Abstract
IAA biosynthetic pathways in a basidiomycetous yeast, Rhodosporidiobolus fluvialis DMKU-CP293, were investigated. The yeast strain showed tryptophan (Trp)-dependent IAA biosynthesis when grown in tryptophan supplemented mineral salt medium. Gas chromatography–mass spectrometry was used to further identify the pathway intermediates of Trp-dependent IAA biosynthesis. The results indicated that the main intermediates produced by R. fluvialis DMKU-CP293 were tryptamine (TAM), indole-3-acetic acid (IAA), and tryptophol (TOL), whereas indole-3-pyruvic acid (IPA) was not found. However, supplementation of IPA to the culture medium resulted in IAA peak detection by high-performance liquid chromatography analysis of the culture supernatant. Key enzymes of three IAA biosynthetic routes, i.e., IPA, IAM and TAM were investigated to clarify the IAA biosynthetic pathways of R. fluvialis DMKU-CP293. Results indicated that the activities of tryptophan aminotransferase, tryptophan 2-monooxygenase, and tryptophan decarboxylase were observed in cell crude extract. Overall results suggested that IAA biosynthetic in this yeast strain mainly occurred via the IPA route. Nevertheless, IAM and TAM pathway might be involved in R. fluvialis DMKU-CP293.
Keywords: Indole-3-acetic acid, IAA biosynthesis, basidiomycetous yeast, Rhodosporidiobolus fluvialis
1. Introduction
Indole-3-acetic acid (IAA) was the first phytohormone to be discovered in auxin class. IAA plays an important role in cell division, elongation, fruit development, and senescence. IAA can be synthesized, not only plants, but also in microorganisms, including bacteria, actinomycetes, filamentous fungi, and yeast [1–3]. IAA can be synthesized via either a Trp-independent pathway or Trp-dependent pathway by sequential conversion of tryptophan to various intermediates. In plants, four Trp-dependent pathways have been proposed, which are indole-3-acetamide (IAM), indole-3-pyruvic acid (IPA), tryptamine (TAM), and indole-3-acetaldoxime (IAOX) pathways [4]. In bacteria, the intermediates determination led to the identification of five different pathways of IAA production using tryptophan as a precursor, which are (1) IAM pathway, in which tryptophan is first converted to IAM by tryptophan-2-monooxygenase and IAM is then converted to IAA by IAM hydrolase activity; (2) IPA pathway, of which the first step is tryptophan conversion to IPA by aminotransferase activity, IPA is then carboxylated to indole-3-acetaldehyde (IAAld) by indole-3-pyruvate decarboxylase (IPDC) activity and the last step is an oxidation of IAAld to IAA; (3) TAM pathway where the first step is decarboxylation of tryptophan to TAM, TAM is then converted to IAAld by amine oxidase and IAAld is converted to IAA as the last step; (4) tryptophan side-chain oxidase pathway where tryptophan is directly converted to IAAld prior to oxidization of IAAld to IAA; (5) indole-3-acetonitrile (IAN) pathway of which intermediates could be directly converted to IAA by nitrilase or converted to IAM prior to conversion of IAM to IAA by nitrile hydratase [5,6]. IAA biosynthetic pathways in bacteria appeared to be more than one pathway for IAA biosynthesis and different intermediates were found. For example, Azospirillum brasilense, simultaneously used IAM, IPA, and IAN pathways for IAA biosynthesis from tryptophan [7]. Recently, studies on IAA producing yeasts, including Basidiomycetous yeasts, such as Cryptococcus flavescens, C. laurentii, C. rajasthanensis, Hannaella sinensis, Pseudozyma aphidis, Rhodosporidiobolus fluvialis, Rhodotorula graminis, Rh. grinbergsii, Rh. paludigena, Rh. terrea, Saitozyma flava, Sporidiobolus ruineniae, Sporobolomyces nylandii, and Trichosporon asahii have been reported [8–11] and Ascomycetous yeasts, such as Aureobasidium pullulans, Candida glabrata, Ca. maltose, Ca. mesorugosa, Ca. metapsilosis, Ca. rugosa, Ca. tropicalis, Cyberlindnera fabianii, Debaryomyces nepalensis, Lodderomyces elongisporus, Metschnikowia koreensis, M. saccharicola, Pichia kudriavzevii, Torulaspora delbrueckii, T. kloeckeriana, and Saccharomyces cerevisiae have been studied [3,9,10,12]. However, only a few studies of yeast IAA biosynthetic pathways have been described. The yeast Rh. paludigena DMKU-RP301 was found to produce IAA via an IPA pathway [13], whereas S. cerevisiae was proven to possess Trp-independent IAA biosynthesis [14]. In addition, Trp-independent IAA biosynthesis was also shown in Ustilago esculenta [15]. To date, an investigation of a Basidiomycetous yeast, R. fluvialis on IAA biosynthesis has not yet been reported. Therefore, the aim of this study was to focus on the clarification of the IAA biosynthetic pathways in R. fluvialis.
2. Materials and methods
2.1. Yeast strain and growth conditions
R. fluvialis DMKU-CP293 (accession number: LC379571) was isolated from corn phylloplane in Thailand using the plating of leaf washings technique [16]. This yeast was identified by the analysis of the D1/D2 region of the large subunit of the rRNA gene sequence, as described by Nutaratat et al. [10] and maintained on YPD agar (gram per liter: yeast extract, 10; peptone, 20; dextrose, 20; agar, 15) at 4 °C.
The inoculum was prepared by cultivation of a loopful of the pure colony into a 250 ml Erlenmeyer flask containing 50 ml YPD medium. The culture was incubated at 30 °C for 16–18 h in the orbital shaking incubator (Jeio Tech, South Korea) at 200 rpm; cells were harvested by centrifugation (4 °C, 10,000×g, 5 min) and washed twice and resuspended in sterile distilled water. The yeast inoculum cells density was estimated by measuring the optical density at 600 nm (OD600) using a spectrophotometer (Genesys 20, Thermo Fisher Scientific, Waltham, MA)
2.2. Tryptophan-dependent and tryptophan-independent pathway analysis for IAA biosynthesis
To investigate whether R. fluvialis DMKU-CP293 employed Trp-dependent or Trp-independent IAA biosynthesis, culture media with and without tryptophan supplements were used. The inoculum was transferred into a 250 ml Erlenmeyer flask containing 50 ml mineral salt (MS) medium (gram per liter: KH2PO4, 1; K2HPO4, 1; NaCl, 1; MgSO4·7H2O, 4; (NH4)2 SO4, 4; Na3C6H5O7·7H2O, 0.5; Glucose, 5), supplemented with or without a 0.1% (w/v) L-tryptophan for investigation of Trp-dependent or Trp-independent IAA biosynthesis, respectively. The yeast inoculum cell density was estimated by measuring the optical density at 600 nm (OD600) using a spectrophotometer and adjusted to 0.2, prior to incubation on an orbital shaker at 200 rpm. The cultures were incubated at 30 °C, and samples of culture broth were taken at daily intervals for 5 days. The culture supernatant was analyzed for IAA using high-performance liquid chromatography (HPLC), equipped with a Cosmosil SC18-MS-II column (Nacalai Tesque, Kyoto, Japan) and UV detector (Agilent Technologies, Santa Clara, CA), methanol: acetic acid: water at the ratio 60: 40: 0.3 v/v/v were used as mobile phase with a flow rate of 0.5 ml/min as described by Nutaratat et al. [17].
2.3. Identification of indole derivatives in the IAA biosynthetic pathway
The inoculum was transferred to MS medium supplemented with 0.1% (v/w) L-tryptophan. All cultures were incubated at 30 °C on an orbital shaker at 200 rpm. Fifty milliliter of the whole culture samples were collected at 1 and 3 days of cultivation and cells were discarded after centrifugation (4 °C, 32,000×g, 5 min). The supernatant was extracted with ethyl acetate, as described by Mujahid et al. [18]. The residue was dissolved in 1 ml methanol prior to GC-MS analysis. The intermediates analysis was carried out on a Shimadzu GC-MS QP2020 (Shimadzu Corporation, Kyoto, Japan) with a HP-5 capillary column system (30 m × 0.25 mm ID, 0.25 mm film thickness). The oven temperature was maintained at 80 °C, for 2 min, then programed to rise from 80 to 200 °C at 10 °C min−1 and then finally programmed to shift from 200 to 280 °C at 20 °C min−1 for 7 min. The mass spectra of intermediates were compared with databases as described by Nutaratat et al. [13].
2.4. Determination of enzyme activity
R. fluvialis DMKU-CP293 cells of 0.1% (w/v) tryptophan supplemented 24 h grown culture were harvested by centrifugation (32,000×g, at 4 °C for 5 min). Cells were washed twice with 50 ml of 0.2 M sodium phosphate buffer (pH 7.2) and resuspended in the same buffer. Cells were lysed by ultrasonication (74 Amp) at 4 °C (Sonics Vibra-Cell CV33, Sonics and Materials Inc., Newtown, CT). After sonication, the lysate was centrifuged (32,000×g, at 4 °C for 10 min) and the clear supernatant was subjected to enzyme activity assay. Tryptophan aminotransferase activity was determined by measuring the formation of IPA, the reaction mixture contained 40 µmol of L-tryptophan, 20 µmol of α-ketoglutarate, 0.1 µmol of pyridoxal-5′-phosphate (PLP), and 0.5 ml of crude cell extract in final volume of 2.5 ml sodium phosphate buffer. The mixture was incubated at 30 °C for 5 min and the reaction was stopped by adding 10% trichloroacetic acid (TCA). The sample was centrifuged at 32,000×g, 4 °C for 5 min and the formation of IPA was analyzed by HPLC as described by Nutaratat et al. [13]. The activity of the other enzyme activities was performed as described by Kulkarni et al. [19]. Tryptophan 2-monooxygenase was assayed by detecting the active conversion of tryptophan to IAM, enzyme assay was carried out in a final volume of 1 ml contained 50 mM sodium phosphate buffer (pH 7.0), 20 mM of L-tryptophan, and 100 μl of crude cell extract. The reaction mixture was incubated at 37 °C for 1 h then the reaction was stopped by adding 100 μl of 5% TCA. The formation of IAM and IAA was analyzed by GC-MS. Tryptophan decarboxylase activity was assayed by studying the active conversion of tryptophan to TAM, the reaction mixture was carried out in final volume of 1 ml which consisted of 20 mM L-tryptophan, 100 μM PLP, 50 mM sodium phosphate buffer (pH 7.0), and 100 μl of crude cell extract. The reaction mixture was incubated at 37 °C for 1 h, after that the reaction was stopped by adding 100 μl of 5% TCA then the formation of TAM and IAA was analyzed by GC-MS. Tryptophan side-chain oxidase activity was assayed by measuring the formation of N-acetyl-α,β-didehydrotryptophanamide, the assay mixture was prepared to final volume of 1 ml containing 50 mM potassium phosphate buffer (pH 6.0), substrate and crude cell extract. The reaction was started by the addition of crude cell extract. The increasing absorbance of N-acetyl-α,β-didehydrotryptophanamide formation was measured at 333 nm. Nitrile hydratase was assayed by monitoring the active conversion of IAN to IAM and nitrilase was examined for the formation of IAM and IAA, the reaction mixtures in 2 ml of final volume consisted of 50 mM sodium phosphate buffer (pH 7.5), 1 mM IAN, and 100 μl of cell crude extract were incubated at 37 °C for 1 h. After that the reaction was stopped by the addition of 100 μl of 5% TCA. The supernatant was extracted twice with ethyl acetate. The formation of IAM confirming of nitrile hydratase activity and the IAA formation confirming of nitrilase activity was observed by GC-MS analysis.
2.5. IAA precursors feeding experiments
The experiments were carried out by feeding the yeast with possible IAA precursors, such as IPA, IAM, TAM, and IAN. The precultured inoculum of the yeast strains was resuspended in MS medium from which 0.1% (w/v) L-tryptophan was individually replaced by 0.1% (w/v) IAA precursors (IPA, IAM, TAM, or IAN). The cultures were incubated at 30 °C on an orbital shaker at 200 rpm, for 1, 2, and 3 days to assure the compounds detection. The supernatants of the cultures fed with IPA, IAM, TAM, or IAN were analyzed for IAA using HPLC.
3. Results and discussion
3.1. Study of IAA biosynthesis in R. fluvialis DMKU-CP293
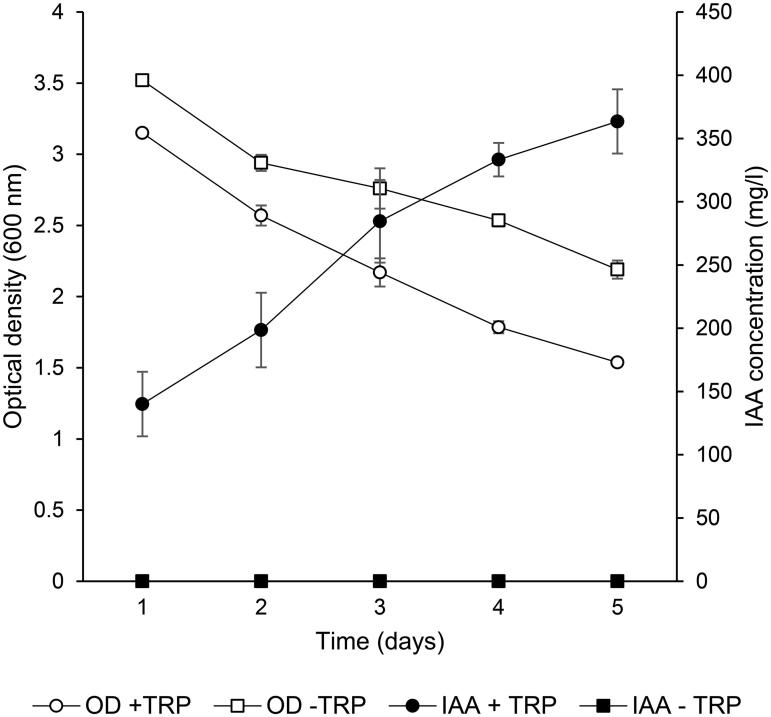
Study of IAA biosynthesis in R. fluvialis DMKU-CP293 was focused because this strain showed higher IAA producing ability (1,061.97 mg IAA/l) when cultivated in YPD medium supplemented with 0.1% L-tryptophan, when compared to A. pullulans and Rh. paludigena DMKU-RP301 those cultivated in the same medium produced 147.4 µg IAA/ml and 321.7 mg IAA/l, respectively [15,17]. In addition, informative report on IAA biosynthesis in R. fluvialis is still required. To investigate tryptophan dependency for IAA biosynthesis of R. fluvialis DMKU-CP293, the culture supernatant of yeast culture grown in MS medium, with or without 0.1% L-tryptophan, was analyzed by HPLC. After 5 days of incubation, yeast stain produced amount (363.49 mg IAA/l) when L-tryptophan was supplemented (Figure 1) whereas in the absence of L-tryptophan in the medium, IAA was not detected. Regarding growth, there was little difference in growth patterns observed when yeast was cultivated in the culture medium, with or without L-tryptophan supplement, indicating that L-tryptophan had no effect on growth. These results implied that R. fluvialis DMKU-CP293 synthesized IAA through a Trp-dependent pathway. Tryptophan has been reported as a main precursor for IAA biosynthesis in yeast, such as C. flavus [15] and Rh. paludigena [13].
Figure 1.
Yeast growth in terms of OD600 (^ with L-tryptophan supplementation, □ without L-tryptophan supplementation) and IAA production (• with L-tryptophan supplementation, ▪ without L-tryptophan supplementation) of R. fluvialis DMKU-CP293 cultivated in MS medium supplemented with 0.1% L-tryptophan and without 0.1% L-tryptophan. Values are presented as means ± standard deviation of triplicate samples.
3.2. IAA intermediates involve with IAA biosynthetic pathway of R. fluvialis DMKU-CP293
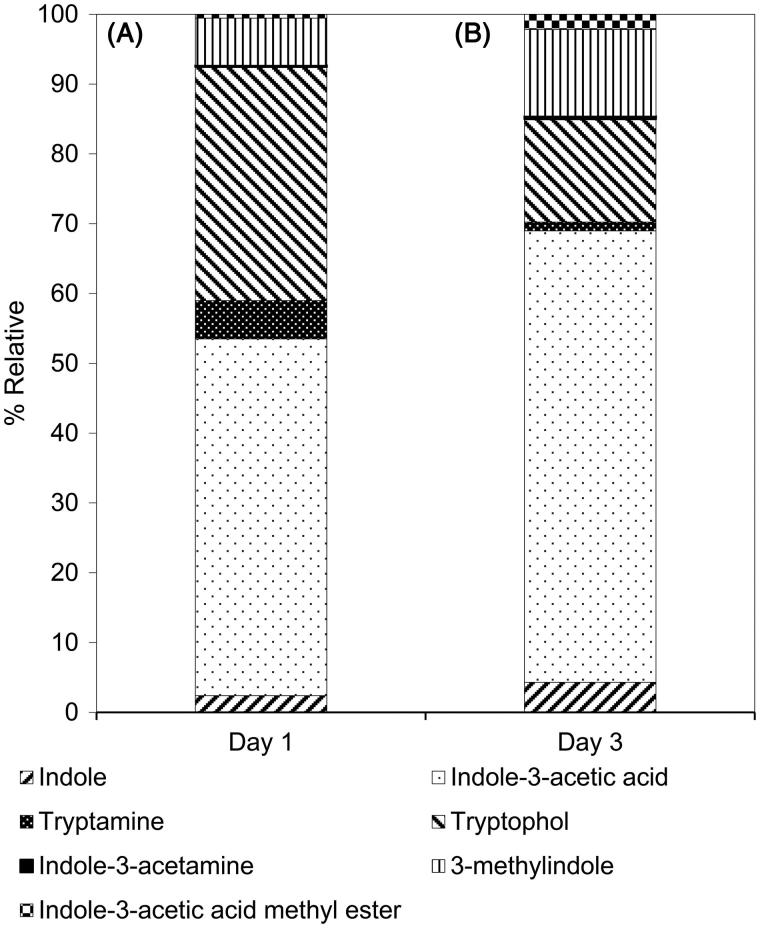
To verify IAA biosynthetic pathway of R. fluvialis DMKU-CP293, IAA intermediates were determined using the GC-MS. Indole derivatives, which were found in the supernatant of yeast cultivated in MS medium supplemented with 0.1% L-tryptophan using glucose as carbon source were: indole, IAA, TAM, TOL, IAM, 3-methylindole and indole-3-acetic acid methyl ester (Figure 2). Indole may serve as the precursor for tryptophan synthesis or, conversely, it may result from tryptophan metabolism similar to IAA, TAM, TOL, and IAM. 3-Methylindole and indole-3-acetic acid methyl ester those may be obtained as a result of IAA degradation.
Figure 2.
Identification of indole derivatives from the culture supernatant of R. fluvialis DMKU-CP293 by GC-MS in MS medium after (A) 1 day and (B) 3 days of incubation.
In the present study, we investigated the pathway(s) that R. fluvialis DMKU-CP293 may use for IAA biosynthesis by determining the pathway intermediates produced by yeast. The results indicated that TAM and IAM, but not IPA, were found in the culture medium of R. fluvialis DMKU-CP293. TOL, which is an aromatic alcohol and is a product of enzymatic reduction of IAAld, was also detected. However, some intermediates of IAA biosynthesis have been reported to be unstable and non-accumulative compounds, such as IPA and IAAld [20]. IPA pathway was shown to be the major route for IAA biosynthesis in yeast or other organisms, such as fungi Ustilago maydis, Tricholoma vaccinum, Piriformospora indica, Aciculosporium take [21–24]; plant Arabidopsis thaliana [25,26] and bacteria Agrobacterium rhizogenes, Pseudomonas savastanoi, Erwinia herbicola, Rhodococcus fascians [27–29]. Nutaratat et al. [13] also reported that the basidiomycetous yeast, Rh. paludigena DMKU-RP301, synthesized IAA via the IPA pathway but IPA and IAAld were not detected. Therefore, we postulated that R. fluvialis DMKU-CP293 may produce IPA but were not detectable due to its instability and its difficulty in isolation [30]. In addition, IPA may be quickly converted to other compounds those were unstable or quickly entered other side-chain pathway of IAA biosynthesis. Overall, investigations of pathway intermediates (Figure 2), suggested the possibility that R. fluvialis DMKU-CP293 used the IPA pathway, IAM pathway and TAM pathway to synthesize IAA from tryptophan.
3.3. Determination of key enzymes involved with IAA biosynthetic pathways
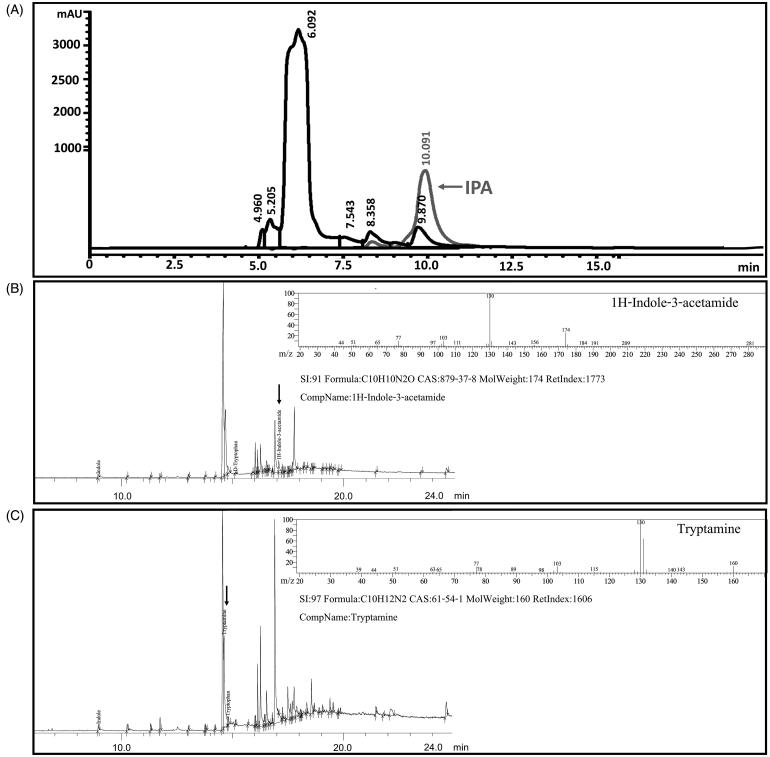
To clarify the pathways of IAA biosynthesis in R. fluvialis DMKU-CP293, activities of the key enzymes involved in the IAA biosynthetic pathway were assayed and the results are summarized in Table 1. Tryptophan aminotransferase (EC 2.6.1.27), the first enzyme that converts tryptophan to IPA, was assayed and the formation of IPA was revealed as a peak with a retention time of 9.870 min corresponding to that of the standard IPA (a retention time of 10.091 min) when analyzed with the HPLC (Figure 3(A)). Finding of tryptophan aminotransferase activity in cell crude extract indicated that the IPA route is probably one of the IAA biosynthetic pathways in this yeast. To determine the key enzyme of the IAM pathway, the active conversion of tryptophan to IAM was investigated by an in vitro assay of the tryptophan 2-monooxygenase activity and IAM was measured by the GC-MS. The formation of IAM was detected to indicate that tryptophan 2-monooxygenase activity present in R. fluvialis DMKU-CP293 (Figure 3(B)). Regarding the TAM pathway, activity of tryptophan decarboxylase was assayed in vitro and TAM in the crude extract was determined by GC-MS. The result showed TAM was detected by GC-MS, tryptophan decarboxylase was revealed in this yeast (Figure 3(C)). To investigate the involvement of the IAN pathway with IAA biosynthesis in this yeast nitrile hydratase and nitrilase activities were assayed but none was found (Table 1). Therefore, the results presented in this study provided evidence that R. fluvialis DMKU-CP293 may use IPA, IAM, and TAM pathways for IAA biosynthesis.
Table 1.
Activity assay for the existence of the enzymes involved with IAA biosynthetic pathway.
| Enzymes | Activity |
|---|---|
| Tryptophan aminotransferase | + |
| Tryptophan 2-monooxygenase | + |
| Tryptophan decarboxylase | + |
| Nitrile hydratase | − |
| Nitrilase | − |
| Tryptophan side-chain oxidase | − |
+: Presence of activity.
−: Absence of activity.
Figure 3.
HPLC and GC-MS chromatograms of key intermediates involved with IAA biosynthesis via IPA (A), IAM (B), and TAM (C) routes.
3.4. Feeding experiments with IAA precursors
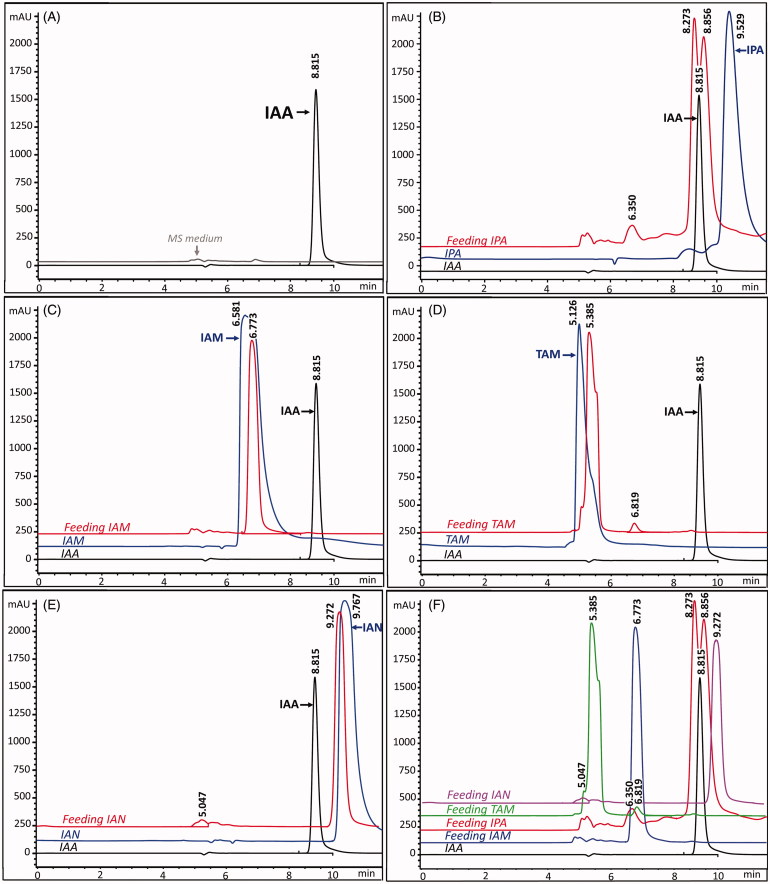
To assure the validity of our results described above, feeding experiments with different types of IAA intermediates were carried out and IAA was detected by HPLC analysis. IPA, IAM, IAN, and TAM were individually added as a precursor. Accumulation of IAA was detected when IPA was added to the medium (Figure 4(B)). However, we did not observe any IAA peak in the medium added with IAN (Figure 4(E)). This result supported the absence of nitrile hydratase and nitrilase activities from the previous determination of the enzyme involved in IAA biosynthetic pathways. The IAN pathway does not involve of IAA biosynthesis pathway in R. fluvialis DMKU-CP293. The culture supernatant of R. fluvialis DMKU-CP293 cultivated in the medium added with IAM was used to determine an existence of the IAM pathway in this yeast. However, the absence of an IAA peak after the HPLC analysis reflected the absence of the IAM pathway in R. fluvialis DMKU-CP293 (Figure 4(C)). This result suggested that R. fluvialis DMKU-CP293 was unable to convert IAM to IAA. Regarding the TAM pathway, TAM was directly converted to IAAld by the activity of amine oxidase [28]. As reported in plants, the level of IAAld was maintained in equilibrium as a form of TOL because IAAld was not stable [31]. Therefore, IAA biosynthesis of R. fluvialis DMKU-CP293 via the TAM pathway was suspected. This was due to a decrease in the TAM amount with longer incubation time (data not shown). To assess the existence of the TAM pathway in this yeast, a TAM feeding experiment was carried out but IAA was not found (Figure 4(D)). From the literature, IPA and TAM are converted to IAA through the IAAld which can be converted to IAA by two alternative enzymatic reactions catalyzed by indole-3-acetaldehyde dehydrogenase (EC. 1.2.1.3) and indole-3-acetaldehyde oxidase (EC. 1.2.3.7) [32]. Alternatively, IPA can also be directly converted to IAA by indole-3-pyruvate monooxygenase (EC:1.14.13.168) [25,33].
Figure 4.
Embedded HPLC chromatogram of standard IAA and culture supernatant of R. fluvialis DMKU-CP293 grown in MS medium (A), standard IAA, standard IPA and culture supernatant with IPA (B); standard IAA, standard IAM, and culture supernatant with IAM (C); standard IAA, standard TAM and culture supernatant with TAM (D), standard IAA, standard IAN and culture supernatant with IAN (E) and standard IAA, culture supernatant IPA, IAM, TAM, and IAN (F) after 3 days of incubation.
In conclusion, R. fluvialis DMKU-CP293 was shown to biosynthesize IAA through Trp-dependent via the IPA route (Figure 5). Based on indole derivatives identified from the culture supernatant using GC-MS, we propose that the possible IAA biosynthetic routes used by this yeast are IPA, IAM and TAM pathways when glucose was used as a carbon source. Overall results obtained from indole derivatives identification, key enzymes activity assay and feeding experiment suggested that R. fluvialis DMKU-CP293 mainly used the IPA pathway for IAA biosynthesis. However, the key enzyme activity assay and IAA intermediates suggested that the IAM and TAM pathways may be involved with IAA biosynthesis in this yeast. Thus, further investigation on the expression of the gene encoding enzymes involved with each pathway possibly confirm multiple routes of IAA biosynthesis in R. fluvialis.
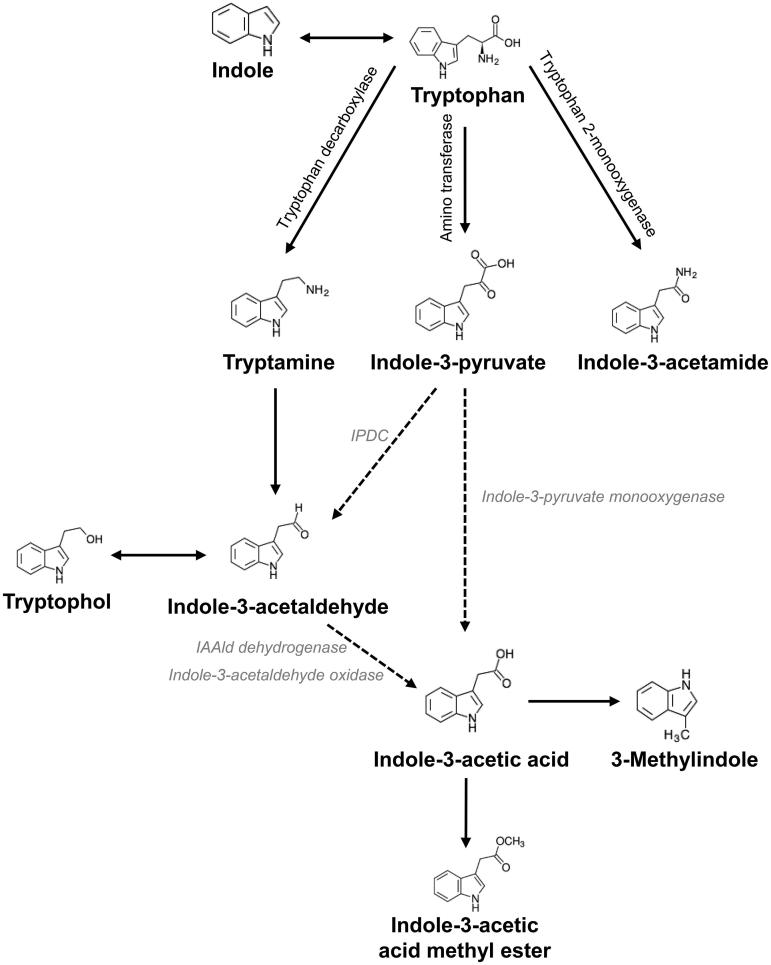
Figure 5.
Proposed tryptophan-dependent IAA biosynthesis pathway and its intermediates in R. fluvialis DMKU-CP293. Arrows with solid lines indicate IAA intermediates confirmed by GC-MS and enzymatic assays in this study, arrows with dashed lines indicate the pathway proposed based on the previous literature [2,18,25].
Funding Statement
This work was supported by the Thailand Research Fund through the TRF Research-Team Promotion Grant (RTA608004) and the Royal Golden Jubilee PhD programme grant no. PHD/0125/2559, Thailand.
Acknowledgments
The authors would like to thank Assist. Prof. Dr. Pakorn Vatana-amorn, Department of Chemistry, Faculty of Science, Kasetsart University, Thailand for his comments and suggestions.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- 1.Libbert E, Risch H. Interactions between plants and epiphytic bacteria regarding their auxin metabolism: v. isolation and identification of the IAA‐producing and destroying bacteria from pea plants. Physiol Plant. 1969;22:51–58. [Google Scholar]
- 2.Ruanpanun P, Tangchitsomkid N, Hyde KD, et al. Actinomycetes and fungi isolated from plant-parasitic nematode infested soils: screening of the effective biocontrol potential, indole-3-acetic acid and siderophore production. World J Microbiol Biotechnol. 2010;26:1569–1578. [Google Scholar]
- 3.Limtong S, Koowadjanakul N. Yeasts from phylloplane and their capability to produce indole-3-acetic acid. World J Microbiol Biotechnol. 2012;28:3323–3335. [DOI] [PubMed] [Google Scholar]
- 4.Mano Y, Nemoto K. The pathway of auxin biosynthesis in plants. J Exp Bot. 2012;63:2853–2872. [DOI] [PubMed] [Google Scholar]
- 5.Spaepen S, Vanderleyden J, Remans R. Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol Rev. 2007;31:425–448. [DOI] [PubMed] [Google Scholar]
- 6.Duca D, Lorv J, Patten CL, et al. Indole-3-acetic acid in plant-microbe interactions. Antonie Van Leeuwenhoek. 2014;106:85–125. [DOI] [PubMed] [Google Scholar]
- 7.Zakharova EA, Shcherbakov AA, Brudnik VV, et al. Biosynthesis of indole-3-acetic acid in Azospirillum brasilense. Insights from quantum chemistry. Eur J Biochem. 1999;259:572–576. [DOI] [PubMed] [Google Scholar]
- 8.Xin G, Glawe D, Doty SL. Characterization of three endophytic, indole-3-acetic acid-producing yeasts occurring in Populus trees. Mycol Res. 2009;113:973–980. [DOI] [PubMed] [Google Scholar]
- 9.Limtong S, Kaewwichian R, Yongmanitchai W, et al. Diversity of culturable yeasts in phylloplane of sugarcane in Thailand and their capability to produce indole-3-acetic acid. World J Microbiol Biotechnol. 2014;30:1785–1796. [DOI] [PubMed] [Google Scholar]
- 10.Nutaratat P, Srisuk N, Arunrattiyakorn P, et al. Plant growth-promoting traits of epiphytic and endophytic yeasts isolated from rice and sugar cane leaves in Thailand. Fungal Biol. 2014;118:683–694. [DOI] [PubMed] [Google Scholar]
- 11.Fu S-F, Sun P-F, Lu H-Y, et al. Plant growth-promoting traits of yeasts isolated from the phyllosphere and rhizosphere of Drosera spatulata Lab. Fungal Biol. 2016;120:433–448. [DOI] [PubMed] [Google Scholar]
- 12.El-Tarabily KA. Suppression of Rhizoctonia solani diseases of sugar beet by antagonistic and plant growth-promoting yeasts. J Appl Microbiol. 2004;96:69–75. [DOI] [PubMed] [Google Scholar]
- 13.Nutaratat P, Srisuk N, Arunrattiyakorn P, et al. Indole-3-acetic acid biosynthetic pathways in the basidiomycetous yeast Rhodosporidium paludigenum. Arch Microbiol. 2016;198:429–437. [DOI] [PubMed] [Google Scholar]
- 14.Rao RP, Hunter A, Kashpur O, et al. Aberrant synthesis of indole-3-acetic acid in Saccharomyces cerevisiae triggers morphogenic transition, a virulence trait of pathogenic fungi. Genetics. 2010;185:211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun P-F, Fang W-T, Shin L-Y, et al. Indole-3-acetic acid-producing yeasts in the phyllosphere of the carnivorous plant Drosera indica L. Plos One. 2014;9:e114196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Surussawadee J, Khunnamwong P, Srisuk N, et al. Papiliotrema siamense f.a., sp. nov., a yeast species isolated from plant leaves. Int J Syst Evol Microbiol. 2014;64:3058–3062. [DOI] [PubMed] [Google Scholar]
- 17.Nutaratat P, Amsri W, Srisuk N, et al. Indole-3-acetic acid production by newly isolated red yeast Rhodosporidium paludigenum. J Gen Appl Microbiol. 2015;61:1–9. [DOI] [PubMed] [Google Scholar]
- 18.Mujahid M, Sasikala C, Ramana CV. Production of indole-3-acetic acid and related indole derivatives from L-tryptophan by Rubrivivax benzoatilyticus JA2. Appl Microbiol Biotechnol. 2011;89:1001–1008. [DOI] [PubMed] [Google Scholar]
- 19.Kulkarni GB, Sanjeevkumar S, Kirankumar B, et al. Indole-3-acetic acid biosynthesis in Fusarium delphinoides strain GPK, a causal agent of wilt in chickpea. Appl Biochem Biotechnol. 2013;169:1292–1305. [DOI] [PubMed] [Google Scholar]
- 20.Ricardo C-L, Campos-Reales N, Elmerich C, et al. Physiological evidence for differently regulated tryptophan-dependent pathways for indole-3-acetic acid synthesis in Azospirillum brasilense. Mol Gen Genet. 2000;264:521–530. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka E, Tanaka C, Ishihara A, et al. Indole-3-acetic acid biosynthesis in Aciculosporium take, a causal agent of witches' broom of bamboo. J Gen Plant Pathol. 2003;69:1–6. [Google Scholar]
- 22.Reineke G, Heinze B, Schirawski J, et al. Indole-3-acetic acid (IAA) biosynthesis in the smut fungus Ustilago maydis and its relevance for increased IAA levels in infected tissue and host tumour formation. Mol Plant Pathol. 2008;9:339–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hilbert M, Voll LM, Ding Y, et al. Indole derivative production by the root endophyte Piriformospora indica is not required for growth promotion but for biotrophic colonization of barley roots. New Phytol. 2012;196:520–534. [DOI] [PubMed] [Google Scholar]
- 24.Krause K, Henke C, Asiimwe T, et al. Biosynthesis and secretion of indole-3-acetic acid and its morphological effects on Tricholoma vaccinum-spruce ectomycorrhiza. Appl Environ Microbiol. 2015;81:7003–7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mashiguchi K, Tanaka K, Sakai T, et al. The main auxin biosynthesis pathway in Arabidopsis. Proc Natl Acad Sci USA. 2011;108:18512–18517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao Y. Auxin biosynthesis. Arabidopsis book. 2014;12:e0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brandi M, Clark EM, Lindow SE. Characterization of the indole-3-acetic acid (IAA) biosynthetic pathway in an epiphytic strain of Erwinia herbicola and IAA production in vitro. Can J Microbiol. 1996;42:586–592. [Google Scholar]
- 28.Spaepen S, Vanderleyden J. Auxin and plant-microbe interactions. Cold Spring Harb Perspect Biol. 2011;3:a001438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stes E, Prinsen E, Holsters M, et al. Plant-derived auxin plays an accessory role in symptom development upon Rhodococcus fascians infection. Plant J. 2012;70:513–527. [DOI] [PubMed] [Google Scholar]
- 30.Koga J, Adachi T, Hidaka H. IAA Biosynthetic pathway from tryptophan via indole-3-pyruvic acid in Enterobacter cloacae. Agric Biol Chem. 1991;55:701–706. [Google Scholar]
- 31.Brown HM, Purves WK. Indole acetaldehyde reductase of Cucumis sativus L: kinetic properties and role in auxin biosynthesis. Plant Physiol. 1980;65:107–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koshiba T, Saito E, Ono N, et al. Purification and properties of flavin- and molybdenum-containing aldehyde oxidase from coleoptiles of maize. Plant Physiol. 1996;110:781–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao Y. Auxin biosynthesis: a simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol Plant. 2012;5:334–338. [DOI] [PMC free article] [PubMed] [Google Scholar]