Abstract
Several factors regulate export of mRNAs from neuronal cell bodies. Using in situ hybridization and RT-PCR, we examined how target interaction influences the distribution of mRNAs expressed in sensory neurons (SNs) of Aplysia maintained in cell culture. Interaction with a synaptic target has two effects on the distribution of mRNA encoding an SN-specific peptide, sensorin: the target affects the accumulation of sensorin mRNA at the axon hillock and the stability of sensorin mRNA exported to distal sites. Synapse formation with motor neuron L7 results in the accumulation of high levels of sensorin mRNA in the axon hillock of the SN and in SN neurites contacting L7. SNs cultured alone or in contact with motor neuron L11, with which no synapses form, show a more uniform distribution of sensorin mRNA in the cytoplasm of the SN cell body, with little expression in neurites. Contact with L7 or L11 had little or no effect on the distribution of two other mRNAs in the cytoplasm of SN cell bodies. Sensorin mRNA exported to SN neurites after 1 d in culture is more stable when the SN contacts L7 compared with SN neurites that contact L11. After removal of the SN cell body, the amounts of sensorin mRNA already exported to the neurites are greater when neurites contact L7 compared with neurites in contact with L11. The results indicate that target interaction and synapse formation regulate both the accumulations of specific mRNAs destined for export and their stability at distant sites.
Keywords: synapse formation, synapse specificity, mRNA expression, mRNA export, mRNA stability, cell culture, Aplysia
During development, the formation of specific synaptic connections in the CNS is established by a sequence of cellular and molecular processes including axonal guidance, recognition of postsynaptic targets, synaptic differentiation, and activity-dependent synaptic stabilization and elimination (for review, see Goodman and Shatz, 1993). Synapse formation and maintenance depend on appropriate expression of many genes and trafficking of the encoded proteins to the proper region of the neuron. Expression of synapse-associated genes may be controlled by an intrinsic differentiation program as well as by extrinsic signals derived from the presynaptic and postsynaptic cells. Local extrinsic signals, diffusible or cell surface molecules, may modulate the expression of these genes and determine either synapse formation or elimination (for review, see Cabelli et al., 1997; Fitzsimonds and Poo, 1998; Sanes et al., 1998; Winberg et al., 1998). How do these signals mediate the long-term maintenance of one set of synaptic connections and the decline in efficacy or elimination of others in the same neuron? One potential mechanism is that intercellular signals modulate the local synthesis and targeting of macrocmolecules required for synapse formation or maintenance.
The export of specific mRNAs into dendrites of vertebrate neurons is well established (Crino and Eberwine, 1996; Knowles et al., 1996;Steward, 1997). Some of these mRNAs are targeted to specific dendritic sites after site-specific activation of synaptic connections (Steward and Worley, 2001). Specific mRNAs, including those encoding neuropeptides and other proteins, are also exported into axons of invertebrate neurons (van Minnen, 1994; Schacher et al., 1999). Isolated axons of invertebrate neurons have the machinery to synthesize proteins (van Minnen et al., 1997; Spencer et al., 2000), including proteins required for synapse formation and the expression of some forms of long-term synaptic plasticity (Casadio et al., 1997;Martin et al., 1997; Sherff and Carew, 1999; Sun et al., 2001; Schacher and Wu, 2002). Thus, local translation of exported mRNAs encoding synapse-specific macromolecules may contribute to the formation or maintenance of specific synapses.
The large identified neurons of Aplysia form specific synapses in dissociated cell culture even when a presynaptic neuron is confronted with several targets (Camardo et al., 1983; Schacher and Montarolo, 1991; Hawver and Schacher, 1993; Casadio et al., 1997;Schacher et al., 1999). Contact with different synaptic targets leads to expression of branch-specific morphological, physiological, and biochemical features in sensory neurons (SNs). These include the formation of varicosities with active zones (Glanzman et al., 1989), expression of electrical excitability and synaptic plasticity (Glanzman et al., 1990; Schacher and Montarolo, 1991; Sun and Schacher, 1996), and expression of high levels of the SN-specific neuropeptide sensorin (Brunet et al., 1991; Santarelli et al., 1996; Casadio et al., 1997). The levels of sensorin mRNA and protein in the SN branches contacting L7 correlate with changes in synaptic efficacy associated with new synapse formation, long-term facilitation with 5-HT, and long-term depression with Phe-Met-Arg-Phe-amide (Santarelli et al., 1996;Sun et al., 2001). Our previous studies also indicated that target interactions influence the export of sensorin mRNA in a branch-specific manner (Schacher et al., 1999).
How does the target regulate levels of specific mRNAs in the distal neurites? In the present study, we have addressed the role of interactions with specific target neurons in regulating the distribution of specific mRNAs in a presynaptic neuron using in situ hybridization and RT-PCR techniques. We found that sensorin mRNA accumulates at the axon hillock and neurites of the SN that is contacting a synaptic partner (L7) compared with SNs contacting a target that fails to induce synapse formation (L11). Sensorin mRNA at distal neurites is more stable when the SN neurites contact L7 compared with SN neurites contacting L11. The results indicate that interaction with a synaptic target can regulate both the distribution of specific mRNAs destined for export and their stability at distal sites.
MATERIALS AND METHODS
Cell culture and electrophysiology. SNs were isolated from pleural ganglia dissected from adult animals (80–100 gm); motor cells L7 and L11 were isolated from juvenile (1–3 gm) abdominal ganglia and maintained in coculture for 1–3 d as described previously (Rayport and Schacher, 1986; Schacher et al., 1999). Individual SNs were plated in the same culture dish at very low density to prevent cells from contacting each other. Cocultures consisted of one SN with one L7 or one L11 (SN-L7 or SN-L11). In some cases, one coculture of each type was plated in the same dish. Because each neuron was isolated from the rest of the nervous system, glial cells typically did not contaminate the cultures. Occasionally, a few glial cells were attached to the cell body of L7 or L11. No glial cells were detected on the distal axons or regenerated neurites.
Standard electrophysiological techniques were used to record the amplitude of the EPSP evoked in L7 or L11 with stimulation of each SN (Schacher and Montarolo, 1991; Schacher et al., 1999). Motor cells (L7 and L11) were impaled with a microelectrode (resistance of 15–20 MΩ) containing 2.0 m K-acetate, 0.5 mKCl, and 10 mm K-HEPES, pH 7.4, and held at −85 mV. Each SN was stimulated with a brief (0.3–0.5 msec) depolarizing pulse to evoke an action potential, using an extracellular electrode placed near the cell body of the SN. The amplitude of the EPSPs evoked in L7 ranged from 1 mV (day 1) to 45 mV (day 3). Action potentials in SNs never evoked EPSPs in L11 (data not shown) (Glanzman et al., 1989;Schacher and Montarolo, 1991).
In situ hybridization. Antisense oligonucleotide probes were designed to cover specific coding portions of the target sequences used successfully in RT-PCR analyses of the mRNAs, synthesized (Genset, La Jolla, CA), lyophilized, and redissolved in sterile distilled water. The specific antisense oligonucleotide probe sequences were as follows: (1) SN-specific protein sensorin: TTAACTCTTCTTGACTCACCAACTGCCTGGAT- AGCA complimentary to nucleotide (nt) 304–339 of its cDNA; (2) neuron-specific isoform of actin: CACTTCATGATGGAGTTGAAGGTGGTCTCGTGGACA complimentary to nt 890–925 of its cDNA; (3) common extracellular region for all isoforms ofAplysia cell adhesion molecule (apCAM): TGGGGCCCTTGTTCACAATGCCATCAGTGCTGGCAC complimentary to nt 1078–1113 of its cDNA. Their sense probes were also designed for use as nonspecific controls. These probes were labeled by 3′ end with biotin-deoxyUTP (dUTP) according to the manufacturer's instructions (Boehringer Mannheim, Indianapolis, IN).
All subsequent reagents and solutions used in the in situhybridization were made with diethyl pyrocarbonate-treated MilliQ water, and care was taken to avoid contamination with RNases. At 1 or 3 d after culture, cells were rinsed briefly in artificial seawater (ASW) and fixed in 0.1 m PBS, pH 7.4, containing 4% paraformaldehyde and 30% sucrose for 1 hr at room temperature. The cultures were washed in 0.1m PBS three times for 10 min at room temperature. The cells were digested with 1 μg/ml of proteinase K in TE buffer (0.1 m Tris-HCl with 1 mmEDTA, pH 8.0) for 20 min at 37°C, then washed with PBS three times for 10 min at room temperature. The cells were post-fixed with 4% paraformaldehyde in PBS for 10 min at room temperature, then washed three times for 10 min at room temperature with PBS. Cells were rinsed in 0.1 m triethanolamine (pH 8.0, plus 0.25% acetic anhydride) for 10 min at room temperature, equilibrated in 50% deionized formamide in 5× SSC (saline sodium citrate, pH 7.2) for 20 min at 42°C, and hybridized overnight at 42°C in hybridization buffer (50% deionized formamide, 5× SSC, 0.02% SDS, 2% blocking reagent) containing 1.5 μg/ml of the biotin-labeled oligonucleotide probes. Unbound probe was washed out with 2× SSC two times for 15 min at 42°C and with 0.1× SSC two times for 15 min at 45°C. After cultures were equilibrated in buffer I (0.1 mTris-HCl and 0.15 m NaCl) for 2 min and in buffer II (0.1 m Tris-HCl with 0.15m NaCl, 0.3% Triton X-100, and 2% normal goat serum) for 30 min at room temperature, cells were incubated in streptavidin-FITC (Invitrogen, Gaithersburg, MD; 1:200, diluted in buffer II) for 4 hr at 4°C. Unbound antibody was washed out with PBS three times for 15 min at room temperature. The hybridization signals were visualized directly with fluorescent microscopy.
The specificity of biotin-labeled antisense oligonucleotide probe was examined by hybridizing with labeled sense probe or excess unlabeled probe, or by omitting probe in hybridization solution as well as RNase pretreatment. All controls were negative. Cells were hybridized under the same conditions to reduce variability in different experimental groups.
To observe the distribution of specific mRNAs in neurites and varicosities of live SNs, the oligonucleotide probes (antisense or sense) were labeled with fluorescent-tagged dUTP (Alexa488, Alexa568, or Alexa594 from Molecular Probes, Eugene, OR) at the 3′ end and injected by pressure into the cell bodies of SNs. The concentration of each probe in the injection electrode was 250 ng/μl dissolved in 0.5m KCl buffered with 10 mm K-HEPES, pH 7.4. In some cases, two oligonucleotides (two antisense probes or one antisense and one sense probe) were coinjected into a single SN. The regenerated neurites and varicosities of SNs were visualized (fluorescent or Nomarski optics) 2–6 hr after injections with a SIT camera (Dage 68; Dage-MTI, Michigan City, IN) attached to a Nikon Diaphot inverted microscope. At earlier time points, staining of regenerated neurites and varicosities was weak because insufficient time had elapsed for fluorescent signal to build up at distal sites.
Dissection of cellular compartments and RT-PCR. Cultures were rinsed with hemolymph-free medium (1:1 by volume of L15 plus Instant Ocean) and allowed to cool to 4°C for ∼20 min. Bovine serum albumin (10 μg/ml) was added to the final rinse to prevent dissected cell bodies and neurites from sticking to the dish surface or the transfer pipettes. In some cultures, the cell body of the SN was removed 1 d before collection. The SN axon survives for several days and maintains synaptic efficacy in the absence of the cell body (Schacher and Wu, 2002). Each sample collected consisted of the SN axon and the cell body and processes of the target cell (L7 or L11). Each sample was transferred to 200 μl of Trizol (Invitrogen), and total RNA was isolated after chloroform extraction and isopropanol precipitation. cDNA from each sample was synthesized by using random hexamers as primers and reverse transcriptase (Superscript II). Duplicate aliquots (2 μl) from each sample were used to amplify the fragments by PCR (35 cycles), using primer sets for two transcripts: (1) SN-specific neuropeptide sensorin (Brunet et al., 1991), AACAGAAACAGTCTTTCCCC and TCTTGACTCACCAACTGCC (nt 43–331) and (2) neuron-specific isoform of actin (DesGroseillers et al., 1994), ACCACCACCTTCAACTCCATC and GGGTAAGAGAAGCAAGAAAG (nt 900–1298). The lengths of the synthesized fragments detected with ethidium bromide staining on 1.2% agarose gels were identical to those predicted from the known sequences in the database. For semi-quantitative comparison of mRNA expression, the number of PCR cycles was adjusted initially to ensure linearity of the fluorescent signals of the fragments amplified by PCR [for methods, see Schacher et al. (1999)]. Each PCR run included amplifications and the detection of specific fragments from known cDNAs of known concentrations (positive controls), amplifications of each fragment from samples of medium collected near cells after removal of the cells from the culture dish, and other negative controls.
Quantification and data analysis. The NIH Scion software package was used to quantify average pixel intensities for hybridized signal of sensorin, actin, and apCAM mRNAs in SN cell bodies, axons, neurites, and varicosities. For examining distribution within the SN cell body, pixel intensity was averaged for six separate 5 × 5 μm squares within the peripheral cytoplasm of each SN cell body. The squares were positioned at the following equivalent clock locations: 12, 2, 4, 6, 8, and 10 (sites 1 through 6). The cytoplasm of the cell body adjacent to the SN axon was positioned at 6 o'clock (position 4), and the signal intensity at the opposite side away from the axon (12 o'clock or position 1) was normalized as 100%. For each cell, the intensities at the other five locations were normalized to that measured at the 12 o'clock location. The overall effect of target interaction was determined by ANOVA, and specific differences for the different target interactions were determined by the Scheffe multiple-comparison tests.
After pressure injection of labeled probes, staining intensity was measured in the axon stump, regenerated neurites, and varicosities of the SNs. Because the staining intensity of neurites and varicosities in SNs contacting L11 declined significantly by 200 μm from the SN axon stump, only structures that regenerated within that distance were measured. The intensity of each neurite was measured at 50 μm intervals. All neurites and their varicosities (sphere-like structures >1.5 μm in diameter) emerging from the SN axon stump were measured. Intensities for each compartment for each SN were averaged to provide an intensity level for that SN.
In the RT-PCR analysis, to quantify differences in the levels of sensorin mRNA expression, we normalized staining intensity for sensorin fragments (percentage) by the signals obtained for actin fragments in the same sample (staining intensity of sensorin/staining intensity of actin × 100). We normalized sensorin levels with actin levels, because the average staining intensity for actin fragments within the compartment (cell body of the target along with the axonal processes and neurites of the SN and the target) was not affected by cell interaction. Scion software package was used to quantify average pixel intensities for PCR fragments in each compartment. On the basis of signals generated from known concentrations of sensorin cDNA (positive controls), a twofold increase in staining intensity for sensorin A fragment equaled approximately a fivefold increase in the starting cDNA levels.
The overall effects of target interaction were determined by ANOVA, and differences between the means ± SEM for the different target interactions were determined by the Scheffe multiple-comparison tests.
RESULTS
The large neurons of Aplysia regenerate and establish stable synaptic connections after 4 d in culture (Montarolo et al., 1986; Rayport and Schacher, 1986; Zhu et al., 1994). We reported previously (Schacher et al., 1999) that an SN-specific mRNA (sensorin) is exported to neurites, and overall levels are regulated by target interaction and synapse formation. In the present study, we examined whether the distribution of three mRNAs, including sensorin, is regulated by interactions with a specific target. We examined whether the distribution of an mRNA encoding a family of membrane proteins (apCAMs) is regulated by target interaction. This family of proteins, homologous to NCAM and fasciclin II, is the major membrane protein inAplysia neurons, accounting for >10% of the total membrane protein (Keller and Schacher, 1990; Mayford et al., 1992). Little or no apCAM mRNA is exported from the cell bodies of Aplysianeurons (Schacher et al., 1999). We also determined whether actin mRNA in SNs is regulated by target interaction, because it is expressed abundantly and is exported from the cell bodies of Aplysianeurons.
Target interaction regulates the distribution of sensorin mRNA in cell bodies of SNs
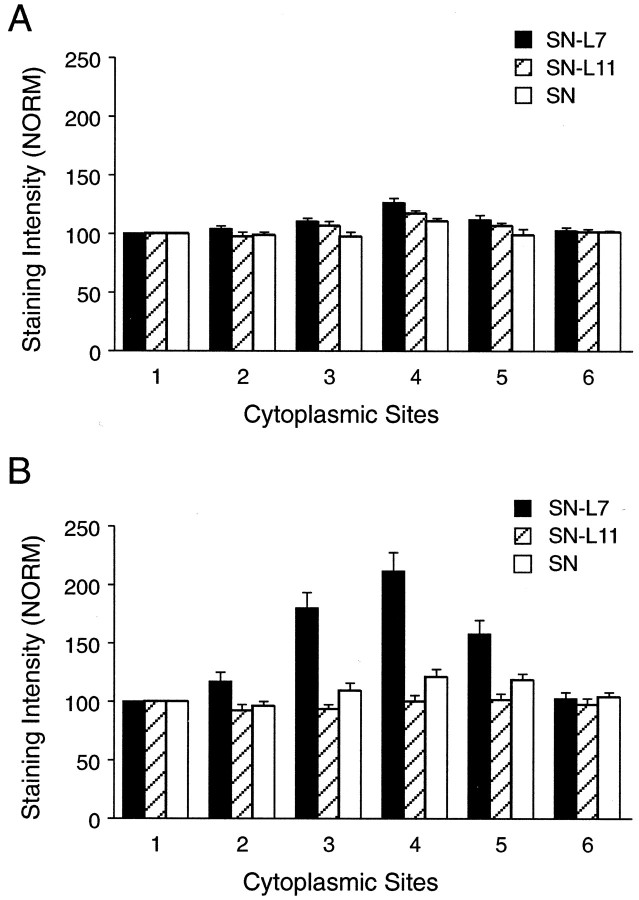
Interaction with a specific target leads to the accumulation of sensorin mRNA in the region of the cytoplasm adjacent to the axon hillock. Using in situ hybridization, we measured the staining intensity of oligonucleotide probes (experimental and control) that recognize sensorin mRNA in six areas of cytoplasm of the SN cell body that were cultured alone or in SNs contacting L7 or L11 (Fig.1). The six areas corresponded to the following clock positions: 12 (position 1 and opposite the axon hillock), 2, 4, 6 (axon hillock), 8, and 10. After day 3 in culture, SNs contacting L7 had intense staining in areas adjacent to the axon hillock (positions 3, 4, and 5) compared with SNs cultured alone or in contact with L11 (Figs. 1B,2B). There was more than a twofold difference in staining intensity near the axon hillock in SNs contacting L7. SNs in contact with target neuron L11 (no synapse formation) show a uniform distribution in cytoplasm of the SN cell body (Figs. 1B, 2B). In contrast, we did not observe obvious differences in staining for sensorin mRNA in the cell bodies of SNs cultured alone or in SNs contacting L7 versus L11 after 1 d in culture (Figs. 1A,2A). In each case, there was a small enhancement in staining near the axon hillock (site 4; p < 0.05). SNs cultured alone showed the same small enhancement at the axon hillock observed on day 3. Contact and synapse formation with motor neuron L7 results in more sensorin mRNA accumulating at the axon hillock of SN cell bodies.
Fig. 1.
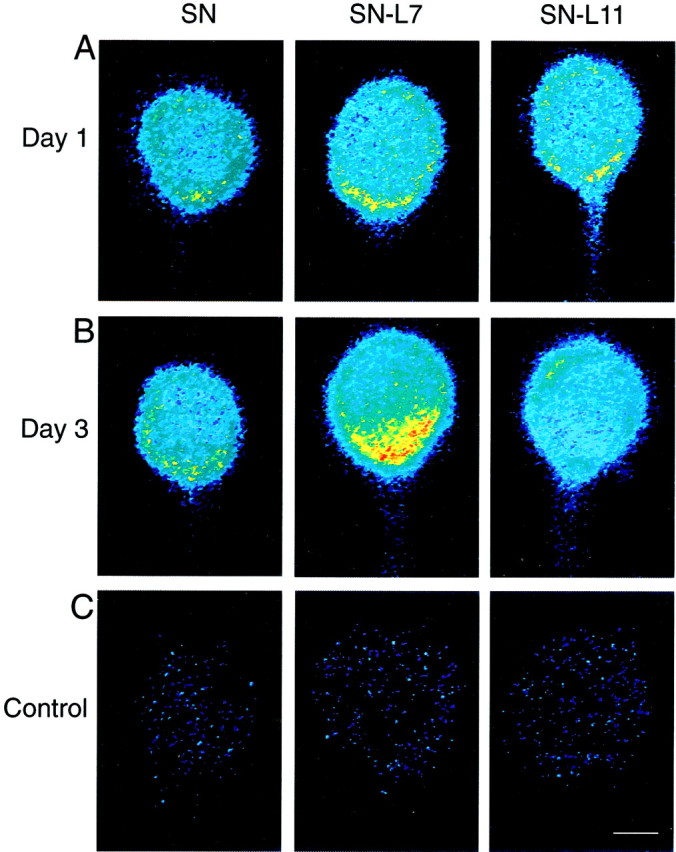
Target interaction regulates the distribution of sensorin mRNA in SN cell bodies. Each micrograph is a pseudocolor representation of fluorescent signals (red is high intensity; blue is low intensity) in SNs cultured alone (SN) and in SNs cocultured with L7 (SN-L7) or with L11 (SN-L11). The low-intensity staining at the outer perimeter of each cell body is an artifact of computer enhancement. Axons emerge from SN cell bodies at approximately the 6 o'clock position. A, After 1 d in culture, there is little difference in the staining intensity within the cytoplasm. There is a small increase in staining near the axon hillock. B, By 3 d there is a significant difference in staining within the cell bodies. Staining intensity is highest where the axon emerges from the SN cell body when the SN contacts L7. Note the uniform staining when SN contacts L11. C, Staining is very weak in SN cell bodies (day 3 in culture) when incubated with control (sense) probes. Scale bar, 25 μm.
Fig. 2.
Interaction with L7 leads to increased accumulation of sensorin mRNA in the axon hillock of the SN. Each graph summarizes relative intensity at five sites in the cytoplasm of SN cell bodies (sites 2-6) compared with site 1, which is the portion of the cytoplasm equivalent to the 12 o'clock position. Sites 2-6are the 2, 4, 6 (axon), 8, and 10 o'clock positions, respectively. At each time point, the height of each bar is the mean ± SEM relative intensity from 10 SNs for each culture condition.A, Interaction of SN with a target after 1 d in culture. The distribution of sensorin mRNA in the cytoplasm of SN cell bodies was not influenced significantly by target interaction (two-factor ANOVA; df = 10, 135; F = 1.34;p > 0.10). There was a small effect of site location on staining intensity (p < 0.01).B, Distribution of sensorin mRNA in SN cell bodies is affected significantly by interaction with L7 in 3 d cultures. An ANOVA (two-factor) indicated an overall significant effect (df = 10,135; F = 11.042; p < 0.001). Interaction with L7 resulted in a significant difference at sites 3 (4 o'clock) and 4 (6 o'clock) compared with SN and SN-L11 cultures (p < 0.01 for both; Scheffe test) and at site 5 (8 o'clock) compared with SN-L11 cultures (p < 0.05).
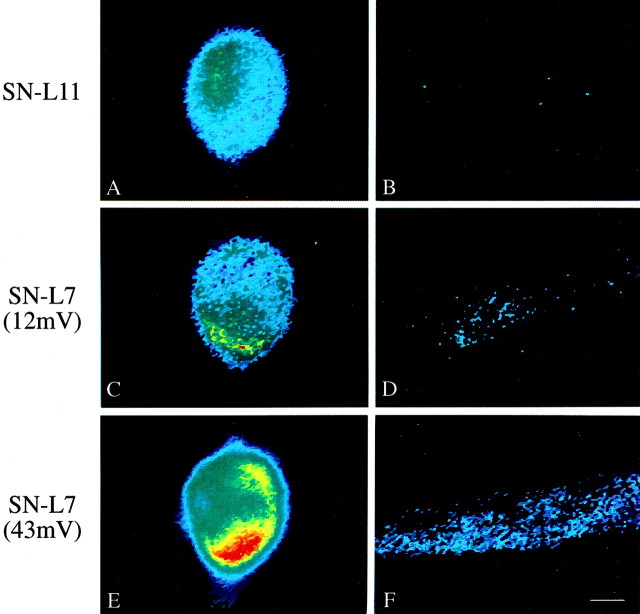
The staining for sensorin mRNA in SN neurites contacting the major processes of L7 is high compared with staining in SN neurites contacting L11 (Fig.3A–D). The major processes of L7 are a preferred site of neurite growth and the formation of varicosities with transmitter release sites (Glanzman et al., 1989). In contrast, SN neurites contacting L11 have few varicosities, and those varicosities fail to develop transmitter release sites (Glanzman et al., 1989; Schacher and Montarolo, 1991). Little staining for sensorin mRNA was detected in SN neurites contacting the major processes of L11 (n = 5 cultures) (Fig. 3A,B). Synapse formation with L7 results in higher levels of sensorin mRNA in SN neurites compared with SN neurites contacting L11. The intensity of staining also correlates with the amplitude of the EPSPs (Fig.3C–F). Low levels of staining are detected when EPSP amplitudes are <20 mV. These results suggest that mRNA export from the SN cell bodies and the stability of the mRNA once it arrives in neurites are regulated by target interaction and synapse formation.
Fig. 3.
SN neurites interacting with L7 on day 3 in culture contain high levels of sensorin mRNA compared with SN neurites interacting with L11. Each micrograph is a pseudocolor representation of fluorescent signals (red is high intensity;blue is low intensity) in SNs cocultured with L11 (SN-L11) or with L7 (SN-L7) after 3 d. The low-intensity staining at the outer perimeter of each cell body is an artifact of computer enhancement. A,B, Staining in the SN cell body (A) is distributed uniformly (Figs. 1, 2) and virtually absent in SN neurites regenerating along the major processes of L11 (center of micrograph from left toright; see D andF). This level of staining was detected in all five SN-L11 cultures. C, D, Stimulation of the SN evoked a 12 mV EPSP on day 3. Staining of cytoplasm near the SN axon is high (C), and modest staining is detected in SN neurites interacting with major processes of L7 (D). This level of staining was observed in all three cultures in which EPSPs were <20 mV. E,F, Stimulation of the SN evoked a 43 mV EPSP on day 3. Staining of cytoplasm near SN axon is high (E), and staining in SN neurites interacting with major processes of L7 (F) is distributed over all of the major processes of L7. This level of staining was detected in all three cultures in which EPSPs were >30 mV. Scale bar, 25 μm.
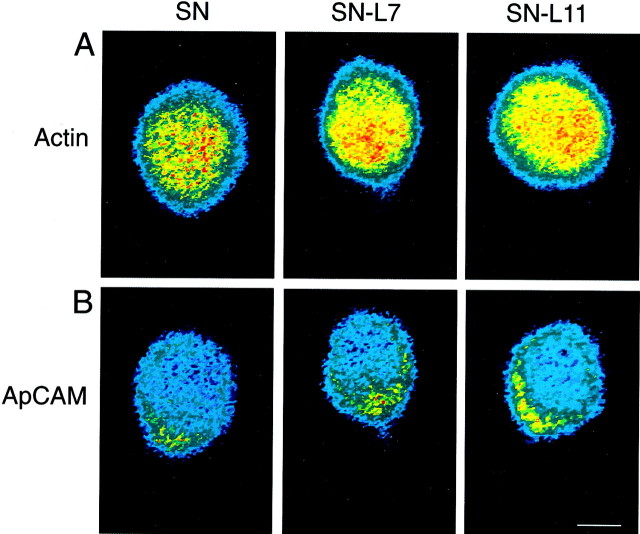
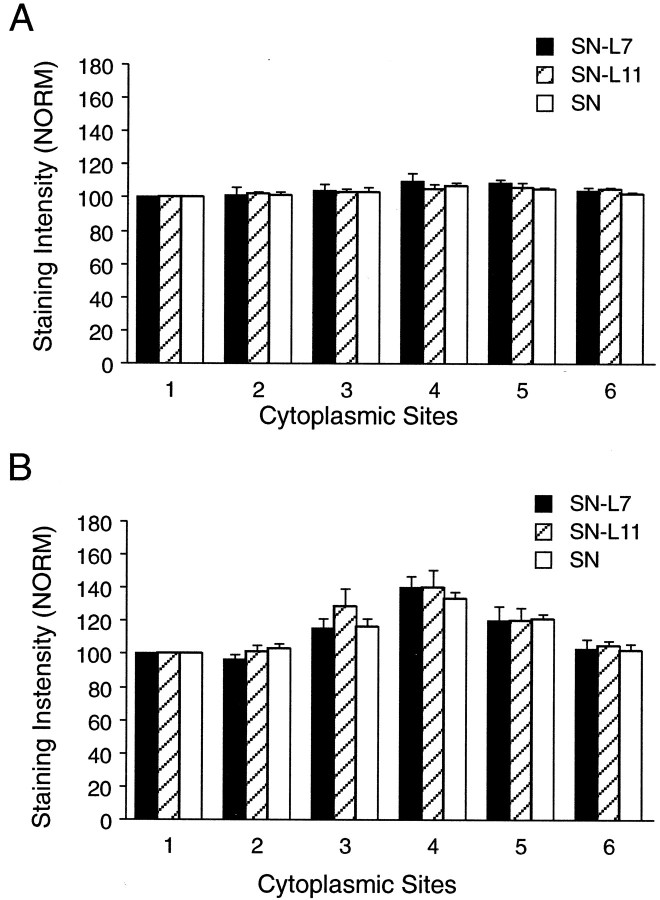
The distribution of two other mRNAs in the cell bodies of SNs was not regulated by target interaction (Figs. 4, 5). Actin mRNA, but not apCAM mRNA, is exported into the axons ofAplysia neurons (Schacher et al., 1999). Actin mRNA was expressed at high levels and was distributed uniformly throughout the cell bodies of all SNs (Figs. 4A, 5A). Target interaction also did not affect the distribution of apCAM mRNA. As was found for sensorin, there was a small increase in staining intensity for apCAM mRNA (∼50%) in the cytoplasm adjacent to the axon hillock of the SN (Figs. 4B, 5B). These results suggest the possibility that mRNAs encoding secretory or membrane proteins accumulate at or near the axon hillock. The distribution of some of these mRNAs within the cytoplasm of neuronal cell bodies is regulated by target interactions.
Fig. 4.
Target interaction does not affect distribution of apCAM and actin mRNAs in SN cell bodies. Each micrograph is a pseudocolor representation of fluorescent signals (redis high intensity; blue is low intensity) in SNs cultured alone (SN) and in SNs cocultured with L7 (SN-L7) or with L11 (SN-L11) after 3 d in culture. Axons emerge from SN cell bodies at approximately the 6 o'clock position. The low-intensity staining at the outer perimeter of each cell body is an artifact of computer enhancement.A, Staining for actin mRNA is relatively high (compared with sensorin or apCAM mRNAs) and distributed uniformly in SN cell bodies for each culture condition. B, Staining for apCAM mRNA is enhanced near the SN axon (6 o'clock position) for each culture condition. Scale bar, 25 μm.
Fig. 5.
Target interaction has no significant effect on distribution of actin or apCAM mRNAs in SN cell bodies after 3 d in culture. Each graph summarizes relative intensity at five sites in the cytoplasm of SN cell bodies (sites2–6) compared with site1, which is the portion of the cytoplasm equivalent to the 12 o'clock position. Sites 2–6 are the 2, 4, 6 (axon), 8, and 10 o'clock positions, respectively. The height of each bar is the mean ± SEM from 10 SNs for each culture condition. A, Distribution of actin mRNA is uniform. An ANOVA (two-factor) indicated no overall significant effect (df = 10, 135; F = 0.593; p > 0.7), including no effect of treatment or site on staining intensity.B, Distribution of apCAM mRNA is not affected by target interaction. An ANOVA (two-factor) indicated no overall significant effect (df = 10, 135; F = 0.48;p > 0.9), but there was a significant effect of site location on staining intensity (p < 0.001).
Target interaction regulates the distribution and stability of mRNA in SN neurites
We next examined whether the mRNAs exported to distal neurites are affected by target interaction. We used two methods: (1) injection of labeled probes into the cell body of SNs and monitoring the subsequent distribution of the labeled probes in live cells or (2) RT-PCR analyses of mRNA in neurites after removal of the SN cell body.
We first tested the probe injection method for monitoring the distribution of mRNA. After injecting labeled probes recognizing an mRNA that is restricted to the cell body (apCAM), fluorescent staining was detected primarily in the cell body (Fig.6A). In contrast, staining was detected in axons and neurites after injecting probes recognizing mRNAs that are exported (sensorin and actin) (Fig.6A,B). Another validation of this method is that labeled antisense probes were detected in the distal processes, whereas labeled sense probes were barely detectable (Fig.7). We therefore applied this technique to evaluate staining intensity for probes recognizing actin or sensorin mRNA in distal SN neurites contacting either L7 or L11. We used two types of cultures: SN contacting a single target and SN contacting two targets.
Fig. 6.
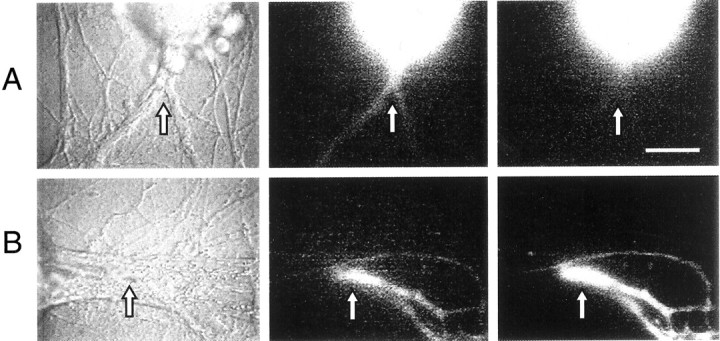
The distribution of specific mRNAs in living cells is comparable with that detected with previous RT-PCR results. A, Sensorin mRNA, but not apCAM mRNA, is exported into the SN axon. Antisense oligos for sensorin and apCAM were coinjected into same SN cell body (left panel). At 3 hr after injection, antisensorin probe (labeled with Molecular Probes Alexa-488 dUTP; middle panel), but not anti-apCAM probe (labeled with Alexa-568 dUTP; right panel), is detected in the bifurcating axon of the SN (arrow). Staining intensity in the SN cell body was comparable for the two probes (<5% difference). B, Both sensorin and actin mRNAs are exported into axons. Antisense probes (Alexa-488 dUTP for sensorin and Alexa-594 dUTP for actin) were coinjected into the SN cell body (not in image). After 3 hr, significant signals for both probes (sensorin in middle panel and actin in right panel) were detected in SN neurites (arrow) contacting L7 axon (extending fromleft to right in center of micrograph) ∼200 μm from the SN cell body (left panel). All oligos contain labeled dUTP at the 3′-end. Scale bar, 25 μm.
Fig. 7.
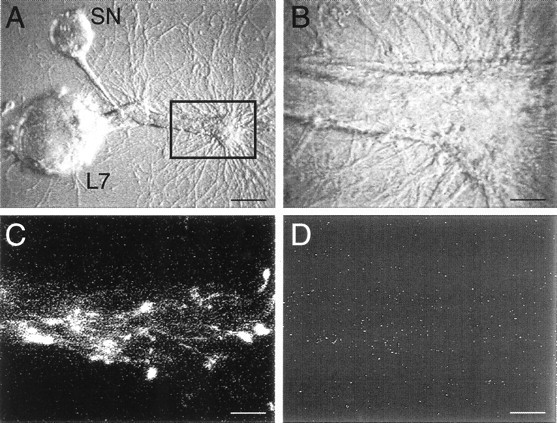
Antisense probes are transported to neurites selectively. A, Micrograph of SN-L7 culture after 3 d. The major processes of L7 extend toward theright. Both sense and antisense probes for sensorin mRNA were coinjected into the cell body of the SN, and fluorescent images were taken 4 hr after injection. The outlined area, which includes an axon stump of L7, is viewed at higher magnifications in B–D. Scale bar, 50 μm.B, High- magnification micrograph of stump and major processes of L7. Fluorescent images from this area are viewed inC and D. Scale bar, 12.5 μm.C, Antisense probe for sensorin mRNA (labeled with Alexa-594) labels neurites and varicosities contacting the major processes of L7. D, Sense probe for sensorin (labeled with Alexa-488) fails to label the same neurites and varicosities. Staining intensity for each probe in the SN cell body was comparable (<5% difference). Scale bars, 12.5 μm.
Staining for probes recognizing sensorin and actin mRNA is stronger in SN neurites contacting L7 compared with staining in SN neurites contacting L11 (Fig. 8). In general, staining in the SN neurites contacting L11 was below levels of detection by 200 μm from the stump of the SN axon (Fig. 8, compareA3 and A4 with B3 andB4). Within this 200 μm zone, the staining intensity of the SN varicosities after normalization with the staining intensity measured in the axon stump (Fig. 8, bottom portions ofA3, A4, B3, and B4) was ∼2.3 fold higher for those contacting L7 versus L11 (n = 6 cultures each; p < 0.01).
Fig. 8.
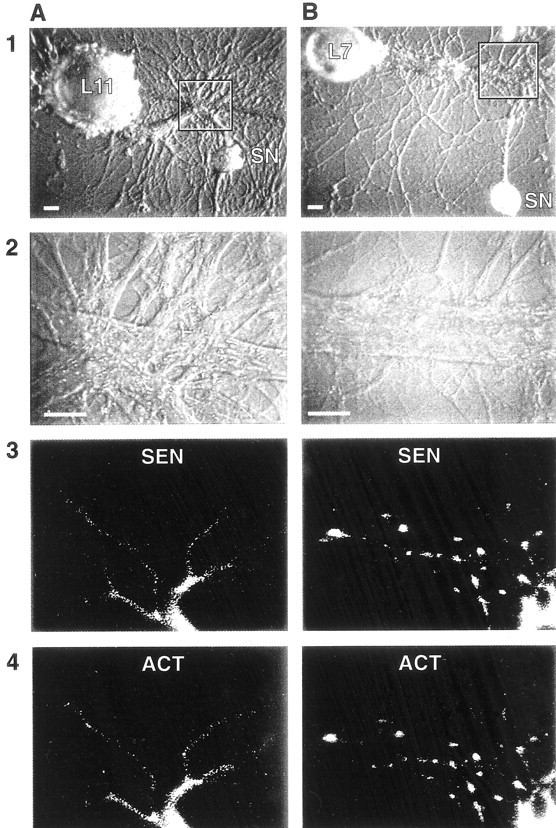
Distribution of both actin and sensorin mRNAs in live SNs is affected by target interaction. SNs cultured with either L11 (A) or L7 (B) after 3 d were coinjected with fluorescent-tagged antisense probes for sensorin (Alexa-488 dUTP; A3, B3) and actin (Alexa-594 dUTP; A4, B4).A1, B1, Nomarski optics image of the cocultures. Areas encompassed by the rectangles are viewed in the remaining images. Scale bars, 30 μm. A2,B2, Nomarski optics image of areas viewed for distribution of tagged probes. Scale bars, 25 μm. A3,A4, B3, B4, Epifluorescent views of staining by antisense probes for sensorin (3) and actin (4) 4 hr after injections. SN neurites and the few varicosities interacting with axon of L11 show weak staining, which diminishes significantly after ∼200 μm from the SN axon stump (bottom portion of micrographs in A3 and A4). The axon stump shows significant staining. By contrast, SN varicosities contacting L7 show significant staining with probes for both sensorin and actin mRNA (B3, B4). Staining intensity of SN neurites and varicosities contacting L7 was 15.8 ± 1.9 and 30.6 ± 2.0, respectively (n = 6 cultures). Staining intensity of SN neurites and varicosities contacting L11 was 9.6 ± 1.2 and 13.5 ± 1.5, respectively. An ANOVA indicated a significant difference in staining when contacting L7 versus L11 (df = 1, 10; F = 20.157; p < 0.001). Staining in neurites (p < 0.03) and varicosities (p < 0.01) was significantly higher when contacting L7 versus L11.
A significant difference in staining was detected as well when probes were injected into the cell body of a single SN in contact with two separate targets (L7 and L11 or two L7s). We compared staining intensity detected in the regenerating neurites emerging from the stumps of bifurcate axons placed near each target (Martin et al., 1997;Schacher et al., 1999). There was approximately a twofold increase in staining intensity of SN neurites and varicosities contacting L7 compared with staining in neurites and varicosities contacting L11 (Fig. 9A). There was no significant difference in staining when the SN contacted two L7 targets (Fig. 9B). These results suggest that the target can regulate the levels of exported mRNAs in SN neurites and varicosities. The differences can be a consequence of target interaction on initial export of mRNA into the regenerated neurites and varicosities or the stability of mRNAs in those cellular compartments once they arrive.
Fig. 9.
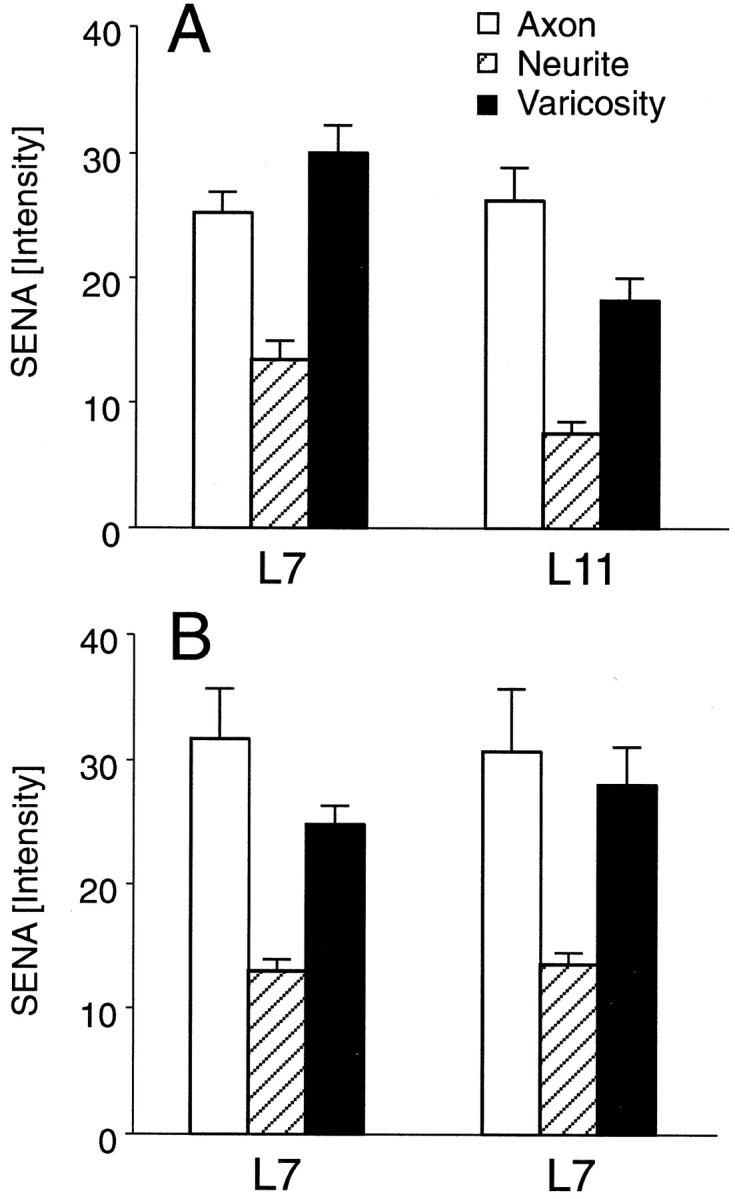
Branch-specific differences in the distribution of sensorin mRNA detected with fluorescent-tagged antisense probe injected into a single SN interacting with multiple contacts. A, Staining (pixel intensity) of SN axons, neurites, and varicosities when cocultured with both L7 and L11 was averaged for five cultures. Staining intensity in the axons was not significantly different. Staining intensity of neurites and varicosities regenerated from SN branch contacting L7 was significantly greater than intensity of neurites and varicosities regenerated from SN branch contacting L11. An ANOVA (two-factor) revealed an overall difference (df = 2, 16;F = 5.279; p < 0.02). Staining in both neurites (p < 0.03) and varicosities (p < 0.02) was significantly higher when they contacted L7 compared with L11. B, Staining of SN axons, neurites, and varicosities when cocultured with two L7 cells was averaged for five cultures. Placement of each SN-L7 interaction in a specific group was determined randomly. Average EPSP amplitudes for the two groups were not significantly different (19.8 ± 3.2 mV vs 18.4 ± 3.0 mV). An ANOVA indicated no significant difference in staining at the two branches (df = 2, 16; F = 0.299; p > 0.7).
To measure stability of mRNA in the distal neurites, we examined the consequences of target interaction on mRNA levels detected in distal neurites after removal of the SN cell body. Previously, we found that the level of sensorin mRNA in SN neurites contacting L7 was not significantly different from the level in SN neurites contacting L11 on day 1 in culture. With further interaction (day 4), there was more sensorin mRNA in neurites contacting L7 versus SN neurites contacting L11 (Schacher et al., 1999). We therefore tested for stability by comparing levels of sensorin mRNA in SN neurites after 1 d in culture with that detected in neurites 1 d after removal of the SN cell body (Fig.10A,B). SN neurites detached from their cell bodies continued to regenerate and form new synapses with L7 (Schacher and Wu, 2002).
Fig. 10.
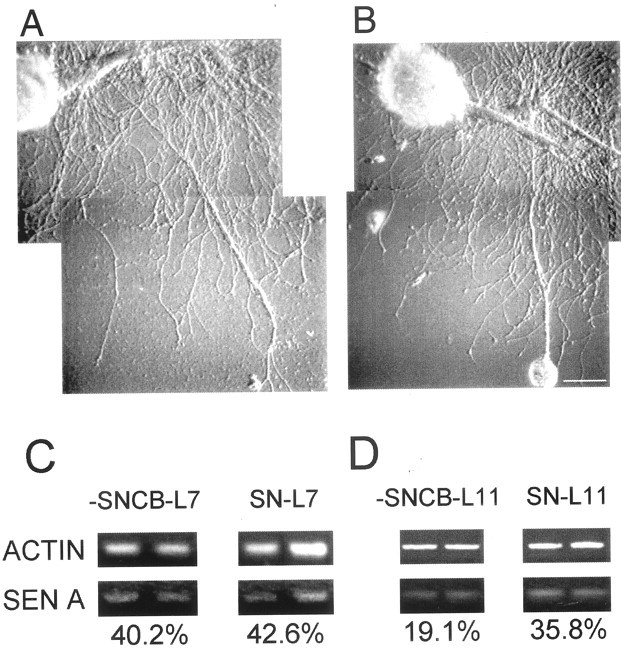
Target influences stability of transported mRNA.A, B, Photographs of SN-L7 cultures 24 hr after dissection of SN cell body (A) and matching controls (B). C, D, Stability of sensorin mRNA in SN axon and neurites is affected by target. RT-PCR fragments for actin and sensorin mRNA were amplified from the neurites of SNs (−SN cell body for 24 hr or control cultures) interacting with targets (L7 or L11) from two sets of SN-L7 and SN-L11 cocultures in each condition. When normalized for actin mRNA levels in the entire culture, levels of sensorin mRNA were not affected in SN-L7 cultures after cell body dissection, whereas there was a significant reduction after dissection when SNs interacted with L11. An ANOVA indicated a significant effect of treatment (df = 3, 18;F = 20.036; p < 0.001). Removal of the SN cell body did not significantly affect sensorin mRNA levels in the neurites contacting each L7 (each cell was selected at random to belong to a specific group; p > 0.7). Removal of the SN cell body resulted in a significant decline in sensorin mRNA in SN neurites contacting L11 (p < 0.01).
Sensorin mRNA exported from the SN cell body is more stable when the SN contacts L7 compared with L11 (Fig. 10C,D). After removal of the SN cell body (24 hr), amounts of sensorin mRNA already exported to the neurites are not significantly lowered when SN neurites contact L7 (Fig. 10C) (n = 5 cultures each). In contrast, sensorin mRNA declines significantly (Fig.10D) (n = 6 cultures each;p < 0.01) in neurites contacting L11. Thus, preexisting sensorin mRNA in SN neurites contacting L11 appears to be less stable than sensorin mRNA in SN neurites contacting L7.
DISCUSSION
Our results indicate that the target modulates the distribution of mRNAs in both the cell body and neurites of the presynaptic neuron. As synapses form and strengthen, mRNA encoding sensorin accumulates near the axon hillock of the SN. Accumulation of sensorin or actin mRNA in SN neurites and varicosities is regulated by the influence of the target on local mRNA stability and transport to SN neurites and varicosities.
The accumulation of sensorin mRNA and to some extent apCAM mRNA at the axon hillock suggests that transcripts encoding proteins destined for distal neurites (Keller and Schacher, 1990; Santarelli et al., 1996) are targeted to sites in the cell body adjacent to the axon hillock (Litman et al., 1993). Such targeting may be mediated by structural sequences within the mRNAs and the potential interaction of the mRNAs with binding proteins associated with the intracellular transport apparatus (Bassell et al., 1994; Litman et al., 1994; Behar et al., 1995; Knowles et al., 1996; Mayford et al., 1996; Kiebler et al., 1999; Zhang et al., 2001). The presence of mRNAs at the axon hillock would facilitate access of the translated products in the axon hillock to the transport apparatus for export to specific distal sites. Because apCAM mRNA is not transported, regulation of its overall expression takes place in the cell body (transcription or translation) and via turnover of existing proteins throughout the cell (Bailey et al., 1992;Zhu et al., 1995; Schacher et al., 1999). In the case of sensorin, the accumulation of its mRNA at the axon hillock may also facilitate the transport of sensorin mRNA to distal synaptic sites (Schacher et al., 1999).
Synapse formation with L7 modulates the targeting of sensorin mRNA to the axon hillock. The consequences of target interaction increase during the first few days in culture. Target interaction might affect accumulation of sensorin mRNA selectively at the axon hillock by regulating splicing events that serve to influence the expression of appropriate mRNA sequences that are required for targeting sensorin mRNA to the axon hillock. Alternatively, target interaction could modulate the expression of selective binding proteins required for mRNA distribution or targeting. Identification of the critical elements for targeting sensorin mRNA will help determine the mechanism mediating its selective accumulation at the axon hillock. The overall effect of this accumulation in the axon hillock is to increase the level of expression of sensorin at distal synaptic sites (Santarelli et al., 1996; Casadio et al., 1997).
The target also regulates the level of sensorin mRNA in SN neurites. As was found with RT-PCR analyses (Schacher et al., 1999, their Fig. 4), staining for sensorin in SN neurites correlated with EPSP amplitude. Because the number of SN varicosities contacting the major processes of L7 increases with EPSP amplitude (Glanzman et al., 1989;Schacher and Montarolo, 1991), the high level of sensorin mRNA staining may reflect the greater density of SN varicosities contacting L7. SN varicosities may also represent a preferred target site where sensorin mRNA accumulates. Sensorin neuropeptide is concentrated at SN varicosities (Santarelli et al., 1996). Translation from the accumulated mRNA in the axon hillock and subsequent transport along with local translation of mRNA at distal sites may contribute to the increase expression of the peptide in varicosities contacting L7. The peptide may have a direct physiological role at SN synapses (Brunet et al., 1991) or some other signaling function that affects the properties of SN or L7 or both.
Target interaction does not appear to influence the distribution of actin mRNA in the cell body of the SN where it is expressed uniformly in the cytoplasm. The uniform expression of actin mRNA throughout the cell body reflects the distribution of the protein itself and suggests that mRNAs encoding this cytoplasmic protein either lack instructions for targeting or contain structural elements that ensure cell body-wide distribution. For example, actin mRNA may contain sequences that prevent its degradation in the cell body, whereas sensorin mRNA may be degraded unless it is protected by binding a molecule that is targeted to the region of the axon hillock (Cooperstock and Lipshitz, 1997). In this scheme, target interaction enhances the expression of a molecule that is targeted to specific sites and serves to stabilize the expression of specific mRNAs that happen to find their way to those sites. This type of mechanism may explain why expression of actin mRNA in SN neurites is enhanced by interaction and synapse formation with L7 and not when the SN neurites contact L11 and fail to form synapses (Glanzman et al., 1989). The local translation of actin mRNA at varicosities may facilitate neuritic growth (Zhang et al., 2001) or the formation of new synapses (Hatada et al., 2000; Schacher and Wu, 2002).
The greater amount of staining of sensorin and actin mRNA in SN neurites and varicosities contacting L7 compared with neurites and varicosities contacting L11 suggests that accumulation at varicosities may be mediated via target-dependent effects on the stability of the mRNA once it arrives at varicosities. That the stability is modulated is supported by the RT-PCR analysis of the level of sensorin mRNA expression remaining in SN neurites 24 hr after removal of the SN cell body. Preexisting sensorin mRNA in the neurites appears to be stable if the SN neurites contact L7 compared with SN neurites that contact L11. A similar mechanism may contribute to the greater staining intensity for actin mRNA in live SN neurites and varicosities when they contact L7 compared with staining intensity in SN neurites and varicosities contacting L11. Because the target neuron has high levels of actin mRNA, we could not test directly the stability of actin mRNA stability in the SN neurites contacting the target. The target might influence expression of mRNA binding proteins that target mRNAs to varicosities and serve to stabilize local mRNA. When contacting L7, proteins that might protect mRNA from degradation are induced and transported to distal neurites of SNs. When contacting L11, SNs may express low levels of these proteins, leading to the degradation of mRNAs at distal sites. These molecules might include those critical for the development or acquisition of the macromolecular complex required for local protein synthesis at distal sites (Tiedge et al., 1999;Kiebler and DesGroseillers, 2000; Steward and Schuman, 2001). Thus the same mechanisms that might affect distribution of some mRNAs in the cell body may contribute to levels of mRNA expression at distal sites.
Footnotes
This research was supported by National Science Foundation Grant IBN-9808938 and National Institutes of Health (NIH) Grant MH-60387, and animals were provided by the National Center for Research Resources for Aplysia at the University of Miami supported by NIH Grant RR 10294. We thank Rachel Yarmolinsky and Eve Vagg for assistance in preparing the figures, and Drs. Kupfermann and Schwartz for comments on this manuscript.
Correspondence should be addressed to Dr. Samuel Schacher, Center for Neurobiology and Behavior, Columbia University College of Physicians and Surgeons, New York State Psychiatric Institute, 1051 Riverside Drive, New York, NY 10032. E-mail:sms2@columbia.edu.
REFERENCES
- 1.Bailey CH, Chen M, Keller F, Kandel ER. Serotonin-mediated endocytosis of apCAM; an early step of learning-related synaptic growth in Aplysia. Science. 1992;256:645–648. doi: 10.1126/science.1585177. [DOI] [PubMed] [Google Scholar]
- 2.Bassell GJ, Zhang H, Kosik KS. Association of polyA mRNA with microtubules in cultured neurons. Neuron. 1994;12:571–582. doi: 10.1016/0896-6273(94)90213-5. [DOI] [PubMed] [Google Scholar]
- 3.Behar L, Marz R, Sadot E, Barg J, Ginzburg I. Cis-acting signals and trans-acting proteins are involved in tau mRNA targeting into neurites of differentiating neuronal cells. Int J Dev Neurosci. 1995;13:113–127. doi: 10.1016/0736-5748(95)00001-w. [DOI] [PubMed] [Google Scholar]
- 4.Brunet J-F, Shapiro E, Foster SA, Kandel ER, Yuichi I. Identification of a peptide specific for Aplysia sensory neurons by PCR-based differential screening. Science. 1991;252:856–859. doi: 10.1126/science.1840700. [DOI] [PubMed] [Google Scholar]
- 5.Cabelli RJ, Shelton DL, Segal RA, Shatz CJ. Blockade of endogenous ligands of trkB inhibits formation of ocular dominance columns. Neuron. 1997;19:63–76. doi: 10.1016/s0896-6273(00)80348-7. [DOI] [PubMed] [Google Scholar]
- 6.Camardo J, Proshansky E, Schacher S. Identified Aplysia neurons form specific chemical synapses in culture. J Neurosci. 1983;3:2614–2620. doi: 10.1523/JNEUROSCI.03-12-02614.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casadio A, Martin KC, Bailey CH, Chen M, Zhu H, Kandel ER. Molecular mechanisms underlying synapse-specific long-term facilitation of Aplysia sensory to motor synapses. Soc Neurosci Abstr. 1997;24:234. doi: 10.1016/s0092-8674(00)80484-5. [DOI] [PubMed] [Google Scholar]
- 8.Cooperstock RL, Lipshitz HD. Control of mRNA stability and translation during Drosophila development. Semin Cell Dev Biol. 1997;8:541–549. doi: 10.1006/scdb.1997.0179. [DOI] [PubMed] [Google Scholar]
- 9.Crino PB, Eberwine J. Molecular characterization of the dendritic growth cone: regulated mRNA transport and local protein synthesis. Neuron. 1996;17:1173–1187. doi: 10.1016/s0896-6273(00)80248-2. [DOI] [PubMed] [Google Scholar]
- 10.DesGroseillers L, Auclair D, Wickham L, Maalouf M. A novel actin cDNA is expressed in the neurons of Aplysia californica. Biochim Biophys Acta. 1994;121:322–324. doi: 10.1016/0167-4781(94)90293-3. [DOI] [PubMed] [Google Scholar]
- 11.Fitzsimonds RM, Poo M-M. Retrograde signaling in the development and modification of synapses. Physiol Rev. 1998;78:143–170. doi: 10.1152/physrev.1998.78.1.143. [DOI] [PubMed] [Google Scholar]
- 12.Glanzman DL, Kandel ER, Schacher S. Identified target motor neuron regulates neurite outgrowth and synapse formation of Aplysia sensory neurons in vitro. Neuron. 1989;3:441–450. doi: 10.1016/0896-6273(89)90203-1. [DOI] [PubMed] [Google Scholar]
- 13.Glanzman DL, Kandel ER, Schacher S. Target-dependent structural changes accompanying long-term synaptic facilitation in Aplysia neurons. Science. 1990;249:799–802. doi: 10.1126/science.2389145. [DOI] [PubMed] [Google Scholar]
- 14.Goodman CS, Shatz CJ. Developmental mechanisms that generate precise patterns of neuronal connectivity. Cell [Suppl] 1993;72:77–98. doi: 10.1016/s0092-8674(05)80030-3. [DOI] [PubMed] [Google Scholar]
- 15. Hatada Y, Wu F, Sun ZY, Schacher S, Goldberg DJ. Presynaptic morphological changes associated with long-term synaptic facilitation are triggered by actin polymerization at pre-existing varicosities. J Neurosci 20 2000. RC82(1–5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hawver D, Schacher S. Selective fasciculation as a mechanism for the formation of specific chemical connections between Aplysia neurons in vitro. J Neurobiol. 1993;24:368–383. doi: 10.1002/neu.480240309. [DOI] [PubMed] [Google Scholar]
- 17.Keller F, Schacher S. Neuron-specific membrane glycoproteins promoting neurite fasciculation in Aplysia californica. J Cell Biol. 1990;111:2637–2650. doi: 10.1083/jcb.111.6.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiebler MA, DesGroseillers L. Molecular insights in mRNA transport and local translation in the mammalian nervous system. Neuron. 2000;25:19–28. doi: 10.1016/s0896-6273(00)80868-5. [DOI] [PubMed] [Google Scholar]
- 19.Kiebler MA, Hemraj I, Verkade P, Kohrman M, Fortes P, Marion RM, Ortin J, Dotti CG. The mammalian Staufen protein localizes to the somatodendritic domain of cultured hippocampal neurons: implications for its involvement in mRNA transport. J Neurosci. 1999;19:288–297. doi: 10.1523/JNEUROSCI.19-01-00288.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knowles RB, Sabry JH, Martone ME, Deerinck TJ, Ellisman MH, Bassell GJ, Kosik KS. Translocation of RNA granules in living neurons. J Neurosci. 1996;16:7812–7820. doi: 10.1523/JNEUROSCI.16-24-07812.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Litman P, Barg J, Rindzoonski L, Ginzburg I. Subcellular localization of tau mRNA in differentiating neuronal cell culture: implications for neuronal polarity. Neuron. 1993;10:627–638. doi: 10.1016/0896-6273(93)90165-n. [DOI] [PubMed] [Google Scholar]
- 22.Litman P, Barg J, Ginzburg I. Microtubules are involved in the localization of tau mRNA in primary neuronal cultures. Neuron. 1994;13:1463–1474. doi: 10.1016/0896-6273(94)90432-4. [DOI] [PubMed] [Google Scholar]
- 23.Martin KC, Casadio A, Zhu H, Yaping E, Rose JC, Chen M, Bailey CH, Kandel ER. Synapse-specific, long-term facilitation of Aplysia sensory to motor synapses: a function for local protein synthesis in memory storage. Cell. 1997;91:927–938. doi: 10.1016/s0092-8674(00)80484-5. [DOI] [PubMed] [Google Scholar]
- 24.Mayford M, Barzilai A, Keller F, Schacher S, Kandel ER. Modulation of an NCAM-related adhesion molecule with long-term synaptic plasticity in Aplysia. Science. 1992;256:638–644. doi: 10.1126/science.1585176. [DOI] [PubMed] [Google Scholar]
- 25.Mayford M, Baranes D, Podsypanina K, Kandel ER. The 3′ UTR of CAMKIIœ mRNA is a cis-acting signal for the localization and translation of mRNA in dendrites. Proc Natl Acad Sci USA. 1996;93:13250–13255. doi: 10.1073/pnas.93.23.13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Montarolo PG, Goelet P, Castellucci VF, Morgan J, Kandel ER, Schacher S. A critical period for macromolecular synthesis in long-term heterosynaptic facilitation in Aplysia. Science. 1986;234:1249–1254. doi: 10.1126/science.3775383. [DOI] [PubMed] [Google Scholar]
- 27.Rayport SG, Schacher S. Synaptic plasticity in vitro: cell culture of identified Aplysia neurons mediating short-term habituation and sensitization. J Neurosci. 1986;6:759–763. doi: 10.1523/JNEUROSCI.06-03-00759.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanes JR, Apel ED, Gautam M, Glass D, Grady RM, Martin PT, Nicholl MC, Yancopoulus GD. Agrin receptors at the skeletal neuromuscular junction. Ann NY Acad Sci. 1998;841:1–13. doi: 10.1111/j.1749-6632.1998.tb10905.x. [DOI] [PubMed] [Google Scholar]
- 29.Santarelli L, Montarolo PG, Schacher S. Neuropeptide localization in varicosities of Aplysia sensory neurons is regulated by target and neuromodulators evoking long-term synaptic plasticity. J Neurobiol. 1996;31:297–308. doi: 10.1002/(SICI)1097-4695(199611)31:3<297::AID-NEU3>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 30.Schacher S, Montarolo PG. Target-dependent structural changes in sensory neurons of Aplysia accompany long-term heterosynaptic inhibition. Neuron. 1991;6:679–690. doi: 10.1016/0896-6273(91)90166-w. [DOI] [PubMed] [Google Scholar]
- 31.Schacher S, Wu F (2002) Synapse formation in the absence of cell bodies requires protein synthesis. J Neurosci, in press. [DOI] [PMC free article] [PubMed]
- 32.Schacher S, Wu F, Panyko JD, Sun ZY, Wang D. Expression and branch-specific export of mRNA are regulated by synapse formation and interaction with specific postsynaptic targets. J Neurosci. 1999;19:6338–6347. doi: 10.1523/JNEUROSCI.19-15-06338.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sherff CM, Carew TJ. Coincident induction of long-term facilitation in Aplysia: cooperativity between cell bodies and remote synapses. Science. 1999;285:1911–1914. doi: 10.1126/science.285.5435.1911. [DOI] [PubMed] [Google Scholar]
- 34.Spencer GE, Syed NI, van Kesteren E, Lukowiak K, Geraerts WP, van Minnen J. Synthesis and functional integration of a neurotransmitter receptor in isolated invertebrate axons. J Neurobiol. 2000;44:72–81. doi: 10.1002/1097-4695(200007)44:1<72::aid-neu7>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 35.Steward O. mRNA localization in neurons: a multipurpose mechanism? Neuron. 1997;18:9–12. doi: 10.1016/s0896-6273(01)80041-6. [DOI] [PubMed] [Google Scholar]
- 36.Steward O, Schuman EM. Protein synthesis at synaptic sites on dendrites. Annu Rev Neurosci. 2001;24:299–325. doi: 10.1146/annurev.neuro.24.1.299. [DOI] [PubMed] [Google Scholar]
- 37.Steward O, Worley PF. A cellular mechanism for targeting newly synthesized mRNAs to synaptic sites on dendrites. Proc Natl Acad Sci USA. 2001;98:7062–7068. doi: 10.1073/pnas.131146398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun Z-Y, Schacher S. Development of short-term heterosynaptic facilitation at Aplysia sensorimotor synapses is accompanied by changes in the functional expression of presynaptic serotonin receptors. J Neurophysiol. 1996;76:2250–2261. doi: 10.1152/jn.1996.76.4.2250. [DOI] [PubMed] [Google Scholar]
- 39.Sun Z-Y, Wu F, Schacher S. Rapid bidirectional modulation of mRNA expression and export accompany long-term facilitation and depression of Aplysia synapses. J Neurobiol. 2001;46:41–47. doi: 10.1002/1097-4695(200101)46:1<41::aid-neu4>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 40.Tiedge H, Bloom FE, Richter D. RNA, whither goest thou. Science. 1999;283:186–188. doi: 10.1126/science.283.5399.186. [DOI] [PubMed] [Google Scholar]
- 41.van Minnen J. Axonal localization of neuropeptide-encoding mRNA in identified neurons of the snail Lymnaea stagnalis. Cell Tissue Res. 1994;276:155–161. doi: 10.1007/BF00354795. [DOI] [PubMed] [Google Scholar]
- 42.van Minnen J, Bergman JJ, van Kesteren ER, Smit AB, Geraerts WPM, Lukowiak K, Hasan SU, Syed NI. De novo protein synthesis in isolated axons of identified neurons. Neuroscience. 1997;80:1–7. doi: 10.1016/s0306-4522(97)00137-1. [DOI] [PubMed] [Google Scholar]
- 43.Winberg ML, Mitchell KJ, Goodman CS. Genetic analysis of the mechanisms controlling target selection: complementary and combinatorial functions of netrins, semaphorins and IgCams. Cell. 1998;93:581–592. doi: 10.1016/s0092-8674(00)81187-3. [DOI] [PubMed] [Google Scholar]
- 44.Zhang HL, Eom T, Oleynikov Y, Shenoy SM, Liebelt DA, Dictenberg JB, Singer RH, Bassell GJ. Neurotrophin-induced transport of a beta-actin mRNP complex increases beta-actin levels and stimulates growth cone motility. Neuron. 2001;31:261–275. doi: 10.1016/s0896-6273(01)00357-9. [DOI] [PubMed] [Google Scholar]
- 45.Zhu H, Wu F, Schacher S. Aplysia cell adhesion molecules and serotonin regulate sensory cell-motor cell interactions during early stages of synapse formation in vitro. J Neurosci. 1994;14:6886–6900. doi: 10.1523/JNEUROSCI.14-11-06886.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu H, Wu F, Schacher S. Changes in expression and distribution of Aplysia cell adhesion molecules can influence synapse formation and elimination in vitro. J Neurosci. 1995;15:4173–4183. doi: 10.1523/JNEUROSCI.15-06-04173.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]