Abstract
Classifying all of the ganglion cells in the mammalian retina has long been a goal of anatomists, physiologists, and cell biologists. The rabbit retinal ganglion cell layer was phenotyped using intrinsic small molecule signals (aspartate, glutamate, glycine, glutamine, GABA, and taurine) and glutamate receptor-gated 1-amino-4-guanidobutane excitation signals as the clustering dimensions for formal classification. Intrinsic signals alone yielded 7 ganglion cell superclasses and 1 amacrine cell superclass; the addition of excitation signals ultimately resolved 14 natural ganglion cell classes and 3 amacrine cell classes. Ganglion cells comprise two-thirds to three-quarters of the cells in the ganglion cell layer and exhibited distinct metabolic, coupling, and excitation phenotypes, as well as characteristic sizes, population fractions, and patterns. Metabolic signatures (mixtures of glutamate, aspartate, glutamine, and GABA) chemically discriminated ganglion from amacrine cells. Coupling signatures reflected heterologous coupling states across ganglion cells: (1) uncoupled, (2) coupled to GABAergic amacrine cells, and (3) coupled to glycinergic amacrine cells. Excitation signatures reflected differential channel permeation rates across classes after AMPA activation. Extraction of unique size and patterning features from the data sets further validated the robustness of the classification. Because the classifications were explicitly blinded to structure, this is strong evidence that molecular phenotype classes are natural classes. Correspondences of molecular phenotype classes to functional classes were inferred from size, coupling, encounter, and physiological attributes. Ganglion cell classes display markedly different ionotropic drives, which may partly explain the physiological brisk–sluggish spectrum of ganglion cell spiking patterns.
Keywords: neuronal classification, molecular phenotyping, 1-amino-4-guanidobutane, AGB, retina, ganglion cells, amacrine cells, patterning
Efforts to assess the classes, numbers, and distributions of retinal ganglion cells have engaged diverse metrics: cell count, size, shape, axon diameter, and physiological encounter rates. Subsets of mammalian ganglion cell classes project to an array of central targets (Fukuda and Stone, 1974;Farmer and Rodieck, 1982; Leventhal et al., 1985; Rodieck and Watanabe, 1993; Pu et al., 1994; Rodieck, 1998), but a unified description of all classes and distributions in the ganglion cell layer has remained elusive. Several summaries of how neuronal typologies might be abstracted have emerged, some accompanied by debates regarding methods, definitions, and results (Rowe and Stone, 1977; Hughes, 1979; Holden, 1981; Rodieck and Brening, 1982; Famiglietti, 1992; Wingate et al., 1992; Cook, 1998; Masland and Raviola, 2000). We have performed a formal multispectral classification (Swain and Davis, 1978) of the rabbit ganglion cell layer using molecular signals as the dimensions of the multispectral space. The attributes of such a classification were precisely articulated by Famiglietti (1992): “. . .the ability to label neurons by neurotransmitter molecules. . .has provided a new means of identifying neurons repeatedly with some reliability. Nevertheless such labels are not unique markers. As a consequence of their very large range of diversity, neurons are likely to be identified and characterized as unique types by quantifying the expression of overlapping sets of markers, rather than by absolutely unique indicators. . . .The plausible hypothesis has been advanced that ‘natural’ cell types emerge as distinct clusters of points in a parametric space. . .”
Although Famiglietti and others (Rodieck and Brening, 1982;Cook, 1998) generally conceived this parametric space to be morphological, mixtures of small molecules do constitute distinctive signatures for many natural classes of retinal cells (Marc et al., 1995, 1998; Kalloniatis et al., 1996). On the basis of previous analyses, we expected that the signatures of natural ganglion cell classes would be poorly separable (Marc, 1999a,b), but when we expanded the set of molecular targets to include aspartate, glutamate, glycine, glutamine, GABA, and taurine signals, strongly heterogeneous multispectral signatures emerged in the ganglion cell layer, suggesting that detailed phenotyping was indeed possible. Excitation signatures based on mapping AMPA-activated 1-amino-4-guanidobutane (AGB) permeation (Marc, 1999b) proved decisive. Fusion of excitation and intrinsic signals permitted the demonstration that the rabbit retinal ganglion cell layer contains no fewer than 14 natural types of ganglion cells, and some are further characterized by cell size, patterning, and population fraction. This separability arises from three forms of molecular phenotype variation: (1) some ganglion cells display different intrinsic glutamatergic metabolite signatures; (2) other cells express different degrees of heterologous coupling with amacrine cells, leading to separable patterns of GABA and glycine leakage into the ganglion cells; and (3) each cell class expresses a characteristic AMPA receptor-mediated drive.
MATERIALS AND METHODS
Terminology. We define five retinal cell types as major cellular divisions traditionally distinguished by position and shape: photoreceptors, bipolar cells, horizontal cells, amacrine cells, and ganglion cells. We define superclasses and classes as statistical groups within a type. Thus we are concerned with discovering the natural classes of the ganglion cell type. A statistical class is defined by its position and dispersion in N-dimensional space. If it is well separated from its fellow classes and cannot be separated further, it may be a natural class. That hypothesis is strengthened if the statistical class can be shown to possess additional features associated with presumed natural classes. In this process, we will see that some natural classes form intermediate statistical groups as superclasses.
Isolated retinal preparations. These methods follow those described in Marc (1999a). Light-adapted adult male and female pigmented rabbits were tranquilized with intramuscular ketamine/xylazine, deeply anesthetized with intraperitoneal urethane in saline, and euthanized by thoracotamy, all in accord with institutional animal care and use guidelines. Both eyes were rapidly removed and hemisected, and large retinal pieces were mounted on cellulose acetate filter discs, then bathed in 35°C Ames medium (Ames and Nesbett, 1981) equilibrated with 95% O2/5% CO2 or HEPES-based modified Ames medium equilibrated with 100% O2. Single 2 × 3 mm retinal chips taken within 2 mm of the visual streak were razor cut from the large pieces and incubated for 10 min at 35°C under gas in 100 μl droplets of Ames or HEPES-based medium containing 5 mm AGB and 25 μl AMPA. All incubations were performed under fluorescent room lighting as described in Marc (1999a). Samples from four rabbits (cases 2366, 2578, 2780, and 2781) were similarly processed and classified (see below). Horizontal sections from retinas of >20 other specimens with and without AGB or agonists were also analyzed. All retinas were fixed by immersing chips in room temperature 1% paraformaldehyde, 2.5% glutaraldehyde, 3% sucrose, 0.01% CaCl2, in 0.1 mphosphate buffer, pH 7.4. All tissue was processed as described previously (Marc et al., 1990).
Specimen preparation and immunocytochemical visualization. Each chip was flat embedded in epoxy resin on a glass slide; a small rectangular sample was scribed from the slide (Stell and Lightfoot, 1975) and sectioned serially at 250 nm onto 12-spot Teflon-coated slides (Cel-Line, Fisher Scientific). The immunocytochemical and IgG production procedures were as described previously (Marc et al., 1990, 1995) using the silver-intensification protocol of Kalloniatis and Fletcher (1993). The samples were serially probed with IgGs targeting aspartate, glutamate, glutamine, glycine, GABA, taurine, and AGB obtained from Signature Immunologics Inc. (Salt Lake City, UT). Primary IgG signals were detected with goat anti-rabbit IgGs adsorbed to 1 nm gold particles (Amersham Biosciences) and visualized with silver intensification. The silvering process was run at 30°C for 240 sec before quenching. All preparations received identical probing and visualization treatments, yielding two orders of magnitude of detection range with differential concentration sensitivity as low as 40 μm based on artificial standards. Single-letter amino acid codes were sometimes used to denote AGB (B), aspartate (D), glutamate (E), glycine (G), glutamine (Q), taurine (τ), and GABA (γ). The selectivities of each of these probes have been characterized extensively. This is particularly critical for the IgG targeting GABA, because we interpret variations in signal strength as reflecting true GABA content arising from different sources. It is important to know the degree to which small signals could reflect contamination. Full cross-reactivity data for the Signature Immunologics YY100 anti-GABA have been obtained for the most plausible contaminating species: alanine, β-alanine, citrulline, cysteine, aspartate, glutamate, glycine, isoleucine, glutathione, lysine, leucine, methionine, asparagine, arginine, ornithine, proline, glutamine, serine, threonine, valine, tryptophan, tyrosine, ATP, ADP, AMP, cAMP, and cGMP, as well as all macromolecules expressed in retina. With the exception of β-alanine, the YY100 IgG is selective for GABA at >10,000:1. The selectivity for β-alanine (the three-carbon analog of GABA) is 80:1. This means that low levels of GABA-like immunoreactivity could be produced by β-alanine only if it were present in 80-fold excess over the inferred GABA level: a 0.1 mm YY100 signal could be produced by 8 mm β-alanine. However, this is an impossible level, because the ganglion cell layer shows little endogenous β-alanine immunoreactivity (<0.5 mm maximum) using our own β-alanine-selective IgG, and the cells in question (those showing the least amount of GABA signal, ∼0.1–1 mm) have no detectable β-alanine immunoreactivity. Thus we are confident that the GABA signal is accurate and uncontaminated. Similar verifications hold for each of the IgGs used in this study.
Image analysis. All images of immunoreactivity were captured as 8-bit 1536 pixel × 1152 line frames under constant flux light with feedback regulation and fixed CCD camera gain and gamma as described previously (Marc, 1999a). Silver visualization produces density-scaled images, and linear image inversion produces intensity-scaled images that are linear with log10(concentration) over a range of 0.05–10 mm. Serial images were aligned to >250 nm root-mean-square error with registration algorithms from PCI Geomatics (Richmond, Hill, Ontario, Canada). Image analysis and morphometry were performed with Image-Pro Plus 2.0 (Media Cybernetics Inc., Silver Spring, MD).
Classification followed these steps: (1) capture of N molecular signal channels, (2) registration of channels, (3) isodata clustering of N channels, (4) theme map generation, (5) histogram and scatter plot exploration, and if necessary, (6) deconvolution and theme map correction. After theme map generation (step 4), univariate and bivariate signal histograms were explored for each emergent class. In cases in which a signal was clearly multimodal, the histogram was deconvolved, and each cell was recoded. This is roughly equivalent to another form of classification based on watershed filtering of N-space histograms to find decision boundaries (Narendra and Goldberg, 1977). Classes completely resolved by clustering were formally statistically separable, and those resolved by deconvolution were formally statistically significant (see below).
Classifications were performed using the isodata algorithm (PCI Geomatics), and data were explored with applications written in IDL (Research Systems Inc., Boulder, CO). An overview of and reference lists for simple classical pattern recognition methods are provided inMarc et al. (1995). Isodata resembles the simpler K-means clustering method but adds heuristic splitting of high variance classes and merging of highly overlapping classes (Ball and Hall, 1967). Statistical separability indicates that the means and covariances for a set of N-dimensional data allow classification of a sample of those data into distinct classes. The probability of error (pe) in classification is estimated from the transformed divergences of the classes assuming equal a priori probability densities. Cell classes described as separable have pe ≤ 0.01. Separable classes are also inherently statistically significant classes. Natural classes need not be inherently separable by clustering and could have signature overlaps greater than required for pe ≤ 0.01 yet still be statistically significant classes. In the separations of class 5 from 9, and class 10 from 11, we used deconvolution methods [previously detailed in Marc et al. (1995)] to detect signals arising from distinct classes that clustering failed to resolve completely. These cells were reassigned as classes associated with a mode of a bimodal distribution by thresholding above and below the mode crossover point.
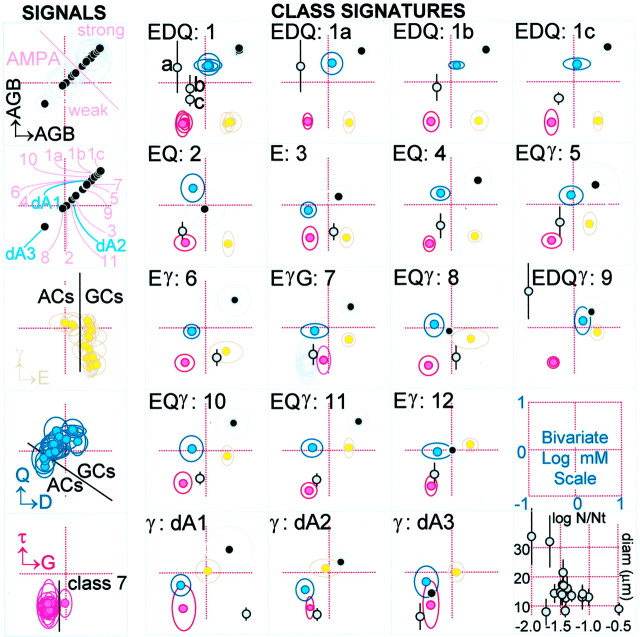
The multidimensional signatures of classes were visualized as selected rgb maps (encoding three molecular signals as red, green, and blue signals, respectively) and as superimposed bivariate 2N-plots. Single bivariate plots (Marc et al., 1995) display the area of concentration space occupied by a class in a two-dimensional chemical space. 2N-plots superimpose several bivariate plots, capturing most of the features discriminating cell groupings in an N-space. This method was inspired by the parallel coordinate space described by Inselberg and Dimsdale (1990). Pairs of signals were displayed as class means bounded by 2 SD margins. The x–y axes spanned 0.1–10 mm with logarithmic scaling, and the [x,y] pairs were color coded: [AGB, AGB] gray; [E, γ] orange; [D, Q] cyan; and [G, τ] magenta.
Average cell diameters and cell spacings were acquired with the object utilities in ImagePro Plus 2.0 (Media Cybernetics). Cell sizes in each sample were measured only from glutamate signals in individual 250 nm sections, so cell size estimates contain no registration errors and require no use of dissector methods. Some classified cells often appeared to form regular arrays, and these were numerically characterized by their conformity ratios (CRs; also known as regularity indices): the ratio of the class mean nearest-neighbor distance to its SD. The significance of the deviation of a conformity ratio from that predicted for a random pattern was determined from significance charts in Cook (1996).
Agents and sources. Ames medium either was purchased from Sigma (St. Louis, MO) or made according to Ames and Nesbett (1981) and modified as needed by equimolar Na+replacement with 15 mm HEPES and reduction of NaHCO3 to 1 mm. AGB and AMPA were added without adjustment. All solutions were made before each preparation. AGB (agmatine sulfate) was obtained from Sigma, and AMPA was from Research Biochemicals International (Natick, MA).
Figure preparation. All images are digital, assembled from the raw data captured by CCD camera (see Image analysis above). Selected frames of raw Tagged Image Format (*.tif) files were extracted for display, each sharpened by unsharp masking, and after entire images were assembled as a single figure, contrasts were adjusted with linear remapping to correct for out-of-gamut effects during printing. All final images were prepared in Adobe PhotoShop 5.0. Combinations of registered channels were viewed as true-color rgb mappings, as described previously (Marc et al., 1995).
RESULTS
GABA signals demonstrate intrinsic heterogeneity in the ganglion cell layer
A common error in immunocytochemistry is the imputation, via imagery, of binary states for small molecules: present versus absent. This is almost never true for amino acids because absolute contents vary across cell types. The error is often technical, arising from nonlinear and excessive amplification during enzyme-linked visualization or saturation of signals (fluorescence or absorption) during image capture or later processing. Silver visualization of immunogold signals on thin sections offers unparalleled signal differentiation. A spectrum of GABA signals, displayed in a single 250 nm section through the plane of the ganglion cell layer in the central streak of the rabbit retina (Fig.1A) and ranging from <50 μm to ∼10 mm, characterizes different compartments. The basic reasons for this differentiation are that some ganglion cells contain no GABA; displaced starburst amacrine cells contain GAD65 (Brandon and Criswell, 1995) and synthesize high GABA levels; Müller cells import GABA via a high-affinity GAT-3 transporter (Johnson et al., 1996) and metabolize it, yet have a persistent low level; and many ganglion cells engage in heterologous dye coupling with amacrine cells (Xin and Bloomfield, 1997), and GABA could clearly leak into ganglion cells. The concentration scaling of this image reveals that Müller cells appear to contain ∼85 μm GABA (uncorrected for fixation loss) and that some large neuronal cells clearly contain measurably less than that, whereas others contain much more. Some cells in the ganglion cell layer contain roughly the same amount of GABA as the Müller cells and are nearly invisible against the glial background (Fig. 1A, ellipse). An example of the sensitivity of the silvering method is also indicated, where a clear difference between Müller cell and ganglion cell labeling can be detected with as little as 40 μmcalculated difference between the two compartments (Fig.1A, box). Müller cell signals do vary across preparations, primarily because the rate of GABA conversion to succinate semi-aldehyde is an explicitly aerobic reaction. This field was chosen for its slightly higher-than-average Müller cell signal, which highlights the range of ganglion cell GABA signals. All of the signal diversity in Figure 1 is caused by intercellular variations in GABA content; none can be caused by corruption of the anti-GABA IgG signal with any known targets. This same pattern of GABA signals has been observed in >150 rabbit retinas, at all loci, with variation in cell proportions. Similarly, fascicles of ganglion cell axons just beneath the layer of somas contain streaks of strong GABA signal (Fig. 1B) that correspond to glutamate immunoreactive axons (Fig. 1C). Regardless of whether the GABA signal in ganglion cells arises from coupling leakage or synthesis, it appears in axons as well.
Fig. 1.
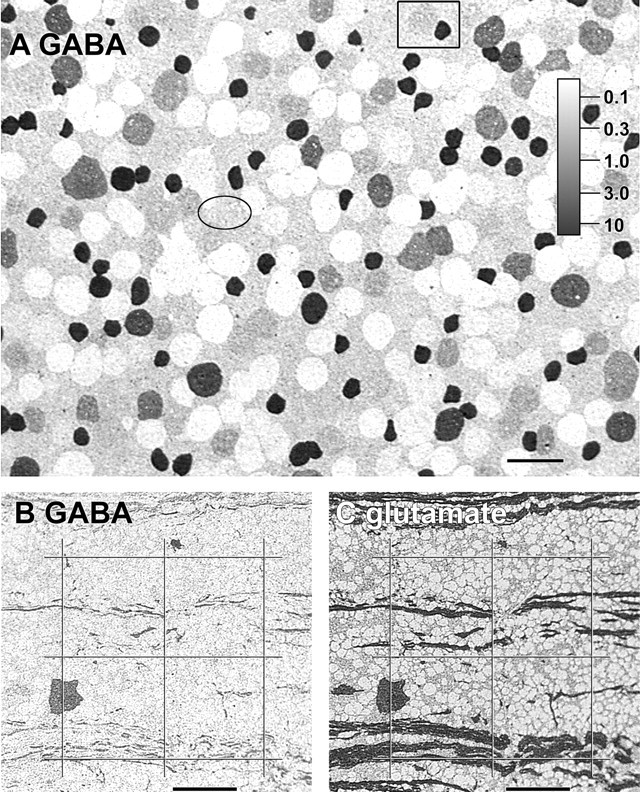
The diversity of GABA signals in the ganglion cell layer. A, Differential GABA content of cells in the ganglion cell layer of the rabbit retinal area centralis visualized by quantitative GABA mapping on a 250 nm section. Scaling was determined from artificial standards. Cellular contents range from <50 μm to ∼10 mm. Some ganglion cells are literally invisible in the array because they have the same GABA content as the surrounding Müller cells (ellipse). Other cells can be differentiated from the Müller cells by having contents roughly 40 μm higher (box); a strongly immunoreactive starburst amacrine cell is also contained in the box. Image is density scaled. Scale bar, 25 μm.B, C, Precision registered images of GABA and glutamate immunoreactivity in serial 250 nm sections through the optic fiber layer just proximal to the ganglion cell somas. Streaks of GABA immunoreactivity represent axons of varying caliber, and all GABA-immunoreactive axons are contained in fascicles of glutamate-immunoreactive axons. The grid overlay permits tracking of common points in both images. Images are density scaled. Scale bars, 25 μm.
Full classification of the ganglion cell layer
The notion that one might be able to formally phenotype the ganglion cell layer by applying pattern recognition algorithms to sets of molecular signatures (Marc et al., 1995) gains credence from images probed for excitation signals (Fig.2A) and intrinsic small molecules (Fig. 2B–H) in a comprehensive data set obtained ∼2 mm below the visual streak. Aspartate levels vary across cell types (Fig. 2B), even among cells displaying similar glutamate signals (Fig.2C). This variation is not random, and strong aspartate signals are biased toward large cells. Glutamine contents (Fig.2E,F) vary in a pattern similar but not identical to aspartate. The high glutamine contents of Müller cells form a background against which many neurons are invisible, but masking out Müller cells with their own signature class (Marc et al., 1990, 1995) exposes the full spectrum of neuronal glutamine contents. Glycine signals in the ganglion cell layer are weak (Fig. 2D), never exceeding a few hundred micromolar and never reaching the 5–10 mm levels achieved by glycinergic amacrine cells (Fig. 2D,inset). Nevertheless, certain cells always display glycine content higher than their neighbors. As in most vertebrates, taurine signals of the ganglion cell layer are restricted to Müller cells (Fig. 2G). Although none of these additional signals alone exhibits differential patterning as dramatic as that of GABA (Fig.2H), separation improves with every dimension that adds even small correlations. Use of these intrinsic signals alone never permitted a complete segregation of all likely natural classes, but isodata clustering in five normal and four excitation-mapped retinas resolved eight neuronal superclasses, labeled according to their dominant molecular signals: one amacrine cell and seven ganglion cell superclasses. At the simplest level, we can resolve two global N-dimensional clusters that distinguish amacrine and ganglion cells as fully separable types, the details of which will be discussed below. The superclasses of ganglion cells arise in part from fully separating or detecting strong three-dimensional modes of metabolic signatures dominated by glutamate (E) and characterized by increasing glutamine (Q) and aspartate (D) content: superclasses E, EQ, and EDQ. These basic superclass signatures also are separable from their counterparts that also possess a GABA signal: superclasses Eγ, EQγ, and EDQγ. As seen below, however, the EQγ and EDQγ modes do not completely separate, because a bridging cell population overlaps the two, although it more appropriately fits in the EQγ superclass. Two variations on this simple theme emerge. First, one population of cells in the Eγ classification space further separates by evidencing coupling to glycinergic amacrine cells: superclass EγG. Finally, some members of the Eγ and EQγ classification spaces contain GABA levels that are indistinguishable from those of bona fide amacrine cells. Several superclasses are composites of natural classes.
Fig. 2.
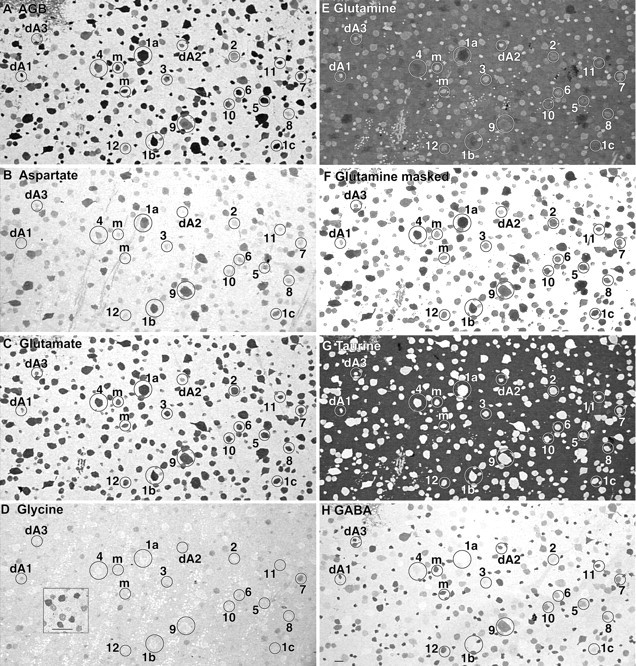
Excitation and intrinsic molecular signals in the ganglion cell layer visualized from an array of seven serial 250 nm sections (sequence: D G Q τ γ B E). All cells are embedded in a matrix of Müller cell cytoplasm. The reticulated patch at thetop left of A and Hrepresents intrusion of the inner plexiform layer into the plane. Sets of the same circled cells appear in every image: labels 1–12 indicate ganglion cell classes, labels dA1–3 indicate amacrine cell classes, andm indicates misplaced amacrine cells. All images are density scaled. Scale bar, 25 μm. A displays AMPA-activated AGB excitation signals, and subsequent panels represent intrinsic signals: B, aspartate; C, glutamate; D, glycine; E, glutamine;F, glutamine with Müller cells masked;G, taurine; H, GABA. Theinset in the glycine image (D) represents an array of glycine-immunoreactive amacrine cells from the same specimen. Ganglion cells contain a spectrum of glutamine signals (E, F), and many cells, such as classes 5 and 9, closely match the glutamine contents of Müller cells. Some classes are significantly higher (class 1a), whereas others are substantially lower (classes 3, dA1, dA2, and dA3). InG, the black taurine background is Müller cell cytoplasm, revealing that taurine levels in ganglion cells are virtually zero.
The addition of an excitation signal proved decisive, enabling a robust estimate of the minimum number of ganglion cell natural classes and some of their morphological attributes. AGB is an organic cation that permeates glutamate-gated ion channels, accumulates in activated neurons, and can be detected with quantitative immunochemical protocols (Marc, 1999a). AGB mapping thus reports recent excitation history. We have shown previously that activation of ionotropic glutamate receptors in the ganglion cell layer yields stable, heterogeneous patterns of AGB signals, implying intrinsic differences in the AMPA receptor properties of ganglion cells. We chose to analyze the excitation patterns generated by 25 μm AMPA in the ganglion cell layer in combination with the signals that permit detection of the eight superclasses. Four normal adult rabbit retinas were incubated for 10 min in Ames medium containing 5 mm AGB and 25 μm AMPA. In all samples, the ganglion cell layer displayed a spectrum of AMPA-driven responses (Fig.2A). These signal differences must arise from variations in either the numbers or types of ionotropic glutamate receptors expressed by neurons and not simply cell size variation (Marc, 1999a). Both the very largest and smallest of cells show the strongest AMPA responsivity, whereas both large and small cell types also show very weak responses.
Inclusion of the AMPA-driven excitation signal in the AGB signal in the classification set resulted in enhanced differentiation of the ganglion cell layer, revealing molecular phenotype classes visualized as rgb maps (Fig. 3A), theme maps (Fig. 3B), and signature arrays (Fig.4) of the constituent cells. Because the 15-color theme map is complex, it is practical to decompose it into its constituent cell patterns in Figure 5. The molecular phenotype data suggest the existence of at least 14 natural classes of ganglion cells and 3 natural classes of displaced amacrine cells. In general, rgb maps suggest the existence of multiple cells classes by revealing a spectrum of hues that correspond to a spectrum of molecular mixtures. Visual examination of such maps is not a proof, however, and classification is essential, revealing both classes and their signature spaces.
Fig. 3.
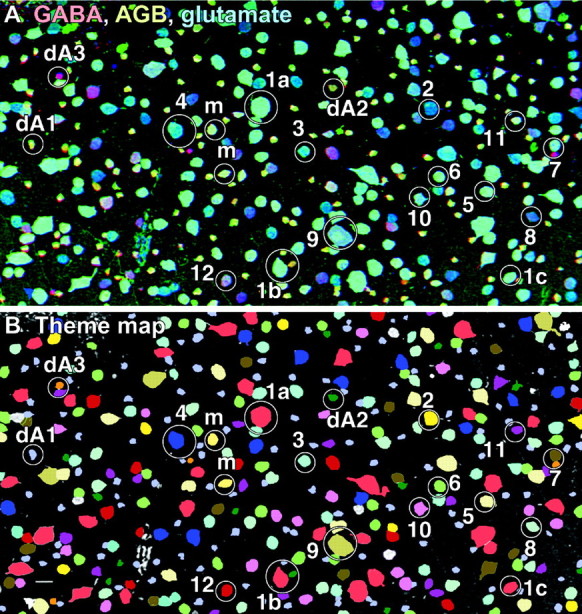
A summary rgb triplet and theme map of the ganglion cell layer. A, One of 35 possible rgb triplet mappings visualized from a registered array of 7 serial 250 nm sections: GABA → red, AGB → green, glutamate → blue. Cells exhibit a vast spectrum of hues associated with their varying intrinsic GABA contents and AMPA-activated AGB excitation signals. For example, class 1 cells (1a, 1b, 1c) are all bright cyan, reflecting strong responses to AMPA and negligible GABA content. At another limit one finds mauve class 12 cells with weak AMPA responses and high GABA content. Class dA1 starburst amacrine cells are yellowish, with high AMPA responses and high GABA content and low glutamate content. Image is intensity scaled. Scale bar, 25 μm. B, The theme map displays the results of isodata clustering and deconvolution to extract all distinct neuronal molecular phenotypes in the ganglion cell layer. The class color code can be read directly from the image:1, light red; 2,yellow; 3, cyan;4, dark blue; 5,light tan; 6, bright green; 7, olive;8, sea green; 9,dark tan; 10, magenta;11, dark purple; 12,dark red; dA1, lavender;dA2, dark green; dA3,orange; misplaced amacrine cells, m,lemon yellow. Symbols as in Figure 2. Image is indexed. Scale bar, 25 μm.
Fig. 4.
Bivariate signal plots (left column) and class signature N-plots (right four columns) that characterize classes of cells in the ganglion cell layer (case 2366). Each molecular signal or signature plot is placed on the same quantitative bivariate log millimolar scale that maps directly to pixel intensity. Each colored spotis the bivariate mean for a pair of values surrounded by a 2 SD border. In x,y axis notation,black is AGB AGB, gold isEγ, cyan is DQ,magenta is Gτ. Gray symbols map log population fraction (x = log N/Nt, where N = number of cells in class, Nt = total cells in all classes) against mean cell diameter ± 2 SD (y) on the scale in the bottom left quadrant. Left signal column, The AGB AGB signal plots (top left two frames, black symbols) reveal that the AMPA response spectrum of classes is large and index the positions in response space for each class. The Eγ signal plot shows the separation of amacrine cells from ganglion cells, especially on the E vector, and the overlap of some ganglion cells into the γ signal space of true amacrine cells. The DQ plot similarly shows that amacrine cells occupy the lowest portion of the signal space. This bivariate space alone is insufficient to completely segregate amacrine cells but is part of a complete separation when combined with the Eγ space. The Gτ plot reveals that class 7 ganglion cells separate on the G vector. The bottom right plot demonstrates that the large cells are the rarest, small cells are the most abundant, and all other cells are clustered between.Right four signature columns, The class signature plots are N-plots, encoding the same five signal pairs (AGB AGB, Eγ, DQ, Gτ, fraction-size) for each of 15 classes.
Fig. 5.
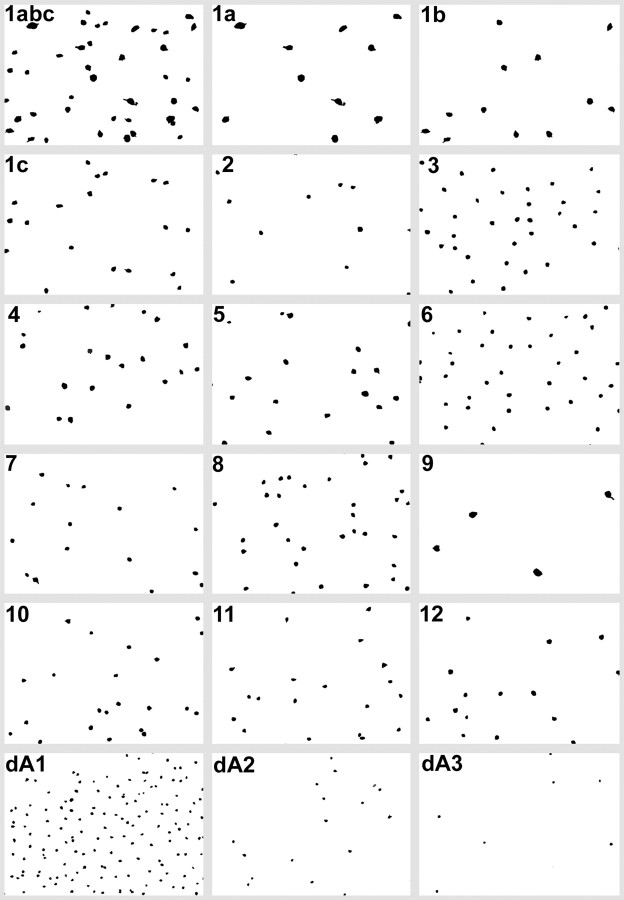
Separated patterns of cell classes derived from the registered array of seven serial 250 nm sections. Class 1 contains a mixture of cell sizes that are separable by deconvolution into classes 1a, 1b, and 1c. Class pairs 9 and 5 and 10 and 11 were not completely separated by the isodata algorithm and were resolved by deconvolution of their GABA signals.
Amacrine and ganglion cells are separate cell types in signature space: amacrine cells are notably deficient in glutamate, aspartate, and glutamine signals, and they show enhanced GABA signals, identical to GABA immunoreactive cells of the amacrine cell layer proper. The classification decision boundaries for amacrine–ganglion cell discrimination are shown on the Eγ and DQ bivariate signal channel summary plots (Fig. 4, left column) and the univariate glutamate signal histogram of the ganglion cell layer (Fig.6A). Although ganglion cells express various GABA signals that may overlap into the amacrine cell range, they are nevertheless completely separable from amacrine cells in EDQ (glutamate–aspartate–glutamine) space.
Fig. 6.
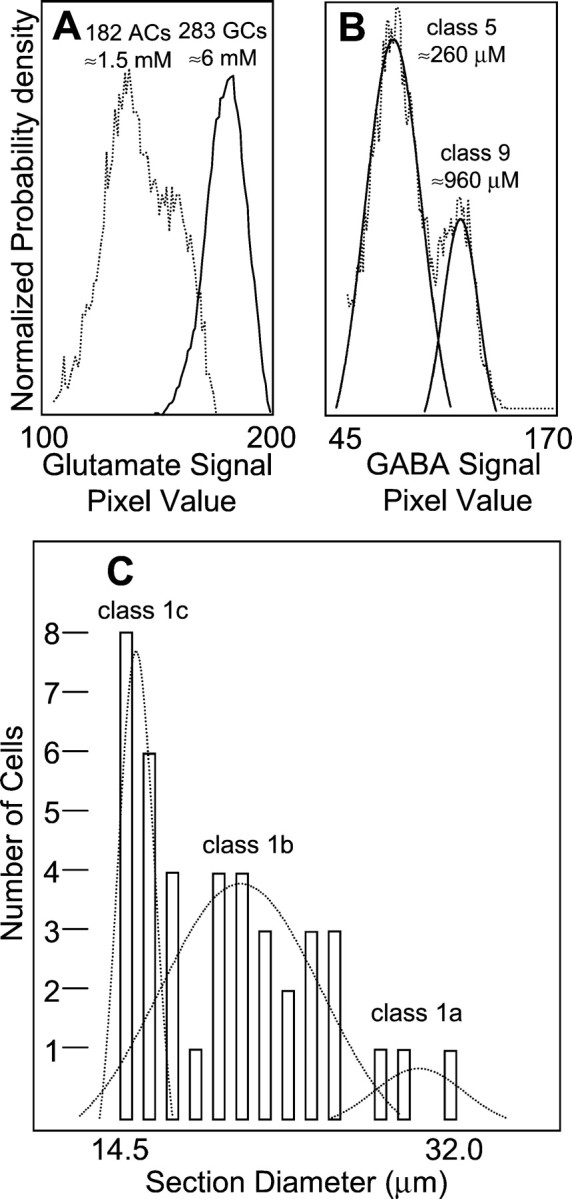
Univariate histograms and histogram deconvolution.A, Peak normalized probability density profiles for glutamate signals in displaced amacrine cells (dotted line) and ganglion cells (solid line).B, Deconvolution of the GABA signal histogram (dotted line) for superclass EDQγ into Gaussian components representing class 5 (EQγ) and class 9 EDQγ ganglion cells (solid lines). C, Deconvolution of the cell size histogram for superclass EDQ, class 1 ganglion cells into three components of identical shape and varying peak height (dotted lines).
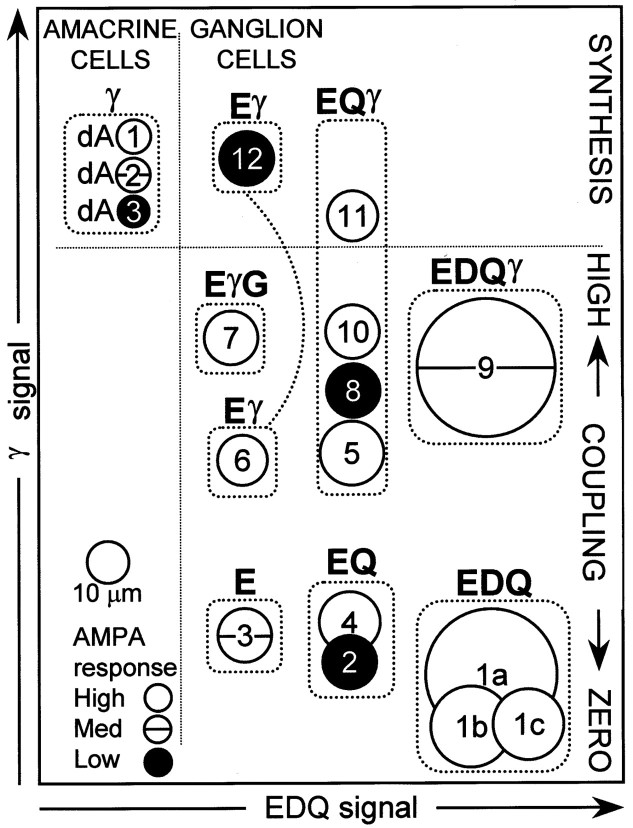
If molecular phenotype classes are natural classes, they must eventually map onto functional and morphological classes of cells. As an interim naming strategy, we chose to label classes in ordinal sequence by increasing GABA signals: i.e., class 1 cells contain no detectable GABA and class 12 cells have high GABA contents, with other classes ordered between. The relationships among superclasses and classes are summarized in Figure 7 and below.
Fig. 7.
A summary hierarchy of superclasses and classes encoded as a metabolic signature (EDQ signal), a coupling/synthesis signature (γ signal), an excitation signature (white = high, horizontal line= medium, black = low), and a size signature (disc diameter).
Superclass EDQ → class 1
This superclass contains a single molecular phenotype, class 1 ganglion cells, characterized by strong DQ signals, no GABA coupling, and three cell-size groups displaying nearly identical AMPA-driven AGB signals. Classes 1a, 1b, and 1c were extracted by deconvolution of cell-size histograms (discussed below in Structural correlates of classes).
Superclass EDQγ → class 9
This group resembled superclass EDQ but also showed significant GABA content and slightly weaker AMPA responses. Another distinct molecular class that did not separate fully with the isodata algorithm also initially contaminated it. Overlapping modes of GABA, aspartate, and glutamine signals suggested that one class of cells bridged N-space between superclasses EQγ and EDQγ, likely because of differing degree of coupling. This hypothesis was tested by deconvolving the univariate histogram of GABA signals for superclass EDQγ into Gaussian modes (Fig. 6B) and recoding each cell by modal membership. These operations yielded two unique size classes: class 5 (medium-sized cells), which properly belonged in superclass EQγ, and class 9 ganglion cells (the largest neurons in the retina) as the sole member of superclass EDQγ. Because classification and deconvolution were blind to structure, the discovery of different size classes is an independent validation of the separation method. Failure to completely separate using clustering algorithms is a common problem in pattern recognition because natural classes need not respect arbitrary separability criteria.
Superclass EQ → classes 2 and 4
With high Q but low D signals, this superclass encompassed classes of low (class 2) and high (class 4) AMPA responsivity.
Superclass EQγ → classes 5, 8, 10, and 11
Superclass EQγ represents the high end of a continuum of GABA content in ganglion cells. In particular, classes 11 and 12 represent high and low AMPA response classes of ganglion cell subsets with GABA contents that overlap those of bona fide GABAergic amacrine cells. However, their absolute glutamate, aspartate, and glutamine values place them outside the amacrine cell molecular phenotype. Classes 8 and 10 represent low and high AMPA response classes of ganglion cell subsets in which GABA levels do not reach those of amacrine cells but that have much higher GABA signals than any other superclass. Class 5 cells were originally mixed with class 9 cells in superclass EDQγ but were resolved by deconvolution.
Superclass E → class 3
Superclass E contains class 3 ganglion cells with negligible GABA signals, weak DQ signals, and medium AMPA responsivity.
Superclass Eγ → classes 6 and 12
Superclass Eγ contains cells with both low and high GABA contents, modest DQ signals, and it expresses either high (class 6) or low (class 12) AMPA responsivity.
Superclass EγG → class 7
This superclass contained only a single population of GABA+ and glycine+ ganglion cells with medium-strength AMPA responses.
Superclass γ → classes dA1, dA2, and dA3
This amacrine cell superclass contained three distinctive molecular phenotypes. Class dA1 (displaced amacrine cells) is composed entirely of the highly AMPA-responsive ON-center starburst amacrine cell cohort and represents 85% of superclass γ. The remainder is composed of a class (dA2) with phenotype and size that resemble starburst amacrine cells but is significantly less responsive to AMPA, and class dA3, a very small cell type displaying no AMPA response at all. It is possible that class dA2 cells are a frequent misplaced variety or true dA1 starburst amacrine cells that, for some reason, have weaker AMPA responses. Class dA3 cells are smaller than any other element and may not be amacrine cells at all, although they have high GABA content. They closely resemble very small cells detected in (and excluded from) the ganglion cell cohort by Oyster et al. (1981) and would fit in the microneuron category of Wong and Hughes (1987), being much larger than microglia.
Stability of classes
The superclasses can easily be found in every retina examined, but can all the classes be extracted? AGB mapping is a physiologicalin vitro experiment, and assessment of variability of the AMPA-induced AGB signal across cell types and individual retinas is pivotal in validating classifications. Each preparation must yield arrays of nearly flawless thin horizontal sections that can be precisely registered, are flat enough to provide some hundreds of ganglion cells in a high-resolution field, and behave similarly in response to physiological stimulation. Four individual retinas (cases 2366, 2578, 2780, and 2781) were prepared and treated identically, using 10 min of 25 μm AMPA activation in the presence 5 mm AGB. All were processed identically as serially probed 250 nm horizontal sections and analyzed as described in Materials and Methods. Each represented a locus 2–4 mm below the visual streak. Figure 8 displays both GABA and AMPA-activated AGB signals from an example of every cell class from each retina. GABA content and AMPA activation are fairly stable across retinas, although there are clear variations in the amount of basal GABA and AGB signals in the Müller cells. The ordinal ranking of AMPA responsivity in ganglion cells classes is consistent and suggests that AMPA responses are stable under our defined experimental conditions. Two of the retinas yielded 14 ganglion cell classes and two yielded 13, the latter lacking detectable examples of class 10. Examination of five other retinas prepared at different AMPA activation levels revealed one case in which class 7 was absent and two more in which class 10 cells could not be found. We have no obvious explanation for this except that the efficacy of heterologous coupling with amacrine cells may vary. Decreases in glycine coupling would collapse class 7 into class 6; a decrease in GABA coupling could disperse class 10 into classes 5, 4 or maybe even 3 if glutamine signal changed as well; increases in GABA coupling would seem less likely, because contamination of the small classes 11 and 12 would be noticeable. On the whole, separation into 14 classes in chemical space seems to most reasonably account for the ganglion cell population. In every preparation, some cells appear to be bona fide misplaced amacrine cells, and ∼2–5% of the small ganglion cells could not be classified, either because of section defects or disappearance from the end of a series, or simply because the signature of a single cell did not fit a single class well enough. Such cells could be damaged cells or true additional classes with sparse distributions.
Fig. 8.
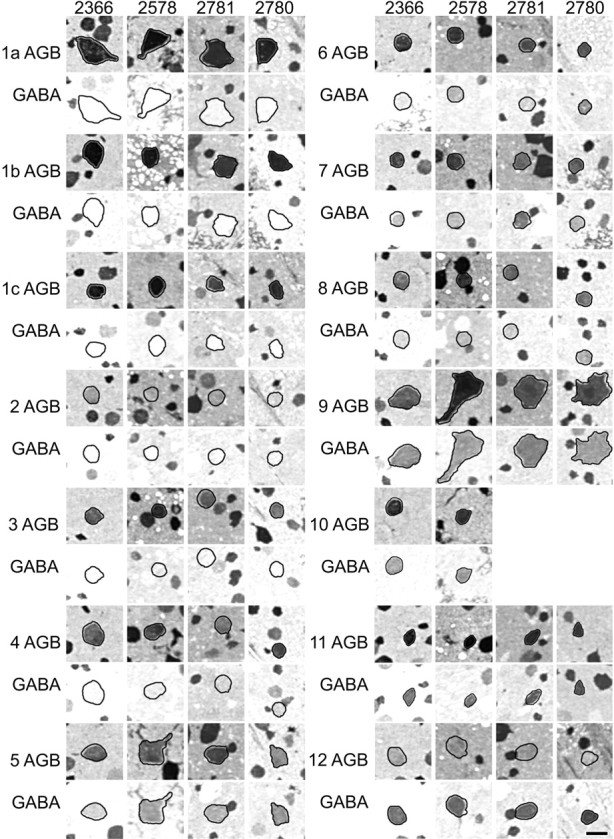
An array of GABA and AGB signals in four retinas. All panels were captured under identical conditions from horizontal 250 nm sections after registration and classification of cells. The outline of each cell was captured in a registered taurine channel (Fig. 2G) and used to track its location in both AGB and GABA channels. AGB signals were activated by exposure to 25 μm AMPA as described in Results. Class 10 cells were not resolved in cases2780 and 2781. Images are density scaled. Scale bar, 25 μm.
Structural correlates of classes
Do molecular phenotype classes represent stable natural classes? They do for some other cell types. Horizontal cells (Marc et al., 1995;Marc, 1999b) and starburst amacrine cells (Marc, 1999b) can be extracted via their distinctive signatures, and other cell types display strong superclasses; e.g., mammalian cone ON-center and OFF-center bipolar cells have distinctive signatures (Kalloniatis et al., 1996). This does not prove that all natural classes should have characteristic signatures, even when excitation signals are added to the data. Independent tests of identity are needed. The classification methods that we used were blind to size differences because they were pixel based and not object based. Thus size and patterning extracted from the classifications become test statistics.
The mean diameter of every cell in the glutamate channel of dataset 2366 was determined, and the values for all classes were compared pair wise [Student's t test (Table1); true diameter measures from single sections are better approximated by the mode or supremum, but for spheres randomly sampled, the true diameter ≈ measured mean × shrinkage correction × 4/π]. The mean suffices for tests of distribution differences. Most classes were significantly different in size from many other classes. For example, extraction of molecular phenotype classes 2 and 4 from superclass EQ permits the demonstration that they are also members of statistically different size groups. Superclass EDQγ was also initially contaminated, and deconvolution exposed molecular phenotype classes that are vastly different size groups. The only groups that did not completely segregate internally based on size were classes within superclasses EQγ and γ. Even so, superclass γ still separates from all other superclasses in terms of size, and superclass EQγ segregated partially. On balance, the extraction of 12 ganglion cell and 3 amacrine cell molecular phenotype classes reveals the existence of underlying structural correlates, which the pixel-based pattern recognition that was applied could not have detected, a priori.
Table 1.
Classification results for 465 cells from retina 2366
| Superclass | Class | Cell type | % Cells | % GCs or ACs | Soma size1-a (μm ± 1 SD) | Size different from class (p < 0.05 ttest) | CR1-b |
|---|---|---|---|---|---|---|---|
| EDQ | 1a | GC | 1.7 | 2.8 | 31.7 ± 4.7 | All but 9 | 2.9 |
| EDQ | 1b | GC | 3.7 | 6.0 | 20.2 ± 2.2 | All | 2.4 |
| EDQ | 1c | GC | 3.4 | 5.7 | 17.4 ± 1.0 | All | 1.8 |
| EQ | 2 | GC | 2.6 | 4.2 | 13.4 ± 1.5 | 1, 4, 5, 9, dA | 2.6 |
| EQ | 4 | GC | 5.0 | 7.7 | 15.2 ± 2.4 | All but 5 | 2.7 |
| E | 3 | GC | 7.5 | 12.2 | 13.2 ± 1.4 | 1, 4, 5, 6, 9, 11, dA | 3.8 |
| Eγ | 6 | γ-Coupled GC | 9.2 | 15.0 | 12.4 ± 1.3 | 1, 3, 4, 5, 8, 9, 12, dA | 3.1 |
| EγG | 7 | γG-coupled GC | 3.4 | 5.6 | 13.0 ± 1.8 | 1, 4, 5, 9, dA | 2.1 |
| EDQγ | 9 | γ-Coupled GC | 0.9 | 1.4 | 34.0 ± 2.1 | All but 1a | 3.0 |
| EQγ | 5 | γ-Coupled GC | 4.3 | 7.0 | 16.2 ± 2.5 | All but 4 | 2.1 |
| EQγ | 8 | γ-Coupled GC | 7.5 | 12.2 | 13.4 ± 1.1 | 1, 4, 5, 6, 9, 11, dA | 2.1 |
| EQγ | 10 | γ-Coupled GC | 5.2 | 8.4 | 13.0 ± 1.2 | 1, 4, 5, 9, 11, dA | 1.9 |
| EQγ | 11 | γ+ GC | 4.1 | 6.6 | 12.3 ± 1.1 | 1, 3, 4, 5, 8, 9, 10, dA | 2.0 |
| EQγ | 12 | γ+ GC | 3.2 | 5.2 | 13.9 ± 1.9 | 1, 4, 5, 6, 9, 11, dA | 2.9 |
| γ | dA1 | Starburst ACs | 32.7 | 85.4 | 8.0 ± 1.0 | All but dA | 2.3 |
| γ | dA2 | γ+ AC | 3.9 | 10.1 | 8.3 ± 1.1 | All but dA | 1.5 |
| γ | dA3 | γ+ AC | 1.7 | 4.5 | 6.9 ± 1.0 | All but dA | 3.5 |
True soma size ≈ measured mean × shrinkage correction × 4/τ.
Significant CR values (classes 3, 6, and 14).
Class 1 (superclass EDQ) separated from all other classes based on size, except for class 9. There was, however, cause to suspect that molecular phenotype class 1 was not an ultimate, natural class: (1) visual examination showed it to be more heterogeneous than any other molecular phenotype class; (2) the class diameter variance was greater than any other class; and (3) the diameter histogram was clearly multimodal. We thus attempted to decompose the histogram into the most probable component Gaussians with variances forced to match the mean variances of all other single classes (Fig. 6C). This assumption forces the shape of all Gaussians to be as broad as bona fide natural classes. Only the sum of three such Gaussians—no more, no less—fits the envelope of the histogram with a correlation coefficient of >0.8. Fits of 1, 2, or 4 or more Gaussians led to correlation coefficients of <0.4. Furthermore, model distributions with more adjustable parameters (e.g., Weibull functions, etc.) did no better. Arguably, three natural populations comprise molecular phenotype class 1, and they were extracted simply by segmenting the population at the two minima in the histogram. This resulted in three classes (1a, 1b, and 1c) that still differed in size from all other cell types with the exception that classes 1a and 9 were not different, although they are fully separable in molecular phenotype space. Taken together, 80% of the 136 possible size comparisons among the classes are significantly different at p ≤ 0.05 (Table 1). This would not be possible had not the underlying size classes and molecular phenotypes been the same natural classes.
This array of size groupings allows a reconstitution of the theoretical ganglion cell size histogram with higher resolution of its underlying structure than previously possible. The cell-size Gaussian for each class was calculated from its mean diameter and SD and weighted by the frequency of the class (Fig. 9). Viewed on both logarithmic and linear probability density ordinates, the envelope of all the individual classes resembles rabbit ganglion cell layer density histograms acquired from whole mounts when the displaced amacrine cells are included (Vaney, 1980; Tancred, 1981; Vaney et al., 1981; Rowe and Dreher, 1982). Several points emerge from inspection of the histograms. First, amacrine and ganglion cells are separable by size. Second, six cell classes dominate the shape of the ganglion cell size histogram: 1a, 1b, 1c, 3, 6, and 8. All other ganglion cell classes are submerged beneath the envelope and are undetectable by simple population counting and sizing. Third, no deconvolution of the total cell size histogram of the ganglion cell layer can extract the correct number of types, regardless of the underlying fundamental shapes chosen.
Fig. 9.
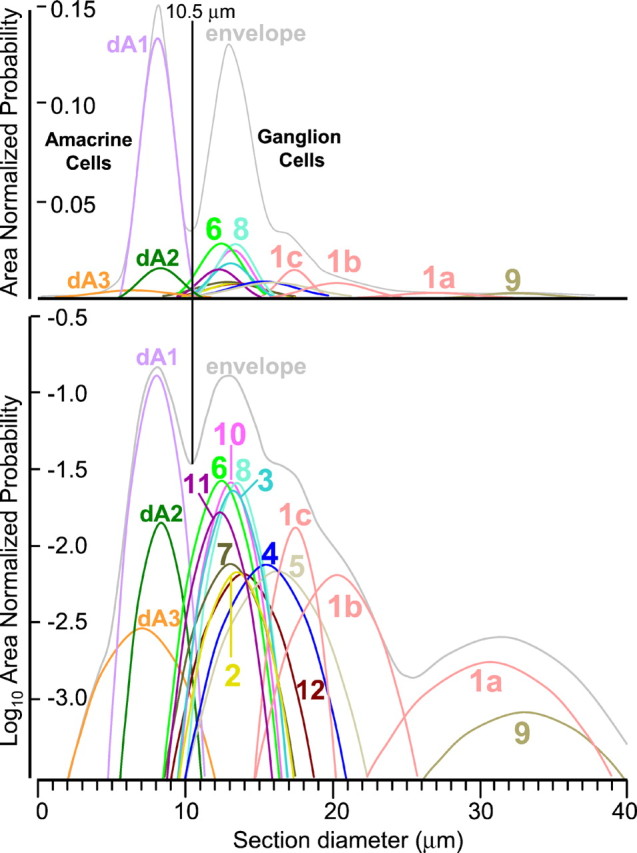
Reconstruction of the ganglion cell layer size histogram. The mean, population density, and variance of each class of ganglion cell in case 2366 was used to control the position, height, and width of a Gaussian distribution for that class. All classes were summed to create the envelope distribution and plotted on linear (A) or logarithmic (B) ordinates. The linear envelope strongly resembles typical soma size or soma area histograms plotted on linear ordinates when starburst amacrine cells are included. The logarithmic envelope and Gaussians demonstrate that the smooth peak of the envelope belies the complex mixture of cell types comprising the ganglion cell layer. At this eccentricity (∼2 mm below the streak), all cells with mean diameters <8 μm are amacrine cells on the basis of signatures.
Molecular phenotyping also uncovers hidden patterning. Quantitative signatures must be extracted from serial thin sections of nearly perfect horizontal orientations, strongly limiting cell numbers. Given that there are so many different classes, any one class will occur with low frequency, and the number of sampled cells in one comprehensive data set is usually lower than practical for robust tests of patterning. We cannot combine sets from other loci or retinas, and each horizontal section array must stand alone. Even so, three classes occurred with high enough frequencies in a single patch to test patterning by measuring the CR (also known as the regularity index) and testing significance (Cook, 1996). All three, (classes 3, 6, and dA1) were patterned significantly at p < 0.01 (Fig.10, Table 1). Classes were extracted by signatures alone, so this demonstrates that molecular phenotyping uncovers patterned classes, which would again be implausible were not the underlying elements natural classes. We expected to see better patterning among the remaining classes, but this is partly a consequence of the limited numbers of cells. If we presumed that the patterning precision of each class was replicated over an area subtending 100 cells of that class, then 10 of 18 classes would have been statistically patterned. In addition, some patterns might represent mixed “subclasses.” For example, we know that ON-OFF direction-selective (DS) ganglion cells exist as four vector subclasses (Oyster and Barlow, 1967) but that they are not morphologically distinguishable (Amthor et al., 1989b). Any class that contains the ON-OFF DS cells should be represented by a mixture of patterns (a “mixed” pattern). Furthermore, it is not clear that somatic positions of all ganglion cell types must be well patterned, because the territory of dendritic coverage is the critical tiling unit (Wässle et al., 1981), and cells with wide, sparse, asymmetrical dendritic arbors, such as many type “W” ganglion cells, are likely to be tiled with rather asymmetrical Dirichlet (Voronoi) domains. In addition, some cells in a single functional class, such as ON DS ganglion cells (ignoring vector subtypes), can be immediate neighbors (He and Masland, 1998), which will strongly break any patterning statistics based on somatic spacings alone. This will again yield mixed patterns.
Fig. 10.
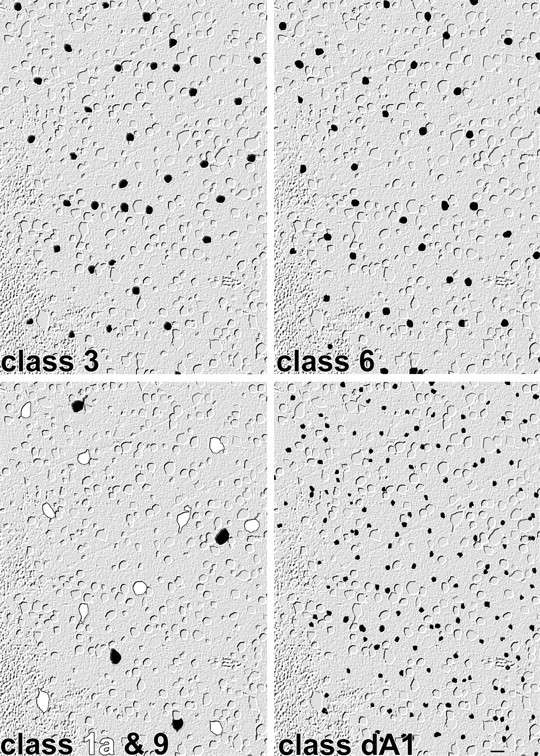
Superposition of class 3, class 6, class 1a (white), class 9, and class dA1 cells on the array of all cells in the ganglion cell layer. Classes 3, 6, and dA1 are significantly patterned (p < 0.01). Classes 1a and 9 cells are too rare to test in this data set. Indexed cell class image is superimposed on a digital “phase” image of the glutamate channel. Scale bar, 25 μm.
Finally, the fractions of some cell classes exposed by classification roughly correspond to previously determined groups. Classes 1a and 9 clearly fit within the size group for α ganglion cells (Peichl et al., 1987) alone and are also the rarest of types, comprising but 1–2% of all cells in the ganglion cell layer in all preparations. Conversely, the well known starburst amacrine cells were the dominant cell class comprising 20–35% of the ganglion cell layer depending on eccentricity. As pointed out by Hughes (1985), the ganglion cell size spectrum in rabbit is quite unimodal, and the remaining populations are difficult to correlate with known types.
Attributes of different ganglion cell classes: coupling and excitatory drive
Classification allows exploration of important structural and functional properties of these cell types. Roughly three domains of GABA signals can be assigned to ganglion cells (Fig. 7). Classes 1a, 1b, 1c, 2, 3, and 4 have little or no detectable GABA signals. This is consistent with arguments that certain mammalian ganglion cells such as a subset of α cells are not tracer coupled to amacrine cells (Xin and Bloomfield, 1997). A spectrum of classes including high population density class 8 cells, large class 9 cells, medium class 5 cells, and small class 10 cells show significant GABA signals, suggesting varying degrees of steady-state leakage of GABA from coupled amacrine cells. Class 7 in particular stands out by having measurable glycine and GABA signals. Finally, ganglion cell classes 11 and 12 contain GABA signals indistinguishable from those of GABAergic amacrine cells and might synthesize GABA directly. As previously argued, however, class 11 and 12 cells possess a glutamate phenotype that places them with ganglion cells and not amacrine cells. Furthermore, GABA immunoreactive axons are abundant in the rabbit optic nerve but always map to glutamate immunoreactive axons (Fig.1B).
The most important feature of these classes, however, is likely to be the fact that they vary greatly in their responses to AMPA activation (Figs. 2A, 8), and the variation has no correlation with size or GABA content. Some of the smallest (class dA1) and largest (classes 1a, 1b, and 1c) cells of the ganglion cell layer show extremely high AMPA responsivity, so cell volume cannot be the mechanism underlying high signals. Similarly, one of the smallest (class 8) and one of the largest (class 12) ganglion cells have weak AMPA responses. We presume that class 9 ganglion cells correspond to OFF-α cells because they could be no other than an α type based on size, and their basal dendrites can be traced into the distal half of the inner plexiform layer (Fig. 11). However, it is clear that in every preparation, the AMPA-driven signals of class 9 ganglion cells are slightly weaker than those of class 1a ganglion cells.
Fig. 11.
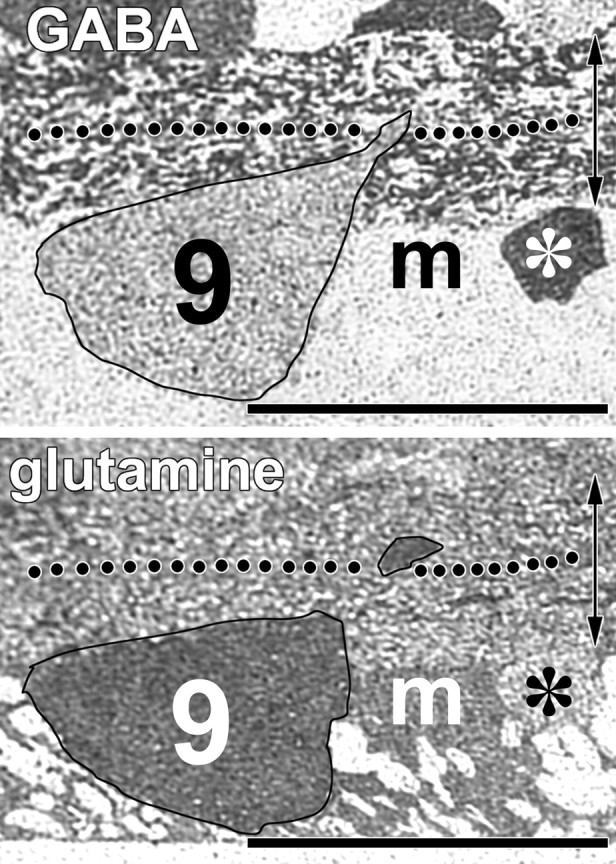
Identification of class 9 ganglion cells as probable OFF α ganglion cells in semi-serial 250 nm vertical sections. The top panel shows the characteristic moderate GABA signal of a class 9 ganglion cell embedded in the Müller cell foot pieces (m), along with a strongly immunoreactive nearby starburst amacrine cell (asterisk). A large-caliber proximal dendrite clearly passes the midway point (dots) of the inner plexiform layer (double-headed arrow). Bottom panel, The glutamine signal validates the identity of the cell as a member of superclass EDQγ, and its dimensions require that it be a member of the α size group. The dendrite continues into the distal inner plexiform layer. Scale bars, 25 μm.
DISCUSSION
Metabolic signatures
The molecular phenotypes of ganglion cells distinguish them from all other retinal neurons (Sherry and Ulshafer, 1992;Kalloniatis et al., 1994, 1996; Marc et al., 1995, 1998). Classification shows that EDQ and EDQγ cells are large ganglion cells, although no biological reason for this specialization is obvious. Of the remaining cells, EQ+EQγ superclasses contain larger ganglion cells than the E+Eγ superclasses. We have documented the association of the EDQ phenotype with large ganglion cells in >30 individual rabbit retinas; it appears to be a reliable index of identity. Amacrine cells possess distinctively weaker EDQ signatures than the true ganglion cells, and this, rather than their GABA signals alone, enables separation of the γ superclass. We hypothesize that signatures are tools for molecular profiling of cell types in all tissues, in all states.
Coupling signatures
One-half of all ganglion cells in the rabbit retina contain nontrivial GABA signals. Where does this GABA come from? A major source must be heterologous coupling between ganglion cells and conventional GABAergic amacrine cells. The evidence for amacrine–ganglion cell coupling in the mammalian retina is unequivocal (Rodieck and Haun, 1991; Vaney, 1991; Dacey and Brace, 1992; Penn et al., 1994; Jacoby et al., 1996; Stafford and Dacey, 1997; Xin and Bloomfield, 1997). Most amacrine cells are GABA immunoreactive, and GABA-immunoreactive dendrites comprise ∼80% of the mass of the inner plexiform layer (Marc, 1992, 1999b; Marc et al., 1995, 1998). GABA is a 97 Da molecule and would clearly leak from GABA in amacrine cells (containing ∼10 mm GABA) through gap junctions at nearly diffusion-limited rates into ganglion cells. A similar phenomenon would explain glycine-rich class 7 ganglion cells. Ganglion cells may possess other sources of GABA signals, such as transport or synthesis, but the existence of a coupling leak for many cell classes is certain, and we are satisfied that coupling patterns must shape molecular phenotypes of ganglion cells. Why do not all ganglion cells coupled to amacrine cells eventually equilibrate to the same level as their source cells? A GABA sink must exist in ganglion cells. All eukaryotic cells are probably capable of metabolizing GABA (Michal, 1999), so it would not be surprising if a GABA leak from a “10 mm” amacrine cell yielded a stable steady-state 0.5 mm GABA level in a ganglion cell that slowly metabolizes it. Coupling efficiency between ganglion cells and amacrine cells might also vary. OFF-center α ganglion cells display heterologous coupling to amacrine cells (Mills and Xia, 2000) but can appear uncoupled, likely because of changes in adaptation state (Hu and Bloomfield, 2000,2001). We hypothesize that coupling signatures can be used to uncover patterns of dendritic cofasciculation and costratification and to track adaptation events.
Excitation signatures
As reported previously, the responses of retinal neurons to glutamate agonists are not identical (Marc, 1999a,b), and ganglion cells show stereotyped differences in AGB permeation after activation with kainate, AMPA, NMDA, and glutamate itself. These differences are not explained by cell size but must arise from cell-specific variations in receptor affinity, macroscopic open-time kinetics, and/or number. Rabbit retinal ganglion cell signal integration is dominated by AMPA and NMDA receptors, with no significant kainate receptor involvement (Marc, 1999c). Differences in AMPA-activated responses should reflect the behaviors of AMPA receptors under native glutamate stimulation, and we predict that glutamate drive from a given bipolar cell targeting different ganglion cell classes may evoke different effects based on AMPA receptor properties. Cells with high and low AMPA responsivity should have fast and slow integration times, respectively. We hypothesize that AMPA receptor type, and the absence or presence of GluR2-edited subunits in particular, may control the brisk–sluggish spectrum of ganglion cell responses.
Correlation with physiological classes
The apparent numbers of molecular phenotype classes and physiological classes in rabbit are similar (Oyster et al., 1971;Amthor et al., 1989a,b,). Given data on coupling patterns (Xin and Bloomfield, 1997; Mills and Xia, 2000; Hu and Bloomfield, 2001), the tabulation of morphologies of physiological classes (Amthor et al., 1989a,b), encounter frequencies (Oyster et al., 1971), and soma size histograms (Provis, 1979; Vaney, 1980; Oyster et al., 1981; Vaney et al., 1981), we can construct provisional matches between physiological and molecular phenotype classes (Table2). We have made the matches as specific as possible and offer them as a target for argument and experiment.
Table 2.
Possible correlations between physiological and molecular phenotype classes
| Physiological class (1, 2) | Size (1, 2) | AC coupling | Possible class, patterning, and coupling features | AMPA response |
|---|---|---|---|---|
| Concentric brisk linear | ||||
| ON sustained (β) | S | ? | 3 well-patterned uncoupled | Medium |
| OFF sustained (β) | S | ? | 6 well-patterned γ coupled | High |
| ON transient | M | ? | 1b uncoupled | High |
| OFF transient | M | ? | 5 γ coupled | High |
| Concentric brisk nonlinear | ||||
| Transient ON (α) | L | No (3) | 1a uncoupled | High |
| Transient OFF (α) | L | Yes (3, 4) | 9 γ coupled | Medium |
| Concentric sluggish linear | ||||
| ON sustained | ? | ? | 2 sparse uncoupled | Low |
| OFF sustained | S | ? | 8 mixed pattern γ coupled | Low |
| Concentric sluggish nonlinear | ||||
| ON transient | M | ? | 12 mixed pattern γ coupled/γ+ | Low |
| OFF transient | M | ? | 12 mixed pattern γ coupled/γ+ | Low |
| Complex | ||||
| Local edge detector | S | ? | 7 γG coupled | High |
| Uniformity detector | M-L | ? | 5 γ coupled or 1b uncoupled | High |
| Orientation-selective (two subclasses) | S | ? | 11 mixed pattern γ coupled/γ+ | |
| ON DS (three vector subclasses) | M-L | No (6) | 4 mixed pattern uncoupled | High |
| ON-OFF DS (four vector subclasses) | S | No (5, 6) | 1c mixed pattern uncoupled | High |
| Yes (5, 6) | 10 mixed pattern γ coupled | High |
Total: 15 classes (21 including vector and orientation types). References: (1) Amthor et al., 1989a; (2) Amthor et al., 1989b; (3) Hu and Bloomfield, 2001; (4) Mills and Xia, 2000; (5) Vaney 1994a, b; (6)Xin and Bloomfield, 1997.
Concentric brisk nonlinear transient cells
Classes 1a and 9 are the α ganglion cells of the rabbit retina (Peichl et al., 1987). Considering dye-coupling patterns (Mills and Xia, 2000; Hu and Bloomfield, 2001) and our tracing of dendrites (Fig.11), class 1a cells are ON-center and class 9 cells are OFF-center, corresponding directly to the concentric brisk nonlinear transient cells of Amthor et al. (1989a) and the Y cells of carnivores.
Concentric brisk linear sustained cells
A presumed X-like, β-like class is one of the complex problems in rabbit retina. Rabbit β homologs have small somas (Amthor et al., 1989a), excluding candidate classes 1b and 1c. Brisk sustained cells are a common cell type (Levick, 1967; Oyster et al., 1971;Amthor et al., 1989a) and should have good AMPA responses. The well patterned, frequent small cells of classes 3 and 6 seem the best candidates. There are conceptual problems: class 6 is γ coupled, whereas ferret β ganglion cells (Penn et al., 1994) and primate midget ganglion cells (Dacey and Brace, 1992) show no amacrine cell coupling. At present, we ignore this conflict. By analogy with coupling patterns of ON and OFF α cells, we propose class 3 as the ON-center and class 6 as the OFF-center classes.
Concentric brisk linear transient cells
Amthor et al. (1989a) distinguished these medium-sized cells from α cells with caveats. Equating briskness and high AMPA responsivity, the best matches are classes 1b (ON) and 5 (OFF).
Concentric sluggish cells
Equating sluggishness with low AMPA responsivity and matching sizes and encounter rates, the small sustained sluggish cells could be members of class 2 or 8. The patterning of class 8 suggests that the population could be mixed and may contain both ON and OFF varieties. Transient sluggish cells are of medium size, matching best with class 12 ganglion cells.
ON-OFF DS cells
The ON-OFF DS cell class harbors four vector types (Oyster and Barlow, 1967) and lies in the small-to-medium soma size spectrum (Amthor et al., 1989b). Because vector type might display a well patterned distribution (Vaney, 1994b) and a single class contains two or more vector types, that class might show mixed distribution patterns. Some ON-OFF DS cells are coupled to amacrine cells and others are not (Vaney, 1994a,b; Xin and Bloomfield, 1997). We match small–medium class 10 and medium-sized 1b cells as the coupled and uncoupled sets of ON-OFF DS cells.
ON DS cells
The vector types of ON DS cells project to the medial terminal nucleus of the rabbit accessory optic system and are medium–large ganglion cells (Oyster et al., 1981). Xin and Bloomfield (1997) suggest that this cell is not coupled. Class 4 seems the best fit for ON DS ganglion cells.
Orientation-selective cells
These cells, with two orientation-preference subclasses, are among the smallest cells in soma size (Amthor et al., 1989b). Of all the small cells, they appear to have the most elongate, polygonal somas, corresponding well to the mixed-pattern, γ-coupled class 11 ganglion cells.
Local edge detector cells
With complex physiologies and small somas, local edge detector (LED) cells also have small, compact dendritic arbors. Xin and Bloomfield (1997) illustrated small “narrow field” cells much like LED cells injected by Amthor et al. (1989b), and they were clearly coupled to amacrine cells. The only remaining group that seems to fit is class 7, but Xin and Bloomfield (1997) speculated that the cell was coupled only to a single type of amacrine cell, and class 7 is apparently both γ and G coupled. Furthermore, if all ganglion cells have coverage factors of at least 1, the class 7 distribution (Fig.2D) does not suffice. LED cells are reported to comprise nearly 19% of the cells in the streak, although this value appears to drop to ∼2% in the periphery (Oyster et al., 1971). LED cells may be embedded in another class. The match of class 7 cells is quite uncertain.
Uniformity detectors
A rare, medium-to-large cell, inhibited by any stimulus, uniformity detectors do not nicely fit anywhere. The size match corresponds to classes 5 and 1b.
Other cells
There is room for more functional–morphological classes as long as they each comprise <1 or 2% of the cohort.
Summary
Rabbit retinal ganglion cells contain mixtures of free small molecules that, when combined with additional excitation markers, allow specification of a molecular phenotype for each cell and a comprehensive classification of the entire layer. The classification appears robust and serves, we believe, as (1) a step toward completeness in describing this population of visual neurons and (2) a bridge for unification of physiological and morphological classifications.
Footnotes
This work was supported by National Institutes of Health Grant EY02576 (R.E.M.) and a Jules and Doris Stein Research to Prevent Blindness Professorship (R.E.M.). R.E.M. is a principal of Signature Immunologics Inc.
Correspondence should be addressed to Robert E. Marc, John Moran Eye Center, University of Utah, 75 North Medical Drive, Salt Lake City, UT 84132. E-mail: robert.marc@hsc.utah.edu.
REFERENCES
- 1.Ames A, Nesbett FB., III In vitro retina as an experimental model of the central nervous system. J Neurochem. 1981;37:867–877. doi: 10.1111/j.1471-4159.1981.tb04473.x. [DOI] [PubMed] [Google Scholar]
- 2.Amthor FR, Takahashi ES, Oyster CW. Morphologies of rabbit retinal ganglion cells with concentric receptive fields. J Comp Neurol. 1989a;280:72–96. doi: 10.1002/cne.902800107. [DOI] [PubMed] [Google Scholar]
- 3.Amthor FR, Takahashi ES, Oyster CW. Morphologies of rabbit retinal ganglion cells with complex receptive fields. J Comp Neurol. 1989b;280:97–121. doi: 10.1002/cne.902800108. [DOI] [PubMed] [Google Scholar]
- 4.Ball GB, Hall DJ. Clustering technique for summarizing multivariate data. Behav Sci. 1967;12:153–155. doi: 10.1002/bs.3830120210. [DOI] [PubMed] [Google Scholar]
- 5.Brandon C, Criswell MH. Displaced starburst amacrine cells of the rabbit retina contain the 67-kDa isoform, but not the 65-kDa isoform, of glutamate decarboxylase. Vis Neurosci. 1995;12:1053–1061. doi: 10.1017/s0952523800006714. [DOI] [PubMed] [Google Scholar]
- 6.Cook JE. Spatial properties of retinal mosaics: an empirical evaluation of some existing measures. Vis Neurosci. 1996;13:15–30. doi: 10.1017/s0952523800007094. [DOI] [PubMed] [Google Scholar]
- 7.Cook JE. Getting to grips with neuronal diversity: what is a neuronal type? In: Chalupa L, Finlay B, editors. Development and organization of the retina. Plenum; New York: 1998. pp. 91–120. [Google Scholar]
- 8.Dacey DM, Brace S. A coupled network for parasol but not midget ganglion cells in the primate retina. Vis Neurosci. 1992;9:279–290. doi: 10.1017/s0952523800010695. [DOI] [PubMed] [Google Scholar]
- 9.Famiglietti EV. New metrics for analysis of dendritic branching patterns demonstrating similarities and differences in ON and ON-OFF directionally selective retinal ganglion cells. J Comp Neurol. 1992;324:295–321. doi: 10.1002/cne.903240302. [DOI] [PubMed] [Google Scholar]
- 10.Farmer SG, Rodieck RW. Ganglion cells of the cat accessory optic system: morphology and retinal topography. J Comp Neurol. 1982;205:190–198. doi: 10.1002/cne.902050210. [DOI] [PubMed] [Google Scholar]
- 11.Fukuda Y, Stone J. Retinal distribution and central projections of Y-, X- and W-cells of the cat's retina. J Neurophysiol. 1974;37:749–772. doi: 10.1152/jn.1974.37.4.749. [DOI] [PubMed] [Google Scholar]
- 12.He S, Masland RH. ON direction-selective ganglion cells in the rabbit retina: dendritic morphology and pattern of fasciculation. Vis Neurosci. 1998;15:369–375. doi: 10.1017/s095252389815215x. [DOI] [PubMed] [Google Scholar]
- 13.Holden AL. Classifying and comparing retinal ganglion cells. Brain Behav Evol. 1981;18:188–193. doi: 10.1159/000121786. [DOI] [PubMed] [Google Scholar]
- 14.Hu EH, Bloomfield SA. Modulation of the tracer coupling pattern of α-ganglion cells in the rabbit retina. Invest Ophthalmol Vis Sci. 2000;41:S936. [Google Scholar]
- 15.Hu EH, Bloomfield SA. Comparison of the tracer coupling pattern and correlated activity of α-ganglion cells in the rabbit retina. Invest Ophthalmol Vis Sci. 2001;42:S677. [Google Scholar]
- 16.Hughes A. A rose by any other name: on naming of neurones by Rowe and Stone. Brain Behav Evol. 1979;16:52–64. doi: 10.1159/000121823. [DOI] [PubMed] [Google Scholar]
- 17.Hughes A. New perspectives in retinal organization. Prog Retinal Res. 1985;4:243–313. [Google Scholar]
- 18.Inselberg A, Dimsdale B. Parallel coordinates: a tool for visualizing multi-dimensional geometry. Proc 1st IEEE Conf Visualization. 1990;1:361–375. [Google Scholar]
- 19.Jacoby R, Stafford D, Kouyama N, Marshak D. Synaptic inputs to ON parasol ganglion cells in the primate retina. J Neurosci. 1996;16:8041–8056. doi: 10.1523/JNEUROSCI.16-24-08041.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson J, Chen TK, Rickman DW, Evans C, Brecha N. Multiple γ-aminobutyric acid plasma membrane transporters (GAT-1, GAT-2, GAT-3) in the rat retina. J Comp Neurol. 1996;375:212–224. doi: 10.1002/(SICI)1096-9861(19961111)375:2<212::AID-CNE3>3.0.CO;2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalloniatis M, Fletcher E. Immunocytochemical localization of amino acid neurotransmitters in the chicken retina. J Comp Neurol. 1993;336:174–193. doi: 10.1002/cne.903360203. [DOI] [PubMed] [Google Scholar]
- 22.Kalloniatis M, Tomisich G, Marc RE. Neurochemical signatures revealed by glutamine labeling in the chicken retina. Vis Neurosci. 1994;11:793–804. doi: 10.1017/s0952523800003096. [DOI] [PubMed] [Google Scholar]
- 23.Kalloniatis M, Marc RE, Murry RF. Amino acid signatures in the primate retina. J Neurosci. 1996;16:6807–6829. doi: 10.1523/JNEUROSCI.16-21-06807.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leventhal AG, Rodieck RW, Dreher B. Central projections of cat retinal ganglion cells. J Neurophysiol. 1985;237:216–226. doi: 10.1002/cne.902370206. [DOI] [PubMed] [Google Scholar]
- 25.Levick WR. Receptive fields and trigger features of ganglion cells in the visual streak of the rabbit's retina. J Physiol (Lond) 1967;188:285–307. doi: 10.1113/jphysiol.1967.sp008140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marc RE. The structure of GABAergic circuits in ectotherm retinas. In: Mize R, Marc RE, Sillito A, editors. GABA in the retina and central visual system. Elsevier; Amsterdam: 1992. pp. 61–92. [Google Scholar]
- 27.Marc RE. Mapping glutamatergic drive in the vertebrate retina with a channel-permeant organic cation. J Comp Neurol. 1999a;407:47–64. doi: 10.1002/(sici)1096-9861(19990428)407:1<47::aid-cne4>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 28.Marc RE. Kainate activation of horizontal, bipolar, amacrine and ganglion cells in the rabbit retina. J Comp Neurol. 1999b;407:65–76. [PubMed] [Google Scholar]
- 29.Marc RE. Subtypes of OFF-center bipolar cells possess different ionotropic glutamate receptor channel properties. Invest Ophthalmol Vis Sci. 1999c;40:S790. [Google Scholar]
- 30.Marc RE, Liu WLS, Kalloniatis M, Raiguel S, Van Haesendonck E. Patterns of glutamate immunoreactivity in the goldfish retina. J Neurosci. 1990;10:4006–4034. doi: 10.1523/JNEUROSCI.10-12-04006.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marc RE, Basinger SF, Murry RF. Pattern recognition of amino acid signatures in retinal neurons. J Neurosci. 1995;15:5106–5129. doi: 10.1523/JNEUROSCI.15-07-05106.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marc RE, Murry R, Fisher SK, Linberg K, Lewis G, Kalloniatis M. Amino acid signatures in the normal cat retina. Invest Ophthalmol Vis Sci. 1998;39:1685–1693. [PubMed] [Google Scholar]
- 33.Masland RH, Raviola E. Confronting complexity: strategies for understanding the microcircuitry of the retina. Annu Rev Neurosci. 2000;23:249–284. doi: 10.1146/annurev.neuro.23.1.249. [DOI] [PubMed] [Google Scholar]
- 34.Michal G. Amino acids and derivatives. In: Michal G, editor. Biochemical pathways. Wiley; New York: 1999. pp. 46–67. [Google Scholar]
- 35.Mills SL, Xia XB. The kinetics of tracer coupling between OFF α ganglion cells in the rabbit retina. Invest Ophthalmol Vis Sci. 2000;41:s936. [Google Scholar]
- 36.Narendra PM, Goldberg M. A non-parametric clustering scheme for Landsat. Pattern Recogn. 1977;9:207–215. [Google Scholar]
- 37.Oyster CW, Barlow HB. Direction-selective units in rabbit retina: distribution of preferred directions. Science. 1967;155:841–842. doi: 10.1126/science.155.3764.841. [DOI] [PubMed] [Google Scholar]
- 38.Oyster CW, Takahashi E, Levick WR. Information processing in the rabbit visual system. Doc Ophthalmol. 1971;30:162–204. doi: 10.1007/BF00142519. [DOI] [PubMed] [Google Scholar]
- 39.Oyster CW, Takahashi ES, Hurst DC. Density, soma size, and regional distribution of rabbit retinal ganglion cells. J Neurosci. 1981;1:1331–1346. doi: 10.1523/JNEUROSCI.01-12-01331.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peichl L, Buhl EH, Boycott BB. Alpha ganglion cells in the rabbit retina. J Comp Neurol. 1987;263:25–41. doi: 10.1002/cne.902630103. [DOI] [PubMed] [Google Scholar]
- 41.Penn AA, Wong ROL, Shatz CJ. Neuronal coupling in the developing mammalian retina. J Neurosci. 1994;14:3805–3815. doi: 10.1523/JNEUROSCI.14-06-03805.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Provis JM. The distribution and size of ganglion cells in the retina of the pigmented rabbit: a quantitative analysis. J Comp Neurol. 1979;185:121–137. doi: 10.1002/cne.901850108. [DOI] [PubMed] [Google Scholar]
- 43.Pu M, Berson DM, Pan T. Structure and function of retinal ganglion cells innervating the cat's geniculate wing: an in vitro study. J Neurosci. 1994;14:4338–4358. doi: 10.1523/JNEUROSCI.14-07-04338.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodieck RW. The first steps in seeing. Sinauer; Sunderland, MA: 1998. [Google Scholar]
- 45.Rodieck RW, Brening RK. On classifying retinal ganglion cells by numerical methods. Brain Behav Evol. 1982;21:42–46. doi: 10.1159/000121614. [DOI] [PubMed] [Google Scholar]
- 46.Rodieck RW, Haun TJ. Parasol retinal ganglion cells in macaques connect intracellularly to other parasol ganglion cells, and to amacrine cells found in the ganglion cell layer. Soc Neurosci Abstr. 1991;17:1375. [Google Scholar]
- 47.Rodieck RW, Watanabe M. Survey of the morphology of macaque retinal ganglion cells that project to the pretectum, superior colliculus, and parvicellular laminae of the lateral geniculate nucleus. J Comp Neurol. 1993;338:289–303. doi: 10.1002/cne.903380211. [DOI] [PubMed] [Google Scholar]
- 48.Rowe MH, Dreher B. Retinal W-cell projections to the medial interlaminar nucleus in the cat: implications for ganglion cell classification. J Comp Neurol. 1982;204:117. doi: 10.1002/cne.902040203. [DOI] [PubMed] [Google Scholar]
- 49.Rowe MH, Stone J. Naming of neurones: classification and naming of cat retinal ganglion cells. Brain Behav Evol. 1977;14:185–216. doi: 10.1159/000125660. [DOI] [PubMed] [Google Scholar]
- 50.Sherry DM, Ulshafer RJ. Neurotransmitter-specific identification and characterization of neurons in the all-cone retina of Anolis carolinesis. II. Glutamate and aspartate. Vis Neurosci. 1992;9:313–323. doi: 10.1017/s0952523800010725. [DOI] [PubMed] [Google Scholar]
- 51.Stafford DK, Dacey DM. Physiology of the A1 amacrine: a spiking, axon-bearing interneuron of the macaque monkey retina. Vis Neurosci. 1997;14:507–522. doi: 10.1017/s0952523800012165. [DOI] [PubMed] [Google Scholar]
- 52.Stell WK, Lightfoot DO. Color-specific interconnections of cones and horizontal cells in the retina of the goldfish. J Comp Neurol. 1975;159:473–502. doi: 10.1002/cne.901590404. [DOI] [PubMed] [Google Scholar]
- 53.Swain PH, Davis SM. Remote sensing: a quantitative approach. McGraw-Hill; New York: 1978. [Google Scholar]
- 54.Tancred E. The distribution and sizes of ganglion cells in the retinas of five Australian marsupials. J Comp Neurol. 1981;196:585–603. doi: 10.1002/cne.901960406. [DOI] [PubMed] [Google Scholar]
- 55.Vaney DI. A quantitative comparison between the ganglion cell populations and axonal outflows of the visual streak and periphery of the rabbit retina. J Comp Neurol. 1980;189:215–233. doi: 10.1002/cne.901890202. [DOI] [PubMed] [Google Scholar]
- 56.Vaney DI. Many diverse types of retinal neurons show tracer coupling when injected with biocytin or Neurobiotin. Neurosci Lett. 1991;125:187–190. doi: 10.1016/0304-3940(91)90024-n. [DOI] [PubMed] [Google Scholar]
- 57.Vaney DI. Patterns of neuronal coupling in the retina. Prog Retinal Eye Res. 1994a;13:301–355. [Google Scholar]
- 58.Vaney DI. Territorial organization of direction-selective ganglion cells in rabbit retina. J Neurosci. 1994b;14:6301–6316. doi: 10.1523/JNEUROSCI.14-11-06301.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vaney DI, Peichl L, Boycott BB. Matching populations of amacrine cells in the inner nuclear and ganglion cell layers of the rabbit retina. J Comp Neurol. 1981;199:373–391. doi: 10.1002/cne.901990305. [DOI] [PubMed] [Google Scholar]
- 60.Wässle H, Peichl L, Boycott BB. Dendritic territories of cat retinal ganglion cells. Nature. 1981;292:344–345. doi: 10.1038/292344a0. [DOI] [PubMed] [Google Scholar]
- 61.Wingate RJT, Fitzgibbon T, Thompson ID. Lucifer yellow, retrograde tracers and fractal analysis characterize adult ferret retinal ganglion cells. J Comp Neurol. 1992;323:449–474. doi: 10.1002/cne.903230402. [DOI] [PubMed] [Google Scholar]
- 62.Wong ROL, Hughes A. Developing neuronal populations of the cat retinal ganglion cell layer. J Comp Neurol. 1987;262:473–495. doi: 10.1002/cne.902620403. [DOI] [PubMed] [Google Scholar]
- 63.Xin D, Bloomfield SA. Tracer coupling pattern of amacrine and ganglion cells in the rabbit retina. J Comp Neurol. 1997;383:512–528. doi: 10.1002/(sici)1096-9861(19970714)383:4<512::aid-cne8>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]