Abstract
Risk for onset of alcoholism is related to genetic differences in acute alcohol withdrawal liability. We previously mapped a locus responsible for 26% of the genetic variance in acute alcohol withdrawal convulsion liability to a >35 centimorgan (cM) interval of murine chromosome 4. Here, we narrow the position of this locus to a <1 cM interval (∼1.8 megabase, containing 15 genes and/or predicted genes) using a combination of novel, interval-specific congenic strains and recombinant progeny testing. We report the development of a small-donor-segment congenic strain, which confirms capture of a gene affecting alcohol withdrawal within the <1 cM interval. We also confirm a pentobarbital withdrawal locus within this interval, suggesting that the same gene may influence predisposition to physiological dependence on alcohol and a barbiturate. This congenic strain will be invaluable for determining whether this interval also harbors a gene(s) underlying other quantitative trait loci mapped to chromosome 4, including loci affecting voluntary alcohol consumption, alcohol-induced ataxia, physical dependence after chronic alcohol exposure, and seizure response to pentylenetetrazol or an audiogenic stimulus. To date, Mpdz, which encodes the multiple PSD95/DLG/ZO-1 (PDZ) domain protein (MPDZ), is the only gene within the interval shown to have allelic variants that differ in coding sequence and/or expression. Sequence analysis of 15 standard inbred mouse strains identifies six Mpdz haplotypes that predict three MPDZ protein variants. These analyses, and evidence using interval-specific congenic lines, show that alcohol withdrawal severity is genetically correlated with MPDZ status, indicating that MPDZ variants may influence alcohol withdrawal liability.
Keywords: quantitative trait locus, recombinant progeny testing, interval-specific congenic strain, PDZ domain, ethanol, barbiturate, physiological dependence, convulsion, seizure, C57BL/6J, DBA/2J
Alcoholism and alcohol abuse are common disorders with lifetime prevalences of 10–17% (Regier et al., 1990). A genetic contribution to alcoholism is supported by adoption studies that demonstrate an increased risk for severe alcohol-related problems in children of alcoholics who were adopted out, even if they had been raised without knowledge of their biological parents' problems (Schuckit et al., 1972; Cadoret et al., 1980). Four large twin studies published in the 1990s substantiate the conclusion that alcoholism is >50% heritable (Goldman, 1993; Reich et al., 1999). Previous studies show that risk for onset of alcoholism is associated with genetic differences in acute alcohol withdrawal liability. Family history-positive men report greater withdrawal effects, measured 3–8 hr after administration of 1 gm/kg ethanol, than family history-negative men (McCaul et al., 1991). Sons of alcoholics also report greater hangover symptoms, which are thought to represent an acute withdrawal syndrome, than sons of nonalcoholics (Newlin and Pretorius, 1990).
Although no animal model exactly duplicates clinically defined alcoholism, models for specific factors, such as the withdrawal syndrome, are useful to identify potential genetic determinants of liability in humans. The DBA/2J (D2) and C57BL/6J (B6) mouse strains are the most widely studied genetic models of severe and mild acute alcohol withdrawal, respectively. We previously used populations derived from these progenitor strains to identify markers for genetic variation in degree of physiological dependence on alcohol. Among the many signs of physiological dependence, withdrawal convulsions are a particularly useful index, because they are displayed in all species tested, including humans (Friedman, 1980). Quantitative trait locus (QTL) analyses identified three genomic regions on mouse chromosomes 1, 4, and 11, each of which contains a gene(s) that influences acute alcohol withdrawal liability (Buck et al., 1997). We subsequently mapped a QTL affecting pentobarbital (PB) withdrawal liability to mouse chromosome 1 and two suggestive QTLs on chromosomes 4 and 11 (Buck et al., 1999). This convergence of QTLs makes it tempting to speculate that these genomic regions contain one or more “withdrawal genes” and shows the cumulative power of QTL mapping to detect multiple effects of the same gene, a condition (if true) called pleiotropism.
A major challenge encountered in subsequent work (e.g., to investigate potential pleiotropic effects of QTLs or to identify the specific genes underlying QTLs) is to attain higher-resolution mapping of QTLs, which are initially mapped to large [10–35 centimorgans (cM)] confidence intervals containing hundreds of genes, any one (or more) of which could be responsible for the QTL association. This previously daunting task has been made feasible through the implementation of strategies to narrow QTL regions to ≤1 cM. One of the most powerful strategies for fine mapping uses interval-specific congenic strains (Darvasi, 1997,1998; Lyons et al., 2000; Cicila et al., 2001). Here, we used a variant of this strategy, sometimes called recombinant progeny testing, in combination with classic congenic analyses to map QTLs affecting physiological dependence on alcohol and PB to the same <1 cM interval of murine chromosome 4.
MATERIALS AND METHODS
Animals. The D2.B6-D4Mit142 conventional congenic strain was developed and bred at the Veterinary Medical Unit at the Portland Veterans Affairs Medical Center. The series of interval-specific congenic lines (ISCL1–ISCL5) and the small-donor-segment (SDS) congenic strain (D2.B6-Mpdz) were developed and bred in our colony at the Department of Comparative Medicine at Oregon Health and Science University. The 129/J, A/HeJ, AKR/J, BALB/cJ, C3H/HeJ, C57BL/6J, C57BR/cdJ, C57L/J, CBA/J, CE/J, DBA/1J, DBA/2J, PL/J, SJL/J, and SWR/J inbred strains were bred in our colony from mice originally purchased from The Jackson Laboratory (Bar Harbor, ME). All procedures were approved by the Veterans Affairs and/or Oregon Health and Science University Institutional Animal Care and Use Committees in accordance with United States Department of Agriculture and United States Public Health Service guidelines.
D2.B6-D4Mit142 congenic strain development. The B6 strain was crossed with D2 mice to yield B6D2 F1 animals, which were backcrossed to D2 mice. Selection of breeders for this and subsequent backcrosses was based on genotyping at four markers within and flanking the chromosome 4 QTL (between D4Mit111 at 21.9 cM and D4Mit327 at 42.5 cM). In this manner, 10 generations of backcrossing (N1–N10) were performed. This was followed by an intercross between N10 animals to isolate the donor homozygotes, which constituted the finished D2.B6-D4Mit142 congenic strain. Genotyping was performed to identify the boundary regions of the introgressed region from the donor (B6) strain, which comprised ∼2% of the genome.
Genotyping. DNA for genotyping mice for congenic development and higher-resolution QTL mapping was extracted from tail biopsy material using the Puregene DNA isolation kit (Gentra Biosystems, Minneapolis, MN) according to the manufacturer's instructions. PCR amplification and gel electrophoresis were performed as in previous work (Buck et al., 1999) using markers from the MIT microsatellite marker set first described by Dietrich et al. (1992)(www.informatics.jax.org/searches/marker_form.shtml).
Development of a series of interval-specific congenic lines for fine-mapping Alcw2. Conventional congenic mice were crossed to D2 mice to yield F1(D2.B6-D4Mit142× D2) animals, which were backcrossed to D2 mice. Individual progeny were genotyped using between 3 and 12 MIT and single nucleotide polymorphism (SNP) markers within or flanking the acute alcohol withdrawal QTL on mouse chromosome 4 (referred to as Alcw2) to identify recombinant mice and define the boundaries of the introgressed region. Individual recombinant mice were backcrossed to D2 mice, resulting in multiple offspring with the same recombination (replicated recombinant progeny, also referred to as an ISCL). At the same time that recombinations in the previous generation were being replicated, additional recombinants were sought in two subsequent backcross generations in an ever-narrowing QTL interval and replicated as needed. A total of five ISCLs were developed and replicated for phenotype analysis. The congenic line in which the large starting introgressed region is intact is referred to as ISCL1. Four additional interval-specific lines, in which one or two recombinations within the original introgressed interval are maintained, are referred to as ISCL2–ISCL5. Because our original F2 mapping study indicated dominance of the B6 allele (Buck et al., 1997), we tested for Alcw2 capture by phenotypic comparisons of mice from ISCL1–ISCL5 that were heterozygous (B6D2) within their introgressed regions with their D2D2 littermates (one-tailed unpairedt test). Identification of the ISCLs that showed the QTL effect on phenotype scores (i.e., less severe withdrawal in B6D2 congenic mice than in D2D2 littermates) and the ISCLs that did not defined the critical genomic interval required for the QTL effect. A total of 492 mice from ISCL1–ISCL5, representing 258 mice containing the donor B6 region and 234 D2D2 littermates, were phenotyped over a period of ∼15 months.
Development of the D2.B6-Mpdz congenic strain. A final intercross using ISCL5 animals was performed to isolate the donor homozygotes, which constituted the finished SDS congenic strain (D2.B6-Mpdz) with a <1 cM introgressed region spanningD4Mit80 and Mpdz.
Alcohol and PB withdrawal phenotyping. Genetic variation in withdrawal was examined by monitoring changes in handling-induced convulsion (HIC) scores, a sensitive index of alcohol and PB withdrawal severity (Goldstein and Pal, 1971; Crabbe et al., 1991). Adult mice were scored twice for baseline HICs immediately before administration of ethanol (4 gm/kg, i.p.), scored hourly between 2 and 12 hr after ethanol administration, or before administration of PB (60 mg/kg, i.p.), and scored hourly between 1 and 8 hr after PB injection. Details of the methods and scoring system have been published previously (Crabbe et al., 1991). Individual mice and different inbred strains can differ in baseline (predrug) HIC scores. Therefore, to assess alcohol withdrawal severity, scores were first computed as the area under the curve (AUC) between 4 and 12 hr after alcohol administration. Separate animals were used to assess PB withdrawal severity, and withdrawal scores were computed as the AUC between 2 and 8 hr after PB administration. The average baseline HIC score for each animal was then subtracted from its AUC score to calculate a corrected alcohol withdrawal score (or PB withdrawal score). These are the same variables as used in the initial QTL mapping studies (Buck et al., 1997, 1999).
Statistical analysis. All statistical calculations were performed using SYSTAT (SPSS, Chicago, IL). One-way ANOVA and Tukey's honestly significant difference post hoccomparisons were used to compare D2.B6-D4Mit142, D2, and B6 strains for acute alcohol and PB withdrawal severity. One-tailedt tests were used to compare ISCLs with their D2D2 littermates for alcohol withdrawal severity. One-tailed ttests were also used to compare the D2.B6-Mpdz strain with the D2 background strain for alcohol withdrawal and PB withdrawal severity.
Fisher's method of combining p values from independent experiments testing the same hypothesis (Sokal and Rohlf, 1981) was used to calculate combined p values for the alcohol and PB withdrawal QTLs detected in the present congenic studies; thep values were combined with those from previous work using BXD recombinant inbred strains, B6D2 F2mice, and selectively bred lines (Buck et al., 1997, 1999). This method is more conservative than adding logarithm of the odds (LOD) scores. From these combined p values, asymptotic LOD scores were estimated as LOD = −log10(p). Guidelines recommended by Lander and Kruglyak (1995) were used for interpreting and reporting confirmation of the alcohol and PB withdrawal QTLs. The relationship between withdrawal severity and three multiple PSD95/DLG/ZO-1 (MPDZ) variants in 15 standard inbred strains was assessed by one-way ANOVA and Tukey's honestly significant differencepost hoc comparisons.
Candidate gene sequencing.Poly(A)+ RNA was isolated from mouse brain with MicroFast-Track (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. cDNA was reverse-transcribed using oligo(dT) primer and SuperScript II RNaseH reverse transcriptase (Invitrogen). The protein coding regions of 12 candidate genes that map to the midregion of mouse chromosome 4 (www.informatics.jax.org) were sequenced using the D2 and B6 progenitor strains. The coding region of Mpdz was also sequenced using 13 additional standard inbred mouse strains for haplotype and protein variant analyses. Two independent PCR products from each strain were gel-purified and sequenced on an ABI 377 automated system (Applied Biosystems, Foster City, CA) using gene-specific primers. For each gene, overlapping fragments were aligned using MacVector 6.5.3 and AssemblyLIGN 1.0.9c (Oxford Molecular/GCG Inc., Madison, WI). Open reading frames and the predicted protein sequences were compared using BLASTP, a search program designed to compare protein sequences against the National Center for Biotechnology Information protein databases (www.ncbi.nlm.nih.gov/blast/). Additional analysis focused on three MPDZ protein variants. Secondary structure was predicted with PeptideStructure (Seq-Web version 1.2, WI Package 10.1; GCG Inc.) using published algorithms (Chou and Fasman, 1978; Garnier et al., 1978). The positions of 13 PDZ domains were predicted with the Pfam protein domain finding software (http://www.sanger.ac.uk./Software/Pfam/search.shtml).
Development of Mpdz and Ptprd SNP markers. Sequence analysis of Mpdz identified a polymorphism at position 5299, which is located within an NlaIII site (CATG) in the B6 allele. This restriction site is absent in the D2 allele. A primer set (5′-GGAGACTGATGCAAGGGGAC and 5′-ACTTTAGCAGGGCAGCGAC) was designed to amplify a 90 bp product, spanning this polymorphism, from genomic DNA.NlaIII cuts the B6 product into 57 and 33 bp fragments, but the D2 product remains intact.
Sequence analysis of Ptprd identified a polymorphism at position 2100. In the B6 allele, this position is located within aBsrI restriction site (CCCAGT). In the D2 allele, the polymorphism is located within an NciI restriction site (CC C/G GG). A primer set (5′-GGACTCCAGAAAGAGCAGC and 5′-GGTGTTTGAAAGTTAAGGCG) was used to amplify a 96 bp product, spanning this SNP, from genomic DNA. BsrI digests the B6 product into 35 and 61 bp fragments, but the D2 product remains intact. Conversely,NciI digests the D2 allele into 36 and 60 bp fragments, whereas the B6 product remains intact. The Mpdz andPtprd products were analyzed by agarose gel electrophoresis with ethidium bromide staining.
RESULTS
D2.B6-D4Mit142 congenic strain
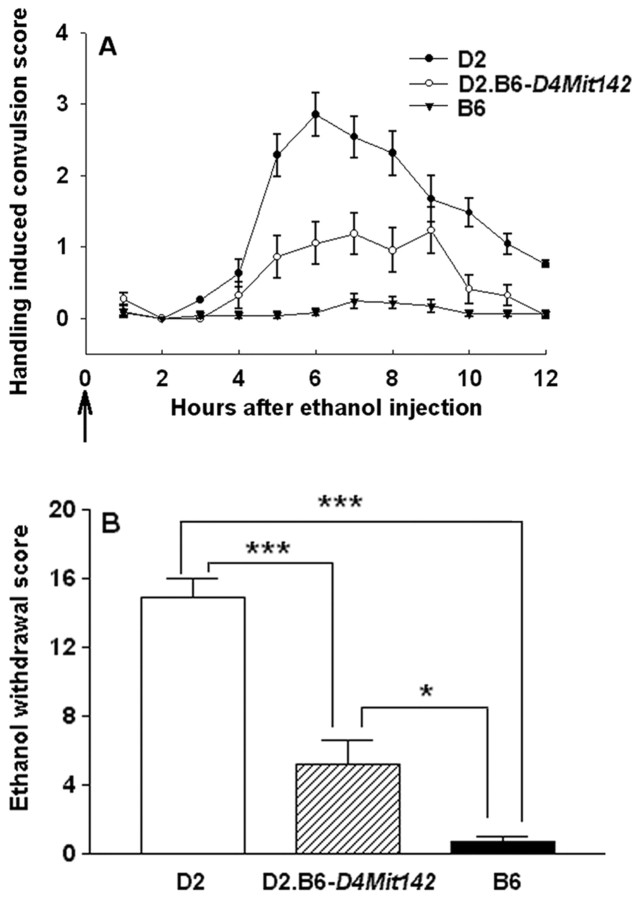
Genotypic analysis of the conventional congenic strain (B6.D2-D4Mit142, so referred to because the markerD4Mit142 at 37.5 cM was associated with the acute alcohol withdrawal QTL mapped to chromosome 4 in the initial mapping work) showed that the introgressed interval of chromosome 4 spannedD4Mit192 (6.3 cM) and D4Mit327 (42.5 cM), whereasD4Mit263 (3.2 cM) and D4Mit245 (42.5 cM) were excluded from the introgressed region. The finished D2.B6-D4Mit142 strain qualifies as an inbred strain (Bailey, 1981) and is ∼98% background (D2) strain genome, with the exception of the introgressed region from the donor (B6) strain. Assessment of the D2.B6-D4Mit142 congenic D2 and B6 strains demonstrated a significant effect of the introgressed genomic region on alcohol withdrawal severity (Fig. 1). The congenic strain showed less severe acute alcohol withdrawal than D2 mice (p < 0.001), confirming that a locus (gene) influencing physiological dependence on alcohol is contained within the introgressed donor interval. Gender-specific comparisons confirmed the strain differences in alcohol withdrawal severity (males:F(2,65) = 60.5, p = 7.1 × 10−6; females:F(2,51) = 25.1, p = 6.9 × 10−5). Alcohol withdrawal scores were significantly lower in D2.B6-D4Mit142 congenic mice than in same-sex D2 mice, emphasizing that the QTL was evident in both genders (males: p < 0.001; females:p = 0.003).
Fig. 1.
The D2.B6-D4Mit142 congenic strain captures a gene influencing acute alcohol withdrawal severity in the introgressed donor (B6) region. A, Alcohol withdrawal was indexed using the HIC in the D2 background strain (closed circles), D2.B6-D4Mit142 congenic strain (open circles), and B6 donor strain (triangles). The mice were scored twice for baseline HICs immediately before administration of 4 gm/kg ethanol (arrow marks ethanol injection at time 0) and hourly between 2 and 12 hr after alcohol administration. After 3–4 hr, convulsion scores increase above baseline, indicating a state of withdrawal hyperexcitability, which peaks ∼6–8 hr after alcohol administration. Data represent the strain mean ± SEM (n = 22–40 mice per strain). B, The mean ± SEM for alcohol withdrawal severity (corrected AUC) for the D2.B6-D4Mit142, D2, and B6 strains. A significant effect of strain on alcohol withdrawal severity was identified (F(2,119) = 79.1; p= 2.4 × 10−9). Tukey's post hoc strain comparisons: ***p < 0.001; *p < 0.05.
Higher-resolution mapping of Alcw2
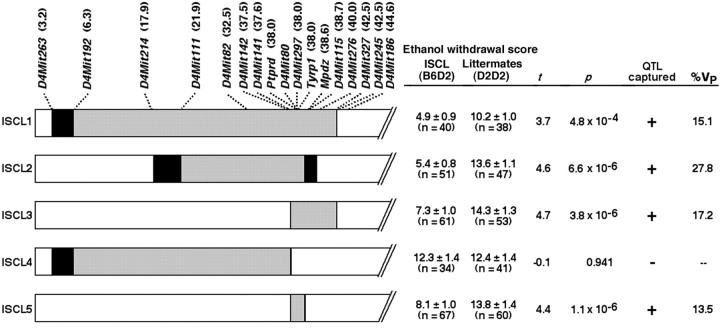
B6D2 F2 analysis previously indicated dominance of B6 alleles for the chromosome 4 locus (Buck et al., 1997), suggesting that direct comparison of backcross progeny that were heterozygous (B6D2) within the introgressed region and their D2D2 littermates could efficiently detect “capture” of the gene(s) underlying the chromosome 4 locus (referred to as Alcw2). This was confirmed by showing that backcross progeny that were heterozygous for the starting introgressed interval (ISCL1) showed less severe acute alcohol withdrawal than their D2D2 littermates (p = 4.8 × 10−4) (Fig.2). The ISCL1 data indicated that a QTL responsible for ∼15.1% of the phenotypic variance in alcohol withdrawal severity is contained within the introgressed interval spanning 6.3–42.5 cM. Genotyping progeny from a D2.B6-D4Mit142 × D2 backcross and two subsequent backcrosses also identified individual recombinant mice that were used to derive four additional interval-specific congenic lines, ISCL2–ISCL5 (Fig. 2). ISCL2 mice showed a strong protective QTL effect for alcohol withdrawal compared with their D2D2 littermates, indicating that a B6 protective allele (gene) was contained in the 16.7 cM introgressed region spanning D4Mit111 and Mpdz. The QTL effect was also evident in ISCL3, with a 4.8 cM introgressed region spanning D4Mit80 and D4Mit327. In contrast, ISCL4 mice, with an introgressed region spanningD4Mit192 and D4Mit141, did not differ from their D2D2 littermates in alcohol withdrawal liability. We therefore concluded that the critical interval spans D4Mit80 (the position of this marker is given as 37.7 cM, but its actual position is distal to Ptprd at 38.0 cM) and Mpdz at 38.6 cM. The critical region is a 1.78 megabase (Mb) interval that contains 15 genes and/or predicted genes; these data were generated through use of the Celera Discovery System and Celera Genomics' associated databases (Celera, Rockville, MD). Moreover, the proximal boundary lies within the interval between D4Mit80 and Ptprd(∼37.7–38.0 cM, see above), and the distal boundary lies within the 0.1 cM interval between Mpdz and D4Mit115(38.6–38.7 cM). Development and testing of ISCL5, with an introgressed interval corresponding to the critical <1 cM interval, confirmed that this interval shows a strong protective QTL effect for alcohol withdrawal. Moreover, the percentage of phenotypic variance accounted for by the introgressed interval in ISCL5 (13.5%) is similar to that accounted for by ISCL1 (15.1%). However, ISCL2 potentially accounted for a greater percentage of the phenotypic variance (27.8%). To test whether ISCL2 actually accounted for a greater percentage of the phenotypic variations than other ISCLs that also showed the protective effect or whether this apparent difference was caused by chance, we compared the alcohol withdrawal scores for ISCL2 with those of ISCL1, ISCL3, and ISCL5. To maximize the power of our analysis, the alcohol withdrawal scores of the B6D2 heterozygote animals of ISCL2 were compared with the B6D2 heterozygote mice of ISCL1, ISCL3, and ISCL5 combined using an unpaired t test (two-tailed). Alcohol withdrawal scores were not significantly different between the two groups (t = −1.6; p = 0.11), suggesting that the full Alcw2 effect may be captured by ISCL1, ISCL2, ISCL3, and ISCL5.
Fig. 2.
Left, Five interval-specific congenic lines (ISCL1–ISCL5) were developed and tested to attain higher-resolution mapping of Alcw2. The MIT and gene markers examined and their positions (in centimorgans, shown inparentheses next to each marker) are indicated attop. For each ISCL, the interval-specific donor region is shown in gray and is heterozygous B6D2. Chromosomal regions homozygous for the background (D2) allele are shown inwhite, and the boundaries between the B6 and D2 regions are shown in black. In some cases (e.g., for the proximal and the distal boundaries of ISCL5), the boundary region is so small (<0.1 cM) that it appears as a black line.Right, Acute alcohol withdrawal scores (mean ± SEM, corrected AUC) are given for the ISCL1–ISCL5 progeny (B6D2) and their D2D2 littermates. Alcohol withdrawal severity was significantly less severe in ISCL (B6D2) mice than their D2D2 littermates for ISCL1, ISCL2, ISCL3, and ISCL5. The percentage of phenotypic variance (%Vp) accounted for in each ISCL is also given.
SDS congenic testing confirms a PB withdrawal QTL and maps it to the same interval as Alcw2
A final intercross using ISCL5 mice was performed to generate donor strain homozygotes, resulting in a finished SDS congenic strain with an introgressed region limited to the <1 cM QTL interval. Demonstration that the SDS congenic strain (D2.B6-Mpdz) had less severe alcohol withdrawal than D2 mice (p = 5 × 10−5) confirmed that a gene(s) underlying Alcw2 was captured in the introgressed interval (Fig. 3A). This congenic study, together with previous work (Buck et al., 1997), provides still more evidence for Alcw2 (combined p = 2.2 × 10−10; LOD = 8.8).
Fig. 3.
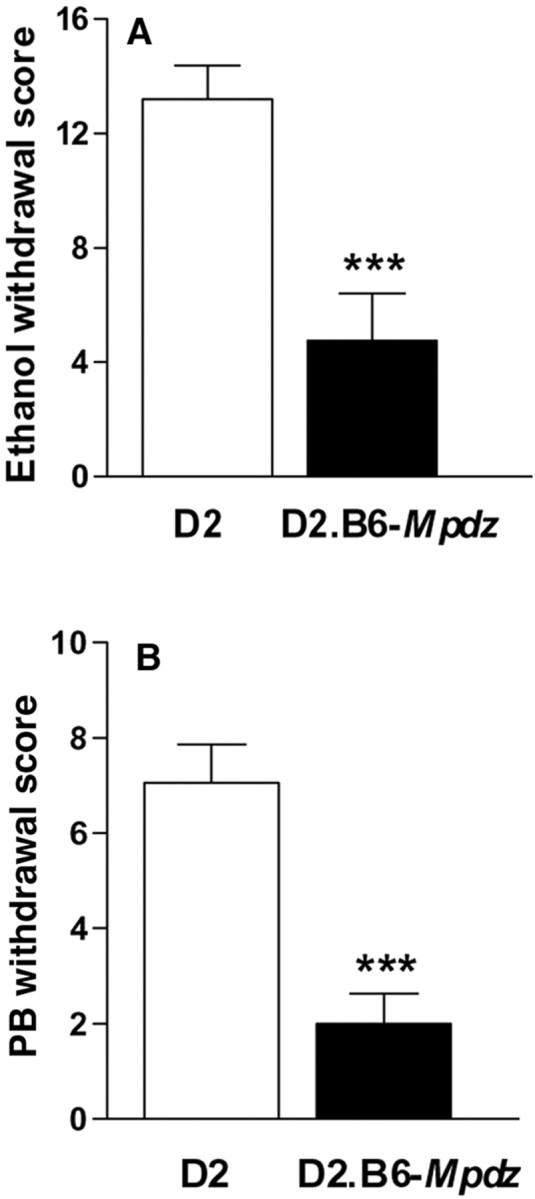
Confirmation that the D2.B6-Mpdz SDS congenic strain captures a gene(s) that influences acute alcohol withdrawal and acute PB withdrawal severity. A, Data represent the alcohol withdrawal scores (corrected AUC, mean ± SEM) for D2.B6-Mpdz congenic and D2 strains (***p = 5 × 10−5;t = 4.3; df = 20). B, PB withdrawal scores (corrected AUC, mean ± SEM) for the D2.B6-Mpdz congenic and D2 strains (***p = 2 × 10−4;t = 3.8; df = 45).
Acute PB withdrawal was also less severe in D2B6-Mpdz mice than in D2 mice (p = 2 × 10−4) (Fig. 3B). The present congenic studies, together with previous work reporting suggestive linkage (Buck et al., 1999), confirm a QTL for acute PB withdrawal liability on chromosome 4 (combined p = 4.1 × 10−7; LOD = 5.6). This QTL,Pbw2, now exceeds the guidelines recommended by Lander and Kruglyak (1995) for significant linkage.
Candidate genes
The critical DNA sequence change(s) underlying Alcw2and Pbw2 could be a result of changes in the protein-coding region that change the encoded protein or of changes in the regulatory region of the gene that influence expression of the gene. Here, we sequenced the protein-coding regions of 12 candidate genes using the D2 and B6 progenitor strains. These genes were chosen for our initial analysis for four reasons: (1) Based on public and private (Celera) mouse databases, these genes map in or near the Alcw2/Pbw2interval on mouse chromosome 4; (2) based on public and private (Celera) human databases, these genes map in or near the syntenic regions of human chromosomes 9p24-p22 and 1p32-p31; (3) these genes are expressed in brain regions implicated in ethanol withdrawal and convulsions; and (4) they have potential known biological relevance to alcohol action. A summary of our sequencing results to date is given in Table 1, and the sequences have been deposited in GenBank. Eight of the genes examined were not polymorphic, two had single polymorphisms, and two had two or more polymorphisms in their protein-coding regions. As part of our fine-mapping effort, we have eliminated three genes that contain coding-region sequence differences between the D2 and B6 progenitor strains (i.e.,Ptprd, Sh3d2a, and Ppt) from the QTL interval. Thus, our sequence comparisons have (1) facilitated our fine-mapping efforts, (2) eliminated three genes that contain coding-region sequence differences between the D2 and B6 alleles from the QTL interval, and (3) identified Mpdz as a viable candidate gene for this QTL. In gene expression profiling analyses, the genes known to map within the Alcw2/Pbw2 interval were expressed at approximately background levels (e.g., Mpdz) or were not differentially expressed between D2 versus B6 mice during alcohol withdrawal (Daniels and Buck, 2002) (G. M. Daniels and K. J. Buck, unpublished data) or during acute PB withdrawal or in naive mice (Daniels and Buck, unpublished data).
Table 1.
Sequence comparisons between B6 and D2 alleles for 12 genes in or near the QTL interval
| Chromosome: centimorgan position | Gene | Description | Proposed function | Coding length (bp) | Δ DNA sequence | Δ Protein sequence |
|---|---|---|---|---|---|---|
| 4:38.0 | Ptprd | Protein tyrosine receptor phosphatase | Brain-specific receptor phosphatase | 3762 | 7 | 0 |
| 4:syn | Rpll8A-rs | Ribosomal protein L18a-related sequence | Protein translation | 567 | 0 | 0 |
| 4:syn | Rps26-rs | Ribosomal protein S26- related sequence | Protein translation | 348 | 0 | 0 |
| 4:38.6 | Mpdz | Multiple PDZ protein | Synaptic clustering, signal transduction efficiency | 6168 | 18 | 10 |
| 4:38.6 | Nfib | Nuclear factor I/B | Brain-selective transcription factor | 1410 | 0 | 0 |
| 4:38.9 | Adfp | Adipose differentiation-related protein | Potential constituent of lipid vesicle membrane | 1381 | 0 | 0 |
| 4:38-41.6 | Sh3d2a | SH3 domain protein 2A | Tyrosine kinase receptor-mediated signal transduction | 1054 | 1 | 0 |
| 4:syn | Ppt | Palmitoyl-protein thioesterase | Thioester cleavage, implicated in seizures | 924 | 1 | 0 |
| 4:syn | Hpca | Hippocalcin | Hippocampal Ca2+ binding protein | 579 | 0 | 0 |
| 4:syn | Gabrd | GABAA receptor, δ subunit | Benzodiazepine insensitive GABAAreceptors | 1350 | 0 | 0 |
| 4:45.8 | Nfia | Nuclear transcription factor I/A | Brain-selective transcription factor | 1530 | 0 | 0 |
| 4:46.8 | Pde4b | Phosphodiesterase 4B | cAMP cleavage | 2166 | 0 | 0 |
It should be pointed out that even higher-resolution mapping of this QTL is currently being pursued, along with continued testing of viable candidate genes that remain within the ever-narrowing QTL interval. However, to date, Mpdz is the only viable candidate gene within the QTL interval shown to differ in gene expression and/or protein-coding sequence between the progenitor D2 and B6 alleles. Sequence analysis identified 18 polymorphisms between the D2 and B6Mpdz alleles, 10 of which are predicted to change amino acid residues in the MPDZ protein (Fig. 4). These include nonsynonymous substitutions in two PDZ domains (V1338M in PDZ8; H1767N in PDZ11).
Fig. 4.
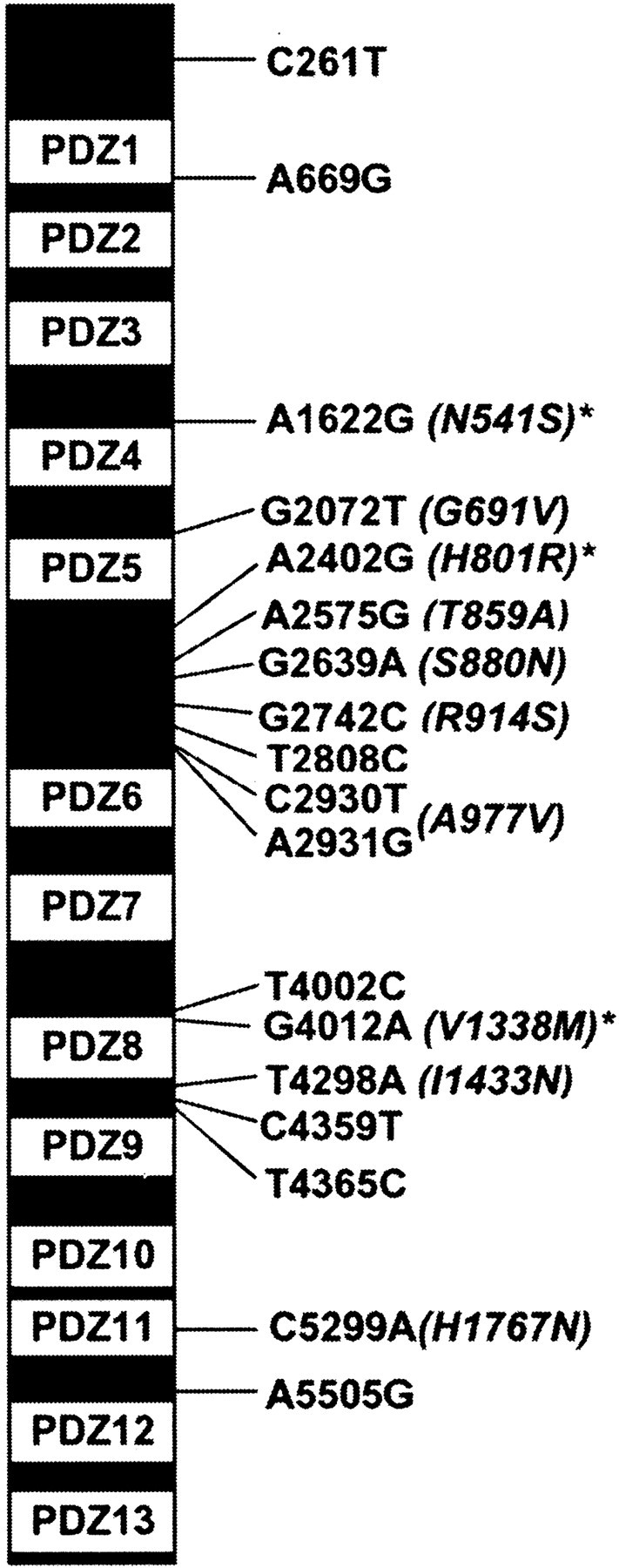
The positions of 18 nucleotide residues within the 6154 bp mouse Mpdz coding sequence that are polymorphic between the B6 and the D2 strains are indicated. The B6 nucleic acid residue is given first, then its position, followed by the D2 residue (e.g., A1622G). Strain differences in the predicted amino acid sequence are given in parentheses, with the B6 amino acid residue given first, followed by its position and the D2 residue (e.g., N541S). The locations of the 13 PDZ domains are indicated. *Difference in predicted secondary structure.
Mpdz encodes an MPDZ protein that contains 13 PDZ domains, named for three prototypical proteins: the mammalian postsynaptic density protein (PSD-95), the Drosophiladisks–large septate junction protein (DLG), and the mammalian epithelial tight-junction protein zona occludens (ZO-1). Functionally, PDZ domain proteins act as scaffolding proteins that assemble supramolecular complexes within the cell and in this way aid in localizing and coupling components of specific signal transduction pathways at specific subcellular locations. This results in an enhanced rate and fidelity of signal transduction in specific pathways (Sheng and Sala, 2001). MPDZ interacts with 5-HT2 receptors (Ullmer et al., 1998; Becamel et al., 2001), suggesting a mechanism by which MPDZ variants may contribute to genetic differences in alcohol withdrawal liability (see Discussion).
Haplotype analysis
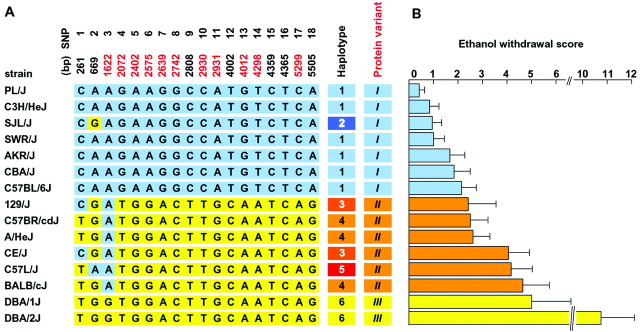
Analysis of 15 inbred mouse strains identified six Mpdzhaplotypes that predict three distinct protein variants (MPDZ1–MPDZ3) (Fig. 5A). The B6 strain has haplotype 1 and, along with other strains with haplotypes 1 and 2, encodes MPDZ1. The D2 and D1 strains have haplotype 6 and encode MPDZ3. The strains with haplotypes 3, 4, or 5 encode MPDZ2, which is nearly identical to MPDZ3 except at amino acid residue 541 between PDZ3 and PDZ4. To maximize the power of our analysis to assess the potential relationship between MPDZ and alcohol withdrawal severity, the 15 inbred strains were divided into three groups based on MPDZ protein status (Fig. 5B). For each group, we calculated a mean ± SEM alcohol withdrawal score based on published strain means (Metten and Crabbe, 1994) and found that alcohol withdrawal severity was significantly associated with MPDZ protein status (F(2,12) = 17.6; p = 0.0003). Because alcohol withdrawal severity is a quantitative trait influenced by at least three loci (genes) (Buck et al., 1997), it is not surprising that the phenotypic withdrawal data do not fall into three clear groups when only one of these loci is taken into account. Nevertheless, inbred strains encoding MPDZ1 protein (Fig. 5, blue bars) showed significantly less severe ethanol withdrawal [ethanol withdrawal score = 1.3 ± 0.2 (mean ± SEM)] than strains with MPDZ2 (Fig. 5, orange bars; 3.4 ± 0.4; p = 0.04) or MPDZ3 (Fig. 5, yellow bars; 7.9 ± 2.9; p = 0.0002). Alcohol withdrawal severity was also different among strains with MPDZ2 versus MPDZ3 (p = 0.006).
Fig. 5.
A, Mpdz haplotypes and protein variants in 15 standard inbred mouse strains. The SNPs shared with the B6 strain are shown in blue, whereas those shared with the D2 strain are shown in yellow. The SNP positions are shown at top, and those that predict variation in the protein sequence are shown in red. Themiddle columns designate the six Mpdzhaplotypes (1–6) and the three MPDZ protein variants (roman characters). B, Mean ± SEM acute ethanol withdrawal scores for 15 standard inbred mouse strains (phenotypic data are from Metten and Crabbe, 1994). The horizontal barsshown in blue are for the strains encoding MPDZ1,orange for strains encoding MPDZ2, andyellow for the strains encoding MPDZ3.
Using the same standard inbred strains, we also assessed the potential relationship between PB withdrawal severity and MPDZ status. To maximize the power of our analysis to assess the potential relationship between MPDZ and PB withdrawal severity, the 15 inbred strains were divided into three groups based on MPDZ protein status. For each group, we calculated a mean ± SEM PB withdrawal score based on published strain means (Metten and Crabbe, 1994). The AKR/J strain, which encodes MPDZ1, was an outlier among the seven inbred strains in this group (Q test; Dean and Dixon, 1951). AKR was considered an outlier, and its mean value was excluded from the group becauseQexp exceeded theQcrit value for the 90% confidence level, where Qexp = [(variance excluding outlier)/(variance including outlier)] = 0.69, andQcrit, the critical value for the rejection quotient Q, was 0.51 for the group of seven strain means. Because PB withdrawal severity is a quantitative trait influenced by more than one locus (gene) (Buck et al., 1999), neither the identification of an outlier strain nor the finding that the phenotypic data do not fall perfectly into three clear groups is surprising, when only one of these loci is taken into account. Our results indicate that PB withdrawal severity is significantly associated with MPDZ protein status (F(2,11) = 10.26; p = 0.003). Inbred strains encoding the MPDZ1 protein showed significantly less severe PB withdrawal [PB withdrawal score = 1.1 ± 0.2 (mean ± SEM)] than strains with the MPDZ2 (2.4 ± 0.4;p = 0.04) or MPDZ3 (4.0 ± 0.9; p= 0.003) variants. A trend for differential PB withdrawal severity was detected among strains with MPDZ2 versus MPDZ3 (p = 0.09; two-tailed).
These data are consistent with Mpdz being the gene that underlies Alcw2 and/or Pbw2 and identifyMpdz as a promising candidate gene. However, it is also possible that the polymorphisms identified in Mpdz are merely in linkage disequilibrium with the polymorphism(s) that actually underlies Alcw2 and/or Pbw2. More definitive confirmation that Mpdz (and other promising candidate genes if they arise) is involved in withdrawal from alcohol and/or PB will most likely require verification using D2/B6 bacterial artificial chromosome (BAC) transgenics for Mpdz (see Discussion).
DISCUSSION
Identification of influential genes in humans is problematic for complex conditions with polygenic or heterogeneous inheritance (e.g., substance abuse) (Lander and Botstein, 1989). Diagnostic criteria for physiological dependence on sedative–hypnotic substances, such as barbiturates and ethanol, include two or more of the following withdrawal signs, developing within several hours to a few days after cessation of drug use: autonomic hyperactivity, tremor, insomnia, nausea, hallucinations, psychomotor agitation, anxiety, and seizures (DSM-IV, 1994). Recent efforts have therefore focused on the genetic dissection of complex traits using animal models as a way to identify important genes for study in humans. The present studies were performed to fine-map QTLs for genetic variation in liability for alcohol and PB withdrawal and identify promising candidate genes.
Genomic scans have proved extremely useful for determining the rough chromosomal location of QTLs. However, the imprecision with which genome screens localize QTLs presents difficulties in further studying the gene(s) responsible for the phenotypic effects and for assessing potential pleiotropism. Theoretical and practical analyses suggest that QTLs can be mapped to confidence intervals of ∼10–35 cM using standard genetic crosses. Congenic strains offer a much more precise QTL localization, establishing definitive limits for a QTL-containing region. In the present studies, we used novel, interval-specific congenic strains and recombinant progeny testing to reduce the interval of alcohol and PB withdrawal QTLs to a <1 cM region of murine chromosome 4. Our results emphasize the possibility of direct comparison between recombinant progeny, particularly in cases in which a QTL shows dominance for the donor allele. This variation of the congenic mapping strategy proposed by Darvasi (1997, 1998) provides an efficient, powerful alternative for fine-mapping of valuable complex phenotypes, including complex behavioral traits.
To date, we have reduced the size of the QTL interval to <1 cM (∼1.8 Mb) and reduced the number of potential candidate genes from several hundred to ∼15. This is a major step toward identification of the actual gene or genes underlying Alcw2 and Pbw2, as well as other QTLs that may be within this interval. Additional QTLs are detected in the midregion of chromosome 4 for initial sensitivity and rapid tolerance to ethanol-induced ataxia (Gallaher et al., 1996), ethanol-induced hypothermia (Crabbe et al., 1996), predisposition to physical dependence after chronic ethanol exposure (Buck et al., 2002), seizure response to pentylenetetrazol (Ferraro et al., 1999), and audiogenic seizure severity (Neumann and Collins, 1991). This convergence of QTLs makes it tempting to speculate that this chromosomal region contains a gene or genes with pleiotropic effects on multiple behaviors of interest. However, many genes reside in each QTL interval, and additional work is required to prove that a single gene is responsible. If most or all of these QTLs actually reflect the influence of a common gene, the nature of these phenotypes suggests that the locus may be involved in controlling alcohol and PB withdrawal severity, as well as mediating withdrawal convulsions per se and additional behavioral responses to ethanol.
Among the candidate genes examined to date for differential expression (Daniels and Buck, 2002) (Daniels and Buck, unpublished data) and protein-coding sequence, Mpdz is a promising candidate gene for Alcw2 and Pbw2. The mouse Mpdzgene and its human (MPDZ, previously referred to asMUPP1) and rat homologs encode an MPDZ. MPDZ is the numerical leader among PDZ domain proteins, containing 13 of these interaction domains. Known interaction partners for MPDZ include 5-HT2C, 5-HT2A, and 5-HT2B receptors; cKIT (a membrane tyrosine kinase receptor); and p75 (a neurotrophin receptor that modulates the binding of nerve growth factor to the functional high-affinity receptor tyrosine kinase, Trk) (Ullmer et al., 1998; Mancini et al., 2000;Becamel et al., 2001). Interaction between MPDZ and 5-HT2 receptors could potentially contribute to genetic differences in alcohol and PB withdrawal liability. Serotonin-mediated neurotransmission has long been implicated in alcoholism (LeMarquand et al., 1994a) and the acquisition of tolerance to PB and ethanol and cross-tolerance between these two drugs (Frankel et al., 1975; Khanna et al., 1980). 5-HT2receptor subtypes have been implicated in convulsions and ethanol withdrawal-induced anxiety in both human and nonhuman animal studies (LeMarquand et al., 1994a,b), and 5-HT2agonists and antagonists affect convulsion severity and ethanol withdrawal-induced anxiety (Prather et al., 1991; Semenova and Ticku, 1992; Lal et al., 1993; Gatch et al., 2000). 5-HT2 receptors also mediate the enhanced response to 5-HT in mice treated with PB (Banks et al., 1988) at approximately the same dose we used to induce acute PB withdrawal (65 vs 60 mg/kg, respectively). PB also augments 5-HT-mediated inhibition of Purkinje cells, which is thought to be mediated by 5-HT2 receptors (Strahlendorf et al., 1988). It is tempting to speculate that genetic variation in the severity of withdrawal from ethanol and PB between D2- and B6-derived mice may be related to genetic variation in MPDZ that affects 5-HT2 receptor expression and/or function. 5-HT2 receptors are increased in D2 mice compared with B6 mice in the cerebral cortex, and the increase in D2 mice is correlated with an age-dependent susceptibility to audiogenic convulsions displayed by the D2 strain (Jazrawi and Horton, 1989). Together, these studies suggest a plausible mechanism by which genetic variation in MPDZ might contribute to genetic differences in predisposition to alcohol and/or PB withdrawal convulsions.
In summary, our results fine-map QTLs on mouse chromosome 4 involved in acute alcohol withdrawal and PB withdrawal and assess polymorphisms in several genes in or near the Alcw2/Pbw2 interval. Our results identify Mpdz as a promising candidate gene, but additional work is necessary to establish with certainty thatMpdz is a withdrawal gene. Until then, it must be kept in mind that another gene within the QTL interval in linkage disequilibrium with Mpdz may ultimately be shown to contribute all or part of the QTL effect. We are therefore continuing our fine-mapping efforts to further reduce the QTL interval and, correspondingly, the number of genes in the interval. Candidate genes identified by QTL mapping or other methods must always be subjected to rigorous evaluation before they can be accepted. More definitive confirmation that Mpdz (and other promising candidate genes if they emerge) is involved in withdrawal from alcohol and/or PB will require direct testing. Nadeau and Frankel (2000) suggest that allele swap should be the formal proof of identity. Certainly, if the allele swap produced the expected phenotypic changes, this would be a powerful argument. However, in simpler systems such as bacteria, allele swapping has not always produced the expected phenotypic results because of genetic background (epistatic) effects (Malke et al., 2000). Thus, it would appear that we should also consider additional means of “proof” to test the involvement of Mpdz in alcohol and PB withdrawal. The approach we are currently pursuing will use BAC transgenics in which one or more B6 BACs are microinjected into separate D2 embryos. This approach is especially attractive for candidate genes for the chromosome 4 QTL, because the QTL (and therefore the underlying gene) shows dominance for the B6 allele. We may therefore be able to detect the QTL effect (i.e., a decrease in alcohol withdrawal severity vs the background strain) even in the presence of the background D2 allele. In the future, we will also take advantage of advances in transgenic technology on the horizon for allele swap, despite the limitations of this approach and, whenever appropriate, use additional means of proof discussed by Belknap et al. (2001). In the end, proof that a gene underlies a QTL will rely on the careful assembly of evidence that leads to only one reasonable conclusion.
Footnotes
This work was supported by United States Public Health Service Grants P50 AA10760, RO1 AA06243, RO1 DA05228, and RO1 AA11114; by the Department of Veterans Affairs; and by the Deutsche Forschungsgemeinschaft (Fe 524/1-1). We gratefully acknowledge Brooks Rademacher, Stephen Cross, Laurie O'Toole, and Melinda Helms for excellent technical assistance. We also thank Dr. Pamela Metten for providing the behavioral data for the standard inbred mouse strains.
Genbank accession numbers: Mpdz: AF326526–AF326544;Ptprd: AF326559, AF326560; Sh3d2a:AF326561, AF326562; Ppt: AF326557, AF326558;Hpca: AF326551, AF326552; Nfia1:AF326553, AF326554; Nfib: AY035852, AY035853;Gabrd: AF326546, AF326564; Pde4b:AF326555, AF326556; Adfp: AY035850, AY035851.
Correspondence should be addressed to Dr. Kari J. Buck, Veterans Affairs Medical Center, Research Service (mail code R&D40), 3710 Southwest US Veterans Hospital Road, Portland, OR 97201. E-mail:buckk@ohsu.edu.
C. Fehr's present address: Department of Psychiatry, University of Mainz, Mainz, Germany.
REFERENCES
- 1.Bailey DW. Recombinant inbred strains and bilineal congenic strains. In: Foster HL, Small JD, Fox JG, editors. The mouse in biomedical research. Academic; New York: 1981. pp. 223–239. [Google Scholar]
- 2.Banks WS, Kastin AJ, Trentman TL, Haynes HS, Johnson BG, Galina ZH. Mediation of serotonin-induced analgesia by the 5-HT2 receptor in the pentobarbital anesthetized mouse model. Brain Res Bull. 1988;21:887–891. doi: 10.1016/0361-9230(88)90022-6. [DOI] [PubMed] [Google Scholar]
- 3.Becamel C, Figge A, Poliak S, Dumuis A, Peles E, Bockaert J, Lubbert H, Ullmer C. Interaction of serotonin 5-hydroxytryptamine type 2C receptors with PDZ10 of the multi-PDZ domain protein MUPP1. J Biol Chem. 2001;276:12974–12982. doi: 10.1074/jbc.M008089200. [DOI] [PubMed] [Google Scholar]
- 4.Belknap JK, Hitzemann R, Crabbe JC, Phillips TJ, Buck KJ, Williams RW. QTL analysis and genome-wide mutagenesis in mice: complementary genetic approaches to the dissection of complex traits. Behav Genet. 2001;31:5–15. doi: 10.1023/a:1010249607128. [DOI] [PubMed] [Google Scholar]
- 5.Buck KJ, Metten P, Belknap JK, Crabbe JC. Quantitative trait loci involved in genetic predisposition to acute alcohol withdrawal in mice. J Neurosci. 1997;17:3946–3955. doi: 10.1523/JNEUROSCI.17-10-03946.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buck KJ, Metten P, Belknap JK, Crabbe JC. Quantitative trait loci affecting risk for pentobarbital withdrawal map near alcohol withdrawal loci on mouse chromosomes 1, 4, and 11. Mamm Genome. 1999;10:431–437. doi: 10.1007/s003359901018. [DOI] [PubMed] [Google Scholar]
- 7.Buck KJ, Rademacher BLS, Metten P, Crabbe JC. Mapping murine loci for physical dependence on ethanol. Psychopharmacology. 2002;160:398–407. doi: 10.1007/s00213-001-0988-8. [DOI] [PubMed] [Google Scholar]
- 8.Cadoret RJ, Cain CA, Grove WM. Development of alcoholism in adoptees raised apart from alcoholic biologic relatives. Arch Gen Psychiatry. 1980;37:561–563. doi: 10.1001/archpsyc.1980.01780180075008. [DOI] [PubMed] [Google Scholar]
- 9.Chou PY, Fasman GD. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- 10.Cicila GT, Garrett MR, Lee SJ, Liu J, Dene H, Rapp JP. High-resolution mapping of the blood pressure QTL on chromosome 7 using dahl rat congenic strains. Genomics. 2001;72:51–60. doi: 10.1006/geno.2000.6442. [DOI] [PubMed] [Google Scholar]
- 11.Crabbe JC, Merrill C, Belknap JK. Acute dependence on depressant drugs is determined by common genes in mice. J Pharmacol Exp Ther. 1991;257:663–667. [PubMed] [Google Scholar]
- 12.Crabbe JC, Phillips TJ, Gallaher EJ, Crawshaw LI, Mitchell SR. Common genetic determinants of the ataxic and hypothermic effects of ethanol in BXD/Ty recombinant inbred mice: genetic correlations and quantitative trait loci. J Pharmacol Exp Ther. 1996;277:624–632. [PubMed] [Google Scholar]
- 13.Daniels GM, Buck KJ. Expression profiling identifies strain-specific changes associated with ethanol withdrawal in mice. Genes Brain Behav. 2002;1:35–45. doi: 10.1046/j.1601-1848.2001.00008.x. [DOI] [PubMed] [Google Scholar]
- 14.Darvasi A. Interval-specific congenic strains (ISCS): an experimental design for mapping a QTL into a 1 centiMorgan interval. Mamm Genome. 1997;8:163–167. doi: 10.1007/s003359900382. [DOI] [PubMed] [Google Scholar]
- 15.Darvasi A. Experimental strategies for the genetic dissection of complex traits in animal models. Nat Genet. 1998;18:19–24. doi: 10.1038/ng0198-19. [DOI] [PubMed] [Google Scholar]
- 16.Dean RB, Dixon WJ. Simplified statistics for small numbers of observations. Anal Chem. 1951;23:636–638. [Google Scholar]
- 17.Dietrich W, Katz H, Lincoln SE, Shin H, Friedman J, Dracopoli N, Lander ES. A genetic map of the mouse suitable for typing intraspecific crosses. Genetics. 1992;131:423–447. doi: 10.1093/genetics/131.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DSM-IV. Diagnostic and statistical manual of mental disorders, Ed 4 (First MB, ed). American Psychiatric Association; Washington, DC: 1994. [Google Scholar]
- 19.Ferraro TN, Golden GT, Smith GG, St Jean P, Schork NJ, Mulholland N, Ballas C, Schill J, Buono RJ, Berrettini WH. Mapping loci for pentylenetetrazol-induced seizure susceptibility in mice. J Neurosci. 1999;19:6733–6739. doi: 10.1523/JNEUROSCI.19-16-06733.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frankel D, Khanna JM, LeBlanc AE, Kalant H. Effect of p-chlorophenylalanine on the acquisition of tolerance to ethanol and pentobarbital. Psychopharmacology. 1975;44:247–252. doi: 10.1007/BF00428901. [DOI] [PubMed] [Google Scholar]
- 21.Friedman HJ. Assessment of physical dependence on and withdrawal from ethanol in animals. In: Rigter H, Crabbe JC, editors. Alcohol tolerance and dependence. Elsevier; Amsterdam: 1980. pp. 93–121. [Google Scholar]
- 22.Gallaher EJ, Jones GE, Belknap JK, Crabbe JC. Identification of genetic markers for initial sensitivity and rapid tolerance to ethanol-induced ataxia using quantitative trait locus analysis in BXD recombinant inbred mice. J Pharmacol Exp Ther. 1996;277:604–612. [PubMed] [Google Scholar]
- 23.Garnier J, Osguthorpe DJ, Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978;120:97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- 24.Gatch MB, Wallis CJ, Lal H. Effects of ritanserin on ethanol withdrawal-induced anxiety in rats. Alcohol. 2000;21:11–17. doi: 10.1016/s0741-8329(99)00095-6. [DOI] [PubMed] [Google Scholar]
- 25.Goldman D. Recent developments in alcoholism: genetic transmission. Recent Dev Alcohol. 1993;11:231–248. [PubMed] [Google Scholar]
- 26.Goldstein DB, Pal N. Alcohol dependence produced in mice by inhalation of ethanol: grading the withdrawal system. Science. 1971;172:288–290. doi: 10.1126/science.172.3980.288. [DOI] [PubMed] [Google Scholar]
- 27.Jazrawi SP, Horton RW. 5-HT2 receptor binding and 5-HT uptake in mouse brain: developmental changes and the relationship to audiogenic seizure susceptibility in DBA/2J mice. Brain Res Dev Brain Res. 1989;45:257–263. doi: 10.1016/0165-3806(89)90043-6. [DOI] [PubMed] [Google Scholar]
- 28.Khanna JM, Le AD, Kalant H, LeBlanc AE. Role of serotonin (5-HT) in tolerance to ethanol and barbiturates. Adv Exp Med Biol. 1980;126:181–195. doi: 10.1007/978-1-4684-3632-7_17. [DOI] [PubMed] [Google Scholar]
- 29.Lal H, Prather PL, Rezazadeh SM. Potential role of 5-HT1C and/or 5-HT2 receptors in the mianserin-induced prevention of anxiogenic behaviors occurring during ethanol withdrawal. Alcohol Clin Exp Res. 1993;17:411–417. doi: 10.1111/j.1530-0277.1993.tb00785.x. [DOI] [PubMed] [Google Scholar]
- 30.Lander ES, Botstein D. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989;121:185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lander ES, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 32.LeMarquand D, Pihl RO, Benkelfat C. Serotonin and alcohol intake, abuse, and dependence: clinical evidence. Biol Psychiatry. 1994a;36:326–327. doi: 10.1016/0006-3223(94)90630-0. [DOI] [PubMed] [Google Scholar]
- 33.LeMarquand D, Pihl RO, Benkelfat C. Serotonin and alcohol intake, abuse, and dependence: findings of animal studies. Biol Psychiatry. 1994b;36:395–421. doi: 10.1016/0006-3223(94)91215-7. [DOI] [PubMed] [Google Scholar]
- 34.Lyons PA, Armitage N, Argentina F, Denny P, Hill NJ, Lord CJ, Wilusz MB, Peterson LB, Wicker LS, Todd JA. Congenic mapping of the type 1 diabetes locus, Idd3, to a 780-kb region of mouse chromosome 3: identification of a candidate segment of ancestral DNA by haplotype mapping. Genome Res. 2000;10:446–453. doi: 10.1101/gr.10.4.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malke H, Steiner K, Gase K, Frank C. Expression and regulation of the streptokinase gene. Methods. 2000;21:111–124. doi: 10.1006/meth.2000.0982. [DOI] [PubMed] [Google Scholar]
- 36.Mancini A, Koch A, Stefan M, Niemann H, Tamura T. The direct association of the multiple PDZ domain containing proteins (MUPP1) with the human cKit C-terminus is regulated by tyrosine kinase activity. FEBS Lett. 2000;482:54–58. doi: 10.1016/s0014-5793(00)02036-6. [DOI] [PubMed] [Google Scholar]
- 37.McCaul ME, Turkkan JS, Svikis DS, Bigelow GE. Alcohol and secobarbital effects as a function of familial alcoholism: extended intoxication and increased withdrawal effects. Alcohol Clin Exp Res. 1991;15:94–101. doi: 10.1111/j.1530-0277.1991.tb00524.x. [DOI] [PubMed] [Google Scholar]
- 38.Metten P, Crabbe JC. Common genetic determinants of severity of withdrawal from ethanol, pentobarbital and diazepam in inbred mice. Behav Pharmacol. 1994;5:533–547. doi: 10.1097/00008877-199408000-00014. [DOI] [PubMed] [Google Scholar]
- 39.Nadeau JH, Frankel WN. The roads from phenotypic variation to gene discovery: mutagenesis versus QTLs. Nat Genet. 2000;25:381–384. doi: 10.1038/78051. [DOI] [PubMed] [Google Scholar]
- 40.Neumann PE, Collins RL. Genetic dissection of susceptibility to audiogenic seizures in inbred mice. Proc Natl Acad Sci USA. 1991;88:5408–5412. doi: 10.1073/pnas.88.12.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Newlin DB, Pretorius MB. Sons of alcoholics report greater hangover symptoms than sons of nonalcoholics: a pilot study. Alcohol Clin Exp Res. 1990;14:713–716. doi: 10.1111/j.1530-0277.1990.tb01231.x. [DOI] [PubMed] [Google Scholar]
- 42.Prather PL, Rezazadeh SM, Lan H. Mianserin in the treatment of ethanol withdrawal in the rat: prevention of behaviors indicative of anxiety. Psychopharmacol Bull. 1991;27:285–289. [PubMed] [Google Scholar]
- 43.Regier DA, Farmer ME, Rae DS, Locke BZ, Keith SJ, Judd LL, Goodwin FK. Comorbidity of mental disorders with alcohol and other drug abuse. results from the Epidemiologic Catchment Area (ECA) Study. JAMA. 1990;264:2511–2518. [PubMed] [Google Scholar]
- 44.Reich T, Hinrichs A, Culverhouse R, Bierut L. Genetic studies of alcoholism and substance dependence. Am J Hum Genet. 1999;65:599–605. doi: 10.1086/302561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schuckit MA, Goodwin DA, Winokur G. A study of alcoholism in half siblings. Am J Psychiatry. 1972;128:1132–1136. doi: 10.1176/ajp.128.9.1132. [DOI] [PubMed] [Google Scholar]
- 46.Semenova TP, Ticku MK. Effects of 5-HT receptor antagonists on seizure susceptibility and locomotor activity in DBA/2 mice. Brain Res. 1992;588:229–236. doi: 10.1016/0006-8993(92)91580-8. [DOI] [PubMed] [Google Scholar]
- 47.Sheng M, Sala C. PDZ domains and the organization of supramolecular complexes. Annu Rev Neurosci. 2001;24:1–29. doi: 10.1146/annurev.neuro.24.1.1. [DOI] [PubMed] [Google Scholar]
- 48.Sokal RR, Rohlf FJ. Biometry. Freeman; San Francisco: 1981. [Google Scholar]
- 49.Strahlendorf JC, Lee M, Netzeband JG, Strahlendorf HK. Pentobarbital augments serotonin-mediated inhibition of cerebellar Purkinje cells. Neuroscience. 1988;27:107–115. doi: 10.1016/0306-4522(88)90222-9. [DOI] [PubMed] [Google Scholar]
- 50.Ullmer C, Schmuck K, Figge A, Lubbert H. Cloning and characterization of MUPP1, a novel PDZ domain protein. FEBS Lett. 1998;424:63–68. doi: 10.1016/s0014-5793(98)00141-0. [DOI] [PubMed] [Google Scholar]