membranes and to be involved in the recruitment of the COPI coat ontrans-Golgi network membranes. ARF1 is also required for the binding of the AP-1 adaptor proteins of clathrin-coated vesicles to Golgi membranes. Additionally, ARF6 appears to regulate some steps of the endocytic pathway by recycling target proteins from endosomal compartments to plasma membrane (D'Souza-Schorey et al., 1995; Peters et al., 1995). Interestingly, tamalin possesses four YXXL consensus motifs, which interact with μ subunits of the clathrin adaptor complexes (Ohno et al., 1995; Mellman, 1996; Heilker et al., 1999). Cytohesin-2 has been shown to activate both ARF1 and ARF6 and colocalize with ARF proteins (Chardin et al., 1996; Frank et al., 1998; Lee and Pohajdak, 2000). It is therefore possible that tamalin may serve as a key element that links the ARF-GEF with the intracellular trafficking machinery and regulate intracellular transport of mGluRs.
Recently, several PDZ domain-containing proteins that interact with NMDA and AMPA receptors have been shown to bind specifically to proteins involved in signal transduction processes of small GTP-binding proteins. For example, PSD-95 and synapse-associated protein-102 bind to synaptic GTPase-activating protein (GAP), a novel rasGAP found only at excitatory synapses (Kim et al., 1998). Citron, a rhoGAP (Furuyashiki et al., 1999; Zhang et al., 1999), and Kalirin, a rhoGEF (Penzes et al., 2001), also bind to the PSD-95 family members and are localized at excitatory synapses. These findings suggest that the PDZ domain-containing proteins may play a key role in connecting glutamate receptors to small GTP-binding proteins that regulate intracellular protein transport and synaptic organization.
Footnotes
This work was supported in part by research grants from the Ministry of Education, Science and Culture of Japan. We thank Bert Vogelstein for providing adenoviral recombination vectors and Haruhiko Bito for a gift of the enolase promoter and technical advice. We are grateful to Atsushi Nishimune and Satoshi Kaneko for technical advice and Kumlesh K. Dev for careful reading of this manuscript.
Correspondence should be addressed to Shigetada Nakanishi, Department of Biological Sciences, Kyoto University Faculty of Medicine, Yoshida, Sakyo-ku, Kyoto 606-8501, Japan. E-mail:snakanis@phy.med.kyoto-u.ac.jp.
REFERENCES
- 1.Abe T, Sugihara H, Nawa H, Shigemoto R, Mizuno N, Nakanishi S. Molecular characterization of a novel metabotropic glutamate receptor mGluR5 coupled to inositol phosphate/Ca2+ signal transduction. J Biol Chem. 1992;267:13361–13368. [PubMed] [Google Scholar]
- 2.Aramori I, Nakanishi S. Signal transduction and pharmacological characteristics of a metabotropic glutamate receptor, mGluR1, in transfected CHO cells. Neuron. 1992;8:757–765. doi: 10.1016/0896-6273(92)90096-v. [DOI] [PubMed] [Google Scholar]
- 3.Bito H, Deisseroth K, Tsien RW. CREB phosphorylation and dephosphorylation: a Ca2+- and stimulus duration-dependent switch for hippocampal gene expression. Cell. 1996;87:1203–1214. doi: 10.1016/s0092-8674(00)81816-4. [DOI] [PubMed] [Google Scholar]
- 4.Bliss TVP, Collingridge GL. A synaptic model of memory: long-term potentiation in the hippocampus. Nature. 1993;361:31–39. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- 5.Boudin H, Doan A, Xia J, Shigemoto R, Huganir RL, Worley P, Craig AM. Presynaptic clustering of mGluR7a requires the PICK1 PDZ domain binding site. Neuron. 2000;28:485–497. doi: 10.1016/s0896-6273(00)00127-6. [DOI] [PubMed] [Google Scholar]
- 6.Chardin P, Paris S, Antonny B, Robineau S, Beraud-Dufour S, Jackson CL, Chabre M. A human exchange factor for ARF contains Sec7- and pleckstrin-homology domains. Nature. 1996;384:481–484. doi: 10.1038/384481a0. [DOI] [PubMed] [Google Scholar]
- 7.Chavrier P, Goud B. The role of ARF and Rab GTPases in membrane transport. Curr Opin Cell Biol. 1999;11:466–475. doi: 10.1016/S0955-0674(99)80067-2. [DOI] [PubMed] [Google Scholar]
- 8.Cho KO, Hunt CA, Kennedy MB. The rat brain postsynaptic density fraction contains a homolog of the Drosophila discs-large tumor suppressor protein. Neuron. 1992;9:929–942. doi: 10.1016/0896-6273(92)90245-9. [DOI] [PubMed] [Google Scholar]
- 9.Choi DW, Rothman SM. The role of glutamate neurotoxicity in hypoxic-ischemic neuronal death. Annu Rev Neurosci. 1990;13:171–182. doi: 10.1146/annurev.ne.13.030190.001131. [DOI] [PubMed] [Google Scholar]
- 10.Craven SE, Bredt DS. PDZ proteins organize synaptic signaling pathways. Cell. 1998;93:495–498. doi: 10.1016/s0092-8674(00)81179-4. [DOI] [PubMed] [Google Scholar]
- 11.D'Souza-Schorey C, Li G, Colombo MI, Stahl PD. A regulatory role for ARF6 in receptor-mediated endocytosis. Science. 1995;267:1175–1178. doi: 10.1126/science.7855600. [DOI] [PubMed] [Google Scholar]
- 12.Dev KK, Nishimune A, Henley JM, Nakanishi S. The protein kinase C alpha binding protein PICK1 interacts with short but not long form alternative splice variants of AMPA receptor subunits. Neuropharmacology. 1999;38:635–644. doi: 10.1016/s0028-3908(98)00230-5. [DOI] [PubMed] [Google Scholar]
- 13.Dev KK, Nakajima Y, Kitano J, Braithwaite SP, Henley JM, Nakanishi S. PICK1 interacts with and regulates PKC phosphorylation of mGLUR7. J Neurosci. 2000;20:7252–7257. doi: 10.1523/JNEUROSCI.20-19-07252.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dixon B, Sahely B, Liu L, Pohajdak B. Cloning a cDNA from human NK/T cells which codes for an unusual leucine zipper containing protein. Biochim Biophys Acta. 1993;1216:321–324. doi: 10.1016/0167-4781(93)90165-a. [DOI] [PubMed] [Google Scholar]
- 15.Dong H, O'Brien RJ, Fung ET, Lanahan AA, Worley PF, Huganir RL. GRIP: a synaptic PDZ domain-containing protein that interacts with AMPA receptors. Nature. 1997;386:279–284. doi: 10.1038/386279a0. [DOI] [PubMed] [Google Scholar]
- 16.Dunah AW, Luo J, Wang YH, Yasuda RP, Wolfe BB. Subunit composition of N-methyl-d-aspartate receptors in the central nervous system that contain the NR2D subunit. Mol Pharmacol. 1998;53:429–437. doi: 10.1124/mol.53.3.429. [DOI] [PubMed] [Google Scholar]
- 17.El Far O, Airas J, Wischmeyer E, Nehring RB, Karschin A, Betz H. Interaction of the C-terminal tail region of the metabotropic glutamate receptor 7 with the protein kinase C substrate PICK1. Eur J Neurosci. 2000;12:4215–4221. doi: 10.1046/j.1460-9568.2000.01309.x. [DOI] [PubMed] [Google Scholar]
- 18.Frank S, Upender S, Hansen SH, Casanova JE. ARNO is a guanine nucleotide exchange factor for ADP-ribosylation factor 6. J Biol Chem. 1998;273:23–27. doi: 10.1074/jbc.273.1.23. [DOI] [PubMed] [Google Scholar]
- 19.Furuyashiki T, Fujisawa K, Fujita A, Madaule P, Uchino S, Mishina M, Bito H, Narumiya S. Citron, a Rho-target, interacts with PSD-95/SAP-90 at glutamatergic synapses in the thalamus. J Neurosci. 1999;19:109–118. doi: 10.1523/JNEUROSCI.19-01-00109.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garner CC, Nash J, Huganir RL. PDZ domains in synapse assembly and signaling. Trends Cell Biol. 2000;10:274–280. doi: 10.1016/s0962-8924(00)01783-9. [DOI] [PubMed] [Google Scholar]
- 21.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracyclin-responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He TC, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci USA. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heilker R, Spiess M, Crottet P. Recognition of sorting signals by clathrin adaptors. BioEssays. 1999;21:558–567. doi: 10.1002/(SICI)1521-1878(199907)21:7<558::AID-BIES4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 24.Jackson CL, Casanova JE. Turning on ARF: the Sec7 family of guanine-nucleotide-exchange factors. Trends Cell Biol. 2000;10:60–67. doi: 10.1016/s0962-8924(99)01699-2. [DOI] [PubMed] [Google Scholar]
- 25.Jackson TR, Kearns BG, Theibert AB. Cytohesins and centaurins: mediators of PI 3-kinase-regulated Arf signaling. Trends Biochem Sci. 2000;25:489–495. doi: 10.1016/s0968-0004(00)01644-3. [DOI] [PubMed] [Google Scholar]
- 26.Kawabata S, Tsutsumi R, Kohara A, Yamaguchi T, Nakanishi S, Okada M. Control of calcium oscillations by phosphorylation of metabotropic glutamate receptors. Nature. 1996;383:89–92. doi: 10.1038/383089a0. [DOI] [PubMed] [Google Scholar]
- 27.Kim JH, Liao D, Lau LF, Huganir RL. SynGAP: a synaptic RasGAP that associates with the PSD-95/SAP90 protein family. Neuron. 1998;20:683–691. doi: 10.1016/s0896-6273(00)81008-9. [DOI] [PubMed] [Google Scholar]
- 28.Kornau HC, Schenker LT, Kennedy MB, Seeburg PH. Domain interaction between NMDA receptor subunits and the postsynaptic density protein PSD-95. Science. 1995;269:1737–1740. doi: 10.1126/science.7569905. [DOI] [PubMed] [Google Scholar]
- 29.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee SY, Pohajdak B. N-terminal targeting of guanine nucleotide exchange factors (GEF) for ADP ribosylation factors (ARF) to the Golgi. J Cell Sci. 2000;113:1883–1889. doi: 10.1242/jcs.113.11.1883. [DOI] [PubMed] [Google Scholar]
- 31.Martin LJ, Blackstone CD, Huganir RL, Price DL. Cellular localization of a metabotropic glutamate receptor in rat brain. Neuron. 1992;9:259–270. doi: 10.1016/0896-6273(92)90165-a. [DOI] [PubMed] [Google Scholar]
- 32.Maximov A, Südhof TC, Bezprozvanny I. Association of neuronal calcium channels with modular adaptor proteins. J Biol Chem. 1999;274:24453–24456. doi: 10.1074/jbc.274.35.24453. [DOI] [PubMed] [Google Scholar]
- 33.Mellman I. Endocytosis and molecular sorting. Annu Rev Cell Dev Biol. 1996;12:575–625. doi: 10.1146/annurev.cellbio.12.1.575. [DOI] [PubMed] [Google Scholar]
- 34.Moss J, Vaughan M. Molecules in the ARF orbit. J Biol Chem. 1998;273:21431–21434. doi: 10.1074/jbc.273.34.21431. [DOI] [PubMed] [Google Scholar]
- 35.Naisbitt S, Kim E, Tu JC, Xiao B, Sala C, Valtschanoff J, Weinberg RJ, Worley PF, Sheng M. Shank, a novel family of postsynaptic density proteins that binds to the NMDA receptor/PSD-95/GKAP complex and cortactin. Neuron. 1999;23:569–582. doi: 10.1016/s0896-6273(00)80809-0. [DOI] [PubMed] [Google Scholar]
- 36.Nakanishi S. Molecular diversity of glutamate receptors and implications for brain function. Science. 1992;258:597–603. doi: 10.1126/science.1329206. [DOI] [PubMed] [Google Scholar]
- 37.Nakanishi S, Masu M. Molecular diversity and functions of glutamate receptors. Annu Rev Biophys Biomol Struct. 1994;23:319–348. doi: 10.1146/annurev.bb.23.060194.001535. [DOI] [PubMed] [Google Scholar]
- 38.Nevrivy DJ, Peterson VJ, Avram D, Ishmael JE, Hansen SG, Dowell P, Hruby DE, Dawson MI, Leid M. Interaction of GRASP, a protein encoded by a novel retinoic acid-induced gene, with members of the cytohesin family of guanine nucleotide exchange factors. J Biol Chem. 2000;275:16827–16836. doi: 10.1074/jbc.275.22.16827. [DOI] [PubMed] [Google Scholar]
- 39.Niethammer M, Kim E, Sheng M. Interaction between the C terminus of NMDA receptor subunits and multiple members of the PSD-95 family of membrane-associated guanylate kinases. J Neurosci. 1996;16:2157–2163. doi: 10.1523/JNEUROSCI.16-07-02157.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohno H, Stewart J, Fournier MC, Bosshart H, Rhee I, Miyatake S, Saito T, Gallusser A, Kirchhausen T, Bonifacino JS. Interaction of tyrosine-based sorting signals with clathrin-associated proteins. Science. 1995;269:1872–1875. doi: 10.1126/science.7569928. [DOI] [PubMed] [Google Scholar]
- 41.Penzes P, Johnson RC, Sattler R, Zhang X, Huganir RL, Kambampati V, Mains RE, Eipper BA. The neuronal Rho-GEF Kalirin-7 interacts with PDZ domain-containing proteins and regulates dendritic morphogenesis. Neuron. 2001;29:229–242. doi: 10.1016/s0896-6273(01)00193-3. [DOI] [PubMed] [Google Scholar]
- 42.Peters PJ, Hsu VW, Ooi CE, Finazzi D, Teal SB, Oorschot V, Donaldson JG, Klausner RD. Overexpression of wild-type and mutant ARF1 and ARF6: distinct perturbations of nonoverlapping membrane compartments. J Cell Biol. 1995;128:1003–1017. doi: 10.1083/jcb.128.6.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pin JP, Duvoisin R. The metabotropic glutamate receptors: structure and functions. Neuropharmacology. 1995;34:1–26. doi: 10.1016/0028-3908(94)00129-g. [DOI] [PubMed] [Google Scholar]
- 44.Roth MG. Lipid regulators of membrane traffic through the Golgi complex. Trends Cell Biol. 1999;9:174–179. doi: 10.1016/s0962-8924(99)01535-4. [DOI] [PubMed] [Google Scholar]
- 45.Sakimura K, Kushiya E, Takahashi Y, Suzuki Y. The structure and expression of neuron-specific enolase gene. Gene. 1987;60:103–113. doi: 10.1016/0378-1119(87)90218-6. [DOI] [PubMed] [Google Scholar]
- 46.Sheng M, Sala C. PDZ domains and the organization of supramolecular complexes. Annu Rev Neurosci. 2001;24:1–29. doi: 10.1146/annurev.neuro.24.1.1. [DOI] [PubMed] [Google Scholar]
- 47.Shigemoto R, Mizuno N. Metabotropic glutamate receptors–immunocytochemical and in situ hybridization analyses. In: Ottersen OP, Storm-Mathisen J, editors. Handbook of chemical neuroanatomy, Vol 18: glutamate. Elsevier; New York: 2000. pp. 63–98. [Google Scholar]
- 48.Shigemoto R, Nomura S, Ohishi H, Sugihara H, Nakanishi S, Mizuno N. Immunohistochemical localization of a metabotropic glutamate receptor, mGluR5, in the rat brain. Neurosci Lett. 1993;163:53–57. doi: 10.1016/0304-3940(93)90227-c. [DOI] [PubMed] [Google Scholar]
- 49.Shigemoto R, Kinoshita A, Wada E, Nomura S, Ohishi H, Takada M, Flor PJ, Neki A, Abe T, Nakanishi S, Mizuno N. Differential presynaptic localization of metabotropic glutamate receptor subtypes in the rat hippocampus. J Neurosci. 1997;17:7503–7522. doi: 10.1523/JNEUROSCI.17-19-07503.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sillevis Smitt P, Kinoshita A, De Leeuw B, Moll W, Coesmans M, Jaarsma D, Henzen-Logmans S, Vecht C, De Zeeuw C, Sekiyama N, Nakanishi S, Shigemoto R. Paraneoplastic cerebellar ataxia due to autoantibodies against a glutamate receptor. N Engl J Med. 2000;342:21–27. doi: 10.1056/NEJM200001063420104. [DOI] [PubMed] [Google Scholar]
- 51.Songyang Z, Fanning AS, Fu C, Xu J, Marfatia SM, Chishti AH, Crompton A, Chan AC, Anderson JM, Cantley LC. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science. 1997;275:73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- 52.Srivastava S, Osten P, Vilim FS, Khatri L, Inman G, States B, Daly C, DeSouza S, Abagyan R, Valtschanoff JG, Weinberg RJ, Ziff EB. Novel anchorage of GluR2/3 to the postsynaptic density by the AMPA receptor-binding protein ABP. Neuron. 1998;21:581–591. doi: 10.1016/s0896-6273(00)80568-1. [DOI] [PubMed] [Google Scholar]
- 53.Tanabe Y, Masu M, Ishii T, Shigemoto R, Nakanishi S. A family of metabotropic glutamate receptors. Neuron. 1992;8:169–179. doi: 10.1016/0896-6273(92)90118-w. [DOI] [PubMed] [Google Scholar]
- 54.Tu JC, Xiao B, Naisbitt S, Yuan JP, Petralia RS, Brakeman P, Doan A, Aakalu VK, Lanahan AA, Sheng M, Worley PF. Coupling of mGluR/Homer and PSD-95 complexes by the Shank family of postsynaptic density proteins. Neuron. 1999;23:583–592. doi: 10.1016/s0896-6273(00)80810-7. [DOI] [PubMed] [Google Scholar]
- 55.Watanabe D, Inokawa H, Hashimoto K, Suzuki N, Kano M, Shigemoto R, Hirano T, Toyama K, Kaneko S, Yokoi M, Moriyoshi K, Suzuki M, Kobayashi K, Nagatsu T, Kreitman RJ, Pastan I, Nakanishi S. Ablation of cerebellar Golgi cells disrupts synaptic integration involving GABA inhibition and NMDA receptor activation in motor coordination. Cell. 1998;95:17–27. doi: 10.1016/s0092-8674(00)81779-1. [DOI] [PubMed] [Google Scholar]
- 56.Xia J, Zhang X, Staudinger J, Huganir RL. Clustering of AMPA receptors by the synaptic PDZ domain-containing protein PICK1. Neuron. 1999;22:179–187. doi: 10.1016/s0896-6273(00)80689-3. [DOI] [PubMed] [Google Scholar]
- 57.Ye B, Liao D, Zhang X, Zhang P, Dong H, Huganir RL. GRASP-1: a neuronal Ras GEF associated with the AMPA receptor/GRIP complex. Neuron. 2000;26:603–617. doi: 10.1016/s0896-6273(00)81198-8. [DOI] [PubMed] [Google Scholar]
- 58.Yoshida Y, Hamada H. Adenovirus-mediated inducible gene expression through tetracyclin-controllable transactivator with nuclear localization signal. Biochem Biophys Res Commun. 1997;230:426–430. doi: 10.1006/bbrc.1996.5975. [DOI] [PubMed] [Google Scholar]
- 59.Zhang W, Vazquez L, Apperson M, Kennedy MB. Citron binds to PSD-95 at glutamatergic synapses on inhibitory neurons in the hippocampus. J Neurosci. 1999;19:96–108. doi: 10.1523/JNEUROSCI.19-01-00096.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]