ABSTRACT
Shigella is the major cause of bacillary dysentery worldwide, especially in developing countries. There are several virulence factors essential for the organism to be virulent which are generally present in the virulence plasmid and on chromosomal pathogenicity islands. The present study was undertaken to determine the virulence gene profile of Shigella spp isolated from a clinical specimen and to study their significant association with common clinical symptoms and antimicrobial resistance. Sixty Shigella whole genome sequences, including 22 S. flexneri, 14 S. sonnei, 17 S. boydii and 7 S. dysenteriae were analyzed for the presence of virulence genes. The gene found predominantly in this study were ipaH (90%) followed by sigA (83%), and lpfA (78%) respectively. The virulence genes were significantly higher in S. flexneri, particularly in serotype 2 compared to S. sonnei. Interestingly, a significant association was observed between sigA gene and fever whereas sepA and sigA were found to be associated with diarrhea. Among the studied Shigella isolates, the presence of virulence genes was found higher in isolates resistant to more than three antibiotic classes. The present work revealed the varying incidence of virulence determinants among different Shigella serogroups and shows their contribution to disease severity.
KEYWORDS: Shigella spp, virulence, sepA, sigA, lpfA, ipaH
Introduction
Shigella is a pathogen restricted to humans and a common cause of diarrhea in developing countries. Among the four Shigella serogroups, S. flexneri is the most prevailing species in developing nations followed by S. sonnei, while S. boydii and S. dysenteriae are less frequently isolated [1]. The spectrum of the disease varies from mild to severe infections.
Although shigellosis is self-limiting, use of antibiotics reduces the duration of illness and thus reduces the person to person transmission. Recently, WHO has recommended ciprofloxacin as the first choice for treating dysentery in adults and children, and azithromycin, cefixime or ceftriaxone as the second choice. Trimethoprim-sulfamethoxazole can be considered as an alternative second-line drug but generally is recommended only when the susceptibility is known or based on the local surveillance data [2].
Several virulence factors have been reported to be associated with the pathogenesis of shigellosis, which may be located either in the chromosome or plasmid. The virulence inv plasmid is an essential virulence determinant of Shigella spp which encodes the molecular machinery necessary for tissue invasion and intracellular survival [3]. The key factor for Shigella pathogenesis is the ability to invade and colonize intestinal epithelial cells [4]. The epithelial cell penetration and host response modification for cell to cell dissemination are generally mediated by an invasion-associated locus (ial), which is located on a plasmid and the invasion plasmid antigen H (ipaH) genes present in both plasmid and chromosome [5].
In addition, Shigella spp also produces distinct enterotoxins genes such as ShET-1, which encodes set1A and set1B genes located on the chromosome are found to be responsible for watery diarrhea. ShET-2, encoded by sen gene that are located on the virulence plasmid are believed to be involved in the invasion process [5,6]. Another important virulence factor related to S. dysenteriae is the presence of stx toxin which is released only during cell lysis [7].
There are very few studies that exist worldwide on the molecular characterization of Shigella virulence factors, and such reports are scarce in India. The present study was undertaken to determine the virulence gene profile of Shigella spp isolated in India and to study their association with common clinical symptoms and antimicrobial resistance.
Materials and methods
Isolate identification and serotyping
Sixty Shigella isolates were included in this study. These isolates were non-duplicate stool specimen obtained from patients suspected with enteric infection, collected as a part of routine diagnostics at the department of clinical microbiology, Christian Medical College, Vellore, India during the year 2011–2017. The stool specimen was inoculated on to the selective media such as deoxycholate citrate agar (DCA), xylose lysine deoxycholate agar (XLD) and on the MacConkey agar. The colonies that exhibit the characteristics of Shigella spp were selected and identified to the species level by biochemical tests [8] and serotypes were determined with commercially available polyclonal and monoclonal-specific antisera (Denka Seiken, Tokyo, Japan). Patient details were obtained through an electronic database maintained in the hospital and included for further analysis.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing of isolates was performed using Kirby-Bauer disc diffusion method against ampicillin (10 µg), trimethoprim/sulfamethoxazole (1.25/23.75 µg), nalidixic acid (30 µg), norfloxacin (10 µg), cefotaxime (30 µg) and cefixime (5 µg). The results were interpreted using breakpoints endorsed by the Clinical Laboratory Standards Institute (CLSI) guidelines 2017 [9]. Quality control strains used were Escherichia coli ATCC 35218 and Escherichia coli ATCC 25922 for the antibiotics tested.
Whole genome sequencing (WGS)
Genomic DNA was extracted using the QiaSymphony DNA extraction platform (Qiagen) as per manufacturer’s instruction. The WGS was performed using Ion Torrent (PGM, Life technologies) with 400-bp read chemistry (Life Technologies, Carlsbad, CA) as described earlier [10]. The genome data was assembled de novo using AssemblerSPAdes v.5.0.0.0 embedded in Torrent suite server v.5.0.5. Sequences were annotated using PATRIC, the bacterial bioinformatics database and analysis resource (http://www.patricbrc.org) [11], and the NCBI Prokaryotic Genome Automatic Annotation Pipeline (PGAAP) (http://www.ncbi.nlm. nih.gov/genomes/static/Pipeline.html) [12].
The whole genome data was analyzed for the presence of virulence genes using an open access tool, VirulenceFinder 1.5 (https://cge.cbs.dtu.dk//services/VirulenceFinder/) with the 90% threshold for identity and with 60% of minimum length coverage, where reads were mapped to the reference sequence in the database [13]. Species-specific virulence genes were also analyzed through PATRIC database. These whole genome shotgun sequences were deposited in DDBJ/ENA/GenBank. This study also includes 20 Shigella genomes retrieved from our previous study for analysis [10].
Statistical analysis
Comparisons of variables were derived using two-tailed Chi-squared test. A p-value of <0.05 was considered statistically significant. The significance levels for important virulence genes were determined between the two predominant serogroups (S. flexneri and S. sonnei) and for the most common clinical symptoms of Shigella.
Results
Sixty Shigella isolates, including S. flexneri (n = 22), S. sonnei (n = 14), S. boydii (n = 17) and S. dysenteriae (n = 7) were analyzed in this study.
Antimicrobial susceptibility
Of the isolates tested, 63% were multi-drug resistant (MDR), resistant to three or more antibiotic classes. Mostly, the isolates were resistant to ampicillin (70%), trimethoprim/sulfamethoxazole (90%) and nalidixic acid (72%). While 33%, 27% and 20% resistance was observed for norfloxacin, cefotaxime and cefixime, respectively. One isolate was susceptible to all tested antimicrobials.
Analysis of WGS results
The prevalence of virulence genes among different Shigella serogroups is shown in Table 1. All isolates were positive for ipaH gene. The virulence genes found predominantly in the study isolates other than ipaH were sigA (83%), and lpfA (78%). The enterotoxin gene such as ShET-1 (28% set1A, 23% set1B) and ShET-2 (72% senB) were identified. The ShET-1 gene was mostly seen in S. flexneri isolates. The genes including virF, iha, sepA, ipaD, ial, gad, celb, capU and pic were also identified in 63%, 40%, 27%, 63%, 57%, 5%, 15%, 57% and 18% of isolates, respectively. A high heterogeneity in the combination of virulence genes was observed.
Table 1.
Distribution of virulence genes among 60 Shigella isolates n (%).
| virF | iha | sepA | set1A | set1B | senB | ipaD | ipaH | ial | gad | celb | capU | sigA | pic | lpfA | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S. dysenteriae (n = 7) | 5 (100) | 5 (100) | 0 (0) | 0 (0) | 1 (14) | 7 (100) | 7 (100) | 7 (100) | 7 (100) | 0 (0) | 0 (0) | 7 (100) | 7 (100) | 0 (0) | 7 (100) |
| S. boydii (n = 17) | 17 (100) | 17 (100) | 0 (0) | 1 (6) | 1 (6) | 17 (100) | 16 (94) | 17 (100) | 15 (88) | 1 (6) | 0 (0) | 15 (88) | 14 (82) | 0 (0) | 7 (41) |
| S. flexneri (n = 22) | 16 (73) | 0 (0) | 16 (73) | 15 (68) | 12 (54) | 5 (23) | 15 (68) | 22 (100) | 7 (32) | 0 (0) | 0 (0) | 11 (50) | 15 (68) | 11 (50) | 21 (95) |
| S. sonnei (n = 14) | 0 (7) | 0 (0) | 0 (0) | 1 (7) | 1 (7) | 14 (100) | 0 (0) | 14 (100) | 5 (36) | 2 (14) | 9 (64) | 1 (7) | 14 (100) | 0 (0) | 14 (100) |
| Flexneri vs Sonnei (p-value) | <0.001 | NS | <0.001 | <0.003 | <0.028 | <0.001 | <0.001 | NS | NS | NS | <0.001 | <0.015 | NS | <0.014 | NS |
| Total | 38 (63) | 24 (40) | 16 (27) | 17 (28) | 14 (23) | 43 (72) | 38 (63) | 60 (100) | 34 (57) | 3 (5) | 9 (15) | 34 (57) | 50 (83) | 11 (18) | 47 (78) |
NS – not significant, p < 0.05 was considered significant
The presence of virulence genes was compared in the most prevalent serogroups. The virulence genes were significantly higher (p < 0.005) in S. flexneri isolates when compared to S. sonnei. (Table 1). Most of the virulence genes were found in all four serogroups. However, the sepA and pic genes were identified only in S. flexneri isolates, while gad gene was found in S. boydii and S. sonnei isolates. Similarly, iha gene was seen only in S. dysenteriae and S. boydii isolates. Whereas, S. sonnei harbored the celb gene but all other serogroups were negative for this gene. While 3 S. flexneri isolates were negative for all gene except ipaH and lpfA genes. The distribution of virulence genes among S. flexneri serotypes were given in the Table 2.
Table 2.
Prevalence of virulence genes among S. flexneri serotypes studied n (%).
| Virulence genes | S. flexneri 1 (n = 3) | S. flexneri 2 (n = 11) | S. flexneri 4 (n = 3) | S. flexneri (untypeable) (n = 5) |
|---|---|---|---|---|
| virF | 2 (67) | 10 (91) | 2 (67) | 2 (40) |
| iha | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| sepA | 2 (67) | 9 (82) | 3 (100) | 2 (40) |
| set1A | 0 (0) | 10 (91) | 3 (100) | 2 (40) |
| set1B | 0 (0) | 8 (73) | 2 (67) | 2 (40 |
| senB | 2 (67) | 1 (9) | 1 (33) | 1 (20) |
| ipaD | 1 (33) | 10 (91) | 2 (67) | 2 (40) |
| ipaH | 3 (100) | 11 (100) | 3 (100) | 5 (100) |
| ial | 2 (67) | 3 (27) | 0 (0) | 2 (40) |
| gad | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| celb | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| capU | 2 (67) | 6 (54) | 2 (67) | 1 (20) |
| sigA | 0 (0) | 10 (91) | 3 (100) | 2 (40) |
| pic | 0 (0) | 6 (54) | 2 (67) | 3 (60) |
| lpfA | 3 (100) | 10 (91) | 3 (100) | 5 (100) |
Additionally, common clinical features of Shigella infection (fever, abdominal pain, vomiting, diarrhea and hospitalization) were compared with the presence of common virulence genes to study their association (Table 3). The most common complaint was fever (38%), followed by diarrhea (33%), vomiting (8%) and abdominal pain (7%). Remarkably, 45% of the patients had a history of hospitalization in this study. A significant association was observed for ipaH with the presence of abdominal pain and vomiting, and similarly for the sigA gene with fever. Whereas sepA and sigA were found to be associated with diarrhea (p < 0.005). The other symptoms were, however, not significantly associated with any of the virulence genes. Also, analyzing the occurrence of clinical manifestations among the different Shigella species showed that the symptoms were not associated with any serogroup in the present study (data not shown). Among the studied Shigella isolates, the presence of virulence genes was found to be higher in isolates resistant to more than three antibiotic classes (Figure 1). The accession numbers of the isolates are provided in Table 4.
Table 3.
Association of common virulence genes with certain symptoms of shigellosis.
| Fever |
Abdominal pain |
p-value |
Diarrhea |
Vomiting |
Hospitalization |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Virulence genes | Yes | No | p-value | Yes | No | p-value | Yes | No | p-value | Yes | No | p-value | Yes | No | p-value | |
| sepA | Positive | 4 | 19 | NS | 2 | 2 | NS | 2 | 14 | p< 0.05* | 0 | 5 | NS | 6 | 21 | NS |
| Negative | 12 | 25 | 14 | 42 | 18 | 26 | 16 | 39 | 10 | 23 | ||||||
| set1A | Positive | 5 | 18 | NS | 1 | 3 | NS | 4 | 16 | NS | 1 | 4 | NS | 9 | 18 | NS |
| Negative | 12 | 25 | 16 | 40 | 13 | 27 | 16 | 39 | 8 | 25 | ||||||
| set1B | Positive | 3 | 20 | NS | 0 | 4 | NS | 2 | 18 | NS | 1 | 4 | NS | 5 | 22 | NS |
| Negative | 11 | 26 | 14 | 42 | 12 | 28 | 13 | 42 | 9 | 24 | ||||||
| senB | Positive | 19 | 4 | NS | 4 | 0 | NS | 17 | 3 | NS | 5 | 0 | NS | 21 | 6 | NS |
| Negative | 24 | 13 | 39 | 17 | 26 | 14 | 38 | 17 | 22 | 11 | ||||||
| ipaH | Positive | 23 | 37 | NS | 4 | 56 | p< 0.05* | 20 | 40 | NS | 5 | 55 | p< 0.05* | 27 | 33 | NS |
| Negative | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||
| ial | Positive | 13 | 10 | NS | 2 | 2 | NS | 11 | 9 | NS | 3 | 2 | NS | 17 | 10 | NS |
| Negative | 21 | 16 | 32 | 24 | 23 | 17 | 31 | 24 | 17 | 16 | ||||||
| sigA | Positive | 16 | 34 | p< 0.05* | 3 | 1 | NS | 13 | 37 | p< 0.05* | 4 | 1 | NS | 22 | 5 | NS |
| Negative | 7 | 3 | 47 | 9 | 7 | 3 | 46 | 9 | 28 | 5 | ||||||
| pic | Positive | 2 | 21 | NS | 0 | 4 | NS | 1 | 19 | NS | 0 | 5 | NS | 4 | 23 | NS |
| Negative | 9 | 28 | 11 | 45 | 10 | 30 | 11 | 44 | 7 | 26 | ||||||
NS – not significant, * significant difference
Figure 1.
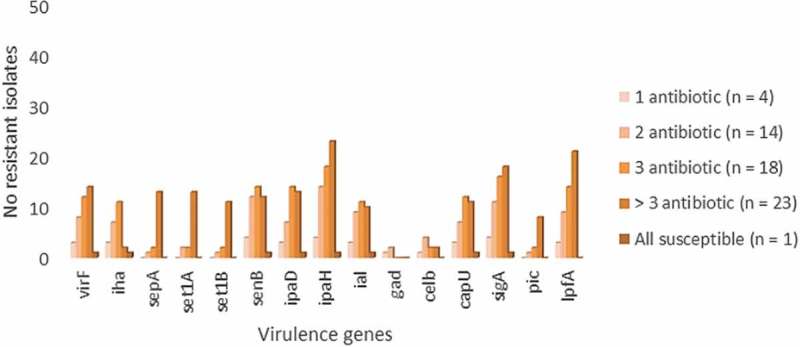
Distribution of virulence genes with varying antimicrobial resistance pattern among the study isolates.
Table 4.
Details of the Shigella isolates analyzed in this study.
| Isolate ID | year | Age/Sex | Organism | AST pattern | Accession |
|---|---|---|---|---|---|
| FC1882 | 2014 | 47/F | S. boydii | SXT-NAL | *MDDI00000000 |
| FC1764 | 2014 | 21/F | S. boydii | AMP-SXT | *MDDH00000000 |
| FC1661 | 2014 | 3/F | S. boydii | SXT-NAL-FIX | *MDGW00000000 |
| FC2833 | 2014 | 1/F | S. boydii | ALL SUSCEPTIBLE | *MDJL00000000 |
| FC1567 | 2012 | 1/M | S. boydii | AMP-SXT-NAL | *MIIV00000000 |
| FC2117 | 2012 | 8/M | S. boydii | AMP-SXT | *MINP00000000 |
| FC2125 | 2012 | 2/M | S. boydii | SXT-NAL-NX | *MINQ00000000 |
| FC2175 | 2012 | 24/M | S. boydii | SXT | *MINR00000000 |
| FC2710 | 2014 | 0/M | S. boydii | AMP-SXT-NAL (MS) | *MINU00000000 |
| FC1139 | 2011 | 1/M | S. flexneri | AMP-SXT | *MECX00000000 |
| FC1172 | 2011 | 3/M | S. flexneri | AMP-SXT-NAL-NX (MS) | *MDJI00000000 |
| FC1056 | 2015 | 75/F | S. dysenteriae | NAL-TAX | *MECW00000000 |
| FC1708 | 2012 | 2/M | S. dysenteriae | SXT | *MIIX00000000 |
| FC1737 | 2013 | 3/M | S. dysenteriae | NAL | *MIIY00000000 |
| FC2531 | 2013 | 74/F | S. dysenteriae | AMP-NAL-TAX | *MINS00000000 |
| FC2541 | 2013 | 31/F | S. dysenteriae | AMP-NAL-TAX | *MINT00000000 |
| FC2383 | 2014 | 26/F | S. boydii | AMP-SXT-NAL | *MDJK00000000 |
| FC1544 | 2014 | 2/M | S. dysenteriae | AMP-SXT-NAL | *MECT00000000 |
| FC3196 | 2015 | 60/M | S. dysenteriae | AMP-SXT-NAL | *MINV00000000 |
| FC288 | 2016 | 1/F | S. sonnei | AMP-SXT-NAL-NX | NGWI00000000 |
| FC1373 | 2016 | 4/F | S. sonnei | AMP-SXT-NAL-NX | NGWH00000000 |
| FC1417 | 2016 | 24/M | S. flexneri 4 | AMP-SXT-NAL-NX-TAX-FIX | NGWG00000000 |
| FC1846 | 2014 | 1/M | S. boydii | AMP-SXT-NAL-TAX-FIX | NGWF00000000 |
| FC2615 | 2015 | 5/M | S. boydii | AMP-SXT-NAL | NGWE00000000 |
| FC906 | 2015 | 0/M | S. flexneri 2 | AMP-SXT-NAL-NX-TAX-FIX | NGWD00000000 |
| FC1182 | 2015 | 1/M | S. flexneri 1 | AMP-SXT-NAL | NGWC00000000 |
| FC1772 | 2014 | 0/M | S. sonnei | AMP-SXT-NAL-NX-TAX-FIX | NGWB00000000 |
| FC1659 | 2015 | 3/M | S. flexneri 2 | SXT-NAL | NGWA00000000 |
| FC470 | 2015 | 6/M | S. flexneri 2 | AMP-SXT-NAL-NX-TAX-FIX | NGVZ00000000 |
| FC1247 | 2015 | 3/F | S. flexneri 2 | AMP-SXT-NAL-NX-TAX-FIX | NGVY00000000 |
| FC1607 | 2015 | 21/F | S. flexneri 4 | AMP-SXT-NAL-NX-TAX-FIX | NGVX00000000 |
| FC1481 | 2015 | 1/M | S. flexneri 4 | AMP-SXT-NAL-NX-TAX-FIX | NGVW00000000 |
| FC3278 | 2015 | 3/F | S. sonnei | AMP-SXT-NAL | NMYB00000000 |
| FC1244 | 2015 | 65/F | S. sonnei | SXT-NAL | NMYA00000000 |
| FC3433 | 2015 | 4/F | S. flexneri 2 | AMP-SXT-NAL-TAX | NMXZ00000000 |
| FC653 | 2017 | 9/M | S. sonnei | AMP-SXT | NMXY00000000 |
| FC1170 | 2015 | 45/F | S. flexneri 2 | AMP-SXT-NAL | NMXX00000000 |
| FC1824 | 2014 | 45/M | S. flexneri 2 | AMP-SXT-NAL | NMXW00000000 |
| FC601 | 2017 | 18/F | S. flexneri 1 | AMP-SXT | NMXV00000000 |
| FC3209 | 2016 | 1/M | S. sonnei | SXT-NAL-NX | NMXU00000000 |
| FC666 | 2017 | 12/M | S. boydii | SXT | NMXT00000000 |
| FC1747 | 2016 | 48/m | S. sonnei | SXT-NAL | NMXS00000000 |
| FC15 | 2017 | 69/F | S. sonnei | AMP-SXT-NAL-NX-TAX-FIX | NMXR00000000 |
| FC401 | 2017 | 2/M | S. flexneri 1 | AMP-SXT-NAL-NX | NMXQ00000000 |
| FC420 | 2017 | 2/M | S. flexneri 2 | AMP-SXT-NAL-NX | NMXP00000000 |
| FC248 | 2017 | 20/F | S. flexneri | AMP-SXT-NAL-NX | NMXO00000000 |
| FC1642 | 2017 | 1/F | S. boydii | SXT | PDYE00000000 |
| FC1655 | 2017 | 2/M | S. boydii | AMP-SXT-TAX-FIX | PDYD00000000 |
| FC1676 | 2017 | 3/F | S. boydii | AMP-SXT | PDYC00000000 |
| FC1706 | 2017 | 31/F | S. sonnei | SXT | PDYB00000000 |
| FC1628 | 2017 | 11/M | S. sonnei | SXT | PDYA00000000 |
| FC1667 | 2017 | 31/F | S. sonnei | NAL | PDXZ00000000 |
| FC1717 | 2017 | 1/M | S. boydii | AMP-SXT | PDXY00000000 |
| FC1653 | 2017 | 2/F | S. sonnei | SXT | PDXX00000000 |
| FC1677 | 2013 | 1/M | S. sonnei | AMP-SXT-NAL-TAX-FIX | PDXW00000000 |
| FC1405 | 2011 | 4/M | S. flexneri | AMP-SXT-NAL-NX | PDXV00000000 |
| FC2101 | 2016 | 2/M | S. flexneri 2 | AMP-SXT-NAL-TAX-FIX | PDXU00000000 |
| FC2414 | 2016 | 0/M | S. flexneri 2 | AMP-SXT-NX | PDXT00000000 |
| FC1954 | 2016 | 0/M | S. flexneri 2 | AMP-SXT-NAL-NX | PDXS00000000 |
| FC1180 | 2011 | 3/M | S. flexneri | AMP-SXT-NAL-NX (MS) | *MDJJ00000000 |
AMP – ampicillin, SXT – trimethoprim/sulfamethoxazole, NAL – nalidixic acid, NX – norfloxacin, TAX – cefotaxime, FIX – cefixime.* isolates from previous study [10]
Discussion
Among the pathogens causing dysentery, Shigella continues to be the major etiological agent, especially in developing countries. The pathogenicity of Shigella is associated with the presence of several virulence determinants that are associated with invasion of the colonic epithelium and cell to cell dissemination [5]. The present study investigated the virulence attributes in all four serogroups of Shigella.
Increased resistance is sometimes associated with increased virulence in bacteria [14]. Several studies support this hypothesis that highly pathogenic bacterial strains would need antibiotic treatment to reduce the severity of symptoms and are probably subjected to the selective pressure of antibiotic exposure. Similarly, the association of virulence and antimicrobial resistance among the study isolates revealed that the number of virulence genes was higher in isolates resistant to three or more antimicrobials which in concordance with the previous study [15]. This could be due to the increased prevalence of MDR Shigella strains. Further proves the fact that virulence and antimicrobial resistance are well associated.
An earlier study showed that S. flexneri 2a was found to possess more virulence factors than other serogroups [16]. Similarly, on the analysis of 60 Shigella whole genome sequences of different serogroups, S. flexneri was found associated with an increased number of virulence genes particularly serotype 2a. Among the invasion-associated genes, ipaH was predominantly seen followed by ipaD and ial genes. Both ial and ipaH were reported to be responsible for epithelial cell penetration and cell to cell dissemination [17]. These genes were seen in all serogroups except S. sonnei, in which ipaD was absent.
The ipaH gene is generally present as multiple copies, seven on chromosomes, of which five are located in ipaH-islands which are acquired through phage-mediated lateral gene transfer and five on large plasmids [6,18]. The presence of the ipaH gene in all the studied isolates could be due to these high copy numbers. Besides, an association of ipaH gene with symptoms, abdominal pain and vomiting was observed in the present study which is in concordance with the previous report where the ipaH gene was found to be associated with fever, vomiting and dehydration in the infected children [6]. Further, ipaD, is part of the Ipa family required for Shigella invasion, plays an important role in Type III secretion systems (T3SSs), which are the common virulence factors among Gram-negative bacteria. In Shigella, T3SS is assembled when the environmental conditions are appropriate for invasion, secretion is initiated only after its contact to the host cell [19].
There is another gene, virF located on the virulence plasmid (pINV) which activates the transcription of icsA and virB genes, prompting the full expression of Shigella invasion program [20]. This gene was identified in 38 of the 60 Shigella isolates tested. Also, the absence of the virF gene in 22 isolates showed the lack of virulence plasmid, which could be due to the loss of plasmid during the culture.
Toxins are some of the major virulence factors produced by bacteria. stx is the toxin gene, exclusively produced by S. dysenteriae type 1 and rarely by other Shigella serogroups. The role of stx in shigellosis is still not clear as this is not essential for invasion [21]. However, the present study does not include S. dysenteriae type 1 and hence the gene was not identified in the study isolates.
Shigella spp also produces an enterotoxin (set1A/B) and sen) and exotoxin called Shiga toxin [7]. Cruz et al. reported that set1B subunit belongs to ShET-1 is the potential aggravating factor for dehydration in shigellosis and detected only in S. flexneri isolates [6]. In this study, set1A/B was predominantly seen among the S. flexneri isolates. Whereas, sen gene which was known to be responsible for inducing bloody diarrhea was identified in all serogroups in concurrence with the previous study by Lluque et al. [16].
Furthermore, all set1A/B positive isolates also presented with sigA gene, which is a Shigella IgA-like protease homolog (sigA) that belongs to the members of cytotoxic class 1 serine protease autotransporters of Enterobacteriaceae (SPATEs). This class also includes plasmid-encoded toxin gene (pet) and secreted autotransporter toxin gene (sat) [22]. In this study, sigA was identified in all four Shigella serogroups while sat and pet genes were absent in the study isolates. Besides the symptom fever was significantly associated with the presence of sigA gene. However, the class 2 members are non-cytotoxic and include the protein involved in mucosal colonization (pic) and tissue invasion (sepA). The pic and sepA genes were more prevalent in S. flexneri isolates, which is in concordance with the previous report [23].
Moreover, the set1A/B gene is reported to be present in the complementary strand of the pic gene. However, the presence of set1A or set1B in the absence of pic, absence of set1A/B genes and the presence of all three genes are observed in the study isolates. The combination of set1A + set1B + pic was present in 8 isolates. The differences observed between the presence of these genes have previously been reported in the studies [16,18,24]. The probable reason for this scenario could be due to inactive/truncated pic gene that results in the absence of set1A/B genes [16,18].
Notably, the presence of both sepA and sigA genes significantly correlated with the presence of diarrhea. A comparable result was observed for sepA gene with diarrhea in enteroaggregative E. coli in another study [22]. A recent study showed that the sen gene had a significant association with hospitalization and bloody diarrhea [5]. Similarly, the presence of sen gene was found higher in patients with hospitalization in the present study.
The limitation of the present study is the lack of invasion efficiency analysis to ensure whether the clinical isolates retained their virulence plasmid and truly invasive. Also, further study is needed to confirm the virulence gene expressions with other techniques like quantitative RT-PCR or Western blots analysis.
In conclusion, the present work revealed the heterogeneity of virulence determinants in Shigella serogroups. S. flexneri was found to have more numbers of virulence genes. The study revealed that particular symptoms were found to be associated with the presence of certain virulence factors. These findings enhance our understanding on the contribution of virulence genes in disease severity. Although, a larger sample size with clinical metadata and outcome is necessary to provide a greater insight into it.
Funding Statement
The study was funded by Indian Council of Medical Research (ICMR), New Delhi (Ref. No: AMR/TF/55/13ECDII dated 23/10/2013).
Acknowledgments
The authors gratefully acknowledge the Institutional Review Board of the Christian Medical College, Vellore (83-i/11/13) for approving the study and providing lab space and facilities.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Nuesch-Inderbinen M, Heini N, Zurfluh K, et al. Shigella antimicrobial drug resistance mechanisms, 2004–2014. Emerg Infect Dis. 2016;22:1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kotloff KL. Shigella infection in children and adults: a formidable foe. Lancet Glob Health. 2017;5:e1166–7. [DOI] [PubMed] [Google Scholar]
- [3].Schroeder GN, Hilbi H. Molecular pathogenesis of Shigella spp.: controlling host cell signaling, invasion, and death by type III secretion. Clin Microbiol Rev. 2008;21:134–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Sansonetti PJ. Rupture, invasion and inflammatory destruction of the intestinal barrier by Shigella, making sense of prokaryote–eukaryote cross-talks. FEMS Microbiol Rev. 2001;25:3–14. [DOI] [PubMed] [Google Scholar]
- [5].Yaghoubi S, Ranjbar R, Dallal MM, et al. Profiling of virulence-associated factors in Shigella species isolated from acute pediatric diarrheal samples in Tehran, Iran. Osong Public Health Res Perspect. 2017;8:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Cruz CB, Souza MC, Serra PT, et al. Virulence factors associated with pediatric shigellosis in Brazilian Amazon. BioMed Res Int. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Nogrady N, Kiraly M, Borbas K, et al. Antimicrobial resistance and genetic characteristics of integron-carrier shigellae isolated in Hungary (1998–2008). J Med Microbiol. 2013;62:1545–1551. [DOI] [PubMed] [Google Scholar]
- [8].Versalovic J, Carroll KC, Funke G, et al. Manual of clinical microbiology. 10th ed. Washington DC: American Society for Microbiology; 2010. [Google Scholar]
- [9].Clinical and Laboratory Standards Institute (CLSI) Performance standards for antimicrobial susceptibility testing In: Twenty-seventh Informational Supplement M100-S27. Wayne, PA: CLSI; 2017:32-41. [Google Scholar]
- [10].Dhiviya Prabaa MS, Naveen Kumar DR, Yesurajan IF, et al. Identification of nonserotypeable Shigella spp. using genome sequencing: a step forward. Future Sci OA. 2017;3:FSO229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wattam AR, Abraham D, Dalay O, et al. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 2013;42:D581–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Tatusova T, DiCuccio M, Badretdin A, et al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44:6614–6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Joensen KG, Scheutz F, Lund O, et al. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J Clin Microbiol. 2014;52:1501–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Roux D, Danilchanka O, Guillard T, et al. Fitness cost of antibiotic susceptibility during bacterial infection. Sci Transl Med. 2015;7:297ra114. [DOI] [PubMed] [Google Scholar]
- [15].Medeiros PH, Lima AA, Guedes MM, et al. Molecular characterization of virulence and antimicrobial resistance profile of Shigella species isolated from children with moderate to severe diarrhea in northeastern Brazil. Diagn Microbiol Infect Dis. 2018;90:198–205. [DOI] [PubMed] [Google Scholar]
- [16].Lluque A, Mosquito S, Gomes C, et al. Virulence factors and mechanisms of antimicrobial resistance in Shigella strains from periurban areas of Lima (Peru). Int J Med Microbiol. 2015;305:480–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Thong KL, Hoe SL, Puthucheary SD, et al. Detection of virulence genes in Malaysian Shigella species by multiplex PCR assay. BMC Infect Dis. 2005;5:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Yang F, Yang J, Zhang X, et al. Genome dynamics and diversity of Shigella species, the etiologic agents of bacillary dysentery. Nucleic Acids Res. 2005;33:6445–6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Roehrich AD, Guillossou E, Blocker AJ, et al. Shigella IpaD has a dual role: signal transduction from the type III secretion system needle tip and intracellular secretion regulation. Mol Microbiol. 2013;87:690–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Di Martino ML, Falconi M, Micheli G, et al. The multifaceted activity of the VirF regulatory protein in the Shigella lifestyle. Front Mol Biosci. 2016;3:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Casabonne C, Gonzalez A, Aquili V, et al. Prevalence and virulence genes of Shigella spp. isolated from patients with diarrhea in Rosario, Argentina. Jpn J Infect Dis. 2016;69:477–481. [DOI] [PubMed] [Google Scholar]
- [22].Boisen N, Scheutz F, Rasko DA, et al. Genomic characterization of enteroaggregative Escherichia coli from children in Mali. J Infect Dis. 2011;205:431–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Nave HH, Mansouri S, Emaneini M, et al. Distribution of genes encoding virulence factors and molecular analysis of Shigella spp. isolated from patients with diarrhea in Kerman, Iran. Microb Pathog. 2016;92:68–71. [DOI] [PubMed] [Google Scholar]
- [24].Durand D, Contreras CA, Mosquito S, et al. pic gene of enteroaggregative Escherichia coli and its association with diarrhea in Peruvian children. Pathog Dis. 2016;74:6. [DOI] [PMC free article] [PubMed] [Google Scholar]


