Abstract
Taste buds are a heterogeneous population of cells exhibiting diverse morphological and biochemical characteristics. Because taste buds arise from multiple progenitors, the different types of taste cells may represent distinct lineages. The present study was undertaken to determine the following: (1) how many progenitors contribute to a taste bud, and (2) whether the specific subpopulation of serotonin-immunoreactive (IR) taste cells are related by lineage to a restricted set of progenitor cells. These questions were addressed using cell lineage analysis of taste buds from H253 X-inactivation mosaic mice. After random X-inactivation of the lacZtransgene, the tongue of hemizygous female mice displays discrete patches of epithelial cells, which are either β-galactosidase (β-gal) positive or β-gal negative. By analyzing the proportion of the two differently stained cell populations in taste buds located at the boundary between positive and negative epithelial patches, we can determine the minimum number of progenitors that may contribute to the formation of a taste bud. The presence of taste buds containing only 6–12% labeled cells indicates that at least eight progenitors contribute to an average taste bud of 55 cells, assuming progenitors contribute equally to the cell population. Cell lineage analysis of serotonin-IR taste cells in such mixed taste buds suggests that this subpopulation likely arises from only one to two progenitors and often is related by lineage. Thus, at least some of the cell types in a taste bud represent distinct lineages of cells and are not merely phenotypic stages as a cell progresses from a young to a mature state.
Keywords: gustatory, development, basal cell, cell lineage, mosaic analysis, type III cell, serotonin
Taste buds, the sensory endorgans for the sense of taste, consist of ∼50–75 spindle-shaped neuroepithelial cells (Finger and Simon, 2000). Taste bud cells are heterogeneous in terms of ultrastructure as well as immunocytochemical profiles (Lindemann, 1996; Finger and Simon, 2000). Based on ultrastructural characteristics, several different subclasses of intragemmal taste cells have been identified: basal cells, type I (dark) cells, type II (light) cells, and type III (intermediate) cells (Farbman, 1965a; Murray, 1969, 1973; Takeda, 1977; Takeda et al., 1981; Farbman et al., 1985; Kinnamon et al., 1985; Delay et al., 1986; Yee et al., 2001).
The significance of the different taste cell types, both in terms of function and cell lineage, is controversial. Like other epithelial cells, taste cells are continually replaced by a proliferative basal cell population (Beidler and Smallman, 1965; Conger and Wells, 1969;Farbman, 1980). Consequently, cells in each taste bud vary in age. Thus, phenotypic differences may reflect the life history of the cell in a taste bud. Conversely, phenotypic differences may reflect functional diversity. For example, some investigators suggest that type III cells are the primary receptor cells of the taste bud, whereas type I cells play a supportive or glia-like role (Murray and Murray, 1971;Murray, 1986; Lindemann, 1996; Lawton et al., 2000). Two conflicting views exist in terms of cell lineage: (1) different taste cell types represent separate lineages that maintain a stable phenotype throughout the lifespan of the cells (Farbman, 1965a,b; Fujimoto and Murray, 1970, 1971, 1980; Pumplin et al., 1997), or (2) the taste cells change substantially with age, and the similarity in phenotype is associated with the acquisition of different functions by cells belonging to a common lineage (Delay et al., 1986). Even if separate lineages exist, some characteristics might appear only under certain conditions or states of maturation but could nonetheless be restricted to particular types of taste cells. For example, only some taste cells have the ability to accumulate serotonin, and these are all type III cells (Yee et al., 2001). However, not all type III cells accumulate serotonin. Whether this capability is related to lineage, functional status, or maturational state of the cell is unknown.
Previous analysis (Stone et al., 1995) of lineage complexity of X-inactivation mosaic tongue tissues has shown that multiple progenitors give rise to individual taste buds. Thus, different cell lineages may exist within a single taste bud. The present study was undertaken to investigate whether a subpopulation of taste cells defined by the serotonin phenotype is a distinct sublineage arising from developmental mechanisms or alternatively consists of taste cells undergoing a serotonergic stage of their life cycle. This question was addressed using cell lineage analysis, which is able to retrospectively identify genealogical relationships among cells arising from common ancestry.
MATERIALS AND METHODS
Mice. The mice used in these studies were X-inactivation mosaic females hemizygous for an X-linkedlacZ marker (Tam and Tan, 1992). This H253 line of mice carries ∼14 tandem copies of an 8.9 kb fragment containing the promoter of the mouse housekeeping gene 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase, linked to theEscherichia coli lacZ gene (Gautier et al., 1989). All studies reported herein were undertaken with the approval of the University of Colorado Institutional Animal Care and Use Committee and in accord with the guidelines of the Society for Neuroscience.
Because the transgenic marker is linked to the X chromosome, natural mosaics can be produced by mating male H253 mice to wild-type females. This mating results in female progeny that are hemizygous for thelacZ marker, with the transgene present only on the paternally inherited X chromosome. During development of mammalian females, one of the two X chromosomes is randomly inactivated in each cell (Lyon, 1961). Inactivation in mouse ectodermal and mesodermal lineages is virtually completed by embryonic day 9.5 (E9.5) (Tan et al., 1993); therefore, all taste cells in the emerging tongue primordia at E11.5 (Paulson et al., 1985; Kaufman, 1992) would have a 50:50 composition, with one-half of the cells expressing the lacZgene product β-galactosidase (β-gal).
Lingual tissue from transgenic male mice was used as a positive control for evaluation of β-gal expression in cells containing an active transgene. X-Inactivation does not occur in males; thus, in H253 transgenic males, all cells should be β-gal positive (β-gal+).
Simvastatin treatment. There is some variability in the expression of β-galactosidase in adult mouse tongue tissue (data not shown). To decrease this variability and to increase the production of β-galactosidase in cells retaining the active marker, Simvastatin was administered orally (1 mg/100 ml drinking water for 30–40 d) to both male transgenic mice and female mosaic mice. Simvastatin lowers cholesterol, a negative inhibitor of HMG-CoA reductase (Goldstein and Brown, 1990). Lower cholesterol, therefore, results in an increase in the production of HMG-CoA reductase and in H253 mice, an increase in expression of the lacZ gene driven by the HMG-CoAreductase promoter.
Tissue preparation for analysis of lingual epithelium and taste buds from male H253 transgenic mice. Lingual tissue from male H253 mice was examined to determine the pattern of β-gal staining in transgenic male mice. Anesthetized mice (0.1 cc, i.p., 50 mg/ml, Nembutal sodium solution; Abbott Laboratories, Chicago, IL) were perfused transcardially with 4% paraformaldehyde in 0.1m phosphate buffer, and the tongues were removed. Fresh fixative was used to postfix lingual tissue for 45 min, after which it was cryoprotected in 20% sucrose in 0.1m phosphate buffer overnight. The following morning, the tongue tissue was cryosectioned at 50–200 μm thickness. Resultant sections were washed three times, for 20 min each (3 × 20 min), in washing buffer (2 mmMgCl2, 5 mm EGTA, 0.01% Na desoxycholate, and 0.02% Nonidet P-40 in 0.1 mphosphate buffer) and then incubated overnight at 37°C in X-gal solution: 0.1% 4-chloro-5-bromo-3-indolyl-β-d-galactopyranoside (X-gal) (Sigma, St, Louis, MO), 5 mmK3Fe(CN)6, and 5 mmK4Fe(CN)6 · 6H2O, in washing buffer. Stained sections were washed in 0.1 mphosphate buffer 3 × 10 min, rinsed in distilled water for 10 min, and dehydrated in graded ethanols, followed by acetone and put into infiltration solution (Historesin Plus basic resin with activator; catalog #70-2224-861; Jung) overnight. The following morning, sections were placed into embedding solution (infiltration solution plus hardener) poured into molds.
Serial sections of 5–10 μm thickness were cut from polymerized plastic blocks after removal from the molds. These sections were counterstained with nuclear fast red, dehydrated through graded ethanols, cleared with Histoclear, coverslipped with Permount, and examined.
Numerical analysis of taste bud composition. To estimate the number of progenitors contributing to individual taste buds, β-gal+ and β-gal-negative (β-gal−) cells were counted in individual taste buds containing few β-gal+ cells. Hemizygous female transgenic mice were anesthetized and perfused by the same procedure described for male H253 mice. Postfixation of tongue tissue and cryoprotection also were identical to the procedures described for male mice. After cryoprotection, regions containing the circumvallate papillae, fungiform papillae, or palatal taste buds were cryosectioned at 50–70 μm thickness. Cryosectioning was done so that taste buds could be viewed longitudinally. Most counts were done using circumvallate taste buds because of the high concentration of taste buds in these papillae. Plastic sections of X-gal-stained material were produced as above for tissue from male tongues.
Image processing and analysis of mixed taste bud counts.Using both photomicrographs and mounted tissue sections, β-gal+ (blue plus red) and β-gal− (red) intragemmal nuclei were counted in serial sections through entire taste buds. Inclusion of perigemmal cells in these counts was avoided because of the lack of β-gal activity in this cell population, even in nonmosaic mice (see Results, Lingual epithelium and taste buds from male H253 mice). Several features were used to distinguish between the nuclei of intragemmal taste cells and perigemmal or epithelial cell nuclei. Taste cell nuclei are more elongate and thinner than nuclei of surrounding epithelial cells; in general, taste cells and their nuclei are oriented approximately perpendicular to the surface of the crypt. In addition, in nuclear fast red counterstained sections, the cytoplasmic region of intragemmal taste cells was lighter than that of surrounding epithelial cells. This staining difference resulted in a distinct boundary around most taste buds. Although elongated and oriented similarly to taste cells, perigemmal cells contained less cytoplasm and were thinner than taste cells. These differences were obvious in central sections of taste buds in which the perigemmal cells were lateral to intragemmal cells. However, in the first and last sections of a taste bud, the distinction between intragemmal and perigemmal cells was less clear. Therefore, some perigemmal cells may have been included in the counts. The width of β-gal+ nuclei ranged in size from 2.5 to 7.5 μm at their widest point (n = 9), and the width of β-gal− nuclei was similar, ranging in size from 2 to 8 μm (n = 67). Because the nuclei were similar in size, the sampling method used was unbiased. However, some nuclei may have been counted twice; serial sections were 5–10 μm in thickness, resulting in larger nuclei being present in two sections.
Analysis of the number of taste bud progenitors. To provide a rough estimate of the minimal number of progenitor cells contributing to individual taste buds, the composition of mixed taste buds in the overall population was determined. These values can be compared with the proportions expected from a binomial distribution if two assumptions are made: (1) progenitors contribute equally to the taste bud population, and (2) different taste cells have approximately the same lifespan. If these assumptions are correct, then the proportions of β-gal-expressing and -nonexpressing cells in individual taste buds will resemble a binomial distribution. For example, if two progenitors contribute equally to a taste bud population, every mixed taste bud would be expected to contain ∼50% β-gal+ cells; three progenitors would result in mixed taste buds containing 33 or 66% β-gal+ cells, etc. The lowest percentage of β-gal+ cells in mixed taste buds would be 50% if there were two progenitors but would be 33% if there were three progenitors (Fig. 1). Thus, the lower the contribution by β-gal+ cells in a taste bud, the greater the predicted number of progenitors for that bud.
Fig. 1.
The expected results of two or three progenitors giving rise to cells in a taste bud if the progenitors contribute equally to the cell population. Dark shadingindicates β-gal+ cells, and white indicates β-gal− cells.
Eleven taste buds of the hundreds examined in the three mosaic mice exhibiting circumvallate mosaicism were selected for scoring because they contained both β-gal+ and β-gal− cells, and they had a low number of β-gal+ cells. Eight of the samples were from circumvallate papillae, two were from fungiform papillae, and one was from a palatal taste bud. In the fungiform and palatal samples, taste buds were more scattered and generally contained many β-gal+ cells and therefore were not included in the present analysis. Initial analysis of the circumvallate taste buds revealed a wide variation in the number of β-gal+ cells. Subsequent analysis was focused on those with few β-gal+ cells. The number of taste buds containing few β-gal− cells in a mostly β-gal+ taste bud was not determined because it is difficult to detect the few unstained cells amid a blue sea of β-gal+ cells.
Serotonin cell lineage analysis. Taste cells recognized by anti-serotonin antibodies [5-HT immunoreactive (IR)] after pretreatment with the serotonin precursor 5-hydroxy-l-tryptophan (5-HTP) were chosen for initial taste cell lineage analysis because the relatively few (5–10) 5-HT-IR cells in mouse taste buds (Uchida, 1985; Fujimoto et al., 1987;Kim and Roper, 1995) facilitates statistical analysis.
Tissue preparation for 5-HT-IR cell lineage analysis. The cell lineage of 5-HT-IR taste cells was determined using mosaic analysis of taste buds double labeled with anti-β-galactosidase antibodies and anti-serotonin antibodies, followed by fluorescent-labeled secondary antibodies. Mice were treated with Simvastatin as described previously and then injected intraperitoneally with 5-HTP (80 mg/kg body weight; catalog #3753; Sigma). One hour after 5-HTP injection, mice were anesthetized and perfused transcardially as above. In some cases, perfusion with fixative was preceded by 10–15 ml of 0.9% NaCl (Baxter, McGaw Park, IL). After perfusion, tongues were removed and placed in fresh fixative for 45–60 min. Postfixation was followed by cryoprotection in 20% sucrose in 0.1m phosphate buffer overnight. On the next day, 40–50 μm cryosections were collected in 0.1 mPBS.
Immunocytochemistry. Two protocols were used for immunolabeling three sets of sections. The first set of sections was double labeled with anti-β-gal and anti-5-HT antisera. The second set of sections was triple labeled with β-gal antibodies, 5-HT antibodies, and 4′-6-diamidin-2-phenylindol-dihydrochlorid (DAPI) (a fluorescent nuclear marker). A third set of sections was triple labeled with both antibodies and propidium iodide (a different nuclear marker). The first two sets of sections were treated as follows. Cryosections were washed in PBS (3 × 10 min), placed into blocking solution [1% bovine serum albumin (BSA), 1% normal horse serum, and 0.3% Triton X-100 in PBS] for 1–2 hr, and then incubated in a mixture of the polyclonal, primary antisera: goat anti-serotonin (1:200 dilution; catalog #108072; Incstar, Stillwater, MN) and rabbit anti-β-galactosidase (1:1000 dilution; catalog #38952; Cappel, Cochranville, PA) in blocking solution. Taste cells labeled with the anti-serotonin antibodies are referred to as 5-HT-IR in this paper, although no attempt was made in this study to determine exactly the chemical nature of the substance recognized by the anti-serotonin antibody. Primary antibody incubation lasted for 36–48 hr, and then sections were washed in PBS 3 × 10 min and incubated for 2–18 hr in a mixture of secondary antibodies: FITC donkey anti-goat (1:100 dilution; catalog #27384; Jackson ImmunoResearch, West Grove, PA) and Lissamine rhodamine donkey anti-rabbit (1:100 dilution; catalog #27162; Jackson ImmunoResearch), followed by washes in PBS 3 × 10 min. The second set of sections was then incubated in DAPI (0.33 gm/ml; catalog #12930720-31; Boehringer Mannheim, Indianapolis, IN) for 2 min and washed 3 × 10 min in PBS. A third set of cryosections was triple labeled with anti-β-gal, anti-5-HT antisera, and propidium iodide. These were treated similarly to double-labeled and triple-labeled DAPI sections with the following exceptions: sections were incubated in PBS containing 2 mg/ml BSA (catalog #A-2153; Sigma) and 10% donkey serum (DS) (catalog #017-000-121; Jackson ImmunoResearch) for 1–2 hr before primary antibody application, which consisted of PBS, BSA, DS, rabbit anti-β-gal (1:1000; catalog #55976; Cappel), and goat anti-5-HT (1:200; catalog #20079; DiaSorin, Stillwater, MN). Sections were left in primary antibody for 2 d at 4°C, washed 3 × 10 min in PBS, and incubated overnight at 4°C in secondary antisera: FITC donkey anti-rabbit (1:100; catalog #711-095-152; Jackson ImmunoResearch) and Cy5 donkey anti-goat (1:100; catalog #705-175-147; Jackson ImmunoResearch). After secondary incubation, sections were washed 3 × 10 min in PBS and placed into 0.1 m PBS containing 10 mg/ml MgCl2 and 250 μg/ml RNase A (Sigma) for 30 min at 35°C. After washing again in PBS, tissue was placed in 0.5 μg/ml propidium iodide (Sigma) in 0.1 m phosphate buffer for 1 min, followed by 3 × 10 min in PBS. Sections then were mounted on slides and coverslipped with Fluoromount G (Southern Biotechnology, Alabaster, AL). All experiments in each set included a negative control consisting of sections treated identically to the experimental sections except for a lack of primary antisera exposure. No inappropriate cross-reactivity by the secondary antisera was present in any of the sections.
Image processing and analysis. The circumvallate papillae of three mosaic mice were sectioned and double labeled with antibodies to β-gal and 5-HT, followed by the appropriate fluorescently tagged secondary antibodies. Taste buds with few β-gal-IR cells were chosen for analysis (n = 16 buds) to increase the probability that the β-gal-IR cells were derived from a single, or few, progenitor cells. The 16 taste buds were analyzed by confocal microscopy. Two sets of confocal images were collected with a Bio-Rad (Hercules, CA) 600 laser scanning confocal microscope equipped with a helium–xenon laser and K1 and K2 filter blocks for simultaneous analysis of FITC and Lissamine rhodamine fluorescence. Two Z-series consisting of three to six images with 3–5 μm steps between images were collected for each taste bud. One Z-series was taken to detect 5-HT-IR and the other to detect β-gal-IR. The parameters for each Z-series of a pair were identical so that the β-gal-IR images and the 5-HT-IR images could be merged, with Photoshop software (Adobe Systems, San Jose, CA), into a series of double-labeled color images. The number of β-gal-IR nuclei, 5-HT-IR cells, and double-labeled (β-gal-IR and 5-HT-IR) cells were counted for the portions of each taste bud contained within the Z-series. Because of a nuclear localization signal associated with the β-gal transgene, the β-gal-IR is nuclear, whereas 5-HT-IR is cytoplasmic. Accordingly, it was easier to locate and identify 5-HT-IR taste cells because of the larger volume occupied by immunoreactive cytoplasm compared with the nucleus. Thus, to avoid biased sampling, 5-HT-IR cells were counted only if the nucleus was visible. In addition, eight of the taste buds were counterstained with the nuclear stain DAPI and examined with aZeiss (Oberkochen, Germany) standard microscope equipped for epifluorescence. Lissamine–rhodamine (β-gal-IR nuclei), fluorescein (5-HT-IR cells), and DAPI (all nuclei) labeling were viewed and photographed separately. The resulting color slides were digitally scanned, and the images were combined using Photoshop software to produce tri-color images. Both images and microscopic viewing were used to count the number of DAPI-labeled nuclei, β-gal-IR nuclei, and 5-HT-IR cells in sections through taste buds.
Circumvallate and foliate papillae were collected from five additional mice, sectioned, and triple labeled with antibodies to β-gal and 5-HT, followed by their appropriate secondary antibodies and then propidium iodide. These sections also were analyzed by confocal microscopy. Two sets of confocal images were collected for each selected region, one to detect β-gal (FITC secondary antibody) and propidium iodide (red fluorescent) labeling and one to detect 5-HT-IR (Cy5 secondary antibody). Each set consisted of a series of images collected 3–5 μm apart, and the parameters for each set of a pair were identical as described for β-gal, 5-HT-IR double-labeled sections. Thus, triple-labeled images were obtained by merging two images collected in a single focal plane using Photoshop software. Merged images of single confocal sections were used to count β-gal-IR cells, 5-HT-IR cells, double-labeled cells, and nuclei (stained with propidium iodide) in selected taste buds (n = 16).
Taste buds from all three sets of sections (double labeled and triple labeled with DAPI or propidium iodide; n = 32 total buds) were used to determine whether the 5-HT-IR cells in individual taste buds arise from one or multiple progenitors. Sections triple labeled with both antibodies and propidium iodide (n = 16) were used to study the cell lineage of 5-HT-IR cells.
Statistical analysis. Counts of labeled nuclei obtained from tri-color images were used to test whether 5-HT-IR taste cells are related by lineage. The final numerical analysis was done using sections labeled with both antisera and propidium iodide because all three labels were identifiable with confocal microscopy (n = 16; see Table 2). The null hypothesis tested was that the presence of β-gal-IR and 5-HT-IR are independent events. For this analysis, the expected distribution of propidium iodide-labeled nuclei, 5-HT-IR cells with nuclei, β-gal-IR nuclei, and double-labeled nuclei (5-HT-IR and β-gal-IR) was compared by χ2 analysis with the actual distribution of labeled nuclei. To determine the expected distribution of independently labeled nuclei, the following calculations were made:Dp =pDX * N, whereDp is the expected number of double-labeled cells in a taste bud,pD is the probability of double-labeled cells in that taste bud, and N is the total number of cells sampled, assuming independent events;pD =pβX *ps, wherepβ is the probability of β-gal+ cells in a taste bud, and ps is the probability of 5-HT-IR cells; and pβ= nβ/N ps =ns/N, wherenβ is the total number of β-gal+ cells in a taste bud, and ns is the total number of 5-HT-IR cells in that taste bud.
Table 2.
Cell lineage analysis of 5-HT-IR taste cells in buds triple labeled with 5-HT antibodies, β-gal antibodies, and propidium iodide
| Total number cells (N) | Total β-gal-IR cells | β-gal-IR only cells | Double-labeled cells | 5-HT-IR only cells | Total 5-HT-IR cells | p value | |
|---|---|---|---|---|---|---|---|
| TB 1 | 11 | 4 | 0 | 4 | 0 | 4 | 0.01 |
| TB 2 | 9 | 6 | 3 | 3 | 0 | 3 | 0.52 |
| TB 3 | 8 | 3 | 3 | 0 | 2 | 2 | 0.66 |
| TB 4 | 10 | 4 | 3 | 1 | 1 | 2 | 0.99 |
| TB 5 | 13 | 3 | 1 | 2 | 1 | 3 | 0.24 |
| TB 6 | 8 | 5 | 0 | 5 | 0 | 5 | 0.05 |
| TB 7 | 15 | 2 | 0 | 2 | 1 | 3 | 0.03 |
| TB 8 | 16 | 3 | 3 | 0 | 3 | 3 | 0.84 |
| TB 9 | 16 | 2 | 1 | 1 | 2 | 3 | 0.70 |
| TB 10 | 16 | 5 | 3 | 2 | 0 | 2 | 0.17 |
| TB 11 | 20 | 7 | 2 | 5 | 0 | 5 | 0.006 |
| TB 12 | 14 | 5 | 3 | 2 | 0 | 2 | 0.24 |
| TB 13 | 17 | 6 | 2 | 4 | 0 | 4 | 0.02 |
| TB 14 | 24 | 12 | 6 | 6 | 1 | 7 | 0.17 |
| TB 15 | 11 | 5 | 2 | 3 | 0 | 3 | 0.17 |
| TB 16 | 10 | 4 | 3 | 1 | 1 | 2 | 0.99 |
β-gal-IR cells, 5-HT-IR cells, cells double labeled for β-gal and 5-HT, and propidium iodide-labeled cells were counted in single confocal sections of 16 taste buds. The distribution of these counts was compared with that expected if β-gal-IR and 5-HT-IR labeled cells independently of each other (see Materials and Methods). χ2 analysis was used to compare the distributions. The resultant p value for each taste bud is listed in the far right column. A p value of 0.05 or less indicated that the null hypothesis (that the labels are independent of each other) should be rejected at the 95% confidence level. Samples with only one β-gal+ cell were excluded from analysis because such taste buds provide no information about lineage. N was obtained by counting nuclei stained with propidium iodide in a specified region. TB, Taste bud.
The above values were determined for a series of sections through each taste bud using actual counts of the total number of β-gal-IR cells, the total number of 5-HT-IR cells, and the total number of propidium iodide-labeled cells in serial confocal images through sections of taste buds. Values for the probability of single-labeled cells were calculated by subtracting the expected number of double-labeled cells from the total number of either 5-HT-IR cells or β-gal-IR cells. The calculated values for 5-HT-IR only cells, β-gal-IR only cells, and double-labeled cells were then compared by χ2 analysis, with the actual counts shown in Table 2.
RESULTS
Lingual epithelium and taste buds from male H253 mice
To ensure that the relevant cells constitutively express reductase-driven lacZ, we examined the lingual epithelium of male transgenic mice treated with Simvistatin. Although not all cells in the adult male transgenic mice expressed β-gal, the tongue tissue displayed uniform β-gal activity in all cells of the cornified, granular, and spinous layers of the lingual epithelium (Fig.2). In contrast, the basal layer showed little or no β-gal activity. All of the intragemmal cells of the taste bud strongly expressed β-gal but not the perigemmal cells surrounding the taste bud (Fig. 2).
Fig. 2.
Light microscopic image of lingual tissue from an adult male H253 mouse treated with Simvistatin. Tissue was sectioned, stained with X-gal, and then counterstained with nuclear fast red. Blue, nuclear precipitate indicates β-gal activity. Basal cells in nontaste bud-bearing epithelium lack X-gal staining (arrow), as do perigemmal cells surrounding the taste bud (arrowhead). Scale bar, 20 μm.
Proportions of β-gal+ cells in mixed taste buds
For lineage analysis, only taste buds with few (2–18) β-gal+ cells were used. The presence of few β-gal+ cells in a taste bud increases the probability that the β-gal+ cells arise from the same progenitor. Cell lineage questions were addressed by using immunocytochemistry to identify taste cells labeled by 5-HT antibodies (5-HT-IR), β-gal antibodies, or both and then analyzing the relationship between 5-HT immunoreactivity and β-gal immunoreactivity. If 5-HT-IR cells arise from a single progenitor, either all cells would be double labeled (with 5-HT-IR and β-gal-IR) or all single labeled (5-HT-IR only). In contrast, if a taste bud contains both 5-HT-IR, β-gal-IR double-labeled cells and single-labeled 5-HT-IR cells, then 5-HT-IR cells arise from multiple progenitors, because β-gal+ and β-gal− cells must have arisen from different progenitors. A correlation analysis of the relative incidence of β-gal-IR and 5-HT-IR cells was performed to test whether 5-HT-IR cells are related by lineage (see Materials and Methods, Statistical Analysis).
Many taste buds of female hemizygous transgenic mice were found to contain fewer than 50% β-gal+ cells. The lowest contribution of β-gal+ cells in a circumvallate taste bud was 6%, although more commonly, 8–13% β-gal+ cells was found (Table 1, Fig.3). In a taste bud that contains an average of 55 cells, 8–13% implies the presence of four to seven labeled cells. If every progenitor in the taste bud gives rise to equal numbers of progeny cells, one labeled cell in a pool of 14 progenitors will produce a taste bud containing four labeled cells (8% of the total population). By a similar argument, one labeled cell in a pool of eight progenitors would produce a taste bud exhibiting seven labeled cells (13% of total population).
Table 1.
Distribution of the percentage of β-gal+ cells in eight circumvallate taste buds
| % β-gal+ cells | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 18 |
| β-gal+/N | 4/66 | 4/48 | 6/59 | 6/49 | 4/30 | 11/62 | |||||
| 4/40 | 10/82 | ||||||||||
| # Taste buds | 1 | 0 | 1 | 0 | 2 | 0 | 2 | 1 | 0 | 0 | 1 |
Nuclei in serial sections through the entire individual taste buds were counted. In some cases, a few cells in the taste bud might have been missed because of unclear taste bud boundaries. Counts did not include perigemmal cells. β-gal+/N, Actual count of β-gal+ cells per total number of cells in that taste bud.
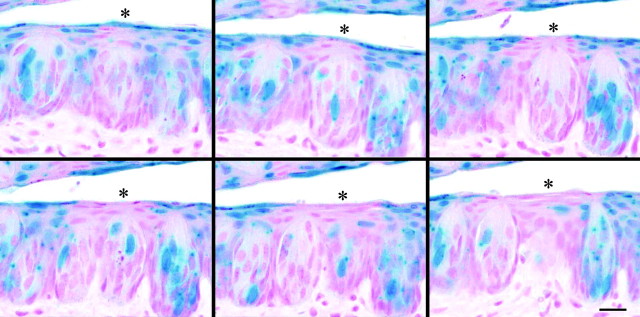
Fig. 3.
Light microscopic images of 5 μm serial sections through circumvallate taste buds from an H253 mosaic mouse. The adult female mouse was treated with Simvistatin, and the tissue section was stained with X-gal solution as described in Materials and Methods, embedded in Historesin, resectioned, and stained with nuclear fast red. Asterisks indicate a taste bud with few β-gal+ cells (blue). Scale bar, 20 μm.
In addition to the circumvallate taste buds, two fungiform taste buds were analyzed. The lowest contribution of β-gal+ cells in the two fungiform taste buds was 31%, suggesting that at least two progenitor cells may be required to populate the whole taste bud (data not shown). However, because only two samples were analyzed, no definitive statement on the size of progenitor pool may be made for the fungiform taste bud.
Cell lineage analysis of taste cells labeled with serotonin antibodies
To resolve the issue of whether different morphological or histochemical types of taste cells represent independent cell lineages or instead a particular developmental stage of a single taste cell lineage, we examined the relationship between 5-HT-IR taste cells [a phenotypic subpopulation of type III cells (Yee et al., 2001)] and β-gal-IR nuclei in H253 transgenic, mosaic mice. Thirty-two taste buds were analyzed (from sections double labeled with both antibodies and from triple-labeled DAPI and triple-labeled propidium iodide sections), and the outcome was grouped under three categories: 5-HT-IR and β-gal-IR, 5-HT-IR only, and β-gal-IR only.
Both 5-HT-IR only cells and 5-HT-IR/β-gal-IR double-labeled cells were present in the majority of taste buds studied (Table2 shows results from triple-labeled propidium iodide sections; Fig. 4). The presence of both 5-HT-IR only cells and double-labeled cells in the same taste bud indicates that more than one progenitor gave rise to the 5-HT-IR cells in that taste bud because β-gal+ cells and β-gal− cells cannot arise from the same progenitor. Furthermore, based on the binomial distribution, because approximately one-half of the taste buds contain both 5-HT-IR only cells and double-labeled cells (15 of 32), it is likely that two progenitors contribute to the 5-HT-IR cells in individual taste buds. However, the number of taste buds with only double-labeled cells (n = 12) is approximately twice the number expected on the basis of binomial statistics with two progenitors (n = 15). This suggests that, in some taste buds, only one progenitor gave rise to the entire 5-HT-IR subpopulation.
Fig. 4.

Confocal image of circumvallate taste buds from an H253 mosaic mouse, triple labeled for β-gal-IR (magenta), 5-HT-IR (green), and propidium iodide (blue). This figure was obtained by merging two confocal images using Photoshop software as described in Materials and Methods. The Photoshop filter “dust and scratches” was used to remove artifactual speckles from the merged image (pixel value was 3). Note that both 5-HT-IR only cells (green) and double-labeled cells (green and magenta) are present in individual taste buds (e.g., bud indicated by asterisk). This indicates that more than one progenitor gave rise to the 5-HT-IR cells in those buds. Scale bar, 20 μm.
Sections triple labeled for 5-HT-IR, β-gal, and propidium iodide (Fig. 4) were analyzed in more detail to determine whether there was a lineage relationship between 5-HT-IR taste cells. Counts from 16 taste buds are presented in Table 2. The idea that 5-HT-IR taste cells are related by lineage is supported by χ2analysis of the actual and expected distributions of 5-HT-IR, β-gal-IR, and propidium iodide-labeled nuclei. In five of the 16 taste buds (Table 2), the distribution of β-gal-IR cells, 5-HT-IR cells, double-labeled cells, and nonimmunoreactive cells differed significantly from the distribution expected if β-gal antibodies and 5-HT antibodies labeled taste cells independently (p < 0.001) (Table 2). Thus, 5-HT-IR cells within a taste bud show lineage relationships and are more likely to share a common progenitor with each other than with non-5-HT-IR cells in a taste bud.
DISCUSSION
The key conclusions of this study are as follows: (1) at least eight progenitor cells contribute to each circumvallate taste bud, based on the assumption that all progenitor cells make equal contribution, and (2) amine-accumulating (5-HT-IR) cells in a taste bud are likely to be clonal descendants of a subset of progenitors. Thus, the different cell types in a mature taste bud are not merely phenotypic, temporal stages of a common taste cell type but likely represent distinct cell lineages that are independent of one another once they have undergone terminal division from the basal cell population. Furthermore, these findings indicate that at least some of the proliferative cells contributing to a taste bud have a limited repertoire of progeny, i.e., particular basal cells may generate only one or two of the three cell types in a taste bud.
Analysis of taste bud progenitor number
Individual taste buds arise from several progenitors. In circumvallate taste buds from mosaic mice, 8–13% of taste cells are β-gal+ in taste buds with the fewest β-gal+ cells. From our counts, ∼55 intragemmal cells are present in an average mouse circumvallate taste bud. Eight to 13% translate into approximately four to eight β-gal+ cells in an average taste bud consisting of 55 cells. If progenitors contribute equally, these proportions would result from a relatively large progenitor pool, e.g., eight total progenitors consisting of seven β-gal− and one β-gal+ cell [producing seven cells each, for a total of 56 cells, with seven (12.5%) being β-gal+]. This fairly large number of progenitors per taste bud correlates reasonably well with the number of cells present in early taste bud placodes that give rise to fungiform papillae and their taste buds. These placodal cells represent, or contain, the progenitor pool for incipient taste buds (Farbman and Mbiene, 1991). In E14 mice, at the earliest stages of molecular differentiation of the incipient taste bud, the fungiform taste placodes consist of ∼10–12 cells exhibiting sonic hedgehog mRNA (Hall et al., 1999).
However, the architecture of β-gal+ patches in the lingual epithelium suggests that a single β-gal+ progenitor would occur only rarely in consort with numerous β-gal- progenitors. In mosaic mice, the borders between β-gal+ and β-gal− patches are generally distinct, arguing against significant tangential migration that could otherwise account for only one β-gal+ progenitor in a small area (Stone et al., 1995). Also, patch sizes in the lingual epithelium are relatively large, usually ∼50 μm, with patch sizes up to 1 mm not uncommon (Stone et al., 1995). Thus, the likelihood of a patch boundary coinciding with a taste bud is low; the likelihood of only a single lineage-marked cell falling within the progenitor pool is lower still. The relative incidence of taste buds with low numbers of β-gal-labeled cells suggests that, in some taste buds, progenitors contribute unequally. In support of this idea, we found occasional taste buds that contained one to two β-gal+ cells (e.g., a palatal taste bud that appeared to contain only one β-gal+ cell), a situation that would require the unlikely contribution of 25–55 progenitors if they all contributed equally. Taste buds with few labeled cells could occur if progenitors give rise to different numbers of progeny or if they take turns producing taste cells. For example, if four progenitors were to contribute to a taste bud and one of these divided at one-third the rate of the others, then it would give rise to only 10% of the final population. Taste cells with different lifespans also could result in a taste bud with few β-gal+ cells. Farbman (1969, 1980) reported that dark (type I) cells have a lifespan of ∼9 d, whereas light (type II) cells live somewhat longer. Thus, the β-gal+ cells in taste buds with the fewest positive cells may be dark cells if dark and light cell progenitors divide at the same rate and dark cells die more quickly. Alternatively, progenitors producing cells with longer lifespans may divide more slowly and contribute cells to the bud less often.
5-HT-IR taste cell lineage analysis
Our mosaic analysis of 5-HT-IR taste cells indicates that, within a taste bud, 5-HT-accumulating taste cells tend to be related by lineage, and, in individual taste buds, one or two progenitors give rise to this subpopulation. Several cell types exist in a taste bud, and controversy exists as to whether different cell types represent distinct lineages or whether one cell type grades into another as the taste cell matures (Delay et al., 1986). This controversy primarily centers around type I (dark), type II (light), and type III (intermediate) cells defined by ultrastructural characteristics. Cells that display 5-HT-IR belong to the type III class of taste cells (Takeda and Kitao, 1980; Uchida, 1985; Fujimoto et al., 1987; Kim and Roper, 1995; Yee et al., 2001), although not all type III cells exhibit 5-HT immunoreactivity (Yee et al., 2001). The cell lineage study presented here indicates that one class of type III cell, the 5-HT-IR cells, exhibit lineage relationships within the taste bud and therefore do not represent one stage of an age-related continuum of cells progressing from type I to type III to type II (dark to intermediate to light), as suggested previously.
The lineage relatedness among amine-accumulating type III cells suggests that at least some proliferative basal cells are restricted in terms of their ability to generate different taste cell types. Based on studies of other tissues, it is likely that more than one source of proliferating cells contributes to the taste bud cell population. In both the epidermis and the olfactory epithelium, two mitotically active basal cell populations exist [epidermis (Barrandon and Green, 1987;Potten and Morris, 1988) and olfactory epithelium (Graziadei and Metcalf, 1971; Graziadei and Monti Graziadei, 1978, 1979; Mackay-Sim and Kittel, 1991; Huard et al., 1998)]. One type of basal cell, the stem cell, has a long cell cycle and, by asymmetric division, maintains both a stem cell population and a second proliferative population. This second proliferative group of basal cells, like a transit-amplifying population, divides a limited number of times and then differentiates. This general scheme also has been proposed for intestinal epithelium (for review, see Gordon and Hermiston, 1994). Furthermore, studies of lingual epithelial basal cells indicate that more than one type of basal cell gives rise to the general lingual epithelium (Fukuda et al., 1978).
Two dividing populations may ultimately contribute to taste buds, with basal cells remote from the taste bud serving as the stem cell population and more proximate taste bud basal cells (and possibly perigemmal cells) serving as a lineage-restricted pool of transit-amplifying cells. The idea that both peripheral cells (perigemmal or surrounding cells) and basal cells within the taste bud contribute to the taste bud is consistent with reports of mitotic activity in both cell populations (Beidler and Smallman, 1965; Conger and Wells, 1969; Murray and Murray, 1971; Farbman, 1980; Delay et al., 1986). The lineage-restricted taste bud basal cells would likely include the mammalian achaete-scute homolog 1 (Mash1)-positive basal cells identified by Seta et al. (1999) and the BMP-4 (bone morphogenetic protein)-expressing basal cells identified by Yee and Finger (2001). Both of these markers are expressed by a specific subset of postmitotic taste cells, as well as the proliferative basal cells. This persistence of expression in only some types of taste cells may indicate that some basal cells only give rise to certain types of taste cells. Thus, for example, one population of basal cells might generate the neural-like type III cells, whereas a different set of basal cells gives rise to the more glial-like type I cells.
This lineage restriction could occur in one of two ways: either the original embryonic progenitor population is restricted or else the embryonic progenitors give rise to multipotent stem cells, which produce a lineage-restricted pool of proliferative basal cells (Fig.5). In the first of these models, the original embryonic progenitor cells give rise to basal cells, which are restricted in terms of their proliferative capabilities from the outset. Thus, a particular basal cell might generate only type III taste cells, whereas other basal cells would generate type II or type I taste cells (Fig. 5A). Conversely, the embryonic progenitors may give rise to multipotent stem cells, which continuously give rise to a set of lineage-restricted proliferative cells intimately associated with the taste buds. These gemmal proliferative cells then would give rise to the different types of taste cells (Fig.5B). That is, proliferative cells near the taste bud (e.g., taste bud basal cells) may give rise to only one type of taste cell (type I, II, or III), whereas multipotent epithelial stem cells are capable of generating all types of epithelial cells, including the lineage-restricted gemmal proliferative cells. Our results are consonant with either model. Thus, additional study will be necessary to determine whether multipotent stem cells are present in association with adult taste buds.
Fig. 5.
Schematic diagram showing possible lineage relationships in taste buds. A, In this scheme, the embryonic progenitor cells give rise to lineage-restricted basal cells that generate the different types of taste cells.B, In this scheme, the embryonic progenitors give rise to multipotent epithelial stem cells that generate lineage-restricted basal cells.
Footnotes
This work was supported by the National Institutes of Health (National Institute on Deafness and Other Communication Disorders Grant P01 DC00244) and the National Health and Medical Research Council (NHMRC) of Australia. P.P.L.T. is an NMHRC Senior Principal Research Fellow. We are grateful to John Caldwell, Joan Hooper, John Kinnamon, David Koeller, and Robert Lasher for critical discussions regarding this project. We also thank Karl Anderson, Bärbel Böttger, and Robin Michaels for technical assistance. Simvastatin was graciously supplied by Merck (Rahway, NJ). We especially appreciate Linda Barlow's comments on previous versions of this manuscript.
Correspondence to be addressed to Leslie M. Stone, Department of Biomedical Sciences, Colorado State University, Fort Collins, CO 80523. E-mail: lstone@lamar.colostate.edu.
REFERENCES
- 1.Barlow LA, Northcutt RG. Embryonic origin of amphibian taste buds. Dev Biol. 1995;169:273–285. doi: 10.1006/dbio.1995.1143. [DOI] [PubMed] [Google Scholar]
- 2.Barrandon Y, Green H. Cell migration is essential for sustained growth of keratinocyte colonies: the roles of transforming growth factor-alpha and epidermal growth factor. Cell. 1987;50:1131–1137. doi: 10.1016/0092-8674(87)90179-6. [DOI] [PubMed] [Google Scholar]
- 3.Beidler LM, Smallman RL. Renewal of cells within taste buds. J Cell Biol. 1965;27:263–272. doi: 10.1083/jcb.27.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conger AD, Wells MA. Radiation and aging effect on taste structure and function. Radiat Res. 1969;37:31–49. [PubMed] [Google Scholar]
- 5.Delay RJ, Kinnamon JC, Roper SD. Ultrastructure of mouse vallate taste buds. II. Cell types and cell lineage. J Comp Neurol. 1986;253:242–252. doi: 10.1002/cne.902530210. [DOI] [PubMed] [Google Scholar]
- 6.Farbman AI. Fine structure of the taste bud. J Ultrastruct Res. 1965a;12:328–350. doi: 10.1016/s0022-5320(65)80103-4. [DOI] [PubMed] [Google Scholar]
- 7.Farbman AI. Electron microscope study of developing taste buds in rat fungiform papilla. Dev Biol. 1965b;11:55–68. doi: 10.1016/0012-1606(65)90040-0. [DOI] [PubMed] [Google Scholar]
- 8.Farbman AI. Fine structure of degenerating taste buds after denervation. J Embryol Exp Morphol. 1969;22:55–68. [PubMed] [Google Scholar]
- 9.Farbman AI. Development of the taste bud. In: Beidler L, editor. Handbook of sensory physiology, Vol 4, Chemical senses II, Taste. Springer; Berlin: 1971. pp. 51–61. [Google Scholar]
- 10.Farbman AI. Renewal of taste bud cells in rat circumvallate papillae. Cell Tissue Kinet. 1980;13:349–357. doi: 10.1111/j.1365-2184.1980.tb00474.x. [DOI] [PubMed] [Google Scholar]
- 11.Farbman AI, Mbiene JP. Early development and innervation of taste bud-bearing papillae on the rat tongue. J Comp Neurol. 1991;304:172–186. doi: 10.1002/cne.903040203. [DOI] [PubMed] [Google Scholar]
- 12.Farbman AI, Hellekant G, Nelson A. Structure of taste buds in foliate papillae of the rhesus monkey Macaca mulatta. Am J Anat. 1985;172:41–56. doi: 10.1002/aja.1001720104. [DOI] [PubMed] [Google Scholar]
- 13.Finger TE, Simon SA. Cell biology of taste epithelium. In: Finger TE, Silver WL, Restrepo D, editors. The neurobiology of taste and smell, Ed 2. Wiley; New York: 2000. pp. 287–314. [Google Scholar]
- 14.Fujimoto S, Murray RG. Fine structure of degeneration and regeneration in denervated rabbit vallate taste buds. Anat Rec. 1970;168:393–413. doi: 10.1002/ar.1091680306. [DOI] [PubMed] [Google Scholar]
- 15.Fujimoto S, Ueda H, Kagawa H. Immunocytochemistry on the localization of 5-hydroxytryptamine in monkey and rabbit taste buds. Acta Anat. 1987;128:80–83. doi: 10.1159/000146320. [DOI] [PubMed] [Google Scholar]
- 16.Fukuda M, Okamura K, Fujita S, Böhm N, Rohrbach R, Sandritter W. The different stem cell populations in mouse epidermis and lingual epithelium. Pathol Res Pract. 1978;163:205–227. doi: 10.1016/S0344-0338(78)80015-6. [DOI] [PubMed] [Google Scholar]
- 17.Gautier C, Mehtali M, Lathe R. A ubiquitous mammalian expression vector, pHMG, based on a housekeeping gene promoter. Nucleic Acids Res. 1989;17:8389. doi: 10.1093/nar/17.20.8389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldstein JL, Brown MS. Regulation of the mevalonate pathway. Nature. 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 19.Gordon JI, Hermiston ML. Differentiation and self-renewal in the mouse gastrointestinal epithelium. Curr Opin Cell Biol. 1994;6:795–803. doi: 10.1016/0955-0674(94)90047-7. [DOI] [PubMed] [Google Scholar]
- 20.Graziadei PPC, Metcalf JF. Autoradiographic and ultrastructural observations on the frog's olfactory mucosa. Z Zellforsch Mikrosk Anat. 1971;116:305–318. doi: 10.1007/BF00330630. [DOI] [PubMed] [Google Scholar]
- 21.Graziadei PPC, Monti Graziadei GA. Continuous nerve cell renewal in the olfactory system. In: Jacobson M, editor. Handbook of sensory physiology, Vol IX, Development of sensory systems. Springer; Berlin: 1978. pp. 55–83. [Google Scholar]
- 22.Graziadei PPC, Monti Graziadei GA. Neurogenesis and neuron regeneration in the olfactory system of mammals. I. Morphological aspects of differentiation and structural organization of the olfactory sensory neurons. J Neurocytol. 1979;8:1–18. doi: 10.1007/BF01206454. [DOI] [PubMed] [Google Scholar]
- 23.Hall JM, Hooper JE, Finger TE. Expression of sonic hedgehog, patched, and Gli1 in developing taste papillae of the mouse. J Comp Neurol. 1999;406:143–155. doi: 10.1002/(sici)1096-9861(19990405)406:2<143::aid-cne1>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 24.Huard JM, Youngentob SL, Goldstein BJ, Luskin MB, Schwob JE. Adult olfactory epithelium contains multipotent progenitors that give rise to neurons and non-neural cells. J Comp Neurol. 1998;400:469–486. [PubMed] [Google Scholar]
- 25.Kaufman MH. The atlas of mouse development. Academic; London: 1992. [Google Scholar]
- 26.Kim D-J, Roper SD. Localization of serotonin in taste buds: a comparative study in four vertebrates. J Comp Neurol. 1995;353:364–370. doi: 10.1002/cne.903530304. [DOI] [PubMed] [Google Scholar]
- 27.Kinnamon JC, Taylor BJ, Delay RJ, Roper SD. Ultrastructure of mouse vallate taste buds. I. Taste cells and their associated synapses. J Comp Neurol. 1985;235:48–60. doi: 10.1002/cne.902350105. [DOI] [PubMed] [Google Scholar]
- 28.Lawton DM, Furness DN, Lindemann B, Hackney CM. Localization of the glutamate-aspartate transporter, GLAST, in rat taste buds. Eur J Neurosci. 2000;12:3163–3171. doi: 10.1046/j.1460-9568.2000.00207.x. [DOI] [PubMed] [Google Scholar]
- 29.Lindemann B. Taste reception. Physiol Rev. 1996;76:719–766. doi: 10.1152/physrev.1996.76.3.719. [DOI] [PubMed] [Google Scholar]
- 30.Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus). Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 31.Mackay-Sim A, Kittel P. Cell dynamics in the adult mouse olfactory epithelium: a quantitative autoradiographic study. J Neurosci. 1991;11:979–984. doi: 10.1523/JNEUROSCI.11-04-00979.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murray RG. Cell types in rabbit taste buds. In: Pfaffman C, editor. Olfaction and taste III. Rockefeller UP; New York: 1969. pp. 331–334. [Google Scholar]
- 33.Murray RG. The ultrastructure of taste buds. In: Friedmann I, editor. The ultrastructure of sensory organs. Elsevier; New York: 1973. pp. 1–81. [Google Scholar]
- 34.Murray RG. The mammalian taste bud type III cell: a critical analysis. J Ultrastruct Mol Struct Res. 1986;95:175–188. doi: 10.1016/0889-1605(86)90039-x. [DOI] [PubMed] [Google Scholar]
- 35.Murray RG, Murray A. Relations and possible significance of taste bud cells. Contrib Sens Physiol. 1971;5:47–95. doi: 10.1016/b978-0-12-151805-9.50008-0. [DOI] [PubMed] [Google Scholar]
- 36.Paulson RB, Hayes TG, Sucheston ME. Scanning electron microscope study of tongue development in the CD-1 mouse fetus. J Craniofac Genet Dev Biol. 1985;5:59–73. [PubMed] [Google Scholar]
- 37.Potten CS, Morris RJ. Epithelial stem cells in vivo. J Cell Sci [Suppl] 1988;10:45–62. doi: 10.1242/jcs.1988.supplement_10.4. [DOI] [PubMed] [Google Scholar]
- 38.Pumplin DW, Yu C, Smith DV. Light and dark cells of rat vallate taste buds are morphologically distinct cell types. J Comp Neurol. 1997;378:389–410. doi: 10.1002/(sici)1096-9861(19970217)378:3<389::aid-cne7>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 39.Seta Y, Toyono T, Takeda S, Toyoshima K. Expression of Mash1 in basal cells of rat circumvallate taste buds is dependent upon gustatory innervation. FEBS Lett. 1999;444:43–46. doi: 10.1016/s0014-5793(99)00023-x. [DOI] [PubMed] [Google Scholar]
- 40.Stone LM, Finger TE, Tam PPL, Tan S-S. Taste receptor cells arise from local epithelium, not neurogenic ectoderm. Proc Natl Acad Sci USA. 1995;92:1916–1920. doi: 10.1073/pnas.92.6.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takeda M. Uptake of 5-hydroxytryptophan by gustatory cells in the mouse taste bud. Arch Histol Jpn. 1977;40:243–250. doi: 10.1679/aohc1950.40.243. [DOI] [PubMed] [Google Scholar]
- 42.Takeda M, Kitao K. Effect of monoamines on the taste buds in the mouse. Cell Tissue Res. 1980;210:71–78. doi: 10.1007/BF00232142. [DOI] [PubMed] [Google Scholar]
- 43.Takeda M, Shishido Y, Kitao K, Suzuki Y. Biogenic monoamines in developing taste buds of mouse circumvallate papillae. Arch Histol Jpn. 1981;44:485–495. doi: 10.1679/aohc1950.44.485. [DOI] [PubMed] [Google Scholar]
- 44.Tam PPL, Tan S-S. The somitogenetic potential of cells in the primitive streak and the tail bud of the organogenesis-stage mouse embryos. Development. 1992;115:703–715. doi: 10.1242/dev.115.3.703. [DOI] [PubMed] [Google Scholar]
- 45.Tan S-S, Williams EA, Tam PPL. X-chromosome inactivation occurs at different times in different tissues of the post-implantation mouse embryo. Nat Genet. 1993;3:170–174. doi: 10.1038/ng0293-170. [DOI] [PubMed] [Google Scholar]
- 46.Uchida T. Serotonin-like immunoreactivity in the taste bud of the mouse circumvallate papilla. Jpn J Oral Biol. 1985;27:132–139. [Google Scholar]
- 47.Yee C, Finger TE. Bone morphogenetic protein 4 (BMP-4) is present in taste cells of adult mice. Chem Senses. 2001;26:1123. [Google Scholar]
- 48.Yee C, Yang R, Böttger B, Finger TE, Kinnamon JC. “Type III” cells of rat taste buds: immunohistochemical and ultrastructural studies of neuron specific enolase, protein gene product 9.5 and serotonin. J Comp Neurol. 2001;440:97–108. doi: 10.1002/cne.1372. [DOI] [PubMed] [Google Scholar]