Abstract
The transcription of the immediate-early genes Arcand Homer 1a (H1a) is dynamically regulated in response to synaptic activity; their protein products function at the postsynaptic sites of excitatory synapses. Previous studies demonstrate a role for Arc in the maintenance of long-term potentiation and in memory consolidation processes and indicate a role for H1a in modifying glutamatergic signaling pathways. Using double-label fluorescence in situ hybridization, we demonstrate that Arc andH1a RNA expression is induced strongly in the same neurons of rat hippocampus and neocortex after exploration of a novel environment. These findings support the view that novel experience activates a cell-specific genomic program and that Arcand H1a may function in concert in the structural and functional modifications of dendrites that lead to long-term changes in synaptic efficacy.
Keywords: memory, learning, transcription, hippocampus, cortex, immediate-early, gene, Arc, Homer 1, dendrite, plasticity
Research over the past few years has identified several proteins localized to the neuronal soma and dendrites that are rapidly and dynamically regulated by synaptic activity. These “effector” immediate-early genes (IEGs) have a number of cellular functions capable of modifying synaptic function (Lanahan and Worley, 1998). Two such effector IEGs are Arc(activity regulated cytoskeletal-associated protein, also known asArg3.1) (Link et al., 1995; Lyford et al., 1995) andHomer 1a (H1a) (Brakeman et al., 1997).Arc mRNA and protein can be selectively targeted to active regions of the dendritic arbor (Steward and Worley, 2001). Moreover, disrupting Arc protein expression in the hippocampus impairs the maintenance of long-term potentiation (LTP) and the consolidation of memory for spatial experience (Guzowski et al., 2000).H1a interacts with several proteins within the postsynaptic density (PSD) and may play an important role in modifying glutamatergic signaling pathways (Xiao et al., 2000).
As more is learned about the individual functions of different IEGs, it is becoming increasingly important to determine which genes function in concert, as part of the same plasticity mechanisms, within the same neurons. Here, we use fluorescence in situ hybridization (FISH) to demonstrate that the transcription of Arc andH1a is dramatically upregulated in the same hippocampal and neocortical neurons of rats after exploration of a novel environment. These findings raise the possibility that Arc andH1a might function in concert as part of an activity-dependent genomic program to induce and stabilize long-term changes in synaptic efficacy in neural networks encoding memory for specific experiences.
MATERIALS AND METHODS
Subjects, apparatus, and behavior. The subjects were male Sprague Dawley rats (Harlan Sprague Dawley, Indianapolis, IN), weighing 250–275 gm at arrival. Maximal electroconvulsive shock (MECS) was induced using a constant-current generator (ECT unit; Ugo Basile, Comerio, Italy) (Cole et al., 1990). For behavioral experiments, the rats were handled daily for 1 week before training to habituate them to the handling procedures. For the Arc/H1abehavioral time course experiment (see Figs. 2, 3), each rat (n = 3 per time point) sampled a novel environment for 5 min. The environment was a box divided into nine 400 cm2 grids surrounded by 30-cm-high walls. The box was positioned on a table in a room with ambient lighting, thus allowing the rats access to both local and distant visual cues. Each rat was picked up and released into the center of a different grid square every 15 sec, on a semirandom schedule. This procedure was used to ensure that the rats sampled the entire environment. At the end of the 5 min session, the rat was returned to its home cage until it was killed at the assigned time after exploration. Because rats have a strong tendency to explore novel environments, the rats (n = 6) used to establish the correlation betweenArc cytoplasmic and H1a intranuclear foci (INF) staining (see Fig. 4) were allowed to explore freely for 6 min a novel open box (61 × 61 cm with 24-cm-high sides), returned to their cages in the colony room, and killed 26 min later. All rats explored the novel environment completely, as evidenced by their multiple crossings of each floor grid (data not shown).
Fig. 2.
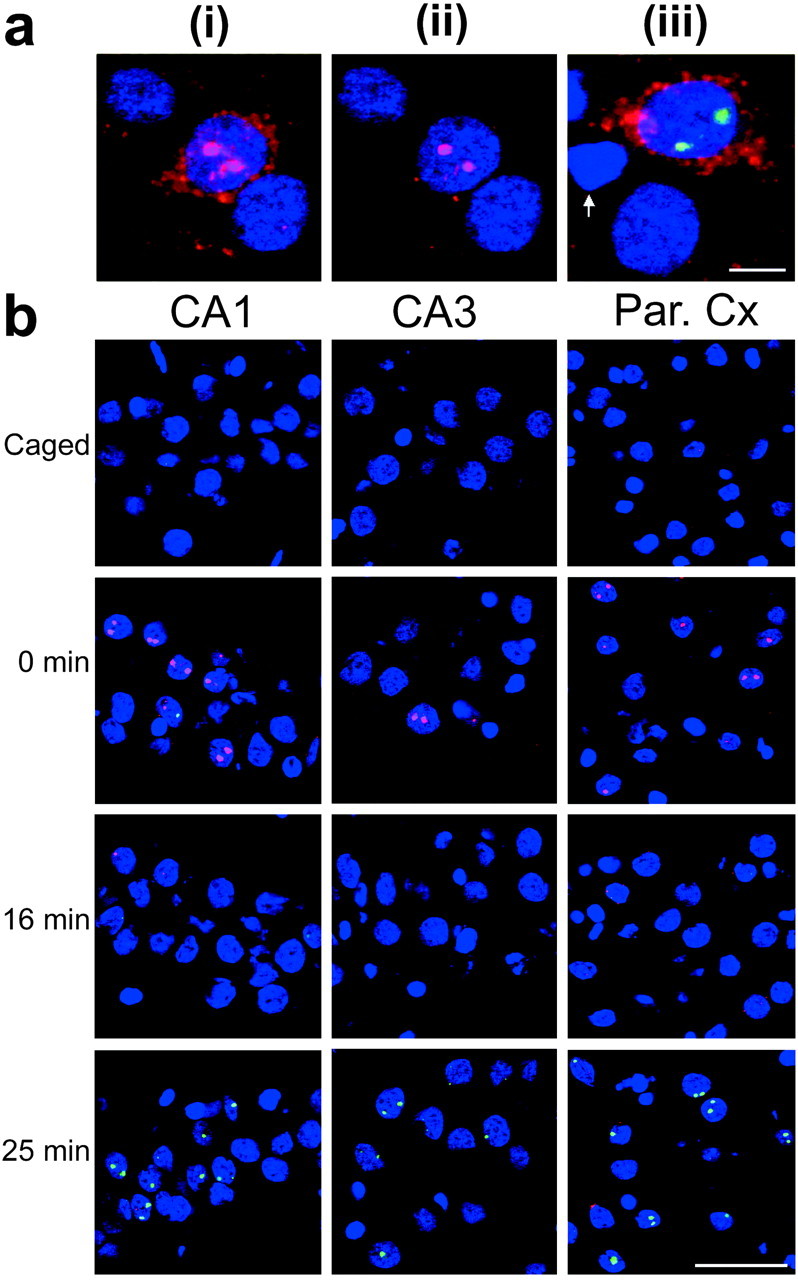
Dynamic appearance of Arc andH1a INF in CA1, CA3, and parietal cortical neurons after behavioral experience. a, Optimizing confocal microscope settings for the detection of either Arc cytoplasmic RNA labeling or Arc INF. i andii represent the same field imaged with different confocal settings: i, Clear Arccytoplasmic RNA labeling is achieved when the PMT offset is low;ii, unobstructed image of Arc INF is achieved by increasing the PMT offset; iii, an image optimized for detection of both Arc cytoplasmic labeling (red) and H1a INF (green). The white arrow points to a putative glial cell nuclei. b, Rats were exposed to a novel environment for 5 min and killed at the indicated time (in minutes) after removal (n = 3/group). Double-label FISH for Arc and H1a was performed on coronal brain sections as described in Materials and Methods. In these images, the confocal microscope settings were optimized for the detection of INF; the Arc signal is indicated inred, the H1a signal is indicated ingreen, and nuclei are indicated in blue. Note the predominance of the Arc INF at 0 min andH1a INF at 25 min in each brain region. Par. Cx, Parietal cortex. Scale bar, 50 μm.
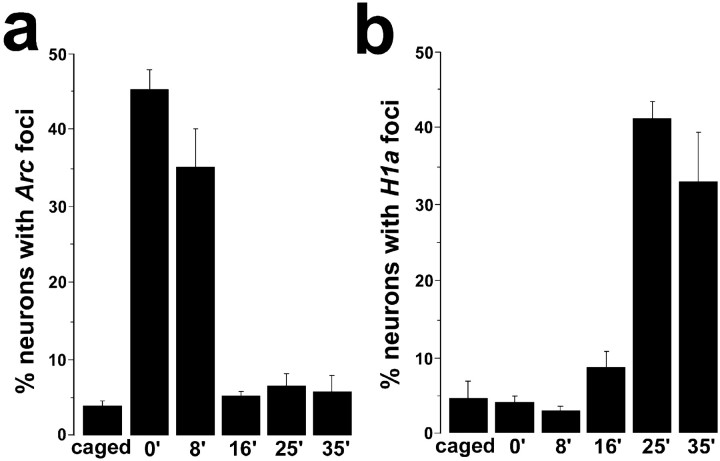
Fig. 3.
Experience-dependent appearance ofArc INF in CA1 neurons is rapid, transient, and does not coincide in time with the delayed appearance of H1a INF. Confocal Z-stacks of the CA1 region were collected fromArc/H1a double-label slides from rats killed at different delays after behavioral exploration (see Results and Fig. 2). The percentage of counted nuclei positive for Arc(a) or H1a(b) INF is indicated for each time point. Note that compared with the caged control group, the percentage of neurons with Arc INF was significantly higher only in the 0 and 8 min groups, whereas the percentage of neurons with H1aINF was significantly higher only in the 25 and 35 min groups.
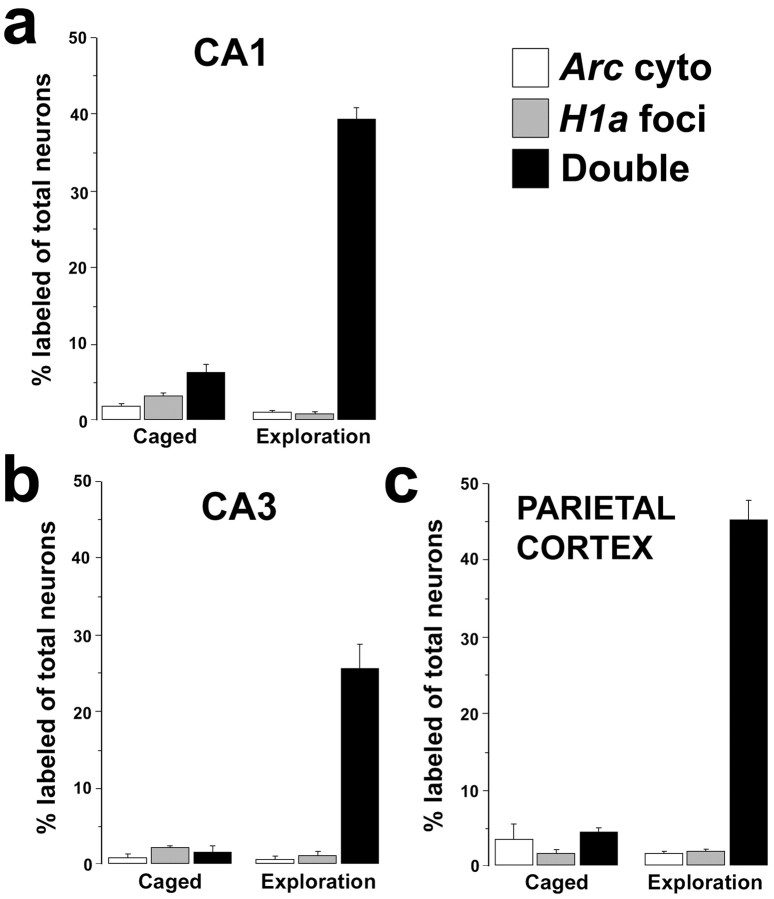
Fig. 4.
Exploration of a novel environment induces the coincident expression of Arc and H1a in single neurons of the rat hippocampus and parietal cortex. One group of rats was exposed to a novel environment for 6 min, returned to their home cages in the colony room for 26 min, and then killed (exploration group; n = 6). A separate group of rats was killed directly from their home cages (caged control group;n = 3). Double-label FISH for Arcand H1a was performed on coronal brain sections, and image stacks were collected and analyzed from CA1, CA3, and parietal cortical regions. The percentage of neurons in a respective brain region expressing Arc cytoplasmic RNA staining only (white bars), H1a INF only (gray bars), or both (black bars) is shown for rats from both groups. Note that the only staining class population that changed with exploration (relative to the caged controls) was the Arc cytoplasmic+/H1aINF+ double-label class (black bars). Arc cyto, Arc cytoplasmic.
FISH. After decapitation with a rodent guillotine, the brains were removed rapidly, quick-frozen in isopentane (approximately −50°C), and then stored at −70°C until being sectioned on a cryostat. Twenty-micrometer-thick sections were mounted on slides such that all groups were represented on each slide. Digoxigenin- or fluorescein-labeled riboprobes were generated using commercial transcription kits (MaxiScript; Ambion, Austin, TX) and RNA labeling mixes (Roche Products, Hertforshire, UK). The plasmid used to generate the Arc antisense and sense riboprobes contained a full-length cDNA (3.0 kbp) of the Arc transcript (Lyford et al., 1995). The H1a antisense riboprobe was generated using an H1a cDNA clone and was directed to the 4.4 kb 3′ untranslated region (UTR) of the H1a mRNA (Brakeman et al., 1997). Single- or double-label FISH was performed as described in detail previously (Guzowski et al., 1999; Guzowski and Worley, 2001;Bottai et al., 2002). In Arc/H1a double-labeling studies, digoxigenin-labeled Arc riboprobe was detected with anti-digoxigenin–HRP (Roche Products) and a cyanine-3 substrate kit (CY3 DirectFISH; PerkinElmer Life Sciences, Emeryville, CA). After detection of the Arc riboprobe, the slides were treated with 2% H2O2 to quench residual HRP activity. Fluorescein-labeled H1a probe was then detected with anti-fluorescein HRP (Roche Products) and a cyanine-5 substrate kit (CY5 DirectFISH; PerkinElmer Life Sciences). Nuclei were counterstained with either 4′,6′-diamidino-2-phenylindole (DAPI; Vector Laboratories, Burlingame, CA) (see Fig. 1) or YOYO-1 (quinolinium,1,1′-[1,3-propanediylbis [(dimethyliminio)-3,1-propanediyl]]bis[4-[(3-methyl-2(3H)-benzoxazolylidene)methyl]]-tetraiodide; Molecular Probes, Eugene, OR). The specificity of the labeling was confirmed by two control conditions. Some slides were hybridized withArc and H1a sense riboprobes; on other slides the riboprobe was omitted. For both control conditions, the remaining detection steps were performed without modification from the standard procedure.
Fig. 1.
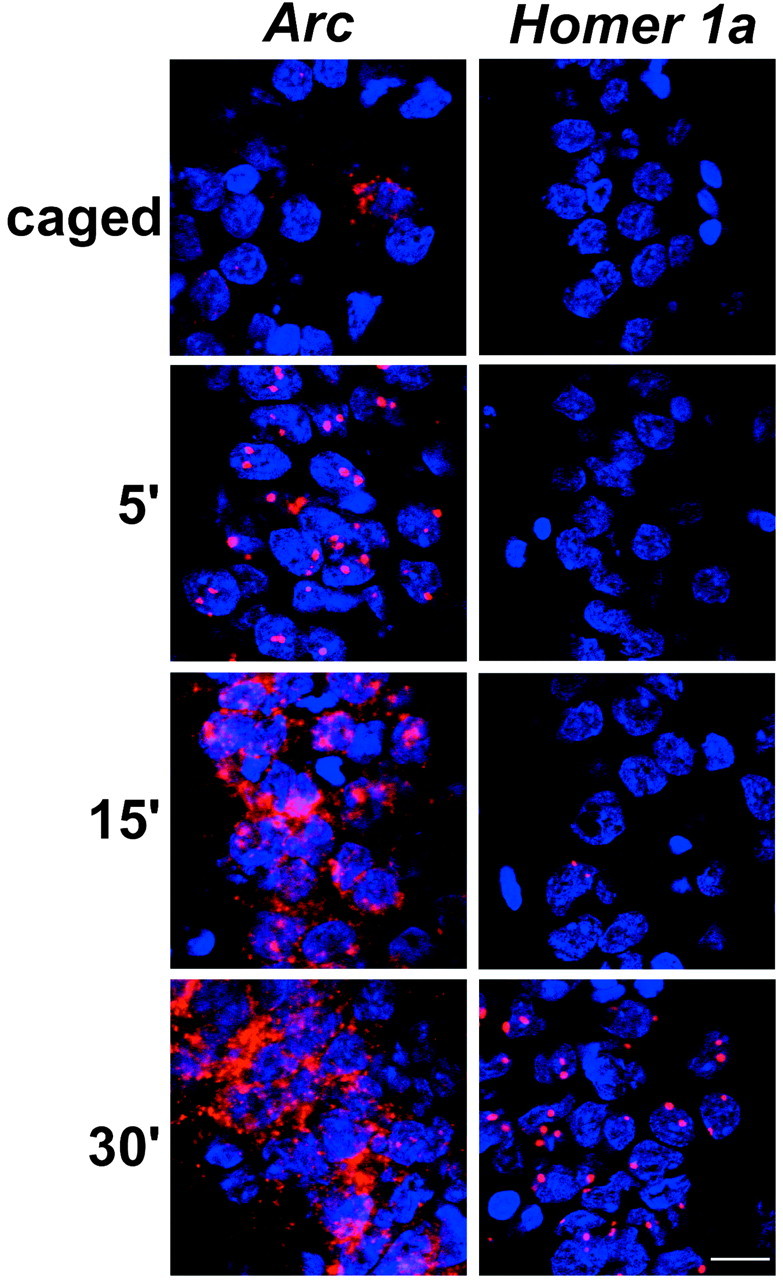
Distinct temporal profiles of Arcand H1a RNA appearance in CA1 neurons after MECS. Rats were killed at the indicated time (in minutes) after MECS.Arc and H1a RNAs were detected with digoxigenin-label antisense riboprobes as described in Materials and Methods. IEG RNAs were detected with CY3 (red), and nuclei were counterstained with DAPI (blue). Scale bar, 20 μm.
Image acquisition and analysis. Images were acquired using a Leica (Nussloch, Germany) TCS-4D confocal microscope equipped with a krypton–argon laser. Photomultiplier tube (PMT) assignments, pinhole size, and contrast values were kept constant for each brain region within a slide. Depending on the required analysis, the settings were adjusted to optimize either the appearance of cytoplasmic labeling or that of INF (see Fig. 2a). Optimizing for INF was achieved by increasing the offset of the PMT. Figure 2a,iii, shows an image optimized for the appearance ofArc cytoplasmic signal and H1a INF. Z-stacks of either 1- or 2-μm-thick optical sections were acquired with either a 40× oil or 20× objective lens, respectively. Images for the behavioral time course analysis of Arc and H1aINF were collected at 20× magnification (one Z-stack per slide in two different slides, totaling 104–170 cells per area per slide). For the Arc/H1a correlation study (see Fig. 4), cell counts for each rat varied between 79 and 124 for CA1, 43 and 54 for CA3, and 73 and 127 for the parietal cortex.
Only putative neurons were included in the analyses. Putative glial-cell nuclei were identified based on their small size (∼5 μm in diameter) and bright, uniform nuclear counterstaining (see Fig.2a, iii). Furthermore, these cells did not express Arc or H1a, consistent with a previous report that Arc was expressed predominantly, if not exclusively, in excitatory neurons (Cirelli and Tononi, 2000). Z-stacks were analyzed using MetaMorph software (Universal Imaging Corporation, West Chester, PA). First, neuronal nuclei present in the median planes (representing 20% of the stack thickness) were identified and outlined. Then nuclei were characterized for the presence ofArc and H1a INF (see Fig. 3) or Arccytoplasmic labeling and H1a INF (see Fig. 4). The results were expressed as a percentage of the total neuronal nuclei analyzed per stack. To prevent bias, the experimenter was unaware of the relationship between the images and the behavioral conditions they represented. The median planes criterion reduced the likelihood of analyzing partial nuclei, which could yield false negative results. This approach is essentially an optical dissector technique that minimizes sampling errors attributable to partial cells and stereological concerns, because variations in cell volumes do not influence sampling frequencies (West, 1993).
Statistical analyses. The main effect of treatment (e.g., caged/exploration or time after the end of behavioral testing) was evaluated by ANOVA. When the main effect was significant at the α = 0.05 level, additional comparisons between groups were conducted with Fisher post hoc tests (Statview software; Statview, Berkeley, CA).
RESULTS
Previous characterization of Homer 1 transcriptional regulation in mice demonstrated that the appearance of the Homer 1 RNA signal after MECS was dependent on the location of the riboprobe along the primary transcript (Bottai et al., 2002). Here, we examined the time course of the appearance of the signal from an antisense riboprobe specific to the 3′UTR of rat H1a. This probe distinguishes synaptic activity-dependent H1atranscripts from the constitutive Homer 1 forms (Xiao et al., 2000). These results were compared with those obtained using an antisense riboprobe generated from the entire Arc cDNA. Rats received MECS to activate IEG expression in the hippocampus and cortex. As reported previously (Guzowski et al., 1999), the Arc RNA signal was detected in INF in ∼95% of the neurons (see Materials and Methods) in the hippocampus (Fig. 1,left) and the parietal cortex within 5 min of MECS. IEG INF appear as either one or two (usually two) distinct areas of intense fluorescent staining within neuronal nuclei (Fig. 1). INF indicate the genomic sites of IEG RNA synthesis (Guzowski et al., 1999; Bottai et al., 2002). By 30 min, the Arc signal was prominent in the cytoplasm, and the proportion of INF-positive cells was similar to baseline (Fig. 1, left). H1a RNA was also detected in most hippocampal (Fig. 1, right) and parietal cortical neurons. However, H1a INF did not appear until 30 min after MECS (Fig. 1, right). The delayed appearance of the H1a INF is explained by the relative position of the 3′UTR riboprobe along the primary transcript (∼40 kb from the transcript start site) (Bottai et al., 2002) and the limited elongation rate of RNA polymerase II (∼1.4 kb/min) (Femino et al., 1998). These data show that with a strong stimulus, both Arc andH1a can be induced in the majority of the hippocampal and cortical neurons.
Next, we examined the kinetics of Arc and H1a RNA appearance after open field exploration (a behavior that induces place-cell activity in hippocampal neurons). Rats were exposed to a novel environment for 5 min and then killed at 0, 8, 16, 25, or 35 min after exploration (n = 3/group). Rats that were killed after a delay were returned to their home cage in the colony room between exploration and being killed. The brains were processed forArc and H1a double-label FISH, and confocal images were acquired for qualitative (Fig.2) or quantitative (Fig.3) analysis. The dynamics ofArc and H1a transcription were similar in the hippocampal CA1 and CA3 regions and in the parietal cortex (Fig.2b). In each region, the greatest proportion ofArc INF-positive cells was seen in the group killed immediately after exploration. Furthermore, the proportion ofArc INF-positive cells returned to control levels within 16 min after exploration. In contrast, an increase in the proportion ofH1a INF-positive cells above control levels was not seen until 25 min after exploration.
Quantitative analysis of Z-stacks from CA1 confirmed the above observations for both Arc (Fig. 3a) andH1a (Fig. 3b). For Arc cell counts, there was a significant effect of time of death (overall ANOVA;F(5,12) = 51.34; p < 0.0001), and post hoc comparisons revealed significant differences between the caged and 0 min groups, caged and 8 min groups, and 8 min and 16 min groups (p < 0.0001 for all three comparisons). The proportions of ArcINF-positive cells in the 16, 25, and 35 min groups were not statistically different from the caged group. In contrast, the percentage of H1a INF-positive CA1 neurons was comparable with that of caged controls until 25 min after exploration (Fig.3b) (overall ANOVA; F(5,12)= 26.97; p < 0.0001). Post hoc comparisons revealed significant differences between the caged and 25 min groups and the caged and 35 min groups (p < 0.0001), whereas those between the caged and 0, 8, and 16 min groups did not reveal a significant effect. In addition, the percentage ofH1a INF-positive neurons in the 25 min postexploration group was significantly higher than that of the 16 min group (p < 0.0001).
The finding that Arc and H1a were expressed in a similar proportion of hippocampal and parietal cortical neurons of rats that had explored a novel environment is consistent with the idea that these genes might be dynamically regulated in the same neurons. To test this possibility directly, the correspondence of neurons containing Arc cytoplasmic labeling andH1a INF in the hippocampus and cortex of caged control rats was compared with rats that had explored a novel environment 30 min before being killed. Neurons were scored as containing Arccytoplasmic labeling only, H1a INF only, or bothArc cytoplasmic labeling and H1a INF (Fig.2a, iii). In the hippocampus (CA1 and CA3 regions) and the parietal cortex of the rats from the exploration group, the correspondence of Arc cytoplasmic labeling andH1a INF was >90% (Fig. 4) (95% for CA1, 94% for CA3, and 93% for the parietal cortex).
DISCUSSION
The current findings demonstrate that transcription of the IEGsArc and H1a is dynamically regulated by physiological activity in the same hippocampal and cortical neurons. Double-label FISH revealed distinct temporal and spatial patterns ofArc and H1a RNA appearance after MECS (Fig. 1) and behavioral experience (Figs. 2-4). The highest proportion ofArc INF-positive cells was observed in the rats killed immediately after exploration (Figs. 2, 3), indicating rapid induction of Arc transcription. This period of transcription is very brief, because the proportion of Arc INF-positive cells returned to control levels within 16 min after exploration, andArc mRNA was prominent within the cytoplasm by 30 min (Figs.2-4) (Guzowski et al., 1999). In contrast, the proportion ofH1a INF-positive cells detected with the 3′UTR riboprobe did not change until 25 min after exploration. At that time, the proportion of H1a INF-positive cells was highest and was similar to that of Arc INF-positive neurons from the rats killed immediately after exploration (Figs. 2, 3). Double-label FISH performed on brain sections of rats that had explored a novel environment 30 min before being killed revealed >90% correspondence of neurons positive for cytoplasmic Arc mRNA and H1a INF (Fig. 4). Thus, with a discrete behavioral stimulus of known onset and limited duration, transcriptional activation of Arc andH1a is coincident in single neurons of the hippocampus and parietal cortex.
The coincident transcription of Arc and H1a may be regulated by the mitogen-activated protein kinase (MAPK)/extracellular signal-regulated protein kinase (ERK) cascade. The finding that a MAPK/ERK inhibitor blocked forskolin-inducedArc transcription in cultured hippocampal neurons (Waltereit et al., 2001) is consistent with this hypothesis. In addition, pretreatment with MAPK kinase (MEK) inhibitors blocked LTP and prevented increases in ERK2 and cAMP response element-binding protein phosphorylation as well as increases in Arc RNA expression caused by the local infusion of BDNF in the dentate gyrus of intact rats (Ying et al., 2002). Similarly, MEK inhibitors blocked glutamate-induced increases in H1a transcription in cultured cerebellar granule cells (Sato et al., 2001). Thus, the coincident transcription of Arc and H1a demonstrated here could result from the activation of the MAPK/ERK pathway rather than from the activation of separate, parallel pathways acting on the promoters of these genes. However, it must be noted that althoughArc and H1a may use the same mechanisms for transcriptional activation, H1a is subject to additional levels of activity-dependent regulation, including the use of an alternative transcription termination signal and conversion of an intronic sequence (in constitutive forms) to an exonic sequence (in IEG forms) (Bottai et al., 2002).
The distinct temporal patterns of Arc and H1a INF are explained by the different architectures of the transcription units for these genes (Arc mRNA is derived from a limited primary transcript; ∼3.5 kb with two small introns; GenBank accession numberAF177701), whereas H1a mRNA is derived from a primary transcript spanning ∼50 kb (Bottai et al., 2002). Thus, the time course of appearance of H1a INF is dependent on the position of the riboprobe along the primary transcript: Riboprobes to either exon 1 or intron 1 of Homer 1 detect H1a INF with induction kinetics indistinguishable from that of ArcINF (Bottai et al., 2002), whereas those to the 3′UTR detectH1a INF with much delayed kinetics (Figs. 1-4) (Bottai et al., 2002).
The coincident expression of Arc and H1a seen after experience (Fig. 4) and the temporally offset appearance/disappearance of Arc and H1a INF (Fig.3) enable an important modification of the cellular analysis of temporal activity by FISH (catFISH) brain imaging method (Guzowski et al., 1999, 2001). The power of catFISH is its ability to identify, at a single-cell level, neuronal ensembles activated by two distinct behavioral experiences within an animal, a property that distinguishes catFISH from other imaging methods. In the original method,Arc INF indicate cells active in the 5–10 min preceding death, whereas the cytoplasmic Arc signal indicates cells active ∼30 min before death. Although catFISH has been used successfully to determine neural activity for two experiences (Guzowski et al., 1999), quantifying the cytoplasmic IEG RNA signal can be difficult when many cells are activated in a region with a high cell density, such as the pyramidal cell layers of the hippocampus. However, the combined use of Arc and 3′UTR H1a riboprobes in double-label FISH circumvents this problem by exploiting the transcription rate of RNA polymerase II (∼1.4 kb/min) (Femino et al., 1998) and the natural “genomic timers” of cellular activation afforded by the dissimilar gene structures of Arc andH1a (Bottai et al., 2002). With this approach (Arc/H1a catFISH), the activity history of many neurons can be distinguished based solely on strong intranuclear signals:Arc INF indicate cells activated by an experience shortly before (<15 min) death, whereas H1a INF indicate cells activated at least 25 min earlier. The exclusive use of intranuclear signals greatly facilitates manual analysis and makes catFISH amenable to computer automation for large-scale investigations of neural population interactions during learning and memory.
Several lines of evidence suggest that both Arc andH1a are involved in synaptic plasticity. First, the expression of both genes is increased dramatically in the dentate gyrus after the induction of perforant-path LTP (Link et al., 1995; Lyford et al., 1995; Brakeman et al., 1997) and in the insular cortex after the LTP-inducing stimulation of the amygdala (Jones et al., 1999). Moreover, inhibiting hippocampal Arc protein expression disrupts the maintenance of perforant-path LTP and impairs memory consolidation (Guzowski et al., 2000). Biochemical evidence indicates that Arc may be a component of NMDA receptor complexes (Plath et al., 2001). The Homer 1 gene is alternatively spliced from a large primary transcript to form two constitutively expressed forms (Homer 1b and Homer 1c) and two synaptic activity-regulated IEG forms (H1a andania3) (Xiao et al., 1998, 2000; Bottai et al., 2002). Homer 1b/c proteins are implicated in coupling group 1 metabotropic glutamate receptors (mGluR) with IP3 and ryanodine receptors and in coupling either of these intracellular receptors with the NMDA receptor-associated PSD-95 complex (Xiao et al., 2000). It has been hypothesized that Homer 1 IEG forms, which lack the C-terminal coiled-coil domain of the constitutive forms, act as natural antagonists to Homer 1b/c forms to modify glutamatergic signaling pathways (Xiao et al., 2000). Consistent with this hypothesis,H1a transgene expression in Purkinje neurons alters mGluR-induced Ca2+ release from intracellular stores (Tu et al., 1998).
Arc transcription has been shown previously to be activated in CA1 neurons in an environmental context-specific manner (Guzowski et al., 1999), as observed for place-cell firing activity in hippocampal neurons. The striking concordance of Arc and H1acoincident expression shown here indicates that, like Arc,H1a is induced in the neural networks engaged in information processing. The fact that both Arc and H1a are dynamically expressed in the same neurons by experience and are localized to the postsynaptic density suggests that these genes may function in concert to modify synaptic efficacy in the hippocampal and neocortical networks responsible for encoding the memory of discrete experiences.
Footnotes
This work was supported by National Institutes of Health Grants MH60123 (J.F.G.), AG09219 (C.A.B. and P.F.W.), MH01565 (B.L.M.), and AG18230 (C.A.B., J.F.G., B.L.M., and P.F.W.). We thank Beth Takacs for assistance with the behavioral training of the rats.
Correspondence should be addressed to Dr. J. F. Guzowski, Department of Neurosciences, Basic Medical Sciences Building, Room 145, University of New Mexico, Health Sciences Center, Albuquerque, NM 87131-5223. E-mail: jguzowski@salud.unm.edu.
REFERENCES
- 1.Bottai D, Guzowski JF, Schwarz MK, Kang SH, Xiao B, Lanahan A, Worley PF, Seeburg PH. Synaptic activity-induced conversion of intronic to exonic sequence in Homer 1 immediate-early gene expression. J Neurosci. 2002;22:167–175. doi: 10.1523/JNEUROSCI.22-01-00167.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brakeman PR, Lanahan AA, O'Brien R, Roche K, Barnes CA, Huganir RL, Worley PF. Homer: a protein that selectively binds metabotropic glutamate receptors. Nature. 1997;386:284–288. doi: 10.1038/386284a0. [DOI] [PubMed] [Google Scholar]
- 3.Cirelli C, Tononi G. Differential expression of plasticity-related genes in waking and sleep and their regulation by the noradrenergic system. J Neurosci. 2000;20:9187–9194. doi: 10.1523/JNEUROSCI.20-24-09187.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cole AJ, Abu-Shakra S, Saffen DW, Baraban JM, Worley PF. Rapid rise in transcription factor mRNAs in rat brain after electroshock-induced seizures. J Neurochem. 1990;55:1920–1927. doi: 10.1111/j.1471-4159.1990.tb05777.x. [DOI] [PubMed] [Google Scholar]
- 5.Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 6.Guzowski JF, Worley PF. Cellular compartment analysis of temporal activity by fluorescence in situ hybridization (catFISH). In: Taylor GP, editor. Current protocols in neuroscience. Wiley; New York: 2001. pp. 1.8.1–1.8.16. [DOI] [PubMed] [Google Scholar]
- 7.Guzowski JF, McNaughton BL, Barnes CA, Worley PF. Environment-specific expression of the immediate-early gene Arc in hippocampal neuronal ensembles. Nat Neurosci. 1999;2:1120–1124. doi: 10.1038/16046. [DOI] [PubMed] [Google Scholar]
- 8.Guzowski JF, Lyford GL, Stevenson GD, Houston FP, McGaugh JL, Worley PF, Barnes CA. Inhibition of activity-dependent arc protein expression in the rat hippocampus impairs the maintenance of long-term potentiation and the consolidation of long-term memory. J Neurosci. 2000;20:3993–4001. doi: 10.1523/JNEUROSCI.20-11-03993.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guzowski JF, McNaughton BL, Barnes CA, Worley PF. Imaging neural activity with temporal and cellular resolution using FISH. Curr Opin Neurobiol. 2001;11:579–584. doi: 10.1016/s0959-4388(00)00252-x. [DOI] [PubMed] [Google Scholar]
- 10. Jones MW, French PJ, Bliss TV, Rosenblum K. Molecular mechanisms of long-term potentiation in the insular cortex in vivo. J Neurosci 19 1999. RC36:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lanahan A, Worley P. Immediate-early genes and synaptic function. Neurobiol Learn Mem. 1998;70:37–43. doi: 10.1006/nlme.1998.3836. [DOI] [PubMed] [Google Scholar]
- 12.Link W, Konietsko U, Kauselmann G, Krug M, Schwanke B, Frey U, Kuhl D. Somatodendritic expression of an immediate-early gene is regulated by synaptic activity. Proc Natl Acad Sci USA. 1995;92:5734–5738. doi: 10.1073/pnas.92.12.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lyford GL, Yamagata K, Kaufmann WE, Barnes CA, Sanders LK, Copeland NG, Gilbert DJ, Jenkins NA, Lanahan AA, Worley PF. Arc, a growth factor and activity-regulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron. 1995;14:433–445. doi: 10.1016/0896-6273(95)90299-6. [DOI] [PubMed] [Google Scholar]
- 14.Plath N, Ohana O, Dammermann B, Waltereit R, Husi H, Blanquet V, Wurst W, Bosi M, Grant SG, Kuhl D. Aberrant LTP in Arg3.1/Arc knockout animals. Soc Neurosci Abstr. 2001;27:611.12. [Google Scholar]
- 15.Sato M, Suzuki K, Nakanishi S. NMDA receptor stimulation and brain-derived neurotrophic factor upregulate homer 1a mRNA via the mitogen-activated protein kinase cascade in cultured cerebellar granule cells. J Neurosci. 2001;21:3797–3805. doi: 10.1523/JNEUROSCI.21-11-03797.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steward O, Worley PF. Selective targeting of newly synthesized Arc mRNA to active synapses requires NMDA receptor activation. Neuron. 2001;30:227–240. doi: 10.1016/s0896-6273(01)00275-6. [DOI] [PubMed] [Google Scholar]
- 17.Tu JC, Xiao B, Yuan JP, Lanahan AA, Leoffert K, Li M, Linden DJ, Worley PF. Homer binds a novel proline-rich motif and links group 1 metabotropic glutamate receptors with IP3 receptors. Neuron. 1998;21:717–726. doi: 10.1016/s0896-6273(00)80589-9. [DOI] [PubMed] [Google Scholar]
- 18.Waltereit R, Dammermann B, Wulff P, Scafidi J, Staubli U, Kauselmann G, Bundman M, Kuhl D. Arg3.1/Arc mRNA induction by Ca2+ and cAMP requires protein kinase A and mitogen-activated protein kinase/extracellular regulated kinase activation. J Neurosci. 2001;21:5484–5493. doi: 10.1523/JNEUROSCI.21-15-05484.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.West MJ. New stereological methods for counting neurons. Neurobiol Aging. 1993;14:275–285. doi: 10.1016/0197-4580(93)90112-o. [DOI] [PubMed] [Google Scholar]
- 20.Xiao B, Tu JC, Petralia RS, Yuan JP, Doan A, Breder CD, Ruggiero A, Lanahan AA, Wenthold RJ, Worley PF. Homer regulates the association of group 1 metabotropic glutamate receptors with multivalent complexes of homer-related, synaptic proteins. Neuron. 1998;21:707–716. doi: 10.1016/s0896-6273(00)80588-7. [DOI] [PubMed] [Google Scholar]
- 21.Xiao B, Tu JC, Worley PF. Homer: a link between neural activity and glutamate receptor function. Curr Opin Neurobiol. 2000;10:370–374. doi: 10.1016/s0959-4388(00)00087-8. [DOI] [PubMed] [Google Scholar]
- 22.Ying SW, Futter M, Rosenblum K, Webber MJ, Hunt SP, Bliss TV, Bramham CR. Brain-derived neurotrophic factor induces long-term potentiation in intact adult hippocampus: requirement for ERK activation coupled to CREB and upregulation of Arc synthesis. J Neurosci. 2002;22:1532–1540. doi: 10.1523/JNEUROSCI.22-05-01532.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]