Abstract
Drug abuse (DA) or drug addiction is a complicated brain disorder which is commonly considered as neurobiological impairments caused by both genetic factors and environmental effects. Among DA-related targets, G protein-coupled receptors (GPCRs) play an important role in DA therapy. However, only 52 GPCRs have been published with crystal structures in the recent two decades. In the effort to overcome the limitations of crystal structure and conformational diversity of GPCRs, we built homology models and performed conformational searches by molecular dynamics (MD) simulation. To accelerate and facilitate the drug abuse research, we construct a DA-related GPCR-specific chemogenomics knowledgebase (KB) (DAKB-GPCRs) for its research that can be implemented with our established and novel chemogenomics tools as well as algorithms for data analysis and visualization. Our established TargetHunter and HTDocking tools, as well as our novel tools that include target classification and Spider Plot, are compiled into the platform. Our DAKB-GPCRs provides the following results for a query compound: (1) blood−brain barrier (BBB) plot via our BBB predictor, (2) docking scores via HTDocking, (3) similarity score via TargetHunter, (4) target classification via machine learning methods that utilize both docking scores and similarity scores, and (5) a drug−target interaction network via Spider Plot.
Graphical Abstract

INTRODUCTION
Drug abuse (DA) is a complicated neurological disorder. The main symptoms of DA and addiction involve compulsive behaviors such as drug craving, seeking, persistent use, and relapses despite serious adverse consequences. In 2017, there were approximate 30.5 million people of age 12 or older in the U.S. who had used illicit drugs or abused psychotherapeutic medications (e.g., pain relievers, stimulants, or tranquilizers), representing 11.2% of the population.1 According to Drug Facts,2 the total overall costs of substance abuse in the U.S. exceed $740 billion annually, including approximately $193 billion for illicit drugs,3,4 $300 billion for tobacco,5,6 and $249 billion for alcohol7 (Figure S1). Thus, research for DA prevention and treatment is a high priority.
DA is a chronic disease with strong genetic and environ-mental influences that can often lead to patient relapse. The basic mechanisms of DA are well accepted that abused substances can disrupt activity of normal nerves cells by interacting with receptors in the brain and activating the downstream signaling pathways, which comprise a host of kinases and transcription factors. Specifically, there is a broad range of G-protein coupled receptors (GPCRs) related to the DA and central nervous system (CNS) side effects, including opioid receptors, cannabinoid receptors, and serotonin receptors.8 Further, it is estimated that genetic factors play a significant role, approximately 40−60% of the total risk.8,9
To accelerate and facilitate the drug abuse research, we constructed a self-service online library named GPCR-specific chemogenomics knowledgebase (KB) for DA research (DAKB-GPCRs) that contains information about drug abuse-related protein targets, small molecules, as well as tools and algorithms for computational data analyses and visualization on those data. In our established drug DA-KB8,10 regarding protein targets of abused drugs, we found that 86 out of 258 proteins (33.3%) are GPCRs including serotonin receptors (5HT 1A-5HT 7),11 adenosine receptors (A1, A2a, A2b, and A3),12 cannabinoid receptors (CB1 and CB2),13 acetylcholine receptors (M1-M5),14 C-X-C motif chemokine receptors (CXCR1−CXCR4), dopamine receptors (D1−D5),15 histamine receptors (H1−H4), melanocortin receptors (MC1−MC5),16 opioid receptors (Mu, Delta, and Kappa),17 glutamate receptors (metabotropic glutamate receptor 1−8),18 etc.8 More details can be found in our previous publication,8 our published DAKB website (https://www.cbligand.org/CDAR/) and Table S1. Moreover, in the past 18 years, only 52 GPCRs have been published with crystal structures or cryo-EM structures,19 in which only 29 GPCRs are related to DA (34% out of 86 GPCRs) (Figure S1). Recently, homology modeling with a sequence identity of 30% or greater,20 and/or multiple conformations-based docking, is becoming a powerful tool and strategy for structural study and drug discovery.21
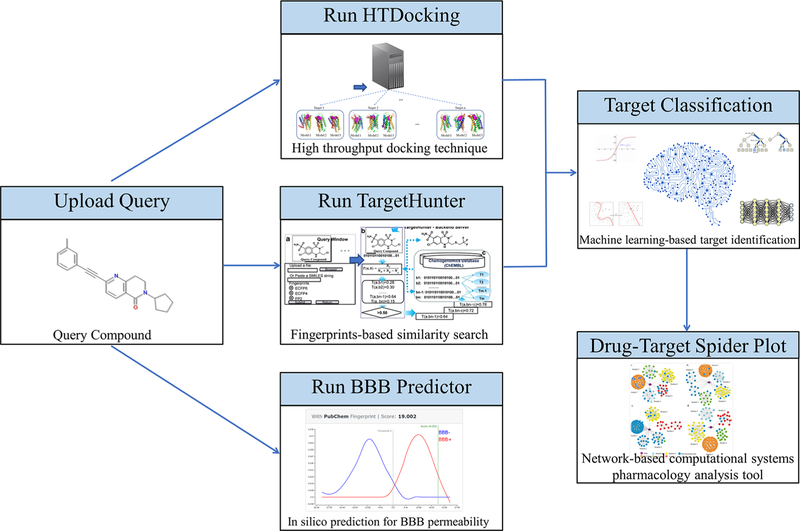
On the basis of our previously established molecular information databases8 and developed computing algorithms,22−24 we constructed a GPCR domain-specific drug abuse knowledgebase (DAKB-GPCRs, Figure 1). To achieve this, we have carried out a literature search to collect DA-related GPCR structures and corresponding drugs.8 Specifically, as discussed in the beginning, there were only a small percentage of GPCR crystal structures available, but we built the homology models for those GPCRs without crystal structure(s) based on published GPCR crystal structures for the use of molecular modeling study. To further increase the diversity of the protein conformation as well as optimize the accuracy of the homology models, molecular dynamics (MD) simulation was used to sample conformations of each GPCR, followed by computational prescreening validation for multiple conformation-based docking. Last but not least, we integrated established tools and algorithms (such as TargetHunter, HTDocking, NGL, ANN) and developed new tools (such as Spider Plot) for data visualization and analyses.
Figure 1.
Overall workflow of DAKB-GPCRs.
MATERIALS AND METHODS
Genes/Proteins.
Genes/proteins that related to drug abuse were collected from public databases such as Ensembl,25 UniProt,26 KEGG,27 GPCRdb,28 and the NCBI Protein Database.29 Available crystal structures or cryo-EM structures of GPCRs were retrieved from the Protein Data Bank (PDB) (https://www.rcsb.org/).
Drugs and Chemicals.
The ChEMBL database (version 23)30 was used in our work. The experimental data for each small molecule against its respective target proteins were collected using text mining techniques and cleaned up by manual inspection. Bioactivity data from different resources were normalized using the same standard. The number of small molecule agents for each GPCR is listed in Table S1. Especially, small molecules with IC50 lower than 1 μM toward a GPCR target were regarded as the active compounds, while those larger than 10 μM were considered as inactive compounds. A training data set that consisted of both active and inactive compounds of each GPCR will be used for prescreening and similarity search using TargetHunter.31
Homology Models.
The sequences of the human GPCRs were collected from the UniProtKB/Swiss-Prot (https://www.uniprot.org/uniprot/) website. We will first truncate some residues from the N terminal and some residues from the C terminal. Moreover, the third intracellular loop (ICL3, between helix 5 and helix 6) has a long-flexible sequence, we only keep ∼15 residues. Then, Modeller 9.1832 was used to construct the homology models: (a) searching and selecting template(s) for the target protein, (b) conducting sequence alignment between target and template(s), (c) adjusting the sequence alignment using the residue tables from GPCRdb (http://gpcrdb.org/residue/residuetable), and (d) building and evaluating the homology models. After generating the 3D models of protein targets, SYBYL-X 1.333 was adopted to carry out the energy minimization. Then the best three models after energy minimization with the root-mean-square deviation (RMSD) of 2−3 Å was selected for further molecular dynamics simulation. This established protocol can be found in our previous publications.34–36
Molecular Dynamics (MD) Simulation for Conformational Sampling.
A pioneer paper entitled “The Biological Functions of Low-Frequency Phonons”37 and a series of following research works on biomacromolecules from dynamic point of view38 suggest that low-frequency collective motions do exist in proteins and DNA.39–42 By studying the low-frequency internal motions,40 many important biological functions in proteins and DNA and their dynamic mechanisms, such as shifting between active and inactive states,43 cooperative effects,44 and allosteric transition,38 can be revealed. Therefore, to further understand the action mechanisms of biomacromolecules, we should consider not only the static structural information (such as crystal structure or cryo-EM structure) but also the dynamical information acquired from molecular dynamic (MD) simulation.45
In order to obtain the diverse conformations of each GPCR for multiple conformations-based docking, we carried out the MD simulations without ligand for either crystal structures or homology models. First, special caution was applied to His residues, because His was ionized at pH 7.40. VEGA ZZ 2.4.046 and PROPKA 3.147 were applied to predict the pK values of His and other residues. Sequentially, the VMD program was used to embed the receptor into a periodic and pre-equilibrated structure of 1-palmytoyl-2-oleoyl-sn-glycero-3-phosphatidylcholine (POPC). Lipid molecules within 3 Å of the receptor were eliminated. We then inserted these into a water box (TIP3P48 water model, 67 Å × 67 Å × 74 Å) with eliminating the waters molecules within 3 Å of the receptor. Two minimizations were carried out. Each minimization was carried out in 50 000 steps. The first minimization was performed with the fixed protein, and the second minimization was carried out with the flexible protein. Then 1.0 ns of MD for heating from 0 to 300 K and equilibration was performed. Finally, the NAMD package (version 2.9b1)49 using the CHARMM2750 force field was applied to the MD simulation. The particle mesh Ewald (PME)51 method (with a 12 Å nonbonded cutoff and a grid spacing of 1 Å per grid point in each dimension) was used to calculate the electrostatics. A smooth cutoff (switching radius 10 Å, cutoff radius 12 Å) was used to calculate the van der Waals energies. The temperature and pressure were kept constant using a Langevin thermostat (310 K) and Langevin barostat (1 atm), respectively. The time step of the MD simulation was set to 1 fs. In order to select reasonable conformations of each homology model or crystal structure, we first performed 5 ns MD simulation for the protein with explicit water and lipid. During 5 ns MD simulation, we fixed the Cα atoms of seven transmembrane domains of the protein and made the side chains flexible. Then, we made the whole protein flexible and performed another 5 ns MD simulation. We selected ten conformations with the lowest energy and the RMSD over 2−3 Å to perform secondary energy minimization by SYBYL. This established protocol can be found in our previous publications.34−36
Prescreening.
Ten models of each GPCR that obtained after MD simulation were utilized to perform the prescreening against its training data set (see Drugs and Chemicals). The training data set of each GPCR included both the active and inactive compounds (Table S1). The docking program HTDocking (see below) was applied to constructed receptor−ligand complexes.
Database Infrastructure.
A query compound can be submitted using JSME Molecular Editor v2017–03-01.52 DAKB-GPCRs was implemented on the basis of our established molecular database prototype CBID (https://www.cbligand.org/cbid/) using the SQLite database management system (https://sqlite.org/) and Kestrel HTTP server (https://github.com/aspnet/KestrelHttpServer) with the Apache HTTP server (https://httpd.apache.org/) as its reverse proxy server. The overview of our design for DAKB-GPCRs is depicted in Figure 1.
HTDocking.
DAKB-GPCRs adopts our online high-throughput molecular docking technique—HTDocking,22,23,53 for identifying possible interactions between protein targets and small molecules. There are three different conformations for each GPCRs that selected from MD sampling and validated by prescreening. For each query compound, HTDocking will automatically dock it into three different conformations and generate docking scores. A higher docking score indicates that the protein is more likely to be the candidate target of the queried small molecule. idock54 can provide up to 9 predicted binding affinity values (ΔG values) from different docking poses for each compound in a binding pocket of a protein. In our HTDocking program, we only consider the best binding affinity value which is further transformed as a docking score. The docking score = −log10(eΔG*4184/8.314/310.15).
TargetHunter.
DAKB-GPCRs integrates our online target-identification service—TargetHunter,31 for predicting the potential off-targets for submitted compounds. TargetHunter exploits an important principle of medicinal chemistry: compounds with structural similarities often have similar physicochemical properties and biological profiles. For each query compound, TargetHunter calculates the similarity (from 0.0 to 1.0, totally different to 100% similar) with its known active compound’s data set that was collected from Drugs and Chemicals (see above).
Machine Learning-Based Target Classification.
For each GPCR, a data set consisting of three docking scores and one similarity score for each known compound were trained to build the target classification models using established machine learning (ML) algorithms. The compound data set collected for each target (as discussed in Drugs and Chemicals) was used for training and testing the classification models. The size of the data sets can be found in Table S1. Molecular docking scores and molecular fingerprint similarity scores were computed using the protocols discussed in HTDocking and TargetHunter, respectively. Four ML algorithms were adopted by our classification models: logistic regression,55 support vector machine (SVM),56 random forest (RF),57 and artificial neural networks (ANN).58 The training and the classification were implemented in Python using scikit-learn (https://scikit-learn.org/).
Spider Plot.
Based on the target classification, our online tool Spider Plot visualizes the molecule−protein interaction network. The average docking scores are displayed as connection labels and the protein targets on which the query compound is active are displayed as circular nodes. By default, a green node denotes a target with high similarity score (>0.7) comparing the best matched known compound and the query compound while a pink one denotes otherwise. With Spider Plot, the colors, font sizes, node sizes, border widths, and node shapes as well as the layout can be completely customized, and the entire network graph can be exported as an image file on particular browsers.
Blood−Brain Barrier (BBB) Predictor.
The BBB predictor22,23 was integrated into DAKB-GPCRs. This predictor was built by applying the support vector machine (SVM) and LiCABEDS59,60 algorithms on four types of fingerprints of 1593 reported compounds.61 It predicts whether or not a query compound can move across the BBB to the central nervous system (CNS).62 The BBB predictor is also available for access from https://www.cbligand.org/BBB/.
Software Requirements.
The DAKB-GPCRs website is compatible with modern web browsers (such as Chrome, Firefox, Microsoft Edge, and Safari) provided JavaScript and cookies are enabled. We recommend the latest release version of these web browsers for better rendering.
RESULTS AND DISCUSSION
Overview of Our Platform.
Our platform intrinsically works on a set of conformations of GPCR homology models/ crystal structures and their relevant compounds. The compounds serve as the training data set where both active and inactive compounds are included for docking and similarity calculation. The platform makes use of three conformations for each GPCR so as to obtain multiple docking scores for training the machine learning models. The predicted values of BBB penetration22 and the potential GPCR targets on which the submitted query compound is active are provided in the output. As shown in Figure 1, The platform first queues a task for the query compound and afterward computes three docking scores via HTDocking22 and a molecular similarity score via TargetHunter31 for each GPCR target. Subsequently a target classification is performed by our ensembled machine learning algorithms featuring docking scores and similarity score. The classification results are then passed to Spider Plot for visualization of the drug-targets interaction network.
Statistical Analysis of Multiple Conformations-Based Docking.
Ten models of each GPCR with the lowest energy and the RMSD over 2−3 Å (compared to the original homology model/crystal structure) obtained after MD simulation were utilized to perform the prescreening against its training data set (see Drugs and Chemicals). Three conformations of each GPCR with the best ROC curve were selected and integrated into our platform. Here, we take adrenoceptor alpha 1d (ADRA1D) as an example, which was constructed using beta-2 adrenergic receptor (PDB ID 3SN6, sequence identity between helixes 39%), beta-1 adrenergic receptor (PDB ID 2Y00, sequence identity between helixes 39%) and dopamine 3 receptor (PDB ID 3PBL, sequence identity between helixes 40%). Figure S2 shows the curves of its best three conformations. Figure S2A shows the statistical results of Model 1 of ADRA1D: the docking score of 6.15 was chosen as the best threshold, because the docking scores of 71% (1−0.29) inactive compounds were lower than 6.15, while the docking scores of 78% (0.78) active compounds were higher than 6.15. The threshold of docking score of other two models of ADRA1D were 6.59 (Figure S2B) and 6.71 (Figure S2C), respectively. We can find that the true positive rate (TPR) and false positive rate (FPR) of model 2 are 71% and 63%, while those of model 3 are 79% and 63%. The similar results can be found for other GPCRs in our platform, indicating the protocol of multiple conformations-based docking is reasonable.
Evaluation of the Machine Learning Classification Models.
A 10-fold cross validation was used to assess the predictive capability of all the models. The performance and evaluation for those classifications are listed in Figure S3. The performance of the ANN models was not robust in this case; thus, those models were eliminated. Therefore, for each protein target, the final prediction was determined by the classifications from the three selected models and their confidence levels.
Case Study.
To elaborate how the DAKB-GPCRs website can be used to facilitate research, we use a published compound (CHEMBL1779871, a positive allosteric modulator of human mGluR5) to showcase the functionalities step by step.
Home Page.
The DAKB-GPCRs can be accessed from https://www.cbligand.org/dakb-gpcrs/. On the top of the home page (Figure S4A) is the navigation bar which contains the HOME button, the ALL TASKS button and the HELP button. Clicking on the HOME button brings the user back to the home page while the ALL TASKS button directs the user to the task list page. The HELP button navigates the user to the User Guide page which provides a step-by-step guide for using DAKB-GPCRs.
Task List Page.
The task list page displays all the user-submitted tasks in a table layout (Figure S4B). Completed tasks are shown as Finished in the status column, and ongoing tasks are shown as Running. The user can click on the task name to access the detailed information on the input compound as well as the output of the computation task. The user can initiate a new task by clicking on the Create a new task button on the upper right corner of the page.
Start a New Task Using Structure Query.
The user can submit a query compound either by drawing its 2D structure with JSME Molecular Editor or by uploading a chemical file with the upload button.63 A meaningful name for the task is also required. Clicking on the Create Task button will initiate a new task. Afterward, a background job worker which periodically monitors the task queue will start to allocate computation resources and dispatch the computation task. In Figure S4C, the structure of CHEMBL1779871 in SMILES format was uploaded.
Detailed Task Page.
After submitting the request, the website will automatically direct users to the detailed task page (Figure S4D). On this page, you can see the task information as well as the progress bar on the top showing the real time progress of computation for target prediction. Following the task information is the section of ligand information which includes 2D/3D structures, multiple structure files of the query compound, and various computed molecular fingerprints.64
BBB Page.
The visualized results from the BBB predictor can be found on the BBB page (Figure S4E).
Output Page for Target Prediction.
The output page can be accessed by clicking on the See Detailed Output button from the detailed task page. On the output page, each block presents three docking scores by HTDocking computed against three protein target models and a similarity score by TargetHunter (Figure S4F and S4G). The classification result is presented suggesting whether the query compound is an active or inactive compound for this protein target, and the confidence level of the prediction is also provided. When clicking on the download button, the user can download the docking results of the submitted compound. The most similar compound within the compound library of a protein target can be seen from the Best Match field. Its comparison with the query compound is shown in a popup window (Figure S4H) after clicking the compound ID. When clicking on the target name, a window that consists of additional resource for the protein target (Figure S4I) will be shown, which includes a 3D interactive visualization,65,66 multiple links to other database websites, and models for direct downloading.
The SPIDER PLOT button on the top of the page leads user to the drug−target network plotting tool for data visualization and analysis (Figure S4J). The most promising predictions (with similarity score > 0.70) for the query compound are shown as green nodes connected with a solid line while other predictions are shown as purple nodes connected with a dashed line. The name and structure of the query compound are placed in the center of the plot and the average docking scores are displayed as connection labels. By clicking on a node, a toolbox will appear for users to customize the colors, font sizes, node sizes, and border widths. Also, the entire network graph can be exported as an image file on particular browsers. As illustrated in Figure S4J, GRM5 was predicted as the most promising target for CHEMBL1779871, which was consistent with the bioactivity data where CHEMBL1779871 is an active compound for GRM5 (EC50 = 7.5 nM).
Prediction Accuracy.
As the statistics show in Table S1, the overall accuracy of our machine learning based prediction was 90% on the testing set containing 178 812 compounds collected in the method discussed in the Drugs and Chemicals section. As a comparison, the overall accuracy using just docking scores with an optimized cutoff value on the same testing set was ranged between 61% and 62%, while the overall accuracy using just similarity score was 93%. In the final output of DAKB-GPCRs all the four scores and the machine learning prediction are provided for the users’ reference.
Future Perspective.
In the past decade, more and more prediction methods and computational tools were developed and released as publicly accessible websites with user-friendly interfaces.67–70 Many of which are practically useful and have increasing impacts on medical science.71 We are aware of the importance of user experience and as such in our future work, to further improve the user-friendliness, we shall make efforts to add more flexibility for users to manipulate the findings according to their needs and more customizations to our visualizing tool Spider Plot.
CONCLUSIONS
To facilitate the DA research, we constructed an integrated online computing platform for drug abuse researchers and scientists who may have no experience in programming, statistics or in silico drug design. This is an optimal approach for tailoring medicinal chemistry drug design and customizing combination therapy in a precision systems pharmacology manner. To our knowledge, no such domain-specific database is available for the proposed computational applications. Our platform is the first web-based service that integrates DA-related genes, proteins, and drugs for DA research. State-of-the-art computational chemistry/chemoinformatics and machine learning algorithms established in our lab have been implemented for this chemogenomics database, which will help characterize the features of genes, proteins, and drugs in DA study. It will also facilitate new information exchange and data sharing of knowledge among relevant scientific communities.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Shifan Ma, Guanxing Hu, Xiaomeng Xu, and Xiaole Yang for collecting drug abuse-related targets and small molecules for each GPCR. The authors thank Si Chen, Yu Zhang, and Nan Wu for preparing the images of the website. The authors thank Yan Zhang for wiring a program that can automatically calculate the ROC curve. The authors thank Weiwei Lin for proofreading.
Funding
This work was supported by the funding to the Xie laboratory from the National Institutes of Health, National Institute on Drug Abuse (P30 DA035778A1), National Institutes of Health (R01 DA025612), and the Department of Defense (W81XWH-16-1-0490).
The authors declare the following competing financial interest(s): X.-Q.X. is the Founder of ID4Pharma and serves as a consultant for Oxis Biotech. The other authors have no relationships to disclose. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jcim.8b0062310.1021/acs.jcim.8b00623.
Figures S1−S4 and Table S1 mentioned in the text (DOCX)
This web-based tool is freely available at https://www.cbligand.org/dakb-gpcrs.
REFERENCES
- (1).NIDA Nationwide Trends https://www.drugabuse.gov/publications/drugfacts/nationwide-trends (January 10).
- (2).NIDA Costs of Substance Abuse https://www.drugabuse.gov/related-topics/trends-statistics (18 December).
- (3).National Drug Intelligence Center (NDIC). National Drug Threat Assessment; 2017.
- (4).Birnbaum HG; White AG; Schiller M; Waldman T; Cleveland JM; Roland CL Societal Costs of Prescription Opioid Abuse, Dependence, and Misuse in the United States. Pain Med 2011, 12, 657−667. [DOI] [PubMed] [Google Scholar]
- (5).Surgeon General of the United States. The Health Consequences of Smoking—50 Years of Progress: A Report of the Surgeon General; January 2014.
- (6).Xu X; Bishop EE; Kennedy SM; Simpson SA; Pechacek TF Annual Healthcare Spending Attributable to Cigarette Smoking An Update. Am. J. Prev. Med 2015, 48, 326–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).CDC https://www.cdc.gov/features/costsofdrinking/ (April 21).
- (8).Xie X-Q; Wang L; Ouyang Q; Fang C; Su W; Liu H Chemogenomics knowledgebased polypharmacology analyses of drug abuse related G-protein coupled receptors and their ligands. Front. Pharmacol 2014, 5, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Kendler KS; Schmitt E; Aggen SH; Prescott CAJA Genetic and environmental influences on alcohol, caffeine, cannabis, and nicotine use from early adolescence to middle adulthood. Arch. Gen. Psychiatry 2008, 65, 674–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Xie X-Q; Wang L; Wang J; Xie Z; Yang P; Ouyang Q In Silico Chemogenomics Knowledgebase and Computational System Neuropharmacology Approach for Cannabinoid Drug Research. In Neuropathology of Drug Addictions and Substance Misuse; Elsevier: 2016; pp 183–195. [Google Scholar]
- (11).LeMarquand D; Pihl RO; Benkelfat CJ Serotonin and alcohol intake, abuse, and dependence: findings of animal studies. Biol. Psychiatry 1994, 36, 395–421. [DOI] [PubMed] [Google Scholar]
- (12).Ballesteros-Yáñez I; Castillo CA; Merighi S; Gessi S The Role of Adenosine Receptors in Psychostimulant Addiction. Front. Pharmacol 2018, 8, 985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Panlilio LV; Goldberg SR; Justinova Z Therapeutics, Cannabinoid abuse and addiction: clinical and preclinical findings. Clin. Pharmacol. Ther 2015, 97, 616–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Fink Jensen A; Fedorova I; Wörtwein G; Woldbye DP; Rasmussen T; Thomsen M; Bolwig TG; Knitowski KM; McKinzie DL; Yamada M Role for M5 muscarinic acetylcholine receptors in cocaine addiction. J. Neurosci. Res 2003, 74, 91–96. [DOI] [PubMed] [Google Scholar]
- (15).Beaulieu J-M; Gainetdinov RR The physiology, signaling, and pharmacology of dopamine receptors. Pharma. Rev 2011, 63, 182–217. [DOI] [PubMed] [Google Scholar]
- (16).Olney JJ; Navarro M; Thiele TE Targeting central melanocortin receptors: a promising novel approach for treating alcohol abuse disorders. Front. Neurosci 2014, 8, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Di Chiara G; Alan North R Neurobiology of opiate abuse. Trends Pharmacol. Sci 1992, 13, 185–193. [DOI] [PubMed] [Google Scholar]
- (18).Kenny PJ; Markou A The ups and downs of addiction: role of metabotropic glutamate receptors. Trends Pharmacol. Sci 2004, 25, 265–272. [DOI] [PubMed] [Google Scholar]
- (19).Xiang J; Chun E; Liu C; Jing L; Al-Sahouri Z; Zhu L; Liu W Successful Strategies to Determine High-Resolution Structures of GPCRs. Trends Pharmacol. Sci 2016, 37, 1055–1069. [DOI] [PubMed] [Google Scholar]
- (20).Chothia C; Lesk AM The Relation between the Divergence of Sequence and Structure in Proteins. EMBO J 1986, 5, 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Kitchen DB; Decornez H; Furr JR; Bajorath J Docking and scoring in virtual screening for drug discovery: Methods and applications. Nat. Rev. Drug Discovery 2004, 3, 935–949. [DOI] [PubMed] [Google Scholar]
- (22).Liu HB; Wang LR; Lv ML; Pei RR; Li PB; Pei Z; Wang YG; Su WW; Xie XQ AlzPlatform: An Alzheimer’s Disease Domain-Specific Chemogenornics Knowledgebase for Poly-pharmacology and Target Identification Research. J. Chem. Inf. Model 2014, 54, 1050–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Zhang Y; Wang LR; Feng ZW; Cheng HZ; McGuire TF; Ding YH; Cheng T; Gao YD; Xie XQ StemCellCKB: An Integrated Stem Cell-Specific Chemogenomics KnowledgeBase for Target Identification and Systems-Pharmacology Research. J. Chem. Inf. Model 2016, 56, 1995–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Zhang H; Ma S; Feng Z; Wang D; Li C; Cao Y; Chen X; Liu A; Zhu Z; Zhang J; Zhang G; Chai Y; Wang L; Xie X-Q Cardiovascular disease chemogenomics knowledgebase-guided target identification and drug synergy mechanism study of an herbal formula. Sci. Rep 2016, 6, 33963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Zerbino DR; Achuthan P; Akanni W; Amode MR; Barrell D; Bhai J; Billis K; Cummins C; Gall A; Giron CG; Gil L; Gordon L; Haggerty L; Haskell E; Hourlier T; Izuogu OG; Janacek SH; Juettemann T; To JK; Laird MR; Lavidas I; Liu Z; Loveland JE; Maurel T; McLaren W; Moore B; Mudge J; Murphy DN; Newman V; Nuhn M; Ogeh D; Ong CK; Parker A; Patricio M; Riat HS; Schuilenburg H; Sheppard D; Sparrow H; Taylor K; Thormann A; Vullo A; Walts B; Zadissa A; Frankish A; Hunt SE; Kostadima M; Langridge N; Martin FJ; Muffato M; Perry E; Ruffier M; Staines DM; Trevanion SJ; Aken BL; Cunningham F; Yates A; Flicek P Ensembl 2018. Nucleic Acids Res 2018, 46, D754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).The Uniprot Consortium. UniProt: the universal protein knowledgebase. Nucleic Acids Res 2017, 45, D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Kanehisa M; Goto S KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000, 28, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Isberg V; Mordalski S; Munk C; Rataj K; Harpsoe K; Hauser AS; Vroling B; Bojarski AJ; Vriend G; Gloriam DE GPCRdb: an information system for G protein-coupled receptors. Nucleic Acids Res 2017, 45, 2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Coordinators NR Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 2018, 46, D8–D13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Gaulton A; Bellis LJ; Bento AP; Chambers J; Davies M; Hersey A; Light Y; McGlinchey S; Michalovich D; Al-Lazikani B; Overington JP ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res 2012, 40, D1100–D1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Wang L; Ma C; Wipf P; Liu H; Su W; Xie XQ TargetHunter: an in silico target identification tool for predicting therapeutic potential of small organic molecules based on chemogenomic database. AAPS J 2013, 15, 395–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Webb B; Sali A Comparative Protein Structure Modeling Using MODELLER. Curr. Protocols Bioinf 2016, 54, 5.6.1–5.6.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33). SYBYL-X, v1.3; Tripos International, 2010.
- (34).Feng Z; Alqarni MH; Yang P; Tong Q; Chowdhury A; Wang L; Xie XQ Modeling, molecular dynamics simulation, and mutation validation for structure of cannabinoid receptor 2 based on known crystal structures of GPCRs. J. Chem. Inf. Model 2014, 54, 2483–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Feng ZW; Pearce LV; Xu XM; Yang XL; Yang P; Blumberg PM; Xie XQ Structural Insight into Tetrameric hTRPV1 from Homology Modeling, Molecular Docking, Molecular Dynamics Simulation, Virtual Screening, and Bioassay Validations. J. Chem. Inf. Model 2015, 55, 572–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Hu J; Feng Z; Ma S; Zhang Y; Tong Q; Alqarni MH; Gou X; Xie XQ Difference and Influence of Inactive and Active States of Cannabinoid Receptor Subtype CB2: From Conformation to Drug Discovery. J. Chem. Inf. Model 2016, 56, 1152–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Chen K-C; Chen N-Y The biological functions of low-frequency phonons. Sci. Sin 1977, 20, 447–457. [Google Scholar]
- (38).Chou K; Chen N; Forsen SJCS The biological functions of low-frequency phonons. 2 Cooperative effects. Chem. Scr 1981, 18, 126–132. [Google Scholar]
- (39).Chou KC; Mao B Collective motion in DNA and its role in drug intercalation. Biopolymers 1988, 27, 1795–1815. [DOI] [PubMed] [Google Scholar]
- (40).Chou K-C Low-frequency collective motion in biomacromolecules and its biological functions. Biophys. Chem 1988, 30, 3–48. [DOI] [PubMed] [Google Scholar]
- (41).Chou K-C; Maggiora GM; Mao B Quasi-continuum models of twist-like and accordion-like low-frequency motions in DNA. Biophys. J 1989, 56, 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Martel P Biophysical aspects of neutron scattering from vibrational modes of proteins. Prog. Biophys. Mol. Biol 1992, 57, 129–179. [DOI] [PubMed] [Google Scholar]
- (43).Wang J-F; Gong K; Wei D-Q; Li Y-X; Chou K-C Design; Selection, Molecular dynamics studies on the interactions of PTP1B with inhibitors: from the first phosphate-binding site to the second one. Protein Eng., Des. Sel 2009, 22, 349–355. [DOI] [PubMed] [Google Scholar]
- (44).Chou K-C Low-frequency resonance and cooperativity of hemoglobin. Trends Biochem. Sci 1989, 14, 212. [DOI] [PubMed] [Google Scholar]
- (45).Yang J; Li J-H; Wang J; Zhang C-Y Molecular modeling of BAD complex resided in a mitochondrion integrating glycolysis and apoptosis. J. Theor. Biol 2010, 266, 231–241. [DOI] [PubMed] [Google Scholar]
- (46).Pedretti A; Villa L; Vistoli G Modelling, VEGA: a versatile program to convert, handle and visualize molecular structure on Windows-based PCs. J. Mol. Graphics Modell 2002, 21, 47–49. [DOI] [PubMed] [Google Scholar]
- (47).Søndergaard CR; Olsson MH; Rostkowski M; Jensen JH Computation, Improved treatment of ligands and coupling effects in empirical calculation and rationalization of p K a values. J. Chem. Theory Comput 2011, 7, 2284–2295. [DOI] [PubMed] [Google Scholar]
- (48).Jorgensen WL; Chandrasekhar J; Madura JD; Impey RW; Klein ML Comparison of simple potential functions for simulating liquid water. J. Chem. Phys 1983, 79, 926–935. [Google Scholar]
- (49).Kalé L; Skeel R; Bhandarkar M; Brunner R; Gursoy A; Krawetz N; Phillips J; Shinozaki A; Varadarajan K; Schulten K NAMD2: Greater Scalability for Parallel Molecular Dynamics. J. Comput. Phys 1999, 151, 283–312. [Google Scholar]
- (50).Brooks BR; Brooks CL 3rd; Mackerell AD Jr.; Nilsson L; Petrella RJ; Roux B; Won Y; Archontis G; Bartels C; Boresch S; Caflisch A; Caves L; Cui Q; Dinner AR; Feig M; Fischer S; Gao J; Hodoscek M; Im W; Kuczera K; Lazaridis T; Ma J; Ovchinnikov V; Paci E; Pastor RW; Post CB; Pu JZ; Schaefer M; Tidor B; Venable RM; Woodcock HL; Wu X; Yang W; York DM; Karplus M CHARMM: the biomolecular simulation program. J. Comput. Chem 2009, 30, 1545–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Essmann U; Perera L; Berkowitz ML; Darden T; Lee H; Pedersen LG A smooth particle mesh Ewald method. J. Chem. Phys 1995, 103, 8577–8593. [Google Scholar]
- (52).Ertl P Molecular structure input on the web. J. Cheminf 2010, 2, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Xu XM; Ma SF; Feng ZW; Hu GX; Wang LR; Xie XQ Chemogenomics knowledgebase and systems pharmacology for hallucinogen target identification-Salvinorin A as a case study. J. Mol. Graphics Modell 2016, 70, 284–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Li H; Leung K-S; Wong M-H idock: A multithreaded virtual screening tool for flexible ligand docking. In 2012 IEEE Symposium on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB); IEEE: 2012; pp 77–84. [Google Scholar]
- (55).Cox DR The Regression-Analysis of Binary Sequences. J. Roy Stat Soc. B 1958, 20, 215–242. [Google Scholar]
- (56).Cortes C; Vapnik V Support-Vector Networks. Mach. Learn 1995, 20, 273. [Google Scholar]
- (57).Ho TK The random subspace method for constructing decision forests. IEEE Trans. Pattern Anal. Mahine Intell 1998, 20, 832–844. [Google Scholar]
- (58).Burden F; Winkler D Bayesian regularization of neural networks. Methods Mol. Biol 2008, 458, 23. [DOI] [PubMed] [Google Scholar]
- (59).Ma C; Wang L; Xie X-Q Modeling, Ligand classifier of adaptively boosting ensemble decision stumps (LiCABEDS) and its application on modeling ligand functionality for 5HT-subtype GPCR families. J. Chem. Inf. Model 2011, 51, 521−531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Ma C; Wang L; Yang P; Myint KZ; Xie X-Q Modeling, LiCABEDS II. Modeling of ligand selectivity for G-protein-coupled cannabinoid receptors. J. Chem. Inf. Model 2013, 53, 11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Zhao YH; Abraham MH; Ibrahim A; Fish PV; Cole S; Lewis ML; de Groot MJ; Reynolds DP Modeling, Predicting penetration across the blood-brain barrier from simple descriptors and fragmentation schemes. J. Chem. Inf. Model 2007, 47, 170–175. [DOI] [PubMed] [Google Scholar]
- (62).Kousik SM; Napier TC; Carvey PM The effects of psychostimulant drugs on blood brain barrier function and neuro-inflammation. Front. Pharmacol 2012, 3, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Bienfait B; Ertl P JSME: a free molecule editor in JavaScript. J. Cheminf 2013, 5, 24–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).O’Boyle NM; Banck M; James CA; Morley C; Vandermeersch T; Hutchison GR Open Babel: An open chemical toolbox. J. Cheminf 2011, 3, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Rose AS; Bradley AR; Valasatava Y; Duarte JM; Prlić A; Rose PW NGL viewer: web-based molecular graphics for large complexes. Bioinformatics 2018, 34, 3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Rose AS; Hildebrand PW NGL Viewer: a web application for molecular visualization. Nucleic Acids Res 2015, 43, W576–W579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Chou K-C; Shen H-B Recent advances in developing web-servers for predicting protein attributes. Nat. Sci 2009, 1, 63. [Google Scholar]
- (68).Xiao X; Wang P; Chou KC GPCR CA: A cellular automaton image approach for predicting G protein−coupled receptor functional classes. J. Comput. Chem 2009, 30, 1414–1423. [DOI] [PubMed] [Google Scholar]
- (69).Xiao X; Min J-L; Lin W-Z; Liu Z; Cheng X; Chou K-C Dynamics, iDrug-Target: predicting the interactions between drug compounds and target proteins in cellular networking via benchmark dataset optimization approach. J. Biomol. Struct. Dyn 2015, 33, 2221–2233. [DOI] [PubMed] [Google Scholar]
- (70).Liu B; Wang S; Long R; Chou K-C iRSpot-EL: identify recombination spots with an ensemble learning approach. Bioinformatics 2017, 33, 35–41. [DOI] [PubMed] [Google Scholar]
- (71).Chou K-C Impacts of bioinformatics to medicinal chemistry. Med. Chem 2015, 11, 218–234. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.