Abstract
Background and Aims
Cardamine occulta (Brassicaceae) is an octoploid weedy species (2n = 8x = 64) originated in Eastern Asia. It has been introduced to other continents including Europe and considered to be an invasive species. Despite its wide distribution, the polyploid origin of C. occulta remained unexplored. The feasibility of comparative chromosome painting (CCP) in crucifers allowed us to elucidate the origin and genome evolution in Cardamine species. We aimed to investigate the genome structure of C. occulta in comparison with its tetraploid (2n = 4x = 32, C. kokaiensis and C. scutata) and octoploid (2n = 8x = 64, C. dentipetala) relatives.
Methods
Genomic in situ hybridization (GISH) and large-scale CCP were applied to uncover the parental genomes and chromosome composition of the investigated Cardamine species.
Key Results
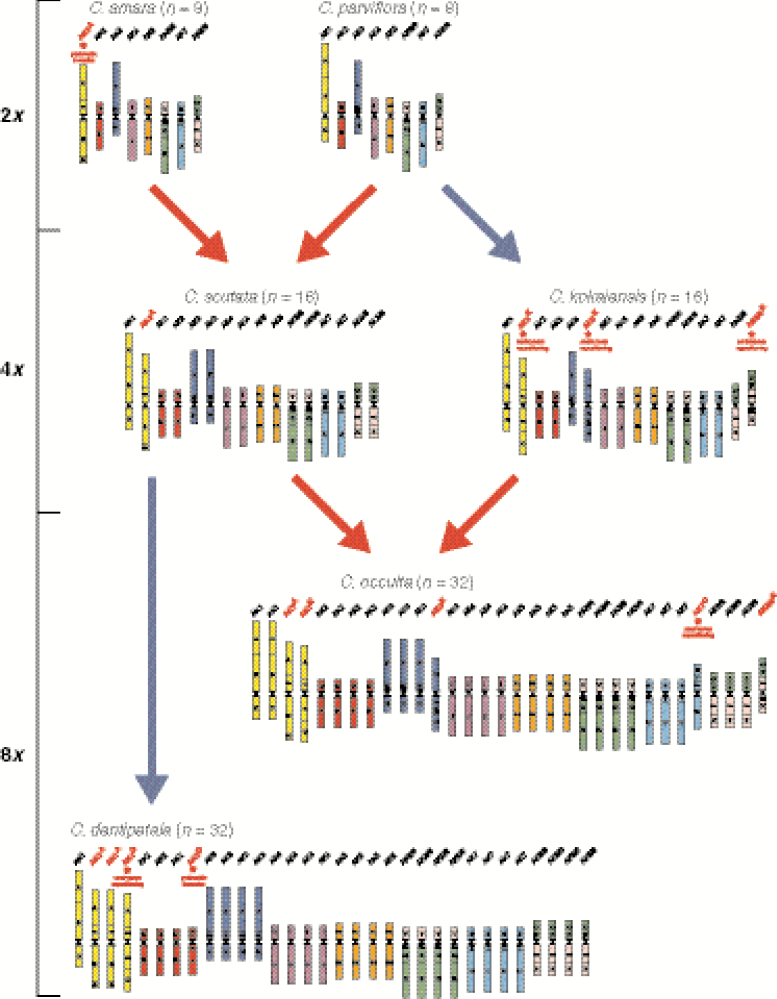
All investigated species descended from a common ancestral Cardamine genome (n = 8), structurally resembling the Ancestral Crucifer Karyotype (n = 8), but differentiated by a translocation between chromosomes AK6 and AK8. Allotetraploid C. scutata originated by hybridization between two diploid species, C. parviflora and C. amara (2n = 2x = 16). By contrast, C. kokaiensis has an autotetraploid origin from a parental genome related to C. parviflora. Interestingly, octoploid C. occulta probably originated through hybridization between the tetraploids C. scutata and C. kokaiensis. The octoploid genome of C. dentipetala probably originated from C. scutata via autopolyploidization. Except for five species-specific centromere repositionings and one pericentric inversion post-dating the polyploidization events, the parental subgenomes remained stable in the tetra- and octoploids.
Conclusions
Comparative genome structure, origin and evolutionary history was reconstructed in C. occulta and related species. For the first time, whole-genome cytogenomic maps were established for octoploid plants. Post-polyploid evolution in Asian Cardamine polyploids has not been associated with descending dysploidy and intergenomic rearrangements. The combination of different parental (sub)genomes adapted to distinct habitats provides an evolutionary advantage to newly formed polyploids by occupying new ecological niches.
Keywords: Allopolyploidy, autopolyploidy, Asian Cardamine, Brassicaceae, centromere repositioning, chromosome rearrangements, comparative chromosome painting, diploidization, genome collinearity, GISH (genomic in situ hybridization), hybridization, invasive species
INTRODUCTION
The genus Cardamine L. (bittercress) is one of the largest genera of the family Brassicaceae, distributed on all continents except Antarctica. It is treated as a member of the tribe Cardamineae Dumort., which is presumably a monophyletic group comprising 12 genera and ~340 species (Al-Shehbaz, 2012). Cardamine is the largest genus of the tribe, comprising more than 200 species, although the number of recognized species partially depends on the taxonomic treatment of several complicated groups (Lihová and Marhold, 2006). The genus shows a high incidence of polyploidy: 58 % of analysed taxa (species and subspecies) are entirely neopolyploid (tetraploids being slightly more abundant than other ploidy levels), 10 % are both diploid and neopolyploid, and 32 % are diploid (Kučera et al., 2005). The genus also comprises species with the highest known chromosome number in the family, C. diphylla (Michx.) Alph.Wood and C. concatenata (Michx.) O.Schwarz (both 2n = 256). Spontaneous interspecific hybridization has been reported to occur quite frequently between several Cardamine species (reviewed by Lihová and Marhold, 2006; Marhold et al., 2018). Hence, hybridization and polyploidy are crucial driving forces of speciation in Cardamine (Marhold and Lihová, 2006).
Eastern Asian Cardamine populations, currently classified as C. occulta Hornem. (Marhold et al., 2016), were for most of the last century treated as part of the widely circumscribed species C. flexuosa With. or as C. hirsuta subsp. flexuosa (With.) Forbes & Hemsl. (Matsumura, 1912; Ohwi, 1953; Ohwi and Kitagawa, 1992; Zhou et al., 2001; Al-Shehbaz et al., 2006). These populations were found to be distinct from the European species C. flexuosa only recently, based on phylogenetic analyses using the internal transcribed spacer (ITS) region of rDNA and the trnL–trnF region of cpDNA (Lihová et al., 2006a). Differences were found also in the ploidy level. While C. flexuosa, as originally described from Europe, is tetraploid (2n = 4x = 32; Kučera et al., 2005; Mandáková et al., 2014) with identified parental diploid species C. amara L. and C. hirsuta L., C. occulta is uniformly octoploid (2n = 8x = 64) throughout its distribution area (Lihová et al., 2006a; Šlenker et al., 2018). It is primarily a weed of rice paddies, orchards, flower beds and various other kinds of anthropogenic vegetation (Kudoh et al., 1993; Yatsu et al., 2003; these studies reported the species as C. flexuosa). From our field experience, we are not aware of any localities in truly natural vegetation. Therefore, it was hypothesized by Lihová et al. (2006a) that the origin and successful expansion of C. occulta have been associated with the establishment of irrigated man-made habitats. Cardamine occulta probably originated in Eastern Asia and was subsequently introduced to other continents almost certainly due to the human activities. Apart from Eastern Asia, C. occulta currently occurs in North (Post et al., 2009; Al-Shehbaz et al., 2010) and Central America (Rankin Rodríguez and Greuter, 2009, as C. flexuosa subsp. debilis O. E. Schulz), Australia (Thompson, 1996, as C. aff. flexuosa), New Zealand (Heenan, 2017), and Hawaii (K. Marhold, unpubl. data). Most recently, C. occulta was introduced also to Europe (first two occurrences recorded in 1977 in Italy and in 1993 in Spain), where it is now spreading and the reports are increasing from many countries (Marhold et al., 2016; Maiorov, 2018). Consequently, C. occulta has become the most widespread Cardamine species.
According to Lihová et al. (2006a), the closest relatives of C. occulta include the Eastern Asian C. scutata Thunb., C. fallax (O.E. Schulz) Nakai and the Japanese endemics C. dentipetala Matsum., C. longifructus Ohwi, and C. niigatensis H. Hara. All of them occur in natural habitats, mostly moist locations at edges of rivulets or in forests. Cardamine scutata is sometimes found also in paddy fields, when they are ill-drained after the rice-cultivation periods. It is a tetraploid species, and based on cpDNA and ITS sequences published by Lihová et al. (2006a) and Carlsen et al. (2009), Shimizu-Inatsugi et al. (2017) suggested diploid C. amara and C. parviflora L. as the parental species of C. scutata. There is almost continuous morphological variation between C. scutata and C. niigatensis (also tetraploid; Lihová and Kučera, 2007) when they co-occur, indicating their close relationships or even conspecificity. The other two endemics, C. dentipetala and C. longifructus, were of unknown ploidy level and they have been either synonymized with C. scutata (Zhou et al., 2001; Al-Shehbaz et al., 2006) or considered to be conspecific (C. longifructus being a variety of C. dentipetala; Ohwi, 1965; Ohwi and Kitagawa, 1992). Cardamine fallax was inferred to be hexaploid, and based on DNA sequence data, it was hypothesized that this species or its diploid progenitors may have contributed to the origin of C. occulta (Lihová et al., 2006a). Most recently, a new tetraploid species, closely related to C. occulta, was described from Japan and easternmost China, namely C. kokaiensis Yahara et al. (Šlenker et al., 2018). Diploid species are very scarce in Eastern Asia (Kučera et al., 2005), and apart from the above-mentioned C. amara and C. parviflora, the only diploid relatives that grow in the area of these Eastern Asian polyploids are C. impatiens L. and C. hirsuta.
In the present study, using up-to-date molecular cytogenetic approaches, we analysed the genome origin and stucture in Cardamine polyploid species that have been poorly explored so far: the invasive octoploid species C. occulta and its Asian relatives.
MATERIAL AND METHODS
Plant material
Multiple populations of C. occulta, and those of relevant diploids (C. amara, C. hirsuta, C. impatiens, C. parviflora) and related polyploids (C. dentipetala, C. kokaiensis, C. scutata) were sampled for this study in Eastern Asia and Europe (Table 1). Voucher specimens are deposited in SAV. The plants were either collected in the wild (C. amara and C. impatiens) or grown in a growth chamber from seeds from plants collected in the wild. Young inflorescences of the analysed plants were collected and fixed in freshly prepared fixative (ethanol/acetic acid, 3: 1) overnight, transferred to 70 % ethanol and stored at −20 °C until used. For genomic in situ hybridization (GISH) in polyploids, leaves of C. amara, C. hirsuta, C. impatiens, C. kokaiensis, C. parviflora and C. scutata were dried in silica gel.
Table 1.
List of population samples analysed in the study.
| Locality code | Locality and collection details |
|---|---|
| Cardamine dentipetala | |
| JP168 | Japan, Honshu, Kyoto Pref., Uji City, Shirakawa, 100 m, 34.881°N, 135.819°E, coll. KM, 6 Apr 2011 |
| Cardamine kokaiensis | |
| CH15/2014 | China, Zhejiang Province, Linan County, Xitianmu Town, Renshanling Village (临安西天目乡仁山岭村), 169 m, 30.239°N, 119.466°E, coll. KM & YZ, 11 Apr 2014 |
| JP149 | Japan, Honshu, Ibaraki Pref., Jyoso-shi, Araigimachi, 10 m, 36.019°N, 140.002°E, coll. HK & KM, 18 Apr 2011 |
| Cardamine occulta | |
| CH02/2014 | China, Zhejiang Province, Anji County, Bamboo Museum Garden (安吉竹博园), 33 m, 30.592°N, 119.656°E, coll. KM & YZ, 15 Apr 2014 |
| CH17/2014 | China, Zhejiang Province, Ninghai County, Chalu Town, Qianhoulou Village (宁海市岔路镇前后娄村), 43 m, 29.195°N, 121.304°E, coll. KM & YZ, 17 Apr 2014 |
| CH18/2014 | China, Zhejiang Province, Linhai County, Kuocang Mountains (括苍山), 79 m, 28.839°N, 120.981°E, coll. KM, YZ & Ming Jiang 蒋明, 18 Apr 2014 |
| JP146 | Japan, Honshu, Mie Pref., Kitamuro-gun, Kihoku-cho, Kiinagashima-ku, Shikooku, 10 m, 34.222°N, 136.326°E, coll. HK, 16 Apr 2011 |
| JP157 | Japan, Kyushu, Fukuoka Pref., Akama, Taku, 11 m, 33.807°N, 130.573°E, coll. KM & J. Sugisaka, 24 Apr 2011 |
| JP170 | Japan, Honshu, Kyoto Pref., Kyoto City, Inari, 60 m, 34.967°N, 135.775°E, coll. KM, 7 Apr 2011 |
| JP173 | Japan, Honshu, Gifu Pref., Nakatsugawa-shi, Sakamoto, 300 m, 35.481°N, 137.481°E, coll. HK & KM, 10 May 2011 |
| JP174 | Japan, Honshu, Nagano Pref., Ueda-shi, Sanada-cho, Ohata, 620 m, 36.428°N, 138.298°E, coll. HK & KM, 16 May 2011 |
| JP175 | Japan, Honshu, Gunma Pref., Agatsuma-gun, Nakanojyo-cho, Nagaishi, 360 m, 36.598°N, 138.863°E, coll. HK & KM, 16 May 2011 |
| JP177 | Japan, Honshu, Gunma Pref., between Shirasawa-mura and Katashina-Mura, Tone, 610 m, 36.683°N, 139.2°E, coll. HK & KM, 17 May 2011 |
| JP178 | Japan, Honshu, Niigata Pref., Minamiuonnma-shi, Osawa, 197 m, 37.015°N, 138.829°E, coll. HK & KM, 17 May 2011 |
| JP181 | Japan, Honshu, Nagano Pref., Minamiazumi-gun, Hotaka-cho, Ariake, 570 m, 36.382°N, 137.857°E, coll. HK & KM, 17 May 2011 |
| JP191 | Japan, Honshu, Niigata Pref., Ohno, Itoigawa city, 52 m, 37.008°N, 137.872°E, coll. KM, T. Kawagoe & J. Sugisaka, 1 Jun 2011 |
| SVC | Spain, Cantabria, San Vicente de la Barquera, 4 m, 43.383°N, 4.398°W, coll. M. Lysák, 9 Jun 2011 |
| Cardamine scutata | |
| JP169 | Japan, Honshu, Kyoto Pref., Uji City, Shirakawa, 100 m, 34.881°N, 135.819°E, coll. KM, 6 Apr 2011 |
| Cardamine amara | |
| URN | Switzerland, Canton Uri, Urnerboden, 46.887°N, 8.901°E, coll. T. Mandáková et al., May 2010 |
| Cardamine hirsuta | |
| OXF | UK, Oxford (Hay and Tsiantis, 2006) |
| Cardamine impatiens | |
| KRE | Slovakia, District Žiar nad Hronom, Kremnica, 720 m, 48.707°N, 18.939°E, coll. KM, 15 Aug 2013 |
| Cardamine parviflora | |
| ZBR | Czech Republic, Hodonín, Zbrod, forest Doubrava, 168 m, 48.888°N, 17.065°E, coll. J. Kučera, May 2013 |
Name abbreviations: HK, Hiroshi Kudoh; KM, Karol Marhold; YZ, Yunpeng Zhao 赵云鹏.
Chromosome preparations
Chromosome spreads from fixed young flower buds containing immature anthers were prepared according to published protocols (Lysak and Mandáková, 2013; Mandáková and Lysak, 2016a). Suitable slides were post-fixed in freshly prepared 4 % formaldehyde in distilled water for 10 min and put into a rack to air-dry. Preparations were kept in a dust-free box at room temperature until used.
DNA probes
The Arabidopsis-type telomere repeat (TTTAGGG)n was prepared according to Ijdo et al. (1991). The C. amara-specific 290-bp Carcen tandem repeat was prepared based on details given by Mandáková et al. (2013) and used as a (peri)centromeric probe. For comparative chromosome painting (CCP), 674 chromosome-specific BAC clones of Arabidopsis thaliana grouped into contigs according to eight chromosomes and 22 genomic blocks (GBs) of the Ancestral Crucifer Karyotype (ACK; Lysak et al., 2016) were used. To determine and characterize species-specific chromosome rearrangements, after initial CCP experiments, some BAC contigs were split into smaller subcontigs. Genomic DNA (gDNA) of C. amara, C. hirsuta, C. impatiens, C. kokaiensis, C. parviflora and C. scutata were extracted from the silica gel-dried leaves using a Qiagen DNA isolation kit and used as GISH probes. All DNA probes were labelled with biotin-dUTP, digoxigenin-dUTP or Cy3-dUTP by nick translation as described by Mandáková and Lysak (2016b). Probes were pooled to follow the design of a given experiment, ethanol precipitated, dried and dissolved in 20 µL of 50 % formamide and 10 % dextran sulphate in 2× SSC per slide.
In situ hybridization and microscopy
For fluorescence in situ hybridization (FISH) and CCP, 20 µL of the probe was pipetted on a chromosome-containing slide and immediately denatured on a hot plate at 80 °C for 2 min. For GISH, 20 µL of the probe was denatured in an Eppendorf tube at 90 °C for 10 min, placed on ice for 10 min, pipetted on a slide and denatured on a hot plate at 80 °C for 2 min. Hybridization was carried out in a moist chamber at 37 °C overnight. Post-hybridization washing was performed in 20 % formamide in 2× SSC at 42 °C. The immunodetection of hapten-labelled probes was performed as described by Mandáková and Lysak (2016b) as follows: biotin-dUTP was detected by avidin–Texas Red (Vector Laboratories) and amplified by goat anti-avidin–biotin (Vector Laboratories) and avidin–Texas Red; digoxigenin-dUTP was detected by mouse anti-digoxigenin (Jackson Immuno Research) and goat anti-mouse–Alexa Fluor 488 (Invitrogen). Chromosomes were counterstained with 2 μg mL−1 DAPI in Vectashield. The preparations were photographed using a Zeiss Axioimager Z2 epifluorescence microscope with a CoolCube camera (MetaSystems). Images were acquired separately for all four fluorochromes using appropriate excitation and emission filters (AHF Analysentechnik). The four monochromatic images were pseudocoloured, merged and cropped using Photoshop CS (Adobe Systems) and ImageJ (National Institutes of Health).
RESULTS
Parental diploid genomes – Cardamine amara and C. parviflora
Four Cardamine species were tested as potential diploid parental genomes of the studied Asian polyploids: C. amara, C. hirsuta, C. impatiens and C. parviflora. Labelled gDNA of the four diploids was used in different combinations for GISH experiments. Only gDNA of C. amara and C. parviflora hybridized to chromosomes of the investigated polyploids (C. dentipetala, C. kokaiensis, C. occulta and C. scutata; more details provided below). Therefore, C. amara and C. parviflora, as potential parental diploid genomes of the Asian Cardamine polyploids, were analysed cytogenetically in detail.
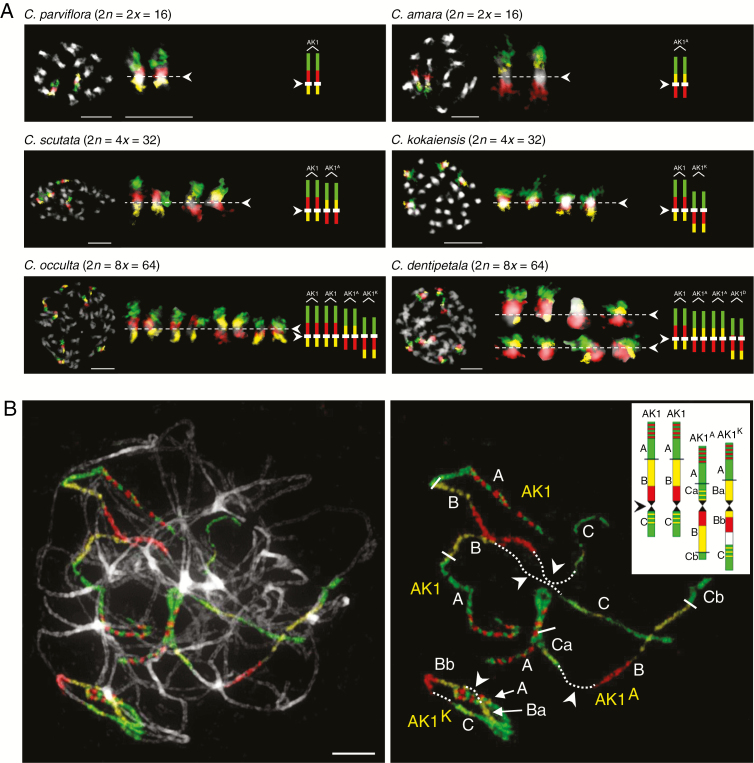
Both analysed accessions of C. amara and C. parviflora were diploid with eight chromosome pairs (2n = 2x = 16). Interstitial telomeric repeats (ITRs) were observed at all pericentromere regions in C. parviflora, but not in C. amara. As expected, the C. amara-specific tandem repeat Carcen localized to all centromeres in C. amara, but not to chromosomes of C. parviflora (Supplementary Data Figure S1). To analyse the genome structure of the species we used CCP with chromosome-specific A. thaliana BAC contigs, representing 22 conserved GBs of the ACK (n = 8; Lysak et al., 2016), on mitotic and meiotic (pachytene) chromosomes.
In C. parviflora, six out of eight chromosome pairs (AK1–AK5 and AK7) retained the ancestral structure as in the ACK. The two remaining chromosomes originated by a reciprocal translocation involving the bottom arm of AK6 and the major part (~6 Mb in the A. thaliana genome) of the bottom arm of chromosome AK8 (~6.6 Mb). The resulting translocation chromosomes AK6/8 and AK8/6 consist of genomic blocks V+Wa+Q+X and O+P+Wb+X, respectively (Fig. 1). The chromosome size expressed as length of A. thaliana BAC contigs in megabases is as follows: AK1 – 17, AK2 – 8.5, AK3 – 13.7, AK4 – 9.6, AK5 – 10.4, AK6/8 – 12.4, AK7 – 12.2, and AK8/6 – 10.6.
Fig. 1.
Genome structure and origin of Asian Cardamine polyploids and their diploid progenitors. Comparative cytogenetic maps are based on the comparison with the Ancestral Crucifer Karyotype (ACK; Lysak et al., 2016) comprising eight chromosomes (AK1–AK8) and 22 genomic blocks. Cardamine chromosomes follow the ‘ancestral colours’ of AK chromosomes and capital letters refer to genomic blocks (A–X). Chromosome rearrangements are indicated by red asterisks, while reshuffled chromosomes are highlighted by red headings. Red and blue arrows represent allo- and autopolyploid events, respectively.
A comparative cytogenomic map of C. amara was reconstructed by us previously (Mandáková et al., 2013) and further specified in the present study. Whereas the genome of C. amara almost completely resembles that of C. parviflora, including the translocation chromosomes AK6/8 and AK8/6, it differs by a ~13-Mb pericentric inversion on chromosome AK1 (AK1A). The breakpoints occurred between GBs A and B (between BAC clones T29M8 and F6F9), and within block C (between BAC clones F9I5 and F6D8). The resulting chromosome AK1A consists of GBs A+Ca+B+Cb (Fig. 1). The absence of AK1A in other Cardamine species (Mandáková et al., 2013, 2014, 2016) suggests that the inversion is probably specific for C. amara.
The allotetraploid origin and conserved subgenome structure in Cardamine scutata
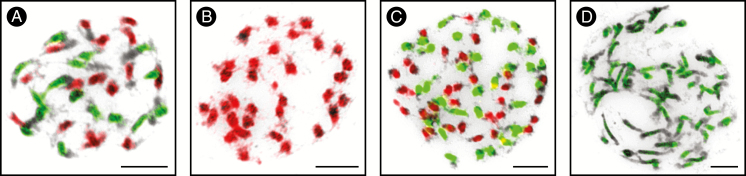
Cardamine scutata was confirmed to be a tetraploid species (2n = 4x = 32). To determine the origin of C. scutata, we carried out GISH with gDNA of four diploid species (see above) in different combinations, and FISH of a telomeric repeat and Carcen probed to mitotic chromosomes of C. scutata. Only gDNA of C. parviflora and C. amara hybridized to C. scutata chromosomes, each to one half of its chromosome complement (Fig. 2A). FISH signals revealed 16 chromosomes bearing ITRs and the other 16 chromosomes containing the C. amara-specific Carcen repeat (Supplementary Data Figure S1). Thus, GISH and FISH of tandem repeats strongly suggested an allotetraploid origin of C. scutata from parental genomes closely related to C. amara and C. parviflora.
Fig. 2.
Genomic in situ hybridization (GISH) in Asian Cardamine polyploids. (A) GISH revealed 16 chromosomes of C. amara (green fluorescence) and 16 chromosomes of C. parviflora (red fluorescence) within a mitotic chromosome complement of the allotetraploid C. scutata (2n = 32). (B) Mitotic chromosomes of the autotetraploid C. kokaiensis (2n = 32) entirelly labelled by gDNA of C. parviflora (red). (C) GISH with gDNA of C. kokaiensis (red) and C. scutata (green) revealed two 32-chromosomal subgenomes in the allooctoploid C. occulta (2n = 64). (D) Mitotic chromosomes of the autooctoploid C. dentipetala (2n = 64) hybridized with gDNA of C. scutata (green). Chromosomes were counterstained by DAPI and inverted in Adobe Photoshop. Scale bars = 10 µm.
Knowing the structure of C. amara and C. parviflora chromosomes, corresponding painting probes were hybridized to chromosomes of C. scutata. The unambiguous identification of all 22 genomic blocks in four and two genomic copies on mitotic and pachytene chromosomes, respectively, confirmed the tetraploid origin of the species. All 16 chromosome pairs were structurally identical to chromosomes of C. amara and C. parviflora, including the diagnostic C. amara-specific chromosome AK1A (Fig. 3A). The parental chromosome signatures further supported the origin of C. scutata by hybridization between genomes closely related to those of diploid C. amara and C. parviflora (Fig. 1).
Fig. 3.
Comparative chromosome painting (CCP) with probes for AK1. (A) CCP revealed the structure of AK1 homeologues within mitotic chromosome complements of Cardamine parviflora, C. amara, C. scutata, C. kokaiensis, C. occulta and C. dentipetala. (B) An example of fine-scale CCP on pachytene chromosomes of C. occulta. Arrowheads point to centromeres. Red, green and yellow colours correspond to fluorescent signals of Texas Red, Alexa 488 and Cy3, respectively. Chromosomes were counterstained by DAPI and cut out using Adobe Photoshop. Scale bars = 10 µm.
The autotetraploid origin of Cardamine kokaiensis followed by three centromere repositionings
Both investigated populations of C. kokaiensis were found to be tetraploid (2n = 4x = 32). The uniform GISH labelling of all 32 chromosomes of C. kokaiensis only by gDNA of C. parviflora (Fig. 2B), and ITRs identified in all pericentromeres strongly suggested an autotetraploid origin of C. kokaiensis from a genome related to C. parviflora.
CCP revealed that the genome of C. kokaiensis is composed of eight C. parviflora-like chromosomes present in four copies. However, although three chromosomes have retained the ancestral organization/colinearity of genomic blocks, their centromeres have relocated. Centromere repositionings were identified in one chromosome pair of AK1 (AK1K), AK3 (AK3K) and AK8/6 (AK8/6K). The AK1 centromere originally located between GBs B and C (between BAC clones F12K21 and F2J6) shifted ~2.8 Mb within block B (between F26F24 and T1K7). The AK3 centromere moved by ~2.3 Mb from the border of GBs G and F (between MWL2 and F16J10) into block F (between MRC8 and K26M9). The AK8/6 centromere originally located between GBs P and Wb (between T3H13 and K26M13) repositioned ~1.1 Mb into block Wb (between MTI20 and K18B18). Both Chinese and Japanese populations of C. kokaiensis exhibited the identical genome structure (Fig. 1).
The allooctoploid origin and complex genome structure of Cardamine occulta
All analysed plants of C. occulta had 64 chromosomes (2n = 8x = 64). The definite identification of all 22 genomic blocks in 16 and eight copies on mitotic and meiotic chromosomes, respectively, confirmed the expected octoploid status of C. occulta (Fig. 1).
Each of the labelled gDNA of C. kokaiensis and C. scutata hybridized to one half of C. occulta chromosomes (Fig. 2C). Telomeric probe and Carcen localized to ~48 and 16 centromeres, respectively. Interstitial telomeric signals thus marked 32 chromosomes originating from C. kokaiensis (i.e. 16 + 16 C. parviflora-like chromosomes forming the complement of C. kokaiensis) and 16 chromosomes of C. scutata (i.e. 16 C. parviflora-like chromosomes). By contrast, Carcen delimited 16 chromosomes originating from C. scutata (i.e. 16 C. amara-like chromosomes from the allotetraploid complement of C. scutata) (Supplementary Data Figure S1). Thus, GISH and tandem repeats suggested an allopolyploid origin of C. occulta from parental genomes closely related to C. kokaiensis and C. scutata.
CCP with a painting probe designed according to the structure of AK1 on pachytene chromosomes in C. occulta revealed two homeologous chromosomes resembling those of C. parviflora, one C. amara-specific AK1A and one homeologue of C. kokaiensis-specific AK1K (Fig. 3). Accordingly, the AK1 and AK1A homeologues correspond to the C. scutata subgenome, whereas homeologues AK1 and AK1K are indicative of the C. kokaiensis subgenome. Cardamine kokaiensis-specific chromosomes AK3K and AK8/6K, characterized by repositioned centromeres, were identified within one subgenome of C. occulta too. Therefore, the CCP data strongly suggest an allooctoploid origin of C. occulta through a merger of unreduced gametes of C. scutata and C. kokaiensis (Fig. 1). The C. occulta genome almost completely matches those of C. scutata and C. kokaiensis, differing only by a centromere repositioning on one out of four AK7 homeologues (AK7O). The centromere moved by ~1.4 Mb from its original position between GBs T and U (between BAC clones F18A5 and T6K21) into block U (between F1C12 and F13K14) (Fig. 1).
To detect whether the genome structure and particularly the C. occulta-specific centromere repositioning on AK7O are stable throughout the distribution area, we analysed three Chinese, three Japanese and one Spanish population of C. occulta. Detailed inspection using CCP, GISH and FISH of tandem repeats did not reveal any population-specific chromosome repatterning and proved the structural stability of the C. occulta genome.
The autooctoploid Cardamine dentipetala structurally resembling C. scutata which experienced two chromosome rearrangements
Similarly to C. occulta, C. dentipetala was found to be octoploid (2n = 8x = 64). We observed uniform labelling of all 64 chromosomes by gDNA of C. scutata (Fig. 2D), and hybridization signals of the telomeric probe and Carcen mirroring the pattern of C. scutata.
The evidence of an autopolyploid origin of C. dentipetala from a genome related to C. scutata (Fig. 1) was further supported by CCP. Structurally, the C. dentipetala genome resembles that of C. scutata including two homeologues of AK1A. The genomes of C. dentipetala and C. scutata differ by a C. dentipetala-specific centromere repositioning on one chromosome pair of AK1 (AK1D) and a ~3.2-Mb pericentric inversion on one homeologue of AK2 (AK2D). The AK1 centromere originally located between GBs B and C (between BAC clones F12K21 and F2J6) was shifted by ~2.4 Mb into block B (between F21K23 and F1K23; Fig. 3A). The pericentric inversion occurred within blocks D (between BAC clones F19C14 and F20B16) and E (between F17O7 and F14O23). The resulting chromosome AK2D consists of GBs Da+Ea+Db+Eb.
Rearrangements shuffled chromosomes from acro- to (sub)metacentric
We identified cross-species collinearity of numerous Cardamine chromosomes differentiated by centromere position (centromere repositioning). Three C. kokaiensis-specific centromere repositionings were revealed, and observed also in its polyploid descendant – in the allooctoploid genome of C. occulta. Furthermore, one C. occulta-specific centromere repositioning and one C. dentipetala-specific centromere repositioning were identified. In addition to the centromere repositionings, a C. amara-specific pericentric inversion was transfered in the allotetraploid C. scutata, allooctoploid C. occulta and autooctoploid C. dentipetala. Furthermore, a C. dentipetala-specific pericentric inversion was found. Interestingly, the inferred intrachromosomal rearrangements shaped the Cardamine chromosomes from acro- to (sub)metacentric as shown by the increased mean centromeric index (CI, length of the short arm to the total chromosome length × 100). CI values of the ancestral chromosomes AK1, AK2, AK3, AK7 and AK8/6 were 27, 27, 23.3, 19.7 and 35.8 %, respectively, vs. 34.9 % of AK1A, 78.9 % of AK1K, 84.8 % of AK1D, 60.4 % of AK2D, 98.6 % of AK3K, 67.1 % of AK7O and 89 % of AK8/6K. These events also made the karyotypes more symmetric: the average CI of the ancestral Cardamine genome and the karyotype of C. parviflora was 27.2 %, vs. 34.9, 31, 38.4, 36.2 and 34 % in C. amara, C. scutata, C. kokaiensis, C. occulta and C. dentipetala, respectively.
DISCUSSION
The power of GISH and CCP approaches to elucidate the origin and structure of polyploid genomes
Discerning the origin of hybrid and polyploid plant species as well as cytogenetic analysis of introgressed chromosomes and chromosome segments, and intergenomic translocations is feasible using GISH (Schwarzacher et al., 1989). The use of GISH in plant cytogenetic studies has been extensively reviewed (Raina and Rani, 2001; D’Hont, 2005; Chester et al., 2010; Silva and Souza, 2013; Younis et al., 2015; Ramzan et al., 2017). GISH represents an effective and low-cost analytical method based on differential hybridization of genome-specific dispersed repeats, providing that parental species are not extinct and their genomes are sufficiently divergent. GISH is not only useful in identifying parental chromosome complements, but at the same time has the capacity to reveal intergenomic translocations. Cruciferous plants, like some other plant groups with small genomes, show a particular chromosome organization characterized by most repetitive DNA sequences forming pericentromeric heterochromatin. Consequently, GISH probes preferentially hybridize to the pericentromeric regions, leaving euchromatic chromosome arms practically unlabelled (Raina and Rani, 2001). Despite these technical impediments, GISH in crucifer cytogenetics is growing in popularity and is largely confined to Brassica crop species (reviewed by Lysak and Lexer, 2006; Snowdon, 2007). Ali et al. (2004) modified the GISH protocol and were able to detect homogenously labelled parental chromosomes in the allotetraploid genome of Arabidopsis suecica. GISH was used to reveal the origin of allopolyploid Boechera (Kantama et al., 2007) and Australian Lepidium species (Dierschke et al., 2009) as well as Cardamine allopolyploid taxa (Mandáková et al., 2013, 2014). Alternatively, genome-specific centromeric repeats and (retro)transposons can be used as GISH-like probes to distinguish parental genomes, as shown in A. suecica (Comai et al., 2003) or Brassica allotetraploids (Lim et al., 2007; Alix et al., 2008).
Although the GISH technique enables easy identification of parental chromosome complements, it is not able to distinguish between individual chromosomes within a hybrid/polyploid genome. Apart from chromosome-specific repeats (e.g. rDNA and other tandem repeats), labelling of large chromosome segments or whole chromosomes in repeat-rich plant genomes using chromosome-specific painting probes has been a long-standing problem (reviewed by Schubert et al., 2001). The first large-scale painting of a plant chromosome was achieved by Lysak et al. (2001) in A. thaliana. Like most Brassicaceae species, A. thaliana is favoured for chromosome painting (CP) due to its small genome size, low amount of repetitive DNA (~15 %) clustered mainly in the pericentromeric regions, and the public availability of assembled chromosome-specific BAC libraries (Arabidopsis Genome Initiative, 2000). In the last decade, CP has been successfully applied to other Brassicaceae species (CCP), and allowed unprecedented analyses of the cruciferous genome evolution at the chromosomal level (Lysak et al., 2005, 2006; Mandáková and Lysak, 2008; Mandáková et al., 2010a, 2010b, 2012, 2013, 2015a, 2015b, 2016, 2017a, 2017b, 2017c; Hay et al., 2014; Geiser et al., 2016). Besides Brassicaceae, CP has been successfully applied in the grass genus Brachypodium (Idziak et al., 2011; Betekhtin et al., 2014). Nevertheless, the B. distachyon-specific BAC pools could not be used as probes for CCP outside the genus, and Brassicaceae remains to date the only plant family with established family-wide CCP. CCP in crucifers thus provides a unique type of data about genome evolution in plants.
An alternative chromosome panting method is based on collections of short oligo probes (about 45 nt in length) designed from any known sequence (oligopainting). A highly specific collection of short oligo probes enables detection of intended targets in (large) repeat-rich plant genomes. Oligopainting was developed by Beliveau et al. (2012) and successfully applied in Aquilegia (Filiault et al., 2018), Cucumis (Lou et al., 2014; Han et al., 2015), rice (Hou et al., 2018), Solanum (Braz et al., 2018) strawberry (Qu et al., 2017) and wheat (Du et al., 2017).
Independent auto- and allopolyploid origins of the Asian Cardamine species
Chromosome numbers and ploidy levels of C. kokaiensis (2n = 4x = 32), C. occulta (2n = 8x = 64) and C. scutata (2n = 4x = 32) were previously reported by Lihová et al. (2006a) and Šlenker et al. (2018), and confirmed in the present study. Chromosome number was newly determined in C. dentipetala (2n = 64). As x = 8 is the most common base chromosome number in the genus Cardamine (Kučera et al., 2005, and references therein; Kiefer et al., 2014), the tetraploid status of C. scutata and C. kokaiensis, and octoploid nature of C. occulta and C. dentipetala are self-evident. Our CCP data demonstrated that genomes of C. scutata and C. kokaiensis consist of 22 duplicated genomic blocks, whereas a two-fold number of GBs was identified in C. occulta and C. dentipetala. This unequivocally supports their tetra- and octoploid origins, respectively.
Despite the previous thorough phylogenetic effort of Lihová et al. (2006a), the origin and parental genomes of the Asian Cardamine polyploids remained uncertain. In the present study, GISH was used as a primary tool for the identification of the parental (sub)genomes contributing to the origin of Cardamine polyploids. GISH data were further supported by the hybridization patterns of Carcen and telomeric repeats, and most importantly by parental-specific chromosome alterations revealed by CCP. Chromosomal rearrangements show only a low tendency towards convergent evolution and are generally considered to be phylogenetically informative rare genomic changes (Rokas and Holand, 2000). Therefore, CCP in the mustard family has the power to uncover parental genomes, resolve conflicting phylogenetic signals or elucidate poorly supported tree topologies (Mandáková and Lysak, 2008; Mandáková et al., 2010a, 2010b, 2013, 2014, 2017c).
Contrasting patterns of genome origin among different polyploid Cardamine species have been revealed in our study. The tetraploid genome of C. scutata probably originated by hybridization between two differentiated diploid genomes closely related to two Eurasian species, C. parviflora and C. amara. By contrast, an autotetraploid origin of C. kokaiensis from a parental genome related to C. parviflora was suggested. The octoploid C. occulta probably arose through hybridization between the two tetraploids. The octoploid genome of C. dentipetala structurally mirrors C. scutata and probably originated from C. scutata via autopolyploidization. The cytogenetic evidence for the parental (sub)genomes of C. kokaiensis, C. occulta and C. scutata also explain differences in relative genome size values recently reported for these polyploids (Šlenker et al., 2018). Significant differences in monoploid genome sizes, even between species of the same ploidy level, were observed for C. flexuosa, C. occulta, C. scutata and C. kokaiensis (C. dentipetala was not included in that study) and attributed to their assumed different polyploid origins. Indeed, taking into account different genome sizes of the diploid progenitors and assuming additivity of genome sizes in the polyploids, the observed genome size data perfectly fit the scenarios of auto- and allopolyploid origins as revealed here by GISH and CCP. Because the taxonomic evaluation of the assumed narrow endemics C. niigatensis (tetraploid) and C. longifructus (ploidy level unknown) remains uncertain (see Introduction), whether they are distinct from C. scutata and C. dentipetala, respectively, or not, we can speculate only on if and how they were involved in the polyploid origins inferred here. Additionally, the hexaploid C. fallax (not available for the present study) might have a similar parentage as C. occulta as deduced from the shared ITS ribotypes and cpDNA haplotypes (Lihová et al., 2006a), or even participated as an intermediary in the origin of this octoploid. Some pieces in the complex mosaic of the polyploid evolution in Asian Cardamine species could thus be still missing. If this is the case, we might also consider that some of the post-polyploid intrachromosomal rearrangements inferred in C. dentipetala, C. kokaiensis and C. occulta (see also below) may have occurred already in some of these unsampled (or even extinct) taxa, representing potential middle steps in the reconstructed polyploid pathways.
The identical genome structure, including C. occulta-specific repositioning of AK7O, revealed in nine different populations of C. occulta from Asia (China and Japan) and Europe (Spain), suggests a single origin of the allooctoploid. Similarly, identical centromere repositionings of AK1K, AK3K and AK8/6K identified in both Chinese and Japanese populations argues for a single origin of C. kokaiensis. Considering the ease of GISH and the strength of hybridization signals (Fig. 2), as well as the structural stasis of the parental subgenomes (Fig. 1), C. dentipetala, C. kokaiensis, C. occulta and C. scutata may be regarded as relatively young polyploids.
Multiple and independent auto- and allopolyploidization events are not rare in the genus Cardamine (Marhold and Lihová, 2006; Carlsen et al., 2009), with 68 % of taxa being entirely neopolyploid or having both diploid and polyploid cytotypes (Kučera et al., 2005). Spontaneous interspecific hybridization has been frequently reported to occur between several Cardamine species (Marhold et al., 2002, 2018; Lihová et al., 2006a, b; Carlsen et al., 2009). For example, the widespread species C. flexuosa was recently shown to be an allotetraploid (2n = 4x = 32) originating from two diploid species, C. amara and C. hirsuta (2n = 2x = 16; Mandáková et al., 2014). Furthermore, recent cytogenetic and molecular studies (Mandáková et al., 2013; Zozomová-Lihová et al., 2014a, b) provided several lines of evidence that C. schulzii is a trigenomic hybrid species comprising two cytotypes (2n = 5x = 38 and 2x = 6x = 46), both containing subgenomes derived from C. amara, C. rivularis (both 2n = 2x = 16) and C. pratensis (2n = 4x = 30). Accordingly, hybridization and polyploidy have been significant driving forces of diversification and speciation in the genus Cardamine.
Post-polyploid genome stasis in Cardamine
Evolution of the post-polyploid genome in the investigated polyploid Cardamine species has not been associated with descending dysploidy. However, a growing body of data suggests that descending dysploidy is not rare in Cardamine. Lawrence (1931) was the first to suspect that the hypotetraploid chromosome number (2n = 30) in C. pratensis might have resulted from chromosome fusion in otherwise tetraploid species (2n = 4x = 32). The origin of the fusion chromosome was recently confirmed by Mandáková et al. (2013) as a nested chromosome insertion between two non-homeologous chromosomes accompanied by centromere elimination. Recently, Mandáková et al. (2016) demonstrated that the North American species C. cordifolia bearing a triploid-like chromosome number (2n = 24) is not a triploid hybrid but a diploidized tetraploid. In C. cordifolia, the ancestral tetraploid chromosome number was reduced (from 2n = 32 to 24) through four terminal chromosome translocations between non-homeologous chromosomes coupled with elimination of four centromeres.
Given the largely conserved chromosome collinearity shared among the parental diploid Cardamine species and the polyploids, individual subgenomes exhibit considerable stasis within the polyploid genomes. No interchromosomal rearrangements, such as reciprocal translocations, were observed. This is surprising, considering the possibility of intergenomic repatterning via homeologous recombination as a result of almost complete interspecies genome collinearity between the parental species (Fig. 1). Similarly, the overall stability of parental subgenomes was observed in tetraploid C. flexuosa (2n = 4x = 32). In C. flexuosa, only single intergenomic translocation between two homeologues has occured since its allopolyploid origin from structurally highly collinear diploid genomes of C. amara and C. hirsuta (Mandáková et al., 2014). Reciprocal translocation between two homeologous chromosomes in C. flexuosa may indicate the start of diploidization of the allopolyploid genome. The absence of intergenomic rearrangements in the Asian Cardamine polyploids suggests their more recent origin or slower tempo of post-polyploid diploidization compared to C. flexuosa. A robust phylogenetic analysis of Cardamine with estimation of divergence times will be essential to resolve this question.
Whereas the genome of C. scutata completely mirrors the parental genomes of C. amara and C. parviflora, and the remaining Cardamine polyploid genomes are largely collinear, two essential post-polyploid intrachromosomal rearrangements were identified as mechanisms shaping the chromosome structure in C. kokaiensis, C. occulta and C. dentipetala: pericentric inversion and centromere repositioning. In particular, the origins of the AK1K, AK1D, AK3K, AK7O and AK8/6K chromosomes are unique in the context of genome evolution in Brassicaceae. Whereas these chromosomes have retained the ancestral organization/collinearity of genomic blocks, their centromeres have relocated. Interestingly, in all cases, centromere repositioning shaped the chromosomes from acro- to (sub)metacentric. The only other examples where centromere repositioning in crucifers are inferred are in the telocentric homeologues of AK3 and AK8 in Cardamine rivularis (Mandáková et al., 2013) and Neslia paniculata (Camelineae; Lysak et al., 2006), respectively.
Cardamineae-specific reciprocal translocation
CCP with A. thaliana painting probes arranged according to the structure of ACK revealed that the analysed Cardamine species share six ancestral chromosomes with ACK (AK1–AK5 and AK7), whereas two chromosomes (AK6 and AK8) participated in a reciprocal translocation event. As the two translocation chromosomes AK6/8 and AK8/6 were also found in complements of other Cardamine species (Mandáková et al., 2013, 2014, 2016; Hay et al., 2014), Armoracia rusticana (Mandáková and Lysak, 2019), Leavenworthia alabamica (Haudry et al., 2013), Nasturtium officinale and N. microphyllum (Mandáková and Lysak, 2019), but not in other Brassicaceae species outside the tribe Cardamineae, the AK6–AK8 translocation probably occured prior to the diversification of Cardamineae. The Cardamineae-specific AK6–AK8 translocation thus corroborates the monophyly of the tribe and represents a unique taxonomic marker delimiting the tribe. Cardamineae genomes descended from the ancestral Cardamineae genome (n = 8) containing chromosomes AK1–AK5, AK6/8, AK7 and AK8/6 (Supplementary Data Figure S2) and exhibit remarkable genome stasis over millions of years of their independent evolution. The data suggest that the infratribal diversification in the Cardamineae was not caused by or associated with gross chromosome alternations. We hypothesize that the AK6–AK8 translocation may confer a selective adaptive advantage to their carriers. As the tribe comprises some important vegetable, weedy and model species, the established ancestral Cardamineae genome provides useful genomic resource for this crucifer group.
Speciation through polyploidization in Cardamine has been associated with ecological niche separation between the parental species and the newly formed polyploid
Hybrid/polyploid establishment and persistence imply that the population has become isolated from both of its parental taxa. This can be achieved either by chromosomal rearrangements and subsequent postzygotic genetic isolation, which have been observed in newly formed hybrids. Alternatively, ecological divergence, e.g. spread into new ecological niches not occupied by the parents, results in spatial isolation and gene flow restriction. Other ecological barriers, such as temporal or pollinator divergence, can contribute to reproductive isolation as well (Gross and Rieseberg, 2005; Hegarty and Hiscock, 2005).
The role of human-induced environmental disturbance for creating new habitats available for hybrids/polyploids is also widely recognized, and apparently was crucial also for the origin of several Brassicaceae hybrids and allopolyploids, such as Cardamine × insueta and C. schulzii (Mandáková et al., 2013) or Rorippa × armoracioides (Bleeker, 2003). The absence of gross chromosome alterations in the studied Asian Cardamine tetra- and octoploids compared to their parental species implies that postzygotic genetic isolation did not play a major role in the speciation events. More probably, the origin and successful spreading of C. occulta have been associated with the establishment of suitable irrigated human-made habitats.
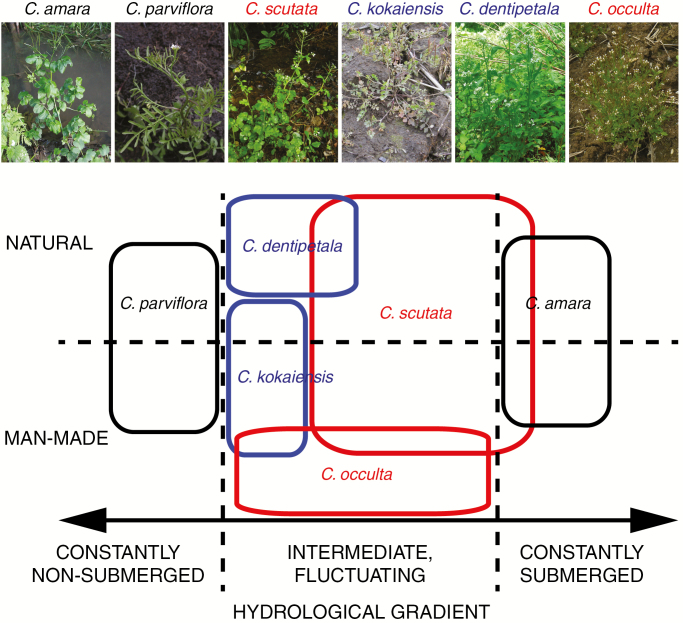
From an ecological point of view, the diversification of the genus Cardamine is characterized by niche separation along the hydrological gradients, ranging from dry habitats to constantly waterlogged habitats, including moist but non-submerged or periodically submerged habitats as intermediates (Lihová and Marhold, 2006). The habitats of the four polyploids examined in this study are characterized by the mode of water fluctuation (Fig. 4). Cardamine scutata grows in habitats where below-ground parts are waterlogged for most of the year, and its ecological distribution extends from natural habitats to human-made habitats surrounding rice paddy fields (Kudoh, 2017). Cardamine dentipetala occurs in similar habitats to those for C. scutata, but less submerged and its distribution is restricted to natural habitats. Cardamine kokaiensis inhabits seasonally flooded habitats, such as river floodplains and paddy fields, and grows ephemerally as annuals by utilizing non-flooded periods (Šlenker et al., 2018). Interestingly, C. occulta primarily occurs in paddy fields and surrounding human-made habitats (Kudoh et al., 1993; Kudoh, 2017). The recurrent speciation through polyploidization in Cardamine has been reported to be associated with ecological niche separation between the parental species and the newly formed polyploids (Shimizu-Inatsugi et al., 2017). In fact, growth experiments in which hydrological conditions were manipulated (i.e. water-logged, un-submerged, and fluctuating conditions) showed that C. scutata grew well across all conditions, but parental diploids, C. amara and C. parviflora, were specialized to waterlogged and non-submerged conditions, respectively (Shimizu-Inatsugi et al., 2017). Although future evaluation is required, the new combinations originating from allo- and autopolyploidization between the parental genomes may have enhanced the establishment of the studied polyploids by exploring novel niches along the hydrological gradient.
Fig. 4.
Schematic diagram representing niche separation among the six studied Cardamine species along the hydrological gradient (a horizontal arrow separated by vertical broken lines) in natural and human-made habitats (vertically arranged and separated by a horizontal broken line). Black, blue and red rectangles represent habitat ranges of the parental diploids, autopolyploids and allopolyploids, respectively.
From the biogeographical perspective, it is worth mentioning that from the revealed diploid parental species, C. parviflora is almost cosmopolitan, occurring throughout Eurasia (Lihová et al., 2006a, and references therein), while C. amara is currently absent from Eastern Asia (it occurs throughout Europe and its easternmost occurrence is reported from West Siberia; Malyschev and Peschkova, 1994). Nevertheless, there are several other polyploid Cardamine perennial species that morphologically closely resemble C. amara, strongly suggesting it as at least one of the parental species [most prominent examples being C. torrentis Nakai, C. amariformis Nakai and C. valida (Takeda) Nakai; Lihová et al., 2010]. Therefore, it seems reasonable to expect that diploid C. amara had a much wider distribution area in the past, also reaching easternmost Asia, and participated in multiple polyploidization events there. Furthermore, the tetraploids C. scutata and C. kokaiensis grow sympatrically in Eastern Asia, although the area of the latter species is at least currently fragmented and much more restricted (Šlenker et al., 2018). Hybridization between them resulted in the widespread and invasive C. occulta colonizing all continents. Finally, the occurrence of the octoploid C. dentipetala seems to be tightly connected with that of its parent, C. scutata, as it grows sympatrically with or in close proximity to the latter species, in line with its inferred autopolyploid origin. They also share similar habitat preferences, but C. dentipetala exhibits a shifted and narrower ecological niche (Fig. 4).
CONCLUSIONS
The present cytogenetic study provides unequivocal evidence of the auto- and allopolyploid origins of Asian Cardamine polyploid species, identifies their parental genomes and presents their genome structures. The comparative cytogenetic maps demonstrate that post-polyploid genome evolution in the Cardamine polyploids was not associated with descending dysploidy or gross chromosome rearrangements. The data serve as a basis for future detailed genomic studies, which should address the invasive allooctoploid C. occulta, the most widespread Cardamine species in the world.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Figure S1: In situ localization of Arabidopsis-type telomere repeat (TTTAGGG)n and C. amara-specific 290-bp Carcen tandem repeat on chromosomes of C. amara, C. parviflora, C. scutata and C. occulta. Interstitial telomeric repeats (ITRs) were observed at all pericentromere regions in C. parviflora, but not in C. amara. As expected, the C. amara-specific tandem repeat Carcen localized to all centromeres in C. amara, but not to chromosomes of C. parviflora. In allotetraploid C. scutata, FISH signals revealed 16 chromosomes bearing ITRs and the other 16 chromosomes containing the Carcen repeat. In allo-octoploid C. occulta, the telomeric probe and Carcen localized to ~48 and 16 centromeres, respectively. Interstitial telomeric signals thus marked 32 chromosomes originating from C. kokaiensis (i.e. 16 + 16 C. parviflora-like chromosomes forming the complement of C. kokaiensis) and 16 chromosomes of C. scutata (i.e. 16 C. parviflora-like chromosomes). By contrast, Carcen delimited 16 chromosomes originating from C. scutata (i.e. 16 C. amara-like chromosomes from the allotetraploid complement of C. scutata). Figure S2: Structure of the ancestral Cardamine genome.
ACKNOWLEDGMENTS
This work was supported by a research grant from the Czech Science Foundation (grant nos. 16-10809S and 17-13029S), the Central European Institute of Technology, CEITEC 2020 project (grant no. LQ1601), and the project of the Vedecká Grantová Agentúra MŠVVaŠ SR a SAV, Slovak Republic (grant no. 2/0133/17). Core Facility Plant Sciences of CEITEC MU is acknowledged for the cultivation of experimental plants used in this paper.
LITERATURE CITED
- Ali HB, Lysak MA, Schubert I. 2004. Genomic in situ hybridization in plants with small genomes is feasible and elucidates the chromosomal parentage in interspecific Arabidopsis hybrids. Genome 47: 954–960. [DOI] [PubMed] [Google Scholar]
- Alix K, Joets J, Ryder CD, et al. . 2008. The CACTA transposon Bot1 played a major role in Brassica genome divergence and gene proliferation. Plant Journal 56: 1030–1044. [DOI] [PubMed] [Google Scholar]
- Al-Shehbaz IA. 2012. A generic and tribal synopsis of the Brassicaceae (Cruciferae). Taxon 61: 931–954. [Google Scholar]
- Al-Shehbaz IA, Arai K, Ohba H. 2006. Cardamine. In: Iwatsuki K, Boufford DE, Ohba H, eds. Flora of Japan, Vol. IIa, Angiospermae, Dicotyledoneae, Archichlamydeae. Tokyo: Kodansha, 482–490. [Google Scholar]
- Al-Shehbaz IA, Marhold K, Lihová J. 2010. Cardamine Linnaeus. In: Flora of North America Editorial Committee, Flora of North America: North of Mexico, Volume 7, Magnoliophyta: Salicaceae to Brassicaceae. New York: Oxford University Press, 464–484. [Google Scholar]
- Arabidopsis Genome Initiative. 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815. [DOI] [PubMed] [Google Scholar]
- Beliveau BJ, Joycea EF, Apostolopoulosa N, et al. . 2012. Versatile design and synthesis platform for visualizing genomes with Oligopaint FISH probes. Proceedings of the National Academy of Sciences of the United States of America 109: 21301–21306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betekhtin A, Jenkins G, Hasterok R. 2014. Reconstructing the evolution of Brachypodium genomes using comparative chromosome painting. PLoS One 9: e115108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleeker W. 2003. Hybridization and Rorippa austriaca (Brassicaceae) invasion in Germany. Molecular Ecology 12: 1831–1841. [DOI] [PubMed] [Google Scholar]
- Braz GT, He L, Zhao H, et al. . 2018. Comparative oligo-FISH mapping: an efficient and powerful methodology to reveal karyotypic and chromosomal evolution. Genetics 208: 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsen T, Bleeker W, Hurka H, Elven R, Brochmann C. 2009. Biogeography and phylogeny of Cardamine (Brassicaceae). Annals of the Missouri Botanical Garden 96: 215–236. [Google Scholar]
- Chester M, Leitch AR, Soltis PS, Soltis DE. 2010. Review of the application of modern cytogenetic methods (FISH/GISH) to the study of reticulation (polyploidy/hybridisation). Genes 1: 166–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L, Tyagi AP, Lysak MA. 2003. FISH analysis of meiosis in Arabidopsis allopolyploids. Chromosome Research 11: 217–226. [DOI] [PubMed] [Google Scholar]
- D’Hont A. 2005. Unraveling the genome structure of polyploids using FISH and GISH; examples of sugarcane and banana. Cytogenetic and Genome Research 109: 27–33. [DOI] [PubMed] [Google Scholar]
- Dierschke T, Mandáková T, Lysak MA, Mummenhoff K. 2009. A bicontinental origin of polyploid Australian/New Zealand Lepidium species (Brassicaceae)? Evidence from genomic in situ hybridization. Annals of Botany 204: 681–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du P, Zhuang L, Wang Y, et al. . 2017. Development of oligonucleotides and multiplex probes for quick and accurate identification of wheat and Thinopyrum bessarabicum chromosomes. Genome 60: 93–103. [DOI] [PubMed] [Google Scholar]
- Filiault DL, Ballerini ES, Mandáková T, et al. . 2018. The Aquilegia genome provides insight into adaptive radiation and reveals an extraordinarily polymorphic chromosome with a unique history. eLife 7: e36426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser C, Mandáková T, Arrigo N, Lysak MA, Parisod C. 2016. Repeated whole-genome duplication, karyotype reshuffling and biased retention of stress-responding genes in Buckler Mustards. Plant Cell 28: 17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross BL, Rieseberg LH. 2005. The ecological genetics of homoploid hybrid speciation. Journal of Heredity 96: 241–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Zhang T, Thammapichai P, Weng Y, Jiang J. 2015. Chromosome-specific painting in Cucumis species using bulked oligonucleotides. Genetics 200: 771–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haudry A, Platts AE, Vello E, et al. . 2013. An atlas of over 90,000 conserved non-coding sequences yields detailed insight into crucifer regulatory regions. Nature Genetics 45: 891–898. [DOI] [PubMed] [Google Scholar]
- Hay AS, Pieper B, Cooke E, et al. . 2014. Cardamine hirsuta: a versatile genetic system for comparative studies. Plant Journal 78: 1–15. [DOI] [PubMed] [Google Scholar]
- Hay A, Tsiantis M. 2006. The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nature Genetics 38: 942–947. [DOI] [PubMed] [Google Scholar]
- Heenan PB. 2017. A taxonomic revision of Cardamine L. (Brassicaceae) in New Zealand. Phytotaxa 330: 1–154. [Google Scholar]
- Hegarty MJ, Hiscock SJ. 2005. Hybrid speciation in plants: new insights from molecular studies. New Phytologist 165: 411–423. [DOI] [PubMed] [Google Scholar]
- Hou L, Xu M, Zhang Z, et al. . 2018. Chromosome painting and its applications in cultivated and wild rice. BMC Plant Biology 18: 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idziak D, Betekhtin A, Wolny E, et al. . 2011. Painting the chromosomes of Brachypodium - current status and future prospects. Chromosoma 120: 469–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ijdo JW, Wells RA, Baldini A, Reeders ST. 1991. Improved telomere detection using a telomere repeat probe (TTAGGG)n generated by PCR. Nucleic Acids Research 19: 4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantama L, Sharbel TF, Schranz ME, Mitchell-Olds T, de Vries S, de Jong H. 2007. Diploid apomicts of the Boechera holboellii complex display large-scale chromosome substitutions and aberrant chromosomes. Proceedings of the National Academy of Sciences of the United States of America 104: 14026–14031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer M, Schmickl R, German DA, et al. . 2014. BrassiBase: Introduction to a novel knowledge database on Brassicaceae evolution. Plant and Cell Physiology 55: e3. [DOI] [PubMed] [Google Scholar]
- Kučera J, Valko I, Marhold K. 2005. On-line database of the chromosome numbers of the genus Cardamine (Brassicaceae). Biologia 60: 473–476. Database available online at http://www.cardamine.sav.sk/www/index.php?lang=en [Google Scholar]
- Kudoh H. 2017. Biology of the weedy species of the genus Cardamine (Brassicaceae) in Japan. Journal of Weed Science and Technology 62: 175–183 (in Japanese with English summary). [Google Scholar]
- Kudoh H, Ishiguri Y, Kawano S. 1993. Phenotypic variability in life history traits and phenology of field populations of Cardamine flexuosa and C. fallax (Cruciferae) in Honshu, Japan. Plant Species Biology 8: 7–20. [Google Scholar]
- Lawrence WJC. 1931. The chromosome constitution of Cardamine pratensis and Verbascum phoeniceum. Genetica 13: 183–208. [Google Scholar]
- Lihová J, Kučera J. 2007. [Reports]. In: Marhold K, ed., IAPT/IOPB Chromosome data reports 4. Taxon 56: 1269, E1–E2. [Google Scholar]
- Lihová J, Marhold K. 2006. Phylogenetic and diversity patterns in Cardamine (Brassicaceae) – a genus with conspicuous polyploid and reticulate evolution. In: Sharma AK, Sharma A, eds. Plant genome: Biodiversity and evolution, Volume 1C, Phanerogams (Angiosperms - Dicotyledons). Enfield: Science Publishers, Inc, 149–186. [Google Scholar]
- Lihová J, Marhold K, Kudoh H, Koch MA. 2006a Worldwide phylogeny and biogeography of Cardamine flexuosa (Brassicaceae) and its relatives. American Journal of Botany 93: 1206–1221. [DOI] [PubMed] [Google Scholar]
- Lihová J, Shimizu KK, Marhold K. 2006b Allopolyploid origin of Cardamine asarifolia (Brassicaceae): incongruence between plastid and nuclear ribosomal DNA sequences solved by a single-copy nuclear gene. Molecular Phylogenetics and Evolution 39: 759–786. [DOI] [PubMed] [Google Scholar]
- Lihová J, Kudoh H, Marhold K. 2010. Morphometric studies of polyploid Cardamine species (Brassicaceae) from Japan: solving a long-standing taxonomic and nomenclatural controversy. Australian Systematic Botany 23: 94–111. [Google Scholar]
- Lim KB, Yang TJ, Hwang YJ, et al. . 2007. Characterization of the centromere and peri-centromere retrotransposons in Brassica rapa and their distribution in related Brassica species. Plant Journal 49: 173–183. [DOI] [PubMed] [Google Scholar]
- Lou Q, Zhang Y, He Y, et al. . 2014. Single‐copy genebased chromosome painting in cucumber and its application for chromosome rearrangement analysis in Cucumis. The Plant Journal 78: 169–179. [DOI] [PubMed] [Google Scholar]
- Lysak MA, Lexer C. 2006. Towards the era of comparative evolutionary genomics in Brassicaceae. Plant Systematics and Evolution 259: 175–198. [Google Scholar]
- Lysak MA, Mandáková T. 2013. Analysis of plant meiotic chromosomes by chromosome painting. In: Clifton NJ, ed. Methods in molecular biology. New York: Humana Press, 13–24. [DOI] [PubMed] [Google Scholar]
- Lysak MA, Fransz PF, Ali HBM, Schubert I. 2001. Chromosome painting in Arabidopsis thaliana. Plant Journal 28: 689–697. [DOI] [PubMed] [Google Scholar]
- Lysak MA, Koch M, Pecinka A, Schubert I. 2005. Chromosome triplication found across the tribe Brassiceae. Genome Research 15: 516–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak MA, Berr A, Pecinka A, Schmidt R, McBreen K, Schubert I. 2006. Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proceedings of the National Academy of Sciences of the United States of America 103: 5224–5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak MA, Mandáková T, Schranz ME. 2016. Comparative paleogenomics of crucifers: ancestral genomic blocks revisited. Current Opinion in Plant Biology 30: 108–115. [DOI] [PubMed] [Google Scholar]
- Maiorov SR. 2018. Melkotsvetkovye serdechniki sektsii Pteroneuron (DC.) Rouy et Fouc. (Cardamine L., Cruciferae) vo flore evropeiskoi Rossii. Phytodiversity of Eastern Europe 12: 6–17. [Google Scholar]
- Malyschev LI, Peschkova GA. 1994. Flora Sibiri, vol. 7 Novosibirsk: Nauka. [Google Scholar]
- Mandáková T, Lysak MA. 2008. Chromosomal phylogeny and karyotype evolution in x = 7 crucifer species (Brassicaceae). Plant Cell 20: 2559–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandáková T, Lysak MA. 2016a Chromosome preparation for cytogenetic analyses in Arabidopsis. Current Protocols in Plant Biology 1: 43–51. [DOI] [PubMed] [Google Scholar]
- Mandáková T, Lysak MA. 2016b Painting of Arabidopsis chromosomes with chromosome-specific BAC clones. Current Protocols in Plant Biology 1: 359–371. [DOI] [PubMed] [Google Scholar]
- Mandáková T, Lysak MA. 2019. Healthy leaves and roots: comparative structure of horseradish and watercress genomes. Plant Physiology 179: 66–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandáková T, Joly S, Krzywinski M, Mummenhoff K, Lysak MA. 2010a Fast diploidization in close mesopolyploid relatives of Arabidopsis. Plant Cell 22: 2277–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandáková T, Heenan PB, Lysak MA. 2010b Island species radiation and karyotypic stasis in Pachycladon. BMC Evolutionary Biology 10: 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandáková T, Mummenhoff K, Al-Shehbaz IA, Mucina L, Muehlhausee A, Lysak MA. 2012. Whole-genome triplication and species radiation in the southern African tribe Heliophileae (Brassicaceae). Taxon 61: 989–1000. [Google Scholar]
- Mandáková T, Shimizu Inatsugi R, Zozomová-Lihová J, et al. . 2013. The more the merrier: recent hybridization and polyploidy in Cardamine. Plant Cell 25: 3280–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandáková T, Marhold K, Lysak MA. 2014. The widespread crucifer species Cardamine flexuosa is an allotetraploid with a conserved subgenomic structure. New Phytologist 201: 982–992. [DOI] [PubMed] [Google Scholar]
- Mandáková T, Schranz ME, Sharbel TF, de Jong H, Lysak MA. 2015a Karyotype evolution in apomictic Boechera and the origin of the aberrant chromosomes. Plant Journal 82: 785–793. [DOI] [PubMed] [Google Scholar]
- Mandáková T, Singh V, Kraemer U, Lysak MA. 2015b Genome structure of the heavy metal hyperaccumulator Noccaea caerulescens and its stability on metalliferous and non-metalliferous soils. Plant Physiology 169: 674–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandáková T, Gloss AD, Whiteman NK, Lysak MA. 2016. How diploidization turned a tetraploid into a pseudotriploid. American Journal of Botany 103: 1187–1196. [DOI] [PubMed] [Google Scholar]
- Mandáková T, Hloušková P, German D, Lysak MA. 2017a Monophyletic origin and evolution of the largest crucifer genomes. Plant Physiology 174: 2062–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandáková T, Li Z, Barker MS, Lysak MA. 2017b Diverse genome organization following 13 independent mesopolyploid events in Brassicaceae contrasts with convergent patterns of gene retention. Plant Journal 91: 3–21. [DOI] [PubMed] [Google Scholar]
- Mandáková T, Pouch M, Harmannová K, Zhan SH, Mayrose I, Lysak MA. 2017c Multi-speed genome diploidization and diversification after an ancient allopolyploidization. Molecular Ecology 26: 1–18. [DOI] [PubMed] [Google Scholar]
- Marhold K, Lihová J. 2006. Polyploidy, hybridization and reticulate evolution: lessons from the Brassicaceae. Plant Systematics and Evolution 259: 143–174. [Google Scholar]
- Marhold K, Lihová J, Perný M, Grupe R, Neuffer B. 2002. Natural hybridization in Cardamine (Brassicaceae) in the Pyrenees: evidence from morphological and molecular data. Botanical Journal of the Linnean Society 139: 275–294. [Google Scholar]
- Marhold K, Šlenker M, Kudoh H, Zozomová-Lihová J. 2016. Cardamine occulta, the correct species name for invasive Asian plants previously classified as C. flexuosa, and its occurrence in Europe. PhytoKeys 62: 57–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhold K, Šlenker M, Zozomová-Lihová J. 2018. Polyploidy and hybridization in the Mediterranean and neighbouring northern areas: examples from the genus Cardamine (Brassicaceae). Biologia Serbica 40: 47–59. [Google Scholar]
- Matsumura J. 1912. Index plantarum japonicarum, Vol. 2 Tokioni: Marunzen. [Google Scholar]
- Ohwi J. 1953. Flora of Japan. Tokyo: Shibundo. [Google Scholar]
- Ohwi J. 1965. Flora of Japan. Revised edition. Tokyo: Shibundo. [Google Scholar]
- Ohwi J, Kitagawa M. 1992. New Flora of Japan. Tokyo: Shibundo Co., Ltd. Publishers. [Google Scholar]
- Post AR, Krings A, Xiang QY, Sosinski BR, Neal JC. 2009. Lectotypification of Cardamine flexuosa (Brassicaceae). Journal of the Botanical Research Institute of Texas 3: 227–230. [Google Scholar]
- Qu M, Li K, Han Y, Chen L, Li Z, Han Y. 2017. Integrated karyotyping of woodland strawberry (Fragaria vesca) with oligopaint FISH probes. Cytogenetic and Genome Research 153: 158–164. [DOI] [PubMed] [Google Scholar]
- Raina SN, Rani V. 2001. GISH technology in plant genome research. Methods in Cell Science 23: 83–104. [PubMed] [Google Scholar]
- Ramzan F, Younis A, Lim KB. 2017. Application of genomic in situ hybridization in horticultural science. International Journal of Genetics 2017: 7561909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin Rodríguez R, Greuter W. 2009. Brassicaceae. In: Greuter W, Rankin Rodríguez R, eds. Flora de la República de Cuba, serie A, Plantas Vasculares, Fascículo 15. Ruggell: AR Gantner Verlag KG, 1–51. [Google Scholar]
- Rokas A, Holland PW. 2000. Rare genomic changes as a tool for phylogenetics. Trends in Ecology and Evolution 15: 454–459. [DOI] [PubMed] [Google Scholar]
- Shimizu-Inatsugi R, Terada A, Hirose K, Kudoh H, Sese J, Shimizu KK. 2017. Plant adaptive radiation mediated by polyploid plasticity in transcriptomes. Molecular Ecology 26: 193–207. [DOI] [PubMed] [Google Scholar]
- Schubert I, Fransz PF, Fuchs J, de Jong JH. 2001. Chromosome painting in plants. Methods in Cell Science 23: 57–69. [PubMed] [Google Scholar]
- Schwarzacher T, Leitch AR, Bennett MD, Heslop-Harrison JS. 1989. In situ localization of parental genomes in a wide hybrid. Annals of Botany 64: 315–324. [Google Scholar]
- Silva GS, Souza MM. 2013. Genomic in situ hybridization in plants. Genetics and Molecular Research 12: 2953–2965. [DOI] [PubMed] [Google Scholar]
- Snowdon RJ. 2007. Cytogenetics and genome analysis in Brassica crops. Chromosome Research 15: 85–95. [DOI] [PubMed] [Google Scholar]
- Šlenker M, Zozomová-Lihová J, Mandáková T, et al. . 2018. Morphology and genome size of the widespread weed Cardamine occulta: how it differs from cleistogamic C. kokaiensis and other closely related taxa in Europe and Asia. Botanical Journal of the Linnean Society 187: 456–482. [Google Scholar]
- Thompson IR. 1996. Cardamine. In: Walsh NG, Entwisle TJ, eds. Flora of Victoria, vol. 3 Melbourne: Inkata Press, 434–442. [Google Scholar]
- Yatsu Y, Kachi N, Kudoh H. 2003. Ecological distribution and phenology of an invasive species, Cardamine hisuta L., and its native counterpart, Cardamine flexuosa With., in central Japan. Plant Species Biology 18: 35–42. [Google Scholar]
- Younis A, Ramzan F, Hwang YJ, Lim KB. 2015. FISH and GISH: molecular cytogenetic tools and their applications in ornamental plants. Plant Cell Reports 34: 1477–1488. [DOI] [PubMed] [Google Scholar]
- Zhou TY, Lu LL, Yang G, Al-Shehbaz IA. 2001. Brassicaceae. In: Wu ZY, Raven PH, eds. Flora of China, vol. 8 Beijing & St. Louis: Science Press & Missouri Botanical Garden Press, 1–193. [Google Scholar]
- Zozomová-Lihová J, Krak K, et al. . 2014. b Multiple hybridization events in Cardamine (Brassicaceae) during the last 150 years: revisiting a textbook example of neoallopolyploidy. Annals of Botany 113: 817–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zozomová-Lihová J, Mandáková T, Mummenhoff K, Kovaříková A, Lysak MA, Kovařík A. 2014a When fathers are instant loosers: homogenisation of rDNA loci in recently formed Cardamine schulzii trigenomic allopolyploid. New Phytologist 203: 1096–1108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.