Abstract
The Rho family of small GTPases, key regulators of the actin cytoskeleton in eukaryotic cells from yeast to human, is implicated in the control of neuronal morphology. Guanine nucleotide exchange factors (GEFs) are upstream positive regulators of Rho GTPases and integrate extracellular signaling for appropriate activation of Rho GTPases at specific subcellular regions. Here we describe the identification of a novel Dbl family GEF for Rho GTPases in Homo sapiens andMus musculus. It contains a tandem Dbl homology–pleckstrin homology domain and FERM domain, characteristic of the plasma membrane proteins linker. This gene, termed FERM domain including RhoGEF (FIR), was abundantly expressed in brain, lung, and testis, as well as embryonic hippocampal and cortical neurons. FIR was found to activate the biochemical pathway specific for Rac1 but not for RhoA or Cdc42. Ectopic expression of FIR in the cortical neurons resulted in significantly shortened neurites and excessive growth cones, presumably mediated by Rac1. These results suggest that FIR may regulate neurite remodeling by mediating the signaling pathways from membrane proteins to Rac.
Keywords: Rac1, Rho guanine nucleotide exchange factor, FERM domain, cytoskeleton, neurite outgrowth, neuronal morphology
The Rho GTPases are key regulators of the actin cytoskeleton in eukaryotic cells from yeast to human (Hall, 1998) and mediate the morphological changes that can be observed during neuronal development and plasticity, such as growth of neurites, axon guidance, and dendrite elaboration (Luo et al., 1994; Threadgill et al., 1997; Yamashita et al., 1999; Li et al., 2000). Each member of the archetypal trio of the Rho GTPases, RhoA, Rac1, and Cdc42, has been found to regulate distinct actin filament-containing structures. RhoA regulates the formation of focal adhesions and subsequent assembly of stress fibers, and Rac1 regulates the formation of membrane lamellae, whereas Cdc42 triggers the outgrowth of peripheral spike-like protrusions known as filopodia (van Aelst and D'Souza-Schorey, 1997;Hall, 1998; Richnau and Aspenstrom, 2001). The potential of the Rho GTPases to function as signaling switches resides in their ability to cycle between active, GTP-bound states and inactive, GDP-bound states. These cyclings are regulated by a variety of intracellular proteins. GTPase-activating proteins stimulate the intrinsic GTP hydrolysis of the GTPases, thus inactivating their targets (Lamarche and Hall, 1994; Whitehead et al., 1997; Zalcman et al., 1999). Guanine nucleotide exchange factors (GEFs) promote the exchange of GDP for GTP, thereby activating GTPases. The Rho GEFs comprise enzymes with Dbl homology (DH) domain (Mackay and Hall, 1998) and are Rho GTPase-specific exchange factors (Whitehead et al., 1997). These proteins are characterized by a DH domain sharing ∼250 amino acids (aa), followed immediately by a pleckstrin homology (PH) domain (Stam and Collard, 1999). They often contain multiple protein motifs, such as Src homology domains 2 and 3 and PDZ (postsynaptic density 95/Discs large/zona occludens 1) domains, most of which are involved in intracellular signal transduction (Matsuo et al., 2002). GEFs from the Dbl family are thought to integrate extracellular signaling for appropriate activation of Rho GTPases at specific subcellular regions. Whereas in Caenorhabditis elegans and Drosophila some GEFs from the Dbl family have been shown to play essential roles in neurite genesis (Steven et al., 1998; Awasaki et al., 2000; Bateman et al., 2000; Liebl et al., 2000; Newsome et al., 2000), in vertebrates, little is known about regulations of neurite outgrowth by GEFs (Kunda et al., 2001;Penzes et al., 2001; Matsuo et al., 2002).
To further understand the molecular mechanisms underlying the activation of Rho GTPases and biological phenomena that the related signaling pathway regulates, especially in neurons, we tried to identify a novel GEF using a consensus sequence for DH domains of Rho guanine nucleotide exchange factors to search DNA databases. Here we describe the identification of a novel GEF for Rho GTPases, including FERM domain in Homo sapiens and Mus musculus. These proteins, termed FERM domain including RhoGEF (FIR), were found to activate the biochemical pathway specific for Rac1. Ectopic expression of FIR in cortical neurons resulted in morphological changes.
MATERIALS AND METHODS
RNA isolation and Northern blot analysis. Total RNA derived from 6-week-old adult mice was extracted from various tissues by the acid guanidium-thiocyanate–phenol chloroform method. Total RNA (20 μg/lane) was separated by electrophoresis on 1.0% agarose–formide gels and transferred overnight onto polyvinylidene difluoride membrane (Millipore, Bedford, MA). The membrane was prehybridized for 1 hr at 65°C in hybridization buffer (0.9m NaCl and 90 mm sodium citrate, pH 7.0) containing 5× Denhardt's solution, 0.5% SDS, and heat-denatured salmon sperm DNA (100 ng/ml). The cDNA probe specific for mouse FIR mRNA was radiolabeled with [32P]dCTP (NZ522; PerkinElmer Life Sciences, Emeryville, CA) by a random labeling kit according to the instructions of the manufacturer (Takara Shuzo, Shiga, Japan). After hybridization overnight at 65°C in hybridization buffer containing radiolabeled cDNA probe (5 ng/ml), the membrane was washed with 2× SSC containing 0.5% SDS, followed by 0.2× SSC containing 0.5% SDS, each for 30 min at 65°C. Then, the filters were exposed to x-ray films (Fujifilm, Tokyo, Japan) and subjected to autoradiography.
Reverse transcription-PCR. Total RNA (5 μg) was reverse-transcribed using oligo-dT by reverse transcriptase from Moloney murine leukemia virus (Invitrogen, San Diego, CA). For PCR amplification, specific oligonucleotide primer pairs (10 pmol each) were incubated with 1 μl of cDNA template in a 20 μl PCR reaction mixture containing 1.5 mmMgCl2, 25 mm KCl, 10 mm Tris, pH 9.2, mixed deoxynucleotides (1 mm each), and 1 U of Taqpolymerase. The sequences of primers used were as follows: mouse FIR sense primer, 5′-AATTGACGGAGCTACAGCGA-3′ and mouse FIR antisense primer, 5′-GACGTGAGATTTGAATTGGA-3′ (product length, 801 bp); mouse glial fibrillary acidic protein (GFAP) sense primer, 5′-TAGACAGGAGGCAGATGAAGCCACC-3′ and mouse GFAP antisense primer, 5′-GTCGTTAGCTTCGTGCTTGGCTTGG-3′ (product length, 337 bp); and, as an internal control, mouse β-actin sense primer, 5′- TCCTCCCTGGAGAAGAGCTA-3′ and mouse β-actin antisense primer, 5′-TCCTGCTTGCTGATCCACAT-3′ (product length, 403 bp). Dilutions of the cDNAs were amplified for 35 cycles at 94°C for 30 sec, 60°C for 30 sec, and 72°C for 30 sec. The amplified PCR products were analyzed by 1.2% agarose gel electrophoresis and ethidium bromide staining. The product for β-actin mRNA served as the internal standard. All of the products were assayed in the linear response range of the reverse transcription (RT)-PCR amplification process; the cycle number used was determined by finding the midpoint of linear amplification on a sigmoid curve for amplification products with cycle numbers of 24–40 plotted against band density. The identity of each PCR product was confirmed by subcloning the amplified cDNAs into the pGEM-T vector (Promega, Madison, WI) and sequencing.
Plasmid constructs. The full-length FIR (FL-FIR) cDNA (KIAA0793; gifts from Dr. T. Nagase, Kazusa DNA Research Institute, Kisarazu, Japan) was digested with SalI andXbaI and subcloned into pEGFP plasmid (Clontech, Palo Alto, CA), which produces the N-terminally green fluorescent protein (GFP)-tagged protein under the control of cytomegalovirus promoter. N-terminal truncated FIR (ΔN-FIR; aa 346–1055) was generated by digestion of the full-length FIR with SacI andXhoI and subcloned into pEGFP plasmid. The 720 bp fragment encoding the DH domain of FIR (aa 516–755) and the 942 bp fragment encoding the DH and PH domains of FIR (aa 540–853) were subcloned into the EcoRI and XhoI sites of pGEX-5X (Amersham Biosciences, Arlington Heights, IL). Wild-type Rac1, Cdc42, and RhoA were N-terminally hemagglutinin (HA) tagged and subcloned into the pcDNA3 (Invitrogen). Rac1–61L, Rac1–17N, and RhoA-19N in pEF-BOS and wild-type Rac1 and RhoA in pGEX-4T to produce glutathioneS-transferase (GST) fusion proteins inEscherichia were gifts from Dr. A. Hall (Department of Biochemistry and Molecular Biology, University College London, London, UK). Wild-type Cdc42 was subcloned into pGEX-5X (Amersham Biosciences). The construct for Rac-binding domain of human PAK2 (PAK2-RBD; aa 66–147) in PGEX-4T was made as described previously (Akasaki et al., 1999). The construct for GST fusion to the RhoA binding domain of Rhotekin (GST-RBD) was a gift from Dr. M. A. Schwartz (Department of Cell Biology, The Scripps Research Institute, La Jolla, CA), and neural Wiskott-Aldrich syndrome protein (NWASP)-Cdc42/Rac1 interactive binding (CRIB) in pEF-BOS was from Dr. T. Takenawa (Division of Biochemistry, Institute of Medical Science, University of Tokyo, Tokyo, Japan).
Expression and purification of recombinant proteins.Bacterially expressed recombinant RhoA, Rac1, Cdc42, and FIR proteins were purified as described previously (Chuang et al., 1995).Escherichia strain DH5α transformed with the vectors was treated for 3 hr at 37°C with 0.1 mmisopropyl-thio-β-D-galactoside to induce the expression of each protein, which was purified through a glutathione-Sepharose 4B column.
In vitro nucleotide exchange assay. Purified Rho proteins were used directly for the3H-loaded GDP dissociation assays as described previously (Horii et al., 1994; Chuang et al., 1995). Briefly, 4 μg of each Rho GTPase was incubated with 10 μm [3H]GDP (PerkinElmer Life Sciences) for 25 min at room temperature, and GST-fused DH domain or DH–PH domain of FIR was added to the assay mixture. At the indicated time, an aliquot of the reaction sample was removed and passed through nitrocellulose filters (IPVH 000; Millipore). The filters were washed and used for scintillation counting. GST protein or the buffer was used as a control.
Interaction of FIR with Rho GTPases. Binding of FIR to nucleotide-free Rho GTPases was determined according to the procedure mentioned by Penzes et al. (2001). Briefly, GST fusion proteins of Rho GTPases purified from Escherichia were depleted of bound nucleotide by incubation with 10 mm EDTA. HEK293 cells expressing GFP-ΔN-FIR, which contains DH and PH domains of FIR, were lysed in binding buffer (40 mm Tris-HCl, pH 7.5, and 50 mm NaCl containing 1% Triton X-100). For each binding reaction, 5 μg of GST-GTPase bound to 25 μl of glutathione-Sepharose beads was mixed with an aliquot of cell extract containing 1 mg of protein for 2 hr at 4°C. The beads were washed with binding buffer, and bound proteins were analyzed by SDS-PAGE and Western blotting using the monoclonal anti-GFP antibody (Santa Cruz Biotechnology, Santa Cruz, CA).
Pull-down GTPase assay. In vivoRac1/Cdc42 activation assay was performed according to the method we described previously (Akasaki et al., 1999). HEK293T cells were transfected using Lipofectamine2000 (Invitrogen, Gaithersburg, MD), cultured for 48 hr, and lysed [in 20 mm HEPES, pH 7.4, 150 mmNaCl, 2% Nonidet P-40, 20% glycerol, 8 mm EGTA, 8 mm EDTA, 80 μmp-amidinophenylmethanesulfonyl fluoride (hydrochloride), 100 μg/ml aprotinin, and 200 μg/ml each of leupeptin, chymostatin, and pepstatin A]. Cell lysates were clarified by centrifugation, and the supernatant was incubated with 20 μg of GST-PAK2 protein immobilized on glutathione-Sepharose beads for 3 min. Beads were washed with washing buffer (20 mmHEPES, pH 7.4, 142.5 mm NaCl, 1% Nonidet P-40, 10% glycerol, 4 mm EGTA, and 4 mm EDTA), and bound GTP-Rho proteins were detected by Western blotting with the anti-HA monoclonal antibody (Boehringer Mannheim, Mannheim, Germany). In vivo RhoA activity assay was performed according to the method reported by Schwartz and his colleagues using a GST fusion to the RhoA binding domain of Rhotekin (GST-RBD) (Ren et al., 1999). To detect endogenous Rac1 activation by FIR, COS-7 cells were cultured for 24 hr after transfection, followed by the pull-down assay.
In vivo nucleotide labeling. Transfected HEK293T cells were cultured for 24 hr, serum starved in DMEM medium, labeled with [32P]orthophosphate (100 μCi/ml; PerkinElmer Life Sciences) for 4 hr, and disrupted in lysis buffer (50 mm Tris-HCl, pH 7.5, 20 mmMgCl2, 150 mm NaCl, 0.5% Nonidet-P40, 1 mm sodium orthovanadate, 1 mm PMSF, 25 μg/ml leupeptin, and 25 μg/ml aprotinin). Lysates were immunoprecipitated with the anti-HA monoclonal antibody for 2 hr using protein Sepharose G beads (Amersham Biosciences). Immunoprecipitates were washed three times in lysis buffer and twice in washing buffer (50 mmTris-HCl, pH 7.5, 20 mmMgCl2, and 500 mm NaCl) and finally were resuspended in 1 mKH2PO4, pH 3.4. Bound nucleotides were released by heating at 68°C and fractionated using polyethyleneimine thin-layer chromatography plates. Radioactive spots, located by autoradiography, were scraped off the plates and counted in a scintillation counter (Yamashita et al., 1999).
Cell cultures and transient transfections.NIH3T3cells and COS-7cells were cultured in DMEM containing 10% fetal bovine serum (Sigma, St. Louis, MO), penicillin, and streptomycin. For immunocytochemistry, cells grown on chamber slides for 3 d to 40–60% confluence were transfected with 0.15 μg of plasmid DNA per 1 cm2 and 0.25 μl/cm2 Lipofectamine2000 in complete serum-free medium for 5 hr, after which they were washed and fed with growth medium for 24 hr. Then, the medium was replaced with DMEM without serum for 16 hr. Cells fixed in 4.0% formaldehyde in PBS (50 mm NaPi, pH7.5, and 150 mmNaCl) for 10 min at room temperature were permeabilized and stained.
Neuronal cultures and transfections. Cerebral cortex from embryonic day 18 rat was digested with papain for 30 min at 37°C, followed by dissociation. Dissociated neurons were plated on the dishes precoated with poly-l-lysine in DMEM containing 10% fetal bovine serum (Sigma), penicillin, and streptomycin. After culturing for 1 d, the medium was replaced with DMEM with B-27 supplement. Two days after dissociation, cultures from rat embryo were transfected using Lipofectamine2000. The neurons were fixed 24 hr after transfection in 4.0% formaldehyde for 20 min, permeabilized, and blocked in PBS containing 5% normal goat serum, 0.1% bovine serum albumin, and 0.1% Triton X-100 for 30 min. The cultures were incubated overnight with the monoclonal antibody to class III β-tubulin (Research Diagnostics, Flanders, NJ), followed by Alexa Fluor-conjugated secondary antibody (Molecular Probes, Eugene, OR). Morphological features were quantified using LSM510 software version 2.02 (Zeiss, Oberkochen, Germany). Pictures of randomly selected fields were taken at low magnification, and the total neurite length as well as the length of the longest process on each individual neuron in the field was measured after it was traced using the computer program; at least 40 neurons from three independent cultures were analyzed.
Dissociated cultures of hippocampus from embryonic day 18 mouse were performed by the same procedure mentioned above. The hippocampal and the cortical neurons were maintained for 1 d after dissociation, and total RNA was isolated from them to perform RT-PCR.
RESULTS
Identification of a novel FERM domain including guanine nucleotide exchange factor for Rho GTPases
We first intended to identify candidate GEFs for Rho GTPases, which contain domains previously implicated in signal transduction, and searched DNA databases using a consensus amino acid sequence of the DH domain of known GEFs for Rho GTPases (Boguski and McCormick, 1993;Fukuhara et al., 1999). A number of yet uncharacterized proteins containing putative DH-like domains were detected. Subsequently, we analyzed their DNA sequences, and their expected translational products suggested that many of them encode putative GEFs for Rho GTPases. We were interested in one of them, KIAA0793, human cDNA clone (GenBank accession number AB018336). The plasmid construct for KIAA0793 was a gift from Dr. T. Nagase, and its nucleotide sequence was confirmed. The cDNA is 3165 bp long and encodes a putative protein consisting of 1055 amino acids. The proposed initiating ATG conforms to a Kozak consensus sequence (Kozak, 1986), and there is a stop codon just upstream of this ATG in frame (data not shown). The encoded putative protein is predicted to have a core molecular mass of 117 kDa and has highly homologous domains implicated in signal transduction (Fig. 1). We also detected possible homolog of KIAA0793 in Mus musculus (GenBank accession number BC009153), whose putative open reading frame consists of 1065 amino acids (Fig. 1A). The putative protein in mouse shows 83% identity and 97% similarity with that encoded by KIAA0793 (Fig. 1A). Each of these putative proteins has one highly conserved DH domain and two PH domains. In N-terminal region, they contain an interesting structure exhibiting homology to the FERM domain (band 4.1 homology domain) of ERM proteins (ezrin, radixin, and moesin). The FERM domain is known to associate with plasma membrane proteins such as CD44 (Bretscher et al., 1997; Tsukita et al., 1997;Vaheri et al., 1997) (Fig. 1B). As well established, a tandem of DH and PH domains are responsible for the nucleotide exchange activity of Rho GTPases, and, therefore, we tentatively named these newly detected molecules as FERM domain including RhoGEF (FIR), which might represent a novel exchange factor for Rho GTPases.
Fig. 1.
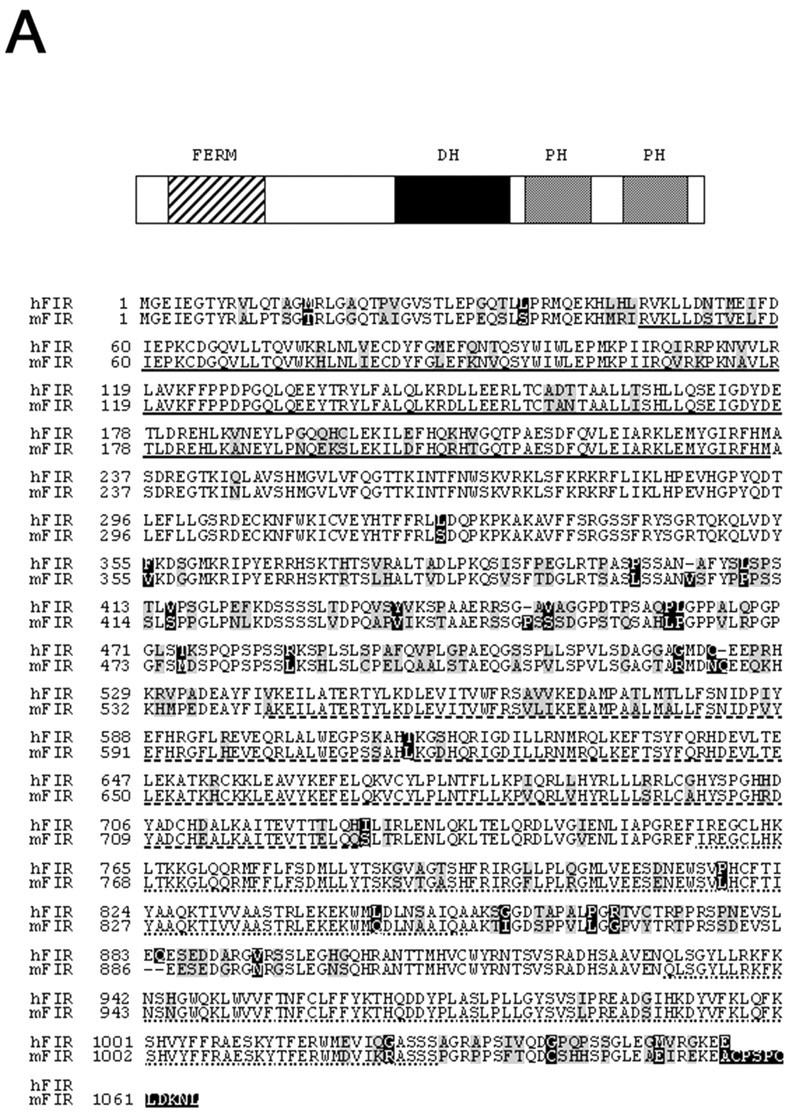

FIR contains several domains in signal transduction. A, Schematic structure and deduced amino acid sequence of human and mouse FIR. The sequences of human and mouse FIR were optimally aligned on the basis of residue identity (nonboxed) and similarity (gray box). Black boxes are nonconserved residues. FERM domain, Single underline; DH domain,interrupted line; two PH domains, dotted lines. B, C, Sequence comparison of FIR with ERM proteins, Dbl, and pleckstrin (B) and other Rac-GEFs (C). Black boxes, Identical residues; gray boxes, similar residues to FIR. hFIR, Human FIR; mFIR, mouse FIR; hSOS1, human SOS1; hTiam1, human Tiam1; hVav1, human Vav1.
Tissue distribution of FIR and the expression in neurons
We determined the tissue distribution of mouse FIR mRNA to gain insight into possible functional roles. A 800 bp fragment corresponding to mouse FIR (nucleotides 2311–3110) was used to specifically detect FIR mRNA. Northern blot analysis of a panel of tissues from adult mouse revealed an ∼8 kb mRNA species, with high levels of expression in brain, lung, and testis (Fig.2A). Low levels of expression could be found in heart and kidney. We next examined the expression of FIR mRNA in primary neuronal cultures (1 d after dissociation) derived from embryonic day 18 mice by RT-PCR. In both cortical and hippocampal neurons, the signals corresponding to FIR mRNA were detected (Fig. 2B). These results suggest that FIR may play some roles in neuronal cells in brain.
Fig. 2.

Tissue distribution of FIR and the expression in neurons. A, Northern blot analysis of a panel of tissues from adult mouse. Ribosomal RNA was used as a standard.B, The expression of mouse FIR mRNA in primary cultured embryonic day 18 hippocampal and cortical neurons was detected by RT-PCR. No signal for GFAP mRNA was found in cortical or hippocampal neurons. M, Molecular weight marker; N, no cDNA; C, cortical neurons; H, hippocampal neurons; W, adult mouse whole brain.
FIR activates Rac1 but not Cdc42 or RhoA
To determine the specific Rho GTPases on which FIR can catalyze GDP–GTP exchange, we used an in vitro assay that measured the ability of FIR to induce the dissociation of3H-labeled GDP from RhoA, Rac1, or Cdc42. As shown in Figure 3A, the isolated DH domain promoted nucleotide exchange of Rac1 but not on RhoA or Cdc42. A fused protein of DH and PH domains had the same effects on Rac1 as DH domain itself, suggesting that PH domain did not enhance nucleotide exchange of Rac1.
Fig. 3.
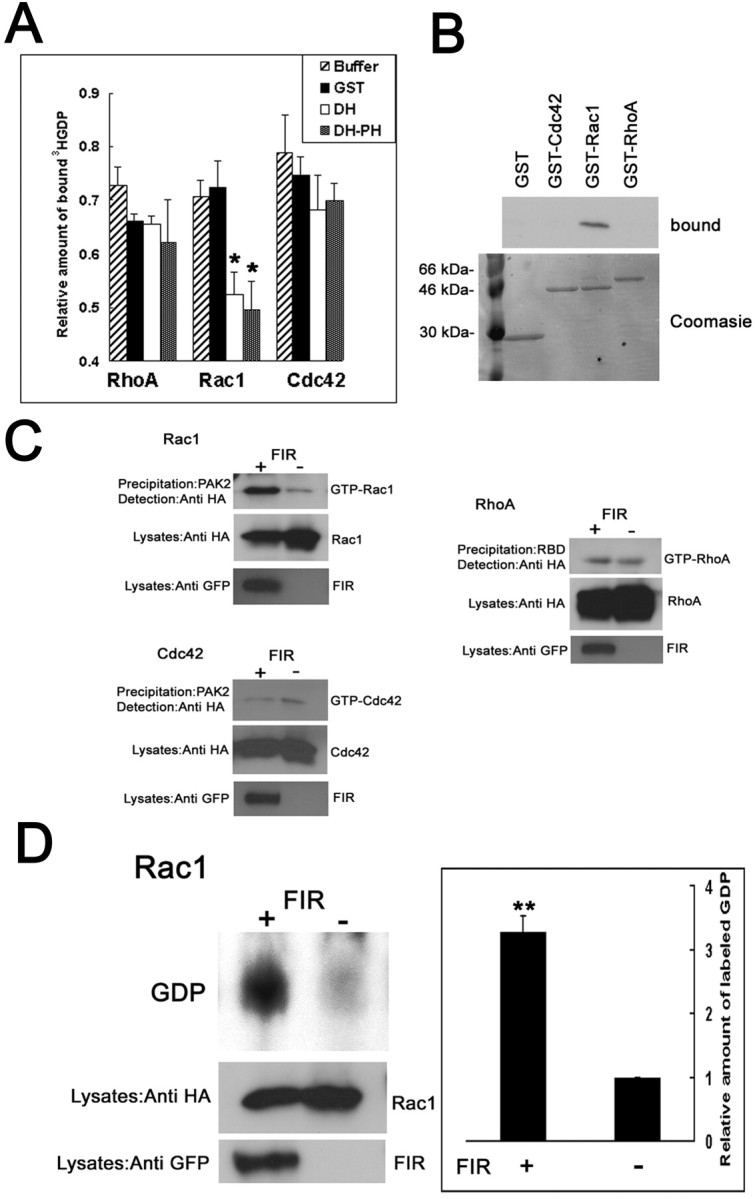
FIR activates Rac1 but not Cdc42 or RhoA.A, In vitro exchange activity of DH domain of FIR. The ability of DH or DH–PH domain of FIR to induce the dissociation of 3H-labeled GDP from RhoA, Rac1, or Cdc42 in 30 min was measured. GST protein or the incubation buffer was used as a control. The graph represents the average ± SE of relative amount of initial [3H]GDP remaining bound from three individual experiments. *p < 0.05;t test, compared with the control. B, Binding of the FIR with nucleotide-depleted Rho GTPases. Lysates (1 mg of protein) from the HEK293 cells transfected with GFP-ΔN-FIR were incubated with 5 μg of GST-fused Rho GTPases. Bound proteins were analyzed by Western blotting with the anti-GFP monoclonal antibody. Precipitated GST fusion proteins were visualized with Coomassie blue (bottom). C, Pull-down GTPase activity assays. The activity for Rho GTPases in HEK293T cells transiently cotransfected with HA-tagged RhoA and with or without FL-FIR was detected by affinity precipitation using GST fusions of their effectors. The amounts of Rho GTPases in the lysates are shown in the middle panels. FIR expression was confirmed with the anti-GFP antibody (bottom). D, Precipitation of [32P]GDP-Rac1. After32P labeling, HEK293 cells transfected with HA-tagged Rac1 and with or without FIR were immunoprecipitated with the anti-HA antibody. [32P]GDP was quantified by scraping the thin-layer plates and counting. Values represent relative amount of radioactivity and are expressed as means ± SEM of four experiments. **p < 0.005; t test, compared with the control vector. Expression of Rac1 (middle) or FIR (bottom) in the lysates was determined.
Next, we assessed binding of FIR with nucleotide-depleted Rho GTPases. HEK293 cells were transfected with the GFP-tagged N-terminal truncated form of FIR, which consists of DH and PH domains without FERM domain. Purified nucleotide-depleted GST fusion proteins of Cdc42, Rac1, and RhoA were incubated with the lysates from the transfected HEK293 cells. The results show that FIR interacted with nucleotide-depleted Rac1 but not with RhoA or Cdc42 (Fig. 3B). These data support the notion that FIR acts on Rac1.
To examine whether FIR acts as a Rac1-specific GEF also in vivo, we measured the activity for Rac1, RhoA, or Cdc42 in HEK293T cells transiently transfected with or without FIR construct by affinity precipitation. Because Rho GTPases in the GTP-bound state bind to their downstream effectors, GST fusions of these effectors can be used to capture active Rho GTPases from cell lysates. Thus, a GST fusion to the Rac1/Cdc42 binding domain of PAK (GST-PAK2) was used to specifically precipitate GTP-bound Rac1 or Cdc42 from cell extracts (Akasaki et al., 1999). RhoA activity was examined using a GST fusion to the RhoA binding domain of Rhotekin (GST-RBD) (Ren et al., 1999). This assay revealed that extracts of HEK293T cells transfected with Rac1 and FIR contained increased amount of GTP-Rac1 compared with the control cells without FIR expression (Fig. 3B), although the levels of expression of Rac1 were comparable. As expected, no or little activation of RhoA or Cdc42 could be found (Fig.3C).
We further confirmed Rac1 activation by FIR by radioactive in vivo nucleotide labeling. HEK293T cells transiently transfected with HA-tagged Rac1 with or without FIR were32P-labeled, and the radioactivity associated with Rac1 was determined by immunoprecipitation with the anti-HA antibody, followed by thin-layer chromatography. The radioactivity precipitated with Rac1 comigrated with a GDP standard. Because Rho proteins have a high intrinsic GTPase activity, this assay precludes the detection of radiolabeled GTP and reflects the nucleotide exchange rates of the GTPases (Yamashita et al., 1999). Massive32P-labeled GDP associated with Rac1 was coprecipitated in the presence of FIR, whereas levels were low in the absence of FIR (Fig. 3D). Together, these results demonstrate that the catalytic domain of FIR acts specifically on Rac1 both in vitro and in vivo. In fact, amino acid sequence of DH domain of FIR is highly similar to that of other Rac-GEFs, such as SOS1, Tiam1, and Vav1 (Fig. 1C). Similarity of the putative DH domain of human FIR with that of human SOS1, Tiam1, or Vav1 is 63.2, 65.6, or 61.5%, respectively.
Effects of FIR on the actin cytoskeleton
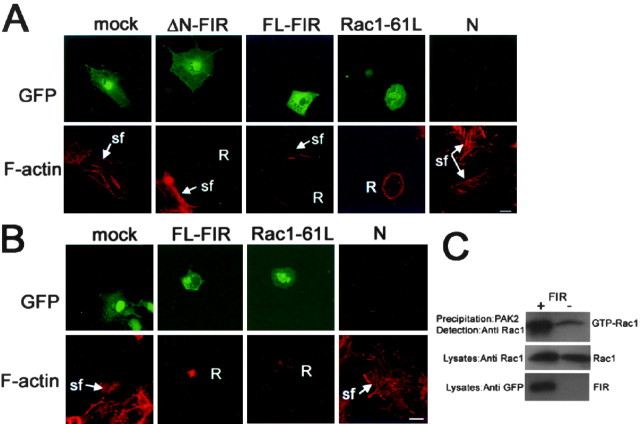
We next examined the effects of FIR on the morphology of fibroblasts, in which RhoA, Rac1, and Cdc42 each elicits distinct morphological changes. Specifically, RhoA induces stress fiber formation, Cdc42 induces filopodia extension, and Rac1 induces lamellipodia formation and membrane ruffling (Hall, 1998). We transfected NIH3T3 fibroblasts with the plasmid for GFP-fused FIR or GFP. After serum starvation for 16 hr, we detected the expression of FIR by GFP autofluorescence and examined the actin structures by staining F-actin with rhodamine-phalloidin. GFP-FL-FIR-transfected cells displayed lamellipodia and ruffles, indicative of Rac (Hall, 1998; Penzes et al., 2000) (Fig.4A). Stress fiber formation was significantly suppressed in the cells expressing FIR compared with the control cells expressing GFP. These changes in actin cytoskeleton induced by FIR were observed in the cells transfected with the dominant active form of Rac1 (Rac1–61L), consistent with our biochemical data that show activation of Rac1 by FIR. Next, we made a GFP fusion to N-terminal truncated form of FIR, in which FERM domain was deleted, to assess whether FERM domain was not necessary to induce these changes in actin structures. N-terminal truncated FIR also induced morphological changes characteristic of the activation of Rac1. Interestingly, the cells expressing full-length FIR fusion protein, which contains FERM domain, showed a punctate pattern similar to that observed previously for ERM proteins fused to GFP (Mangeat et al., 1999; Olsson et al., 1999), whereas GFP signals for N-terminal truncated FIR were diffuse in the cytoplasm (Fig.4A). Signals for N-terminal truncated FIR were seen in the nucleus, as is the case with Rac1, although function of Rac1 in the nucleus has remained to be elucidated (Kraynov et al., 2000). In contrast, the full-length FIR was mainly localized in the cytoplasm, suggesting that FERM domain may regulate the subcellular localization of FIR.
Fig. 4.
Effects of FIR on the actin cytoskeleton.A, NIH3T3 cells transfected with the full-length FIR and N-terminal truncated FIR displayed lamellipodia and ruffles, indicative of Rac. Scale bar, 50 μm. B, COS-7 cells transfected with FIR showed similar phenotypic changes induced by Rac1-61L Scale bar, 50 μm. C, Endogenous Rac1 activation by FIR in COS-7 cells was detected using pull-down Rac1 activation assay.N, Nontransfected cells; R, ruffles;sf, stress fibers.
These alterations of the actin structures by expression of FIR were also found in COS-7 cells (Fig.5B). Pull-down Rac1 activation assay revealed that endogenous Rac1 was activated in COS-7 cells transiently transfected with FIR compared with the control cells (Fig.5C). Overall, these results show that FIR regulates the structure of actin cytoskeleton presumably through activation of Rac1.
Fig. 5.
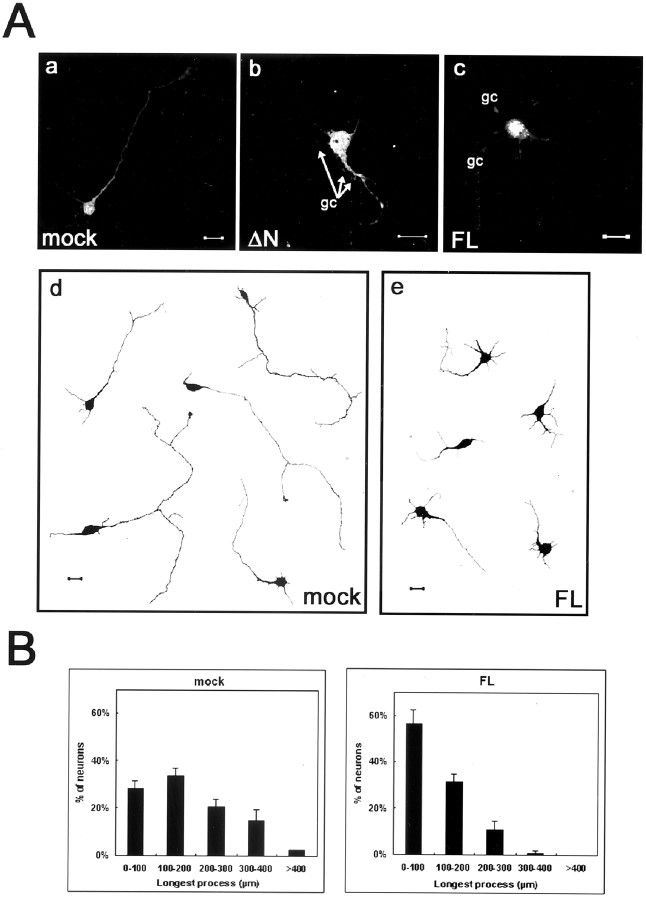
FIR regulates neuronal process length.A, Cortical neurons (2 d after dissociation) were transfected with the empty pEGFP vector (a), pEGFP-ΔN-FIR (b), or pEGFP-FL-FIR (c). Transfected cells were detected by GFP autofluorescence. Neurons overexpressing FL-FIR or ΔN-FIR display multiple lateral growth cones extending from neurites and growth cones on their cell soma. Computer-assisted tracing of β-tubulin III-labeled neurites of control GFP (c)- and FL-FIR (d)-expressing neurons revealed apparent difference between two groups. Scale bar, 20 μm. B, The length of the longest process in each FL-FIR- or GFP-expressing neurons was analyzed. Histograms of lengths of the longest process of transfected neurons. For B, error bars are the SE for three experiments, each containing 40–50 neurons. Overexpression of FIR affects the length of neurites. FL, Full-length;gc, growth cones.
FIR regulates neuronal morphology
Because FIR mRNA was expressed in cortical and hippocampal neurons in the developmental stages as well as the adult brain from mice, it is suggested that FIR may play some roles in neuronal function. Therefore, we established an assay to directly address the role of FIR in regulating neuronal morphology. Dissociated culture of embryonic 18 d rat cortical neurons were transfected with GFP or GFP-fused FIR and cultured in the defined medium for 48 hr, and then morphological changes were assessed. Neurons overexpressing the full-length or N-terminal truncated FIR often displayed multiple lateral growth cones extending from neurites compared with those transfected with GFP (Fig. 5A). A large fraction of neurons overexpressing the full-length or N-terminal truncated FIR had growth cones on their cell soma. These alternations in cortical neurons are consistent with the previous finding in which the cortical neurons were transfected with Rac1-specific GEF (Penzes et al., 2001). Similar to the finding in NIH3T3 cells, cortical neurons overexpressing the full-length FIR, which contains FERM domain, showed the punctated pattern, characteristic of ERM proteins.
Next, we evaluated the effects of overexpression of FIR by measuring the total neurite length per neuron and the length of the longest neurite per neuron visualized by autofluorescence. The average length of the longest neurite per full-length FIR-expressing neuron was significantly shorter than that from the GFP control (Fig.5Ad,Ae,B). The average length of the longest neurite per neuron expressing GFP-fused N-terminal truncated FIR was not significantly different from that expressing full-length FIR (data not shown). Exactly the same results were obtained when the total neurite length per neuron was measured (data not shown). Thus, regulation of neurite outgrowth by overexpression of FIR in embryonic cortical neurons may be attributable to DH and PH domains of FIR rather than its FERM domain.
Neurite remodeling by FIR is dependent on activation of Rac1
The effects of FIR on the neuronal morphology may be attributable to Rac1 activation. To test this hypothesis, we used mammalian expression vectors for constitutive active and dominant negative forms of Rac1 (Ridley and Hall, 1992; Nobes and Hall, 1995). Cortical neurons were transfected with constitutive active Rac1 (Rac1–61L). The average length of the longest neurite per neuron expressing Rac1–61L was significantly shorter than the control (Figs.5B, 6A), demonstrating that the effect of Rac1–61L was comparable with that of FIR. On the other hand, ectopic expression of the dominant negative mutant of Rac1 (Rac1–17N) blocked the effect of FIR with regard to the neurite outgrowth (Figs. 5B, 6A). We further tested whether RhoA or Cdc42 was involved in FIR signaling. However, cotransfection of dominant negative RhoA (RhoA-19N) did not modify the effect of FIR (Figs. 5B, 6B). Coexpression of FIR with CRIB domain of NWASP, which was used to competitively inhibit Cdc42 (Miki et al., 1998), also failed to affect the morphology induced by FIR (Figs. 5B,6B). These data demonstrate that FIR regulates neurite remodeling of the dissociated cortical neurons through activation of Rac1.
Fig. 6.
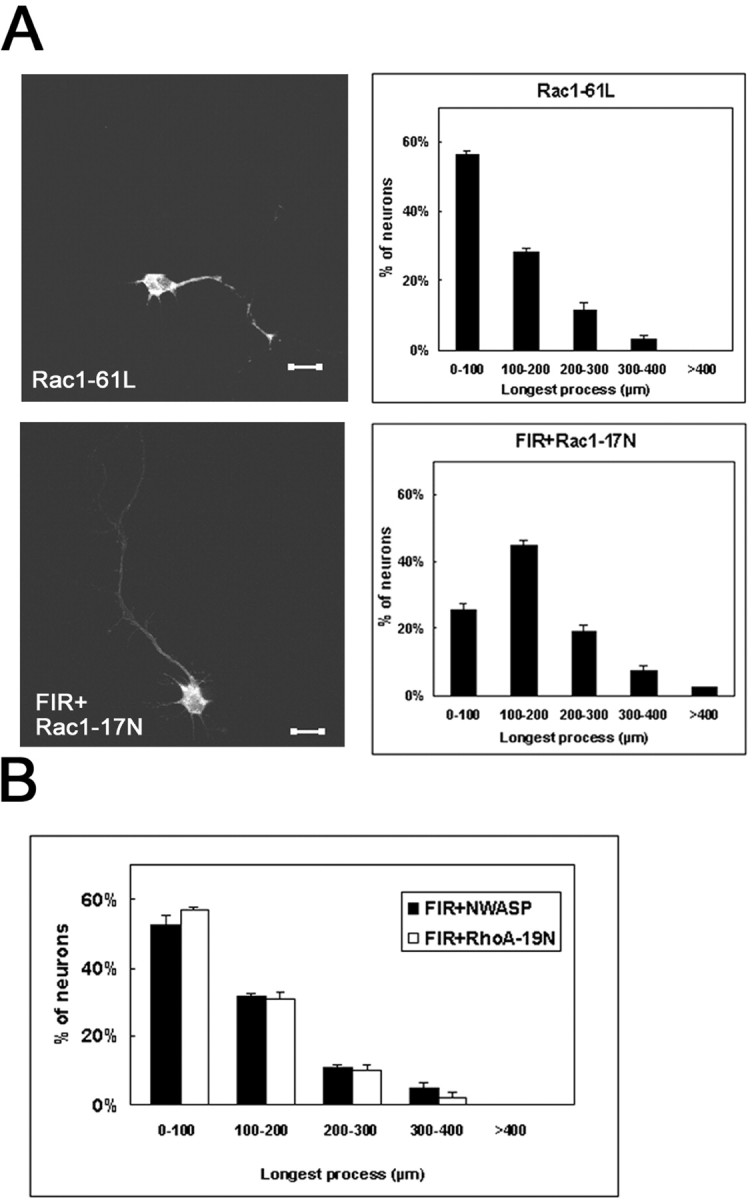
Neurite remodeling by FIR is involved in Rac1 signaling. A, Cortical neurons expressing Rac1-61L (top) or Rac1-17N plus FIR (bottom). Transfected cells were detected by GFP autofluorescence.B, The length of the longest process in each neuron expressing FIR plus NWASP or FIR plus RhoA-19N was analyzed. Histograms of the length of the longest process of the transfected neuron. ForA and B, error bars are SE from three experiments, each containing 40–50 neurons.
DISCUSSION
The GEFs from the Dbl family are multifunctional molecules that transduce diverse intracellular signals leading to the activation of Rho GTPases. The tandem of DH and PH domains shared by all members of this family represent the structural module responsible for catalyzing the GDP–GTP exchange reaction of Rho GTPases. Recent progress in genomic, genetic, structural, and biochemical studies have implicated GEFs from Dbl family members in diverse biological processes, including growth and development, tissue organization, and neuronal axon guidance. In the nervous system, Rho GTPases are essential for establishing highly asymmetrical neuronal forms and might adjust the shape of neurites in differentiated neurons (Zhai et al., 2001). This notion is substantiated by the facts that expression of dominant inactive forms of Rac1 and Cdc42 caused defects in axon guidance and cell migration in C. elegans, Drosophila, and mouse (Luo, 2000). Among many regulators of Rho GTPases, Trio and ephexin, GEFs for Rho GTPases, have emerged recently as key factors for axon guidance (Bateman et al., 2000; Shamah et al., 2001). Trio and ephexin associate with the receptor phosphatase LAR and EphA, respectively, and transduce the signal from these receptors to Rho GTPases. However, because at least these GEFs seem to mediate signals from the specific receptors and there are much more guidance receptors, identified so far, whose molecular signals remain to be elucidated, new molecules will be added in the course of elucidation of the mechanisms of neuronal navigation in the future. We identified a previously uncharacterized GEF for Rac and named FERM domain including RhoGEF (FIR), which would be implicated in axon guidance, because FIR mRNA is expressed in cortical and hippocampal neurons during the developmental stages. FIR shares mild homology with CDEP (chondrocyte-derived ezrin-like domain-containing protein), which has a FERM domain at the N terminus and a DH domain followed by two PH domains in the C-terminal region (Koyano et al., 1997, 2001). However, CDEP was reported to act on Rho in vitro, and the distribution pattern was different from FIR, suggesting distinct function of CDEP. To clearly elucidate the specific roles of these GEFs, it will be necessary to find the interactors of FIR to find how FIR is regulated in the cells.
Previous reports have shown diverse effects of Rho GTPases on neuronal morphology (Luo et al., 1994; Li et al., 2000). Activated Rac1 inhibited axonal outgrowth of both Drosophila sensory and mouse Purkinje neurons (Luo et al., 1994; Luo et al., 1997), suppressed axonal formation in Xenopus retinal ganglion cells (Ruchhoeft et al., 1999), and elicited growth cone collapse in embryonic chick dorsal root ganglion neurons (Jin and Strittmatter, 1997). Penzes et al. (2001) showed shortened axons and excessive growth cones of rat cortical neurons, mediated by Rac1-specific catalytic domain of RhoGEF protein, Kalirin-9. The inhibition of neurite elongation and morphological changes by overexpression of FIR in rat cortical neurons through Rac1 activation are consistent with these previous reports showing Rac1 phenotype. However, the effects of any DH domain might depend on the complement of Rho GTPases present in any given neuron at a particular time (Penzes et al., 2001). In addition, the action of Rho GTPases might vary with cell type and developmental stage. For example, we found that axonal outgrowth was facilitated through inactivation of RhoA and inhibited by activation of RhoA in other cells, such as embryonic chick ciliary neurons, cerebellar granule neurons, and hippocampal neurons (Yamashita et al., 1999;Neumann et al., 2002; Yamashita et al., 2002), that are seemingly contradictory. Therefore, FIR may mediate diverse actions that are dependent on the cell context.
Many Rho GTPase-specific GEFs contain multiple protein motifs involved in intracellular signal transduction, such as Src homology domains and PDZ domains (Matsuo et al., 2002). FIR includes the FERM domain. ERM proteins, collectively composed of ezrin, radixin, and moesin, are a group of closely related membrane cytoskeleton linkers that regulate cell adhesion and cortical morphogenesis (Mangeat et al., 1999). The N-terminal membrane binding domain, the so-called FERM domain, of FERM has been shown to associate with several membrane associated proteins, such as hyaluronan receptor CD44 (Tsukita et al., 1994), intercellular adhesion molecule (ICAM-1), and ICAM-2 (Heiska et al., 1998). Therefore, considering the fact that Rho regulators play important roles in neuronal navigation, it is possible that FIR transmits signals from plasma membrane receptors for guidance molecules to cytoskeletal reorganization through interaction of FERM domain with the receptors. In fact, Max-1, a recently identified cytoplasmic protein that has FERM domain, was shown to be involved in netrin-induced axonal guidance by modulating the Unc5 receptor signaling pathway (Huang et al., 2002). In addition, FERM proteins themselves are also known as upstream regulators of RhoA, and their FERM domain can bind directly with PH domain of Dbl (Mangeat et al., 1999; Tsukita and Yonemura, 1999;Louvet-Vallee, 2000). Thus, the FERM domain of FIR may elicit Rho signal through binding to Dbl, independent of the internal DH domain. Alternatively, it is possible that FERM domain and the internal PH domains might mutually interact intramolecularly and regulate additional transductional functions for the cytoskeleton. Detailed structure–function analyses of FIR should help to elucidate the precise mechanism of FIR in regulating cell morphology, especially in neurons.
Footnotes
We thank Yumiko Hara and Akemi Arakawa for participating in this work.
Correspondence should be addressed to Dr. Toshihide Yamashita, Department of Anatomy and Neuroscience, Osaka University Graduate School of Medicine, 2-2 Yamadaoka, Suita, Osaka 565-0871, Japan. E-mail: tyama@anat2.med.osaka-u.ac.jp.
REFERENCES
- 1.Akasaki T, Koga H, Sumimoto H. Phosphoinositide 3-kinase-dependent and -independent activation of the small GTPase Rac2 in human neutrophils. J Biol Chem. 1999;274:18055–18059. doi: 10.1074/jbc.274.25.18055. [DOI] [PubMed] [Google Scholar]
- 2.Awasaki T, Saito M, Sone M, Suzuki E, Sakai R, Ito K, Hama C. The Drosophila trio plays an essential role in patterning of axons by regulating their directional extension. Neuron. 2000;26:119–131. doi: 10.1016/s0896-6273(00)81143-5. [DOI] [PubMed] [Google Scholar]
- 3.Bateman J, Shu H, Van Vactor D. The guanine nucleotide exchange factor trio mediates axonal development in the Drosophila embryo. Neuron. 2000;26:93–106. doi: 10.1016/s0896-6273(00)81141-1. [DOI] [PubMed] [Google Scholar]
- 4.Boguski MS, McCormick F. Proteins regulating Ras and its relatives. Nature. 1993;366:643–654. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- 5.Bretscher A, Reczek D, Berryman M. Ezrin: a protein requiring conformational activation to link microfilaments to the plasma membrane in the assembly of cell surface structures. J Cell Sci. 1997;110:3011–3018. doi: 10.1242/jcs.110.24.3011. [DOI] [PubMed] [Google Scholar]
- 6.Chuang TH, Xu X, Kaartinen V, Heisterkamp N, Groffen J, Bokoch GM. Abr and Bcr are multifunctional regulators of the Rho GTP-binding protein family. Proc Natl Acad Sci USA. 1995;92:10282–10286. doi: 10.1073/pnas.92.22.10282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fukuhara S, Murga C, Zohar M, Igishi T, Gutkind JS. A novel PDZ domain containing guanine nucleotide exchange factor links heterotrimeric G proteins to Rho. J Biol Chem. 1999;274:5868–5879. doi: 10.1074/jbc.274.9.5868. [DOI] [PubMed] [Google Scholar]
- 8.Hall A. Identification of two distinct mechanisms of phagocytosis controlled by different Rho GTPases. Science. 1998;279:509–514. doi: 10.1126/science.282.5394.1717. [DOI] [PubMed] [Google Scholar]
- 9.Heiska L, Alfthan A, Gronholm M, Vilja P, Vaheri A, Carpen O. Association of ezrin with intercellular adhesion molecule-1 and -2 (ICAM-1 and ICAM-2). Regulation by phosphatidylinositol 4, 5-bisphosphate. J Biol Chem. 1998;273:21893–21900. doi: 10.1074/jbc.273.34.21893. [DOI] [PubMed] [Google Scholar]
- 10.Horii Y, Beeler JF, Sakaguchi K, Tachibana M, Miki T. A novel oncogene, ost, encodes a guanine nucleotide exchange factor that potentially links Rho and Rac signaling pathways. EMBO J. 1994;13:4776–4786. doi: 10.1002/j.1460-2075.1994.tb06803.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang X, Cheng HJ, Tessier-Lavigne M, Jin Y. Max-1, a novel PH/MyTH4/FERM domain cytoplasmic protein implicated in netrin-mediated axon repulsion. Neuron. 2002;34:563–576. doi: 10.1016/s0896-6273(02)00672-4. [DOI] [PubMed] [Google Scholar]
- 12.Jin Z, Strittmatter SM. Rac1 mediates collapsin-1-induced growth cone collapse. J Neurosci. 1997;17:6256–6263. doi: 10.1523/JNEUROSCI.17-16-06256.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koyano Y, Kawamoto T, Shen M, Yan W, Noshiro M, Fujii K, Kato Y. Molecular cloning and characterization of CDEP, a novel human protein containing the ezrin-like domain of the band 4.1 superfamily and the Dbl homology domain of Rho guanine nucleotide exchange factors. Biochem Biophys Res Commun. 1997;241:369–375. doi: 10.1006/bbrc.1997.7826. [DOI] [PubMed] [Google Scholar]
- 14.Koyano Y, Kawamoto T, Kikuchi A, Shen M, Kuruta Y, Tsutsumi S, Fujimoto K, Noshiro M, Fujii K, Kato Y. Chondrocyte-derived ezrin-like domain containing protein (CDEP), a rho guanine nucleotide exchange factor, is inducible in chondrocytes by parathyroid hormone and cyclic AMP and has transforming activity in NIH3T3 cells. Osteoarthritis Cartilage. 2001;9:S64–S68. doi: 10.1053/joca.2001.0446. [DOI] [PubMed] [Google Scholar]
- 15.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;31:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 16.Kraynov VS, Chamberlain C, Bokoch GM, Schwartz MA, Slabaugh S, Hahn KM. Localized Rac activation dynamics visualized in living cells. Science. 2000;290:333–337. doi: 10.1126/science.290.5490.333. [DOI] [PubMed] [Google Scholar]
- 17.Kunda P, Paglini G, Quiroga S, Kosik K, Caceres A. Evidence for the involvement of Tiam1 in axon formation. J Neurosci. 2001;21:2361–2372. doi: 10.1523/JNEUROSCI.21-07-02361.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamarche N, Hall A. GAPs for rho-related GTPases. Trends Genet. 1994;10:436–440. doi: 10.1016/0168-9525(94)90114-7. [DOI] [PubMed] [Google Scholar]
- 19.Li Z, van Aelst L, Cline HT. Rho GTPases regulate distinct aspects of dendritic arbor growth in Xenopus central neurons in vivo. Nat Neurosci. 2000;3:217–225. doi: 10.1038/72920. [DOI] [PubMed] [Google Scholar]
- 20.Liebl EC, Forsthoefel DJ, Franco LS, Sample SH, Hess JE, Cowger JA, Chandler MP, Shupert AM, Seeger MA. Dosage-sensitive, reciprocal genetic interactions between the Abl tyrosine kinase and the putative GEF trio reveal trio's role in axon pathfinding. Neuron. 2000;26:107–118. doi: 10.1016/s0896-6273(00)81142-3. [DOI] [PubMed] [Google Scholar]
- 21.Louvet-Vallee S. ERM proteins: from cellular architecture to cell signaling. Biol Cell. 2000;92:305–316. doi: 10.1016/s0248-4900(00)01078-9. [DOI] [PubMed] [Google Scholar]
- 22.Luo L. Rho GTPases in neuronal morphogenesis. Nat Rev Neurosci. 2000;1:260–264. doi: 10.1038/35044547. [DOI] [PubMed] [Google Scholar]
- 23.Luo L, Liao YJ, Jan LY. Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes Dev. 1994;8:1787–1803. doi: 10.1101/gad.8.15.1787. [DOI] [PubMed] [Google Scholar]
- 24.Luo L, Jan LY, Jan YN. Rho family GTP-binding proteins in growth cone signalling. Curr Opin Neurobiol. 1997;7:81–86. doi: 10.1016/s0959-4388(97)80124-9. [DOI] [PubMed] [Google Scholar]
- 25.Mackay DG, Hall A. Rho GTPases. J Biol Chem. 1998;273:20685–20688. doi: 10.1074/jbc.273.33.20685. [DOI] [PubMed] [Google Scholar]
- 26.Mangeat P, Roy C, Martin M. ERM proteins in cell adhesion and membrane dynamics. Trends Cell Biol. 1999;9:187–192. doi: 10.1016/s0962-8924(99)01544-5. [DOI] [PubMed] [Google Scholar]
- 27.Matsuo N, Hoshino M, Yoshizawa M, Nabeshima Y. Characterization of STEF, a guanine nucleotide exchange factor for Rac1, required for neurite growth. J Biol Chem. 2002;277:2860–2868. doi: 10.1074/jbc.M106186200. [DOI] [PubMed] [Google Scholar]
- 28.Miki H, Sasaki T, Takai Y, Takenawa T. Induction of filopodium formation by a WASP-related actin-depolymerizing protein N-WASP. Nature. 1998;391:93–96. doi: 10.1038/34208. [DOI] [PubMed] [Google Scholar]
- 29.Neumann H, Schweigreiter R, Yamashita T, Rosenkranz K, Wekerle H, Barde YA. Tumor necrosis factor inhibits neurite outgrowth and branching of hippocampal neurons by a rho-dependent mechanism. J Neurosci. 2002;22:854–862. doi: 10.1523/JNEUROSCI.22-03-00854.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newsome TP, Schmidt S, Dietzl G, Keleman K, Asling B, Debant A, Dickson BJ. Trio combines with dock to regulate Pak activity during photoreceptor axon pathfinding in Drosophila. Cell. 2000;101:283–294. doi: 10.1016/s0092-8674(00)80838-7. [DOI] [PubMed] [Google Scholar]
- 31.Nobes CD, Hall A. Rho, Rac, and Cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 32.Olsson P, Korhonen L, Mercer EA, Lindholm D. MIR is a novel ERM-like protein that interacts with myosin regulatory light chain and inhibits neurite outgrowth. J Biol Chem. 1999;274:36288–36292. doi: 10.1074/jbc.274.51.36288. [DOI] [PubMed] [Google Scholar]
- 33.Penzes P, Johnson RC, Alam MR, Kambampati V, Mains RE, Eipper BA. An isoform of kalirin, a brain-specific GDP/GTP exchange factor, is enriched in the postsynaptic density fraction. J Biol Chem. 2000;275:6395–6403. doi: 10.1074/jbc.275.9.6395. [DOI] [PubMed] [Google Scholar]
- 34.Penzes P, Johnson RC, Kambampati V, Mains RE, Eipper BA. Distinct roles for the two Rho GDP/GTP exchange factor domains of kalirin in regulation of neurite growth and neuronal morphology. J Neurosci. 2001;21:8426–8434. doi: 10.1523/JNEUROSCI.21-21-08426.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ren XD, Kiosses WB, Schwartz MA. Regulation of the small GTP-binding protein Rho by cell adhesion and the cytoskeleton. EMBO J. 1999;18:578–585. doi: 10.1093/emboj/18.3.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richnau N, Aspenstrom P. Rich, a rho GTPase-activating protein domain-containing protein involved in signaling by Cdc42 and Rac1. J Biol Chem. 2001;276:35060–35070. doi: 10.1074/jbc.M103540200. [DOI] [PubMed] [Google Scholar]
- 37.Ridley AJ, Hall A. The small GTP-binding protein regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 1992;70:389–399. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- 38.Ruchhoeft ML, Ohnuma S, McNeill L, Holt CE, Harris WA. The neuronal architecture of Xenopus retinal ganglion cells is sculpted by rho-family GTPases in vivo. J Neurosci. 1999;19:8454–8463. doi: 10.1523/JNEUROSCI.19-19-08454.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shamah SM, Lin MZ, Goldberg JL, Estrach S, Sahin M, Hu L, Bazalakova M, Neve RL, Corfas G, Debant A, Greenberg ME. EphA receptors regulate growth cone dynamics through the novel guanine nucleotide exchange factor ephexin. Cell. 2001;105:233–244. doi: 10.1016/s0092-8674(01)00314-2. [DOI] [PubMed] [Google Scholar]
- 40.Stam JC, Collard JG. The DH protein family, exchange factors for Rho-like GTPases. Prog Mol Subcell Biol. 1999;22:51–83. doi: 10.1007/978-3-642-58591-3_4. [DOI] [PubMed] [Google Scholar]
- 41.Steven R, Kubiseski TJ, Zheng H, Kulkarni S, Mancillas J, Ruiz Morales A, Hogue CW, Pawson T, Culotti J. UNC-73 activates the Rac GTPase and is required for cell and growth cone migrations in C. elegans. Cell. 1998;92:785–795. doi: 10.1016/s0092-8674(00)81406-3. [DOI] [PubMed] [Google Scholar]
- 42.Threadgill R, Bobb K, Ghosh A. Regulation of dendritic growth and remodeling by Rho, Rac, and Cdc42. Neuron. 1997;19:625–634. doi: 10.1016/s0896-6273(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 43.Tsukita S, Yonemura S. Cortical actin organization: lessons from ERM (ezrin/radixin/moesin) proteins. J Biol Chem. 1999;274:34507–34510. doi: 10.1074/jbc.274.49.34507. [DOI] [PubMed] [Google Scholar]
- 44.Tsukita S, Oishi K, Sato N, Sagara J, Kawai A, Tsukita S. ERM family members as molecular linkers between the cell surface glycoprotein CD44 and actin-based cytoskeletons. J Cell Biol. 1994;126:391–401. doi: 10.1083/jcb.126.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsukita S, Yonemura S, Tsukita S. ERM proteins: head-to-tail regulation of actin-plasma membrane interaction. Trends Biochem Sci. 1997;22:53–58. doi: 10.1016/s0968-0004(96)10071-2. [DOI] [PubMed] [Google Scholar]
- 46.Vaheri A, Carpen O, Heiska L, Helander TS, Jaaskelainen J, Majander-Nordenswan P, Sainio M, Timonen T, Turunen O. The ezrin protein family: membrane-cytoskeleton interactions and disease associations. Curr Opin Cell Biol. 1997;9:659–666. doi: 10.1016/s0955-0674(97)80119-6. [DOI] [PubMed] [Google Scholar]
- 47.van Aelst L, D'Souza-Schorey C. Rho GTPases and signaling networks. Genes Dev. 1997;11:2295–2322. doi: 10.1101/gad.11.18.2295. [DOI] [PubMed] [Google Scholar]
- 48.Whitehead IP, Campbell S, Rossman KL, Der CJ. Dbl family proteins. Biochim Biophys Acta. 1997;1332:F1–F23. doi: 10.1016/s0304-419x(96)00040-6. [DOI] [PubMed] [Google Scholar]
- 49.Yamashita T, Tucker KL, Barde YA. Neurotrophin binding to the p75 receptor modulates Rho activity and axonal outgrowth. Neuron. 1999;24:585–593. doi: 10.1016/s0896-6273(00)81114-9. [DOI] [PubMed] [Google Scholar]
- 50.Yamashita T, Higuchi H, Tohyama M. The p75 receptor transduces the signal from myelin-associated glycoprotein to Rho. J Cell Biol. 2002;157:565–570. doi: 10.1083/jcb.200202010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zalcman G, Dorseuil O, Garcia-Ranea JA, Gacon G, Camonis J. RhoGAPs and RhoGDIs, (His)stories of two families. Prog Mol Subcell Biol. 1999;22:85–113. doi: 10.1007/978-3-642-58591-3_5. [DOI] [PubMed] [Google Scholar]
- 52.Zhai J, Lin H, Shamim M, Schlaepfer WW, Canete-Soler R. Identification of a novel interaction of 14–3-3 with p190RhoGEF. J Biol Chem. 2001;276:41318–41324. doi: 10.1074/jbc.M107709200. [DOI] [PubMed] [Google Scholar]