Abstract
Ethanol has complex but similar effects on behavior in mammals and the fruit fly Drosophila melanogaster. In addition, genetic and pharmacological approaches have implicated the cAMP pathway in the regulation of ethanol-induced behaviors in both flies and rodents. Here we examine the neuroanatomical loci that modulate ethanol sensitivity in Drosophila by targeting the expression of an inhibitor of cAMP-dependent protein kinase (PKA) to specific regions in the fly's brain. Expression of the inhibitor in most brain regions or in muscle has no effect on behavior. In contrast, inhibition of PKA in a relatively small number of cells, possibly neurosecretory cells, in the fly's brain is sufficient to decrease sensitivity to the incoordinating effects of ethanol. Additional brain areas are, however, also involved. The mushroom bodies, brain structures where cAMP signaling is required for olfactory classical conditioning, are dispensable for the regulation of ethanol sensitivity. Finally, different behavioral effects of ethanol, motor incoordination and sedation, appear to be regulated by PKA function in distinct brain regions. We conclude that the regulation of ethanol-induced behaviors by PKA involves complex interactions among groups of cells that mediate either increased or reduced sensitivity to the acute intoxicating effects of ethanol.
Keywords: Drosophila, ethanol, PKA, neuroanatomy, mushroom bodies, postural control, locomotion
Alcohol is among the most widely abused drugs in the world, yet the mechanisms by which it acts are only partially understood. Both rodents and the fruit fly Drosophila melanogaster show behavioral responses to acute ethanol exposure that are remarkably similar to human behaviors. Increasing doses of ethanol elicit hyperactivity, then ataxia or incoordination, and finally sedation (Singh and Heberlein, 2000; Parr et al., 2001). Importantly, some of the mechanisms that regulate these behavioral responses also appear to be conserved. For example, genetic and pharmacological manipulations that disrupt dopaminergic systems reduce ethanol-induced locomotor activation in both rodents (Phillips and Shen, 1996) and flies (Bainton et al., 2000).
Although ethanol does not act through a specific receptor, it affects the function of certain cell surface proteins, including several ion channels (Harris, 1999). In addition, some intracellular signaling pathways, such as the cAMP pathway, are also affected by ethanol (Diamond and Gordon, 1997; Tabakoff and Hoffman, 1998). A genetic screen for Drosophila mutants with increased ethanol sensitivity identified amnesiac (Moore et al., 1998), a gene encoding a putative neuropeptide believed to activate the cAMP pathway (Feany and Quinn, 1995). Consistent with this, flies with mutations in the calcium/calmodulin-sensitive adenylyl cyclase rutabagashow increased ethanol sensitivity. In contrast, a mutation in thepka-RII gene, encoding a regulatory subunit of cAMP-dependent protein kinase (PKA), causes decreased ethanol sensitivity (Park et al., 2000). Genetic manipulations of the cAMP pathway in mice have been shown recently to alter ethanol sensitivity as well (Thiele et al., 2000; Wand et al., 2001).
A complete understanding of the mechanisms by which ethanol alters behavior requires knowledge of not only the molecules, but also the neuronal circuits that mediate these effects. In Drosophila, specific groups of neurons can be manipulated using the GAL4/UAS binary expression system (Brand and Perrimon, 1993). This targeted expression approach has been used to map neuroanatomical loci underlying behaviors such as learning and memory (Connolly et al., 1996; Zars et al., 2000a,b), courtship behavior (Ferveur et al., 1995; O'Dell et al., 1995; Joiner and Griffith, 1999), and locomotion (Martin et al., 1999;Gatti et al., 2000). In addition, chemical ablation of the mushroom bodies (MBs), prominent central brain structures, has demonstrated their importance in olfactory and courtship conditioning (de Belle and Heisenberg, 1994; McBride et al., 1999).
We used the GAL4/UAS system to drive expression of a transgene that inhibits PKA activity in restricted brain regions and measured the flies' sensitivity to ethanol. We find that inhibition of PKA in discrete brain regions alters the flies' sensitivity to the acute intoxicating effects of ethanol. We postulate that normal ethanol responsiveness is achieved by a complex balance between loci that increase and reduce the flies' sensitivity to the incoordinating effects of ethanol. In addition, different brain regions seem to regulate distinct aspects of intoxication, such as postural control and sedation. Chemical ablation of the mushroom bodies had no effect, suggesting that the mechanisms that regulate ethanol sensitivity and olfactory conditioning, although sharing molecular components, rely on separable neural structures for their manifestation.
MATERIALS AND METHODS
Drosophila stocks and genetics. P[GAL4] lines 201Y (chromosome II), c522 (III), c107 (I), c747 (II), and c290 (II) as well as additional P[GAL4] lines were obtained from K. Kaiser (University of Glasgow, Scotland, UK) (Yang et al., 1995; Manseau et al., 1997). 3A4 and P[GAL4]MHC82 (III) were obtained from G. Davis (University of California, San Francisco, CA); MHC82 contains the myosin heavy chain promoter fused to GAL4 (Davis et al., 1998). Other P[GAL4] lines were obtained from L. Griffith (Brandeis University, Waltham, MA; MJ lines; see below), C. O'Kane (University of Cambridge, Cambridge, UK; OK lines; see below), and K. Ito (National Institute for Basic Biology, Okazaki, Japan). hsGAL4 flies were obtained from the Bloomington Stock Center at Indiana University (Bloomington, IN). UAS-PKAinh(III) (also called BDK33) flies were obtained from D. Kalderon (Columbia University, New York, NY). These flies carry a transgene coding for the Drosophila type I regulatory subunit of PKA with mutated cAMP-binding sites: Gly-196 and -321 were replaced by Glu and Asp, respectively (Li et al., 1995; D. Kalderon, personal communication). The UAS-PKAm-inhtransgene (II) (Kiger and O'Shea, 2001) contains, in addition to the mutations carried by UAS-PKAinh, mutations in Arg-91 and -92, which were replaced with Gly to abolish binding to the PKA catalytic subunit. The UAS-PKAc transgene (II) (Kiger et al., 1999) encodes a FLAG-tagged Drosophila PKA catalytic subunit. All lines are homozygous viable but were used as heterozygotes or hemizygotes in behavioral assays. Lines used in behavioral experiments (with the exception of P[GAL4]MHC82) were outcrossed for five generations to a w1118 stock isogenic for chromosomes II and III. Flies were raised on standard cornmeal and molasses food at 25°C and 70% relative humidity in constant light.
For behavioral testing, flies carrying both the P[GAL4] and UAS insertions were generated by crossing P[GAL4] virgin females to UAS males. As controls, P[GAL4] or UAS heterozygotes or hemizygotes were generated by crossing males to virgin females carrying attached X chromosomes in the w1118 genetic background. All experiments were performed with 2- to 4-d-old males, ∼110 for the inebriometer and 20 for the locomotor tracking system. All genotypes were tested on multiple days.
P[GAL4] screen. Fifty-nine P[GAL4] lines with diverse expression patterns in the brain were initially screened by crossing to UAS-PKAinh. Of these, 27 were not tested behaviorally because of lethality, low viability, or other defects, such as unexpanded wings. The remaining 32 lines were tested in the inebriometer in the presence of UAS-PKAinh. Lines with increased mean elution time (MET) in the presence of UAS-PKAinh as well as a few control lines were also tested with UAS-PKAm-inh. An additional 34 P[GAL4] lines driving UAS-PKAinh or UAS-PKAm-inh were screened. These were chosen on the basis of expression in specific structures, such as the fan-shaped body, ellipsoid body, mushroom bodies, pars intercerebralis (PI), or other. Expression patterns listed in Table 1 derive from the following published information: All c and Y lines, www.flytrap.org; c161, 78Y, 7Y, 64Y, c561, c105, c481, c346, c819, and c232, Renn et al. (1999); 78Y and 7Y, Martin et al. (1999); c309, c747, and OK348,Connolly et al. (1996); MJ126a and MJ162a, Joiner and Griffith (1999); MZ423, Ito et al. (1997); c302 and c739, Yang et al. (1995); MHC82,Davis et al. (1998); 17D, Martin et al. (1998); and Okt30, Feb204, Feb170, Kurs6, Jan129, Mai301, Jan229, Kurs21, Mai179, Kurs58, and Sep54, Siegmund and Korge (2001).
Table 1.
List of GAL4 lines used to drive expression of PKAinh
| Major site of expression | P[GAL4] lines |
|---|---|
| Lines showing normal behavior when driving PKAinh | |
| Mushroom bodies | c309, c747, c772, c739, c758,1-a MJ162a, c7651-a |
| Ellipsoid body | c346,1-a64Y,1-a c747, c772, c739, c758,1-a c767, c41,1-ac119,1-a c491, c7651-a |
| Fan-shaped body | c739, c453,1-a 104Y, OK348,1-ac687,1-ac7651-a |
| Central complex small field | 7Y, 78Y |
| Dorsal giant interneurons | c346,1-a MZ423 |
| PI neurons/other ring gland projecting neurons | c309, c747, c772, c767, c229,1-aOkt30,1-a,1-bFeb204,1-a,1-bKurs58,1-a,1-bJan129,1-a,1-bMai301,1-a,1-bJan229,1-a,1-bKurs21,1-a,1-b Mai179,1-a,1-bKurs61-a,1-b |
| Antennal lobes | c747, c772, c739, c758,1-a,1-bMJ162a |
| Unknown CNS expression | OK086, OK028,1-aOK062,1-aOK144D,1-a OK144L, OK165,1-a OK168, OK175,1-aOK309,1-a 3A4, c123a |
| Muscle | MHC82 |
| No CNS expression | c290 |
| Lines showing resistance when driving PKAinh and/or PKAm-inh | |
| Mushroom bodies | 121Y, 17D, c302, c584 |
| Ellipsoid body | c819, c81,1-ac105,1-ac161,1-ac217,1-ac338,1-a c401a, c481,1-a c628, MJ126a,1-a c302, c232, c561, c584 |
| Fan-shaped body | c159b,1-ac259,1-a 121Y, c584, c61 |
| PI neurons/other ring gland projecting neurons | MJ126a,1-aFeb170,1-a,1-b 121Y, c548 |
| Antennal lobes | c584 |
P[GAL4], UAS-PKAinh, P[GAL4] + UAS-PKAinh, and in some cases, P[GAL4] + UAS-PKAm-inh males were tested in the inebriometer. Lines were classified as normal with PKAinh if the MET of P[GAL4] + UAS-PKAinh was within 5 min of the MET of controls and resistant with PKAinh and PKAm-inhif the METs of P[GAL4] + UAS-PKAinh and P[GAL4] + UAS-PKAm-inh were more than 5 min increased compared with controls. The major sites of expression were determined from the references listed in Materials and Methods. In some cases, expression patterns were confirmed in P[GAL4] + UAS-lacZ males, and in all cases, these were consistent with published patterns.
n = 1 or 2; in all other cases, n = 3 or more.
Larval expression pattern.
Behavioral assays. Inebriometer assays were performed as described previously (Moore et al., 1998; Singh and Heberlein, 2000). Inebriometers were equilibrated to 20°C and an ethanol/humidified air mixture of ∼50/40 U. Flies were equilibrated for 1 hr at 20°C before being loaded into the inebriometer. Elution was quantified in 3 min intervals using a monitor (Trikinetics, Waltham, MA), and mean elution times were calculated from the resulting distribution. One-way ANOVAs were performed in Statistica (StatSoft, Tulsa, OK). To maintain an experiment-wide error rate of α = 0.05, the adjusted error rates were p = 0.0167 for the 3 subsequent planned pair-wise comparisons in Figures 1 and 3. In Figure 5, two-way ANOVA was performed in Statistica (StatSoft) with post hocNewman–Keuls testing.
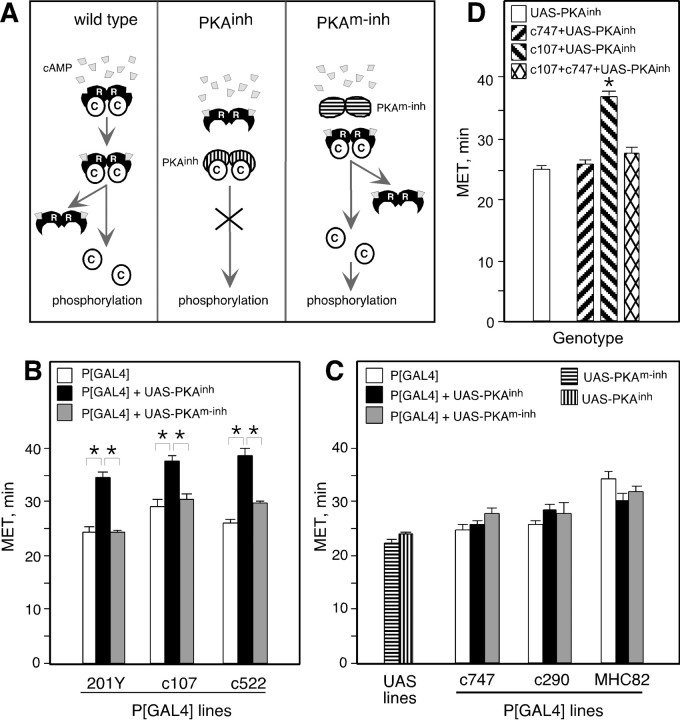
Fig. 1.
Expression of PKAinh in specific brain regions alters ethanol sensitivity in the inebriometer. A, Inactive PKA holoenzymes consist of two regulatory (R) and two catalytic (C) subunits. Binding of cAMP to R subunits results in their dissociation from C and hence C activation. PKAinh lacks cAMP binding ability and remains bound to C even on increases in cellular cAMP, thereby inhibiting C activation. PKAm-inh is unable to bind cAMP or C and therefore should not have an inhibitory effect. B, PKAinh expression under the control of 201Y, c107, and c522 resulted in increased MET. One-way ANOVA revealed a significant effect of genotype for all three P[GAL4] lines: 201Y (F(2,24) = 24.2; p< 0.0001), c107 (F(2,15) = 14.8;p < 0.0001), and c522 (F(2,15) = 28.1; p< 0.0001). Pair-wise planned comparisons, with the criticalp value adjusted to α = 0.0167, revealed significant differences between P[GAL4]+UAS-PKAinhand both P[GAL4] and P[GAL4]+UAS-PKAm-inh(p < 0.001 for all comparisons). Pairwise planned comparisons did not reveal significant differences between P[GAL4] and P[GAL4]+UAS-PKAm-inh: 201Y (p = 0.966), c107 (p = 0.346), and c522 (p = 0.061). For each P[GAL4] line,n is the same for P[GAL4], P[GAL4]+UAS-PKAinh, and P[GAL4]+UAS-PKAm-inh: 201Y (n= 9), c107 (n = 6), c522 (n = 6), c747 (n = 8), c290 (n = 5), MHC82 (n = 7), UAS-PKAinh(n = 59), and UAS-PKAm-inh(n = 20). Asterisks denote significant differences. In all figures, error bars indicate SEM. In this and subsequent figures, flies were heterozygous for autosomal insertions and hemizygous for X-linked insertions (see Materials and Methods for chromosomal location of insertions).C, The MET of c747, c290, and MHC82 P[GAL4] lines was not altered by the presence of UAS-PKAinh or UAS-PKAm-inh. One-way ANOVA comparing P[GAL4], P[GAL4]+UAS-PKAinh, and P[GAL4]+UAS-PKAm-inh revealed no significant effect of genotype for c290 (F(2,12) = 1.35; p = 0.30) and MHC82 (F(2,18) = 2.93; p= 0.078). For c747, there was a weak effect of genotype (F(2,21) = 4.0; p = 0.038). Planned pair-wise comparisons, with the criticalp value adjusted to α = 0.0167, revealed a marginally significant difference between c747+UAS-PKAm-inh and c747 (p = 0.012) but not between c747+UAS-PKAm-inh and c747+UAS-PKAinh (p = 0.061) or between c747+UAS-PKAinh and c747 (p = 0.459). D, Expression of PKAinh under the control of c107 and c747 resulted in normal MET. t tests with the criticalp value adjusted to α = 0.025 revealed no significant difference between c107+c747+UAS-PKAinhand c747+UAS-PKAinh (p = 0.05) but did reveal a significant difference between c107+c747+UAS-PKAinh and c107+UAS-PKAinh (p < 0.0001). The UAS-PKAinh MET is fromB. n = 10 for c747+UAS-PKAinh and c107+UAS-PKAinh; n = 8 for c107+c747+UAS-PKAinh.
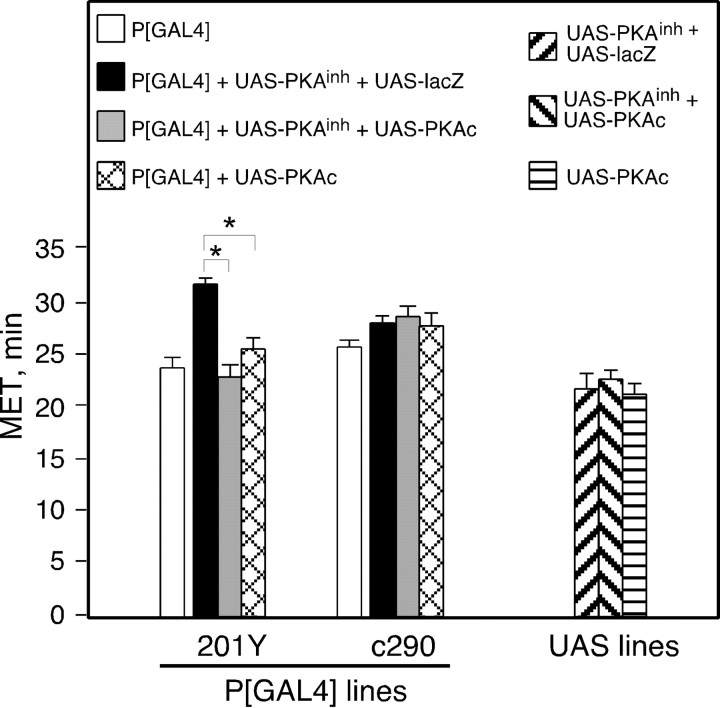
Fig. 3.
PKAc coexpression suppresses PKAinh-induced ethanol resistance. Flies of the indicated genotypes were tested in the inebriometer; METs are shown. Simultaneous expression of PKAinh and lacZ under the control of 201Y resulted in an increase in MET. This increase in MET was suppressed by coexpression of PKAc with PKAinh. Expression of PKAc alone did not alter MET, nor did the expression of any of the transgenes under the control of c290. The values for P[GAL4] were obtained from the experiments shown in Figure 1 and are displayed for comparison. One-way ANOVA revealed no significant effect of genotype for c290 (F(2,12) = 0.45;p = 0.647). A significant effect of genotype was observed for 201Y (F(2,12) = 45.5;p < 0.0001). Planned pair-wise comparisons with the critical p value adjusted to α = 0.0167 revealed a significant difference between 201Y+UAS-lacZ+UAS-PKAinh and both 201Y+UAS-PKAc+UAS-PKAinh and 201Y+UAS-PKAc (p < 0.0001 for both comparisons) but not between 201Y+UAS-PKAc+UAS-PKAinh and 201Y+UAS-PKAc (p = 0.019) (n = 4 for each UAS genotype; n = 5 for all others).Asterisks denote significant differences.
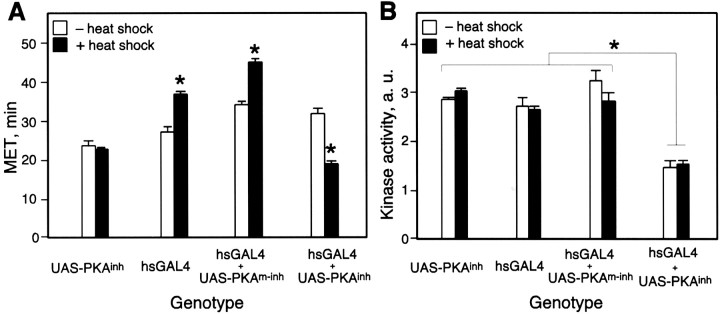
Fig. 5.
Heat shock-induced expression of PKAinh results in increased ethanol sensitivity. Male flies of the indicated genotypes were grown at 25°C. Flies were either heat-shocked for 1 hr at 37°C or kept at 25°C. Twenty-four hr after the heat shock, flies were tested in the inebriometer, or extracts were prepared for kinase activity assays. A, PKAinh-expressing flies are more sensitive to ethanol after heat shock. Two-way ANOVA revealed a significant effect of genotype (F(3,24) = 94.5;p < 0.0001), a significant effect of heat shock treatment (F(1,24) = 4.6;p < 0.05), and a significant interaction between genotype and treatment (F(3,24) = 53.5;p < 0.0001). Newman–Keuls post hoctesting revealed no significant difference between UAS-PKAinh with or without heat shock (p = 0.6) but did reveal significant differences for hsGAL4, hsGAL4+UAS-PKAm-inh, and hsGAL4+UAS-PKAinh with and without heat shock (p < 0.001 in all cases). Newman–Keuls tests also revealed significant differences between heat-shocked hsGAL4+UAS-PKAinh and heat-shocked UAS-PKAinh (p = 0.02), hsGAL4 (p = 0.0001), and hsGAL4+UAS-PKAm-inh (p = 0.0001). B, PKAinh expression results in reduced PKA activity. Two-way ANOVA revealed a significant effect of genotype (F(3.16) = 55.1;p < 0.0001) but no effect of heat shock (F(1,16) = 0.5; p = 0.5) and no interaction between genotype and heat shock (F(3,16) = 1.9; p = 0.2). Newman–Keuls post hoc testing revealed significant differences between hsGAL4+UAS-PKAinh(with or without heat shock) and all other genotypes (p < 0.001 in all cases). The only other significant difference was between hsGAL+UAS-PKAm-inh (without heat shock) and hsGAL4 (with heat shock) (p = 0.046;n = 3 for each genotype and condition).Asterisks denote significant differences.
Locomotor tracking assays were performed as described previously (Scholz et al., 2000), with an ethanol/humidified air mixture of 40/25 U. Flies were exposed to humidified air for 6 min, filmed in air for 2 min, and then filmed in the presence of ethanol vapor for an additional 18 min. Ten second movie clips were captured on an Apple G4 computer using Adobe Premiere. The flies' locomotion was tracked using the dynamic image analysis system (DIAS) (Solltech, Oakdale, IA). Average speed over the interval was calculated by dividing the flies' total path length by time. One-way ANOVAs across each of 16 time points were performed in Statistica (StatSoft). To maintain an experiment-wise error rate of α = 0.05, the critical p value was adjusted to α = 0.003. For time points showing a main effect of genotype, post hoc Newman–Keuls tests were performed to determine whether P[GAL4]+UAS-PKAinh was significantly different from both P[GAL4] and UAS controls at α = 0.05.
Ethanol absorption. Absorption assays were performed as described previously (Moore et al., 1998). Thirty flies of each genotype were exposed in triplicate to an ethanol/humidified air mixture of 50/45 U for 30 min. The alcohol concentration in extracts was measured using an alcohol dehydrogenase assay (Sigma, St. Louis, MO). To calculate the ethanol concentration in the flies, we estimated the volume of one fly to be 2 μl. The values for 201Y+UAS-PKAinh and 201Y were corrected for the total amount of protein, because 201Y+UAS-PKAinh flies are slightly smaller than 201Y flies. Protein was measured using Coomassie blue reagent (Pierce, Rockford, IL).
Histology. Staining for β-galactosidase expression in CNSs of 5- to 7-d-old P[GAL4]/UAS-lacZ males was done as described previously (Scholz et al., 2000). Samples were incubated in 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal; Labscientific, Livingston, NJ) solution at 37°C for 20–30 min (c747), 1–2 hr (c522 and c107) or overnight (c290, 201Y, and c747). Multiple specimens were observed for each genotype.
For anti-tau immunohistochemistry, larval brains were dissected in cold PBS, fixed in 4% paraformaldehyde for 50 min at room temperature, and washed in PBS with 0.3% Triton X-100. Specimens were incubated with anti-tau antibody (Sigma) diluted 1:500, and with a secondary HRP-coupled goat anti-mouse antibody (Vector Laboratories, Burlingame, CA) diluted 1:100. The Vectastain Elite ABC kit was used for DAB staining (Vector).
PKA activity assay. Fifty male UAS-PKAinh, hs-GAL4, hs-GAL4+UAS-PKAinh, or hs-GAL4+UAS-PKAm-inh flies were reared at 25°C and collected under CO2 anesthesia as for a behavioral assay. One day after collection, flies were heat-shocked for 1 hr at 37°C in a water bath and then returned to 25°C. Non-heat-shocked controls were kept at 25°C. Twenty-four hours after the beginning of heat shock, flies were transferred to Eppendorf tubes with brief CO2 anesthesia and frozen by placing the tubes on dry ice. Whole flies were homogenized in 600 μl of buffer (in mm: 10 sodium phosphate, pH 6.8, 1 EDTA, 0.5 EGTA, 2.5 β-mercaptoethanol, 25 benzamidine, and 1 PMSF) and centrifuged for 5 min at 14,000 rpm; the pellet was discarded. Protein concentration in the extracts was measured with Coomassie blue reagent (Pierce). Eight micrograms of total protein were assayed with the Colorimetric PKA assay kit, Spinzyme format (Pierce) according to the manufacturer's instructions. Each extract was assayed in duplicate, and the experiment was repeated three times. Two-way ANOVA was performed in Statistica (StatSoft) with post hocNewman–Keuls testing.
Hydroxyurea ablation of mushroom bodies. Hydroxyurea ablation was performed according to the method of de Belle and Heisenberg (1994). 201Y virgin females were crossed to UAS-lacZ or UAS-lacZ;UAS-PKAinh males. Eggs were collected on apple juice plates at 25°C in 1 hr intervals and kept at 25°C for 23.5 hr (this time was determined empirically to result in the most efficient MB ablation). The newly hatched first instar larvae were transferred to a microcentrifuge tube containing a paste of heat-killed yeast with or without 50 mg/ml hydroxyurea (Sigma) for 4 hr at 25°C. At this time, larvae were washed and transferred to regular food bottles; adult males, 2–4 d old, were tested in the inebriometer. Males eluting from each inebriometer run were collected and stained for β-galactosidase expression (between 20 and 50 from hydroxyurea-treated groups and ∼10 from each control group). Ablated, partially ablated, and unablated mushroom bodies were observed. For 201Y+UAS-lacZ, the percent complete ablation values observed in individual inebriometer runs were 58, 78, 82, and 85%, with an average of 76%. For 201Y+UAS-lacZ+UAS-PKAinh, the percent complete ablation values were 68, 70, 84, and 95, with an average of 79%, which is not significantly different from the percent ablation seen in 201Y+UAS-lacZ flies (Student's t test). No significant correlation was found between percent ablation and MET: 201Y+UAS-lacZ, p = 0.604; 201Y+UAS-lacZ+UAS-PKAinh,p = 0.328. No mushroom body ablation was observed in untreated flies. METs were subjected to two-way ANOVA using Statistica (StatSoft).
RESULTS
PKA inhibitor expression in specific brain regions decreases ethanol sensitivity
A collection of Drosophila strains, in which the transcription factor GAL4 is expressed in various discrete regions of the CNS under the control of endogenous enhancers, was used to express a PKA inhibitor in a spatially restricted manner. Inactive PKA holoenzyme consists of a dimer of regulatory subunits bound to two catalytic subunits. PKA activation occurs when cAMP binds to regulatory subunits, causing dissociation of the holoenzyme and release of active catalytic subunits (Taylor et al., 1990). The PKA inhibitory transgene PKAinh (Li et al., 1995) encodes aDrosophila type I regulatory subunit with mutated cAMP binding sites; it therefore remains bound to the endogenous catalytic subunit when cAMP levels rise, inhibiting activation in a dominant manner (Fig. 1A). As a control we used a transgene encoding an inactive PKA inhibitor, PKAm-inh, which is identical to PKAinh with the exception of two additional mutations in amino acids needed for binding to the catalytic subunit (Kiger and O'Shea, 2001). Both transgenes are positioned downstream of UAS sites that allow transcriptional regulation by GAL4. As shown below, expression of PKAinh or PKAm-inh had the expected effect on PKA catalytic activity.
Flies carrying UAS-PKAinh and individual P[GAL4] insertions were screened for sensitivity to ethanol in the inebriometer. This apparatus consists of a 4-foot-long vertical column fitted with obliquely oriented mesh baffles on which sober flies will stand (Cohan and Hoffman, 1986; Weber, 1988). When exposed to ethanol vapor, flies become uncoordinated, lose postural control, and fall through the column. The MET of a population of flies is a measure of their sensitivity to ethanol (increased MET equals decreased sensitivity).
Of 64 P[GAL4] lines tested, 42 displayed normal ethanol sensitivity in the presence of UAS-PKAinh (Table1). These included lines with expression in a variety of structures, including the MBs, ellipsoid body (EB), fan-shaped body (FSB), central complex small-field neurons, and antennal lobes (ALs). These structures are composed of multiple different cell types (Hanesch et al., 1989; Crittenden et al., 1998;Renn et al., 1999), and GAL4 expression in the lines tested may be restricted to only certain subsets of cells. Therefore, we cannot completely rule out the possibility that PKA signaling in these structures contributes to the modulation of ethanol sensitivity. An additional 19 P[GAL4] lines resulted in altered MET in the presence of either PKAinh or PKAm-inh, presumably because of an effect of protein overexpression (Table 1). In total, then, 61 of 64 lines screened did not result in a specific effect of PKAinh expression on ethanol sensitivity.
In contrast, expression of PKAinh under the control of three P[GAL4] lines, 201Y, c107, and c522, led to a decrease in ethanol sensitivity (resistance) in the presence of UAS-PKAinh but not UAS-PKAm-inh (Fig. 1B). The lack of effect of UAS-PKAm-inhindicated that the inhibitory activity of PKAinh (the ability to bind to the PKA catalytic subunit) is required for the altered behavior. Thus, expression of the inhibitor in a subset of CNS cells caused a specific reduction in ethanol sensitivity.
Three examples of lines with normal sensitivity when driving expression of PKAinh are shown in Figure 1C. Expression of PKAinhin postembryonic muscle, under the control of P[GAL4]MHC82 (Davis et al., 1998), did not affect ethanol sensitivity (Fig. 1C). Line c290 shows little or no expression in the CNS, whereas c747 is expressed broadly and at high levels in the CNS (Fig.2).
Fig. 2.
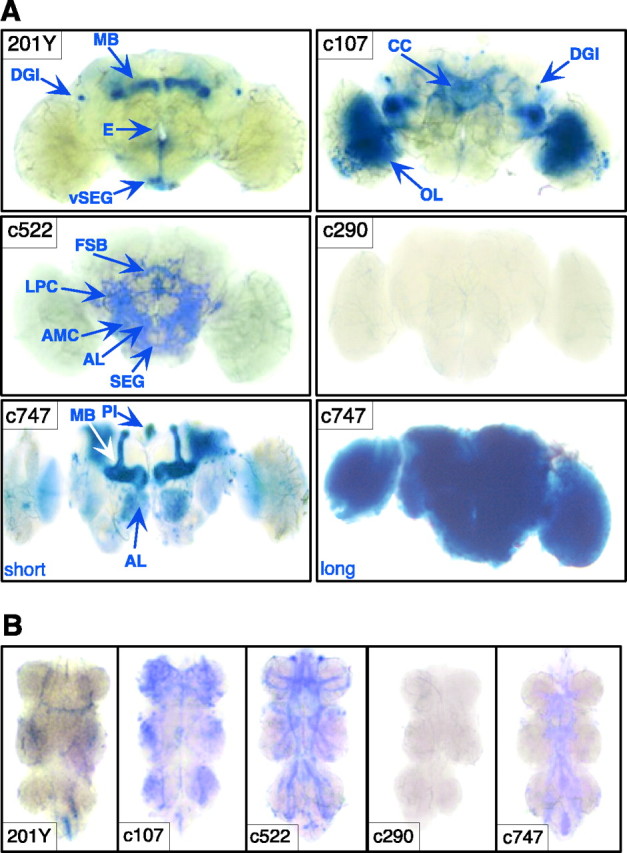
Expression patterns of P[GAL4] lines. β-Galactosidase expression under the control of each P[GAL4] was visualized by staining whole-mount nervous systems with X-gal. Some planes are out of focus in the photographs. A, In the brains of 201Y males, expression is observed in a subset of MB neurons, in the DGIs, and in neurons in the PI, vSEG, and lateral protocerebrum. PI neurons are easily dissociated from the brain during dissection and are not observed in this sample (but see Fig.4D). In c107, expression is seen in small-field neurons projecting to the FSB and EB of the central complex (CC), in the DGIs and optic lobes (OL), and elsewhere. c522 is expressed in parts of the central complex (subsets of FSB and EB neurons), in the ALs, antenno-mechanosensory center (AMC), and SEG. c290 has little expression in the CNS. c747 is widely and highly expressed. A short (20–30 min) exposure in X-gal solution reveals high levels of expression in the MBs, PI, EB, AL, and OL (Zars et al., 2000a). Longer X-gal exposure (overnight) reveals widespread expression. E, Esophagus;LPC, lateral protocerebrum. B, Ventral nerve cord (VNC) expression is observed in all P[GAL4] lines except c290. 201Y has very limited expression in a small number of cell bodies in the abdominal ganglion, two longitudinal fibers, and one transverse fiber. c107, c522, and c747 are expressed in thoracic and abdominal neuromeres. c107 expression is seen in a median longitudinal tract of the VNC, which appears to either give rise to or derive from tangles of fibers at each of the three levels of the thoracic ganglia. There is additional staining in the abdominal ganglion of the VNC. In c522, prominent longitudinal tracts in the VNC, which are continuous with tracts in the ventral part of the brain, resemble the median tract of the dorsal cervical fasciculus and the dorsal lateral tract of the ventral cervical fasciculus described byPower (1948). In addition, transverse fiber tracts are also observed, as well as additional cell bodies. In c747, some tracts appear to overlap with those observed in c522, specifically the median tract of the dorsal cervical fasciculus.
Given the widespread expression of GAL4 in line c747, the normal ethanol sensitivity of c747+UAS-PKAinh was surprising. We reasoned that if expression of PKAinh in the c747 cells was truly inert, simultaneous expression of PKAinh under the control of both c107 and c747 should result in resistance to ethanol, the phenotype displayed by c107+UAS-PKAinh flies. However, expression of PKAinh under the control of both c107 and c747 resulted in a MET similar to that of c747+UAS-PKAinh flies; i.e., the lack of effect of line c747 was epistatic to the ethanol resistance caused by line c107 (Fig. 1D). One possible explanation for this result is that the normal behavior displayed by c747+UAS-PKAinh represents the balanced sum of increased and decreased sensitivity caused by PKAinh expression in different sets of cells. Adding expression in c107 cells would not tip the balance toward resistance if the GAL4-expressing cells in c107 are already present in c747. That is, c107 may express GAL4 in resistance-causing cells that are also present in c747; in c107, however, these cells would not be counterbalanced by sensitivity-causing cells. It is also possible that the expression of PKAinh in certain c747 cells clamps ethanol sensitivity at a relatively normal level, and that these cells are epistatic to the resistance-causing cells of line c107; these cells may also mask resistance-causing cells in c747. Regardless of the exact mechanism, these data clearly show that broad PKAinh expression under the control of line c747 is not behaviorally inert but, rather, the result of complex interactions between different populations of cells. It is therefore important to exercise caution when drawing conclusions about neurons (or brain regions) not involved in a behavior of interest based on a lack of effect of a particular manipulation.
Decreased sensitivity to ethanol is not attributable to altered pharmacokinetics
Because reduced ethanol sensitivity could be caused by impaired ethanol absorption (Singh and Heberlein, 2000), we measured the ethanol content in P[GAL4] flies in the absence and presence of UAS-PKAinh after a 30 min exposure to ethanol vapor (Table 2). Expression of PKAinh did not affect the levels of absorbed ethanol in these flies. Thus, the altered behavior is attributable to functional differences in ethanol responsiveness rather than changed pharmacokinetics.
Table 2.
PKAinh expression does not alter ethanol absorption
| Transgenes | [Ethanol] |
|---|---|
| 201Y | 21.4 ± 0.8 |
| 201Y + UAS-PKAinh | 23.7 ± 1.8 |
| c107 | 34.2 ± 1.2 |
| c107 + UAS-PKAinh | 32.8 ± 0.4 |
| c522 | 30.1 ± 0.5 |
| c522 + UAS-PKAinh | 29.0 ± 0.9 |
| c747 | 35.7 ± 1.9 |
| c747 + UAS-PKAinh | 33.6 ± 1.7 |
| c290 | 32.8 ± 0.1 |
| c290 + UAS-PKAinh | 32.0 ± 0.1 |
| MHC82 | 36.4 ± 0.2 |
| MHC82 + UAS-PKAinh | 38.9 ± 1.1 |
| UAS-PKAinh | 31.8 ± 0.7 |
Ethanol concentration after a 30 min exposure to ethanol vapor is shown. Concentration values are millimolar, except for 201Y + UAS-PKAinh and 201Y, for which values represent millimoles of ethanol per microgram of protein (see Materials and Methods). No significant differences were seen between P[GAL4] and P[GAL4] + UAS-PKAinh flies (Student's t test;n = 3).
Expression patterns of P[GAL4] lines
Expression of GAL4 was determined using a reporter transgene, UAS-lacZ, encoding β-galactosidase. The expression pattern of the UAS-lacZ reporter was found to be indistinguishable in P[GAL4] and P[GAL4]+UAS-PKAinh flies on analysis at the light microscope level (data not shown). Thus, PKAinh expression does not alter the gross morphology of the structures in which it is expressed, although it could have effects at the ultrastructural level.
Of the three P[GAL4] lines that caused a specific change in ethanol sensitivity in the presence of UAS-PKAinh, two lines, c107 and c522, showed restricted expression in the CNS, whereas 201Y expression was even more limited (Fig. 2). The most prominent expression in c107 is in the optic lobes, small-field neurons of the central complex projecting to the EB and FSB (Renn et al., 1999), and the dorsal giant interneurons (DGIs), a bilateral pair of neurons with characteristic dorsally looping projections (Ito et al., 1997) (Fig. 2A). Line c522 is expressed in the antennal lobes, the antenno-mechanosensory center, subesophageal ganglion (SEG), central complex, and lateral protocerebrum (Zars et al., 2000b) (Fig. 2A). Central complex expression in c522 includes intrinsic neurons of the FSB and a small number of EB intrinsic neurons. Lines c107 and c522 are also expressed in the ventral nerve cord (VNC) (Fig. 2B). Line 201Y, used extensively in previous behavioral (O'Dell et al., 1995; Connolly et al., 1996; Martin et al., 1998; Joiner and Griffith, 1999; Zars et al., 2000a; McGuire et al., 2001) and neuroanatomical (Yang et al., 1995;Tettamanti et al., 1997; Armstrong et al., 1998; Kraft et al., 1998;Lee et al., 2000) studies, is expressed in a subset of MB neurons that project to the cores of the α and β lobes and broadly in γ lobe neurons (Fig. 2A). In addition, 201Y is expressed in the DGIs (K. Ito, personal communication), in ∼13 neurons of the PI projecting through the median bundle, and in 6 neurons located in the ventral SEG (see below). There is also very restricted and low expression in the VNC (Fig. 2B).
It is unlikely that PKAinh expression in the optic lobes of c107 affects ethanol sensitivity, because flies with severely abnormal optic lobes attributable to the small optic lobes mutation (sol1) are normal (Scholz et al., 2000). As mentioned above, small-field neurons of c107 project onto the EB and FSB, whereas expression of c522 includes intrinsic neurons of both of these structures, providing a possible functional link between the two P[GAL4] lines. As described above, however, PKAinh expression in 25 P[GAL4] lines with EB expression and 11 lines with FSB expression did not result in a specific effect of the inhibitor on behavior (Table 1). Therefore, it is unclear whether PKA signaling in these structures plays a role in modulating ethanol sensitivity. Because of the more limited expression of 201Y, we focused most subsequent experiments on this line.
Effect of PKAinh is suppressed by overexpression of catalytic subunit
As discussed above, the effect of PKAinh expression on ethanol sensitivity appears to require the binding of PKAinhto catalytic subunits, because PKAm-inh, which lacks this function, is inert. If this is correct, overexpression of the PKA catalytic subunit (PKAc) should counteract PKAinh and therefore suppress its effect on ethanol sensitivity. Indeed, PKAinh-induced defects in wing development are suppressed by coexpression of PKAc (Kiger et al., 1999). We therefore expressed both UAS-PKAinh and UAS-PKAc simultaneously under the control of line 201Y. As shown in Figure 3, coexpression of PKAc did suppress the PKAinh-induced reduction in ethanol sensitivity. This effect was not caused by the presence of two UAS transgenes, which may reveal limiting amounts of GAL4, because coexpression of an inert transgene, UAS-lacZ, with UAS-PKAinh did not cause phenotypic suppression (Fig. 3). Overexpression of PKAc alone with the 201Y driver did not alter behavior, suggesting that ethanol sensitivity was not affected by enhanced PKA function in the 201Y-expressing cells. Expression of any combination of transgenes with the control line c290 had no effect on ethanol sensitivity. These data strongly suggest that inhibition of PKA function in the 201Y-expressing cells decreases ethanol sensitivity, and that PKAinh acts by sequestering endogenous PKA catalytic subunits.
Mushroom bodies are not involved in the regulation of ethanol sensitivity
As has been documented previously (O'Dell et al., 1995; Yang et al., 1995; Connolly et al., 1996; Tettamanti et al., 1997; Armstrong et al., 1998; Kraft et al., 1998; Martin et al., 1998; Joiner and Griffith, 1999; Lee et al., 2000; Zars et al., 2000a,b; McGuire et al., 2001), 201Y is expressed in the mushroom bodies (Fig.2A), structures in the fly's brain that play an important role in olfactory classical conditioning. Interestingly, several olfactory learning and memory mutants, such asamnesiac (amn), rutabaga(rut), and the cell adhesion molecule fasciclin II (fasII), show altered ethanol sensitivity (Moore et al., 1998; Cheng et al., 2001). rutand fasII are preferentially expressed in the MBs, along with the catalytic and regulatory subunits of PKA (for review, seeRoman and Davis, 2001); amn is expressed in two prominent neurons that project onto the MB axons (Waddell et al., 2000). We were therefore interested in determining whether the MBs are required for normal ethanol sensitivity and whether the expression of PKAinh in the MBs is responsible for the altered sensitivity seen in 201Y+UAS-PKAinh flies.
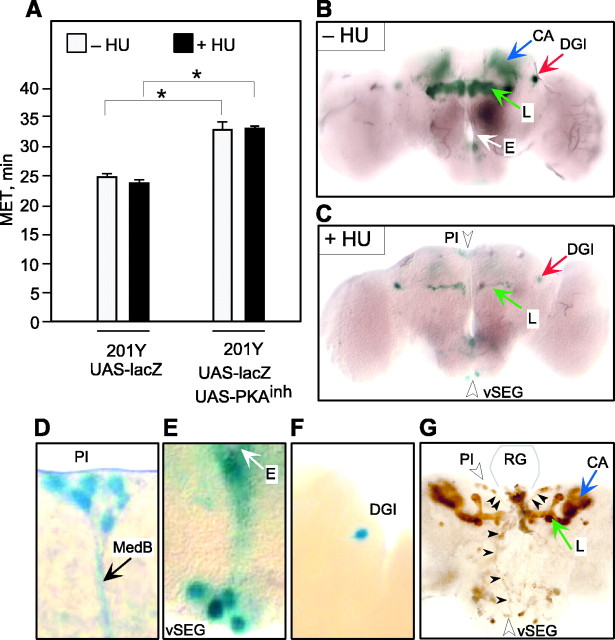
During the first few hours of larval life, the only proliferating neuroblasts are four that give rise to the MBs and one that contributes to the antennal lobes (Truman and Bate, 1987; Prokop and Technau, 1991,1994; Ito and Hotta, 1992). The MBs can thus be selectively ablated by feeding hydroxyurea to newly hatched larvae, a manipulation that has been shown to impair olfactory and courtship conditioning (de Belle and Heisenberg, 1994; McBride et al., 1999). To determine whether MB ablation alters ethanol sensitivity, we fed 201Y+UAS-lacZ larvae yeast paste with or without hydroxyurea and tested the resulting adults in the inebriometer; MB ablation was assessed by examining β-galactosidase expression (Fig. 4; see Materials and Methods). As reported previously (Armstrong et al., 1998), hydroxyurea treatment eliminated most MB neurons in which 201Y is expressed (Fig. 4C); a few γ lobe neurons remain, probably those arising during embryogenesis. Also consistent with previous results (Armstrong et al., 1998), hydroxyurea treatment spared neurons outside of the mushroom bodies, such as the DGIs (Fig.4C). Flies lacking the MBs showed normal sensitivity in the inebriometer (Fig. 4A). Thus, unlike olfactory learning, in which the MBs play a central role, ethanol sensitivity does not appear to be regulated by these structures.
Fig. 4.
Ablation of mushroom bodies does not alter ethanol sensitivity. A, The MET in the inebriometer of hydroxyurea-treated (+HU) and mock-treated (−HU) flies is shown. METs were subjected to two-way ANOVA revealing a significant main effect of genotype (F(1,12) = 148.7; p< 0.001) but no effect of hydroxyurea treatment (F(1,12) = 3.3; p = 0.093) and no significant interaction (F(1,12) = 0.62; p= 0.446). These data indicate that ablation of the mushroom bodies had no effect in either genotype. (n = 4 for all groups). Asterisks denote significant differences.B, C, Males eluting from each inebriometer run were collected, and their brains were dissected and stained with X-gal. Examples of an unablated brain (−HU) and ablated brain (+HU) are shown. HU treatment ablated all but a few gamma lobe neurons (L) but spared other neurons such as the DGIs. D–F, Close-up view of the expression of P[GAL4] outside the MBs in 201Y. D, Expression in the PI neurons, which extend axons through the median bundle (MedB). E, Expression in the vSEG neurons. Note axons extending from the cell bodies toward the esophagus (E). F, Expression in the DGI neurons. G, Expression of the microtubule-binding protein tau under the control of 201Y in third instar larvae. Expression is observed in MB lobes (L) and the cell bodies and axons (arrowheads) of PI and vSEG neurons. These axons project to the base of the ring gland (RG, the outline of the unstained gland, based on a phase-contrast image, gray lines).
To determine whether expression of PKAinhin MBs contributes to the reduced sensitivity of 201Y+UAS-PKAinh flies, we treated larvae of this genotype with hydroxyurea as described above. These flies also carried UAS-lacZ to monitor MB ablation. Decreased ethanol sensitivity of 201Y+UAS-PKAinh flies remained even after mushroom body ablation (Fig. 4A). We conclude from these results that PKA inhibition in neurons other than those forming the MBs is responsible for the decreased ethanol sensitivity of 201Y+UAS-PKAinh flies. This is consistent with our finding that expression of PKAinhin MB neurons under the control of c747 (and five additional lines) did not alter MET (Fig. 1B, Table 1).
Because few neurons outside of the MBs express GAL4 in 201Y, this limits the number of neurons that cause the altered inebriometer phenotype. These include the PI neurons (Fig. 4D), the ventral SEG neurons (Fig. 4E), the DGIs (Fig.4F), and a very small number of other neurons in the protocerebrum and abdominal ganglion. Diffuse staining near the esophagus (Fig. 4E) is attributable to expression in the axons and nerve endings of PI and SEG neurosecretory cells known to project to this area (Rajashekhar and Singh, 1994; Shiga et al., 2000). We do not believe that PKAinh expression in the DGIs is responsible for the altered behavior, because three lines with DGI expression did not cause altered ethanol sensitivity when driving PKAinh (Table 1; data not shown). We also tested P[GAL4] lines with PI and vSEG expression: 8 lines with adult expression in PI neurons and 10 lines with expression in larval PI and SEG neurosecretory cells projecting to the ring gland did not result in altered ethanol sensitivity (Table 1). However, PI and SEG neurons are heterogeneous; there are at least three different subtypes of larval PI neurons and two subtypes of SEG neurons based on their projection patterns (Siegmund and Korge, 2001). In addition, different PI neurons in the blowfly Calliphora express different neuropeptides (Duve and Thorpe, 1980, 1981, 1983; Duve et al., 1983). This leaves open the possibility that a specific subset of these neurosecretory cells expresses GAL4 in 201Y but not in the other GAL4 lines tested.
Acute PKA inhibition in the adult fly increases ethanol sensitivity
Previous experiments had shown that global reduction of PKA activity throughout development (by reducing the dose of the PKA catalytic subunit) or acute pharmacological inhibition of PKA in the adult led to increased ethanol sensitivity (Moore et al., 1998). Yet we were unable to find a GAL4 line that mimicked that effect when driving PKAinh. To test the effect of expression of PKAinh throughout the fly, we used a heat shock-inducible GAL4 transgene, hsGAL4, in which GAL4 is under the control of the hsp70 promoter. Flies carrying the hsGAL4 transgene and either UAS-PKAinh or UAS-PKAm-inh were heat-shocked for 1 hr and tested in the inebriometer the following day. Under these conditions, heat shock alone did not affect ethanol sensitivity, because the MET of UAS-PKAinh flies was unchanged by heat shock (Fig.5A). Heat shock-induced expression of GAL4 (in hsGAL4 flies) or GAL4 and PKAm-inh (in hsGAL4+UAS-PKAm-inh flies) resulted in a significant increase in ethanol resistance (Fig. 5A), probably a nonspecific effect of protein overexpression. However, heat shock-induced expression of PKAinh (in hsGAL4+UAS-PKAinh flies) resulted in increased sensitivity to ethanol (Fig. 5A). Thus, in contrast to restricted expression of PKAinh, short-term ubiquitous expression of the inhibitor in adult flies led to increased ethanol sensitivity, an observation that is consistent with previous pharmacological experiments (Moore et al., 1998).
To determine whether expression of PKAinhinhibited PKA activity as expected, we measured cAMP-stimulated PKA activity in extracts of flies expressing either PKAinh or PKAm-inh under the control of hsGAL4. PKA activity was strongly reduced by expression of PKAinh but not PKAm-inh (Fig. 5B). However, the degree of PKA inhibition was the same in the presence or absence of heat shock. These data suggest that there is considerable “leaky” expression of GAL4 even in the absence of heat shock, an observation that has been made before with transgenes expressed under the control of heat-inducible promoters (for example, see Grotewiel et al., 1998;Cheng et al., 2001). However, as shown above, this leaky expression of PKAinh did not result in altered ethanol sensitivity, whereas heat-induced expression of the inhibitor led to strongly increased ethanol sensitivity (Fig. 5A). There are several possible explanations for this observation. For example, the assay used to detect kinase activity may not be sensitive enough to detect subtle differences in activity between heat-shocked and non-heat-shocked flies. Alternatively, heat induction of PKAinh above baseline levels may occur in only a few cells, an effect that would not be detected when assaying kinase activity in extracts of whole flies. Although it has not been reported, it is possible that PKA inhibition may alter the flies' ability to cope with the heat shock itself, subsequently altering the flies' response to ethanol. Finally, PKA activity may be reduced transiently between heat shock and testing, altering nervous system susceptibility to ethanol. Extracts prepared 4 hr after the heat shock did not reveal such a difference (data not shown), but this does not preclude an effect at other time points.
In summary, we have shown that PKAinh and PKAm-inh display the expected effects on cAMP-stimulated kinase activity, and that ubiquitous expression of PKAinh resulted in increased ethanol sensitivity, similar to that observed with genetic and pharmacological manipulations that globally reduce cAMP signaling (Moore et al., 1998).
PKA inhibition in different regions has distinct effects on ethanol-induced locomotor activity
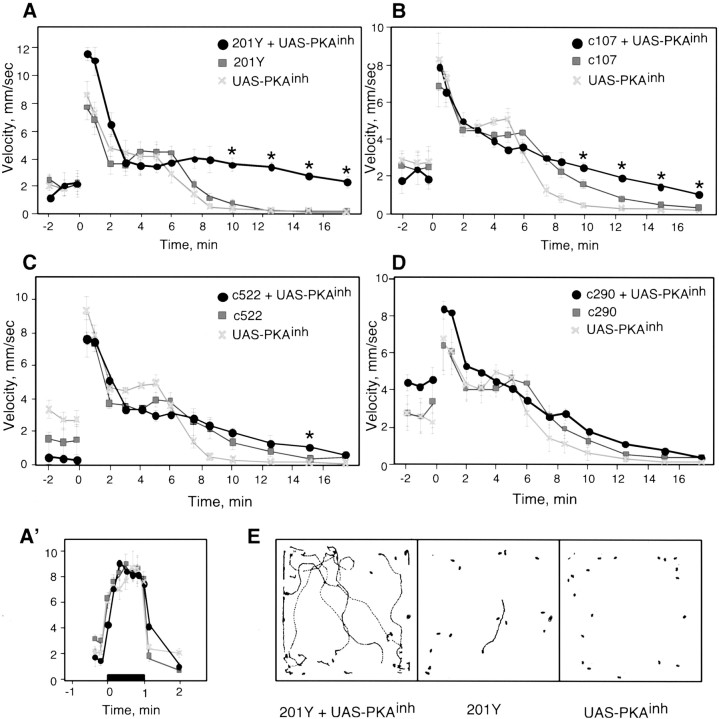
To further analyze the ways in which PKAinh expression affects ethanol sensitivity, we examined the flies' response to ethanol in a different behavioral assay. The locomotor video-tracking system allows the monitoring of horizontal walking activity of groups of flies as they are exposed continuously to ethanol vapor of defined concentrations (Scholz et al., 2000). Immediately after the start of ethanol exposure, flies showed a transient increase in locomotor velocity (Fig.6); this is probably a startle response to a novel odor (F. Wolf and U. Heberlein, unpublished observations). All P[GAL4]+UAS-PKAinh flies tested had robust startle responses, indicating that locomotor function in these flies is unaffected by PKAinh expression (Fig. 6). With continued exposure to the relatively high ethanol concentration used in these experiments, locomotor activity decreased gradually until the flies became completely sedated, at which point they became resistant to arousal by mechanical stimulation. When compared with 201Y and UAS-PKAinhcontrols, 201Y+UAS-PKAinh flies were resistant to the locomotor-depressant effects of ethanol (Fig.6A). This effect can be seen clearly in examples of locomotor traces obtained 12.5 min after the start of ethanol exposure (Fig. 6E); whereas control flies were nearly completely immobile, most 201Y+UAS-PKAinhflies remained active. Because the kinetics of ethanol absorption are indistinguishable in control and experimental flies (Table 1), these data show that higher levels of ethanol are needed to sedate flies with targeted inactivation of PKA in 201Y-expressing cells. We note that baseline locomotion levels (before the start of ethanol exposure) are somewhat variable, because flies are still in the process of adapting to their new environment (Fig. 6). These differences in spontaneous locomotion, however, do not affect the extent of the response to ethanol (F. W. Wolf and U. Heberlein, unpublished observations).
Fig. 6.
PKA inhibition differentially affects the ethanol-induced locomotor activity pattern. The average velocity of a population of 20 flies is shown as a function of time. Ethanol exposure started at time 0. A, 201Y and UAS-PKAinh flies were relatively calm in air (−2–0 min), startled on exposure to alcohol (0–2 min), remained relatively active until ∼6 min, but were strongly sedated by ∼8.5 min of ethanol exposure. 201Y+UAS-PKAinh flies behaved similarly at early time points but remained active during the later time points. One-way ANOVA across each time point, with the criticalp value adjusted to α = 0.003, revealed a significant effect of genotype at 10 min (F(2,9) = 34.5; p< 0.0001), 12.5 min (F(2,9) = 130.8;p < 0.0001), 15 min (F(2,9) = 74.3; p< 0.0001), and 17.5 min (F(2,9) = 27.7; p < 0.0005). Post hocNewman–Keuls testing showed that at each starredtime-point, P[GAL4]+UAS-PKAinh flies were different from both P[GAL4] and UAS-PKAinhcontrols (p < 0.001 for all comparisons).A′, 201Y, 201Y+UAS-PKAinh and 201Y+UAS-PKAm-inh flies were exposed to air, followed by an ethanol/air dose of 40/25 U beginning at time 0 for 1 min. Consecutive 10 sec clips were analyzed beginning 20 sec before the onset of ethanol and ending 20 sec after ethanol exposure was terminated; an additional 10 sec was analyzed 1 min after the end of exposure. The startle response was similar in all three genotypes (n = 3 for each genotype). B, c107+UAS-PKAinh flies are also resistant to the locomotor-depressant effects of ethanol compared with controls. A significant effect of genotype was seen at 7.5 min (F(2,9) = 18.1; p< 0.001), 10 min (F(2,9) = 30.9;p < 0.0001), 12.5 min (F(2,9) = 47.0; p< 0.0001), 15 min (F(2,9) = 65.3;p < 0.0001), and 17.5 min (F(2,9) = 37.7; p< 0.0001). Newman–Keuls post hoc tests showed that c107+UAS-PKAinh flies were significantly different from both c107 and UAS-PKAinh controls at 10, 12.5, 15, and 17.5 min (p < 0.01 for all comparisons). C, c522+UAS-PKAinhflies have normal sensitivity to the locomotor depressant effects of ethanol. A significant effect of genotype was observed only at 15 min (F(2,9) = 12.1; p = 0.0028). In Newman–Keuls post hoc tests, c522+UAS-PKAinh flies were significantly different from both c522 and UAS-PKAinh controls (p < 0.01) at this time point.D, c290+UAS-PKAinh flies were also similar to controls. In A–D, n = 4 for all genotypes. E, Examples of locomotor traces of 20 flies corresponding to a 10 sec period at 12.5 min of ethanol exposure. Although almost all 201Y and UAS-PKAinh flies were completely sedated at 12.5 min, 201Y+UAS-PKAinhflies were still active at this time. Asterisks denote significant differences.
c107+UAS-PKAinh flies were also resistant to ethanol-induced sedation, although to a lesser degree than 201Y+UAS-PKAinh flies (Fig.6B). In contrast, both c522+UAS-PKAinh and c290+UAS-PKAinh flies did not differ significantly from controls, despite their different inebriometer phenotypes (Figs. 1, 6C,D). Thus, although PKAinh expression under the control of 201Y, c107, and c522 resulted in decreased ethanol sensitivity in the inebriometer, only 201Y and c107 showed an altered response to the locomotor-depressant effects of ethanol.
We suggest that distinct components of ethanol sensitivity are differentially modulated by cAMP signaling in different neurons in the flies' CNS. Signaling in the 201Y and c107 cells regulates sensitivity to the effects of ethanol on both motor coordination (postural control) and sedation, whereas signaling in the c522 cells appears to only modulate sensitivity to the effect of ethanol on postural control.
DISCUSSION
A role for cAMP signaling in the modulation of acute sensitivity to ethanol in Drosophila has been demonstrated previously (Moore et al., 1998). Here we show that several neuroanatomical loci interact in complex ways to regulate normal responsiveness to acute ethanol exposure, yet inhibition of PKA in just a few cells in the brain, possibly neurosecretory cells, is sufficient to reduce the flies' sensitivity to the effects of ethanol on postural control. Using a different behavioral assay that measures the locomotor-depressant effects of ethanol, we show that cAMP signaling in different populations of neurons regulates distinct aspects of the acute behavioral response to ethanol. In addition, we show that despite the regulation of olfactory learning and ethanol sensitivity by a common set of genes, these two complex Drosophila behaviors are controlled by distinct neural circuits.
Neuroanatomy of ethanol sensitivity
Of the neurons we have identified as candidates for mediating the resistance to ethanol intoxication, the PI and ventral SEG (vSEG) neurons are the most interesting. These resemble known neuropeptide-expressing neurosecretory cells that extend axons toward the esophagus, where they intermingle with the projections from the neurosecretory PI neurons (Rajashekhar and Singh, 1994; Shiga et al., 2000). Both PI and vSEG neurons also send projections to the hypocerebral ganglion–corpus cardiacum complex, a neuroendocrine organ that is an adult derivative of the larval ring gland (Thomsen, 1969;Nassel, 1993; Shiga et al., 2000). Interestingly, an enhancer–trap insertion in the neuropeptide-encoding amnesiac gene,amnchpd, shows strong β−galactosidase expression in the ring gland (data not shown), suggesting a possible association between the PI and vSEG neurons and neuropeptides modulating ethanol sensitivity.
Ethanol sensitivity and the cAMP pathway
Several conditions known or expected to decrease the function of the cAMP pathway in the whole fly were shown previously to increase the flies' ethanol sensitivity in the inebriometer. These include mutations in amn, rut, and the catalytic subunit of PKA, as well as acute pharmacological inhibition of PKA (Moore et al., 1998). Similarly, ubiquitous expression of PKAinh in the adult fly under the control of hsGAL4, as shown here, resulted in increased ethanol sensitivity. In contrast, restricted inhibition of PKA under the control of specific GAL4 lines caused reduced ethanol sensitivity. Several mechanisms may contribute to this apparent paradox. First, the different phenotypes may be the consequence of cAMP pathway requirements in different populations of CNS cells. Although we identified cells that confer reduced sensitivity to ethanol, we failed to find those that confer increased sensitivity. The existence of the latter cells is, however, inferred from the fact that ubiquitous expression of PKAinh results in increased ethanol sensitivity. If so, cells conferring increased sensitivity likely act downstream in the neural circuits regulating the effects of ethanol, because they mask the effect of cells that confer increased resistance. This is consistent with the result obtained with coexpression of the PKAinh with both c107 and c747, in which the resistant phenotype obtained with c107 was masked by simultaneously expressing PKAinh in the c747 neurons.
It is also possible that the specific effect of PKA inhibition on ethanol sensitivity depends on the timing of such inhibition. Perhaps inhibition in the adult fly, such as that expected to be caused by feeding flies a PKA inhibitor and that caused by heat shock induction of PKAinh, results in increased sensitivity, whereas expression under the control of c107, c522, and 201Y during development results in reduced sensitivity. These three lines all exhibit GAL4 expression in the CNS in third instar larvae (Fig. 4G; data not shown).
Finally, it is possible that quantitative differences between the degree of PKA inhibition in the three GAL4 lines that cause ethanol resistance, c107, c522, and 201Y, and the experimental conditions that result in increased ethanol sensitivity are responsible for the dichotomy. Strong and global reductions in PKA activity, for example inpka-C1 null mutants or by expressing PKAinh pan-neurally, result in lethality attributable to developmental defects (Lane and Kalderon, 1993; data not shown), precluding behavioral analyses. It is therefore possible that the degree of PKA inhibition achieved in the c107, c522, or 201Y cells is greater than that obtained with other GAL4 lines, but flies survive because expression is restricted in these lines.
The regulation of ethanol sensitivity by cAMP signaling is also complex in mammals. As in flies with a mutation in PKA-RII, cAMP-stimulated PKA activity is greatly reduced in mice with a targeted disruption in the PKA-RIIβ gene (Park et al., 2000; Thiele et al., 2000); like the PKA-RII mutant flies, these mice show decreased sensitivity to the sedating effects of ethanol. Interestingly, mice expressing a transgene similar to PKAinh in the forebrain, which therefore have decreased PKA activity in these brain regions, display increased ethanol sensitivity (Wand et al., 2001). Mice heterozygous for a deletion of a Gsa gene are also more sensitive to ethanol. Thus, as with flies, different genetic manipulations that decrease cAMP signaling result in opposite effects on ethanol sensitivity.
Regulation of different ethanol-induced behaviors
Exposure of both mammals and flies to varying concentrations of ethanol has distinct behavioral consequences. In Drosophila, these can be separated using different assays, such as the inebriometer and the locomotor tracking system. Using these assays, we have shown that expression of PKAinh under the control of c107, c522, and 201Y, which in all three cases resulted in similar resistance to the incoordinating effects of ethanol in the inebriometer, altered sensitivity to the locomotor-depressant effects of ethanol to varying degrees. Thus, PKA inhibition in different sets of neurons affects distinct aspects of the acute behavioral effects of ethanol. The neural circuitry controlling different aspects of ethanol sensitivity has not been studied in mammals, but different inbred mouse strains, as well as mice with targeted disruption of the 5-HT1B receptor, show different relative sensitivities to acute ethanol administration in different behavioral assays, such as the stationary dowel, rotarod, and loss of righting reflex tests (Crabbe et al., 1994; Boehm et al., 2000; Browman and Crabbe, 2000). Together, the data from flies and mice suggest that different aspects of ethanol sensitivity are both genetically and neuroanatomically separable.
Ethanol sensitivity and olfactory conditioning
Flies learn to avoid an odor that has been paired previously with an electric shock, a fact that they can remember for hours to days, depending on the training paradigm (for review, see Dubnau and Tully, 1998). cAMP signaling plays a central role in this classical conditioning paradigm (for review, see Davis, 1996; Dubnau and Tully, 1998). Interestingly, the genetic overlap between olfactory conditioning and ethanol sensitivity includes not only rut,amn, and pka-C but also the cell adhesion molecule fasciclin II (Cheng et al., 2001).
Because the mechanisms that regulate olfactory learning and ethanol sensitivity appear to share molecular components, we investigated whether some of the same neuroanatomical structures might be involved in both behaviors. Multiple different lines of experimentation, including hydroxyurea-induced MB ablation (de Belle and Heisenberg, 1994), have shown that these structures play a central role in olfactory learning and memory (for review, see Roman and Davis, 2001). In contrast, our experiments show that MBs are not necessary for proper regulation of ethanol sensitivity measured in the inebriometer.
amn, although not expressed in the MBs, is expressed in the dorsal paired medial (DPM) interneurons, which project to the neuropil containing MB axons; preventing neurotransmission of these neurons impairs olfactory learning (Waddell et al., 2000). It seems unlikely that amn expressed in the DPM neurons plays a role in regulating ethanol sensitivity, because ablation of the presumed DPM targets, the MBs, has no effect. Thus, amn expressed elsewhere in the nervous system (Waddell et al., 2000) must modulate ethanol sensitivity. As discussed above, an interesting possibility is the neuroendocrine hypocerebral ganglion–corpus cardiacum complex. Consistent with distinct roles for amn in ethanol sensitivity and olfactory learning is the finding that the ethanol sensitivity defect of amn can be rescued by ubiquitous expression of an amn transgene in the adult fly, a protocol that fails to restore normal learning (Moore et al., 1998; DeZazzo et al., 1999).
These observations show that despite the overlap among the genes involved in regulating ethanol sensitivity and olfactory conditioning, the neural circuitry underlying these behaviors is separable. It is therefore important to determine not only the identity of genes involved in the behavior of interest but also the neuroanatomical circuits that regulate its manifestations. The ability to manipulate subsets of neurons in the fly's nervous system allows dissection of neuroanatomical loci regulating specific behaviors, in this case different aspects of ethanol sensitivity, allowing exploration of both the molecular and neural circuitry underlying behavior.
Footnotes
This work was supported by an award from the McKnight Foundation for Neuroscience (U.H.), National Institutes of Health Grant AA10035 (U.H.), and the Medical Scientist Training Program (A.R.R.). We are grateful to Kim Kaiser, Dan Kalderon, Grae Davis, Leslie Griffith, Thomas Siegmund, Gunter Korge, and Cahir O'Kane for providing fly lines used in this study. Kei Ito generously shared unpublished lines and knowledge of fly neuroanatomy. This manuscript benefited from the comments of Linus Tsai, Adrian Rothenfluh, Doug Guarnieri, Henrike Scholz, Fred Wolf, and Ammon Corl. We thank Linus Tsai for outcrossing many of the lines used as well as for thoughtful suggestions throughout the project.
Correspondence should be addressed to Ulrike Heberlein, 513 Parnassus Avenue, S-1332, San Francisco, CA 94143-0452. E-mail:ulrike@itsa.ucsf.edu.
REFERENCES
- 1.Armstrong JD, de Belle JS, Wang Z, Kaiser K. Metamorphosis of the mushroom bodies; large-scale rearrangements of the neural substrates for associative learning and memory in Drosophila. Learn Mem. 1998;5:102–114. [PMC free article] [PubMed] [Google Scholar]
- 2.Bainton RJ, Tsai LT-Y, Singh CM, Moore MS, Neckameyer WS, Heberlein U. Dopamine modulates acute responses to cocaine, nicotine, and ethanol in Drosophila. Curr Biol. 2000;10:187–194. doi: 10.1016/s0960-9822(00)00336-5. [DOI] [PubMed] [Google Scholar]
- 3.Boehm SL, II, Schafer GL, Phillips TJ, Browman KE, Crabbe JC. Sensitivity to ethanol-induced motor incoordination in 5-HT(1B) receptor null mutant mice is task-dependent: implications for behavioral assessment of genetically altered mice. Behav Neurosci. 2000;114:401–409. [PubMed] [Google Scholar]
- 4.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 5.Browman KE, Crabbe JC. Quantitative trait loci affecting ethanol sensitivity in BXD recombinant inbred mice. Alcohol Clin Exp Res. 2000;24:17–23. [PubMed] [Google Scholar]
- 6.Cheng Y, Endo K, Wu K, Rodan AR, Heberlein U, Davis RL. Drosophila fasciclin II is required for the formation of odor memories and for normal sensitivity to alcohol. Cell. 2001;105:757–768. doi: 10.1016/s0092-8674(01)00386-5. [DOI] [PubMed] [Google Scholar]
- 7.Cohan FM, Hoffman AA. Genetic divergence under uniform selection. II. Different responses to selection for knockdown resistance to ethanol among Drosophila melanogaster populations and their replicate lines. Genetics. 1986;114:145–163. doi: 10.1093/genetics/114.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Connolly JB, Roberts IJH, Armstrong JD, Kaiser K, Forte M, Tully T, O'Kane CJ. Associative learning disrupted by impaired Gs signaling in Drosophila mushroom bodies. Science. 1996;274:2104–2107. doi: 10.1126/science.274.5295.2104. [DOI] [PubMed] [Google Scholar]
- 9.Crabbe JC, Belknap JK, Buck KJ. Genetic animal models of alcohol and drug abuse. Science. 1994;264:1715–1723. doi: 10.1126/science.8209252. [DOI] [PubMed] [Google Scholar]
- 10.Crittenden JR, Skoulakis EM, Han KA, Kalderon D, Davis RL. Tripartite mushroom body architecture revealed by antigenic markers. Learn Mem. 1998;5:38–51. [PMC free article] [PubMed] [Google Scholar]
- 11.Davis GW, DiAntonio A, Petersen SA, Goodman CS. Postsynaptic PKA controls quantal size and reveals a retrograde signal that regulates presynaptic transmitter release in Drosophila. Neuron. 1998;20:305–315. doi: 10.1016/s0896-6273(00)80458-4. [DOI] [PubMed] [Google Scholar]
- 12.Davis RL. Physiology and biochemistry of Drosophila learning mutants. Physiol Rev. 1996;76:299–317. doi: 10.1152/physrev.1996.76.2.299. [DOI] [PubMed] [Google Scholar]
- 13.de Belle JS, Heisenberg M. Associative odor learning in Drosophila abolished by chemical ablation of mushroom bodies. Science. 1994;263:692–695. doi: 10.1126/science.8303280. [DOI] [PubMed] [Google Scholar]
- 14.DeZazzo J, Xia S, Christensen J, Velinzon K, Tully T. Developmental expression of an amn(+) transgene rescues the mutant memory defect of amnesiac adults. J Neurosci. 1999;19:8740–8746. doi: 10.1523/JNEUROSCI.19-20-08740.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diamond I, Gordon AS. Cellular and molecular neuroscience of alcoholism. Physiol Rev. 1997;77:1–20. doi: 10.1152/physrev.1997.77.1.1. [DOI] [PubMed] [Google Scholar]
- 16.Dubnau J, Tully T. Gene discovery in Drosophila: new insights for learning and memory. Annu Rev Neurosci. 1998;21:407–444. doi: 10.1146/annurev.neuro.21.1.407. [DOI] [PubMed] [Google Scholar]
- 17.Duve H, Thorpe A. Localisation of pancreatic polypeptide (PP)-like immunoreactive material in neurones of the brain of the blowfly, Calliphora erythrocephala (Diptera). Cell Tissue Res. 1980;210:101–109. doi: 10.1007/BF00232146. [DOI] [PubMed] [Google Scholar]
- 18.Duve H, Thorpe A. Gastrin/cholecystokinin (CCK)-like immunoreactive neurones in the brain of the blowfly, Calliphora erythrocephala (Diptera). Gen Comp Endocrinol. 1981;43:381–391. doi: 10.1016/0016-6480(81)90298-7. [DOI] [PubMed] [Google Scholar]
- 19.Duve H, Thorpe A. Immunocytochemical identification of alpha-endorphin-like material in neurones of the brain and corpus cardiacum of the blowfly, Calliphora vomitoria (Diptera). Cell Tissue Res. 1983;233:415–426. doi: 10.1007/BF00238307. [DOI] [PubMed] [Google Scholar]
- 20.Duve H, Thorpe A, Strausfeld NJ. Cobalt-immunocytochemical identification of peptidergic neurons in Calliphora innervating central and peripheral targets. J Neurocytol. 1983;12:847–861. doi: 10.1007/BF01258155. [DOI] [PubMed] [Google Scholar]
- 21.Feany MB, Quinn WG. A neuropeptide gene defined by the Drosophila memory mutant amnesiac. Science. 1995;268:869–873. doi: 10.1126/science.7754370. [DOI] [PubMed] [Google Scholar]
- 22.Ferveur JF, Störtkuhl KF, Stocker RF, Greenspan RJ. Genetic feminization of brain structures and changed sexual orientation in male Drosophila. Science. 1995;267:902–905. doi: 10.1126/science.7846534. [DOI] [PubMed] [Google Scholar]
- 23.Gatti S, Ferveur J-F, Martin J-R. Genetic identification of neurons controlling a sexually dimorphic behaviour. Curr Biol. 2000;10:667–670. doi: 10.1016/s0960-9822(00)00517-0. [DOI] [PubMed] [Google Scholar]
- 24.Grotewiel MS, Beck CDO, Wu K, Zhu X, Davis RL. Integrin-mediated short-term memory in Drosophila. Nature. 1998;391:455–461. doi: 10.1038/35079. [DOI] [PubMed] [Google Scholar]
- 25.Hanesch U, Fishbach K-F, Heisenberg M. Neuronal architecture of the central complex in Drosophila melanogaster. Cell Tissue Res. 1989;257:343–366. [Google Scholar]
- 26.Harris RA. Ethanol actions on multiple ion channels: which are important? Alcohol Clin Exp Res. 1999;23:1563–1570. [PubMed] [Google Scholar]
- 27.Ito K, Hotta Y. Proliferation pattern of postembryonic neuroblasts in the brain of Drosophila melanogaster. Dev Biol. 1992;149:134–148. doi: 10.1016/0012-1606(92)90270-q. [DOI] [PubMed] [Google Scholar]
- 28.Ito K, Sass H, Urban J, Hofbauer A, Schneuwly S. GAL4-responsive UAS-tau as a tool for studying the anatomy and development of the Drosophila central nervous system. Cell Tissue Res. 1997;290:1–10. doi: 10.1007/s004410050901. [DOI] [PubMed] [Google Scholar]
- 29.Joiner MA, Griffith LC. Mapping of the anatomical circuit of CaM kinase-dependent courtship conditioning in Drosophila. Learn Mem. 1999;6:177–192. [PMC free article] [PubMed] [Google Scholar]
- 30.Kiger JA, Jr, O'Shea C. Genetic evidence for a protein kinase A/cubitus interruptus complex that facilitates processing of cubitus interruptus in Drosophila. Genetics. 2001;158:1157–1166. doi: 10.1093/genetics/158.3.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kiger JA, Jr, Eklund JL, Younger SH, O'Kane CJ. Transgenic inhibitors identify two roles for protein kinase A in Drosophila development. Genetics. 1999;152:281–290. doi: 10.1093/genetics/152.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kraft R, Levine RB, Restifo LL. The steroid hormone 20-hydroxyecdysone enhances neurite growth of Drosophila mushroom body neurons isolated during metamorphosis. J Neurosci. 1998;18:8886–8899. doi: 10.1523/JNEUROSCI.18-21-08886.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lane ME, Kalderon D. Genetic investigation of cAMP-dependent protein kinase function in Drosophila development. Genes Dev. 1993;7:1229–1243. doi: 10.1101/gad.7.7a.1229. [DOI] [PubMed] [Google Scholar]
- 34.Lee T, Winter C, Marticke SS, Lee A, Luo L. Essential roles of Drosophila RhoA in the regulation of neuroblast proliferation and dendritic but not axonal morphogenesis. Neuron. 2000;25:307–316. doi: 10.1016/s0896-6273(00)80896-x. [DOI] [PubMed] [Google Scholar]
- 35.Li W, Ohlmeyer JT, Lane ME, Kalderon D. Function of protein kinase A in hedgehog signal transduction and Drosophila imaginal disc development. Cell. 1995;80:553–562. doi: 10.1016/0092-8674(95)90509-x. [DOI] [PubMed] [Google Scholar]
- 36.Manseau L, Baradaran A, Brower D, Budhu A, Elefant F, Phan H, Philp AV, Yang M, Glover D, Kaiser K, Palter K, Selleck S. GAL4 enhancer traps expressed in the embryo, larval brain, imaginal discs, and ovary of Drosophila. Dev Dyn. 1997;209:310–322. doi: 10.1002/(SICI)1097-0177(199707)209:3<310::AID-AJA6>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 37.Martin JR, Ernst R, Heisenberg M. Mushroom bodies suppress locomotor activity in Drosophila melanogaster. Learn Mem. 1998;5:179–191. [PMC free article] [PubMed] [Google Scholar]
- 38.Martin JR, Raabe T, Heisenberg M. Central complex substructures are required for the maintenance of locomotor activity in Drosophila melanogaster. J Comp Physiol [A] 1999;185:277–288. doi: 10.1007/s003590050387. [DOI] [PubMed] [Google Scholar]
- 39.McBride SM, Giuliani G, Choi C, Krause P, Correale D, Watson K, Baker G, Siwicki KK. Mushroom body ablation impairs short-term memory and long-term memory of courtship conditioning in Drosophila melanogaster. Neuron. 1999;24:967–977. doi: 10.1016/s0896-6273(00)81043-0. [DOI] [PubMed] [Google Scholar]
- 40.McGuire SE, Le PT, Davis RL. The role of Drosophila mushroom body signaling in olfactory memory. Science. 2001;293:1330–1333. doi: 10.1126/science.1062622. [DOI] [PubMed] [Google Scholar]
- 41.Moore MS, DeZazzo J, Luk AY, Tully T, Singh SM, Heberlein U. Ethanol intoxication in Drosophila: genetic and pharmacological evidence for regulation by the cAMP signaling pathway. Cell. 1998;93:997–1007. doi: 10.1016/s0092-8674(00)81205-2. [DOI] [PubMed] [Google Scholar]
- 42.Nassel DR. Neuropeptides in the insect brain: a review. Cell Tissue Res. 1993;273:1–29. doi: 10.1007/BF00304608. [DOI] [PubMed] [Google Scholar]
- 43.O'Dell KM, Armstrong JD, Yang MY, Kaiser K. Functional dissection of the Drosophila mushroom bodies by selective feminization of genetically defined subcompartments. Neuron. 1995;15:55–61. doi: 10.1016/0896-6273(95)90064-0. [DOI] [PubMed] [Google Scholar]
- 44.Park SK, Sedore SA, Cronmiller C, Hirsh J. PKA-RII-deficient Drosophila are viable but show developmental, circadian and drug response phenotypes. J Biol Chem. 2000;275:20588–20596. doi: 10.1074/jbc.M002460200. [DOI] [PubMed] [Google Scholar]
- 45.Parr J, Large A, Wang X, Fowler SC, Ratzlaff KL, Ruden DM. The inebri-actometer: a device for measuring the locomotor activity of Drosophila exposed to ethanol vapor. J Neurosci Methods. 2001;107:93–99. doi: 10.1016/s0165-0270(01)00357-0. [DOI] [PubMed] [Google Scholar]
- 46.Phillips TJ, Shen EH. Neurochemical bases of locomotion and ethanol stimulant effects. Int Rev Neurobiol. 1996;39:243–282. doi: 10.1016/s0074-7742(08)60669-8. [DOI] [PubMed] [Google Scholar]
- 47.Power ME. The thoracico-abdominal system of an adult insect, Drosophila melanogaster. J Comp Neurol. 1948;88:347–409. doi: 10.1002/cne.900880303. [DOI] [PubMed] [Google Scholar]
- 48.Prokop A, Technau GM. The origin of postembryonic neuroblasts in the ventral nerve cord of Drosophila melanogaster. Development. 1991;111:79–88. doi: 10.1242/dev.111.1.79. [DOI] [PubMed] [Google Scholar]
- 49.Prokop A, Technau GM. Normal function of the mushroom body defect gene of Drosophila is required for the regulation of the number and proliferation of neuroblasts. Dev Biol. 1994;161:321–327. doi: 10.1006/dbio.1994.1034. [DOI] [PubMed] [Google Scholar]
- 50.Rajashekhar KP, Singh RN. Neuroarchitecture of the tritocerebrum of Drosophila melanogaster. J Comp Neurol. 1994;349:633–645. doi: 10.1002/cne.903490410. [DOI] [PubMed] [Google Scholar]
- 51.Renn SCP, Armstrong JD, Yang M, Wang Z, An X, Kaiser K, Taghert PH. Genetic analysis of the Drosophila ellipsoid body neuropil: organization and development of the central complex. J Neurobiol. 1999;41:189–207. [PubMed] [Google Scholar]
- 52.Roman G, Davis RL. Molecular biology and anatomy of Drosophila olfactory associative learning. BioEssays. 2001;23:571–581. doi: 10.1002/bies.1083. [DOI] [PubMed] [Google Scholar]
- 53.Scholz H, Ramond J, Singh CM, Heberlein U. Functional ethanol tolerance in Drosophila. Neuron. 2000;28:261–271. doi: 10.1016/s0896-6273(00)00101-x. [DOI] [PubMed] [Google Scholar]
- 54.Shiga S, Toyoda I, Numata H. Neurons projecting to the retrocerebral complex of the adult blow fly, Protophormia terraenovae. Cell Tissue Res. 2000;299:427–439. doi: 10.1007/s004419900110. [DOI] [PubMed] [Google Scholar]
- 55.Siegmund T, Korge G. Innervation of the ring gland of Drosophila melanogaster. J Comp Neurol. 2001;431:481–491. doi: 10.1002/1096-9861(20010319)431:4<481::aid-cne1084>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 56.Singh CM, Heberlein U. Genetic control of acute ethanol-induced behaviors in Drosophila. Alcohol Clin Exp Res. 2000;24:1127–1136. [PubMed] [Google Scholar]
- 57.Tabakoff B, Hoffman PL. Adenylyl cyclases and alcohol. Adv Second Messenger Phosphoprotein Res. 1998;32:173–193. doi: 10.1016/s1040-7952(98)80011-6. [DOI] [PubMed] [Google Scholar]
- 58.Taylor SS, Buechler JA, Yonemoto W. cAMP-dependent protein kinase: framework for a diverse family of regulatory enzymes. Annu Rev Biochem. 1990;59:971–1005. doi: 10.1146/annurev.bi.59.070190.004543. [DOI] [PubMed] [Google Scholar]
- 59.Tettamanti M, Armstrong JD, Endo K, Yang MY, Furukubo-Tokunaga K, Kaiser K, Reichert H. Early development of the Drosophila mushroom bodies, brain centres for associative learning and memory. Dev Genes Evol. 1997;207:242–252. doi: 10.1007/s004270050112. [DOI] [PubMed] [Google Scholar]
- 60. Thiele TE, Willis B, Stadler J, Reynolds JG, Bernstein IL, McKnight GS. . High ethanol consumption and low sensitivity to ethanol-induced sedation in protein kinase A-mutant mice. J Neurosci 20 2000. RC75:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thomsen M. The neurosecretory system of the adult Calliphora erythrocephala. IV. A histological study of the corpus cardiacum and its connections with the nervous system. Z Zellforsch Mikrosk Anat. 1969;94:205–219. doi: 10.1007/BF00339357. [DOI] [PubMed] [Google Scholar]
- 62.Truman JW, Bate M. Spatial and temporal patterns of neurogenesis in the central nervous system of Drosophila melanogaster. Dev Biol. 1987;125:145–157. doi: 10.1016/0012-1606(88)90067-x. [DOI] [PubMed] [Google Scholar]
- 63.Waddell S, Armstrong JD, Kitamoto T, Kaiser K, Quinn WG. The amnesiac gene product is expressed in two neurons in the Drosophila brain that are critical for memory. Cell. 2000;103:805–813. doi: 10.1016/s0092-8674(00)00183-5. [DOI] [PubMed] [Google Scholar]
- 64.Wand G, Levine M, Zweifel L, Schwindinger W, Abel T. The cAMP-protein kinase A signal transduction pathway modulates ethanol consumption and sedative effects of ethanol. J Neurosci. 2001;21:5297–5303. doi: 10.1523/JNEUROSCI.21-14-05297.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weber KE. An apparatus for measurement of resistance to gas-phase reagents. Drosoph Inf Serv. 1988;67:91–93. [Google Scholar]
- 66.Yang MY, Armstrong JD, Vilinsky I, Strausfeld NJ, Kaiser K. Subdivision of the Drosophila mushroom bodies by enhancer-trap expression patterns. Neuron. 1995;15:45–54. doi: 10.1016/0896-6273(95)90063-2. [DOI] [PubMed] [Google Scholar]
- 67.Zars T, Fischer M, Schulz R, Heisenberg M. Localization of a short-term memory in Drosophila. Science. 2000a;288:672–675. doi: 10.1126/science.288.5466.672. [DOI] [PubMed] [Google Scholar]
- 68.Zars T, Wolf R, Davis R, Heisenberg M. Tissue-specific expression of a type I adenylyl cyclase rescues the rutabaga mutant memory defect: in search of the engram. Learn Mem. 2000b;7:18–31. doi: 10.1101/lm.7.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]