Abstract
We used serial analysis of gene expression to identify new NGF-responsive immediate early genes (IEGs) with potential roles in neuronal differentiation. Among those identified was MafK, a small Maf family basic region and leucine zipper transcriptional repressor and coactivator expressed in immature neurons. NGF treatment elevates the levels of both MafK transcripts and protein. In contrast, there is no effect on expression of the closely related MafG. Unlike many other NGF-responsive IEGs, MafK regulation shows selectivity and is unresponsive to epidermal growth factor, depolarization, or cAMP derivatives. Inhibitor studies indicate that NGF-promoted MafK regulation is mediated by an atypical isoform of PKC but not by mitogen-activated kinase kinase, phospholipase Cγ, or phosphoinositide 3′-kinase. Interference with MafK expression or activity by small interfering RNA and dominant negative strategies, respectively, suppresses NGF-promoted outgrowth and maintenance of neurites by PC12 cells and neurite outgrowth by immature telencephalic neurons. Our findings support a role for MafK as a novel regulator of neuronal differentiation.
Keywords: MafK, NGF, immediate early gene, transcription factor, differentiation, PC12 cell
Research spanning the past decade or so has provided vast insight regarding the intracellular signaling pathways that are activated by binding of neurotrophic factors to their tyrosine kinase receptors (Friedman and Greene, 1999; Kaplan and Miller, 2000). These pathways include those shared with many receptor tyrosine kinases such as those activated by phosphoinositide 3′-kinase (PI3K), phospholipase Cγ (PLCγ), and Ras (Marshall, 1995), as well as less studied mechanisms that include activation of atypical forms of PKC (Wooten et al., 2000). Less well characterized are the transcriptional regulators that are downstream of such signaling cascades and that play major roles in mediating neurotrophin actions such as promotion of neurite outgrowth. Studies of transcriptional responses to neurotrophins indicate that there is a temporal and probably causally linked progression of changes in gene expression that occur in response to neurotrophin stimulation. As first established with NGF-responsive PC12 cells (Greene and Tischler, 1976), these commence with the rapidly elevated expression of immediate early genes (many of which are transcription factors; Curran and Morgan, 1985;Greenberg et al., 1985) and progress to changes in gene expression that are apparent only after several days of neurotrophin exposure (Leonard et al., 1987). Although a number of such genes have been described, the complexity of the NGF mechanism suggests that many more remain to be recognized, including immediate early genes.
In an effort to obtain a comprehensive and quantitative view of changes in gene expression that underlie neurotrophin actions, we have applied serial analysis of gene expression (SAGE; Velculescu et al., 1995;Velculescu et al., 2000; Yamamoto et al., 2001) to analysis of PC12 cells. An initial comparison of gene expression in cells either untreated with NGF or exposed to the factor for 9 d revealed a total of >22,000 distinct transcripts of which ∼4% responded to NGF by changes in expression of sixfold or more (Angelastro et al., 2000). In the present study, we applied SAGE to detect NGF-responsive immediate early genes (IEGs) with the aim in part of identifying transcription factors that might in turn regulate expression of late genes that are involved in neuronal differentiation and neurite outgrowth. We focused our analysis on PC12 cell cultures exposed to NGF for 1 hr in complete growth medium, that is, under the same conditions for which NGF-promoted regulation of IEGs such as c-fos was first discovered (Curran and Morgan, 1985; Greenberg et al., 1985). In addition to detecting previously reported NGF-dependent IEGs, we find rapid elevation of MafK, a member of the Maf transcription factor family that is present in immature neurons during development, but without previously known function there. Our experiments indicate that NGF-dependent regulation of MafK is mediated by a mechanism dependent on activity of an atypical form of PKC. We find that MafK plays a role in neuronal outgrowth and maintenance in that these are promoted by overexpression of wild-type MafK and markedly reduced by expression of a MafK dominant negative or by small interfering RNA (siRNA)-mediated downregulation of MafK in PC12 cells and immature neurons.
MATERIALS AND METHODS
Reagents
Anisomycin and 12-O-tetradecanoyl phorbol-13-acetate (TPA) were purchased from Sigma (St. Louis, MO). Actinomycin D and PKC inhibitors bisindolylmaleimide I (GF 109203X) and 2-[1(3-dimethylaminopropyl)-5-methoxyindol-3-yl]-3-(1H-indol-3yl)maleimide (Go6983) were purchased from Calbiochem (San Diego, CA). 2-(4-Morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002) was purchased from Biomol (Plymouth Meeting, PA). Mitogen-activated kinase kinase (MEK) inhibitor 1,4-diamino-2,3-dicyano-1,4-bis(2-aminophenylthio)butadiene (UO126) was purchased from Alexis (San Diego, CA). siRNA duplexes were synthesized by Dharmacon Research (Lafayette, CO). The MafK target sequence (AA_N19) was AAG GAG GAG GUA ACU CGG CUG, was based on our rat MafK sequence, and was designed according to guidelines provided by the supplier. This sequence showed no substantial similarity to other sequences present in the National Center for Biotechnology Information (NCBI) database. The indifferent siRNA target sequence was AAG AAG CAG GAG ACC AUC GAG and was kindly provided by Dr. Carol M. Troy (Columbia University).
Cell culture
PC12 cells. These were grown as described previously on collagen-coated dishes in RPMI 1640 medium supplemented with 10% heat-inactivated horse serum and 5% fetal bovine serum (Greene et al., 1998) Human recombinant NGF was a generous gift from Genentech (San Francisco, CA).
Dissociated culture of telencephalic cells. Embryonic day 14 (E14) embryos from two Sprague Dawley rats were used for each preparation, which was modified from the protocol of Li et al. (1998). Telencephalons were harvested and trypsinized (0.05% trypsin and 0.53 mm EDTA; Invitrogen, Carlsbad, CA) for 30 min. The dissociated cells were centrifuged and resuspended in DMEM containing 6–7% FBS, 20 ng/ml basic FGF (bFGF), and 10 ng/ml epidermal growth factor (EGF) (Laywell et al., 1999) and plated on poly-l-lysine-coated 24-well dishes at 3–5 × 105 cells per well. The next day, the medium was removed, and the cells were refed with DMEM containing 20 ng/ml bFGF and 10 ng/ml EGF. The initial presence of FBS inhibits trypsin and along with bFGF and EGF permits cell division to be maintained. The removal of FBS on the second day slows cell division of non-neuronal lineage cells, but bFGF and EGF allow cell division and neuronal differentiation of progenitor cells to continue.
SAGE
SAGE libraries from 1 hr NGF-treated PC12 cells were prepared and sequenced as described previously (Angelastro et al., 2000). To match SAGE tags with transcripts of known proteins, tags were initially analyzed with the NCBI rat SAGE tag to the gene-mapping database (ftp://ncbi.nlm.nih.gov/pub/sage/map/Rn/Nla3), which matches possible 14 mer tags with known rat genes and expressed sequence tags (ESTs). With the use of sequences present in the NCBI Unigene rat database (http://www.ncbi.nlm.nih.gov/UniGene/Rn.Home.html), potential matches were further scrutinized to determine whether there was a match at the 15th base and to determine whether the matched sequence was at the most 3′ end of a known rat transcript or EST. We considered only cases in which a clear poly(A) tail and a polyadenylation signal were present at the 3′ end of the transcript or EST. Appropriate ESTs (i.e., those in a Unigene cluster in which the tag matched an EST according to the above criteria) were further analyzed by an advanced BLAST search for matches with known genes. If there was a high match between an EST and a known gene from another species, the corresponding tag was identified as originating from a rat ortholog of the known gene.
Semiquantitative and quantitative PCR
Total cellular RNA was isolated as described previously (Angelastro et al., 2000), and 5 μg was used for reverse transcription with 5′-T30NN-3′ primer using Superscript II RNase H reverse transcriptase according to the manufacturer's specifications (Invitrogen). Semiquantitative PCR was performed under the following conditions for 25 cycles: denaturation at 95°C for 30 sec, annealing at 50°C for 1 min, and extension at 72°C for 2 min, using platinum Taq polymerase (Invitrogen). Quantitative real-time PCRs were performed as described by Troy et al. (2001). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used to normalize input cDNA. Products generated during quantitative and semiquantitative PCR were run on agarose gels, excised, and sequenced to verify the identity of the originating transcript. Forward and reverse primer pairs used for quantitative and semiquantitative PCR were MafK: MA_F, 5′-CAA AGA TAC AAA AGC AGT CAC GG-3′, and MA_R, 5′-AAG TGA GTT TTC TGT CTT GTT CCT-3′; and GAPDH: GDP_F, 5′-GAAACCTGCCAAGTAGATGA-3′, and GDP_R, 5′-TCTCTCTTGCTCTCAGTATCC-3′.
Constructs
The rat MafK open reading frame was cloned using PC12 cell cDNA as a template and primers based on the known mouse sequence (GenBank accession number NM 010757) that were as follows [XbaI andSalI sites (underlined) were incorporated into the primers to facilitate subcloning]: MK_cds_F, 5′- TCT AGA AAG CGC TTG TGA AAG AGT GC-3′; and MK_cds_R, 5′- GTC GAC TGA GGA ATC TGT GCC AGG GA-3′. A SalI–XbaI fragment containing the MafK open reading frame was subcloned into the pCMS-enhanced green fluorescent protein (eGFP) vector (Clontech, Palo Alto, CA).
The dominant-negative p18 NF-E2 mutant of rat MafK was constructed by overlap extension using PCR (Kotkow and Orkin, 1995). Mutant oligonucleotide primers used in the PCR were as follows: P18MF1, 5′-GCA CAC TC G CCG CGG CTG GCT ACG C-3′; and P18MR1, 5′-GCG T AG CCA GCC GCG GCG AGT GTG C-3′ (mutant residues are underlined). cDNAs encoding the mutant p18 were subcloned into theEcoRI–XbaI site of the pCMS-eGFP vector.
Western immunoblotting
PC12 cells were harvested in Laemmli sample buffer, and protein concentration was measured by the Bradford assay (Bio-Rad, Hercules, CA). After electrophoresis on a 4–20% Tris-glycine gel (Invitrogen), the protein were transferred to a Protran nitrocellulose membrane (Schleicher & Schuell, Keene, NH). Immune complexes were visualized using an enhanced chemiluminescence detection kit (Amersham Biosciences, Piscataway, NJ). The following antisera were used: MafK, 1:200 dilution (Santa Cruz Biotechnology, Santa Cruz, CA); extracellular signal-regulated kinase 1 (ERK-1), catalog #sc-094, 1:10,000 dilution (Santa Cruz Biotechnology); MafG, 1:200 dilution, kindly provided by Dr. Volker Blank (McGill University, Montreal, Quebec, Canada) (Blank et al., 1997); and actin monoclonal antibody, kindly provided by Dr. James Lessard (Children's Hospital Medical Center, Cincinnati, OH) (Lessard, 1988). In some cases, as indicated, blots were coprobed with a mixture of ERK-1 and MafK antisera. Densitometry was performed with NIH Image 1.62 software.
Immunohistochemistry
Fluorescence immunohistochemistry was performed as described previously (Angelastro et al., 2001). Briefly, PC12 cells were fixed in 4% formaldehyde for 10 min and then, after washing with PBS, exposed to 10% nonimmune goat serum and 0.2% Triton X-100 in PBS for 30 min at room temperature. Staining for MafK, MafG, and neurofilament proteins was performed overnight at room temperature with antisera dilutions of 1:200 in PBS containing 10% nonimmune goat serum and 0.2% Triton X-100, followed by 1 hr exposure to Alexa fluorescence secondary goat anti-rabbit or anti-mouse IgG (1:4000 dilution). Neurofilament antibody was a generous gift from Dr. Ronald Liem (Columbia University) (Kaplan et al., 1991). Staining for green fluorescence protein was as above and used either monoclonal antibody (Sigma; 1:500) or polyclonal antiserum (Clontech; 1:1000).
Transient transfections
PC12 cells were plated at 80% confluency on 24-well rat tail collagen-coated plates in 500 μl of complete medium the day before transfection. Transient transfection with expression vectors was achieved by using the cationic lipid LipofectAMINE 2000, following the manufacturer's instructions (Invitrogen). Empty vector (pCMS-eGFP; Clontech) and the wild-type MafK expression vector were used at 1 μg/well; the MafK dominant negative expression vector was used at 10 μg/well. For transfection of siRNA, LipofectAMINE 2000 was diluted in serum and antibiotic-free DMEM (3 μl/50 μl) and incubated for 5 min. Empty pCMS-eGFP vector (1 μg) and siRNA (80 pmol) were mixed in DMEM (50 μl) and added to the LipofectAMINE mixture, and incubation was continued for an additional 20 min before addition to cultures and transfection as above. For neuritogenesis assays, PC12 cells were treated with NGF (50 ng/ml) 2 d after transfection and then monitored at various times thereafter for proportions of cells with processes longer than two cell body diameters (∼20 μm). For neurite stability assays, PC12 cells were pretreated with NGF for 5 d, transfected, and scored at various times thereafter for proportions of neurite-bearing cells. For dissociated E14 telencephalic cultures, the cells were transfected the day after plating with 2.0 μg of pCMS-eGFP vector or the pCMS-eGFP constructs mentioned above and 6.6 μl of ExGene500 (Fermentas, Hanover, MD) according to the manufacture's directions.
RESULTS
NGF rapidly elevates MafK expression
A SAGE library was generated and analyzed as described previously (Angelastro et al., 2000) using PC12 cells exposed for 1 hr to NGF. In total, 27,721 tags were sequenced, which represented 3575 unique transcripts whose corresponding tags were found at least twice. These data were then compared with those derived from a library of NGF-untreated cells (Angelastro et al., 2000) comprising a total of 74,880 SAGE tags representing 16,662 unique transcripts. This comparison revealed 213 transcripts that underwent a fivefold or greater change in expression in response to the 1 hr treatment with NGF. When the regulated tags were analyzed (see Materials and Methods) for correspondence with known genes, a number were found to encode known transcription factors (Table 1). Most of these transcription factors have been previously reported to be immediate early genes whose expressions are rapidly elevated in response to NGF. Although serum response factor has been found to respond to other growth factors (Spencer and Misra, 1996), to our knowledge, it has not been previously reported to be NGF-responsive. The detection of such known IEGs in our analysis supports the quality and reliability of our SAGE libraries.
Table 1.
Identification by SAGE of transcription factors that show elevated levels of transcripts in PC12 cells at 1 hr of NGF exposure
| Tag sequence | Number of tags | Identified gene | Accession number | Reference | ||
|---|---|---|---|---|---|---|
| 9 d NGF | Naive | 1 hr NGF | ||||
| GGATATGTGGT | 1 | 0 | 116 (348) | Zif268/NGFI-A | NM_012551 | Milbrandt, 1987 |
| GCCCCCTTCCA | 3 | 0 | 37 (111) | JunB | X54686 | Bartel et al., 1989 |
| GTCACATCCTC | 2 | 0 | 17 (51) | Serum response factor | NM_020493 | Sheng et al., 1988 |
| TGGAAAGTGAA | 0 | 1 | 13 (39) | c-Fos | X06769 | Greenberg et al., 1985 |
| TAGCTGAAAAA | 0 | 0 | 13 (39) | nur77/NGFI-B | U17254 | Milbrandt, 1988 |
| TTTGAGGGGTA | 0 | 0 | 10 (30) | TIS11 | AB025017 | Varnum et al., 1991 |
| TTTGTTTGTTT | 1 | 0 | 7 (21) | c-Jun | X17163 | Wu et al., 1989 |
| TCGCCGTGACT | 0 | 0 | 5 (15) | MafK | AF461686 | |
SAGE tags are given along with corresponding genes and accession numbers. The number of tags indicates the frequency a given tag was detected in analyses of 74,880 tags from untreated PC12 cells and 82,743 tags from PC12 cells treated with NGF for 9 d (both fromAngelastro et al., 2000) and from the current analysis of 27,721 tags from PC12 cells exposed to NGF for 1 hr. Values in parentheses are the calculated numbers of tags that would be anticipated if the number of tags from the 1 hr NGF library was equal to that for the untreated library. Examples of references for regulation by NGF are given.
One transcription factor not previously reported as rapidly responding to NGF or other growth factors is the Maf family member MafK (Fujiwara et al., 1993; Motohashi et al., 1997). Analysis of rat EST and the GenBank databases indicated that the regulated tag TCGCCGTGACT corresponded to a transcript encoding the rat ortholog of murine MafK (accession number NM_010757). Cloning of a cDNA encoding rat MafK (accession number AF461686) revealed that the rat and mouse coding sequences were 95% identical, and that the corresponding proteins were 87% identical.
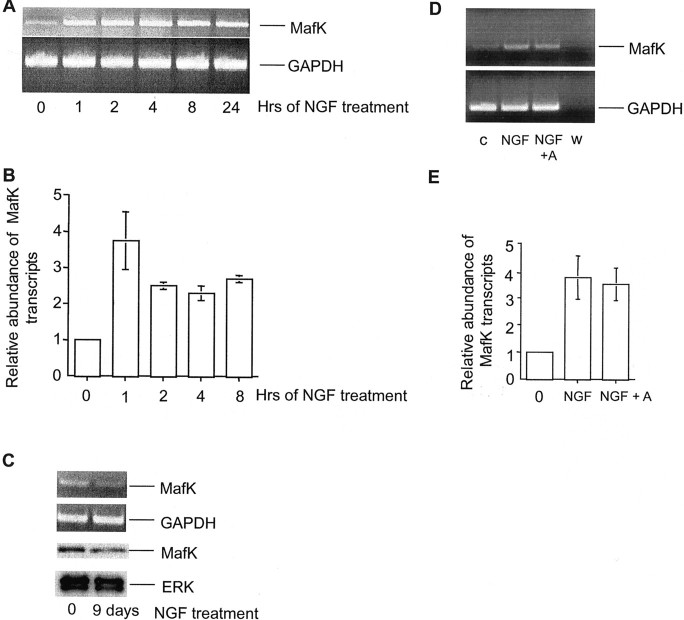
To confirm rapid regulation of MafK expression by NGF, we first performed semiquantitative reverse transcription (RT)-PCR (Fig.1A) as well as real-time PCR (Fig. 1B). These showed significant elevation of MafK transcripts within 1 hr of NGF exposure and indicated that expression remained elevated for at least 24 hr. By 9 d of NGF exposure, at which time the cells had attained a neuronal phenotype, MafK transcript levels had returned to near those in nontreated cells (Fig. 1C). This is consistent with the absence of tags corresponding to MafK in our SAGE analysis of long-term (9 d) NGF-treated cells (Table 1) and with the report by Motohashi et al. (1998) that 5 d of NGF treatment does not affect PC12 cell levels of MafK transcripts. The absence of SAGE tags for MafK in our previous SAGE analyses indicates that the relative abundance of this transcript is <0.001% in naive and NGF-primed cells and that its detection would require analysis of a larger number of SAGE tags.
Fig. 1.
MafK is an immediate early gene upregulated by NGF in PC12 cells. A, B, Time dependence of effect of NGF on MafK mRNA levels in PC12 cells. A, PC12 cells were treated with NGF for the indicated times, and then total RNA was isolated and converted to cDNA by RT-PCR. Semiquantitative PCRs were performed with 25 cycles for MafK and 20 cycles for GAPDH.B, RT reactions were subjected to quantitative real-time PCR. The relative abundances of MafK transcripts were measured as described in Materials and Methods and were normalized to GAPDH mRNA. Data represent mean values, and error bars indicate SE for measurements from four independent experiments. C, MafK mRNA and protein levels in PC12 cells before and after 9 d of NGF treatment. Cells were harvested, and total RNA or total protein was isolated and subjected to RT-PCR (top 2 panels) or immunoblotting (bottom 2 panels), respectively. Immunoblots were coprobed with MafK and ERK antisera. Comparable results were achieved in three independent experiments. D, E, Effect of the protein synthesis inhibitor anisomycin on regulation of MafK mRNA by NGF. PC12 cells were treated for 1 hr with NGF, and total RNA was isolated and subjected to the RT reaction. Anisomycin (A; 10 μm), when used, was added to cultures 1 hr before NGF. Semiquantitative (D) and quantitative real-time (E) PCRs were performed. Sample wcontained water instead of the RT reaction. Data in Erepresent mean values, and error bars indicate SE for measurements from four independent experiments. c, Control, no additive.
The data in Figure 1, D and E, show that the rapid NGF-dependent elevation of MafK transcripts was not affected by the presence of the protein synthesis inhibitor anisomycin. Thus, the effect NGF on MafK expression does not appear to be mediated byde novo synthesis of regulatory proteins.
We next examined the effect of NGF on levels of MafK protein by Western immunoblotting. This revealed that MafK protein levels increase by 1 hr of NGF treatment, are maximally increased by 2 hr, and remain elevated for at least 24 hr (Fig.2A). Similarly to the behavior found with MafK transcripts, the increase in MafK protein was not sustained at 9 d of NGF exposure (Fig. 1C). The rapid effect of NGF on MafK protein levels was blocked by actinomycin D, suggesting a requirement for gene transcription (Fig.2B). This observation, together with the rapidity of the effect and its blockade by anisomycin, suggests that MafK is an NGF-responsive immediate early gene.
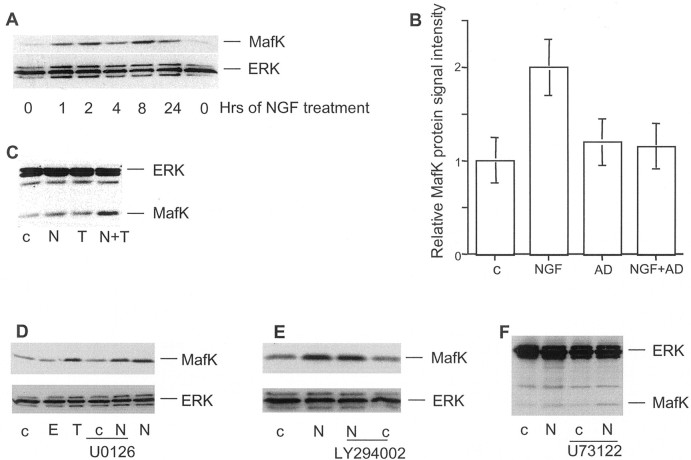
Fig. 2.
Mechanisms of MafK protein regulation by NGF. A, Time dependence of effect of NGF on MafK protein levels in PC12 cells. Total cellular protein isolated from PC12 cells was analyzed by Western immunoblotting with ERK and MafK antisera. Comparable results were achieved in two or three independent experiments. B, Effect of the transcriptional inhibitor actinomycin D on regulation of MafK protein by NGF. Cells were pretreated for 90 min with 10 μm actinomycin D (AD) and then for 2 hr with NGF. Total cell protein was subjected to Western immunoblotting with ERK and MafK antisera. Band intensities were determined by densitometry (using shorter times of exposure than shown and for which bands were not at a level of saturation), and values were normalized to ERK signals. Values are mean ± SEM (n = 9). C, Effect of NGF and TPA on MafK protein levels. PC12 cells were treated with no additive (c) or NGF, 50 nm TPA, or both for 2 hr, and then Western immunoblotting was performed with ERK and MafK antisera. Comparable results were achieved in two or three independent experiments. D–F, Effect of signal transduction inhibitors and activators on NGF-promoted MafK levels. PC12 cells were treated for 2 hr with no additive (c), EGF (E), 50 nm TPA (T), or NGF (N) as indicated. Inhibitors (MEK inhibitor U1026, PI3K inhibitor LY294002, and PLCγ inhibitor U73122) were added 1 hr before NGF where indicated. Total cellular protein was isolated and subjected to Western immunoblotting with ERK and MafK antisera. Comparable results were achieved in two or three independent experiments.
The effect of NGF on MafK is selective
To assess the specificity of the effect on MafK, we examined the expression of MafG, a closely related member of the Maf family (Shavit et al., 1998). Western immunoblots revealed little if any effect of NGF on levels of MafG protein after 2 hr of treatment (Fig.3E).
Fig. 3.
Role of PKC in NGF-promoted regulation of MafK elevation and lack of effect of NGF MafG levels.A–C, PKC inhibitors GF 109203X and Go6983 inhibit NGF-promoted MafK elevation at concentrations that block novel and atypical PKC isoforms. PC12 cells were pretreated for 1 hr with 10 nm to 10 μm GF 109203X and 10 nmto 1 μm Go6983 where indicated and then with NGF for 2 hr (in the continued presence of the inhibitors). Cells were harvested, and total cellular protein was subjected to Western coimmunoblotting with ERK and MafK antisera. Comparable results were achieved in two or three independent experiments. D, Depletion of classic and novel forms of PKC by pretreatment with 50 nm TPA does not inhibit NGF-promoted MafK regulation. PC12 cell cultures were pretreated for 2 d with 10 nm to 1 μmTPA before 2 hr of NGF treatment. Cells were collected, and total cellular protein was subjected to Western coimmunoblotting with ERK and MafK antisera. Comparable results were achieved in two or three independent experiments. E, The same membrane that was used in A was stripped and reprobed with actin antibody and MafG antiserum. Comparable results were achieved in two or three independent experiments. c, Control, no additive;N, NGF.
To explore the selectivity of MafK regulation, PC12 cell cultures were treated with 100 ng/ml EGF, 50 nm TPA, depolarizing (46 mm) levels of K+, 1 μm ionomycin, or 100 μm8-(4-chlorophenylthio)-cAMP for 2 hr. Of these, only TPA led to increased MafK protein expression (Fig. 2C; data not shown). The TPA effect appeared to be approximately additive with that of NGF (Fig. 2C). Thus, unlike many NGF-sensitive IEGs that also respond to EGF and other extracellular stimulants, MafK appears to be relatively selective in its response.
NGF regulates MafK expression via an atypical PKC
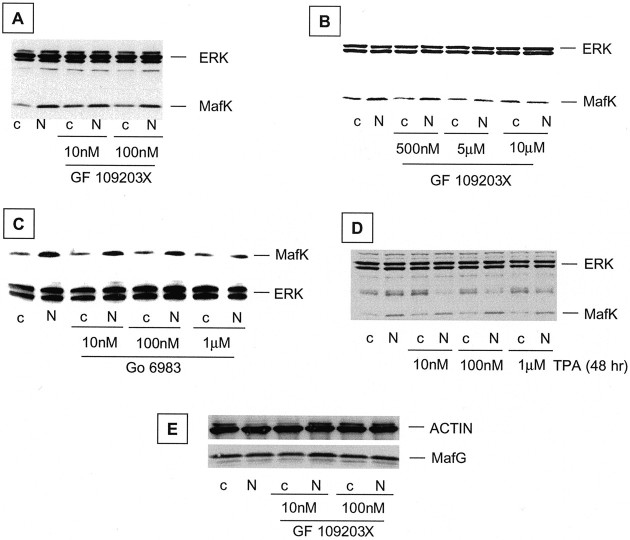
We next used inhibitors of specific signaling pathways to uncover the mechanism by which NGF regulates MafK expression. The MEK inhibitor U-1026 (10 μm) (Fig. 2D) (Favata et al., 1998), the PI3K inhibitor LY294002 (50 μm) (Fig. 2E) (Vlahos et al., 1994), and the PLCγ inhibitor U-73122 (5 μm) (Fig.2F) (Bleasdale et al., 1989) were each without effect on basal or NGF-stimulated MafK levels (although they blocked other cellular responses to NGF as anticipated). An additional group of signaling molecules that have been implicated in responses to NGF are various isoforms of PKC (Wooten et al., 1997). PC12 cells were exposed to NGF in the presence of various concentrations (10 nm to 10 μm) of the PKC inhibitors GF 109203X (Martiny-Baron et al., 1993) and Go6983 (Douglas et al., 1999) and assessed for expression of MafK protein with or without 2 hr of NGF treatment. There was no effect on NGF-promoted elevation of MafK at 10–100 nm GF 109203X (Fig.3A). These concentrations should inhibit TPA-dependent “classical” PKC isoforms in intact cells (Martiny-Baron et al., 1993, Uberall et al., 1997, 1999). In contrast, there was partial inhibition at 500 nm and near-complete inhibition at 5–10 μm GF 109203X (Fig. 3B). The latter concentrations inhibit both the TPA-dependent “novel” and the TPA-independent “atypical” forms of PKC (Martiny-Baron et al., 1993, Uberall et al., 1997, 1999; Gerstin et al., 1998; Zheng et al., 2000; Tsuji et al., 2001). Similarly, there was little effect of Go6983 at 10 and 100 nm (which block classical PKC activity), whereas 1 μm, which inhibits both novel and atypical PKC forms (Gschwendt et al., 1996; Douglas et al., 1999), effectively suppressed the NGF-dependent MafK protein response (Fig. 3C). To distinguish between the roles of the novel and atypical PKC subfamilies, cultures were pretreated with 10 nm to 1 μm TPA for 2 d to deplete the TPA-dependent classical and novel isoforms (Roivainen et al., 1993; Wooten et al., 1994) and then exposed to NGF for 2 hr. As shown in Figure 3D, this pretreatment did not diminish the ability of NGF to promote elevation of MafK protein. Together, these findings favor a mechanism by which rapid NGF-dependent elevation of MafK is mediated by an atypical form of PKC.
MafK participates in NGF-promoted neuritogenesis
MafK is expressed in the embryonic and mature nervous systems and on this basis has been suggested to play a role in neuronal development (Motohashi et al., 1996; Motohashi et al., 1997; Shavit et al., 1998;Onodera et al., 2000). We used several different approaches to evaluate the potential role of MafK in NGF-promoted neuritogenesis. First, we constructed expression vectors of wild-type rat MafK and a mutant form in which three conserved amino acids (Lys-59, Asn-60, and Arg-61) in the basic domain were changed to Ala residues (Kotkow and Orkin, 1995). The latter construct encodes a mutant protein that (along with its homodimers and heterodimers) is unable to bind DNA and that therefore acts as a dominant negative mutant. For instance, in murine erythroleukemia cells, this mutant reduced MafK-promoted globin gene expression (Kotkow and Orkin, 1995). The expression vectors along with an empty control vector (all of which also express eGFP) were transfected into PC12 cells, and 2 d later, the cultures were exposed to NGF. At various times, the transfected cells (identified by GFP expression) were scored for the presence of neurites. Transfection with wild-type MafK alone did not cause neurite outgrowth in the absence of NGF (data not shown). Expression of the transfected protein was verified by immunofluorescence staining, which showed greatly elevated levels of MafK signal in nuclei of transfected cells (data not shown). In the presence of NGF, cells transfected with wild-type MafK exhibited a significantly faster rate of neurite generation than cells transfected with the empty vector (Fig.4A). In contrast, cells transfected with the dominant negative (d/n) form of MafK showed a markedly reduced rate of neurite genesis when compared with control-transfected cells (Fig. 4A).
Fig. 4.
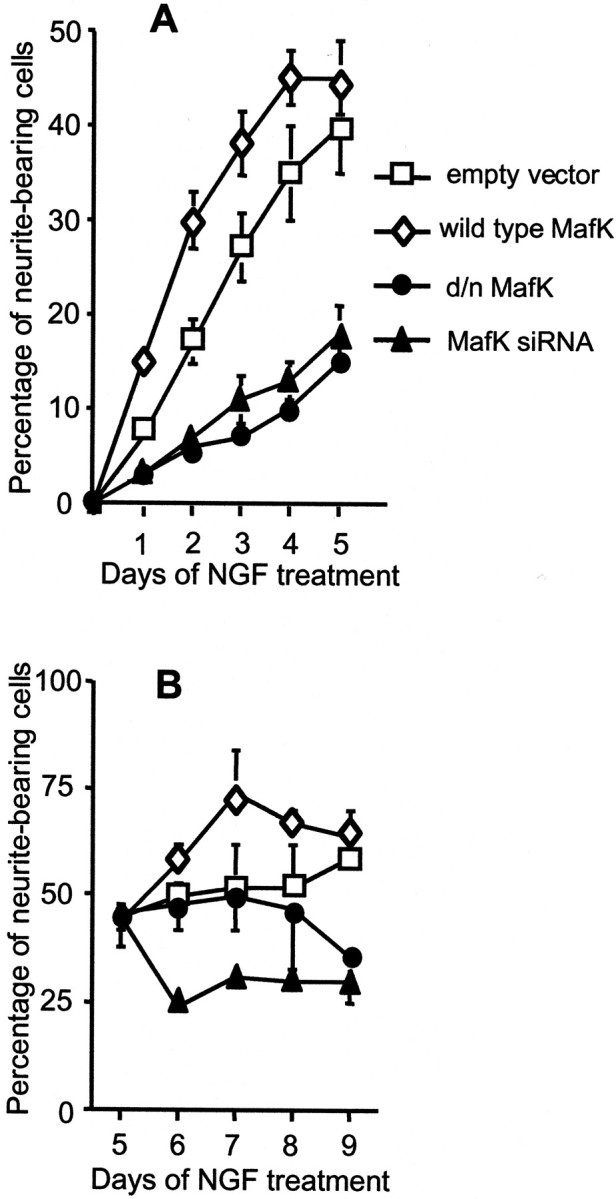
Functional role of MafK in NGF-promoted neurite genesis (A) and maintenance (B). A, PC12 cells were transfected with the indicated constructs as described in Materials and Methods. Two days later, the cells were treated with NGF. GFP-positive cells were scored for neurite outgrowth on the following 5 d. Data are mean ± SEM (n = 4–12 independent experiments, in each of which at least 200 cells were scored).B, PC12 cells were treated with NGF for 5 d and then transfected with the indicated constructs. Proportions of GFP-expressing cells were scored for the presence of neurites on the following 4 d. Data are mean ± SEM (n = 4 independent experiments, in each of which at least 200 cells were scored).
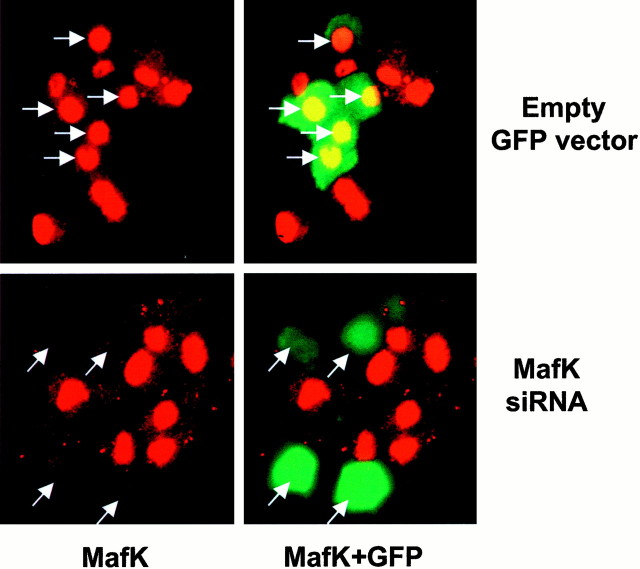
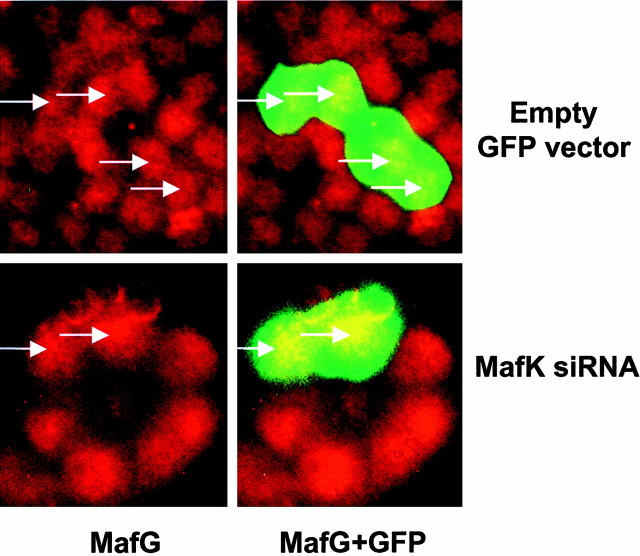
As an alternative strategy to assess the importance of MafK in neurite genesis, we used a siRNA construct designed to specifically reduce the cellular levels of MafK transcripts. Figure5 shows that cells cotransfected with the siRNA and a construct expressing eGFP showed little or no immunostaining for MafK. This effect was evident by 24 hr after transfection and persisted for 3 d. By 5 d after transfection, the suppression was only partial; by 7 d, control- and siRNA-transfected cells were indistinguishable (data not shown). To control for the specificity of the siRNA, transfected cells were also immunostained for the closely related family member MafG. As shown in Figure 6, the MafK siRNA had no detectable effect on expression of MafG. Such selectivity is consistent with previous reports regarding the specificity of siRNA constructs in mammalian cells (Elbashir et al., 2001). When cells transfected with the MafK siRNA construct were assessed for response to NGF, it was observed that the rate of neurite genesis was greatly diminished in comparison with control cells and to a degree comparable with that observed with the d/n MafK construct (Fig. 4A). In contrast, an indifferent siRNA construct (which shows no homology with known rat sequences) did not reduce neurite outgrowth (data not shown), thus indicating that the actions of the MafK-directed siRNA are not attributable to nonspecific interference with growth of neurites.
Fig. 5.
Depletion of MafK by exposure to MafK siRNA. PC12 cells were cotransfected with empty GFP-expressing vector with or without MafK siRNA. Two days after transfection, cell cultures were immunostained with antiserum against MafK and visualized by fluorescence microscopy for MafK (red) or GFP (green). Left panels, Staining for MafK alone; right panels, same fields with merged images for MafK and GFP. Arrows indicate positions of GFP-expressing cells.
Fig. 6.
MafK siRNA does not affect expression of MafG. The experiment was performed as in Figure 5, except that the cells were stained with MafG antiserum rather than with MafK antiserum.Arrows indicate positions of GFP-expressing cells.
We next wished to determine whether MafK plays a role in stability of existing neurites. PC12 cells were pretreated with NGF for 5 d to promote neurite outgrowth. The cells were then transfected with expression vectors containing wild-type or dominant negative MafK or with the MafK siRNA, and the transfected cells (as well as those transfected with an empty vector) were assessed over the next 4 d for the presence of neurites (Fig. 4B). Expression of excess wild-type MafK elevated the rate of neurite outgrowth, whereas in contrast, the MafK siRNA led to loss of neurites. The dominant negative construct also caused a decrease in neurite numbers, but this was delayed by several days compared with the effect of the MafK siRNA, possibly because of the need for accumulation of sufficient levels of the mutant protein. Together, these data support a major role for MafK in NGF-promoted generation and maintenance of neurites.
To determine whether MafK may also play a role in neurite outgrowth by normal immature neurons (in which it is expressed), we next examined the effects of our constructs on cells in cultures of rat E14 telencephalon that contain neural progenitor cells and immature neurons. We reasoned that in such cultures, many of the immature neurons would be generating processes for the first time, and thus the neuritogenesis would be in some ways comparable with the de novo generation of neurites by NGF-treated PC12 cells. The cultures were transfected with empty eGFP vector, wild-type MafK, d/n MafK, or MafK siRNA and 30 hr later were assessed for expression of GFP, the neuronal marker neurofilament M (NFM), and neurites. Overexpression of wild-type MafK elevated the proportion of NFM-positive cells with neurites, whereas d/n MafK and MafK siRNA both decreased the number of NFM-positive cells with neurites by approximately twofold (Fig. 7). Staining with MafK antiserum verified expression of MafK in nuclei of neurite-bearing cells in control cultures and overexpression of wild-type MafK and depletion of endogenous MafK by MafK siRNA in transfected cells (data not shown).
Fig. 7.
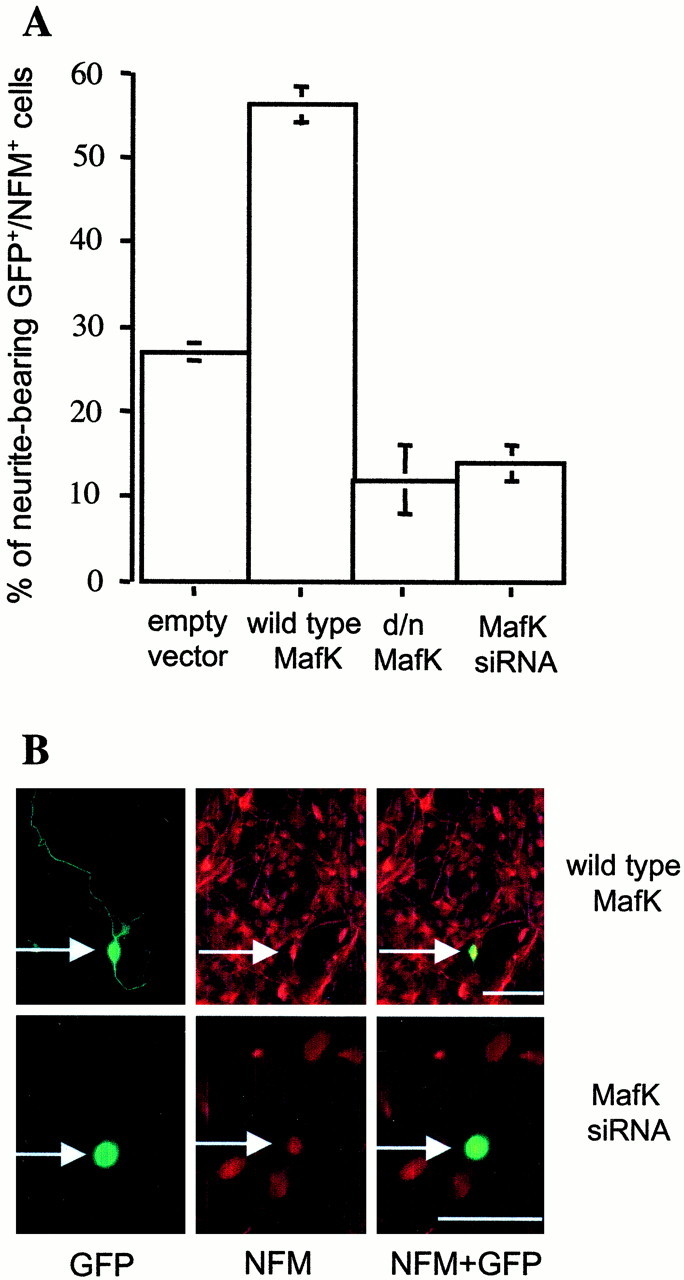
Functional role of MafK in neurite outgrowth by telencephalic neurons. Cells cultured from E14 rat telencephalon were transfected with empty vector, wild-type MafK, d/n MafK, or MafK siRNA plus empty vector. Thirty hours after transfection, cultures were immunostained with antisera against GFP (green) and NFM (red). A, Percentage of neurite-bearing GFP- and NFM-positive cells. Values are averages of two counts representing >50 cells each. Error bars indicate range of counts. Comparable results were achieved in this and an independent experiment by scoring the proportion of neurite-bearing GFP-positive cells. B, Examples of morphology of NFM-positive cells transfected with wild-type MafK and MafK siRNA. Left panels, Staining for GFP; middle panels, staining for NFM; right panels, merged images.Arrows indicate the same cell (left toright) stained with GFP and NFM. Scale bars, 50 μm.
DISCUSSION
MafK is an NGF-regulated IEG
Because of the importance of transcriptional regulation in the NGF mechanism (Greene et al., 1982; Segal and Greenberg, 1996), we aimed to identify and functionally characterize new NGF-responsive early genes. Through application of SAGE technology, we found that MafK transcripts and protein are elevated within 1–2 hr of NGF exposure. Such behavior in many ways resembles that of previously described NGF-regulated IEGs (Herschman, 1991; Segal and Greenberg, 1996). For instance, upregulation of MafK transcripts occurs within 1 hr, is independent of protein synthesis, and is blocked by inhibition of transcription. Also, as do at least several NGF-regulated IEG transcription factors (Sassone-Corsi et al., 1989; Schlingensiepen et al., 1994; Qu et al., 1998; Levkovitz et al., 2001) MafK appears to play an important role in promotion of neurite outgrowth. On the other hand, MafK also displayed characteristics that distinguish it from other IEGs. Thus, there was a considerable level of basal MafK expression without NGF. Furthermore, unlike many NGF-responsive IEGs whose levels are transiently elevated over a time course of hours, MafK remains upregulated for at least 1 d after NGF exposure and returns to baseline levels only after longer-term NGF exposure.
An additional important distinction between MafK and other described NGF-responsive IEGs regards specificity. It has been noted most NGF-responsive IEGs also respond to a variety of other growth factors and extracellular signals in both neuronal and non-neuronal backgrounds (Herschman, 1991; Hill and Treisman, 1995). This has raised the question of how specificity of the NGF response is achieved, which has in turn raised the possibility that additional IEGs remained to be discovered that are specific to NGF and perhaps other agents that promote neuronal differentiation. We found that in contrast to many IEGs, MafK does not respond to EGF (to which PC12 cells respond but not with neuronal differentiation), elevated K+, a calcium ionophore, or a cAMP derivative. This observation indicates that there is at least a degree of specificity to the MafK response and thus supports the possibility that MafK regulation represents one of the ways by which specificity of neurotrophic factor signaling is achieved. The urokinase plasminogen activator receptor is another example of an IEG that is differentially induced by NGF compared with EGF and that plays a required role in NGF-promoted neuritogenesis (Farias-Eisner et al., 2000).
Regulation of MafK by NGF appears to be mediated by an atypical PKC
Application of well characterized inhibitors appeared to rule out MEK, PI3K, and PLCγ as mediators of NGF-promoted MafK elevation. These molecules participate in regulation of many NGF-responsive IEGs examined to date (Segal and Greenberg, 1996). We also evaluated involvement of a PKC-dependent mechanism. PC12 cells express multiple PKC isoforms, including those in the classical, novel, and atypical families (Quest, 1996; Wooten et al., 1997; Moscat and Diaz-Meco, 2000). Although acute TPA treatment increased MafK levels, this appeared to be additive with NGF and to be mediated by classical or novel PKC isoforms (which are TPA-sensitive). In contrast, our findings with PKC inhibitors and TPA pretreatment support the hypothesis that regulation of MafK by NGF is mediated by an atypical PKC. In conformity with this, it was reported that NGF promotes transient activation of atypical PKC isoforms ι and ζ in PC12 cells (Coleman and Wooten, 1994; Wooten et al., 2000) and that downregulation of PKC-ζ with an antisense oligonucleotide attenuates NGF-induced neurite outgrowth (Coleman and Wooten, 1994). Moreover, overexpression of atypical PKC-ι and -ζ, as in the case of MafK seen here, potentiates NGF-promoted neurite outgrowth (Wooten et al., 1999). Finally, recent evidence indicates that NGF activates atypical PKCs by an src-dependent mechanism that would not appear to include MEK, PI3K, or PLCγ (Wooten et al., 2001).
It has long been clear that there are elements of the NGF signaling pathway that are not mediated by ras/ERK-, PI3K-, or PLCγ-dependent mechanisms and that might in part provide a degree of specificity to neurotrophin actions (Greene and Kaplan, 1995; Peng et al., 1995). The indications that MafK is regulated by NGF via an atypical PKC and by a mechanism that is not blocked by inhibitors of the aforementioned signaling molecules makes it a potential and attractive candidate in this regard.
MafK plays a role in neurite outgrowth and maintenance
Because of its rapid response to NGF, regulation via atypical PKCs that appear to be required for NGF-mediated differentiation, identification as a transcription factor, and presence in the developing nervous system, we evaluated whether MafK plays a role in NGF-promoted neurite outgrowth as well as in outgrowth of neurites by immature neurons. Overexpression of MafK elevated the rate of neuritogenesis by NGF-treated PC12 cells and immature neurons cultured from E14 rat brain telencephalon. In contrast, overexpression of a dominant negative MafK or downregulation of MafK by siRNA significantly reduced neurite production in both systems. In addition, interference with MafK expression decreased the stability of preexisting PC12 cell neurites.
The success of siRNA in downregulating MafK was particularly encouraging with respect to the use of this technique in vertebrate neuronal cells. Although the transfection efficiency in our cultures was too low to permit direct quantitative evaluation of effects on MafK protein, immunostaining indicated reduction of MafK expression to undetectable levels. In contrast, there was no detectable effect on the closely related MafG protein. Moreover, an irrelevant siRNA did not block neurite outgrowth, thus indicating that this approach does nonspecifically interfere with neuritogenesis. It was significant that the same outcomes were achieved with the dominant negative and siRNA approaches.
Our findings are consistent with current knowledge about MafK. Maf proteins are structurally similar to the founding family member v-Maf, the transforming component of avian musculoaponeurotic fibrosarcoma virus AS42 (Nishizawa et al., 1989). Maf family members share a relatively well conserved basic region and leucine zipper (b-Zip) motif that promotes DNA binding and dimer formation (Motohashi et al., 1997). In contrast to large Mafs, small Mafs (MafF, MafG and MafK) lack transcriptional activation domains (Motohashi et al., 1997, 2000) and thus function either as heterodimeric partners for non-Maf b-Zip proteins to form transcription-activating complexes or as homodimers or heterodimers with other small Mafs to form transcriptional repressors (Motohashi et al., 1997). It has been suggested that the propensity of MafK to form either transcription-activating or repressor complexes depends on the relative ratio between available small Mafs and alternative b-Zip partners (Motohashi et al., 2000; Yoh et al., 2001). In this context, it may be relevant that MafK can heterodimerize with partners such as c-Fos and c-Jun (Motohashi et al., 1997), which are also IEG targets of NGF.
Various evidence implicates MafK as a required factor in erythroid cell differentiation (Motohashi et al., 1997, 2000). However, little else is known about its potential role in other tissue or cell types. Developmental expression of murine MafK and MafG has been described previously (Motohashi et al., 1996; Shavit et al., 1998; Katsuoka et al., 2000). Of particular pertinence here, MafK was strongly expressed in neuronal cells at E13 and was broadly expressed in postnatal neurons (Motohashi et al., 1996). Significantly, neuronal expression of MafK is driven by a different promoter than in mesodermal and cardiac tissues (Motohashi et al., 1996; Katsuoka et al., 2000). As for MafG, expression is present throughout most of the embryo at E7.5–E14.5, including brain (Shavit et al., 1998).
MafK null mice showed no discernable phenotype (Kotkow and Orkin, 1996; Shavit et al., 1998), whereas mafG nulls showed, in addition to mild thrombocytopenia, mild deficits in restricted motor skills (Shavit et al., 1998). Animals null for both genes exhibited perinatal lethality (Onodera et al., 2000), and the status of their nervous system was not reported. However,mafG−/−:: mafK−/+animals do survive and exhibit a neurological phenotype that is greatly exaggerated in comparison with mafG nulls. This includes hind leg clasping and severe motor ataxia with intermittently spastic hindlimbs (Onodera et al., 2000). Together with the expression data, these findings indicate a role for MafK in neuronal differentiation and function in vivo and thus support the relevance of ourin vitro studies.
The observation that loss of MafK in mice shows no phenotype on its own, but that loss of one copy greatly exacerbates the neurological phenotype of mafG null mice, indicates that loss of MafK may be partially compensated by MafG and vice versa. This may explain in part why interference with MafK expression and activity does not totally abolish neurite outgrowth. In this context, it is intriguing that NGF regulates MafK but not MafG expression.
A final point of potential importance is that besides NGF, TPA also rapidly elevates mafK expression. This indicates that in addition to atypical PKC isoform(s), MafK is subject to regulation by one or more classical or novel PKCs. Thus, a variety of physiological circumstances that activate PKCs may also induce MafK. In this respect, it is of interest that classical and novel PKC activation by TPA, like MafK overexpression, significantly potentiates NGF-promoted neuritogenesis (Burstein et al., 1982; Hall et al., 1988).
Footnotes
This work was supported in part by grants from the National Institute of Neurological Disorders and Stroke, National Institutes of Health. We thank Claudine Bitel for outstanding technical assistance.
Correspondence should be addressed to Lloyd A. Greene, Department of Pathology, Columbia University College of Physicians and Surgeons, 630 West 168th Street, New York, NY 10032. E-mail: Lag3@columbia.edu.
Dr. Töröcsik's present address: Department of Medical Biochemistry, Semmelweis University, Budapest, Hungary. On leave from the Department of Biology, University Medical School of Pecs, Pecs, Hungary.
REFERENCES
- 1.Angelastro JM, Klimaschewski L, Tang S, Vitolo OV, Weissman TA, Donlin LT, Shelanski ML, Greene LA. Identification of diverse nerve growth factor-regulated genes by serial analysis of gene expression (SAGE) profiling. Proc Natl Acad Sci USA. 2000;97:10424–10429. doi: 10.1073/pnas.97.19.10424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angelastro JM, Moon NY, Liu DX, Yang AS, Greene LA, Franke TF. Characterization of a novel isoform of caspase-9 that inhibits apoptosis. J Biol Chem. 2001;276:12190–12200. doi: 10.1074/jbc.M009523200. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP, Sheng M, Lau LF, Greenberg ME. Growth factors and membrane depolarization activate distinct programs of early response gene expression: dissociation of fos and jun induction. Genes Dev. 1989;3:304–313. doi: 10.1101/gad.3.3.304. [DOI] [PubMed] [Google Scholar]
- 4.Blank V, Kim MJ, Andrews NC. Human MafG is a functional partner for p45 NF-E2 in activating globin gene expression. Blood. 1997;89:3925–3935. [PubMed] [Google Scholar]
- 5.Bleasdale JE, Bundy GL, Bunting S, Fitzpatrick FA, Huff RM, Sun FF, Pike JE. Inhibition of phospholipase C dependent processes by U-73, 122. Adv Prostaglandin Thromboxane Leukot Res. 1989;19:590–593. [PubMed] [Google Scholar]
- 6.Burstein DE, Blumberg PM, Greene LA. Nerve growth factor-induced neuronal differentiation of PC12 pheochromocytoma cells: lack of inhibition by a tumor promoter. Brain Res. 1982;247:115–119. doi: 10.1016/0006-8993(82)91033-2. [DOI] [PubMed] [Google Scholar]
- 7.Coleman ES, Wooten MW. Nerve growth factor-induced differentiation of PC12 cells employs the PMA-insensitive protein kinase C-zeta isoform. J Mol Neurosci. 1994;5:39–57. doi: 10.1007/BF02736693. [DOI] [PubMed] [Google Scholar]
- 8.Curran T, Morgan JI. Superinduction of c-fos by nerve growth factor in the presence of peripherally active benzodiazepines. Science. 1985;229:1265–1268. doi: 10.1126/science.4035354. [DOI] [PubMed] [Google Scholar]
- 9.Douglas DN, Fink HS, Ridgway ND, Cook HW, Byers DM. Myristoylated alanine-rich C-kinase substrate is phosphorylated and translocated by a phorbol ester-insensitive and calcium-independent protein kinase C isoform in C6 glioma cell membranes. Biochim Biophys Acta. 1999;1448:439–449. doi: 10.1016/s0167-4889(98)00161-x. [DOI] [PubMed] [Google Scholar]
- 10.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 11.Farias-Eisner R, Vician L, Silver A, Reddy S, Rabbani SA, Herschman HR. The urokinase plasminogen activator receptor (UPAR) is preferentially induced by nerve growth factor in PC12 pheochromocytoma cells and is required for NGF-driven differentiation. J Neurosci. 2000;20:230–239. doi: 10.1523/JNEUROSCI.20-01-00230.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Favata MF, Horiuchi KY, Manos EJ, Daulerio AJ, Stradley DA, Feeser WS, Van Dyk DE, Pitts WJ, Earl RA, Hobbs F, Copeland RA, Magolda RL, Scherle PA, Trzaskos JM. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J Biol Chem. 1998;273:18623–18632. doi: 10.1074/jbc.273.29.18623. [DOI] [PubMed] [Google Scholar]
- 13.Friedman WJ, Greene LA. Neurotrophin signaling via Trks and p75. Exp Cell Res. 1999;253:131–142. doi: 10.1006/excr.1999.4705. [DOI] [PubMed] [Google Scholar]
- 14.Fujiwara KT, Kataoka K, Nishizawa M. Two new members of the maf oncogene family, mafK, and mafF, encode nuclear b-Zip proteins lacking putative trans-activator domain. Oncogene. 1993;8:2371–2380. [PubMed] [Google Scholar]
- 15.Gerstin EH, Jr, McMahon T, Dadgar J, Messing RO. Protein kinase C delta mediates ethanol-induced up-regulation of L-type calcium channels. J Biol Chem. 1998;273:16409–16414. doi: 10.1074/jbc.273.26.16409. [DOI] [PubMed] [Google Scholar]
- 16.Gschwendt M, Dietrich S, Rennecke J, Kittstein W, Müller H-J, Johannes F-J. Inhibition of protein kinase C mu by various inhibitors. Differentiation from protein kinase c isoenzymes. FEBS Lett. 1996;392:77–80. doi: 10.1016/0014-5793(96)00785-5. [DOI] [PubMed] [Google Scholar]
- 17.Greenberg ME, Greene LA, Ziff EB. Nerve growth factor and epidermal growth factor induce rapid transient changes in proto-oncogene transcription in PC12 cells. J Biol Chem. 1985;260:14101–14110. [PubMed] [Google Scholar]
- 18.Greene LA, Kaplan DR. Early events in neurotrophin signalling via Trk and p75 receptors. Curr Opin Neurobiol. 1995;5:579–587. doi: 10.1016/0959-4388(95)80062-x. [DOI] [PubMed] [Google Scholar]
- 19.Greene LA, Tischler AS. Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc Natl Acad Sci USA. 1976;73:2424–2428. doi: 10.1073/pnas.73.7.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greene LA, Burstein DE, Black MM. The role of transcription-dependent priming in nerve growth factor promoted neurite outgrowth. Dev Biol. 1982;91:305–316. doi: 10.1016/0012-1606(82)90037-9. [DOI] [PubMed] [Google Scholar]
- 21.Greene LA, Farinelli SE, Cunningham ME, Park DS (1998) Culture and experimental use of the PC12 rat pheochromocytoma cell line. In: Culturing nerve cells (Banker G, Goslin K, eds), Ed 2, pp 161–188. Cambridge MA: MIT.
- 22.Hall FL, Fernyhough P, Ishii DN, Vulliet PR. Suppression of nerve growth factor-directed neurite outgrowth in PC12 cells by sphingosine, an inhibitor of protein kinase C. J Biol Chem. 1988;263:4460–4466. [PubMed] [Google Scholar]
- 23.Herschman HR. Primary response genes induced by growth factors and tumor promoters. Annu Rev Biochem. 1991;60:281–319. doi: 10.1146/annurev.bi.60.070191.001433. [DOI] [PubMed] [Google Scholar]
- 24.Hill CS, Treisman R. Transcriptional regulation by extracellular signals: mechanisms and specificity. Cell. 1995;80:199–211. doi: 10.1016/0092-8674(95)90403-4. [DOI] [PubMed] [Google Scholar]
- 25.Kaplan DR, Miller FD. Neurotrophin signal transduction in the nervous system. Curr Opin Neurobiol. 2000;10:381–391. doi: 10.1016/s0959-4388(00)00092-1. [DOI] [PubMed] [Google Scholar]
- 26.Kaplan MP, Chin SS, Macioce P, Srinawasan J, Hashim G, Liem RK. Characterization of a panel of neurofilament antibodies recognizing N-terminal epitopes. J Neurosci Res. 1991;30:545–554. doi: 10.1002/jnr.490300312. [DOI] [PubMed] [Google Scholar]
- 27.Katsuoka F, Motohashi H, Onodera K, Suwabe N, Engel JD, Yamamoto M. One enhancer mediates mafK transcriptional activation in both hematopoietic and cardiac muscle cells. EMBO J. 2000;19:2980–2991. doi: 10.1093/emboj/19.12.2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kotkow KJ, Orkin SH. Dependence of globin gene expression in mouse erythroleukemia cells on the NF-E2 heterodimer. Mol Cell Biol. 1995;15:4640–4647. doi: 10.1128/mcb.15.8.4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kotkow KJ, Orkin SH. Complexity of the erythroid transcription factor NF-E2 as revealed by gene targeting of the mouse p18 NF-E2 locus. Proc Natl Acad Sci USA. 1996;93:3514–3518. doi: 10.1073/pnas.93.8.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laywell ED, Kukekov VG, Steindler DA. Multipotent neurospheres can be derived from forebrain subependymal zone and spinal cord of adult mice after protracted postmortem intervals. Exp Neurol. 1999;156:430–433. doi: 10.1006/exnr.1999.7029. [DOI] [PubMed] [Google Scholar]
- 31.Leonard DG, Ziff EB, Greene LA. Identification and characterization of mRNAs regulated by nerve growth factor in PC12 cells. Mol Cell Biol. 1987;7:3156–3167. doi: 10.1128/mcb.7.9.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lessard JL. Two monoclonal antibodies to actin: one muscle selective and one generally reactive. Cell Motil Cytoskeleton. 1988;10:349–362. doi: 10.1002/cm.970100302. [DOI] [PubMed] [Google Scholar]
- 33.Levkovitz Y, O'Donovan KJ, Baraban JM. Blockade of NGF-induced neurite outgrowth by a dominant-negative inhibitor of the egr family of transcription regulatory factors. J Neurosci. 2001;21:45–52. doi: 10.1523/JNEUROSCI.21-01-00045.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li W, Cogswell CA, LoTurco JJ. Neuronal differentiation of precursors in the neocortical ventricular zone is triggered by BMP. J Neurosci. 1998;18:8853–8862. doi: 10.1523/JNEUROSCI.18-21-08853.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marshall CJ. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 36.Martiny-Baron G, Kazanietz MG, Mischak H, Blumberg PM, Kochs G, Hug H, Marme D, Schachtele C. Selective inhibition of protein kinase C isozymes by the indolocarbazole Go 6976. J Biol Chem. 1993;268:9194–9197. [PubMed] [Google Scholar]
- 37.Milbrandt J. A nerve growth factor-induced gene encodes a possible transcriptional regulatory factor. Science. 1987;238:797–799. doi: 10.1126/science.3672127. [DOI] [PubMed] [Google Scholar]
- 38.Milbrandt J. Nerve growth factor induces a gene homologous to the glucocorticoid receptor gene. Neuron. 1988;1:183–188. doi: 10.1016/0896-6273(88)90138-9. [DOI] [PubMed] [Google Scholar]
- 39.Moscat J, Diaz-Meco MT. The atypical protein kinase Cs. Functional specificity mediated by specific protein adapters. EMBO Rep. 2000;1:399–403. doi: 10.1093/embo-reports/kvd098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Motohashi H, Igarashi K, Onodera K, Takahashi S, Ohtani H, Nakafuku M, Nishizawa M, Engel JD, Yamamoto M. Mesodermal- vs. neuronal-specific expression of MafK is elicited by different promoters. Genes Cells. 1996;1:223–238. doi: 10.1046/j.1365-2443.1996.d01-230.x. [DOI] [PubMed] [Google Scholar]
- 41.Motohashi H, Shavit JA, Igarashi K, Yamamoto M, Engel JD. The world according to Maf. Nucleic Acids Res. 1997;25:2953–2959. doi: 10.1093/nar/25.15.2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Motohashi H, Ohta J, Engel JD, Yamamoto M. A core region of the mafK gene IN promoter directs neurone-specific transcription in vivo. Genes Cells. 1998;3:671–684. doi: 10.1046/j.1365-2443.1998.00222.x. [DOI] [PubMed] [Google Scholar]
- 43.Motohashi H, Katsuoka F, Shavit JA, Engel JD, Yamamoto M. Positive or negative MARE-dependent transcriptional regulation is determined by the abundance of small Maf proteins. Cell. 2000;103:865–875. doi: 10.1016/s0092-8674(00)00190-2. [DOI] [PubMed] [Google Scholar]
- 44.Nishizawa M, Kataoka K, Goto N, Fujiwara KT, Kawai S. v-maf, a viral oncogene that encodes a “leucine zipper” motif. Proc Natl Acad Sci USA. 1989;86:7711–7715. doi: 10.1073/pnas.86.20.7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Onodera K, Shavit JA, Motohashi H, Yamamoto M, Engel JD. Perinatal synthetic lethality and hematopoietic defects in compound mafG:: mafK mutant mice. EMBO J. 2000;19:1335–1345. doi: 10.1093/emboj/19.6.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peng X, Greene LA, Kaplan DR, Stephens RM. Deletion of a conserved juxtamembrane sequence in Trk abolishes NGF-promoted neuritogenesis. Neuron. 1995;15:395–406. doi: 10.1016/0896-6273(95)90043-8. [DOI] [PubMed] [Google Scholar]
- 47.Qu Z, Wolfraim LA, Svaren J, Ehrengruber MU, Davidson N, Milbrandt J. The transcriptional corepressor NAB2 inhibits NGF-induced differentiation of PC12 cells. J Cell Biol. 1998;142:1075–1082. doi: 10.1083/jcb.142.4.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quest AF. Regulation of protein kinase C: a tale of lipids and proteins. Enzyme Protein. 1996;49:231–261. doi: 10.1159/000468635. [DOI] [PubMed] [Google Scholar]
- 49.Roivainen R, McMahon T, Messing RO. Protein kinase C isozymes that mediate enhancement of neurite outgrowth by ethanol and phorbol esters in PC12 cells. Brain Res. 1993;624:85–93. doi: 10.1016/0006-8993(93)90063-s. [DOI] [PubMed] [Google Scholar]
- 50.Sassone-Corsi P, Der CJ, Verma IM. ras-induced neuronal differentiation of PC12 cells: possible involvement of fos and jun. Mol Cell Biol. 1989;9:3174–3183. doi: 10.1128/mcb.9.8.3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schlingensiepen KH, Wollnik F, Kunst M, Schlingensiepen R, Herdegen T, Brysch W. The role of Jun transcription factor expression and phosphorylation in neuronal differentiation, neuronal cell death, and plastic adaptations in vivo. Cell Mol Neurobiol. 1994;14:487–505. doi: 10.1007/BF02088833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Segal RA, Greenberg ME. Intracellular signaling pathways activated by neurotrophic factors. Annu Rev Neurosci. 1996;19:463–489. doi: 10.1146/annurev.ne.19.030196.002335. [DOI] [PubMed] [Google Scholar]
- 53.Shavit JA, Motohashi H, Onodera K, Akasaka J, Yamamoto M, Engel JD. Impaired megakaryopoiesis and behavioral defects in mafG-null mutant mice. Genes Dev. 1998;12:2164–2174. doi: 10.1101/gad.12.14.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sheng M, Dougan ST, McFadden G, Greenberg ME. Calcium and growth factor pathways of c-fos transcriptional activation require distinct upstream regulatory sequences. Mol Cell Biol. 1988;8:2787–2796. doi: 10.1128/mcb.8.7.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spencer JA, Misra RP. Expression of the serum response factor gene is regulated by serum response factor binding sites. J Biol Chem. 1996;271:16535–16543. doi: 10.1074/jbc.271.28.16535. [DOI] [PubMed] [Google Scholar]
- 56.Troy CM, Rabacchi SA, Hohl JB, Angelastro JM, Greene LA, Shelanski ML. Death in the balance: alternative participation of the caspase-2 and -9 pathways in neuronal death induced by nerve growth factor deprivation. J Neurosci. 2001;21:5007–5016. doi: 10.1523/JNEUROSCI.21-14-05007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsuji M, Inanami O, Kuwabara M. Induction of neurite outgrowth in PC12 cells by alpha-phenyl-N-tert-butylnitron through activation of protein kinase C and the Ras-extracellular signal-regulated kinase pathway. J Biol Chem. 2001;276:32779–32785. doi: 10.1074/jbc.M101403200. [DOI] [PubMed] [Google Scholar]
- 58.Uberall F, Giselbrecht S, Hellbert K, Fresser F, Bauer B, Gschwendt M, Grunicke HH, Baier G. Conventional PKC-alpha, novel PKC-epsilon and PKC-theta, but not atypical PKC-lambda are MARCKS kinases in intact NIH 3T3 fibroblasts. J Biol Chem. 1997;272:4072–4078. doi: 10.1074/jbc.272.7.4072. [DOI] [PubMed] [Google Scholar]
- 59.Uberall F, Hellbert K, Kampfer S, Maly K, Villunger A, Spitaler M, Mwanjewe J, Baier-Bitterlich G, Baier G, Grunicke HH. Evidence that atypical protein kinase C-lambda and atypical protein kinase C-zeta participate in Ras-mediated reorganization of the F-actin cytoskeleton. J Cell Biol. 1999;144:413–425. doi: 10.1083/jcb.144.3.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Varnum BC, Ma QF, Chi TH, Fletcher B, Herschman HR. The TIS11 primary response gene is a member of a gene family that encodes proteins with a highly conserved sequence containing an unusual Cys-His repeat. Mol Cell Biol. 1991;11:1754–1758. doi: 10.1128/mcb.11.3.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 62.Velculescu VE, Vogelstein B, Kinzler KW. Analysing uncharted transcriptomes with SAGE. Trends Genet. 2000;16:423–425. doi: 10.1016/s0168-9525(00)02114-4. [DOI] [PubMed] [Google Scholar]
- 63.Vlahos CJ, Matter WF, Hui KY, Brown RF. A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002). J Biol Chem. 1994;269:5241–5248. [PubMed] [Google Scholar]
- 64.Wooten MW, Zhou G, Seibenhener ML, Coleman ES. A role for zeta protein kinase C in nerve growth factor-induced differentiation of PC12 cells. Cell Growth Differ. 1994;5:395–403. [PubMed] [Google Scholar]
- 65.Wooten MW, Zhou G, Wooten MC, Seibenhener ML. Transport of protein kinase C isoforms to the nucleus of PC12 cells by nerve growth factor: association of atypical zeta-PKC with the nuclear matrix. J Neurosci Res. 1997;49:393–403. doi: 10.1002/(sici)1097-4547(19970815)49:4<393::aid-jnr1>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 66.Wooten MW, Seibenhener ML, Zhou G, Vandenplas ML, Tan TH. Overexpression of atypical PKC in PC12 cells enhances NGF-responsiveness and survival through an NF-kappaB dependent pathway. Cell Death Differ. 1999;6:753–764. doi: 10.1038/sj.cdd.4400548. [DOI] [PubMed] [Google Scholar]
- 67.Wooten MW, Seibenhener ML, Neidigh KB, Vandenplas ML. Mapping of atypical protein kinase C within the nerve growth factor signaling cascade: relationship to differentiation and survival of PC12 cells. Mol Cell Biol. 2000;20:4494–4504. doi: 10.1128/mcb.20.13.4494-4504.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wooten MW, Seibenhener ML, Mamidipudi V, Diaz-Meco MT, Barker PA, Moscat J. The atypical protein kinase C-interacting protein p62 is a scaffold for NF-kappaB activation by nerve growth factor. J Biol Chem. 2001;276:7709–7712. doi: 10.1074/jbc.C000869200. [DOI] [PubMed] [Google Scholar]
- 69.Wu BY, Fodor EJ, Edwards RH, Rutter WJ. Nerve growth factor induces the proto-oncogene c-jun in PC12 cells. J Biol Chem. 1989;264:9000–9003. [PubMed] [Google Scholar]
- 70.Yamamoto M, Wakatsuki T, Hada A, Ryo A. Use of serial analysis of gene expression (SAGE) technology. J Immunol Methods. 2001;250:45–66. doi: 10.1016/s0022-1759(01)00305-2. [DOI] [PubMed] [Google Scholar]
- 71.Yoh K, Sugawara T, Motohashi H, Takahama Y, Koyama A, Yamamoto M, Takahashi S. Transgenic over-expression of MafK suppresses T cell proliferation and function in vivo. Genes Cells. 2001;6:1055–1066. doi: 10.1046/j.1365-2443.2001.00489.x. [DOI] [PubMed] [Google Scholar]
- 72.Zheng WH, Kar S, Quirion R. Stimulation of protein kinase C modulates insulin-like growth factor-1-induced akt activation in PC12 cells. J Biol Chem. 2000;275:13377–13785. doi: 10.1074/jbc.275.18.13377. [DOI] [PubMed] [Google Scholar]