Abstract
In Drosophila, a number of key processes such as emergence from the pupal case, locomotor activity, feeding, olfaction, and aspects of mating behavior are under circadian regulation. Although we have a basic understanding of how the molecular oscillations take place, a clear link between gene regulation and downstream biological processes is still missing. To identify clock-controlled output genes, we have used an oligonucleotide-based high-density array that interrogates gene expression changes on a whole genome level. We found genes regulating various physiological processes to be under circadian transcriptional regulation, ranging from protein stability and degradation, signal transduction, heme metabolism, detoxification, and immunity. By comparing rhythmically expressed genes in the fly head and body, we found that the clock has adapted its output functions to the needs of each particular tissue, implying that tissue-specific regulation is superimposed on clock control of gene expression. Finally, taking full advantage of the fly as a model system, we have identified and characterized a cycling potassium channel protein as a key step in linking the transcriptional feedback loop to rhythmic locomotor behavior.
Keywords: oligonucleotide arrays, Drosophila, circadian outputs, slowpoke, locomotor behavior, gating
Circadian behaviors take place at regular intervals because of the action of a cell autonomous clock, which marks time even in the absence of environmental information. This molecular clock relies on negative feedback loops, in which transactivators drive the transcription of repressor proteins, which in turn block transcription at their own promoters (Young and Kay, 2001). It is important to note that this oscillation at the mRNA level is only the first step toward sustainable molecular rhythms, which are accomplished by introducing a number of “delays” affecting message stability (Suri et al., 1998) and protein stability (Price et al., 1998) or controlling subcellular localization (Saez and Young, 1996).
Although we have a relatively good understanding of how these molecular oscillations are generated in Drosophila, a clear link between the oscillator and the downstream biological processes under clock control is still missing. Previous attempts to describe the extent of circadian transcriptional regulation have identified a handful of clock-controlled “output” genes. A cDNA library screen identified ∼20 oscillating mRNAs, most of them with unknown functions (Van Gelder et al., 1995). Alternative approaches included the screening of subtractive cDNA libraries, which retrievedcircadian regulated gene-1 (Rouyer et al., 1997) andtakeout (Sarov-Blat et al., 2000) differential display, which identified vrille (vri) (Blau and Young, 1999), and luminescence-based enhancer trap screens that uncovered additional clock-controlled genes (ccgs), among thosenumb and a putative nicotinamide adenine dinucleotide kinase (Stempfl et al., 2002) and regular (Scully et al., 2002).
To carry out a more global examination of clock-controlled transcription, we used high-density oligonucleotide-based arrays. This approach has been used successfully in Arabidopsis and mammals, uncovering an astonishing range of physiological processes under circadian control (Harmer et al., 2000; Panda et al., 2002;Storch et al., 2002). Recently three groups used a similar strategy to identify the extent of clock-controlled transcription in flies.McDonald and Rosbash (2001) described 134 genes that oscillate under free-running [constant dark (DD)] conditions and display altered levels in the Clkjrk mutant background. Claridge-Chang et al. (2001), on the other hand, analyzed cycling profiles under driven [light/dark (LD) cycles] and free-running (DD) conditions in different genetic backgrounds. They identified 158 cycling genes, some of which are involved in learning and memory, vision, olfaction, locomotion, detoxification, and metabolism. Ueda et al. (2002) reported a very striking observation. Of 712 genes cycling in the fly head in LD, only 115 were also cycling in DD, and conversely, they identified 341 genes that were considered cycling only in DD.
To gain deeper insight into clock-controlled processes in the whole fly, we probed high-density arrays with RNA isolated from both head and body fractions. We found that although the number of cycling genes is similar between the two samples, only a small proportion of genes cycles in both structures, indicating that the clock controls different aspects of physiology throughout the organism. As is the case in mammals, we identified a large group of genes that although expressed to mid-high levels in both fractions are cycling in only one of them. Finally, taking advantage of the fly as a model genomics organism, we have characterized a rhythmic potassium channel as a key step in linking the transcriptional feedback loop to rhythmic locomotor behavior.
MATERIALS AND METHODS
Fly stocks. Time courses for the DNA chipping experiments were performed using y w as wild-type control and y w;;Clkjrk(Allada et al., 1998) as the mutant with impaired clock function. Theslo 4 (Atkinson et al., 1991) stock was kindly provided by Dr. N. Atkinson (University of Texas, Austin, TX). The stI slo I stock (Elkins et al., 1986) was a gift from the Bloomington Stock Center. slo I was generated by ethyl methane sulfonate mutagenesis and slo 4 was generated by gamma irradiation; the latter is a result of a chromosome inversion with one breakpoint within this gene (Atkinson et al., 2000).slo 4 mutants have been shown to carry a null allele by a number of approaches, and it fails to complement the slo I mutation in electrophysiological studies in the dorsal longitudinal flight muscle (Atkinson et al., 1991). No molecular characterization of the slo I mutant is available.
RNA and protein time courses. Newly eclosed wild-type (y w) or mutant (y w;;Clkjrk) flies were entrained for 5 d to a 12 hr LD regime under constant temperature (25°C). Time courses were performed under either LD or DD conditions. Samples were collected every 4 hr for 2 consecutive days and immediately frozen in dry ice. Frozen flies were shaken and sieved to generate two fractions: head (without antennas) and body (decapitated, and without legs or wings). Heads and bodies were kept apart for RNA/protein extraction. For the DD time courses, flies were entrained to LD cycles during 5 d and then released into constant darkness; samples were taken during the first and second day in DD.
RNA extraction and hybridization. Total RNA was prepared from fly head or body homogenized in Trizol reagent (Invitrogen). RNA was purified using RNeasy kit (Qiagen). Total RNA (7.5 μg) was labeled for hybridization according to manufacturer's instructions (Affymetrix). For Northern analyses, 25 μg of total RNA was loaded into a 1.2% formaldehyde agarose gel and transferred to nylon (Hybond N, Amersham). expressed sequence tags used as probes were obtained from ResGen (Invitrogen) and correspond to GH27053 (CT38753), LP 11415 (CT8171), AT29985 (CT18196), GH13172 (CT33647), and LD06553 (CT12127).
DNA chip experiments. cDNA synthesis, biotin labeling of cRNA, and hybridization of the chips were performed as recommended by the manufacturer (Affymetrix). Cel to text file condensation was performed using the MAS 4 algorithm in the Microarray Suite v.4 (Affymetrix) and visualized using the GeneSpring (Silicon Genetics) software package. To identify clock-controlled genes, we used a statistical program, COSOPT, which we have described in detail elsewhere (Harmer et al., 2000; Panda et al., 2002). Finally, we have constructed a publicly accessible databasehttp://expression.gnf.org/circadian, where users can query for circadian-regulated genes in the fly head and body.
Locomotor behavior analysis. Newly eclosed flies were entrained to 12 hr LD cycles for 3 d, and adult males were placed in glass tubes and monitored for activity with infrared detectors and a computerized data collection system (Trikinetics) (Hamblen-Coyle et al., 1992). Activity was monitored in LD conditions for 3–4 d, when the flies were released into DD at least for 1 week. Data were analyzed using the Clocklab software package (Actimetrics, Evanston, IL) as inYang and Sehgal (2001). Only those flies that were alive 2 d after analysis were taken into account. Periodogram analysis of flies that were scored as arrhythmic in Table 4 produced no strong peak that was statistically significant (p < 0.05). Those flies that showed recognizable daily onsets and offsets of activity that was not consolidated, thus resulting in periodograms that displayed weakly significant periods, were classified as weakly rhythmic (Yang and Sehgal, 2001) in Table 4 and were not taken into account for average period calculations. Overall activity was calculated by averaging equivalent bins during LD or DD cycles for each fly and then taking an average of all the flies within each genotype.
Table 4.
A null mutation in the slowpoke potassium channel does not dramatically change overall activity levels
| Rhythmicity (%) LD | DD | DD period (analyzed) | Activity (total counts per day) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| R | WR | AR | R | WR | AR | LD | DD | ||
| CS (70) | 92.9 | 5.7 | 1.4 | 75.7 | 8.6 | 15.7 | 23.58 (51) | 940 | 1055 |
| y w(72) | 87.5 | 9.7 | 2.8 | 87.5 | 12.5 | 0.0 | 23.46 (75) | 633 | 863 |
| slo I (59) | 39.0 | 27.1 | 33.9 | 55.9 | 25.4 | 18.6 | 24.19 (33) | 874 | 795 |
| slo 4 (78) | 6.4 | 24.4 | 69.2 | 0.0 | 29.5 | 70.5 | 791 | 665 | |
| slo 4/slo I (81) | 34.6 | 30.9 | 34.6 | 23.5 | 29.6 | 46.9 | 23.10 (19) | 1152 | 731 |
| Clkjrk(46) | 30.4 | 30.4 | 39.1 | 8.7 | 39.1 | 52.2 | 24.56 (3) | 1076 | 1491 |
| per 0 (35) | 60.0 | 31.4 | 8.6 | 0.0 | 37.1 | 62.9 | 695 | 1141 | |
Number of flies analyzed within each genotype is expressed in parentheses. Actograms were analyzed as described in Materials and Methods. R, WR, and AR stand for strongly rhythmic, weakly rhythmic and arrhythmic flies.
Western blots. Time courses were performed as described. Approximately 10 μl of wild-type and mutant fly heads was loaded into precast 4–15% acrylamide gels (Bio-Rad). Gels were transferred in 15% methanol Tris-glycine buffer onto Nitrobind-supported nitrocellulose (Osmonics). Primary antibodies used were rabbit IgG anti-SLO (1:500) and mouse monoclonal IgG anti-hsp70 (1:2000; Sigma). Washes were performed in TBS (for SLO) or TBS 0.5% Tween (for hsp70). Appropriate secondary antibodies were detected using ECF (Amersham Biosciences).
RESULTS
Clock-controlled transcription throughout the day
We monitored steady-state RNA levels using DNA GeneChips (Affymetrix) spanning the entire Drosophila genome. Temporal profiling of 13,500 probe sets was performed with a 4 hr time resolution during 2 consecutive days under entrained and free-running conditions. Each sample was hybridized to two DNA GeneChips to test the reproducibility of the technique (Harmer et al., 2000). The mean hybridization signal strength and the SEM for each probe set was calculated from the duplicate hybridizations.
To identify the cyclic mRNAs among the pool of expressed genes, we used COSOPT, an analytical algorithm that we developed for detection and statistical characterization of rhythmic gene expression in gene array experiments. Briefly, data were fit to 100,000 cosine test waves, and the significance of each fit was then determined empirically by temporally randomizing the data sets; those genes with traces that fit a cosine wave with a period between 20 and 28 hr were scored as cycling with a given probability (Panda et al., 2002). In the LD y whead time course, COSOPT scored 120 genes as cycling withp < 0.01 (Table 1). This may be an underestimation of the total number of cycling genes because previously characterized clock components such as per (which cycles with p = 0.015) and Clk(p = 0.036) fall outside of this category. We chose a conservative p value for selecting clock-controlled genes for further study. Storch et al. (2002) used a similar criterion to minimize the overall number of genes in the array selected as cycling while maximizing the number of “guide genes” (known ccgs) present, which further validates our method. Internal validation of this approach toward the identification of novel output genes came from the observation that previously characterized cycling genes (such astim, vri, to, and even low amplitude ones such as cry) displayed a circadian pattern of gene expression in our array experiment (data not shown). We also detecteddreg-5 (Van Gelder et al., 1995) with some element of low-amplitude cycling, although it was not identified as such by our algorithm. Recently, an enhancer trap-based screen retrieved four additional ccgs (Stempfl et al., 2002). Of those, COSOPT identified CG2207, CG13432, and CG3779 as cycling in the fly head, although none of them reached the significance level of p < 0.01. Interestingly, CG6145 appears to cycle robustly in the fly body (see below and Table 3).
Table 1.
COSOPT assigned a p value to each putative cycling gene
| Cycling genes | |||
|---|---|---|---|
| p < 0.01 | p < 0.025 | p < 0.05 | |
| Head | 120 | 478 | 1206 |
| Body | 177 | 524 | 1144 |
Table 3.
List of cycling genes in fly bodies (p < 0.01), as in Table 2
| Gene ID | Function | LD experiment | |
|---|---|---|---|
| p-β | Phase | ||
| Defense/immunity | |||
| LysX | Lysozyme X | 9.50E-03 | 17.55 |
| Lectin-galC1 | Galactose-specific C-type lectin | 3.60E-03 | 9.00 |
| CT8705 | PGRP-SC1b, peptidoglycan recognition protein | 1.87E-03 | 22.20 |
| Phas1 | Eukaryotic-initiation-factor-4E binding protein | 9.76E-03 | 23.00 |
| Ag5r | Antigen 5-related | 2.10E-03 | 21.31 |
| Chit | Chitinase-like | 6.84E-03 | 7.60 |
| CT5624 | Chitinase-like | 3.01E-03 | 17.25 |
| CT30310 | Drosomycin-like | 9.84E-03 | 15.12 |
| Idgf4 | Imaginal Disc Growth Factor 4, chitin binding protein | 2.20E-03 | 3.20 |
| CT29102 | Tep 4, complement-like protein | 3.51E-03 | 5.40 |
| Development | |||
| CT13185 | dorso-ventral patterning in oogenesis | 8.53E-03 | 4.90 |
| Detoxification | |||
| cyp9b2 | 4.91E-03 | 4.70 | |
| CT20826 | Cyp18a1 | 4.07E-03 | 19.34 |
| Cyp6gl | 1.86E-03 | 8.03 | |
| CT38747 | Glutathione transferase | 2.29E-03 | 5.90 |
| GstD1 | GlutathioneS-transferase B1 | 5.67E-03 | 7.30 |
| CT35150 | GlutathioneS-transferase-like | 4.46E-03 | 7.50 |
| CT35071 | UDP-glucuronosyltransferase | 9.59E-03 | 4.90 |
| Electron transfer | |||
| Cyt-A5 | Cytochrome A5-related | 7.50E-03 | 7.90 |
| Ligand binding/carrier | |||
| CT3751 | Triglyceride binding | 6.09E-03 | 6.50 |
| Mp20 | Muscle protein 2 | 8.59E-03 | 18.85 |
| Dbi | Diazepam-binding inhibitor | 7.95E-03 | 8.60 |
| PebIII | Ejaculatory bulb protein III | 4.14E-03 | 19.62 |
| CT33980 | Neural Lazarillo | 9.36E-03 | 19.72 |
| CT31326 | Antennal binding protein X-like | 6.24E-03 | 3.90 |
| CT32778 | Odorant binding protein | 2.78E-03 | 5.40 |
| CT21061 | Fatty acid binding protein-like | 1.94E-03 | 9.00 |
| Metabolism | |||
| CT16885 | Galactokinase | 1.46E-03 | 8.50 |
| wun | Phosphatidate phosphatase | 6.99E-03 | 19.01 |
| CT16187 | 3-Hydroxyisobutyryl-CoA hydrolase | 6.41E-03 | 0.50 |
| CT24308 | Threonine dehydratase | 6.48E-03 | 7.90 |
| CT6492 | Maltase L-like | 2.28E-03 | 6.80 |
| Hmgs | Hydroxymethylglutaryl-CoA synthase | 6.40E-03 | 23.06 |
| LvpH | Larval visceral protein H | 4.80E-03 | 7.50 |
| Pepck | Phosphoenolpyruvate carboxykinase | 6.50E-03 | 11.00 |
| Scu | 3-Hydroxyacyl-CoA dehydrogenase | 9.57E-03 | 8.90 |
| CT17038 | Malate dehydrogenase | 3.07E-03 | 8.10 |
| CT36683 | α-Amylase-like | 1.74E-03 | 8.40 |
| CT6532 | Maltase H-like | 4.49E-03 | 7.30 |
| CT22063 | Triacylglycerol lipase-like | 9.88E-03 | 7.60 |
| CT22069 | Hexokinase | 2.84E-03 | 8.00 |
| CT21171 | Carbonate dehydratase-like | 6.81E-03 | 4.70 |
| CT12913 | Ubiquinone biosynthesis | 6.51E-03 | 21.39 |
| CT34308 | UDP-glucose epimerase | 5.17E-03 | 8.50 |
| CT18319 | Pyrroline 5-carboxylate reductase-like | 6.39E-03 | 7.30 |
| CT33098 | α-glucosidase II | 3.93E-03 | 7.40 |
| CT19053 | Carbonate dehydratase-like | 5.82E-03 | 16.25 |
| CT41369 | 2.57E-03 | 4.70 | |
| CT28913 | Glucosylceramidase-like | 7.84E-03 | 9.45 |
| RfaBp | Retinoid- and fatty-acid binding protein | 2.28E-03 | 8.40 |
| RfaBp | Retinoid- and fatty-acid binding protein | 1.50E-03 | 9.60 |
| CT26924 | Glucose dehydrogenase-like | 6.49E-03 | 7.50 |
| α-Est5 | Carboxylesterase | 8.47E-03 | 10.00 |
| pug | Formate–tetrahydrofolate ligase | 8.23E-03 | 7.60 |
| α-Est7 | α-Esterase-7 | 9.35E-03 | 9.70 |
| ade5 | Phosphoribosylaminoimidazole carboxylase; EC:4.1.1.21 | 3.38E-03 | 9.50 |
| Pdh | Photoreceptor dehydrogenase | 9.23E-03 | 6.90 |
| CT30991 | IMP cyclohydrolase | 4.38E-03 | 9.80 |
| CT24026 | α-Galactosidase | 7.24E-03 | 8.90 |
| CT9666 | Transaldolase | 3.14E-03 | 8.50 |
| CT39259 | Oxidoreductase | 3.46E-03 | 3.10 |
| CT41283 | Cholinephosphate cytidylyltransferase | 6.56E-03 | 0.42 |
| CT33239 | N-acetyl transferase | 9.11E-03 | 15.44 |
| CT31087 | rRNA methyltransferase | 4.32E-03 | 13.20 |
| CT34977 | Epoxide hydrolase | 5.71E-03 | 6.30 |
| Muscle contraction | |||
| Mhc | Myosin II heavy chain | 6.81E-03 | 17.56 |
| TpnC73F | Troponin C at 73F | 6.66E-03 | 17.79 |
| Proteases | |||
| CT15463 | Zinc carboxypeptidase | 7.73E-03 | 21.92 |
| CT14806 | Vitellogenic carboxypeptidase | 7.54E-03 | 6.80 |
| CT14378 | Carboxypeptidase-like (inactive) | 3.89E-03 | 1.50 |
| CT4209 | Chymotrypsin | 3.30E-03 | 22.58 |
| CT37183 | Serine protease inhibitor | 1.66E-03 | 9.00 |
| CT25448 | Chymotrypsin | 8.12E-03 | 21.11 |
| CT8699 | Serine protease-like | 7.10E-03 | 22.52 |
| CT19724 | Serine protease | 6.97E-03 | 20.04 |
| CT37173 | Serine protease-like | 1.98E-03 | 14.30 |
| CT3996 | Cathepsin | 4.41E-03 | 9.30 |
| CT20780 | Cathepsin L | 5.44E-03 | 13.40 |
| Ser99Da | Serine protease 1 | 9.03E-03 | 23.59 |
| Ser99Db | Serine protease 2 | 9.91E-03 | 23.18 |
| CT29408 | Serine protease | 4.45E-03 | 22.77 |
| γ-Try | γ-Trypsin | 2.77E-03 | 20.66 |
| Protein synthesis/folding | |||
| CT25442 | Ribosomal protein K11-like | 7.71E-03 | 20.20 |
| CT17486 | Chaperone | 2.69E-03 | 19.57 |
| Signal transduction | |||
| pp1-13C | Ser/Thr phosphatase | 9.91E-03 | 5.80 |
| CkII-β | Casein kinase II β subunit | 7.15E-03 | 16.77 |
| cry | Cryptochrome | 5.77E-03 | 4.10 |
| Lk6 | Heat shock construct of Kidd | 4.74E-03 | 5.20 |
| CT22109 | Accessory gland peptide 36DE | 9.84E-03 | 17.93 |
| Stress response | |||
| Cat | Catalase | 1.53E-03 | 8.80 |
| CT33074 | Takeout | 4.71E-03 | 0.65 |
| CT9894 | Heat shock protein 70 | 6.26E-03 | 23.78 |
| Structural proteins | |||
| Gel | Gelsolin | 6.95E-03 | 7.80 |
| Msp-300 | Muscle-specific protein 3 | 8.02E-03 | 16.42 |
| CT33880 | Cuticle protein-like | 9.68E-03 | 17.33 |
| Glt | Glutactin | 4.72E-03 | 9.10 |
| CT41348 | A-band-protein-225 | 6.88E-03 | 17.09 |
| CT39472 | Peritrophin-15a | 4.86E-03 | 20.12 |
| Transcription | |||
| CT15944 | 7.39E-03 | 17.21 | |
| CT6009 | 9.47E-03 | 13.50 | |
| tim | Timeless | 3.98E-03 | 17.53 |
| Transporter | |||
| Vha13 | Hydrogen-transporting two-sector ATPase | 6.26E-03 | 21.66 |
| BY8473 | ATP-binding cassette transporter | 1.48E-03 | 22.77 |
| CT27832 | Sodium/phosphate cotransporter | 4.17E-03 | 17.16 |
| CT13124 | zetaCOP | 5.41E-03 | 20.56 |
| CT14766 | ATP-binding cassette transporter | 9.08E-03 | 18.53 |
| CT30783 | Sugar transporter-like | 6.99E-03 | 8.00 |
| CT10168 | Sodium/phosphate cotransporter | 6.97E-03 | 18.12 |
| CT19169 | Sugar transporter | 6.06E-03 | 6.70 |
| CT33284 | 4.70E-03 | 8.90 | |
| CT15971 | Amino-acid permease-like | 9.52E-03 | 17.98 |
| Unknown function | |||
| CT22723 | Senescence marker protein-3 | 3.39E-03 | 8.90 |
| Mst57Dc | Male-specific RNA 57Dc | 6.37E-03 | 16.93 |
| Gip | Gip-like | 8.12E-03 | 9.80 |
| CT26166 | Kisir | 4.69E-03 | 21.50 |
| CT19307 | 7.73E-03 | 20.48 | |
| CT21613 | 8.81E-03 | 17.97 | |
| CT35864 | 9.38E-03 | 14.41 | |
| CT14230 | 6.06E-03 | 6.50 | |
| CT14296 | 2.49E-03 | 6.60 | |
| CT23898 | 2.59E-03 | 9.30 | |
| CT23894 | 8.82E-04 | 7.50 | |
| CT21019 | 7.01E-03 | 5.70 | |
| CT9093 | 8.76E-03 | 18.52 | |
| CT35241 | 6.39E-03 | 9.30 | |
| CT25774 | 7.17E-03 | 8.00 | |
| CT27394 | 3.05E-03 | 9.00 | |
| CT35953 | 8.23E-03 | 3.00 | |
| CT34370 | 9.41E-03 | 17.70 | |
| CT22505 | 6.20E-03 | 7.60 | |
| CT12127 | 1.59E-03 | 7.70 | |
| CT35796 | 5.00E-03 | 17.12 | |
| CT26922 | 4.52E-03 | 8.90 | |
| CT33647 | 3.27E-03 | 0.00 | |
| CT27834 | 2.31E-03 | 7.80 | |
| CT18564 | 2.24E-03 | 23.50 | |
| CT35055 | 8.36E-03 | 7.50 | |
| CT35089 | 9.38E-03 | 5.00 | |
| CT8227 | 5.83E-03 | 20.29 | |
| CT18118 | 9.50E-03 | 20.02 | |
| CT42569 | 1.54E-03 | 22.14 | |
| CT40493 | 9.81E-03 | 9.00 | |
| CT33419 | 9.49E-03 | 1.95 | |
| CT34875 | 9.46E-03 | 15.36 | |
| CT32604 | 9.65E-03 | 19.09 | |
| CT39634 | 6.22E-03 | 8.40 | |
| CT22395 | 5.73E-03 | 21.98 | |
| CT33153 | 8.49E-03 | 5.20 | |
| CT12789 | 5.68E-03 | 21.28 | |
| CT1789 | 6.56E-03 | 2.80 | |
| CT40966 | 7.27E-03 | 6.00 | |
| CT36877 | 6.76E-03 | 23.39 | |
| CT26804 | 3.60E-03 | 21.31 | |
| CT26802 | 4.05E-03 | 21.61 | |
| CT28427 | 2.07E-03 | 5.40 | |
| CT38785 | 9.84E-03 | 2.50 | |
| CT34717 | 4.17E-03 | 5.70 | |
| CT18134 | 3.68E-03 | 9.90 | |
| CT33073 | 5.64E-03 | 13.90 | |
| CT37044 | 4.08E-03 | 3.30 | |
| CT33083 | 8.21E-03 | 9.30 | |
| CT29612 | 9.10E-03 | 20.14 | |
| CT20261 | 4.25E-03 | 7.80 | |
| CT34939 | 1.41E-03 | 9.40 | |
| CT34936 | 8.21E-03 | 11.00 | |
| CT37419 | 8.18E-03 | 13.90 | |
| CT36793 | 3.98E-03 | 16.56 | |
| CT18390 | 3.53E-03 | 1.30 | |
| CT9987 | 9.65E-03 | 7.70 | |
| CT29500 | 4.76E-03 | 21.63 | |
| CT35842 | 6.17E-03 | 8.60 | |
| CT32316 | 7.80E-03 | 3.40 | |
| CT16297 | 9.32E-03 | 17.56 | |
| CT21565 | 9.20E-03 | 18.83 | |
| CT42272 | 5.07E-03 | 6.20 | |
To address type 1 and type 2 errors in our analysis, 1000 Gaussian random temporal patterns of 13 time points (analogous to 48 hr of 4 hr sampling) with no rhythmic component were analyzed with COSOPT using the same criteria applied to our experimental data (p < 0.01; no multiple-measurement correction). COSOPT identified two cycling curves when it should have detected none, indicating that the false positive rate under these conditions is 0.2%. If we applied this percentage to the 14,000 genes assessed in the experiment, we would have 28 false positives. If it is more appropriately applied to only the ∼5000 genes that are detectable, the number of false positives becomes 10. Determining the rate of false negatives is a more complex process because it imposes a priori assumptions on the quality of the data. We ran these simulations under two hypothetical conditions: a worse case scenario in which the signal/noise ratio is 1 (signal and noise are equal in magnitude), and a more likely scenario in which this ratio equals 2. When we analyzed 1000 cosine waves of amplitude equal to 1 SD above the noise level, COSOPT detected 445 (implying that the false negative rate under these conditions is 55%); interestingly, when the amplitude equals 2 SDs above the noise (still a conservative assumption), the false negative rate becomes negligible. COSOPT identified 997 of 1000 (that is, 0.3% error rate) of the test cosine waves, providing an exceptionally high degree of confidence that our approach is valid (data not shown).
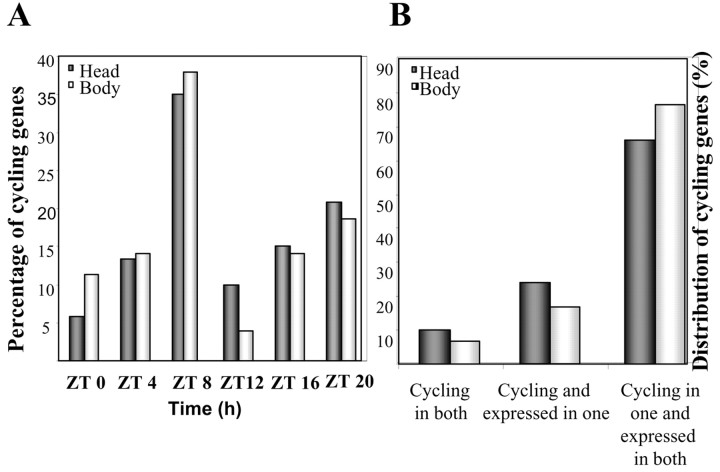
Cycling genes were grouped in clusters according to the phase of peak expression. The phase distribution is shown in Figure1A. This approach allowed the identification of several novel phases of peak expression [Zeitgeber time 4 (ZT4), ZT8, ZT12] and, more importantly, made possible a direct comparison between the different genes within the same experiment. All phases were similarly represented with the exception of ZT8, and to a lesser extent ZT20, where a larger proportion of the genes reach maximum levels of expression. This is similar to what has been reported by Ueda et al. (2002) and contrasts with the findings of Claridge-Chang et al. (2001), although this could be attributable partially to differences in the cluster analysis that was performed.
Fig. 1.
A, Output genes peak throughout the day in the fly head and body. Genes were grouped according to the phase calculated by COSOPT; each cluster represents genes peaking at the specified time ± 2 hr. ZT0 refers to the time when lights were switched on. Cluster size is represented as percentage of the total number of cycling genes. B, The clock controls different subsets of genes in head and body fractions. Distribution of genes cycling in both tissues, cycling and expressed in only one, and expressed to mid-high levels in both but cycling in one is displayed.
One of the hallmarks of clock-controlled activity is the persistence of the rhythms in the absence of environmental cues. To confirm the circadian nature of the newly identified target genes, we performed an independent experiment under free-running conditions. We tested which proportion of the novel outputs showed reliable cycling profiles under constant darkness. One of the limitations of this analysis in DD is the “dampening” of the amplitude of the oscillation, which could partially result from the desynchronization of individual cells in the absence of resetting environmental cues, or it could reflect a direct effect of light boosting the amplitude of the oscillations (Hardin, 1994). Approximately 50% of the genes identified in the LD experiment were found to cycle robustly in DD as well (Table2). Interestingly, the phase predicted by COSOPT for each individual cycling gene is very similar in LD and DD.
Table 2.
Cycling genes in fly heads (p < 0.01) are categorized by known or predicted function
| Gene ID | Function | LD | DD | Levels inClkjrk | ||
|---|---|---|---|---|---|---|
| p-β | Phase | p-β | Phase | |||
| Cell adhesion | ||||||
| CT35785 | 8.3E-03 | 21.3 | ||||
| Trn | tartan | 7.7E-03 | 9.7 | |||
| Metabolism | ||||||
| CT1116 | Succinyl CoA transferase | 4.6E-03 | 4.2 | |||
| CT22171 | Isocitrate dehydrogenase (NADP+) | 4.6E-03 | 20.6 | 2.4E-01 | 21.1 | |
| CT34968 | 3-OH isobutyrate dehydrogenase | 4.2E-03 | 3.2 | 3.1E-02 | 2.5 | Up |
| CT12987 | Short-branched chain acyl CoA dehydrogenase | 4.8E-03 | 0.5 | 9.1E-02 | 21.1 | |
| CT41571 | Aldehyde dehydrogenase | 9.5E-03 | 18.5 | 8.4E-02 | 22.0 | |
| CT1187 | Choline phosphate cytidil transferase | 9.9E-03 | 8.8 | 2.3E-02 | 8.8 | |
| CT11757 | Anon 23-Da | 4.0E-03 | 14.6 | |||
| Hmgs | Hydroxymethylglutaryl-CoA synthase | 6.9E-03 | 21.6 | |||
| CT16527 | Cholesterol O acyl transferase | 8.0E-03 | 7.8 | |||
| CT40163 | Angiotensin-converting enzyme | 2.3E-03 | 13.4 | |||
| CT12411 | Oxido reductase | 7.5E-03 | 8.4 | |||
| Development | ||||||
| CT27880 | Yellow-d2 | 5.5E-03 | 8.8 | 5.0E-03 | 15.0 | |
| Idgf1 | Imaginal Disc Growth Factor 1 | 6.0E-03 | 20.7 | Up | ||
| mbc | Myoblast city | 5.0E-03 | 11.7 | Up | ||
| Detoxification | ||||||
| cyp6a21 | Cytochrome P450 6a21 | 3.8E-03 | 21.2 | 6.8E-03 | 2.2 | |
| Ugt35b | UDP-glycosyltransferase 35b | 4.0E-03 | 3.7 | 9.8E-03 | 3.0 | |
| CT38753 | GlutathioneS-transferase | 4.9E-03 | 5.6 | |||
| CT38747 | GlutathioneS-transferase | 7.5E-03 | 5.1 | Down | ||
| cyp4e3 | Cytochrome P450 4e3 | 3.3E-03 | 9.0 | 6.4E-03 | 14.2 | Down |
| cyp6d5 | Cytochrome P450 6d5 | 6.0E-03 | 21.6 | |||
| cyp6a17 | Cytochrome P450 6a17 | 9.4E-03 | 2.9 | Up | ||
| CT16545 | Glutathione transferase | 2.9E-03 | 7.8 | 1.6E-01 | 2.9 | |
| cyp6a2 | Cytochrome P450 6a2 | 3.1E-03 | 7.9 | 1.5E-02 | 13.0 | Down |
| cyp18 | Cytochrome P450 18 | 6.9E-03 | 17.1 | 6.9E-03 | 15.7 | |
| Heme metabolism | ||||||
| Alas | 5-Aminolevulinate synthase | 1.0E-02 | 22.5 | |||
| CT34507 | Heme oxygenase-like | 8.8E-03 | 9.1 | 1.3E-02 | 14.2 | |
| Ligand binding/carrier | ||||||
| CT6764 | Glucan α 1,4 glucosidase | 4.2E-03 | 18.4 | 1.0E-01 | 22.5 | |
| glob1 | Globin 1 | 6.7E-03 | 20.1 | 1.2E-01 | 21.2 | |
| CT33362 | 7.6E-03 | 20.4 | 2.4E-02 | 22.0 | ||
| CT28495 | 2.8E-03 | 13.9 | 2.9E-02 | 21.9 | ||
| Neurotransmission | ||||||
| ple | Tyrosine 3-monooxygenase | 4.5E-03 | 2.4 | 6.6E-03 | 1.6 | |
| Eaat2 | Excitatory amino acid transporter 2 | 4.4E-03 | 13.2 | |||
| Dat | DopamineN-acetyltransferase | 5.2E-03 | 9.5 | |||
| CT42497 | Glycine transporter | 5.6E-03 | 6.9 | 5.8E-03 | 13.1 | |
| b | Black, glutamate decarboxylase | 7.7E-03 | 16.2 | Down | ||
| Photoreceptor | ||||||
| cry | Cryptochrome | 3.6E-03 | 1.1 | |||
| Protease inhibitors | ||||||
| serpin | Serine protease inhibitor | 8.3E-03 | 20.0 | |||
| CT37177 | 9.3E-03 | 14.6 | Up | |||
| CT37195 | 8.2E-03 | 10.4 | Up | |||
| Protein synthesis/degradation | ||||||
| CT18196 | Ubiquitin thiolesterase | 2.2E-03 | 13.1 | 7.3E-04 | 16.5 | |
| pros26 | Proteasome 26 kDa protein | 4.9E-03 | 8.7 | 1.7E-02 | 8.4 | |
| pros26.4 | Proteasome 26S subunit subunit 4 ATPase | 7.1E-03 | 7.9 | |||
| CT28749 | 26S proteasome regulatory subunit p39A | 6.2E-03 | 8.9 | 1.4E-02 | 10.7 | |
| CT7240 | Translation initiation factor (eIF-5) | 7.4E-03 | 8.5 | 2.9E-02 | 9.5 | |
| CT39962 | tRNA synthase | 6.5E-03 | 20.8 | |||
| Protein folding? | ||||||
| CT16169 | Heat shock protein 27-like | 7.4E-03 | 21.9 | |||
| Cnx99A | Calnexin 99A | 2.8E-03 | 17.4 | 6.6E-02 | 19.2 | |
| Transcription | ||||||
| CT30663 | 7.0E-03 | 11.6 | ||||
| tim | timeless | 4.7E-03 | 16.4 | 4.6E-03 | 17.4 | |
| Mad | Mothers against dpp | 5.0E-03 | 14.5 | |||
| vri | vrille | 5.5E-03 | 13.6 | 1.6E-03 | 15.4 | |
| CT31519 | 5.1E-03 | 2.8 | 3.4E-01 | 7.5 | ||
| CT19628 | 6.3E-03 | 16.6 | ||||
| CT17296 | 4.8E-03 | 14.1 | 1.1E-02 | 17.4 | ||
| CT18631 | TfII A-L | 8.4E-03 | 9.3 | |||
| 140up | Upstream of RpII14 | 9.1E-03 | 17.5 | 9.1E-02 | 15.4 | |
| Signal transduction | ||||||
| Akap200 | Protein kinase A anchoring protein | 6.2E-03 | 15.6 | |||
| Pk61C | Protein kinase 61C | 2.9E-03 | 10.8 | 2.3E-02 | 14.1 | |
| Pka-C3 | Ser/Thr kinase | 4.9E-03 | 15.3 | 5.7E-02 | 7.6 | |
| CT35755 | 9.1E-03 | 21.5 | 1.9E-01 | 4.4 | ||
| CT27512 | Ras small monomeric GTPase | 8.1E-03 | 9.3 | 1.2E-03 | 12.4 | |
| Slob | Slowpoke binding protein | 3.0E-03 | 18.0 | 2.2E-03 | 16.3 | |
| CT34849 | Chd64 | 9.8E-03 | 5.5 | |||
| Stress response | ||||||
| CT25938 | Superoxide dismutase (Cu-Zn) | 3.3E-03 | 8.6 | |||
| Hsp22 | Heat shock protein 22 | 7.8E-03 | 17.6 | |||
| to | Takeout | 8.3E-03 | 21.7 | 5.3E-02 | 2.1 | |
| Structural proteins | ||||||
| dlp | Heparin sulfate proteoglycan | 7.4E-03 | 11.2 | |||
| Sulf1 | N-acetylglucosamine-6-sultase | 9.0E-03 | 8.8 | 1.3E-02 | 9.4 | |
| βTub56D | βTubulin56D | 8.7E-03 | 10.0 | 1.4E-01 | 22.9 | Up |
| CT31310 | 8.9E-03 | 6.2 | Down | |||
| CT31613 | Thrombospondin | 2.3E-03 | 10.3 | 2.4E-02 | 11.9 | |
| CT15377 | 9.1E-03 | 5.2 | 2.3E-02 | 3.0 | ||
| Cpn | Calphotin | 8.9E-03 | 7.2 | 3.0E-02 | 9.9 | Up |
| capu | Cappuccino | 1.0E-02 | 18.5 | |||
| Transporters | ||||||
| CT42567 | High-affinity inorganic phosphate:sodium transporter | 3.8E-03 | 0.2 | 4.6E-02 | 22.9 | Up |
| CT30701 | Glucose transporter | 8.2E-03 | 7.7 | |||
| trpl | Trp-like | 6.7E-03 | 9.8 | |||
| CT16777 | 8.6E-03 | 8.6 | 1.3E-02 | 13.9 | ||
| Unknown function | ||||||
| CT12588 | 8.9E-03 | 23.5 | 2.9E-02 | 17.9 | ||
| CT31141 | 9.9E-03 | 15.2 | ||||
| CT28701 | 7.7E-03 | 8.3 | ||||
| CT34631 | 8.3E-03 | 8.9 | Up | |||
| CT6802 | 7.4E-03 | 9.4 | ||||
| CT32204 | 3.9E-03 | 7.1 | 2.4E-03 | 7.7 | ||
| CT16557 | 9.3E-03 | 8.8 | 2.1E-03 | 16.2 | ||
| CT16503 | 7.9E-03 | 2.2 | 7.2E-03 | 4.9 | ||
| CT33900 | 7.9E-03 | 18.9 | ||||
| CT22515 | 9.9E-03 | 14.1 | ||||
| CT33647 | 4.1E-03 | 1.0 | 7.5E-03 | 19.6 | ||
| CT18564 | 9.9E-03 | 21.8 | ||||
| CT25838 | 5.2E-03 | 8.7 | ||||
| CT30256 | 8.0E-03 | 20.5 | ||||
| CT33484 | 3.6E-03 | 18.4 | 6.1E-03 | 21.0 | ||
| CT32008 | 6.9E-03 | 8.3 | ||||
| CT28709 | 8.7E-03 | 7.9 | ||||
| CT31865 | 2.3E-03 | 9.2 | 1.2E-03 | 14.3 | ||
| CT32600 | 7.9E-03 | 22.7 | 1.0E-01 | 22.0 | ||
| CT15908 | 4.8E-03 | 8.8 | 2.4E-03 | 13.2 | ||
| CT32262 | 2.3E-03 | 8.3 | 1.0E-03 | 10.2 | ||
| CT34135 | 9.5E-03 | 5.5 | 7.1E-02 | 16.3 | ||
| CT34439 | 9.6E-03 | 3.3 | 1.7E-01 | 15.7 | ||
| CT10556 | 4.4E-03 | 8.7 | Up | |||
| CT42539 | 5.5E-03 | 8.5 | ||||
| CT35916 | 9.6E-03 | 5.9 | 1.1E-02 | 7.5 | ||
| CT33073 | 4.6E-03 | 7.2 | 3.8E-03 | 10.0 | ||
| CT37044 | 2.4E-03 | 20.6 | 7.0E-03 | 0.9 | ||
| CT29508 | 8.8E-03 | 19.1 | 1.4E-01 | 1.3 | ||
| CT29612 | 2.7E-03 | 19.5 | 3.4E-03 | 19.3 | ||
| CT35582 | 2.7E-03 | 8.5 | 1.0E-03 | 14.4 | ||
| CT32596 | 8.9E-03 | 10.8 | ||||
| CT26517 | 1.0E-02 | 4.8 | 3.1E-01 | 17.4 | ||
| CT29500 | 4.7E-03 | 21.4 | 1.8E-02 | 2.3 | ||
| CT16351 | 4.8E-03 | 15.5 | ||||
| CT24597 | 9.5E-03 | 4.5 | 2.8E-01 | 16.1 | ||
| CT26954 | 9.3E-03 | 8.7 | ||||
| CT25972 | 8.5E-03 | 15.9 | ||||
Gene ID lists either the gene name (if known) or the Celera Transcript (CT) number, as well as the associated p value and phase under entrained (LD) and free-running (DD) conditions. The last column displays the genes that were found to change significantly in the y w;;Clkjrk background.
To examine the cycling profiles of the newly identified output genes in a behaviorally arrhythmic background, we entrained y w;;Clkjrk and performed a time course with a 4 hr time resolution during 2 consecutive days in LD; this resolution was chosen to properly compare genes peaking at different phases in the wild-type condition with their counterparts in the mutant background. We found that 116 of the 120 identified output genes ceased to cycle in this mutant background, commensurate with the role that the CLOCK protein plays in sustaining the molecular oscillations. The remaining four genes were scored by COSOPT as cycling with p < 0.01. Closer inspection of the individual traces highlighted the direct effect of light and light/dark transitions on gene expression: three of the mRNAs appeared to slowly build up during the day and decrease during the night, whereas others appeared to be acutely driven by light and decreased thereafter (data not shown). To determine whether the levels of expression changed in the mutant background, we calculated the arithmetic average difference (subtracting the average level of expression in y w;;Clkjrk from that in wild type) and corresponding SD. By this criteria we found that 5 genes were downregulated and 11 were upregulated (p < 0.025) in the y w;;Clkjrkmutant background (as indicated in the last column of Table 2).
The clock regulates gene expression in a tissue-specific manner
To provide a more comprehensive view of clock control in a whole organism, we analyzed cyclic gene expression under entrained conditions using male bodies as the source of total RNA and compared these transcripts with those derived from heads. COSOPT identified 177 genes that cycle within a period between 20 and 28 hr with p< 0.01 (Table 1); the data are summarized in Figure 1, Aand B, and Table 3. As in the mouse (Panda et al., 2002), only a small proportion of cycling transcripts was found to overlap between the two tissues (12 genes), including some previously identified clock components (such astim, vri, and cry). The analysis of the remaining genes revealed an even more interesting feature: a large number of genes that cycle solely in fly heads are still expressed at medium to high levels in bodies, pointing to differential transcriptional regulation in this subset of clock outputs (Fig.1B). This finding is also in agreement with what has been found in the mouse (Panda et al., 2002; Storch et al., 2002).
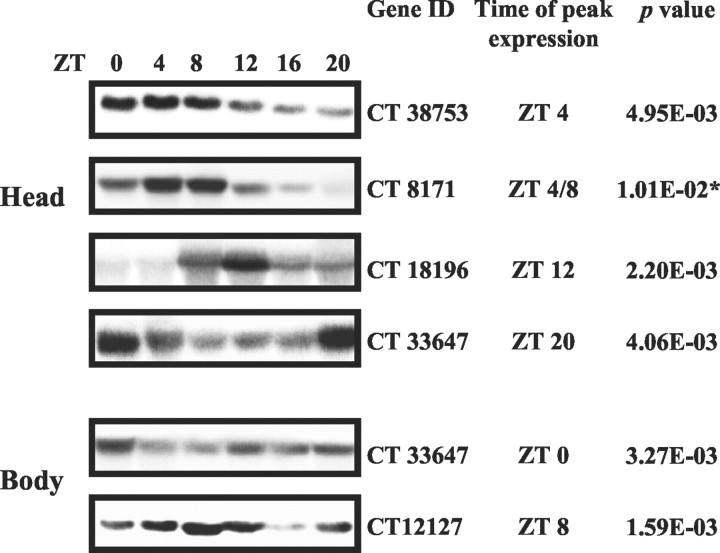
To independently confirm the cyclic pattern of a subset of output genes, we performed Northern blot analysis on separate time courses. We chose genes spanning a range of p values; all were found to oscillate with a phase concordant to that predicted by the microarray experiments (Fig. 2).
Fig. 2.
Northern blot analysis of several clock-controlled genes. Northern blot analysis of independent head and body time courses confirms both the cyclic nature of the candidates tested and their respective phases of expression. Gene ID refers to the cDNA used as probes. The right columns indicate the expected peak time and the corresponding p value predicted by the array experiment, respectively. Theasterisk indicates a gene that is outside of thep < 0.01 cutoff and still appears rhythmic. According to our experiments, the CT18196 transcript has smaller size than predicted.
Circadian transcriptional regulation of physiology
Protein stability
Although cyclic transcription is a signature of clock activity, important levels of regulation take place downstream. Several genes involved in various aspects of protein stability were under rhythmic transcriptional control. We identified three genes cycling in the fly head with a peak at ZT8 that are part of the 26S proteasome complex (Fig. 3A). pros26 encodes a multicatalytic endopeptidase (Saville and Belote, 1993).pros26.4 and rpn9 are part of the 19S regulatory complex and correspond to subunit 4 of the AAA-ATPase and are required for assembly and stability of the proteasome. In Drosophila, different proteasome subunits are expressed throughout development, possibly to control specific processes such as cell division or morphogenesis (Haass and Kloetzel, 1989). We also identified a putative de-ubiquitinating enzyme, the homolog of the mouse Usp8, with peak expression at ZT12 (Fig. 3A). The cyclic pattern of CG5798 was confirmed independently by Northern blot analysis (Fig. 2). De-ubiquitinating enzymes remove the polyubiquitin chain from conjugated proteins before their degradation by the proteasome. These enzymes either may regulate degradation by the proteasome or may be involved in ubiquitination processes regulating subcellular localization. In summary, these data suggest that temporal regulation of the proteosome may be important in Drosophilaphysiology.
Fig. 3.
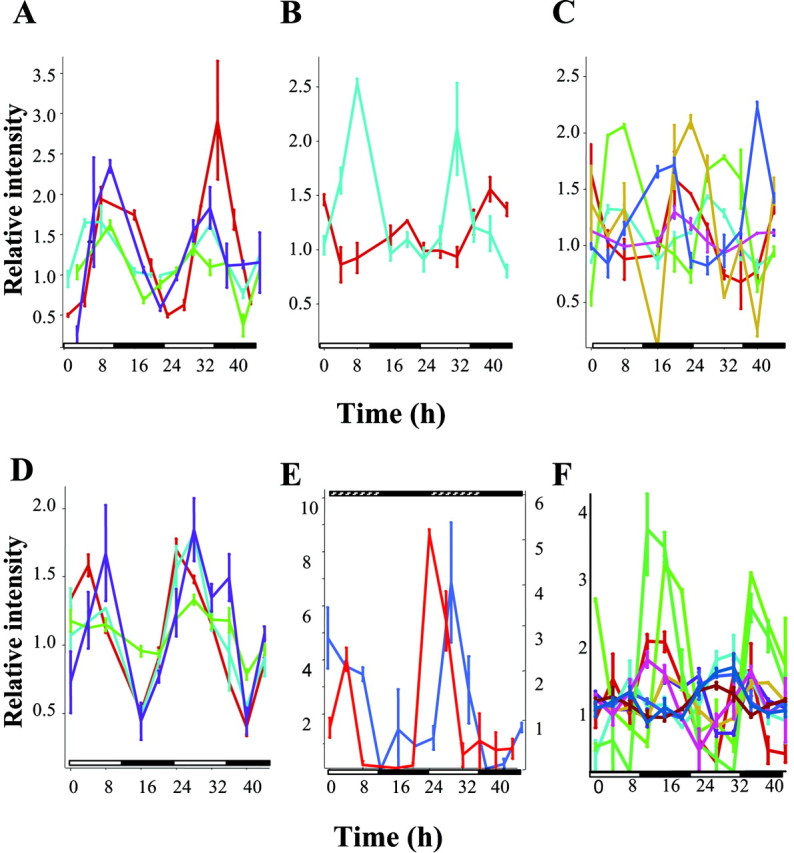
Cyclic patterns of clock-controlled genes in the fly head and body under entrained conditions. Complementary RNA samples were prepared as described and hybridized to duplicate DNA GeneChips. Data were normalized such that the mean expression level for each particular gene over the course of all time points equals 1. The average signal strength at each time point was then expressed as a ratio over the median signal strength for that particular gene. Representative traces of cycling transcripts implicated in various physiologies and metabolic pathways are shown inA–F. A, Clock control of protein stability. pros26 (light blue),pros26.4 (green),rpn9 (purple), and ubiquitin thiolesterase (red). B, Heme metabolism. The gene encoding alas is in red; heme-oxygenase is in light blue. C, Genes implicated in detoxification are under circadian regulation. Phase I cytochrome P450s are colored as follows: cyp4e3 (green), cyp6a2 (light blue), cyp6a17 (brown),cyp6a21 (red), cyp6d5 (dark blue), and cyp18 (pink). D, Phase II genes:ugt35b in red, GST3 inpurple, and two uncharacterized GSTs (CT38753 and CT38747) in light blue and green, respectively. (E) Neurotransmission.ple expression under entrained and free-running conditions is depicted in red and blue, respectively. F, Immunity. A number of genes involved in different aspects of innate immunity are under circadian control and are indicated as follows: in recognition and phagocytosis: Agr5 (light brown), a peptidoglycan protein (light green), lectin galC (aqua), Idgf4 (brown), CT29102 (deep blue); antimicrobial peptides: lysX (red) and CT 30310 (pink); Chitinase-like molecules: Chit (light blue) and CT5624 (dark green). Relative intensities ± SEM at each time point are shown.White and black boxes on theabscissa represent the duration of the light and dark periods. Hatched boxes (E) indicate subjective day.
Heme metabolism
The observation that the expression of both the rate-limiting enzyme in heme biosynthesis, δ-aminolevulinate synthase,alas-, and the rate-limiting enzyme of heme degradation, heme oxygenase, cycle in Drosophila heads suggests that heme metabolism is tightly regulated by the clock (with a peak at ZT20 and ZT8, respectively) (Fig. 3B). Heme, or heme-containing proteins, is involved in respiration, oxygen transport, detoxification, and signal transduction processes, but as a chelator of iron it may promote deleterious cellular effects such as oxidative membrane damage. Thus, maintaining a proper balance between heme biosynthetic and degradative pathways is crucial for cellular homeostasis (Ryter and Tyrrell, 2000). Although alas has been found to cycle in the mouse liver, heme oxygenase has not been observed to cycle (Kornmann et al., 2001).
Detoxification/olfaction
Heme is also required for P450 function. In insects, P450 enzymes are thought to be involved in the biosynthetic pathways of ecdysteroids and juvenile hormones, and as such play a role in insect growth, development, and reproduction as well as in the metabolism of natural plant products and insecticides, resulting in bioactivation or detoxification (Feyereisen, 1999). This biotransformation process has been described as occurring in two phases. The initial compound can be transformed into a more reactive species (usually via redox reactions catalyzed by cytochrome P450s), whereas in the second phase, highly polar groups such as UDP-glucuronosyl or glutathione are added either to the products of the first phase or in some cases directly to the toxic chemicals. The enzymes involved in this second-stage phase are UDP-glucuronosyl transferases (UGTs) and gluthathioneS-transferases (GSTs). Products of phase II are highly hydrophilic, can no longer cross membranes, and are eliminated by secretion (Wang et al., 1999). We found that six different cytochrome P450 genes, cyp4e3, cyp6a2, cyp6a17,cyp6a21, cyp6d5, and cyp18, cycle with different phases in heads (ZT8, 4, 0, 20, 20, and 16, respectively) (Fig. 3C) and cyp6g1, cyp9b2, andcyp18a1 cycle with different phases in the body (Table 3). The only functionally characterized enzyme thus far iscyp6a2, which is involved in the metabolism of organophosphorus insecticides. Interestingly, we also found thatugt35b and several GSTs also cycle in fly heads (“phase II” enzymes) (Fig. 3D), implying that multiple steps in the biotransformation process appear to be under circadian control.
In Drosophila, ugt35b is preferentially expressed in the third antennal segment where most of the olfactory sensilla are found (Wang et al., 1999), although it can be found in the fly head as well. Given that olfaction is known to be under circadian control (Krishnan et al., 1999) and olfactory UGTs have been found to play a role in odorant signal termination (Lazard et al., 1991), we can speculate that ugt35b could potentially be a link between them.
Neurotransmission
Another gene that appears to be under circadian control isple, implicating neurotransmission as a clock-controlled process. ple codes for the tyrosine 3-monooxygenase (also known as tyrosine hydroxylase), the first and rate-limiting enzyme in dopamine biosynthesis. Dopamine is an intermediate in cuticular sclerotization and also functions as a neurotransmitter in the fly nervous system. It has been shown to modulate certain forms of learning (such as female sexual receptivity and habituation), as well as motor neuron activity and neuromuscular function in larva (Neckameyer, 1998;Cooper and Neckameyer, 1999) and to exert circadian control over reflexive locomotion in decapitated flies as well (Andretic and Hirsh, 2000). We found that ple cycles with high amplitude, peaking at ZT4 under both entrained and free-running conditions (Fig.3E). ple expression falls below the level of detection in y w;;Clkjrk flies (a mutation that impairs clock function) (Allada et al., 1998).ple represents one of the several examples of homologous genes cycling in both flies and mammals (Panda et al., 2002), further stressing the significance of our observation. The oscillation ofple expression is unlikely to relate to its role in the head cuticle because the expression is undetectable in the body. Rather, the dramatic oscillation may contribute to circadian regulation of behavior.
Immunity
Drosophila is able to mount a rapid immune reaction in response to the infection by a pathogen. In fact, two distinct pathways are triggered depending on the nature of the pathogen involved, namely Gram-negative bacteria or fungi (Khush et al., 2001). We found a number of genes that play a role in different aspects of the immune response that cycle throughout the day in the fly body (Fig. 3F, Table 3). These genes are involved in microbial recognition and phagocytosis (such as PGR-SC1b, ldgf4, Tep4, Agr5, and lectin-GalC), or they encode antimicrobial peptides (such as lysX and CT30310); others have unknown function but have been found to be induced specifically by infection (Phas1) (De Gregorio et al., 2001).
A potassium channel is an output of the clock involved in sustained rhythmic behavior
The availability of a more complete description of clock-controlled genes enabled the selection of several candidates for the control of locomotor behavior. One of these candidates was slowpoke binding protein (slob), which binds to the Ca2+-dependent voltage-gated potassium channel slowpoke (slo) (Schopperle et al., 1998). A mutation in this channel causes behavioral defects (Atkinson et al., 2000) and an altered mating song, also a hallmark of certain clock components (Peixoto and Hall, 1998). slowpoke participates in the repolarization of the action potential in flight muscles and in motoneurons (Elkins et al., 1986; Gho and Mallart, 1986). SLOB has been shown to modulate SLO activity per se, and through the formation of a complex with the ζ isoform of 14-3-3 protein that acts downstream in several signaling pathways (Zhou et al., 1999).
We found that slob mRNA cycled robustly in fly heads in LD and DD (Fig.4A,B, Table 2), consistent with recent reports (Claridge-Chang et al., 2001;McDonald and Rosbash, 2001; Ueda et al., 2002). This pattern was lost in the y w;;Clkjrk mutant background. Although slo was not detected as cycling by COSOPT because of its low level of expression, we noticed that it appeared to cycle in phase with slob in both LD and DD (Fig.4A,B). The cycling ofslo was investigated by RT-PCR analysis (data not shown), and the protein was shown to cycle and peak at ZT20 by Western blot (Fig. 4C). The slo spatial expression pattern has been studied extensively (Becker et al., 1995) and has revealed that the slo mRNA is widely expressed in the adult brain. Furthermore, SLO protein has been localized both to neuronal cell bodies as well as to the neuronal projections.
Fig. 4.
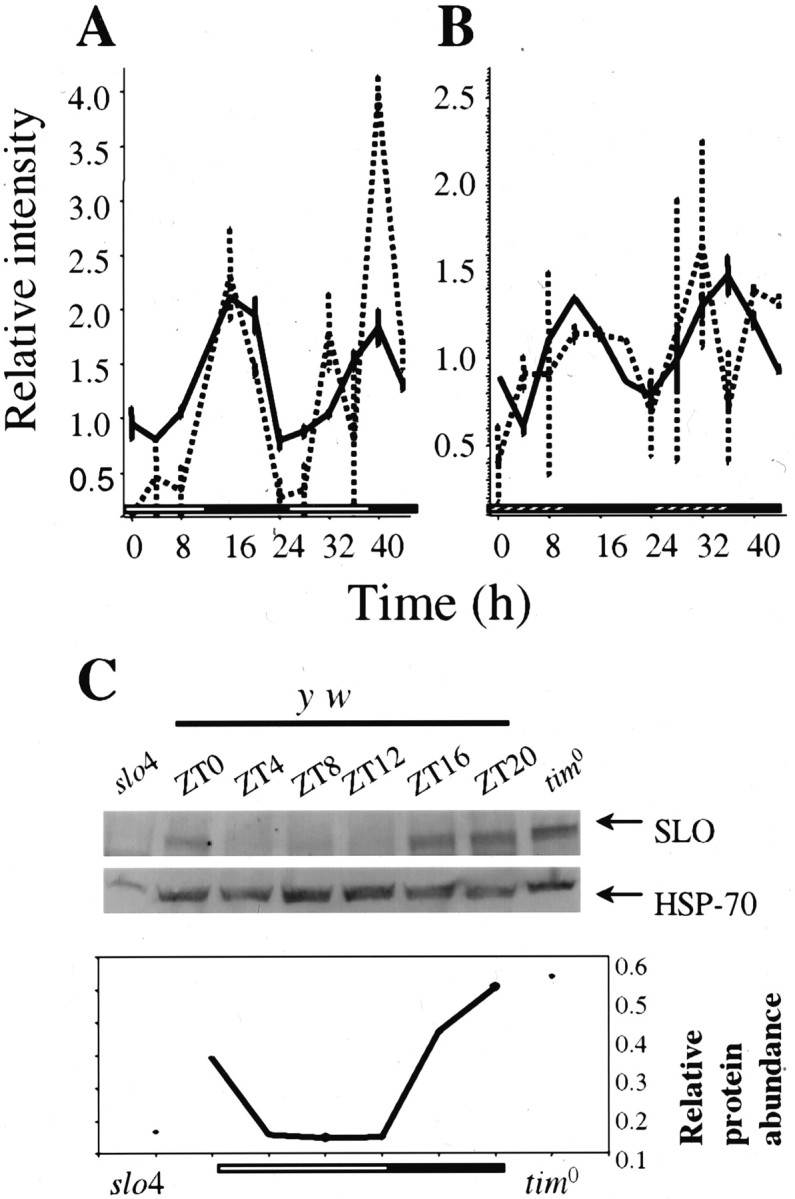
slo and slob cycle in LD and DD in wild-type flies. Shown is circadian pattern of expression of slob (gray) andslo (black) under entrained (A) and free-running (B) conditions. Relative intensities ± SEM at each time point are shown. C, Representative example of a Western blot to detect the SLO protein under LD conditions. Flies were entrained and collected as described. The first and last lanes correspond to protein extracts collected at ZT16 fromslo4 and tim0 flies, respectively. The graph indicates the quantification of SLO levels throughout the day (normalized to HSP-70).
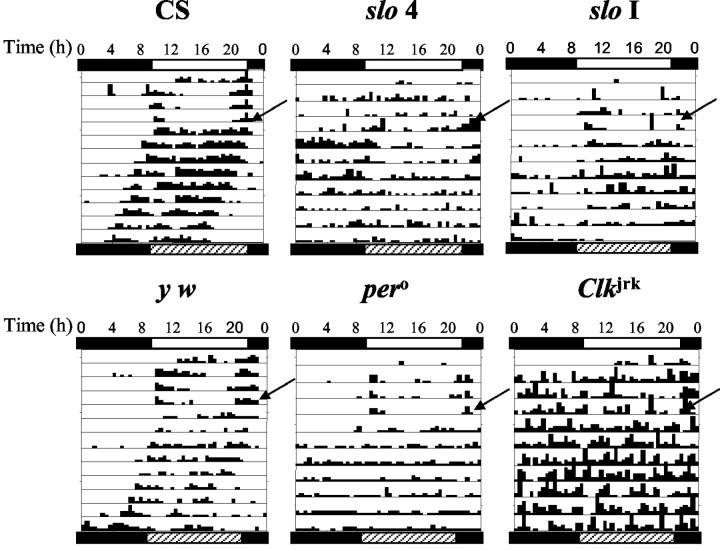
Prompted by the speculation that SLO might be involved in circadian control of activity, we examined the locomotor activity in twoslo mutants, slo I and slo 4. Wild-type flies show increased locomotor activity near dawn and dusk and remain quiescent the rest of the day (Fig.5) (Hamblen-Coyle et al., 1992). These bursts of activity do not merely follow the next temporal transition, but instead anticipate it. Figure 5 shows individual representative actograms of wild-type and mutant flies. Wild-type Canton S (CS) and y w are rhythmic in LD and in DD, where the endogenous period becomes apparent. peroand Clkjrk mutants, which have defects in core clock components, behave differently under entrained conditions. Although pero flies still look mostly rhythmic in LD,Clkjrk is often not (Fig. 5) (Wheeler et al., 1993; Allada et al., 1998). This apparent rhythmicity in pero flies is caused by the so-called “startle effect,” an immediate behavioral response to the light/dark transitions. We found that most of the slo 4 mutant flies display weak rhythms (defined as lacking a consolidated peak in the periodogram analysis) or no rhythms at all in LD. As expected, the lack of rhythmicity persisted under free-running conditions (Fig. 5, Table 4). Surprisingly, this arrythmicity is comparable to, if not worse than, the one displayed by Clkjrk.slo I mutants, on the other hand, displayed a milder phenotype, with only 40–55% of rhythmic flies in LD and DD, respectively, which is commensurate with a hypomorphic slomutation (as opposed to a true null, as is the case for slo4). Given the nature of the slo 4 mutation and the difference in the strength of the phenotype observed betweenslo I and slo 4 mutants, we tested slo4/slo I trans-heterozygotes to rule out the possibility that other loci (also affected by the chromosomal inversion) could be contributing to the observed phenotype. We found that the slo 4/slo I mutants show a somewhat intermediate phenotype (especially obvious in DD) between that ofslo 4 and slo I (Table 4). We also tested a small number of slo4 heterozygotes (slo 4/+) and found that most are either strongly or weakly rhythmic; no arrhythmic flies were found (data not shown). This argues against an effect exerted by the other putative loci.
Fig. 5.
slowpoke is required for sustained rhythmic behavior. Representative actograms of wild-type CS andy w flies (left) and the mutantsper0, Clkjrk, and arrhythmicslo I and slo 4 flies are displayed. Flies were entrained for 5 d before the onset of the experiment. During the experiments, flies were kept in LD for 3–4 d and then switched to DD and monitored for at least another week. Rhythmicity and total activity in LD and DD conditions were determined using the Clocklab software package.
To determine whether this mutation caused a general decrease in motility, which by itself could result in arrhythmicity, we quantified the total locomotor activity displayed by the different genotypes under LD and DD conditions. Although wild-type flies appear to be slightly more active under constant darkness, both slo mutants are impervious to the lighting regimen. More importantly, the overall levels of activity are not different from those of the wild-type flies (Table 4). We superimposed the actograms of wild-type, slo4, and slo I mutant flies because the average activity plots are known to reveal features not apparent when individual flies are inspected. This analysis revealed that the most striking difference is the impaired anticipation of the transitions in theslo 4 (null) mutant flies, indicating that the temporal gating that consolidates behavior around dawn and dusk is absent in flies lacking slo function (Fig.6).
Fig. 6.
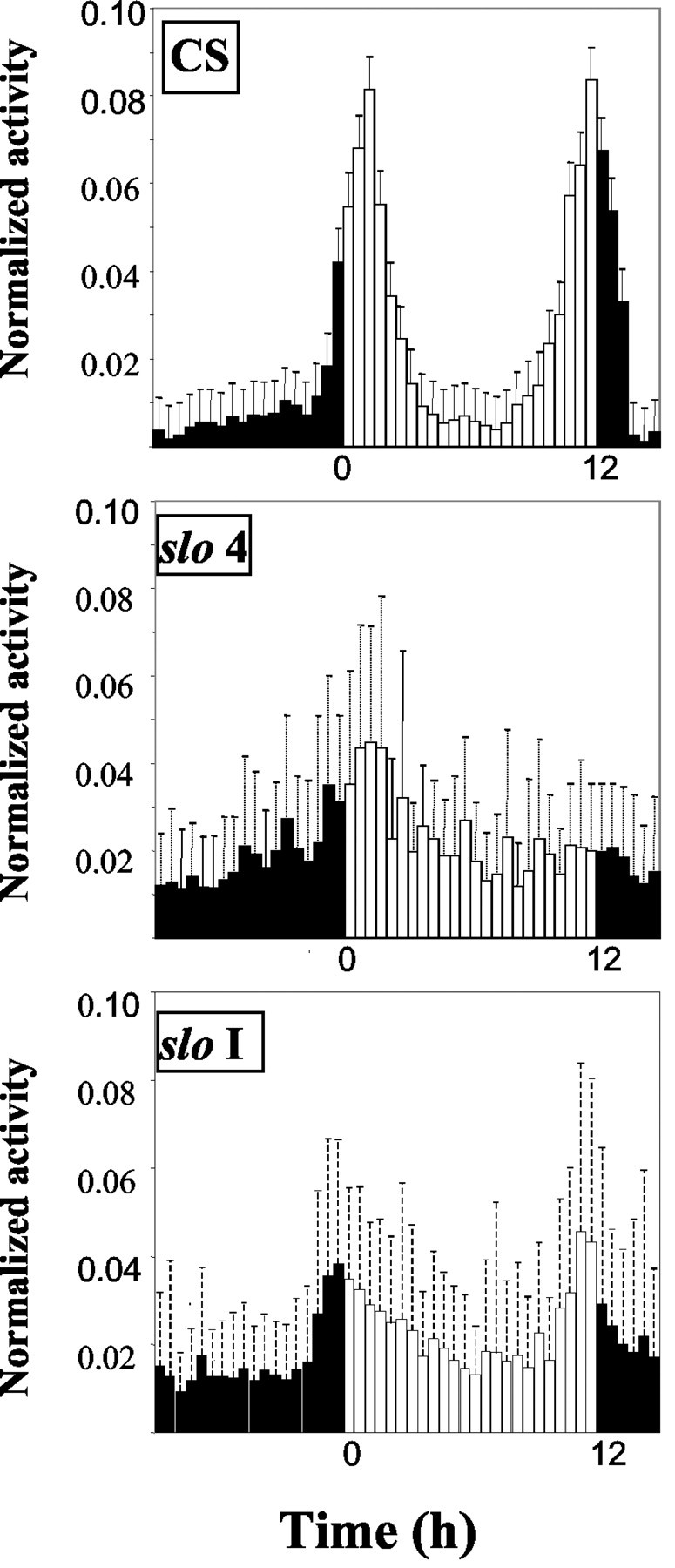
Mutations in slowpoke disrupt the consolidation of activity around the transitions. Average activity plots for CS, slo 4, and slo I mutant flies are shown. Activity records of the LD portion of the experiment for 53 wild-type, 28 slo 4, and 56 slo I flies were used for the analysis. To superimpose the separate animal records, the levels of activity were normalized per fly per day. Each vertical bar represents the mean ± SD.
DISCUSSION
Although a description of the core circadian oscillator exists in flies and mammals, a connection between the molecular basis of rhythmicity and physiology under clock control has been lacking. Because many clock mutants are transcription factors, a central approach in linking core mechanism to physiology is to examine mRNAs for rhythmic expression. To accomplish this on a whole genome scale, we and others have examined steady-state mRNA levels using high-density oligonucleotide arrays for circadian patterns of expression in the fly head and body. This analysis has identified several genes known to be rhythmically expressed, as well as several hundred genes of known and unknown function that are likewise under clock control. We found that a number of aspects of fly physiology ranging from basic cellular metabolism to neurotransmission, stress resistance, and detoxification appear to be under the control of the biological clock. Importantly, as in the mouse, few genes cycled in both heads and bodies, suggesting that tissue specificity is an important component of circadian transcriptional regulation. Furthermore, several cycling genes in the fly were also cycling in the mouse, suggesting that these were conserved output mediators, or perhaps even core clock components.
Recently, three groups reported the use of a similar strategy to identify output genes in Drosophila heads (Claridge-Chang et al., 2001; McDonald and Rosbash, 2001; Ueda et al., 2002). Each study identified >100 transcripts that cycle within a 24 hr period in the fly head, and the expression of these transcripts was affected in arrhythmic backgrounds. Similarly to our work, Claridge-Chang et al. (2001) confirmed a subset of the cycling genes by performing Northern blots on independent time courses. Instead, Ueda et al. (2002) used quantitative RT-PCR to confirm a number of cyclers. A direct comparison of the four datasets yields a surprising result: the overlap among lists represents a small fraction of the total number of cycling genes (14, among those the previously characterized clock components). Is this a reflection of differences in the experimental approaches, genotypes used, or variability of the techniques used? The four experiments have two major aspects in common: the use of total RNA prepared from fly heads and Affymetrix Genechips, although different genotypes have been used (y w, CS, cn bw, and w). Jackson and Schroeder (2001) proposed that the use of different methods to identify the cycling genes is responsible for the variability observed. We favor that hypothesis, coupled with the subtly different experimental designs used in each case [we performed independent analysis on 48-hr-long LD and DD time courses as did Ueda et al. (2002); meanwhile Claridge-Chang et al. (2001) concatenated three independent time courses, each representing the last day of entrainment and the first in free-running conditions (LD + DD), andMcDonald and Rosbash (2001) only looked at a single day in DD)].
In that regard, it appears particularly informative to contrast our observations with those of Ueda et al. (2002) given the similarity in experimental approaches. If we grouped the cycling genes identified by COSOPT (with p < 0.01) in the LD and DD fly head experiments as in Ueda et al. (2002), class I (LD and DD), class II (only LD), and class III (only DD) subsets would contain 24, 96, and 321 genes, respectively. The overlap between the two datasets is high, ranging from ∼40% in class I to 20% in class II genes. Conversely, the overlap between the genes that cycle exclusively in DD is minimal, which might allude to problems in the detection of low-amplitude cyclers or the dampening of the signal, which is characteristic of gene expression under free-running conditions. Nevertheless, the ultimate comparison that will address the limits of the experimental approach is to use a single algorithm to analyze all datasets. As in Ueda et al. (2002), we found a large proportion of cycling genes in DD that were not identified as such in the LD experiment. A direct effect of light on gene expression (by masking or suppression) could account for such an observation.
The important question to ask then is the following: how many of these transcripts are truly cycling? The answer can be derived only from independent observations. In that sense it is encouraging to find that ∼67 of the cycling genes were identified in at least two datasets, which together with those that have been shown to cycle by independent means brings the number of confirmed cyclers to >80 in the fly head. Our results have also shown that looking into different tissues will help address the issue of which proportion of the genome is under clock control, because there is very minimal overlap between the ones that cycle in the head and the body, as is the case in mammals (Panda et al., 2002; Storch et al., 2002).
Microarray experiments are extremely powerful in their scope and should be taken as a starting point to delve into the specifics of different aspects of physiology that appear to be under control of the clock. We identified several genes potentially linked to behavior. Follow-up of one of them, slo, implicates it as a central regulator of locomotor activity, because a null mutation (slo 4) in this locus results in behavioral arrhymicity without a major change in total activity levels. Several scenarios could account for these observations. A mutation in slocould cause arrhythmicity if it directly affects the output pathway controlling behavior by affecting the excitability of the neurons that control it, although if such were the case we would expect hyperkinetic or hypokinetic flies. Alternatively, the mutation could act at the level of the pacemaker neurons by reducing the synchronous firing between the lateral neurons, which would also cause the observed lack of behavioral rhythmicity. slowpoke could also be “gating” (McWatters et al., 2000) fly locomotor activity that would be regulated by additional unidentified components. The observation that slo 4 mutants lack the consolidation of behavior around dawn and dusk clearly favors this hypothesis, although additional work will be required to rule out other plausible scenarios, such as its involvement in the light input pathway that conveys environmental information to the clock or the core oscillator itself.
The notion that a potassium channel is involved in the generation of rhythmic activity was proposed a number of years ago after the analysis of membrane conductance changes in isolated retinal neurons of the mollusk Bulla (McMahon and Block, 1987; Michel et al., 1993). This observation, together with the finding that potassium currents are under circadian regulation in the mouse (D. McMahon, unpublished observations) and that expression of Kcnma1, the slowpokemouse ortholog, cycles (Panda et al., 2002), strongly suggests that this mechanism of control of rhythmic activity could play a role in more complex organisms as well.
Footnotes
This work was supported by National Institutes of Health Grant MH51573 to S.A.K. We thank Karen Wager-Smith, Alejandro Schinder, and Jeff Hall for invaluable discussions; Stacey Harmer and Trey Sato for helpful comments on this manuscript; Camilo Orozco for assistance with data analysis; Andrew Su for his assistance with data visualization; Nigel Atkinson (University of Texas, Austin, TX) for providingslo 4 mutant flies; and the Bloomington Stock Center for the slo I flies. We are also grateful to Dr. I Levitan (University of Pennsylvania, Philadelphia, PA) for the anti-slo antibodies. We also thank Doug McMahon for sharing unpublished observations and Hiroki Ueda for sharing raw data.
Correspondence should be addressed to Steve A. Kay, The Scripps Research Institute, ICND-216, 10550 North Torrey Pines Road, La Jolla, CA, 92037. E-mail: stevek@scripps.edu.
M. Fernanda Ceriani's present address: Instituto de Investigaciones Bioquímicas, Fundación Instituto Leloir, Avenida Patricias Argentinas 435, 1405 Buenos Aires, Argentina.
REFERENCES
- 1.Allada R, White NE, So WV, Hall JC, Rosbash M. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93:791–804. doi: 10.1016/s0092-8674(00)81440-3. [DOI] [PubMed] [Google Scholar]
- 2.Andretic R, Hirsh J. Circadian modulation of dopamine receptor responsiveness in Drosophila melanogaster. Proc Natl Acad Sci USA. 2000;97:1873–1878. doi: 10.1073/pnas.97.4.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atkinson NS, Robertson GA, Ganetzky B. A component of calcium-activated potassium channels encoded by the Drosophila slo locus. Science. 1991;253:551–555. doi: 10.1126/science.1857984. [DOI] [PubMed] [Google Scholar]
- 4.Atkinson NS, Brenner R, Chang W, Wilbur J, Larimer JL, Yu J. Molecular separation of two behavioral phenotypes by a mutation affecting the promoters of a Ca-activated K channel. J Neurosci. 2000;20:2988–2993. doi: 10.1523/JNEUROSCI.20-08-02988.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker MN, Brenner R, Atkinson NS. Tissue-specific expression of a Drosophila calcium-activated potassium channel. J Neurosci. 1995;15:6250–6259. doi: 10.1523/JNEUROSCI.15-09-06250.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blau J, Young MW. Cycling vrille expression is required for a functional Drosophila clock. Cell. 1999;99:661–671. doi: 10.1016/s0092-8674(00)81554-8. [DOI] [PubMed] [Google Scholar]
- 7.Claridge-Chang A, Wijnen H, Naef F, Boothroyd C, Rajewsky N, Young MW. Circadian regulation of gene expression systems in the Drosophila head. Neuron. 2001;32:657–671. doi: 10.1016/s0896-6273(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 8.Cooper RL, Neckameyer WS. Dopaminergic modulation of motor neuron activity and neuromuscular function in Drosophila melanogaster. Comp Biochem Physiol B Biochem Mol Biol. 1999;122:199–210. doi: 10.1016/s0305-0491(98)10160-8. [DOI] [PubMed] [Google Scholar]
- 9.De Gregorio E, Spellman PT, Rubin GM, Lemaitre B. Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proc Natl Acad Sci USA. 2001;98:12590–12595. doi: 10.1073/pnas.221458698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elkins T, Ganetzky B, Wu CF. A Drosophila mutation that eliminates a calcium-dependent potassium current. Proc Natl Acad Sci USA. 1986;83:8415–8419. doi: 10.1073/pnas.83.21.8415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feyereisen R. Insect P450 enzymes. Annu Rev Entomol. 1999;44:507–533. doi: 10.1146/annurev.ento.44.1.507. [DOI] [PubMed] [Google Scholar]
- 12.Gho M, Mallart A. Two distinct calcium-activated potassium currents in larval muscle fibres of Drosophila melanogaster. Pflügers Arch. 1986;407:526–533. doi: 10.1007/BF00657511. [DOI] [PubMed] [Google Scholar]
- 13.Haass C, Kloetzel PM. The Drosophila proteasome undergoes changes in its subunit pattern during development. Exp Cell Res. 1989;180:243–252. doi: 10.1016/0014-4827(89)90228-0. [DOI] [PubMed] [Google Scholar]
- 14.Hamblen-Coyle MJ, Wheeler DA, Rutila JE, Rosbash M, Hall JC. Behavior of period-altered rhythm mutants of Drosophila in light:dark cycles. J Insect Behav. 1992;5:417–446. [Google Scholar]
- 15.Hardin PE. Analysis of period mRNA cycling in Drosophila head and body tissues indicates that body oscillators behave differently from head oscillators. Mol Cell Biol. 1994;14:7211–7218. doi: 10.1128/mcb.14.11.7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harmer SL, Hogenesch JB, Straume M, Chang H-S, Han B, Zhu T, Wang X, Kreps JA, Kay SA. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- 17.Jackson FR, Schroeder AJ. A timely expression profile. Dev Cell. 2001;1:730–731. doi: 10.1016/s1534-5807(01)00100-9. [DOI] [PubMed] [Google Scholar]
- 18.Khush RS, Leulier F, Lemaitre B. Drosophila immunity: two paths to NF-kappaB. Trends Immunol. 2001;22:260–264. doi: 10.1016/s1471-4906(01)01887-7. [DOI] [PubMed] [Google Scholar]
- 19.Kornmann B, Preitner N, Rifat D, Fleury-Olela F, Schibler U. Analysis of circadian liver gene expression by ADDER, a highly sensitive method for the display of differentially expressed mRNAs. Nucleic Acids Res. 2001;29:E51–1. doi: 10.1093/nar/29.11.e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krishnan B, Dryer SE, Hardin PE. Circadian rhythms in olfactory responses of Drosophila melanogaster. Nature. 1999;400:375–378. doi: 10.1038/22566. [DOI] [PubMed] [Google Scholar]
- 21.Lazard D, Zupko K, Poria Y, Nef P, Lazarovits J, Horn S, Khen M, Lancet D. Odorant signal termination by olfactory UDP glucuronosyl transferase. Nature. 1991;349:790–793. doi: 10.1038/349790a0. [DOI] [PubMed] [Google Scholar]
- 22.McDonald MJ, Rosbash M. Microarray analysis and organization of circadian gene expression in Drosophila. Cell. 2001;107:567–578. doi: 10.1016/s0092-8674(01)00545-1. [DOI] [PubMed] [Google Scholar]
- 23.McMahon DG, Block GD. The Bulla circadian pacemaker I. Pacemaker neuron membrane potential controls phase through a calcium-dependent mechanism. J Comp Physiol [A] 1987;161:335–346. doi: 10.1007/BF00603959. [DOI] [PubMed] [Google Scholar]
- 24.McWatters HG, Bastow RM, Hall A, Millar AJ. The ELF3 zeitnehmer regulates light signalling to the circadian clock. Nature. 2000;408:716–720. doi: 10.1038/35047079. [DOI] [PubMed] [Google Scholar]
- 25.Michel S, Geusz ME, Zaritsky JJ, Block GD. Circadian rhythm in membrane conductance expressed in isolated neurons. Science. 1993;259:239–241. doi: 10.1126/science.8421785. [DOI] [PubMed] [Google Scholar]
- 26.Neckameyer WS. Dopamine and mushroom bodies in Drosophila: experience-dependent and -independent aspects of sexual behavior. Learn Mem. 1998;5:157–165. [PMC free article] [PubMed] [Google Scholar]
- 27.Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 28.Peixoto AA, Hall JC. Analysis of temperature-sensitive mutants reveals new genes involved in the courtship song of Drosophila. Genetics. 1998;148:827–838. doi: 10.1093/genetics/148.2.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Price JL, Blau J, Rothenfluh A, Abodeely M, Kloss B, Young MW. Double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell. 1998;94:83–95. doi: 10.1016/s0092-8674(00)81224-6. [DOI] [PubMed] [Google Scholar]
- 30.Rouyer F, Rachidi M, Pikielny C, Rosbash M. A new gene encoding a putative transcription factor regulated by the Drosophila circadian clock. EMBO J. 1997;16:3944–3954. doi: 10.1093/emboj/16.13.3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ryter SW, Tyrrell RM. The heme synthesis and degradation pathways: role in oxidant sensitivity. Heme oxygenase has both pro- and antioxidant properties. Free Radic Biol Med. 2000;28:289–309. doi: 10.1016/s0891-5849(99)00223-3. [DOI] [PubMed] [Google Scholar]
- 32.Saez L, Young MW. Regulation of nuclear entry of the Drosophila clock proteins Period and Timeless. Neuron. 1996;17:911–920. doi: 10.1016/s0896-6273(00)80222-6. [DOI] [PubMed] [Google Scholar]
- 33.Sarov-Blat L, So WV, Liu L, Rosbash M. The Drosophila takeout gene is a novel molecular link between circadian rhythms and feeding behavior. Cell. 2000;101:647–656. doi: 10.1016/s0092-8674(00)80876-4. [DOI] [PubMed] [Google Scholar]
- 34.Saville KJ, Belote JM. Identification of an essential gene, l(3)73Ai, with a dominant temperature-sensitive lethal allele, encoding a Drosophila proteasome subunit. Proc Natl Acad Sci USA. 1993;90:8842–8846. doi: 10.1073/pnas.90.19.8842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schopperle WM, Holmqvist MH, Zhou Y, Wang J, Wang Z, Griffith LC, Keselman I, Kusinitz F, Dagan D, Levitan IB. Slob, a novel protein that interacts with the Slowpoke calcium-dependent potassium channel. Neuron. 1998;20:565–573. doi: 10.1016/s0896-6273(00)80995-2. [DOI] [PubMed] [Google Scholar]
- 36.Scully AL, Zelhof AC, Kay SA. A P element with a novel fusion of reporters identifies regular, a C(2)H(2) zinc-finger gene downstream of the circadian clock. Mol Cell Neurosci. 2002;19:501–514. doi: 10.1006/mcne.2001.1091. [DOI] [PubMed] [Google Scholar]
- 37.Stempfl T, Vogel M, Szabo G, Wulbeck C, Liu J, Hall JC, Stanewsky R. Identification of circadian-clock-regulated enhancers and genes of Drosophila melanogaster by transposon mobilization and luciferase reporting of cyclical gene expression. Genetics. 2002;160:571–593. doi: 10.1093/genetics/160.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 39.Suri V, Qian Z, Hall J, Rosbash M. Evidence that the TIM light response is relevant to light-induced phase shifts in Drosophila melanogaster. Neuron. 1998;21:225–234. doi: 10.1016/s0896-6273(00)80529-2. [DOI] [PubMed] [Google Scholar]
- 40.Ueda HR, Matsumoto A, Kawamura M, Iino M, Tanimura T, Hashimoto S. Genome-wide transcriptional orchestration of circadian rhythms in Drosophila. J Biol Chem. 2002;277:14048–14052. doi: 10.1074/jbc.C100765200. [DOI] [PubMed] [Google Scholar]
- 41.Van Gelder RN, Bae H, Palazzolo MJ, Krasnow MA. Extent and character of circadian gene expression in Drosophila melanogaster: identification of twenty oscillating mRNAs in the fly head. Curr Biol. 1995;5:1424–1436. doi: 10.1016/s0960-9822(95)00280-6. [DOI] [PubMed] [Google Scholar]
- 42.Wang Q, Hasan G, Pikielny CW. Preferential expression of biotransformation enzymes in the olfactory organs of Drosophila melanogaster, the antennae. J Biol Chem. 1999;274:10309–10315. doi: 10.1074/jbc.274.15.10309. [DOI] [PubMed] [Google Scholar]
- 43.Wheeler DA, Hamblen-Coyle MJ, Dushay MS, Hall JC. Behavior in light-dark cycles of Drosophila mutants that are arrhythmic, blind, or both. J Biol Rhythms. 1993;8:67–94. doi: 10.1177/074873049300800106. [DOI] [PubMed] [Google Scholar]
- 44.Yang Z, Sehgal A. Role of molecular oscillations in generating behavioral rhythms in Drosophila. Neuron. 2001;29:453–467. doi: 10.1016/s0896-6273(01)00218-5. [DOI] [PubMed] [Google Scholar]
- 45.Young MW, Kay SA. Time zones: a comparative genetics of circadian clocks. Nat Rev Genet. 2001;2:702–715. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- 46.Zhou Y, Schopperle WM, Murrey H, Jaramillo A, Dagan D, Griffith LC, Levitan IB. A dynamically regulated 14–3-3, Slob, and Slowpoke potassium channel complex in Drosophila presynaptic nerve terminals. Neuron. 1999;22:809–818. doi: 10.1016/s0896-6273(00)80739-4. [DOI] [PubMed] [Google Scholar]