Abstract
The first several months of life are a critical period for neuronal plasticity in the visual cortex during which anatomic and physiological development depends on visual experience. In cats, electrophysiologically assessed neuronal plasticity is minimal until ∼3 weeks, peaks at 5 weeks, gradually declines to low levels at 20 weeks, and disappears at ∼1 year of age (Daw, 1994). Rearing in darkness slows the entire time course of this critical period, such that at 5 weeks of age, normal cats are more plastic than dark-reared cats, whereas at 20 weeks, dark-reared cats are more plastic (Mower, 1991; Beaver et al., 2001). Thus, a stringent criterion is that genes that are important for plasticity in visual cortex will show differences in expression between normal rearing and dark rearing that are of opposite direction in young versus older animals. The present study reports the identification by differential display PCR of Munc13-3, a mammalian homolog of the Caenorhabditis elegans “uncoordinated” gene (unc-13), as a candidate gene for critical-period neuronal plasticity, the expression of which is regulated according to this criterion specifically in visual cortex and not in frontal cortex. Other members of the Munc13 family (Munc13-1 and Munc13-2) do not meet this criterion in visual cortex, indicating that Munc13-3 is the only family member that is regulated by age and dark rearing in the same manner as physiological plasticity during the visual cortical critical period.
Keywords: visual cortex, Munc13, critical period, dark rearing, differential display PCR, neuronal plasticity
The neonatal visual cortex is a highly plastic structure, the development of which is guided by visual experience during early postnatal life. The clearest example of such environmental effects on visual cortical development is to rear cats with one eye sutured closed, a condition that leads to dramatic anatomic and physiological abnormalities (for review, see Sherman and Spear, 1982; Daw, 1994). In normal development, sensitivity to monocular deprivation is limited to a “critical period,” which in cats begins several weeks after birth, peaks at ∼5–6 weeks, gradually declines to low levels at 20 weeks, and disappears at ∼1 year of age (Daw, 1994).
Rearing in total darkness extends this critical period and prolongs neuronal plasticity far beyond its normal age limits (Cynader and Mitchell, 1980; Mower et al., 1981). Dark rearing therefore provides a means to dissociate changes associated with the state of visual cortical plasticity from changes associated with the general maturation of the animal. It is an ideal system for isolating changes in gene expression involved in controlling neuronal plasticity. Electrophysiological results indicate that the effect of dark rearing is to slow the entire time course of the critical period, such that at young ages (5 weeks), normal cats are more plastic than dark-reared cats, whereas at later ages (20 weeks), dark-reared cats are more plastic (Mower, 1991; Beaver et al., 2001). Thus, a stringent criterion is that genes that are important for plasticity in visual cortex will show differences in expression between normal and dark-reared cats that are of opposite direction in young versus older animals. The present study reports the identification by differential display (dd) PCR of Munc13-3, a mammalian homolog of the Caenorhabditis elegans“uncoordinated” gene (unc-13), as a candidate gene for critical-period neuronal plasticity, the expression of which is regulated according to this criterion in visual cortex.
MATERIALS AND METHODS
Animals. Cats were reared in a normal 12 hr light/dark cycle or in complete darkness from birth to 5 or 20 weeks of age. For each of the four age and rearing conditions [normal (N) 5-week-old, dark-reared (D) 5-week-old, N20-week-old, D20-week-old], two animals were used in ddPCR to reduce interanimal variability and false positives. For Northern blots, these eight cats and additional ones were used. Additional animals included one cat at each age and rearing condition that were used to compare Munc13-3 regulation in visual cortex, frontal cortex, and cerebellum in animals not used in the original ddPCR. In addition, two normal cats at 20 weeks were used to compare the regional distribution of the different Munc13 genes (see below) in total RNA from visual cortex, frontal cortex, and cerebellum. The animals were killed by an overdose of sodium pentobarbital (75 mg/kg, i.p.). These procedures conform to the guidelines of the National Institutes of Health and were approved by the Institutional Animal Care and Use Committee. Fresh brain regions [visual cortex (all of area 17 and possibly a small part of 18), frontal cortex (the anterior quarter of the cerebral hemisphere), and cerebellum (entire structure)] were dissected, immediately frozen in liquid nitrogen, and stored at −80°C until used for extraction of total RNA. RNA was extracted, and its amount and integrity were determined as described previously (Rosen et al., 1992). DNA contamination was removed from the RNA (MessageClean; GenHunter Corporation, Nashville, TN), and cDNA was synthesized (SMART; Clontech, Palo Alto, CA or RNAimage; GenHunter Corp., Nashville, TN).
Differential display PCR. PCR on cDNA generated from each independent RNA sample was done using the appropriate oligo-dT-X (oligo-dT-C, dT-G, or dT-A) primer and 80 arbitrary primers as provided in a commercially available kit (RNAimage; GenHunter Corporation). The radiolabeled PCR products (33P-dATP) were size-separated on a standard DNA sequencing gel (6% acrylamide, 8m urea) to generate eight lanes (two cats in each of four age and rearing conditions) for each arbitrary primer (see Fig.1). The gel was dried and processed for film autoradiography. PCR products showing appropriate differential expression caused by age and rearing were recovered by cutting from the gel. The PCR products were amplified in a PCR reaction using the same primers as in the ddPCR, then cloned for use in sequencing and as probes for Northern blots (see below). Because ddPCR bands frequently contain more than one gene fragment, multiple clones from each ddPCR band were sequenced. ddPCR fragments are generally in the range of 150–600 bp. For smaller fragments, we used the PCR-TRAP cloning system (GenHunter Corporation) and for larger fragments (>450 bp), we used the TOPO TA cloning system (Invitrogen, Carlsbad, CA). The resultant sequences were run against gene and expressed sequence tag (EST) databases to determine similarity to known genes. 5′ rapid amplification of cDNA ends (RACE) PCR and cloning (SMART; Clontech) were used to generate additional sequences for ddPCR fragments.
Fig. 1.
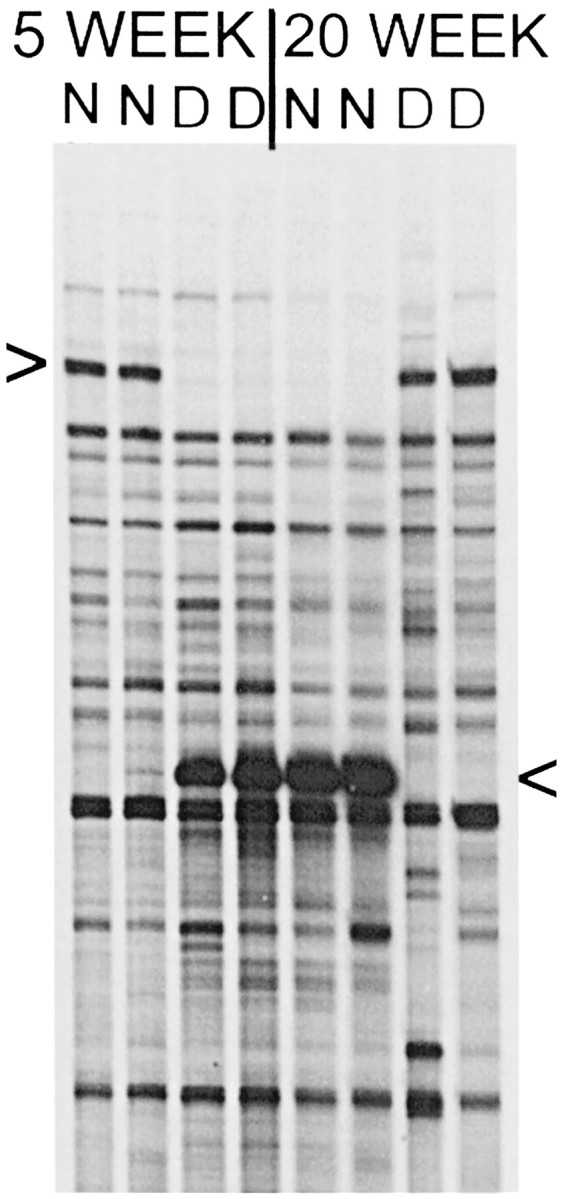
Candidate gene fragments identified by ddPCR. Portion of a ddPCR sequencing gel showing a ddPCR product that is expressed more highly in normal (N) cat visual cortex at 5 weeks and in dark-reared (D) cat visual cortex at 20 weeks (top arrowhead). Below it is seen as a different band that is expressed more highly in D cats at 5 weeks and N cats at 20 weeks (bottom arrowhead). In this case, both patterns of differentially expressed bands were detected by a single oligo dT-X/arbitrary primer pair. Independently isolated RNA samples from two cats at each age and rearing condition were run together (8 lanes) to help screen out false positives. The ddPCR analysis was also repeated with the same primer pair, and it confirmed reproducibility (data not shown).
Northern analysis. Probes used for Northern blotting included (1) the cat ddPCR fragment (503 bp) obtained from the screen; (2) a 2094 bp human retinal est (GenBank AA016047) found to have high homology to the cat ddPCR fragment (see below); (3) full-length rat cDNAs for Munc13-1, Munc13-2, and Munc13-3 (kindly provided by Dr. N. Brose, Max-Planck-Institute for Experimental Medicine, Göttingen, Germany); and (4) segments generated by PCR of cat cortical total RNA and the full-length rat Munc 13 cDNAs using primers designed from the rat sequences to yield 5′ probes specific to the different Munc13 genes (Munc13-1: bases 1202–1780; Munc13-2: bases 881–1398; Munc13-3: bases 747–1277). In both rats and cats, the resultant 5′ cDNA segments for the Munc13 family members were the predicted size (∼500 bp), and their sequences were the same between rats and cats (98–99% base pair identity). BLAST searching confirmed that the designed 5′ segments matched specifically with the appropriate Munc13 family members. Results obtained with the full-length rat Munc13-1/2/3 probes, and the designed 5′ subtype-specific probes were the same in terms of differences in expression caused by age and rearing and regional distribution (see Fig. 3b,c).
Fig. 3.
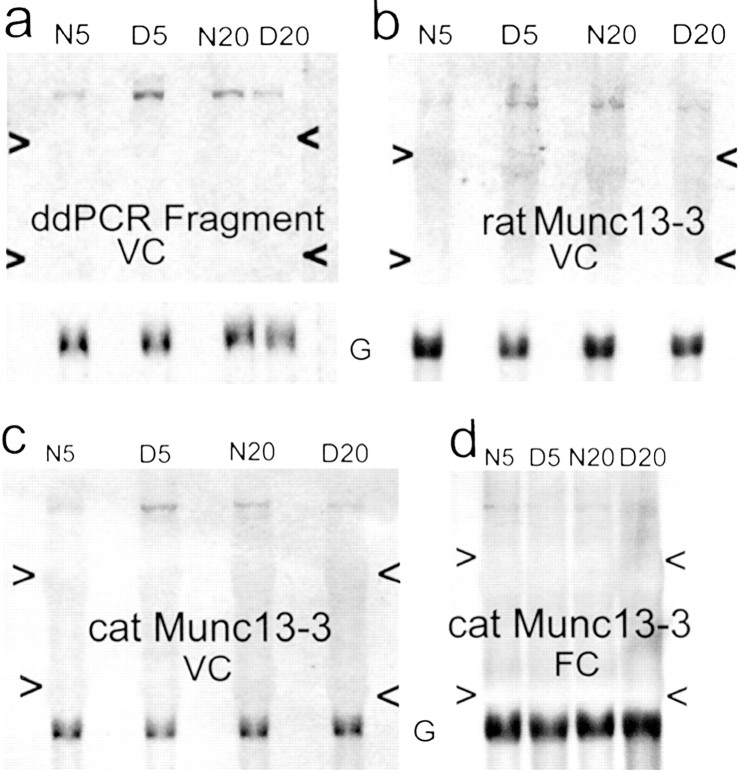
Differential expression of Munc13-3. Northern blots confirming differential expression of the candidate gene Munc13-3. Total RNA from a 5 week normal (N5), a 5 week dark-reared (D5), a 20 week normal (N20), and a 20 week dark-reared (D20) cat visual cortex (a–c) or frontal cortex (d) was loaded in each blot. Blots were either rehybridized (a, b) with GAPDH (G) or simultaneously hybridized (c, d) with a Munc13-3 probe and GAPDH.a, Filter of visual cortical (VC) total RNA hybridized with the 3′ cat Munc13-3 gene fragment recovered from ddPCR; b, filter of VC total RNA hybridized with a full-length rat Munc13-3 probe; c, filter of VC total RNA hybridized with a probe designed for the 5′ end of the cat Munc13-3 gene (see Materials and Methods); d, filter of frontal cortical (FC) total RNA hybridized with the same 5′ cat Munc13-3 probe. All probes identified a band of appropriate size (∼7 kb) for Munc13-3, based on the rat sequence. Arrowheadsindicate 28S and 18S rRNA.
Filters with lanes from all experimental conditions and with lanes from different brain regions (see Figs. 3, 4) were hybridized (stringency: 0.1× SSC, 0.1× SDS, 42°C) using these probes according to our standard procedures (Rosen et al., 1992). All filters were either rehybridized with a probe to GAPDH (after stripping) or simultaneously hybridized with a Munc13 probe and GAPDH to correct for loading errors. GAPDH levels in cat visual cortex are not affected by age and dark rearing during the visual cortical critical period (McCormack et al., 1992; Rosen et al., 1992). For GAPDH hybridization, a human cDNA clone inserted into pBR322 vector was obtained from the American Type Culture Collection (Manassas, VA) (ATCC 57090). The probe was synthesized from an 800 bp XbaI-PstI fragment of the cDNA. The relative intensity of signals in Northern blots was determined by densitometric scanning. In each blot, Munc13 signals were corrected against GAPDH mRNA and expressed as a percentage of the maximal corrected signal for that blot. Results from the three animals in each group were averaged.
Fig. 4.
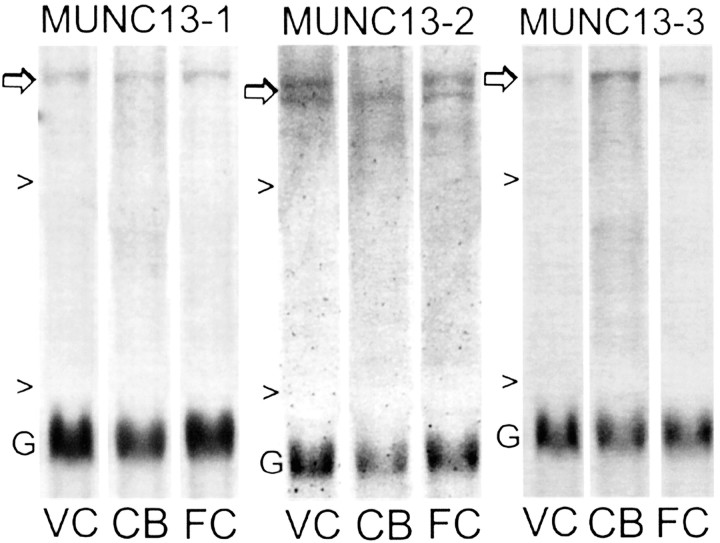
Differential regional distribution of Munc13 mRNAs. Portions of Northern blots showing regional differences in expression of Munc13 family members (Munc13-1, Munc13-2, Munc13-3) in two neocortical structures [visual cortex (VC) and frontal cortex (FC)] and cerebellum (CB) of normal 20 week cats. Arrows indicate the Munc13 band of interest, and arrowheads indicate 28S and 18S rRNA. Blots were simultaneously hybridized with the Munc13 probe and with the probe to GAPDH (G). Probes were 5′ cDNAs specific for each Munc family member, and they share 98–99% sequence identity between rats and cats (see Materials and Methods).
RESULTS
Characterization of a candidate plasticity gene
We have performed a ddPCR screening of theoretically 96% (GenHunter Corp.) of the expressed genes in the visual cortex of normal (N) and dark-reared (D) cats at 5 and 20 weeks to directly identify candidate plasticity genes that show opposite direction differences in expression because of age and dark rearing. A manageable set of candidate plasticity genes (∼25) has been identified. Figure1 shows ddPCR results for one different primer pair (240 primer pairs in total were used), and it indicates two distinct patterns of differential expression. One pattern is an elevation in N cats at 5 weeks and in D cats at 20 weeks. Genes with this pattern of expression (“plasticity” profile) are increased in expression by age and rearing condition combinations in which neuronal plasticity is highest. The other pattern is the opposite: elevated expression in D cats at 5 weeks and in N cats at 20 weeks. These genes (“antiplasticity” profile) show decreased levels of expression in age and rearing conditions in which plasticity is highest and elevated expression when neuronal plasticity is reduced. The finding of these inverse patterns of expression fits with current thinking that a balance between activator and suppressor genes controls neural plasticity (Abel et al., 1998).
Cloning and sequencing of the ddPCR fragments in the upper and lower bands of Figure 1 revealed several mitochondrial gene fragments and an unknown gene. Northern blotting confirmed differential expression of the mitochondrial genes, some showing the plasticity and others the antiplasticity expression profile (Yang et al., 2001). The unknown gene showed the antiplasticity profile (Figs. 1, bottom band, 3). A database search with the 503 bp unknown cat gene fragment revealed high sequence identity (80%) with an EST cDNA clone from human retina. To identify the gene, we first sequenced 2094 bases of the human EST, which database searching indicated is located on chromosome 15, and found a region of high sequence identity (87%) with rat Munc13-3 (Fig.2). Northern blotting with the cat ddPCR fragment (3′ end of Munc13-3), the full-length rat Munc13-3 sequence, and a PCR segment from the 5′ end of the cat Munc13-3 gene (see Materials and Methods) all showed the same antiplasticity profile of differential expression caused by age and rearing condition (Fig.3a–c). Northern blotting with the human EST also showed the same antiplasticity profile (data not shown). This opposite direction pattern of differences caused by age and rearing was not observed in frontal cortex (Fig. 3d) or cerebellum (data not shown), and in both structures there was a slight downregulation caused by dark rearing at both ages.
Fig. 2.
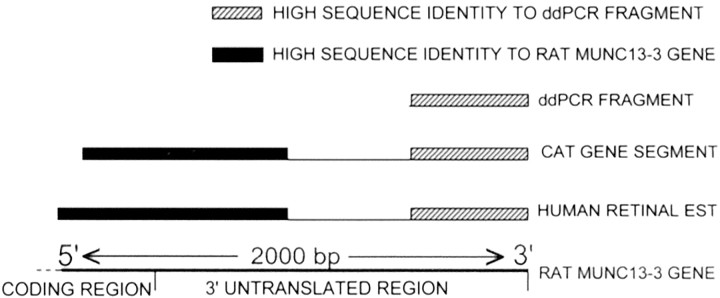
Sequence of the human EST and cat gene. Schematic results from sequencing ∼2 kb of the human retinal EST fragment and of the cat gene. High sequence identity to the 503 bp ddPCR fragment was found in both the human (80%) and cat (96%) gene segments in their 3′ ends. The ddPCR fragment had no significant identity with rat Munc13-3 based on pairwise BLAST (National Center for Biotechnology Information). In the 5′ portion of the cat and human gene segments, regions of high sequence identity to Munc13-3 were found in both the human EST (87%) and the cat gene segment (89%).
5′ RACE PCR was used to generate and sequence part (1852 bases) of the cat gene. High sequence identity (85%) was found between the resultant cat candidate gene partial sequence and the human EST throughout their available lengths. The 5′ portion of this cat partial sequence also showed high identity (89%) with the 3′ region of rat Munc13-3 (Fig.2). The cat sequence has been submitted to GenBank (AF465212); the human Munc13-3-like sequence was identical to that recently submitted by another laboratory (AK054981). On the basis of the predicted amino acid sequence, the 5′ portion of the cat gene fragment encoded 94 amino acids of rat Munc13-3. There was a 98% match between the predicted cat amino acid sequence and that of rat Munc13-3 protein, suggesting their identity.
Differential regulation of Munc13 family members
Northern blotting with probes specific to rat Munc13-3 and the other members of its gene family, Munc13-1 and Munc13-2 (Brose et al., 1995), revealed differential regional distributions of the family members, as shown in Figure 4. The bands detected by each probe in cat visual cortex were of appropriate size for Munc13 mRNAs (∼7 kb) on the basis of the rat sequences. Munc 13-1 was expressed about equally in visual cortex, frontal cortex, and cerebellum. Munc13-2 was expressed as a doublet, which could represent an alternative splice variant of Munc13-2 that has a wide tissue distribution (Song et al., 1998). The larger band was expressed in both visual and frontal cortex (slightly higher in visual cortex) but was not detected in cerebellum. The smaller band was also expressed at higher levels in neocortical structures but was also detected in cerebellum. Munc13-3 was expressed most highly in cerebellum and was also present at lower levels in frontal and visual cortex (slightly higher in frontal cortex). These differential patterns of regional expression of the Munc13 genes are consistent with findings in rodents (Augustin et al., 1999), lending confidence to the specificity of the probes.
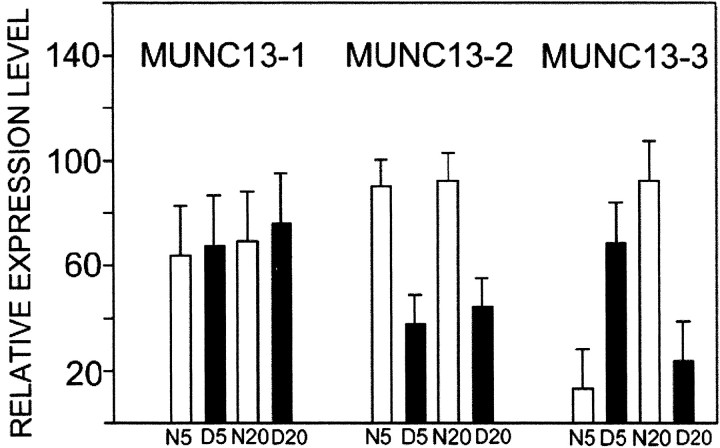
Northern blotting with probes specific to Munc13-3 and the other members of its gene family, Munc13-1 and Munc13-2 (Brose et al., 1995), revealed differential regulation of the family members by age and rearing condition, as summarized in Figure5. Munc13-3 showed the antiplasticity profile, being elevated fourfold to fivefold in dark-reared cats at 5 weeks and in normal cats at 20 weeks. ANOVA showed a significant statistical interaction (ANOVA,F(1,11) = 17.14;p = 0.003), indicating that the effect of dark rearing depended on age. Munc13-3 expression was also regulated by development and increased sevenfold between 5 and 20 weeks in normal cat visual cortex (but decreased between these ages in dark-reared cat visual cortex). The other Munc13 genes did not show opposite direction differences in expression level caused by age and rearing condition. Munc13-1 showed no statistically significant differences caused by age or rearing condition. Munc13-2 was elevated (twofold) in normal visual cortex at both ages (ANOVA, F(1,11) = 22.22; p = 0.002). The expression profile of Munc13-2 therefore suggests upregulation and downregulation by neuronal activity level per se. Munc13-3, conversely, shows an expression profile that mirrors the physiological effects of age and dark rearing on critical-period neuronal plasticity in visual cortex.
Fig. 5.
Differential regulation of Munc13 family member expression by age and dark rearing. Histograms showing relative levels of expression of Munc13-1, Munc13-2, and Munc13-3 in visual cortex of N5, D5, N20, and D20 cat visual cortex (n = 3 at each age and rearing condition). The relative intensity of signals in Northern blots was determined by densitometric scanning. In each blot, Munc13 signals were normalized against GAPDH mRNA and expressed as a percentage of the maximal corrected signal for that blot. Results from the three animals in each group were averaged. Error bars indicate mean and SEM.
DISCUSSION
Plasticity-related genes
Differential gene screening has identified a candidate gene for critical-period neuroplasticity in the visual cortex, and combined sequence and Northern blot data indicate that the gene is Munc13-3. The effects of dark rearing on Munc13-3 gene expression matched its effects on the physiological plasticity of visual cortical neurons, indicating a relationship between expression of these genes and neuronal plasticity. This bidirectional regulation of Munc13-3 gene expression across the age and rearing combinations examined is difficult to explain in terms of the level of neuronal activity. Dark rearing reduces the responsiveness of visual cortical neurons in both young and older cats (Imbert and Buisseret, 1975; Mower et al., 1981), and spontaneous activity is lower in young dark-reared animals and comparable in older dark-reared animals (Imbert and Buisseret, 1975;Czepita et al., 1994). Therefore, the overall level of activity (including visual responses and spontaneous activity) in light-reared animals is higher than in dark-reared animals at all ages. If the expression of Munc 13-3 reflected neuronal activity level, it would be expected that dark rearing would have the same effect on expression level in both young and old cats. Rather, Munc13-3 shows an expression profile that mirrors the effects of age and dark rearing on critical-period neuronal plasticity in visual cortex. The expression profile of Munc13-2, conversely, indicates unidirectional downregulation by decreases in neuronal activity.
Several studies have manipulated neuronal activity levels to identify genes that may be important for neuronal plasticity, and this approach has proved fruitful in identifying candidate effector genes involved in the response of neurons to electrical (Yamagata et al., 1993) and chemical (Nedivi et al., 1993; Qian et al., 1993) seizures or blockade of neuronal activity in the visual system (Corriveau et al., 1998). This approach has yielded important information on a number of genes that are responsive to visual input, regulated during the critical period, and involved in structural and functional plastic responses (Nedivi et al., 1996, 1998; Corriveau et al., 1998, 1999; Lanahan and Worley, 1998; Huh et al., 2000).
The gene screening criterion used in the present study, based on physiological properties of the visual cortical critical period, offers a promising alternative approach to identifying genes important for neuronal plasticity. In addition to the present gene screening, a number of molecules consistent with the criterion of opposite direction differences caused by age and dark rearing have been identified with other biochemical procedures. Such molecules include the NR2A subunit of the NMDA receptor (Chen et al., 2000), basal and induced levels of cAMP (Reid et al., 1996), stimulated phosphoinositide turnover (Dudek and Bear, 1989), and the immediate early gene product Fos (Mower and Kaplan, 1999). The identification of neurotransmitter receptor, second messenger system, and gene transcription regulation molecules indicates a signaling pathway from the cell surface to the nucleus. Recently, a candidate effector molecule for neuronal plasticity identified by differential gene screening after chemical seizures was proven to show opposite direction regulation caused by age and dark rearing (Lee and Nedivi, 2002). The changes in expression of Munc13-3 found here could also indicate a change in the nuclear output of a long-term plasticity effector molecule.
Munc13 genes and neuronal plasticity
A number of considerations indicate that the gene identified to show the antiplasticity profile of regulation in the present screen of cat visual cortex is Munc13-3. The available sequence indicated a high homology with rat Munc13-3 specifically. Probes derived from cat, rat, and human for Munc13-3 all yielded the same expression pattern. The regional pattern of expression and the regulation of expression by age and dark rearing yielded distinct results for the candidate gene Munc13-3 and two closely related family members, Munc13-1 and Munc13-2. Munc13-1, 2, and 3 are a family of three brain-specific proteins in mammals with homology to C. elegans unc-13 (Brose et al., 1995). The most abundant isoform in rat brain, Munc13-1, is expressed in all regions of the rat CNS, but Munc13-2 and 13-3 are most highly expressed in the forebrain and hindbrain, respectively (Augustin et al., 1999a). The present findings on regional distribution in cat are consistent with those in the rat, the major difference being that there is higher expression of Munc13-3 outside the cerebellum in cats than has been described in rodents. There are several possible reasons for this difference. One possibility is procedural. The present results are based on Northern blots, whereas the rat data are based on in situ hybridization. Optimizing the high Munc13-3 in situ hybridization signal that is concentrated in the cell dense granule layer of the cerebellum would result in an underestimation of expression in cerebral cortex, in which the signal would be spread across the entire cortical thickness. Another possibility is a species difference, such that there are differences in the regional distribution and the interaction of Munc13 family members between rats and cats. Direct comparative studies with the same methods in different species will resolve this issue. Nevertheless, the facts that both full-length and designed 5′ end Munc13 subtype-specific probes labeled bands of appropriate size for Munc13 genes on the basis of the rat sequences and that each subtype-specific probe indicated a different regional distribution and a different regulation profile caused by age and rearing condition indicates that all three Munc13s are transcribed in cat visual cortex.
All three Munc13 genes are phorbol ester receptors that bind syntaxin, are specifically targeted to presynaptic active zones, and play an essential role in synaptic vesicle release (Betz et al., 1997, 1998). Munc13-1 deletion mouse mutants die near birth, but primary hippocampal cultures from these animals indicate an inability of docked vesicles to fuse in response to physiological stimuli, and this effect is evident in glutamatergic but not GABAergic neurons (Augustin et al., 1999b). Munc13-3 mutants thrive and show grossly normal brain morphology, but there are impairments of synaptic transmission at cerebellar synapses and of motor learning (Augustin et al., 2001). Munc13-3 mutants show enhanced paired pulse facilitation at parallel fiber–Purkinje cell synapses, which is indicative of decreased neurotransmitter release probability (Zucker, 1989), suggesting that normally Munc13-3 functions to increase release probability and decrease paired pulse facilitation. Mutation of brain-derived neurotrophic factor, a molecule that promotes neuronal plasticity, conversely, results in decreased paired pulse facilitation at parallel fiber–Purkinje cell synapses (Carter et al., 2002). Thus, the data in cerebellum may suggest that Munc13-3 acts in a manner consistent with the antiplasticity role proposed for it in visual cortical critical-period neuronal plasticity by the present results. Firm conclusions must await better determination of the function of Munc13-3, particularly in visual cortex. Overall, growing evidence indicates a role of Munc13 genes in presynaptic transmission and plasticity, and the present results extend this evidence by implicating Munc13-3 as a candidate plasticity gene for critical-period neuroplasticity of the visual cortex. The visual cortical critical period involves opposite processes, probably with overlapping but distinct temporal profiles. One process maximizes adaptive responsiveness to external input, and the other promotes crystallization of semi-immutable mature response properties. Identification and characterization of genes showing plasticity and antiplasticity profiles is a promising new direction for insight into these interactive developmental mechanisms.
Footnotes
This work was supported by National Science Foundation (NSF) Grant 0090777 and NSF EPSCoR Grant EPS-9874764. We thank Dr. N. Brose (Max-Planck-Institute for Experimental Medicine, Göttingen, Germany) for kindly providing the full-length Munc13-1/2/3 cDNAs.
Correspondence should be addressed to Dr. George D. Mower, Department of Anatomical Sciences and Neurobiology, University of Louisville School of Medicine, 500 South Preston Street, Louisville, KY 40202. E-mail: george.mower@louisville.edu.
REFERENCES
- 1.Abel T, Martin KC, Bartsch D, Kandel ER. Memory suppressor genes: inhibitory constraints on the storage of long-term memory. Science. 1998;279:338–341. doi: 10.1126/science.279.5349.338. [DOI] [PubMed] [Google Scholar]
- 2.Augustin I, Betz A, Herrmann C, Jo T, Brose N. Differential expression of two novel Munc13 proteins in rat brain. Biochem J. 1999a;337:363–371. [PMC free article] [PubMed] [Google Scholar]
- 3.Augustin I, Rosenmund C, Sudhof TC, Brose N. Munc13-1 is essential for fusion competence of glutamatergic synaptic vesicles. Nature. 1999b;400:457–461. doi: 10.1038/22768. [DOI] [PubMed] [Google Scholar]
- 4.Augustin I, Korte S, Rickmann M, Kretzschmar HA, Sudhof TC, Herms JW, Brose N. The cerebellum-specific Munc13 isoform Munc13-3 regulates cerebellar synaptic transmission and motor learning in mice. J Neurosci. 2001;21:10–17. doi: 10.1523/JNEUROSCI.21-01-00010.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beaver CJ, Ji Q-H, Daw NW. Layer differences in the effect of monocular vision in light and dark reared kittens. Vis Neurosci. 2001;18:811–820. doi: 10.1017/s0952523801185147. [DOI] [PubMed] [Google Scholar]
- 6.Betz A, Okamoto M, Benseler F, Brose N. Direct interaction of the rat unc-13 homologue Munc13-1 with the N terminus of syntaxin. J Biol Chem. 1997;272:2520–2526. doi: 10.1074/jbc.272.4.2520. [DOI] [PubMed] [Google Scholar]
- 7.Betz A, Ashery U, Rickmann M, Augustin I, Neher E, Sudhof TC, Rettig J, Brose N. Munc13-1 is a presynaptic phorbol ester receptor that enhances neurotransmitter release. Neuron. 1998;21:123–136. doi: 10.1016/s0896-6273(00)80520-6. [DOI] [PubMed] [Google Scholar]
- 8.Brose N, Hofmann K, Hata Y, Sudhof TC. Mammalian homologues of Caenorhabditis elegans unc-13 gene define novel family of C2-domain proteins. J Biol Chem. 1995;270:25273–25280. doi: 10.1074/jbc.270.42.25273. [DOI] [PubMed] [Google Scholar]
- 9.Carter AR, Chen C, Schwartz PM, Segal RA. Brain-derived neurotrophic factor modulates cerebellar plasticity and synaptic ultrastructure. J Neurosci. 2002;22:1316–1327. doi: 10.1523/JNEUROSCI.22-04-01316.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen L, Cooper NG, Mower GD. Developmental changes in the expression of NMDA receptor subunits (NR1, NR2A, NR2B) in the cat visual cortex and the effects of dark rearing. Mol Brain Res. 2000;78:196–200. doi: 10.1016/s0169-328x(00)00076-0. [DOI] [PubMed] [Google Scholar]
- 11.Corriveau RA, Huh GS, Shatz CJ. Regulation of class I MHC gene expression in the developing and mature CNS by neural activity. Neuron. 1998;21:505–520. doi: 10.1016/s0896-6273(00)80562-0. [DOI] [PubMed] [Google Scholar]
- 12.Corriveau RA, Shatz CJ, Nedivi E. Dynamic regulation of cpg15 during activity-dependent synaptic development in the mammalian visual system. J Neurosci. 1999;19:7999–8008. doi: 10.1523/JNEUROSCI.19-18-07999.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cynader M, Mitchell DE. Prolonged sensitivity to monocular deprivation in dark reared cats. J Neurophysiol. 1980;43:1026–1040. doi: 10.1152/jn.1980.43.4.1026. [DOI] [PubMed] [Google Scholar]
- 14.Czepita D, Reid SN, Daw NW. Effect of longer periods of dark rearing on NMDA receptors in cat visual cortex. J Neurophysiol. 1994;72:1220–1226. doi: 10.1152/jn.1994.72.3.1220. [DOI] [PubMed] [Google Scholar]
- 15.Daw NW. Mechanisms of plasticity in the visual cortex. The Friedenwald Lecture. Invest Ophthalmol Vis Sci. 1994;35:4168–4179. [PubMed] [Google Scholar]
- 16.Dudek SM, Bear MF. A biochemical correlate of the critical period for synaptic modification in the visual cortex. Science. 1989;246:673–675. doi: 10.1126/science.2573152. [DOI] [PubMed] [Google Scholar]
- 17.Huh GS, Boulanger LM, Du H, Riquelme PA, Brotz TM, Shatz CJ. Functional requirement for class I MHC in CNS development and plasticity. Science. 2000;290:2155–2159. doi: 10.1126/science.290.5499.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imbert M, Buisseret P. Receptive field characteristics and plastic properties of visual cortical cells in kittens reared with or without visual experience. Exp Brain Res. 1975;22:25–36. doi: 10.1007/BF00235409. [DOI] [PubMed] [Google Scholar]
- 19.Lanahan A, Worley P. Immediate-early genes and synaptic function. Neurobiol Learn Mem. 1998;70:37–43. doi: 10.1006/nlme.1998.3836. [DOI] [PubMed] [Google Scholar]
- 20.Lee WC, Nedivi E. Extended plasticity of visual cortex in dark-reared animals may result from prolonged expression of cpg15-like genes. J Neurosci. 2002;22:1807–1815. doi: 10.1523/JNEUROSCI.22-05-01807.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCormack MA, Rosen KM, Villa-Komaroff L, Mower GD. Changes in immediate early gene expression during postnatal development of cat cortex and cerebellum. Mol Brain Res. 1992;12:215–223. doi: 10.1016/0169-328x(92)90087-r. [DOI] [PubMed] [Google Scholar]
- 22.Mower GD. The effect of dark rearing on the time course of the critical period in cat visual cortex. Dev Brain Res. 1991;58:151–158. doi: 10.1016/0165-3806(91)90001-y. [DOI] [PubMed] [Google Scholar]
- 23.Mower GD, Kaplan IV. Fos expression during the critical period in visual cortex: differences between normal and dark reared cats. Mol Brain Res. 1999;64:264–269. doi: 10.1016/s0169-328x(98)00333-7. [DOI] [PubMed] [Google Scholar]
- 24.Mower GD, Berry D, Burchfiel JL, Duffy FH. Comparison of the effects of dark rearing and binocular suture on development and plasticity of cat visual cortex. Brain Res. 1981;220:255–267. doi: 10.1016/0006-8993(81)91216-6. [DOI] [PubMed] [Google Scholar]
- 25.Nedivi E, Hevroni D, Naot D, Israeli D, Citrl Y. Numerous candidate plasticity related genes revealed by differential cDNA cloning. Nature. 1993;363:718–722. doi: 10.1038/363718a0. [DOI] [PubMed] [Google Scholar]
- 26.Nedivi E, Fieldust S, Theill LE, Hevron D. A set of genes expressed in response to light in the adult cerebral cortex and regulated during development. Proc Natl Acad Sci USA. 1996;93:2048–2053. doi: 10.1073/pnas.93.5.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nedivi E, Wu GY, Cline HT. Promotion of dendritic growth by CPG15, an activity-induced signaling molecule. Science. 1998;281:1863–1866. doi: 10.1126/science.281.5384.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qian Z, Gilbert ME, Colicos MA, Kandel ER, Kuhl D. Tissue-plasminogen activator is induced as an immediate-early gene during seizure, kindling and long-term potentiation. Nature. 1993;361:453–457. doi: 10.1038/361453a0. [DOI] [PubMed] [Google Scholar]
- 29.Reid SN, Daw NW, Gregory DS, Flavin H. cAMP levels increased by activation of metabotropic glutamate receptors correlate with visual plasticity. J Neurosci. 1996;16:7619–7626. doi: 10.1523/JNEUROSCI.16-23-07619.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosen KM, McCormack MA, Villa-Komaroff L, Mower GD. Brief visual experience induces immediate early gene expression in the cat visual cortex. Proc Natl Acad Sci USA. 1992;89:5437–5441. doi: 10.1073/pnas.89.12.5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sherman SM, Spear PD. Organization of visual pathways in normal and visually deprived cats. Physiol Rev. 1982;62:738–855. doi: 10.1152/physrev.1982.62.2.738. [DOI] [PubMed] [Google Scholar]
- 32.Song Y, Ailenberg M, Silverman M. Cloning of a novel gene in the human kidney homologous to rat munc13s: its potential role in diabetic nephropathy. Kidney Int. 1998;53:1689–1695. doi: 10.1046/j.1523-1755.1998.00942.x. [DOI] [PubMed] [Google Scholar]
- 33.Yamagata K, Andreasson KI, Kaufmann WE, Barnes CA, Worley PF. Expression of a mitogen-inducible cyclooxygenase in brain neurons: regulation by synaptic activity and glucocorticoids. Neuron. 1993;11:371–386. doi: 10.1016/0896-6273(93)90192-t. [DOI] [PubMed] [Google Scholar]
- 34.Yang C, Silver B, Ellis SR, Mower GD. Bidirectional regulation of mitochondrial gene expression during developmental neuroplasticity of visual cortex. Biochem Biophys Res Commun. 2001;287:1070–1074. doi: 10.1006/bbrc.2001.5706. [DOI] [PubMed] [Google Scholar]
- 35.Zucker RS. Short-term synaptic plasticity. Annu Rev Neurosci. 1989;12:13–31. doi: 10.1146/annurev.ne.12.030189.000305. [DOI] [PubMed] [Google Scholar]