Abstract
Sensory neurons can respond to dynamic stimuli with temporally precise firing events. In the lateral geniculate nucleus (LGN) of the thalamus, we found previously that when a flickering visual stimulus was repeated, individual cells fired action potentials at the same time in every trial to within 1 msec. We now show that these precise firing events are also reproducible across cells of the same class. Therefore, the mechanisms for producing precise timing must be conserved within a cell class. Our results further suggest that cortical neurons would require only a few generic processing mechanisms to extract the fine temporal information available in their LGN inputs.
Keywords: reliability, precision, synchrony, timing, coding, thalamus, LGN, vision
Lateral geniculate nucleus (LGN) relay cells can be remarkably deterministic, with reliable and temporally precise responses controlled primarily by the visual stimulus and altered very little by noise (Reinagel and Reid, 2000). Similar results have been found for many other visual neurons (Bair et al., 1994; Berry et al., 1997; de Ruyter van Steveninck et al., 1997;Buračas et al., 1998). Yet the reliability of each LGN neuron does not imply similarity of different neurons. Reliable, precise firing events could be determined by the idiosyncrasies of each LGN cell and the retinal circuit providing its input (such as connectivity, synaptic strengths, dendritic branching, and balance of conductances). Indeed, heterogeneity of precise firing events might be expected because LGN neurons within a class, and their retinal inputs, vary significantly in other temporal properties, such as the average time courses of visual responses (Victor, 1987, 1988; Cai et al., 1997;Wolfe and Palmer, 1998).
Precise temporal patterns of firing in the LGN carry rich information about time-varying visual stimuli, such as a strong diffuse flicker; however, whether this information is transmitted to the cortex depends on the nature of the cortical responses to the LGN input. If cortical cells discriminated LGN spike times to a precision of only 8 msec, they could use <50% of the available information; but with 1 msec precision, they could use >90% of the available information (Reinagel and Reid, 2000). Moreover, if cortical targets could distinguish different sequences of spikes, up to ∼20% more information could be extracted from some LGN cells (Reinagel and Reid, 2000). This raises the question of whether cortical cells are sensitive to either precise spike timing or spike interval patterns.
The synapse between LGN neurons and their targets in layer 4 of the primary visual cortex can transmit LGN spikes with a temporal precision of ∼1 msec in vivo (Tanaka, 1983; Reid and Alonso, 1995). In addition, this thalamocortical transmission is sensitive to temporal patterns: the efficacy of an LGN spike in driving a cortical target to fire depends on the time since the previous spike, and even on the time of the spike before that (Usrey et al., 2000). More generally, cortical synapses are modulated in a complex but consistent manner by the temporal pattern of input spikes (Markram and Tsodyks, 1996; Tsodyks and Markram, 1997; Varela et al., 1997; Nelson and Abbott, 2001), and cortical neurons produce spikes reproducibly when injected with large time-varying currents (Mainen and Sejnowski, 1995). What was not known, however, was whether the precise timing of visual responses in the LGN was a reproducible phenomenon or an arbitrary property of each cell. Therefore, we compared the responses of different LGN neurons to determine whether precise firing events were conserved across cells of a class, specifically ON and OFF X cells.
MATERIALS AND METHODS
Experimental procedures
Physiology. Individual neurons were recorded in the LGN of cats anesthetized with sodium pentothal according to standard methods, as described by Reinagel and Reid (2000). Cells were classified by conventional physiological tests, including spatiotemporal receptive field mapping (Reid et al., 1997) and a modified null test to differentiate X and Y cells (Hochstein and Shapley, 1976). Experimental data were recorded only for X cells in this study. We included in our analysis all 12 well isolated cells from which we recorded responses to 128 repeats of this visual stimulus: seven ON X cells and five OFF X cells recorded in four different experimental animals.
Visual stimuli. The visual stimulus was a spatially uniform wide-field illumination that was modulated in time (diffuse flicker). The time course of modulation was a random sequence of luminance values drawn from the distribution measured in a natural stimulus (Reinagel and Reid, 2000). The stimulus was displayed on a computer monitor with photopic mean luminance (>10 cd/m2; eyes dilated) and a refresh rate of 128 Hz. Because the luminance value was chosen independently for each time frame, the power spectrum of the stimulus was flat between 0 and 64 Hz (white noise).
Analysis
Identification of firing events. We presented the same dynamic stimulus repeatedly for 128 trials and accumulated a peristimulus time histogram (PSTH), which represents the probability of firing as a function of time. We divided this PSTH into discrete firing events using the method of Berry et al. (1997). Briefly, the PSTH was smoothed by a Gaussian filter whose width was determined by the trial-to-trial jitter of spike timing, as computed from the autocorrelation between two trials. In this smoothed PSTH, minima that were deep compared with adjacent maxima were taken as boundaries between distinct events. These boundaries were then used to divide the original, unsmoothed PSTH into discrete firing events, such that all spikes were assigned to one and only one event. We excluded from additional analysis any event that was represented by <10 spikes over all of the trials. We defined the time of the event as the mean of the best-fit Gaussian to the peak in the unsmoothed PSTH and the precision of the event as the width (2ς) of this Gaussian.
Alignment of events between cells. To compare event timing across cells, we created an event train for each cell by representing every peak as a single event, as defined above. We matched corresponding events of different cells by a method originally developed to compare the spike trains of a single cell in different trials (Victor and Purpura, 1997). This elegant and efficient algorithm is based on a method used for aligning homologous genetic sequences. The optimal alignment of two event trains is defined in terms of the shortest path to convert one into the other by means of three elementary operations: deleting a spike, adding a spike, or shifting a spike in time. The optimal alignment depends on one free parameter, the “cost” of shifting a spike. This parameter defines how far we may shift an event to align it with an event of the other cell. We chose this cost parameter such that events separated by more than four times the average width of events were considered distinct events. Many cell pairs had a small systematic latency difference that affected all events. We corrected for latency before aligning the events of the two cells, based on time of the peak in the cross-correlation between the PSTHs of the two cells. This latency correction ensured that the mean difference in event time was 0 in all cases. The mean latency correction was 2.9 msec (range, 0.3–6.2).
Normalized time difference. For each aligned peak, we defined the normalized time difference as the difference in peak times between the two cells divided by the average width of the two peaks. Thus, if the Gaussian fits to the aligned PSTH peaks of two cells are defined by (μ1,ς1) and (μ2,ς2) and the width of each peak is defined as 2ς, then the normalized time difference of the paired firing event is given by: (μ1 − μ2)/ς1 + ς2).
RESULTS
Precise firing events happen at the same times for different cells
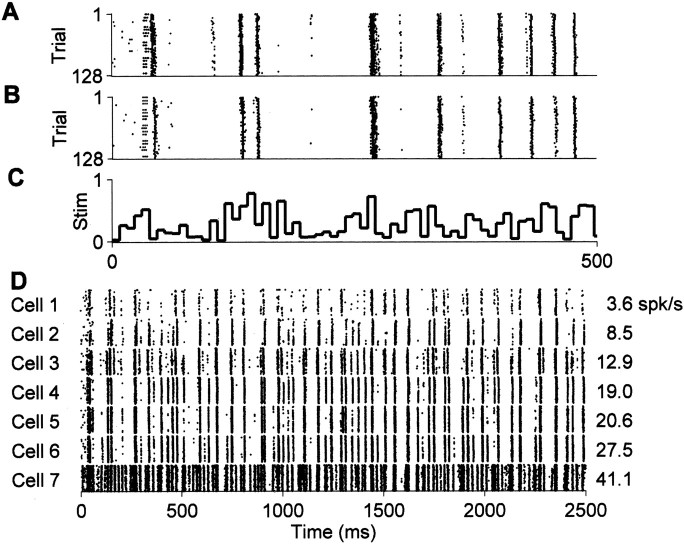
Our finding can be readily observed by direct inspection of the data. When the same dynamic visual stimulus was presented repeatedly, individual neurons responded with action potentials at the same precise times in every trial (Fig.1A). When the identical stimulus was used to study different cells, even in different animals, the precise spike times were remarkably similar from cell to cell (Fig.1, compare A and B). This similarity was found to a greater or lesser degree for all cells of the same class (Fig.1D). The remainder of our discussion quantifies this observation for a small sample data set.
Fig. 1.
Precise firing events happen at the same times in different cells. A, Responses of an LGN neuron (ON-center X cell) to 128 repeats of the same dynamic visual stimulus. Each row represents a single trial, with each action potential represented by a single point; thehorizontal axis represents time (the first 500 msec of an 8 sec trial is shown; the time axis is shared withB and C). B, Responses of another ON-center X cell to the same stimulus, recorded in a different animal. C, Luminance time course of the visual stimulus (Stim) for the responses shown in A andB. Spike events typically occurred between three and four frames after a dark-to-light transition in the stimulus (frame duration, 7.8 msec). D, Responses of all ON-center X cells for which we recorded at least 128 trials with this stimulus (7 cells from 4 different animals). For each cell, 128 repeats are shown for the first 2500 msec of the 8 sec trial. The spike times are aligned relative to the stimulus onset, without correction for latency differences among the cells. The mean firing rate of each cell for the entire trial is indicated at the right. Cells 6 and 4 are expanded in A and B, respectively.
Precision of firing events of individual cells
The first step in quantifying the similarity of responses across cells was to identify the times and precisions of the firing events in each cell. Each peak in the PSTH was fitted by a Gaussian curve whose width (2ς) we take as a measure of the trial-to-trial variability of the event time (Fig.2A). In the cell shown, the peak widths ranged from 0.7 to 4.0 msec, with an average width of 2ς = 2.1 msec (Fig. 2B). The distribution of peak widths was similar for all cells (Fig. 2C), despite a >10-fold range of the overall firing rate. The average width ranged from 1 to 4 msec, and all cells had peaks of width of <1 msec. This result is consistent with our previous finding that the temporal precision required to extract all of the information in individual spike trains is 1–2 msec for all cells, regardless of the firing rate (Reinagel and Reid, 2000).
Fig. 2.
Precision of firing events across trials for individual cells. A, PSTH for the cell shown in Figure1A (average number of spikes in each 1 msec bin; 500 msec of the 8 sec trial is shown). The width of each peak (2ς, in milliseconds) is indicated above. Peaks with <10 spikes were not analyzed. B, Distribution of peak widths for the cell shown in A. The mean and SD are indicated by thepoint and line above the histogram (2.1 ± 0.9 msec; n = 179 peaks).C, Mean peak width for each cell recorded with this stimulus: the seven ON-center X cells shown in Figure1D (white bars) and five OFF-center X cells (black bars). Error bars indicate the SD over peaks (n = 85–329 peaks). Cell 6 is shown in A andB.
Peak timing differs by less than peak widths
The peaks in the firing rates of two cells are often aligned to within a fraction of the widths of the peaks (Fig.3A, PSTH). To formalize this observation, we used Victor and Purpura's (1997)method to identify the corresponding peaks of two cells. We allowed for corresponding peaks to be separated up to four times the average peak width for the two cells (for a maximum distance of ∼8 msec). In the example shown, 159 of 176 (90%) peaks in one cell were aligned to 1 of the 179 peaks in the other cell. These peaks were aligned much more precisely than required by our cutoff criterion (Fig. 3B,black bars vs dashed lines). Most (94%) were displaced by less than the average peak width of the two cells (1.8 msec). The magnitudes of peaks tended to scale with the average firing rate of the cell, but not always in a simple way (for example, the second event from the right in Fig. 3A).
Fig. 3.
Peak timing differs by less than peak widths.A, PSTHs for cells 6 (black) and 4 (red). A small systematic difference in latency (1.6 msec) was removed before this analysis. The mean times of the analyzed peaks for both cells are indicated by the tick marks above. Numbers above the paired events indicate the normalized time difference between the two cells, in units of peak widths. B, Histogram of absolute time differences for all paired events for these two cells (black bars; n = 159). Because of the latency correction, the mean time difference was 0 by construction.Dashed lines indicate the minimum and maximum time difference we allowed (4 times the average peak width, or 7.4 msec for this cell pair). The blue curve shows the average distribution obtained from time-shifted controls (averaged over 31 different time shifts with an average of n = 52 peaks matched by chance). C, Distribution of normalized time differences for all paired events of this cell pair. Theblue curve shows the distribution of normalized time differences for randomly matched events in time-shifted controls. The SD of each distribution is given at the right; this is a measure of the overall alignment precision for a pair of responses. As a control for the uncertainty in our estimate of event times, we did the same calculation for a single cell, comparing one-half of the data set to the other half. This yielded a much narrower distribution of normalized time differences (SD = 0.09). D, The SD of the normalized time difference for all 21 possible pairings of the seven ON cells (white bars; gray bar is for pair shown in A–C) and all 10 pairings of the five OFF cells (black bars). Results for time-shifted controls are shown as blue points.
To measure alignment on a peak-by-peak basis, we normalized the time difference between cells by the average width of the two matched peaks. A value of <1 indicates that the average spike time was more precise from cell to cell than the timing of individual spikes from trial to trial within each cell. For the cell pair shown, the normalized time difference was 0.02 ± 0.51 peak widths (Fig.3C) (Norm'd Δt of example peaks shown in Fig. 3A). In other words, 68% of peaks were aligned within 0.51 peak width, and 95% were aligned within 1.02 peak widths. The SD of the normalized time difference is a measure of how well any of the peaks of any two cells were aligned (summarized for the population in Fig. 3D).
All pairs of cells of the same class had a substantial number of peaks in common. In any pair, the cell with the lower firing rate had fewer peaks, but those peaks tended to be aligned with peaks of the other cell. For ON cell pairs, 80–100% of the peaks (of the cell with fewer peaks) were aligned; 44–85% were aligned for OFF cell pairs. When we measured the quality of the alignment, 16 of 21 ON cell pairs (Fig.3D, white bars) and 5 of 10 OFF cell pairs (black bars) had an SD of ≤1 for the normalized time difference. Thus, for these cell pairs, most events were aligned even more precisely than the trial-to-trial precision of either cell.
As a control, for each cell pair we compared the responses of the two cells to different segments of the visual stimulus. In these time-shifted controls, considerably fewer peaks were aligned by chance, and these were equally likely to be shifted by any amount of time within the range allowed (Fig. 3B, blue curve). Therefore, the normalized time difference (our measure of the precision of alignment) also had a broad distribution (Fig. 3C,blue curve). For all cell pairs, the true alignment of peaks (Fig. 3D, bars) was much more precise than for the time-shifted controls (Fig. 3D, blue dots).
DISCUSSION
Cells in the LGN can respond with precise firing events whose widths are on the order of milliseconds. Here we have shown that the timing of these events is conserved among cells of a given class, even in different animals. LGN cells of a class differed in firing rate, absolute latency, the number of peaks, and the heights of peaks, without altering the relative timing or temporal precision of these firing events. The striking invariance of event timing suggests that this may be a functionally important feature of LGN responses.
Origins of conserved patterns
It is likely that event times are at least as well conserved in retinal ganglion cells, which provide the feedforward input to the LGN neurons we studied. To the extent that intrinsic properties or circuit connectivity vary in the retina and LGN, those physiological parameters are apparently not important in determining the precise timing of firing events. In the retina, simultaneously recorded cells of the same class were found to transmit redundant information about a full-field visual stimulus (Warland et al., 1997). Keat et al. (2001) presented a 20 parameter model that can predict the precise firing events of individual retinal ganglion cells or LGN neurons. In some cases, two or more cells of the same class were analyzed, and the parameters that fit the cells were similar. These results are consistent with our findings.
Generality of results
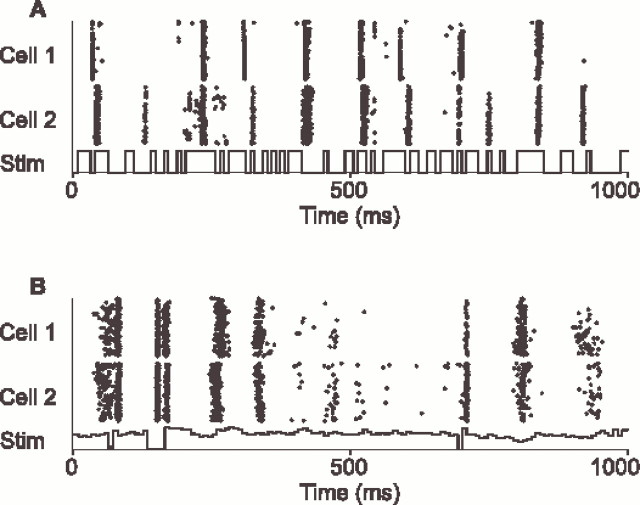
In this study, we found that precisely timed firing events were conserved within broad classes of cells (ON and OFF center X cells). We have shown the analysis of responses to a spatially uniform random flicker (white noise) with a natural distribution of luminance values. We found qualitatively similar results for other spatially uniform stimuli: a binary black–white random flicker (Reid et al., 1997) (Fig.4A) and a temporally nonwhite stimulus whose luminance time course was taken from a recorded natural scene (van Hateren, 1997) (Fig. 4B). It remains to be seen whether a similar conservation of firing events is found within other cell types in the LGN and whether this result is restricted to spatially uniform stimuli.
Fig. 4.
Examples of conserved firing events for other stimuli. A, Responses of two OFF X cells recorded in two different animals. The visual stimulus (Stim) was a wide-field binary flicker with the luminance time course shown beneath.B, Responses of another two OFF X cells (cells 8 and 9 of Fig. 2C), recorded sequentially in the same animal. The stimulus was a spatially uniform field modulated with a luminance time course recorded from nature, shown beneath. In bothA and B, spike times are shown relative to the stimulus time, with no correction for differences in absolute latency. For each cell, spike times are shown for 128 repeats of the stimulus for 1 sec in the middle of an 8 sec trial.
Another example of conserved firing patterns has been reported recently for the motion-sensitive neuron H1 of the fly. In that study, an information-theoretic measure was used to quantify the similarity between H1 neurons in different flies. The visual information was measured from the responses of each H1 cell separately (such that each cell could have a unique neural code) and compared with the information in the pooled responses of several H1 cells (such that all cells were constrained to use a common code). In the case of H1, 70% of the information was found to be universal for the cell class; that is, it did not depend on knowing which H1 neuron produced a given response (Schneidman et al., 2001). We performed the same analysis and found that most of the visual information in our LGN responses was universal for a given class (75% of information was universal for ON X cells, 78% was universal for OFF X cells; data not shown).
Consequences for downstream neurons
Our stimuli subtended 5–10 receptive field diameters, not the entire visual field. This might resemble the situation in which an identical visual stimulus abruptly covers a local neighborhood of receptive fields, such as when an object moves across part of the visual field or when the animal makes a saccade across an edge between uniform regions of an image. Our result suggests that in this situation, the local population of LGN neurons could signal this event with a temporally precise and nearly synchronous response, regardless of whether they share common retinal ganglion cell inputs. A convergence of several synchronous spikes would be a highly effective input to cortical targets (Alonso et al., 1996; Usrey et al., 2000).
Apart from of the issue of synchrony, there is the question of whether reproducible temporal patterns in the LGN are important to downstream neurons. It was known that single neurons produce reliable patterns of spikes (Bair et al., 1994; Berry et al., 1997; de Ruyter van Steveninck et al., 1997; Buračas et al., 1998; Reinagel and Reid, 2000) and that dynamic synapses respond to a given pattern of spikes in a reproducible way (Markram and Tsodyks, 1996; Abbott et al., 1997;Dobrunz et al., 1997; Tsodyks and Markram, 1997; Varela et al., 1997;Dobrunz and Stevens, 1999). The fact that spiking patterns are reproducible across the LGN cell class now raises the question of whether the dynamics of thalamocortical synapses are also stereotyped for the presynaptic and postsynaptic cell class, as is the case among cortical interneurons (Gupta et al., 2000). If so, cortical targets of a given class could also have consistent temporal responses to equivalent visual stimuli and precisely synchronized responses to simultaneous large-field stimuli.
In conclusion, we have argued previously that precise temporal patterns of firing in the LGN are visually driven and that they could provide a rich source of visual information to the cortex. Our finding that this complexity can be reduced to just a few types of temporal patterns lends much greater plausibility to the hypothesis that the patterns might be decoded in the cortex and might play an important role in vision.
Footnotes
This work was supported by National Eye Institute Grants R01 EY10115, R01 EY12815, and P30 EY12196. We thank Christine Couture for expert technical assistance and Sergey Yurgenson for software support.
Correspondence should be addressed to Dr. R. Clay Reid, Department of Neurobiology, Harvard Medical School, 220 Longwood Avenue, Boston, MA 02115. E-mail clay_reid@hms.harvard.edu.
REFERENCES
- 1.Abbott LF, Varela JA, Sen K, Nelson SB. Synaptic depression and cortical gain control. Science. 1997;275:220–224. doi: 10.1126/science.275.5297.221. [DOI] [PubMed] [Google Scholar]
- 2.Alonso JM, Usrey WM, Reid RC. Precisely correlated firing in cells of the lateral geniculate nucleus. Nature. 1996;383:815–819. doi: 10.1038/383815a0. [DOI] [PubMed] [Google Scholar]
- 3.Bair W, Koch C, Newsome W, Britten K. Power spectrum analysis of bursting cells in area MT in the behaving monkey. J Neurosci. 1994;14:2870–2892. doi: 10.1523/JNEUROSCI.14-05-02870.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berry MJ, Warland DK, Meister M. The structure and precision of retinal spike trains. Proc Natl Acad Sci USA. 1997;94:5411–5416. doi: 10.1073/pnas.94.10.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buračas GT, Zador AM, DeWeese MR, Albright TD. Efficient discrimination of temporal patterns by motion-sensitive neurons in primate visual cortex. Neuron. 1998;20:959–969. doi: 10.1016/s0896-6273(00)80477-8. [DOI] [PubMed] [Google Scholar]
- 6.Cai D, DeAngelis GC, Freeman RD. Spatiotemporal receptive field organization in the lateral geniculate nucleus of cats and kittens. J Neurophysiol. 1997;78:1045–1061. doi: 10.1152/jn.1997.78.2.1045. [DOI] [PubMed] [Google Scholar]
- 7.de Ruyter van Steveninck RR, Lewen GD, Strong SP, Koberle R, Bialek W. Reproducibility and variability in neural spike trains. Science. 1997;275:1805–1808. doi: 10.1126/science.275.5307.1805. [DOI] [PubMed] [Google Scholar]
- 8.Dobrunz LE, Stevens CF. Response of hippocampal synapses to natural stimulation patterns. Neuron. 1999;22:157–166. doi: 10.1016/s0896-6273(00)80687-x. [DOI] [PubMed] [Google Scholar]
- 9.Dobrunz LE, Huang EP, Stevens CF. Very short-term plasticity in hippocampal synapses. Proc Natl Acad Sci USA. 1997;94:14843–18487. doi: 10.1073/pnas.94.26.14843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta A, Wang Y, Markram H. Organizing principles for a diversity of GABAergic interneurons and synapses in the neocortex. Science. 2000;287:273–278. doi: 10.1126/science.287.5451.273. [DOI] [PubMed] [Google Scholar]
- 11.Hochstein S, Shapley RM. Quantitative analysis of retinal ganglion cell classifications. J Physiol (Lond) 1976;262:237–264. doi: 10.1113/jphysiol.1976.sp011594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keat J, Reinagel P, Reid RC, Meister M. Predicting every spike: a model for the responses of visual neurons. Neuron. 2001;30:803–817. doi: 10.1016/s0896-6273(01)00322-1. [DOI] [PubMed] [Google Scholar]
- 13.Mainen ZF, Sejnowski TJ. Reliability of spike timing in neocortical neurons. Science. 1995;268:1503–1506. doi: 10.1126/science.7770778. [DOI] [PubMed] [Google Scholar]
- 14.Markram H, Tsodyks M. Redistribution of synaptic efficacy between neocortical pyramidal neurons. Nature. 1996;382:807–810. doi: 10.1038/382807a0. [DOI] [PubMed] [Google Scholar]
- 15. Nelson SB, Abbott LF. Temporal dynamics of biological synapses. The handbook of brain theory and neural networks Arbib M. 2002. MIT; Cambridge, MA, in press. [Google Scholar]
- 16.Reid RC, Alonso JM. Specificity of monosynaptic connections from thalamus to visual cortex. Nature. 1995;378:281–284. doi: 10.1038/378281a0. [DOI] [PubMed] [Google Scholar]
- 17.Reid RC, Victor JD, Shapley RM. The use of m-sequences in the analysis of visual neurons: linear receptive field properties. Vis Neurosci. 1997;14:1015–1027. doi: 10.1017/s0952523800011743. [DOI] [PubMed] [Google Scholar]
- 18.Reinagel P, Reid RC. Temporal coding of visual information in the thalamus. J Neurosci. 2000;20:5392–5400. doi: 10.1523/JNEUROSCI.20-14-05392.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneidman E, Brenner N, Tishby N, de Ruyter van Steveninck RR, Bialek W. Universality and individuality in a neural code. In: Leen TK, Dietterich TG, Tresp V, editors. Advances in neural information processing systems 13. MIT; Cambridge, MA: 2001. pp. 159–165. [Google Scholar]
- 20.Tanaka K. Cross-correlation analysis of geniculostriate neuronal relationships in cats. J Neurophysiol. 1983;49:1303–1318. doi: 10.1152/jn.1983.49.6.1303. [DOI] [PubMed] [Google Scholar]
- 21.Tsodyks MV, Markram H. The neural code between neocortical pyramidal neurons depends on neurotransmitter release probability. Proc Natl Acad Sci USA. 1997;94:719–723. doi: 10.1073/pnas.94.2.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Usrey WM, Alonso JM, Reid RC. Synaptic interactions between thalamic inputs to simple cells in cat visual cortex. J Neurosci. 2000;20:5461–5467. doi: 10.1523/JNEUROSCI.20-14-05461.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Hateren JH. Processing of natural time series of intensities by the visual system of the blowfly. Vision Res. 1997;37:3407–3416. doi: 10.1016/s0042-6989(97)00105-3. [DOI] [PubMed] [Google Scholar]
- 24.Varela JA, Sen K, Gibson J, Fost J, Abbott LF, Nelson SB. A quantitative description of short-term plasticity at excitatory synapses in layer 2/3 of rat primary visual cortex. J Neurosci. 1997;17:7926–7940. doi: 10.1523/JNEUROSCI.17-20-07926.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Victor JD. The dynamics of the cat retinal X cell centre. J Physiol (Lond) 1987;386:219–246. doi: 10.1113/jphysiol.1987.sp016531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Victor JD. The dynamics of the cat retinal Y cell subunit. J Physiol (Lond) 1988;405:289–320. doi: 10.1113/jphysiol.1988.sp017334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Victor J, Purpura K. Metric-space analysis of spike trains: theory, algorithms and application. Netw Comput Neural Syst. 1997;8:127–164. [Google Scholar]
- 28.Warland DK, Reinagel P, Meister M. Decoding visual information from a population of retinal ganglion cells. J Neurophysiol. 1997;78:2336–2350. doi: 10.1152/jn.1997.78.5.2336. [DOI] [PubMed] [Google Scholar]
- 29.Wolfe J, Palmer LA. Temporal diversity in the lateral geniculate nucleus of cat. Vis Neurosci. 1998;15:653–675. doi: 10.1017/s0952523898154068. [DOI] [PubMed] [Google Scholar]