Abstract
The chemoattractant stromal cell-derived factor-1 (SDF-1) and its receptor CXC chemokine receptor 4 (CXCR4) are key modulators of immune function. In the developing brain, SDF-1 is crucial for neuronal guidance; however, cerebral functions of SDF-1/CXCR4 in adulthood are unclear. Here, we examine the cellular expression of SDF-1 isoforms and CXCR4 in the brain of mice receiving systemic lipopolysaccharide (LPS) or permanent focal cerebral ischemia. CXCR4 mRNA was constitutively expressed in cortical and hippocampal neurons and ependymal cells. Hippocampal neurons targeted the CXCR4 receptor to their somatodendritic and axonal compartments. In cortex and hippocampus, CXCR4-expressing neurons exhibited an overlapping distribution with neurons expressing SDF-1 transcripts. Although neurons synthesized SDF-1α mRNA, the SDF-1β isoform was selectively expressed by endothelial cells of cerebral microvessels. LPS stimulation dramatically decreased endothelial SDF-1β mRNA expression throughout the forebrain but did not affect neuronal SDF-1α. After focal cerebral ischemia, SDF-1β expression was selectively increased in endothelial cells of penumbral blood vessels and decreased in endothelial cells of nonlesioned brain areas. In the penumbra, SDF-1β upregulation was associated with a concomitant infiltration of CXCR4-expressing peripheral blood cells, including macrophages. Neuronal SDF-1α was transiently downregulated and neuronal CXCR4 was transiently upregulated in the nonlesioned cerebral cortex in response to ischemia. Although endothelial SDF-1β may control cerebral infiltration of CXCR4-carrying leukocytes during cerebral ischemia, the neuronal SDF-1α/CXCR4 system may contribute to ischemia-induced neuronal plasticity. Thus, the isoform-specific regulation of SDF-1 expression modulates neurotransmission and cerebral infiltration via distinct CXCR4-dependent pathways.
Keywords: CXC chemokine, stromal cell-derived factor-1, CXCR4, focal cerebral ischemia, lipopolysaccharide, in situhybridization, immunocytochemistry, antibody
Chemokines are small proteins with chemotactic effects on leukocytes. Among chemokines expressed in brain, stromal cell-derived factor-1 (SDF-1) attracted much attention because it was shown to be crucial for cerebellar development (Ma et al., 1998;Zou et al., 1998; Klein et al., 2001; Lu et al., 2001). The isoforms SDF-1α and SDF-1β arise by alternative splicing from the SDF-1 gene. They share an identical 5′-untranslated and coding region but differ at the 3′-end (Tashiro et al., 1993; Nagasawa et al., 1994;Shirozu et al., 1995). A rat SDF-1γ mRNA has been described, which is similar to SDF-1β but contains a 2.5 kb insertion after codon 89 (Gleichmann et al., 2000). The SDF-1α and SDF-1β proteins are identical except four C-terminal amino acids and represent endogenous ligands of the CXC chemokine receptor 4 (CXCR4), which also functions as coreceptor for human immunodeficiency virus (HIV-1) in lymphocytes (Bleul et al., 1996b; Feng et al., 1996; Oberlin et al., 1996). CXCR4-mediated apoptosis in neuronal cultures induced by HIV-1 glycoprotein gp120 and SDF-1 indicates a potential role of this receptor in neurodegeneration (Hesselgesser et al., 1998; Kaul and Lipton, 1999). Microglia-enhanced SDF-1-stimulated release of TNFα and glutamate from astrocytes in vitro was proposed as a novel excitotoxic pathomechanism in neurodegenerative diseases (Bezzi et al., 2001). Because SDF-1 modulates synaptic transmission via neuronal CXCR4 receptors in the developing cerebellum (Limatola et al., 2000), SDF-1 may exert physiological functions also in other brain regions.
Our understanding of the role of the SDF-1/CXCR4 receptor system in the adult brain has been hampered, however, by the lack of information about the cellular expression of CXCR4 and SDF-1 isoforms. Previous contradictory studies described SDF-1 expression either as widespread in neurons and oligodendrocytes (Gleichmann et al., 2000) or as restricted to distinct neuronal populations and endothelial cells (Tham et al., 2001). Moreover, the differential cellular expression, regulation, and function of SDF-1 isoforms has not been characterized in the brain.
SDF-1 is involved in inflammation and wound healing (Nanki et al., 2000; Fedyk et al., 2001). SDF-1 triggers adhesion of lymphocytes to activated endothelial cells in vitro (Kantele et al., 2000). Most likely, endothelial SDF-1 plays a crucial role in trans-endothelial migration of leukocytes (Campbell et al., 1998; Pablos et al., 1999; Grabovsky et al., 2000). This raises the question to what extent SDF-1 in the brain may exert chemotactic effects on monocytes and lymphocytes after endothelial activation by systemic LPS-induced immune challenge (Bleul et al., 1996a; Quan et al., 1997; Lebel et al., 2000; Rivest et al., 2000). The potential excitotoxic and chemotactic activities of SDF-1 may also be relevant in focal cerebral ischemia, a condition involving disruption of the blood–brain barrier (BBB), excitotoxic neurodegeneration, and glial activation (Stoll et al., 1998).
Here, we established the cellular expression patterns of SDF-1 isoforms and CXCR4 in the adult mouse brain by in situ hybridization. The subcellular distribution of CXCR4 was characterized by confocal microscopy using a novel antiserum. We provide evidence for differential expression of SDF-1 isoforms in endothelial and neuronal cells. Furthermore, our results suggest that isoform-specific regulation of SDF-1α and SDF-1β selectively modulates CXCR4-dependent neurotransmission and cerebral leukocyte infiltration.
MATERIALS AND METHODS
Generation of anti-peptide antisera
Rabbit polyclonal antisera were generated against the C-terminal portion of the mouse CXCR4 receptor (residues 338–359, KGKRGGHSSVSTESESSSFHSS), which is identical in mouse, rat, and human. The peptide was coupled to keyhole limpet hemocyanin, and the conjugate was emulsified with Freund's adjuvant and injected into three rabbits (2144–2146) for immunization as described (Schreff et al., 2000). Two of the three antisera (2144 and 2145) developed a titer against their immunizing peptide and yielded essentially identical results during antibody characterization. Antiserum 2144 was subjected to immunoaffinity purification and used throughout this study.
Western blot analysis
The mouse CXCR4 cDNA (kind gift of Dr. Martin Lipp and Dr. Reinhold Förster, Max Delbrück Center for Molecular Medicine, Berlin, Germany) was tagged at its N terminus with the T7 epitope tag sequence MASMTGGQQMG and subcloned into pcDNA3.1 expression vector (Invitrogen, Groningen, The Netherlands) as described (Koch et al., 2001). Cell membranes were prepared from human embryonic kidney (HEK) 293 cells stably transfected with the CXCR4 receptor, as well as from mouse brain, and solubilized in lysis buffer (150 mmNaCl, 5 mm EDTA, 3 mm EGTA, 20 mmHEPES, pH 7.4, containing 4 mg/ml dodecyl-β-maltoside, and proteinase inhibitors: 1 mm phenylmethylsulfonylfluoride, 1 μm pepstatin, 10 μg/ml leupeptin, and 2 μg/ml aprotinin). The lysate was cleared and subjected to immunoprecipitation with anti-CXCR4 antibodies. Alternatively, glycoproteins were partially purified using wheat germ lectin agarose (WGA; Vector Laboratories, Burlingame, CA). Beads were washed and eluted with SDS-sample buffer for 20 min at 60°C. Either crude membrane proteins (100 μg per lane) or WGA extracts purified from 500 μg membrane proteins were subjected to 10% SDS-PAGE and immunoblotted onto nitrocellulose. Blots were incubated with 1 μg/ml affinity-purified anti-CXCR4 (2144) or rat monoclonal anti-CXCR4 antibody clone 2B11, which is directed against the N terminus (Forster et al., 1998), overnight at 4°C. Bound primary antibodies were then detected using peroxidase-conjugated secondary antibodies and enhanced chemiluminescence. For absorption controls, anti-CXCR4 antibody was preincubated with 10 μg/ml of its cognate peptide for 2 hr at room temperature.
Immunocytochemistry
Wild-type HEK 293 cells or HEK 293 cells stably transfected with T7-tagged CXCR4 receptor were grown on coverslips overnight. For internalization studies, cells were treated with SDF-1α in concentrations ranging from 1 to 1000 ng/ml for 30 min and fixed with 4% paraformaldehyde and 0.2% picric acid in 0.1 mphosphate buffer, pH 7.4, for 1 hr at room temperature. Cells were washed and stained with affinity-purified anti-CXCR4 or anti-T7 antibodies (Gramsch Laboratories) as described (Pfeiffer et al., 2001). Bound primary antibodies were detected with biotinylated anti-rabbit IgG antibodies (Vector) followed by cyanine 3.18 (Cy3)-conjugated streptavidin (Amersham).
Immunohistochemistry
Male NMRI mice or male Wistar rats were deeply anesthetized with chloral hydrate and transcardially perfused with Tyrode's solution followed by Zamboni's fixative containing 4% paraformaldehyde and 0.2% picric acid in 0.1 m phosphate buffer, pH 7.4. Brains were rapidly dissected and postfixed in the same fixative for 2 hr at room temperature. Tissue was cryoprotected by immersion in 30% sucrose for 48 hr at 4°C before sectioning using a freezing microtome. Free-floating sections (30–40 μm) were washed and incubated with 1 μg/ml affinity-purified anti-CXCR4 antibodies. Staining of primary antibody was detected using Cy3-conjugated anti-rabbit antibody or the tyramine amplification procedure as described (Schulz et al., 1998). For dual immunofluorescence of CXCR4 and GFAP, sections were first stained for CXCR4 as described above and subsequently labeled with guinea pig anti-GFAP antibody (1:2000) (Progen, Heidelberg, Germany), which was visualized with a Cy5-conjugated secondary antibody. For immunocytochemical controls, the primary antibody was omitted, replaced by preimmune sera, or absorbed with several concentrations ranging from 1 to 10 μg/ml of homologous or heterologous peptides for 2 hr at room temperature. Specimens were examined using a Leica TCS-NT laser scanning confocal microscope (Heidelberg, Germany) equipped with a krypton/argon laser. Cy3 was imaged with 568 nm excitation and 570–630 nm bandpass emission filters, and Cy5 was imaged with 647 nm excitation and 665 nm longpass emission filters.
Probes used in in situ hybridization
The expression of the SDF-1 isoforms was characterized using riboprobes derived from three partial mouse cDNAs. The first probe (called SDF-1) was complementary to nucleotides (nt) 11–273 of SDF-1α and SDF-1β (GI:12025675 and GI:12025674). This corresponds to parts of the 5′-untranslated as well as the coding region (nt 82–351 of GI:12025675). The nt 19–262 of this probe are also 96% identical to rat SDF-1α, SDF-1β, and SDF-1γ. The second probe (SDF-1α) was complementary to nt 1005–1780 of GI:12025675, which are specific for mouse SDF-1α. The mouse SDF-1α cDNA (Tashiro et al., 1993) was kindly provided by Dr. T. Honjo and Dr. K. Tashiro (Kyoto University, Kyoto, Japan). The third probe (SDF-1β) was complementary to nt 366–1186 of SDF-1β (GI:12025674) and exhibited no similarity with SDF-1α. In the rat, a SDF-1γ mRNA has recently been described, which is similar to SDF-1β but contains a 2.5 kb insertion after codon 89 (Gleichmann et al., 2000). Although a similar isoform has not been described so far in mice, our SDF-1β probe would be expected to hybridize also to such a putative mouse SDF-1γ mRNA. For simplicity, we refer to the detected mRNA as SDF-1β mRNA. The SDF-1 and SDF-1β cDNAs as well as the cDNAs of the rat CXCR4 (U90610, nt 1–1050), the β polypeptide of the mouse complement component 1q (C1q; nt 35–1041) (Wood et al., 1988), mouse GFAP (nt 189–1054) (Lewis et al., 1984), rat pre-prosomatostatin (nt 32–307) (Goodman et al., 1985), and mouse interleukin-1 β (IL-1β; nt 205–724) (Telford et al., 1986) were amplified by RT-PCR. C1q was used as marker for both brain microglial cells and macrophages (Schwaeble et al., 1995; Schafer et al., 2000). All described cDNAs were cloned into the pGEM-Teasy vector (Promega) and subjected to double-strand DNA sequencing. A riboprobe for glutamate acid decarboxylase was used previously (Stumm et al., 2001). For the mouse CXCR4 receptor (Heesen et al., 1996; Nagasawa et al., 1996), the cDNA of Dr. M. Lipp and Dr. R. Förster was used. Riboprobes in antisense and sense orientation were generated from the linearized vector constructs by in vitro transcription (Melton et al., 1984) using [35S]-UTP (1000 Ci/mmol; 15 μm concentration in the transcription reaction) or digoxigenin-UTP (DIG) (Roche Diagnostics, Mannheim, Germany) as label. The probes were subjected to mild alkaline hydrolysis (Angerer et al., 1987) and purified using P-30 spin columns (Bio-Rad).
In situ hybridization histochemistry
Radioactive in situ hybridization was performed as described previously (Stumm et al., 2001). Fixation of slide-mounted frozen sections (14 μm thickness) was performed in phosphate-buffered 4% paraformaldehyde for 60 min. After washing in PBS, the slides were incubated for 10 min in 0.4% Triton X-100, rinsed in distilled water, and incubated for 10 min in 0.1 mtriethanolamine, pH 8.0 (Sigma, Deisenhofen, Germany) containing 0.25% v/v acetic anhydrate (Sigma). After washing in PBS, the sections were dehydrated in 50 and 70% 2-propanol, air dried, and stored at −20°C. Riboprobes were diluted in hybridization buffer [3× sodium chloride/sodium citrate (SSC), 50 mmNaPO4, 20 mmdithiothreitol, 1 × Denhardt's solution, 0.25 gm/l yeast tRNA, 10% dextran sulfate, and 50% formamide] to 50,000 dpm/μl. Hybridization was performed for 14 hr at 60°C. The slides were washed in 2× SSC and 1× SSC before a 30 min treatment with 1 U/ml RNase T1 and 20 μg/ml RNase A (Roche Diagnostics) at 37°C in 10 mm Tris, pH 8.0, 0.5 mNaCl, 1 mm EDTA. Slides were extensively washed in 1× and 0.2× SSC at room temperature and subsequently in 0.2× SSC for 60 min at 60°C. After washing in water, the tissue was dehydrated in 50 and 70% 2-propanol. Slides were exposed together with [14C] standards (ARC, St. Louis, MO) to x-ray film for 2–5 d. For high-power bright- and dark-field microscopic analysis, autoradiographic detection of35S was performed with NTB-2 nuclear emulsion (Eastman Kodak, Rochester, NY). Exposure times were 10–42 d. Cresyl violet was used as counterstain.
Double in situhybridization histochemistry
The visualization of two different RNA transcripts in the same tissue section was performed by combining radioactive and nonradioactive in situ hybridization essentially as described (Stumm et al., 2001). Digoxigenin-labeled riboprobes (see above) were diluted in radioactive hybridization solution to 1 μg/ml, hybridized, and washed as above. To detect nonradioactive hybrids, slides were equilibrated in buffer 1 (0.1 mmaleic acid, 150 mm NaCl, pH 7.5) containing 0.05% Tween 20 (Merck, Darmstadt, Germany). After blocking for 1 hr in blocking buffer (buffer 1 containing 2% blocking reagent; Roche), alkaline phosphatase-conjugated anti-DIG Fab fragments (Roche) were applied overnight at a concentration of 0.5 U/ml blocking buffer. Slides were washed twice for 30 min in buffer 1 and equilibrated in buffer 2 (100 mm Tris, 100 mm NaCl, 50 mmMgCl2, pH 9.4, 0.05% Tween 20) before a 16 hr color reaction using 0.2 mm5-bromo-4-chloro-3-indolyl phosphate and 0.2 mmnitroblue tetrazolium salt (Roche). The reaction was stopped by washing the slides in distilled water. The35S-labeled probes were detected by K5 photo-emulsion (Ilford); exposure times were 14–60 d.
Combination of immunohistochemistry and in situ hybridization
Immunocytochemistry for von Willebrand factor (vWF), an established marker of cerebral vascular endothelial cells (Yamamoto et al., 1998), was combined with radioactive in situhybridization for SDF-1 and SDF-1β mRNAs. Before hybridization, the formalin-fixed sections were heated to 92–95°C for 15 min in 0.01m citrate buffer, pH 6.0. Hybridization and washing were performed as described above. After washing, sections were blocked by incubation in PBS containing 5% bovine serum albumin (BSA) for 30 min. A rabbit anti-human vWF antibody (Dako, Glostrup, Denmark) was diluted 1:1000 in PBS containing 2% BSA and applied overnight. For negative controls, the primary antibody was omitted. After several washes in distilled water followed by rinsing in PBS, a biotinylated secondary antibody (Dianova, Hamburg, Germany) was applied for 1 hr. After another series of washes, sections were incubated for 45 min with the ABC reagents (Vectastain ABC Kit, Vector) followed by diaminobenzidine reaction (0.125 μg/ml diaminobenzidine) for 10 min at room temperature. Autoradiographic detection of35S was performed with NTB-2 nuclear emulsion (Eastman Kodak).
Mapping of SDF-1, SDF-1α, SDF-1β, and CXCR4 mRNA expression in the brain
Coronal brain sections of two male NMRI mice were cut at 14 μm. At intervals of 500 μm, cresyl violet staining and in situ hybridization analysis of the SDF-1, SDF-1α, SDF-1β, and CXCR4 mRNAs was performed on adjacent sections. Cerebral structures were identified, and abbreviations were assigned according to the mouse brain atlas (Franklin and Paxinos, 1997).
Animal experiments
For all animal procedures, ethical approval was sought before the experiments according to the requirements of the German National Act on the Use of Experimental Animals. Male virus- and pathogen-free NMRI mice (30 gm, Tierzucht Schönwalde or Charles River, Germany) and male Wistar rats (300 gm, Tierzucht Schönwalde) were used.
Lipopolysaccharide challenge. NMRI mice were intraperitoneally injected (8:30–9:30 A.M.) with lipopolysaccharide (LPS) from Escherichia coli (50 μg, total volume 500 μl; Sigma-Aldrich L3129, Schnelldorf, Germany) suspended in sterile saline. Five groups of three LPS-injected mice each were killed by cervical dislocation and decapitation after 0.5, 1, 3, 6, and 24 hr. As a control group, four mice were injected with 500 μl sterile saline and killed after 3 hr. Brains and spleens were removed and processed for quantitative in situ hybridization. Changes in cerebral SDF-1 and CXCR4 mRNA expression were analyzed by comparison of the five LPS-treated groups with the control group. To verify the efficacy of the LPS stimulus, we analyzed IL-1β mRNA levels in the spleens of all groups by in situ hybridization. Expression of IL-1β was dramatically elevated in all animals 0.5–3 hr after LPS and was virtually absent 24 hr after LPS as well as in the control group, which is in excellent agreement with previous studies.
Animal model of focal cerebral ischemia. Permanent middle cerebral artery occlusion (MCAO) was performed in male NMRI mice according to Welsh et al. (1987). After the mice were anesthetized by intraperitoneal injection of tribromoethanol (600 mg/kg), a hole was drilled into the skull to expose the middle cerebral artery. The stem of the middle cerebral artery and both branches were permanently occluded by electrocoagulation. Body temperature was maintained at 37 ± 1°C with a heating lamp during the surgical procedure. After MCAO, mice were kept at an environmental temperature of 30°C for 2 hr and then transferred to their home cages. At different time points after ischemia, the animals were deeply anesthetized with halothane, and the brains were quickly removed and frozen in isopentane at −30°C.
Three independent experiments were performed to determine ischemia-related changes in SDF-1 and CXCR4 mRNA expression by quantitative in situ hybridization. In experiment 1, three naive animals were compared with the MCAO groups, which consisted of three animals at 6 hr and 1 d as well as four animals at 2 and 4 d. In experiments 2 and 3, the possible effects of surgery (skull trepanation) were analyzed 1 and 2 d after treatment. At either time point three MCAO-treated, three sham-operated, and three naive animals were compared with each other. For double in situ hybridization, two animals were prepared 2 d after MCAO and after sham operation, respectively.
Image analysis
Film autoradiograms of three sections from each animal were digitized on an illumination screen using NIH Image 1.62. For calibration, density measurements of the film background and the [14C] standards were obtained and plotted against the tissue radioactivity equivalents (nanoCuries per gram tissue; American Radiolabeled Chemicals, St. Louis, MO). A threshold level was set to exclude pixel values below background density from the measurement. The density measurements were expressed as integrated optical density (IOD) (i.e., average optical density × proportional area). The regions of interest were the entire brain section in the LPS experiment and three manually selected areas in the ischemia experiments: the infarct including the directly adjacent area, the ipsilateral area, in which neurodegeneration was not observed, and the entire contralateral hemisphere. To obtain the mean value of a region of interest in a single animal, measurements of three sections were averaged.
Statistical analysis
One-way ANOVA followed by Dunnett's multiple comparison test was used in the LPS experiment and in the ischemia-experiment 1. In Ischemia-experiments 2 and 3, the groups were compared with each other using ANOVA followed by Newman–Keuls multiple comparison test.p values <0.05 were considered statistically significant.
RESULTS
Cell-specific expression of SDF-1 splice variants
To analyze the differential cellular expression of SDF-1 splice variants in the brain, two riboprobes were used for in situhybridization that were specific for either the SDF-1α or the SDF-1β mRNA. As shown in x-ray autoradiograms of the septal area, SDF-1α mRNA was expressed by distinct neuronal structures in the cortex and claustrum (Fig.1A, arrows). In contrast, SDF-1β was expressed in numerous cells that exhibited a homogenous distribution throughout the brain (Fig.1B). A riboprobe that recognized all SDF-1 isoforms (SDF-1 mRNA) showed a combined expression pattern of the SDF-1α and SDF-1β mRNAs (Fig. 1C). Low-power analysis of emulsion-coated serial sections revealed that SDF-1β mRNA-expressing cells were not selectively localized in any specific structures of the white or gray matter (Fig. 1D–G). At high-power magnification it was apparent that SDF-1β mRNA-expressing cells contained small round or small oval-shaped nuclei (Fig.2D, inset). These cells had a rod-like shape without processes and were aligned in clusters of three to five cells, which suggests that SDF-1β is expressed in the cerebral endothelium (Fig. 2E). To test this hypothesis, a combination of immunohistochemistry for vWF, which is an established marker for cerebral endothelial cells, and in situ hybridization for SDF-1β mRNA was used. This approach clearly revealed that SDF-1β was expressed by vWF-immunoreactive (ir) endothelial cells of small but not large cerebral blood vessels (Fig. 2F). SDF-1β transcripts were not detected in neurons (Fig. 2D). The possible expression of the SDF-1 gene in glia was analyzed by combining radioactive in situ hybridization for all SDF-1 mRNAs and nonisotopic in situ hybridization for GFAP and C1q, established markers for astrocytes and microglial cells. However, SDF-1 expression was not observed in these glial populations (Fig.2B,C). The use of the riboprobe for all SDF-1 transcripts allowed the visualization of a SDF-1 splice variant in neurons (Fig. 2A). Hybridization with the SDF-1α-selective probe revealed that the neuronally generated SDF-1 transcripts consisted of the α-isoform. SDF-1α mRNA was also detected in meningeal cells and occasionally in the endothelium of large parenchymal blood vessels (data not shown). These findings provide evidence for cell-specific splicing of SDF-1 transcripts in the mouse brain. Although SDF-1β mRNA is selectively generated in endothelial cells, SDF-1α mRNA is expressed predominantly by neurons. SDF-1 expression in astrocytes and microglial cells was below the detection limit.
Fig. 1.
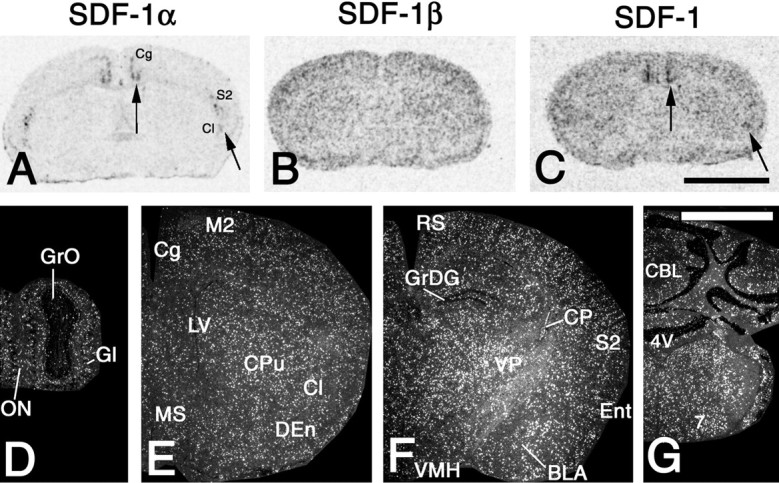
Cerebral expression patterns of SDF-1 isoforms. In situ hybridization was performed with riboprobes specific for SDF-1α (A), SDF-1β (B, D–G), and a riboprobe detecting all SDF-1 isoforms (C). A–C, X-ray autoradiograms of coronal brain sections at the striatal level.A, Arrows, SDF-1α mRNA is expressed in neuronal structures of the cingulate (Cg) and the secondary somatosensory cortex (S2) as well as the claustrum (Cl). Specific hybridization signals for SDF-1α are undetectable in other structures of the brain parenchyma. Note that some SDF-1α expression occurs in the meninges. B, SDF-1β mRNA is evenly expressed throughout the brain and not related to specific structures.C, The probe for all SDF-1 isoforms shows an expression pattern that reflects the simultaneous detection of SDF-1α (C, arrows) and SDF-1β mRNAs.D–G, The left hemisphere of coronal sections after hybridization with the SDF-1β-selective probe is shown at the level of the olfactory bulb (D), the medial septal nucleus (MS) (E), the ventromedial hypothalamic nucleus (VMH) (F), and the facial nucleus (7) (G).D–G, Note that SDF-1β mRNA-expressing cells do not show any specific clustering in structures of gray or white matter.Gl, Glomerular layer of the olfactory bulb;GrO, granule layer of the olfactory bulb;ON, olfactory nerve layer; M2, secondary motor cortex; LV, lateral ventricle; CPu, caudate putamen; DEn, dorsal endopiriform nucleus;RS, retrosplenial cortex; GrDG, granule layer of the dentate gyrus; CP, choroid plexus;VP, ventral posterolateral and posteromedial thalamic nuclei; Ent, entorhinal cortex; BLA, basolateral amygdaloid nucleus, anterior part; CBL, cerebellum; 4V, fourth ventricle.D–G, Exposure time, 21 d. Scale bars: A–C, 3 mm; G, 2 mm.
Fig. 2.
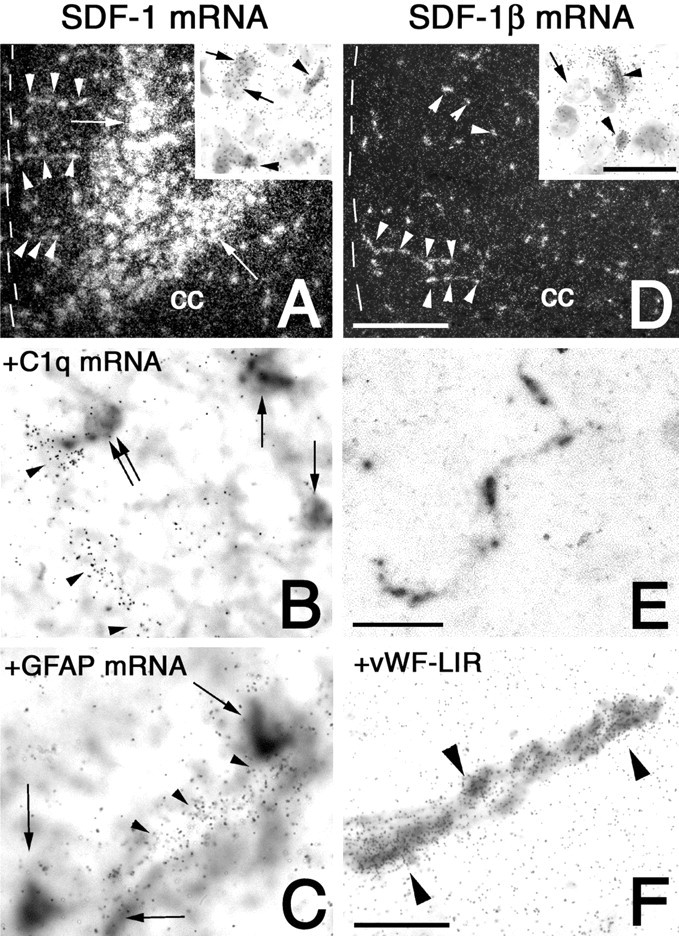
Demonstration of neuronal SDF-1 expression and endothelial SDF-1β expression by in situ hybridization with a riboprobe detecting all SDF-1 splice variants (A–C) and a riboprobe specific for SDF-1β mRNA (D–F). A, D, Dark-field micrographs of coronal sections of the cingulate cortex reveal the mRNA expression of SDF-1 isoforms by neuronal cells in laminas V and VI (A, arrows) and the absence of SDF-1β mRNA from neurons in the cingulate cortex (D). The corpus callosum (cc) and the meningeal surface (dotted line) are indicated for orientation. SDF-1 and SDF-1β mRNAs are expressed in vascular structures (A, D,arrowheads) running perpendicularly to the meningeal surface. A, Inset, High-power bright-field magnifications showing SDF-1 mRNA expression by neuronal cells with large round nuclei (arrows) as well as endothelial cells with small round and small oval nuclei (arrowheads). D, Inset, SDF-1β mRNA is present in endothelial cells with small round and small oval nuclei (arrowheads) but not in a neuron with large round nucleus (arrow). E, Nonradioactive in situ hybridization showing the rod-shaped morphology of SDF-1β-expressing endothelial cells of a microvessel. F, Combination of immunohistochemistry for vWF (detected as black reaction product) and in situ hybridization for SDF-1β mRNA (detected asgrains) identifies specific SDF-1β expression by vWF-ir endothelial cells (arrowheads). B,C, Black reaction products, microglial cells (B), and astrocytes (C) are identified by nonradioactive in situ hybridization for C1q and GFAP, respectively. Co-hybridized 35S-labeled riboprobes to SDF-1 (detected asgrains) reveal SDF-1 expression by endothelial cells (arrowheads in B, C) and the absence of SDF-1 expression from microglial cells (arrows inB) and astrocytes (arrows inC). B, Double arrow, Note the juxtavascular localization of the SDF-1 mRNA-negative microglial cell. Exposure times: A, 42 d; D, 21 d; B, C, F, 28 d. Scale bars: A, D, 400 μm;insets, 25 μm; B, C,F, 30 μm; E, 100 μm.
In a brain-mapping study using the probe detecting all SDF-1 isoforms, we clearly identified neurons expressing SDF-1 mRNA in the cerebral cortex (Fig. 3B–D), claustrum (Fig. 3B), granular layer of the dentate gyrus (Fig.3C,D), basolateral nucleus of the amygdala (Fig.3C,D), and the amygdalohippocampal area (Fig.3D). In the cortical areas, high neuronal SDF-1 mRNA levels were localized to the deep laminae of the prelimbic and infralimbic cortex and areas 1 and 2 of the cingulate cortex (laminas V and VI), as well as in the insular and entorhinal cortex. In the secondary motor, secondary somatosensory, and secondary visual as well as the ventral part of the auditory cortex, SDF-1 mRNA-expressing neurons were seen in the deepest layer.
Fig. 3.

Cerebral expression patterns of CXCR4 mRNA and neuronal and non-neuronal SDF-1 isoforms. Dark-field micrographs of coronal sections hybridized with riboprobes detecting all SDF-1 transcripts (A–F) or CXCR4 (G–L). A–F, In addition to moderate SDF-1 mRNA levels in endothelial cells throughout the brain, high SDF-1 mRNA levels are present in neuronal structures of the forebrain (B–D). G, In the olfactory bulb, CXCR4 mRNA expression is abundant in the glomerular layer (Gl). B–D,H–J, SDF-1 and CXCR4 mRNA-expressing neurons are located in areas 1 and 2 (Cg1, Cg2) of the cingulate cortex (B, H), in the insular cortex (In) (B,H), in the lateral entorhinal and ectorhinal cortex (LEnt, Ect) (C,D, I, J), and in the claustrum (Cl) (B,H). B–D, In the secondary motor cortex (M2) (B, C) and secondary somatosensory cortex (S2) (B,C), the medial aspect of the secondary visual cortex (V2) (D) and the ventral auditory cortex (AuV) (D), cortex neurons expressing SDF-1 mRNA are present, whereas CXCR4 mRNA-expressing neurons are mostly absent (H–J). Lower neuronal SDF-1 mRNA levels and fewer CXCR4 mRNA-expressing neurons are present in the retrosplenial (RS) (C, I) than in the cingulate cortex (B, H). C,D, Cerebrocortical neurons expressing SDF-1 mRNA are frequently located in the deep cortical layers. B,H, Neurons expressing CXCR4 mRNA but none expressing SDF-1 mRNAs are present in the dorsal endopiriform nucleus (DEn) (H). Note SDF-1 mRNA expression in the choroid plexus (CP) of the lateral ventricle (B, C) and CXCR4 mRNA expression in the subfornical organ (SFO) (H) as well as the ependymal layers of the lateral ventricle (LV) (H–J), the dorsal and ventral parts of the third ventricle (D3V, 3V) (H–J), and the fourth ventricle (4V) (K). In the amygdala, neuronal SDF-1 mRNA is selectively located in the anterior (BLA) (C) and posterior (BLP) (D) parts of the basolateral nucleus and in the amygdalohippocampal area (AHi) (D), whereas only few CXCR4 mRNA-expressing neurons without any clear assignment to subnuclei are detectable (I, J). In the hippocampal formation, numerous CXCR4 mRNA-expressing neurons are present in the moleculare lacunosum and molecular layers (Mol) (I, J) as well as the polymorphic layer (PoDG) (I,J), whereas SDF-1 mRNA is expressed only in the granular layer (GrDG) (C,D). K, L, In the cerebellum (CBL), CXCR4 mRNA is expressed in the Purkinje cell layer. K, L, In the brainstem, neuronal CXCR4 mRNA expression cannot be identified. Exposure time, 42 d. 7n, Facial nerve. Scale bar (shown in A for A–L): 2 mm.
Expression of the CXCR4 receptor in mouse brain
To examine the distribution of CXCR4-expressing cells in the brain, we first performed in situ hybridization (Fig.3G–L). In contrast to the pancerebral endothelial SDF-1β mRNA expression, the expression of CXCR4 mRNA was restricted to distinct neuronal cell groups, the ependymal layer of the four ventricles, and cells in the Purkinje cell layer of the cerebellum (Fig. 3G–L). Neuronal populations expressing CXCR4 mRNA were localized to the cerebral cortex, hippocampal formation, and amygdala. In the cerebral cortex, scattered CXCR4 mRNA-expressing neurons were found in the prelimbic and infralimbic areas as well as the cingulate, insular, and entorhinal areas (Fig.3H–J). Thus, neuronal CXCR4 and neuronal SDF-1 revealed a closely overlapping distribution in these regions. The highest levels of CXCR4 mRNA were detected in the glomerular layer of the olfactory bulb and in the hippocampal formation (Fig.3G,I,J).
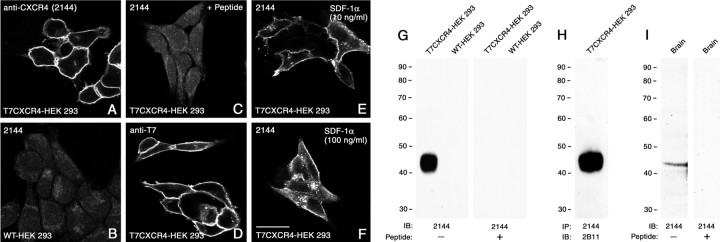
To further characterize the subcellular distribution of the CXCR4 receptor protein, we generated an anti-peptide antibody directed against the cytoplasmic tail of the CXCR4 receptor. Antibodies were affinity purified and further characterized using immunofluorescent staining of stably transfected HEK 293 cells. When wild-type HEK 293 cells or T7-tagged CXCR4-transfected cells were stained, the anti-CXCR4 antibodies yielded prominent immunofluorescence at the plasma membrane only in HEK 293 cells bearing the CXCR4 receptor but not in wild-type cells (Fig.4A,B). This staining was virtually identical to that seen with anti-T7 tag antibodies and completely blocked by preincubation of the antiserum with homologous peptide (Fig. 4C,D). After exposure of CXCR4-expressing cells with SDF-1α, a proportion of immunoreactive CXCR4 receptors underwent a dose-dependent redistribution from the plasma membrane into vesicle-like structures within the cytoplasm (Fig.4E,F). In Western blot assays, the anti-CXCR4 antibody detected a broad band migrating at 45 kDa only in membrane extracts from CXCR4-expressing cells but not in wild-type cells (Fig. 4G). Essentially identical results were obtained when CXCR4 receptors were first immunoprecipitated with our C-terminal rabbit anti-CXCR4 antibody (2144) and then immunoblotted with N-terminal rat monoclonal anti-CXCR4 antibody (2B11) (Fig.4H). The anti-CXCR4 antibody (2144) also detected a single 45 kDa band in membrane extracts from mouse brain (Fig.4I). The CXCR4-ir bands were no longer detected when the antiserum was preincubated with its immunizing peptide (Fig.4G,I).
Fig. 4.
Characterization of the anti-CXCR4 antiserum using stably transfected HEK 293 cells. Immunofluorescent labeling and confocal imaging of wild-type HEK 293 cells (B) and HEK 293 cells transfected with a construct coding for a T7-tagged CXCR4 receptor (A, C–F) are shown. Cells were either not treated (A–D) or exposed to 10 or 100 ng/ml SDF-1α for 30 min (E,F), fixed, and stained with the anti-CXCR4 antiserum (2144) (A–C, E,F) or anti-T7 antibody (D).G–I, Western blot analysis of CXCR4-LIR in transfected HEK 293 cells and mouse brain. G, I, Membrane preparations from wild-type HEK 293 cells, T7CXCR4-expressing HEK 293 cells, or mouse brain were immunoblotted (IB) with the anti-CXCR4 antibody (2144).H, CXCR4 receptors were immunoprecipitated (IP) using the C-terminal rabbit anti-CXCR4 antibody (2144) and immunoblotted with the N-terminal rat anti-CXCR4 antibody (2B11) as described in Materials and Methods. For absorption controls, the anti-CXCR4 antiserum was preincubated with 10 μg/ml of its immunizing peptide (C, G, I). Note that anti-CXCR4 antiserum yielded prominent immunofluorescence localized at the level of the plasma membrane only in CXCR4 T7tag-expressing HEK 293 cells but not in wild-type cells. This staining was essentially identical to that seen with the anti-T7 antibody and completely abolished by preincubation with homologous peptide. SDF-1α induced a dose-dependent internalization of the CXCR4 receptor. Note that anti-CXCR4 antiserum detected a single 45 kDa band in both transfected cells and mouse brain. WT, Wild-type. Ordinate represents migration of protein molecular weight markers (Mr × 10−3). Scale bar (A–F): 15 μm.
In brain sections of adult mice, high levels of CXCR4-like-immunoreactivity (LIR) were detected in brain regions expressing high levels of CXCR4 mRNA, including the olfactory bulb, the hippocampal formation, and the ependymal layer of the four ventricles (Fig. 5). CXCR4-ir axon terminals were occasionally detected in regions where CXCR4 mRNA was not detectable, e.g., in the accumbens shell (Fig. 5B). In contrast to in situ hybridization, immunocytochemistry did not label neurons in the claustrum and dorsal endopiriform nucleus. All immunostainings were completely abolished by preabsorption of the CXCR4 antibody with homologous but not with heterologous peptides (Fig.5C).
Fig. 5.
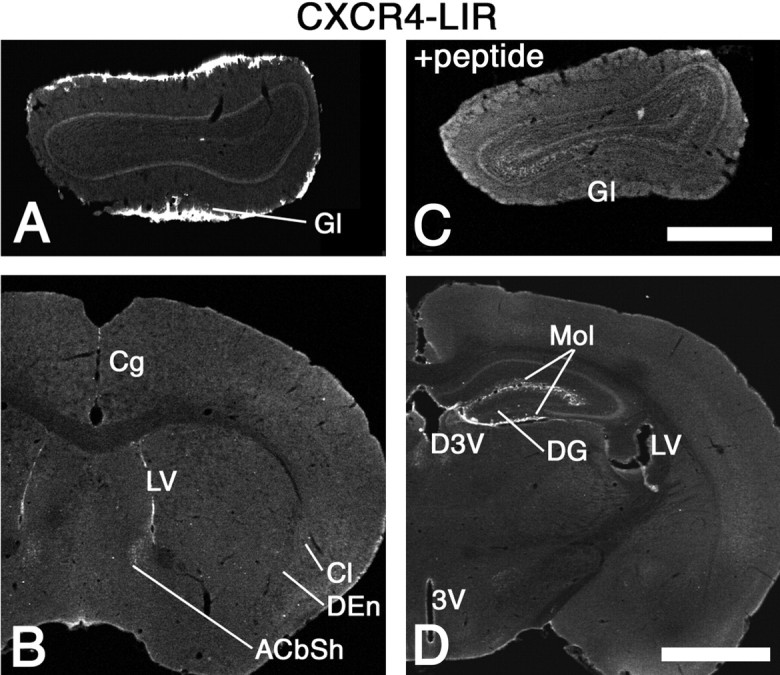
Distribution of CXCR4-LIR in the forebrain. Low-power confocal images of coronal brain sections stained by fluorescent immunohistochemistry. Strong CXCR4-LIR is present in the glomerular layer of the olfactory bulb (Gl) (A) and the ependymal layer of the lateral ventricle (LV) (B,D) as well as the ventral and dorsal parts of the third ventricle (3V, D3V) (D). The molecular layer of the dentate gyrus and the lacunosum moleculare layer of the hippocampus (Mol) (D) exhibit strong CXCR4-LIR. Note the presence of CXCR4-ir fibers in the shell region of the accumbens nucleus (ACbSh) (B) and the virtual absence of CXCR4-ir perikarya from the claustrum and the dorsal endopiriform nucleus (Cl, DEn) (B). C, CXCR4-LIR is completely neutralized by preabsorption with the peptide used for immunization. Scale bars: A, C, 700 μm;B, D, 1.2 mm.
The potential hippocampal targets of the SDF-1-producing dentate gyrus granule cells and entorhinal neurons were then characterized in detail. In the hippocampus, CXCR4 mRNA-expressing neurons were detected most frequently in the lacunosum moleculare layer, sparsely in the oriens layer, and very rarely in the pyramidal cell layer and the stratum radiatum (Fig. 6A). In the dentate gyrus, numerous CXCR4 mRNA-expressing neurons were localized in the molecular layer (Fig. 6A). In addition, numerous CXCR4-expressing neurons, some of which coexpressed glutamic acid decarboxylase (data not shown), were detected in the polymorphic layer as well as at the border of the granular and polymorphic layers (Fig. 6A). The distribution patterns of CXCR4-ir neuronal perikarya and CXCR4 mRNA-expressing neurons in the hippocampal formation were virtually identical (Fig.6A,B). The only exception were several neurons in the polymorphic layer (hilar neurons) that exhibited high CXCR4 mRNA levels, coexpressed the mRNA of pre-prosomatostatin (data not shown), but exhibited no detectable CXCR4-LIR at their somatodendritic domain (Fig. 6A,B). Because hilar somatostatinergic neurons are known to project to the outer molecular layer, this discrepancy could be explained by the selective targeting of the CXCR4 receptor to their axonal compartment in the molecular layer. In fact, CXCR4-ir axons running perpendicularly to the granular layer were observed in the granular layer and in the inner molecular layer (Fig. 6C, arrowheads). These axons may contribute to the dense network of CXCR4-ir fibers in the outer molecular layer (Fig. 6B). In the CXCR4-ir neurons at the border of the granular and polymorphic layers, CXCR4 receptors were restricted to the somata and proximal dendrites (Fig.6C, asterisk). In neurons of the lacunosum moleculare layer and the molecular layer, CXCR4-LIR was present at the plasma membrane of somata and dendrites (Fig.6C,D, arrows) as well as axons (Fig.6C,D,G,arrowheads). In the outer molecular layer, CXCR4-ir neurons were frequently orientated parallel to the hippocampal fissure. In both the molecular and the lacunosum moleculare layers, immunoreactive CXCR4 receptors were found in spine-free neurons (Fig. 6F) and neurons with spine-like protrusions at their cell bodies and dendrites (Fig. 6E,G,arrows).
Fig. 6.
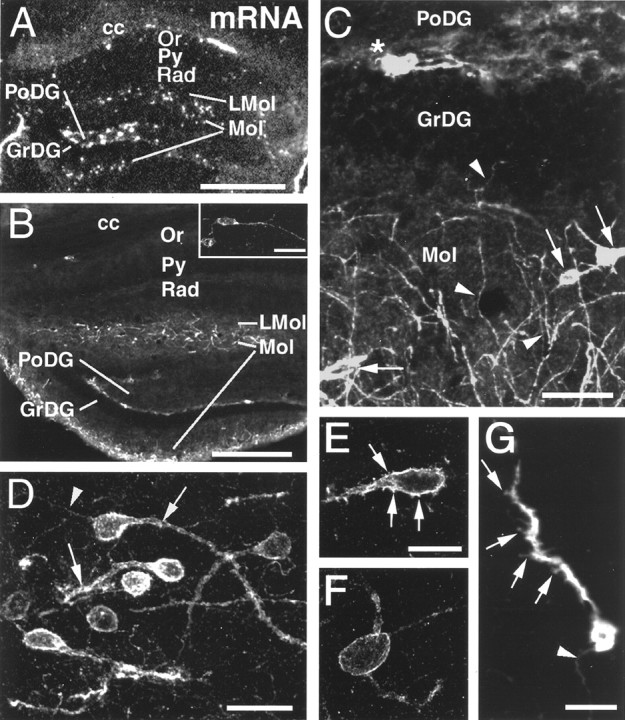
CXCR4 mRNA expression and cellular localization of CXCR4-LIR in the hippocampal formation. Coronal sections are shown in a dark-field micrograph after in situ hybridization (A) and in confocal images after fluorescent immunohistochemistry (B–G). A, B, Low-power micrographs comparing the distribution of CXCR4 mRNA (A) and CXCR4-LIR (B); note corpus callosum (cc) for orientation. In the oriens layer (Or), few CXCR4 mRNA-expressing and CXCR4-LIR neurons are detectable (shown at higher magnification in B), but none are detectable in the pyramidal cell layer (Py), stratum radiatum (Rad), or granular layer (GrDG). In the lacunosum moleculare layer (LMol), in the molecular layer (Mol), and at the border of the granular and polymorphic layers (PoDG), numerous CXCR4 mRNA-expressing and CXCR4-ir neurons are detected. Within the polymorphic layer, neurons express high CXCR4 mRNA levels but exhibit no detectable somatodendritic CXCR4-LIR.C–G, High-power confocal images demonstrating the subcellular targeting of CXCR4-LIR. C, In neurons of the molecular layer, CXCR4-LIR is somatodendritic (C,arrows) and axonal (C,arrowheads). Note the perpendicular orientation of several CXCR4-ir axons to the granular layer (C,arrowheads in GrDG andMol). At the border of the granular and polymorphic layers, CXCR4-LIR is restricted to neuronal perikarya and their proximal dendrites (C, asterisk).D, In the lacunosum moleculare layer, CXCR4-LIR typically exhibits somatodendritic (D,arrows) and axonal targeting (D,arrowhead) in parallel orientation to the hippocampal fissure. E–G, CXCR4-ir neurons in the molecular and lacunosum moleculare layers either exhibit CXCR4-ir spine-like protrusions at their cell bodies and dendrites (E,G, arrows) or exhibit a spine-free morphology (F). G, Note the CXCR4-ir axon (arrowhead). Scale bars: A, 400 μm; B, 300 μm; inset inB, 30 μm; C, 40 μm; D, 20 μm; E, F, 10 μm; G, 15 μm.
Cell-specific regulation of SDF-1 expression after systemic LPS administration
To test whether endothelial and neuronal SDF-1 isoforms may be differently regulated in the brain, we applied LPS systemically at a dose known to activate the cerebral endothelium and analyzed expression levels of endothelial SDF-1β and neuronal SDF-1α transcripts. Quantitative analysis of x-ray autoradiograms of hybridized brain sections (Fig.7E,F) revealed that the expression levels of SDF-1β mRNA at 3, 6, and 24 hr after LPS administration were decreased to 34, 36, and 53% (p < 0.05) of the levels seen in saline-injected animals, respectively (Table1). The downregulation was of similar intensity in cortex, caudate putamen, basal forebrain, and choroid plexus. Qualitative analysis at high power confirmed that at 3 hr after LPS stimulation the endothelial SDF-1β mRNA expression was decreased to levels near the detection limit (Fig.7G,H). Hybridization with the probe detecting all SDF-1 isoforms showed a LPS-induced decrease in endothelial SDF-1 expression similar to the SDF-1β-selective probe (Fig. 7A–D). Hybridization with the probe to SDF-1α or the probe to all SDF-1 isoforms revealed that LPS application did not affect SDF-1 mRNA expression in neurons of cortex or claustrum (Fig.7A–D). Thus, systemic LPS stimulation selectively affects endothelial SDF-1β expression but does not alter neuronal SDF-1α levels.
Fig. 7.
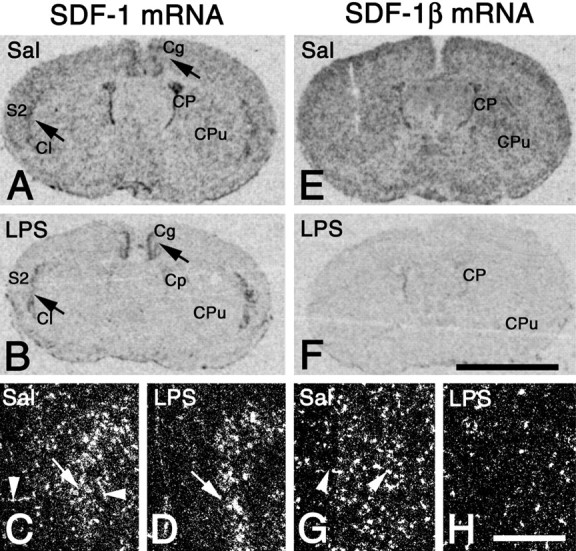
LPS-induced decrease in endothelial but not neuronal SDF-1 mRNA expression revealed by in situhybridization with a riboprobe detecting all SDF-1 isoforms (A–D) and a riboprobe specific for SDF-1β mRNA (E–H). Coronal brain sections 3 hr after LPS (LPS) (B, D,F, H) or saline application (Sal) (A, C,E, G) are shown as x-ray autoradiograms of the whole brain (A, B,E, F) and as dark-field micrographs of the cingulate cortex (C, D,G, H). A,B, After LPS application, SDF-1 mRNA expression is dramatically reduced as compared with saline-injected animals. Note that the LPS-induced decrease in SDF-1 mRNA levels is most obvious where neuronally expressed SDF-1 isoforms are absent, e.g., in the caudate putamen (CPu) and choroid plexus (CP). SDF-1 expression in neuronal structures (A, B, arrows) of the cingulate and secondary somatosensory cortex (Cg, S2) as well as the claustrum (Cl) is unchanged after LPS stimulation as compared with the saline-injected control.C, D, SDF-1 expression in the cingulate cortex is decreased by LPS in endothelial cells (arrowheads) but not neurons (arrows).E, F, Throughout the brain, SDF-1β mRNA levels are strongly reduced after LPS stimulation as compared with the saline-injected control. Note the pronounced downregulation of SDF-1β in endothelial cells after LPS injection (H) as compared with the control (G, arrowheads). Exposure times:A, B, 3 d; E,F, 2 d; C, D,G, H, 21 d. Scale bars:A, B, E, F, 3 mm; C, D, G,H, 550 μm.
Table 1.
SDF-1β mRNA levels in the mouse brain after peripheral LPS administration
| 3 hr saline | 0.5 hr LPS | 1 hr LPS | 3 hr LPS | 6 hr LPS | 24 hr LPS | |
|---|---|---|---|---|---|---|
| SDF-1β mRNA | 381 ± 36 | 289 ± 26 | 275 ± 44 | 133 ± 82* (34%) | 138 ± 92* (36%) | 203 ± 57*(53%) |
SDF-1β mRNA levels of whole-brain sections expressed in nanoCuries per gram ± SD (mean values of four animals in the group 3 hr after saline injection and three animals per LPS-treated group). Bold values express measurements as percentage of the group 3 hr after saline injection. *p < 0.05 versus the saline-injected group. One-way ANOVA followed by Dunnett's multiple comparison test.
CXCR4 mRNA levels analyzed in the ependymal layer of the lateral ventricles and neurons in the cingulate cortex were not changed after LPS stimulation as compared with the saline-injected group. There was no evidence for an infiltration of the brain by CXCR4 mRNA-expressing leukocytes in any of the investigated regions (data not shown).
Adaptive plasticity of the SDF-1/CXCR4 receptor system after focal ischemia
We then investigated adaptive changes of endothelial and neuronal SDF-1 mRNA expression in relation to the presence of CXCR4 mRNA-expressing infiltrates and regulation of neuronal CXCR4 mRNA levels after focal cerebral ischemia. A comparative analysis of the primary infarcted area, the area at the border of the infarcted and nonlesioned tissues (border zone), and the ipsilateral area without detectable neurodegeneration (ipsilateral nonlesioned area) as well as the contralateral hemisphere was performed (Fig.8G). For simplicity during quantitative image analysis, the border zone and the primary infarct were treated as one region.
Fig. 8.
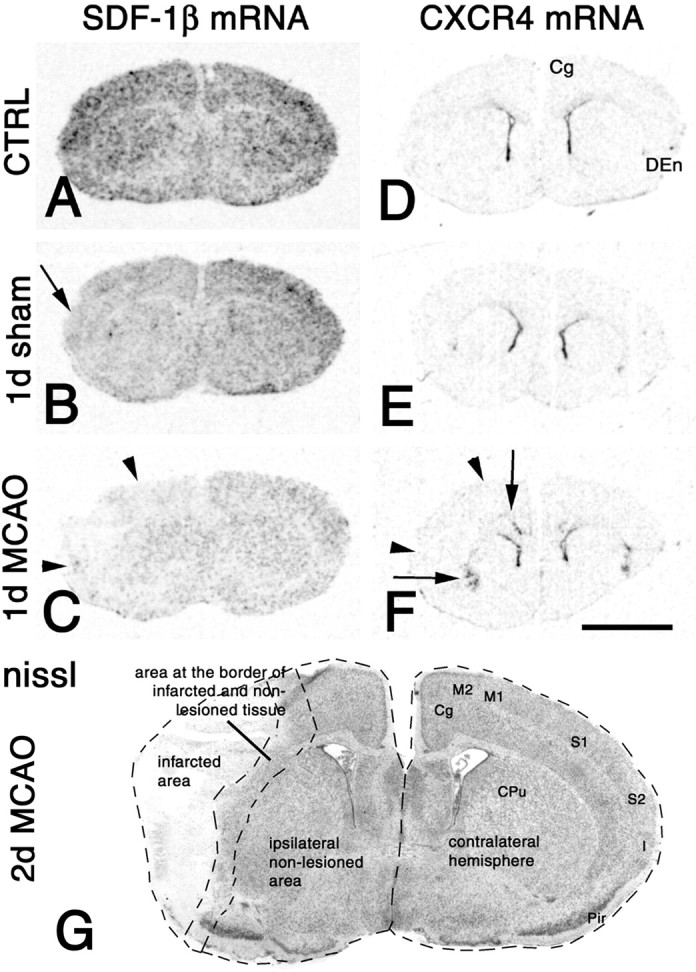
Patterns of middle cerebral artery occlusion (MCAO)-induced changes in SDF-1β (A–C) and CXCR4 (D–F) mRNA expression. A–F, Low-power micrographs of x-ray autoradiograms of hybridized coronal sections from naive mice (CTRL) (A, D) and mice 1 d after sham operation (B, E) or MCAO (C, F). The lateral borders of the infarct are indicated by arrowheads(C, F). C, After MCAO, SDF-1β mRNA levels are decreased throughout the brain as compared with the sham operation (B) or the naive control (A). The decrease in SDF-1β mRNA expression is most pronounced within the infarct. C, In the nonlesioned brain areas, SDF-1β expression is decreased more strongly in the ipsilateral than in the contralateral hemisphere.B, Note that sham operation induced a decrease in SDF-1β mRNA expression throughout the brain, which increases with proximity to the trepanation site (B,arrow). F, In the hemisphere ipsilateral to MCAO, there is a strong increase in CXCR4 mRNA expression in lamina VI of the cingulate cortex and the dorsal endopiriform nucleus (F, arrows) as compared with naive animals or sham operations (D, E). A similar but less pronounced increase is observed in cingulate cortex and dorsal endopiriform nucleus of the contralateral hemisphere.G, Low-power micrograph of a cresyl violet (nissl)-stained coronal section through a mouse forebrain 2 d after MCAO at bregma +0.14 mm. Delineated bydotted lines are the infarcted area, which includes the primary (S1) and secondary (S2) somatosensory cortex, the insular cortex (I), and the dorsal parts of the piriform cortex (Pir). The cingulate (Cg) and secondary motor (M2) cortices are nonlesioned cerebrocortical areas. The primary motor cortex (M1), the lateral aspect of the caudate putamen (CPu), and parts of the piriform cortex contribute to an area at the border of nonlesioned and infarcted tissue. The largest part of the caudate putamen (medial aspect) is not infarcted. Exposure times:A–C, 2 d; D–F, 3 d. Scale bar (shown in F for A–F): 3 mm.
Changes in endothelial SDF-1β mRNA expression
As shown for 1 d after MCAO (Fig.8A,C), there was a downregulation of SDF-1β mRNA expression to different extents in the four regions as compared with controls. The decrease in SDF-1β mRNA levels was most pronounced in the primary infarct, most likely due to degeneration of cells. However, SDF-1β expression was also decreased in the nonlesioned areas. This downregulation was more pronounced in the ipsilateral than in the contralateral hemisphere (Fig. 8C). Quantitative analysis of SDF-1β mRNA expression revealed that MCAO induced a decrease from 6 hr to 4 d in all regions as compared with control animals. In addition, the quantitative analysis revealed an inverse relationship of lesion-distance and extent of SDF-1β downregulation (Table 2). All changes in endothelial SDF-1 mRNA expression were similar, when examined with the SDF-1β−selective probe or the probe detecting all SDF-1 isoforms (Figs. 8, 9). Notably, also the sham operation induced a decrease in SDF-1β mRNA expression 1 d and 2 d after treatment as compared with control animals (Table3, Fig. 8A,B). Comparison of ischemic and sham operated animals at 1 d and 2 d revealed significantly lower SDF-1β mRNA levels after MCAO than after sham operation (Table 3). These findings strongly suggest, that skull trepanation decreases SDF-1β expression in the brain and that focal ischemia induces a reduction in endothelial SDF-1β mRNA expression independently from trepanation.
Table 2.
Influence of MCAO on SDF-1β mRNA levels in the mouse brain
| Control | 6 hr MCAO | 1 d MCAO | 2 d MCAO | 4 d MCAO | |
|---|---|---|---|---|---|
| Infarct and border zone | 318 ± 49 | 6.7 ± 3.6* (3%) | 7.6 ± 3.6* (3%) | 21.7 ± 13.6* (7%) | 49.2 ± 19.5* (16%) |
| Ipsilateral nonlesioned area | 175 ± 39.3 | 33.2 ± 26.6* (19%) | 25.2 ± 28.1* (15%) | 34.3 ± 31.5* (20%) | 48.5 ± 14.8* (28%) |
| Contralateral hemisphere | 233 ± 35.7 | 69.0 ± 26.3* (30%) | 88.3 ± 67.0* (38%) | 96.8 ± 46.0* (42%) | 104 ± 38.2* (45%) |
Measurements are mean SDF-1β mRNA levels in nanoCuries per gram ± SD (three naive animals as control group, three animals at 6 hr and 1 d after MCAO, four animals at 2 d and 4 d after MCAO, respectively). The regions of interest in animals receiving MCAO were the infarct including the border zone, the ipsilateral area where no neurodegeneration was observed (ipsilateral nonlesioned area), and the entire hemisphere contralateral to MCAO. In the controls, the three corresponding regions were analyzed. Bold values express measurements after MCAO as percentage of controls. *p< 0.05 versus the control animals; one-way ANOVA followed by Dunnett's multiple comparison test.
Fig. 9.
Dark-field micrographs of the brain hemisphere ipsilateral to middle cerebral artery occlusion at various time points after surgery (B, D,E–H) and of the corresponding brain hemisphere of control mice (CRTL) (A,C). In situ hybridization was performed with a riboprobe detecting all SDF-1 isoforms (A,B, E, F) and a riboprobe for CXCR4 mRNA (C, D,G, H). A,B, E, F, SDF-1 mRNA expression by neurons in the cingulate cortex is strongly decreased at 6 hr (B) and unchanged at 2 and 4 d after MCAO (E, F) as compared with the control (A). After 6 hr and 2 d, endothelial SDF-1 mRNA expression is at the detection limit within the infarct and strongly decreased in the nonlesioned area (B,E) as compared with the control (A). Note the very strong induction of SDF-1 mRNA expression in the border zone of the infarct after 2 d (E, arrowhead). F, After 4 d a moderate to strong SDF-1 mRNA expression is detectable within the infarct and its border zone (F,arrowhead), whereas non-neuronal SDF-1 mRNA expression in the nonlesioned area is moderately decreased as compared with the control (A). C, D,G, H, After 6 hr and 2 d but not after 4 d, neuronal CXCR4 mRNA expression is strongly increased in the dorsal endopiriform nucleus (D, G,asterisks) and lamina VI of the cingulate cortex (D, G, arrows) as compared with the control (C). G, Note the induction of CXCR4 mRNA expression in the superficial layers of the cingulate cortex after 2 d. G, H, In the border zone of the infarct (G,arrowhead), a massive accumulation of CXCR4 mRNA-expressing cells is detectable after 2 and 4 d.H, After 4 d, numerous CXCR4 mRNA-expressing cells are also present within the infarcted core. Exposure times:A, B, E, F, 21 d; C, D, G,H, 42 d. Scale bar (shown in H forA–H): 3 mm.
Table 3.
Relative influence of sham operation and MCAO on SDF-1β mRNA levels in the mouse brain
| Control 1 | 1 d sham | 1 d MCAO | Control 2 | 2 d sham | 2 d MCAO | |
|---|---|---|---|---|---|---|
| Infarct and border zone | 248 ± 7.5 | 126 ± 31.0* (50%) | 13.3 ± 6.2*3-151 (5%) | 495 ± 29.4 | 279 ± 34.9* (56%) | 61 ± 77*3-151 (12%) |
| Ipsilateral nonlesioned area | 187 ± 14.3 | 122 ± 14.5* (65%) | 70.8 ± 27.6*3-151 (37%) | 442 ± 40.6 | 318 ± 13.8* (71%) | 225 ± 40.0*3-151 (50%) |
| Contralateral hemisphere | 234 ± 15.4 | 183 ± 11.0* (78%) | 117.9 ± 31.0*3-151 (50%) | 465 ± 42.1 | 351 ± 26.5* (75%) | 281 ± 51.3* (60%) |
Mean SDF-1β mRNA levels (expressed in nanoCuries per gram ± SD; calculated from three animals per group) are shown for two independent experiments. In both experiments, naive mice were used as control groups (control 1, control 2). In the first experiment, animals 1 d after sham operation and 1 d after MCAO were analyzed. In the second experiment, animals 2 d after sham operation and 2 d after MCAO were analyzed. The regions of interest in animals receiving MCAO were the infarct including the border zone, the ipsilateral area where no neurodegeneration was observed (ipsilateral nonlesioned area), and the entire hemisphere contralateral to MCAO. In the controls and sham-operated animals, the three corresponding regions were analyzed. Bold values express SDF-1β mRNA levels after MCAO as percentage of controls. *p < 0.05 versus the control animals;
F3-151: p < 0.05 versus the sham-operated animals. One-way ANOVA followed by Newman–Keuls multiple comparison test.
Recruitment of CXCR4-expressing cells to the lesioned brain
The spatial relationship of SDF-1- and CXCR4-expressing cells in the ischemic brain was analyzed at low-power under dark-field illumination (Fig. 9). There were virtually no peripheral CXCR4-expressing cells infiltrating the ischemic brain after 6 hr or 1 d (Fig. 9D, Fig. 8F), when endothelial SDF-1 expression was strongly decreased in all analyzed regions (Figs. 9A,B, 8A,C). Surprisingly, after 2 d numerous CXCR4 mRNA-expressing cells were detected in the border-zone of the infarct (Fig. 9G,arrow-head). In close spatial and temporal overlap with the accumulation of CXCR4-expressing cells a strong focal induction of SDF-1 mRNA expression was observed in single vascular structures (Fig.9E,G, arrow-heads). In contrast, in the infarcted core and the nonlesioned areas SDF-1 expression was still strongly decreased at 2 d after MCAO as compared with the control (Fig. 9A,E). After 4 d numerous CXCR4-expressing cells were present in both the primary infarct and the border-zone, which was in correspondence with a moderate SDF-1 mRNA expression in both regions (Fig.9F,H). It should be noted, that there was no evidence for peripheral CXCR4-expressing cells infiltrating the nonlesioned areas at all stages after cerebral ischemia.
Transient changes of neuronal SDF-1 and neuronal CXCR4 mRNA expression
In addition to the changes of non-neuronal SDF-1β and CXCR4 expression, there were pronounced changes of neuronal SDF-1 and CXCR4 expression in nonlesioned brain regions after focal ischemia (Figs.8,9). Sham operation had no effect on neuronal SDF-1 (not shown) or CXCR4 expression (Fig. 8D,E). An ischemia-induced upregulation of neuronal CXCR4 mRNA expression was seen in the dorsal endopiriform nucleus (Fig. 9D,G, asterisks) and the cingulate cortex (Fig. 9D,G, arrows) from 6 hr to 2 d after MCAO. Neuronal CXCR4 levels in both structures were similar to controls after 4 d (Fig.9C,H). At 6 hr and 1 d the increased CXCR4 expression in the cingulate cortex was restricted to lamina VI (Figs.9D, 8F). After 2 d CXCR4 mRNA expression was induced in laminae II/III and VI (Fig. 9G). Neuronal SDF-1 mRNA expression in the cingulate cortex was decreased in lamina V and VI 6 hr and 1 d after MCAO (Fig. 9A,B) but virtually unchanged at later time-points (Fig.9A,E,F). All described changes of neuronal SDF-1 and CXCR4 mRNA expression were also seen in the contralateral hemisphere but were less pronounced than in the ipsilateral hemisphere (Fig.8F).
Characterization of SDF-1- and CXCR4-expressing cells in the border zone
Combinations of radioactive in situ hybridization and immunocytochemistry or nonisotopic in situ hybridization were applied to further characterize SDF-1- and CXCR4-expressing cells in the border zone after 2 d. The vascular SDF-1 upregulation was detected in vWF-ir endothelial cells using the probe to all SDF-1 isoforms (data not shown) as well as the SDF-1β−selective probe (Fig. 10A,arrowheads). SDF-1 isoforms were not detected by either probe in reactive astrocytes (data not shown) and large reactive microglial cells (Fig. 10B,asterisks), which were identified by their strong GFAP and C1q mRNA expression, respectively. CXCR4 mRNA was detectable in a small subset of C1q-positive cells (∼30 cells per section). These cells were small, without processes, and restricted to the border zone (Fig.10C, arrow). Most of the C1q-positive cells that were devoid of any CXCR4 labeling were larger, exhibited swollen processes, and were frequent in the entire ipsilateral hemisphere (Fig.10C, asterisk). Because C1q detects both macrophages and microglial cells in the brain, the smaller C1q-positive cells expressing CXCR4 most likely represent infiltrating macrophages, whereas the larger CXCR4-negative cells are resident activated microglial cells. Notably, the cells coexpressing CXCR4 and C1q mRNAs were only a subset (∼10%) of CXCR4 mRNA-expressing cells in the border zone. Double in situ hybridization and double immunohistochemistry revealed that most of the CXCR4 mRNA-expressing cells were negative for GFAP (Fig.10D,E,E′). A minor population (< 1%) of reactive astrocytes in the border zone expressed CXCR4 mRNA (data not shown). These findings strongly suggest that resident astrocytes and microglial cells do not upregulate CXCR4 expression during activation after cerebral ischemia. CXCR4-expressing cells in the border zone 2 d after MCAO most likely represent infiltrating peripheral blood cells, including macrophages.
Fig. 10.
High-resolution analysis of the cellular expression of SDF-1 isoforms and CXCR4 mRNA in the border zone 2 d after MCAO. Hybridized 35S-labeled riboprobes (35S) to SDF-1β (A), SDF-1 (B), and CXCR4 (C,D) mRNAs are detected as grains; immunocytochemistry for vWF (A) and co-hybridized DIG-labeled riboprobes (DIG) to C1q (B,C) and GFAP (D) mRNAs are detected as black reaction products. A,Arrowheads, Strong SDF-1β mRNA expression is detected specifically in vWF-ir endothelial cells. B, SDF-1 mRNA is absent from activated C1q mRNA-expressing microglial cells/macrophages (B, asterisks) but expressed by endothelial cells (B,arrowheads). Note the absence of SDF-1 mRNA expression from a juxtavascular C1q mRNA-expressing cell (B,arrow). C, CXCR4 mRNA is absent from a large C1q mRNA-expressing activated microglial cell (C,asterisk) but present in a small presumed C1q mRNA-expressing macrophage (C, arrow). Note C1q mRNA-negative CXCR4 mRNA-expressing cells (C,arrowheads). D, Detection of numerous CXCR4 mRNA-expressing cells (D, arrows) in the presence of CXCR4 mRNA-negative GFAP-positive activated astrocytes (D, asterisks). E,E′, Double immunohistochemistry for CXCR4 (E) and GFAP (E′) demonstrating the absence of CXCR4-LIR (arrows) on activated astrocytes (asterisks). Scale bar:A–D, 40 μm; E,E′, 20 μm.
DISCUSSION
In the present study, we provide the first in situ hybridization evidence for cell- and tissue-specific expression of SDF-1 splice variants in the mouse brain using a pan-SDF-1 probe, a SDF-1α-selective probe, and a SDF-1β−selective probe, which does not detect SDF-1α but may hybridize to a putative mouse SDF-1γ isoform as well. We show that in the adult mouse brain SDF-1β is selectively generated in the endothelium of cerebral microvessels but not in neurons, whereas SDF-1α is generated predominantly in distinct neuronal populations and meningeal cells. After systemic LPS administration, cerebro-endothelial SDF-1β was selectively downregulated, whereas neuronal SDF-1α remained unchanged. After focal ischemia, endothelial SDF-1β showed a persistent downregulation in the nonlesioned brain, and neuronal SDF-1α showed a transient downregulation in the nonlesioned brain. In the penumbra, SDF-1β was selectively upregulated in the endothelium of single blood vessels. Thus, alternative splicing may represent the primary mechanism by which the expression of SDF-1 in the CNS is regulated in a tissue- and stimulus-specific manner.
We observed a striking mismatch between the widespread endothelial SDF-1β expression and the restricted CXCR4 expression yet a close overlap between the distinct neuronal SDF-1α and neuronal CXCR4 expression, indicating that SDF-1α and CXCR4 constitute a functional receptor/ligand system in neurons. The pronounced gene regulation of neuronal SDF-1 and neuronal CXCR4 after focal ischemia indicates a potential role of SDF-1/CXCR4 in adaptive neuronal plasticity. The lesion-induced upregulation of endothelial SDF-1β and the concomitant appearance of CXCR4-expressing cells 2 d after ischemia indicate that cerebro-endothelial SDF-1β may represent a chemoattractant for peripheral blood cells.
Conserved expression of SDF-1 and CXCR4 in the adult mouse and rat brain
Using highly sensitive isotopic in situhybridization, we showed the expression of SDF-1 in neurons of the cerebral cortex, dentate gyrus, basolateral amygdala, and the amygdalohippocampal area as well as endothelial cells of the adult mouse brain. Our findings are in line with previous reports showing the expression of SDF-1 in distinct neuronal populations and endothelial but not glial cells of the adult rat brain (Tham et al., 2001). Another study reported widespread expression of SDF-1 in neurons and oligodendrocytes but not in astrocytes using nonisotopic in situ hybridization (Gleichmann et al., 2000). However, the findings by Tham et al. (2001) and our own observations in mice and rats indicate conserved SDF-1/CXCR4 expression patterns in rodent brain and do not support a widespread distribution of SDF-1 in neuronal or glial cells.
Using species-specific riboprobes and our antiserum, we identified CXCR4-expressing cells in the ventricular ependyma, olfactory bulb, cerebral cortex, hippocampus, amygdala, and cerebellum in rat (data not shown) and mouse. In the cerebral cortex and hippocampus, CXCR4 is expressed exclusively in neurons. The axonal and somatodendritic targeting of CXCR4-LIR suggests presynaptic and postsynaptic functions of this receptor. In a recent in vitro study, Bezzi et al. (2001) implicated SDF-1α in glutamate release from cultured astrocytes. The proposed mechanism involves CXCR4-mediated TNFα release from activated microglia and astrocytes. However, in the adult nonlesioned and ischemia-lesioned brain, CXCR4 expression in the vast majority of identified astrocytes and microglial cells was below the detection limit. Nevertheless, the possibility cannot be excluded that glial CXCR4 expression levels are differently regulated during development or in astrocyte cultures (Wong et al., 1996; Klein et al., 1999; Lazarini et al., 2000; Bezzi et al., 2001).
CXCR4-mediated neuromodulation and plasticity
SDF-1 has recently been shown to modulate synaptic transmission in the developing cerebellum (Limatola et al., 2000). The codistribution of neurons expressing SDF-1α mRNA and neurons expressing CXCR4 in the adult cerebral cortex and hippocampus suggests that SDF-1 may exert neuromodulatory actions also in these brain regions. Prominent CXCR4-LIR was detected in spiny and aspiny neuronal somata, in dendrites and axons of the moleculare lacunosum, and in molecular layers of hippocampus and dentate gyrus. Neuronal SDF-1 mRNA is highly expressed in the granule cells of the dentate gyrus and in the entorhinal area of the cortex. Although SDF-1 synthesized in granule cells is likely to interact locally with CXCR4-expressing interneurons of the polymorphic layer of the dentate gyrus, SDF-1 synthesized in the entorhinal cortex may be present in perforant path projections that terminate in the molecular layer of the dentate gyrus.
Cerebral ischemia evoked dramatic changes in neuronal SDF-1 and CXCR4 mRNA expression in noninfarcted areas. The strong decrease in neuronal SDF-1 mRNA expression in cerebrocortical neurons was most prominent at 6 hr after MCAO and may be related to the acute excitotoxic phase and peri-infarct depolarizations. In contrast, neuronal CXCR4 mRNA levels in the cerebral cortex and the dorsal endopiriform nucleus were persistently upregulated from 6 hr to 2 d after MCAO. This upregulation may be related to lesion-induced neuronal adaptations rather than acute excitotoxicity. However, pro-apoptotic effects of CXCR4 in neurons must also be considered (Kaul and Lipton, 1999). Together, our findings point to a potential role of the SDF-1/CXCR4 system in neuronal plasticity and repair in the adult brain.
Endothelial-specific functions of SDF-1β
We found a remarkable mismatch between the pancerebral endothelial distribution of SDF-1β mRNA and the restricted cerebral expression of CXCR4. The simplest explanation for this finding is that circulating leukocytes expressing CXCR4 but not cerebral resident cells are likely targets for endothelial SDF-1β. Recently, SDF-1 has been shown to augment tethering and firm adhesion of leukocytes to TNFα-activated endothelial cells (Grabovsky et al., 2000). SDF-1 modulates the interaction of endothelial adhesion molecules including intercellular adhesion molecule-1 (ICAM-1) and vascular cell adhesion molecule-1 (VCAM-1) with leukocyte integrins (Campbell et al., 1998; Grabovsky et al., 2000; Kantele et al., 2000). In the cerebral endothelium, constitutive expression of ICAM-1 and VCAM-1 is low but strongly upregulated by systemic LPS or TNFα administration (Henninger et al., 1997). Interestingly, the massive LPS-induced downregulation of endothelial SDF-1β mRNA expression observed in the present study exhibited a similar time course as the VCAM-1/ICAM-1 upregulation reported by Henninger et al. (1997). Thus, balanced expression of SDF-1β and endothelial adhesion molecules in cerebral endothelial cells may be crucial for preservation of BBB integrity during systemic infection. This is in line with the absence of CXCR4-expressing leukocyte infiltrates in the brain after systemic LPS. However, other chemokines including monocyte chemoattractant protein 1 (MCP-1) may selectively target leukocytes to specific brain regions during systemic infection. MCP-1 mRNA expression is most potently induced in all circumventricular organs and choroid plexus after systemic LPS (Thibeault et al., 2001).
Systemic LPS application may regulate cerebral SDF-1 expression via direct Toll-like receptor-mediated mechanisms or via an array of indirect pathways. Direct treatment of cultured neurons, astrocytes, and microglial cells with LPS in vitro can induce either downregulation or upregulation of SDF-1 expression (Ohtani et al., 1998; Bajetto et al., 1999). In contrast, in vivo LPS application selectively downregulates endothelial but not neuronal SDF-1, suggesting that neuronal SDF-1α expression is mostly resistant to systemic LPS.
Like systemic LPS, focal ischemia caused a decrease in endothelial SDF-1β expression throughout the forebrain. Unlike systemic LPS, the MCAO-induced decrease in SDF-1β expression was dependent on the distance to the lesion site. At later stages after ischemia, we detected a strong induction of SDF-1β expression in single vascular structures and a local infiltration of CXCR4-expressing peripheral blood cells in the border zone of the infarct. Thus, focal endothelial SDF-1β upregulation may guide peripheral CXCR4-expressing cells to the lesioned site and is likely to contribute to local post-ischemic inflammation and later reorganization of the infarct.
CXCR4-expressing infiltrates were not observed in brain regions with persistent endothelial SDF-1β downregulation. Thus, the ischemia-induced decrease in endothelial SDF-1β mRNA expression in nonlesioned areas could reflect a protective mechanism that prevents SDF-1-induced leukocyte attraction and platelet activation in brain microvessels (Forster et al., 1998; Kowalska et al., 2000). This is particular relevant under conditions of impaired blood flow and ischemia-induced upregulation of adhesion molecules, including ICAM-1, P-selectin, and E-selectins in endothelial cells (Dirnagl et al., 1999). SDF-1β mRNA expression in the brain appears to be sensitive to even small brain lesions, as indicated by the effect of the sham operation. However, the SDF-1β downregulation after sham operation was only moderate, and neuronal SDF-1α and CXCR4 expression were not affected, indicating that ischemia caused a SDF-1β downregulation that extended far beyond the effects of surgical stress. Moreover, the particular upregulation of SDF-1β and infiltration of CXCR4-positive cells were undetectable in sham-operated animals.
CONCLUSION
In the present study, we have established the cellular expression patterns of SDF-1 isoforms and the CXCR4 receptor in the adult mouse brain demonstrating that SDF-1β is selectively generated in endothelial cells of microvessels, whereas SDF-1α is predominantly generated in distinct neuronal populations. We provide evidence for isoform-specific regulation of SDF-1 gene expression after systemic LPS administration and focal ischemia. The sensitive regulation of endothelial SDF-1β suggests that the SDF-1/CXCR4 system is a promising target for pharmacological intervention under conditions of impaired blood–brain barrier integrity.
Footnotes
This work was supported by Volkswagen Stiftung Grant I/75172 (V.H., S.S.). We thank Dr. Martin Lipp and Dr. Reinhold Förster (Max Delbrück Center for Molecular Medicine, Berlin, Germany) for kindly providing mouse CXCR4 cDNA and rat monoclonal anti-CXCR4 antibody clone 2B11. We also thank Dr. Tasuku Honjo and Dr. Kei Tashiro (Kyoto University, Kyoto, Japan) for kindly providing mouse SDF-1α cDNA. Excellent technical assistance by Diana Gericke, Evelyn Kahl, Dana Mayer, and Nancy Schlawin is gratefully acknowledged.
Correspondence should be addressed to Stefan Schulz, Institut für Pharmakologie und Toxikologie, Otto-von-Guericke-Universität Magdeburg, Leipziger Strasse 44, 39120 Magdeburg, Germany. E-mail:Stefan.Schulz@medizin.uni-magdeburg.de.
REFERENCES
- 1.Angerer LM, Cox KH, Angerer RC. Demonstration of tissue-specific gene expression by in situ hybridization. Methods Enzymol. 1987;152:649–661. doi: 10.1016/0076-6879(87)52071-7. [DOI] [PubMed] [Google Scholar]
- 2.Bajetto A, Bonavia R, Barbero S, Piccioli P, Costa A, Florio T, Schettini G. Glial and neuronal cells express functional chemokine receptor CXCR4 and its natural ligand stromal cell-derived factor 1. J Neurochem. 1999;73:2348–2357. doi: 10.1046/j.1471-4159.1999.0732348.x. [DOI] [PubMed] [Google Scholar]
- 3.Bezzi P, Domercq M, Brambilla L, Galli R, Schols D, De Clercq E, Vescovi A, Bagetta G, Kollias G, Meldolesi J, Volterra A. CXCR4-activated astrocyte glutamate release via TNFalpha: amplification by microglia triggers neurotoxicity. Nat Neurosci. 2001;4:702–710. doi: 10.1038/89490. [DOI] [PubMed] [Google Scholar]
- 4.Bleul CC, Fuhlbrigge RC, Casasnovas JM, Aiuti A, Springer TA. A highly efficacious lymphocyte chemoattractant, stromal cell-derived factor 1 (SDF-1). J Exp Med. 1996a;184:1101–1109. doi: 10.1084/jem.184.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bleul CC, Farzan M, Choe H, Parolin C, Clark-Lewis I, Sodroski J, Springer TA. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature. 1996b;382:829–833. doi: 10.1038/382829a0. [DOI] [PubMed] [Google Scholar]
- 6.Campbell JJ, Hedrick J, Zlotnik A, Siani MA, Thompson DA, Butcher EC. Chemokines and the arrest of lymphocytes rolling under flow conditions. Science. 1998;279:381–384. doi: 10.1126/science.279.5349.381. [DOI] [PubMed] [Google Scholar]
- 7.Dirnagl U, Iadecola C, Moskowitz MA. Pathobiology of ischaemic stroke: an integrated view. Trends Neurosci. 1999;22:391–397. doi: 10.1016/s0166-2236(99)01401-0. [DOI] [PubMed] [Google Scholar]
- 8.Fedyk ER, Jones D, Critchley HO, Phipps RP, Blieden TM, Springer TA. Expression of stromal-derived factor-1 is decreased by IL-1 and TNF and in dermal wound healing. J Immunol. 2001;166:5749–5754. doi: 10.4049/jimmunol.166.9.5749. [DOI] [PubMed] [Google Scholar]
- 9.Feng Y, Broder CC, Kennedy PE, Berger EA. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 10.Forster R, Kremmer E, Schubel A, Breitfeld D, Kleinschmidt A, Nerl C, Bernhardt G, Lipp M. Intracellular and surface expression of the HIV-1 coreceptor CXCR4/fusin on various leukocyte subsets: rapid internalization and recycling during activation. J Immunol. 1998;160:1522–1531. [PubMed] [Google Scholar]
- 11.Franklin KBJ, Paxinos G. The mouse brain in stereotaxic coordinates. Academic; San Diego, CA: 1997. [Google Scholar]
- 12.Gleichmann M, Gillen C, Czardybon M, Bosse F, Greiner-Petter R, Auer J, Muller HW. Cloning and characterization of SDF-1gamma, a novel SDF-1 chemokine transcript with developmentally regulated expression in the nervous system. Eur J Neurosci. 2000;12:1857–1866. doi: 10.1046/j.1460-9568.2000.00048.x. [DOI] [PubMed] [Google Scholar]
- 13.Goodman RH, Montminy MR, Low MJ, Habener JF. Biosynthesis of rat preprosomatostatin. Adv Exp Med Biol. 1985;188:31–47. doi: 10.1007/978-1-4615-7886-4_3. [DOI] [PubMed] [Google Scholar]
- 14.Grabovsky V, Feigelson S, Chen C, Bleijs DA, Peled A, Cinamon G, Baleux F, Arenzana-Seisdedos F, Lapidot T, van Kooyk Y, Lobb RR, Alon R. Subsecond induction of alpha4 integrin clustering by immobilized chemokines stimulates leukocyte tethering and rolling on endothelial vascular cell adhesion molecule 1 under flow conditions. J Exp Med. 2000;192:495–506. doi: 10.1084/jem.192.4.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heesen M, Berman MA, Benson JD, Gerard C, Dorf ME. Cloning of the mouse fusin gene, homologue to a human HIV-1 co-factor. J Immunol. 1996;157:5455–5460. [PubMed] [Google Scholar]
- 16.Henninger DD, Panes J, Eppihimer M, Russell J, Gerritsen M, Anderson DC, Granger DN. Cytokine-induced VCAM-1 and ICAM-1 expression in different organs of the mouse. J Immunol. 1997;158:1825–1832. [PubMed] [Google Scholar]
- 17.Hesselgesser J, Taub D, Baskar P, Greenberg M, Hoxie J, Kolson DL, Horuk R. Neuronal apoptosis induced by HIV-1 gp120 and the chemokine SDF-1 alpha is mediated by the chemokine receptor CXCR4. Curr Biol. 1998;8:595–598. doi: 10.1016/s0960-9822(98)70230-1. [DOI] [PubMed] [Google Scholar]
- 18.Kantele JM, Kurk S, Jutila MA. Effects of continuous exposure to stromal cell-derived factor-1 alpha on T cell rolling and tight adhesion to monolayers of activated endothelial cells. J Immunol. 2000;164:5035–5040. doi: 10.4049/jimmunol.164.10.5035. [DOI] [PubMed] [Google Scholar]
- 19.Kaul M, Lipton SA. Chemokines and activated macrophages in HIV gp120-induced neuronal apoptosis. Proc Natl Acad Sci USA. 1999;96:8212–8216. doi: 10.1073/pnas.96.14.8212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klein RS, Williams KC, Alvarez-Hernandez X, Westmoreland S, Force T, Lackner AA, Luster AD. Chemokine receptor expression and signaling in macaque and human fetal neurons and astrocytes: implications for the neuropathogenesis of AIDS. J Immunol. 1999;163:1636–1646. [PubMed] [Google Scholar]
- 21.Klein RS, Rubin JB, Gibson HD, DeHaan EN, Alvarez-Hernandez X, Segal RA, Luster AD. SDF-1 alpha induces chemotaxis and enhances Sonic hedgehog-induced proliferation of cerebellar granule cells. Development. 2001;128:1971–1981. doi: 10.1242/dev.128.11.1971. [DOI] [PubMed] [Google Scholar]
- 22.Koch T, Schulz S, Pfeiffer M, Klutzny M, Schroder H, Kahl E, Hollt V. C-terminal splice variants of the mouse mu-opioid receptor differ in morphine-induced internalization and receptor resensitization. J Biol Chem. 2001;276:31408–31414. doi: 10.1074/jbc.M100305200. [DOI] [PubMed] [Google Scholar]
- 23.Kowalska MA, Ratajczak MZ, Majka M, Jin J, Kunapuli S, Brass L, Poncz M. Stromal cell-derived factor-1 and macrophage-derived chemokine: 2 chemokines that activate platelets. Blood. 2000;96:50–57. [PubMed] [Google Scholar]
- 24.Lazarini F, Casanova P, Tham TN, De Clercq E, Arenzana-Seisdedos F, Baleux F, Dubois-Dalcq M. Differential signalling of the chemokine receptor CXCR4 by stromal cell-derived factor 1 and the HIV glycoprotein in rat neurons and astrocytes. Eur J Neurosci. 2000;12:117–125. doi: 10.1046/j.1460-9568.2000.00894.x. [DOI] [PubMed] [Google Scholar]
- 25.Lebel E, Vallieres L, Rivest S. Selective involvement of interleukin-6 in the transcriptional activation of the suppressor of cytokine signaling-3 in the brain during systemic immune challenges. Endocrinology. 2000;141:3749–3763. doi: 10.1210/endo.141.10.7695. [DOI] [PubMed] [Google Scholar]
- 26.Lewis SA, Balcarek JM, Krek V, Shelanski M, Cowan NJ. Sequence of a cDNA clone encoding mouse glial fibrillary acidic protein: structural conservation of intermediate filaments. Proc Natl Acad Sci USA. 1984;81:2743–2746. doi: 10.1073/pnas.81.9.2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Limatola C, Giovannelli A, Maggi L, Ragozzino D, Castellani L, Ciotti MT, Vacca F, Mercanti D, Santoni A, Eusebi F. SDF-1alpha-mediated modulation of synaptic transmission in rat cerebellum. Eur J Neurosci. 2000;12:2497–2504. doi: 10.1046/j.1460-9568.2000.00139.x. [DOI] [PubMed] [Google Scholar]
- 28.Lu Q, Sun EE, Klein RS, Flanagan JG. Ephrin-B reverse signaling is mediated by a novel PDZ-RGS protein and selectively inhibits G protein-coupled chemoattraction. Cell. 2001;105:69–79. doi: 10.1016/s0092-8674(01)00297-5. [DOI] [PubMed] [Google Scholar]
- 29.Ma Q, Jones D, Borghesani PR, Segal RA, Nagasawa T, Kishimoto T, Bronson RT, Springer TA. Impaired B-lymphopoiesis, myelopoiesis, and derailed cerebellar neuron migration in CXCR4- and SDF-1-deficient mice. Proc Natl Acad Sci USA. 1998;95:9448–9453. doi: 10.1073/pnas.95.16.9448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melton DA, Krieg PA, Rebagliati MR, Maniatis T, Zinn K, Green MR. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984;12:7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagasawa T, Kikutani H, Kishimoto T. Molecular cloning and structure of a pre-B-cell growth-stimulating factor. Proc Natl Acad Sci USA. 1994;91:2305–2309. doi: 10.1073/pnas.91.6.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagasawa T, Nakajima T, Tachibana K, Iizasa H, Bleul CC, Yoshie O, Matsushima K, Yoshida N, Springer TA, Kishimoto T. Molecular cloning and characterization of a murine pre-B-cell growth-stimulating factor/stromal cell-derived factor 1 receptor, a murine homolog of the human immunodeficiency virus 1 entry coreceptor fusin. Proc Natl Acad Sci USA. 1996;93:14726–14729. doi: 10.1073/pnas.93.25.14726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nanki T, Hayashida K, El-Gabalawy HS, Suson S, Shi K, Girschick HJ, Yavuz S, Lipsky PE. Stromal cell-derived factor-1-CXC chemokine receptor 4 interactions play a central role in CD4+ T cell accumulation in rheumatoid arthritis synovium. J Immunol. 2000;165:6590–6598. doi: 10.4049/jimmunol.165.11.6590. [DOI] [PubMed] [Google Scholar]
- 34.Oberlin E, Amara A, Bachelerie F, Bessia C, Virelizier JL, Arenzana-Seisdedos F, Schwartz O, Heard JM, Clark-Lewis I, Legler DF, Loetscher M, Baggiolini M, Moser B. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature. 1996;382:833–835. doi: 10.1038/382833a0. [DOI] [PubMed] [Google Scholar]
- 35.Ohtani Y, Minami M, Kawaguchi N, Nishiyori A, Yamamoto J, Takami S, Satoh M. Expression of stromal cell-derived factor-1 and CXCR4 chemokine receptor mRNAs in cultured rat glial and neuronal cells. Neurosci Lett. 1998;249:163–166. doi: 10.1016/s0304-3940(98)00425-x. [DOI] [PubMed] [Google Scholar]
- 36.Pablos JL, Amara A, Bouloc A, Santiago B, Caruz A, Galindo M, Delaunay T, Virelizier JL, Arenzana-Seisdedos F. Stromal-cell derived factor is expressed by dendritic cells and endothelium in human skin. Am J Pathol. 1999;155:1577–1586. doi: 10.1016/S0002-9440(10)65474-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pfeiffer M, Koch T, Schroder H, Klutzny M, Kirscht S, Kreienkamp HJ, Hollt V, Schulz S. Homo- and heterodimerization of somatostatin receptor subtypes. Inactivation of sst(3) receptor function by heterodimerization with sst(2A). J Biol Chem. 2001;276:14027–14036. doi: 10.1074/jbc.M006084200. [DOI] [PubMed] [Google Scholar]
- 38.Quan N, Whiteside M, Kim L, Herkenham M. Induction of inhibitory factor kappaBalpha mRNA in the central nervous system after peripheral lipopolysaccharide administration: an in situ hybridization histochemistry study in the rat. Proc Natl Acad Sci USA. 1997;94:10985–10990. doi: 10.1073/pnas.94.20.10985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rivest S, Lacroix S, Vallieres L, Nadeau S, Zhang J, Laflamme N. How the blood talks to the brain parenchyma and the paraventricular nucleus of the hypothalamus during systemic inflammatory and infectious stimuli. Proc Soc Exp Biol Med. 2000;223:22–38. doi: 10.1046/j.1525-1373.2000.22304.x. [DOI] [PubMed] [Google Scholar]
- 40.Schafer MK, Schwaeble WJ, Post C, Salvati P, Calabresi M, Sim RB, Petry F, Loos M, Weihe E. Complement C1q is dramatically up-regulated in brain microglia in response to transient global cerebral ischemia. J Immunol. 2000;164:5446–5452. doi: 10.4049/jimmunol.164.10.5446. [DOI] [PubMed] [Google Scholar]
- 41.Schreff M, Schulz S, Handel M, Keilhoff G, Braun H, Pereira G, Klutzny M, Schmidt H, Wolf G, Hollt V. Distribution, targeting, and internalization of the sst4 somatostatin receptor in rat brain. J Neurosci. 2000;20:3785–3797. doi: 10.1523/JNEUROSCI.20-10-03785.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schulz S, Schreff M, Schmidt H, Handel M, Przewlocki R, Hollt V. Immunocytochemical localization of somatostatin receptor sst2A in the rat spinal cord and dorsal root ganglia. Eur J Neurosci. 1998;10:3700–3708. doi: 10.1046/j.1460-9568.1998.00386.x. [DOI] [PubMed] [Google Scholar]
- 43.Schwaeble W, Schafer MK, Petry F, Fink T, Knebel D, Weihe E, Loos M. Follicular dendritic cells, interdigitating cells, and cells of the monocyte-macrophage lineage are the C1q-producing sources in the spleen. Identification of specific cell types by in situ hybridization and immunohistochemical analysis. J Immunol. 1995;155:4971–4978. [PubMed] [Google Scholar]
- 44.Shirozu M, Nakano T, Inazawa J, Tashiro K, Tada H, Shinohara T, Honjo T. Structure and chromosomal localization of the human stromal cell-derived factor 1 (SDF1) gene. Genomics. 1995;28:495–500. doi: 10.1006/geno.1995.1180. [DOI] [PubMed] [Google Scholar]
- 45.Stoll G, Jander S, Schroeter M. Inflammation and glial responses in ischemic brain lesions. Prog Neurobiol. 1998;56:149–171. doi: 10.1016/s0301-0082(98)00034-3. [DOI] [PubMed] [Google Scholar]
- 46.Stumm R, Culmsee C, Schafer MK, Krieglstein J, Weihe E. Adaptive plasticity in tachykinin and tachykinin receptor expression after focal cerebral ischemia is differentially linked to GABAergic and glutamatergic cerebrocortical circuits and cerebrovenular endothelium. J Neurosci. 2001;21:798–811. doi: 10.1523/JNEUROSCI.21-03-00798.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tashiro K, Tada H, Heilker R, Shirozu M, Nakano T, Honjo T. Signal sequence trap: a cloning strategy for secreted proteins and type I membrane proteins. Science. 1993;261:600–603. doi: 10.1126/science.8342023. [DOI] [PubMed] [Google Scholar]
- 48.Telford JL, Macchia G, Massone A, Carinci V, Palla E, Melli M. The murine interleukin 1 beta gene: structure and evolution. Nucleic Acids Res. 1986;14:9955–9963. doi: 10.1093/nar/14.24.9955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tham TN, Lazarini F, Franceschini IA, Lachapelle F, Amara A, Dubois-Dalcq M. Developmental pattern of expression of the alpha chemokine stromal cell-derived factor 1 in the rat central nervous system. Eur J Neurosci. 2001;13:845–856. doi: 10.1046/j.0953-816x.2000.01451.x. [DOI] [PubMed] [Google Scholar]
- 50.Thibeault I, Laflamme N, Rivest S. Regulation of the gene encoding the monocyte chemoattractant protein 1 (MCP-1) in the mouse and rat brain in response to circulating LPS and proinflammatory cytokines. J Comp Neurol. 2001;434:461–477. doi: 10.1002/cne.1187. [DOI] [PubMed] [Google Scholar]
- 51.Welsh FA, Sakamoto T, McKee AE, Sims RE. Effect of lactacidosis on pyridine nucleotide stability during ischemia in mouse brain. J Neurochem. 1987;49:846–851. doi: 10.1111/j.1471-4159.1987.tb00971.x. [DOI] [PubMed] [Google Scholar]
- 52.Wong ML, Xin WW, Duman RS. Rat LCR1: cloning and cellular distribution of a putative chemokine receptor in brain. Mol Psychiatry. 1996;1:133–140. [PubMed] [Google Scholar]
- 53.Wood L, Pulaski S, Vogeli G. cDNA clones coding for the complete murine B chain of complement C1q: nucleotide and derived amino acid sequences. Immunol Lett. 1988;17:115–119. doi: 10.1016/0165-2478(88)90078-8. [DOI] [PubMed] [Google Scholar]
- 54.Yamamoto K, de Waard V, Fearns C, Loskutoff DJ. Tissue distribution and regulation of murine von Willebrand factor gene expression in vivo. Blood. 1998;92:2791–2801. [PubMed] [Google Scholar]
- 55.Zou YR, Kottmann AH, Kuroda M, Taniuchi I, Littman DR. Function of the chemokine receptor CXCR4 in haematopoiesis and in cerebellar development. Nature. 1998;393:595–599. doi: 10.1038/31269. [DOI] [PubMed] [Google Scholar]