Abstract
A new species, Pythium subutonaiense, isolated from aquatic environments (lake) in China is being described based on morphological characters and molecular evidence. The isolates grew at temperatures between 5 °C and 38 °C, and the optimum temperature was 30 °C, with a radial growth rate of 17.6 mm at 25 °C per day. It is homothallic and characterized by globose to sub-globose shaped and mostly terminal or sometimes catenulate hyphal swellings, filamentous non-inflated sporangia, and smooth oogonia with hypogynous and monoclinous antheridia that contained one plerotic oospore. In phylogenetic analysis, inferred based on the internal transcribed spacer region of the ribosomal RNA gene and mitochondrial cytochrome c oxidase subunit 1 gene, the new species formed a distinct lineage in Pythium clade B. Differences between the new species and phylogenetically related and morphologically similar species are discussed.
Keywords: Oomycote, phylogenetic analysis, Pythium clade B, taxonomy
1. Introduction
Pythium [1] (Pythiaceae, Pythiales), typified with P. monospermum Pringsh., is characterized by hyaline and coenocytic hyphae without septa, various shaped sporangia, and the development of zoospores in a vesicle which is formed at the tip of a discharge tube derived from a sporangium [2]. Pythium spp. are cosmopolitan and represent a range of functional groups, such as saprophytes in natural environments, plant and animal pathogens, and biological control agents protecting against pathogenic fungi [3]. Following recent taxonomic revisions [4,5] and discoveries (e.g., Refs. [6–11]), more than 140 species are currently recorded in the genus Pythium [11].
During studies on Pythium species diversity in southern China, one new species of Pythium, P. utonaiense was isolated. Phylogenetic analysis was performed using the internal transcribed spacer (ITS) regions of the ribosomal RNA and mitochondrial cytochrome c oxidase subunit 1 (COI) genes. Combined with the morphological characters, a new species is described in this study. Differences between the new species and phylogenetically related and/or morphologically similar species are also provided.
2. Materials and methods
2.1. Isolation
The samples were recovered from surface water of the lake in Nanjing, Jiangsu Province of China using flowers of Bougainvillea glabra Choisy as baits to isolate Pythium species [12]. The isolation procedure followed the method described by Benard and Punja [13]. Pieces of tissue 5–10 mm were cut from the baits, washed in tap water and superficially dried on a paper towel, and plated on V8 juice agar (V8A) containing rifampicin (50 mg L−1), phenamacril (5 mg L−1), ampicillin (50 mg L−1), and pentachloronitrobenzene (50 mg L−1) and incubated at 25 °C for 2–3 days. When mycelial growth was observed, purification was carried out twice by transferring a single hyphal tip of colonies onto V8A. Two isolates (Chen 220 and Chen 229) of undescribed Pythium species were recovered and deposited in the herbarium of the College of Plant Protection, Nanjing Agricultural University (NJAU).
2.2. Morphology and growth rate
Colony patterns of two P. subutonaiense isolates were examined after incubation for 3 days at 25 °C in corn meal agar (CMA), potato carrot agar (PCA), and V8A media. Sporulation was induced using a modification of the method described by Chang [14]. Agar blocks (8 mm × 8 mm × 3 mm) were cut from the mycelia fronts of 3-day-old V8A cultures and placed in 10% clarified and sterile V8 juice (one block per dish) and incubated in darkness at 25 °C for 48–72 h. Once the mycelium has reached 4–5 cm in diameter, the V8 juice was pipetted out and the mycelia mats were then rinsed with sterile distilled water (SDW) three times at 20 min intervals. The rinsed mycelia mats were then submerged in SDW. The plates containing SDW were incubated at 25 °C for 24–48 h prior to examining for release of zoospores. Fifty measurements were taken for each morphological feature, such as sporangia, oogonia, and oospores. The cardinal temperatures of growth rates, on PCA media [2], were measured at 24 h of incubation for each isolate at 5–40 °C, with intervals of 5 °C. When no growth was observed, the intervals were reduced from 5 to 2 or 1 °C and the culture was returned to room temperature to check growth revival.
2.3. DNA extraction and sequencing
A cetyl trimethylammonium bromide rapid plant genome extraction kit (Demeter Biotechnologies Co., Ltd., Beijing, China) was used to extract total genomic DNA from purified isolates, and performed the polymerase chain reaction (PCR) according to the study by Chen and Cui [15]. A small piece of dried fungal specimen (about 30 mg) was grounded to powder with liquid nitrogen, transferred to a 1.5-mL centrifuge tube and resuspended in 0.4 mL of lysis buffer (PL), and incubated at 65 °C water bath for 60 min. Then, 0.4 mL phenol–chloroform (24:1) was added to each tube and the suspension was shaken vigorously. After centrifugation at 13,000 rpm for 5 min, 0.3 mL supernatant was transferred to a new tube and mixed with 0.45 mL binding buffer (PQ). The mixture was later transferred to an adsorbing column (AC) for a centrifugation at 13,000 rpm for 0.5 min. Then, 0.5 mL inhibitor removal fluid (IR) was added to the AC for a centrifugation at 12,000 rpm for 0.5 min. After washing twice with 0.5 mL washing buffer (WB), removing the AC to a clean centrifuge tube, and adding 100 μL elution buffer in the middle of adsorbed film, the genome DNA was eluted in 100 µL elution buffer (EB). The ITS region was amplified with the primers: ITS4 and ITS5 [16]. The COI gene was amplified with the primers: OomCoxI-Levlo (CYTCHGGRTGWCCRAAAAACCAAA) and OomCoxI-Levup (TCAWCWMGATGGCTTTTTTCAAC) [17]. The PCR procedure for ITS was as follows: initial denaturation at 95 °C for 3 min, followed by 35 cycles at 94 °C for 40 s, 54 °C for 45 s and 72 °C for 1 min, and a final extension of 72 °C for 10 min. The PCR procedure for COI was as follows: initial denaturation at 94 °C for 2–5 min, followed by 35 cycles at 94 °C for 30 s, 52 °C for 30 s and 72 °C for 1–2 min, and a final extension of 72 °C for 5–10 min [18]. The PCR products were purified and sequenced in Genscript Company (Nanjing, China) with the same primers.
2.4. Phylogenetic analysis
Sequences generated in this study were aligned with additional sequences downloaded from GenBank (Table 1) using ClustalX [19] and BioEdit [20]. Sequence alignment was deposited at TreeBase (http://purl.org/phylo/treebase; submission ID S22483).
Table 1.
A list of species, cultures, and GenBank accession numbers of sequences used in this study.
| Species name | Sample no. | Locality | GenBank accession no. |
|
|---|---|---|---|---|
| ITS | COI | |||
| Pythium adhaerens | CBS 520.74 | The Netherlands | HQ643415 | HQ708462 |
| P. afertile | Lev 2066 | Canada | HQ643416 | HQ708463 |
| P. angustatum | CBS 522.74 | The Netherlands | HQ643437 | HQ708484 |
| P. apleroticum | CBS 772.81 | The Netherlands | HQ643444 | HQ708491 |
| P. aquatile | CBS 215.80 | United Kingdom | HQ643445 | HQ708492 |
| P. aristosporum | CBS 263.38 | Canada | HQ643447 | HQ708494 |
| P. arrhenomanes | CBS 324.62 | USA | HQ643452 | HQ708499 |
| P. biforme | UZ00796 | Japan | KJ995584 | KJ995590 |
| P. brachiatum | UZ00736 | Japan | KJ995581 | KJ995593 |
| P. brachiatum | UZ00746 | Japan | KJ995583 | KJ995594 |
| P. capillosum | CBS 222.94 | France | HQ643483 | HQ708529 |
| P. catenulatum | CBS 226.94 | France | HQ643490 | HQ708536 |
| P. chondricola | CBS 203.85 | The Netherlands | HQ643498 | HQ708544 |
| P. coloratum | CBS 154.64 | Australia | HQ643501 | HQ708547 |
| P. conidiophorum | CBS 223.88 | United Kingdom | HQ643509 | HQ708555 |
| P. contiguanum | CBS 221.94 | Algeria | HQ643514 | HQ708560 |
| P. diclinum | CBS 526.74 | The Netherlands | HQ643523 | HQ708569 |
| P. dissimile | CBS 155.64 | Australia | HQ643526 | HQ708572 |
| P. dissotocum | CBS 166.68 | USA | HQ643528 | HQ708574 |
| P. flevoense | CBS 234.72 | The Netherlands | HQ643538 | HQ708582 |
| P. folliculosum | CBS 220.94 | Switzerland | HQ643540 | HQ708584 |
| P. graminicola | CBS 327.62 | Jamaica | HQ643545 | HQ708589 |
| P. inflatum | CBS 168.68 | USA | HQ643566 | HQ708610 |
| P. kashmirense | CBS 122908 | India | HQ643671 | HQ708715 |
| P. lutarium | CBS 222.88 | United Kingdom | HQ643682 | HQ708726 |
| P. myriotylum | CBS 254.70 | Israel | HQ643701 | HQ708745 |
| P. oopapillum | BR 632 | – | FJ655174 | FJ655178 |
| P. pachycaule | CBS22494 | France | HQ643726 | HQ708767 |
| P. pectinolyticum | CBS 122643 | France | HQ643739 | HQ708780 |
| P. periilum | CBS 169.68 | USA | HQ643740 | HQ708781 |
| P. phragmitis | CBS 117104 | Germany | HQ643746 | HQ708787 |
| P. plurisporium | CBS 100530 | USA | HQ643749 | HQ708790 |
| P. pyrilobum | CBS 158.64 | Australia | HQ643755 | HQ708796 |
| P. rhizo-oryzae | CBS 119169 | India | HQ643757 | HQ708798 |
| P. salpingophorum | BR 1024 | United Kingdom | HQ643770 | HQ708811 |
| P. scleroteichum | CBS 294.37 | USA | HQ643771 | HQ708812 |
| P. subutonaiense | Chen 220a | China | MG654703 | MG674169 |
| P. subutonaiense | Chen 229a | China | MG654704 | MG674170 |
| P. sukuiense | CBS 110030 | Taiwan | HQ643836 | HQ708877 |
| P. sulcatum | CBS 603.73 | USA | HQ643837 | HQ708878 |
| P. tardicrescens | Lev 1534 | USA | HQ643855 | HQ708896 |
| P. torulosum | CBS 316.33 | The Netherlands | HQ643859 | HQ708900 |
| P. tracheiphilum | CBS 323.65 | Italy | HQ643862 | HQ708903 |
| P. utonaiense | UZ00758 | Japan | KJ995586 | KJ995588 |
| P. utonaiense | UZ00769 | Japan | KJ995587 | KJ995589 |
| P. vanterpoolii | CBS 295.37 | United Kingdom | HQ643952 | HQ708993 |
| P. volutum | CBS 699.83 | Japan | HQ643971 | HQ709012 |
| P. zingiberis | CBS 216.82 | Japan | HQ643973 | HQ709014 |
New sequences determined in the present study.
Phylogenetic analysis was done as in the study by Chen and Cui [15]. Maximum parsimony (MP) analysis was applied to the combined dataset of ITS and COI sequences. Pythium adhaerens Sparrow and P. chondricola De Cock were used as outgroups [7]. The tree construction procedure was performed in PAUP* version 4.0b10 [21]. All characters were equally weighted and gaps were treated as missing data. Trees were inferred using the heuristic search option with TBR branch swapping and 1000 random sequence additions. Max-trees were set to 5000, branches of zero length were collapsed and all parsimonious trees were saved. Clade robustness was assessed using a bootstrap analysis with 1000 replicates [22]. Descriptive tree statistics tree length (TL), consistency index (CI), retention index (RI), rescaled consistency index (RC), and homoplasy index (HI) were calculated for each maximum parsimonious tree generated.
MrModeltest 2.3 [23] was used to determine the best-fit evolution model for the dataset for Bayesian inference (BI). BI was calculated with MrBayes3.1.2 [24] with a general time reversible (GTR) model of DNA substitution and an invgamma distribution rate variation across sites. Four Markov chains were run for 2 million generations until the split deviation frequency value <0.01, and sampled every 100th generations. The burn-in was set to discard the first 25% of the trees. A majority rule consensus tree of all remaining trees was calculated. Phylogenetic trees were visualized using Treeview [25]. Branches that received bootstrap support for maximum likelihood (BS), maximum parsimony (MP), and Bayesian posterior probabilities (BPP) greater than or equal to 75% (MP) and 0.95 (BPP) were considered as significantly supported, respectively.
3. Results
3.1. Isolation
Water samples were collected from two lakes, respectively in Zixiahu Park and Soul Valley Temple in China in 2016. The two lakes are located in national park of Dr. Sun Yat-sen’s mausoleum of southern China. Two surface water samples were collected at the edge of the lake. Two isolates of the new species, P. subutonaiense, were respectively obtained from two water samples and isolated from flowers of Bougainvillea glabra as bait on V8A.
3.2. Morphology and growth rate
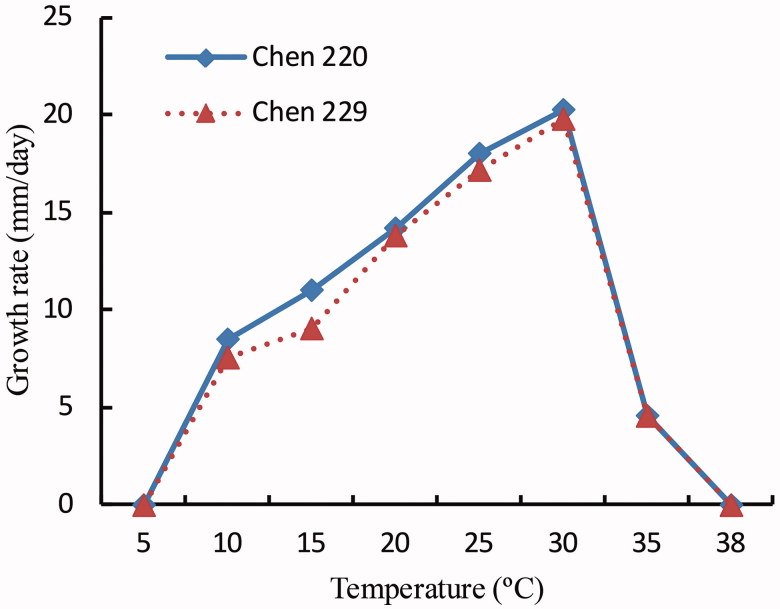
Both two isolates of P. subutonaiense showed similar colony patterns and growth temperature results (Figure 1). They were rosette pattern on PCA, stellate pattern on V8A, and without a special pattern on CMA. The isolates showed maximum growth at 30 °C, and no growth at 5 °C and 38 °C. The growth rate was 17.6 mm per day at 25 °C.
Figure 1.
Mecelial growth rate of P. subutonaiense isolates Chen 220 and Chen 229 on potato carrot agar at different temperatures.
The morphology of asexual and sexual structures was also similar between the two isolates. Hyphal swellings were frequently observed, and zoospores were rarely observed in both isolates. The sexual structures were abundantly produced on V8A. Oogonia were smooth-walled, but sometimes had one outstanding slender projection. Antheridia were hypogynous and monoclinous, produced one or two per oogonium. Oospores were one per oogonium.
P. subutonaiense differs other Pythium species from homothallic sexuality, globose to sub-globose shaped, mostly terminal or sometimes catenulate hyphal swellings, filamentous non-inflated sporangia, and smooth oogonia, hypogynous, and monoclinous antheridia with slender antheridial cells and plerotic oospores.
Further differences between the new species and other related ones are listed in Table 2.
Table 2.
Morphological description of Pythium subutonaiense and the most closely related species.
| P. subutonaiense (Chen 218) | P. brachiatum | P. capillosum | P. flevoense | P. utonaiense | |
|---|---|---|---|---|---|
| Width of hyphae (μm) | Up to 5 | Up to 5.5 | 3–5 | Up to 6 | Up to 5 |
| Hyphal swellings | Present | Absent | Absent | Absent | Absent |
| Sporangia or inflated structures | Filamentous non-inflated | Filamentous slightly inflated | Filamentous, non-inflated or slightly inflated | Filamentous | Filamentous non-inflated |
| Oogonia (μm) | 17.5–22.5 (av. 20.8), intercalary or rarely terminal | 13.3–34.4 (av. 22.7), intercalary or terminal, sometimes in chain | 16–35.2 (av. 23.09), mostly intercalary, rarely terminal | 17–20 (av. 19), mostly terminal | 10–23.3 (av. 17.5), terminal, rarely intercalary |
| Antheridia | Hypogynous or monoclinous | Monoclinous or diclinous or rarely lacking | Mostly diclinous, rarely monoclinous | Diclinous | Lacking |
| Oospores | Plerotic | Plerotic, occasionally aplerotic | Plerotic or aplerotic | Aplerotic, occasionally nearly plerotic | Plerotic |
| Oospore wall thickness (μm) | 0.5–2 | 0.3–3.2 | 2–3.5 | 2–3 | 0.3–3.1 |
| Cardinal temperature | Min 5 °C, optimum 30 °C and max 38 °C | Min 4 °C, optimum 25 °C and max 30 °C | Min 1–2 °C, optimum 25 °C and max 40 °C | Min under 5 °C, optimum 25 °C and max over 35 °C | Min 4 °C, optimum 30 °C and max 40 °C |
| Daily growth rates on PCA at 25 °C (mm) | 17.6/per day | 14/per day | 7/per day | 7–10/per day | 23.5/per day |
| Reference | This study | [7] | [27] | [28] | [7] |
3.3. Molecular phylogeny
Two ITS and two COI sequences were newly generated for this study and their accession numbers are available in GenBank (Table 1). BLAST analyses of ITS and COI sequences of the two isolates, described here as P. subutonaiense, showed the best phylogenetic matches were with species of Pythium clade B.
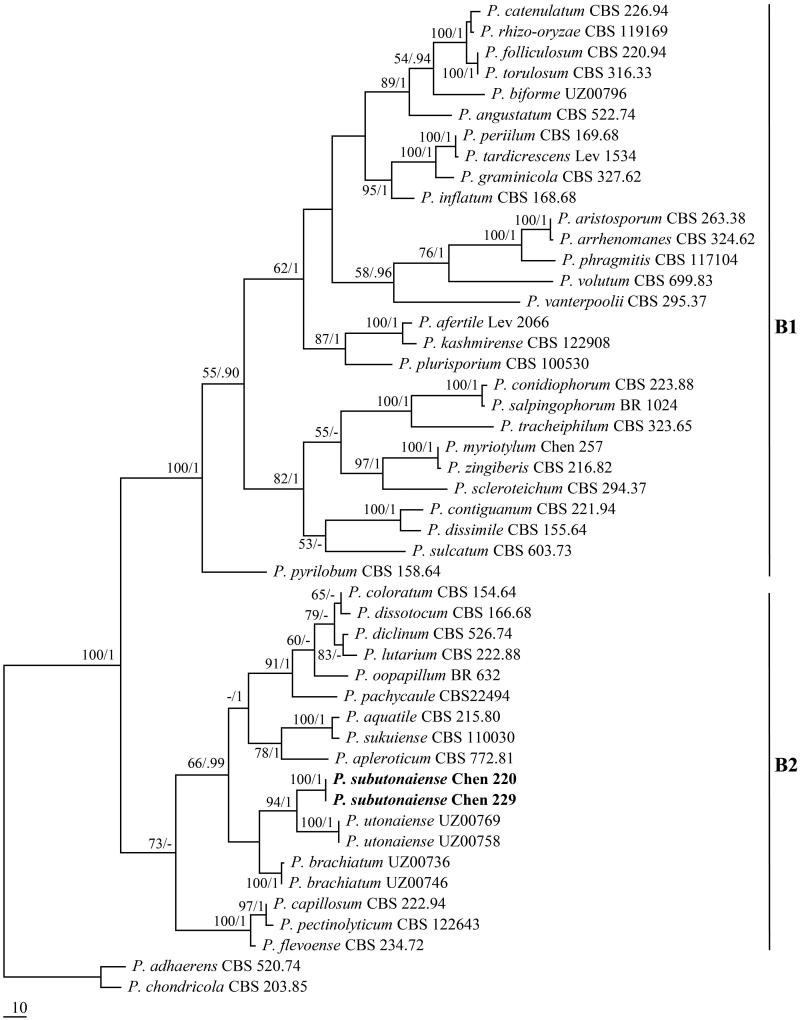
The combined ITS + COI dataset included sequences from 48 oomycetes representing 45 taxa. The dataset had an aligned length of 1563 characters, of which 1111 characters are constant, 64 are variable and parsimony-uninformative, and 388 are parsimony-informative. Maximum parsimony analysis yielded three equally parsimonious trees (TL = 1369, CI = 0.455, RI = 0.757, RC = 0.344, HI = 0.545). Best model for the combined ITS + COI sequences dataset estimated and applied in the Bayesian analysis: GTR + I + G, lset nst = 6, rates = invgamma; prset statefreqpr = dirichlet (1,1,1,1). Bayesian analysis resulted in the same topology with an average standard deviation of split frequencies = 0.006409. The sequences of Pythium species in clade B clustered together with strong supports (100% ML, 1 BPP, Figure 2). The phylogeny inferred from the ITS + COI dataset showed that the two newly sequenced isolates formed a lineage within Pythium subclade B2 with full statistical supports (100% MP, 1 BPP) and clustered with P. utonaiense [7].
Figure 2.
Phylogeny of Pythium clade B species inferred from ITS + COI dataset.
3.4. Taxonomy
P. subutonaiense: Jia J. Chen and X.B. Zheng, sp. nov. (Figure 3).
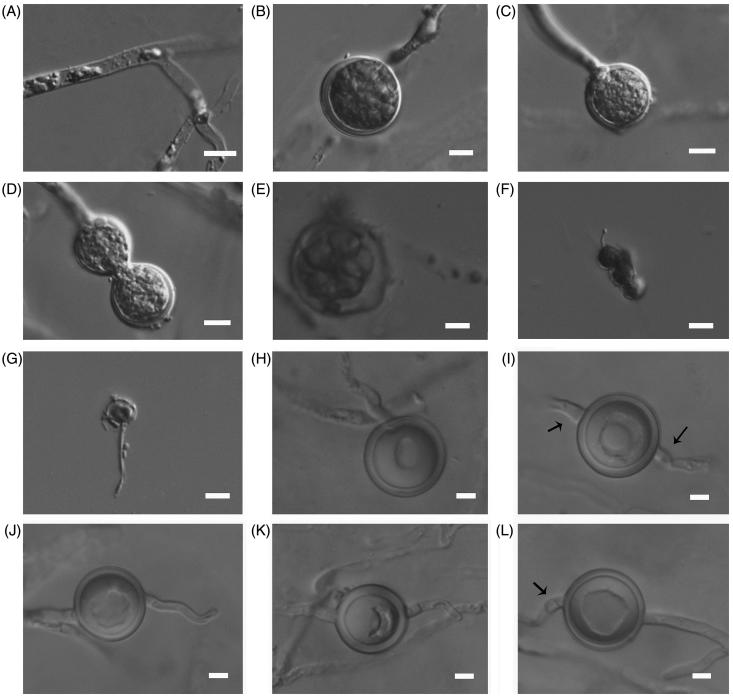
Figure 3.
Asexual and sexual reproductive bodies of P. subutonaiense (Chen 220). (A) Mycelia; (B,C) Globose, terminal hyphal swellings; (D) Catenulate hyphal swellings; (E) A vesicle with zoospores; (F,G) Germinated encysted zoospore; (H) Terminal oogonium with plerotic oospore; (I) Intercalary oogonium with a plerotic oospore; (J,K) Terminal oogonia with a projection; (L) Plerotic oospore and a hypogynous antheridium. Arrows indicate antheridia are hypogynous. Bars A–F = 10 μm, G–L = 5 μm.
MycoBank no.: MB 824731.
Type.—China. Jiangsu Prov., Nanjing, Zixiahu Park, from lake water, November 1, 2016, J.J. Chen, Chen 220 (NJAU, holotype).
Etymology—Subutonaiense (Lat.) referring to the species is somewhat similar to P. utonaiense [7].
Cardinal temperatures: minimum 5 °C, optimum 30 °C, maximum 38 °C. Main hyphae hyaline, aseptate, up to 5.0 µm wide. Hyphal swellings globose to sub-globose, mostly terminal or sometimes catenulate, 20–37.5 (mean 26.5) μm in diameter. Sporangia filamentous non-inflated, rarely giving rise to vesicles containing zoospores. Zoospores formed in SDW at room temperature, and encysted zoospores 5–7.5 μm (mean 6.5 μm) in diameter. Homothallic; oogonia globose, smooth or with one outstanding slender projection, intercalary, rarely terminal, 17.5–22.5 µm (mean 20.8 µm) in diameter. Antheridia hypogynous and monoclinous; antheridial stalks unbranched; antheridial cells slender. Oospore one per oogonium, plerotic, globose, 15.5–20.5 μm (mean 18.3 µm) in diameter, wall up to 0.5–2 (mean 1.6) µm thick.
Additional specimens examined.—CHINA. Jiangsu Prov., Nanjing, Soul Valley Temple, from lake water, November 1, 2016, J.J. Chen, Chen 229 (paratype, NJAU).
4. Discussion
P. subutonaiense is characterized by aquatic habit, homothallic sexuality, globose to sub-globose shaped, mostly terminal or sometimes catenulate hyphal swellings, filamentous non-inflated sporangia, and smooth oogonia, hypogynous, and monoclinous antheridia with slender antheridial cells, and plerotic oospores.
According to Lévesque and de Cock [26], Pythium could be split into 11 clades (A–K), of which the species in subclade B2 are characterized by filamentous non-inflated to slightly inflated sporangia, smooth oogonia mostly smaller than 30 mm in diameter, and a moderate growth rate (mostly 10–20 mm per day). Phylogenetic analysis based on ITS and COI sequences indicated P. subutonaiense belongs to Pythium subclade B2 with full statistical supports. P. subutonaiense shares several morphological characteristics with other Pythium subclade B2 species, such as filamentous sporangia, smooth oogonia, and moderate growth rate. However, P. subutonaiense can be readily distinguished in having globose to sub-globose shaped and mostly terminal or sometimes catenulate hyphal swellings.
P. subutonaiense has a closer relationship with P. utonaiense according to the ITS and COI-based phylogeny (Figure 2). P. utonaiense resembles P. subutonaiense in having aquatic habit, filamentous non-inflated sporangia, and plerotic oospores, but it is distinguished in its absence of hyphal swellings, faster growth rate, and the lack of antheridia [7] (Table 2).
P. brachiatum [7] is similar to P. subutonaiense by sharing aquatic habit and the formation of projections on oogonia, but the former species has slightly slower growth rate, lower optimum growth temperature, filamentous slightly inflated sporangia, and catenulate oogonia (Table 2). In addition, P. brachiatum is distant from P. subutonaiense in the ITS + COI sequence-based phylogeny [7] (Figure 2).
Both P. capillosum [27] and P. flevoense [28] have oogonia with a finger-like projection, and they somehow resemble P. subutonaiense; however, except for the lack of hyphal swellings, the two species can be distinguished from P. subutonaiense by slower growth rate, and thicker oospore wall [28] (Table 2). Besides, these three species clustered in different lineages in the phylogenetic analysis.
Funding Statement
The research was supported by the National Natural Science Foundation of China (31601618), and the Fundamental Research Funds for the Central Universities (KJQN201738) and the Natural Science Foundation of Jiangsu Province (BK20160737).
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- 1.Pringsheim N. Beiträge zur Morphology and Systematik der Algen. 2. Die Saprolegníeen. Jb Wiss Bot. 1858;1:284–306. [Google Scholar]
- 2.van der Plaäts-Niterink AJ. Monograph of the genus Pythium. Stud Mycol. 1981;21:1–242. [Google Scholar]
- 3.Ali-Shtayeh MS, Saleh ASF. Isolation of Pythium acanthicum, P. oligandrum, and P. periplocum from soil and evaluation of their mycoparasitic activity and biocontrol efficacy against selected phytopathogenic Pythium species. Mycopathologia. 1999;145:143–153. [Google Scholar]
- 4.Bala K, Robideau GP, Lévesque CA, et al. Phytopythium sindhum Lodhi. Shahzad & Levesque, sp. nov. Persoonia. 2010;24:136–137. [Google Scholar]
- 5.Uzuhashi S, Tojo M, Kakishima M. Phylogeny of the genus Pythium and description of new genera. Mycoscience. 2010;51:337–365. [Google Scholar]
- 6.Bouket AC, Arzanlou M, Tojo M, et al. Pythium kandovanense sp. nov., a fungus-like eukaryotic micro-organism (Stramenopila, Pythiales) isolated from snow-covered ryegrass leaves. Int J Syst Evol Microbiol. 2015;65:2500–2506. [DOI] [PubMed] [Google Scholar]
- 7.Uzuhashi S, Okada G, Ohkuma M. Four new Pythium species from aquatic environments in Japan. Antonie Van Leeuwenhoek. 2015;107:375–391. [DOI] [PubMed] [Google Scholar]
- 8.Abrinbana M, Badali F, Abdollahzadeh J. Molecular and morphological characterization of three new species of Pythium from Iran: P. ershadii, P. pyrioosporum, and P. urmianum. Mycologia. 2016;108:1175–1188. [DOI] [PubMed] [Google Scholar]
- 9.Ueta S, Tojo M. Pythium barbulae sp. nov. isolated from the moss, Barbula unguiculata; morphology, molecular phylogeny and pathogenicity. Mycoscience. 2016;57:11–19. [Google Scholar]
- 10.Chen J-J, Lü LI, Ye W-W, et al. Pythium cedri sp. nov. (Pythiaceae, Pythiales) from southern China based on morphological and molecular characters. Phytotaxa. 2017;309:135–142. [Google Scholar]
- 11.Veterano ST, Coffua LS, Mena-Ali JI, et al. Pythium yorkensis sp. nov., a potential soybean pathogen from southeastern Pennsylvania. Plant Pathol. 2018;67:619–625. [Google Scholar]
- 12.Bush EA, Hong CX, Stromberg EL. Fluctuations of Phytophthora and Pythium spp. in components of a recycling irrigation system. Plant Dis. 2003;87:1500–1506. [DOI] [PubMed] [Google Scholar]
- 13.Benard D, Punja ZK. Role of Pythium species in cavity spot development on carrots in British Columbia. Can J Plant Pathol. 1995;17:31–45. [Google Scholar]
- 14.Chang HS. Phytophthora species associated with strawberry fruit rot in Taiwan. Bot Bull Acad Sin. 1988;29:61–67. [Google Scholar]
- 15.Chen JJ, Cui BK. Studies on Wrightoporia from China 3. Wrightoporia subavellanea sp. nov. based on morphological characters and rDNA sequence data. Phytotaxa. 2014;175:225–234. [Google Scholar]
- 16.White TJ, Bruns T, Lee S, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics In: Innis MA, editor. PCR protocols: a guide to methods and applications. San Diego: Academic Press; 1990. p. 315–322. [Google Scholar]
- 17.Robideau GP, de Cock AWAM, Coffey MD, et al. DNA barcoding of oomycetes with cytochrome c oxidase subunit I and internal transcribed spacer. Mol Ecol Resour. 2011;11:1002–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blair JE, Coffey MD, Park SY, et al. A multi-locus phylogeny for Phytophthora utilizing markers derived from complete genome sequences. Fungal Genet Biol. 2008;45:266–277. [DOI] [PubMed] [Google Scholar]
- 19.Thompson JD, Gibson TJ, Plewniak F, et al. The Clustal_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 21.Swofford DL. PAUP*: phylogenetic analysis using parsimony (*and other methods). version 4.0b10. Sunderland: Sinauer Associates; 2002. [Google Scholar]
- 22.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. [DOI] [PubMed] [Google Scholar]
- 23.Nylander JAA. MrModeltest v2. Program distributed by the author. Uppsala, Sweden: Evolutionary Biology Centre, Uppsala University; 2004. [Google Scholar]
- 24.Ronquist F, Huelsenbeck JP. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. [DOI] [PubMed] [Google Scholar]
- 25.Page RMD. Treeview: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. [DOI] [PubMed] [Google Scholar]
- 26.Lévesque CA, de Cock AWAM. Molecular phylogeny and taxonomy of the genus Pythium. Mycol Res. 2004;108:1363–1383. [DOI] [PubMed] [Google Scholar]
- 27.van der Plaäts-Niterink AJ. The occurrence of Pythium in the Netherlands: III. Pythium flevoense sp.nov. Acta Bot Neerl. 1972;21:633–639. [Google Scholar]
- 28.Paul B. A new species of Pythium with filamentous sporangia from Algeria. Trans Br Mycol Soc. 1987;89:195–198. [Google Scholar]