Abstract
Myosin-1c plays an essential role in adaptation of hair-cell mechanoelectrical transduction. To mediate adaptation, myosin-1c must interact directly or indirectly with other components of the transduction apparatus, including the mechanically gated transduction channel. As a first step toward identifying myosin-1c receptors, we used recombinant myosin-1c fragments to identify specific binding sites in hair cells and to biochemically characterize their interaction with myosin-1c. Myosin-1c fragments bound to tips of hair-cell stereocilia, the location of transduction and adaptation. Surprisingly, this interaction did not depend on the C-terminal tail of myosin-1c, proposed previously to be the receptor-binding site of the molecule. Instead, the interaction of myosin-1c with stereociliary receptors depended on its calmodulin-binding IQ domains. This interaction was blocked by calmodulin, which probably bound to a previously unoccupied IQ domain of myosin-1c. The calcium-sensitive binding of calmodulin to myosin-1c may therefore modulate the interaction of the adaptation motor with other components of the transduction apparatus.
Keywords: Key words or phrases: adaptation, calmodulin, hair cell, hair bundle, Myo1c, myosin-1c, myosin Iβ, IQ domain, stereocilia, transduction
Situated at the apical surface of the hair cell, the mechanosensitive hair bundle is positioned to transduce mechanical stimuli into electrical signals. Bundle displacements, which can result from head movements or sound, open mechanically gated, cation-selective transduction channels that are located at the tips of stereocilia (Hudspeth, 1989). Hair-cell mechanoelectrical transduction is exceptionally sensitive; hair cells can detect bundle displacements of atomic dimensions. Such sensitivity requires the ability of the cell to adapt to prolonged stimuli. In the prevailing model of transduction, one component of adaptation is driven by the relocation of the transduction apparatus along the stereociliary length in response to changes in both Ca2+concentration and tension in the gating spring (Holt and Corey, 2000). This repositioning is powered by clustered myosin-1c molecules (Myo1c; formerly known as myr 2 and myosin-Iβ), which form the adaptation motor (Gillespie and Corey, 1997; Holt et al., 2002).
As an adaptation-motor myosin, Myo1c must associate directly or indirectly with other components of the transduction apparatus, including the transduction channel itself. To date, however, no proteins have been identified (except actin and calmodulin) that bind to any vertebrate myosin-I. Like other unconventional myosins, Myo1c contains an actin- and ATP-binding globular head, a neck region of tandem calmodulin-binding IQ motifs, and a tail domain, suggested to bind intracellular receptors. In its neck, Myo1c has four IQ motifs; the first two conform well to the IQ consensus [IQX3RGX3R (Rhoads and Friedberg, 1997)], whereas the third conforms less well (Sherr et al., 1993; Metcalf et al., 1994; Reizes et al., 1994; Solc et al., 1994). These three IQ domains bind calmodulin with relatively high affinity; a newly recognized fourth IQ motif binds calmodulin weakly (P. G. Gillespie and J. L. Cyr, unpublished observations). Under cellular conditions, all four calmodulin-binding sites are unlikely to be simultaneously occupied by calmodulin (Gillespie and Cyr, unpublished observations).
Specific receptors should couple Myo1c to the rest of the transduction apparatus. (We use the term “Myo1c receptor” to refer to specific intracellular binding sites for Myo1c.) To examine the interaction of Myo1c with its hair-cell receptors, we used recombinant Myo1c fragments to localize these receptors and biochemically characterize the interaction. We demonstrate here that the neck region of Myo1c suffices for binding to receptors at the tips of stereocilia. Because binding to stereociliary receptors requires Myo1c to be partially free of calmodulin, intracellular calcium levels (by modulating the number of calmodulin molecules bound to Myo1c) should control access of Myo1c to its receptors.
MATERIALS AND METHODS
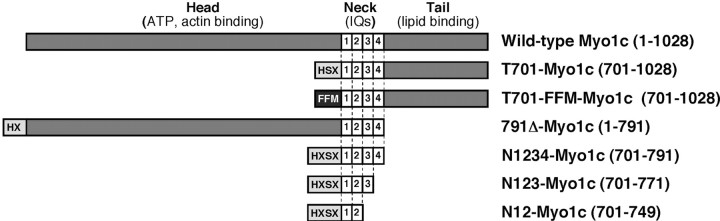
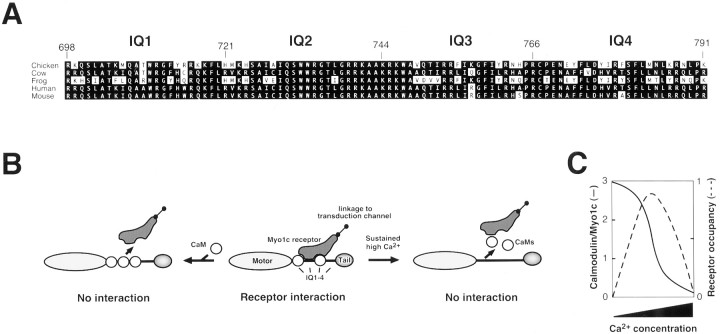
Construction of Myo1c baculoviruses. For expression in Sf9 insect cells, cDNA encoding residues 701 to 1028 of bullfrog Myo1c was amplified by PCR, an N-terminal Xpress tag (DLYDDDDK) for antibody detection was added, and the insert was cloned into the baculovirus transfer vector pBacGus-2cp (Novagen, Madison, WI). The recombinant protein HSX-T701 (frog Myo1c tail, residues 701–1028; hereafter called T701) also contained an His6 tag for Ni2+-nitrilotriacetic acid purification, an S tag (LETAAAKFERQHMN), and the Xpress tag at its N terminus (Fig. 1, tags abbreviatedHSX). 791Δ-Myo1c, a construct with the head and neck regions (amino acids 1–791) but lacking the tail, was constructed from bullfrog Myo1c using similar methods (Fig. 1).
Fig. 1.
Recombinant Myo1c constructs. Myo-1c fragments used in these studies. Shaded regions represent Myo1c sequences, and N-terminal boxes depict epitope tags used for purification and detection. Amino acid residues are indicated inparentheses.
Four additional deletion constructs, each encoding portions of the IQ-containing neck region of Myo1c, were generated by PCR from T701 and were cloned into the pBlueBacHis2A baculovirus transfer vector (Invitrogen, Carlsbad, CA). The final constructs were named N1 (residues 701–722), N12 (residues 701–749), N123 (residues 701–771), and N1234 (residues 701–791), indicating the presence of IQ domains 1–4 (Fig. 1); each encoded Myo1c fragments with 84 additional N-terminal residues, including the His6 tag, Xpress tag, and S tag, as well as a second Xpress tag.
To append alternative epitope tags at the N terminus of T701, the His6, S, and Xpress tag sequences were excised from pBacGus-T701, and then phosphorylated complementary oligonucleotides were used to generate double-stranded DNA inserts encoding FLAG tags (DYKDDDDK) and a c-myc tag (EQKLISEEDL). Two FLAG cassettes and one c-myc cassette were ligated to the N terminus of the T701 Myo1c sequences (abbreviated FFM). This region of the clone was sequenced to confirm epitope tag assembly.
Virus stocks were obtained and manipulated using standard methods (O'Reilly et al., 1994). Xenopus laeviscalmodulin was expressed using the pVL1393 baculovirus transfer vector (courtesy of Dr. James Sellers, National Institutes of Health, Bethesda, MD).
Myo1c fragment expression and purification. Myo1c fragments (T701, N1234, N123, N12, and N1) were coexpressed withXenopus calmodulin in Sf9 cells using methods described previously (Gillespie et al., 1999). Recombinant proteins were purified by high-speed centrifugation of an Sf9 cell extract and Ni2+-nitrilotriacetic acid chromatography in solutions containing 1–2 mm EGTA. In some cases, fragments were further purified using gel filtration on a 25 ml Superdex 200 HR 10/30 column with an AKTA-FPLC system (Amersham Biosciences, Piscataway, NJ). To obtain better yields, N12 was purified in the presence of Ca2+, substituting 25 μm CaCl2 for EGTA in all solutions. Eluate protein concentrations were determined by Coomassie blue dye binding (Bradford, 1976).
Cell pellets of Sf9 cells coexpressing FFM-T701 and calmodulin were resuspended, lysed, and centrifuged as indicated for T701. Before purification on a 1 ml anti-FLAG affinity column (M2-agarose; Sigma, St. Louis, MO), the salt concentration of the high-speed supernatant was reduced to 150 mm. The column was washed with 150 mm NaCl and 100 mm Tris at pH 7.5, and recombinant protein was eluted with 100 μg/ml 3× FLAG peptide (Sigma). Eluted protein was exchanged into 25 mm Tris at pH 8, 0.5 mm MgCl2, 300 mmNaCl, 0.5 mm EGTA, 2.5 mm 2-mercaptoethanol, 0.2 mm phenylmethylsulfonyl fluoride, 1 μmleupeptin, and 1 μm pepstatin using size-selective filtration units (MicroCon 30 filtration unit; Millipore, Bedford, MA).
HSX-GST expression and purification. The tandem His6, S, and Xpress tag sequences were amplified by PCR from pBacGus-T701 and fused to the N terminus of glutathioneS-transferase (GST) in the pET-HTG vector (Jensen et al., 1995). Recombinant protein was expressed in BL21(DE3) Escherichia coli cells and was purified on a Ni2+-nitrilotriacetic acid column as described for the Myo1c protein fragments. Use of the His6 tag for purification ensured that the epitope tags were accessible and intact.
Labeling sacculi and isolated hair cells with recombinant proteins. Bullfrog sacculi were dissected in frog standard saline (in mm: 110 NaCl, 2 KCl, 4 CaCl2, 2 MgCl2, 3d-glucose, and 10 HEPES, pH 7.25); otolithic membranes were manually removed without protease treatment. Sacculi were fixed in 3% formaldehyde in cold PBS (in mm: 137 NaCl, 2.7 KCl, 4.3 Na2HPO4, and 1.4 KH2PO4, pH 7.4) for 20–25 min and washed in PBS. Sacculi were permeabilized for 1 hr with 0.1% Sarkosyl in PBS; this and all subsequent steps were performed at room temperature. Detergent was removed by washing one time 10 min with PBS and two times for 10 min each with 25 mm HEPES, pH 7.5. Nonspecific binding sites were blocked for 1.5 hr in 5 mg/ml bovine serum albumin (fraction V; Calbiochem, La Jolla, CA) in 25 mm HEPES, pH 7.5. Sacculi were incubated overnight with purified Myo1c fragments or with control protein at ∼20 μg/ml (corresponding to 230 nm for T701) in a solution of 5 mg/ml bovine serum albumin, 25 mm HEPES, pH 7.5, and 1 mmEGTA (block solution), supplemented with 400 mmNaCl. After washing three times for 10 min each with 25 mm HEPES, pH 7.5, 1 mmEGTA, and 0.1% Tween 20 (HET), sacculi were incubated with 7.5 μg/ml anti-Xpress tag antibody (Invitrogen) in block solution for 2 hr. Unbound antibody was removed by washing three times for 10 min each in HET. Bound antibody and F-actin were detected by incubation with 13 μg/ml Alexa 488-goat anti-mouse antibody (Molecular Probes, Eugene, OR) and 66 nm Alexa 568-phalloidin (Molecular Probes) in block solution for 2 hr. Sacculi were washed three times for 10 min each in HET and one time for 10 min in 25 mm HEPES, pH 7.5, and 1 mmEGTA and were mounted with Vectashield (Vector Laboratories, Burlingame, CA). Fluorescently labeled samples were viewed on a Nikon (Tokyo, Japan) Eclipse TE300 inverted microscope with a Bio-Rad (Hercules, CA) MRC 1024 confocal imaging system. Images were processed with Scion Image (version 1.62; Scion Corp., Frederick, MD) and Photoshop (versions 5.03 and 6.0; Adobe Systems, San Jose, CA).
Binding with FFM-T701 was performed as described for T701. Bound probe was detected with an anti-myc antibody (9E10 ascites fluid) at a dilution of 1:500. All other steps were identical.
T701 binding was unaltered by the presence of 1 m NaCl or by treating sacculi with 50 μg/ml type XXIV protease (Sigma) at room temperature for 20 min to facilitate otolithic membrane removal. To prevent binding of the motor domain to stereociliary actin when labeling with 791Δ-Myo1c, we included 5 mm ATP, with or without 1 mm MgCl2. For calmodulin blocking experiments, we coincubated 230 nm T701 and 15 μm calmodulin with fixed and permeabilized sacculi. Alternatively, sacculi were preincubated with 15 μmcalmodulin overnight, washed to remove excess calmodulin, and incubated for 2 hr with T701. For IQ peptide-blocking experiments, sacculi were coincubated with 230 nm T701 and 25 μmpeptide. To control for sacculus-to-sacculus variability, each experimental condition used sacculi from several different bullfrogs.
Hair bundles, imbedded in agarose, were isolated as described previously (Gillespie and Hudspeth, 1991). Unfixed bundles were permeabilized with 0.1% saponin in PBS for 1 hr and then were labeled with T701 as described above.
Bullfrog saccular hair cells were enzymatically isolated (Lumpkin and Hudspeth, 1995) and allowed to settle on conconavalin A-coated coverslips. Cells were fixed for 1 hr in 3% formaldehyde in PBS, washed in PBS, and permeabilized in 0.05% Triton X-100 in PBS for 15 min. After permeabilization, hair cells were processed as described for sacculi.
Other methods. Immunolabeling of bullfrog sacculi with the anti-Myo1c antibody R-4280 (rafMIβ) was performed as described previously (Hasson et al., 1997). Bovine brain calmodulin was purified using isoelectric precipitation, followed by phenyl-Sepharose (Sigma) chromatography (Gopalakrishna and Anderson, 1982) and quantified assuming 330 μm/A276nm. IQ1 (residues 698–720; CRKHSIATFLQARWRGYHQRQKFL), IQ2 (residues 721–743; CHMKHSAVEIQSWWRGTIGRRKAA), and IQ3 (residues 744–766; CKRKWAVDVVRRFIKGFIYRNQPR) peptides were synthesized by Genemed Synthesis Inc. (South San Francisco, CA) with added cysteine residues at their N termini. Sequence alignments were performed with MegAlign Software (version 3.16; DNA Star Inc., Madison, WI).
RESULTS
Myo1c localization
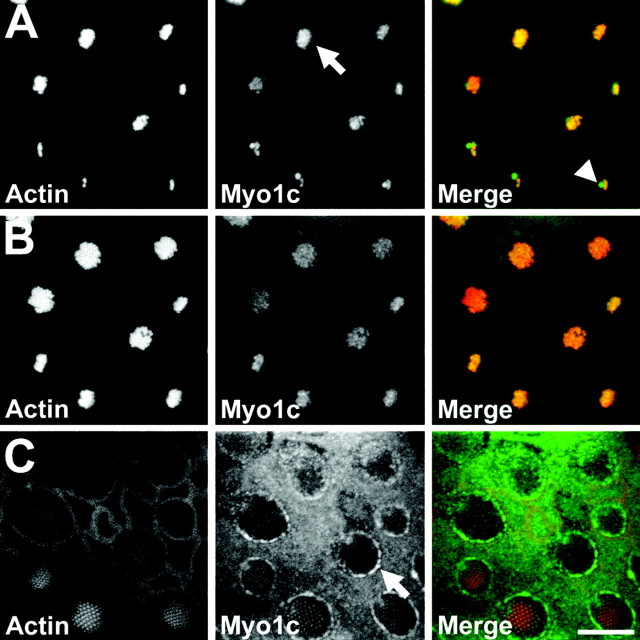
Reasoning that its receptors would be found close to hair-cell Myo1c, we first localized this myosin in the bullfrog sacculus using Myo1c-selective antibodies. The sensory epithelium of a bullfrog sacculus, optimized for detection of ground-borne vibrations and low-frequency sound, contains ∼2500 hair cells, each encircled by nonsensory supporting cells. In this receptor organ, Myo1c was present in both cell types (Gillespie et al., 1993; Hasson et al., 1997) (Fig.2). Within hair cells, Myo1c immunoreactivity was found in both the soma and hair bundle. Bundle Myo1c was primarily restricted to the most distal one-third of the stereocilia and was concentrated at stereociliary tips (Fig.2A,B). Little Myo1c was present where the stereocilia taper and insert into the cuticular plate, an actin meshwork beneath the bundle. Myo1c immunoreactivity was also present in the kinocilium, an axonemal cilium located at the tall edge of the hair bundle, and in the kinociliary bulb (Fig.2A). In the soma, Myo1c was prominent in the pericuticular necklace, a vesicle-rich compartment located between the cuticular plate and the circumferential actin belt (Fig.2C). Supporting cells (Fig. 2C) and epithelial cells surrounding the sensory epithelium also contained Myo1c immunoreactivity at their apical surfaces.
Fig. 2.
Localization of Myo1c in the bullfrog sacculus by immunofluorescence. Fluorescence images of a sacculus at three optical-section planes (see Fig. 3O for schematic).Left column, Actin (phalloidin); middle column, Myo1c (R-4280 antibody); right column, combined actin (red) and endogenous Myo1c (green). A, At the tops of hair bundles, Myo1c immunoreactivity is enriched at stereociliary tips (arrow), the kinocilium, and the kinociliary bulb (arrowhead). B, In sterociliary shafts, endogenous Myo1c labeling is present but reduced compared with tip labeling. C, At the apical surface of the sacculus, endogenous Myo1c labeling is strongly enriched along the hair-cell pericuticular necklace (arrow). Scale bar, 10 μm.
Binding of Myo1c fragment T701 to hair-cell receptors
To examine the interaction of Myo1c with hair-cell proteins, we initially assumed that it bound to intracellular receptors through its C-terminal tail domain. Accordingly, we generated T701, an N-terminal deletion construct that removed the Myo1c head domain and replaced it with epitope tag sequences for purification and detection (Fig. 1). After coexpression with calmodulin, T701 contained approximately two calmodulins at room temperature in the presence of EGTA (Gillespie and Cyr, unpublished observations).
To detect Myo1c receptors in hair cells, we incubated fixed and permeabilized sacculi or isolated hair cells with T701 in the presence of EGTA and then visualized Myo1c bound to its receptors by antibody labeling and confocal microscopy (Fig.3). T701 bound to receptors in hair cells at the tips of stereocilia, in the kinocilium, and in the kinociliary bulb (Fig. 3A–C, J). In addition, T701 bound to the shafts of the stereocilia in the upper two-thirds of the bundle (Fig. 3B,J). Smaller hair bundles were often more intensely labeled than neighboring larger hair bundles. At the level of the cuticular plate, we detected Myo1c receptors in the pericuticular necklace (Fig. 3C), although often only in hair cells with larger hair bundles and somas. Within the hair-cell soma, binding was usually absent from the cuticular plate but was present throughout the cytoplasm in regions that contained actin (Fig. 3J). We observed low levels of T701 binding at the apical surfaces of supporting and nonsensory epithelial cells (Fig.3C). T701 retained its ability to bind to hair-cell receptors when NaCl was as high as 1 m. Labeling with the detection antibodies alone was negligible (data not shown).
Fig. 3.
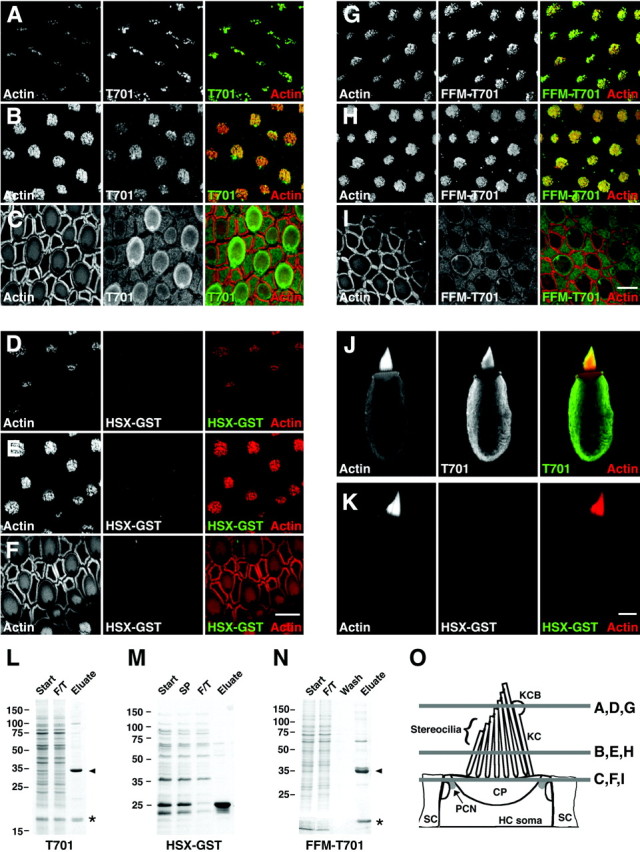
Binding of Myo1c-calmodulin probes to hair-cell receptors. A–I, Left columns, Actin (phalloidin); middlecolumns, epitope-tagged Myo1c or control probe; right columns, combined actin (red) and receptors (green). A–C, T701 binding at stereociliary tips, stereociliary shafts, and pericuticular necklace, respectively. D–F, HSX-GST (negative control) binding at stereociliary tips, stereociliary shafts, and pericuticular necklace, respectively. G–I, Binding of FFM-T701 to hair-cell receptors at stereociliary tips, stereociliary shafts, and pericuticular necklace, respectively. Pericuticular necklace binding is reduced in this particular sample. J, Labeling of isolated hair cell with T701. This cell did not have an intact kinocilium. K, Labeling of an isolated hair cell with HSX-GST. L–N, Coomassie blue-stained SDS-PAGE gels of purified T701, HSX-GST, and FFM-T701. Start, Starting material; SP, soluble proteins;F/T, column flow through; Wash, column wash; Eluate, column eluate. Arrowhead, Myo1c fragments; asterisk, copurifying calmodulin.O, Locations of saccular confocal cross sections shown in A–I. KCB, Kinociliary bulb;KC, kinocilium; HC, hair cell;SC, supporting cell; CP, cuticular plate;PCN, pericuticular necklace. Scale bars: 10 μm (F applies to A–F; Iapplies to G–I; K applies toJ, K).
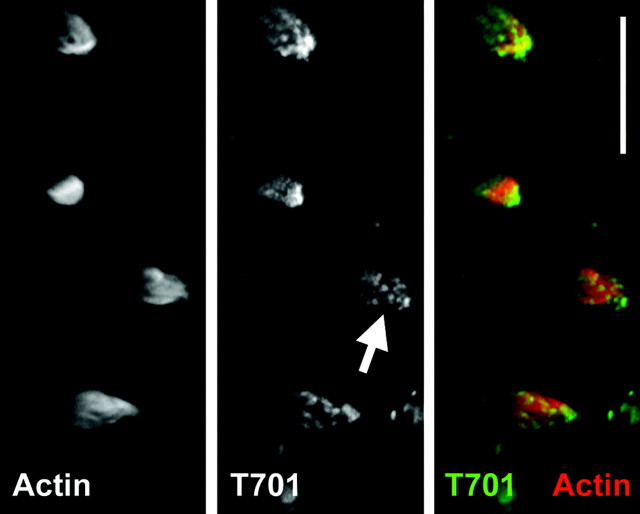
We observed a similar labeling pattern in isolated hair bundles, permeabilized with saponin but unfixed (Gillespie and Hudspeth, 1991). As seen with fixed hair cells, T701 labeling in isolated bundles was strong and focused near stereociliary tips, in the kinocilium, and in the kinociliary bulb (Fig. 4).
Fig. 4.
Binding of Myo1c–calmodulin probes to isolated, unfixed hair bundles. Hair bundles were isolated in agarose, permeabilized with saponin, and then labeled with phalloidin and T701. No fixation was used. Left, Actin (phalloidin);middle, T701-Myo1c; right, combined actin (red) and T701 (green). Hair-bundle labeling by T701 was pronounced, particularly at stereociliary tips (arrow). Scale bar, 10 μm.
To unambiguously verify the specificity of T701 binding to saccular receptors, we probed sacculi with two additional recombinant proteins. One protein, HSX-GST, which included identical epitope tags fused to GST (Fig. 3M), did not label sacculi or isolated hair cells (Fig. 3D–F, K). The second construct, FFM-T701, which contained the Myo1c neck and tail regions with alternative epitope tags (Fig. 1), labeled Myo1c receptors in an identical manner to that obtained with T701 (Fig.3G–I). In addition, coincubation with a twofold excess of FFM-T701 partially blocked T701 binding (data not shown). Collectively, these data confirmed the specificity of T701 labeling and showed that the binding domain was contained within Myo1c neck and tail sequences.
The Myo1c binding domain
We used other deletion constructs (Fig. 1) to delineate the region of Myo1c that was responsible for binding to tips of stereocilia. Surprisingly, a construct that contained the Myo1c head and neck regions (791Δ-Myo1c; residues 1–791) also bound to stereociliary receptors (data not shown). This result focused our attention on the neck region (resides 701–791) with its four calmodulin-binding IQ domains.
To determine the role of the individual IQ domains in binding, we generated epitope-tagged Myo1c fragments that contained all four IQ domains (N1234), the first three IQ domains (N123), the first and second IQ domains (N12), or only the first IQ domain (N1). As anticipated, N1234 bound to Myo1c receptors in the presence of EGTA, yielding a pattern identical to that seen with T701 (Fig.5A). In addition, purified N123 and N12 (Fig. 5D) labeled Myo1c receptors (Fig.5B,C), narrowing the interaction domain down to the first two or three IQ motifs. N12 labeling of stereociliary tips was appreciably less intense, however, than that seen with T701, N1234, or N123 (Fig. 5A–C), suggesting an important role for IQ3. Conversely, stereociliary shafts of smaller hair bundles were more strongly labeled with N12 than with the other epitope-tagged Myo1c fragments (data not shown). These subtle differences in labeling were reproducible and did not reflect sacculus-to-sacculus variability. Although we never observed hair-cell labeling with N1, this probe readily aggregated, complicating interpretation of the negative result. Finally, excess (25 μm) IQ1, IQ2, or IQ3 synthetic peptides did not interfere with T701 binding (Fig. 5E, and data not shown), suggesting that single IQ domains cannot suffice for binding.
Fig. 5.
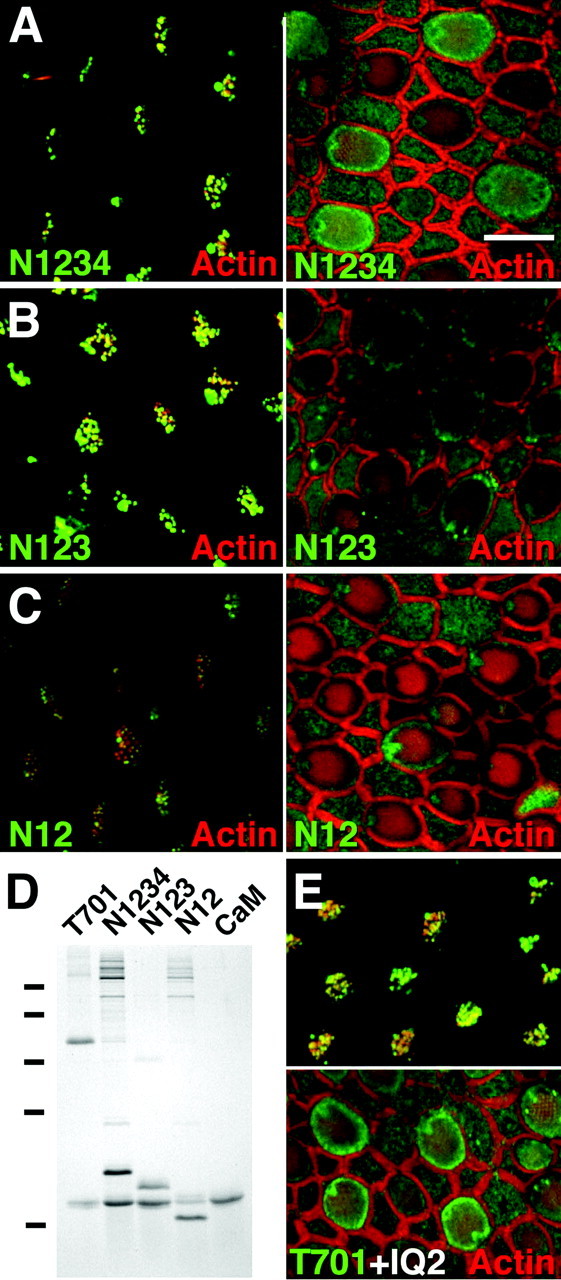
Binding of Myo1c neck fragments to hair-cell receptors. A–C, Confocal sections of sacculi at hair-bundle tips (left) and pericuticular necklace (right). Green, N1234, N123, or N12 binding; red, actin. N12 labeling of stereociliary tips was diminished compared with N1234 or N123. D, Coomassie-stained SDS-PAGE gel showing purified complexes of calmodulin (CaM) with T701, N1234, N123, and N12. Molecular mass markers of 75, 50, 35, 25, and 15 kDa are indicated to theleft. E, IQ2 peptide (25 μm) does not affect T701 (230 nm) labeling.Top, Sterociliary tips; bottom, pericuticular necklace. Scale bar: A–C, E, 10 μm.
Calmodulin and T701 labeling
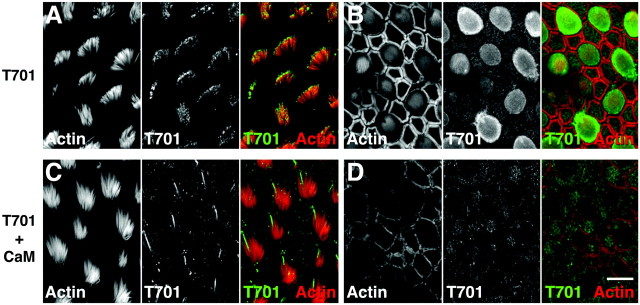
Given the presence of IQ domains in the region of Myo1c responsible for receptor interaction, we investigated whether binding of Myo1c to its receptors was modulated by calmodulin. A 70-fold molar excess of calmodulin completely eliminated binding of T701 to stereociliary tips and the pericuticular necklace (Fig.6C,D); under these conditions, T701 has at least three bound calmodulin molecules (Gillespie and Cyr, unpublished observations). Because calmodulin did not abolish kinocilium or kinociliary bulb labeling (Fig.6C), hair cells may contain more than one type of Myo1c receptor. When we preincubated sacculi with calmodulin and then washed to remove unbound protein, binding of T701 was unaffected (data not shown). Calmodulin must therefore block labeling by either weakly interacting with Myo1c receptors or, more likely, saturating the IQ domains of T701 and preventing binding of the Myo1c probe to its receptors.
Fig. 6.
Calmodulin blocks Myo1c–receptor interaction.Left columns, Actin (phalloidin); middle columns, Myo1c receptors (T701); right columns, combined actin (red) and Myo1c receptors (green). A, B, Labeling of actin and Myo1c receptors (with 230 nm T701) at the tips of stereocilia and pericuticular necklace, respectively.C, D, Simultaneous addition of calmodulin (15 μm) blocks binding of T701 (230 nm) to stereociliary tips and pericuticular necklace but not the kinocilium and kinociliary bulb. Scale bar: A–D, 10 μm.
Ca2+ modulates calmodulin binding to most IQ domains (Jurado et al., 1999). At room temperature, elevated Ca2+ causes the release of nearly all calmodulins from T701 (Gillespie and Cyr, unpublished observations). Under similar elevated-Ca2+ conditions, interaction of T701 with its receptors was blocked completely in some experiments but retained in others (data not shown).
DISCUSSION
Receptor localization using tagged ligands
Myo1c is the only molecule known to be part of the mechanotransduction apparatus in hair-cell stereocilia (Holt et al., 2002). By detecting Myo1c receptors and defining the region of Myo1c responsible for the interaction, we initiated the next step in defining the chain of protein–protein interactions that constitute the transduction complex. We located and characterized Myo1c receptorsin situ using an approach that combines the manipulability of classic ligand–receptor biochemistry with the specificity of immunolocalization (Pardue et al., 1981; Welsh et al., 1982; Knowles and McCulloch, 1992). After adding epitope tags to recombinant fragments of Myo1c, we allowed these tagged probes to bind to receptors in fixed and permeabilized cells, and then we localized bound probes by standard immunocytochemical methods. Radioisotope- or enzyme-tagged ligands are commonly used to report receptor location in cells (Aruffo et al., 1990; Flanagan and Leder, 1990; Devos et al., 1996;Kolodkin et al., 1997; Muller et al., 1997). Our variation, using immunodetection of epitope-tagged protein, allows us to achieve the sensitivity and specificity necessary for detecting receptors of low abundance in the hair cell.
Several conditions must be met for a tagged ligand to accurately report the location and properties of its cognate receptor. First, a subset of the receptors must be free of endogenous ligand and remain capable of binding the probe, even after fixation and permeabilization. Second, the tagged form of the ligand must be biologically active and remain so during incubation with the tissue. The strong and specific binding we observed with epitope-tagged Myo1c indicates that these conditions prevailed in our experiments. Tagged Myo1c bound to the tips of stereocilia, despite the presence of endogenous Myo1c in these locations, indicating that unoccupied receptors were present. Myo1c binding was retained even after stringent fixation and permeabilization conditions. Admittedly, our results were obtained with exogenously applied proteins, and it is not clear whether similar binding would be seen if the Myo1c constructs were expressed in hair cells. Nevertheless, our results demonstrated the feasibility of using a tagged Myo1c to identify the locations and binding properties of its receptors.
The neck domain of Myo1c mediates receptor binding
We make two important assumptions about the probe–receptor interaction described here: (1) tagged Myo1c fragments mimic endogenous Myo1c, and (2) binding sites represent endogenous receptors. Supporting the former point, hydrodynamic properties of T701-Myo1c are similar to those of wild-type Myo1c, after accounting for the ∼85 kDa head domain (Gillespie and Cyr, unpublished observations). Supporting the latter point, receptors for Myo1c probes reside in the same subcellular compartments as endogenous Myo1c, including the tips of stereocilia and the pericuticular necklace. Because active transduction channels (Hudspeth, 1982) and the adaptation mechanism (Assad et al., 1989) are located at tips of stereocilia, receptors located there could couple Myo1c to the rest of the transduction apparatus. Admittedly, these receptors could localize Myo1c to tips of the stereocilia for other purposes, a possibility that cannot be eliminated until the receptors are identified.
Using our in situ binding assay, we narrowed down the region of Myo1c required for receptor binding. Other unconventional myosins have protein–protein interaction motifs in their tails, which presumably modulate the interaction of the motor with intracellular proteins (Mermall et al., 1998; Beningo et al., 2000; Kussel-Andermann et al., 2000a,b; Berg et al., 2001; Jung et al., 2001; Lapierre et al., 2001). By analogy, we expected the tail of Myo1c to be involved in receptor binding. Our unexpected result was that Myo1c interacts with its receptors not through its tail but through its calmodulin-binding IQ domains. This observation suggested the exciting possibility that the interaction of Myo1c with the transduction complex might be modulated by calmodulin.
Calmodulin modulates the Myo1c–receptor interaction
Coincubation of T701 and a large molar excess of calmodulin blocked the interaction of T701 with stereociliary and pericuticular necklace receptor sites. Calmodulin might have blocked labeling by either binding to receptors, thereby competing with Myo1c, or binding to Myo1c itself, interfering with its ability to interact with the receptor. Our data do not support the former hypothesis: we found calmodulin to be ineffective at blocking labeling if applied to the tissue before applying the probe. This suggests the alternative, that excess calmodulin prevents interaction by binding to Myo1c, presumably by binding to previously unoccupied IQ domains.
Which IQ domains mediate the negative effects of calmodulin binding? The N12 fragment of Myo1c labeled stereociliary receptors, indicating that IQ1 and IQ2 are sufficient for interaction. Consistent with an essential role for IQ2, a comparison of the Myo1c IQ domain sequences from several species showed a striking conservation in IQ2 and part of IQ3. From residues 729–748, the amino acid sequence is 100% conserved, with the single exception of a Leu to Ile substitution at residue 737 in the bullfrog protein (Fig.7A). The high degree of sequence identity of the IQ2 region between species is a characteristic that is shared neither with the other three IQ domains nor between the four Myo1c IQ domains of any single species. Because calmodulin binds to widely divergent IQ domains (Rhoads and Friedberg, 1997), the high sequence conservation in IQ2 reinforces the suggestion that this domain interacts with proteins other than calmodulin, such as hair-cell Myo1c receptors.
Fig. 7.
Interaction of Myo1c with receptors.A, Alignment of amino acid sequences of the Myo1c neck region from indicated species, with IQ domains indicated. Note the sequence identity of residues 729–748, spanning IQ2 and the beginning of IQ3. B, Model for interaction of Myo1c with receptors. If IQ domains 1–3 are occupied by calmodulin, the receptor cannot bind (left). Calmodulin dissociation from IQ2 permits receptor interaction (middle). If calmodulin dissociates from all IQ domains, receptors cannot bind (right). Calmodulin-bound IQ3 and possibly IQ1 may contribute to the binding site. C, Predicted effects of Ca2+ concentration on the occupancy of Myo1c IQs 1–3 with calmodulin and hence binding to Myo1c receptors.
Does calmodulin also contribute to the binding site for Myo1c receptors? N123 bound hair-cell receptors more avidly than did N12. Because N123 consistently had approximately twofold more calmodulin associated with it than did N12 (Fig. 5D), we infer that calmodulin occupied the IQ3 site and that this calmodulin probably enhanced receptor binding. In addition, a large excess of IQ2 peptide did not block T701 labeling (Fig. 5E), consistent with the suggestion that the receptor-binding site on Myo1c encompasses more than IQ2. We suggest that calmodulin-free IQ2 contributes significantly to binding of Myo1c with its receptors and that calmodulin-bound IQ3 (and, perhaps, IQ1) enhances this interaction. If calmodulin binds to IQ2, however, the receptor interaction should be blocked sterically (Fig. 7B,C). Calmodulin therefore appears to have a dual role, enhancing and inhibiting Myo1c–receptor interaction under different conditions.
The C-terminal 30 kDa of Myo1c was not required for interaction with Myo1c receptors. This protein domain is enriched in basic amino acids, which presumably mediate its interaction with acidic phospholipids (Reizes et al., 1994). The sole role of the tail therefore may be to localize Myo1c to particular membrane compartments; the IQ domains would then associate specifically with nearby receptors.
Myo1c receptors
We were able to draw several conclusions about these hair-cell Myo1c receptors. First, the receptors were not endogenous calmodulin. Although localization of the Myo1c binding domain to the neck region and block of binding by calmodulin raises this possibility, three observations strongly refute this conclusion. (1) Within hair cells, the distribution of calmodulin does not match that of Myo1c receptors (Shepherd et al., 1989; Pack and Slepecky, 1995; Ogata and Slepecky, 1998; Nakazawa, 2001). (2) High concentrations of free IQ peptides, which should bind to endogenous calmodulin, had no effect on Myo1c binding. (3) Myo1c fragments bound to stereociliary receptors in unfixed and permeabilized hair bundles; under these conditions, free calmodulin and other small soluble proteins should diffuse out of the stereocilia. Our second conclusion was that the receptors were not membrane lipids. Although Myo1c binds to acidic phospholipids through its tail (Reizes et al., 1994) and perhaps its neck (Swanljung-Collins and Collins, 1992), not only should the stringent detergent conditions used for permeabilization have extracted most lipids, but the high ionic strength (as high as 1 m NaCl) used for probe binding should have prevented interaction with any remaining phospholipids (Reizes et al., 1994). A third conclusion was that, as expected for a component of the transduction apparatus, Myo1c receptors are directly or indirectly associated with the membrane or cytoskeleton of stereocilia; unfixed, detergent-permeabilized hair bundles retained T701 binding during overnight incubation. Finally, we concluded that Myo1c receptors were not Myo1c itself. Although Myo1c apparently can self-associate under some in vitro conditions, this behavior has not been observed under the conditions of the sacculus-labeling experiments (Cyr and Gillespie, unpublished observations). The most plausible interpretation of our tagged-Myo1c binding experiments is that the recombinant Myo1c fragments interact with so far unidentified receptors that serve to restrict Myo1c to specific hair-cell locations.
We observed Myo1c receptors in a number of saccular sites besides the stereocilia, including the kinocilium, the apical surfaces of the supporting cells, and the pericuticular necklace. This last location may be a special case; as with several other stereociliary proteins, we speculate that Myo1c receptors are localized to the pericuticular necklace because this subcellular region serves as precursor compartment for proteins bound for the stereocilia (Hasson et al., 1997; Dumont et al., 2001). Myo1c receptors in other locations outside the stereocilia are colocalized well with endogenous Myo1c itself and much less so with actin, suggesting that other compartments use both Myo1c and its receptors for other, as yet unknown, roles.
Our discovery that Myo1c–receptor interactions are mediated by IQ domains provides new avenues for molecular identification of the receptors. Although inner-ear Myo1c receptors may be too scarce for conclusive identification by protein sequencing, Myo1c receptors could be purified from alternative, more abundant tissue sources. The presence of these Myo1c receptors could then be confirmed in hair cells and their properties could be characterized. The role of calmodulin in receptor binding complicates biochemical and molecular identification efforts, however. If not removed, free calmodulin in tissue extracts will block receptor interactions with Myo1c. Furthermore, expression-screening approaches, like two-hybrid methods, may require experimental control of calmodulin binding and Ca2+ concentration.
Consequences for Myo1c function in hair cells
Calcium and calmodulin regulation of Myo1c–receptor binding provides the transduction process an effective mechanism to readily modulate properties of the adaptation motor. With a free-calmodulin concentration of 35 μm in the hair bundle (Walker et al., 1993), IQ domains 1–3 should be fully saturated by calmodulin when Ca2+ is low (Gillespie and Cyr, unpublished observations). Under these conditions, Myo1c should not interact with its receptors. When the Ca2+concentration is high, as it should be near the transduction channel (Lumpkin and Hudspeth, 1998) and hence the adaptation motor, calmodulin molecules should dissociate from Myo1c, permitting interaction of Myo1c with its receptors (Fig. 7B,C). In addition, if Ca2+ remains elevated and all calmodulin molecules dissociate, receptor interaction may be disrupted (Fig. 7B,C). Although clearly important for coupling Myo1c to other transduction components, whether this mode of regulation could directly contribute to adaptation (Gillespie and Corey, 1997) depends on the kinetics of Ca2+ regulation of the Myo1c-receptor interaction. Although our data do not directly address this point, we suspect that the proposed Ca2+-dependent decoupling of Myo1c from its receptors will be relatively slow and hence will not play a role on the time scale of adaptation (microseconds to hundreds of milliseconds). Nevertheless, by controlling Myo1c–receptor interactions, Ca2+ entering a hair cell through transduction channels could optimally couple the recruitment of Myo1c molecules into the adaptation motor to transduction–channel activity.
Footnotes
This work was supported by National Institutes of Health Grant DC02368. We thank Dr. Stefan Heller for sharing unpublished sequence information. We appreciate the technical assistance of Dr. Kevin Nusser and Weiyi Zhao. In addition, we are indebted to Dr. Susan Gillespie for technical assistance and for her insightful comments on this manuscript.
Correspondence should be addressed to Dr. Peter G. Gillespie, Oregon Hearing Research Center and Vollum Institute, L335A/Oregon Health and Science University, 3181 SW Sam Jackson Park Road, Portland, OR 97201. E-mail: gillespp@ohsu.edu.
J. Cyr's present address: Department of Otolaryngology and Sensory Neuroscience Research Center, West Virginia University School of Medicine, Morgantown, WV 26506-9303.
REFERENCES
- 1.Aruffo A, Stamenkovic I, Melnick M, Underhill CB, Seed B. CD44 is the principal cell surface receptor for hyaluronate. Cell. 1990;61:1303–1313. doi: 10.1016/0092-8674(90)90694-a. [DOI] [PubMed] [Google Scholar]
- 2.Assad JA, Hacohen N, Corey DP. Voltage dependence of adaptation and active bundle movement in bullfrog saccular hair cells. Proc Natl Acad Sci USA. 1989;86:2918–2922. doi: 10.1073/pnas.86.8.2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beningo KA, Lillie SH, Brown SS. The yeast kinesin-related protein Smy1p exerts its effects on the class V myosin Myo2p via a physical interaction. Mol Biol Cell. 2000;11:691–702. doi: 10.1091/mbc.11.2.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berg JS, Powell BC, Cheney RE. A millennial myosin census. Mol Biol Cell. 2001;12:780–794. doi: 10.1091/mbc.12.4.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 6.Devos R, Richards JG, Campfield LA, Tartaglia LA, Guisez Y, van der Heyden J, Travernier J, Plaetinck G, Burn P. OB protein binds specifically to the choroid plexus of mice and rats. Proc Natl Acad Sci USA. 1996;93:5668–5673. doi: 10.1073/pnas.93.11.5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dumont RA, Lins U, Filoteo AG, Penniston JT, Kachar B, Gillespie PG. Plasma membrane Ca2+-ATPase isoform 2a is the PMCA of hair bundles. J Neurosci. 2001;21:5066–5078. doi: 10.1523/JNEUROSCI.21-14-05066.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flanagan JG, Leder P. The kit ligand: a cell surface molecule altered in steel mutant fibroblasts. Cell. 1990;63:185–194. doi: 10.1016/0092-8674(90)90299-t. [DOI] [PubMed] [Google Scholar]
- 9.Gillespie PG, Corey DP. Myosin and adaptation by hair cells. Neuron. 1997;19:955–958. doi: 10.1016/s0896-6273(00)80387-6. [DOI] [PubMed] [Google Scholar]
- 10.Gillespie PG, Hudspeth AJ. High-purity isolation of bullfrog hair bundles and subcellular and topological localization of constituent proteins. J Cell Biol. 1991;112:625–640. doi: 10.1083/jcb.112.4.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gillespie PG, Wagner MC, Hudspeth AJ. Identification of a 120 kd hair-bundle myosin located near stereociliary tips. Neuron. 1993;11:581–594. doi: 10.1016/0896-6273(93)90071-x. [DOI] [PubMed] [Google Scholar]
- 12.Gillespie PG, Gillespie SK, Mercer JA, Shah K, Shokat KM. Engineering of the myosin-Iβ nucleotide-binding pocket to create selective sensitivity to N6-modified ADP analogs. J Biol Chem. 1999;274:31373–31381. doi: 10.1074/jbc.274.44.31373. [DOI] [PubMed] [Google Scholar]
- 13.Gopalakrishna R, Anderson WB. Ca2+-induced hydrophobic site on calmodulin: application for purification of calmodulin by phenyl-Sepharose affinity chromatography. Biochem Biophys Res Commun. 1982;104:830–836. doi: 10.1016/0006-291x(82)90712-4. [DOI] [PubMed] [Google Scholar]
- 14.Hasson T, Gillespie PG, Garcia JA, MacDonald RB, Zhao Y, Yee AG, Corey DP. Unconventional myosins in inner-ear sensory epithelia. J Cell Biol. 1997;137:1287–1307. doi: 10.1083/jcb.137.6.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holt JR, Corey DP. Two mechanisms for transducer adaptation in vertebrate hair cells. Proc Natl Acad Sci USA. 2000;97:11730–11735. doi: 10.1073/pnas.97.22.11730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holt JR, Gillespie SKH, Provance DW, Shah K, Shokat KM, Corey DP, Mercer JA, Gillespie PG. A chemical-genetic strategy implicates myosin-1c in adaptation by hair cells. Cell. 2002;108:371–381. doi: 10.1016/s0092-8674(02)00629-3. [DOI] [PubMed] [Google Scholar]
- 17.Hudspeth AJ. Extracellular current flow and the site of transduction by vertebrate hair cells. J Neurosci. 1982;2:1–10. doi: 10.1523/JNEUROSCI.02-01-00001.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hudspeth AJ. How the ear's works work. Nature. 1989;341:397–404. doi: 10.1038/341397a0. [DOI] [PubMed] [Google Scholar]
- 19.Jensen TH, Jensen A, Kjems J. Tools for the production and purification of full-length, N- or C-terminal 32P-labeled protein, applied to HIV-1 Gag and Rev. Gene. 1995;162:235–237. doi: 10.1016/0378-1119(95)00328-4. [DOI] [PubMed] [Google Scholar]
- 20.Jung G, Remmert K, Wu X, Volosky JM, Hammer JA., III The Dictyostelium CARMIL protein links capping protein and the Arp2/3 complex to type I myosins through their SH3 domains. J Cell Biol. 2001;153:1479–1497. doi: 10.1083/jcb.153.7.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jurado LA, Chockalingam PS, Jarrett HW. Apocalmodulin. Physiol Rev. 1999;79:661–682. doi: 10.1152/physrev.1999.79.3.661. [DOI] [PubMed] [Google Scholar]
- 22.Knowles GC, McCulloch CA. Simultaneous localization and quantification of relative G and F actin content: optimization of fluorescence labeling methods. J Histochem Cytochem. 1992;40:1605–1612. doi: 10.1177/40.10.1527379. [DOI] [PubMed] [Google Scholar]
- 23.Kolodkin AL, Levengood DV, Rowe EG, Tai YT, Giger RJ, Ginty DD. Neuropilin is a semaphorin III receptor. Cell. 1997;90:753–762. doi: 10.1016/s0092-8674(00)80535-8. [DOI] [PubMed] [Google Scholar]
- 24.Kussel-Andermann P, El-Amraoui A, Safieddine S, Hardelin JP, Nouaille S, Camonis J, Petit C. Unconventional myosin VIIA is a novel A-kinase-anchoring protein. J Biol Chem. 2000a;275:29654–29659. doi: 10.1074/jbc.M004393200. [DOI] [PubMed] [Google Scholar]
- 25.Kussel-Andermann P, El-Amraoui A, Safieddine S, Nouaille S, Perfettini I, Lecuit M, Cossart P, Wolfrum U, Petit C. Vezatin, a novel transmembrane protein, bridges myosin VIIA to the cadherin-catenins complex. EMBO J. 2000b;19:6020–6029. doi: 10.1093/emboj/19.22.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lapierre LA, Kumar R, Hales CM, Navarre J, Bhartur SG, Burnette JO, Provance DW, Jr, Mercer JA, Bähler M, Goldenring JR. Myosin Vb is associated with plasma membrane recycling systems. Mol Biol Cell. 2001;12:1843–1857. doi: 10.1091/mbc.12.6.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lumpkin EA, Hudspeth AJ. Detection of Ca2+ entry through mechanosensitive channels localizes the site of mechanoelectrical transduction in hair cells. Proc Natl Acad Sci USA. 1995;92:10297–10301. doi: 10.1073/pnas.92.22.10297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lumpkin EA, Hudspeth AJ. Regulation of free Ca2+ concentration in hair-cell stereocilia. J Neurosci. 1998;18:6300–6318. doi: 10.1523/JNEUROSCI.18-16-06300.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mermall V, Post PL, Mooseker MS. Unconventional myosins in cell movement, membrane traffic, and signal transduction. Science. 1998;279:527–533. doi: 10.1126/science.279.5350.527. [DOI] [PubMed] [Google Scholar]
- 30.Metcalf AB, Chelliah Y, Hudspeth AJ. Molecular cloning of a myosin Iβ isozyme that may mediate adaptation by hair cells of the bullfrog's internal ear. Proc Natl Acad Sci USA. 1994;91:11821–11825. doi: 10.1073/pnas.91.25.11821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muller U, Wang D, Denda S, Meneses JJ, Pedersen RA, Reichardt LF. Integrin alpha8beta1 is critically important for epithelial-mesenchymal interactions during kidney morphogenesis. Cell. 1997;88:603–613. doi: 10.1016/s0092-8674(00)81903-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakazawa K. Ultrastructural localization of calmodulin in gerbil cochlea by immunogold electron microscopy. Hear Res. 2001;151:133–140. doi: 10.1016/s0378-5955(00)00220-3. [DOI] [PubMed] [Google Scholar]
- 33.O'Reilly DR, Miller LK, Luckow VA. Baculovirus expression vectors: a laboratory manual. Oxford UP; New York: 1994. [Google Scholar]
- 34.Ogata Y, Slepecky NB. Immunocytochemical localization of calmodulin in the vestibular end-organs of the gerbil. J Vestib Res. 1998;8:209–216. [PubMed] [Google Scholar]
- 35.Pack AK, Slepecky NB. Cytoskeletal and calcium-binding proteins in the mammalian organ of Corti: cell type-specific proteins displaying longitudinal and radial gradients. Hear Res. 1995;91:119–135. doi: 10.1016/0378-5955(95)00173-5. [DOI] [PubMed] [Google Scholar]
- 36.Pardue RL, Kaetzel MA, Hahn SH, Brinkley BR, Dedman JR. The identification of calmodulin-binding sites on mitochondria in cultured 3T3 cells. Cell. 1981;23:533–542. doi: 10.1016/0092-8674(81)90149-5. [DOI] [PubMed] [Google Scholar]
- 37.Reizes O, Barylko B, Li C, Sudhof TC, Albanesi JP. Domain structure of a mammalian myosin Iβ. Proc Natl Acad Sci USA. 1994;91:6349–6353. doi: 10.1073/pnas.91.14.6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rhoads AR, Friedberg F. Sequence motifs for calmodulin recognition. FASEB J. 1997;11:331–340. doi: 10.1096/fasebj.11.5.9141499. [DOI] [PubMed] [Google Scholar]
- 39.Shepherd GMG, Barres BA, Corey DP. “Bundle-blot” purification and initial protein characterization of hair cell stereocilia. Proc Natl Acad Sci USA. 1989;86:4973–4977. doi: 10.1073/pnas.86.13.4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sherr EH, Joyce MP, Greene LA. Mammalian myosin Iα, Iβ, and Iγ: new widely expressed genes of the myosin I family. J Cell Biol. 1993;120:1405–1416. doi: 10.1083/jcb.120.6.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Solc CK, Derfler BH, Duyk GM, Corey DP. Molecular cloning of myosins from bullfrog saccular macula: a candidate for the hair cell adaptation motor. Auditory Neurosci. 1994;1:63–75. [Google Scholar]
- 42.Swanljung-Collins H, Collins JH. Phosphorylation of brush border myosin I by protein kinase C is regulated by Ca2+-stimulated binding of myosin I to phosphatidylserine concerted with calmodulin dissociation. J Biol Chem. 1992;267:3445–3454. [PubMed] [Google Scholar]
- 43.Walker RG, Hudspeth AJ, Gillespie PG. Calmodulin and calmodulin-binding proteins in hair bundles. Proc Natl Acad Sci USA. 1993;90:2807–2811. doi: 10.1073/pnas.90.7.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Welsh MJ, Aster JC, Ireland M, Alcala J, Maisel H. Calmodulin binds to chick lens gap junction protein in a calcium-independent manner. Science. 1982;216:642–644. doi: 10.1126/science.6280283. [DOI] [PubMed] [Google Scholar]