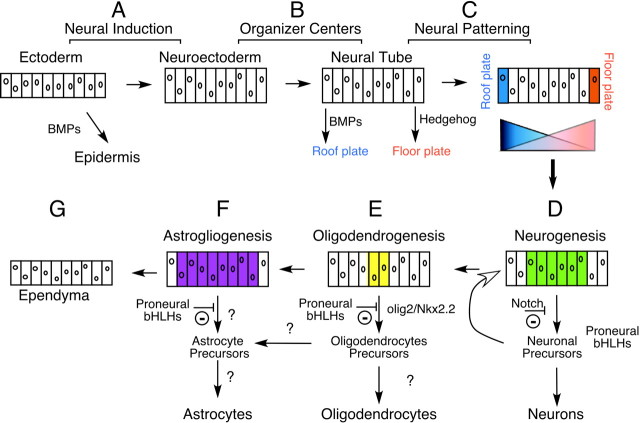
How the differentiated cell types that comprise the vertebrate CNS are generated during development is one of the central questions in developmental neurobiology. Considerable progress recently made in addressing this question has led to a rudimentary understanding of the molecular mechanisms that enable cells to acquire a neural fate in embryos. The purpose of this review is to discuss how the molecular mechanisms promoting neurogenesis in embryos might compare to those used in adult neural stem cells. To make this comparison, a focus will be placed on the nature of the progenitor cell populations and how these progenitors are instructed to become differentiated cell types.
From ectoderm to a neuroepithelium: default differentiation?
The adult vertebrate CNS has traditionally been subdivided into four major cell types: the neurons, the myelin-forming oligodendrocytes, the astrocytes, and the ependymal lining of the central lumen. All of these cell types are generated during development from a common source, the neuroepithelial cells that arise in early embryos in the form of the neural tube. The developmental events leading up to the formation of neuroepithelial cells involve inductive events that underlie axis determination. Significantly, neuroepithelial cells seem to form from cells that avoid a variety of instructive signals that induce non-neural fates, including the bone morphogenetic proteins (BMPs), which induce epidermal differentiation around the start of gastrulation (Wilson and Edlund, 2001) (Fig.1A). As a result, neuroepithelial cells probably represent a default, ground state, perhaps explaining both their ability to generate a variety of cell types, and the ease with which they form in culture from embryonic stem cells (Tropepe et al., 2001).
Fig. 1.
Diagram showing the embryonic origins of the progenitor cells in the developing CNS and some of the factors that promote the differentiation of these progenitors into neural cell types. See text for details.
The neuroepithelial cells within the neural tube give rise to differentiated neural cell types, producing neurons first, and glia at later stages. During neurogenesis, neurons are produced from almost all regions of the neuroepithelium except for a few specialized areas such as the optic stalk. It is unlikely, however, that neurogenesis represents a default pathway of differentiation, because both the onset and period of neurogenesis among neuroepithelial cells varies greatly depending on their location along the neuraxis. Moreover, given the diversity of neurons that comprise any given region of the CNS, one might expect a corresponding diversity of genetic programs that promote neuronal differentiation. However, the current view is that a core genetic program involving basic helix-loop-helix (bHLH) transcription factors is required for the differentiation of neuroepithelial cells into neurons, regardless of where and when they form. What seems to vary is how these bHLH proteins are activated within neuroepithelial cells at different points of the neuraxes by patterning genes. To illustrate this point, the following discussion will focus on the spinal cord where perhaps the most is known about how neuroepithelial cells are patterned, and how this patterning subsequently promotes neurogenesis, thus leading to the generation of specific classes of neurons.
Neurogenesis in the spinal cord
Patterning of the developing spinal cord begins at neural plate stages of development via inductive interactions that create organizing centers at the dorsal and ventral poles of the neural tube (Fig.1B, Floor plate, Roof plate) (Jessell, 2000). These specialized neuroepithelial cells generate signals that induce, often in a concentration-dependent manner, the expression of patterning genes in neighboring neuroepithelial cells (Fig. 1C). In the spinal cord, these patterning genes encode homeodomain transcription factors, and their expression patterns divide the cells in the neuroepithelium into different zones along the dorsoventral axis of the neural tube (Lee and Pfaff, 2001). These patterning genes are thought to specify neuronal subtype identity and, in addition, when and for how long neurons of a particular type will be generated during neurogenesis. This latter function likely depends on interactions between the patterning genes and a family of bHLH transcription factors referred to collectively as the proneural proteins.
Proneural proteins as obligatory factors in neuronal differentiation
The vertebrate proneural bHLH genes fall into two families based on homology to bHLH genes required for neural cell differentiation inDrosophila; those related to the Drosophila Achaete-Scute genes such as Mash1, and those related to Drosophila atonal, such as theneurogenins, the NeuroD-like, and theATH genes (Brunet and Ghysen, 1999). As transcriptional activators, these proteins have been proposed to have multiple functions during neurogenesis, including a role in neuronal subtype specification. However, the most relevant function to this discussion is their proposed role in promoting the differentiation of neural precursor cells into neurons during embryonic development (Fig. 1D).
Proneural bHLH proteins are likely to be the critical factor in causing the neuroepithelial cells to become neurons, which they do by exiting the cell cycle, delaminating out of the epithelium, and activating the expression of a large panel of genes indicative of generic neuronal differentiation. At the point where cells in the neuroepithelium make this decision, a region just outside of the ventricular zone, they express one or several of the proneural bHLH genes: the exact gene expressed varies depending on time and place. When eliminated by targeted mutation, loss of specific proneural bHLH genes results in specific deletion of neuronal elements (Schwab et al., 2000; Olson et al., 2001; Wang et al., 2001). However, the loss of neurons is likely to be much more severe when multiple members are simultaneously eliminated, a result of genetic redundancy as also found for the proneural bHLH genes in Drosophila (Scardigli et al., 2001). From the loss-of-function analysis, it is clear that the neural bHLH proteins are required for neuronal differentiation in specific cases, but because of functional redundancy it is not straightforward experimentally to ask whether all neuronal differentiation is driven by proneural gene action. However, we also know that the proneural proteins are potent inducers of neuronal differentiation when ectopically expressed (Lee et al., 1995). In addition, all neurons express a generic set of neuronal genes that are activated by the proneural bHLH proteins in ectopic expression experiments. Finally, proneural proteins seem to be key in promoting cell cycle exit, an event shared by all neurons when they undergo terminal differentiation (Farah et al., 2000). Thus, a reasonable assumption is that the proneural proteins are responsible for promoting neuronal differentiation, regardless of when and where a neuron forms in the CNS.
The proneural bHLH cascade in neuronal determination
One can view the activation of proneural bHLH genes as setting in motion an irreversible set of events that result in terminal neuronal differentiation. As a consequence, one would like to know how these neuronal switch genes are activated within the neuroepithelial cells during neurogenesis in the embryo, or for that matter within neural stem cells when neurogenesis occurs in the adult. Indeed, this question may have several answers, and the one that applies to embryos may be different from that for the adult. To explain why this might be the case, one needs to consider in detail how the bHLH genes that promote neuronal differentiation are activated in neurogenic epithelium. In all cases examined thus far, the key factor seems to be a bHLH cascade that mediates both neuronal determination and differentiation. The distinction between determination and differentiation is an important one and dates back to a model first proposed by Weintraub (1993) in his analysis of bHLH proteins that promote myogenesis.
In the Weintraub (1993) model, mesodermal cells are converted into dividing muscle precursors, the myoblasts, via the action of myogenic bHLH proteins such as MyoD (Weintraub, 1993). Because the myoblast “state” is largely maintained by MyoD expression, and MyoD maintains its expression by autoactivation, this state is easily reversible by simply disrupting the positive feedback loop between MyoD and its own transcription. Indeed, if MyoD activity is inhibited in myoblasts, they apparently convert back to a multipotential mesodermal cell. Alternatively, myoblasts are converted into muscle cells when the level and/or activity of MyoD is sufficiently high to activate downstream target genes, including bHLH proteins such as myogenin, which results in exit from the cell cycle, and terminal differentiation. This model explains a distinction that embryologists have long made between the determination of myogenic cells, a state inherently plastic and easily reversible particularly in culture, and terminal differentiation, a less reversible state that leads to muscle.
In a similar manner, the proneural bHLH genes that promote neuronal differentiation are likely to be activated by other proneural bHLH proteins that are expressed in neurogenic epithelium and act as neuronal determination genes (Fig. 1D, green shading). Experiments show that when their activity is sufficiently high in neuroepithelial cells, these bHLH proteins activate the expression of downstream differentiation bHLH genes, which then act to promote exit from the cell cycle and neuronal differentiation (Ma et al., 1996). Conversely, other experiments show that when the activity and expression of the determinative bHLH genes is inhibited, neuroepithelial cells seem to revert back to a ground state where they have the option to divide and perhaps become a neuron at a later time, or serve as the source of progenitor cells for various glia. Indeed, regulation of the determinative bHLH proteins seems to be a critical event for maintaining a proper balance between the need to generate cells that undergo terminal neuronal differentiation and the need to retain neuroepithelial cells in a progenitor mode, thus creating the progenitor cells for later-born neurons or for glia (Nieto et al., 2001). This balance allows for protracted neurogenesis and the proliferation of neuronal progenitor cells, a prerequisite for the use of neuronal birth date as an important mechanism for neuronal subtype specification (Perron and Harris, 2000). The Notch signaling pathway is well established as one means for maintaining this balance by inhibiting the activity of the proneural proteins (Fig.1D), but one imagines that other mechanisms are also used in this respect (Lewis, 1998; Lyden et al., 1999). Artificially elevating the activity of bHLH proteins seems to short-circuit the process, resulting in enhanced neuronal differentiation, but at the expense of a depleted precursor pool.
Neural patterning and the control of neurogenesis
Although we do not yet know whether all regions of neurogenic epithelium use a bHLH cascade to mediate neuronal determination, we do know that neural patterning genes activate the bHLH cascade in cases where neurogenesis has been studied in detail within the developing spinal cord. In ventral spinal cord, the first neurons to be generated are motor neurons, and their generation is correlated with the early expression of a determinative bHLH protein, Ngn2, within a narrow ventral domain of neuroepithelium. This region of the neuroepithelium is patterned by the expression of a key transcription factor, called Olig2, induced within the ventral neuroepithelium by hedgehog signaling (Fig. 1C). Significantly, one function of Olig2 in the generation of motor neurons is to induce Ngn2 expression, thus setting in motion the bHLH cascade (Mizuguchi et al., 2001;Novitch et al., 2001). When ectopically expressed in the embryonic spinal cord, Olig2 induces ectopic motor neuron differentiation, and does so in part by inducing ectopic and precocious expression ofNgn2. Significantly, ectopic Olig2 expression seems to recapitulate the normal balance between determination and differentiation discussed above, because only a fraction of theOlig2 expressing cells go on to form motorneurons over a given period of time. As expected, expressing high levels ofNgn2 along with Olig2 short circuits the determination phase, resulting in a very high level of motor neuron differentiation among the transgenic cells.
A similar link has also been made between patterning genes and bHLH cascades that operates in the dorsal neuroepithelium of the spinal cord to promote the generation of certain classes of dorsal interneurons (Gowan et al., 2001). In this case, neighboring domains of neuroepithelium producing these different classes of interneurons appear to do so, in part, by promoting the expression of distinct members of the proneural bHLH family. Genetic evidence suggests that these distinct neurogenic domains of neuroepithelial cells are formed by matching a given patterning transcription factor and a given proneural bHLH protein required for neuronal determination. If correct, this interpretation supports the view that patterning genes are key factors in activating a core bHLH cascade within neuroepithelial cells at each point along the neuraxis. The diversity in programs of neurogenesis that occur along the neuraxis presumably arises therefore by the way different patterning genes engage the bHLH cascade.
Converting neuroepithelial cells into glia
Neuroepithelial cells initially form neurons, but gradually switch over and produce different forms of glia. Rather than arising from separate populations of progenitor cells, the glial precursors are likely to be the neuroepithelial cells that were kept back from neuronal differentiation during the determinative phase of neurogenesis. One fate open to these cells is to form oligodendrocyte precursors (OPCs) that arise within a very restricted domain of the neural tube along the dorsoventral axis. As is the case with neurogenesis, a distinct genetic program has recently emerged as critical in promoting the differentiation of neuroepithelial cells into OPC, regardless of position along the neuraxis (Sun et al., 2001; Zhou et al., 2001). Surprisingly, this genetic program requiresOlig2, the gene required for promoting motor neuron differentiation at early stages. However, once this process is complete, Olig2 expression in the neuroepithelium begins to overlap with that of another patterning gene called Nkx2.2. Genetic studies suggest that when this overlap occurs, the neuroepithelium gives rise to OPCs, which subsequently delaminate out of the neuroepithelium, disperse within the white matter, and undergo a terminal differentiation program of myelination. A speculative analogy is that Olig2 and Nkx2.2 act as a genetic switch during the determination of OPCs, much the same way that proneural bHLH proteins determine neuronal precursors, as described above. This may account for the plasticity of OPCs in culture, because by analogy with determined neuronal precursors, factors that inhibit the action of Olig2 and Nkx2.2 will revert these cells back to the neuroepithelial ground state, whereas those that promote their activity will result in terminal oligodendrocyte differentiation into myelinating cells. The plasticity of these determined states seems to explain, for example, why they can be easily overcome by other determination factors. Misexpression of the proneural proteins in neuroepithelial cells inhibits OPC formation by activating neuronal determination; conversely, inhibition of proneural gene function by Olig2 and Nkx2.2 may be a factor in promoting the formation of OPCs (Sun et al., 2001;Zhou et al., 2001).
In contrast with OPCs, the genetic programs required for generating other forms of glia in the CNS are less clear, and in some cases may involve default pathways of differentiation that are available to neuroepithelial cells that have not formed neurons or oligodendrocytes (Fig. 1E). Radial glia cells found throughout the embryonic nervous system or the Müller glial cells found in the retina have been classified as distinct differentiated cell types, but in fact are remarkably similar to neuroepithelial cells by a number of criteria: the most important one being that they remain part of the neuroepithelium. The formation of these cells is stimulated when neurogenesis is inhibited by Notch, suggesting to some investigators that their relationship with neuroepithelial cells may be quite direct (Gaiano et al., 2000). Default differentiation may also apply to the astrocytes, a catch-all term for what appears to be a fairly plastic and heterogeneous population of cells. How these cells arise from the neuroepithelium is not entirely clear, but one emerging theme is that inhibiting neurogenesis or oligodendrocyte differentiation seems to stimulate their formation, like that of radial glia. For example, inhibition of proneural bHLH function via the Notch pathway is a potent means of “inducing” astrocyte differentiation (Tanigaki et al., 2001). Similarly, developing neuroepithelium gives rise precociously to astrocytes when proneural genes are eliminated by mutation (Nieto et al., 2001). In this sense, the embryonic origins of astrocytes are analogous to those of embryonic fibroblasts within the mesodermal lineage: a cell type distinguished more by the fact that it has not differentiated into a more specialized cell such as muscle, fat, cartilage, or bone cells, rather than the fact that it requires a distinct program of differentiation. Finally, a population of neuroepithelial cells left over after neurogenesis and gliogenesis form the ependyma, the epithelial lining of the central canal (Fig.1G). As direct remnants of the embryonic neuroepithelial cells, they are in the best position to retain the “code” of transcription factor gene expression that was used embryonically to pattern neurogenesis within this epithelium.
Is neurogenesis in embryos mechanistically similar to that in the adult?
How related is neurogenesis in embryos to that occurring in the adult? Several points of comparison are worth discussing, based on what is known about neurogenesis in embryos, although the answer to this question is obviously a problem for the future. One point is that the source of new neurons in the adult is unlikely to be a direct counterpart to the neuroepithelial cells that generate neurons in the embryo. The obvious requirement that stem cells divide makes such terminally differentiated cells such as neurons or oligodendrocytes unlikely sources of neural stem cells. Cell types retained within the neuroepithelium of the adult nervous system, such as ependyma or the so-called radial glia cells, are more likely sources based on their similarity to embryonic neuroepithelial cells from which they are closely derived (Alvarez-Buylla et al., 2001; Tamamaki et al., 2001). Indeed, as direct remnants of the embryonic neuroepithelium, these cells conceivably retain or continue to respond to the patterning signals that promote neurogenesis in the embryo. Again, a critical issue for these cells is whether they are capable of the cell division required of stem cells. Finally, another potential source is astrocytes, based on fact that they continue to undergo cell division and the idea that their differentiation is primarily a default pathway, leaving open the possibility that if the appropriate genes are activated, (i.e., the bHLH cascade), they can be coaxed into neuronal differentiation. Thus, the neurons generated in the adult may have several sources, some of which are close to but not directly equivalent to the embryonic neuroepithelium.
A second point of comparison is the whether the bHLH cascade, so critical to neurogenesis in the embryo, is also required in the adult. For reasons discussed above, a reasonable assumption is that it will be, because at present, alternative mechanisms for generating neurons do not currently exist. If this assumption is correct, then how might the bHLH cascade be activated during adult neurogenesis, and is this mechanism different or the same as that used in embryos? In embryos, a key factor in generating diverse patterns of neurogenesis that occur in the various regions of the developing CNS is the interactions that occur between bHLH cascade and the myriad of transcription factors that pattern the embryonic neuroepithelium. These interactions are critical both for neuronal cell determination and differentiation. Thus, one key question is whether the bHLH cascade is used in adult stem cells using this same mechanism, indicating that the patterning genes expressed in embryonic neuroepithelium will also be expressed by adult stem cells. The alternative scenario is that the constellation of patterning genes that are normally used in the embryo for promoting neurogenesis are absent in adult stem cells, which instead use an alternative means to activate the bHLH cascade. One possibility, for example, is that adult stem cells generate neurons by mechanisms that activate the differentiation proneural genes directly, thus bypassing the determination bHLH proteins. If this latter scenario is true, adult neural stem cells may be more specialized and only generate a limited range of neuronal subtypes because they are unable to activate the bHLH cascade in the same diverse ways used by embryonic cells.
Based on what we know about neurogenesis in embryos, one would like to know how the bHLH cascade is used by adult neural stem cells and how this mechanism compares to that used in embryos. This comparison may not only provide additional insights into how neurogenesis is mediated in the adult, but perhaps suggest strategies that might can be used to increase the neurogenic potential of neural stem cells in terms of their ability to undergo neuronal determination and differentiation.
Footnotes
Correspondence should be addressed to Chris Kintner at the above address. E-mail: kintner@salk.edu.
REFERENCES
- 1.Alvarez-Buylla A, Garcia-Verdugo JM, Tramontin AD. A unified hypothesis on the lineage of neural stem cells. Nat Rev Neurosci. 2001;2:287–293. doi: 10.1038/35067582. [DOI] [PubMed] [Google Scholar]
- 2.Brunet JF, Ghysen A. Deconstructing cell determination: proneural genes and neuronal identity. Bioessays. 1999;21:313–318. doi: 10.1002/(SICI)1521-1878(199904)21:4<313::AID-BIES7>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 3.Farah MH, Olson JM, Sucic HB, Hume RI, Tapscott SJ, Turner DL. Generation of neurons by transient expression of neural bHLH proteins in mammalian cells. Development. 2000;127:693–702. doi: 10.1242/dev.127.4.693. [DOI] [PubMed] [Google Scholar]
- 4.Gaiano N, Nye JS, Fishell G. Radial glial identity is promoted by Notch1 signaling in the murine forebrain. Neuron. 2000;26:395–404. doi: 10.1016/s0896-6273(00)81172-1. [DOI] [PubMed] [Google Scholar]
- 5.Gowan K, Helms AW, Hunsaker TL, Collisson T, Ebert PJ, Odom R, Johnson JE. Crossinhibitory activities of Ngn1 and Math1 allow specification of distinct dorsal interneurons. Neuron. 2001;31:219–232. doi: 10.1016/s0896-6273(01)00367-1. [DOI] [PubMed] [Google Scholar]
- 6.Jessell TM. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- 7.Lee JE, Hollenberg SM, Snider L, Turner DL, Lipnick N, Weintraub H. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix-loop-helix protein. Science. 1995;268:836–844. doi: 10.1126/science.7754368. [DOI] [PubMed] [Google Scholar]
- 8.Lee SK, Pfaff SL. Transcriptional networks regulating neuronal identity in the developing spinal cord. Nat Neurosci. 2001;4:1183–1191. doi: 10.1038/nn750. [DOI] [PubMed] [Google Scholar]
- 9.Lewis J. Notch signalling and the control of cell fate choices in vertebrates. Semin Cell Dev Biol. 1998;9:583–589. doi: 10.1006/scdb.1998.0266. [DOI] [PubMed] [Google Scholar]
- 10.Lyden D, Young AZ, Zagzag D, Yan W, Gerald W, O'Reilly R, Bader BL, Hynes RO, Zhuang Y, Manova K, Benezra R. Id1 and Id3 are required for neurogenesis, angiogenesis and vascularization of tumour xenografts. Nature. 1999;401:670–677. doi: 10.1038/44334. [DOI] [PubMed] [Google Scholar]
- 11.Ma Q, Kintner C, Anderson DJ. Identification of neurogenin, a vertebrate neuronal determination gene. Cell. 1996;87:43–52. doi: 10.1016/s0092-8674(00)81321-5. [DOI] [PubMed] [Google Scholar]
- 12.Mizuguchi R, Sugimori M, Takebayashi H, Kosako H, Nagao M, Yoshida S, Nabeshima Y, Shimamura K, Nakafuku M. Combinatorial roles of olig2 and neurogenin2 in the coordinated induction of pan-neuronal and subtype-specific properties of motoneurons. Neuron. 2001;31:757–771. doi: 10.1016/s0896-6273(01)00413-5. [DOI] [PubMed] [Google Scholar]
- 13.Nieto M, Schuurmans C, Brz O, Guillemot F. Neural bHLH genes control the neuronal versus glial fate decision in cortical progenitors. Neuron. 2001;29:401–413. doi: 10.1016/s0896-6273(01)00214-8. [DOI] [PubMed] [Google Scholar]
- 14.Novitch BG, Chen AI, Jessell TM. Coordinate regulation of motor neuron subtype identity and pan-neuronal properties by the bHLH repressor Olig2. Neuron. 2001;31:773–789. doi: 10.1016/s0896-6273(01)00407-x. [DOI] [PubMed] [Google Scholar]
- 15.Olson JM, Asakura A, Snider L, Hawkes R, Strand A, Stoeck J, Hallahan A, Pritchard J, Tapscott SJ (2001) NeuroD2 is necessary for development, survival of central nervous system neurons Dev Biol 234:174–187. [DOI] [PubMed]
- 16.Perron M, Harris WA. Determination of vertebrate retinal progenitor cell fate by the Notch pathway and basic helix-loop-helix transcription factors. Cell Mol Life Sci. 2000;57:215–223. doi: 10.1007/PL00000685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scardigli R, Schuurmans C, Gradwohl G, Guillemot F. Crossregulation between Neurogenin2 and pathways specifying neuronal identity in the spinal cord. Neuron. 2001;31:203–217. doi: 10.1016/s0896-6273(01)00358-0. [DOI] [PubMed] [Google Scholar]
- 18.Schwab MH, Bartholomae A, Heimrich B, Feldmeyer D, Druffel-Augustin S, Goebbels S, Naya FJ, Zhao S, Frotscher M, Tsai MJ, Nave KA. Neuronal basic helix-loop-helix proteins (NEX and BETA2/Neuro D) regulate terminal granule cell differentiation in the hippocampus. J Neurosci. 2000;20:3714–3724. doi: 10.1523/JNEUROSCI.20-10-03714.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun T, Echelard Y, Lu R, Yuk D, Kaing S, Stiles CD, Rowitch DH. Olig bHLH proteins interact with homeodomain proteins to regulate cell fate acquisition in progenitors of the ventral neural tube. Curr Biol. 2001;11:1413–1420. doi: 10.1016/s0960-9822(01)00441-9. [DOI] [PubMed] [Google Scholar]
- 20.Tamamaki N, Nakamura K, Okamoto K, Kaneko T. Radial glia is a progenitor of neocortical neurons in the developing cerebral cortex. Neurosci Res. 2001;41:51–60. doi: 10.1016/s0168-0102(01)00259-0. [DOI] [PubMed] [Google Scholar]
- 21.Tanigaki K, Nogaki F, Takahashi J, Tashiro K, Kurooka H, Honjo T. Notch1, Notch3 instructively restrict bFGF-responsive multipotent neural progenitor cells to an astroglial fate. Neuron. 2001;29:45–55. doi: 10.1016/s0896-6273(01)00179-9. [DOI] [PubMed] [Google Scholar]
- 22.Tropepe V, Hitoshi S, Sirard C, Mak TW, Rossant J, van der Kooy D. Direct neural fate specification from embryonic stem cells: a primitive mammalian neural stem cell stage acquired through a default mechanism. Neuron. 2001;30:65–78. doi: 10.1016/s0896-6273(01)00263-x. [DOI] [PubMed] [Google Scholar]
- 23.Wang SW, Kim BS, Ding K, Wang H, Sun D, Johnson RL, Klein WH, Gan L. Requirement for math5 in the development of retinal ganglion cells. Genes Dev. 2001;15:24–29. doi: 10.1101/gad.855301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weintraub H. The MyoD family and myogenesis: redundancy, networks, and thresholds. Cell. 1993;75:1241–1244. doi: 10.1016/0092-8674(93)90610-3. [DOI] [PubMed] [Google Scholar]
- 25.Wilson SI, Edlund T. Neural induction: toward a unifying mechanism. Nat Neurosci. 2001;4:1161–1168. doi: 10.1038/nn747. [DOI] [PubMed] [Google Scholar]
- 26.Zhou Q, Choi G, Anderson DJ. The bHLH transcription factor Olig2 promotes oligodendrocyte differentiation in collaboration with Nkx2.2. Neuron. 2001;31:791–807. doi: 10.1016/s0896-6273(01)00414-7. [DOI] [PubMed] [Google Scholar]