Abstract
Background: The forkhead box O3 (FOXO3) and p27Kip1 are two important genes in breast cancer progression. In the present study we analyzed the effect of simultaneous FOXO3 silencing and p27Kip1 activation on breast cancer cell survival and the potential targets of these changes in cancer molecular pathways.
Materials and methods: The present study involved the cloning of FOXO3a shRNA and p27Kip1 genes under the control of the bidirectional survivin promoter to down- and up-regulate FOXO3 and p27Kip1 genes, respectively. After transfection of the recombinant expression vector into the breast cancer cell line, the inhibition of cell growth was assessed by MTS and flow cytometry assays. Following the extraction of total mRNA and protein, the expression of target genes was evaluated by qPCR and Western blotting in both treated and untreated cell lines. Then, the downstream protein responses were examined by 2 D electrophoresis. The differentially expressed proteins were also identified by mass spectrometry.
Results: Rates of cell proliferation were significantly inhibited in the transfected cell line 72 h post-transfection. Proteomic profiling of the cell line resulted in the identification of seven novel protein markers in breast cancer responsive to these changes in expression of FOXO3 and p27Kip1. The changes in expression of these markers suggested that certain signaling pathways contribute to the development of breast cancer.
Conclusion: Simultaneous silencing of FOXO3 and activation of p27Kip1 in MDA-MB-231 cells caused alterations in the expression level of several genes involved in apoptosis, cell proliferation, cell cycle control, tissue invasion, drug resistance, and metastasis. It seems that the identified genes might serve as useful biomarkers for breast cancer.
Keywords: Breast cancer, cell signaling, FOXO3a, gene therapy, P27Kip1, proteomics
Introduction
Breast cancer is an important global health concern and the second leading cause of cancer-related mortality among women (1). Omics studies have revealed several pathways and genes related to breast cancer (2), such as p27Kip1 and forkhead box O3 (FOXO3). The p27Kip1 protein interacts with cyclin E and cyclin-dependent kinase 2 (CDK2). In addition, the p27Kip1 gene arrests the progression of the cell cycle and leads to apoptosis, tumor suppression, and cell adhesion (3). Poor prognosis in patients with colorectal, gastric, pulmonary, and breast cancers is associated with a low expression level of the p27Kip1 gene (4).
The FOXO proteins, a subgroup of forkhead transcription factors, are involved in apoptosis, differentiation, stress response, cell cycle control, and cell metabolism (5). Overall, the FOXO3 gene is involved in many signaling pathways and makes major contributions to several processes, including cancer development (6). Therefore, the identification of FOXO3 inhibitors presents a promising strategy for future anticancer drugs (7). Proteomic analysis is generally applied in breast cancer serving two ends, namely the identification of novel molecular markers for breast tumor profiling and determination of the intracellular signaling pathways contributing to the growth of breast cancer cells (8). To our knowledge, there is no information on the effect of simultaneously decreasing FOXO3 gene and increasing p27Kip1 gene expression, respectively, to inhibit breast cancer. With this background in mind, the present study aimed to analyze the effect of the FOXO3 and p27Kip1 genes on the control of the breast cancer MDA-MB-231 cell line, using a bidirectional survivin promoter. Moreover, the changes in the protein profile of the transfected cell line were investigated in order to find cancer molecular markers.
Materials and methods
Cell line, plasmid constructs, and transfection
The MDA-MB-231 cell line adopted in this study was supplied by the Pasteur Institute (Tehran, Iran). Cells were cultured in a high-glucose Dulbecco’s modified Eagle’s medium (ThermoFisher scientific, Waltham, MA, USA), supplemented with penicillin-streptomycin (100 units/mL) and fetal bovine serum (10% FBS; Gibco BI102-100) in a damp atmosphere (5% CO2) at 37 °C. The cells were transfected with a plasmid construct designed in a bidirectional promoter (dual targeting of FOXO3a shRNA and simultaneous induction of p27Kip1 gene).
In addition, an empty plasmid of pcDNA3.1+ expression vector with EGFP was used as the control (Figure 1). The vectors were purchased from BioMatic, Toronto, Ontario, Canada. To verify the identity of the inserted sequences, all plasmids were examined through direct sequence analysis. Twenty-four hours before transfecting DNAs into the cells, some of the cells were applied in the growth medium without antibiotics. Transfection was performed as recommended by the manufacturer using Lipofectamine 2000 (Invitrogen, Waltham, MA, USA).
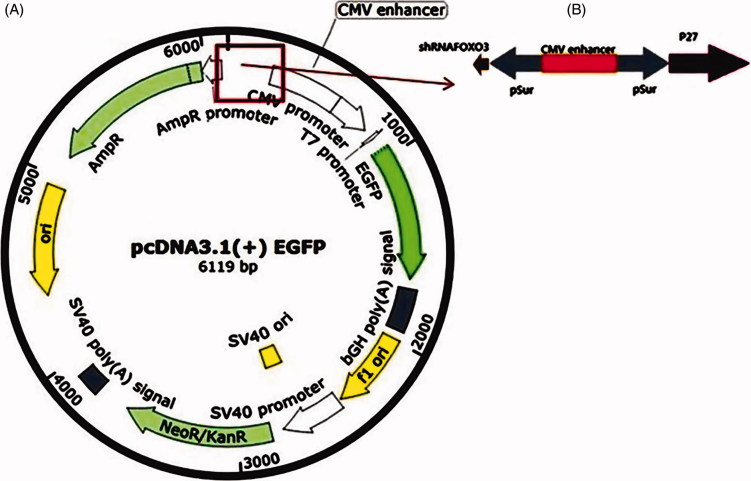
Figure 1.
(A) Map of pcDNA3.1(+) EGFP; (B) pSur (shFOXO3-p27Kip1) vector with a survivin promoter.
Cell viability assay
The viability of the cells (2–5 × 103/well) was tested in a 96-well plate in a final volume of 200 μL/well, incubated in a CO2 incubator (5% CO2) at 37 °C overnight. Then, 20 μL/well of MTS reagent (Promega, Fitchburg, WI, USA) were added to each well and incubated for 4 h at 37 °C. The plate was shaken briefly, and then the absorbance of the treated and untreated cells was measured at 490 nm 24 h, 48 h, and 72 h after transfection, using a plate reader (BioTek, Winooski, VT, USA).
Quantitative real-time polymerase chain reaction assay
The extraction of total RNA was accomplished using TRIzol solution (Invitrogen). Total RNA (2 µg) was used for complementary DNA synthesis as recommended by the manufacturer (Thermo Scientific, Vilnius, Lithuania). Quantitative real-time reverse transcriptase polymerase chain reaction (qPCR) assay was performed using ABI 7300 RT-PCR systems (Applied Biosystems, Foster City, CA, USA) to determine the relative expression of the p27Kip1 and FOXO3a genes (Table 1). The threshold cycle (Ct) value was estimated, and the REST 2009 software was used to assess the amplification plots. In addition, the comparative Ct (2−ΔΔCt) method was applied to analyze the data (8).
Table 1.
Primers used in this study.
| Primer | Sequence (5’→3’) |
|---|---|
| FOXO3a (R) | GACTATGCAGTGACAGGTTGTG |
| FOXO3a (F) | TCTACGAGTGGATGGTGCGTT |
| P27 (R) | TCCATTCCATGAAGTCAGCG |
| P27 (F) | GAGCAGACGCCCAAGAAGC |
| GAPDH (R) | AGGCAGGGATGATGTGGAGAG |
| GAPDH (F) | ATCGTGGAACTCAGTACCACA |
Apoptosis assay
To identify apoptotic cells, the FITC Annexin V Detection Kit with propidium iodide (PI) (BioLegend, San Diego, CA, USA) was exclusively developed. After seeding the cells (2 × 105 cells/mL) in tissue culture plates, they were incubated overnight, and then treated after 24 h, 48 h, and 72 h. In the next stage, the cells were washed with a cell-staining buffer, and then re-suspended in the annexin V binding buffer at a concentration of 0.25–1.0 × 107 cells/mL. Subsequently, the PI solution (10 μL) and FITC annexin V (5 μL) were added. The cells were vortexed and incubated at 25 °C for 15 min in darkness. Then, the annexin V binding buffer (400 μL) was added to the tubes. Finally, a BD Accuri™ C6 cytometer (Seattle, WA, USA) was used to perform flow cytometry.
Protein extraction and immunoblotting
A protein lysis buffer (100 mM Tris-HCl, 150 mM NaCl, 5% SDS, 1 mM dithiothreitol, 5 mM EDTA, pH of 7.4, 5% sodium deoxycholate, and 10% glycerol) was used for lysing the harvested cells collected via trypsinization. A protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO,USA) mixture was added to 1 mM of PMSF. The total protein in the resultant supernatant was used for Western blotting analysis. After separating the total protein using polyacrylamide gel electrophoresis, it was transferred to a nitrocellulose membrane, which was blocked with bovine serum albumin (5% w/v bovine serum albumin; Sigma-Aldrich) in TBST. Furthermore, the primary antibody was used against FOXO3a and p27Kip1 antibodies (Santa Cruz, Santa Cruz, CA, USA). Visualization of the bands was accomplished by means of the Enhanced Chemiluminescence Detection Kit (Parstous, Tehran, Tehran, Iran) following the manufacturer’s protocol. In addition, the National Institutes of Health ImageJ program (Bethesda, MD, USA) was used to determine the band density.
Two-dimensional gel electrophoresis
Cell pellets were resuspended by adding an adequate amount of 2 D lysis buffer, containing 2 M thiourea, 1% dithiothreitol, 4% CHAPS, 7 M urea, 0.001% bromophenol blue, and 0.5% immobilized pH gradient (IPG) buffer (pH of 3–10; GE Healthcare, USA), followed by extracting the total protein. In addition, a 2 D Quant Kit (GE Healthcare) was used to measure the protein level with bovine serum albumin as the standard. To this end, after rehydrating the IPG strips (7 cm 3–10 NL, Immobilone Dry Strip; GE Healthcare) at a pH gradient of 3–10, 300 µg of each protein sample was loaded on the IPG strip. The 2 D electrophoresis was carried out based on our previously published method (9). For staining the gels, a colloidal Coomassie Brilliant Blue dye was used as described by Candiano et al. (10). A scanner (GE Healthcare Life Sciences, Chicago, IL, USA) was also employed to scan the stained gels. The analysis of the protein spots in the gel images was performed by the ImageMaster 2 D Platinum 6.0 (GE Healthcare Life Sciences). Finally, the intensities of the treated and untreated gels were compared, and their ratios were determined.
Protein identification via liquid chromatography-mass spectrometry
After cutting the protein spots into 1-mm3 pieces, they were removed from the stained gel manually and added to tubes. The pieces were destained with an equal solution of ammonium bicarbonate and acetonitrile. Then, they were digested with trypsin (Promega, USA) at 37 °C for a minimum of 16 h. Trifluoroacetic acid (TFA) 10% was also added to prevent digestion. Extraction of the gel pieces was accomplished using an equal solution of 0.1% TFA and acetonitrile (50 μL).
Supernatants from the pooled samples were dried, using Speed-Vac centrifugation (Thermo Scientific, Waltham, MA, USA). Then, 2% acetonitrile (10 μL) and 0.1% TFA were used for dissolving the samples. The Q-TRAP nLC-MS/MS system (Applied Biosystems) was used for all analyses, as described (11). Mass data were collected and analyzed using the Analyst software (Applied Biosystems). For finding the SwissProt-Trembl subdatabase, the MS/MS lists were searched, and Mascot 3 (version 2.2., Matrix Science, Boston, MA, USA) was used to blast the data lists against databases.
Data analysis
Data were analyzed in Prism, version 6.0 (GraphPad Software Inc, San Diego, CA, USA) using two-way ANOVA and Student’s t test. The data were also presented as mean ± standard error of mean of at least three independent analyses. A p values less than 0.05 was considered statistically significant.
Results
Combinatorial effects of induced p27Kip1 and shFOXO3a on cell viability
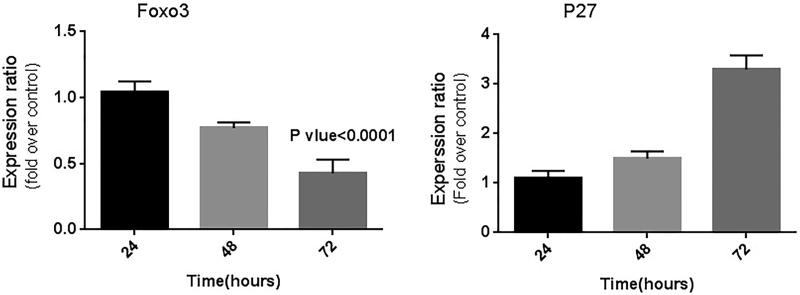
The p27Kip1 mRNA expression level was significantly higher, and FOXO3a mRNA expression significantly lower, in treated cells 72 h post-transfection in comparison with that found in controls (P < 0.05). Analysis of the qPCR results showed a 0.41-fold change in FOXO3a gene expression (down-regulation) and more than a three-fold change in p27Kip1 gene expression (up-regulation) 72 h after transfection (Figure 2).
Figure 2.
Relative expression of FOXO3 and p27Kip1 genes in the MDA-MB-231 cell line after transfection was significantly down-regulated and up-regulated, respectively, 72 h after transfection. Expression of the genes was analyzed by the REST program (P < 0.05).
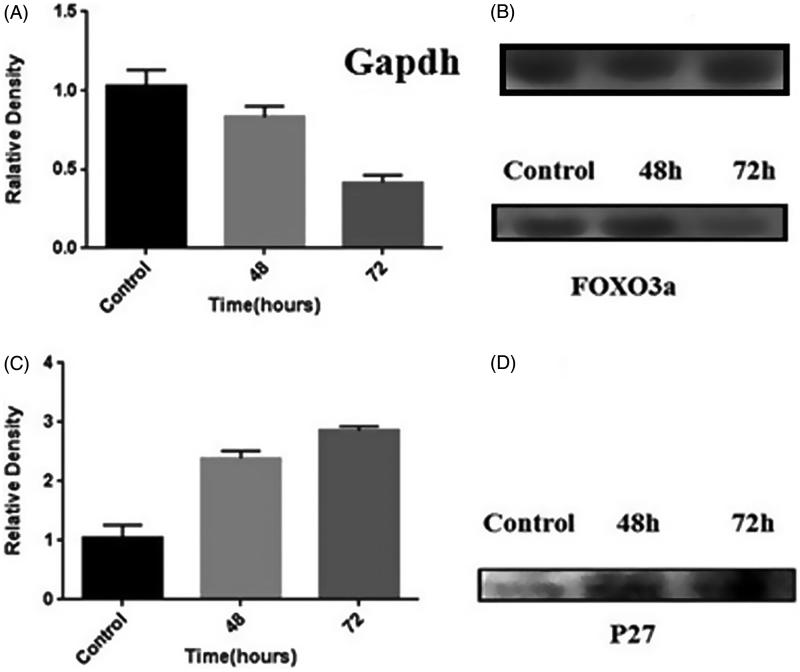
FOXO3 expression decreased (60%), while p27Kip1 expression increased 72 h after transfection, compared to the control expression (Figure 3).
Figure 3.
Relative band densitometry of Western blotting data, showing: (A) the incidence of the highest down-regulation of FOXO3a gene 72 h after transfection; (B) reduction of FOXO3a gene expression in the cells treated with the construct after 72 h, compared to the control; (C) the highest up-regulation of P27Kip1 gene in MDA-MB-231 cells occurring 72 h post-transfection; and (D) an increase in P27Kip1 expression in the cells treated with the construct 72 h after transfection, compared to the control. Implementation of quantification in three repeats (GAPDH was used as the control).
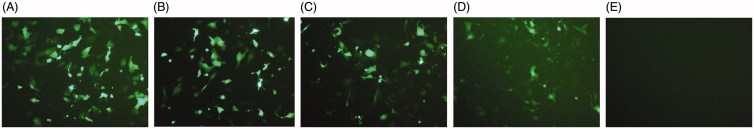
The expression of construct in MDA-MB-231 significantly reduced the growth of the cell line (P < 0.05) (Figure 4).
Figure 4.
Microscopic morphology of MDA-MB-231 cells under a fluorescent microscope after transfection for 24 h, 48 h, and 72 h with: (A) an empty vector; (B) a vector containing the genes after 24 h, (C) 48 h, (D) 72 h; and (E) negative signal of control cells without transfection.
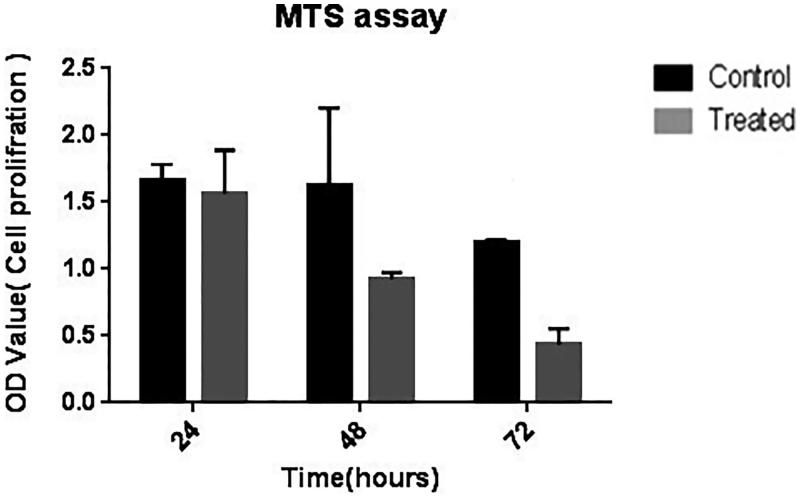
The effect of the construct on the rate of cell proliferation was not statistically significant 24 h post-transfection. However, it was significantly inhibited at 72 h after treatment (P < 0.05; Figure 5).
Figure 5.
Inhibition of MDA-MB-231 cell proliferation after 24 h, 48 h, and 72 h of treatment in wells (in triplicate) using MTS assay. Data are expressed as the mean cell viability percentage versus that of the control.
Flow cytometry
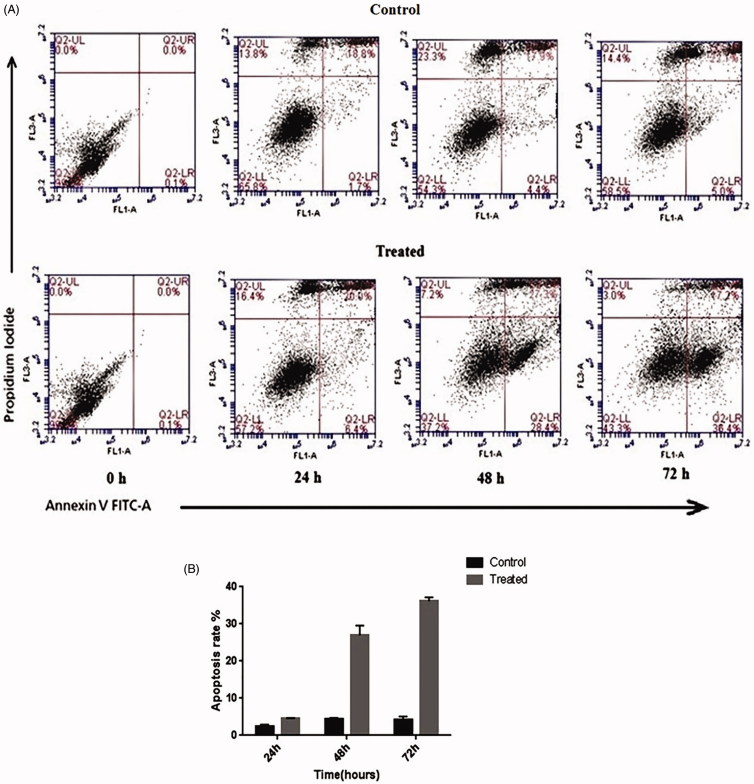
Apoptotic cells included both early and late apoptosis (Annexin V+/PI- and Annexin V+/PI+, respectively), while Annexin V and PI were negative for viable cells. In the controls, the majority of the cells (99.9%) were viable and non-apoptotic at baseline. However, early apoptotic cells showed a significant increase after treatment, compared to the untreated cells after 72 h (from 5.0% to 36.4%; Annexin V+/PI–; Figures 6(A, B)).
Figure 6.
Apoptosis detection via flow cytometry. A: use of Annexin V-FITC staining in the treated and untreated MDA-MB-231 cells after 24 h, 48 h, and 72 h. The upper and lower right quadrants (Annexin V+/PI + and Annexin V+/PI−, respectively) indicate late and early apoptosis, respectively (10.4% of cells died by lipofectamine, as shown in Supplementary Figure 1, available online). B: Apoptosis in the treated and control MDA-MB-231 cells 24 h, 48 h, and 72 h post-transfection. Data were presented as mean ± SD (P < 0.05).
Effect of overexpression p27Kip1 and FOXO3a shRNA on downstream proteins
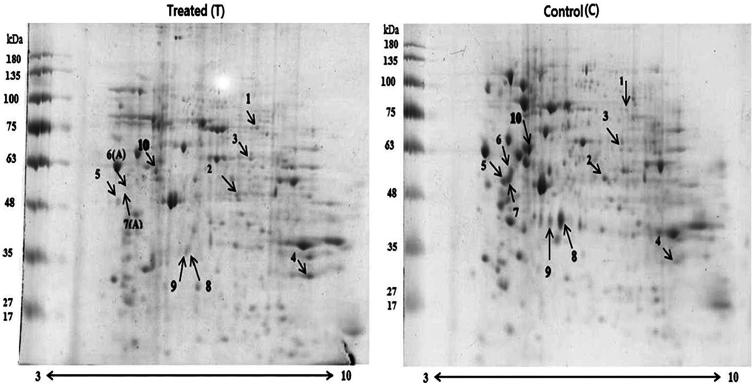
To identify the downstream responsive proteins associated with the changes in p27Kip1 and shFOXO3a expression at 72 h after transfection, the treated and untreated samples were compared by 2 D gel electrophoresis. There was differential expression of 10 protein spots in the breast cancer cell line (Figure 7). Out of the 10 proteins, 3 and 7 proteins showed up-regulated and down-regulated expression, respectively (Figure 7). Table 2 presents the mass spectrometry (MS) information of the appeared spots. Succinate dehydrogenase (SDH; T1/C1), albumin (ALB; T3/C3), and serine/threonine-protein kinase NLK (NLK; T4/C4) were significantly up-regulated, while tubulin alpha (TUBA1C; T2/C2), tubulin beta chain (TUBB; T10/C10), β-actin (ACTB) with two isoforms (T8/C8 and T9/C9), and vimentin (VIM) with three isoforms (T5/C5, T6/C6, and T7/C7) were significantly down-regulated (Figure 7). The theoretical and experimental masses and the isoelectric points of these differentially expressed proteins were also nearly the same, verifying the identified MS results (Table 2). Gene ontology (GO) analysis of 10 differentially expressed proteins consisted of protein–protein interactions using the GeneMANIA (https://genemania.org) and KEGG (https://www.genome.jp/kegg/kegg2.html) databases (Table 3). Based on the result of two-way ANOVA, after the transfection of genes in MDA-MB-231 cells for 72 h, FOXO3 gene expression showed the greatest reduction (P < 0.05), while P27Kip1 gene showed the highest increase, compared to the control (Figure 7). The 2 D gel electrophoresis of the plasmid construct and empty vector-transfected MDA-MB-231 cells indicated changes in the expression level. Out of the collected 25 spots, 10 spots (indicated by arrows, Figure 7) that had the highest scores were selected and confirmed to be involved in breast cancer.
Figure 7.
Two-dimensional gel electrophoresis of MDA-MB-231 cell line in treated and untreated cells, showing the increased expression of tubulin beta chain, vimentin (three isoforms), and β-actin (two isoforms) and decreased expression of succinate dehydrogenase complex, albumin, and NLK. Protein spots 6 and 7 were absent in the treated cells. The experiment was performed in two biological repeats. A = absence.
Table 2.
Protein spots collected from two-dimensional gels in MDA-MB-231 cell line transfected with the plasmid construct in comparison with the control (empty vector) after 72 h.
| Spot ID | Accession | Name of protein | Gene | Theoretical MW (kDa)/PI | Experimental MW (kDa)/PI | Ratio | Peptide | Score |
|---|---|---|---|---|---|---|---|---|
| 1 | P31040 | Succinate dehydrogenase | SDHA | 72.6/7.39 | 72.2/7.8 | 2.1 (up-regulated) | VGSVLQEGGK | 2.65 |
| 2 | Q9BQE3 | α-Tubulin | TUBA1C | 51/6.0 | 49.9/7.1 | 0.2 (down- regulated) | AVFVDLEPTVIDEVR; EIIDLVLDR; EDAANNYAR; DVNAAIATIK; TIGGGDDSFNTFFSETGAGK; LSVDYGKK; LSVDYGK | 12.49 |
| 3 | P02768 | Albumin | ALB | 69.3/6.28 | 65.1/6.5 | 1.2 (up-regulated) | KVPQVSTPTLVEVSR; AEFAEVSK; LVTDLTK | 2.49 |
| 4 | Q9UBE8 | Serine/threonine-protein kinase NLK | NLK | 8.13/58.28 | 34..6/8.7 | 4.5 (up-regulated) | TQEVVTQYYRAPEILMGSRHYSNAIDIWSVGIFAELLGR; PNVFQNLVSCK; ICDFGLARVEELDESR | 2.6 |
| 5 | P08670 | Vimentin | VIM | 53.65/5.12 | 49.8/5.5 | 0.3 (down-regulated) | QDVDNASLAR; QVQSLTEVDALK; VELQELNDR; QQYESVAAK; EEAENTLQSFR; ETNLDSLPLVDTHSK; EYQDLLNVK; LQEEMLQR; | 19.82 |
| 6 | P08670 | Vimentin | VIM | 53.65/5.12 | 52.5/5.7 | a/p (down-regulated) | EEENFAVEAANYQDTIGR; ALDIEIATYR; EKLQEELQR; QVQSLTEVDALK; QDVDNASLAR; VESLQEEIAFLK; VELQELNDR; EEAENTLQSFR; ETNLDSLPLVDTHSK; QQYESVAAK; LQEEMLQR; LLEGEESR; DNLAEDIR; QVDQLTNDK | 35.71 |
| 7 | P08670 | Vimentin | VIM | 53.65/5.12 | 50.4/5.6 | a/p (down-regulated) | VELQELNDR; FADLSEAANR; QYESVAAKLGDLYEEEMR; EAENTLQSFR; ALDIEIATYR; LQEEMLQR; QESTEYR; ETNLDSLPLVDTHSK; QVQSLTEVDALK; LGDLYEEER; DNLAEDIR; LLEGEESR; LQDEIQNKEEAR; FANYIDK | 27.58 |
| 8 | P60709 | β-Actin | ACTB | 41.7/5.48 | 40.5/6.5 | 0.2 (down-regulated) | QEYDESGPSIVHR;GYSFTTTAER; DLTDYLMK; APEEHPVLLTEAPLNPK; DSYVGDEAQSKR; DSYVGDEAQSK; EITALAPSTK; DLTDYLK; DLYANTVLSGGTTYPGIADR | 17.02 |
| 9 | P60709 | β-Actin | ACTB | 41.7/5.48 | 40.89/6.7 | 0.1 (down-regulated) | QEYDESGPSIVHR;GYSFTTTAER; DLTDYLMK; APEEHPVLLTEAPLNPK; DSYVGDEAQSKR; DSYVGDEAQSK; EITALAPSTK; DLTDYLK; DLYANTVLSGGTTYPGIADR | 10.72 |
| 10 | P07437 | Tubulin beta chain | TUBB | 51.2/5 | 49.6/5.8 | 0.3 (down-regulated) | INTFSVVPSPK; TAVDIPPR; ISVYYNEATGGK; NAADPR; LAVNVPFPR | 46.38 |
a/p: absence/presence; MW: molecular weight.
Table 3.
Protein–protein interactions of all proteins for collecting the data.
| Gene | |||||||
|---|---|---|---|---|---|---|---|
| Vim | ACTB | TUBA1C | TUBB | SDHA | NLK | ALB | |
| Expand | |||||||
| Physical interaction | SERPINH, MAGOH, EIFA3, SPRM4, KRT18, UPP1, ITGB4, BASP1, PPL, URGDP, XPNPEP3 | CFL1, CFL2, PFN1, CCT8, TCP1, CCT3, CCT4, RUVBL1, MYH11 | TUBA4B, TUBB6, TUBB, TUBA4A, TUBB4A, TUBB4B, TUBB3, TUBB2A, TUBB6, TUBB2B, GAPDH, | THBS1, CLIP1, TCP11L1, TUBB2A, YBX1, TUBA1A, TUBA1B, TUBA1C, TUBA1A, TUBA4B, PNKD, TNPO2, DNAJA4 | SLC2A14, CPNE1, RAB14, LRRC8A, HIGD1A, SDHAF2, SDHAF2, SDHC, SDHB, UBC | MYB, RANGAP1, ZHX3, MAP3K7, TAB1, HIPK2, LEF1, TP53, TCF7L2 | F2, HP, ITIH1, AMBP, AHSG, APOA2, RBP4, FCGRT |
| Co-expression | SERPINH, TTN | CFL1, PFN1 | TUBB6, TUBB, TUBA4A, TUBB4B, TUBB3, TUBB2A, TUBB6, TUBB2B, GAPDH, TUBA1A, TUBA1B, TBCB, PFDN1, STMN1 | TUBAB, TUBB2A, TUBG2, YBX1, TUBA1B, TUBA1C, TUBA4A, TUBB3, TUBB4B, TUBG1 | RAB14, HIGD1A, SDHC, SDHB, NDUFV1, UBC, ALAD, UQCRC1 | — | F2, HP, ITH1, GC, AMBP, AFM, AHSG, AFP, APO2, RBP4, SLCO1B1, SLCO1B3 |
| Co-localization | SERPINHI | — | STMN1, TBCB, TUBA1B, TUBA1A, TUBB2A | — | NDUFV1, UQCRC1, NDUFS1 | RANGAP1, MAP3K7, TAB1 | F2, HP, ITH1, GC, AMBP, AHSG, APO2, RBP4, SLCO1B1, SLCO1B3 |
| Gene interaction | — | — | — | — | — | TP53 | — |
| Role | Proliferation, tissue invasion, and metastasis | Cell mobility, endocytosis, cell adhesion, division, drug resistance, and metastasis | Cellular mobility | Cellular mobility | Tumor suppressor | Inhibit cellular proliferation, induce apoptosis | Stabilizing DNA, regulation calcium, antioxidant properties |
Discussion
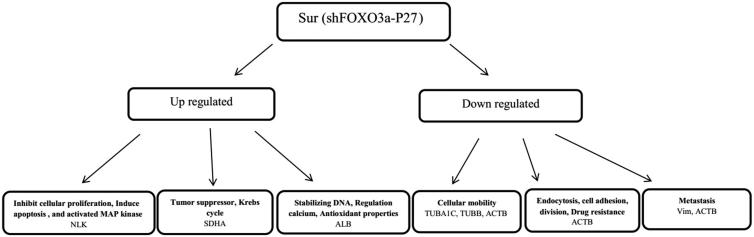
There are studies regarding the independent effect of P27Kip1 and FOXO3a genes on the inhibition of breast cancer; however, they have reported a poor prognosis (12,13). The present study involved the design of a bidirectional expression vector suppressing FOXO3a and inducing p27Kip1 expression. Our results showed that the synergistic effects of p27Kip1 and shFOXO3a induced 36.4% apoptosis in cells. However, the overexpression of p27Kip1 alone has been reported to induce 12.73% apoptosis (12). Recently, proteomic methods have been used as a strategy for the detection of protein biomarkers (14). Expression of seven proteins changed after FOXO3a suppression and p27Kip1 induction (Figure 8). These proteins are all involved in apoptosis, metastasis, drug resistance, proliferation, and cell cycle control.
Figure 8.
Effect of simultaneous FOXO3a suppression and P27Kip1 induction on protein expression in treated cells.
Three up-regulated proteins included SDHA, albumin (with cell source), and NLK, while four down-regulated proteins included TUBA1C, TUBB, ACTB with two isoforms, and VIM with three isoforms. The SDH is identified as a mitochondrial enzyme, involved in the Krebs cycle and electron transfer chain, and contributes to tumor suppression. This enzyme has four subunits, namely SDHA, SDHB, SDHC, and SDHD (15). Research over the past decades has shown an inverse relationship between albumin level and severity/risk of the progression of different cancers, including pulmonary, gastric, ovarian, and breast cancers. There are various mechanisms for the anticancer effects of albumin, such as DNA stabilization and duplication, calcium preservation and regulation, and antioxidant properties. In a study performed by Xiao-Jing Du et al., the growth of human cancer cells, such as breast cancer cells, was inhibited by high concentrations of albumin (16). The main cause of the reduction of the albumin level, especially in cancer patients, is the inhibition of albumin-regulating gene by tumor necrosis factor which results in mRNA expression reduction of approximately 90% in the liver (17). NLK, which belongs to the mitogen-activated protein kinase family, is a vital regulator of various cancer types. In a study conducted by Lv et al., NLK inhibited non-small-cell lung cancer through the Wnt signaling pathway (18). In addition, Huang et al. showed that NLK inhibits cellular proliferation in breast cancer and induces apoptosis. Similar studies confirm that NLK is a tumor suppressor gene in ovarian cancer (19). The theoretical molecular weight of NLK is about 58 kDa, while its experimental molecular weight is about 35 kDa, which can be attributed to the degradation of this protein. Actin and its proteins have a major functional and structural role in cell motility, endocytosis, cell adhesion and division, and maintenance of cell morphology. The expression of ACTB increases in a metastatic state (20). Accordingly, Chunmei Guo et al. reported that ACTB increased in many cancers of the liver, kidneys, colon, stomach, pancreas, esophagus, lung, breast, prostate, and ovaries (21). These tumors are associated with increased drug resistance and metastasis, especially in MDA-MB-231 cells (21).
Tubulins consist of microtubules, with two alpha and beta subunits (approximate weight of 50 kDa). They play a large role in cellular activities, such as cellular mobility. Dysfunction of the microtubule network causes disturbance in apoptosis and differentiation (20). In addition, VIM is one of the most filamentous proteins, expressed in various types of cancers, such as prostate, gastrointestinal, central nervous system, pulmonary, and breast cancers. In a study carried out by Satelli, it was shown that the increased invasive/migratory potential of cancer cells is related to VIM overexpression (22). The VIM is generally considered as an indicator of poor prognosis (22).
In conclusion, the simultaneous silencing of FOXO3 and activation of p27Kip1 in MDA-MB-231 cells caused alterations in the expression level of several genes which are involved in cell apoptosis, cell proliferation, cell cycle control, tissue invasion, drug resistance, and metastasis. Therefore, it seems that these genes may serve as useful biomarkers for breast cancer.
Supplementary Material
Biographies
Sabah Mayahi is a Ph.D. student of Molecular Medicine at Golestan University of Medical Sciences, Gorgan, Iran. She works in the Department of Parasitology and Mycology, Faculty of Medicine, Mazandaran University of Medical Sciences.
Dr. Masood Golalipour is an Associate Professor of Molecular Genetics at the Department of Genetic and member of the Cellular and Molecular Research Center of Golestan University of Medical Sciences.
Dr. Ahad Yamch is an Assistant Professor of Genetic Engineering at the Department of Biotechnology, Gorgan University of Agricultural Sciences and Natural Resources.
Dr. Gagan Deep Jhingan completed his Masters and Ph.D. in Biotechnology from Jawaharlal Nehru University. He has worked as Wellcome/DBT Early Career Fellow at National Institute of Immunology, New Delhi. He has specialization in the field of proteomics and is responsible for the development and management of Vproteomics platform technology.
Majid Shahbazi is a Professor of Genetics working on target therapy and Director of Cellular and Molecular Research Center; head of the Department of Molecular Medicine, School of Advanced Technologies in Medicine of Golestan University of Medical Sciences; also CEO of AryaTinaGene biopharmaceutical company.
Funding Statement
This work was supported by the Golestan University of Medical Sciences and Health Services (Grant number: 940714170 ) and AryaTinagene biopharmaceutical company.
Acknowledgements
We thank the Golestan University of Medical Sciences and Arya Tina Gene (ATG) Biopharmaceutical Company, Gorgan, Iran.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- 1.Abdulkareem IH. Aetio-pathogenesis of breast cancer. Niger Med J. 2013;54:371–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sadeghi H, Golalipour M, Yamchi A, Farazmandfar T, Shahbazi M. CDC25A pathway toward tumorigenesis: molecular targets of CDC25A in cell-cycle regulation. J Cell Biochem. 2019;120:2919–28. [DOI] [PubMed] [Google Scholar]
- 3.Sgambato A, Cittadini A, Faraglia B, Weinstein IB. Multiple functions of p27(Kip1) and its alterations in tumor cells: a review. J Cell Physiol. 2000;183:18–27. [DOI] [PubMed] [Google Scholar]
- 4.Kudoh S, Kumaravel TS, Kuramavel B, Eguchi M, Asaoku H, Dohy H, et al. Protein expression of cell cycle regulator, p27Kip1, correlates with histopathological grade of non-Hodgkin's lymphoma. Jpn J Cancer Res. 1999;90:1262–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Obsil T, Obsilova V. Structure/function relationships underlying regulation of FOXO transcription factors. Oncogene 2008;27:2263–75. [DOI] [PubMed] [Google Scholar]
- 6.Fu Z, Tindall DJ. FOXOs, cancer and regulation of apoptosis. Oncogene 2008;27:2312–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor S, Lam M, Pararasa C, Brown JE, Carmichael AR, Griffiths HR. Evaluating the evidence for targeting FOXO3a in breast cancer: a systematic review. Cancer Cell Int. 2015;15:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Golalipour M, Mahjoubi F, Sanati MH, Alimoghaddam K, Kamran A. Gene dosage is not responsible for the upregulation of MRP1 gene expression in adult leukemia patients. Arch Med Res. 2007;38:297–304. [DOI] [PubMed] [Google Scholar]
- 9.Yamchi A, Ben C, Rossignol M, Zareie SR, Mirlohi A, Sayed-Tabatabaei BE, et al. Proteomics analysis of Medicago truncatula response to infection by the phytopathogenic bacterium Ralstonia solanacearum points to jasmonate and salicylate defence pathways. Cell Microbiol. 2018;20:e12796. [DOI] [PubMed] [Google Scholar]
- 10.Candiano G, Bruschi M, Musante L, Santucci L, Ghiggeri GM, Carnemolla B, et al. Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis. 2004;25:1327–33. [DOI] [PubMed] [Google Scholar]
- 11.Boudart G, Jamet E, Rossignol M, Lafitte C, Borderies G, Jauneau A, et al. Cell wall proteins in apoplastic fluids of Arabidopsis thaliana rosettes: identification by mass spectrometry and bioinformatics. Proteomics. 2005;5:212–21. [DOI] [PubMed] [Google Scholar]
- 12.Jiang D, Wang X, Liu X, Li F. Gene delivery of cyclin-dependent kinase inhibitors p21Waf1 and p27Kip1 suppresses proliferation of MCF-7 breast cancer cells in vitro. Breast Cancer. 2014;21:614–23. [DOI] [PubMed] [Google Scholar]
- 13.Storz P, Doppler H, Copland JA, Simpson KJ, Toker A. FOXO3a promotes tumor cell invasion through the induction of matrix metalloproteinases. Mol Cell Biol. 2009;29:4906–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gamez-Pozo A, Trilla-Fuertes L, Berges-Soria J, Selevsek N, Lopez-Vacas R, Diaz-Almiron M, et al. Functional proteomics outlines the complexity of breast cancer molecular subtypes. Sci Rep 2017;7:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bardella C, Pollard PJ, Tomlinson I. SDH mutations in cancer. Biochim Biophys Acta. 2011;1807:1432–43. [DOI] [PubMed] [Google Scholar]
- 16.Du XJ, Tang LL, Mao YP, Sun Y, Zeng MS, Kang TB, et al. The pretreatment albumin to globulin ratio has predictive value for long-term mortality in nasopharyngeal carcinoma. PLoS One 2014;9:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seaton KA. concentration controls cancer. J Natl Med Assoc 2001;93:490–3. [PMC free article] [PubMed] [Google Scholar]
- 18.Lv L, Wan C, Chen B, Li M, Liu Y, Ni T, et al. Nemo-like kinase (NLK) inhibits the progression of NSCLC via negatively modulating WNT signaling pathway. J Cell Biochem. 2014;115:81–92. [DOI] [PubMed] [Google Scholar]
- 19.Huang Y, Jiang Y, Lu W, Zhang Y. Nemo-like kinase associated with proliferation and apoptosis by c-Myb degradation in breast cancer. PLoS One 2013;8:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee S, Terry D, Hurst DR, Welch DR, Sang QX. Protein signatures in human MDA-MB-231 breast cancer cells indicating a more invasive phenotype following knockdown of human endometase/matrilysin-2 by siRNA. J Cancer. 2011;2:165–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo C, Liu S, Wang J, Sun MZ, Greenaway FT. ACTB in cancer. Clin Chim Acta. 2013;417:39–44. [DOI] [PubMed] [Google Scholar]
- 22.Satelli A, Li S. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell Mol Life Sci. 2011;68:3033–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.