Abstract
IgA is prominently secreted at mucosal surfaces and coats a fraction of the commensal microbiota, a process that is critical for intestinal homeostasis. However, the mechanisms of IgA induction and the molecular targets of these antibodies remain poorly understood, particularly in humans. Here we demonstrate that microbiota from a subset of human individuals encode two protein ‘superantigens’ expressed on the surface of commensal bacteria of the family Lachnospiraceae such as Ruminococcus gnavus that bind IgA variable regions and stimulate potent IgA responses in mice. These superantigens stimulate B cells expressing human VH3 or murine VH5/6/7 variable regions and subsequently bind their antibodies, allowing these organisms to become highly coated with IgA in vivo. These findings demonstrate a previously unappreciated role for commensal superantigens in host-microbiota interactions. Furthermore, as superantigen-expressing strains show a strikingly uneven distribution across human populations, they should be systematically considered in studies evaluating human B cell responses and microbiota during homeostasis and disease.
One Sentence Summary:
Identification of bacterial superantigens in the human intestinal microbiota that induce and bind IgA.
Introduction
At mammalian mucosal surfaces, a homeostatic barrier consisting of immunoglobulin A (IgA) antibodies, mucus, and antimicrobial peptides serves as a first line of defense against a complex array of antigens from the commensal microbiota, diet, and occasional enteric pathogens (1). IgA is produced by plasma cells (PCs) that reside predominantly in the small intestinal (SI) lamina propria (LP), a large fraction of which notably arise in the absence of inflammation or immunization. These cells differentiate via T cell-dependent or –independent mechanisms in mucosa-associated secondary lymphoid tissues such as Peyer’s patches (PPs) or mesenteric lymph nodes (mLNs) (2–4). However, despite their abundance, the specificity and function of IgA antibodies remain poorly understood.
The specificity of IgA has been investigated using bacterial flow cytometry and 16S ribosomal RNA (rRNA) gene sequencing to visualize and identify members of the microbiota that are endogenously coated with IgA. These studies have revealed that a taxonomically distinct subset of microbiota in humans and mice is coated with IgA in vivo, whereas other microbes are not (2, 5–9). Notably, IgA-coated microbiota have been associated with inflammation and disease in humans with inflammatory bowel diseases or kwashiorkor malnutrition (5, 8, 10). Analysis of monoclonal antibodies (mAbs) derived from single murine SI IgA PCs has demonstrated that these cells typically produce polyreactive antibodies that individually bind multiple microbial taxa (11). Individual polyreactive IgA antibodies can bind to multiple structurally diverse antigens including lipopolysaccharides, flagellins, DNA, and bacterial glycans in vitro (11). However, the microbiota-derived antigens recognized by IgA in vivo remain unknown. Moreover, microbial communities across human populations are exceptionally diverse (12), and it remains possible that distinct patterns of IgA reactivity might be observed in individuals with divergent microbial communities.
The functional consequences of IgA binding to microbiota remain poorly understood, and generalizable effects on commensal fitness in vivo have not been documented. While numerous hypotheses have suggested that IgA may have detrimental effects on microbes by neutralizing surface determinants, promoting agglutination or enchained growth, altering gene expression, or by various other mechanisms (13–16), the constitutive presence of IgA-coated commensals at steady state suggests that any negative effects of IgA binding are not generally sufficient to drive extinction. By contrast, one recent study suggested that Bacteroides fragilis might express certain polysaccharides that attract IgA antibodies to enhance its colonization (17); however, the extent to which this phenomenon reflects active attraction of antibodies versus secondary changes in capsular composition that alter antibody accessibility to surface antigens remains unclear. Furthermore, members of the Bacteroides are not major targets of IgA in vivo (2). Thus, it remains unknown whether commensals might express factors that allow them to attract or evade IgA antibodies. Identification of such factors might shed critical light on the enigmatic functions of IgA.
Whereas most microbial antigens stimulate only a tiny fraction of lymphocytes bearing specific B or T cell receptors, certain pathogenic microorganisms express superantigens that can stimulate large fractions of lymphocytes by binding to particular variable region gene families. While numerous T cell superantigens have been characterized (18), only a handful of B cell superantigens have been described including staphylococcal protein A (SpA) and peptrostreptococcal protein L (Protein L) (19). Due to their highly potent immunostimulatory properties, these proteins have been associated exclusively with pathogenic organisms. Although Staphylococcus aureus can colonize the human nasopharynx and gastrointestinal tract, this represents the key risk factor for invasive disease (20, 21), enabling this organism to become one of the deadliest known pathogens (22). Expression of the SpA superantigen is required for S. aureus persistence and virulence owing to its ability to stimulate non-specific antibody responses and block the effector functions of the IgG Fcγ region (23–26). Notably, however, superantigens expressed by indigenous commensal, non-pathogenic bacteria have not been identified.
Here we report the identification and characterization of a new class of B cell superantigens expressed by commensal bacteria of the family Lachnospiraceae that induce and bind a substantial fraction of IgA antibodies, thereby profoundly influencing intestinal B cell responses and IgA coating of commensals. These organisms and their superantigens are widespread, yet strikingly unevenly distributed across and within human populations around the world. Thus, superantigen-mediated stimulation and attraction of host IgA antibodies may represent a major mechanism of host-microbiota interaction in humans.
Results
A subset of humans harbor microbiota with B cell superantigen-type properties
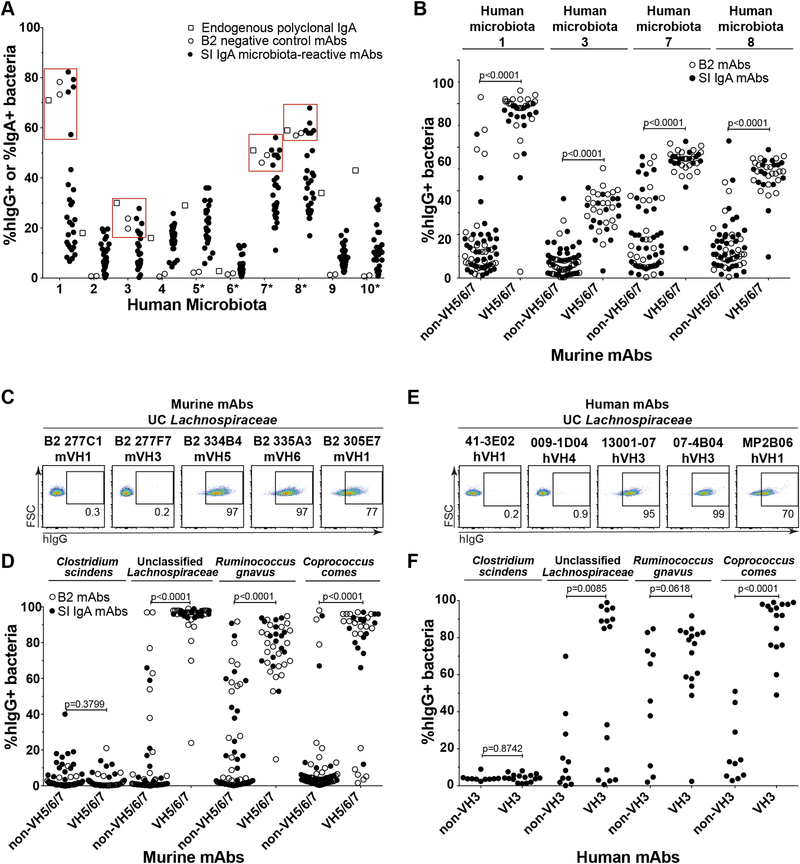
Our previous study of antibodies cloned from single murine SI LP IgA PCs indicated that these mAbs are commonly polyreactive and individually bind multiple bacterial taxa to coat a broad but defined subset of microbiota (11). To extend these observations, we tested whether murine IgA-derived mAbs might also bind human microbiota. We screened a panel of ~27 microbiota-reactive SI IgA mAbs, as well as two non-reactive negative control mAbs from naïve B2 cells (Data file S1), by bacterial flow cytometry against fecal microbiota from ten human infants that had been engrafted into germ-free mice as part of a separate study (27). We found that six out of ten individuals showed a reactivity pattern consistent with that observed in mice, as expected, indicating that polyreactive murine IgA antibodies indeed bind human microbiota (Fig. 1A). However, four of ten individuals showed an atypical pattern in which the two negative control mAbs as well as a subset of SI IgA mAbs bound to microbiota with very high intensity (Fig. 1A). Notably, the mAb-binding microbes from these individuals were also highly coated with endogenous polyclonal IgA (Fig. 1A). While our cohort included both healthy and cow’s milk allergic infant donors, this binding property did not segregate with allergic status (Fig 1A). To further characterize this unusual pattern of reactivity, we examined binding of a larger panel of 53 SI IgA and 47 naïve B2-derived mAbs against these four individuals’ microbiota (Data file S1). This analysis revealed that virtually all mAbs that expressed VH5, VH6, or VH7 (VH5/6/7)-family variable regions, regardless of their SI IgA or naïve B2 origin, bound strongly to these individuals’ microbiota (Fig. 1B). This binding pattern was reminiscent of superantigens such as SpA which, in addition to antibody Fc regions, binds particular variable region gene families irrespective of other variable elements (24, 28). In contrast to human microbiota, we did not observe this superantigen-type binding in various mouse strains from several different colonies (fig. S1A), although it remains possible that the relevant microbes might be present in other strains or colonies of mice not examined here. These data indicate that a subset of humans harbor microbiota that exhibit superantigen-type binding to antibodies expressing murine VH5/6/7 variable regions.
Fig. 1. A subset of humans harbor microbiota with superantigen-type binding to monoclonal antibodies expressing murine VH5/6/7 and human VH3 variable regions.
(A) Bacterial flow cytometry analysis indicating the percent of fecal bacteria from ten human donors grafted into germ-free mice that were coated with endogenous polyclonal IgA (open squares), or various negative control B2 mAbs (open circles) or microbiota-reactive SI IgA mAbs (closed circles) expressed with a human IgG1 backbone and detected with anti-hIgG reagents. Each circle represents a distinct mAb. Red squares highlight the four individuals with superantigen-type reactivity. Asterisks denote microbiota from cow’s milk allergic individuals, and the remaining samples were from healthy individuals. Data compiled from four independent experiments. (B) Bacterial flow cytometry analysis of the four human individuals’ microbiota from panel 1A that showed superantigen-type reactivity with a larger panel of 53 murine SI IgA-derived (closed circles) or 47 naïve B2-derived mAbs (open circles), grouped by heavy chain variable gene usage as indicated. (C) Representative flow cytometry plots and (D) Summary of bacterial flow cytometry analysis of indicated strains cultured in vitro and stained with 53 SI IgA or 47 B2-derived mAbs, grouped by heavy chain variable gene usage as indicated. Data compiled from three independent experiments. (E) Representative flow cytometry plots and (F) Summary bacterial flow cytometry analysis of indicated strains cultured in vitro and stained with 26 fully human anti-influenza mAbs, grouped by heavy chain variable gene usage as indicated. Data compiled from three independent experiments. P values in panels (B-F) calculated by unpaired t test.
To further characterize the nature of this atypical pattern of reactivity, we purified superantigen-expressing microbiota from one individual using a VH5 mAb (11), and isolated a strain of unclassified (UC) Lachnospiraceae, a member of the phylum Firmicutes and order Clostridiales, in pure culture. This strain showed strong binding to mAbs expressing VH5/6/7 variable regions in vitro (Fig. 1C–D). By contrast, non-VH5/6/7 mAbs generally lacked this reactivity, although a few appeared to bind with somewhat reduced intensity (Fig. 1C–D). Based on the phylogeny of this organism, we obtained and screened nine related strains for superantigen-type reactivity (fig. S1B). We identified two additional strains within the family Lachnospiraceae that showed highly similar superantigen-type binding patterns to the original isolate: Ruminococcus gnavus and Coprococcus comes (Fig. 1D and S1B). By contrast, several other Lachnospiraceae members including Clostridium scindens, Clostridium nexile, and Dorea formicigenerans lacked this binding pattern, as did all members of the Firmicutes analyzed outside of this family (Fig. 1D and S1B). These data indicate that expression of antibody-binding superantigens is frequent but not ubiquitous among different strains of the family Lachnospiraceae.
As the superantigen-type binding was identified in human but not mouse microbiota (Fig. 1A and S1A), we reasoned that the superantigens would likely also bind to human antibodies. Indeed, staining with a panel of 26 fully human mAbs derived from anti-influenza responses revealed that strains of Lachnospiraceae exhibited superantigen-type binding to human VH3 variable regions (Fig. 1E–F, Data file S1). Similar to our observations with mouse mAbs, non-VH3 antibodies usually did not show reactivity, although a handful of mAbs bound with lower intensity. We conclude that multiple members of the family Lachnospiraceae express superantigens that bind human VH3 and murine VH5/6/7 variable regions. Of note, these patterns are strikingly similar to the variable region-binding patterns exhibited by known superantigens such as SpA (23, 24, 29, 30). However, these proteins lack the characteristic Fcγ binding activity of SpA, as both reactive and non-reactive mAbs used in our study were expressed on a common human IgG1 backbone (Fig. 1C–F). Thus, Lachnospiraceae superantigens exhibit similar but distinct binding properties from those described for SpA and other known B cell superantigens.
Identification of the superantigens
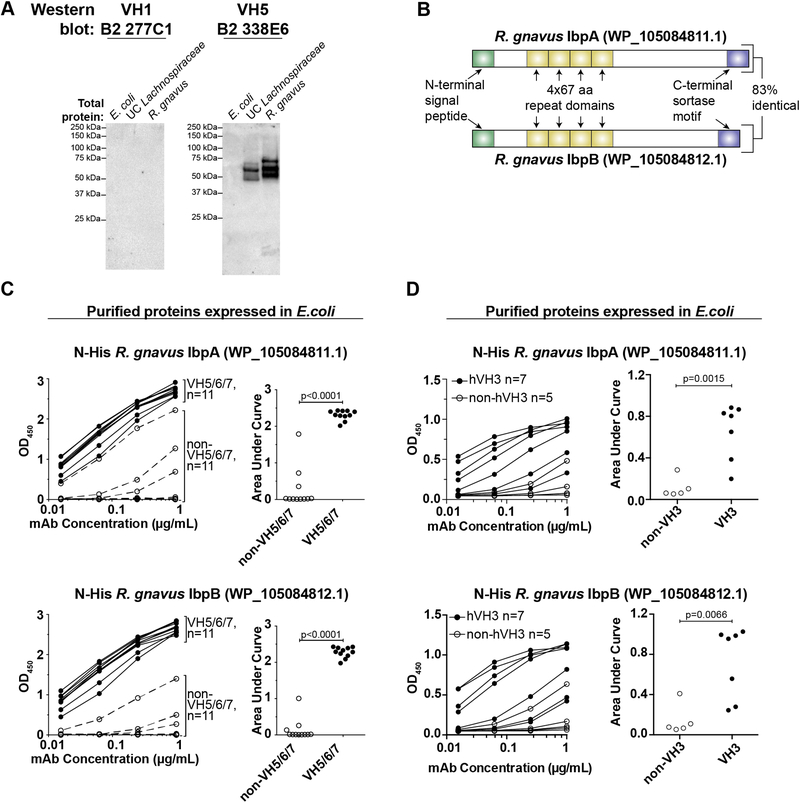
We next sought to identify the superantigen(s). As a first step, we extracted total protein from the UC Lachnospiraceae or R. gnavus superantigen-expressing strains or a control E. coli strain, and probed the lysates by western blot with VH1 or VH5 mAbs. While no reactivity was detected with the VH1 mAb, as expected, the VH5 mAb identified three reactive bands of 50–75 kDa in the superantigen-expressing strains (Fig. 2A). We reasoned that the superantigens would likely be cell wall-anchored proteins, as our flow cytometry analyses suggested surface localization (Fig. 1C–F). Cell wall-targeted proteins in gram-positive bacteria possess well-characterized signals that direct sorting to this compartment, namely an N-terminal signal peptide and a C-terminal LPXTG sortase motif (31). Additionally, all known B cell superantigens including SpA and Protein L contain a series of 50–100 amino acid repeat domains that bind immunoglobulins and allow potent cross-linking of receptors on the B cell surface (23). By computational analysis of the publicly available R. gnavus genome sequenced for the human microbiome project ((32); GenBank NZ_AAYG00000000.2), we identified ~17 predicted cell wall-targeted proteins. However, none of these proteins possessed a repeat-domain structure consistent with known superantigens. Further, cloning each of these proteins into E. coli followed by expression and purification with an N-terminal His tag failed to identify any factors that bound to VH5/6/7-family mAbs. Moreover, immunoprecipitation from R. gnavus lysates with a VH5 mAb followed by mass spectrometry analysis failed to identify any likely candidates or any proteins with predicted cell surface localization (fig. S2A–B).
Fig. 2. Identification of the superantigens.
(A) Western blot analysis of total protein lysates from indicated strains probed with non-superantigen-reactive VH1 mAb 277C1 or superantigen-reactive VH5 mAb 338E6. Representative of >3 independent experiments. (B) Diagram of the two superantigens. aa, amino acid. (C) ELISA analysis of purified R. gnavus proteins expressed in E. coli with an N-terminal His tag. Proteins were coated on plates and probed for reactivity against the indicated dose titration of 11 VH5/6/7 or 11 non-VH5/6/7 mAbs from naïve B2 cells, as indicated (left panels) or (D) against 7 human VH3 and 5 non-VH3 antibodies. Right panels summarize the area under the curve for each antibody shown in the left panels. Representative of two independent experiments. P values calculated by unpaired t test.
During the course of these studies, a new R. gnavus genome was published (33), which resolved a number of additional proteins that were absent from the 2007 genome by utilizing Pacific Biosciences’ long-read sequencing platform (34). Among these we found two uncharacterized ‘hypothetical’ proteins that bore features of known superantigens, including an N-terminal signal peptide, C-terminal sortase motif, four repeat domains of 67 amino acids each, and a predicted molecular weight consistent with that observed by western blot (Fig. 2B; NCBI reference sequences WP_105084811.1 and WP_105084812.1). Reanalysis of mass spectrometry data against this new genome indicated that the two proteins were highly enriched after VH5 mAb immunoprecipitation (fig. S2B). The two proteins were located directly adjacent to one another in the genome and showed substantial homology (Fig. 2B; ~83% sequence identity), suggesting that they arose by gene duplication. Notably, these proteins had no sequence homology to previously characterized superantigens or other known proteins.
To test whether these factors encode the mAb-binding superantigens, we cloned and expressed them with an N-terminal His tag in E. coli and analyzed the recombinant purified proteins by western blot and enzyme-linked immunosorbent assay (ELISA). Both proteins bound strongly to mAbs expressing murine VH5/6/7 or human VH3 variable regions but rarely to mAbs expressing other variable regions, and they closely recapitulated the binding patterns observed by flow cytometry against whole bacteria (Fig. 2C–D, S2C, 1C). These two proteins therefore account for the two lower molecular weight bands observed by western blot analysis of total lysates (Fig. 2A). The upper band likely represents a mobility shift of the same proteins due to residual attached peptidoglycan, a common feature of sortase-anchored surface proteins (35), as mass spectrometry analysis of this band demonstrated the presence of the two superantigens but no other putative surface proteins (fig. S2B). Based on these analyses, we have designated these proteins as immunoglobulin binding protein A (IbpA; WP_105084811.1) and IbpB (WP_105084812.1). We conclude that R. gnavus expresses IbpA and IbpB, two homologous immunoglobulin-binding superantigens, on its cell surface.
IbpA and IbpB stimulate murine and human B cells in vitro
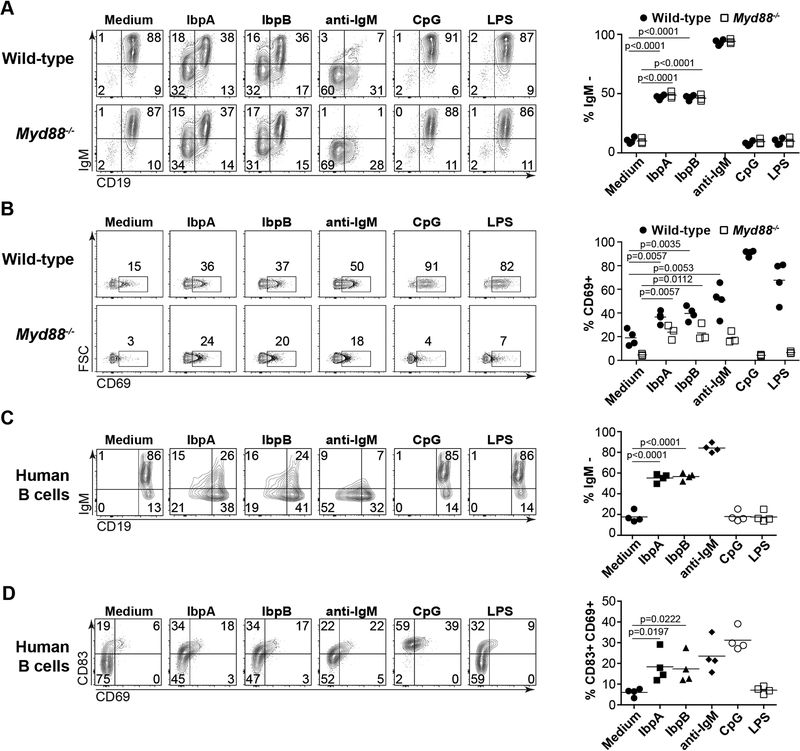
We next sought to determine whether IbpA and IbpB were capable of directly stimulating B cells in vitro. We found that a fraction of about 30% of murine B cells downregulated surface IgM and CD19 (Fig. 3A) and upregulated CD69 (Fig. 3B) after six hours of incubation in vitro with plate-bound IbpA or IbpB, consistent with potent BCR-mediated cellular activation, and similar to that found in a majority of B cells after anti-IgM stimulation. To further exclude the possibility that this activation resulted in part from contamination (e.g. LPS) during protein purification after expression in E. coli, we assessed the ability of IbpA and IbpB to activate B cells isolated from Myd88−/− mice that lack the ability to respond to LPS. IbpA and IbpB activated Myd88−/− B cells to a similar extent as wild-type B cells (Fig. 3A–B). In contrast to Ibps, purified LPS or CpG DNA upregulated CD69 but failed to downregulate IgM and CD19 and this response was lost in Myd88−/− cells, as expected (Fig. 3A–B). Direct quantification of LPS contamination in Ibp protein preparations revealed a concentration of less than 5 EU/mL. We conclude that IbpA and IbpB can directly activate a fraction of murine B cells in vitro, likely by binding the BCR.
Fig. 3: IbpA and IbpB activate murine and human B cells in vitro.
(A) Representative flow cytometry analyses and summary plots depicting purified murine B cells from wild-type or Myd88−/− mice 6 hours after in vitro incubation with indicated stimuli and analyzed for surface expression of CD19 and IgM or (B) CD69. Data pooled from two independent experiments. (C) Representative flow cytometry analyses and summary plots depicting purified human B cells 6 hours after in vitro incubation with indicated stimuli and analyzed for surface expression of CD19 and IgM or (D) CD83 and CD69. Each data point represents one human sample. Samples were obtained from three individuals and cells from one individual were isolated and analyzed in two independent experiments. Data pooled from two independent experiments. P values calculated by unpaired t test.
Using a similar assay, we next assessed whether IbpA and IbpB could activate human B cells purified from the peripheral blood of three individuals. IbpA and IbpB individually activated a large fraction of human B cells in all individuals, as indicated by downregulation of surface IgM and CD19 (Fig. 3C) and upregulation of CD83 and CD69 (Fig. 3D) after six hours of stimulation in vitro. Purified LPS or CpG DNA failed to elicit a similar pattern of response, as expected (Fig. 3C–D). Together, these data suggest that both IbpA and IbpB can stimulate a significant fraction of B cells in both mice and humans in vitro.
Superantigen-expressing strains induce and bind IgA antibodies in vivo
To determine whether these factors function as superantigens in vivo, we examined germ-free (GF) controls or three groups of gnotobiotic mice colonized with: 1) A control strain of Proteobacteria (UC Pseudomonadaceae) isolated from the fecal IgA+ fraction of an SPF mouse, which is bound by polyreactive IgA antibodies in vivo but lacks superantigen-type binding activity (11); 2) The UC Lachnospiraceae superantigen-expressing strain; and 3) The R. gnavus superantigen-expressing strain. Four weeks after colonization, fecal bacteria from all three groups were coated with IgA in vivo, as expected (fig. S3A). mAb staining demonstrated that fecal bacteria from mice colonized with the superantigen-expressing strains but not the UC Pseudomonadaceae bound a VH5 mAb, as expected, whereas none of the strains bound a control VH1 mAb (fig. S3B). In addition, bacteria from all three groups were bound to a lesser extent by a polyreactive VH3 antibody (fig. S3B). These data indicate that the superantigen-expressing strains can become IgA-coated by both superantigen-type binding interactions as well as low-affinity natural polyreactive antibodies, although the relative contributions of each to their IgA coating in vivo remain unclear.
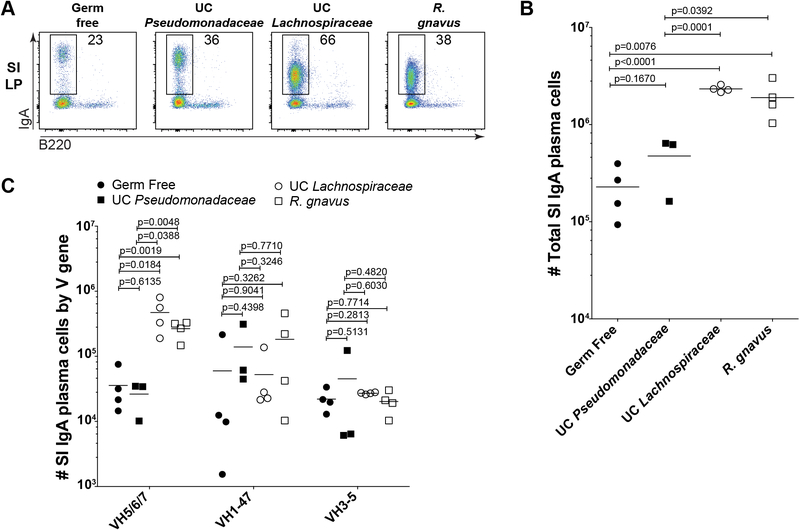
Strikingly, recipients of both superantigen-expressing strains showed a 5-fold and 10-fold expansion of SI IgA PCs over recipients of UC Pseudomonadaceae or GF controls, respectively (Fig. 4A–B). By contrast, the UC Pseudomonadaceae-colonized controls showed only a minor induction over GF (Fig. 4A–B). Intriguingly, SI IgA PCs in mice colonized with superantigen-expressing strains also showed reduced surface IgA staining relative to GF or UC Pseudomonadaceae-colonized controls (Fig. 4A). While the mechanisms underlying this decreased surface IgA expression remain unclear, this observation further suggests that these commensal strains elicit different immune responses. In line with the observed increase in SI IgA PCs, luminal free IgA levels were dramatically increased in the SI and colon of mice colonized with either superantigen-expressing strain relative to UC Pseudomonadaceae or GF controls (fig. S3C). Colonic LP IgA PCs showed a modest increase over GF in all groups that was not statistically significant (fig. S3D). The observed increase in SI IgA PCs could be associated with preferential induction of T cell-dependent (TD) IgA responses by these organisms rather than superantigen-driven interactions (7, 11). However, repertoire analysis indicated that SI IgA PCs in all groups showed very low levels of somatic hypermutation (fig. S3E). Further, mLN or PP germinal center B cells, T follicular helper cells, IgA class-switched B cells, or CCR6+ IgD+ precursors were similar in all groups to GF controls (fig. S3F–G) (36). While these data suggest that TI responses may be predominantly involved in the response to these superantigens, they do not rule out the possibility that both TI and TD responses are involved. Notably, the responses elicited by these organisms appear distinct from those induced by segmented filamentous bacteria, a strong inducer of IgA that lacks superantigens, in which preferential stimulation of TD responses was reported (37). We conclude that superantigen-expressing strains stimulate substantial SI IgA PC accumulation in monocolonized mice.
Fig. 4. Superantigen-expressing strains stimulate and attract IgA in vivo.
(A) Representative flow cytometry plots gated on FSChi Lineage- cells or (B) absolute number summaries of SI LP IgA PCs from GF or gnotobiotic mice four weeks after monocolonization with indicated bacterial strains. Data compiled from three independent experiments. (C) Absolute number of SI IgA PCs expressing indicated variable gene families. Calculated from the total cell number shown in panel (B) multiplied by the percent of the repertoire expressing indicated variable region genes as determined by DNA sequencing. One-way ANOVA p values were 0.0048 for VH5/6/7, 0.5156 for VH1–47, and 0.6958 for VH3–5. P values in panels (B-C) calculated by unpaired t test.
To determine whether the expanded SI IgAs preferentially included superantigen-reactive variable regions, we analyzed the immunoglobulin repertoire of sorted SI IgA PCs from each group of mice. Strikingly, both superantigen-expressing strains stimulated substantial accumulation of IgA PCs expressing VH5/6/7 gene families (Fig. 4C). By contrast, cells expressing VH1–47 or VH3–5 gene families were detected at similar numbers in all groups (Fig. 4C), indicating that, as expected, the observed IgA induction preferentially involved the VH5/6/7 gene families. We conclude that superantigen-expressing UC Lachnospiraceae and R. gnavus polyclonally stimulate differentiation of IgA PCs expressing VH5/6/7 variable regions and bind their secreted antibodies, thereby increasing both free and bacteria-bound IgA in vivo.
R. gnavus and its superantigens are prevalent across human metagenomes
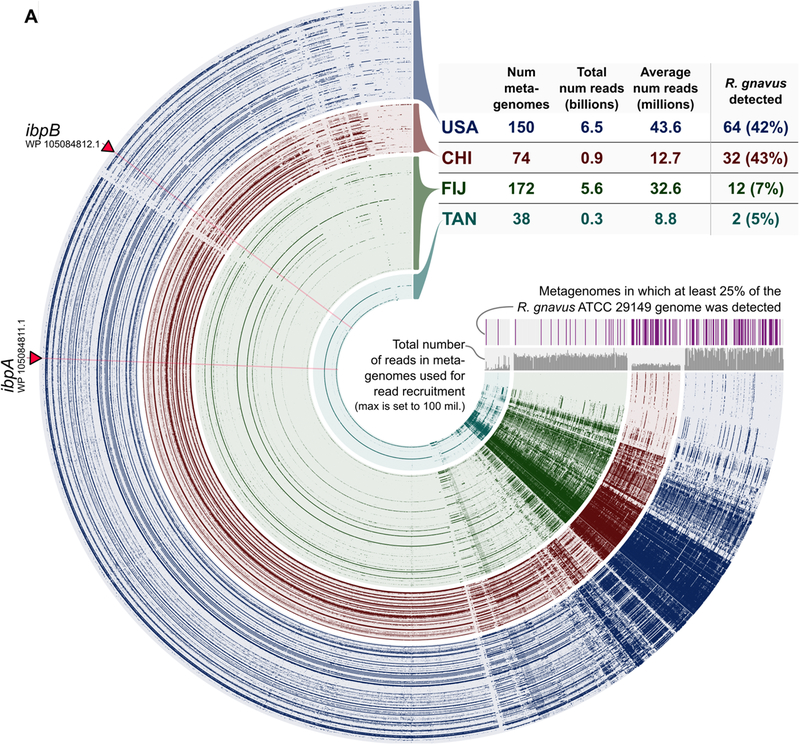
We previously observed microbiota with superantigen-type binding in four of ten human individuals (Fig. 1A–B); however, this analysis represented a limited cohort whose microbial communities were engrafted into GF mice. Having identified the genes encoding the superantigens, we therefore sought to determine their distribution across 434 healthy human metagenomes using publicly available data (Fig. 5, Table S1, Data file S2) (38–41). Metagenomic read recruitment analysis revealed that R. gnavus and its superantigens were present in 42% of individuals from the USA and 43% from China, but rare among those from Fiji or Tanzania (5% and 9%, respectively; Fig. 5, Table S1). Importantly, the two superantigens appeared to be core genes of R. gnavus, as both were detected whenever R. gnavus was present but not in its absence and their coverage matched the rest of the genome (Fig. 5, Table S1, Data file S3). These results likely underestimate the true prevalence of this class of superantigens, as related superantigen-expressing microbes such as the UC Lachnospiraceae and Coprococcus comes identified here (Fig. 1C–F) may also contribute. However, a characterization of their prevalence will only be possible when full genome sequences for these strains become available. We conclude that superantigen-expressing strains of commensal bacteria are widespread, likely present in billions of individuals worldwide, but variably distributed within and between human populations.
Fig. 5. Distribution of R. gnavus and its superantigens across human metagenomes.
(A) Dendrogram alignment of the R. gnavus ATCC 29149 genome to 434 human metagenomes. Each spoke represents one gene in the R. gnavus genome and each layer represents an individual human metagenome. The two superantigen genes are labeled. Intensity represents coverage of the open reading frame in the metagenome.
Discussion
We describe here the first B cell superantigens expressed by commensal bacteria from the human gut microbiota. These factors are widespread but unevenly distributed across human populations and therefore may represent a common and fundamental mechanism of host-microbe interaction in the intestinal mucosa. We identified the superantigenic factors at the biochemical and molecular level, and demonstrated that they represent a novel class of proteins unrelated to other known B cell superantigens. Our data suggest that these proteins induce and bind IgA antibodies of the human VH3 and murine VH5/6/7 gene families with high affinity in vivo.
Ruminococcus was reported to be the defining taxon of one of three ‘enterotypes’ that characterize human microbiota diversity across individuals (12). While the precise classification of these enterotypes remains somewhat controversial (42), it is clear that many bacterial species including Ruminococcus are unevenly distributed across human individuals in the same geographic area and between populations in different locations (43). While distinct microbial communities have been associated with a variety of diseases including obesity, diabetes, autoimmunity, cancer, and inflammatory bowel diseases (44), in most cases it remains unclear how these communities mechanistically contribute to pathology in these complex disorders. Notably, our findings suggest that the Ruminococcus enterotype is associated with a distinct immunological phenotype characterized by the presence of B cell superantigens. As the superantigen-targeted VH3 gene family is expressed by 50–60% of human B cells (24), it is possible that these factors could significantly affect B cell responses in carriers and therefore alter the course of various diseases involving B cells and/or antibodies. Notably, a number of recent studies have identified enrichment of R. gnavus in the microbiota of humans with allergic disease (45), lupus nephritis (46), spondyloarthritis (47), and inflammatory bowel disease (48, 49). Whether these relationships are correlative or causative remains unknown; however, our results suggest that detailed further investigations are warranted.
Recent studies have suggested that IgA coating may mark disease-associated members of the microbiota in inflammatory bowel diseases, autoimmunity, and kwashiorkor malnutrition (5, 8, 10). Although bona fide pathogens are likely to become IgA-coated in vivo, such studies are complicated by the presence of a variety of innocuous commensals that are also coated with IgA during homeostasis (2). Our findings emphasize that it will be essential to determine the presence of superantigen-expressing Lachnospiraceae strains in future studies of human IgA-coated microbiota, as these organisms may alter not only their own IgA coating but also the magnitude of IgA responses to other bacteria, particularly those that can be bound by polyreactive human VH3 antibodies (11).
Further studies of the impact of B cell superantigens on host and microbiota are warranted and will depend on the establishment of methods for genetic manipulation of these commensal organisms. In particular, it remains unclear why these organisms would express superantigens. Intriguingly, while bacterial genomes are exceptionally plastic and readily adapt in order to maximize fitness (50), our analysis of wild R. gnavus strains across hundreds of human metagenomes indicated that the ibp superantigens are core components of the R. gnavus genome. Moreover, the presence of two proteins with highly similar functions suggests strong evolutionary pressure to express these factors. However, the precise mechanisms by which these superantigens impact the fitness of the commensals and/or the host remain unknown, and it is possible that they might have either beneficial or detrimental effects. For example, these organisms might attract IgA in order to promote intra- or inter-species agglutination, which could facilitate cooperative physiology (51), horizontal gene transfer (52), or antagonistic interactions via Type VI secretion or other mechanisms (53). In addition, superantigen-mediated stimulation could amplify the host IgA response, which could lead to either enhanced or diminished ability to mount protective immune responses to pathogens. Future studies should assess whether Lachnospiraceae superantigens alter the human IgA repertoire in particular physiological conditions, such as during spontaneous colonization in infants, or in dysbiotic conditions during which the abundance of these strains is altered.
Our study has several important limitations. First, although we identified the superantigenic factors expressed by R. gnavus, the nature of the superantigenic reactivity observed in other strains of Lachnospiraceae remains unknown and it is possible that this is achieved through either conserved or divergent mechanisms. Second, although we documented binding and activation of human B cells in vitro, we did not prove that these factors stimulate or modulate human immunity in vivo and this remains an important area of future investigation. Lastly, although we frequently identified the presence of the superantigen genes in human metagenomes, we did not determine whether these genes are expressed and functional in all individuals.
In summary, we report that commensal bacteria of the family Lachnospiraceae express potent, conserved B cell superantigens that stimulate IgA production and bacterial coating. Our observations are distinct from known mechanisms of IgA responses to commensals including polyreactive recognition by natural IgA antibodies (11) or high-affinity responses to pathogens or vaccines (54), and highlight an unexpected mechanism of host-commensal interaction previously associated only with pathogens. Our findings are in apparent contrast with well-known strategies for evasion of antibodies by bacteria (55), and suggest instead that certain commensal bacteria express factors that attract host antibodies.
Materials and Methods
Study design
The objective of this study was to assess the mechanisms of antibody recognition of the human microbiota and to define the molecular factors that regulate IgA targeting. All data, including outliers, were reported in relevant figures. Sample sizes were determined by reagent availability and were not pre-specified. Randomization and blinding were not performed. Information regarding experimental replicates is reported in the figure legends.
Mice
All mouse studies were performed according to guidelines approved by the University of Chicago Institutional Animal Care and Use Committee. Myd88−/− mice were purchased from The Jackson Laboratory. For gnotobiotic experiments, mice were housed in isolators in the University of Chicago Gnotobiotic facility. For non-gnotobiotic experiments, mice were housed under specific pathogen-free conditions at the University of Chicago. In most experiments, mice were of the C57BL/6 strain background. In some experiments, fecal samples were collected and analyzed from six week old male mice of C57BL/6, BALB/c, NOD, or C3H strains two days after arrival from The Jackson Laboratory, Taconic Biosciences, or Charles River Laboratories. Colonization of mice with fecal samples from healthy or cow’s milk allergic infants was performed in the University of Chicago Gnotobiotic Animal Research Facility. C3H/HeN mice were colonized with ten individual human infants’ microbiota and maintained in individual gnototobiotic isolators, one per donor as part of a separate study (27). The ten individuals were Italian infants of mixed gender; informed consent was obtained and experiments were approved by the University of Chicago Institutional Review Board. Detailed information on each individual will be reported separately (27). Here, fecal samples from colonized mice were collected and analyzed for mAb reactivity. Experiments involving monocolonized gnotobiotic mice were performed in individual gnotobiotic isolators. Mice were 9–24 weeks old, with ages and genders randomly distributed across each experimental group and were analyzed 4 weeks after colonization.
Bacterial strains and culture
Ruminococcus gnavus (29149), Coprococcus comes (27758), Dorea formicigenerans (27755), Clostridium nexile (27757), Clostridium scindens (35704), and Clostridium ramosum (25582) were purchased from the ATCC. The UC Lachnospiraceae strain was isolated as described below. Listeria monocytogenes was a gift from Bana Jabri and Lactobacillus rhamnosus was a gift from John Alverdy. Microbial strains were grown on Schaedler blood agar (BD) or chopped meat carbohydrate broth (BD) in an anaerobic jar at 37°C with the exception of Candidatus Arthromitus (SFB), which was propagated in monocolonized mice in gnotobiotic isolators and analyzed by staining of fecal samples.
mAb staining of fecal and cultured bacteria
For staining of fecal bacteria, 1–2 fecal pellets were homogenized by adding 1 mL sterile PBS and vortexing horizontally for 5 mins. Debris was pelleted by centrifugation at 400g for 5 mins, and the supernatant was filtered through a sterile 70 μm strainer (Fisher) and transferred to a new 1.5 mL tube. Cells were centrifuged for 5 mins at 8000g to pellet bacteria and the supernatant was discarded. The bacterial pellet was resuspended in 3 mL PBS 0.25% BSA with SYTO BC (1:7500; Life Technologies) for 15 mins on ice.
For staining of cultured bacteria, 5 mL cultures were grown overnight under anaerobic conditions at 37°C and pelleted by centrifugation at 4700g for 15 mins. Cells were resuspended in 1 mL PBS 0.25% BSA with SYTO BC (1:7500) per mL of original culture and incubated for 15 mins on ice.
For staining, 50 μL of bacterial suspension was transferred to a 96-well v-bottom plate. 50 μL 2X mAb master mix was added for a final concentration of 10 μg/mL and incubated for 20 mins on ice. Cells were washed with PBS 0.25% BSA and centrifuged for 15 mins at 4700g. Supernatant was discarded and cells were resuspended in PBS 0.25% BSA 10% normal goat serum (Jackson Immunoresearch) for 10 mins on ice. 50 μL of 2X goat anti-hIgG-biotin (1:400 final concentration; Southern Biotech) and goat anti-mouse IgA-PE (1:800 final concentration; Southern Biotech) was then added and incubated for 20 mins on ice. For staining of cultured bacteria, the anti-mouse IgA reagent was not included. Cells were washed and centrifuged for 15 mins at 4700g and the supernatant was discarded. Cells were resuspended in 100 μL PBS 0.25% BSA with streptavidin-APC (1:800; Biolegend) and incubated for 20 mins on ice. Cells were washed with PBS 0.25% BSA and centrifuged 20 mins at 4700g. Supernatant was discarded and cells were resuspended in 200 μL PBS 0.25% BSA with DAPI (0.1 μg/mL; Life Technologies) prior to flow cytometry. Flow cytometry was performed with a low forward and side scatter threshold on an LSRII cytometer (BD) to facilitate bacterial detection.
For purification and culture of mAb-binding bacteria, fecal pellets from mice colonized with Human Microbiota 1 (Fig. 1A) were homogenized and processed as described above. 250 μL of bacterial suspension was added to a new 1.5 mL tube and 250 μL 2X mAb B2 334B4 in PBS 0.25% BSA was added (final concentration 10 μg/mL) and incubated for 20 mins on ice. Cells were washed and centrifuged for 5 mins at 8000g, then resuspended in 250 μL PBS 0.25% BSA 10% goat serum (Jackson Immunoresearch) for 10 mins on ice. 250 μL of 2X anti-human IgG-biotin (1:400 final concentration) in PBS 0.25% BSA was added and incubated for 20 mins on ice. Cells were washed and centrifuged for 5 mins at 8000g, then resuspended in 500 μL of streptavidin-APC (1:800 final concentration) in PBS 0.25% BSA for 20 mins on ice. Cells were washed and centrifuged for 5 mins at 8000g, then resuspended in 500 μL PBS 0.25% BSA with anti-APC MACS beads (1:50; Miltenyi) for 20 mins on ice. Cells were washed and centrifuged for 5 mins at 8000g, then resuspended in 1 mL PBS 0.2% BSA and run on an autoMACS separator using the posselds program followed by a qrinse. The positive fraction was then run a second time using the posseld2 program. Eluate was serially diluted and plated on Schaedler blood agar (BD) under anaerobic conditions at 37°C. Two days later, individual colonies were restreaked to obtain pure cultures. Two days later, these cultures were screened for VH5/6/7 mAb reactivity using the staining protocol described above.
Bacterial protein extraction
Saturated 5 mL cultures grown in chopped meat carbohydrate broth under anaerobic conditions at 37°C were pelleted by centrifugation at 4700g for 15 mins. The pellet was resuspended in 800 μL lysis buffer from the NoviPure Microbial Protein Kit (Qiagen) and transferred to a bead beating tube. Cells were lysed by bead beating for 10 mins with tubes taped horizontally on a vortex at room temperature. Tubes were centrifuged for one minute at 12,000g and supernatant was transferred to a new tube. Trichloroacetic acid (Sigma) was added to 10% v/v and proteins were precipitated by incubation for 30 mins on ice. Tubes were centrifuged for 10 mins at 16,000g and the supernatant was removed by aspiration. The pellet was washed by resuspension in 1 mL ice cold acetone and was centrifuged 10 mins at 16,000g. Supernatant was removed by aspiration and the pellet was air dried for several seconds. The pellet was then resuspended in 0.5M Tris-HCl 4% SDS with Halt protease inhibitors (Thermo) and was solubilized by incubating for 30 mins at room temperature followed by 3 mins at 100°C. Protein was quantified by nanodrop.
Coomassie gels and Western blots
20 μg of total protein was mixed with 2X Laemmli buffer (Biorad) and 2.5% v/v beta-mercaptoethanol (Sigma) and heated at 70°C for 10 mins. The prep was left to cool for 2–3 mins and was loaded onto a 10% precast polyacrylamide gel (Biorad) and run for 1 hour at 100V.
For Coomassie staining, gels were transferred to nanopure water and incubated for 15 mins. Water was drained and replaced with Coomassie stain (Biorad) for 45 mins at room temperature. Gels were rinsed with water and then destained overnight with Coomassie destaining solution (Biorad).
For Western blot, PVDF membranes (Biorad) were dipped in 100% ethanol then membrane, gel, sponges, and filter paper were incubated in Tris-Glycine 20% methanol (Biorad) for 15 mins at room temperature. Transfer cassettes were assembled and protein was transferred to the membrane with an electrophoretic transfer apparatus (Biorad) at 65V for two hours. After transfer, the membrane was rinsed briefly in TBS-T (Thermo) and blocked in TBS-T 5% milk for 30 mins at room temperature. Membranes were incubated overnight at 4°C with 8 mL mAb at 1 μg/mL in TBS-T 5% milk. The next day, the membrane was rinsed three times for 5 mins each with TBS-T, then incubated with 8 mL goat anti-human IgG-HRP (1:5000; Southern Biotech) in TBS-T 5% milk for 1 hour at room temperature. The membrane was washed three times with 5 mL TBS-T for 5 mins each, then incubated for 5 mins with 7 mL Clarity HRP substrate (Biorad). The blot was removed from solution and imaged with a ChemiDoc system (Biorad).
Superantigen cloning and protein expression in E. coli
DNA from bacterial isolates was prepared using the PureLink Microbiome DNA Purification Kit (Thermo). For expression, proteins were cloned and expressed without their N-terminal signal peptide or C terminal sorting signal. R. gnavus gene ibpA (WP_105084811.1) was purchased as a gBlock from IDT. The ibpB (WP_105084812.1) gene was cloned from genomic DNA using primers tatgcatcatcatcatcatcacgtgGCAGAGCCAGTGGAAAAG and gctcgaatatcatcgatctcgagcgtcaATTTTTCTTCTTATCTTCTTTCTTAGTATCC with an initial denaturation of 98°C for 30s followed by five cycles of 98°C for 10s, 61°C for 30s, and 72°C for 50s. This was followed by 35 cycles of 98°C for 10s and 72°C for 80s. DNA bands were visualized by agarose gel electrophoresis and were cut and purified using a QIAQuick Gel Extraction Kit (Qiagen). Fragments were incubated with EcoRI-digested Champion pET302 N-His vector (Thermo) and assembled by HiFi assembly (NEB). This mixture was transformed into XL10 Gold ultracompetent cells and individual colonies were sequenced with forward primer TAATACGACTCACTATAGGG and reverse primer TAGTTATTGCTCAGCGGTGG. Colonies with appropriate inserts were grown in 35 mL cultures in LB with ampicillin and plasmids purified using the HiSpeed Plasmid Midi Kit (Qiagen). Plasmids were then transformed into NiCo21 (DE3) competent E. coli (NEB) for protein expression. 20 mL cultures were grown overnight in MagicMedia E. coli expression media (Thermo) and His-Tagged proteins were purified under native conditions using the Ni-NTA spin kit (Qiagen). Protein was quantified by nanodrop.
ELISA analysis of mAb reactivity
R. gnavus proteins WP_105084811.1 and WP_105084812.1 were expressed in E. coli and purified as described above. Protein was then coated overnight at room temperature with 10 μg/mL, 50 μL/well in carbonate buffer (Bethyl) onto 96-well ELISA plates (Thermo). The next day, plates were washed four times with TBS-T and incubated one hour with 150 μL/well TBS 1% BSA blocking buffer at room temperature. Plates were washed four times with TBS-T then incubated one hour with mAbs at 1 μg/mL or four 1:4 serial dilutions in blocking buffer (50 μL/well) at room temperature. Plates were washed four times with TBS-T then incubated one hour at room temperature with 75 μL/well goat anti-human IgG-HRP (1:5000; Southern Biotech) diluted in TBS-T 1% BSA. Plates were washed four times with TBS-T then 100 μL/well TMB HRP substrate (Bethyl) was added and incubated for approximately five minutes. The reaction was quenched with 100 μL 0.18 M H2SO4 and the absorbance at 450 nm was read using an ELISA plate reader (Biotek).
Immunoprecipitation and mass spectrometry
mAbs were buffer exchanged into PBS using an Amicon 30K centrifugal filter and linked to agarose beads using the Pierce Direct IP Kit (Thermo). 1000 μg protein lysate was precleared by mixing with Pierce Control Agarose Resin according to the manufacturer’s protocol, and subsequently incubated with mAb-linked beads overnight at 4°C with rotation. The next day, the beads were washed three times with IP/Lysis buffer and once with conditioning buffer as described by the manufacturer. Proteins were eluted in provided glycine pH 2.8 buffer and neutralized with 1M Tris. Eluate from ten immunoprecipitations was pooled and concentrated using a 0.5 mL Amicon 30K ultracentrifugal filter. The concentrate was then processed for SDS-PAGE as described above under Coomassie gels and Western Blots.
For mass spectrometry analysis, a SDS-PAGE gel run as described above was stained using the Pierce Silver Stain for Mass Spectrometry Kit (Thermo) according to the manufacturer’s protocol. Bands were cut from the gel and destained per the manufacturer’s protocol, then placed in 100 μL nanopure H2O and shipped to MS Bioworks, LLC for mass spectrometry analysis. Common contaminants (i.e. keratins or trypsin) were removed from the analysis. Mass spectrometry data were analyzed using Scaffold (Proteome Software).
Human and mouse B cell purification and in vitro stimulation
For purification of untouched murine B cells, splenocytes were incubated with anti-mouse CD43 biotin (S7) and magnetically labeled with Streptavidin MicroBeads (Miltenyi) following the manufacturers recommendations. Cells were separated using an autoMACS Separator with a purity of ≥ 90–95%.
For purification of human B cells, whole blood was carefully overlaid on Ficoll-Paque PLUS density gradient media (GE Healthcare) and centrifuged at room temperature for 30 mins and 400 g without breaks. PBMCs were harvested from the interlayer, washed with PBS and untouched B cells were purified (≥95% purity) using the Human B cell Isolation Kit (STEMCELL Technologies) following the manufacturers recommendations.
B cells were stimulated in 96 well plates for 6 hours at a density of 0.2 ×106 cells/ml in complete RPMI medium supplemented with 10% FCS (Gibco). Final concentrations of soluble stimuli: 100 nM CpG (ODN2006 for human-, ODN1668 for mouse B cells, Invivogen) and 2 μg/ml LPS (Sigma). For stimulation with plate-bound stimuli, high-binding 96 well plates (Corning) were coated with 100 μl of 10 μg/ml IbpA, IbpB and F(ab’)2-goat anti-mouse or F(ab’)2 anti-H IgM (fisher) diluted in carbonyl coating buffer (Bethyl). To minimize endotoxin contamination of IbpA and IbpB produced from NiCo21 (DE3) E. coli lysates, we used High Capacity Endotoxin Removal Spin Columns (Fisher), and monitoring with Chromogenic Endotoxin Quant kit (Fisher) showed < 5 residual EU/mL.
Free IgA ELISAs
Fecal samples were weighed and resuspended at 0.1 mg/mL in PBS with protease inhibitors (Sigma), then homogenized by vortexing horizontally for 5 mins. Debris was pelleted by centrifugation at 400g for 5 mins, and the supernatant was transferred to a new 1.5 mL tube. This tube was centrifuged for 5 mins at 8000g and the supernatant was removed and saved for further analysis at −20°C. For analysis, 96-well ELISA plates (Thermo) were coated with 100 μL/well goat anti-mouse IgA (10 μg/mL; Bethyl) for one hour at room temperature in carbonate buffer (Bethyl). Plates were washed four times with TBS-T, then incubated with 200 μL/well TBS 1% BSA blocking buffer for one hour at room temperature. Plates were then washed four times with TBS-T. Samples (100 μL/well) were analyzed at 1:100 in TBS-T 1% BSA and four consecutive 1:10 serial dilutions and were compared against a standard curve generated using a mouse serum standard (Bethyl). Plates were incubated for one hour at room temperature and were washed four times with TBS-T. 100 μL/well goat anti-mouse IgA-HRP (1:50,000; Bethyl) diluted in TBS-T 1% BSA was added and incubated for one hour at room temperature. Plates were washed four times with TBS-T and then developed with 100 μL/well TMB HRP substrate for approximately 5 mins at room temperature. The colorimetric reaction was quenched with 100 μL/well H2SO4 and the absorbance at 450 nm was read using an ELISA plate reader. Sample concentrations were determined by comparison to the standard curve fit to a four-parameter logistic curve.
Lymphocyte isolation from tissues
Intestines were dissected and fat and Peyer’s patches were removed prior to processing. Intestines were opened by cutting longitudinally and were subsequently cut into ~1 cm pieces and placed into a 50 mL conical tube. Small intestines were cut in half and processed in two separate tubes whereas colons were processed in a single tube. 10 mL pre-warmed RPMI 1% FCS 1 mM EDTA was added to the intestinal pieces and they were incubated for 15 mins at 37°C with shaking. Pieces were collected with a 100 μm cell strainer and the supernatant was filtered into a new 50 mL tube. The pieces were then transferred back into the original 50 mL tube and the strainer was washed and discarded. The intestinal pieces were again incubated with 10 mL mL RPMI 1% FCS 1 mM EDTA for 15 mins at 37°C with shaking and were collected with a 100 μm cell strainer. The supernatant from these first two washes was discarded as it contained predominantly epithelial cells and intraepithelial lymphocytes. Pieces were transferred to the original 50 mL tube and 10 mL pre-warmed RPMI 20% FCS with 0.5 mg/mL collagenase (Roche) and 0.1 mg/mL DNase (Sigma) was added. Pieces were incubated for 30 mins at 37°C with shaking and were collected with a 100 μm cell strainer. The supernatant was collected and the pieces were transferred back into the 50 mL tube. 10 mL pre-warmed RPMI 20% FCS with 0.5 mg/mL collagenase (Roche) and 0.1 mg/mL DNase (Sigma) was added and pieces were again incubated for 30 mins at 37°C with shaking and were collected with the same 100 μm cell strainer. Pieces were mashed using a disposable syringe plunger (BD) and the strainer was washed with 30 mL RPMI. Tubes were centrifuged for 5 mins at 800g and the supernatant was removed by aspiration. Cell pellets from the two SI fractions were combined and they and the one colon fraction were each resuspended in 10 mL 40% Percoll (Sigma) and transferred to a 15 mL conical tube. This tube was centrifuged for 20 mins at 2100 rpm with no brake. Percoll was aspirated and the pellet was washed with 1 mL HBSS 0.25% BSA and centrifuged 5 mins at 800g. The pellet was then resuspended in HBSS 0.25% BSA with anti-CD16/32 (Biolegend) and stained for flow cytometry as described below.
Other tissues (spleens, mLNs, PPs) were dissected and placed in HBSS 0.25% BSA, then transferred to and mashed on a 70 μm cell strainer with the plunger from a disposable 1 mL syringe (BD). The resultant cell suspension was transferred to a new tube and centrifuged for 5 mins at 500g, then resuspended in HBSS 0.25% BSA with Fc block (Biolegend) and stained for flow cytometry as described below.
Lymphocyte flow cytometry
Single cell suspensions were resuspended in HBSS 0.25% BSA with anti-CD16/32 (Fc Block; Biolegend) for 10 mins on ice prior to staining with the following fluorophore or biotin conjugated monoclonal antibodies purchased from Biolegend, eBioscience, or BD unless otherwise indicated (clone in parentheses): B220 (RA3-6B2), CCR6 (29-2L17), CD4 (GK1.5 or RM4-5), CD8 (53–6.7), CD11c (HL3), CD19 (6D5), CD69 (1D4-C5), F4/80 (BM8), Gl7 (GL7), IgD (11–26c.2a), IgM (Il/41), NK1.1 (PK136), TER-119 (TER-119), Tcrb (H57–597), and goat anti-mouse IgA(Southern Biotech). Lineage cocktail for plasma cell staining included CD3, CD4, CD11c, F4/80, NK1.1, Tcrb, and TER-119. Cells were washed and centrifuged for 5 mins at 500g. Supernatant was removed and cells were resuspended in HBSS 0.25% BSA for analysis by flow cytometry using an LSRII cytometer (BD). Cell numbers were quantified using Spherotech counting beads.
Human cells were first incubated with TrueStain FcX (Biolegend) for 20 minutes at 4–8°C and then stained with the following fluorophore-conjugated antibodies: CD19 (HIB1), CD69 (FN50), CD83 (HBI5e) or IgM (MHM-88). In order to avoid cross reactions with monoclonal antibodies used for flow cytometry stainings, in vitro stimulated mouse and human B cells were incubated for 30 min at 4–8°C with high dose human IgG (1mg/ml; Sigma) prior to the addition of fluorophore-conjugated antibodies to block all binding sites of Ibps potentially still bound to the stimulated cells.
IgA repertoire analysis
20,000–60,000 SI IgA PCs (Lineage− B220− IgA+) were sorted from each mouse using a BD AriaII cell sorter. Post-sort purity analysis verified the sorting efficacy of each sample. Cells were centrifuged for 5 mins at 800g, supernatant was removed, and the pellet was resuspended in 1 mL TRIzol (Thermo) for 5 mins at room temperature, then frozen at −80°C for later processing. Samples were thawed on ice and 0.2 mL chloroform was added; samples were shaken vigorously for 20s and then allowed to settle at room temperature for 2–3 mins. Tubes were then centrifuged at 10,000g for 15 mins at 4°C. The aqueous (top) layer was removed with a pipette and transferred to a new sterile RNase-free tube. An equivalent volume of 100% EtOH was added and the sample was mixed. The sample was then loaded onto an RNeasy column (Qiagen) in a collection tube and spun for 15s at 8,000g and flow through was discarded. 700 μL buffer RW1 (Qiagen) was added to the column and it was centrifuged for 15s at 8,000g. The column was transferred to a new collection tube and 500 μL buffer RPE was added and then centrifuged for 15s at 8,000g. Flow through was discarded and 500 μL buffer RPE was added prior to centrifugation at 8,000g for 2 mins. Flow through was discarded and the column was centrifuged for 1 min at 8,000g. The column was transferred to a new 1.5 mL collection tube and 30 μL RNase-free water was added and incubated for 1–2 mins, then centrifuged for 1 min at 8,000g to elute the RNA. cDNA synthesis was performed with the Superscript IV First Strand cDNA synthesis kit (Thermo) using poly-d(T) primers according to the manufacturer’s protocol.
Immunoglobulin heavy chain variable regions were amplified from 2 μL cDNA using Q5 HotStart polymerase (NEB) and forward primer MsVHE GGGAATTCGAGGTGCAGCTGCAGGAGTCTGG and reverse Cα primer GAAAGTTCACGGTGGTTATATCC. Thermal cycling conditions involved an initial denaturation at 98°C for 30s followed by 35 cycles of 98°C for 10s, 64°C for 20s, and 72°C for 20s and a final elongation step of 72°C for 2 mins. PCR fragments were visualized by agarose gel electrophoresis and a ~500 nt band was excised from the gel and purified using the QIAquick gel extraction kit (Qiagen). PCR fragments were cloned into vectors using the NEB PCR cloning kit, transformed into 10-beta competent E. coli (NEB), and plated on LB agar containing ampicillin. Individual colonies were picked, miniprepped, and sequenced using the forward primer ACCTGCCAACCAAAGCGAGAAC. 96 colonies were picked per plate which led to 80–90 sequences per mouse. Sequences were identified and mapped to variable region genes using IMGT (www.imgt.org). For somatic mutation analysis, mutations were identified using IMGT and those found at the beginning or end of the sequence that were introduced by degenerate primers were discarded from analysis.
Characterizing the occurrence of R. gnavus across healthy human gut metagenomes
To estimate the abundance of R. gnavus ATCC 29149 genome and its superantigen genes across healthy human gut metagenomes we used Bowtie2 v2.3.2 (56) with default parameters to recruit reads from publicly available gut metagenomes from healthy individuals. We used anvi’o v5.1(57) to profile short metagenomic reads aligned to the R. gnavus ATCC 29149 genome to estimate coverage and detection statistics per metagenome, and to visualize merged profiles in ‘gene mode’ where the distribution of each gene of a genome is shown independently for accurate estimates of gene-level detection. To avoid overestimating ‘detection’ as a result of non-specific short read recruitment due to genomic regions conserved across multiple populations, we assumed R. gnavus was detected in a given metagenome only if more than 25% of it was covered by at least 1X. We applied the same principle to identify metagenomes in which superantigens were detected.
Statistical analysis
Statistical analysis by unpaired student’s t test or one-way ANOVA was performed using GraphPad Prism.
Supplementary Material
Data file S1. List of mAbs used in this study.
Data file S2. List of metagenomes analyzed.
Data file S3. Metagenomic recruitment analysis of R. gnavus genes including its superantigens.
Data file S4. Primary data.
Fig. S1. mAb reactivity to mouse microbiota and cladogram of individual bacterial strains analyzed.
Fig. S2. Immunoprecipitation and mass spectrometry analysis and Coomassie and western blot analysis of purified proteins.
Fig. S3. Additional analyses of GF or gnotobiotic monocolonized mice.
Table S1. Distribution of superantigens across human metagenomes.
Acknowledgements:
We thank the University of Chicago Flow Cytometry Core for assistance with cell sorting, the University of Chicago DNA Sequencing core for assistance with plasmid minipreps and sequencing, B. Theriault and the University of Chicago Gnotobiotic facility for assistance with gnotobiotic experiments, M. Meisel, B. Jabri, and E. Chang for sharing germ-free mice, P. Wilson and C. Henry for sharing anti-influenza monoclonal antibodies, and P. Wilson and A. Chervonsky for critical reading of the manuscript.
Funding: This work was supported by NIH grants T32GM007281 and F30AI124476 (to J.J.B), a fellowship of the German Research Foundation (DFG) (to CD), grants U01AI125250, R01AI038339, R01AI108643, R01GM106173, and R01HL118092 (to A.B.), and grant P30DK042086 to the University of Chicago Digestive Diseases Research Core Center.
Footnotes
Competing interests: The authors declare no competing interests.
Data and materials availability: All data necessary to understand and interpret the results are tabulated in the main and supplementary figures.
References and Notes:
- 1.Macpherson AJ, Yilmaz B, Limenitakis JP, Ganal-Vonarburg SC, IgA Function in Relation to the Intestinal Microbiota. Annu Rev Immunol 36, 359–381 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Bunker JJ, Flynn TM, Koval JC, Shaw DG, Meisel M, McDonald BD, Ishizuka IE, Dent AL, Wilson PC, Jabri B, Antonopoulos DA, Bendelac A, Innate and Adaptive Humoral Responses Coat Distinct Commensal Bacteria with Immunoglobulin A. Immunity 43, 541–553 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Macpherson AJ, Gatto D, Sainsbury E, Harriman GR, Hengartner H, Zinkernagel RM, A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science 288, 2222–2226 (2000). [DOI] [PubMed] [Google Scholar]
- 4.Bunker JJ, Bendelac A, IgA Responses to Microbiota. Immunity 49, 211–224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kau AL, Planer JD, Liu J, Rao S, Yatsunenko T, Trehan I, Manary MJ, Liu TC, Stappenbeck TS, Maleta KM, Ashorn P, Dewey KG, Houpt ER, Hsieh CS, Gordon JI, Functional characterization of IgA-targeted bacterial taxa from undernourished Malawian children that produce diet-dependent enteropathy. Sci Transl Med 7, 276ra224 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawamoto S, Maruya M, Kato LM, Suda W, Atarashi K, Doi Y, Tsutsui Y, Qin H, Honda K, Okada T, Hattori M, Fagarasan S, Foxp3(+) T cells regulate immunoglobulin a selection and facilitate diversification of bacterial species responsible for immune homeostasis. Immunity 41, 152–165 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Kubinak JL, Petersen C, Stephens WZ, Soto R, Bake E, O’Connell RM, Round JL, MyD88 signaling in T cells directs IgA-mediated control of the microbiota to promote health. Cell Host Microbe 17, 153–163 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L, Degnan PH, Hu J, Peter I, Zhang W, Ruggiero E, Cho JH, Goodman AL, Flavell RA, Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 158, 1000–1010 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Planer JD, Peng Y, Kau AL, Blanton LV, Ndao IM, Tarr PI, Warner BB, Gordon JI, Development of the gut microbiota and mucosal IgA responses in twins and gnotobiotic mice. Nature 534, 263–266 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Viladomiu M, Kivolowitz C, Abdulhamid A, Dogan B, Victorio D, Castellanos JG, Woo V, Teng F, Tran NL, Sczesnak A, Chai C, Kim M, Diehl GE, Ajami NJ, Petrosino JF, Zhou XK, Schwartzman S, Mandl LA, Abramowitz M, Jacob V, Bosworth B, Steinlauf A, Scherl EJ, Wu HJ, Simpson KW, Longman RS, IgA-coated E coli enriched in Crohn’s disease spondyloarthritis promote TH17-dependent inflammation. Sci Transl Med 9, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bunker JJ, Erickson SA, Flynn TM, Henry C, Koval JC, Meisel M, Jabri B, Antonopoulos DA, Wilson PC, Bendelac A, Natural polyreactive IgA antibodies coat the intestinal microbiota. Science 358, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S, Torrents D, Ugarte E, Zoetendal EG, Wang J, Guarner F, Pedersen O, de Vos WM, Brunak S, Dore J, Meta HITC, Antolin M, Artiguenave F, Blottiere HM, Almeida M, Brechot C, Cara C, Chervaux C, Cultrone A, Delorme C, Denariaz G, Dervyn R, Foerstner KU, Friss C, van de Guchte M, Guedon E, Haimet F, Huber W, van Hylckama-Vlieg J, Jamet A, Juste C, Kaci G, Knol J, Lakhdari O, Layec S, Le Roux K, Maguin E, Merieux A, Melo Minardi R, M’Rini C, Muller J, Oozeer R, Parkhill J, Renault P, Rescigno M, Sanchez N, Sunagawa S, Torrejon A, Turner K, Vandemeulebrouck G, Varela E, Winogradsky Y, Zeller G, Weissenbach J, Ehrlich SD, Bork P, Enterotypes of the human gut microbiome. Nature 473, 174–180 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moor K, Diard M, Sellin ME, Felmy B, Wotzka SY, Toska A, Bakkeren E, Arnoldini M, Bansept F, Co AD, Voller T, Minola A, Fernandez-Rodriguez B, Agatic G, Barbieri S, Piccoli L, Casiraghi C, Corti D, Lanzavecchia A, Regoes RR, Loverdo C, Stocker R, Brumley DR, Hardt WD, Slack E, High-avidity IgA protects the intestine by enchaining growing bacteria. Nature 544, 498–502 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Pabst O, New concepts in the generation and functions of IgA. Nat Rev Immunol 12, 821–832 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Williams RC, Gibbons RJ, Inhibition of bacterial adherence by secretory immunoglobulin A: a mechanism of antigen disposal. Science 177, 697–699 (1972). [DOI] [PubMed] [Google Scholar]
- 16.Nakajima A, Vogelzang A, Maruya M, Miyajima M, Murata M, Son A, Kuwahara T, Tsuruyama T, Yamada S, Matsuura M, Nakase H, Peterson DA, Fagarasan S, Suzuki K, IgA regulates the composition and metabolic function of gut microbiota by promoting symbiosis between bacteria. J Exp Med 215, 2019–2034 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donaldson GP, Ladinsky MS, Yu KB, Sanders JG, Yoo BB, Chou WC, Conner ME, Earl AM, Knight R, Bjorkman PJ, Mazmanian SK, Gut microbiota utilize immunoglobulin A for mucosal colonization. Science, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scherer MT, Ignatowicz L, Winslow GM, Kappler JW, Marrack P, Superantigens: bacterial and viral proteins that manipulate the immune system. Annu Rev Cell Biol 9, 101–128 (1993). [DOI] [PubMed] [Google Scholar]
- 19.Fleischer B, Gerlach D, Fuhrmann A, Schmidt KH, Superantigens and pseudosuperantigens of gram-positive cocci. Med Microbiol Immunol 184, 1–8 (1995). [DOI] [PubMed] [Google Scholar]
- 20.Krismer B, Weidenmaier C, Zipperer A, Peschel A, The commensal lifestyle of Staphylococcus aureus and its interactions with the nasal microbiota. Nat Rev Microbiol 15, 675–687 (2017). [DOI] [PubMed] [Google Scholar]
- 21.van Belkum A, Melles DC, Nouwen J, van Leeuwen WB, van Wamel W, Vos MC, Wertheim HF, Verbrugh HA, Co-evolutionary aspects of human colonisation and infection by Staphylococcus aureus. Infect Genet Evol 9, 32–47 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Lowy FD, Staphylococcus aureus infections. N Engl J Med 339, 520–532 (1998). [DOI] [PubMed] [Google Scholar]
- 23.Kim HK, Cheng AG, Kim HY, Missiakas DM, Schneewind O, Nontoxigenic protein A vaccine for methicillin-resistant Staphylococcus aureus infections in mice. J Exp Med 207, 1863–1870 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pauli NT, Kim HK, Falugi F, Huang M, Dulac J, Henry Dunand C, Zheng NY, Kaur K, Andrews SF, Huang Y, DeDent A, Frank KM, Charnot-Katsikas A, Schneewind O, Wilson PC, Staphylococcus aureus infection induces protein A-mediated immune evasion in humans. J Exp Med 211, 2331–2339 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Falugi F, Kim HK, Missiakas DM, Schneewind O, Role of protein A in the evasion of host adaptive immune responses by Staphylococcus aureus. MBio 4, e00575–00513 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun Y, Emolo C, Holtfreter S, Wiles S, Kreiswirth B, Missiakas D, Schneewind O, Staphylococcal protein A contributes to persistent colonization of mice with Staphylococcus aureus. J Bacteriol, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feehley T, Plunkett CH, Bao R, Choi Hong SM, Culleen E, Belda-Ferre P, Campbell E, Aitoro R, Nocerino R, Paparo L, Andrade J, Antonopoulos DA, Berni Canani R, Nagler CR, Healthy infants harbor intestinal bacteria that protect against food allergy. Nat Med 25, 448–453 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Graille M, Stura EA, Corper AL, Sutton BJ, Taussig MJ, Charbonnier JB, Silverman GJ, Crystal structure of a Staphylococcus aureus protein A domain complexed with the Fab fragment of a human IgM antibody: structural basis for recognition of B-cell receptors and superantigen activity. Proc Natl Acad Sci U S A 97, 5399–5404 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ibrahim S, Kaartinen M, Seppala I, Matoso-Ferreira A, Makela O, The alternative binding site for protein A in the Fab fragment of immunoglobulins. Scand J Immunol 37, 257–264 (1993). [DOI] [PubMed] [Google Scholar]
- 30.Young WW Jr., Tamura Y, Wolock DM, Fox JW, Staphylococcal protein A binding to the Fab fragments of mouse monoclonal antibodies. J Immunol 133, 3163–3166 (1984). [PubMed] [Google Scholar]
- 31.Schneewind O, Missiakas DM, Protein secretion and surface display in Gram-positive bacteria. Philos Trans R Soc Lond B Biol Sci 367, 1123–1139 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sudarsanam P, Ley R, Guruge J, Turnbaugh PJ, Mahowald M,, Liep D, Gordon J. (GenBank NZ_AAYG00000000.2, 2007). [Google Scholar]
- 33.Beaulaurier J, Zhu S, Deikus G, Mogno I, Zhang XS, Davis-Richardson A, Canepa R, Triplett EW, Faith JJ, Sebra R, Schadt EE, Fang G, Metagenomic binning and association of plasmids with bacterial host genomes using DNA methylation. Nat Biotechnol 36, 61–69 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J, Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10, 563–569 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Fischetti VA, Jones KF, Scott JR, Size variation of the M protein in group A streptococci. J Exp Med 161, 1384–1401 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reboldi A, Arnon TI, Rodda LB, Atakilit A, Sheppard D, Cyster JG, IgA production requires B cell interaction with subepithelial dendritic cells in Peyer’s patches. Science 352, aaf4822 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lecuyer E, Rakotobe S, Lengline-Garnier H, Lebreton C, Picard M, Juste C, Fritzen R, Eberl G, McCoy KD, Macpherson AJ, Reynaud CA, Cerf-Bensussan N, Gaboriau-Routhiau V, Segmented filamentous bacterium uses secondary and tertiary lymphoid tissues to induce gut IgA and specific T helper 17 cell responses. Immunity 40, 608–620 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Brito IL, Yilmaz S, Huang K, Xu L, Jupiter SD, Jenkins AP, Naisilisili W, Tamminen M, Smillie CS, Wortman JR, Birren BW, Xavier RJ, Blainey PC, Singh AK, Gevers D, Alm EJ, Mobile genes in the human microbiome are structured from global to individual scales. Nature 535, 435–439 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.C. Human Microbiome Project, A framework for human microbiome research. Nature 486, 215–221 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, Peng Y, Zhang D, Jie Z, Wu W, Qin Y, Xue W, Li J, Han L, Lu D, Wu P, Dai Y, Sun X, Li Z, Tang A, Zhong S, Li X, Chen W, Xu R, Wang M, Feng Q, Gong M, Yu J, Zhang Y, Zhang M, Hansen T, Sanchez G, Raes J, Falony G, Okuda S, Almeida M, LeChatelier E, Renault P, Pons N, Batto JM, Zhang Z, Chen H, Yang R, Zheng W, Li S, Yang H, Wang J, Ehrlich SD, Nielsen R, Pedersen O, Kristiansen K, Wang J, A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490, 55–60 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Rampelli S, Schnorr SL, Consolandi C, Turroni S, Severgnini M, Peano C, Brigidi P, Crittenden AN, Henry AG, Candela M, Metagenome Sequencing of the Hadza Hunter-Gatherer Gut Microbiota. Curr Biol 25, 1682–1693 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Knights D, Ward TL, McKinlay CE, Miller H, Gonzalez A, McDonald D, Knight R, Rethinking “enterotypes”. Cell Host Microbe 16, 433–437 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smits SA, Leach J, Sonnenburg ED, Gonzalez CG, Lichtman JS, Reid G, Knight R, Manjurano A, Changalucha J, Elias JE, Dominguez-Bello MG, Sonnenburg JL, Seasonal cycling in the gut microbiome of the Hadza hunter-gatherers of Tanzania. Science 357, 802–806 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Round JL, Palm NW, Causal effects of the microbiota on immune-mediated diseases. Sci Immunol 3, (2018). [DOI] [PubMed] [Google Scholar]
- 45.Chua HH, Chou HC, Tung YL, Chiang BL, Liao CC, Liu HH, Ni YH, Intestinal Dysbiosis Featuring Abundance of Ruminococcus gnavus Associates With Allergic Diseases in Infants. Gastroenterology 154, 154–167 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Azzouz D, Omarbekova A, Heguy A, Schwudke D, Gisch N, Rovin BH, Caricchio R, Buyon JP, Alekseyenko AV, Silverman GJ, Lupus nephritis is linked to disease-activity associated expansions and immunity to a gut commensal. Ann Rheum Dis, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Breban M, Tap J, Leboime A, Said-Nahal R, Langella P, Chiocchia G, Furet JP, Sokol H, Faecal microbiota study reveals specific dysbiosis in spondyloarthritis. Ann Rheum Dis 76, 1614–1622 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Hall AB, Yassour M, Sauk J, Garner A, Jiang X, Arthur T, Lagoudas GK, Vatanen T, Fornelos N, Wilson R, Bertha M, Cohen M, Garber J, Khalili H, Gevers D, Ananthakrishnan AN, Kugathasan S, Lander ES, Blainey P, Vlamakis H, Xavier RJ, Huttenhower C, A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med 9, 103 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Machiels K, Sabino J, Vandermosten L, Joossens M, Arijs I, de Bruyn M, Eeckhaut V, Van Assche G, Ferrante M, Verhaegen J, Van Steen K, Van Immerseel F, Huys G, Verbeke K, Wolthuis A, de Buck Van Overstraeten A, D’Hoore A, Rutgeerts P, Vermeire S, Specific members of the predominant gut microbiota predict pouchitis following colectomy and IPAA in UC. Gut 66, 79–88 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Romero D, Palacios R, Gene amplification and genomic plasticity in prokaryotes. Annu Rev Genet 31, 91–111 (1997). [DOI] [PubMed] [Google Scholar]
- 51.Rakoff-Nahoum S, Foster KR, Comstock LE, The evolution of cooperation within the gut microbiota. Nature 533, 255–259 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu L, Chen X, Skogerbo G, Zhang P, Chen R, He S, Huang DW, The human microbiome: a hot spot of microbial horizontal gene transfer. Genomics 100, 265–270 (2012). [DOI] [PubMed] [Google Scholar]
- 53.Russell AB, Wexler AG, Harding BN, Whitney JC, Bohn AJ, Goo YA, Tran BQ, Barry NA, Zheng H, Peterson SB, Chou S, Gonen T, Goodlett DR, Goodman AL, Mougous JD, A type VI secretion-related pathway in Bacteroidetes mediates interbacterial antagonism. Cell Host Microbe 16, 227–236 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bergqvist P, Stensson A, Hazanov L, Holmberg A, Mattsson J, Mehr R, Bemark M, Lycke NY, Re-utilization of germinal centers in multiple Peyer’s patches results in highly synchronized, oligoclonal, and affinity-matured gut IgA responses. Mucosal Immunol 6, 122–135 (2013). [DOI] [PubMed] [Google Scholar]
- 55.Finlay BB, McFadden G, Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell 124, 767–782 (2006). [DOI] [PubMed] [Google Scholar]
- 56.Langmead B, Salzberg SL, Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eren AM, Esen OC, Quince C, Vineis JH, Morrison HG, Sogin ML, Delmont TO, Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ 3, e1319 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data file S1. List of mAbs used in this study.
Data file S2. List of metagenomes analyzed.
Data file S3. Metagenomic recruitment analysis of R. gnavus genes including its superantigens.
Data file S4. Primary data.
Fig. S1. mAb reactivity to mouse microbiota and cladogram of individual bacterial strains analyzed.
Fig. S2. Immunoprecipitation and mass spectrometry analysis and Coomassie and western blot analysis of purified proteins.
Fig. S3. Additional analyses of GF or gnotobiotic monocolonized mice.
Table S1. Distribution of superantigens across human metagenomes.