Abstract
CASK, an adaptor protein of the plasma membrane, is composed of an N-terminal calcium/calmodulin-dependent protein (CaM) kinase domain, central PSD-95, Dlg, and ZO-1/2 domain (PDZ) and Src homology 3 (SH3) domains, and a C-terminal guanylate kinase sequence. The CaM kinase domain of CASK binds to Mint 1, and the region between the CaM kinase and PDZ domains interacts with Velis, resulting in a tight tripartite complex. CASK, Velis, and Mint 1 are evolutionarily conserved in Caenorhabditis elegans, in which homologous genes (called lin-2, lin-7, and lin-10) are required for vulva development. We now demonstrate that the N-terminal CaM kinase domain of CASK binds to a novel brain-specific adaptor protein called Caskin 1. Caskin 1 and a closely related isoform, Caskin 2, are multidomain proteins containing six N-terminal ankyrin repeats, a single SH3 domain, and two sterile α motif domains followed by a long proline-rich sequence and a short conserved C-terminal domain. Unlike CASK and Mint 1, no Caskin homolog was detected in C. elegans. Immunoprecipitations showed that Caskin 1, like Mint 1, is stably bound to CASK in the brain. Affinity chromatography experiments demonstrated that Caskin 1 coassembles with CASK on the immobilized cytoplasmic tail of neurexin 1, suggesting that CASK and Caskin 1 coat the cytoplasmic tails of neurexins and other cell-surface proteins. Detailed mapping studies revealed that Caskin 1 and Mint 1 bind to the same site on the N-terminal CaM kinase domain of CASK and compete with each other for CASK binding. Our data suggest that in the vertebrate brain, CASK and Velis form alternative tripartite complexes with either Mint 1 or Caskin 1 that may couple CASK to distinct downstream effectors.
Keywords: CASK, synapse, scaffold, lin-2, Mint 1, Caskin, neurexin, syndecan, Velis
An important development in biology over the past 10 years has been the realization that many proteins are composed of similar, relatively small autonomous domains. Adaptor proteins containing multiple such domains often form molecular scaffolds that link proteins into large signaling networks (Tsunoda and Zuker, 1999; Pawson and Nash, 2000). Adaptor proteins appear to be particularly important at the synapse, where large protein complexes are emerging as scaffolds of presynaptic and postsynaptic signaling machines (for review, see Garner et al., 2000; Sheng and Sala, 2001). One of the synaptic scaffolding proteins is CASK, a multidomain protein that was first identified by virtue of its interaction with the intracellular tails of neurexins, which are neuron-specific cell-adhesion molecules (Hata et al., 1996), and as the product of the lin-2 gene in Caenorhabditis elegans, which is essential for vulva development (Hoskins et al., 1996). CASK is composed of an N-terminal calcium/calmodulin-dependent protein (CaM) kinase domain that is catalytically inactive, central PSD-95, Dlg, and ZO-1/2 domain (PDZ) and Src homology 3 (SH3) domains, and a C-terminal guanylate kinase domain that is also catalytically inactive. The domain structure of the C-terminal half of CASK identifies it as a member of the membrane-associated guanylate kinase (MAGUK) family; accordingly,Drosophila CASK was named CamGUK (Dimitratos et al., 1997). Among vertebrate MAGUKs, CASK is most closely related to p55, Dlg2, Dlg3, and Vam-1, which, however, lack the N-terminal CaM kinase domain of CASK (Tseng et al., 2001). In vertebrates, CASK is expressed primarily in the brain but is also present at low levels in all cells (Hata et al., 1996). Brain CASK is localized largely to synapses, in which it is evenly distributed between presynaptic and postsynaptic densities (Hsueh et al., 1998).
As a scaffolding protein, CASK participates in multiple interactions. The central PDZ domain of CASK binds to the cytoplasmic tails of several cell-surface proteins, including neurexins, syndecans, NG2, glycophorins, and junctional adhesion molecules (JAMs) (Hata et al., 1996; Cohen et al., 1998; Hsueh et al., 1998; Martinez-Estrada et al., 2001; T. Biederer and T. C. Südhof, unpublished observations). This interaction presumably targets CASK to synapses in the brain and to tight junctions in epithelia. The N-terminal sequences of CASK participate in an evolutionarily conserved complex with two other PDZ domain proteins, Mint 1 (the vertebrate form of C. elegans lin-10; also known as X11α) and Velis (the vertebrate homologs of C. elegans lin-7; also known as Mals) (Borg et al., 1998b; Butz et al., 1998; Kaech et al., 1998). Mint 1 binds to the CaM kinase domain of CASK, whereas Velis bind to a sequence between the CaM kinase and PDZ domains (Butz et al., 1998). In C. elegans, the lin-2–lin-7–lin-10 complex is required for the correct targeting of epidermal growth factor (EGF) receptors to the basolateral plasma membrane of vulval precursor cells (Kaech et al., 1998). In mice, a transgenic insertion mutant that changes CASK expression causes a cleft palate syndrome, suggesting that CASK also has a role in vertebrate development (Laverty and Wilson, 1998); however, its precise function and mechanism of action are unknown.
In addition to Mint 1 and Velis, CASK binds to calmodulin in a Ca2+-dependent manner via its N-terminal CaM kinase domain (Hata et al., 1996) and to protein 4.1 via a C-terminal sequence (Cohen et al., 1998; Biederer and Südhof, 2001). The resulting CASK–cell-surface protein–protein 4.1 complex nucleates actin assembly and may mediate the attachment of actin microfilaments to the cytoplasmic tails of cell-surface receptors (Biederer and Südhof, 2001). Furthermore, the SH3 domain of CASK binds to Ca2+ channels (Maximov et al., 1999), and the SH3 domains and guanylate kinase domains of CASK and Dlg interact intramolecularly and intermolecularly (Nix et al., 2000). Finally, the guanylate kinase domain of CASK binds to the transcription factor Tbr1-1 and may translocate from the synapse into the nucleus, in which it could function as a coactivator of Tbr-1 in transcription (Hsueh et al., 2000).
The interactions of CASK support the notion that it functions as a scaffolding protein, possibly by binding sequentially to different proteins. Such a function would imply that multiple proteins should bind competitively to the same domains. In the present study, we describe a novel protein called Caskin 1 that binds to the CaM kinase domain of CASK in competition with Mint 1, suggesting that CASK participates in alternative complexes with Mint 1 or Caskin 1.
MATERIALS AND METHODS
Purification of CASKIN. Rat brain proteins were affinity-purified on immobilized glutathioneS-transferase (GST)–CASK1-337 fusion protein essentially as described previously (Butz et al., 1998). Three proteins were detectable on Coomassie-stained gels: Mint 1, a 180 kDa protein called Caskin 1 in this study, and a 225 kDa protein. The 180 kDa and 225 kDa bands were eluted from the gel and digested by trypsin, and tryptic peptides were purified by HPLC and sequenced by Edman degradation essentially as described previously (Hata et al., 1993). The 180 kDa peptide sequences did not match any protein in the database except for KIAA and expressed sequence tag (EST) sequences (see Results), whereas the 225 kDa protein was identified as the rat homolog of NEDD4-like ubiquitin ligase 1 (GenBank accession #BAB13352).
cDNA cloning and sequencing. GenBank searches identified a random human brain cDNA (KIAA1306) and human EST clones that contained the sequences of the peptide fragments. To determine the complete structure of Caskin 1, PCRs were performed with rat brain cDNA and degenerative oligonucleotides based on the human sequences. The products were used to screen a rat brain cDNA library in λZAPII by standard cDNA cloning methods (Sambrook et al., 1989). Fifty positive clones were isolated, and the full-length sequence was assembled from their combined nucleotide sequences and translated into amino acid sequences. Human Caskin 1 (KIAA1306), human Caskin 2 (KIAA1139), and mouse Caskin 2 (AW321773.1, AA030081.1, AA036182.1, AA879547.1, AI466464.1,AA216903.1, AI4253971.1, BI837871, and BG872937) clones were identified in the EST databank using basic local alignment search tool searches with rat Caskin 1 sequence. The cDNA sequences were submitted to GenBank (accession #'s AF451975–AF451978).
Construction and expression of bacterial and eukaryotic expression vectors. Expression vectors for GST fusion proteins were constructed in pGEX–KG (Guan and Dixon, 1991) by standard procedures (Sambrook et al., 1989). The following GST fusion proteins were produced by the indicated vectors (residue numbers in parentheses): Caskin 1: GST–Caskin 11–374, pGEX–Caskin 1NcoI–BglII-4 (1–378); GST–Caskin 1375–471, pGEX–Caskin 1-SS (375–471); GST–Caskin 1374–540, pGEX–Caskin 1–sterile α motif 1 (SAM1) (374–540); GST–Caskin 1539–659, pGEX–Caskin 1–SAM2 (539–659); GST–Caskin 1374–659, pGEX–Caskin 1 BglII–SacI-1 (374–659); CASK: GSTCASK, pGEX–CASK full-length; GST–CASK1–275, pGEX–Cash3–9 (1–275); GST–CASK1–310, pGEX–CASK ΔCaM (1–310); GST–CASK1–337, pGEX–Cash3–13 (1–337); GST–CASK98–337, pGEX–CASKBglII–CaM (98–337); GST–CASK237–337, pGEX–CASK P-CaM (237–337); GST–CASK328–909, pGEX–CASK3–25 (328–909); Mint 1: GST–Mint 1, pGEX–Mint 1 5 (116–432); and Neurexin I: GST–NxI and GST–NxIΔ10 were described previously (Biederer and Südhof, 2000). For maltose-binding protein (MBP)–Caskin 1 fusion protein, theBamHI–HindIII fragment from pGEX–Caskin 1BglII–SacI-1(374–659) was subcloned into the same site of pMalC2 (New England Biolabs, Beverly, MA). A full-length eukaryotic expression vector for Caskin 1 (pCMV–CB16) was constructed by cloning the EcoRI insert from clone pBlue-CB16 into the same site of pCMV5. Bacterial expression of GST fusion proteins and COS cell transfections have been described previously (Okamoto and Südhof, 1997). MBP–Caskin 1 fusion proteins were immobilized on amylose resin and purified by 10 mmmaltose/PBS elution.
GST-pulldown experiments. GST fusion proteins were expressed in Escherichia coli and immobilized on glutathione agarose (Sigma, St. Louis, MO) using standard procedures. Rat forebrains were homogenized in a pestle tissue grinder at slow speed in solubilization buffer (25 mm HEPES–NaOH, pH 7.4, 125 mm K-acetate, 5 mmMgCl2, 0.32 m sucrose, and 1.0% Triton X-100). Proteins solubilized from rat forebrain were incubated with immobilized GST fusion proteins for 14 hr at 4°C under mild agitation, and bound proteins were analyzed after extensive washes with the solubilization buffer as described previously (Butz et al., 1998; Biederer and Südhof, 2000).
Immunoprecipitations. Immunoprecipitations were performed from rat brain homogenates as described previously (Butz et al., 1998). To characterize the CASK–Caskin 1 interaction further, immunoprecipitates were subjected to different washing conditions; after five washes using lysis buffer (20 mmimidazole, pH 6.8, 0.1 m NaCl, 5 mm EDTA, 1 mm DTT, 0.5% Triton X-100, 1 mm PMSF, 10 mg/l leupeptin, 1 mg/l pepstatin A, and 10 mg/l aprotinin), bound immune complexes were treated with lysis buffer supplemented with final concentrations of either 0.2, 0.5, or 1 m NaCl, 0.6m KI, or 0.5% SDS/2% Triton X-100.
Antibodies. Most antibodies have been described previously (Okamoto and Südhof, 1997; Butz et al., 1998). Antibodies against Caskin 1 were raised in rabbits against a peptide corresponding to the last 15 residues of Caskin 1 with additional cysteine at the N terminus (sequence: CSMFDDLADQLDAMLE). CASK and Mint monoclonal antibodies were obtained from Transduction Laboratories (Cincinnati, OH).
RNA blotting experiments. RNA blotting experiments were performed using multiple tissue blots purchased from Clontech (Cambridge, UK). Northern blots were hybridized at high stringency (Hata et al., 1996) with a DNA fragment encoding residues 59–232 of Caskin 1.
Miscellaneous procedures. SDS-PAGE was performed as described previously (Laemmli, 1970). Protein concentrations were determined by commercial protein assay kits obtained from Bio-Rad (Hercules, CA) and Pierce (Rockford, IL) and by comparison of samples run on SDS-PAGE with known amounts of BSA standards analyzed on the same gels. Immunocytochemistry experiments with cultured hippocampal neurons and rat brain sections were performed as described previously (Rosahl et al., 1995; Hsueh et al., 1998).
RESULTS
Identification and cloning of Caskin 1
Affinity purification of rat brain proteins with a GST fusion protein of the N-terminal CaM kinase domain of CASK revealed that in addition to Mint 1 (∼170 kDa), a second, slightly larger protein (∼180 kDa) was captured in relatively large amounts (data not shown;Butz et al., 1998). To identify the 180 kDa protein, we isolated preparative amounts of binding proteins by affinity chromatography on immobilized CASK and obtained sequences of tryptic peptides by Edman degradation. Ten unequivocal peptide sequences were obtained from the 180 kDa protein (1, ATPSPVPS; 2, SQEYLLDEGPAPGTPPK; 3, VLPSGVSHFTP?QK; 4, KPTPQ?LI; 5, DTTDP?GTSPL; 6, ASVPPVPGKPR; 7, EASAALQVR; 8, AGDIITVLEQHPDGR; 9, HGTPPPVSPKPP; 10, AASD?EP). Databank searches did not uncover significant homologies of the peptide sequences with known proteins, suggesting that the 180 kDa protein has not been studied previously. However, we did identify several EST clones and two human KIAA clones (KIAA1306 and KIAA1139; GenBank accession #'s AB037727 andAB032965, respectively) that encode the 180 kDa protein or a closely related homolog. We designed PCR primers based on the human nucleotide and rat peptide sequences and obtained a PCR product with rat brain cDNA that was subsequently used to clone the respective cDNAs from rat and mouse brain cDNA libraries using standard techniques (Okamoto and Südhof, 1997). Because of the tight binding of the 180 kDa protein to CASK, we named it Caskin 1 for CASK interacting protein 1 and called its closely related homolog Caskin 2.
Figure 1 displays alignments of the rat and human Caskin 1 and the mouse and human Caskin 2 sequences. Each Caskin isoform is highly conserved evolutionarily, and the two Caskins are very similar. Databank searches indicated that Caskins are composed of two principal regions: the N-terminal half exhibits a multidomain architecture composed of six ankyrin repeats (Fig. 1,purple), a single SH3 domain (Fig. 1, green), and two SAM domains (Fig. 1, blue). The C-terminal half is made up of a long proline-rich sequence (prolines highlighted inred in Fig. 1 to illustrate their abundance) and a unique conserved C-terminal domain (CTD) (Fig. 1, yellow). The N-terminal halves of Caskins 1 and 2 are very similar, especially in the identified domains. In contrast, the C-terminal regions diverge considerably but exhibit patches of identity that are often organized around proline residues. For example, an unusual sequence (KGPPPPPPKRxSS) that could form a polyproline helix and a protein kinase A phosphorylation site is conserved in all sequences, as is a second protein kinase A consensus site (RRRTLSxP) and another cluster of positively charged amino acids (LTESDTVKRRPKxKE).
Fig. 1.

Sequence analysis of Caskins. Alignment of the primary sequences of rat and human Caskin 1 (rC1 andhC1, respectively) and human and mouse Caskin 2 (hC2 and mC2, respectively). Sequences are identified on the left and numbered on theright. Residues that are identical between Caskins 1 and 2 in at least two of the sequences shown are highlighted by a domain-specific color code: The N-terminal ankyrin repeats are shown inpurple, the SH3 domain is in green, the SAM domains are in blue, and the Caskin-specific C-terminal domain is in yellow. Outside of these defined domains, shared sequences are highlighted in blackexcept for prolines, which are shown in the C-terminal half of the proteins on a red background if conserved among different Caskins and in red typeface if specific for a given Caskin isoform.
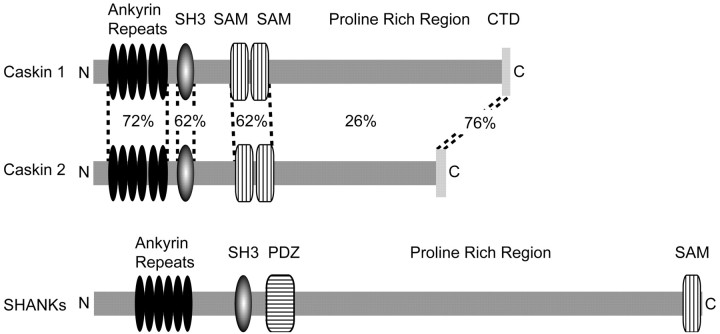
Figure 2 summarizes the domain organization of Caskins. Although the N-terminal ankyrin repeats and the SH3 and SAM domains were identified by standard domain searches, such as that of the conserved domain database of the National Center for Biotechnology Information, these domains are rather atypical. The SH3 domain in particular is only distantly related to other SH3 domains (data not shown); its closest homolog is the SH3 domain of yeast CDC25. The domain organization of Caskins is different from that of other multidomain proteins in the current databases. The structure of Caskins resembles that of Shanks, a family of postsynaptic scaffolding proteins that bind to guanylate kinase-associated protein, glutamate receptor-interacting protein homer, and cortactin (for review, see Sheng and Sala, 2001). Shanks and Caskins both contain N-terminal ankyrin repeats followed by an SH3 domain and a large C-terminal proline-rich sequence. However, in Shanks, the SH3 domain is followed by a PDZ domain that is absent from Caskins. Furthermore, in Shanks, a single SAM domain is positioned at the end of the C terminus, whereas Caskins contain two SAM domains in the center (Fig. 2). Finally, the ankyrin repeats and the SH3 and SAM domains of Caskins and Shanks exhibit little sequence identity and are more related to analogous domains in other proteins than to each other. Thus, it appears that the similar domain organization of Caskins and Shanks does not reflect an evolutionary connection but rather possibly a similar scaffolding function. Databank searches failed to uncover a direct homolog of Caskins in Drosophila or C. elegans, although multiple proteins containing N-terminal ankyrin repeats and SAM domains were observed. Because the nearly completeDrosophila and C. elegans genome sequences are available, this suggests that Caskins, unlike CASK and Mint 1, are not conserved in invertebrates.
Fig. 2.
Domain structures of Caskins. The domain structures of Caskins 1 and 2 are shown schematically and compared with the domain structure of Shanks. Numbers between the Caskin 1 and 2 structures indicate percentage identity between the various domains. The color code used corresponds to that of Figure1.
Tissue distribution of Caskin 1
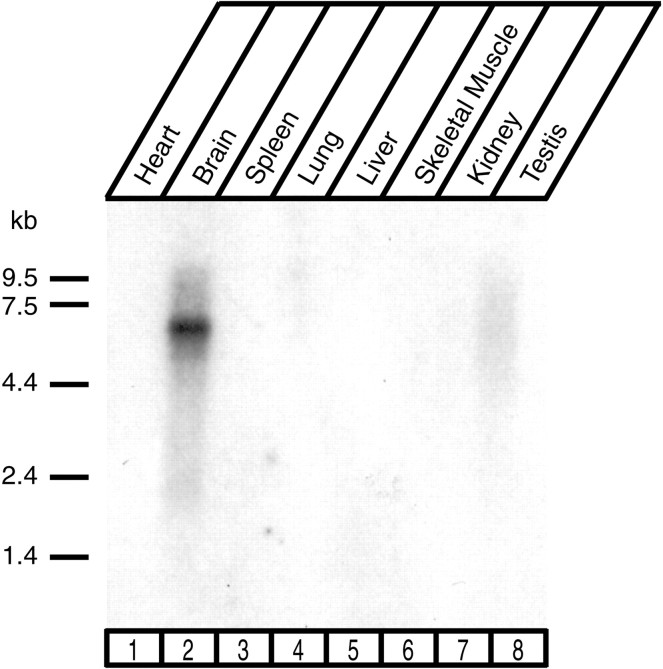
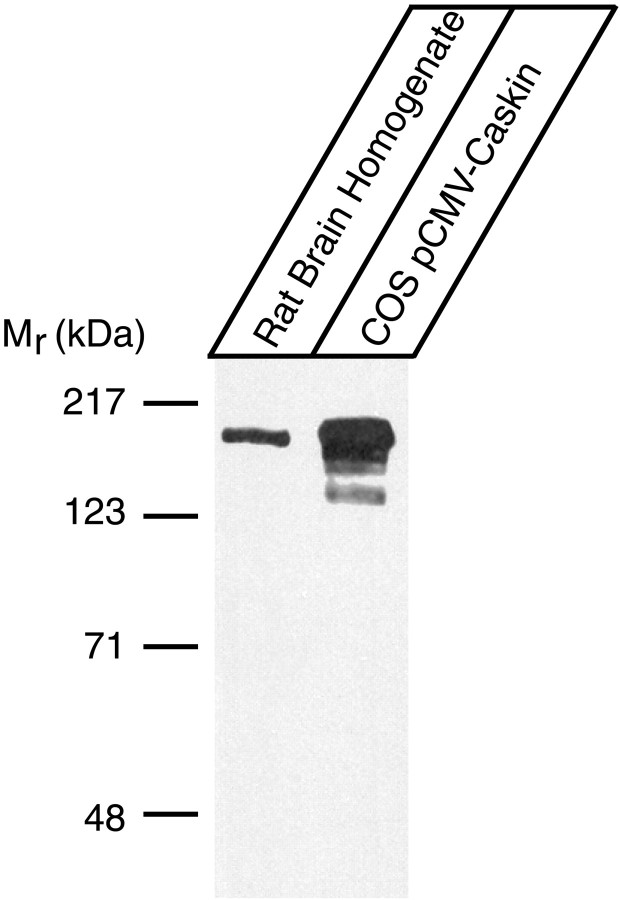
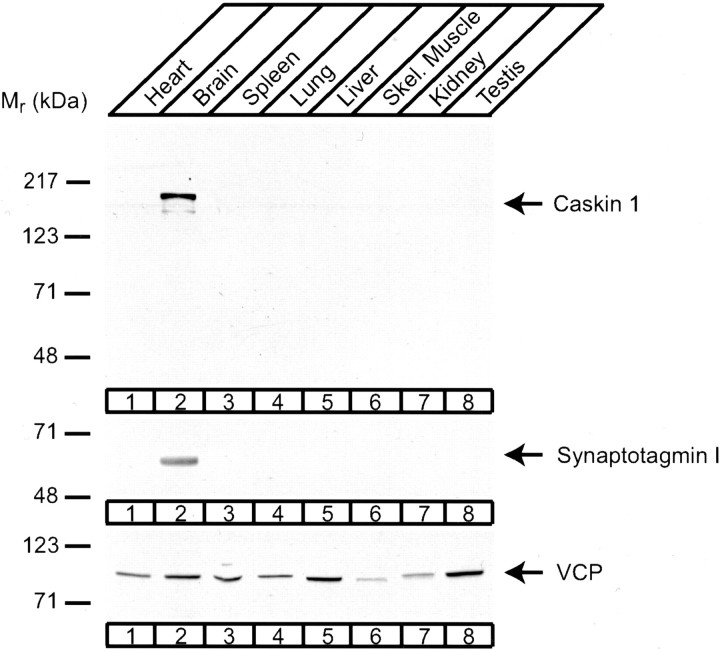
To examine which tissues express Caskin 1, we hybridized a multitissue RNA blot at high stringency with a Caskin 1 cDNA probe. The resulting signal revealed that among the tissues examined, Caskin 1 mRNA was detectable only in the brain (Fig.3). We subsequently raised an antibody against a synthetic peptide corresponding to the C terminus of Caskin 1. Immunoblotting of rat brain homogenates with the antibody uncovered a single major band of ∼180 kDa that was not observed with preimmune serum (Fig. 4 and data not shown). Comparison of this band with the band detected in COS cells transfected with a full-length Caskin 1 expression vector showed that the recombinant Caskin 1 comigrated with the band observed in brain homogenates (Fig. 4). This result suggests that the band observed in the brain corresponds to Caskin 1 and that the cDNA we isolated for Caskin 1 is full-length with respect to the coding region. Immunoblots of different rat tissues with the Caskin 1 antibody confirmed the conclusion from RNA blotting that among all tissues examined, Caskin 1 is expressed at detectable levels only in the brain (Fig.5).
Fig. 3.
RNA blotting analysis of Caskin 1 expression. A blot containing poly(A)-enriched RNA from the indicated rat tissues was probed at high stringency with a probe from the Caskin 1 cDNA.Numbers at left indicate positions of size markers.
Fig. 4.
Characterization of Caskin 1 antibodies. Proteins from total rat brain homogenate and from COS cells transfected with a full-length Caskin 1 expression vector were analyzed by immunoblotting with antibodies raised against a synthetic peptide corresponding to the C-terminal 15 residues of Caskin 1. Signals were visualized by ECL.Numbers at left indicate positions of molecular weight markers. Preimmune serum did not cause a signal (data not shown).
Fig. 5.
Immunoblotting analysis of Caskin 1 expression in adult rat tissues. Total proteins from the indicated tissues (50 μg/lane) were analyzed by immunoblotting with antibodies to Caskin 1 (top), the synaptic vesicle protein synaptotagmin 1 as a control for a brain-specific protein (middle), and vasolin-containing protein (VCP) as a control for a widely expressed protein (bottom). Signals were visualized by ECL. Numbers at leftindicate positions of molecular weight markers.
We used immunocytochemistry experiments to determine the distribution of Caskin 1 in the brain (Fig. 6). Immunoperoxidase labeling of rat brain sections revealed that Caskin 1 was localized primarily to the neuropil and was enriched in synaptic areas. This pattern resembles that of synaptic vesicle proteins, such as rab3A, and is illustrated in Figure 6A,B for the cerebellum. The observed labeling was specific, as evidenced by the lack of staining obtained with preimmune serum (Fig. 6C). To ensure that Caskin 1 is indeed a neuronal protein, we stained cultured hippocampal neurons (Fig. 6D,E). Caskin 1 was highly concentrated in these neurons and was present throughout the cells. Although Caskin was also observed in synapses, we observed no apparent enrichment in synapses in hippocampal neurons (Fig.6E and data not shown). In this regard, Caskin 1 was very similar to proteins such as CASK and syntaxin 1 that were also enriched in synapses in brain sections, but it appeared to be more widely distributed throughout the cells in cultured neurons (data not shown; also see Hsueh et al., 1998).
Fig. 6.
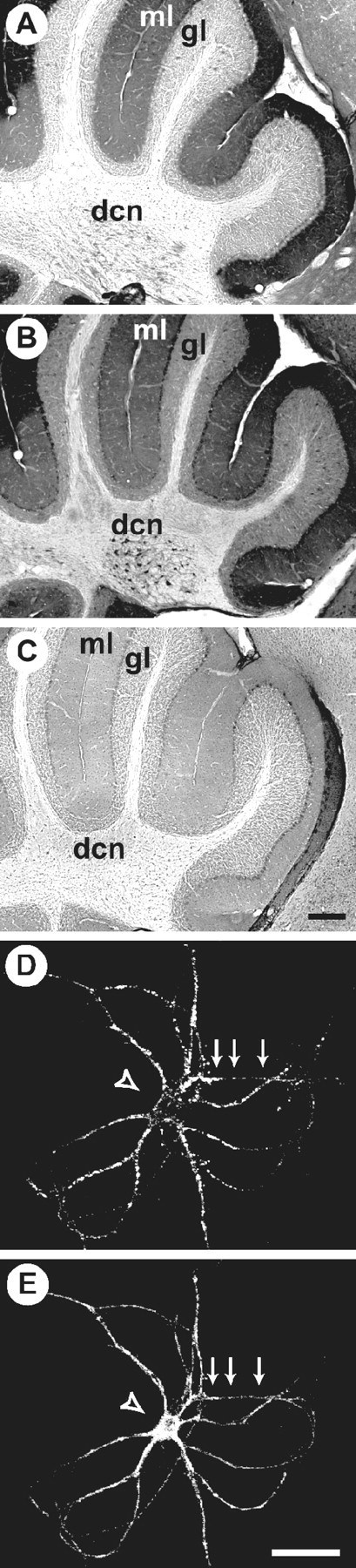
Localization of Caskin 1 analyzed by immunocytochemistry. A–C, Adjacent cryostat sections from rat cerebellum stained with antibodies to Rab3A (A) or Caskin 1 (B) or with Caskin 1 preimmune serum (C). The molecular layer (ml), granule cell layer (gl), and deep cerebellar nuclei (dcn) are identified. Scale bar in C, 0.1 mm (applies to A–C). D,E, Cultured hippocampal neurons double-labeled with a monoclonal antibody to synaptophysin (D) and a polyclonal antibody to Caskin 1 (E). Scale bar inE, 40 μm; (applies to D,E).
Immunoprecipitation of a CASK–Caskin 1 complex from brain homogenates
To obtain independent evidence that Caskin 1 and CASK form a complex in the brain, we performed immunoprecipitations. Proteins from rat brain homogenates were immunoprecipitated with preimmune or anti-Caskin 1 serum and analyzed by immunoblotting. CASK was specifically coimmunoprecipitated with Caskin 1 (Fig.7A). Mint 1 and Munc18 were not detected in the immunoprecipitates, suggesting that Caskin 1 antibodies do not bring down Mint 1 together with CASK. We subsequently washed the immunoprecipitates with solutions of increasing ionic strength (0.2–1.0 m NaCl), a chaotropic agent (0.6 m KI), or a denaturing solution (0.5% SDS in 2% Triton X-100). Figure 7B shows that the CASK–Caskin complex resisted high-salt washes of up to 1 mNaCl and was disrupted only by chaotropic or denaturing agents, suggesting that it is very stable.
Fig. 7.
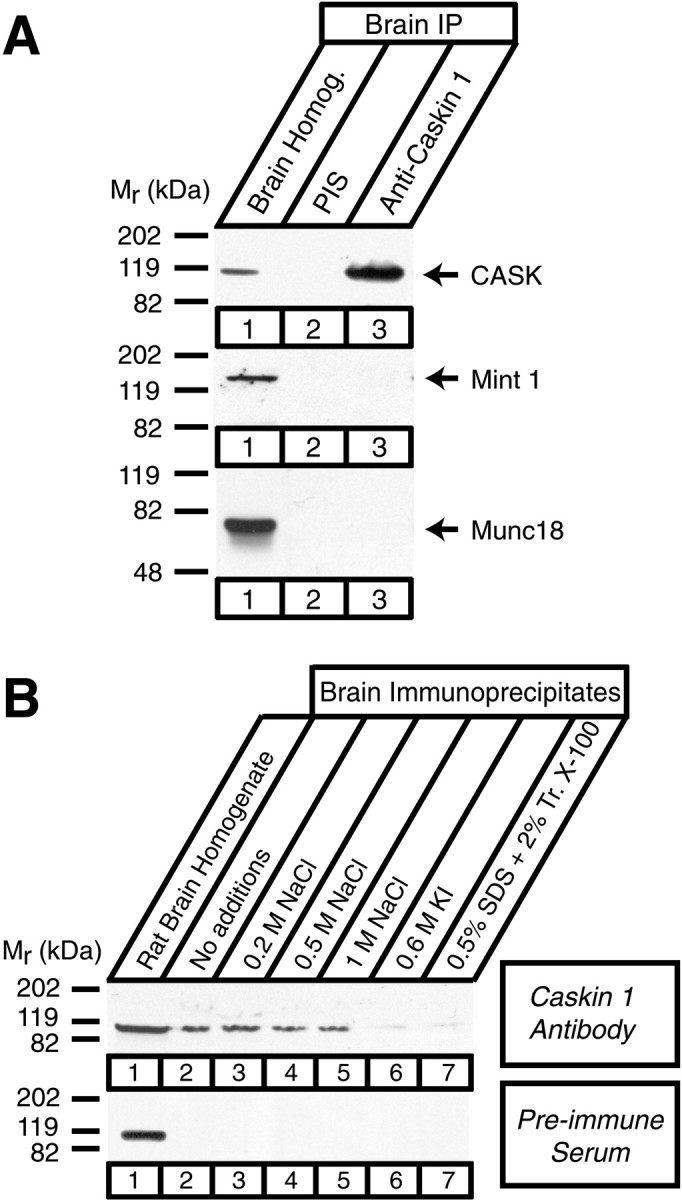
Coimmunoprecipitation of CASK and Caskin 1 from rat brain homogenates. A, Proteins solubilized from rat brains in Triton X-100 (lane 1) were immunoprecipitated with preimmune serum (PIS) (lane 2) and polyclonal antibodies to Caskin 1 (lane 3). Precipitated proteins were analyzed by immunoblotting with monoclonal antibodies to CASK, Mint 1, and Munc18-1 as indicated. IP, immunoprecipitates. B, Immunoprecipitates from rat brain homogenates (lane 1) with Caskin 1 antiserum or preimmune serum were washed in the presence of increasing concentrations of NaCl (lanes 3–5) or of the denaturing agents 0.6 m KI and 0.5% SDS as indicated (lanes 6 and 7) and analyzed by immunoblotting with monoclonal antibodies to CASK. Signals were visualized by ECL.Numbers at left indicate positions of molecular weight markers.
Caskin 1 and Mint 1 bind to the same site on CASK
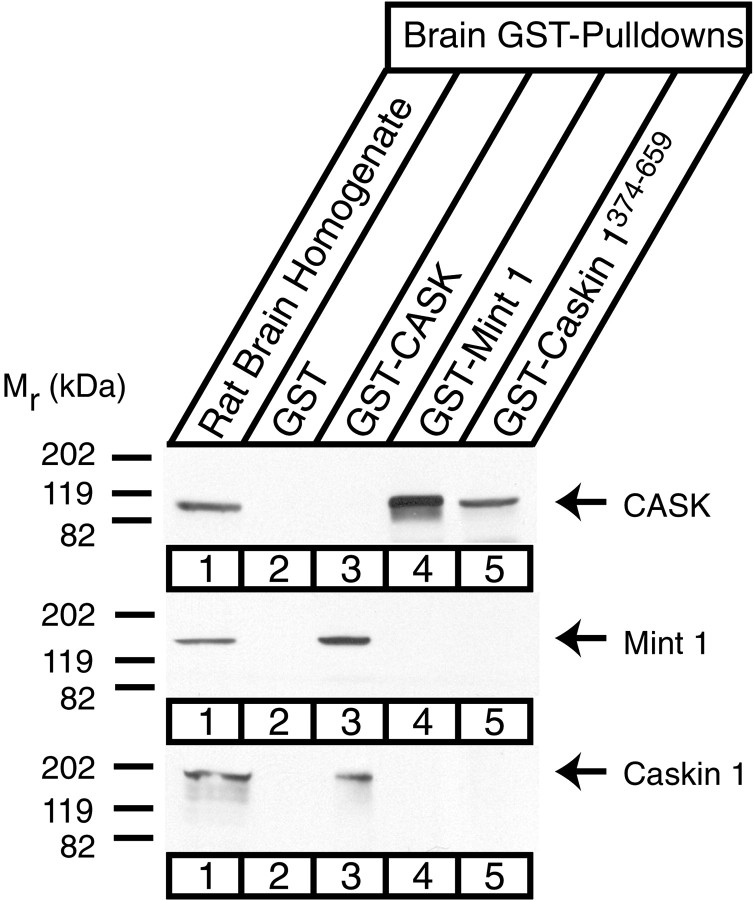
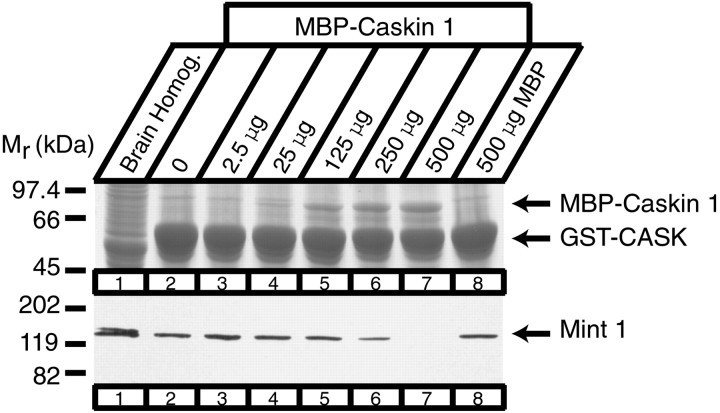
We subsequently used GST pulldowns to examine the interaction of Caskin 1 with CASK. Initial experiments showed that GST–CASK efficiently captured Caskin 1 and Mint 1 from the brain, as expected (Fig. 8). Interestingly, although both GST–Mint 1 and GST–Caskin 1 bound CASK, Mint 1 did not pull down Caskin 1 with CASK, and Caskin 1 did not pull down Mint 1. Because Mint 1 also was not coimmunoprecipitated with CASK by the Caskin antibodies (Fig. 7), this result suggests the possibility that Caskin 1 and Mint 1 bind to the same site on CASK. To test this hypothesis, we expressed a Caskin 1 fusion protein with MBP in bacteria. We subsequently added increasing amounts of purified recombinant MBP–Caskin during GST–CASK pulldown experiments from brain homogenates (Fig.9). The results show that higher Caskin 1 concentrations prevent Mint 1 binding to CASK, suggesting that CASK cannot bind Mint 1 and Caskin 1 at the same time.
Fig. 8.
Confirmation of the CASK–Caskin 1 interaction by GST pulldowns. Proteins from rat brain homogenates (lane 1) were bound to GST alone (lane 2) or to GST fusion proteins of the N-terminal CaM kinase domain of CASK (lane 3) or of the N-terminal sequences of Mint 1 (lane 4) or of Caskin 1 (lane 5). Bound proteins were analyzed by immunoblotting with antibodies to CASK, Mint 1, and Caskin 1 as indicated. Numbers on theleft indicate positions of molecular weight markers.
Fig. 9.
Caskin 1 competes with Mint 1 for CASK binding. Proteins from rat brain homogenates (Homog.) (lane 1) were bound to immobilized GST–CASK1-337 in the presence of increasing concentrations of recombinant MBP–Caskin 1 fusion protein up to 500 μg (lanes 2–7) or of MBP alone at 500 μg (lane 8). Bound proteins were visualized by Coomassie blue staining (top) or immunoblotting with Mint 1 antibodies (bottom). Numbers atleft indicate positions of molecular weight markers.
The N-terminal regions of CASK and Caskin 1 bind to each other
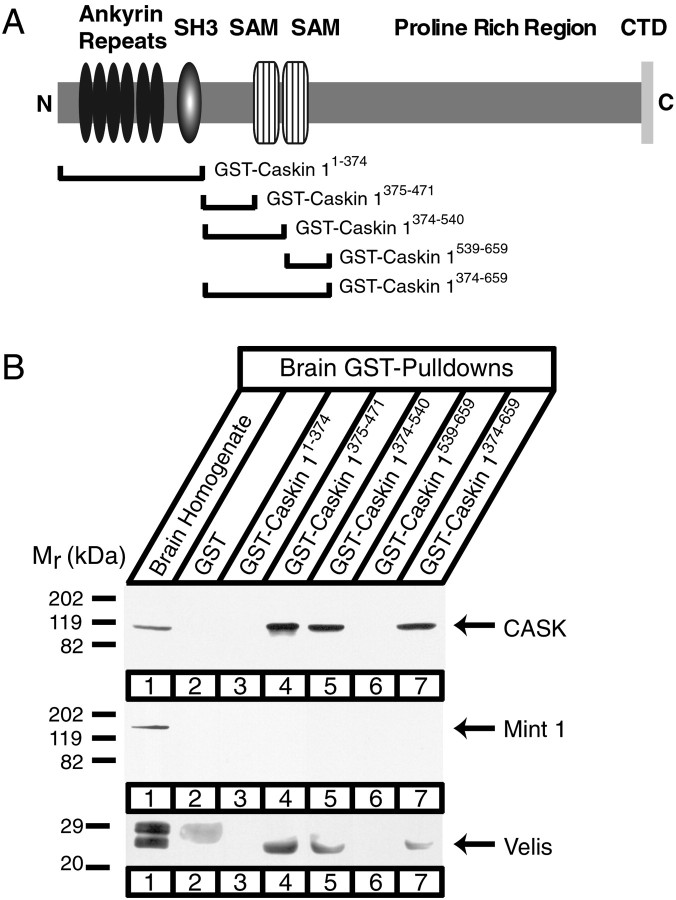
In the next set of experiments, we examined which CASK and Caskin 1 sequences interact with each other. Binding of rat brain CASK to a series of increasingly smaller GST–Caskin 1 fusion proteins uncovered a short sequence in Caskin 1 (residues 375–471) that was fully capable of affinity-purifying CASK from rat brain homogenates (Fig.10). As expected, Mint 1 was not copurified with CASK, but Velis (which bind to a different sequence in CASK) (Butz et al., 1998) were pulled down together with CASK. This indicates that CASK forms a tripartite complex with Caskin 1 and Velis similar to the CASK–Mint 1–Veli complex (Butz et al., 1998). The Caskin 1 sequence that binds to CASK lies between the SH3 domain and the two SAM domains and is relatively poorly conserved between Caskins 1 and 2 (Fig. 1), suggesting that only Caskin 1, and not Caskin 2, binds to CASK, which is similar to the interaction of only Mint 1, and not Mints 2 and 3, with CASK (Butz et al., 1998). This hypothesis was confirmed in experiments that showed that GST–Caskin 2 fusion proteins cannot capture CASK from rat brain homogenates (data not shown).
Fig. 10.
Mapping of the CASK-binding site on Caskin 1 by GST pulldowns. A, Position of GST–Caskin 1 fusion proteins used for pulldowns in B.B, GST pulldowns of rat brain proteins with the indicated Caskin fusion proteins were analyzed by immunoblotting for CASK, Mint 1, and Velis as indicated. Signals were visualized by ECL.Numbers at left indicate positions of molecular weight markers.
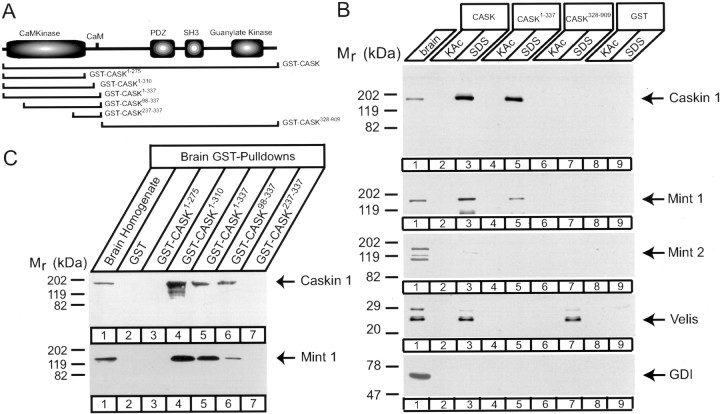
We subsequently studied which CASK sequences bind to Caskin 1 and Mint 1 (Fig. 11). Again, we used a series of GST fusion proteins to elucidate the sequence requirement for binding (Fig. 11A). The first set of experiments showed that like Mint 1, Caskin 1 was efficiently bound by the N-terminal CaM kinase domain of CASK, consistent with competition between the two proteins for binding to CASK (Fig. 11B). Velis, in contrast, were bound by the C-terminal fragment. We subsequently used deletion analysis to determine which sequences are precisely required for Caskin 1 and Mint 1 binding. Figure 11C demonstrates that a C-terminal truncation of CASK, in which the autoinhibitory region and calmodulin-binding site of the CaM kinase homology region of CASK are included (Cask1–310), still binds to both Caskin 1 and Mint 1. In contrast, removal of the autoinhibitory region and calmodulin-binding site of CASK (Cask1–275) abolishes Caskin 1 and Mint 1 binding. Thus, for both Caskin 1 and Mint 1, the sequence of CASK that is C terminal to the actual kinase domain and corresponds to the regulatory sequences in CaM kinase II (including its autophosphorylation site threonine286) (Hata et al., 1996) is essential for binding. N-terminal truncation of the CASK CaM kinase domain demonstrated that Caskin 1 and Mint 1 not only bind to the C-terminal peptide sequence of the domain but also require a larger region. Deletion of the N-terminal 98 residues of CASK strongly diminished Caskin 1 and Mint 1 binding, whereas removal of an additional 139 residues abolished binding (Fig. 11C). Together, these results suggest that the entire CaM kinase domain of CASK is involved in binding relatively short sequences of Caskin 1 and Mint 1. This binding reaction is specific for the catalytically inactive CASK CaM kinase domain, because genuine CaM kinase II was unable to bind to either Caskin 1 or Mint 1 (data not shown).
Fig. 11.
Mapping of the Caskin 1 binding site on CASK.A, Domain structure of CASK and positions of GST–CASK fusion proteins used for pulldowns. B, C, GST pulldowns of proteins in rat brain homogenates with the indicated CASK fusion proteins analyzed by immunoblotting for the proteins identified atright. B, Proteins were eluted with 0.8m K-acetate (KAc) followed by SDS sample buffer, whereas in C, proteins bound to the beads were examined. Note that in CASK, not only the CaM kinase domain but also the region homologous to the autoregulatory sequence of CaM kinase II are required for binding. Also note the separation between the common Mint 1–Caskin 1 and the Veli binding sites on CASK. Signals were visualized by ECL. Numbers at leftindicate positions of molecular weight markers.
Assembly of a CASK–Caskin 1 complex on the immobilized cytoplasmic tails of neurexins
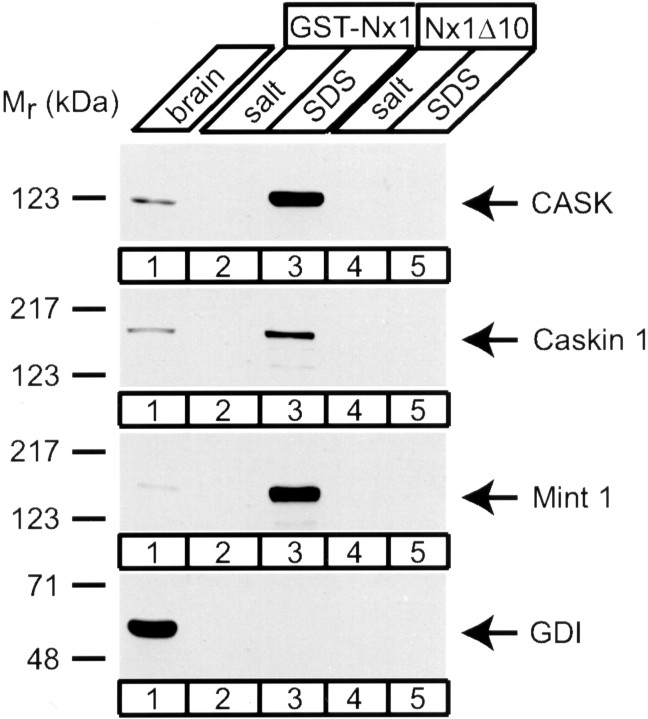
CASK binds to the cytoplasmic tails of several cell-surface proteins, including neurexins, syndecans, and JAMs (Hata et al., 1996;Cohen et al., 1998; Hsueh et al., 1998; Biederer and Südhof, unpublished observations). This reaction can be biochemically reconstituted with the immobilized recombinant tail of neurexin 1, which recruits not only CASK but also Mint 1 and Velis from brain homogenates (Biederer and Südhof, 2000). To test whether Caskin 1 can also be coassembled with CASK on the cytoplasmic tails of neurexins, we used affinity chromatography (Fig.12). Both Caskin 1 and Mint 1 were efficiently bound together with CASK, indicating that the CASK–Caskin 1 complex could be assembled on the plasma membrane. In contrast, GDP dissociation inhibitor (GDI, used as a negative control) remained unbound.
Fig. 12.
Assembly of CASK–Caskin 1 and CASK–Mint 1 complexes on the immobilized cytoplasmic domain of neurexin. Immobilized GST fusion protein of the wild-type cytoplasmic tail of neurexin 1 (GST–NxI) or of mutant cytoplasmic tail lacking the final 10 residues (NxIΔ10) was used for binding reactions with rat brain homogenates and eluted with 0.8 m K-acetate buffer (salt) followed by SDS sample buffer (SDS). Samples were analyzed by immunoblotting for CASK, Mint 1, Caskin 1, and GDI (as a negative control). Note the tight binding of CASK and Caskin 1 to the neurexin tail that requires the C-terminal residues of neurexin.Numbers at left indicate positions of molecular weight markers.
DISCUSSION
In the present study, we purified a novel CASK-binding protein that we named Caskin 1, determined its primary structure from cDNA clones, and studied its interactions with CASK. A closely related second isoform of Caskin 1, called Caskin 2, was also identified and cloned. Our data reveal that Caskin 1 constitutes a multidomain protein that in adult rodents is detected only in brain, in which it binds tightly to CASK. Caskin 1 and 2 are novel proteins that are distinct from other previously reported multidomain proteins, with a structure that suggests an adaptor function. At the N terminus, Caskins contain six ankyrin repeats that in many proteins participate in protein–protein interactions, followed by an atypical SH3 domain and two SAM repeats (Fig. 2). After a long proline-rich region, the two Caskins end in a similar C-terminal sequence (Fig. 1). This sequence may correspond to a novel evolutionarily conserved domain, because a similar sequence is found in an otherwise unrelatedDrosophila protein in GenBank (accession #AAF58176; data not shown). Caskins resemble many other adaptor proteins that usually contain a series of small, autonomously folded binding modules (such as the ankyrin repeats, SAM domains, and SH3 domain in Caskins) and in addition often include extended proline-rich sequences with characteristic signature motifs that may recruit the SH3 or WW domains of other proteins. The domain composition of Caskins is most similar to that of the Shank family of postsynaptic adaptor proteins (for review, see Sheng and Sala, 2001), although the precise number, type, and arrangement of domains differ (Fig. 2). It is generally noticeable that multidomain adaptor proteins appear to be particularly important for synaptic transmission and nuclear transcription, presumably because these reactions are most tightly regulated.
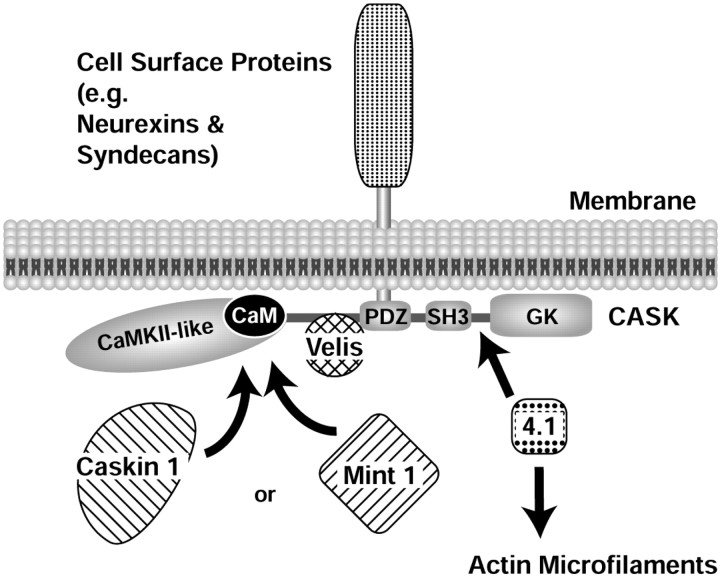
To define the protein network in which Caskin 1 participates, we mapped its interactions with CASK and compared them with those of Mint 1, a previously identified interaction partner of CASK (Butz et al., 1998). We showed that Caskin 1 binds to the N-terminal CaM kinase domain of CASK in direct competition with Mint 1. As a result, CASK participates in two independent and alternative tripartite complexes: the previously characterized CASK–Velis–Mint 1 complex (Borg et al., 1998a,b; Butz et al., 1998) and the new CASK–Velis–Caskin 1 complex. The coimmunoprecipitation of Caskin 1 and Mint 1 with CASK (Fig. 7) and the coassembly of Caskin 1 and Mint 1 with CASK on the immobilized cytoplasmic tail of neurexin 1 (Fig. 12) suggest that both complexes are normally present in the brain and bind to the cytoplasmic tails of neurexins and other cell-surface proteins. Viewed together with previous studies (Hata et al., 1996; Borg et al., 1998b; Butz et al., 1998; Cohen et al., 1998; Hsueh et al., 1998; Biederer and Südhof, 2001), these results show that in the vertebrate brain, CASK forms tight complexes with a multitude of distinct proteins (Fig.13): The N-terminal CaM kinase domain of CASK binds to either Caskin 1 or Mint 1 and interacts with calmodulin; the central Veli-interacting domain binds to Velis; the following PDZ domain interacts with the C-terminal cytoplasmic sequences of cell-surface proteins such as neurexins and syndecans, which anchor the entire complex on the plasma membrane; and finally, the C-terminal region of CASK binds to protein 4.1, which in turn connects to the actin cytoskeleton.
Fig. 13.
Model of the protein–protein interactions mediated by CASK. CASK is recruited to the plasma membrane via interactions of its PDZ domain with the C-terminal cytoplasmic tails of cell-surface proteins, for example neurexins, syndecans, and SynCaM. As a multidomain protein, CASK binds to multiple potential downstream effectors, many of which are also multidomain adaptor proteins containing domains similar to those of CASK. The N-terminal CaM kinase II homology region binds to calmodulin as a function of Ca2+ and to Caskin 1 and Mint 1 independently of Ca2+ (data not shown). The central sequence N-terminal to the PDZ domain binds to Velis, and the C-terminal SH3 and guanylate kinase (GK) domains and connecting sequences bind to protein 4.1, thereby recruiting actin filaments, and to Ca2+ channels (data not shown); in addition, the C-terminal regions may be involved in intramolecular and intermolecular interactions of CASK.
It is striking that except for the cell-surface proteins and calmodulin, all proteins known to bind to CASK are themselves adaptor proteins composed of multiple domains. The two alternative tripartite complexes formed by CASK (CASK–Mint 1– Velis and CASK–Caskin 1–Velis) incorporate a large number of potential interaction domains: multiple PDZ, SAM, and SH3 domains, in addition to ankyrin-like repeats, a guanylate kinase region, and other unique motifs, suggesting that these complexes form the nucleus of multiple high-molecular-weight complexes. The fact that Mint 1 and Caskin 1 are expressed primarily in the brain indicates that the two tripartite complexes are assembled only in the brain. In contrast, CASK and all other interacting proteins are widely expressed outside of the brain, where they presumably participate in other complexes with as yet unidentified interactors for the CASK CaM kinase domain that are ubiquitously expressed. Finally, it is noticeable that Caskin 1 is vertebrate-specific, whereas CASK and all of its other interacting proteins are evolutionarily conserved in invertebrates. The evolutionary derivation of novel proteins by mixing and matching established, phylogenetically old domains represents a quick mechanism of inventing new proteins that contribute to the rapid emergence of molecular complexity during mammalian evolution. The selective presence of Caskin 1 only in vertebrates but not in invertebrates suggests the possibility that Caskin 1 functionally expands CASK by regulating, among others, its interaction with Mint 1.
The functions of the multiple interactions of CASK with presumptive targets are at present unclear. In C. elegans, the mammalian CASK, Mint 1, and Veli proteins correspond to the proteins encoded by the lin-2, lin-7, and lin-10 genes, which are essential for targeting the EGF receptor homolog in vulval precursor cells (for review, seeKim, 1997; Kaech et al., 1998); in addition, the lin-10 gene has a function in the delivery of AMPA-type glutamate receptors to synapses (Rongo et al., 1998). In vertebrates, both CASK and Mint 1 have been associated with a number of possible functions that cannot be reconciled at present into a single biological perspective. For example, Mint 1 was demonstrated to bind to Munc18-1, an essential component of the synaptic vesicle fusion apparatus (Okamoto and Südhof, 1997); it was shown (under the name of X11) to bind to amyloid precursor protein (APP) and to alter APP processing (McLoughlin and Miller, 1996; Borg et al., 1998a) and was proposed (after being renamed mLin-10) to transport NMDA-type (but not AMPA-type) glutamate receptors to synapses in vertebrates (Setou et al., 2000). However, the lack of correspondence to the known role of lin-10 in C. elegans is puzzling. Our results provide a first step toward addressing this question by demonstrating that CASK participates in a complex network of protein–protein interactions in which Mint 1 competes with a vertebrate-specific new protein, Caskin 1. Although thein vivo importance of these interactions remains to be tested, preliminary data suggest that CASK and Mint 1 have distinct functions in vertebrates. CASK mutants in mice have a much more severe phenotype (Laverty and Wilson, 1998) than Mint 1 mutants (Südhof, unpublished observation). This, together with the apparent lack of Caskin conservation in invertebrates, suggests that CASK diversified evolutionarily toward the use of multiple effectors that include Caskin 1. Although much remains to be done to elucidate the precise functions of CASK and its alternative interactions with Caskin 1 and Mint 1, the description of the protein networks in which these proteins participate clearly represents the first necessary step toward addressing this question.
K.T. and T.B. contributed equally to this work.
This study was supported by National Institutes of Health Grant R37-MH52804-06 to T.C.S. and by a fellowship from the Human Frontiers in Science Program to T.B. We thank Dr. C. Slaughter for assistance in protein sequencing, Dr. A. Ho for help with the immunocytochemistry, and the Kazusa DNA Research Institute (Kisarazu, Japan) for providing the KIAA1139 and KIAA1306 cDNA clones.
Correspondence should be addressed to T. C. Südhof, The Center for Basic Neuroscience, Department of Molecular Genetics, and Howard Hughes Medical Institute, The University of Texas Southwestern Medical Center, Dallas, TX 75390. E-mail:Thomas.Sudhof@UTSouthwestern.edu.
S. Butz's present address: Zentrum für Molekularbiologie der Entzündung, Institut für Zellbiologie, Von Esmarchstradße 56, 48149 Münster, Germany.
REFERENCES
- 1.Biederer T, Südhof TC. Mints as adaptors: direct binding to neurexins and recruitment of munc18. J Biol Chem. 2000;275:39803–39806. doi: 10.1074/jbc.C000656200. [DOI] [PubMed] [Google Scholar]
- 2.Biederer T, Südhof TC. Cask and protein 4.1 support F-actin nucleation on neurexins. J Biol Chem. 2001;276:47869–47876. doi: 10.1074/jbc.M105287200. [DOI] [PubMed] [Google Scholar]
- 3.Borg JP, Yang Y, De Taddeo-Borg M, Margolis B, Turner RS. The X11alpha protein slows cellular amyloid precursor protein processing and reduces Abeta40 and Abeta42 secretion. J Biol Chem. 1998a;273:14761–14766. doi: 10.1074/jbc.273.24.14761. [DOI] [PubMed] [Google Scholar]
- 4.Borg JP, Straight SW, Kaech SM, de Taddeo-Borg M, Kroon DE, Karnak D, Turner RS, Kim SK, Margolis B. Identification of an evolutionarily conserved heterotrimeric protein complex involved in protein targeting. J Biol Chem. 1998b;273:31633–31636. doi: 10.1074/jbc.273.48.31633. [DOI] [PubMed] [Google Scholar]
- 5.Butz S, Okamoto M, Südhof TC. A tripartite protein complex with the potential to couple synaptic vesicle exocytosis to cell adhesion in brain. Cell. 1998;94:773–782. doi: 10.1016/s0092-8674(00)81736-5. [DOI] [PubMed] [Google Scholar]
- 6.Cohen AR, Woods DF, Marfatia SM, Walther Z, Chishti AH, Anderson JM, Wood DF. Human CASK/LIN-2 binds syndecan-2 and protein 4.1 and localizes to the basolateral membrane of epithelial cells. J Cell Biol. 1998;142:129–138. doi: 10.1083/jcb.142.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dimitratos SD, Woods DF, Bryant PJ. Camguk, Lin-2, and CASK: novel membrane-associated guanylate kinase homologs that also contain CaM kinase domains. Mech Dev. 1997;63:127–130. doi: 10.1016/s0925-4773(97)00668-0. [DOI] [PubMed] [Google Scholar]
- 8.Garner CC, Kindler S, Gundelfinger ED. Molecular determinants of presynaptic active zones. Curr Opin Neurobiol. 2000;10:321–327. doi: 10.1016/s0959-4388(00)00093-3. [DOI] [PubMed] [Google Scholar]
- 9.Guan KL, Dixon JE. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- 10.Hata Y, Slaughter CA, Südhof TC. Synaptic vesicle fusion complex contains unc-18 homologue bound to syntaxin. Nature. 1993;366:347–351. doi: 10.1038/366347a0. [DOI] [PubMed] [Google Scholar]
- 11.Hata Y, Butz S, Südhof TC. CASK: a novel dlg/PSD95 homologue with an N-terminal calmodulin-dependent protein kinase domain identified by interaction with neurexins. J Neurosci. 1996;16:2488–2494. doi: 10.1523/JNEUROSCI.16-08-02488.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoskins R, Hajnal AF, Harp SA, Kim SK. The C. elegans vulval induction gene lin-2 encodes a member of the MAGUK family of cell junction proteins. Development. 1996;122:97–111. doi: 10.1242/dev.122.1.97. [DOI] [PubMed] [Google Scholar]
- 13.Hsueh YP, Yang C, Kharazia V, Naisbitt S, Cohen AR, Weinberg RJ, Sheng M. Direct interaction of CASK/LIN-2 and syndecan heparan sulfate proteoglycan and their overlapping distribution in neuronal synapses. J Cell Biol. 1998;142:139–151. doi: 10.1083/jcb.142.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsueh YP, Wang TF, Yang FC, Sheng M. Nuclear translocation and transcription regulation by the membrane-associated guanylate kinase CASK/LIN-2. Nature. 2000;404:298–302. doi: 10.1038/35005118. [DOI] [PubMed] [Google Scholar]
- 15.Kaech SM, Whitfield CW, Kim SK. The LIN-2/LIN-7/LIN-10 complex mediates basolateral membrane localization of the C. elegans EGF receptor LET-23 in vulval epithelial cells. Cell. 1998;94:761–771. doi: 10.1016/s0092-8674(00)81735-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim SK. Polarized signaling: basolateral receptor localization in epithelial cells by PDZ-containing proteins. Curr Opin Cell Biol. 1997;9:853–859. doi: 10.1016/s0955-0674(97)80088-9. [DOI] [PubMed] [Google Scholar]
- 17.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 18.Laverty HG, Wilson JB. Murine CASK is disrupted in a sex-linked cleft palate mouse mutant. Genomics. 1998;53:29–41. doi: 10.1006/geno.1998.5479. [DOI] [PubMed] [Google Scholar]
- 19.Martinez-Estrada OM, Villa A, Breviario F, Orsenigo F, Dejana E, Bazzoni G. Association of junctional adhesion molecule with calcium/calmodulin-dependent serine protein kinase (CASK/LIN-2) in human epithelial caco-2 cells. J Biol Chem. 2001;276:9291–9296. doi: 10.1074/jbc.M006991200. [DOI] [PubMed] [Google Scholar]
- 20.Maximov A, Sudhof TC, Bezprozvanny I. Association of neuronal calcium channels with modular adaptor proteins. J Biol Chem. 1999;274:24453–24456. doi: 10.1074/jbc.274.35.24453. [DOI] [PubMed] [Google Scholar]
- 21.McLoughlin DM, Miller CC. The intracellular cytoplasmic domain of the Alzheimer's disease amyloid precursor protein interacts with phosphotyrosine-binding domain proteins in the yeast two-hybrid system. FEBS Lett. 1996;397:197–200. doi: 10.1016/s0014-5793(96)01128-3. [DOI] [PubMed] [Google Scholar]
- 22.Nix SL, Chishti AH, Anderson JM, Walther Z. hCASK and hDlg associate in epithelia, and their src homology 3 and guanylate kinase domains participate in both intramolecular and intermolecular interactions. J Biol Chem. 2000;275:41192–41200. doi: 10.1074/jbc.M002078200. [DOI] [PubMed] [Google Scholar]
- 23.Okamoto M, Südhof TC. Mints, munc18-interacting proteins in synaptic vesicle exocytosis. J Biol Chem. 1997;272:31459–31464. doi: 10.1074/jbc.272.50.31459. [DOI] [PubMed] [Google Scholar]
- 24.Pawson T, Nash P. Protein-protein interactions define specificity in signal transduction. Genes Dev. 2000;14:1027–1047. [PubMed] [Google Scholar]
- 25.Rongo C, Whitfield CW, Rodal A, Kim SK, Kaplan JM. LIN-10 is a shared component of the polarized protein localization pathways in neurons and epithelia. Cell. 1998;94:751–759. doi: 10.1016/s0092-8674(00)81734-1. [DOI] [PubMed] [Google Scholar]
- 26.Rosahl TW, Spillane D, Missler M, Herz J, Selig DK, Wolff JR, Hammer RE, Malenka RC, Südhof TC. Essential functions of synapsins I and II in synaptic vesicle regulation. Nature. 1995;375:488–493. doi: 10.1038/375488a0. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 28.Setou M, Nakagawa T, Seog DH, Hirokawa N. Kinesin superfamily motor protein KIF17 and mLin-10 in NMDA receptor-containing vesicle transport. Science. 2000;288:1796–1802. doi: 10.1126/science.288.5472.1796. [DOI] [PubMed] [Google Scholar]
- 29.Sheng M, Sala C. Pdz domains and the organization of supramolecular complexes. Annu Rev Neurosci. 2001;24:1–29. doi: 10.1146/annurev.neuro.24.1.1. [DOI] [PubMed] [Google Scholar]
- 30.Tseng TC, Marfatia SM, Bryant PJ, Pack S, Zhuang Z, O'Brien JE, Lin L, Hanada T, Chishti AH. VAM-1: a new member of the MAGUK family binds to human Veli-1 through a conserved domain. Biochim Biophys Acta. 2001;1518:249–259. doi: 10.1016/s0167-4781(01)00191-9. [DOI] [PubMed] [Google Scholar]
- 31.Tsunoda S, Zuker CS. The organization of INAD-signaling complexes by a multivalent PDZ domain protein in Drosophila photoreceptor cells ensures sensitivity and speed of signaling. Cell Calcium. 1999;26:165–171. doi: 10.1054/ceca.1999.0070. [DOI] [PubMed] [Google Scholar]