Abstract
Despite the importance of myelinating Schwann cells in health and disease, little is known about the genetic mechanisms underlying their development. The POU domain transcription factor pou3f1 (Tst-1, SCIP, Oct-6) is required for the normal differentiation of myelinating Schwann cells, but its precise role requires identification of the genes that it regulates. Here we report the isolation of six genes whose expression is reduced in the absence of pou3f1. Only one of these genes, the fatty acid transport protein P2, was known previously to be expressed in Schwann cells. The LIM domain proteins cysteine-rich protein-1 (CRP1) and CRP2 are expressed in sciatic nerve and induced by forskolin in cultured Schwann cells, but only CRP2 requires pou3f1 for normal expression. pou3f1 appears to require the claw paw gene product for activation of at least some of its downstream effector genes. Expression of the novel Schwann cell genes after nerve injury suggests that they are myelin related. One of the genes,tramdorin1, encodes a novel amino acid transport protein that is localized to paranodes and incisures. Our results suggest that pou3f1 functions to activate gene expression in the differentiation of myelinating Schwann cells.
Keywords: myelin, pou3f1, Oct-6, Tst-1, SCIP, claw paw, Schwann cells, tramdorin, dendrin, CRP1, CRP2, P2, representational difference analysis
Schwann cells generate myelin in the peripheral nervous system. One myelinating Schwann cell forms a single myelin sheath around a single axon, whereas nonmyelinating Schwann cells typically ensheathe several unmyelinated axons without myelinating any of them. Axons are thought to regulate the differentiation of Schwann cells; presumptive myelinated axons signal Schwann cells into the myelinating lineage, whereas the axons lacking this signal remain associated with nonmyelinating Schwann cells (Mirsky and Jessen, 1996, 1999; Taylor and Suter, 1997; Garbay et al., 2000).
pou3f1 [also known as octomer-6 (Oct-6), Testes-1 (Tst-1), suppressed cAMP-inducible POU (SCIP), Octamer transcription factor-6 (Otf6)] is a member of the POU domain family of transcription factors (for review, see Ryan and Rosenfeld, 1997). pou3f1 is expressed transiently during Schwann cell development (Monuki et al., 1989), specifically in promyelinating Schwann cells, which have formed a 1:1 relationship with axons but have not yet formed a myelin sheath (Zorick et al., 1996;Arroyo et al., 1998). In the absence of pou3f1, the differentiation of myelinating Schwann cells is delayed, transiently arrested at the promyelinating stage (Bermingham et al., 1996; Jaegle et al., 1996).
The role of pou3f1 in Schwann cell differentiation is unknown. It has often been considered to be a repressor of Schwann cell gene expression and/or differentiation. Cotransfection experiments suggest that pou3f1 inhibits the expression of the P0(Mpz), myelin basic protein (Mbp), and p75(low-affinity neurotrophin receptor) genes (Monuki et al., 1990, 1993;He et al., 1991). Furthermore, an N-terminal deletion of pou3f1 (ΔSCIP) acts as a dominant-negative inhibitor of pou3f1 repression of a 1.1 kb rat Mpz (P0) promoter in cotransfection experiments. In transgenic mice, ΔSCIP causes precocious PNS myelination, taken as evidence that pou3f1 represses the progression from promyelinating Schwann cells to myelinating Schwann cells (Weinstein et al., 1995). However, Schwann cells inpou3f1-null mice show delayed differentiation, and their expression of myelin-related mRNAs, including those of Mpzand Mbp, was not higher than in wild-type mice (Bermingham et al., 1996; Jaegle et al., 1996). These results contradict the idea that pou3f1 represses the expression of these genes in Schwann cells. To reconcile these observations, pou3f1 has been hypothesized to activate genes required for Schwann cell differentiation but to repress terminal differentiation (Mirsky and Jessen, 1996; Zorick and Lemke, 1996; Jaegle and Meijer, 1998). In this scheme, pou3f1-null mice lack both functions, whereas ΔSCIP inhibits only the repression function.
Our approach sought to identify target genes of pou3f1 in Schwann cells to better understand the mechanisms by which it controls their differentiation. Representational difference analysis (RDA), a PCR-based technique that permits the isolation of DNA fragments that are present in one DNA sample but absent in another (Lisitsyn and Wigler, 1993; Lisitsyn, 1995), was used with a carrier RNA facilitating the isolation of differentially expressed genes (Erkman et al., 2000; Bermingham et al., 2001). We found six genes that are misexpressed in the sciatic nerves of pou3f1-null mice, and the expression of five of these genes correlates with the expression of other myelin-related genes after nerve injury. Because all of these mRNAs are expressed at lower levels in the sciatic nerves ofpou3f1-null mice, pou3f1 appears to be an activator of gene expression in Schwann cells. The identification of these genes indicates that pou3f1 functions to induce membrane biogenesis, cytoskeletal rearrangement, and amino acid transport during the differentiation of myelinating Schwann cells.
MATERIALS AND METHODS
Isolation of RNA from cultured Schwann cells. Schwann cells were isolated from 3-d-old rat pups (Brockes et al., 1979) and expanded on 10 cm plates coated with poly-l-lysine in DMEM supplemented with 10% FCS, 2 μm forskolin (Porter et al., 1987), and 10 ng/ml recombinant human secreted β isoform of neuregulin (glial growth factor-2). The cells were passaged six times, grown to confluence, and then maintained for 3 d in DMEM and 10% FCS. The cells were maintained for an additional 3 d in DMEM and 10% FCS alone or supplemented with 20 μm forskolin. All of the cultures used in these experiments were essentially free of fibroblasts. RNA was isolated from rat sciatic nerves and Schwann cells by CsCl2 gradient centrifugation (Chirgwin et al., 1979).
Isolation of RNA from sciatic nerves from lesioned rat sciatic nerves. Adult Sprague Dawley rats were anesthetized with 50 mg/kg pentobarbital intraperitoneally, and the sciatic nerves were exposed at the obturator tendon. To prevent axonal regeneration, nerves were doubly ligated and transected between the ligatures. Nerves were crushed by compression with flattened forceps twice, each time for 10 sec. Animals were allowed to survive for various periods of time before being killed by CO2 inhalation. For RNA extraction, several millimeters of nerve adjacent to the lesion site were trimmed off, and the distal nerve stumps were frozen in liquid nitrogen. Where indicated, the distal stumps of crushed nerves were subdivided into proximal and distal segments of equal lengths. Unlesioned nerves were taken from animals of various ages. Total RNA was isolated by CsCl2 gradient centrifugation (Chirgwin et al., 1979).
Isolation of RNA from sciatic nerves from newborn mice.pou3f1TM1Rsd mutant mice (Bermingham et al., 1996) were maintained either on a pure 129Sv genetic background or on a mixed 129/Sv-C57BL/6 genetic background. Newborn mice on the 129/Sv genetic background were euthanized by decapitation. Sciatic nerves were dissected from hindquarters and immediately frozen in liquid nitrogen. Additional tissue was taken for genotyping by PCR (Bermingham et al., 1996). Homozygous mutant and homozygous wild-type nerves were pooled separately in Trizol reagent (Invitrogen, Carlsbad, CA), and RNA and DNA were purified according to the manufacturer's instructions. The DNA was analyzed by PCR to confirm the purity of the samples.
Carrier RNA. The template that lacks sites forDpnII and NlaIII was chosen and generated as follows. A 584 bp BstXI–XbaI fragment ofDrosophila genomic DNA, located 1310 bp 5′ to the transcription initiation site of the Antennapedia(Antp) P1 promoter (Laughon et al., 1986), was cloned into the XbaI and SmaI sites of pBKSII+ (Stratagene, La Jolla, CA). Oligonucleotides encoding an artificial poly(A) tract were introduced adjacent to theAntp sequences. The carrier itself was synthesized in vitro using T7 polymerase (Promega, Madison, WI).
cDNA synthesis. Carrier RNA (150 ng) was added to 1 μg of total RNA; the amount of carrier added was designed to bring the total amount of poly(A+) RNA to 200 ng. Poly(A+) RNA was selected using Oligotex beads (Qiagen, Hilden, CA), and cDNA was synthesized using the Superscript system (Invitrogen); both oligo(dT) and random oligonucleotides were used to prime the first-strand synthesis.
Adaptors/primers. The J, N, and R oligonucleotides were designed by Lisitsyn and Wigler (1995) for use on genomic RDA withBglII and are used here with DpnII. The K, O, and S primers are the corresponding adaptors/primers designed to be used with NlaIII: K Sph 24 (RDA with NlaIII), 5′-GAC AAC CGA CGT CGA CTA TGC ATG-3′; K Sph 12 (RDA withNlaIII), 5′-CAT AGT CGA CGT-3′; O Sph 24 (RDA withNlaIII), 5′-AGG CAA CTG TGC TAT CCG AGC ATG-3′; O Sph 12 (RDA with NlaIII), 5′-CTC GGA TAG CAC-3′; S Sph 24 (RDA withNlaIII), 5′-GGC ACT CTC CAG CCT CTC AGC ATG-3′ and S Sph 12 (RDA with NlaIII), 5′ CTG AGA GGC TGG 3′.
Representational difference analysis. RDA was performed as described previously (Hubank and Schatz, 1994; Lisitsyn and Wigler, 1995; Edman et al., 1997) with the following modifications. For most experiments, carrier mRNA was added to total RNA before the isolation of poly(A+) RNA. cDNAs that were derived from mouse tissue were digested separately with DpnII orNlaIII. DpnII cleaves at the recognition site GATC, and because the four base 5′ overhang that results from its cleavage is identical to that generated by BglII, RDA primers designed previously for experiments that use BglII were used to amplify DpnII-generated fragments. N adaptors (Lisitsyn and Wigler, 1995) were ligated to the DpnII ends, and O primers were ligated to NlaIII ends. Two rounds of PCR were performed with Invitrogen Taq polymerase and buffer, 2 mm MgCl2, and 0.2 mm deoxyNTPs. The first round of PCR consisted of four separate 50 μl reactions run in parallel for five cycles (95°C for 1 min; 72°C for 3 min) and then pooled. The second round of PCR consisted of 16 separate 100 μl reactions run in parallel for 12 cycles and subsequently pooled. Only fragments with two appropriately spaced adaptors (typically 50–600 bp apart) are amplified efficiently. It was determined empirically that amplification remained exponential under these conditions. Excess primers were removed from the PCR products using Qiaquick columns (Qiagen). A small proportion of each of the drivers was digested with DpnII to remove the N adaptors or with NlaIII to remove the O adaptors, after which they were replaced with R or S adaptors, respectively, yielding “tester” DNA. Tester DNA was mixed with an excess of driver that was derived from the other source, denatured, and permitted to anneal in a 5 μl volume at 67°C, under oil, for 24–36 hr, which corresponds to a Cot of ∼300–400. Because single-copy genomic DNA reassociates between Cot 100 and 104, and because ∼3% of the nonrepetitive genome is transcribed (Lewin, 1994), a Cot of 3–300 should be sufficient for cDNA. The reannealed DNA was amplified in four parallel 100 μl reactions for 10 cycles of PCR using R or S primers. This material was phenol extracted, ethanol precipitated, resuspended in 0.2 × echo time, and digested with mung bean nuclease (New England Biolabs, Beverly, MA) to remove the single strands that result from priming at only one end of a template. The resulting material was subjected to a second round of amplification in four parallel 100 μl PCRs for 16 cycles with R or S primers. Difference product 1 (DP1) cDNA was digested withDpnII or NlaIII to remove the R or S adaptors, after which they were replaced with J or K adaptors. This material was used as the tester for a second round of subtraction, after mixing with an excess of driver. The products of the second round of subtraction were subjected to two rounds of PCR amplification to produce the second difference product (DP2). The linkers were removed from the DP2 DNA, after which it was cloned. DpnII fragments were ligated into the BamHI site of pBKSII+(Stratagene), whereas the NlaIII fragments were ligated into the Sph site of pGEM7Zf+ (Promega). Ligations were transfected into DH5α (Invitrogen).
Sequence analysis and rapid amplification of cDNA ends.Twenty-four clones were sequenced for each experiment (96 total). Some clones contained multiple inserts. The sequences were compared with the GenBank nonredundant and expressed sequence tag (EST) databases using the National Center for Biotechnology Information (NCBI) basic local alignment search tool (BLAST) (blastn) program (www.ncbi.nlm. nih.gov/blast) (Altschul et al., 1997). Similar queries were performed using the Celera mouse genome assembly (www.celera.com). The sequence indicated which cDNAs were derived from the same gene and which were duplicates. Based on sequence, some inserts were not analyzed further, such as those corresponding to ribosomal RNAs (rRNAs) that were found in both +/+ and −/− samples. Clones 18 and 138 contained overlapping cDNAs of a novel gene,18-138. 5′ and 3′ rapid amplification of cDNA ends (RACE) was performed with the SmartRACE kit (Clontech, Cambridge, UK), using cDNA that had been synthesized using Superscript II polymerase (Invitrogen) and P0–P2Sprague Dawley rat sciatic nerve RNA.
“Snorthern” blots. cDNA that was amplified by PCR for use as driver in the RDA experiments was used also for Snorthern blots. As described above, the PCR conditions were optimized to ensure that amplification was exponential, and parallel PCRs were pooled to minimize variation from individual samples and obtain sufficient material. Driver DNA (0.5 or 1 μg) was electrophoresed through 3% NuSieve 3:1 agarose (FMC Bioproducts, Rockland, ME) or 4% acrylamide gels and then transferred by conventional Southern blotting or electroblotting onto Hybond N+ membranes (Amersham Biosciences, Arlington Heights, IL).
In situ hybridization. Mice were anesthetized and perfused with formalin. Hindquarters were dissected to expose the sciatic nerve and then stored at 4°C in formalin. Tissues were frozen in a 1:1 mixture of optimal cutting temperature compound (O.C.T.) and Aquamount and sectioned at 20 μm. In situ hybridization was performed as described by Simmons et al. (1989). Single-stranded antisense transcripts were prepared from the following clones: pou3f1, AP700 (Bermingham et al., 1996); LacZ, JBSN3, which consists of 423 bp of sequence starting at the DpnII site at 2344 inV00296; P0, rat cDNA as described by Bermingham et al. (1996); cysteine-rich protein-2 (CRP2), JBSN61B, which contains 231 bp starting at the DpnII site at 171 in D88792; Dendrin, JBSN64, which consists of 361 bp between nucleotides 2856 and 3203 of X96589; tramdorin1 (tramd1), 343 bp between nucleotides 1549 and 1892 of AF512429; and 21-70gene, a 673 bp fragment of a mouse RACE clone that contains the 3′ 222 nt of JBSN70, all of JBSN21, and 114 nt of additional 3′ untranslated region sequence. For 18-138, a 185 bp DpnII RDA fragment (clone JBSN18) and an overlapping 245 bp NlaIII fragment (JBSN 138) were fused at a common BglI site to generate a 333 bp DpnII–NlaIII fragment, 18-138, which was used for in situ hybridization. Emulsion-dipped sections were exposed for 7–17 d, counterstained in 0.001% bisbenzimide (Sigma, St. Louis, MO), and photographed using dark-field optics and either Kodak (Rochester, NY) Ectachrome 400 or 160T film.
RNase protection assays. Single-stranded antisense transcripts were generated with the appropriate RNA polymerase. Equal amounts (10 μg) of total RNA from mouse tissues and rat Schwann cells were incubated with 100,000 cpm of riboprobes. The RNA was denatured at 85°C for 10 min, hybridized overnight at 48°C, and then digested with RNase T1 and RNase A (final concentrations: 1 μg/μl and 40 μg/ml, respectively) for 1 hr at 30°C. The reaction was stopped by adding proteinase K and SDS (final concentrations, 0.28 μg/μl and 0.56%, respectively) for 30 min at 37°C. The RNA was purified by phenol–chloroform extraction and precipitation with LiCl, yeast tRNA, and 100% ethanol. The RNA pellet was resuspended in loading dye and counted, and the protected fragments were separated on a sequencing gel.
Northern blot analysis. Equal amounts (10 μg) of total RNA were electrophoresed in 1% agarose and 2.2 mformaldehyde gels, transferred to nylon membranes (Duralon; Stratagene) in 6× SSC, and ultraviolet cross-linked (0.12 J). Blots were prehybridized, hybridized, and washed using standard techniques; the final stringency of the wash was 0.2× SSC at 65°C for 30 min (Sambrook et al., 1989). The following cDNAs were used as probes: JBSN64, a 361 bp RDA fragment of mouse dendrin for the Northern blot tissue; a 2 kb fragment of mouse dendrin cDNA (kindly provided by Torsten Shultz and Peter Seeburg, Max-Planck Institute, Heidelberg, Germany) for the injury Northern blots; a 1 kb cDNA that corresponds to the 18-138 gene; a 590 bp fragment of mouse CRP2 and a 0.6 kb fragment of mouse CRP1, both kindly provided by S.-F. Yet (Harvard University, Boston, MA), and a 2 kb fragment of mouse tramdorin1 cDNA 1920302. Plasmid inserts were isolated after restriction endonuclease digestion by agarose gel electrophoresis and purified by electroelution. 32P-labeled cDNA probes with specific activities of 2–5 × 109 cpm/μg were prepared by primer extension with random hexamers using the Prim-a-gene kit (Promega) according to the manufacturer's instructions.
Preparation of tramdorin1 antiserum. Peptide ESAKKLQSQDPSPANGTSC, containing amino acids 22–39 near the N terminus of tramdorin1, was coupled to KLH and injected into two rabbits, one of which generated useful antiserum.
Transfections. A 1566 bp BglII–XmnI fragment of EST cDNA 1920302, containing the entire coding region, was cloned between the BamHI and EcoRV sites in the expression plasmid pcDNA3.1 (Invitrogen) and transiently transfected into Cos, HeLa, and 293 cells. The cells were grown in low-glucose DMEM supplemented by 10% fetal bovine serum and antibiotics (100 μg/ml penicillin–streptomycin) in a humidified atmosphere containing 5% CO2 at 37°C. Both Lipofectin (Invitrogen) and plasmid DNA were incubated in Optimen for 30 min at room temperature and then combined for another 15 min. The cells (∼80% confluent) were washed with Optimen and then incubated with the combined Lipofectin/DNA solution for 6 hr at 37°C. After 6 hr, the cells were washed once with HBSS (calcium or magnesium free), incubated for 3 d in DMEM at 37°C, and then replated for immunoblotting or immunostaining.
Immunoblotting. Plates of confluent cells in 100 mm plates were harvested in cold Dulbecco's PBS lacking calcium and magnesium (Invitrogen). The cell pellet was lysed in ice-cold 50 mm Tris, pH 7.0, 1% SDS, and 0.017 mg/ml phenylmethylsulfonyl fluoride (Sigma), followed by a brief sonication on ice with a dismembrator (Fisher Scientific, Houston, TX). Protein concentration was determined using the Bio-Rad (Richmond, CA) kit according to the manufacturer's instructions. For each sample, after a 5–15 min incubation in loading buffer at room temperature, 100 μg of protein lysate was loaded onto a 12% SDS-polyacrylamide gel, electrophoresed, and transferred to an Immobilon-polyvinylidene fluoride membrane (Millipore) over a period of 1 hr, using a semidry transfer unit (Fisher Scientific). The blots were blocked (5% powdered skim milk and 0.5% Tween 20 in Tris-buffered saline) overnight at 4°C and incubated for 24 hr at 4°C in a rabbit antiserum against tramdorin1 (diluted 1:1000). After washing in blocking solution and Tris-buffered saline containing 0.5% Tween 20, blots were visualized by enhanced chemiluminescence (Amersham Biosciences) according to the manufacturer's protocols.
Immunostaining. Transfected cells were plated onto four chamber glass slides (Nalge; Nunc, Roskilde, Denmark) and incubated for 2–3 d to ∼60% confluency. The cells were washed in PBS, fixed in acetone at −20°C for 10 min, and then blocked with 5% fish-skin gelatin in PBS containing 0.1% Triton X-100 for 1 hr at room temperature. Cells were labeled with rabbit anti-tramdorin1 (1:500) and processed as described below. Unfixed rat sciatic nerves were embedded in O.C.T. and immediately frozen in a dry ice–acetone bath. Five-micrometer-thick cryostat sections were thaw mounted on SuperFrost Plus glass slides (Fisher Scientific) and stored at −20°C. Teased nerve fibers were prepared from adult rat sciatic nerves, dried on SuperFrost Plus glass slides overnight at room temperature, and stored at −20°C. Sections and teased fibers were postfixed and permeabilized by immersion in −20°C acetone for 10 min, blocked at room temperature for ≥1 hr in 5% fish-skin gelatin containing 0.5% Triton X-100 in PBS, and incubated 16–48 hr at 4°C with various combinations of primary antibodies: rabbit anti-tramdorin1 (1:500), mouse anti-rat myelin-associated glycoprotein (MAG) (clone 513, 1:100; Boehringer Mannheim, Indianapolis, IN), and mouse anti-lysosome-associated membrane protein (LAMP)1 (1:10; Developmental Hydridoma Bank). After incubating with the primary antibodies, the slides were washed and incubated with the appropriate fluorescein- and rhodamine-conjugated donkey cross-affinity-purified secondary antibodies (diluted 1:100; Jackson ImmunoResearch, West Grove, PA). Slides were mounted with Vectashield (Vector Laboratories, Burlingame, CA), examined by epifluorescence with tetramethylrhodamine isothiocyanate (TRITC) and FITC optics on a Leica (Nussloch, Germany) DMR light microscope, and photographed with a cooled Hamamatsu (Tokyo, Japan) camera or followed by image manipulation with Adobe Systems (San Jose, CA) PhotoShop.
RESULTS
RDA using pou3f1 +/+ and −/− sciatic nerve
To identify target genes of pou3f1 in peripheral nerves, sciatic nerves were isolated from newborn [postnatal day 0 (P0)] pups. Not only do most pou3f1-null mice die at this time (Bermingham et al., 1996; Jaegle et al., 1996), but P0 is near the peak of pou3f1 mRNA expression (Monuki et al., 1989), so the expression of pou3f1 target genes should be maximally affected at this time. The confounding effects of myelin gene expression per se should be minimized at this time, because myelination has not commenced in mice at P0 (Webster, 1993), and the expression of genes whose function or expression correlates with myelination increases primarily after birth (Stahl et al., 1990; Baron et al., 1994). Furthermore, because the maturation of myelinating Schwann cells is delayed in pou3f1-null mice (Bermingham et al., 1996; Jaegle et al., 1996), comparing gene expression after the onset of myelin gene expression would be complicated by the expression of myelin-related genes (i.e., genes whose function or expression correlates with myelination) in the wild-type nerves.
RDA on pou3f1 −/− and +/+ sciatic nerves was performed as described previously (Erkman et al., 2000; Bermingham et al., 2001). Nerves were dissected from newborn pups of heterozygous pou3f1 intercrosses and saved separately for each animal. After the pups were genotyped, the nerves from 7 to 10 pou3f1 +/+ and −/− pups were pooled separately, and total RNA was isolated from each pool. DNA was also isolated from the pooled nerves and used to confirm their genotypes (data not shown). Poly(A+) RNA was isolated from a mixture of 1 μg of total RNA and 150 ng of carrier RNA and used to generate cDNA. cDNAs from +/+ and −/− nerves were digested separately with DpnII or NlaIII, ligated to adaptors, and amplified by PCR to generate driver DNAs (Fig.1A). From each driver DNA, tester DNAs were derived by replacing adaptor sequences, after which four RDA experiments were performed. For each enzyme, −/− cDNA served as driver and +/+ cDNA as tester, to isolate putative pou3f1-activated genes. Separately, +/+ cDNA was used as driver and −/− cDNA as tester to isolate putative pou3f1-repressed genes. Figure1A shows +/+ and −/− drivers, as well as the first and second difference products, DP1 and DP2, generated from one and two rounds of the RDA procedure, respectively. Amplified DP2 DNA was cloned into plasmid vectors and, after transformation, 24 random colonies from each experiment (96 total) were sequenced.
Fig. 1.
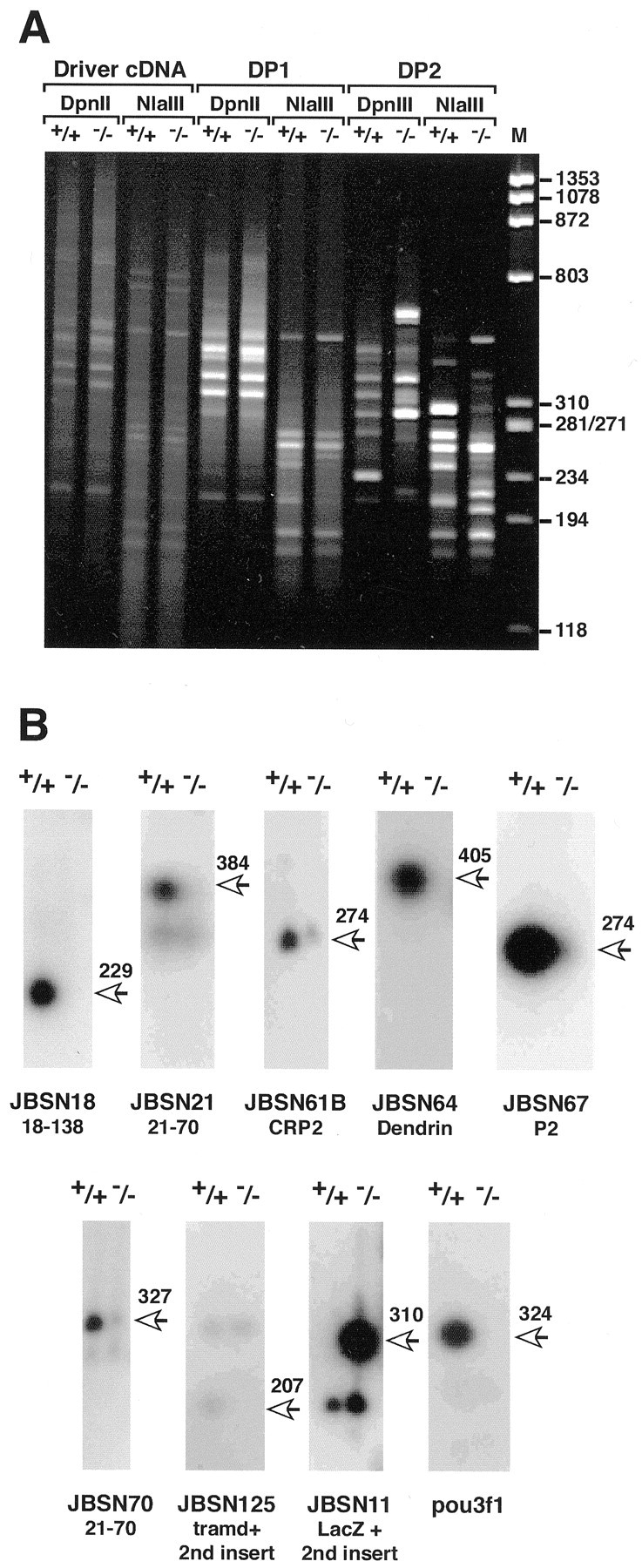
Isolation of genes that are misexpressed in sciatic nerve in the absence of pou3f1. A, Photograph of an ethidium bromide-stained acrylamide gel containing driver, DP1, and DP2 cDNA from P0 pou3f1 +/+ and −/− sciatic nerves. The sizes of the cDNAs can be estimated from the φX174HaeIII digest (lane M). RDA was performed in parallel using cDNAs that were digested with eitherDpnII or NlaIII. Faint bands, presumably attributable to ribosomal RNA or other highly repetitive RNAs, can be seen in the lanes containing the driver cDNA. Note the absence of any band derived from carrier RNA (584 bp). After one round of RDA (DP1 lanes), specific bands, many of similar sizes, begin to appear in both the +/+ and −/−lanes, with either DpnII- orNlaIII-digested cDNA. After two rounds of RDA, the +/+ and −/− lanes contain distinct banding patterns (DP2 lanes). DP2 DNAs were cloned into pBKSII+ for analysis. In DP3 cDNA, only a few bands that correspond to Neo and LacZ increase in intensity relative to DP2 cDNA (data not shown). B, Southern blots of PCR-amplified cDNA (Snorthern blots) from pou3f1 +/+ and −/− sciatic nerves after hybridization to different radiolabeled cDNA fragments generated by RDA. The sizes of bands are shown to theright of each panel and include the 24 bp linkers on the ends of the fragments. All of the misexpressed genes that were confirmed by this analysis are shown, except for Neo. The lacZ probe was made from JBSN11, a plasmid with a double insert. In addition to lacZ, it contains an unknown sequence, which is not differentially expressed, and thus serves as an internal control. JBSN18, JBSN21, JBSN61B, JBSN64, JBSN57, JBSN70, and JBSN125 are activated by pou3f1, because they are expressed at lower levels in cDNA derived from pou3f1 −/− nerves. The pou3f1 probe was derived from a separate RDA experiment and is used here as a control to confirm its absence in −/− cDNA. The names of the genes containing these fragments are given below each pair oflanes.
The sequences were compared to the NCBI nonredundant and EST databases. This analysis revealed, consistent with our previous RDA results (Bermingham et al., 2001), that some clones contained multiple cDNAs, some cDNAs were isolated multiple times, some cDNAs represented different parts of the same gene, and some RDA fragments were homologous to EST clones that correspond to two different genes. A total of 114 RDA cDNA fragments were sequenced. Of these, 23 RDA fragments corresponded to 18S and 28S ribosomal RNA genes; these were found in both the pou3f1 −/− and +/+ samples and were not studied further. Similarly, fragments that correspond to presumed “housekeeping” genes and to known muscle-specific genes such as perinatal myosin heavy chain and troponin C, probably introduced by contaminating muscle tissue in the −/− nerves, also were eliminated. We were left with 78 RDA fragments that corresponded to 51 different candidate pou3f1 target genes. To determine whether these candidate genes were differentially expressed, we performed Southern blot hybridization of 44 of these genes to blots of PCR-amplified cDNA from wild-type/pou3f1 +/+ and mutant/pou3f1 −/− sciatic nerves (Table1). In our hands, these Snorthern blots have proved to be reliable indicators of differential expression (Bermingham et al., 2001) (our unpublished observations).
Table 1.
Genes isolated by RDA of pou3f1 +/+ versus −/− sciatic nerve
| A. Increased expression in +/+ | Isolated–tested–confirmed | Confirmed genes |
|---|---|---|
| DpnII-digested cDNA | 28-26-10 cDNAs | Unknown gene 18–138 (4 copies) |
| 21-19-5 genes | Unknown gene 21–70 (3 copies, 2 different adjacent DpnII fragments) | |
| CRP2 (D88792) | ||
| Dendrin (X96589) | ||
| P2 (S39508) | ||
| NlaIII-digested cDNA | 20-13-4 cDNAs | Unknown gene 18–138 (3 copies) |
| 12-8-2 genes | Unknown gene 125-10:Tramdorin1 | |
| B. Increased expression in −/− | Isolated-tested-confirmed | Confirmed genes |
| DpnII-digested cDNA | 29-20-12 cDNAs | LacZ V00296 (7 copies, 4 different fragments) |
| 14-12-2 genes | pMC1neo U43612(5 copies) | |
| NlaIII-digested cDNA | 38-21-6 cDNAs | LacZV00296 (5 copies, 2 different fragments) |
| 21-14-2 genes | pMC1neo U43612 | |
The results of four independent experiments are summarized using cDNA from pou3f1 +/+ and −/− nerves, digested with eitherDpnII or NlaIII. Clones with increased expression in pou3f1 +/+ sciatic nerve refer to experiments in which tester cDNAs were derived from wild-type sciatic nerve; these genes are putatively upregulated by pou3f1. Likewise, clones with increased expression in pou3f1 −/− sciatic nerve refer to experiments in which tester cDNAs were derived from pou3f1−/− sciatic nerve; these genes are putatively repressed by pou3f1. Not all genes were analyzed (see Results), and for some genes, multiple cDNA fragments were isolated; for each experiment, the number of genes and cDNA fragments that were isolated, tested, and confirmed is indicated. The genes that were shown to be differentially expressed are listed, and in the cases in which multiple independent cDNAs were isolated for these genes (either the same or different sequences corresponding to that gene), their numbers are also indicated.
As summarized in Figure 1B, 11 RDA fragments from eight genes were confirmed to be differentially expressed by Snorthern blots. Hybridization to JBSN18 was seen only in +/+ cDNA; an identical result was seen with JBSN138 (data not shown), which is a different fragment of the same gene (see below). JBSN21 hybridized to a differentially expressed cDNA and, to a lesser degree, with two cDNAs that are not differentially expressed. JBSN61B (CRP2), JBSN64 (dendrin), and JBSN67 (myelin P2) are all differentially expressed, as is JBSN70. Clone JBSN125 contains two inserts, a 215 bp unknown sequence, which is not differentially expressed and serves as an internal control, and a differentially expressed cDNA fragment that is a fragment of tramdorin1(see below). A subclone (125-10) that contains only the smaller cDNA hybridizes only to the differentially expressed fragment (data not shown). pou3f1 itself was not identified in the experiments presented here, presumably because of an insufficient number of clones that were analyzed. However, hybridization of a Snorthern blot to a mouse pou3f1 RDA fragment from brain demonstrates that pou3f1 is present in +/+ but not in −/− cDNA, providing further confirmation of the genotype of the tissue from which the cDNA is derived. P0 and peripheral myelin protein 22 kDa (PMP-22) cDNA fragments were isolated but were not differentially expressed on Snorthern blots, consistent with their apparent normal mRNA expression in pou3f1-null mice (Bermingham et al., 1996). In summary, six genes, representing nine RDA fragments, were expressed at lower levels in the absence of pou3f1 and thus are putatively activated by pou3f1. Two genes, LacZ (Fig. 1B) and Neo(data not shown), were expressed at higher levels inpou3f1-null sciatic nerve. These genes replacepou3f1 in the knock-out mutation and demonstrate that we could have detected repressed genes; the absence of other such genes suggests that pou3f1 is not a repressor in Schwann cells. Finally, these results demonstrate that misexpressed genes can be isolated from small amounts of mutant tissue using an artificial mRNA as a carrier.
Of the six activated genes, only one was known previously to be expressed by Schwann cells: the fatty acid transport protein myelin P2 (Narayanan et al., 1991). Two genes had been identified previously but were not known to be expressed by Schwann cells: the LIM-domain protein CRP2 [smooth muscle LIM (smLIM)] (Weiskirchen et al., 1995; Jain et al., 1996), which binds to components of the actin cytoskeleton (Louis et al., 1997), and dendrin, a protein of unknown function found in dendrites of CNS neurons (Neuner-Jehle et al., 1996; Herb et al., 1997). The subclone 125-10 (from JBSN125) corresponds to numerous EST clones, including AI786604 (I.M.A.G.E. clone 1920302), AI035402 (1380757), andAI005767 (1363993). As described below, we have named this genetramd1. Gene 18-138 is represented by clone JBSN18 (DpnII-derived) and clone JBSN138 (NlaIII-derived) fragments that overlap; the full characterization of this gene will be published elsewhere. RDA clones JBSN21 and JBSN70, which show limited or no homology to sequences currently in the public databases, have been shown by RACE experiments to correspond to adjacent DpnII fragments of a novel putative extracellular matrix gene, 21-70 (data not shown).
Schwann cells express putative pou3f1target genes
To determine whether the cDNAs that appeared to be differentially expressed on Snorthern blots are in fact misexpressed in vivo, in situ hybridization was performed with antisense cRNA probes. Figure2A shows adjacent sections of P0 sciatic nerves frompou3f1 +/+ and −/− mice. As expected, pou3f1 mRNA was expressed in +/+ but not in −/− nerves, LacZ mRNA was expressed in the opposite pattern, and P0 mRNA was abundantly expressed in both genotypes (Bermingham et al., 1996). In contrast to P0 mRNA, P2 mRNA was more abundant in +/+ nerves than in −/− nerves. Dendrin, 18-138, and CRP2 mRNA were all present in +/+ nerve, but the signals in −/− nerve were not above background levels. Minimal expression was detected for 21-70, and no signal was observed using a sense cRNA probe from clone 18-138 or for tramdorin1 (data not shown).
Fig. 2.
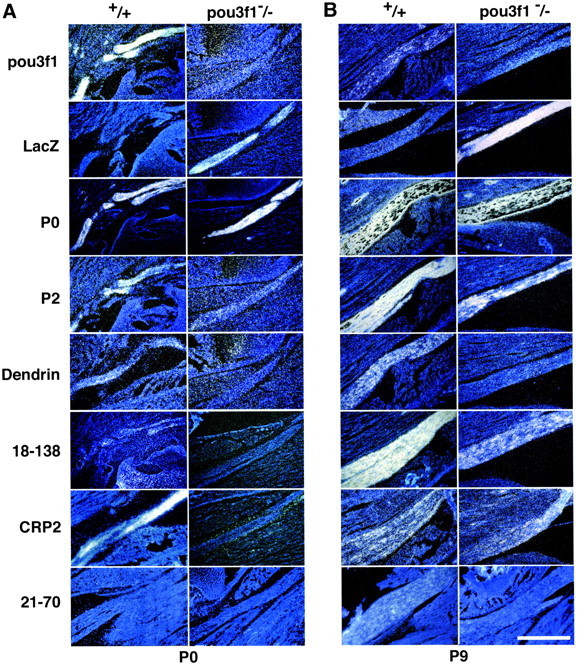
Expression of novel Schwann cell genes in pou3f1 +/+ and −/− sciatic nerves at P0 and P9. Sections of sciatic nerves are shown after in situhybridization with antisense probes for pou3f1, LacZ, P0(Mpz), P2 (P2), dendrin, 18-138, CRP2, and 21-70 mRNA. A, P0 sciatic nerves. Theleft panels show wild-type nerves (+/+); theright panels show knock-out −/− nerves. pou3f1 hybridizes to +/+ but not to −/− nerves, whereas LacZ is the opposite; these results confirm the genotypes of the tissues. Hybridization to mRNA for P0, whose expression is independent of pou3f1, is observed in both +/+ and −/− sciatic nerves and serves to demarcate them. Myelin P2 and CRP2 are expressed in +/+ sciatic nerve, but in pou3f1 mutant nerve, little or no signal is observed. Dendrin and 18-138 hybridization produce faint signals in +/+ and no signals in −/−. At this age, 21-70 mRNAs were not detected. These results demonstrate that differences in expression can be detected by in situ hybridization, and that the genes isolated by RDA are misexpressed in pou3f1 mutant nervesin vivo. B, P9 sciatic nerves. The same probes that were hybridized to P0 sciatic nerves were used to assay expression in P9 sciatic nerves. pou3f1 and LacZ expression confirm the genotypes of the tissues, although by P9, pou3f1 expression is declining, whereas LacZ expression in mutants remains high, as has been observed previously (Jaegle and Meijer, 1998). Myelin P0mRNA expression confirms no significant differences in P0expression in +/+ and pou3f1 −/− mice. At P9, myelin P2and 18-138 expression in mutant sciatic nerve is much stronger than at P0 but remains reduced relative to wild type. Dendrin expression is present in +/+ nerves, but there is little or no signal from mutant nerves. CRP2 expression appears equivalent in +/+ and −/−; at this time, expression is declining in +/+ nerves but is probably increasing in −/− nerves. Faint 21-70 expression can be observed in +/+ nerves but not in −/− nerves. Scale bar, 0.5 mm.
To determine whether these changes persisted in older nerves, we also performed in situ hybridization on sciatic nerves from P9pou3f1 +/+ and −/− mice (Fig. 2B). As expected (Monuki et al., 1989), pou3f1 mRNA expression in P9 +/+ sciatic nerves was reduced from that in P0 +/+ nerves. No pou3f1 mRNA expression was observed in P9 −/− sciatic nerves, whereas LacZ mRNA expression was observed in −/− nerves but not in +/+ nerves, confirming the genotypes of these tissues. The P0 mRNA signal increased from P0 to P9 but was comparable in +/+ and −/− nerves, as has been described previously (Bermingham et al., 1996). Compared with +/+ nerves, the P2 signal was still relatively reduced in P9 −/− nerves but increased compared with P0. Dendrin mRNA was detected in P9 +/+ nerves but not in −/− nerves. The mRNA level of 18-138 was increased in both +/+ and −/− nerves between P0 and P9, but the levels were lower in P9 −/− nerves. Expression of CRP2 mRNA was comparable in P9 +/+ and −/− nerves, but compared with its expression at P0, its expression had decreased in P9 +/+ nerve and increased in P9 −/− nerve (see below). Expression of 21-70 mRNA was faint but greater in P9 +/+ nerve than in P0 +/+ nerve and greater in +/+ than in −/− nerve. Tramdorin1 expression was too faint to be reliably detected (data not shown). These results demonstrate that the genes isolated by RDA are not only expressed in sciatic nerve, but also that their relative expression is altered in the predicted direction in thepou3f1 −/− nerves. However, the pattern of expression between P0 and P9 differed; the expression of P2, CRP2, and 18-138 mRNA in pou3f1 −/− nerves appeared to partially recover, whereas dendrin expression did not.
To evaluate whether the genes that we have identified are important for timely peripheral myelination, we examined their expression inclaw paw (clp) mutant mice (Fig. 3). Likepou3f1-null mice, homozygous clp mice display delayed peripheral myelination, whereas central myelination is unaffected (Henry et al., 1991). Myelin P2 and 18-138 expression is reduced in P1.5clp −/− sciatic nerves but recovered somewhat by P13. Dendrin expression also is reduced in P1.5 clp −/− nerves, but the recovery of its expression by P13 is less obvious. In wild-type nerves, CRP2 expression is robust at P1.5 but is greatly reduced at P13. Inclp mutant nerves, CRP2 activation is reduced, but its expression at P13 is elevated, consistent with the delay in myelination. P0 mRNA expression appears unaffected, although the nerves are thinner in clp mutant mice. These results demonstrate that delayed expression of the genes that we have identified correlates with delayed peripheral myelination in clp as well as pou3f1 mutant mice. In contrast, pou3f1 expression appears normal in P1.5 clp −/− nerves but fails to be downregulated at P13. Therefore, expression of pou3f1 is insufficient for activation of its putative target genes, indicating either that pou3f1 regulates these genes indirectly or that it requires a cofactor to activate them.
Fig. 3.
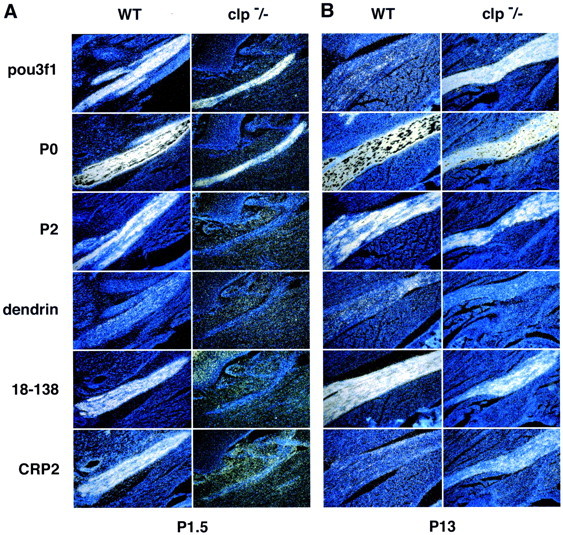
Expression of novel Schwann cell genes inclaw paw mutant sciatic nerves. Adjacent sections of sciatic nerves are shown, after in situ hybridization with antisense probes for pou3f1, P0, P2, CRP2, 18-138, and dendrin mRNA. For each probe, the sections were processed in parallel. A, Sciatic nerves from day P1.5 pups: left, wild type (WT); right,clp/clp. Although nerves are thinner in claw paw mutant mice, pou3f1 and P0 expression is readily apparent. At this stage, expression of myelin P2, 18-138, and CRP2 is robust, whereas dendrin expression is faint. In contrast to pou3f1, all four genes are not significantly expressed in P1.5 clp/clp mutant mice. B, P13 sciatic nerves: pou3f1 expression is minimal in wild-type nerve but remains robust in clp/clp nerve. P13 wild-type expression for P2, dendrin, and 18-138 is comparable with that of P1.5, and expression of these genes appears to recover in P13clp mutant nerves, consistent with the delay in peripheral myelination observed in these mice. CRP2 expression is greater inclp/clp mutant nerve than in wild-type nerve at P13. Note that in wild-type nerves, both CRP2 and pou3f1 are transiently expressed, high at P1.5 and low at P13.
Axotomy results in a prompt decline in the levels of myelin-related mRNAs distal to the lesion, including those of P0, MBP, P2, connexin32, PMP-22, myelin-associated glycoprotein, and periaxin (Poduslo, 1993;Scherer et al., 2001). To determine whether CRP2, tramd1, 18-138, and dendrin are expressed in parallel with other myelin-related genes, we performed Northern blot analysis on rat sciatic nerves collected at various times after crush or transection (Fig.4). For comparison, the blot was also hybridized for CRP1, pou3f1, P0, p75, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The transections were performed to prevent axonal regeneration, whereas nerve crush allows axonal regeneration, and many of the regenerating axons are remyelinated in a proximal-to-distal manner. To better illustrate these points, the distal stumps of crush-injured nerves were divided into proximal (D1) and distal (D2) segments. Tramd1, 18-138, and dendrin mRNA levels paralleled that of P0; their mRNA levels fell after transection and did not increase thereafter, whereas their mRNA levels returned after crush, first in the D1 segment (at 24 d) and then in the D2 segment (at 58 d). The expression of CRP2 mRNA, in contrast, was remarkably similar to that of pou3f1, being low in unlesioned adult nerves and increasing after nerve crush but not transection. These data indicate that tramd1, 18-138, and dendrin are expressed in myelinating Schwann cells and are expressed as part of the program of myelin-related gene expression.
Fig. 4.
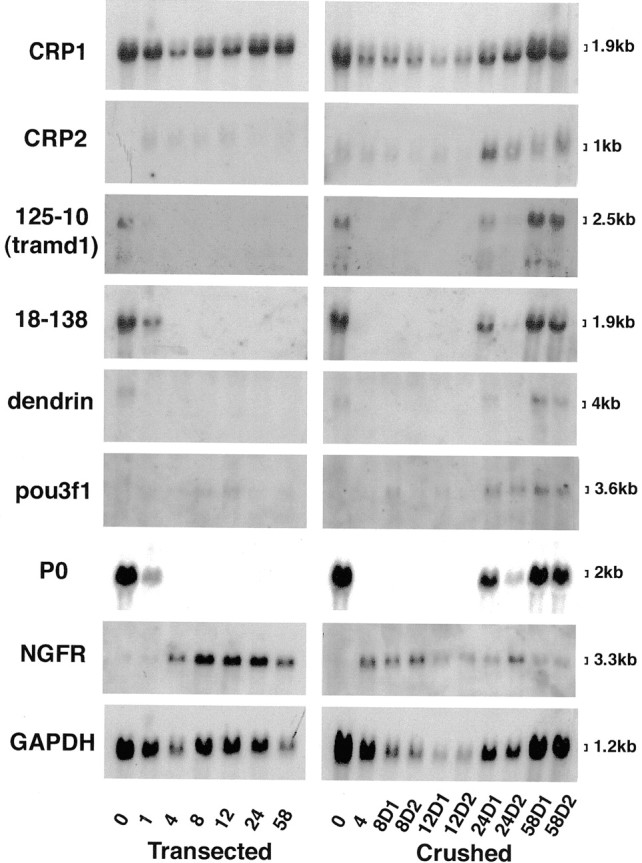
Expression of novel Schwann cell genes after nerve injury. Each lane contains an equal amount (10 μg) of total RNA isolated from the distal stumps of rat sciatic nerves after transection or crush injury. The number of days after these lesions is indicated; the 0 time point is from unlesioned nerves. For the crushed nerves, RNA was made separately from two distal segments, one immediately distal to the injury (D1) and one more distal to the injury (D2). The blots were successively hybridized with radiolabeled cDNA probes for tramd1,18-138, dendrin, CRP2,CRP1, pou3f1, NGF receptor(NGFR), GAPDH, and P0and exposed to film for 14, 5, 14, 14, 7, 14, 14, 4, and 0.15 d, respectively. The steady-state levels of tramd1, 18-138, and dendrin mRNA parallel that of P0, whereas CRP2 mRNA levels parallel that of pou3f1.
Expression of CRP genes in Schwann cells
CRP2 mRNA expression parallels that of pou3f1 both during development as well as after nerve injury. In situhybridization of CRP2 mRNA at P0, P1.5, P9, and P13 (Figs. 2,3) indicates that CRP2 expression is transient, like that of pou3f1 (Monuki et al., 1989). To evaluate the relationship between CRP2 and pou3f1 further, we examined CRP2 mRNA expression in cultured Schwann cells and observed that CRP2 is induced by forskolin (Fig. 5A), as is pou3f1 (Monuki et al., 1989). These results further indicate that CRP2 expression closely mimics that of pou3f1 and that CRP2, like pou3f1, may be expressed in promyelinating Schwann cells (Arroyo et al., 1998). To determine whether CRP2 performs an essential role in Schwann cell myelination, we examined sciatic nerves from P4 and P5 mice that lack the CRP2 gene (kindly provided by S.-F. Yet and M.-E. Lee, Harvard Medical School). We found no differences in myelin from these mice and their wild-type littermates in epoxy sections (data not shown), suggesting that CRP2 is not essential or that other CRP genes may compensate for the absence of CRP2. To test the latter possibility, we examined the expression of CRP1, which is closely related (Liebhaber et al., 1990; Crawford et al., 1994). CRP1 is also induced by forskolin, but unlike CRP2, its expression is not regulated by pou3f1 (Fig. 5B) or nerve injury (Fig. 4). As potential regulators of the actin cytoskeleton, the CRP proteins may act in the changes in Schwann cell morphology that accompany myelination.
Fig. 5.
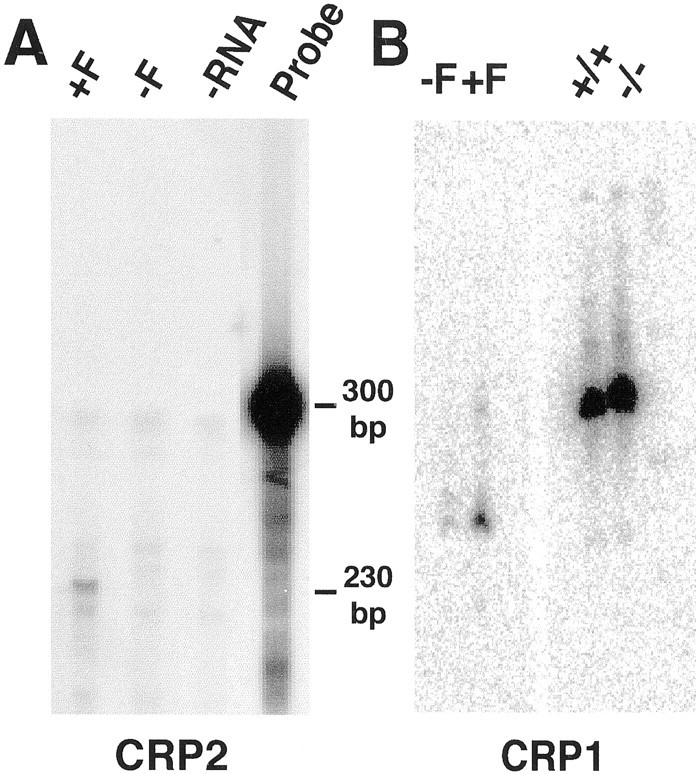
Comparison of CRP1 and CRP2 Schwann cell expression. A, RNase protection demonstrates that forskolin (F) increases the steady-state level of CRP2 mRNA in cultured rat Schwann cells. B, Snorthern blots demonstrate that CRP1 is also induced by forskolin in cultured Schwann cells, but unlike CRP2, it is not misexpressed in P0 nerves in the absence of pou3f1 (compare +/+ with −/−).
Tissue-specific expression of putative pou3f1 target genes
The expression of dendrin, 18-138, and tramdorin1 mRNAs was analyzed on a Northern blot of adult mouse tissues to determine their tissue distribution and the sizes of their transcripts. A dendrin probe hybridized to a 4 kb mRNA in adult cerebrum, as expected (Neuner-Jehle et al., 1996; Herb et al., 1997), and more weakly to a similarly sized mRNA from sciatic nerve (Fig.6A). The 18-138 probe generated a strong hybridization signal with a 1.9 kb mRNA and a weaker signal with a 4.7 kb transcript (Fig. 6B); of the tissues examined, 18-138 mRNA was detected only in sciatic nerve.
Fig. 6.
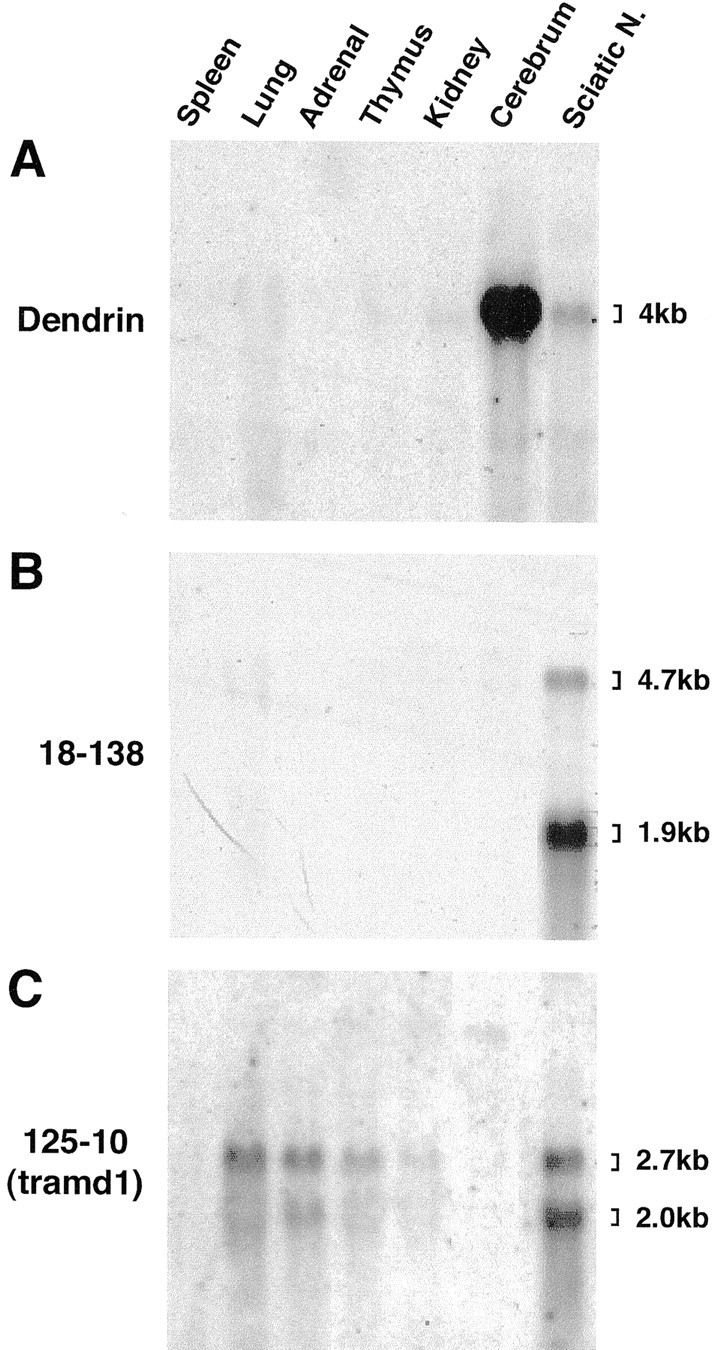
Tissue-specific expression of dendrin, 18-138, and tramdorin1 genes. Each lane contains an equal amount (10 μg) of total RNA isolated from the indicated mouse tissues. The blot was successively hybridized to a radiolabeled probe corresponding to dendrin (A), 18-138 (B), and tramdorin1 (C) and exposed to film for 8, 7, and 14 d, respectively, after each hybridization. The sizes of the transcripts are estimated according to the sizes of 28S and 18S rRNAs (4712 and 1869 nt, respectively) (Hassouna et al., 1984; Raynal et al., 1984). Sciatic N., Sciatic nerve.
The major tramdorin1 transcripts in sciatic nerve were ∼2 and 2.7 kb; the 2.7 kb species is the most prominent tramdorin1 mRNA in lung, adrenal gland, and thymus (Fig. 6C). No expression was observed in the spleen, but a faint band (>6.5 kb) was observed in cerebrum. These results indicate that EST cDNA 1920302, as well as the RACE cDNAs, define a nearly full-length tramd1 mRNA that likely corresponds to the larger 2.7 kb transcript, because they are ∼2.45 kb without their poly(A) tails. Therefore, tramdorin1 expression is neither ubiquitous nor restricted to sciatic nerve. A similar Northern blot analysis of rat tissue showed expression in sciatic nerve but not in lung and thymus (data not shown), suggesting either variation in tramdorin1 expression among species or cross-hybridization of the tramd1 probe to related sequences.
Chromosomal localization of putative pou3f1 target genes
The chromosomal locations of all six putative pou3f1 target genes were examined to ascertain any linkage to known human or mouse peripheral neuropathy loci. Dendrin has been mapped to distal mouse chromosome 15 (Brady et al., 1997), and myelin P2(Pmp2) has been mapped to proximal mouse chromosome 3 (Dietrich et al., 1996). The mouse chromosomal locations of the CRP2 (Csrp2), 18-138, 21-70, and tramd1 genes were determined by interspecific backcross analysis using progeny derived from matings of [(C57BL/6J × Mus spretus)F1 × C57BL/6J] mice (N. A. Jenkins, D. Gilbert, and N. Copeland, unpublished observations). The location of Csrp2 in mice near Myf6 on chromosome 10 confirms its previous mapping to human to 12q21.1 (Weiskirchen et al., 1997) (data not shown). 18-138 resides near the Laglocus on distal mouse chromosome 4, in a region that corresponds to human 1q36. 21-70 is closely linked to the Atm andCsk loci on mouse chromosome 9; the corresponding human locus is on 15q21. The chromosomal locations of these genes were confirmed using Celera and public mouse and human genome databases. Currently, there are no peripheral neuropathy loci for which dendrin, myelin P2, CRP2, 18-138, or 21-70 are candidate loci.
The results of backcross mapping of the tramd1 locus confirm its localization to 11 exons residing between nucleotides 56,775,171 and 56,748,143 on mouse chromosome 11 in the Celera mouse chromosome database (May 5, 2002 update; our unpublished observations; data not shown). The proximal region of mouse chromosome 11 is syntenic with human chromosome 5q31–33, and the homology of mouse tramdorin1 coding exons to 5q sequences in the Celera and public human genome assemblies (Lander et al., 2001; Venter et al., 2001) confirms the mapping in mouse. Although no candidate mouse mutations reside near the tramd1 locus, kindreds with an autosomal recessive form of inherited demyelinating neuropathy have been mapped to this region (LeGuern et al., 1996; Gabreels-Festen et al., 1999;Guilbot et al., 1999a,b), prompting us to characterize this gene further.
cDNAs homologous to clone 125-10 encode a novel gene product, tramdorin1
Rat and mouse cDNAs that correspond to clone 125-10 were isolated by 5′ and 3′ RACE (Frohman, 1993, 1994) and by obtaining and sequencing the homologous EST clone AI786604 (I.M.A.G.E. clone 1920302). As shown in Figure 7A, the combined 5′ and 3′ rat RACE fragments define a 2.5 kb cDNA, as does mouse EST 1920302. The putative full-length rat and mouse cDNAs contain a 1.4 kb open reading frame that contains an ATG codon that closely matches the Kozak consensus for initiator ATGs (Fig. 7B), predicted to encode a protein of 52 kDa. The rat and mouse cDNAs and the hypothetical proteins they encode do not match any genes in the GenBank nonredundant database or the SwissProt database (release 39), although while this manuscript was in preparation, a homologous but not identical gene was cloned (Sagne et al., 2001; see below). Therefore, the cDNAs that we have cloned are derived from a novel gene.
Fig. 7.
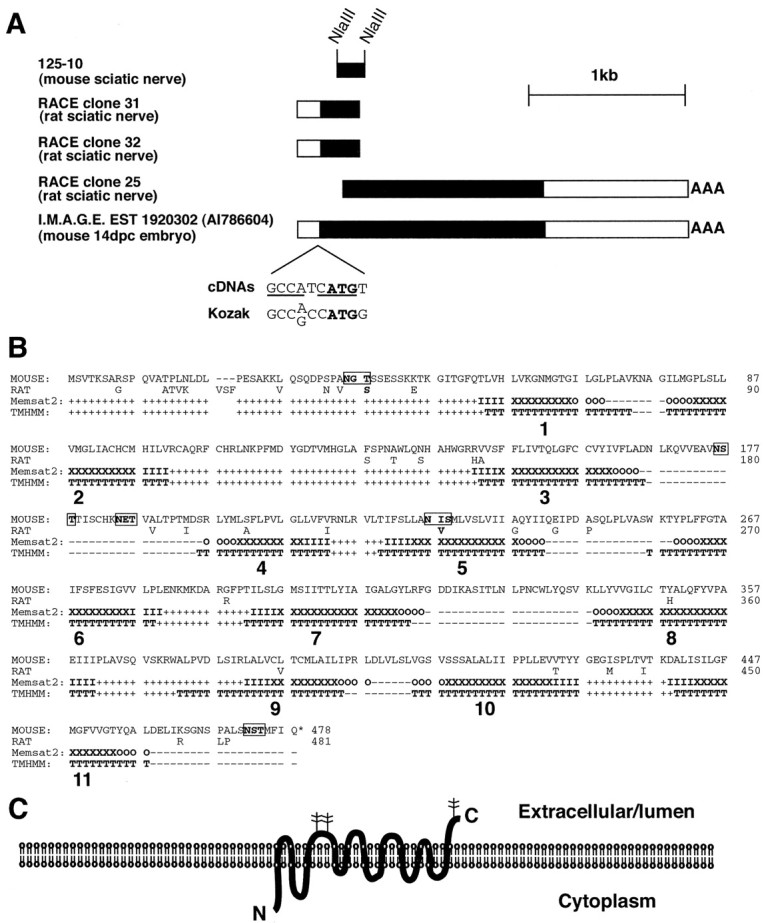
RDA clone 125-10 identifies the gene for a novel protein, tramdorin1. A, cDNAs that correspond to RDA clone 125-10. Five cDNA fragments are shown; they include the RDA fragment 125-10, two independent, nonidentical 5′ RACE rat cDNAs, a 3′ RACE rat cDNA, and a mouse EST cDNA. Of these, the 2.45 kb EST cDNA 1920302 appears to include the entire open reading frame, which is depicted in black. This cDNA appears to be nearly full length, based on the calculated size of tramdmRNA (Figs. 3, 7). The organism and tissue of origin of each cDNA fragment are listed. The open reading frame commences with an ATG initiation codon (in bold) that closely matches the Kozak consensus (Kozak, 1987); nucleotides that are an exact match areunderlined. 14dpc, 14 d postcoitum. B, Amino acid sequences encoded by mouse tramdorin EST cDNA 1920302 and a hypothetical rat tramdorin1 cDNA, derived from 5′ and 3′ RACE sequences, are shown. These sequences have been assigned GenBank accession numbers AF512429 and AF512430, respectively. The mouse cDNA encodes a 478 aa protein, whereas the corresponding rat protein is 481 aa. The amino acid sequence of mouse EST cDNA 1920302 was analyzed using the transmembrane domain prediction programs Memsat2 (McGuffin et al., 2000) and TMHMM (Sonnhammer et al., 1998). Eleven putative transmembrane domains are numbered. Transmembrane domains predicted by TMHMM are marked with capital Ts. For Memsat2, transmembrane domains are marked as follows: O, Outside transmembrane helix cap; X, central transmembrane helix segment; I, inside transmembrane helix cap. For both Memsat2 and TMHMM, predicted cytoplasmic domains are marked by +, whereas noncytoplasmic domains are marked by −. Five consensus glycosylation sites within the protein sequence are shown inbold and are boxed. C, Diagram depicting the predicted topology of tramdorin1 with 11 transmembrane domains, as predicted by Memsat2 and TMHMM. The three extracellular/lumenal glycosylation sites are marked with branched structures. However, the electrophoretic mobility of tramdorin suggests that it is not glycosylated extensively (Fig. 8).
The protein encoded by mouse cDNA 1920302 was predicted by the Memsat2 (PSIPRED) (McGuffin et al., 2000) and transmembrane hidden Markov model (TMHMM) (Sonnhammer et al., 1998) protein structure prediction programs to consist of 11 transmembrane helices, with the N terminus facing the cytoplasm (Figs.7B,C). Although other programs produced differing predictions (data not shown), the following considerations support this topology. First, those programs that predicted 11 transmembrane domain helices more accurately predicted the number and topology of the transmembrane helices of a set of transmembrane proteins of known structure (Tusnady and Simon, 2001). Second, a signal sequence prediction program (Nielsen et al., 1997) indicates that tramdorin1 does not appear to contain a cleaved N-terminal signal sequence that would be expected if the N terminus was lumenal/extracellular. Third, membrane proteins are glycosylated only on their extracytoplasmic faces, and the distribution of glycosylation consensus sites (NXS/T, where X is any amino acid except proline) within the protein supports the proposed topology. Five putative glycosylation sites were found, of which three are predicted to be extracytoplasmic in the structures predicted by Memsat2 and TMHMM. A fourth is not conserved in humans (our unpublished observations), and a fifth is buried in predicted transmembrane helix 5. Fourth, the protein displays homology to the amino acid and anxin permease superfamily of amino acid transport proteins, which are predicted to possess 11 transmembrane domains (Young et al., 1999). However, the related vesicular GABA transporters are thought to consist of 10 transmembrane domains that correspond to predicted transmembrane domains 2–11 (McIntire et al., 1997; Sagne et al., 1997). Regardless of their differing topological predictions, all of the programs indicated that cDNA 1920302 encodes a polytopic transmembrane protein. Therefore, we have named the putative gene product tramdorin, for transmembrane domain rich protein. Thetramdorin1 gene has been assigned the human and mouse gene symbol tramd1. The known proteins that are most closely related to tramdorin1 are two additional tramdorins: tramdorin2 and tramdorin3 (our unpublished observations). While this manuscript was in preparation, tramdorin1 was isolated independently and shown to encode a proton-dependent amino acid transporter (Boll et al., 2002). A recently cloned rat lysosomal amino acid transporter (LYAAT), LYAAT-1 (Sagne et al., 2001), is the rat homolog of tramdorin3; rat tramdorin1 is 67% identical to this protein.
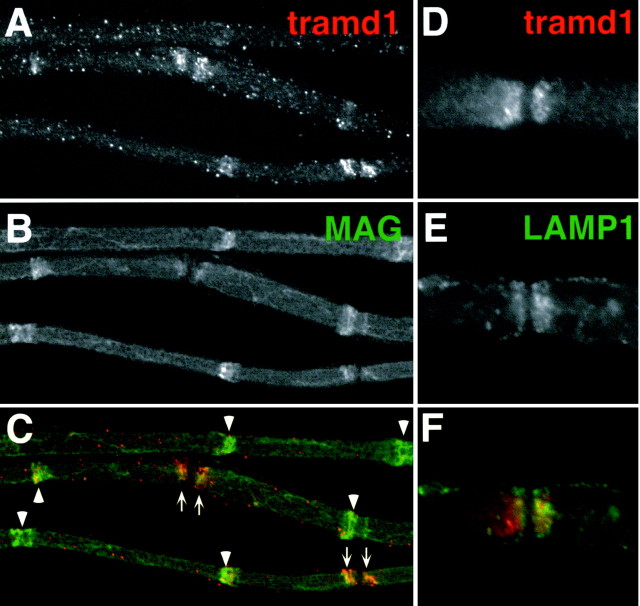
To characterize tramdorin1, an antiserum was raised against a sequence near the N terminus that was divergent among the tramdorin gene family (see Materials and Methods). In immunoblots of adult rat sciatic nerve, the antiserum binds to a single band close to the predicted molecular weight of unglycosylated tramdorin1 (Fig.8A). A band of similar molecular weight is observed on immunoblots of cells transfected with full-length mouse tramdorin1 cDNA (Fig. 8B). Transfected cells were stained with the anti-tramdorin1 antiserum, whereas parental cells were unstained (Fig.8C,D). To localize tramdorin1, we labeled unfixed myelinated fibers from rat sciatic nerves with this antiserum. Tramdorin1 immunoreactivity was observed in the paranodes and in most incisures, as shown by colabeling with a monoclonal antibody against MAG (Fig. 9A–C). To determine whether tramdorin1, like tramdorin3/LYAAT-1, is associated with lysosomes, teased fibers were double labeled for tramdorin1 and LAMP1, a lysosomal marker. Figure 9D–F shows that tramdorin1 and LAMP1 are found in paranodes but do not colocalize. These observations indicate that tramdorin1 is localized to noncompact myelin but is not a component of lysosomes.
Fig. 8.
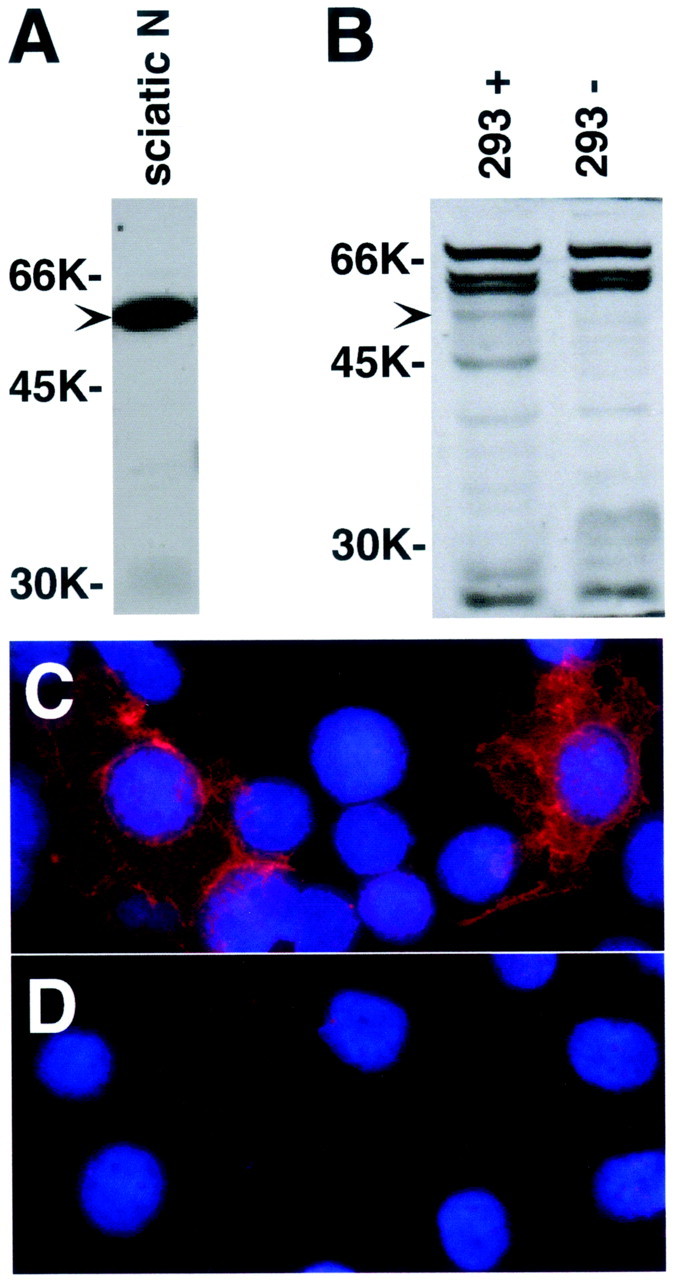
Immunoblot analysis of tramdorin1.A, B, Western blot analysis of anti-tramdorin1 antiserum. Lysates (100 μg) from adult rat sciatic nerve (sciatic N; A) and 293 cells (B) transfected with tramdorin1 expression vector (+) and untransfected 293 cells (−) were used for Western blot analysis of the tramdorin1 antiserum (diluted 1:1000); the blots were developed with chemiluminescence. Arrowheads mark the location of immunoreactive tramdorin protein. C,D, Micrographs showing transfected (C) or untransfected (D) 293 cells viewed by immunofluorescence using the anti-tramdorin1 antiserum. Nuclei were visualized using 4′,6′-diamidino-2-phenylindole.
Fig. 9.
Localization of tramdorin1 in myelinating Schwann cells. Images of unfixed teased fibers from adult rat sciatic nerve, double labeled with a rabbit tramdorin1 antiserum (A,D) (TRITC) and a mouse monoclonal antibody against MAG (B) (FITC) or LAMP1 (E) (FITC) are shown. Tramdorin immunoreactivity is found at paranodes (arrows) and most incisures (arrowheads), colocalizing with MAG (A–C), and in puncta along the outer surface. The failure to detect tramdorin in all incisures is more likely the result of technical problems of antibody penetration than of heterogeneity of expression. At paranodes (D–F), tramdorin1 immunostaining does not appear to colocalize with that of LAMP1, a lysosomal marker. Both markers possess cone-like expression patterns in the paranode, in which the tramdorin expression domain is nested within the LAMP1 expression domain.
DISCUSSION
To identify candidate target genes of pou3f1, we used RDA in conjunction with an artificial carrier mRNA to isolate genes that are differentially expressed in sciatic nerves from newbornpou3f1 −/− pups compared with their +/+ littermates. The sequences of the RDA fragments led to the discovery of six candidate genes, each of which was verified to be misexpressed by Snorthern blots and in situ hybridization. Furthermore, the mRNA levels of five of six of these genes paralleled other myelin-related genes in nerve-injury paradigms. These genes are good candidates to fulfill important functions in generating or stabilizing the myelin sheath or in the metabolism of Schwann cell proteins.
Misexpressed genes in pou3f1 mutant mice
CRP2 is the only putative gene whose mRNA levels parallel those of pou3f1 in developing as well as lesioned adult peripheral nerves and in cultured Schwann cells treated with forskolin. Their parallel expression patterns strongly support the idea that CRP2 is a direct target of pou3f1. In keeping with this suggestion, the CRP2 promoter (Yet et al., 1998) has seven consensus pou3f1-binding sites within 4.8 kb of the transcription start site (data not shown). However, pou3f1 expression is insufficient to activate CRP2 in clp mutant mice, suggesting that if CRP2 is directly activated by pou3f1, this activation also requires the clp gene product. Although absence of CRP2 does not appear to affect Schwann cell myelination, it is a second gene whose expression marks the promyelinating stage of Schwann cell differentiation. The related protein CRP1 is induced in Schwann cells by forskolin treatment, also expressed in adult sciatic nerve, but its expression is not modulated by nerve injury or dependent on pou3f1.
CRP1 and CRP2 (smLIM) are members of the cysteine-rich protein subclass of LIM-only proteins, which consist of two LIM domains, each followed by a glycine-rich region (Liebhaber et al., 1990; Arber et al., 1994;Crawford et al., 1994; Jain et al., 1996). All three members of this subclass are associated with actin fibers, bind to the actin cytoskeletal proteins α-actinin and zyxin (Louis et al., 1997), and are thought to function in the differentiation of the cells that express them. CRP1 and CRP2 are downregulated in transformed cells (Weiskirchen and Bister, 1993; Weiskirchen et al., 1995), and mice that lack the third member of the family, CRP3/MLP, display a disruption of the cytoarchitecture of differentiated muscle cells (Arber et al., 1997). Thus, if CRP2 and CRP1 perform similar functions in Schwann cell differentiation, the loss of CRP2 may be compensated for by CRP1. These proteins may participate in cytoskeletal changes that accompany the transition from a promyelinating Schwann cell to a myelinating Schwann cell (Kidd et al., 1996; Fernandez-Valle et al., 1997; Bernier et al., 1998).
Myelin P2 is a member of the fatty acid binding protein (FABP) family of cytosolic lipid binding proteins (Narayanan et al., 1988), which are thought to function by binding and transporting fatty acids and/or by modulating fatty acid-mediated signal transduction (Glatz et al., 1995; Coe and Bernlohr, 1998). Adipocyte P2 (aP2), the most closely related FABP to myelin P2, activates the nuclear receptor transcription factor peroxisome proliferator activated receptor (PPAR)γ, which activates theaP2 gene in a positive feedback loop that controls lipid metabolism in adipocytes (Coe and Bernlohr, 1998;Hertzel and Bernlohr, 1998). If myelin P2performs an analogous function to a P2 in Schwann cells, then lipid metabolism and perhaps PPAR-mediated gene regulation may be compromised in pou3f1 −/− Schwann cells. In addition, myelin P2 has been shown to bind retinoic acid and retinol (Uyemura et al., 1984), suggesting that it could modulate the activities of retinoid nuclear receptors. The myelin P2 promoter (Narayanan et al., 1991; Bharucha et al., 1993) contains four consensus pou3f1-binding sites (data not shown), located in sequences that are required for forskolin induction of the myelin P2 gene and high-level expression transient transfection assays. Together, these observations suggest that myelin P2 is a target of pou3f1, and that forskolin induction of myelin P2 (Monuki et al., 1989) may be mediated by pou3f1.
As its name suggests, dendrin was originally found in dendrites (Neuner-Jehle et al., 1996; Herb et al., 1997). It is a highly basic protein with putative phosphorylation sites for PKA and protein kinase C, suggesting that phosphorylation regulates its function. It is one of several proteins thought to be translated in dendrites using internal ribosome entry sites (Pinkstaff et al., 2001), suggesting that its translation could be localized to discrete locations in Schwann cells as well. Our data indicate that dendrin is expressed by myelinating Schwann cells, but its function both in dendrites and in Schwann cells remains to be determined.
Tramdorin1/mPAT2 has been shown to function as a proton-dependent transporter of small amino acids (Boll et al., 2002). It is related to yeast vacuolar and rat lysosomal amino acid transport proteins, but unlike tramdorin3/LYAAT-1, tramdorin1 does not appear to be a lysosomal protein. Tramdorin1 lacks a possible lysosomal targeting motif [an acidic residue located near the C terminus, −4 to −6 relative to a dileucine (LL(I,M,V)] (Sandoval et al., 2000) that is present in tramd3/LYAAT-1. Tramdorin1/mPAT2 shows strong specificity for glycine, l-alanine, and l-proline, with greatest inward currents generated by glycine (Boll et al., 2002). Its restricted substrate specificity and localization suggest an unsuspected function of glycine in myelinating Schwann cells.
Regulation of Schwann cell differentiation by pou3f1
Several observations indicate that the genes we have identified are not regulated exclusively by pou3f1. First, pou3f1 is expressed in P1.5 clp mutant sciatic nerves, whereas dendrin, myelin P2, CRP2, and 18-138 are not, suggesting that factor(s) in addition to pou3f1 are required for normal activation of those genes. The gene product that is altered by the clpmutation is unknown; directly or indirectly, this gene product could act in concert with pou3f1 or downstream of it. Second, only the expression of CRP2 appears to parallel that of pou3f1; the other genes normally are expressed after the decline of pou3f1 expression. It is possible that pou3f1 indirectly activates these genes through its activation of the distal enhancer of Krox-20 (Ghislain et al., 2002). In fact, two of the putative pou3f1 target genes are also activated by Krox-20. Dendrin is one of the most strongly activated genes by overexpression of Krox-20 in Schwann cells (Nagarajan et al., 2001), and myelin P2 expression is reduced or absent inkrox-20-null mice (Topilko et al., 1994). However, the transient expression pattern of CRP2 suggests that this gene may be regulated independently of Krox-20. Third, the genes that we have identified appear to be activated slowly in the absence of pou3f1, in conjunction with the delayed onset of myelination (Bermingham et al., 1996; Jaegle et al., 1996). The cause of delayed maturation of myelinating Schwann cells in pou3f1-null mice is unknown, but compensation by another POU protein is possible. In skin, pou3f1 and a member of a different class of POU domain protein, Skn-1, compensate for one another in keratinocyte differentiation (Andersen et al., 1997), indicating that divergent POU domain transcription factors can regulate pou3f1 target genes. The POU domain transcription factor Brn5 is a good candidate for such a compensatory factor, because it is expressed in myelinated Schwann cells (Wu et al., 2001).
Our results suggest that pou3f1 functions primarily as an activator of gene expression in Schwann cells. All six genes that we identified were upregulated by pou3f1. The two repressed genes that we found,Neo and LacZ, indicate that such genes could have been found, and we were able to identify genes that were repressed by forskolin using the same approach (Bermingham et al., 2001). It is possible that repressed genes were missed or that pou3f1 acts as a repressor in Schwann cells at times other than P0 or in other tissues. However, the simplest explanation for our data, and for the phenotype of the pou3f1-null mice, is that pou3f1 is an activator. Because the POU domain of pou3f1 itself can function as an activator (Fyodorov and Deneris, 1996), we suggest that the ΔSCIP transgene (Weinstein et al., 1995) also functions as an activator in vivo, triggering precocious myelination by prematurely activating pou3f1 target genes.
The sample of RDA clones that we analyzed does not contain all of the genes that are misexpressed in the absence of pou3f1. By looking at P0, before most myelin-related genes are highly expressed, we may have reduced the number of genes that we identified. For example, pou3f1 is required for its own downregulation (Jaegle and Meijer, 1998) (Fig. 2), but this effect is not apparent at P0. The Krox-20 distal enhancer has been shown to be activated by pou3f1 (Ghislain et al., 2002), but we did not isolate any Krox-20 clones, perhaps because of confounding effects of its expression from the early, pou3f1-independent enhancer. We did not find other genes that are known or likely to be differentially expressed by myelinating Schwann cells, such asperiaxin, dystroglycan-related protein-2,connexin32, MAG, neurofascin,β4 integrin, and E-cadherin, although most of their cDNAs have appropriately spaced restriction sites forDpnII and NlaIII. Either these genes were missed or their expression does not require pou3f1.
Only a subset of myelin-related genes appears to be downregulated in the absence of pou3f1. The mRNA levels of P0, MBP, PMP-22, and MAG do not appear to be altered significantly in the first 10 d after birth by the absence of pou3f1 (Bermingham et al., 1996) (our unpublished results). Of the genes that were known previously to be expressed by myelinating Schwann cells, only myelin P2 was identified as a putative pou3f1 target gene. The results presented here suggest that myelination involves the coordinated expression of more genes than has been appreciated previously. The use of RDA screens to identify genes that are downregulated in the absence of pou3f1 is a fruitful approach to identify novel genes that may perform important functions in myelination. Until microarrays that contain every mouse or human gene are available, RDA remains a useful way to identify novel genes from specialized tissues, such as sciatic nerve.
Footnotes
This research was supported by a National Alliance for Research on Schizophrenia and Depression Young Investigator Award and National Institutes of Health (NIH) Grants NS40751 (J.R.B.), NS37100 (S.S.S.), and 5O1 NS 34934-11 (M.G.R.). We thank Drs. Mark Sornson and Linda Erkman for insightful discussions during critical phases of this work; Dr. Mark Marchionni for the recombinant glial growth factor; Shaw-Fang Yet, Torsten Shultz, and Peter Seeburg for plasmids; Shaw-Fang Yet and the late Mu-En Lee for CRP2 knock-out mice; Nancy Jenkins, Debra Gilbert, and Neal Copeland for interspecific backcross mapping; Peggy Myer for assistance with the figures; Jamie Pennington, Jill O'Moore, and Ted Xu for excellent technical assistance; and George Carlson for his comments on this manuscript. Photomicroscopy and image analysis by magnetic resonance imaging was made possible by equipment purchased with a grant from the M. J. Murdock Charitable Trust.
Correspondence should be addressed to John R. Bermingham Jr, McLaughlin Research Institute, 1520 23rd Street South, Great Falls, MT 59405. E-mail: jrbjr@po.mri.montana.edu.
T. Whisenhunt's present address: School of Medicine, University of Alabama-Birmingham, Birmingham, AL 35294-0019.
S. O'Connell's present address: Syrrx Inc., 10410 Science Center Drive, San Diego CA 92121.
REFERENCES
- 1.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen B, Weinberg WC, Rennekampff O, McEvilly RJ, Bermingham JR, Jr, Hooshmand F, Vasilyev V, Hansbrough JF, Pittelkow MR, Yuspa SH, Rosenfeld MG. Functions of the POU domain genes Skn-1a/i and Tst-1/Oct-6/SCIP in epidermal differentiation. Genes Dev. 1997;11:1873–1884. doi: 10.1101/gad.11.14.1873. [DOI] [PubMed] [Google Scholar]
- 3.Arber S, Halder G, Caroni P. Muscle LIM protein, a novel essential regulator of myogenesis, promotes myogenic differentiation. Cell. 1994;79:221–231. doi: 10.1016/0092-8674(94)90192-9. [DOI] [PubMed] [Google Scholar]
- 4.Arber S, Hunter JJ, Ross J, Jr, Hongo M, Sansig G, Borg J, Perriard JC, Chien KR, Caroni P. MLP-deficient mice exhibit a disruption of cardiac cytoarchitectural organization, dilated cardiomyopathy, and heart failure. Cell. 1997;88:393–403. doi: 10.1016/s0092-8674(00)81878-4. [DOI] [PubMed] [Google Scholar]
- 5.Arroyo EJ, Bermingham JR, Jr, Rosenfeld MG, Scherer SS. Promyelinating Schwann cells express Tst-1/SCIP/Oct-6. J Neurosci. 1998;18:7891–7902. doi: 10.1523/JNEUROSCI.18-19-07891.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baron P, Shy M, Hondo H, Sessa M, Kamholz J, Pleasure D. Developmental expression of P0 mRNA and P0 protein in the sciatic nerve and the spinal nerve roots of the rat. J Neurocytol. 1994;23:249–257. doi: 10.1007/BF01275529. [DOI] [PubMed] [Google Scholar]
- 7.Bermingham JR, Jr, Scherer SS, O'Connell S, Arroyo E, Kalla KA, Powell FL, Rosenfeld MG. Tst-1/Oct-6/SCIP regulates a unique step in peripheral myelination and is required for normal respiration. Genes Dev. 1996;10:1751–1762. doi: 10.1101/gad.10.14.1751. [DOI] [PubMed] [Google Scholar]
- 8.Bermingham JR, Jr, Shumas S, Whisenhunt T, Rosenfeld MG, Scherer SS. Modification of representational difference analysis applied to the isolation of forskolin-regulated genes from Schwann cells. J Neurosci Res. 2001;63:516–524. doi: 10.1002/jnr.1046. [DOI] [PubMed] [Google Scholar]
- 9.Bernier G, De Repentigny Y, Mathieu M, David S, Kothary R. Dystonin is an essential component of the Schwann cell cytoskeleton at the time of myelination. Development. 1998;125:2135–2148. doi: 10.1242/dev.125.11.2135. [DOI] [PubMed] [Google Scholar]
- 10.Bharucha VA, Peden KW, Subach BR, Narayanan V, Tennekoon GI. Characterization of the cis-acting elements of the mouse myelin P2 promoter. J Neurosci Res. 1993;36:508–519. doi: 10.1002/jnr.490360503. [DOI] [PubMed] [Google Scholar]
- 11.Boll M, Foltz M, Rubio-Aliaga I, Kottra G, Daniel H. Functional characterization of two novel mammalian electrogenic proton-dependent amino acid cotransporters. J Biol Chem. 2002;277:22966–22973. doi: 10.1074/jbc.M200374200. [DOI] [PubMed] [Google Scholar]
- 12.Brady KP, Rowe LB, Her H, Stevens TJ, Eppig J, Sussman DJ, Sikela J, Beier DR. Genetic mapping of 262 loci derived from expressed sequences in a murine interspecific cross using single-strand conformational polymorphism analysis. Genome Res. 1997;7:1085–1093. doi: 10.1101/gr.7.11.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brockes JP, Fields P, Raff MC. Studies on cultured rat Schwann cells. I. Establishment of purified populations from cultures of peripheral nerve. Brain Res. 1979;165:105–118. doi: 10.1016/0006-8993(79)90048-9. [DOI] [PubMed] [Google Scholar]
- 14.Chirgwin JM, Przbyla AE, MacDonald RJ, Rutter RJ. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochem J. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 15.Coe NR, Bernlohr DA. Physiological properties and functions of intracellular fatty acid-binding proteins. Biochim Biophys Acta. 1998;1391:287–306. doi: 10.1016/s0005-2760(97)00205-1. [DOI] [PubMed] [Google Scholar]
- 16.Crawford AW, Pino JD, Beckerle MC. Biochemical and molecular characterization of the chicken cysteine-rich protein, a developmentally regulated LIM-domain protein that is associated with the actin cytoskeleton. J Cell Biol. 1994;124:117–127. doi: 10.1083/jcb.124.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dietrich WF, Miller J, Steen R, Merchant MA, Damron-Boles D, Husain Z, Dredge R, Daly MJ, Ingalls KA, O'Connor TJ, Evans CA, DeAngelis MM, Levinson DM, Kruglyak L, Goodman N, Copeland NG, Jenkins NA, Hawkins TL, Stein L, Page DC. A comprehensive genetic map of the mouse genome. Nature. 1996;380:149–152. doi: 10.1038/380149a0. [DOI] [PubMed] [Google Scholar]
- 18.Edman CF, Prigent SA, Schipper A, Feramisco JR. Identification of ErbB3-stimulated genes using modified representational difference analysis. Biochem J. 1997;323:113–118. doi: 10.1042/bj3230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Erkman L, Yates PA, McLaughlin T, McEvilly RJ, Whisenhunt T, O'Connell SM, Krones AI, Kirby MA, Rapaport DH, Bermingham JR, O'Leary DD, Rosenfeld MG. A POU domain transcription factor-dependent program regulates axon pathfinding in the vertebrate visual system. Neuron. 2000;28:779–792. doi: 10.1016/s0896-6273(00)00153-7. [DOI] [PubMed] [Google Scholar]
- 20.Fernandez-Valle C, Gorman D, Gomez AM, Bunge MB. Actin plays a role in both changes in cell shape and gene-expression associated with Schwann cell myelination. J Neurosci. 1997;17:241–250. doi: 10.1523/JNEUROSCI.17-01-00241.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frohman MA. Rapid amplification of complementary DNA ends for generation of full-length complementary DNAs: thermal RACE. Methods Enzymol. 1993;218:340–356. doi: 10.1016/0076-6879(93)18026-9. [DOI] [PubMed] [Google Scholar]
- 22.Frohman MA. On beyond classic RACE (rapid amplification of cDNA ends). PCR Methods Appl. 1994;4:S40–S58. doi: 10.1101/gr.4.1.s40. [DOI] [PubMed] [Google Scholar]
- 23.Fyodorov D, Deneris E. The POU domain of SCIP/Tst-1/Oct-6 is sufficient for activation of any acetylcholine receptor promoter. Mol Cell Biol. 1996;16:5004–5014. doi: 10.1128/mcb.16.9.5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gabreels-Festen A, van Beersum S, Eshuis L, LeGuern E, Gabreels F, van Engelen B, Mariman E. Study on the gene and phenotypic characterisation of autosomal recessive demyelinating motor and sensory neuropathy (Charcot-Marie-Tooth disease) with a gene locus on chromosome 5q23–q33. J Neurol Neurosurg Psychiatry. 1999;66:569–574. doi: 10.1136/jnnp.66.5.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garbay B, Heape AM, Sargueil F, Cassagne C. Myelin synthesis in the peripheral nervous system. Prog Neurobiol. 2000;61:267–304. doi: 10.1016/s0301-0082(99)00049-0. [DOI] [PubMed] [Google Scholar]
- 26.Ghislain J, Desmarquet-Trin-Dinh C, Jaegle M, Meijer D, Charnay P, Frain M. Characterisation of cis-acting sequences reveals a biphasic, axon-dependent regulation of Krox20 during Schwann cell development. Development. 2002;129:155–166. doi: 10.1242/dev.129.1.155. [DOI] [PubMed] [Google Scholar]
- 27.Glatz JF, Borchers T, Spener F, van der Vusse GJ. Fatty acids in cell signalling: modulation by lipid binding proteins. Prostaglandins Leukot Essent Fatty Acids. 1995;52:121–127. doi: 10.1016/0952-3278(95)90010-1. [DOI] [PubMed] [Google Scholar]
- 28.Guilbot A, Kessali M, Ravise N, Hammadouche T, Bouhouche A, Maisonobe T, Grid D, Brice A, Leguern E. The autosomal recessive form of CMT disease linked to 5q31–q33. Ann NY Acad Sci. 1999a;883:56–59. [PubMed] [Google Scholar]
- 29.Guilbot A, Ravise N, Bouhouche A, Coullin P, Birouk N, Maisonobe T, Kuntzer T, Vial C, Grid D, Brice A, LeGuern E. Genetic, cytogenetic and physical refinement of the autosomal recessive CMT linked to 5q31–q33: exclusion of candidate genes including EGR1. Eur J Hum Genet. 1999b;7:849–859. doi: 10.1038/sj.ejhg.5200382. [DOI] [PubMed] [Google Scholar]
- 30.Hassouna N, Michot B, Bachellerie JP. The complete nucleotide sequence of mouse 28S rRNA gene. Implications for the process of size increase of the large subunit rRNA in higher eukaryotes. Nucleic Acids Res. 1984;12:3563–3583. doi: 10.1093/nar/12.8.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He X, Gerrero R, Simmons DM, Park RE, Lin CJ, Swanson LW, Rosenfeld MG. Tst-1, a member of the POU domain gene family, binds the promoter of the gene encoding the cell surface adhesion molecule P0. Mol Cell Biol. 1991;11:1739–1744. doi: 10.1128/mcb.11.3.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henry EW, Eicher EM, Sidman RL. The mouse mutation claw paw: forelimb deformity and delayed myelination throughout the peripheral nervous system. J Hered. 1991;82:287–294. doi: 10.1093/oxfordjournals.jhered.a111088. [DOI] [PubMed] [Google Scholar]
- 33.Herb A, Wisden W, Catania MV, Marechal D, Dresse A, Seeburg PH. Prominent dendritic localization in forebrain neurons of a novel mRNA and its product, dendrin. Mol Cell Neurosci. 1997;8:367–374. doi: 10.1006/mcne.1996.0594. [DOI] [PubMed] [Google Scholar]
- 34.Hertzel AV, Bernlohr DA. Regulation of adipocyte gene expression by polyunsaturated fatty acids. Mol Cell Biochem. 1998;188:33–39. [PubMed] [Google Scholar]
- 35.Hubank M, Schatz DG. Identifying differences in mRNA expression by representational difference analysis of cDNA. Nucleic Acids Res. 1994;22:5640–5648. doi: 10.1093/nar/22.25.5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jaegle M, Meijer D. Role of Oct-6 in Schwann cell differentiation. Microsc Res Tech. 1998;41:372–378. doi: 10.1002/(SICI)1097-0029(19980601)41:5<372::AID-JEMT4>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 37.Jaegle M, Mandemakers W, Broos L, Zwart R, Karis A, Visser P, Grosveld F, Meijer D. The POU factor Oct-6 and Schwann cell differentiation. Science. 1996;273:507–510. doi: 10.1126/science.273.5274.507. [DOI] [PubMed] [Google Scholar]
- 38.Jain MK, Fujita KP, Hsieh CM, Endege WO, Sibinga NE, Yet SF, Kashiki S, Lee WS, Perrella MA, Haber E, Lee ME. Molecular cloning and characterization of SmLIM, a developmentally regulated LIM protein preferentially expressed in aortic smooth muscle cells. J Biol Chem. 1996;271:10194–10199. doi: 10.1074/jbc.271.17.10194. [DOI] [PubMed] [Google Scholar]
- 39.Kidd G, Andrews SB, Trapp BD. Axons regulate the distribution of Schwann cell microtubules. J Neurosci. 1996;16:946–954. doi: 10.1523/JNEUROSCI.16-03-00946.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 42.Laughon A, Boulet AM, Bermingham JR, Jr, Laymon RA, Scott MP. Structure of transcripts from the homeotic Antennapedia gene of Drosophila melanogaster: two promoters control the major protein-coding region. Mol Cell Biol. 1986;6:4676–4689. doi: 10.1128/mcb.6.12.4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.LeGuern E, Guilbot A, Kessali M, Ravise N, Tassin J, Maisonobe T, Grid D, Brice A. Homozygosity mapping of an autosomal recessive form of demyelinating Charcot-Marie-Tooth disease to chromosome 5q23–q33. Hum Mol Genet. 1996;5:1685–1688. doi: 10.1093/hmg/5.10.1685. [DOI] [PubMed] [Google Scholar]
- 44.Lewin B. Genes V. Oxford UP; New York: 1994. [Google Scholar]
- 45.Liebhaber SA, Emery JG, Urbanek M, Wang XK, Cooke NE. Characterization of a human cDNA encoding a widely expressed and highly conserved cysteine-rich protein with an unusual zinc-finger motif. Nucleic Acids Res. 1990;18:3871–3879. doi: 10.1093/nar/18.13.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lisitsyn NA. Representational difference analysis: finding the differences between genomes. Trends Genet. 1995;11:303–307. doi: 10.1016/s0168-9525(00)89087-3. [DOI] [PubMed] [Google Scholar]
- 47.Lisitsyn NA, Wigler M. Cloning the differences between two complex genomes. Science. 1993;259:946–951. doi: 10.1126/science.8438152. [DOI] [PubMed] [Google Scholar]
- 48.Lisitsyn NA, Wigler M. Representational difference analysis in detection of genetic lesions in cancer. Methods Enzymol. 1995;254:291–304. doi: 10.1016/0076-6879(95)54021-0. [DOI] [PubMed] [Google Scholar]
- 49.Louis HA, Pino JD, Schmeichel KL, Pomies P, Beckerle MC. Comparison of three members of the cysteine-rich protein family reveals functional conservation and divergent patterns of gene expression. J Biol Chem. 1997;272:27484–27491. doi: 10.1074/jbc.272.43.27484. [DOI] [PubMed] [Google Scholar]
- 50.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 51.McIntire SL, Reimer RJ, Schuske K, Edwards RH, Jorgensen EM. Identification and characterization of the vesicular GABA transporter. Nature. 1997;389:870–876. doi: 10.1038/39908. [DOI] [PubMed] [Google Scholar]
- 52.Mirsky R, Jessen KR. Schwann cell development, differentiation and myelination. Curr Opin Neurobiol. 1996;6:89–96. doi: 10.1016/s0959-4388(96)80013-4. [DOI] [PubMed] [Google Scholar]
- 53.Mirsky R, Jessen KR. The neurobiology of Schwann cells. Brain Pathol. 1999;9:293–311. doi: 10.1111/j.1750-3639.1999.tb00228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Monuki ES, Weinmaster G, Kuhn R, Lemke G. SCIP: a glial cell POU domain gene regulated by cyclic AMP. Neuron. 1989;3:783–793. doi: 10.1016/0896-6273(89)90247-x. [DOI] [PubMed] [Google Scholar]
- 55.Monuki ES, Kuhn R, Weinmaster G, Trapp BD, Lemke G. Expression and activity of the POU transcription factor SCIP. Science. 1990;249:1300–1303. doi: 10.1126/science.1975954. [DOI] [PubMed] [Google Scholar]
- 56.Monuki ES, Kuhn R, Lemke G. Repression of the Myelin-P(0) gene by the POU transcription factor SCIP. Mech Dev. 1993;42:15–32. doi: 10.1016/0925-4773(93)90095-f. [DOI] [PubMed] [Google Scholar]
- 57.Nagarajan R, Svaren J, Le N, Araki T, Watson M, Milbrandt J. EGR2 mutations in inherited neuropathies dominant-negatively inhibit myelin gene expression. Neuron. 2001;30:355–368. doi: 10.1016/s0896-6273(01)00282-3. [DOI] [PubMed] [Google Scholar]
- 58.Narayanan V, Barbosa E, Reed R, Tennekoon G. Characterization of a cloned cDNA encoding rabbit myelin P2 protein. J Biol Chem. 1988;263:8332–8337. [PubMed] [Google Scholar]
- 59.Narayanan V, Kaestner KH, Tennekoon GI. Structure of the mouse myelin P2 protein gene. J Neurochem. 1991;57:75–80. doi: 10.1111/j.1471-4159.1991.tb02101.x. [DOI] [PubMed] [Google Scholar]
- 60.Neuner-Jehle M, Denizot JP, Borbely AA, Mallet J. Characterization and sleep deprivation-induced expression modulation of dendrin, a novel dendritic protein in rat brain neurons. J Neurosci Res. 1996;46:138–151. doi: 10.1002/(SICI)1097-4547(19961015)46:2<138::AID-JNR2>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 61.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 62.Pinkstaff JK, Chappell SA, Mauro VP, Edelman GM, Krushel LA. Internal initiation of translation of five dendritically localized neuronal mRNAs. Proc Natl Acad Sci USA. 2001;98:2770–2775. doi: 10.1073/pnas.051623398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Poduslo JF. Regulation of myelin gene expression in the peripheral nervous system. In: Dyck PJ, Thomas PK, Griffin JW, Low PA, Poduslo JF, editors. Peripheral neuropathy. Saunders; Philadelphia: 1993. pp. 282–289. [Google Scholar]
- 64.Porter S, Glaser L, Bunge RP. Release of autocrine growth factor by primary and immortalized Schwann cells. Proc Natl Acad Sci USA. 1987;84:7768–7772. doi: 10.1073/pnas.84.21.7768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Raynal F, Michot B, Bachellerie JP. Complete nucleotide sequence of mouse 18 S rRNA gene: comparison with other available homologs. FEBS Lett. 1984;167:263–268. doi: 10.1016/0014-5793(84)80139-8. [DOI] [PubMed] [Google Scholar]
- 66.Ryan AK, Rosenfeld MG. POU domain family values: flexibility, partnerships, and developmental codes. Genes Dev. 1997;11:1207–1225. doi: 10.1101/gad.11.10.1207. [DOI] [PubMed] [Google Scholar]
- 67.Sagne C, El Mestikawy S, Isambert MF, Hamon M, Henry JP, Giros B, Gasnier B. Cloning of a functional vesicular GABA and glycine transporter by screening of genome databases. FEBS Lett. 1997;417:177–183. doi: 10.1016/s0014-5793(97)01279-9. [DOI] [PubMed] [Google Scholar]
- 68.Sagne C, Agulhon C, Ravassard P, Darmon M, Hamon M, El Mestikawy S, Gasnier B, Giros B. Identification and characterization of a lysosomal transporter for small neutral amino acids. Proc Natl Acad Sci USA. 2001;98:7206–7211. doi: 10.1073/pnas.121183498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sambrook J, Maniatis T, Fritsch EF. Molecular cloning: a laboratory manual, Ed 2. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 70.Sandoval IV, Martinez-Arca S, Valdueza J, Palacios S, Holman GD. Distinct reading of different structural determinants modulates the dileucine-mediated transport steps of the lysosomal membrane protein LIMPII and the insulin-sensitive glucose transporter GLUT4. J Biol Chem. 2000;275:39874–39885. doi: 10.1074/jbc.M006261200. [DOI] [PubMed] [Google Scholar]
- 71.Scherer SS, Salzer JL. Axon-Schwann cell interactions in peripheral nerve regeneration. In: Jessen KR, Richardson WD, editors. Glial cell development. Oxford UP; Oxford: 2001. pp. 299–330. [Google Scholar]
- 72.Simmons D, Arriza JL, Swanson DW. Complete protocol for in situ hybridization in brain and other tissues with radiolabeled single-stranded RNA probes. J Histotechnol. 1989;12:169–181. [Google Scholar]
- 73.Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- 74.Stahl N, Harry J, Popko B. Quantitative analysis of myelin protein gene expression during development in the rat sciatic nerve. Brain Res Mol Brain Res. 1990;8:209–212. doi: 10.1016/0169-328x(90)90018-9. [DOI] [PubMed] [Google Scholar]
- 75.Taylor V, Suter U. Molecular biology of axon-glia interactions in the peripheral nervous system. Prog Nucleic Acid Res Mol Biol. 1997;56:225–256. [PubMed] [Google Scholar]
- 76.Topilko P, Schneider-Maunoury S, Levi G, Baron-Van Evercooren A, Chennoufi AB, Seitanidou T, Babinet C, Charnay P. Krox-20 controls myelination in the peripheral nervous system. Nature. 1994;371:796–799. doi: 10.1038/371796a0. [DOI] [PubMed] [Google Scholar]
- 77.Tusnady GE, Simon I. Topology of membrane proteins. J Chem Inf Comput Sci. 2001;41:364–368. doi: 10.1021/ci0001280. [DOI] [PubMed] [Google Scholar]
- 78.Uyemura K, Yoshimura K, Suzuki M, Kitamura K. Lipid binding activities of the P2 protein in peripheral nerve myelin. Neurochem Res. 1984;9:1509–1514. doi: 10.1007/BF00964676. [DOI] [PubMed] [Google Scholar]
- 79.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 80.Webster HD. Development of peripheral nerve fibers. In: Dyck PJ, Thomas PK, Low PA, Poduslo JF, editors. Peripheral neuropathy. Saunders; Philadelphia: 1993. pp. 243–266. [Google Scholar]
- 81.Weinstein DE, Burrola PG, Lemke G. Premature Schwann cell differentiation and hypermyelination in mice expressing a targeted antagonist of the POU transcription factor SCIP. Mol Cell Neurosci. 1995;6:212–229. doi: 10.1006/mcne.1995.1018. [DOI] [PubMed] [Google Scholar]
- 82.Weiskirchen R, Bister K. Suppression in transformed avian fibroblasts of a gene (crp) encoding a cysteine-rich protein containing LIM domains. Oncogene. 1993;8:2317–2324. [PubMed] [Google Scholar]
- 83.Weiskirchen R, Pino JD, Macalma T, Bister K, Beckerle MC. The cysteine-rich protein family of highly related LIM domain proteins. J Biol Chem. 1995;270:28946–28954. doi: 10.1074/jbc.270.48.28946. [DOI] [PubMed] [Google Scholar]
- 84.Weiskirchen R, Erdel M, Utermann G, Bister K. Cloning, structural analysis, and chromosomal localization of the human CSRP2 gene encoding the LIM domain protein CRP2. Genomics. 1997;44:83–93. doi: 10.1006/geno.1997.4855. [DOI] [PubMed] [Google Scholar]
- 85.Wu R, Jurek M, Sundarababu S, Weinstein DE. The POU gene Brn-5 is induced by neuregulin and is restricted to myelinating Schwann cells. Mol Cell Neurosci. 2001;17:683–695. doi: 10.1006/mcne.2000.0957. [DOI] [PubMed] [Google Scholar]
- 86.Yet SF, Folta SC, Jain MK, Hsieh CM, Maemura K, Layne MD, Zhang D, Marria PB, Yoshizumi M, Chin MT, Perrella MA, Lee ME. Molecular cloning, characterization, and promoter analysis of the mouse Crp2/SmLim gene. Preferential expression of its promoter in the vascular smooth muscle cells of transgenic mice. J Biol Chem. 1998;273:10530–10537. doi: 10.1074/jbc.273.17.10530. [DOI] [PubMed] [Google Scholar]
- 87.Young GB, Jack DL, Smith DW, Saier MH., Jr The amino acid/auxin:proton symport permease family. Biochim Biophys Acta. 1999;1415:306–322. doi: 10.1016/s0005-2736(98)00196-5. [DOI] [PubMed] [Google Scholar]
- 88.Zorick TS, Lemke G. Schwann cell differentiation. Curr Opin Cell Biol. 1996;8:870–876. doi: 10.1016/s0955-0674(96)80090-1. [DOI] [PubMed] [Google Scholar]
- 89.Zorick TS, Syroid DE, Arroyo E, Scherer SS, Lemke G. The transcription factors SCIP and Krox-20 mark distinct stages and cell fates in Schwann cell differentiation. Mol Cell Neurosci. 1996;8:129–145. doi: 10.1006/mcne.1996.0052. [DOI] [PubMed] [Google Scholar]