Abstract
Motoneurons are an essential component of all metazoan nervous systems, but it is unknown whether there is an evolutionarily conserved mechanism for generating motoneurons during neurogenesis. In the vertebrate CNS, HB9/MNR2 transcription factors are specifically expressed in all somatic motoneurons and are necessary to distinguish motoneurons from interneurons, in part by repressing interneuron-specific gene expression. Here, we identify and characterize the single Drosophila ortholog of the HB9/MNR2 gene family. Drosophila HB9 is detected in a subset of motoneurons with ventral muscle targets and in a small group of interneurons, including the well characterized serotonergic interneurons. RNA interference knockdown of HB9 levels leads to defects in motoneuron ventral muscle target recognition, ectopic expression of a marker for dorsally projecting motoneurons (Even-skipped), and defects in serotonergic interneuronal projections. Conversely, ectopic HB9 expression causes an expansion of ventral motoneuron projections and repression of Even-skipped. Thus,Drosophila HB9 is required in a subset of motoneurons and interneurons for establishing proper axon projections but does not have a general role in distinguishing motoneuron and interneuron cell types.
Keywords: motoneuron, interneuron, axonogenesis, HB9, MNR2, even-skipped, serotonergic
The CNS contains three primary cell types: motoneurons, interneurons, and glia. Drosophila genes expressed specifically in all motoneurons have not been described, although a growing number of genes are known to be expressed in subsets of motoneurons. The Even-skipped (Eve) homeodomain transcription factor is expressed in dorsally projecting motoneurons and a subset of interneurons; loss of function and misexpression experiments show that it is necessary and sufficient for dorsal axon projections in motoneurons (Landgraf et al., 1999). The Huckebein (Hkb) zinc finger transcription factor is expressed in a subset of dorsally and ventrally projecting motoneurons and a subset of interneurons; it is required for motoneuronal pathfinding and target recognition (Chu-LaGraff et al., 1995). Islet and Lim3 are LIM (lin-11, isl-1, mec-3) homeodomain transcription factors that are expressed in overlapping subsets of ventrally projecting motoneurons and a subset of interneurons, in which they regulate motoneuronal pathfinding and target recognition (Thor and Thomas, 1997; Thor et al., 1999). Together, these studies have led to the model that the motoneuron population consists of small groups of motoneurons that are each specified by a distinct “combinatorial code” of transcription factors (Thor et al., 1999). It is unknown whether any additional transcription factors promote a general motoneuron identity.
On the basis of recent vertebrate studies, the HB9/MNR2 gene family is a prime candidate for a general determinant of somatic motoneuron cell type. In vertebrates, chick MNR2 and mouse HB9 are expressed in presumptive somatic motoneuron progenitors. Mouse HB9 maintains expression in somatic motoneurons, whereas chick HB9 is expressed only in postmitotic somatic motoneurons. Together, vertebrate HB9/MNR2 transcription factors are expressed in all somatic motoneurons and excluded from interneurons. Chick MNR2 or HB9 misexpression in interneurons causes a decrease in interneuronal markers; in addition, chick HB9 misexpression causes an increase in motoneuron markers (Tanabe et al., 1998). Mice lacking HB9 have somatic motoneurons with a hybrid motoneuron/interneuron fate; they extend motoneurons into the muscle field but transiently express interneuronal markers (Arber et al., 1999; Thaler et al., 1999). Thus, vertebrate HB9/MNR2 genes are expressed specifically in somatic motoneurons and are essential for distinguishing motoneuron/interneuron cell types.
Here, we characterize Drosophila HB9, the sole fly ortholog of the HB9/MNR2 gene family. Drosophila HB9differs from its vertebrate orthologs in several ways: it is not expressed in all somatic motoneurons, it is expressed in a subset of interneurons, and it is required for the proper development of both interneurons and motoneurons.
MATERIALS AND METHODS
Cloning of Drosophila HB9. Basic local alignment search tool (BLAST) searches of the Drosophila genome with the entire coding region of chick HB9 and chick MNR2 protein sequences identified a single related Drosophila gene (CG8254). PCR was used to amplify a 751 nt genomic DNA fragment, which was used to screen a pNB40 cDNA library and obtain one full-length clone; conceptual translation of the cDNA yielded a protein identical to that predicted by the Drosophila genome sequence project.
Antibodies and immunocytochemistry. The peptide NH2-CEQEALQRIRDSREYDSPSPDGMSR-NH2was conjugated to BSA and injected into rabbits. A fusion protein containing HB9 amino acids 203–408 was expressed as a His-tagged protein, purified using nickel-coupled Sepharose beads, and injected into rats and rabbits. Antibody specificity was confirmed by reduced staining in HB9RNAi embryos and correspondence between mRNA and protein patterns. Embryos and larval brains were prepared using standard methods (Chu-LaGraff et al., 1995) and stained with the following primary antibodies and dilutions: rat anti-HB9, 1:1000; rabbit anti-HB9, 1:1000 (fluorescent staining) or 1:10,000 (histochemical staining); mouse anti-Tau, 1:40; mouse anti-Eve, 1:20 (C. Goodman, University of California Berkeley, Berkeley, CA); rat anti-reversed polarity (Repo), 1:1000 (Campbell et al., 1994); rabbit anti-β-galactosidase (β-gal), 1:1000 (Cappel, Cochranville, PA); mouse anti-β-gal, 1:500 (Promega, Madison, WI); rabbit anti-phosphohistone H3, 1:1000 (Upstate Biotechnology, Lake Placid, NY); and mouse anti-fasciclin II (FasII) (C. Goodman). Histochemical stainings were done with Vectastain kits (Vector Laboratories, Burlingame, CA), imaged on a Zeiss (Oberkochen, Germany) Axioplan with a digital color camera, and photomontages were generated in Photoshop (Adobe Systems, San Jose, CA). Fluorescent stainings were done by standard methods (Chu-LaGraff et al., 1995) and imaged on a Bio-Rad (Hercules, CA) Radiance confocal microscope.
RNA interference and misexpression of HB9. For RNA interference (RNAi), a 567 nt ClaI/EcoRI fragment of HB9 (not including the homeodomain) was cloned into pBlueScript SK, and double-stranded RNA was transcribed, concentrated, and injected as described previously (Sullivan et al., 1999). Embryos injected with buffer or another CNS-expressed gene (CG6218) showed no change in HB9 expression or motor axon projections (data not shown). Transgenic UAS-HB9 fly strains were generated using standard P element transformation methods. For each transgene, multiple independent lines were established. Scabrous-Gal4(Sca-Gal4) and a chromosome containing bothpatched-Gal4 and engrailed-Gal4(ptc,en-Gal4) (T. von Ohlen, unpublished results) were crossed independently to UAS-HB9(P1) flies, and collections were taken at 25 and 29°C, respectively. Both lines induce HB9 expression in neuroblasts and ganglion mother cells (GMCs) before endogenous HB9 expression.
RESULTS
Identification and expression of the Drosophila HB9 gene
We cloned a 2.2 kb cDNA (see Materials and Methods) that matches the predicted full-length transcript and intron/exon junctions of the conceptual gene CG8254, located at 66A on the third chromosome. The only conserved protein motif is a homeodomain closely related to the HB9/MNR2 homeodomain family (Fig. 1). Because there are no other Drosophila genes more similar to the HB9/MNR2 gene family, we named this gene Drosophila HB9.
Fig. 1.
Identification and expression of theDrosophila HB9 gene. A–C, HB9 protein is first detected in the gut (asterisk), followed by the brain (arrowhead) and ventral CNS. Left, Anterior. A, B, Lateral views of stage 10 (A) and stage 13 (B) embryos; C, ventral view of a stage 15 embryo.VNC, Ventral nerve cord. D, HB9 protein is detected only in bilateral clusters of ∼50 cells in the third instar larval brain; anterior is up. E, HB9 (red) is coexpressed with the mitotic marker phosphohistone H3 (PPH3; green) in one or two cells per hemisegment; ventral view is of a late stage 10 embryo; anterior is left; the ventral midline is thebottom of each panel. F, Comparison of percentage amino acid identity between HB9/MNR2 proteins in the homeodomain (HD, gray).d, Drosophila; m, mouse;c, chick; AmphiMnx, amphioxus HB9.
Expression of HB9 mRNA and protein is indistinguishable during embryogenesis (data not shown) and is restricted to the gut and CNS (Fig. 1). Endoderm expression is conserved from sea urchins (Bellomonte et al., 1998) to vertebrates (Harrison et al., 1994, 1999), and neuronal expression is conserved from amphioxus (Ferrier et al., 2001) to vertebrates (Harrison et al., 1994, 1999; Saha et al., 1997).Drosophila HB9 CNS expression begins during late stage 10 in ∼12 cells per hemisegment and increases to ∼30 cells per hemisegment at stage 15 and later (Fig. 1C). None of these cells express the glial marker Repo (data not shown), indicating that all HB9 cells are neurons or GMCs. The timing of expression of HB9 and the size of cells expressing HB9 indicate that cells are likely to be postmitotic. To confirm this, we performed a double-labeling of HB9 and phosphohistone H3, a marker for cells undergoing mitosis. We found one or two cells in stereotyped mediolateral positions that express phosphohistone H3 and express HB9 weakly (Fig. 1E); we conclude that HB9 can be expressed just before GMC division but is usually restricted to postmitotic neurons. HB9 is also detected in the third instar larval brain (Fig. 1D).
HB9 is expressed in a subset of ventrally projecting motoneurons
On the basis of vertebrate studies, we expected that HB9 would be expressed specifically in motoneurons. Thus, we double-labeled embryos for HB9 and various motoneuron markers, including Eve (labels all dorsally projecting motoneurons) (Landgraf et al., 1999; Thor et al., 1999), Islet-Tau:myc (labels all ventrally projecting motoneurons) (Landgraf et al., 1999; Thor et al., 1999), and Eagle (labels a subset of ventrally projecting motoneurons) (Isshiki et al., 2001). We found that HB9 was coexpressed widely with Islet in most ventrally projecting neurons, including the well characterized RP1, RP3, RP4, and RP5 motoneurons that project via segmental nerve B (SNb) and segmental nerve D (SNd) to ventral muscles 6, 7, 12, and 13 (Fig.2B,D,F). To help distinguish exactly which Islet+motoneurons are HB9+, we used alim3:lacZ transgene to identify all Islet-positive motoneurons that project via SNb (Thor et al., 1999). We find that HB9 is coexpressed with Lim3 and Islet in the four RP motoneurons, two lateral SNb-projecting motoneurons, and a single ventral neuron that has not been described previously (Fig.2B,D,F). HB9 is not expressed in all ventrally projecting motoneurons, however, because it is not detected in the Eagle+ GW motoneuron that projects to muscle 15 or the Eagle+ motoneuron derived from neuroblast 2–4 that projects via segmental nerve a (SNa) to muscle 8 (Schmid et al., 1999) (data not shown). In addition, HB9 is not expressed in any of the Eve+motoneurons that innervate dorsal muscle targets (Fig.2C,E). Thus, HB9 is not expressed in all motoneurons but rather is restricted to a small subset of motoneurons that includes those projecting via SNb and SNd to ventral muscle targets.
Fig. 2.
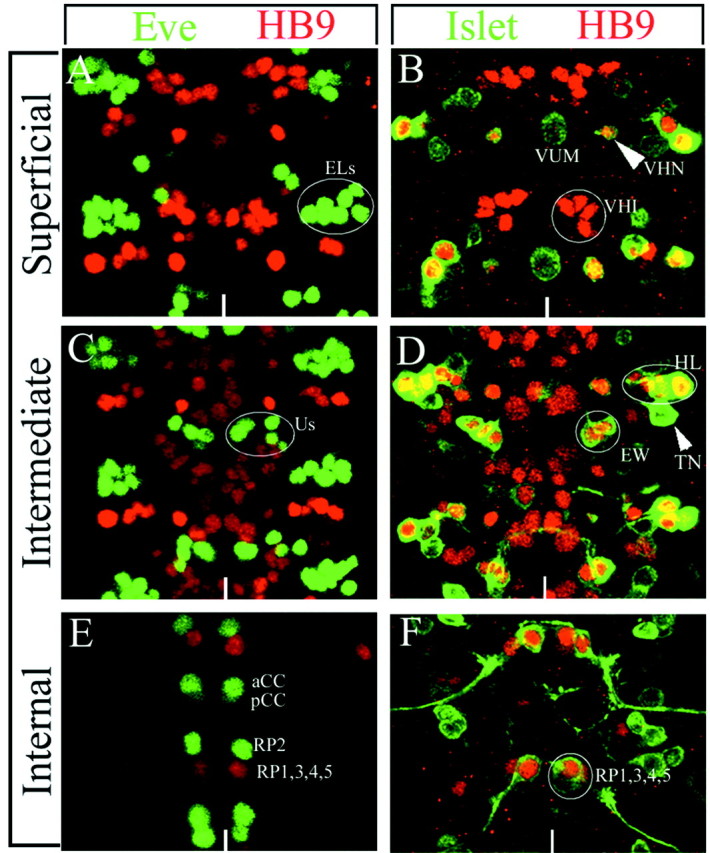
HB9 protein is detected in a subset of motoneurons and interneurons. Localization of HB9 (red) compared with Eve (green) or Islet-tau:myc (green, anti-Tau) is shown. Two segments from a late stage 16 CNS are shown as projections of the superficial layer (A, B), intermediate layer (C, D), or internal layer (E, F) of the CNS. Anterior isup; the ventral midline is indicated by the white line. A, C, E, HB9 is not detected in any Eve+ motoneurons (U1–5,aCC, RP2) or interneurons (ELs, pCC). B, HB9 is detected in six interneurons (VHI neurons) and a single Islet+ neuron (arrowhead, VHN) but not in the Islet+ VUM motoneuron.D, HB9 is detected in the Islet+serotonergic interneurons (EW) and a lateral group of motoneurons (HL) that overlaps partially with a lateral group of Islet+ motoneurons but not in the Islet+ transverse nerve motoneurons (arrowhead, TN). F, HB9 is detected in the Islet+ RP1/3/4/5 motoneurons. Us, U1-5.
HB9 is expressed in a subset of interneurons
To determine whether HB9 expression is limited to motoneurons like its counterparts in vertebrates, we assayed for expression in several identified interneurons. The best-characterized interneurons in the CNS are the serotonergic interneurons derived from neuroblast 7-3, for which there are numerous markers (Isshiki et al., 2001; Novotny et al., 2002). Using several of these markers in combination with HB9, we find that HB9 is expressed in the two serotonergic interneurons (EW1/EW2) and a third lineally related interneuron (EW3); each of these interneurons projects contralaterally across the posterior commissure (Fig. 2D). In addition, we assayed FasII, a transmembrane protein that labels all motoneurons and a few interneurons (Vactor et al., 1993), reasoning that any HB9+, FasII− cells would be interneurons. Only a ventral cluster of six HB9+ cells fails to express FasII (data not shown), indicating that these HB9+ cells are interneurons. In summary, HB9 is expressed in at least nine interneurons (three Islet+ Lim3−Eagle+ EW neurons and a ventral cluster of six Islet−Lim3−Eagle−FasII− neurons).
HB9 regulates motoneuronal gene expression
To determine the function of HB9 in ventrally projecting motoneurons, we used RNAi to knock down the levels of HB9. We found that >90% of injected embryos showed extremely low HB9 protein levels (Fig. 3B); thus, theseHB9RNAi embryos can be considered strong hypomorphs for HB9 expression. Control RNAi injections of buffer or other CNS genes showed no effect on HB9 expression (data not shown).
Fig. 3.
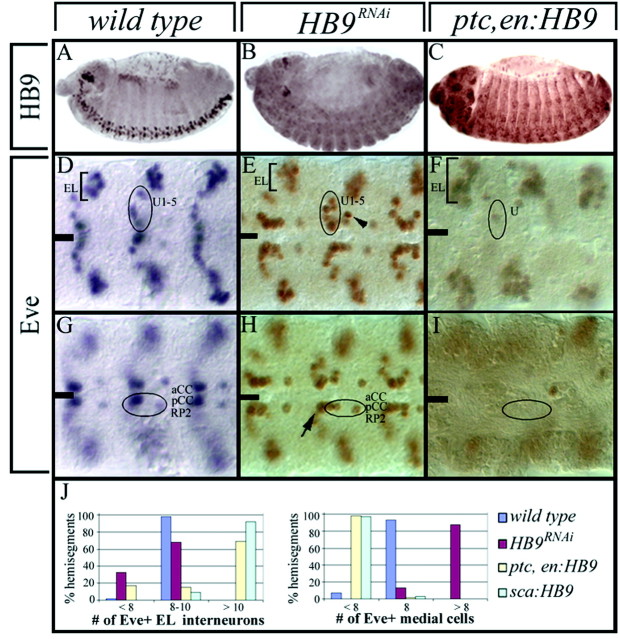
HB9 suppresses expression of the dorsal motoneuron determinant Eve. Wild-type,HB9RNAi, or HB9misexpression (ptc,en-Gal4/UAS-HB9 genotype) stage 16 embryos stained for HB9 (A–C) or Eve (D–I) are shown. Anterior isleft; the ventral midline is indicated by theblack line. A–C, Most HB9 protein is lost in HB9RNAi embryos, whereas all neurons, mesoderm, and epidermal cells express low levels of HB9 protein in HB9 misexpression embryos.D–F, Eve+ interneurons (ELs) are not affected significantly by loss of HB9 but are increased in number by HB9 misexpression. Eve+ U1–5 motoneurons have an extra Eve+ cell nearby (arrowhead) when HB9 is reduced but are almost completely missing after HB9 misexpression. G–I, Eve+ aCC/RP2 motoneurons and pCC interneuron have an extra Eve+ cell nearby (arrow) when HB9 is reduced but are almost completely missing after HB9 misexpression. J, Quantification of the Eve phenotype in wild-type,HB9RNAi, or two HB9misexpression genotypes in stage 16 embryos; Eve medial cells areaCC, pCC, RP2, and the U1–5 motoneurons.
We assayed HB9RNAi embryos for expression of the ventrally projecting motoneuron determinant Islet (Thor and Thomas, 1997) and the dorsally projecting motoneuron determinant Eve (Landgraf et al., 1999). We observed no change in the expression of the number of cells expressing the islet-tau:myc transgene (data not shown), despite strong coexpression of Islet and HB9 in a subset of ventrally projecting motoneurons (Fig. 2D,F). In contrast, HB9RNAi embryos show a derepression of the Eve dorsal motoneuron determinant: there are consistently two ectopic Eve+neurons, one located adjacent to the Eve+dorsally projecting aCC motoneuron (Fig. 3H,arrow) and the other located near the Eve+ dorsally projecting U1–5 motoneurons (Fig. 3E, arrowhead). We reasoned that if loss of HB9 leads to ectopic Eve expression, then perhaps misexpression of HB9 would inhibit Eve expression. We used the Gal4/UAS system (Brand and Perrimon, 1993) to misexpress HB9, using eitherptc,en-Gal4 (which drives HB9 expression in all neuroectoderm and newly formed neuroblasts) or sca-Gal4(which drives HB9 expression in all newly formed neuroblasts and the initial GMCs and neurons in their lineages). Both drivers result in precocious and ubiquitous expression of HB9 throughout the CNS, and both result in a nearly complete elimination of Eve expression in all dorsally projecting motoneurons (aCC, RP2, U1–5) and the pCC intersegmental interneuron (Fig. 3F,I,J). Interestingly, there is an increase in the number of Eve+ EL local interneurons, showing that the inhibition of Eve expression is not a general effect but depends on the cell type in which HB9 is expressed (Fig.3F,I,J). There is no change in Islet expression after misexpression of HB9 (data not shown). We conclude that HB9 restricts Eve expression to a specific subset of motoneurons, in which Eve promotes innervation of dorsal muscle targets (Landgraf et al., 1999). However, we find no role for HB9 in regulating islet gene expression in ventrally projecting motoneurons.
HB9 promotes motoneuron ventral projections and inhibits dorsal projections
HB9 is expressed in ventrally projecting motoneurons, so we assayed HB9RNAi embryos for defects in ventral motoneuron axon outgrowth, pathfinding, and muscle target recognition. We focused on the four HB9+RP1/3/4/5 motoneurons, which project out of the CNS via the SNb to form three distinct synaptic endings between the ventral muscles 7, 6, 13, and 12. In wild-type embryos, we observe three well defined synaptic endings at the 7/6, 6/13, and 13/12 muscle clefts in 90% of the hemisegments assayed (n = 71) (Fig.4A). InHB9RNAi embryos, general CNS morphology is normal (Fig. 4G,H); axons project out of the SNb nerve at approximately the normal time and terminate within the appropriate ventral muscle field but fail to establish synaptic endings at the 7/6, 6/13, and 13/12 muscle clefts in 65% of the hemisegments assayed (n = 72). In the affected segments, axons have expanded growth cones and terminate in an abnormally broad pattern (Fig. 4B, brackets). In addition to defects in ventral muscle target recognition, we observe a slight increase in the thickness of the intersegmental nerve (ISN) innervating dorsal muscles (Fig. 4B), which may reflect ectopic dorsal motor projections from the two extra Eve+ neurons (Fig.3E,H), because it is known that ectopic Eve induces dorsal axon projections (Landgraf et al., 1999). Thus, HB9 is required for ventral muscle target recognition by the well characterized RP motoneurons.
Fig. 4.
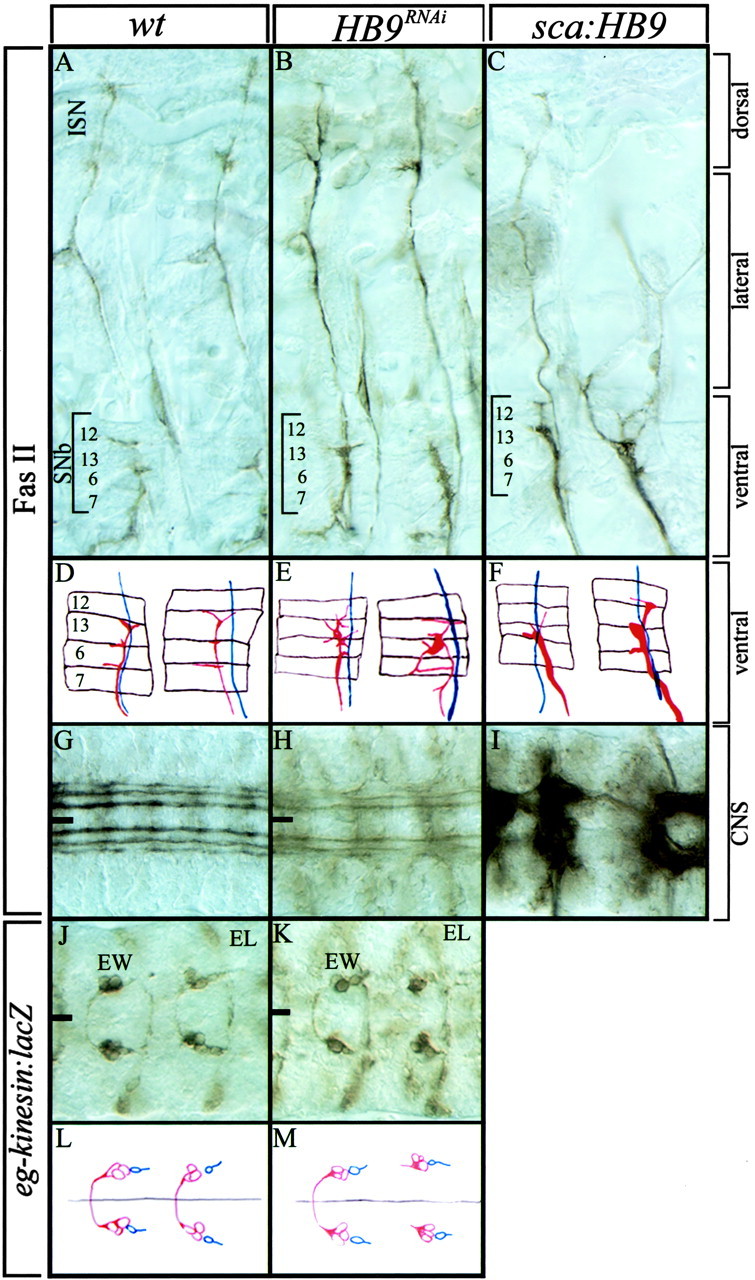
HB9 promotes motoneuron ventral projections and inhibits motoneuron dorsal projections. Wild-type,HB9RNAi, or HB9misexpression (sca-Gal4/UAS-HB9 genotype) early stage 17 embryos stained for FasII (A–I) or β-gal (eagle-kinesin:lacZ genotype) (J–M) are shown. Anterior isleft; the ventral midline is indicated by theblack line; images are a montage of multiple focal planes (A–C, J–K), single focal planes (G–I), or camera lucida tracings (D–F: red, SNb; blue, ISN; brown, muscles 12, 13, 6, and 7 fromtop to bottom) (L–M:red, EW1–EW3 interneurons; blue, GW motoneuron). A–C, Dorsal, lateral, and ventral musculature are indicated with brackets. See Results for details.
To determine whether misexpression of HB9 leads to an increase in ventral muscle target innervation at the expense of dorsal muscle projections (the opposite of the HB9RNAiphenotype), we used scabrous-Gal4 to misexpress HB9 throughout the CNS (but not muscles) and scored motoneuron projections with FasII. Indeed, we observed a striking increase in the thickness of the SNb projections to ventral muscles, with many hemisegments showing the normal synaptic endings in three muscle clefts (43%;n = 58) (Fig. 4C,F). We also observed a clear decrease in the number of motoneuron projections to dorsal muscles (48%; n = 59 hemisegments) (Fig.4C). The loss of dorsal projections is a result of multiple defects, including stalling of the ISN in the ventral muscle field or ISN neurons forming apparent synapses on the more ventral transverse nerve, but we never observed the ISN and the SNb fasciculating together. We conclude that HB9 promotes the targeting of motoneuron projections to ventral muscles and inhibits motoneuronal projections to dorsal muscle targets.
HB9 regulates interneuron axon projections
Drosophila HB9, unlike vertebrate HB9/MNR2 genes, is expressed in a subset of interneurons. To determine whether HB9 plays a role in interneuronal axon targeting similar to its role in motoneurons, we used the eagle-kinesin:lacZ transgene as an axonal marker for the HB9+ EW1/EW2 serotonergic interneurons and the lineally related EW3 interneuron. In wild-type embryos, the EW1–EW3 axons are fasciculated tightly, project anteriorly, turn medially to cross the midline, and synapse within the contralateral neuropil (95%; n = 58 hemisegments) (Fig. 4J,L). In HB9RNAiembryos, the EW1–EW3 neurons project anteriorly as usual but are occasionally defasciculated and fail to cross the midline (26%;n = 61 hemisegments) (Fig.4K,M). No change was observed in the contralateral projections of other interneurons (e.g., the HB9− EL interneurons) (Fig.4J,K), suggesting that EW1–EW3 axon pathfinding defects are not caused indirectly by midline defects. We conclude that HB9 is required in EW1–EW3 interneurons to promote contralateral axon projections. We also find that misexpression of HB9 causes many general defects in the CNS, some of which, such as broken longitudinal connectives (Fig. 4I), can be attributed to interneuronal defects.
DISCUSSION
HB9 function in motoneurons
HB9 is expressed in a subset of ventrally projecting motoneurons but not in their target muscles, and HB9 is required for proper muscle target recognition by these motoneurons. It is possible that the HB9 phenotype is caused by loss of Islet or Lim3 expression, because both Islet and Lim3 are required to establish normal ventral axon projections (Thor and Thomas, 1997; Thor et al., 1999), and HB9 expression overlaps with Islet and Lim3 (Fig. 2; data not shown). However, the HB9 loss and gain of function has no effect on Islet expression (data not shown), and the HB9 and lim3phenotypes are clearly different (lim3 mutants show a rerouting of SNb motoneurons into the SNd, which we do not observe inHB9RNAi embryos). Instead, we favor a model in which HB9, Islet, and Lim3 have independent functions in establishing RP motoneuronal axon projections. lim3 mutants have pathfinding defects in which SNb motoneurons are diverted into the SNd nerve and terminate outside their normal muscle field (Thor and Thomas, 1997; Thor et al., 1999); islet mutants also show pathfinding and fasciculation defects, in addition to target recognition defects (Thor and Thomas, 1997; Thor et al., 1999).HB9 RNAi causes target recognition defects but not fasciculation defects or pathfinding defects.
Misexpression of HB9 is sufficient to promote thickening of ventral motor projections and reduced innervation of dorsal muscles. This phenotype could arise in several ways. First, HB9 could transform interneurons into ventrally projecting motoneurons. We think that this is unlikely, because we have seen no transformation ofapterous- or islet-expressing interneurons into motoneurons after HB9 misexpression (data not shown); moreover, HB9 is normally expressed in some interneurons without turning them into motoneurons. Second, HB9 could transform dorsal motoneurons into ventral motoneurons. This seems unlikely, because dorsal motoneurons are clearly extending in the ISN, and although it is truncated, it is not fused with the SNb nerve. Third, the SNb motoneurons could be slightly defasciculated, which would make them appear thicker than normal. Transmission electron microscopy to count axons in the SNb would be necessary to test this model. Finally, we favor the model that HB9 induces SNa motoneurons to fasciculate into the SNb nerve root and innervate the SNb muscle target field, because misexpression of HB9 leads to the loss of the SNa nerve root in parallel with the thickening of the SNb nerve root (data not shown). The SNa normally extends past the SNb target muscles en route to its own target muscles, so misexpression of HB9 might lead to precocious termination at the SNb target muscles.
HB9 expression and function in interneurons
HB9 is expressed in the serotonergic EW1 and EW2 interneurons and a third lineally related EW3 interneuron and is required to establish their normal contralateral projections. We suspect that HB9 is acting autonomously in these interneurons rather than leading to defects at the midline that block contralateral projections, because HB9 is not expressed in midline cells of the CNS, and other interneurons (ELs) and motoneurons (RPs) show normal contralateral projections. The generation of hb9 mutant clones in the serotonergic interneurons will be necessary to distinguish between a cell autonomous or cell nonautonomous function of HB9 in regulating interneuronal axon projections. Interestingly, many transcription factors known to regulate motoneuron development, including Eve, Islet, Lim3, and Hkb, are also expressed in a subset of interneurons.islet and hkb are both expressed in the serotonergic interneurons, and each is required for proper axon pathfinding and neurotransmitter synthesis (Lundell et al., 1996; Thor and Thomas, 1997); lim3 is required for axon pathfinding of a different subset of interneurons (Thor and Thomas, 1997;Thor et al., 1999). An open question is how interneurons can maintain expression of motoneuronal determinants such as HB9, Islet, Lim3, and Eve without fasciculating with motoneurons or exiting the CNS.
The expression and function of HB9 in interneurons is unexpected, because all known vertebrate HB9/MNR2 genes are expressed only in motoneurons within the CNS. Perhaps there are small groups of HB9/MNR2+ interneurons in vertebrates that have evaded detection; alternatively, Drosophila may have co-opted an ancestral motoneuronal determinant, HB9, for a parallel function in interneurons. Detailed analysis of HB9 expression patterns in additional organisms will be necessary to resolve this question.
Temporal differences of cell fate decisions in flies and vertebrates
We have shown that Drosophila HB9 is expressed primarily in postmitotic neurons, whereas vertebrate HB9 family members are expressed in progenitor cells and in differentiated cells (Tanabe et al., 1998; Arber et al., 1999; Thaler et al., 1999). This difference in the timing of HB9 expression may reflect differences in the timing of motoneuron cell fate commitment in each organism. In mouse and chick embryos, a domain of ventral spinal cord neural precursors goes through a phase in which they generate only motoneurons; during this period, the precursors express HB9. In Drosophila, there are no known precursors that generate solely motoneurons; virtually every motoneuron derives from a terminal cell division that produces one motoneuron and a non-motoneuron sibling cell (Schmid et al., 1999). Thus, it is not surprising that Drosophila motoneuron determinants either are restricted to postmitotic cells (Islet and HB9) or are expressed just before the terminal division but downregulated rapidly in the non-motoneuronal sibling (Eve).
Evolution of motoneuron subtypes
The expression of Drosophila HB9 may help illuminate how Drosophila motoneurons are related evolutionarily to vertebrate motoneurons. It has been proposed that Drosophilamotoneurons fall into two nonoverlapping groups: Islet+ motoneurons that are homologous to vertebrate somatic and visceral motoneurons (Thor and Thomas, 1997) and Eve+ motoneurons that have no vertebrate motoneuron counterpart (Landgraf et al., 1999). TheDrosophila Islet+ motoneurons contain somatic motoneurons projecting to ventral muscle targets and a visceral TMNp motoneuron that projects to the heart (Gorczyca et al., 1994; Thor and Thomas, 1997); the Eve+ motoneurons project to dorsal muscles (Landgraf et al., 1999). Drosophila HB9 expression provides additional support for this model. Drosophila HB9 is expressed in many Islet+ somatic motoneurons but not in the Islet+ TMNp visceral motoneuron, similar to the observed restriction of vertebrate HB9/MNR2 expression to somatic motor neurons but not visceral motor neurons. Our data are consistent with a model in whichDrosophila Islet+HB9+ somatic motoneurons are homologous to vertebrate Islet+HB9+ somatic motoneurons, theDrosophila Islet+ TMNp visceral motoneuron is homologous to vertebrate Islet+ visceral motoneurons, and theDrosophila Eve+ motoneurons have no vertebrate motoneuron counterpart. We note, however, that murine Even-skipped (Evx1/2) CNS expression is restricted to the locally projecting commissural V0 interneurons (Moran-Rivard et al., 2001). Perhaps this class of vertebrate interneurons has unrecognized similarities to Drosophila Eve+motoneurons and/or interneurons.
Note added in proof. A recent article describingDrosophila HB9 expression and mutant phenotype (Broihier and Skeath, 2002) matches the expression and RNAi phenotype described here.
Footnotes
This work was supported by a National Institutes of Health (NIH) Genetics Training Grant (J.O.) and NIH Grant HD-27056 (C.Q.D.). We thank Alice Schmid and Takako Isshiki for technical assistance; S. Thor, S. Certel, and C. Goodman for fly stocks and antibodies; and the Kai Zinn laboratory, the Judith Eisen laboratory, and current members of the Doe laboratory for helpful comments.
Correspondence should be addressed to Chris Q. Doe, Institute of Neuroscience, Institute of Molecular Biology, Howard Hughes Medical Institute, 1254 University of Oregon, Eugene OR 97403-1254. E-mail:cdoe@uoneuro.uoregon.edu.
REFERENCES
- 1.Arber S, Han B, Mendelsohn M, Smith M, Jessell TM, Sockanathan S. Requirement for the homeobox gene Hb9 in the consolidation of motor neuron identity. Neuron. 1999;23:659–674. doi: 10.1016/s0896-6273(01)80026-x. [DOI] [PubMed] [Google Scholar]
- 2.Bellomonte D, Di Bernardo M, Russo R, Caronia G, Spinelli G. Highly restricted expression at the ectoderm-endoderm boundary of PIHbox 9, a sea urchin homeobox gene related to the human HB9 gene. Mech Dev. 1998;74:185–188. doi: 10.1016/s0925-4773(98)00064-1. [DOI] [PubMed] [Google Scholar]
- 3.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 4.Broihier HT, Skeath JB. Drosophila homeodomain protein dHb9 directs neuronal fate via crossrepressive and cell-nonautonomous mechanisms. Neuron. 2002;35:39–50. doi: 10.1016/s0896-6273(02)00743-2. [DOI] [PubMed] [Google Scholar]
- 5.Campbell G, Goring H, Lin T, Spana E, Andersson S, Doe CQ, Tomlinson A. RK2, a glial-specific homeodomain protein required for embryonic nerve cord condensation and viability in Drosophila. Development. 1994;120:2957–2966. doi: 10.1242/dev.120.10.2957. [DOI] [PubMed] [Google Scholar]
- 6.Chu-LaGraff Q, Schmid A, Leidel J, Bronner G, Jackle H, Doe CQ. huckebein specifies aspects of CNS precursor identity required for motoneuron axon pathfinding. Neuron. 1995;15:1041–1051. doi: 10.1016/0896-6273(95)90093-4. [DOI] [PubMed] [Google Scholar]
- 7.Ferrier DE, Brooke NM, Panopoulou G, Holland PW. The Mnx homeobox gene class defined by HB9, MNR2 and amphioxus AmphiMnx. Dev Genes Evol. 2001;211:103–107. doi: 10.1007/s004270000124. [DOI] [PubMed] [Google Scholar]
- 8.Gorczyca MG, Phillis RW, Budnik V. The role of tinman, a mesodermal cell fate gene, in axon pathfinding during the development of the transverse nerve in Drosophila. Development. 1994;120:2143–2152. doi: 10.1242/dev.120.8.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harrison KA, Druey KM, Deguchi Y, Tuscano JM, Kehrl JH. A novel human homeobox gene distantly related to proboscipedia is expressed in lymphoid and pancreatic tissues. J Biol Chem. 1994;269:19968–19975. [PubMed] [Google Scholar]
- 10.Harrison KA, Thaler J, Pfaff SL, Gu H, Kehrl JH. Pancreas dorsal lobe agenesis and abnormal islets of Langerhans in Hlxb9-deficient mice. Nat Genet. 1999;23:71–75. doi: 10.1038/12674. [DOI] [PubMed] [Google Scholar]
- 11.Isshiki T, Pearson B, Holbrook S, Doe CQ. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell. 2001;106:511–521. doi: 10.1016/s0092-8674(01)00465-2. [DOI] [PubMed] [Google Scholar]
- 12.Landgraf M, Roy S, Prokop A, VijayRaghavan K, Bate M. even-skipped determines the dorsal growth of motor axons in Drosophila. Neuron. 1999;22:43–52. doi: 10.1016/s0896-6273(00)80677-7. [DOI] [PubMed] [Google Scholar]
- 13.Lundell MJ, Chu-LaGraff Q, Doe CQ, Hirsh J. The engrailed and huckebein genes are essential for development of serotonin neurons in the Drosophila CNS. Mol Cell Neurosci. 1996;7:46–61. doi: 10.1006/mcne.1996.0004. [DOI] [PubMed] [Google Scholar]
- 14.Moran-Rivard L, Kagawa T, Saueressig H, Gross MK, Burrill J, Goulding M. Evx1 is a postmitotic determinant of v0 interneuron identity in the spinal cord. Neuron. 2001;29:385–399. doi: 10.1016/s0896-6273(01)00213-6. [DOI] [PubMed] [Google Scholar]
- 15.Novotny T, Eiselt R, Urban J. Hunchback is required for the specification of the early sublineage of neuroblast 7–3 in the Drosophila central nervous system. Development. 2002;129:1027–1036. doi: 10.1242/dev.129.4.1027. [DOI] [PubMed] [Google Scholar]
- 16.Saha MS, Miles RR, Grainger RM. Dorsal-ventral patterning during neural induction in Xenopus: assessment of spinal cord regionalization with xHB9, a marker for the motor neuron region. Dev Biol. 1997;187:209–223. doi: 10.1006/dbio.1997.8625. [DOI] [PubMed] [Google Scholar]
- 17.Schmid A, Chiba A, Doe CQ. Clonal analysis of Drosophila embryonic neuroblasts: neural cell types, axon projections and muscle targets. Development. 1999;126:4653–4689. doi: 10.1242/dev.126.21.4653. [DOI] [PubMed] [Google Scholar]
- 18.Sullivan W, Ashburner M, Hawley SR. Drosophila protocols. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1999. [Google Scholar]
- 19.Tanabe Y, William C, Jessell TM. Specification of motor neuron identity by the MNR2 homeodomain protein. Cell. 1998;95:67–80. doi: 10.1016/s0092-8674(00)81783-3. [DOI] [PubMed] [Google Scholar]
- 20.Thaler J, Harrison K, Sharma K, Lettieri K, Kehrl J, Pfaff SL. Active suppression of interneuron programs within developing motor neurons revealed by analysis of homeodomain factor HB9. Neuron. 1999;23:675–687. doi: 10.1016/s0896-6273(01)80027-1. [DOI] [PubMed] [Google Scholar]
- 21.Thor S, Thomas JB. The Drosophila islet gene governs axon pathfinding and neurotransmitter identity. Neuron. 1997;18:397–409. doi: 10.1016/s0896-6273(00)81241-6. [DOI] [PubMed] [Google Scholar]
- 22.Thor S, Andersson SG, Tomlinson A, Thomas JB. A LIM-homeodomain combinatorial code for motor-neuron pathway selection. Nature. 1999;397:76–80. doi: 10.1038/16275. [DOI] [PubMed] [Google Scholar]
- 23.Vactor DV, Sink H, Fambrough D, Tsoo R, Goodman CS. Genes that control neuromuscular specificity in Drosophila. Cell. 1993;73:1137–1153. doi: 10.1016/0092-8674(93)90643-5. [DOI] [PubMed] [Google Scholar]