Abstract
Background and Aims
Compensatory base changes (CBCs) that occur in stems of ribosomal internal transcribed spacer 2 (ITS2) can have important phylogenetic implications because they are not expected to occur within a single species and also affect selection of appropriate DNA substitution models. These effects have been demonstrated when studying ancient lineages. Here we examine these effects to quantify their importance within a more recent lineage by using both DNA- and RNA-specific models.
Methods
We examined the phylogenetic implications of the CBC process by using a comprehensive sampling of ITS2 from ten closely related species of Corydalis. We predicted ITS2 secondary structures by using homology modelling, which was then used for a structure-based alignment. Paired and unpaired regions were analysed separately and in combination by using both RNA-specific substitution models and conventional DNA models. We mapped all base-pair states of CBCs on the phylogenetic tree to infer their evolution and relative timing.
Key Results
Our results indicate that selection acted to increase the thermodynamic stability of the secondary structure. Thus, the unpaired and paired regions did not evolve under a common substitution model. Only two CBCs occurred within the lineage sampled and no striking differences in topology or support for the shared clades were found between trees constructed using DNA- or RNA-specific substitution models.
Conclusions
Although application of RNA-specific substitution models remains preferred over more conventional DNA models, we infer that application of conventional DNA models is unlikely to be problematic when conducting phylogenetic analyses of ITS2 within closely related lineages wherein few CBCs are observed. Each of the two CBCs was found within the same lineages but was not observed within a given species, which supports application of the CBC species concept.
Keywords: Compensatory base change, Corydalis, ITS2, RNA-specific model, secondary structure
INTRODUCTION
Many plant systematics studies have effectively used the internal transcribed spacer (ITS) region of rDNA (Álvarez and Wendel, 2003; Feliner and Rosselló, 2007; Qin et al., 2017). Many of these studies have focused on ITS2 for both DNA barcoding and inferring relationships among recently diverged lineages, mainly because of its high information content and ease of amplification (Chen et al., 2010). Despite a rapid rate of nucleotide substitutions, ITS2 has a highly conserved secondary structure throughout Eukaryota (Hershkovitz and Zimmer, 1996; Schultz et al., 2005; Coleman, 2007, 2015), indicating that functional constraints affect ITS2 evolution. This conserved secondary structure can facilitate sequence alignment between divergent taxa by anchoring their conserved motifs and homologous positions, thus improving the accuracy of phylogenetic reconstructions (Kjer, 1995; Keller et al., 2010; Letsch et al., 2010; Zhang et al., 2015, 2016).
The secondary structure of ITS2 is maintained by hydrogen bonds between complementary base pairs, which form the double-stranded regions (stems) of the ITS2 rDNA molecule (Coleman, 2003, 2007). Only six of the 16 possible base pairs occur frequently and are considered to be stable or relatively stable. These are the Watson–Crick pairs GC/CG and AU/UA and the intermediates UG/GU; the remaining ten base pairs are considered mismatches (Rousset et al., 1991; Savill et al., 2001). Substitutions that occur between stable base pairs often decrease the stability of the stems, which is deleterious to RNA function. Compensatory base changes (CBCs), wherein substitutions on one side of a pair are compensated by substitutions on the other side, can restore stability (Rousset et al., 1991; Wolf et al., 2013).
This co-variation pattern of stem regions violates the assumption of most phylogenetic inference methods that sites are evolving independently of each other (Posada and Crandall, 1998). Some authors have argued that failing to account for CBC substitutions results in the same variation being counted twice and can lead to misleading phylogenetic inferences with strong support (Wheeler and Honeycutt, 1988; Dixon and Hillis, 1993; Galtier, 2004).
Matthias Wolf and colleagues have effectively improved rDNA sequence alignments and tree searches by directly addressing the CBC process. First, they coded the four bases and their structural states on each side of stems by using 12 nucleotide letters (for paired left, paired right, or unpaired; Seibel et al., 2006; Wolf et al., 2014). This coding enables sequences to be aligned using secondary structure with the program 4SALE (Seibel et al., 2006, 2008). The resulting sequence-structure alignment can then be transferred to ProfDistS (Wolf et al., 2008) for neighbour-joining tree construction and to PAUP* and R for parsimony and maximum-likelihood tree construction, respectively (e.g. Markert et al., 2012; Heeg and Wolf, 2015). This integrated approach has been shown to improve both accuracy and robustness of phylogenetic analyses relative to application of standard four-state DNA models (Keller et al., 2010; Wolf, 2015; Buchheim et al., 2017).
Some RNA-specific substitution models have been suggested to account for the base-pair substitution together in both stem sides instead of considering the base states separately in each side. These models can be classified as six-state, seven-state and 16-state models according to their alternative treatments of complementary base pairs (Tillier and Collins, 1998; Savill et al., 2001; Allen and Whelan, 2014). Most widely used programs for phylogenetic inference do not implement these RNA-specific substitution models, though MrBayes (Ronquist et al., 2012) does provide a doublet model. The PHASE package (Jow et al., 2002; Hudelot et al., 2003; Allen and Whelan, 2014) was specifically designed for phylogenetic analyses of RNA and includes RNA-specific substitution models. Application of these RNA models has often demonstrated their superiority over commonly used DNA models based on their shorter inferred branch lengths and higher likelihoods (e.g. Hudelot et al., 2003; Telford et al., 2005; Patiño-Galindo et al., 2018). But other studies have found that using RNA models down-weights phylogenetic signal from stems, thereby effectively up-weighting signal from loops (Letsch et al., 2010; Letsch and Kjer, 2011). The loops are more liable to be saturated by multiple hits along individual branches and/or be misaligned. Both of these problems can result in inaccurate phylogenetic inferences (Letsch et al., 2010; Letsch and Kjer, 2011). These results, wherein using RNA-specific models can be both advantageous as well as disadvantageous for phylogenetic inference, come primarily from studies that have sampled ancient lineages for which saturation and/or misalignment are particular concerns. Few studies have quantified the benefit of RNA-specific models in the context of phylogenetic inferences among closely related species (Marinho et al., 2011; Adebowale et al., 2016).
Another potential use of CBCs in ITS2 is species delimitation. Based on her study of ITS2 among species of unicellular green alga, Coleman (2000, 2009) hypothesized that organisms differing by even a single CBC in ITS2 conserved stems are unable to cross. Müller et al. (2007) corroborated Coleman’s hypothesis based on their large-dataset analyses of fungi and plants. In 93 % of the cases wherein two organisms differed by a CBC in ITS2 they were classified as distinct species (Müller et al., 2007). CBCs in ITS2 were also found to have taxonomic value in some animal lineages. For example, Wolf et al. (2007) asserted that Trichoplax adhaerens, the simplest known animal species, consists of at least two species based on ITS2 CBC evidence. Likewise, three new species identified using ITS2 CBCs that are morphologically indistinguishable from Paramacrobiotus richtersi have been confirmed by using 18S rDNA, physiological and biochemical data (Schill et al., 2010). Based on this evidence, wherein CBCs occur among rather than within species, Wolf et al. (2013) developed a generalized ‘CBC species concept’.
Corydalis (Papaveraceae) species are often characterized by their large and colourful petal spurs. Hence they may have economic value as ornamentals. However, they are not well known to the public because they often bloom in early spring, have a limited geographical distribution, and are short-lived perennials. Some species with tuberous roots, especially species of section Pes-gallinaceus, are important medical plants in East Asia. Jiang et al. (2018) sequenced the ITS region of some species in section Pes-gallinaceus, but found it to be polymorphic. In this study we sampled a clade of 10 Chinese species in section Pes-gallinaceus, including multiple specimens from nine species that were sampled from different geographical regions. We cloned and sequenced the PCR products to identify the ITS2 alleles within polymorphic individuals. We then traced the history of CBC substitutions and tested whether they correspond with species delimitations. Finally, we quantified the effects of alternative substitution models (both conventional DNA models and RNA-specific models) on the inferred phylogeny.
MATERIALS AND METHODS
Taxon sampling
The principles that we used for our sampling procedure in order to effectively study CBC evolution are as follows. First, we sampled multiple closely related species to enable us to trace the step-by-step substitution pattern within a single lineage. Second, we sampled multiple individuals from nine of the ten species (a single individual of Corydalis linjiangensis was sampled) to distinguish between apomorphies among species from variation within individual species. We sampled 35 plants from these ten species in Corydalis section Pes-gallinaceus as delimited by the plastid-based phylogeny inferred from Jiang et al. (2018) (Supplementary Data Table S1). Corydalis huangshanensis, from the sister section (Duplotuber), was used as the outgroup for comparisons between DNA- and RNA-specific models. Eleven additional species from Pérez-Gutiérrez et al. (2015) were added to calibrate node ages for the molecular dating analysis (Supplementary Data Table S1).
DNA extraction, amplification and sequencing
Genomic DNA was extracted from silica gel-dried leaves following a modified CTAB protocol (Porebski et al., 1997) and then purified with Plant DNA Extraction Kits (Tiangen Biotech, Beijing, China). The PCR amplifications were carried out with primer pair ITS2F and ITS3R (Hou et al., 2013). PCR reactions in a 25 μL volume included 40–100 ng of DNA template, 2.5 µL of 2.5 mm of each dNTP, 2.5 µL of 10× PCR buffer, 0.5 µL of 10 µm of each primer, and 0.625 U of Taq polymerase. The PCR program setting was as follows: 94 °C for 4 min; 35 cycles of 94 °C for 30 s, 53 °C for 30 s and 72 °C for 60 s; followed by an extension period of 72 °C for 10 min. We cloned PCR products by using the pUCm-T carrier system. At least eight clones per individual were sequenced with the primer M13 (-48) on an ABI 3730XL sequencer (Applied Biosystems, Foster City, CA, USA).
Secondary-structure prediction and partition
ITS2 boundaries were identified by using hidden Markov models implemented in the ITS2 Ribosomal RNA Database (http://its2.bioapps.biozentrum.uni-wuerzburg.de/;Ankenbrand et al., 2015). The secondary structure (Vienna format) of ITS2 was obtained via homology prediction using the most similar sequence with a modelled structure in the database (Selig et al., 2008). 4SALE (Seibel et al., 2006, 2008) was developed to both align sequences and associate secondary structures simultaneously for subsequent sequence-structure and phylogenetic analyses (e.g. Keller et al., 2010; Markert et al., 2012; Wolf et al., 2014; Heeg and Wolf, 2015). Alternatively, in this study we used 4SALE to generate the consensus secondary structure of our dataset after sequence structures had been aligned and manually refined. This consensus secondary structure provides an accessible and informative visualization of the structural information contained in the alignment (Seibel et al., 2008) (Fig. 1). We partitioned the ITS2 primary sequence into paired and unpaired regions, and analysed them both separately and in combination in order to test whether the following variables differed between them. Nucleotide composition and variation, the ratio of transitions to transversions (Ts/Tv) and genetic distances were calculated using MEGA 7 (Kumar et al., 2016). Parsimony-informative sites were identified using PAUP* 4.0b10 (Swofford, 2003). Levels of homoplasy were compared between paired and unpaired regions using the substitution-saturation test implemented in DAMBE5 (Xia, 2013).
Fig. 1.
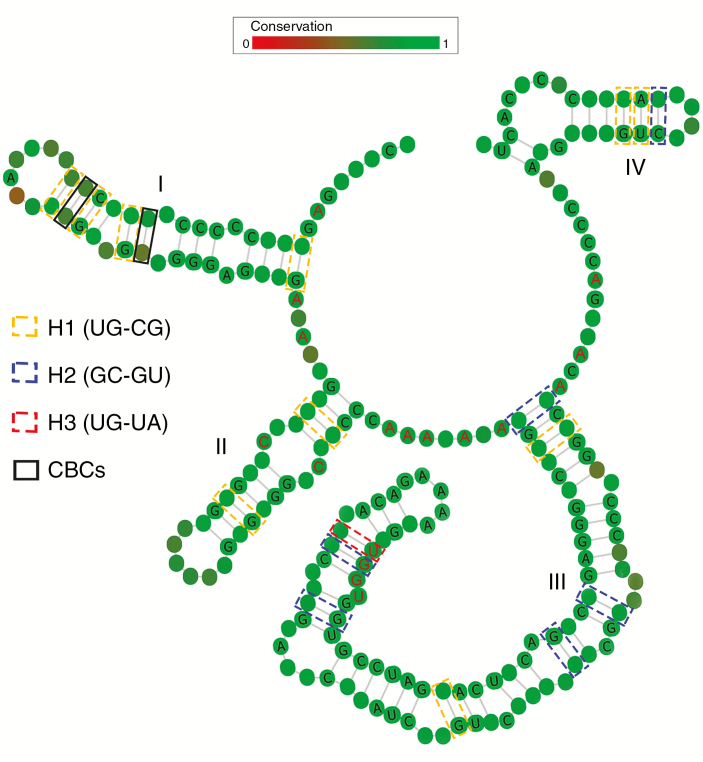
Consensus ITS2 secondary structure derived from Corydalis species. The four stems are labelled I–IV. The pyrimidine–pyrimidine (C) bulge in stem II, the UGGU in stem III and the high A content between stems that are common to nearly all angiosperms are indicated in red colour. CBCs and hemi-CBCs (H1–3) distributed in the stems are highlighted in black solid boxes and coloured dashed boxes, respectively. Degree of conservation over the entire alignment is displayed in colour grades from green (conservative) to red (variable).
Phylogenetic analyses of ITS2 using DNA substitution model
Two sets of alignments and phylogenetic analyses were performed. The first set relied strictly upon DNA sequences and models without reference to RNA secondary structure or RNA-specific models. The second set used both RNA secondary structure and RNA-specific models.
The first set of alignments and phylogenetic analyses were performed as follows. The complete ITS2 nucleotide sequences were aligned using a purely sequence-based method (G-INS-i), which is the most accurate iterative refinement method in MAFFT that does not take into account secondary structure (Katoh and Toh, 2008). Gaps were treated as missing data rather than coded as separate characters (Simmons and Ochoterena, 2000) in order to make the DNA-specific analyses directly comparable to the RNA-specific analyses (PHASE, which was used for the RNA-specific analyses, cannot analyse gap characters). Parsimony analysis was performed using PAUP* 4.0b10 (Swofford, 2003) with the following settings: heuristic search of 1000 random-addition replicates; tree-bisection-reconnection branch swapping and up to 5000 trees saved; 1000 bootstrap replicates with ten heuristic searches per replicate.
Alternative models of nucleotide substitution were examined using jModeltest 2.1.7 (Darriba et al., 2012). The best-fit model selected using the Akaike information criterion (AIC; Akaike, 1974) was GTR+G, which was then used for maximum likelihood (ML) analysis in PhyML 3.0 (Guindon et al., 2010). Non-parametric bootstrap values were then computed from 1000 pseudoreplicates with both nearest-neighbour interchange (NNI) and subtree-pruning-regrafting (SPR) tree searches. Bayesian inference (BI; Yang and Rannala, 1997) was implemented in MrBayes 3.2 (Ronquist et al., 2012) using the GTR+G model and Prset statefreqpr=dirichlet (1,1,1,1), which was selected as the best-fit model by MrModeltest 2.3 (Nylander, 2004). Two independent runs with four Markov chain Monte Carlo (MCMC) chains were each performed for 1 000 000 generations with trees every 100 generations. The initial 3000 sampled trees were discarded as burn-in. Convergence of the two Bayesian MCMC runs was verified by examining effective sample sizes (>200) for each parameter estimate in Tracer 1.6 (http://tree.bio.ed.ac.uk/software/tracer/). We also determined topological convergence based on split frequencies and tree distances by using RWTY (Warren et al., 2017).
Molecular dating
In order to infer ages of CBCs, we used a Bayesian relaxed-clock method as implemented in BEAST 1.7.5 (Drummond et al., 2012). Because of the lack of reliably identified fossils within Corydalis, we used a molecular chronogram of subfamily Fumarioideae (Papaveraceae) estimated by Pérez-Gutiérrez et al. (2015), from which we set the crown age of Corydalis at 37.73 mya, with a 95 % highest posterior density (HPD) of 27–49 mya. Likewise, the crown age of the sister group of Corydalis was set at 33.33 mya (95 % HPD 24–43.5 mya).We performed the BEAST analysis using the GTR+G model, four-category gamma-shaped distribution and a Yule speciation process as a prior to model the tree. The MCMC analysis was run for 10 000 000 generations, sampling every 1000 generations, with a burn-in of 3000 (30 %) trees. The log file was examined using Tracer 1.6 to check that effective sample size was >200 for chain convergence. A final tree generated using TreeAnnotator 1.7.5 (Drummond et al., 2012) was viewed and edited using Figtree 1.5.4 (http://tree.bio.ed.ac.uk/software/figtree/).
Phylogenetic analyses of ITS2 using RNA substitution models
To facilitate comparison of base-pair information and thereby infer the CBC substitution history, we coded each base pairing and transformed them into comparable characters using our modified software RNAconvert from RNAstat (Subbotin et al., 2007). Both softwares can generate a multiple alignment using secondary structure, while RNAconvert can further separate the conversion into a paired partition and an unpaired partition (Supplementary Data Fig. S1). In order to display base-pair variation and thus detect CBCs and hemi-CBCs (one-sided substitutions before CBCs), we aligned paired sites in the transformed matrix using MAFFT and then adjusted manually while referring to the secondary-structure information. Unlike the commonly used CBCAnalyzer (Wolf et al., 2005), this method of structure-guided alignment following base-pair conversion shows both the number of CBCs and the base-pair state of each site (Supplementary Data Fig. S1). More importantly, we can map each base-pair state in the phylogenetic tree to trace the CBC process.
We performed BI analysis using PHASE package 3.0 (Allen and Whelan, 2014). The best-fitting models for BI analyses were estimated using a Perl script (model_selection.pl) from PHASE. This Perl script includes both DNA models (HKY85 and REV) and RNA models (seven RNA seven-state models, wherein the ten mismatch base pairs have a single frequency parameter, and nine RNA 16-state models). We used Allen and Whelan’s (2014) likelihood-correction method to account for different numbers of parameters between the four-, seven-, and 16-state models. The best-fit models (REV+G for unpaired regions and RNA16D+G for paired regions) were then used for the phylogenetic analyses. We used the optimal known ML tree topology from PhyML as the input tree for this procedure. Phylogenetic analyses were performed using the mcmcPHASE program from the PHASE package. The MCMC analysis was run for 1 000 000 generations, sampling every 100 generations, with a burn-in of 3000 (30 %) trees. After convergence had been verified by using Tracer 1.6 (effective sample size >200), a consensus tree with posterior probabilities, base-pair frequencies and substitution rate parameters was generated using the mcmcsummarize program from the PHASE package.
RESULTS
Sequences and secondary-structure analyses
Direct sequencing of ITS amplicons indicated multiple alleles in some individuals. Subsequent cloning confirmed the ITS2 polymorphism in Corydalis. Five individuals sampled from four species each contained three to nine alleles. A total of two to 17 alleles were identified in each ingroup species except C. linjiangensis, for which a single specimen was sampled (Supplementary Data Table S1). The length of ITS2 ranged from 218 to 239 bp, with an average of 133 bp from paired regions and 90 bp from unpaired regions. Nucleotide composition varied between the paired and unpaired regions. For example, the average G+C content in the paired regions (82 %) was greater than that in unpaired regions (54 %), while the adenine content in the unpaired region was 4.4-fold higher than that in the paired region (Table 1). Their substitution pattern was also different. For example, the Ts/Tv ratio was higher in paired than in unpaired regions, while the unpaired regions were more variable than the paired regions. Given that 60 % of the ITS2 bp are involved in stem pairing together with these differences in nucleotide frequencies, Ts/Tv ratios and variation, structural-partition and paired-sites models may be more appropriate for the ITS2 region than standard nucleotide models. The substitution saturation test indicated that neither the paired nor the unpaired ITS2 regions were saturated (Supplementary Data Table S2).
Table 1.
Comparison of sequence characteristics and phylogenetic information between different regions of ITS2
| Category | Paired regions | Unpaired regions | All regions |
|---|---|---|---|
| Mean length (bp) | 133 | 90 | 223 |
| Aligned length | 134 | 119 | 253 |
| A/T/C/G content (%) | 8/10/40/42 | 35/12/42/12 | 19/11/41/30 |
| Ts/Tv | 1.63 | 0.87 | 1.58 |
| MCL Ts/Tv | 19.93 | 0.67 | 1.10 |
| No. of VCs (%) | 65 (48.5 %) | 72 (60.5 %) | 137 (54.2 %) |
| No. of PICs (%) | 42 (31.3 %) | 56 (47.1 %) | 98 (38.7 %) |
MCL, maximum composite likelihood method (Tamura et al., 2007); VC, variable character; PIC, parsimony-informative character.
The consensus ITS2 secondary-structure model had four stems (helices), of which stem III was the longest and had a UGGU motif while stem II contained a pyrimidine–pyrimidine bulge and the loop between stems had a pronounced adenine bias (Fig. 1). All of these are common features of ITS2 among angiosperms (Coleman, 2003). The greater G/C content and GC base pairing in the stem regions help maintain the supporting scaffold.
Model test and substitution analyses of base-pair interaction
There are a total of seven RNA seven-state models (7A–G) and nine RNA 16-state models (16A–F, 16I–K) according to the naming convention of Allen and Whelan (2014), among which the most parameterized models are 7A and 16A, respectively. The remaining 14 models are derived from 7A or 16A with different parameter constraints (Savill et al., 2001; Allen and Whelan, 2014).The best-fit substitution model was REV+G_RNA16D+G (REV+G for unpaired regions and RNA16D+G for paired regions) according to the AIC. The best-fit conventional DNA substitution model (GTR+G) had a higher AIC value than the REV+G_RNA16D+G model (Supplementary Data Table S3), which corroborates our expectation that non-independent base-pair substitutions have occurred in ITS2 within the study lineage.
Early RNA six-state models, which treat mismatch (MM) pairs as missing data, are not considered in Allen and Whelan’s (2014) model-test method. Our base-pair statistics indicate that the total mutability (43.02) and frequency (9.31 %) of MM pairs are greater than those of GU/UG states (4.74/8.06 %; Supplementary Data Table S4), indicating that MM pairs should not be neglected. We therefore adopted Allen and Whelan’s (2014) method and focused on the RNA16 and RNA7 series models in this study.
The best-fit model for paired regions, RNA16D, was developed specifically to account for GU/UG frequencies that are low relative to Watson–Crick base pairs, but still greater than MM (Savill et al., 2001). Compared with the earlier RNA models, this model includes an extra frequency parameter and does not allow simultaneous substitution of both nucleotides in a base pair. For example, the GC base pair can only change at one site at a time to six possible base pairs (GU, GA, GG; CC, UC, AC; Supplementary Data Table S4). The parameter estimates obtained from RNA16D+G can provide insight into the ITS2 substitution process for paired regions. The equilibrium frequency of stable Watson–Crick base pairs is 83 %, with the remaining base pairs being 8 % wobble GU/UG base-pairs and 9 % unstable MM. Substitutions from unstable to stable base pairs always occurred at higher rates. By summing across each row in Supplementary Data Table S4 we obtained the net rate of change from one base pair to the others (‘mutability’). We found that GC/CG had the highest frequency but the lowest mutability. In contrast, MM had the lowest frequency but the highest mutability. These base-pair frequency, mutation rate and mutability results all indicate that natural selection was acting to maintain ITS2 secondary structure. Yet the moderate (9 %) frequency of the MM state indicates that mismatches can be tolerated to some extent and do not completely disrupt the secondary structure.
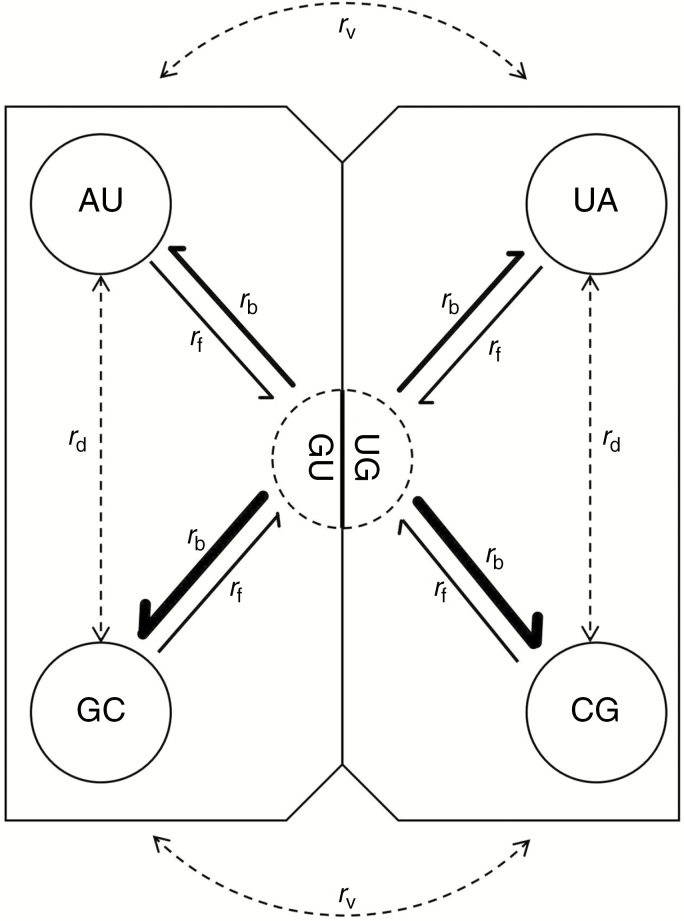
In addition to examining individual rates between pair states, we also binned base pairs following Higgs (2000), which allows double transitions between Watson–Crick pairs (rd), double transversions between Watson–Crick pairs (rv), a single transition from Watson–Crick pairs to GU/UG (forward rates, rf) and a single transition from GU/UG to Watson–Crick pairs (backward rates, rb; Fig. 2). Our results indicate an absence of double substitutions between Watson–Crick pairs (rd/rv = 0), such that CBC substitutions proceeded only by the two-step mutation mechanism via the GU/UG intermediate. But the GU/UG intermediate quickly changed to Watson–Crick pairs, and thus rb is about four times higher than rf (Supplementary Data Table S4).
Fig. 2.
Schematic representation of substitution rate parameters. rd represents double transitions between Watson–Crick pairs; rv represents double transversions between Watson–Crick pairs; rf represents a single transition from Watson–Crick pairs to GU/UG; and rb represents a single transition from GU/UG to Watson–Crick pairs. The thickness of the arrows indicates the substitution rates according to Supplementary Data Table S4.
Analyses of CBC substitution process in a phylogenetic context
A total of 65 ITS2 alleles were observed in the 11 species that we sampled. From these alleles two CBCs and 18 hemi-CBCs were identified (Table 2). Three types of hemi-CBC were found in the stems. The most frequent was UG→CG, followed by GU→GC and GU→AU. Hemi-CBCs were observed in all four stems. The longest stem (III) had the highest number of hemi-CBCs. The two CBCs occurred in stem I.
Table 2.
Type and distribution of CBCs and hemi-CBCs in stems of the consensus ITS2 secondary structure
| Substitution (number) | Stem (length) | Base change (type) | Number of each type in stem/ITS2 | Total number of types in stem |
|---|---|---|---|---|
| Hemi-CBCs (18) | I (13) | UG→CG (H1) | 4/10 | 4 |
| II (10) | UG→CG (H1) | 2/10 | 3 | |
| GU→GC (H2) | 1/7 | |||
| III (36) | UG→CG (H1) | 2/10 | 8 | |
| GU→GC (H2) | 5/7 | |||
| GU→AU (H3) | 1/1 | |||
| IV (8) | UG→CG (H1) | 2/10 | 3 | |
| GU→GC (H2) | 1/7 | |||
| CBCs (2) | I (13) | CG→UA (C1) | 1/1 | 2 |
| UA→CG (C2) | 1/1 |
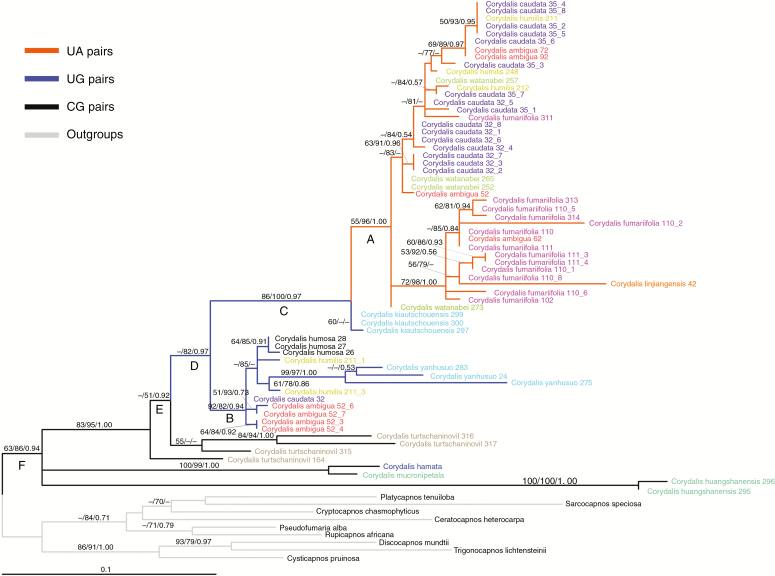
Two CBCs were inferred in the gene tree (Figs 3 and 4). A CBC from CG to UG and then UA is shown in Fig. 3. No individual species was found to have all three of these CBC states. The intermediate UG state is shared by six species, including three species that also have the UA state.
Fig. 3.
A CBC substitution process mapped onto the ITS2 maximum likelihood tree, with different CBC states indicated using different branch colours. Alleles from the same species are assigned the same colour for their terminal names. Numbers on the branches indicate ≥50 % support for MP (bootstrap), ML (bootstrap) and BI (posterior probabilities). ML and BI analyses are based on the GTR+G model. Numbers following a species name represent voucher and clone numbers. Clades A and B share three species with distinct alleles; Clades A–C and F are all supported in MP/ML/BI trees, whereas clades D and E are only supported in ML/BI trees.
Fig. 4.
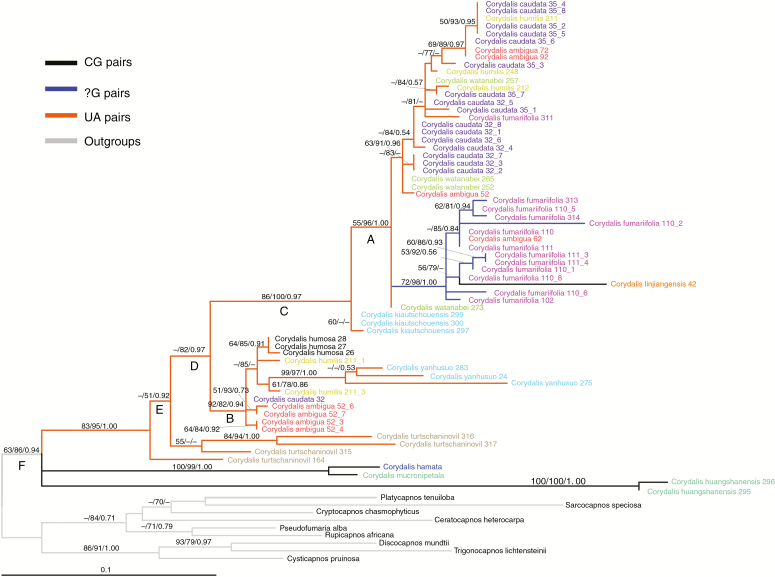
The other CBC substitution process mapped onto the ITS2 ML tree with different CBC states indicated using different branch colours. Alleles from the same species are assigned the same colour for their terminal names. Numbers on the branches indicate ≥50 % support for MP (bootstrap), ML (bootstrap) and BI (posterior probabilities). ML and BI analyses are based on the GTR+G model. Numbers following a species name represent voucher and clone numbers. One side (U) of the expected intermediate UG base pair that was missing in sequences represents an ‘?G’ base pair. Clades A and B share three species with distinct alleles; Clades A–C and F are all supported in MP/ML/BI trees, whereas clades D and E are only supported in ML/BI trees.
A second CBC, from UA to UG and then CG, was also observed (Fig. 4). Unlike the first CBC, the CG state was observed in a single species (C. linjiangensis) within the clade of Corydalis that encompasses the complete CBC. The inferred intermediate UG state was observed in the form of a U-deletion on one side of the stem in six individuals of Corydalis ambigua and Corydalis fumariifolia. The CG state was also observed in three early-derived Corydalis species.
The molecular dating results indicate that the most recent common ancestor of the CG and UG species for the first CBC (Fig. 3) occurred ~ 28.5 mya (95 % HPD 19.6–35.7 mya), the crown age of species with the UG state occurred ~25.3 mya (the 95 % HPD was unavailable from BEAST), and stem and crown nodes for the UA species occurred ~14.9 mya (95 % HPD 10.4–26.1 mya) and ~13.0 mya (95 % HPD 7.7–20.8 mya), respectively (Supplementary Data Fig. S2). Based on the optimal age estimates, we infer that the CBC substitutions were separated by ~10.4–15.5 my.
ITS2 phylogenetic trees derived from DNA/RNA models
The ITS2 matrix was first analysed using conventional four-state DNA models, for which the GTR+G model fitted best. This model was then used in the subsequent ML, BI and BEAST analyses. The maximum parsimony (MP) tree was also constructed for comparison. There were many polytomies in the strict consensus MP tree, but four of the six focal clades (A–F in Fig. 3) were consistently resolved by all optimality criteria applied. The exceptions are clades D and E, which were contradicted in the MP consensus tree because all four individuals of Corydalis turtschaninovii were resolved as a clade sister to clade B with 57 % bootstrap support.
We observed a diversity of ITS2 alleles in 11 Corydalis species. Of the ten species for which two or more alleles were sampled, these alleles were not resolved as exclusive lineages for six species. Alleles from three of these six Corydalis species (C. ambigua, C. caudata and C. humilis) were divided into two clades (A and B in Fig. 3) together with other Corydalis species. Based on these results, wherein alleles from each of three species are polyphyletic and separated into two well-supported clades that are on long branches (Supplementary Data Fig. S2), we hypothesize that these divergent ITS2 alleles from the same species are the result of hybridization. Although the ITS2 gene tree did not fully resolve phylogenetic relationships among the sampled species, it was sufficient to infer the CBCs.
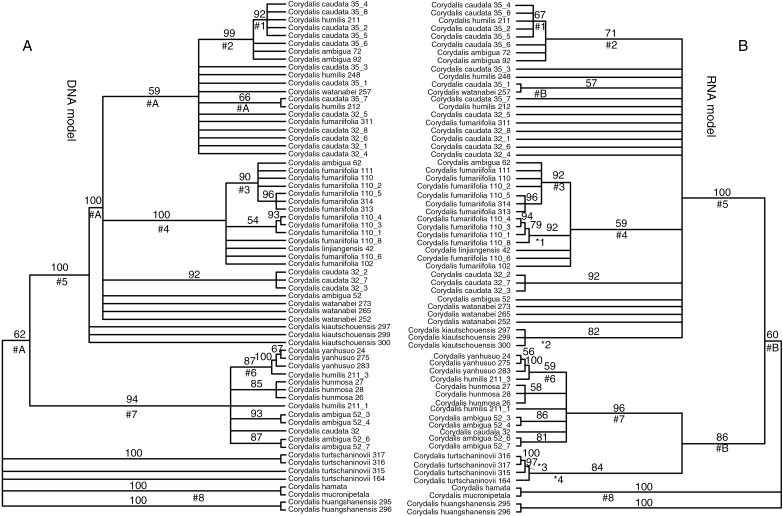
We compared the BI trees produced by the optimal DNA- and RNA-specific alignments and models (Fig. 5). The two trees were generally topologically consistent with each other (the only contradiction is the resolution of C. turtschaninovii), though the DNA tree was generally more resolved and provided higher support for clades consisting of alleles from two or more species. We found 13 different support values for clades above the species level between the two trees, among which four clades (#A) were resolved only in the DNA tree and three clades (#B) were resolved only in the RNA tree. We identified eight identical clades consisting of alleles from two or more species. Four of these clades (#3, #5, #7 and #8) had equal or similar support values, whereas the remaining four clades were more highly supported in the DNA tree. In contrast, the RNA tree was generally more resolved (four unique clades, *1–*4) and provided higher support for clades consisting of alleles from a single species.
Fig. 5.
Comparison of BI trees inferred from different substitution models and alignments. (A) BI tree inferred using the DNA model GTR+G. (B) BI tree inferred using the RNA-specific model REV+G_RNA16D+G. Numbers on the branches indicate the Bayesian posterior probabilities (>50). Corresponding clades of the two trees that shared the same alleles from two or more species are marked with # and number; clades with >50 posterior probability that are resolved only in the DNA or RNA tree are marked with #A and #B, respectively. Additional intraspecific resolution in tree B is indicated using asterisks (*) and numbers. Numbers following a species name represent voucher and clone numbers.
DISCUSSION
Mutational dynamics of ITS2 in terms of secondary structure
Since ITS2 secondary structure is crucial to the process of pre-rRNA maturation (Coleman, 2015), one of our concerns is how selective constraint acts on this functional secondary structure. All ITS2 sequences in our study appear to be functional copies because they all have the conserved ‘four-fingered hand’ form (Coleman, 2003) and have common core motifs that are conserved within angiosperms (Hershkovitz and Zimmer, 1996; Coleman, 2003, 2007). A consensus RNA secondary structure is a prerequisite for phylogenetic analyses that apply RNA models. We found that the paired region had a high G/C content whereas the unpaired region had a high A/C content. In addition, the GC/CG base pair had the highest frequency but the lowest mutability among all base pairs. These observations support the hypothesis that functional rRNA sequences are selected to increase their structural and thermodynamic stability (Higgs, 2000).
Coleman (2003, 2007) showed that nucleotide sequences evolve more slowly in helices II and III than helices I and IV, which is expected given their crucial function in rRNA transcript processing (Coleman, 2015). In our dataset, the two identified CBCs occurred in the more variable helix I, whereas hCBCs were found across all four helices. Despite the greater nucleotide variability in helix I, the CBCs maintained its functional stability. A key feature of the compensatory substitutions is the directionality. For example, we found that UA base pairs changed most frequently to UG base pairs than to UC or UU base pairs (Supplementary Data Table S4). This rate difference can be interpreted as indicating that, when UA changes to CG, it occurs most frequently via a relatively stable UG intermediate (Fig. 2) rather than GU or the unstable UC or UU intermediates.
Phylogenetic implications of CBC
The CBC mutation pattern of paired regions violates the site-independence assumption of typical phylogenetic analyses. Application of RNA-specific models is thus theoretically justified but still largely confined to studies of ancient lineages (e.g. Hudelot et al., 2003; Mallatt et al., 2010; Letsch and Kjer, 2011; Allen and Whelan, 2014; Patiño-Galindo et al., 2018). To date, few studies have critically assessed to what extent this co-variation pattern will affect the phylogenetic inference among recently diverged lineages (Marinho et al., 2011; Adebowale et al., 2016). In this study we inferred the ITS2 gene tree among closely related species by using both DNA and RNA models as well as tracing the CBC process, as suggested by Caisová et al. (2011). The strong nucleotide composition bias between paired and unpaired regions and the higher likelihoods for DNA/RNA models over DNA-only models (Supplementary Data Table S3; but note that this comparison is confounded by our use of different alignments) are in agreement with the previous analyses that allowed RNA loops and stems to evolve under separate models (Telford et al., 2005; Biffin et al., 2007; Allen and Whelan, 2014).
The better fit of RNA-specific models to our data and the three additional clades in the RNA topology both indicate advantages to using this model instead of typical DNA models. Yet the DNA-model-based topology was largely consistent with the RNA-model-based topology and included several clades that were either novel or more highly supported than in the RNA-model-based tree (Fig. 5). Although application of a typical DNA model to RNA stems can violate the site-independence assumption, this assumption is only severely violated in the context of CBCs rather than other substitutions, such as hemi-CBCs. Indeed, of the 20 compensatory changes in paired regions that we identified only two CBCs (Table 2), accounting for no more than 3 % of the variable characters in the entire matrix (Table 1). Therefore, the empirical effect of the site-independence violation is minimal in the context of this study of closely related species, in contrast to the more severe effects that have been demonstrated in phylogenetic analyses of more ancient lineages (Jow et al., 2002; Hudelot et al., 2003; Letsch et al., 2010; Mallatt et al., 2010; Patiño-Galindo et al., 2018). Application of RNA models is useful for improving alignment and inferring the process of molecular evolution. But based on our results we do not consider application of these models to be necessary for effective phylogenetic inference among closely related species using ITS2. The generality of our results and inference should be tested in other empirical studies.
CBC analyses and species delimitation
A highlight of this study is identification of each step of the CBC substitution process in Corydalis. We did not observe the entire CBC process within any individual Corydalis species, which supports the ‘CBC species concept’ hypothesis (Wolf et al., 2013). Although the CBC species concept may be used to help identify distinct species, one should not expect CBCs to differentiate all species from each other. In our study, we generally observed at least two species that shared the same state in a base pair that included a CBC (Figs 3 and 4). Some recent studies in chlorophytes (Caisová et al., 2011, 2013) and Cymatosirales (Samanta et al., 2018) also found that CBCs most often correspond to supra-specific divergence rather than individual species. Likewise, in blowflies CBCs were not found in 33 % of congeneric species pairs (Marinho et al., 2011); CBCs are also absent in four distinct species of Strychnos (Adebowale et al., 2016).
Identification of each stage in a CBC remains problematic in practice. In most phylogenetic studies, wherein a single sequence represents an entire species, low-frequency base-pair states are generally not observed. If taxon sampling is insufficient, some base-pair states will be lost in CBC analyses. Given sufficient intraspecific and interspecific sampling, as in this study, the entire CBC process may be directly inferred within an individual lineage rather than relying upon indirect statistical methods.
Conclusions
In this study we inferred the ITS2 gene tree among closely related species by using both conventional DNA as well as RNA-specific models, and then traced the CBC process on the inferred tree. By doing so we identified just two CBCs, both in the most variable stem (stem I) via GU/UG intermediates, and showed that a pair of CBC substitutions may be separated by ~10.4–15.5 my. Neither of the CBCs occurred within any given species, which is consistent with application of the CBC species concept. ITS2 clearly evolved under secondary-structure constraints within the study lineage. Yet application of conventional DNA models appears unlikely to be problematic when conducting phylogenetic analyses of ITS2 within such closely related lineages, wherein few CBCs are observed. The generality of these results and inferences should be tested in other empirical studies of recently diverged lineages.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Figure S1: workflow illustrating how base-pair information is transformed into an alignment. Figure S2: molecular chronogram showing the divergence times of each substitution in the CBC process. Table S1: list of sample information used in this study. Table S2: substitution-saturation test for different ITS2 partitions. Table S3: comparison of likelihood scores between DNA- and RNA-specific models applied to the ITS2 alignments. Table S4: best-fit substitution rate matrix, mutabilities, base-pair frequencies and substitution rate parameters for the ITS2 paired region in Corydalis, inferred using the RNA16D+G model.
ACKNOWLEDGEMENTS
We thank two anonymous reviewers for several constructive criticisms with which to improve the manuscript. This work was supported by the National Natural Science Foundation of China (grant 81673551) and the Young Scholars Program of Shandong University, Weihai (2017WHWLJH05).
LITERATURE CITED
- Adebowale A, Lamb J, Nicholas A, Naidoo Y. 2016. ITS2 secondary structure for species circumscription: case study in southern African Strychnos L. (Loganiaceae). Genetica 144: 1–12. [DOI] [PubMed] [Google Scholar]
- Akaike H. 1974. A new look at the statistical model identification. IEEE Transactions on Automatic Control 19: 716–723. [Google Scholar]
- Allen JE, Whelan S. 2014. Assessing the state of substitution models describing noncoding RNA evolution. Genome Biology and Evolution 6: 65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez I, Wendel JF. 2003. Ribosomal ITS sequences and plant phylogenetic inference. Molecular Phylogenetics and Evolution 29: 417–434. [DOI] [PubMed] [Google Scholar]
- Ankenbrand MJ, Keller A, Wolf M, Schultz J, Förster F. 2015. ITS2 database V: twice as much. Molecular Biology and Evolution 32: 3030–3032. [DOI] [PubMed] [Google Scholar]
- Biffin E, Harrington MG, Crisp MD, Craven LA, Gadek PA. 2007. Structural partitioning, paired-sites models and evolution of the ITS transcript in Syzygium and Myrtaceae. Molecular Phylogenetics and Evolution 43: 124–139. [DOI] [PubMed] [Google Scholar]
- Buchheim MA, Müller T, Wolf M. 2017. 18S rDNA sequence-structure phylogeny of the Chlorophyceae with special emphasis on the Sphaeropleales. Plant Gene 10: 45–50. [Google Scholar]
- Caisová L, Marin B, Melkonian M. 2011. A close-up view on ITS2 evolution and speciation – a case study in the Ulvophyceae (Chlorophyta, Viridiplantae). BMC Evolutionary Biology 11: 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caisová L, Marin B, Melkonian M. 2013. A consensus secondary structure of ITS2 in the Chlorophyta identified by phylogenetic reconstruction. Protist 164: 482–496. [DOI] [PubMed] [Google Scholar]
- Chen S, Yao H, Han J, et al. . 2010. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS ONE 5: e8613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman AW. 2000. The significance of a coincidence between evolutionary landmarks found in mating affinity and a DNA sequence. Protist 151: 1–9. [DOI] [PubMed] [Google Scholar]
- Coleman AW. 2003. ITS2 is a double-edged tool for eukaryote evolutionary comparisons. Trends in Genetics 19: 370–375. [DOI] [PubMed] [Google Scholar]
- Coleman AW. 2007. Pan-eukaryote ITS2 homologies revealed by RNA secondary structure. Nucleic Acids Research 35: 3322–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman AW. 2009. Is there a molecular key to the level of “biological species” in eukaryotes? A DNA guide. Molecular Phylogenetics and Evolution 50: 197–203. [DOI] [PubMed] [Google Scholar]
- Coleman AW. 2015. Nuclear rRNA transcript processing versus internal transcribed spacer secondary structure. Trends in Genetics 31: 157–163. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9: 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon MT, Hillis DM. 1993. Ribosomal RNA secondary structure: compensatory mutations and implications for phylogenetic analysis. Molecular Biology and Evolution 10: 256–267. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution 29: 1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feliner GN, Rosselló JA. 2007. Better the devil you know? Guidelines for insightful utilization of nrDNA ITS in species-level evolutionary studies in plants. Molecular Phylogenetics and Evolution 44: 911–919. [DOI] [PubMed] [Google Scholar]
- Galtier N. 2004. Sampling properties of the bootstrap support in molecular phylogeny: influence of nonindependence among sites. Systematic Biology 53: 38–46. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML3.0. Systematic Biology 59: 307–321. [DOI] [PubMed] [Google Scholar]
- Heeg JS, Wolf M. 2015. ITS2 and 18S rDNA sequence-structure phylogeny of Chlorella and allies (Chlorophyta, Trebouxiophyceae, Chlorellaceae). Plant Gene 4: 20–28. [Google Scholar]
- Hershkovitz MA, Zimmer EA. 1996. Conservation patterns in angiosperm rDNA ITS2 sequences. Nucleic Acids Research 24: 2857–2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs PG. 2000. RNA secondary structure: physical and computational aspects. Quarterly Reviews of Biophysics 33: 199–253. [DOI] [PubMed] [Google Scholar]
- Hou D, Song J, Shi L, et al. . 2013. Stability and accuracy assessment of identification of traditional Chinese materia medica using DNA barcoding: a case study on Flos Lonicerae Japonicae. BioMed Research International 2013: 549037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudelot C, Gowri-Shankar V, Jow H, Rattray M, Higgs PG. 2003. RNA-based phylogenetic methods: application to mammalian mitochondrial RNA sequences. Molecular Phylogenetics and Evolution 28: 241–252. [DOI] [PubMed] [Google Scholar]
- Jiang L, Li MH, Zhao FX, et al. . 2018. Molecular identification and taxonomic implication of herbal species in genus Corydalis (Papaveraceae). Molecules 23: 1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jow H, Hudelot C, Rattray M, Higgs PG. 2002. Bayesian phylogenetics using an RNA substitution model applied to early mammalian evolution. Molecular Biology and Evolution 19: 1591–1601. [DOI] [PubMed] [Google Scholar]
- Katoh K, Toh H. 2008. Improved accuracy of multiple ncRNA alignment by incorporating structural information into a MAFFT-based framework. BMC Bioinformatics 9: 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Förster F, Müller T, Dandekar T, Schultz J, Wolf M. 2010. Including RNA secondary structures improves accuracy and robustness in reconstruction of phylogenetic trees. Biology Direct 5: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjer KM. 1995. Use of rRNA secondary structure in phylogenetic studies to identify homologous positions: an example of alignment and data presentation from the frogs. Molecular Phylogenetics and Evolution 4: 314–330. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letsch HO, Kjer KM. 2011. Potential pitfalls of modelling ribosomal RNA data in phylogenetic tree reconstruction: evidence from case studies in the Metazoa. BMC Evolutionary Biology 11: 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letsch HO, Kück P, Stocsits RR, Misof B. 2010. The impact of rRNA secondary structure consideration in alignment and tree reconstruction: simulated data and a case study on the phylogeny of hexapods. Molecular Biology and Evolution 27: 2507–2521. [DOI] [PubMed] [Google Scholar]
- Mallatt J, Craig CW, Yoder MJ. 2010. Nearly complete rRNA genes assembled from across the metazoan animals: effects of more taxa, a structure-based alignment, and paired-sites evolutionary models on phylogeny reconstruction. Molecular Phylogenetics and Evolution 55: 1–17. [DOI] [PubMed] [Google Scholar]
- Marinho MAT, Junqueira ACM, Azeredoespin AML. 2011. Evaluation of the internal transcribed spacer 2 (ITS2) as a molecular marker for phylogenetic inference using sequence and secondary structure information in blow flies (Diptera: Calliphoridae). Genetica 139: 1189–1207. [DOI] [PubMed] [Google Scholar]
- Markert SM, Müller T, Koetschan C, Friedl T, Wolf M. 2012. ‘ Y’Scenedesmus (Chlorophyta, Chlorophyceae): the internal transcribed spacer 2 rRNA secondary structure re‐revisited. Plant Biology 14: 987–996. [DOI] [PubMed] [Google Scholar]
- Müller T, Philippi N, Dandekar T, Schultz J, Wolf M. 2007. Distinguishing species. RNA 13: 1469–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander JAA. 2004. MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University; https://github.com/nylander/MrModeltest2. [Google Scholar]
- Patiño-Galindo JÁ, González-Candelas F, Pybus OG. 2018. The effect of RNA substitution models on viroid and RNA virus phylogenies. Genome Biology and Evolution 10: 657–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Gutiérrez MA, Romero-García AT, Fernández MC, Blanca G, Salinas-Bonillo MJ, Suárez-Santiago VN. 2015. Evolutionary history of fumitories (subfamily Fumarioideae, Papaveraceae): an old story shaped by the main geological and climatic events in the Northern Hemisphere. Molecular Phylogenetics and Evolution 88: 75–92. [DOI] [PubMed] [Google Scholar]
- Porebski S, Bailey LG, Baum BR. 1997. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Molecular Biology Reporter 15: 8–15. [Google Scholar]
- Posada D, Crandall KA. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14: 817–818. [DOI] [PubMed] [Google Scholar]
- Qin Y, Li MH, Cao Y, Cao Y, Zhang W. 2017. Molecular thresholds of ITS2 and their implications for molecular evolution and species identification in seed plants. Scientific Reports 7: 17316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, Van Der Mark P, et al. . 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset F, Pélandakis M, Solignac M. 1991. Evolution of compensatory substitutions through G.U intermediate state in Drosophila rRNA. Proceedings of the National Academy of Sciences of the USA 88: 10032–10036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta B, Ehrman JM, Kaczmarska I. 2018. A consensus secondary structure of ITS2 for the diatom order Cymatosirales (Mediophyceae, Bacillariophyta) and reappraisal of the order based on DNA, morphology, and reproduction. Molecular Phylogenetics and Evolution 129: 117–129. [DOI] [PubMed] [Google Scholar]
- Savill NJ, Hoyle DC, Higgs PG. 2001. RNA sequence evolution with secondary structure constraints: comparison of substitution rate models using maximum-likelihood methods. Genetics 157: 399–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schill RO, Förster F, Dandekar T, Wolf M. 2010. Using compensatory base change analysis of internal transcribed spacer 2 secondary structures to identify three new species in Paramacrobiotus (Tardigrada). Organisms Diversity & Evolution 10: 287–296. [Google Scholar]
- Schultz J, Maisel S, Gerlach D, Müller T, Wolf M. 2005. A common core of secondary structure of the internal transcribed spacer 2 (ITS2) throughout the Eukaryota. RNA 11: 361–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibel PN, Müller T, Dandekar T, Schultz J, Wolf M. 2006. 4SALE – a tool for synchronous RNA sequence and secondary structure alignment and editing. BMC Bioinformatics 7: 498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibel PN, Müller T, Dandekar T, Wolf M. J.. 2008. Synchronous visual analysis and editing of RNA sequence and secondary structure alignments using 4SALE. BMC Research Notes 1: 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selig C, Wolf M, Müller T, Dandekar T, Schultz J. 2008. The ITS2 Database II: homology modelling RNA structure for molecular systematics. Nucleic Acids Research 3: D377–D380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons MP, Ochoterena H. 2000. Gaps as characters in sequence-based phylogenetic analyses. Systematic Biology 49: 369–381. [PubMed] [Google Scholar]
- Subbotin SA, Sturhan D, Vovlas N, et al. . 2007. Application of the secondary structure model of rRNA for phylogeny: D2–D3 expansion segments of the LSU gene of plant-parasitic nematodes from the family Hoplolaimidae Filipjev, 1934. Molecular Phylogenetics and Evolution 43: 881–890. [DOI] [PubMed] [Google Scholar]
- Swofford DL. 2003. PAUP*. Phylogenetic analysis using parsimony (*and other methods), version 4.0b10. Sunderland, MA: Sinauer. [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution 24: 1596–1599. [DOI] [PubMed] [Google Scholar]
- Telford MJ, Wise MJ, Gowri-Shankar V. 2005. Consideration of RNA secondary structure significantly improves likelihood-based estimates of phylogeny: examples from the bilateria. Molecular Biology and Evolution 22: 1129–1136. [DOI] [PubMed] [Google Scholar]
- Tillier ER, Collins RA. 1998. High apparent rate of simultaneous compensatory base-pair substitutions in ribosomal RNA. Genetics 148: 1993–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren DL, Geneva AJ, Lanfear R. 2017. RWTY (R We There Yet): an R package for examining convergence of Bayesian phylogenetic analyses. Molecular Biology and Evolution 34: 1016–1020. [DOI] [PubMed] [Google Scholar]
- Wheeler WC, Honeycutt RL. 1988. Paired sequence difference in ribosomal RNAs: evolutionary and phylogenetic implications. Molecular Biology and Evolution 5: 90–96. [DOI] [PubMed] [Google Scholar]
- Wolf M. 2015. ITS so much more. Trends in Genetics 31: 175–176. [DOI] [PubMed] [Google Scholar]
- Wolf M, Friedrich J, Dandekar T, Müller T. 2005. CBCAnalyzer: inferring phylogenies based on compensatory base changes in RNA secondary structures. In Silico Biology 5: 291–294. [PubMed] [Google Scholar]
- Wolf M, Selig C, Müller T, Philippi N, Dandekar T, Schultz J. 2007. Placozoa: at least two. Biologia 62: 641–645. [Google Scholar]
- Wolf M, Ruderisch B, Dandekar T, Schultz J, Müller T. 2008. ProfDistS:(profile-) distance based phylogeny on sequence–structure alignments. Bioinformatics 24: 2401–2402. [DOI] [PubMed] [Google Scholar]
- Wolf M, Chen S, Song J, Ankenbrand M, Müller T. 2013. Compensatory base changes in ITS2 secondary structures correlate with the biological species concept despite intragenomic variability in ITS2 sequences – a proof of concept. PLoS ONE 8: e66726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf M, Koetschan C, Mueller T. 2014. ITS2, 18S, 16S or any other RNA – simply aligning sequences and their individual secondary structures simultaneously by an automatic approach. Gene 546: 145–149. [DOI] [PubMed] [Google Scholar]
- Xia X. 2013. DAMBE5: a comprehensive software package for data analysis in molecular biology and evolution. Molecular Biology and Evolution 30: 1720–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Rannala B. 1997. Bayesian phylogenetic inference using DNA sequences: a Markov chain Monte Carlo method. Molecular Biology and Evolution 14: 717–724. [DOI] [PubMed] [Google Scholar]
- Zhang W, Yuan Y, Yang S, Huang J, Huang L. 2015. ITS2 secondary structure improves discrimination between medicinal “Mu Tong” species when using DNA barcoding. PLoS ONE 10: e0131185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Yang S, Zhao H, Huang L. 2016. Using the ITS2 sequence-structure as a DNA mini-barcode: a case study in authenticating the traditional medicine “Fang Feng”. Biochemical Systematics and Ecology 69: 188–194. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.