Abstract
Ca2+ channel β subunits modify α1 subunit gating properties through direct interactions with intracellular linker domains. In a previous report (Helton and Horne, 2002), we showed that alternative splicing of the β4 subunit had α1 subunit subtype-specific effects on Ca2+ channel activation and fast inactivation. We extend these findings in the present report to include effects on slow inactivation and block by the peptide toxin ω-conotoxin (CTx)-MVIIC. N-terminal deletion and site-directed mutagenesis experiments revealed that the effects of alternative splicing on toxin block and all aspects of gating could be attributed to a proline-rich motif found within N-terminal β4b amino acids 10–20. Interestingly, this motif is conserved within the third postsynaptic density-95 (PSD-95)/Discs large/zona occludens-1 domain of the distantly related membrane-associated guanylate kinase homolog, PSD-95. Sequence identity of ∼30% made possible the building of β4a and β4bthree-dimensional structural models using PSD-95 as the target sequence. The models (1) reveal that alternative splicing of the β4 N terminus results in dramatic differences in surface charge distribution and (2) localize the proline-rich motif of β4b to an extended arm structure that flanks what would be the equivalent of a highly modified PSD-95 carboxylate binding loop. Northern blot analysis revealed a markedly different pattern of distribution for β4a versus β4bin the human CNS. Whereas β4a is distributed throughout evolutionarily older regions of the CNS, β4b is concentrated heavily in the forebrain. These results raise interesting questions about the functional role that alternative splicing of the β4 subunit has played in the evolution of complex neural networks.
Keywords: calcium channel, β4 subunit, PSD-95, alternative splicing, gating, N terminus, ω-CTx-MVIIC
Voltage-gated Ca2+ channels participate in an extensive array of cellular activities including excitation–contraction coupling, transcription, and neurotransmitter release. Neuronal Cav2 channels are assemblies of up to five subunits, α1, α2/δ, β, and γ. The α1 subunit consists of four homologous repeats (I–IV) of six helices (S1–S6) that arrange to form the selectivity filter and pore. The 24 transmembrane helices are connected by a series of alternating intracellular and extracellular loops. These loops are targets for a host of modifying proteins, including β subunits, G-proteins, calmodulin, and syntaxin, as well as the peptide toxins of venomous spiders and marine snails (Catterall, 2000). Interaction of these proteins with α1subunits typically alters the voltage dependency and kinetics of channel gating, which in turn modifies Ca2+ entry into neurons.
Ultimately, gating behavior is determined by the interactions of individual amino acid side chains with the electrostatic forces within their microenvironments. This is especially true for the positively charged S4 helical segments that constitute the voltage sensors in Na+, Ca2+, and K+ channels. Biophysical studies have shown that depolarization disrupts S4 side-chain interactions of Shaker K+ channels to the extent that S4 helices rotate 180° along their axes (Cha et al., 1999; Glauner et al., 1999). This motion likely triggers a cascade of side-chain disruptions that ultimately leads to rotation and separation of the intracellular S6 segments that form the K+ channel gate (Bezanilla, 2000). Such a mechanism is supported by recent studies delineating the conformational changes associated with open and closed states of bacterial two-membrane-spanning K+ channels (Jiang et al., 2002b) and is generally applicable to Na+ and Ca2+ channel gating.
Attempts have been made to assign specific gating functions to individual Ca2+ channel homology domains. Early chimera studies indicated that the IS6 segment was critical for setting the rate of fast inactivation (Zhang et al., 1994); however, substitution of IIS6 and IIIS6 of the Cav2.3 channel into the slow inactivating Cav1.2 channel caused a leftward shift in the voltage dependence of inactivation and increased the rate of Cav1.2 channel inactivation to near Cav2.3 rates (Stotz et al., 2000). Effects on gating have been reported for amino acid substitutions in IS3 (Zhong et al., 2001), the I-II linker (Berrou et al., 2001), IIS6 (Stotz and Zamponi, 2001), extracellular linkers IIIS3-S4 (Lin et al., 1997), and IVS3-S4 (Hans et al., 1999), and IVS6 (Berjukow et al., 2001). Additive effects on Cav1.2 channel inactivation were reported recently for individual IS6, IIS6, IIIS6, and IVS6 substitutions (Shi and Soldatov, 2002). Together, these data support a structural model of α1 subunits in which individual transmembrane segments are interdependently entwined (Horn, 2000).
Our results indicate that this model also applies to β subunit interactions with α1 subunit intracellular linkers. We have shown previously that β4subunit alternative splicing had α1subtype-specific effects on voltage-dependent activation and inactivation (Helton and Horne, 2002). In this report, we extend these findings to include effects on slow inactivation and block by ω-conotoxin (CTx)-MVIIC; we also identify a proline-rich motif in β4b that is responsible for the observed differences in effects.
MATERIALS AND METHODS
Deletion mutants. Truncation of the β4b N terminus in 10 aa increments was performed using PCR and custom oligonucleotide primers [Integrated DNA Technologies (IDT), Coralville, IA]. All nucleotide and amino acid positions for primers and restriction enzymes correspond to the β4b sequence (GenBank accession number U95020). Each forward primer sequence contained an idealized Kozak (1991)sequence and start codon corresponding to the beginning of each of the deleted 10 amino acids as follows: β4bΔ1–10F, 5′-GCCACCATGACCGCGGACGGGCCG; β4b Δ1–20F, 5′-GCCACCATGCAGGTGGCCCGAGGC. Both reactions included a common β4 reverse primer, β4b 732R (5′-TGACGGCCCCACTAACACC). Full-length β4b was used as the template for these reactions. The β4b Δ10–20 deletion mutant was generated with the primer β4bΔ10–20F (5′-GCCACCATGTCCTCCTCCTCCTACGCCAAGAACTCG) paired with β4b 732R using the β4b Δ1–20 mutant as the template. Annealing temperature for PCR was 56°C with Gene Choice Taq DNA polymerase (PGC Scientific, Durham, NC). Correctly sized PCR fragments were cloned into the pT-Advantage vector (Clontech, Palo Alto, CA). PCR-based cycle sequencing (FS chemistry; Applied Biosystems, Foster City, CA) was used with an ABI Prism 310 Genetic Analyzer. The data were analyzed using ABI Prism DNA sequencing software (version 2.12; PerkinElmer Biosystems), and sequence alignments and restriction maps were generated using Lasergene Software (DNA Star, Madison, WI). Correct clones were digested with BamHI (Roche Molecular Biochemicals, Indianapolis, IN), and the corresponding ∼530 bp fragments were ligated into BamHI (nucleotide position 550) -digested β4b in pBluescript II S/K+ (Stratagene, La Jolla, CA). Each β4b deletion mutant was sequenced to confirm correct reading frame and proper N-terminal orientation.
Site-directed mutagenesis. For all β4b site-directed mutants, full-length β4b cDNA (U95020) was used as the template unless otherwise indicated. All site-directed mutagenesis reactions were performed using a QuikChange site-directed mutagenesis kit (Stratagene) and custom forward and reverse compliment oligonucleotide primers (IDT): β4bG10A,D13A, 5′-ACGCCAAGAACGCGACCGCGGCCGGGCCGCAC; β4bP15A,P18A, 5′-GCGGACGGGGCGCACTCCGCCACCTCGCAGGTG; β4bG10A,D13A,P15A,P18A, 5′-ACGCCAAGAACGCGACCGCGGCCGGGGCGCAC (β4bP15A,P18A used as template); β4bG10A,P15A, 5′-TACGCCAAGAACGCGACCGCGGACGGGGCGCACTCCCCCACCTCGCAGGTG; β4bH16A, 5′-ACCGCGGACGGGCCGGCCTCCCCCACCTC; β4bG10A,P18A, 5′-TACGCCAAGAACGCGACCGCGGACGGGCCGCACTCCGCCACCTCGCAGGTG;β4bD13A,P15A, 5′-TACGCCAAGAACGGGACCGCGGCCGGGGCGCACTCCCCCACCTCGCAGGTG; β4bD13A,P18A, 5′-TACGCCAAGAACGGGACCGCGGCCGGGCCGCACTCCGCCACCTCGCAGGTG;β4bH16A, 5′-ACCGCGGACGGGCCGGCCTCCCCCACCCTCG; and β4bT11A,S17A,T19A,S20A, 5′-CAAGAACGGGGCCGCGGACGGGCCGCACGCCCCCGCCGCGCAGGTGGCC. Each of the mutant clones was sequenced to confirm reaction fidelity.
Electrophysiology. cRNAs were synthesized in vitro using an mMessage mMachine RNA transcription kit from Ambion (Austin, TX) (T3 or T7 depending on clone orientation in pBluescript II S/K+). StandardXenopus laevis oocyte expression methods were used to characterize β deletion and site-directed mutants. Briefly, full-length α1, α2/δ-1, and β4 cRNAs were injected in equimolar ratios (5.6 ng of α1A, 2.4 ng of α2/δ-1, and 1.6 ng of β4 in 46 nl) into defolliculated oocytes (stage V–VI). The BI-2 (α1A) and α2/δ-1 clones used in this study were provided by T. Tanabe (Tokyo Medical and Dental University, Tokyo, Japan). Calcium channel currents were recorded 2–4 d after oocyte injection by standard two-electrode voltage clamp using a Warner amplifier (OC-725B) at 20–22°C, and data were collected using pClamp6 software (Axon Instruments, Foster City, CA). Microelectrodes were filled with 3 m KCl, and the resistances of the current and voltage electrodes were 0.3–1.5 MΩ. Data were filtered at 2 kHz and sampled at 10 kHz. Currents were recorded in a chloride-free bath containing (in mm): 5 Ba(OH)2, 5 HEPES, 85 TEA-OH, and 2 KOH, pH adjusted to 7.4 with methanesulfonic acid. In experiments with the peptide toxin ω-CTx-MVIIC (Peptide Institute Inc. Osaka, Japan), the 5 mm Ba2+ solution was supplemented with 0.1 mg/ml cytochrome c to saturate nonspecific peptide binding sites. Cytochrome c at 0.1 mg/ml had no noticeable effect on recorded Ba2+currents. Peptides were reconstituted according to the manufacturer's instructions (100 μm stock solutions in sterile, deionized water). Fresh dilutions of the peptide were made immediately before use. Currents typically ranged between 0.8 and 2.5 μA, and leak currents were between 20 and 100 nA. Data were analyzed using pClamp6 software (Axon Instruments) and Excel 7.0 (Microsoft Corp., Redmond, WA). The leak and capacitive currents were subtracted on line using a standard P/4 protocol. Curve fitting was performed with SigmaPlot version 5.0 (SSPS Inc., Chicago, IL).
Slow inactivation. Oocytes were held at −80 mV for ∼2 min before a 300 msec reference pulse (IR) to 0 mV (β4b) or +10 mV (β4a). AfterIR, the membrane potential was stepped immediately to a conditioning pulse potential ranging from −100 mV to −20 mV (20 mV increments) and held for 5 min. During the conditioning pulse, a 300 msec test pulse (IT) to 0 mV (β4b) or +10 mV (β4a) was applied every 15 sec. Data were normalized as the ratio of the maximum current at time T (IT) divided by the maximum reference current (IR). Data were fit to the double-exponential equationIT/IR=A1e−x/τ1+A2e−x/τ2, where IT equals current at time T,IR equals maximum reference current, x equals time in seconds, and A1 andA2 are components for the time constants τ1 and τ2, respectively. The SEM is shown for each data point unless the values are smaller than the symbol.
Recovery from slow inactivation. Currents were stabilized at −80 mV or −100 mV for ∼2 min before a 300 msec reference pulse (IR) to 0 mV (β4b) or +10 mV (β4a). After a 100 msec step to either −80 mV or −100 mV, the oocytes were held at a conditioning pulse potential of −30 mV for 5 min. Immediately after the conditioning pulse, a 300 msec test pulse was applied (I1), and then the holding potential was stepped back to either −80 mV or −100 mV and 300 msec test pulses (I2–12) were applied at 15 sec intervals starting at time 0 for a total of 3 min. Data were normalized as the ratio of the maximum current at time T (IT) divided by the maximum reference current (IR). Data were fit to the single exponential equationIT/IR =I∞ +Ae−x/τ, whereIT equals current at time point T,IR equals maximum reference current,I∞ equals current remaining at end of protocol, x equals time in seconds, and A is the component for the time constant τ.
Voltage dependence of activation and inactivation. Voltage dependency of activation data was generated from I–Vcurves. Maximal currents were obtained from 300 msec depolarizations from a holding potential of −80 mV to various test potentials (−40 to +10 mV in 5 mV increments). Each individual recording was then normalized, inverted, and fit to the Boltzmann equation %IBa = 1/[1 + exp(−(Vtest −V1/2)/k)], whereVtest equals I–V test potential, Vpre equals prepulse potential, V1/2 equals midpoint of activation or inactivation, and k equals slope factor. The fit curves, V1/2, and kvalues were then averaged and plotted as a function of membrane voltage.
Voltage dependency of inactivation data was obtained from peak currents elicited by a 300 msec maximal current test depolarization after a 20 sec conditioning prepulse to voltages ranging from −80 to +20 mV. Each individual recording was then normalized and fit to the Boltzmann equation %IBa = 1/[1 + exp([Vpre −V1/2]/k)], whereVtest equals I–V test potential, Vpre equals prepulse potential, V1/2 equals midpoint of activation or inactivation, and k equals slope factor. The fit curves, V1/2, and kvalues were then averaged and plotted as a function of prepulse potential.
Pharmacology. Oocytes were held at a potential of −80 mV with maximal currents elicited by 150 msec test pulses to 0 mV (β4b) or +10 mV (β4a) every 15 sec for a total of 10 min. During recordings, oocytes were perfused at a constant rate of ∼0.5 ml/min. Average current sizes for β4a and β4b complexes were 1.9 ± 0.2 μA (4–5 d of incubation) and 2.3 ± 0.3 μA (2–3 d of incubation), respectively. The data were fit to the single-exponential equationIT/IR= I∞ +Ae−x/τ, whereIT equals maximum current at time point T, IR equals maximum current at time point 0, I∞ equals residual current at end of protocol, x equals time in seconds, and A is the component for the time constant, τ. The averaged rate constants (1/τ) for the four ω-CTx-MVIIC concentrations (0.2, 0.6, 2, and 6 μm) were plotted as a log function of their concentration and were fit well by the equation (τ)−1 =kon[Tx] +koff.
Northern blot analysis. A commercially available human neuronal tissue Northern blot [Multiple Tissue Northern (MTN) Blot brain II; Clontech] was probed with a nonspecific β4 subunit probe (β4ΔN; nucleotides 215–1628 plus ∼300 bp of 3′ untranslated). A 32P-labeled β4 subunit probe was made with a nick translation kit (Promega, Madison, WI) using the β4ΔN mutant as the template. The β4ΔN mutant is missing the first coding 147 bp corresponding to the 49 aa N terminus of β4[clone from Helton and Horne (2002)]. The MTN blot was hybridized overnight at 42°C in hybridization buffer [5× SSC, 5% w/v blocking reagent (Roche), 0.1% N-lauroylsarcosine, 0.02% w/v SDS, 50% w/v formamide] plus 100 μg/ml herring sperm DNA (Promega). The probe concentration was 1 million counts/ml. The blot was washed with successive stringency washes (four washes, 15 min each at 37°C) ranging from 2× SSC/0.1% SDS to 0.1× SSC/0.1% SDS. The blot was then exposed to radiographic film for 12 hr at −80°C. One microgram of cRNA for both β4a and β4b was run out on a 1% denaturing formaldehyde gel along with a poly(A)-tailed cRNA mass ladder (RNA Molecular Weight Marker 1; Roche). The β4a cRNA is longer than the β4b cRNA because of the additional ∼400 nt of 5′ untranslated sequence.
Molecular modeling. The sequences for rat postsynaptic density-95 (PSD-95) (DLG4_rat) and human β4 (CACNB4) were obtained (accession numbers P31016 and U95020) from the Swiss-prot database. Amino acids 10–96 of β4b were aligned to residues 303–390 of PSD-95 based on secondary structure prediction (nnPredict) and visual inspection. For β4a, amino acids 50–96 of β4b were aligned to residues 345–390 of PSD-95. Using default parameters, the program MODELLER 6 (Sali and Blundell, 1993) was used to produce 50 models each of β4b and β4a structure based on the solved structure of the third PSD-95/Discs large/zona occludens-1 (PDZ) domain of PSD-95 (1BEF). Five models each were chosen for additional analysis based on the molecular probability density function (PDF) output from MODELLER and stereochemical analysis obtained through Ramachandran output from PROCHECK-NMR (Laskowski et al., 1993). The interactions between different atom types within these models and Cα root mean square deviation (RMSD) comparisons between the models and 1BEF were characterized with ERRAT (Colovos and Yeates, 1993). The β4a and β4b models chosen for comparison had the fewest disallowed residues (Ramachandran), lowest molecular PDF and RMSD values, and highest percentage of residues in acceptable conformations based on ERRAT and PROCHECK-NMR analysis. Models were visualized with the program MOLMOL (Koradi et al., 1996).
RESULTS
Alternative splicing of the β4 subunit affects slow inactivation of Cav2.1 Ca2+ channels
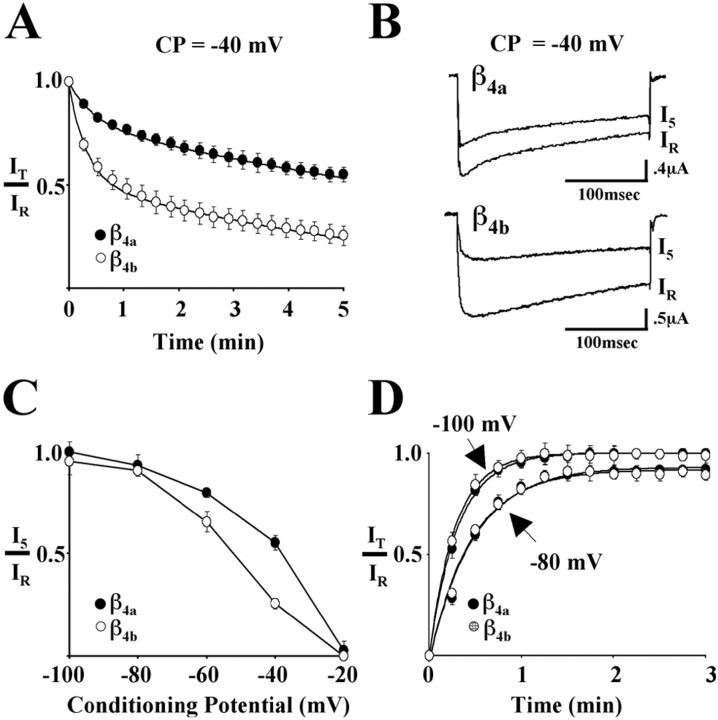
In a previous study (Helton and Horne, 2002), we showed that Cav2.1 complexes containing the longer form of an alternatively spliced β4 subunit N terminus, β4b (49 aa), inactivated at more negative potentials in response to 20 sec conditioning prepulses than complexes containing a shorter form, β4a (15 aa). To determine whether this response extended to slower types of inactivation, we examined in the present study the effects of β4a and β4b on Cav2.1 cumulative inactivation elicited by 5 min conditioning prepulses combined with stimulation at 0.25 Hz. Oocytes were stabilized at −80 mV before a 300 msec reference pulse (IR) to potentials that were predetermined to give peak inward currents (β4b, 0 mV; β4a, +10 mV). The membrane potential was then stepped to and held at the conditioning prepulse potential (ranging from −100 to −20 mV) for 5 min. A 300 msec test pulse (IT) was elicited from the conditioning prepulse potential every 15 sec (I5 equals test pulse at 5 min). The kinetics of entry to slow inactivation for Cav2.1 complexes containing either β4a or β4b at −40 mV is shown in Figure1A. For comparison purposes, we fit the data points for both β4aand β4b to two exponentials (smooth curves in the figure). The time constants for the fast component of entry (τ1) for β4a and β4b were 28.6 ± 2.6 and 18.9 ± 1.2 sec, respectively, and the time constants for the slow component of entry (τ2) were 769 ± 23.6 and 384 ± 14.8 sec, respectively. Overall, theIT/IRratio for Cav2.1 complexes containing β4b decreased to 0.5 in ∼70 sec, whereas those containing β4a required ∼380 sec (data not shown). This indicated that β4b caused a more than fivefold acceleration of the kinetics of slow inactivation. Representative current traces for reference and 5 min test pulses from a conditioning potential of −40 mV for Cav2.1 complexes containing β4a (top) and β4b (bottom) are shown in Figure1B. As seen in the figure and as described in our previous study, Cav2.1 complexes containing β4a underwent open-state fast inactivation faster than did complexes containing β4b. After 5 min at −40 mV, the rate of fast inactivation was unaltered for complexes containing β4a, and slowed only somewhat for complexes containing β4b. The absence of any appreciable tail-current indicated that deactivation was not affected by prolonged depolarization. TheI5/IRratio is plotted against the range of conditioning potentials (−100 to −20 mV) in Figure 1C. The figure illustrates that the voltage dependence of Cav2.1 slow inactivation is shifted to the left for complexes containing β4b relative to those containing β4a. Half-maximal inactivation occurred at approximately −50 mV for complexes containing β4b and −35 mV for β4a. These values are ∼10 mV (β4b) and 5 mV (β4a) more negative than were determined for inactivation in response to 20 sec conditioning prepulses (Helton and Horne, 2002). Figure1D shows that recovery from 5 min of slow inactivation at −30 mV is nearly complete when the membrane potential is stepped back to −80 mV, and that there is no difference in the time course of recovery for Cav2.1 complexes containing either β4a or β4b. Recovery was somewhat faster and more complete when the membrane potential was stepped back to −100 mV. The recovery data at both potentials fit well to single exponentials. The time constants for recovery for β4a and β4b at −80 mV were 28.6 ± 2.0 and 27.8 ± 1.6 sec, respectively, and at −100 mV, 18.9 ± 0.5 and 17.2 ± 0.4 sec, respectively.
Fig. 1.
Effects of β4a and β4bon slow inactivation and recovery from slow inactivation of Cav2.1 Ca2+ channels. Studies were performed with Xenopus oocytes expressing α1A, α2/δ-1, and either β4a or β4b. Reference (IR) and test current (IT) traces were generated by 300 msec step depolarizations from various holding potentials to either 0 mV (β4b) or +10 mV (β4a). Maximum values from 300 msec IR andIT current traces were used to calculateIT/IRwhere indicated. Barium (5 mm) was used as the charge carrier. A, Influence of β4a and β4b on the development of slow inactivation at a conditioning potential (CP) of −40 mV. After a reference pulse (IR) measured from a holding potential of −80 mV, oocytes were held at −40 mV for 5 min. During this time, 300 msec test pulses (IT) were applied every 15 sec. Eachpoint represents the mean value ofIT/IR from 11 (β4a) or 10 (β4b) different recordings. The SEM is shown for each pointunless the values were smaller than the symbol. Thesolid lines represent double-exponential fits to the data. B, Representative reference (IR) and 5 min (I5) current traces from Cav2.1 complexes containing either β4a(top) or β4b (bottom) generated as described in A. C, Voltage dependence of slow inactivation. The ratio ofI5 to IR, generated as in A, plotted as a function of conditioning potential for Cav2.1 complexes containing either β4a or β4b. Data pointsrepresent the means of at least six determinations at a given membrane potential. Lines serve only to connect the data points. D, Influence of β4a and β4b on the time course of recovery from slow inactivation. After a 300 msec reference pulse (IR) measured from a holding potential of either −80 or −100 mV, oocytes were held at −30 mV for 5 min. The membrane potential was then returned to either −80 or −100 mV, and sequential test pulses (IT) were applied at 15 sec intervals for a total of 3 min. Eachpoint represents the mean of at least seven different recordings. Solid lines represent the single-exponential fits of the data.
Alternative splicing of the β4 subunit affects ω-CTx-MVIIC block of Cav2.1 Ca2+channels
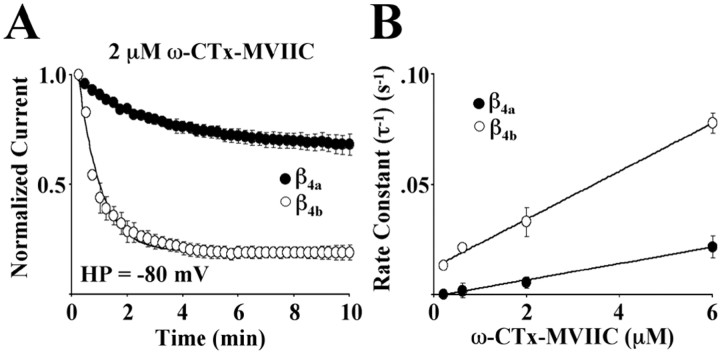
The results to this point indicate that changes in the structure of the β4 subunit N terminus impact α1A subunit structures that are important for many aspects of gating, including activation, open-state inactivation, and fast and slow closed-state inactivation. Given that recent evidence indicates that cytosolic determinants of two-membrane-spanning K+ channel gating are coupled to changes in outer vestibule structure (Perozo et al., 1999; Jiang et al., 2002a,b), we next sought to determine whether alternative splicing of the β4 subunit would affect the block of Cav2.1 channels by a marine snail peptide conotoxin, ω-CTx-MVIIC. Conotoxin interactions with voltage-gated Ca2+ channels are entirely extracellular and occur through binding sites located near H5 (P) helices in several of the six helix transmembrane-spanning motifs (Ellinor et al., 1994). Figure 2A shows the effects of 2 μm ω-CTx-MVIIC on Cav2.1 Ca2+ channel complexes expressed in Xenopus oocytes in the presence of either β4a or β4b. The oocytes were held at −80 mV for 10 min and stimulated every 15 sec. Under these conditions, ω-CTx-MVIIC associated with Cav2.1 complexes containing β4b at a faster rate (τ = 50 ± 0.75 sec) than complexes containing β4a(τ = 200 ± 16 sec). The loss of Ca2+ current resulting from slow inactivation over 10 min at −80 mV (<15%) was subtracted from the data plotted in the figure. Figure 2B demonstrates that as expected for a first-order reaction, the rate constants (τ−1) for toxin block were linearly dependent on toxin concentration as described by the equation (τ)−1 =kon [Tx] +koff. Slopes of linear fits to the data for Cav2.1 complexes containing either β4a and β4b were 3.7 × 10−6m−1 · sec−1 and 1.1 × 10−5m−1 · sec−1, respectively. This indicated that the on-rate (kon) for toxin block was approximately threefold faster for Cav2.1 complexes containing β4b than for those containing β4a.
Fig. 2.
Effects of β4a and β4bon the blockade of Cav2.1 channels by ω-CTx-MVIIC. Studies were performed with Xenopus oocytes expressing α1A, α2/δ-1, and either β4a or β4b.A, Onset and degree of block by a 10 min exposure to 2 μmω-CTx-MVIIC for Cav2.1 subunit combinations at a holding potential (HP) of −80 mV. Each pointrepresents the mean of seven (β4a) or eight (β4b) different recordings. The SEM is shown for each data point unless smaller than thesymbol. Onset of block for both subunit combinations was fit (line) to a single-exponential time course plus a constant. B, The rate constants for the time course of the onset of toxin block were determined from steady-state degree of block from single exponential fits at four different toxin concentrations (0.2, 0.6, 2, and 6 μm) for Cav2.1 complexes containing either β4a or β4b. The averaged rate constants were plotted as a function of toxin concentration (minimum of n = 7, ±SEM). The line represents a linear fit to the data.
The molecular determinants of alternatively spliced β4 subunit differential effects on gating and pharmacology are located within amino acids 10–20 of β4b
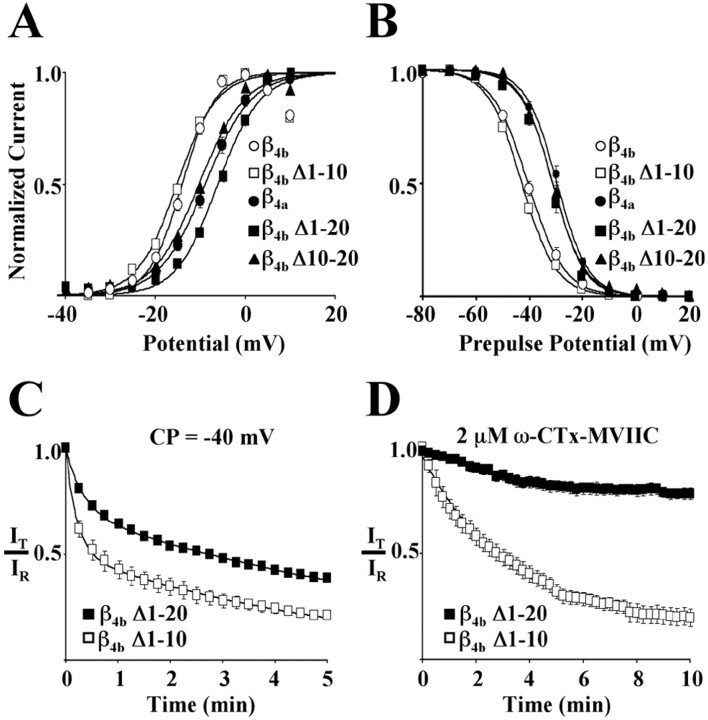
Having characterized many of the functional consequences of alternative splicing of the β4 A domain, we focused next on identifying the key structural determinants underlying the observed differences in effects. It was of particular interest to determine whether or not the effects of alternative splicing on gating and pharmacology could be assigned to separate structural entities. To accomplish this, we first created a series of β4b deletion mutants in which the N terminus was shortened by multiples of 10 aa (β4bΔ1–10 through β4b Δ1–49) and characterized their effects on gating and pharmacology of Cav2.1 complexes. Figure3A–D shows that, relative to full-length β4b, deletion of the first 10 aa (β4b Δ1–10) had no effect on the voltage dependence of activation (Fig. 3A), isochronal (20 sec prepulse) inactivation (Fig. 3B), onset into slow inactivation (Fig. 3C), or susceptibility to block by 2 μm ω-CTx-MVIIC (Fig. 3D). However, when amino acids 1–20 were removed (β4bΔ1–20), both the voltage dependence of activation (Fig.3A) and inactivation (Fig. 3B) of Cav2.1 complexes shifted to more depolarized potentials. As shown in the figure, the acquired gating properties were essentially identical those for Cav2.1 complexes containing β4a. Cav2.1 complexes containing β4b Δ1–20 also had a slower onset into slow inactivation (Fig. 3C) and were less susceptible to block by 2 μm ω-CTx-MVIIC (Fig. 3D). The effects of constructs β4b Δ1–30, β4bΔ1–40, and β4b Δ1–49 were identical to those of β4b Δ1–20 (data not shown). As a first attempt at determining whether the effects of β4b Δ1–20 were simply the result of a decreased size of the β4b N terminus, we reintroduced amino acids 1–10 to the N terminus of β4b Δ1–20 to create the construct β4b Δ10–20. As shown in Figure3A,B, this did not restore theV1/2 of either activation or inactivation to the hyperpolarized potentials characteristic of Cav2.1 complexes containing β4b. Together, these results indicated that the molecular determinants responsible for the observed differences between Cav2.1 complexes containing β4a versus β4b were located in amino acids 10–20 of β4b. Moreover, it was apparent that their influence extended to changes in both gating and pharmacology.
Fig. 3.
Localization of differential effects on Cav2.1 gating and pharmacology to β4bN-terminal amino acids 10–20. The first 10 (β4bΔ1–10), first 20 (β4b Δ1–20), or second 10 (β4b Δ10–20) aa of the N terminus of the β4b subunit were removed using PCR. The deletion mutants as well as β4a or β4b were expressed with α1A and α2δ-1 in Xenopusoocytes. A, Effects of the N-terminal deletion mutants on the voltage dependency of activation of Cav2.1 channels. Plots were derived from averaged I–V data up to +10 mV for each β4 subunit combination. Data pointsrepresent the means of the normalized data at a given membrane potential for a minimum of nine different recordings. Smooth lines represent single Boltzmann fits to the averaged data.B, Normalized, averaged isochronal inactivation curves for Cav2.1 complexes containing the various β4 subunits. Points represent the means of the normalized data at a given membrane potential for a minimum of nine different recordings. Smooth lines represent single Boltzmann fits to the averaged data. C, Effects of β4 N-terminal deletion mutants on the development of slow inactivation at a conditioning potential (CP) of −40 mV. Reference (IR) and test (IT) currents were generated as in Figure 1A. Each point represents the mean value ofIT/IR from 13 (β4b Δ1–10) or nine (β4b Δ1–20) different recordings. The solid lines represent double-exponential fits to the data. D, Onset and degree of block by a 10 min exposure to 2 μm ω-CTx-MVIIC for Cav2.1 complexes containing β4b Δ1–10 or β4b Δ1–20. Data were generated as in Figure2A. Each point represents the average of a minimum of seven recordings. The solid lines represent single-exponential fits to the data.
The β4 A domain is a distant homolog of the third PDZ domain of PSD-95
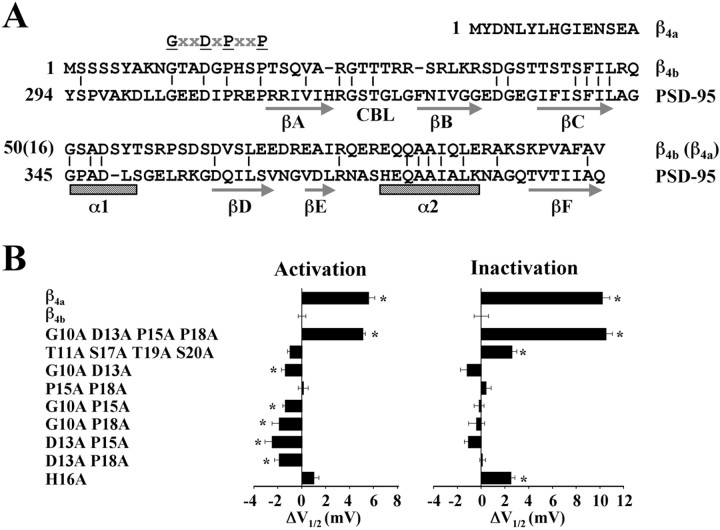
With the results of the deletion experiments highlighting a specific location for the molecular determinants of β4b gating and pharmacology effects, and with the observation that the β1b A domain resembles PDZ domains (Hanlon et al., 1999), we began a systematic comparison of the β4b sequence with similar regions of a number of PDZ domains. Unexpectedly, we found that the entire β4b A domain was weakly homologous to the third PDZ domain of PSD-95 (Fig.4A). Of the 87 aa that have been shown by x-ray crystallography to form the modular PDZ structure of PSD-95 (Doyle et al., 1996), 27 of these (∼31%) are conserved in the β4b sequence. Most importantly, these identities are conserved within key secondary structural elements, such as β-strand C and α-helix 2 of PSD-95. Also of note is the conservation of an RG(S/T)T motif in what would be the equivalent of the carboxylate binding loop (CBL) between β-strands A and B of PSD-95 and the loss of the GLGF motif that is extremely common among PDZ domain subtypes (Bezprozvanny and Maximov, 2001; Harris and Lim, 2001). Four of β4b amino acids 10–20 (G10, D13, P15, and P18) were found in PSD-95. We used these as a starting point for further defining key β4b residues involved in setting Cav2.1 gating parameters.
Fig. 4.
The β4 subunit is a distant homolog of PSD-95. Identification of a conserved GXXDXPXXP motif critical to Cav2.1 gating. A, Amino acid alignment of the A domains of the human spinal cord β4a (amino acids 1–63) and β4b (amino acids 1–97) subunits with the third PDZ domain (amino acids 294–391) of PSD-95. Vertical bars denote identical amino acids between β4b and PSD-95. Important amino acids involved in modulating the leftward shift in the voltage dependence of activation and inactivation of β4b (GXXDXPXXP) are highlighted.Arrows and hatched bars represent predicted α-helices and β-strands of the third PDZ domain of PSD-95, respectively. B, Differences in theV1/2 values of activation and inactivation of β4a, β4b, and β4b N-terminal amino acid mutants. Solid bars represent average V1/2 values of a minimum of nine different recordings for each β4subunit variant. Positive or negative shifts in theV1/2 values (in millivolts) of activation and inactivation of β4a and β4b mutants are relative to the V1/2 values of activation and inactivation of β4b. Currents were generated as described in Figure 3, A and B. The SEM for each bar is shown. Asterisks denote statistical significance (p < 0.05) by a Student's two-sample equal variance t test.
Figure 4B lists a series of site-directed mutants (left), along with their effects on the voltage dependence (V1/2) of activation (middle) and inactivation (right) of Cav2.1 complexes. TheV1/2 values for complexes containing β4a and β4b are included for comparison. Interestingly, the first site-directed β4b mutant tested, G10A, D13A, P15A, P18A, in which all four of the amino acids in common with PSD-95 were altered, displayed activation and inactivation properties similar to that of β4a. To determine whether this was a specific effect, we altered four different amino acids in the β4b 10–20 sequence to create the mutant, T11A, S17A, T19A, S20A. As shown in Figure 4B, Cav2.1 gating properties changed little in response to these mutations. Cav2.1 complexes containing the G10A, D13A, P15A, P18A mutant also had β4a-like slow inactivation and pharmacological properties (data not shown). This indicates that the conserved amino acids are playing a defining role in the gating motif. To delineate the structure in more detail, we subsequently characterized six of the possible G10, D13, P15, P18 amino acid pairs for their effects on gating. Surprisingly, none of the pairs were absolutely essential for maintaining wild-type β4b gating behavior, although small but statistically significant hyperpolarizing effects on activation were noted for five of the six pairs. To complete the alanine substitution study, we created the mutant H16A, which had a small but statistically significant effect on inactivation but not activation.
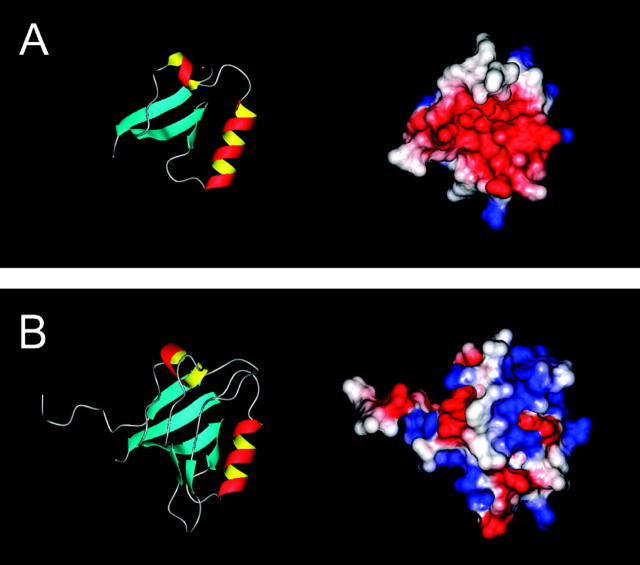
One interpretation of these results is that β4bamino acids 10–20 form a ligand motif that interacts with a binding pocket located somewhere either on the α1Asubunit or on the β4 subunit itself. The affinity of the ligand motif for its receptor site could be defined, for example, by the sum of the interactions of amino acids G10, D13, P15, and P18 with their individual targets. Any given pair may be capable of maintaining a binding interaction under the conditions of our experiments. As a first step toward addressing this possibility, we created three-dimensional structural models of the β4a and β4b A domains (Fig. 5A,B) using the real-space optimization method used in the computer program MODELLER (Sali and Blundell, 1993). The models were initiated using the distance and dihedral angle restraints derived from alignments with portions of the sequence of the third PDZ domain of PSD-95. For β4b, amino acids 10–96 were aligned with amino acids 303–390 of PSD-95. There is 31% sequence identity over this region, which is considered minimally acceptable for this type of comparative modeling (Martí-Renom et al., 2000). For β4a, amino acids 10–49 of β4b were deleted from the alignment. Thus, the models do not include the first 15 aa of β4aand the first 9 aa of β4b. Figure 5A(left) shows that β4a models as a compact structure containing three β-sheets and 2 α-helices. Stereochemical quality assessment of the model using PROCHECK-NMR (Laskowski et al., 1993) identified 41 residues (87.7%) in most favored regions, 5 (10.6%) in additional and generously allowed regions, and 1 (2.1%) in a disallowed region. Calculation of the electrostatic surface potential using MOLMOL (Koradi et al., 1996) reveals that the face of the molecule as oriented in Figure5A, left, contains a pocket of negative charge (red residues) between the two α-helices (Fig. 5A,right). Figure 5B, right andleft, illustrates that the overall effect of alternative splicing to form β4b is to bury the charged pocket beneath three additional β-sheets. Interestingly, the molecule acquires as the result of splicing a positively charged binding pocket (blue residues) in what would be the equivalent of the CBL in PSD-95 (Fig. 4A). The stereochemical quality of the β4b model as shown is not quite as good as that for β4a. PROCHECK-NMR identified 64 residues (79%) in most favored regions, 11 residues (13.6%) in additional allowed regions, and 3 residues (3.7%) each in generously allowed and disallowed regions. Two of the three disallowed residues (R30 and K34) flank what would be the equivalent PSD-95 β-sheet B. Together with the loss of the highly conserved PDZ GLGF sequence, these results are consistent with the notion that through evolution this region of the β4b structure has evolved away from the capacity to bind C-terminal peptide motifs. Of most importance to our present results, however, is the observation that β4b amino acids 10–20 model as an extended arm (pointing to the left in Fig. 5B, right, left) that may serve as a ligand motif. Interestingly, the orientation of the arm appears to be dictated by the isomerization state of proline18 (data not shown).
Fig. 5.
Real-space optimization structural models of the A domains of β4a (A) and β4b (B) based on sequence identities with the third PDZ domain of PSD-95. Ribbon (left) and electrostatic surface potential (right) diagrams were created using MOLMOL (Koradi et al., 1996). For ribbon diagrams, arrows indicate β-strands, and helices indicate α-helices. For surface potential diagrams, red, white, and blue regions indicate negatively charged, hydrophobic, and positively charged amino acids, respectively.
Differential distribution of alternatively spliced β4subunit mRNA
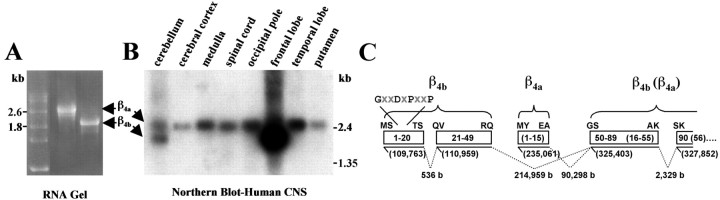
We noted in our previous study (Helton and Horne, 2002) that, based on extensive cDNA library screening, β4awas the predominant alternatively spliced variant of the β4 subunit expressed in human spinal cord. To confirm this observation, we performed a comparative Northern blot analysis using a commercially available multiple tissue Northern blot (Human Brain II; Clontech) and a β4 cDNA probe containing sequence common to both β4a and β4b. The mRNAs for β4aand β4b can be readily distinguished by their distinct migration pattern in agarose-formaldehyde gels (Fig.6A). The results of the Northern blot analysis, shown in Figure 6B, were striking, revealing that not only was β4a the predominant form of β4 subunit in the spinal cord, but also in other “reptilian” portions of the human CNS such as the medulla and putamen. Moreover, β4a was the predominant form of β4 subunit expressed in evolutionarily older regions of the cerebrum, the temporal lobe, and occipital pole. In marked contrast, β4b was highly expressed in the most recent and most highly integrative region of the cerebrum, the frontal lobe. The two forms of the β4 subunit were equally expressed in the cerebellum.
Fig. 6.
Differential distribution of β4a and β4b mRNA in the human CNS. A, Electrophoresis of full-length β4a (left) and β4b (right) cRNAs (includes 5′ and 3′ untranslated) in a 1% agarose formaldehyde denaturing gel. RNA markers (in kilobases) are indicated on the left.B, Northern blot analysis performed with human multiple tissue blot (Human Brain II; Clontech) and a 32P-labeled β4 subunit probe (coding nucleotides 215–1628 plus ∼300 bp 3′ untranslated sequence). Molecular masses on theright correspond to labeled blot markers.C, A human β4 subunit genome map depicting the lengths of intron sequences (b, bases) between alternatively spliced β4a and β4b N-terminal exons and the beginning of exon 2. Solid lines represent exons, and dashed lines represent introns.Numbers in parentheses below solid lines indicate position on chromosome 2. Boxesindicate protein sequence (β4a inparentheses). The first and last two amino acids of each sequence are indicated above each box.
A basic local alignment search tool search of the human genome (Altschul et al., 1990) with β4 sequences revealed that the exons coding for alternatively spliced forms of the β4 subunit A domain are distributed widely on human chromosome 2. Figure 6C shows that, depending on the splice variant, the coding sequence for the β4PDZ domain is contained within three (β4a) or four (β4b) exons spread out over ∼218,000 bases. The coding sequence for the GXXDXPXXP motif is included in the 5′-most exon of a pair of short exons that code for β4b amino acids 1–49. Assembly of the β4b mRNA requires that three RNA segments (536 bases, 214,959 bases, and 2329 bases) be spliced out. The short exon coding for the first 15 aa of β4a lies between the β4b N-terminal exons and the exon coding amino acids 50–89 and 16–55 of β4b and β4a, respectively. By comparison, the third PDZ domain of PSD-95 is encoded by two exons separated by a 200 bp intron (data not shown).
DISCUSSION
We have identified an alternatively spliced proline-rich motif in the Ca2+ channel β4 subunit that has considerable influence over gating of neuronal Cav2.1 Ca2+ channels. Given that the motif also affects extracellular toxin binding, it is likely that the interaction of this motif with its binding site has wide-reaching impact on resting and open-state Ca2+ channel conformations. This notion is supported by recent images of the conformational changes that occur with gating of bacterial two-membrane-spanning K+ channels (Liu et al., 2001; Jiang et al., 2002a,b). Like eukaryotic six-membrane-spanning K+ channels, KcsA and MthK channels are tetramers that pack with fourfold symmetry around a central pore (Doyle et al., 1998; Jiang et al., 2002a). The principal structural elements of KcsA and MthK from N to C terminus include an outer transmembrane helix (M1), a pore helix (P), and an inner transmembrane helix (M2). These correspond to S5, H5, and S6 segments of voltage-gated Ca2+ channels, respectively. In the closed conformation of the KcsA structure, the four M2 helices are straight and arranged such that they form the walls of an inverted teepee that narrows from a 12 Å diameter at its center to a 4 Å pore at its tip (Doyle et al., 1998). After opening, the KcsA M2 helices tilt away from the permeation pathway and rotate about their helical axis (Liu et al., 2001). In MthK, bending and splaying of the inner helices after opening expands the diameter of the pore threefold (Jiang et al., 2002a,b). The nearly 30° bend occurs at a “gating hinge” corresponding to a glycine residue just below the selectivity filter. Applying a radial-outward force on the intracellular aspects of the inner helices places a torque on the gating hinge such that a conformational change is transmitted the full length of the M2 helix. This results in a widening of the outer vestibule [Jiang et al. (2002b), see supplementary information (movie)]. It is has been hypothesized that similar mechanical forces are at work in the gating of voltage-gated Ca2+ channels (Jiang et al., 2002b).
Considered in the context of this mechanical framework, our results suggest that an interaction of the β4b ligand motif with an inner aspect of the Cav2.1 complex either directly or indirectly fine-tunes the torque experienced by α1A S6 segments. In so doing, the interaction alters the conformation of the voltage sensor or gate (or both) as well as the outer vestibule. This would explain why alternative splicing of the β4 subunit would affect both gating and toxin sensitivity. Given the potential for the ligand motif interaction to occur over a wide range (Helton and Horne, 2002), it is not possible from our results to pinpoint which S6 helices might be most affected. And although our ω-CTx-MVIIC binding results might be providing some direction, a previous study has shown that ω-CTx-GVIA binding to α1B is affected by alterations in any of the four P loops that make up the outer vestibule (Ellinor et al., 1994). A case can be made for an indirect effect on the IS6 helix, because the primary α1-β4 subunit interaction occurs on the intracellular loop between homology domains I and II (I-II loop) (Pragnell et al., 1994). This is consistent with a previous study showing that IS6 is a critical determinant of voltage-dependent inactivation in Cav2.1 and Cav2.3 channels (Zhang et al., 1994). However, site-directed mutagenesis and domain-swapping studies have highlighted the equal importance of IIS6, IIIS6, and IVS6 in Ca2+ channel gating (for review, see Stotz and Zamponi, 2001; Shi and Soldatov, 2002), making the case for direct effects on these S6 helices equally plausible. It is interesting that many of the proteins that have evolved to modulate Ca2+ channel gating target α1 I-II (β subunits, Gβγ subunits, protein kinase C) and II-III loops (syntaxin, synaptotagmin, and synaptosomal-associated protein-25). The IS6 and IIS6 (but not IIIS6 or IVS6) helices of Cav2.1, 2.1, and 2.3 Ca2+ channels have glycines in hinge positions comparable with those present in KcsA and MthK (see alignments in Horne et al., 1993).
Despite extensive binding studies with β4subunits, there is currently no evidence to support the notion that the β4b N terminus binds directly to α1A subunits (Walker et al., 1998, 1999). One possible explanation for this is that the interaction is too weak to be detected in solution binding assays. The other possibility is that the α1A subunit is not the primary binding target. The sequence of the ligand motif (GXXDXPXXP) may provide an important clue as to the nature of its own binding site. Proline-rich motifs are common within the primary structures of many ligands important for protein–protein interactions (for review, see Kay et al., 2000; Macias et al., 2002). Src homology 3 (SH3) and WW domains, for example, recognize proline-rich sequences containing a core PXXP, where X denotes any amino acid. These sequences adopt a PPII helix conformation that presents a hydrophobic surface as well as backbone carbonyls that are ideal for hydrogen bonding. Proline-rich ligands bind with low affinity, allowing for rapid modulation and added versatility in signaling pathways. In many respects, the β4bligand motif resembles the serine/threonine proline motifs recognized by group IV WW domains (Sudol and Hunter, 2000). As is the case for the β4b motif, the structural basis for recognition of these motifs is based on the summed contributions of a series of side-chain interactions, none of which is absolutely essential for ligand binding (Verdecia et al., 2000). It is possible that the β4b proline-rich motif binds to its own B domain, which is structurally similar to SH3 and WW domains (Hanlon et al., 1999). This possibility is supported by what is known about related membrane-associated guanylate kinase family proteins in which intramolecular interactions are a key aspect of their functional diversity (Dimitratos et al., 1999).
To date, no specific function has been assigned to the GXXDXPXXP motif of the third PDZ domain of PSD-95. Most attention has been paid to the structure of the carboxylate binding loop to which it is immediately adjacent (Doyle et al., 1996) (Fig. 4, CBL). A partial list of the proteins that interact specifically with the third PDZ domain of PSD-95 include the cell-surface neuroligins (Irie et al., 1997), the microtubule binding protein cysteine-rich interactor of PDZ 3 (Niethammer et al., 1998), the Rho effector protein citron (Zhang et al., 1999), and the β1 adrenergic receptor (Hu et al., 2000). It would be interesting to determine whether the GXXDXPXXP motif of PSD-95 plays a role maintaining the structure of the CBL. Such a role could also be considered for the GXXDXPXXP motif of β4b. As is apparent in the β4b A domain model structure (Figs.4A, 5B), the sequences of β4b corresponding to the hydrophobic CBL and β-strand B of PSD-95 are highly positively charged. It is possible, as is true for some PDZ domains (Cuppen et al., 1998), that this region binds to an internal (as opposed to a C-terminal) negatively charged domain. Interestingly, the immediate 5′ sequence of the II-III loop of Cav2.1 and Cav2.2 Ca2+ channels fits this description, because it is densely packed with glutamate residues. Moreover, several lines of evidence point to the II-III loop as a critical determinant of channel gating and pharmacology. Similar to the effects of β4b on α1A, binding of syntaxin to a “synprint site” just downstream from this region in the II-III loop of α1B shifts theV1/2 of inactivation to more hyperpolarized potentials (Bezprozvanny et al., 2000) and accelerates entry into slow inactivation (Degtiar et al., 2000), and an α1B splice variant that lacks a large portion of the II-III linker region (Δ1) (Kaneko et al., 2002) inactivates at more depolarized potentials and is less sensitive to ω-CTx-MVIIA than is the full-length Cav2.2 variant.
Viewed from the perspective of changing Ca2+ channel function, the differential distribution of β4a and β4b subunit mRNA displayed in Figure6B provides an unexpected snapshot of the evolution of forebrain synapses. It raises the possibility that with the introduction of β4b to the genome, synapses acquired properties that fit better with the overall demand to organize complex neural networks. It could be that with the advent of Cav2.1 complexes that enter more readily into closed inactivation states (Fig. 1A,C) without perceptible gain in time required for recovery (Fig.1D), synapses inherited an enhanced mechanism for synaptic plasticity. In this regard, short-term synaptic depression has been linked to Ca2+ channel inactivation (Forsythe et al., 1998) through mechanisms shown to be β subunit dependent (Patil et al., 1998). This form of short-term synaptic plasticity has been implicated in cortical gain control (Abbott et al., 1997) and low-pass temporal filtering (Fortune and Rose, 2001). In addition, long-lasting Cav2.1 channel inhibition has been identified as the mechanism underlying certain forms of long-term synaptic depression (Robbe et al., 2002). Accordingly, our future studies will be directed toward characterizing the responsiveness of Cav2 Ca2+ channels containing alternatively spliced β4 subunits to changes in more dynamic regulatory inputs, such as neuronal firing frequency and action potential waveform.
Footnotes
This work was supported by National Institutes of Health Grants NS32094 (W.A.H.) and GM5576 (J.C.), by a College of Veterinary Medicine State Research Support grant (W.A.H.), and by an award from the North Carolina State University Keenan Institute (J.C.).
Correspondence should be addressed to William A. Horne, Department of Molecular Biomedical Sciences, North Carolina State University, College of Veterinary Medicine, 4700 Hillsborough Street, Raleigh, NC 27606. E-mail: bill_horne@ncsu.edu.
REFERENCES
- 1.Abbott LF, Varela JA, Sen K, Nelson SB. Synaptic depression and cortical gain control. Science. 1997;275:220–224. doi: 10.1126/science.275.5297.221. [DOI] [PubMed] [Google Scholar]
- 2.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Berjukow S, Marksteiner R, Sokolov S, Weiss RG, Margreiter E, Hering S. Amino acids in segment IVS6 and β-subunit interaction support distinct conformational changes during Cav2.1 inactivation. J Biol Chem. 2001;276:17076–17082. doi: 10.1074/jbc.M010491200. [DOI] [PubMed] [Google Scholar]
- 4.Berrou L, Bernatchez G, Parent L. Molecular determinants of inactivation within the I-II linker of α1E (CaV2.3) calcium channels. Biophys J. 2001;80:215–228. doi: 10.1016/S0006-3495(01)76008-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bezanilla F. The voltage sensor in voltage-dependent ion channels. Physiol Rev. 2000;80:555–592. doi: 10.1152/physrev.2000.80.2.555. [DOI] [PubMed] [Google Scholar]
- 6.Bezprozvanny I, Maximov A. PDZ domains: more than just a glue. Proc Natl Acad Sci USA. 2001;98:787–789. doi: 10.1073/pnas.98.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bezprozvanny I, Zhong P, Scheller RH, Tsien RW. Molecular determinants of the functional interaction between syntaxin and N-type Ca2+ channel gating. Proc Natl Acad Sci USA. 2000;97:13943–13948. doi: 10.1073/pnas.220389697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Catterall WA. Structure and regulation of voltage-gated Ca2+ channels. Annu Rev Cell Dev Biol. 2000;16:521–555. doi: 10.1146/annurev.cellbio.16.1.521. [DOI] [PubMed] [Google Scholar]
- 9.Cha A, Snyder GE, Selvin PR, Bezanilla F. Atomic scale movement of the voltage-sensing region in a potassium channel measured via spectroscopy. Nature. 1999;402:809–813. doi: 10.1038/45552. [DOI] [PubMed] [Google Scholar]
- 10.Colovos C, Yeates TO. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 1993;2:1511–1519. doi: 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cuppen E, Gerrits H, Pepers B, Wieringa B, Hendriks W. PDZ motifs in PTP-BL and RIL bind to internal protein segments in the LIM domain protein RIL. Mol Biol Cell. 1998;9:671–683. doi: 10.1091/mbc.9.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Degtiar VE, Scheller RH, Tsien RW. Syntaxin modulation of slow inactivation of N-type calcium channels. J Neurosci. 2000;20:4355–4367. doi: 10.1523/JNEUROSCI.20-12-04355.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dimitratos SD, Woods DF, Stathakis DG, Bryant PJ. Signaling pathways are focused at specialized regions of the plasma membrane by scaffolding proteins of the MAGUK family. BioEssays. 1999;21:912–921. doi: 10.1002/(SICI)1521-1878(199911)21:11<912::AID-BIES3>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 14.Doyle DA, Lee A, Lewis J, Kim E, Sheng M, MacKinnon R. Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ. Cell. 1996;85:1067–1076. doi: 10.1016/s0092-8674(00)81307-0. [DOI] [PubMed] [Google Scholar]
- 15.Doyle DA, Morais-Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 16.Ellinor PT, Zhang JF, Horne WA, Tsien RW. Structural determinants of the blockade of N-type calcium channels by a peptide neurotoxin. Nature. 1994;372:272–275. doi: 10.1038/372272a0. [DOI] [PubMed] [Google Scholar]
- 17.Forsythe ID, Tsujimoto T, Barnes-Davies M, Cuttle MF, Takahashi T. Inactivation of presynaptic calcium current contributes to synaptic depression at a fast central synapse. Neuron. 1998;20:797–807. doi: 10.1016/s0896-6273(00)81017-x. [DOI] [PubMed] [Google Scholar]
- 18.Fortune ES, Rose GJ. Short-term synaptic plasticity as a temporal filter. Trends Neurosci. 2001;24:381–385. doi: 10.1016/s0166-2236(00)01835-x. [DOI] [PubMed] [Google Scholar]
- 19.Glauner KS, Mannuzzu LM, Gandhi CS, Isacoff EY. Spectroscopic mapping of voltage sensor movement in the Shaker potassium channel. Nature. 1999;402:813–817. doi: 10.1038/45561. [DOI] [PubMed] [Google Scholar]
- 20.Hanlon MR, Berrow NS, Dolphin AC, Wallace BA. Modeling of a voltage-dependent Ca2+ channel β subunit as a basis for understanding its functional properties. FEBS Lett. 1999;445:366–370. doi: 10.1016/s0014-5793(99)00156-8. [DOI] [PubMed] [Google Scholar]
- 21.Hans M, Urrutia A, Deal C, Brust PF, Stauderman K, Ellis SB, Harpold MM, Johnson EC, Williams ME. Structural elements in domain IV that influence biophysical and pharmacological properties of human α1A-containing high-voltage-activated calcium channels. Biophys J. 1999;76:1384–1400. doi: 10.1016/S0006-3495(99)77300-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harris BZ, Lim WA. Mechanism and role of PDZ domains in signaling complex assembly. J Cell Sci. 2001;114:3219–3231. doi: 10.1242/jcs.114.18.3219. [DOI] [PubMed] [Google Scholar]
- 23.Helton TD, Horne WA. Alternative splicing of the β4 subunit has α1 subunit subtype-specific effects on Ca2+ channel gating. J Neurosci. 2002;22:1573–1582. doi: 10.1523/JNEUROSCI.22-05-01573.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horn R. A new twist in the saga of charge movement in voltage-dependent ion channels. Neuron. 2000;25:511–514. doi: 10.1016/s0896-6273(00)81055-7. [DOI] [PubMed] [Google Scholar]
- 25.Horne WA, Ellinor PT, Inman I, Zhou M, Tsien RW, Schwarz TL. Molecular diversity of Ca2+ channel α1 subunits from the marine ray Discopyge ommata. Proc Natl Acad Sci USA. 1993;90:3787–3791. doi: 10.1073/pnas.90.9.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu LA, Tang Y, Miller WE, Cong M, Lau AG, Lefkowitz RJ, Hall RA. β1-adrenergic receptor association with PSD-95: inhibition of receptor internalization and facilitation of β1-adrenergic receptor interaction with N-methyl-d-aspartate receptors. J Biol Chem. 2000;275:38659–38666. doi: 10.1074/jbc.M005938200. [DOI] [PubMed] [Google Scholar]
- 27.Irie M, Hata Y, Takeuchi M, Ichtchenko K, Toyoda A, Hirao K, Takai Y, Rosahl TW, Sudhof TC. Binding of neuroligins to PSD-95. Science. 1997;277:1511–1515. doi: 10.1126/science.277.5331.1511. [DOI] [PubMed] [Google Scholar]
- 28.Jiang Y, Lee A, Chen J, Cadene M, Chait BT, MacKinnon R. Crystal structure and mechanism of a calcium-gated potassium channel. Nature. 2002a;417:515–522. doi: 10.1038/417515a. [DOI] [PubMed] [Google Scholar]
- 29.Jiang Y, Lee A, Chen J, Cadene M, Chait BT, MacKinnon R. The open pore conformation of potassium channels. Nature. 2002b;417:523–526. doi: 10.1038/417523a. [DOI] [PubMed] [Google Scholar]
- 30.Kaneko S, Cooper CB, Nishioka N, Yamasaki H, Suzuki A, Jarvis SE, Akaike A, Satoh M, Zamponi GW. Identification and characterization of novel human Cav2.2 (a1B) calcium channel variants lacking the synaptic protein interaction site. J Neurosci. 2002;22:82–92. doi: 10.1523/JNEUROSCI.22-01-00082.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kay BK, Williamson MP, Sudol M. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 2000;14:231–241. [PubMed] [Google Scholar]
- 32. Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph 14 1996. 51 55:29–32. [DOI] [PubMed] [Google Scholar]
- 33.Kozak M. An analysis of vertebrate mRNA sequences: intimations of translational control. J Cell Biol. 1991;115:887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J Mol Biol. 1993;231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- 35.Lin Z, Haus S, Edgerton J, Lipscombe D. Identification of functionally distinct isoforms of the N-type Ca2+ channel in rat sympathetic ganglia and brain. Neuron. 1997;18:153–166. doi: 10.1016/s0896-6273(01)80054-4. [DOI] [PubMed] [Google Scholar]
- 36.Liu YS, Sompornpisut P, Perozo E. Structure of the KcsA channel intracellular gate in the open state. Nat Struct Biol. 2001;8:883–887. doi: 10.1038/nsb1001-883. [DOI] [PubMed] [Google Scholar]
- 37.Macias MJ, Wiesner S, Sudol M. WW and SH3 domains, two different scaffolds to recognize proline-rich ligands. FEBS Lett. 2002;513:30–37. doi: 10.1016/s0014-5793(01)03290-2. [DOI] [PubMed] [Google Scholar]
- 38.Martí-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 39.Niethammer M, Valtschanoff JG, Kapoor TM, Allison DW, Weinberg TM, Craig AM, Sheng M. CRIPT, a novel postsynaptic protein that binds to the third PDZ domain of PSD-95/SAP90. Neuron. 1998;20:693–707. doi: 10.1016/s0896-6273(00)81009-0. [DOI] [PubMed] [Google Scholar]
- 40.Patil PG, Brody DL, Yue DT. Preferential closed-state inactivation of neuronal calcium channels. Neuron. 1998;20:1027–1038. doi: 10.1016/s0896-6273(00)80483-3. [DOI] [PubMed] [Google Scholar]
- 41.Perozo E, Cortes DM, Cuello LG. Structural rearrangements underlying K+-channel activation gating. Science. 1999;285:73–78. doi: 10.1126/science.285.5424.73. [DOI] [PubMed] [Google Scholar]
- 42.Pragnell M, De Waard M, Mori Y, Tanabe T, Snutch TP, Campbell KP. Calcium channel β-subunit binds to a conserved motif in the I-II cytoplasmic linker of the α1-subunit. Nature. 1994;368:67–70. doi: 10.1038/368067a0. [DOI] [PubMed] [Google Scholar]
- 43.Robbe D, Alonso G, Chaumont S, Bockaert J, Manzoni AJ. Role of P/Q-Ca2+ channels in metabotropic glutamate receptor 2/3-dependent presynaptic long-term depression at nucleus accumbens synapses. J Neurosci. 2002;22:4346–4356. doi: 10.1523/JNEUROSCI.22-11-04346.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 45.Shi C, Soldatov NM. Molecular determinants of voltage-dependent slow inactivation of the Ca2+ channel. J Biol Chem. 2002;277:6813–6821. doi: 10.1074/jbc.M110524200. [DOI] [PubMed] [Google Scholar]
- 46.Stotz SC, Zamponi GW. Identification of inactivation determinants in the domain IIS6 region of high voltage-activated calcium channels. J Biol Chem. 2001;276:33001–33010. doi: 10.1074/jbc.M104387200. [DOI] [PubMed] [Google Scholar]
- 47.Stotz SC, Hamid J, Spaetgens RL, Jarvis SE, Zamponi GW. Fast inactivation of voltage-dependent calcium channels. A hinged-lid mechanism? J Biol Chem. 2000;275:24575–24582. doi: 10.1074/jbc.M000399200. [DOI] [PubMed] [Google Scholar]
- 48.Sudol M, Hunter T. New wrinkles for an old domain. Cell. 2000;103:1001–1004. doi: 10.1016/s0092-8674(00)00203-8. [DOI] [PubMed] [Google Scholar]
- 49.Verdecia MA, Bowman ME, Lu KP, Hunter T, Noel JP. Structural basis for phosphoserine-proline recognition by group IV WW domains. Nat Struct Biol. 2000;7:639–643. doi: 10.1038/77929. [DOI] [PubMed] [Google Scholar]
- 50.Walker D, Bichet D, Campbell KP, De Waard M. A β4 isoform-specific interaction site in the carboxyl-terminal region of the voltage-dependent Ca2+ channel α1A subunit. J Biol Chem. 1998;273:2361–2367. doi: 10.1074/jbc.273.4.2361. [DOI] [PubMed] [Google Scholar]
- 51.Walker D, Bichet D, Geib S, Mori E, Cornet V, Snutch TP, Mori Y, De Waard M. A new β subtype-specific interaction in α1A subunit controls P/Q-type Ca2+ channel activation. J Biol Chem. 1999;274:12383–12390. doi: 10.1074/jbc.274.18.12383. [DOI] [PubMed] [Google Scholar]
- 52.Zhang JF, Ellinor PT, Aldrich RW, Tsien RW. Molecular determinants of voltage-dependent inactivation in calcium channels. Nature. 1994;372:97–100. doi: 10.1038/372097a0. [DOI] [PubMed] [Google Scholar]
- 53.Zhang W, Vazquez L, Apperson M, Kennedy MB. Citron binds to PSD-95 at glutamatergic synapses on inhibitory neurons in the hippocampus. J Neurosci. 1999;19:96–108. doi: 10.1523/JNEUROSCI.19-01-00096.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhong H, Li B, Scheuer T, Catterall WA. Control of gating mode by a single amino acid residue in transmembrane segment IS3 of the N-type Ca2+ channel. Proc Natl Acad Sci USA. 2001;98:4705–4709. doi: 10.1073/pnas.051629098. [DOI] [PMC free article] [PubMed] [Google Scholar]