Abstract
Disruption of the circadian timing system arising from travel between time zones (“jet lag”) and rotational shift work impairs mental and physical performance and severely compromises long-term health. Circadian disruption is more severe during adaptation to advances in local time, because the circadian clock takes much longer to phase advance than delay. The recent identification of mammalian circadian clock genes now makes it possible to examine time zone adjustments from the perspective of molecular events within the suprachiasmatic nucleus (SCN), the principal circadian oscillator. Current models of the clockwork posit interlocked transcriptional/post-translational feedback loops based on the light-sensitive Period (Per) genes and the Cryptochrome (Cry) genes, which are indirectly regulated by light. We show that circadian cycles ofmPer expression in the mouse SCN react rapidly to an advance in the lighting schedule, whereas rhythmic mCry1expression advances more slowly, in parallel to the gradual resetting of the activity–rest cycle. In contrast, during a delay in local time the mPer and mCry cycles react rapidly, completing the 6 hr shift together by the second cycle, in parallel with the activity–rest cycle. These results reveal the potential for dissociation of mPer and mCry expression within the central oscillator during circadian resetting and a differential molecular response of the clock during advance and delay resetting. They highlight the indirect photic regulation ofmCry1 as a potentially rate-limiting factor in behavioral adjustment to time zone transitions.
Keywords: entrainment, period, cryptochrome, circadian, suprachiasmatic nuclei, jet lag
Disruption of circadian timing during jet lag and rotational shift work acutely impairs performance and causes chronic health problems (Siegel et al., 1969; Costa, 1996;Cho, 2001; Rajaratnam and Arendt, 2001; Schernhammer et al., 2001). It arises from conflict between the entraining cycle of light and darkness and the temporal program of the clock (Czeisler and Dijk, 1995) within the hypothalamic suprachiasmatic nuclei (SCN) (Reppert and Weaver, 2001). Usually, discordance is minimal because small phase advances and delays at dawn and dusk synchronize the SCN to solar time (Pittendrigh and Daan, 1976; Aschoff, 1984). Larger shifts to light pulses can occur in experimental subjects “free-running” in continuous dim light. Advances are characterized by gradual resetting, in which the activity–rest cycle takes several days to achieve the steady-state shift, whereas delays are executed rapidly. Resetting to acute time zone transitions also exhibits differential re-entrainment and is limited further by inability of the circadian system to advance by >3 hr/cycle. Consequently, behavioral adaptation to advances in local time and to rotational shift work is slow and progressive, accompanied by temporal disorder of physiology.
Identifying the origin of inertia during resetting has theoretical and practical relevance. Formal analyses indicate, paradoxically, that the phase response curve, an indication of directly light-responsive elements of the oscillator, resets rapidly, within 2 hr (Best et al., 1999). If these formal analyses depict the behavior of the clock accurately, gradual behavioral advances must reflect progressive readjustment of other elements within the circadian timing system. Understanding of circadian readjustment demands, therefore, an analysis of the molecular and cellular events during resetting. Current models propose that the clockwork is a series of interlocked transcriptional/post-translational feedback loops (Dunlap, 1999;Reppert and Weaver, 2001). Alternating activation by BMAL1/CLOCK heterodimers and negative feedback by PER/CRY complexes drives circadian expression of the Period (Per) andCryptochrome (Cry) genes (Shearman et al., 2000). The PER2 protein is pivotal, initiating the subsequent circadian cycle of Bmal1 gene expression, whereas mPer3 is not essential to the clockwork (Bae et al., 2001; Reppert and Weaver, 2001). mCry1 appears to be more important thanmCry2. mCry1 exhibits a more pronounced circadian cycle in the SCN, and a single wild-type mCry1 allele sustains circadian function in an mCry2-deficient background, whereas the reciprocal genotype is arrhythmic (van der Horst et al., 1999).
This model has important implications for understanding adjustment to time zone transitions. Whereas the Per1 and Per2are acutely upregulated by nocturnal light (Shigeyoshi et al., 1997), the Cry genes are not (Okamura et al., 1999; Field et al., 2000). Photic synchronization of Cry/CRY expression is mediated indirectly. Our working hypothesis was that inertia in re-entrainment of circadian behavior arises from the indirect, progressive response to light of SCN Cry. We monitoredmPer and mCry expression in the SCN of mice undergoing acute advance or delays in local time. We show differential molecular responses to advance and delay shifts and highlight the expression of mCry1 in the SCN as a potential rate-limiting factor in behavioral readjustment to time zone transitions.
MATERIALS AND METHODS
All experiments were licensed by the Home Office (United Kingdom). Adult male CD1(ICR) mice (6 weeks; Harlan Olac, Bicester, UK) were housed individually or in groups, with food and water availablead libitum, in light-proof, ventilated chambers, under a 12 hr bright white light (L) (220 μW/cm2), 12 hr dim red light (DR) (<5 μW/cm2) cycle. Activity–rest cycles were monitored with passive infrared movement detectors or running wheels linked to an automated recording system (Dataquest IV; DSI, Frankfurt, Germany). Lights-off was defined as zeitgeber time (ZT) 12. Circadian time (CT) 12 indicated activity onset under continuous DR.
Experiment 1. Groups of animals were killed by cervical dislocation hourly on the baseline day. Others were subjected to an acute advance in the lighting schedule (advanced lights-on) and were killed hourly for 18 hr intervals, commencing at the time of new lights-on, over the first, third, and eighth days after the shift.
Experiment 2. Mice were subjected to a 6 hr delay in the time of lights-on and lights-off and sampled in groups every hour for 44 hr. In both experiments wheel-running was recorded in five additional mice to monitor behavioral re-entrainment.
Frozen brains were processed for in situ hybridization as described previously (Field et al., 2000). Tissues from each sampling day were processed together. The intensity of the hybridization signal in the whole SCN was assessed against a14C calibration curve, using NIH Image software. To compare between days, signal intensity at each time was expressed as a percentage of the daily maximum. At selected times additional brains were obtained for immunocytochemistry by transcardial perfusion fixation with 0.01 m PBS and 4% paraformaldehyde under terminal anesthesia (Kume et al., 1999; Field et al., 2000). NIH Image was used to count the number of immunoreactive nuclei above a thresholded background in representative sections. Differences were assessed by ANOVA and Dunnett's ttests.
RESULTS
Advance resetting is accompanied by dissociation of SCNmPer and mCry expression
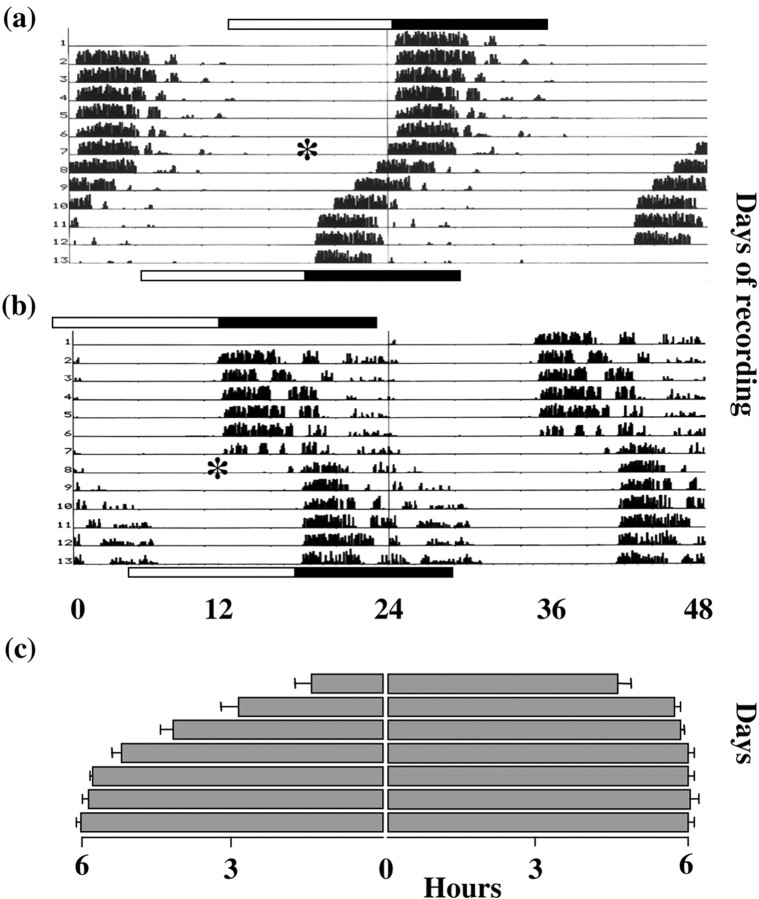
Mice entrained to a 12 hr light/dark cycle (12L:12D) became active at lights-off (Fig. 1a,c). After a 6 hr advance in the photoschedule, the activity–rest cycle exhibited progressive re-entrainment. After 3 d, activity onset had advanced by 4.17 ± 0.25 SEM hr. Stable re-entrainment was only achieved after 1 week.
Fig. 1.
Re-entrainment of mouse activity–rest cycles to shifts of the photoschedule. a, b, Representative double-plotted actograms of mice subjected to 6 hr phase advance or phase delay on days marked by asterisk.Top bars indicate initial L:D cycle; bottom bars below indicate shifted cycle. c, Activity onset (mean + SEM; n = 5) after the 6 hr advance (left panel) or delay (right panel).
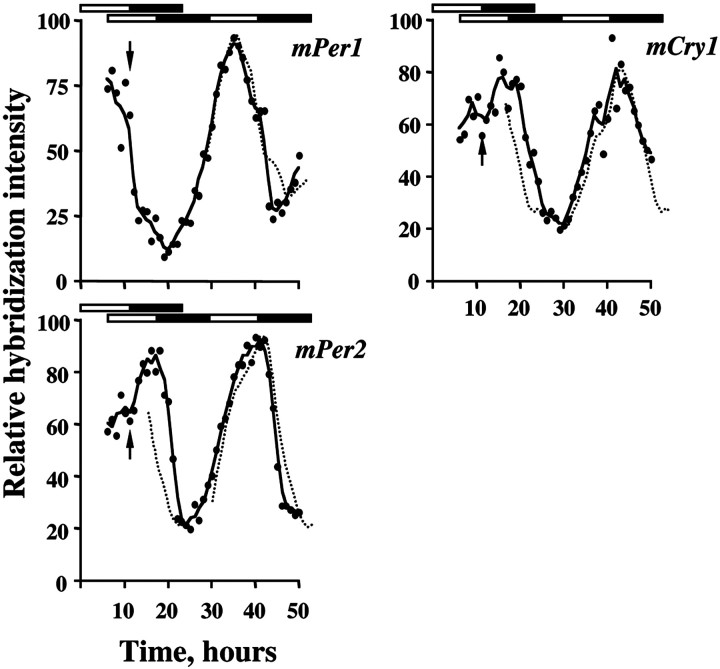
Clock gene expression within the SCN of mice entrained to 12L:12D was equivalent to that reported previously. mPer1 mRNA peaked around ZT06, and mPer2 and mCry1 mRNA around lights-off (ZT12) (Fig. 2a). On day 1 of the advanced schedule, light onset occurred in the middle of the former night, original ZT18. mPer1 mRNA was strongly induced, especially in the ventral SCN (Figs. 2a,3a) (ANOVA over first 6 hr of day 1, treatment and time effects, p < 0.01; interaction p < 0.01). Despite the continuing illumination, however, mPer1 expression returned to control levels within 2–3 hr. For the remainder of day 1, the endogenous cycle of mPer1 expression was not significantly different from that of the control cycle. Acute induction of mPer2 mRNA also occurred on day 1, peaking in the first 4–5 hr of illumination (Figs. 2a, 3a), followed by a decline to control levels (ANOVA treatment effect, and treatment × time interaction,p < 0.01). The spontaneous cycle of mPer2expression for the remainder of day 1 was not significantly different from the control cycle. In contrast to mPer,mCry1 in the SCN was not acutely affected by the advanced lighting (Figs. 2a, 3a). Although there was a trend for mCry1 expression to be advanced slightly, this was not significant.
Fig. 2.
Re-entrainment of SCN gene expression cycles to an advanced lighting schedule. a, Intensity of mRNA hybridization signals for mPer1, mPer2,or mCry1 in SCN before (baseline) and on the first day of the advance phase shift. Baseline data are plotted as observed hourly values (closed circles, mean of 3 mice/hr), and as the three-point moving average (solid line, double-plotted on right as dotted linefor clarity). Data from day 1 of the shift are single-plotted on the right (open circles, solid line), represented by the observed hourly mean (3 mice/hr, for the first 6 hr of light exposure) or as the three point moving average for the hourly means (3 mice/hr) for the subsequent 12 hr. b, Intensity of SCN mRNA hybridization signals before (baseline data, dotted line) and on the third (closed circles, dashed line) and eighth (open circles, solid line) days of the advance phase shift. Plotted as the three point moving average for the hourly means (3 mice/hr).
Fig. 3.
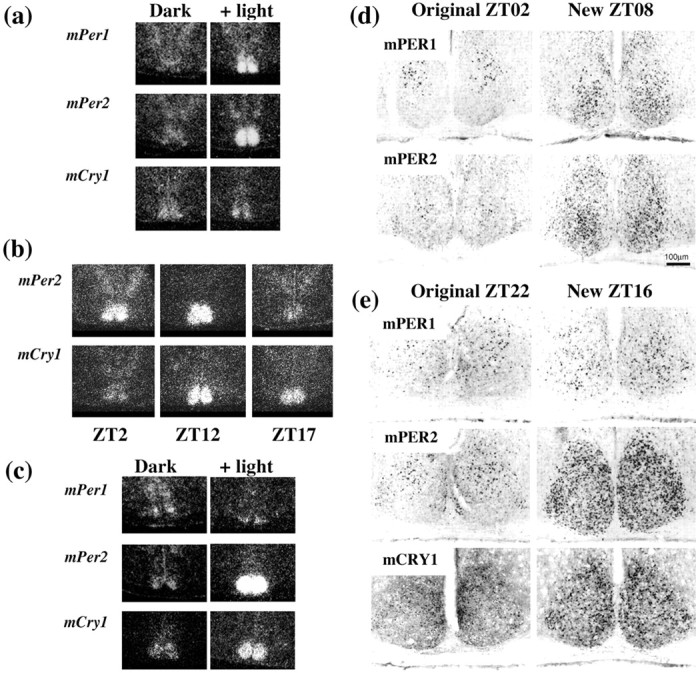
Induction of circadian clock genes in SCN of phase-shifted mice. Representative dark-field autoradiograms ofin situ hybridization. A,mPer and mCry1 expression in mice entrained to 12L:12D and sampled in darkness 1(mPer1) or 3 hr (mPer2, mCry1) after lights-off, or after acute 6 hr phase advance of the L:D cycle. b,mPer2 and mCry1 expression 3 d after a 6 hr advance, sampled 2, 12, or 17 hr after lights-on. Note dissociation of Per and Cry expression. Images are derived from the same animal at each time. c,mPer and mCry1 expression in mice subjected to an acute 6 hr delay of the light/dark cycle. Mice were sampled in darkness at ZT18 or after 6 hr of light exposure between former ZT12 to ZT18. Note coordinate upregulation of bothmPer2 and mCry1 mRNA. Induction tomPer1 was weak and restricted to the ventral SCN. Images for the three mRNAs are derived from the same animal at either time. Representative coronal sections immunostained for mPER or mCRY.d, Mice entrained to 12L:12D, sampled at ZT02, or subjected to an acute 6 hr phase advance to lights-on (now on at original ZT18) and sampled after 8 hr of illumination (new ZT08).e, Mice entrained to 12L:12D, sampled at ZT22, or subjected to an acute 6 hr phase delay (lights now off at original ZT18) and sampled 4 hr after the end of illumination (original ZT22, new ZT16).
Dissociation of mPer and mCry1 expression was again apparent on the third day of the advance. Expression of bothmPer genes filled the entire light phase, in contrast to the limited induction on day 1 (Fig. 2b). Consequently, their rhythmic expression on day 3 was not significantly different from day 8. In contrast, the peak of mCry1 expression on day 3 was in an intermediate position between the baseline day and day 8 (Fig.2b). Moreover, the profile of mCry1 mRNA was distorted insofar as levels remained high for several hours after the peak of expression. There was no clear fall in mCry1 mRNA levels after lights-off, in contrast to that seen in entrained mice. This dissociation of gene cycles, with oppositely phased peaks and troughs, is apparent in Figure 3b. Two hours after lights-on, mPer2 levels were high, whereas mCry1expression was basal in the same SCN. Both genes were highly expressed after 12 hr of illumination. Five hours after lights-off, however,mPer2 levels were basal, whereas mCry1 mRNA remained high. By day 8 the rhythm of mCry1 expression had re-entrained to the advanced lighting cycle, consistent with the completed behavioral response, and was again synchronized to themPer2 cycle. Three-way ANOVA revealed a highly significant interaction between gene, time of day, and day of treatment, confirming that these two genes exhibited differential responses to time zone advances. A similar, highly significant interaction was also observed for the comparison between mPer1 andmCry1 (data not shown), demonstrating that the cycles of expression of these gene products were also dissociated during advance resetting.
The contrast between the rapid response of mPer genes and the gradual advance of mCry1 to the shifted photoschedule is consistent with the model in which mCry is reset through light-dependent changes in mPER2 and thence of mBMAL1. We examined mPER expression in the SCN of control mice at ZT02 or mice sampled at an equivalent time, 8 hr after the advanced light onset on day 1 (n = 3 both groups). Nuclear mPER1 and mPER2 expression in control, nonshifted animals exhibited the very restricted pattern of immunoreactive cells in the dorsal SCN, typical of this phase (Fig.3d) (mPER1 108.6 ± 7.0, mPER2 160.3 ± 25.8, cells per SCN section, mean + SEM). In contrast, mice subjected to the advance expressed significantly higher mPER1-IR (321.7 ± 32.5) and mPER2-IR (618.7 ± 74.5), predominantly in the ventral SCN, confirming translation of the acutely induced mRNAs.
Adjustment to a delay in local time is accompanied by coordinated SCN expression of mCry1 and mPer2
Our working hypothesis predicted that coordinate and rapid readjustment of the Per and Cry genes would accompany rapid behavioral resetting during phase delays. We therefore examined SCN clock gene expression in mice subjected to a 6 hr delay of the photo schedule. The resulting delay in the rhythm of wheel-running was completed within 2 cycles, with little evidence of gradual resetting (Fig. 1b,c).
The delayed photo schedule had little immediate impact on the expression of mPer1 (Figs. 3c,4a). Levels were already falling because the delayed lighting illuminated the original dark phase (ZT12–ZT18). There was limited induction of mRNA in the ventralmost SCN, but less marked than accompanied the advance. This was mirrored by only sporadic mPER1 protein in the ventral SCN 4 hr after the delayed illumination ended (Fig. 3e). Nevertheless, on the following cycle, the profile of mPer1 expression was not significantly different from the projected 6 hr delay of the baseline cycle. The phase delay was complete by the second night (Fig. 4).
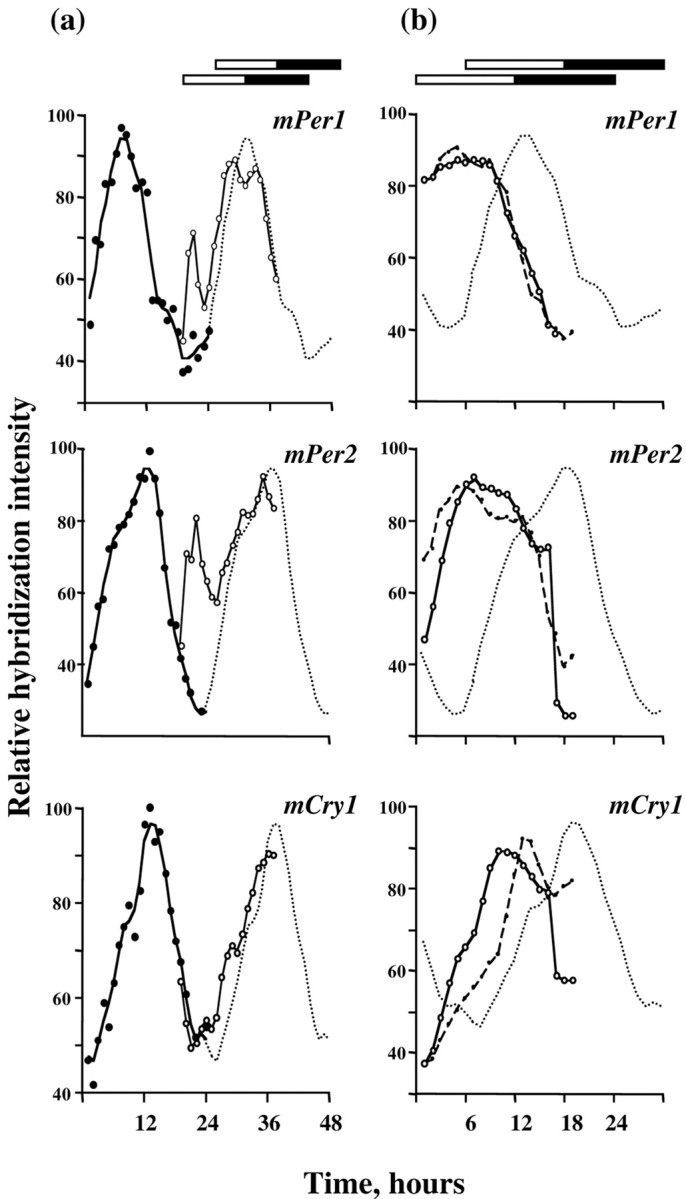
Fig. 4.
Rapid readjustment of rhythmic expression ofmPer and mCry mRNAs in SCN of mice subjected to an acute 6 hr phase delay. Data plotted as hourly means (circles; 3 mice/hr) and as a three-point moving average for the hourly data (solid line). For comparison, the baseline data are plotted in their original phase (dotted line, left panel) and with a predicted 6 hr delay on the second cycle after the shift (dotted line, right panel). Bars depict original (top bar) and shifted photoschedules (bottom bars).Arrow indicates time of exposure to unanticipated nocturnal light.
In contrast to mPer1, the peak of mPer2expression in the SCN was dramatically extended during illumination of the original ZT12 and ZT18 (Figs. 3c, 4). Levels then fell after lights-off. On the second day and night after the shift, the 6 hr phase delay to the expression of mPer2 had been completed. Exposure to a delayed photoschedule also upregulated the expression ofmCry1 in the SCN. On the first cycle of the delay there was a pronounced extension to the mCry1 peak, and levels did not fall until after the new time of lights-off (Figs. 3c, 4). This was mirrored by an extension in mCRY1 expression. In control mice sampled at original ZT22, sporadic mCRY1-IR nuclei were located in the dorsal SCN (98 ± 12 cells per section; n = 3). During the 6 hr delay, mCRY1-IR nuclei were abundant across the SCN, including the ventral retinorecipient regions (389 ± 62;n = 3; p < 0.01 vs controls) (Fig.3e). As for mPers, the rhythm of mCry1on the second cycle after the shift matched that predicted by an acute 6 hr delay of the baseline profile.
The mechanism underlying mCry1 induction is not clear, although the simultaneous and prolonged rise in mPer2 mRNA levels was translated into an extended expression of mPER2 protein. In control mice at ZT22, mPER2-IR nuclei were infrequent and predominantly in the dorsal SCN (mean ± SEM; 68 ± 7 cells per section;n = 3). In mice subjected to the delay, mPER2-IR nuclei were abundant across the SCN (509 ± 25; n = 3;p < 0.01 vs controls) (Fig. 3e).
These results demonstrate that acute phase delays of the circadian system are associated with coordinate shifts of both components of the oscillator, mPer and mCry. This rapidmCry response may underlie the rapid adjustment of behavioral rhythms to delays in local time. It contrasts markedly with gradual advance resetting characterized by dissociation of rapidly adjusting mPer and slowly adjusting mCry1.
DISCUSSION
During phase advances of the SCN clockwork the mPer andmCry genes dissociate as the mPers react rapidly to the new lighting regime, whereas the mCry1 cycle lags behind, advancing in phase with the activity–rest rhythm. Conversely, rapid behavioral responses during delay resetting are accompanied by coordinate, rapid induction of both mPer and mCryexpression in the SCN. The degree of molecular dissociation in the SCN accompanying time zone transitions is therefore a function of the direction of resetting and arises from the differential sensitivity of core circadian genes to light.
The differential speed of advance and delay resetting corresponds directly to previous reports in animals exposed either to an acute shift in the complete lighting cycle (Yamazaki et al., 2000) or to single pulses of light presented during circadian night (Johnson, 1990). Whereas the presence of a complete lighting cycle has the potential to mask gradual resetting of activity onset during delays, it is clear that activity offset, an unmasked index of behavior, also adjusts rapidly. This is equally evident where delays are caused by single or multiple light pulses delivered under free-running conditions (Best et al., 1999).
The rapid readjustment of mPer1 to advanced lighting is consistent with an earlier report in C57BL/6 mice sampled every 6 hr (Sun et al., 1997) and with studies using rats bearing arPer1:: Luc transgene to plot rPer1expression in SCN explants dissected after an advance shift in vivo (Yamazaki et al., 2000). These rapid responses, in the case of the SCN slice observed free-running in vitro, raised the question of where inertia in circadian resetting might arise. Clearly, it is not in SCN Per1 expression. In transgenic ratsrPer1:: Luc expression in peripheral tissues readjusts more slowly, indicating that inertia lies in the periphery not in the SCN. The current study shows, however, that the dynamic responses of SCN mPer expression are not necessarily shared by mCry1. Expression of mPer in the SCN is determined by both circadian time and by the presence of light. This was most apparent on day 1 of the shift with two peaks of expression, one light-induced, the second circadian. By day 3 of the shift the biphasic pattern to mPer expression was lost as the circadian peak had advanced to fuse with the light induction, andmPer levels fell immediately with dark onset, indicating that the circadian advance was completed. In contrast, mCry1levels peaked later and remained high into the dark phase.
We therefore propose an alternative, although not necessarily contradictory, model in which inertia during time zone adjustment occurs within the SCN, and that it arises from the slow reaction of themCry1 gene, a consequence of its indirect photic regulation. This model does not, of course, preclude further elaboration, and the rate of resetting may also be limited by intercommunication between SCN neurons, especially light-responsive and nonresponsive populations. Moreover, lags in signaling between the SCN and dependent peripheral tissues via slow neuroendocrine pathways will also contribute to the observed slow resetting of peripheral tissues (Yamazaki et al., 2000).
Our model is supported by the temporal correlation between behavioral resetting and readjustment of the SCN mCry1 cycle. During both gradual advances and immediate delays, the shift of themCry1 loop matched that for the overt activity–rest rhythm, suggesting that the response of mCry1 in the SCN may be sufficient to explain gradual resetting of locomotor behavior. Although not tested in the current study, it is strongly predicted that progressive advances of mCry, dissociated frommPer expression, would also accompany gradual advance resetting arising from single light pulses presented under free-running conditions. mCRY proteins are the principal mediators of transcriptional suppression within the clockwork and also on clock-controlled genes (Kume et al., 1999). The simplest interpretation of our data is that mCRY proteins gate the molecular and cellular events that determine the circadian timing of activity onset. Mice devoid of functional CRY proteins are unable to express organized circadian activity onsets. Equally, it is clear that light-induced expression of mPer genes is insufficient to determine circadian activity onset.
Photic regulation of mCry is presumably a consequence of altered expression of mPER2, although the relative contribution ofmPer1 and mPer2 to resetting is not known. Antisense oligonucleotides to mPer1 can block resetting (Akiyama et al., 1999), suggesting a necessary role for mPER1. However, mice lacking mPER1 can entrain to a light/dark cycle (Bae et al., 2001;Zheng et al., 2001), insofar as they free-run with a coherent circadian pattern phased to the previous dark interval. Mice with mutations of the mPer2 gene also appear capable of entrainment. However, such mice have a compromised circadian activity–rest pattern when released into continuous darkness, suggesting that the rhythm observed under a light/dark cycle arises from the direct masking effect of light, as it does in mCry knock-out mice (van der Horst et al., 1999). In our delay study, resetting occurred with very littlemPer1 mRNA or protein induction in the SCN, indicating that mPER1 may not be necessary for resetting. For both advance and delay, however, there was a marked induction of mPer2 mRNA and mPER2-IR. Taken with the data on mutant mice and the model proposed byShearman et al. (2000), these findings point to changes ofmPer2/mPER2 as key regulatory points for circadian entrainment.
Why does mCry1 shift rapidly during delays but not during advances? Acute light pulses do not affect mCry expression, although sustained exposure to light clearly can. One obvious difference is that advances follow exposure to light during late subjective night, when mCry1 expression is spontaneously low. Delay resetting with “dusk” light occurs when mCry1levels are high. The molecular substrate for mCry1expression may be easier to sustain once spontaneously active, rather than to activate de novo in a cellular environment that will not support spontaneous (i.e., circadian) mCry1 expression. Further clarification of the protein:DNA interactions drivingmCry1 expression is necessary to address this, although mPER2 is likely to be an important contributor. In delay resetting, mPER2 protein expression is spontaneously high and sustained by exposure to light, and photic activation of mCry1 andmPer2 occurred over the same 6 hr of light exposure (this study), raising the possibility that mPER2 has an effect onmCry1 expression independently of its proposed regulation of mBMAL1. With the advance shifts, mPER2 protein levels are initially low and may take some time to increase in response to light, thereby compromising induction of mCry1.
In conclusion, our results identify mPer genes, in particular mPer2, as critical components of the photic entrainment pathway, whereas mCry1 appears to be a rate-limiting factor in the expression of behavioral resetting. As such, this gene may be an important therapeutic target for management of circadian disorders such as sleep disturbance, jet lag, and shift work.
A.B.R. and M.D.F. contributed equally to this work.
We thank J. Bashford for expert assistance with photomicrography, Profs. H. Okamura (Kobe University, Japan) and S. M. Reppert, (University of Massachusetts, Boston, MA) for plasmid templates, and W. Rashband (National Institutes of Health, Bethesda, MD) for Image software.
Correspondence should be addressed to Michael H. Hastings, Medical Research Council Laboratory of Molecular Biology, Hills Road, Cambridge, CB2 2QH, UK. E-mail: mh105@cus.cam.ac.uk.
REFERENCES
- 1.Akiyama M, Kouzu Y, Takahashi S, Wakamatsu H, Moriya T, Maetani M, Watanabe S, Tei H, Sakaki Y, Shibata S. Inhibition of light- or glutamate-induced mPer1 expression represses the phase shifts into the mouse circadian locomotor and suprachiasmatic firing rhythms. J Neurosci. 1999;19:115–1121. doi: 10.1523/JNEUROSCI.19-03-01115.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aschoff J. Circadian timing. Ann NY Acad Sci. 1984;423:442–468. doi: 10.1111/j.1749-6632.1984.tb23452.x. [DOI] [PubMed] [Google Scholar]
- 3.Bae K, Jin X, Maywood ES, Hastings MH, Reppert SM, Weaver DR. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron. 2001;30:525–536. doi: 10.1016/s0896-6273(01)00302-6. [DOI] [PubMed] [Google Scholar]
- 4.Best JD, Maywood ES, Smith KL, Hastings MH. Rapid resetting of the mammalian circadian clock. J Neurosci. 1999;19:828–835. doi: 10.1523/JNEUROSCI.19-02-00828.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho K. Chronic “jet lag” produces temporal lobe atrophy and spatial cognitive deficits. Nat Neurosci. 2001;4:567–568. doi: 10.1038/88384. [DOI] [PubMed] [Google Scholar]
- 6.Costa G. The impact of shift and night work on health. Appl Ergonomics. 1996;27:9–16. doi: 10.1016/0003-6870(95)00047-x. [DOI] [PubMed] [Google Scholar]
- 7.Czeisler CA, Dijk DJ. Use of bright light to treat maladaptation to night shiftwork and circadian rhythm sleep disorders. J Sleep Res. 1995;4:70–73. doi: 10.1111/j.1365-2869.1995.tb00231.x. [DOI] [PubMed] [Google Scholar]
- 8.Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 9.Field MD, Maywood ES, O'Brien JA, Weaver DR, Reppert SM, Hastings MH. Analysis of clock proteins in mouse SCN demonstrates phylogenetic divergence of the circadian clockwork and resetting mechanisms. Neuron. 2000;25:437–447. doi: 10.1016/s0896-6273(00)80906-x. [DOI] [PubMed] [Google Scholar]
- 10.Johnson CH. An atlas of phase response curves for circadian and circatidal rhythms. Vanderbilt UP; Nashville, TN: 1990. [Google Scholar]
- 11.Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 12.Okamura H, Miyake S, Sumi Y, Yamaguchi S, Yasui A, Muijtjens M, Hoeijmakers JH, van der Horst GT. Photic induction of mPer1 and mPer2 in cry-deficient mice lacking a biological clock. Science. 1999;286:2531–2534. doi: 10.1126/science.286.5449.2531. [DOI] [PubMed] [Google Scholar]
- 13.Pittendrigh CS, Daan S. A functional analysis of circadian pacemakers in nocturnal rodents. J Comp Physiol [A] 1976;106:333–355. [Google Scholar]
- 14.Rajaratnam SMW, Arendt J. Health in a 24-h society. Lancet. 2001;358:999–1005. doi: 10.1016/S0140-6736(01)06108-6. [DOI] [PubMed] [Google Scholar]
- 15.Reppert SM, Weaver DR. Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol. 2001;63:647–676. doi: 10.1146/annurev.physiol.63.1.647. [DOI] [PubMed] [Google Scholar]
- 16.Schernhammer ES, Laden F, Speizer FE, Willett WC, Hunter DJ, Kawachi I, Colditz GA. Rotating night shifts and risk of breast cancer in women participating in the Nurses' Health Study. J Natl Cancer Inst. 2001;93:1563–1568. doi: 10.1093/jnci/93.20.1563. [DOI] [PubMed] [Google Scholar]
- 17.Shearman LP, Sriram S, Weaver DR, Maywood ES, Chaves I, Zheng B, Kume K, Lee CC, van der Horst GT, Hastings MH, Reppert SM. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288:1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- 18.Shigeyoshi Y, Taguchi K, Yamamoto S, Takekida S, Yan L, Tei H, Moriya T, Shibata S, Loros JJ, Dunlap JC, Okamura H. Light-induced resetting of a mammalian circadian clock is associated with rapid induction of the mPer1 transcript. Cell. 1997;91:1043–1053. doi: 10.1016/s0092-8674(00)80494-8. [DOI] [PubMed] [Google Scholar]
- 19.Siegel PV, Gerathwohl SJ, Mohler SR. Time zone effects. Science. 1969;167:47–50. doi: 10.1126/science.164.3885.1249. [DOI] [PubMed] [Google Scholar]
- 20.Sun ZS, Albrecht U, Zhuchenko O, Bailey J, Eichele G, Lee CC. RIGUI, a putative mammalian ortholog of the Drosophila period gene. Cell. 1997;90:1003–1011. doi: 10.1016/s0092-8674(00)80366-9. [DOI] [PubMed] [Google Scholar]
- 21.van der Horst G, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, de Wit J, Verkerk A, Eker AP, van Leenen D, Buijs R, Bootsma D, Hoeijmakers JH, Yasui A. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 22.Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, Block GD, Sakaki Y, Menaker M, Tei H. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288:682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- 23.Zheng B, Albrecht U, Kaasik K, Sage M, Lu W, Vaishnav S, Li Q, Sun ZS, Eichele G, Bradley A, Lee CC. Non-redundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell. 2001;105:683–694. doi: 10.1016/s0092-8674(01)00380-4. [DOI] [PubMed] [Google Scholar]