Abstract
Xenopuslaevis retinas, like retinas from all vertebrate classes, have endogenous circadian clocks that control many aspects of normal retinal physiology occurring in cells throughout all layers of the retina. The localization of the clock(s) that controls these various rhythms remains unclear. One of the best studied rhythmic events is the nocturnal release of melatonin. Photoreceptor layers can synthesize rhythmic melatonin when these cells are in isolation. However, within the intact retina, melatonin is controlled in a complex way, indicating that signals from many parts of the retina may contribute to the production of melatonin rhythmicity. To test this hypothesis, we generated transgenic tadpoles that express different levels of a dominant negative XenopusCLOCK specifically in the retinal photoreceptors. Eyes from these tadpoles continued to produce melatonin at normal levels, but with greatly disrupted rhythmicity, the severity of which correlated with the transgene expression level. These results demonstrate that although many things contribute to melatonin production in vivo, the circadian clock localized in the retinal photoreceptors is necessary for its rhythmicity. Furthermore, these data show that the control of the level of melatonin synthesis is separable from the control of its rhythmicity and may be controlled by different molecular machinery. This type of specific “molecular lesion” allows perturbation of the clock in intact tissues and is valuable for dissection of clock control of tissue-level processes in this and other complex systems.
Keywords: dominant negative CLOCK, transgenic Xenopus, circadian clock, retinal photoreceptor, melatonin rhythm, arylalkylamine N-acetyltransferase (AANAT)
Circadian oscillators exist in many tissues and control timing of local events (Damiola et al., 2000;Stokkan et al., 2001). This type of local circadian control is well established in the vertebrate retina, where many aspects of retinal physiology are regulated by an endogenous retinal circadian clock(s) (for review, see Besharse, 1988; Cahill and Besharse, 1995; Anderson and Green, 2000). This temporal regulation is a fundamental part of the normal function and maintenance of the retina, and disruption of these controls may contribute to retinal dysfunction and degeneration (Fain and Lisman, 1993).
Although retinal rhythms have been demonstrated in vertebrates of all classes, the Xenopus laevis retina has been a particularly valuable model system for these studies because of its large cells and its survivability in culture. In the Xenopusretina, several rhythmic phenomena are under the control of the circadian clock, including release of neuromodulators, gene expression, cone elongation, rod disc shedding (for review, see Besharse, 1988;Anderson and Green, 2000), and visual sensitivity (Manglapus et al., 1999).
Perhaps the most extensively studied rhythm in the retina is melatonin synthesis and release, which peaks during the night (Besharse, 1988;Cahill et al., 1991). Melatonin synthesis is under complex control, including regulation by the circadian clock, acute suppression by light, and suppression by dopamine, which is released from inner retinal cells (for review, see Cahill et al., 1991).
As in all clock systems, very little is known about how these various individual cellular rhythms are organized within the context of the intact tissue. It is unclear how many retinal cell types contain clocks or what the relationships of the various cellular clocks are. The few attempts that have been made to examine this issue have relied primarily on gross lesioning techniques. For example, destruction of the entire inner layers of the retina result in an enriched photoreceptor preparation that still maintains rhythmicity of melatonin production (Cahill and Besharse, 1993). However, within the intact eye, it is likely that other rhythmic processes may also contribute to the generation of the high amplitude melatonin rhythms. For example, dopamine from inner retinal neurons can both acutely suppress melatonin and affect the phase of the melatonin rhythm (Cahill et al., 1991).
The general molecular makeup of circadian clocks appears to be primarily conserved among all animals (for review, see Dunlap, 1999). One well conserved aspect of these clocks is the positive regulation ofperiod gene expression by the transcription factor heterodimer composed of CLOCK and BMAL1/CYCLE (Bae et al., 1998;Darlington et al., 1998; Lee et al., 1998; Glossop et al., 1999), which is necessary for clock function in all cases where it has been tested (Vitaterna et al., 1994; Antoch et al., 1997; King et al., 1997; Allada et al., 1998; Darlington et al., 1998; Rutila et al., 1998; Bunger et al., 2000).
Here we have used a transgenic approach to introduce a mutant form of the Clock gene specifically into photoreceptor cells of theXenopus retina. We demonstrate that expression of this mutant gene results in a very specific “molecular lesion” that ablates circadian clock function only in rods and cones in the context of the intact retina and results in arrhythmic melatonin production. Surprisingly, although melatonin is arrhythmic in these retinas, overall melatonin levels are not altered, demonstrating that the circadian regulation is separable from the general regulation of melatonin synthesis.
MATERIALS AND METHODS
DNA constructs. The transgene IRBP-XCLΔQ-GFP was constructed as follows. Xenopus laevis Clock cDNA (Zhu et al., 2000) (GenBank accession number AF227985) was truncated by PCR at bp 1629 using the following primers: 5′-CCTACAGAAGCATAAAGAAATCAGTGC-3′ and 5′-GA AGATCTT ACCGGTTTTGTTGGCGTTGATGATGGG-TC-3′ (this primer has introduced AgeI and BglII sites at the end, underlined). The truncated Clock cDNA was then digested with EcoRI and BglII and inserted into the EcoRI- and BglII-digested pBluescript II vector (pBS-XCLΔQ). The mouse interphotoreceptor retinoid-binding protein (IRBP) promoter (−155/+101) (Boatright et al., 1997) was digested with BamHI and inserted into the BamHI site of the pBS-XCLΔQ (pIRBP-XCLΔQ). Then pIRBP-XCLΔQ was digested with AgeI and SacII, and the excised fragment was ligated with a pEGFP-1 vector (Clontech, Palo Alto, CA) that was linearized with the same enzymes to produce IRBP-XCLΔQ-green fluorescent protein (GFP).
For transient transfections, pCMV-XCL (wild-type xClock) and pCMV-XBM (wild-type xBmal1) were constructed as follows. pVAX1 (Invitrogen, Carlsbad, CA) was digested with eitherBamHI and XhoI or EcoRI andXhoI, and BamHI- and SalI-digestedxClock or EcoRI- and XhoI-digestedxBmal1cDNA fragments (Anderson et al., 2001) were subcloned into the vector, respectively.
The luciferase reporter gene contains three tandem repeats of a fragment containing an E-box sequence reported from the mousePer1 gene promoter (Gekakis et al., 1998; Hida et al., 2000), which was cloned upstream of the rat prolactin basal promoter (Dr. Richard Day, University of Virginia; unpublished data) and firefly luciferase cDNA (pPer-Ebox).
Luciferase assay. Expression plasmids (0.1 μg each) and the luciferase reporter gene (0.1 μg) were cotransfected into COS-7 cells using Lipofectoamine following the manufacturer's instructions (Invitrogen, Gaithersburg, MD). After 24 hr incubation at 37°C, cells were lysed and examined for luciferase using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI). Luciferase activity was normalized by cotransfecting 25 ng of a control plasmid (pRL-TK, Promega) and measuring Renilla luciferase activity. Luciferase activity was measured using a luminometer (TD-20/20, Turner Designs, Sunnyvale, CA). ANOVA with Tukey-Kramerpost hoc multiple comparison tests (InStat Software, Graphpad, San Diego, CA) were performed to determine whether luciferase activities are significantly different between wild-type and truncated CLOCK.
Transgenesis. Xenopus laevis adults were purchased from NASCO (Fort Atkinson, WI), and eggs and sperm were obtained from the adult frogs. Transgenic tadpoles expressing IRBP-XCLΔQ-GFP were produced using restriction enzyme-mediated integration (REMI) in which the transgene is stably inserted into the sperm genome, followed by fertilization of eggs with the sperm nuclei (Kroll and Amaya, 1996), with the following modifications. IRBP-XCLΔQ-GFP was linearized with SpeI and used as a transgene. The REMI reaction was prepared by mixing 1 μl of linearized plasmid (150 ng/μl) with 4 μl of sperm nuclei (∼4 × 104 nuclei). After a 5 min incubation at room temperature, 1 μl 100 mmMgCl2, 0.5 μl of a 1:10 dilution ofSpeI (10 U/μl), 2 μl oocyte extract (preheated at 80°C for 10 min), and 9 μl sperm dilution buffer (250 mm sucrose, 75 mm KCl, 0.5 mm spermidine trihydrochloride, 0.2 mm spermidine tetrahydrochloride) were added and incubated for 10 min at room temperature. The reaction mix was diluted in MOH (10 mm KPO4, 125 mm Kgluconate, 5 mm NaCl, 0.5 mm MgCl2, 25 mm sucrose, 0.25 mmspermidine, 0.125 mm spermine) to a final concentration of ∼3 sperm nuclei in 5 nl, and then injected into dejellied Xenopus laevis eggs at a rate of 5 nl/sec using a glass needle and a syringe pump (Model 1111, Harvard Apparatus, Holliston, MA). The concentration of the sperm nuclei was empirically determined to result in one functional sperm nucleus being delivered to the majority of eggs. Normally developing embryos were maintained in 12 hr light/dark (LD 12:12) cycles until they reached the appropriate age for analysis (usually 2–3 weeks).
Genotyping. The tail tip was cut from each tadpole, and genomic DNA was isolated using the DNeasy Tissue Kit (Qiagen, Valencia, CA). PCR was then performed using AmpliTaq Gold (PE Applied Biosystems, Foster City, CA) and both GFP-specific and mouse IRBP promoter-specific primers. The sequences of the primers are as follows: GFP primers, 5′-CAAGCTGACCCTGAAGTTCATCTG-3′ and 5′-CGGATCTTGAAGTTCACCTTGATG-3′; IRBP primers, 5′-ATCCCT-ACACAGACATGGCT-3′ and 5′-ATCCCAGAGCCTTGGCTCCT-3′. PCR conditions were as follows: 95°C for 10 min, 30 cycles of 94°C for 40 sec, 55°C for 1 min, 72°C for 1 min; and 72°C for 10 min.
Images. Eyes from transgenic tadpoles were dissected and fixed in phosphate-buffered 4% paraformaldehyde overnight, then infiltrated with 30% sucrose and embedded in O.C.T. compound (Ted Pella, Redding, CA). GFP and phase images were obtained from frozen sections (10 μm thickness) using an inverted fluorescence microscope (IX70, Olympus, Melville, NY).
Flow-through culture. Tadpoles were maintained under LD 12:12 cycles before the experiments. Eyes from entrained 2- to 3-week-old tadpoles were dissected shortly before normal dark onset, and the cornea and lens were removed. Eyecups from independent tadpoles were transferred to a well of a 96-well microtiter plate containing culture medium consisting of 80% defined balanced salts and amino acids (Cahill et al., 1991) and 20% Wolf and Quimby amphibian tissue culture medium (Invitrogen). The medium was supplemented with 100 μm 5-hydroxy-l-tryptophan to enhance melatonin production (Cahill and Besharse, 1990), and the pH of the culture medium was equilibrated with 5% CO2/95% O2 during flow-through culture. Eyecups were then cultured in flow-through chambers as described previously for 3–5 d in constant darkness (DD) (Cahill et al., 1991; Green et al., 1999). Temperature was maintained at 21°C. The culture plates were kept in light-tight chambers, and the medium was continuously delivered with a syringe pump (model 2000, Harvard Apparatus) to each well at a constant rate of 0.2 ml/hr. Superfusates were collected in a fraction collector over 4 hr intervals.
Melatonin measurement. Melatonin levels in the superfusate samples from flow-through culture were determined by radioimmunoassay (RIA) as described previously (Rollag and Niswender, 1976) and validated for measurement of melatonin (ruling out cross-reactivity to other related compounds) in our culture medium (Cahill and Besharse, 1990).
Period analysis. Circadian rhythmicity of melatonin release was evaluated using a fast Fourier transform–nonlinear least squares (FFT-NLLS) estimation method developed by Dr. Martin Straume (National Science Foundation, Center for Biological Timing, University of Virginia) (Plautz et al., 1997). Time series were classified as arrhythmic if no rhythms were returned by the algorithm, i.e., the cosine function could not be fit to the data at a 95% confidence level. Instead of applying any cutoff for the relative amplitude of period as reported in the previous paper (Plautz et al., 1997), we classified eyecups as rhythmic if the relative amplitude of the period was <1 (FFT-NLLS default setting).
Quantitative RT-PCR. After flow-through culture was complete, culture medium was removed, eyecups were homogenized, and total RNA was isolated using Trizol reagent (Invitrogen) following the manufacturer's instructions. RNA was then reverse transcribed into cDNA using Superscript II (Invitrogen) and used as a template for PCR. Real-time quantitative PCR was performed using GeneAmp 5700 Sequence Detection System and SYBR Green Master Mix that includes SYBR Green Dye and AmpliTaq Gold (PE Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. For the quantitation of the transgene expression, GFP-specific primers were used for the PCR reaction (5′-AGCAAAGACCCCAACGAGAA-3′, 5′-GGCGGCGGTCACGA-A-3′). For quantitation of endogenous arylalkylamineN-acetyltransferase (AANAT) expression, the following primers were used: 5′-GGA-AGGCTGGTGGCATTTATC-3′, 5′-CCGGTTTGTGAAAGGTGAG-TG-3′ (Drs. Steven Coon and David Klein, personal communication). Human 18S rRNA primers (PE Applied Biosystems) (5′-CGGCTACCACATCCAAGGAA-3′, 5′-GCTGGAATTACCGCGGCT-3′) were used as an endogenous control for the purpose of normalization. For each experiment, a standard curve was prepared for each primer set using as template dilution series of cDNA from transgenic eyes, where the most concentrated standard was assigned an arbitrary value of 10. The levels of GFP and 18S rRNA levels in each test sample were then determined on the basis of the standard curve. We then normalized the GFP expression levels to the 18S rRNA expression levels for each pair of eyes. The data shown are the averages of three or six independent measurements for each tadpole.
RESULTS
XCLΔQ acts in a dominant negative mannerin vitro
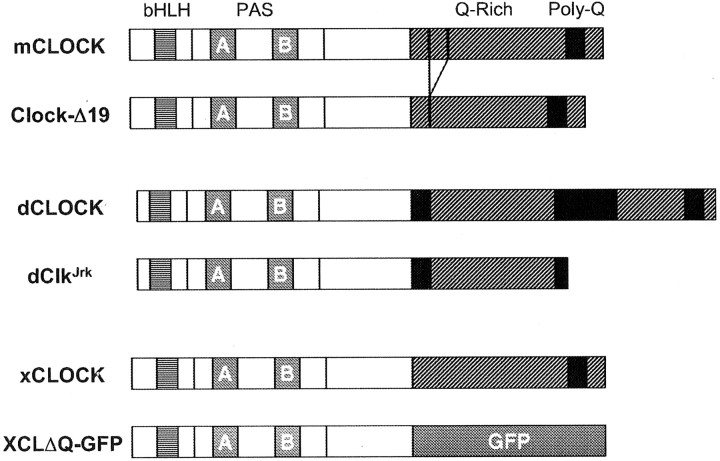
We have previously characterized the Xenopus homolog ofClock (Zhu et al., 2000). Like its orthologs, theXenopus CLOCK protein has a bHLH DNA-binding and protein interaction domain, a PAS protein–protein interaction domain near the N terminus, and a glutamine-rich (Q-rich) domain located at the C terminus that is thought to be a transcription activation domain (Zhu et al., 2000) (Fig. 1). From the structure of this protein, we predicted that an xCLOCK molecule lacking the activation domain would act as a dominant negative, because it would still bind DNA (the E-box element) and the BMAL1 binding partner but would not result in transcriptional activation (Gekakis et al., 1998). This prediction is supported by previous reports in whichClock mutants in both mouse (Clock) andDrosophila (ClkJrk) are semidominant in circadian phenotypes, and each mutation results in a partial deletion of the Q-rich domain at the C terminus (King et al., 1997; Allada et al., 1998) (Fig. 1).
Fig. 1.
Construction of XCLΔQ-GFP, a dominant negative form of xCLOCK. Diagrammatic comparison of XCLΔQ-GFP with normal CLOCK proteins and their known mutant forms in mouse andDrosophila. Hatched and filled regions at the C terminus correspond to the glutamine-rich (Q-rich) and poly-glutamine putative transactivation domains, respectively. PAS-A and PAS-B domains are indicated by Aand B. The mouse Clock mutation (Clock-Δ19) results in the deletion of 51 amino acids of the Q-rich transactivation domain. TheDrosophila dClkJrk mutant has a nonsense mutation that results in truncation of part of the Q-rich domain. In this study, XCLΔQ-GFP was constructed by deleting the Q-rich region from xCLOCK and replacing it with an in-frame GFP cDNA.
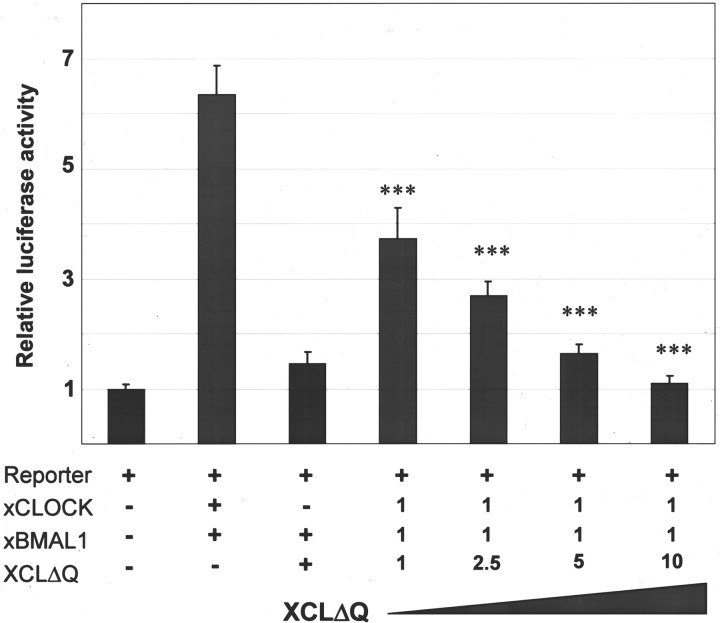
We therefore constructed a truncated version of xClock in which a sequence encoding the Q-rich activation domain was removed and replaced with an in-frame GFP (Fig. 1,XCLΔQ-GFP). To test whether XCLΔQ-GFP has the predicted dominant negative effect in vitro, we performed transient transfection assays using a luciferase reporter gene. We cotransfected COS-7 cells with expression vectors encoding either wild-type xCLOCK (pCMV-XCL) or XCLΔQ-GFP (pCMV-XCLΔQ-GFP) along with xBMAL1 (pCMV-XBM) and a luciferase reporter gene under the control of a basal promoter and with a triple mPer1 E-box cloned upstream (pPer-Ebox) (Gekakis et al., 1998). Although wild-type xCLOCK, along with xBMAL1, activated luciferase activity, no activation was observed when XCLΔQ-GFP and xBMAL1 were coexpressed (Fig.2). In addition, when XCLΔQ-GFP was cotransfected with wild-type xCLOCK and xBMAL1, it inhibited CLOCK/BMAL1-mediated activation of the luciferase activity in a dose-dependent manner (Fig. 2). Even at 1:1:1 ratio, significant inhibition of activation was observed, whereas complete inhibition was seen when 10-fold excess of pCMV-XCLΔQ-GFP was present (p < 0.001) (Fig. 2).
Fig. 2.
XCLΔQ acts as a dominant negative in vitro. Expression plasmids and the luciferase reporter gene (containing three repeats of mPer1 E-box and a basal promoter) were cotransfected into COS-7 cells, and luciferase activities were measured from cell lysates. xCLOCKrefers to a vector encoding wild-type xCLOCK (pCMV-XCL),XCLΔQ refers to a truncated xCLOCK (pCMV-XCLΔQ-GFP), and xBMAL1 refers to wild-type xBMAL1 (pCMV-XBM). In the right four columns, pCMV-XCL and pCMV-XBM were cotransfected with increasing amounts of pCMV-XCLΔQ-GFP. The ratios of the transfected plasmids are indicated by numbers below each column. The data are averages from three independent transfections. ANOVAs with Tukey-Kramer multiple comparison tests were performed to determine significant difference in luciferase activity between wild-type and truncated xCLOCK (***p < 0.001).
IRBP-XCLΔQ-GFP is specifically expressed in the retinal photoreceptor cells
A circadian oscillator located in the photoreceptor layer inXenopus can regulate retinal circadian melatonin rhythms when this layer is cultured without the rest of the retina (Cahill and Besharse, 1993). To investigate the role of the photoreceptor clock in the regulation of melatonin synthesis in vivo (in the intact retina), transgenic studies were performed using the REMI method (Kroll and Amaya, 1996). We designed a transgene that expresses XCLΔQ-GFP in all photoreceptor cells, using the mouse IRBP gene promoter. This promoter drives reporter gene expression in rod and cone photoreceptors in retina and occasionally low level expression in the pineal gland in Xenopus (Fig.3) (data not shown). We have never observed mouse IRBP-driven expression in any other body part or any nonphotoreceptor cells in the retina.
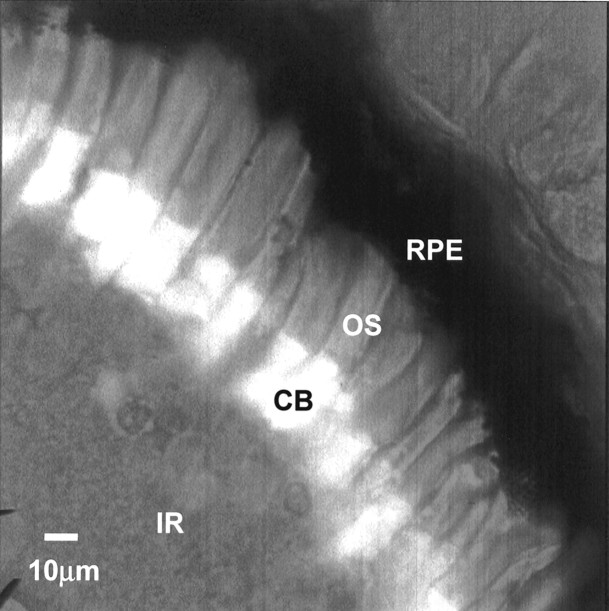
Fig. 3.
XCLΔQ-GFP accumulates in the photoreceptor cell bodies. An image of a section through a transgenic tadpole retina expressing XCLΔQ-GFP under the control of the IRBP promoter. XCLΔQ-GFP is present in the cell bodies and inner segments of all the photoreceptor cells. CB, Photoreceptor cell body;OS, photoreceptor outer segment; IR, inner retina; RPE, retinal pigment epithelium. Note that this is one of the strongest expressors of GFP among many transgenic eyes examined. This is a merged image of GFP signal and phase.
We first tested the IRBP-XCLΔQ-GFP transgene for general translation, stability, and cellular localization by producing transgenicXenopus tadpoles and visualizing the fluorescent XCLΔQ-GFP in the retina. Sections through the retinas of these transgenic tadpoles verified that IRBP-XCLΔQ-GFP was stably expressed in both rod and cone cells and was localized in the cell bodies, including nuclei (Fig. 3). No staining was observed in any other cell types. The cells expressing this transgene looked morphologically normal at the light microscopic level. It is of note that we observed different levels of GFP expression in eyes from different transgenic tadpoles, and in some cases GFP was not expressed at a detectable level, even when transgene was stably incorporated (data not shown).
Transgenic eyecups show abnormal rhythms of melatonin release but normal average melatonin levels
Circadian rhythms of melatonin release in Xenopus can be monitored over time when Xenopus retinas or eyecups are maintained in a flow-through culture system (Cahill et al., 1991). To investigate the involvement of xCLOCK in the regulation of circadian melatonin release, we performed eyecup culture for several days using transgenic tadpoles expressing XCLΔQ-GFP, with wild-type siblings as controls. Embryos were held in cyclic light (LD 12:12) until they reached 2–3 weeks of age, then eyecups were dissected and maintained in flow-through culture in DD for 3–5 d. The transgenic technique results in a mixture of developing embryos, some carrying the transgene and some nontransgenic (wild-type) siblings. The tail of each embryo was saved for PCR analysis to determine the genotype of each animal after the flow-through culture was complete. Therefore, each experiment contained wild-type nontransgenic tadpoles as controls that were obtained from the same set of microinjections and were maintained in exactly the same conditions as the transgenic animals.
Most of the wild-type eyecups showed circadian rhythms of melatonin synthesis (87%; average τ = 24.6 ± 0.6 hr) (Fig.4A, Table1), consistent with previous reports (Cahill and Besharse, 1990, 1993; Cahill et al., 1991; Green et al., 1999). Those few wild-type eyes that were judged arrhythmic showed weak rhythms that damped and were statistically nonrhythmic. In contrast, only 29% of the transgenic eyecups showed circadian expression of melatonin, with periods similar to those of wild-type siblings (Table1). Of the transgenic tadpoles, rhythms were disrupted in 71%, including 8.5% that exhibited rhythms with extremely long periods (average τ = 33.9 hr), and 62.5% showed no significant rhythmicity (Fig. 4B, Table 1). We also compared the average melatonin production levels (averaged over all fractions) of the transgenic and wild-type eyecups, but no significant difference was detected between those two distinct genotypes (Fig.5). It is of note that even the arrhythmic transgenic eyes showed an initial peak of melatonin release during the first night after transfer from LD 12:12 to DD, which is comparable in level to the peak of the wild-type eyes. This peak is most likely the result of melatonin rebound after the previous light period, as has been observed previously in Xenopus retina and chick pineal cells (Zatz, 1991; Green et al., 1999). Although we do not know the mechanism driving this “rebound” phenomenon, this suggests a direct effect of light or light cycles on the amplitude of the melatonin rhythm, and this effect still exists in the transgenic eyes.
Fig. 4.
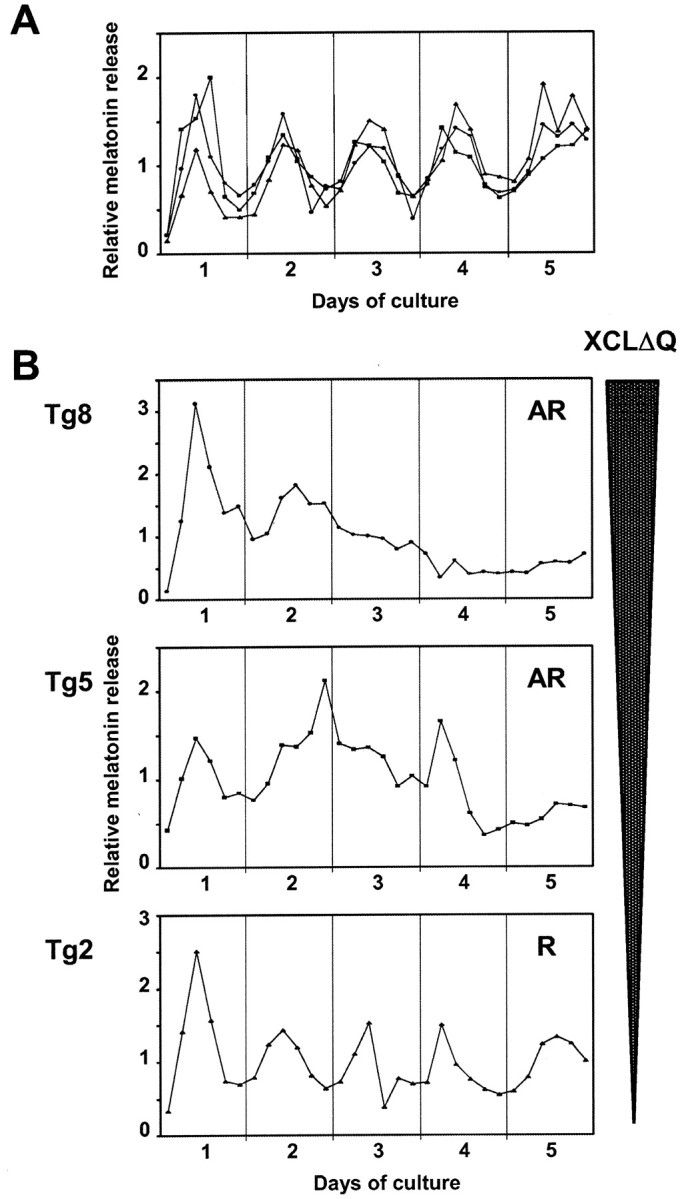
Melatonin release from wild-type and transgenic eyes. Each line represents melatonin release from a single pair of eyes in a flow-through culture for 5 d in constant darkness. Media fractions were collected every 4 hr and assayed for melatonin. Relative melatonin levels (melatonin content in each fraction divided by average) are shown to better allow comparisons between different animals. A, Wild-type eyes;B, patterns of melatonin release from three representative transgenic tadpole eyes are shown (Tg8,Tg5, and Tg2 refer to the numbering used in Fig. 6). The expression level of XCLΔQ was highest inTg8, intermediate in Tg5, and lowest inTg2 (see also Fig. 6). Of these tadpoles, onlyTg2 showed a significant circadian rhythm of melatonin release. Tg5 showed some indication of rhythmic release with long period, although melatonin release was not significantly rhythmic by our criteria. Circadian rhythmicity of melatonin release was analyzed using FFT-NLLS. AR, Arrhythmic; A, rhythmic.
Table 1.
Summary of the melatonin release rhythms
| Number of animals | Period (hr ± SEM) R + LP | % Abnormal | |||||
|---|---|---|---|---|---|---|---|
| Total | R | LP | AR | AR | Total (AR + LP) | ||
| XCLΔQ-GFP | 24 | 7 | 2 | 151-165 | 26.4 ± 1.5 | 62.5 | 71.0 |
| Wild type | 23 | 20 | 1 | 2 | 24.6 ± 0.6 | 8.7 | 13.0 |
Circadian rhythmicity of melatonin release was analyzed using FFT-NLLS. Total % abnormal includes both arrhythmic and rhythmic eyes with long periods (>28 hr). R, rhythmic (period range between 20 and 28 hr); LP, long period (>28 hr); AR, arrhythmic (no significant rhythm at a 95% level of confidence).
F1-165: indicates significant difference between wild-type and transgenic eyes determined by Fisher's exact test (p < 0.001).
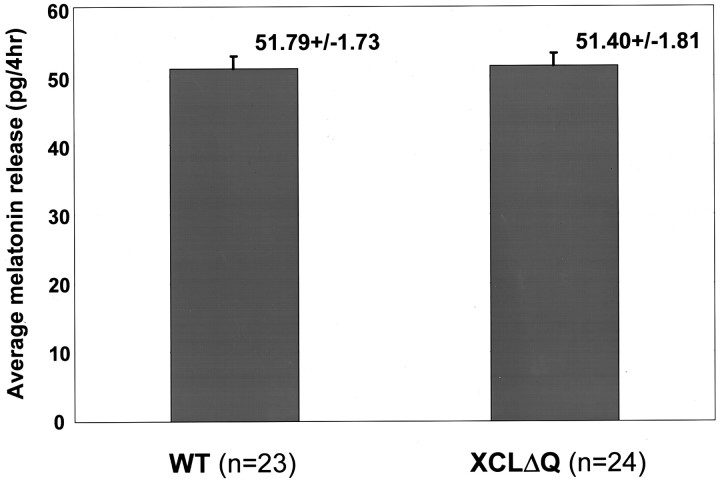
Fig. 5.
Total melatonin levels were not affected in transgenic eyes. Average melatonin release from eyes from transgenic and wild-type tadpoles was calculated from all fractions of all flow-through experiments. Values on the figure are average melatonin content (picograms per 4 hr) ± SEM.
XCLΔQ alters the circadian expression of melatonin in a dose-dependent manner
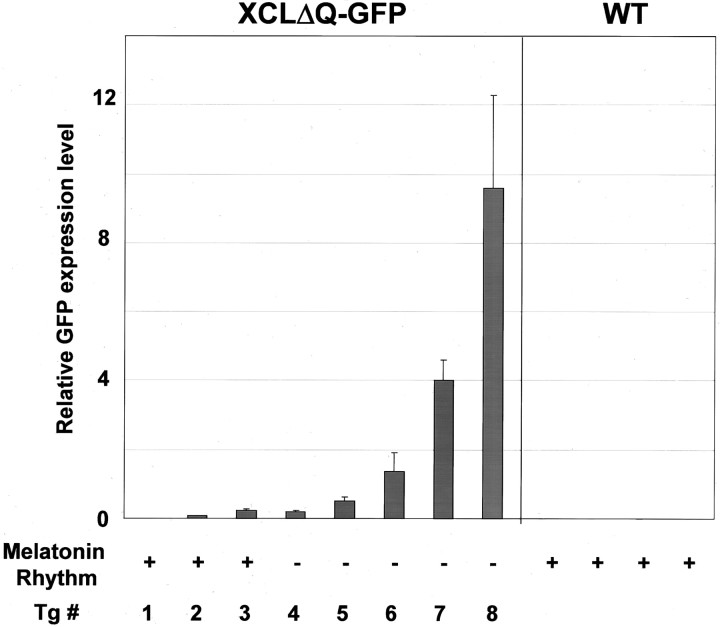
As mentioned above, we observed variability in the phenotypes of retinal melatonin release among individual transgenic tadpoles. This incomplete penetrance is most likely caused by variability in transgene expression levels in individual tadpoles as observed when visualizing GFP fluorescence. To test this possibility, we compared transgene expression levels in individual eyes from some of our tadpoles by quantitative real-time RT-PCR. At the end of the flow-through experiments, total RNA was extracted from the cultured eyecups, and cDNA was synthesized from the RNA for use as a template in quantitative PCR. We averaged the relative expression levels of the transgene from six independent measurements of each sample and found a large range of transgene expression levels among all eyes tested (Fig.6). This variability in expression levels could be attributable to differences in copy numbers of the transgene or its integration sites, or both. These different expression levels correlate well with the melatonin phenotype: only the eyecups with low expression of the transgene had exhibited rhythmic melatonin release, whereas stronger expressors were arrhythmic (Figs.4B, 6). As an example, among the three melatonin profiles from transgenic eyes shown in Figure 4B, the highest expressor (Tg8) was arrhythmic, whereas the lowest (Tg2) showed a statistically significant circadian rhythm of melatonin release. The eyecups that express the transgene at an intermediate level (Tg5) did not produce statistically rhythmic melatonin, although a few peaks can be observed (Figs.4B, 6).
Fig. 6.
Relative expression levels of the XCLΔQ-GFP transgene in the eyes. Quantitative RT-PCR was performed using GFP primers on RNA from each pair of eyes after they were used for the flow-through culture. Relative expression levels of the GFP mRNA were determined and normalized to the endogenous control (18S rRNA). Each value is the average of six independent measurements.Tg1–Tg8 indicate expression levels of the transgene from pairs of eyes from individual transgenic tadpoles. The presence (+) or absence (−) of melatonin rhythmicity in flow-through culture is noted for each tadpole. Tg8,Tg5, and Tg2 correspond to the transgenic eyes that produced the melatonin profiles shown in Figure4B.
XCLΔQ expression does not affect endogenous AANAT mRNA levels
AANAT is one of the melatonin synthetic enzymes, and its activity peaks at night in all species examined (Klein et al., 1997). A recent report describes circadian transcriptional regulation of AANAT in chicken, where an E-box in the chicken AANAT 5′-flanking region was shown to be essential for its transcriptional activation and rhythmic expression (Chong et al., 2000). The CLOCK/BMAL1 heterodimer is capable of directly binding to this E-box and enhancing its transcription in transient transfection assays, suggesting that CLOCK controls melatonin production by directly regulating expression of this enzyme involved in its synthesis (Chong et al., 2000). To examine whether expression of XCLΔQ alters AANAT mRNA levels in our transgenic eyes, we performed quantitative real-time RT-PCR using AANAT-specific primers on some of the transgenic and wild-type eyes used previously for flow-through culture. These eyes had been collected from the flow-through culture in early subjective night (circadian time 13–14). Although we observed variation in the expression levels of XCLΔQ-GFP and a range of melatonin profiles among individuals, there were no significant differences in the AANAT mRNA levels between strong and weak expressors of the transgene, nor between wild-type and transgenic animals (Table 2).
Table 2.
XCLΔQ alters melatonin rhythmicity, but not the endogenous AANAT mRNA levels
| Genotype | Relative mRNA level | Melatonin rhythm | |
|---|---|---|---|
| XCLΔQ-GFP | AANAT (% of WT) | ||
| WT | ND | 100.0 | R |
| Tg | ND | 74.2 ± 10.5 | R |
| Tg | 1.2 ± 0.2 | 72.3 ± 1.8 | R |
| Tg | 1.6 ± 0.1 | 86.5 ± 3.2 | R |
| Tg | 20.1 ± 3.0 | 128.9 ± 7.9 | AR |
| Tg | 91.1 ± 11.6 | 63.5 ± 6.6 | AR |
Quantitative RT-PCR was performed using GFP (for XCLΔQ-GFP mRNA) and Xenopus AANAT primers (for endogenous AANAT mRNA) on RNA from some pairs of eyes after they were analyzed for rhythmicity. Eyes were collected for RNA extraction at the end of the flow-through culture period (circadian time 13–14). Relative expression levels of the GFP and AANAT mRNAs were determined and normalized to the endogenous control (18S rRNA). Each value is the average of three independent measurements (± SEM). ND, Not detectable; R, rhythmic; AR, arrhythmic.
DISCUSSION
In this study, we generated transgenic Xenopus laevis tadpoles that express a truncated xCLOCK (XCLΔQ-GFP) in the photoreceptor cells. This approach demonstrates that it is feasible to specifically manipulate a molecular clock component in a subset of cells within the intact retina in such a way that circadian clock function is compromised. Our data indicate that a circadian clock in the photoreceptors is not only sufficient but also necessary to generate circadian rhythms of melatonin release. However, a functional photoreceptor clock is not necessary to generate normal levels of melatonin.
Although most of the transgenic eyes did not show significant circadian rhythms of melatonin release, some were rhythmic. Because copy numbers and integration sites of the transgene can vary among tadpoles when the REMI method is used, we expected eyes from individual transgenic tadpoles to express XCLΔQ at different levels. In fact, our results from quantitative RT-PCR (Fig. 6) indicate extreme differences in the transgene expression levels. These experiments also demonstrate that those transgenic eyes that were capable of rhythmic melatonin release expressed XCLΔQ-GFP at low levels. In contrast, the arrhythmic eyes expressed the transgene at higher levels (Figs. 4B,6). These data suggest dose effects of the transgene expression, which is consistent with the dose-dependent effect observed in our transient transfection (Fig. 2).
It is of note that average total melatonin secretion levels were not affected by dominant negative xCLOCK (Fig. 6). This indicates that xCLOCK is involved only in regulating melatonin rhythmicity and not absolute levels of melatonin. These data also demonstrate that photoreceptor cells of the transgenic retina are metabolically active and still produce melatonin, and the loss of circadian rhythmicity is not caused by a general toxic effect of XCLΔQ-GFP overexpression. Also, more importantly, these results suggest that there are other mechanisms that control melatonin synthesis and that melatonin synthesis and its rhythmicity are separable events.
Circadian control of melatonin rhythms may occur at several levels. It is known that the melatonin rhythm is regulated, at least in part, by dopamine, another neuromodulator that is expressed during the day. Dopamine acutely suppresses the activity of one of the melatonin synthetic enzymes, AANAT (Iuvone and Besharse, 1986; Boatright et al., 1994), and also causes phase-dependent phase shifts of the circadian clock controlling melatonin rhythms (Cahill et al., 1991). Because melatonin (presumably coming from the photoreceptors) also acutely suppresses dopamine, it has not been known previously which arm of this negative feedback loop (and which cell type) is most critical for driving melatonin rhythms. Our data indicate that the melatonin rhythms are driven by clocks in photoreceptor cells and not simply by rhythmic dopamine.
Other evidence for complex control of melatonin rhythmicity inXenopus comes from studies in which the inner retina was lesioned with detergent treatment (Cahill and Besharse, 1993). Although these photoreceptor layers maintained rhythmic release of melatonin, the period was slightly lengthened. In addition, overall levels of melatonin released were reduced by ∼90% when the retinal pigment epithelial layer was removed, suggesting that these cells also contribute to the regulation of melatonin production. Therefore, although the photoreceptor clocks are both necessary and sufficient for driving melatonin rhythms, these other cell types appear to influence melatonin levels and rhythmicity.
The CLOCK protein has been implicated in transcriptional control of both the core clock mechanism and other downstream rhythmic genes (Jin et al., 1999; Park et al., 2000). A previous report suggests that in chicken, AANAT transcription is regulated through an E-box in the AANAT 5′-flanking sequence (Chong et al., 2000). Transient transfection experiments demonstrated that this E-box could bind both CLOCK/BMAL1 and the closely related MOP4/BMAL1 heterodimers, resulting in activated transcription (Chong et al., 2000). Our present data suggest that inXenopus retina, melatonin rhythmicity is regulated by CLOCK, but probably not through direct CLOCK control of AANAT transcription (Table 2). Because our measurements of AANAT mRNA levels could only be done at a single time point (the end of the flow-through experiment), we cannot rule out the possibility that the XCLΔQ-expressing eyes may have altered AANAT levels at other time points. However, our data that show no change in overall melatonin levels suggest that this is not the case (Fig. 5). In Xenopus retina, AANAT activity is under circadian control (Besharse and Iuvone, 1983), and one possibility is that CLOCK controls circadian NAT activity rhythms inXenopus retina indirectly by regulating some other gene(s). In chicken and Xenopus retina, another melatonin synthetic enzyme, tryptophan hydroxylase, is also expressed in a circadian manner, implying that the transcription of this gene may also be controlled by CLOCK or some other clock-controlled factor (Green et al., 1996; Chong et al., 1998). However, because our culture medium contains excess 5-hydroxy-l-tryptophan (the product of tryptophan hydroxylase) to enhance melatonin levels, any potential effect of this mutation on tryptophan hydroxylase would not be observed in these experiments.
Because our study demonstrated that total melatonin production is not altered in the transgenic eyes, we suggest that the role of CLOCK is not to simply turn on melatonin synthesis at night. If this were the case, then the transgenic animals that are arrhythmic (presumably with disrupted CLOCK-mediated transcriptional activation) would show constitutively low levels of melatonin. Although the reason for the normal levels of melatonin in these eyes is not known, one possibility is that the melatonin synthetic pathway is under both positive and negative transcriptional control, both of which require CLOCK. Alternatively, it is possible that melatonin synthesis is regulated through a negative feedback loop in which CLOCK directly or indirectly activates melatonin synthesis, which eventually feeds back to suppress melatonin output to trough levels. If this were the case, then the transgenic retinas that are missing the activation mechanism may never show the negative feedback and melatonin might remain at average levels. The present data cannot answer this question directly, but this method provides a means for more careful dissection of these control pathways in future studies.
Finally, this study raises the question of whether the circadian clock controlling melatonin release rhythms is localized either in rods or cones or in both photoreceptor cell types. In Xenopus eyes, all of the clock gene homologs identified thus far are expressed in both rod and cone photoreceptor cells (Zhu et al., 2000; Anderson et al., 2001; Zhu and Green, 2001), implying that both cell types have a circadian oscillator(s). Another interesting issue is whether the circadian clock that controls rhythmic melatonin expression regulates other outputs as well. As mentioned above, many physiological rhythms are under the control of the ocular clock(s), although it is not clear whether one circadian oscillator in the photoreceptor cells directs the many different rhythms in the retina, or whether more than one oscillator, located in different retinal cell types, controls different circadian processes. The localization of the clock controlling melatonin synthesis and other rhythmic events can be addressed by using distinct cell type-specific promoters to target XCLΔQ-GFP followed by the assessment of different rhythmic outputs.
In conclusion, we have succeeded in making transgenicXenopus tadpoles in which the xCLOCK function is abolished or altered, specifically in photoreceptor cells in the intact animal. The expression of this dominant negative transgene inXenopus is a valuable tool to selectively inactivate circadian clock function in specific cells and should allow careful dissection of the mechanism of circadian clock function within a complex tissue such as the retina.
Footnotes
This work was supported by grants from the National Institute of Mental Health (MH61461) and the Thomas F. Jeffress and Kate Miller Jeffress Memorial Trust. We thank Barry Knox and Richard Day for providing vectors, and Mark Rollag for melatonin antibody. We are also grateful to Manuel Miranda-Anaya and Xiaorong Liu for generous technical advice, Ignacio Provencio and Ana Maria Castrucci for tremendous help with real-time PCR, and Michael Menaker, Jay Hirsh, Julie Baggs, Carl Strayer, and Cara Constance for critical comments on this manuscript.
Correspondence should be addressed to Dr. Carla B. Green, Department of Biology, University of Virginia, 264 Gilmer Hall, Charlottesville, VA 22904-4328. E-mail: cbg8b@virginia.edu.
REFERENCES
- 1.Allada R, White NE, So WV, Hall JC, Rosbash M. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93:791–804. doi: 10.1016/s0092-8674(00)81440-3. [DOI] [PubMed] [Google Scholar]
- 2.Anderson FE, Green CB. Symphony of rhythms in Xenopus laevis retina. Microsc Res Tech. 2000;50:360–372. doi: 10.1002/1097-0029(20000901)50:5<360::AID-JEMT5>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 3.Anderson FE, Hayasaka N, Green CB. Rhythmic expression of BMAL in the retina of Xenopus Laevis and nonrhythmic expression in other tissues. Invest Ophthalmol Vis Sci. 2001;42:S776. [Google Scholar]
- 4.Antoch MP, Song EJ, Chang AM, Vitaterna MH, Zhao Y, Wilsbacher LD, Sangoram AM, King DP, Pinto LH, Takahashi JS. Functional identification of the mouse circadian Clock gene by transgenic BAC rescue. Cell. 1997;89:655–667. doi: 10.1016/s0092-8674(00)80246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bae K, Lee C, Sidote D, Chuang KY, Edery I. Circadian regulation of a Drosophila homolog of the mammalian Clock gene: PER and TIM function as positive regulators. Mol Cell Biol. 1998;18:6142–6151. doi: 10.1128/mcb.18.10.6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Besharse JC, Iuvone PM, Pierce ME. Regulation of rhythmic photoreceptor metabolism: a role for post-receptoral neurons. Prog Retinal Res. 1988;17:21–61. [Google Scholar]
- 7.Besharse JC, Iuvone PM. Circadian clock in Xenopus eye controlling retinal serotonin N-acetyltransferase. Nature. 1983;305:133–135. doi: 10.1038/305133a0. [DOI] [PubMed] [Google Scholar]
- 8.Boatright JH, Rubim NM, Iuvone PM. Regulation of endogenous dopamine release in amphibian retina by melatonin: the role of GABA. Vis Neurosci. 1994;11:1013–1018. doi: 10.1017/s0952523800003941. [DOI] [PubMed] [Google Scholar]
- 9.Boatright JH, Buono R, Bruno J, Lang RK, Si JS, Shinohara T, Peoples JW, Nickerson JM. The 5′ flanking regions of IRBP and arrestin have promoter activity in primary embryonic chicken retina cell cultures. Exp Eye Res. 1997;64:269–277. doi: 10.1006/exer.1996.0222. [DOI] [PubMed] [Google Scholar]
- 10.Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA. MOP3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103:1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cahill GM, Besharse JC. Circadian regulation of melatonin in the retina of Xenopus laevis: limitation by serotonin availability. J Neurochem. 1990;54:716–719. doi: 10.1111/j.1471-4159.1990.tb01932.x. [DOI] [PubMed] [Google Scholar]
- 12.Cahill GM, Besharse JC. Circadian clock functions localized in Xenopus retinal photoreceptors. Neuron. 1993;10:573–577. doi: 10.1016/0896-6273(93)90160-s. [DOI] [PubMed] [Google Scholar]
- 13.Cahill GM, Besharse JC. Circadian rhythmicity in vertebrate retinas: regulation by a photoreceptor oscillator. Prog Retin Eye Res. 1995;14:267–291. [Google Scholar]
- 14.Cahill GM, Grace MS, Besharse JC. Rhythmic regulation of retinal melatonin: metabolic pathways, neurochemical mechanisms, and the ocular circadian clock. Cell Mol Neurobiol. 1991;11:529–560. doi: 10.1007/BF00734814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chong NW, Cassone VM, Bernard M, Klein DC, Iuvone PM. Circadian expression of tryptophan hydroxylase mRNA in the chicken retina. Brain Res Mol Brain Res. 1998;61:243–250. doi: 10.1016/s0169-328x(98)00219-8. [DOI] [PubMed] [Google Scholar]
- 16.Chong NW, Bernard M, Klein DC. Characterization of the chicken serotonin N-acetyltransferase gene. Activation via clock gene heterodimer/E box interaction. J Biol Chem. 2000;275:32991–32998. doi: 10.1074/jbc.M005671200. [DOI] [PubMed] [Google Scholar]
- 17.Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Darlington TK, Wager-Smith K, Ceriani MF, Staknis D, Gekakis N, Steeves TDL, Weitz CJ, Takahashi JS, Kay SA. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science. 1998;280:1599–1603. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- 19.Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 20.Fain GL, Lisman JE. Photoreceptor degeneration in vitamin A deprivation and retinitis pigmentosa: the equivalent light hypothesis. Exp Eye Res. 1993;57:335–340. doi: 10.1006/exer.1993.1132. [DOI] [PubMed] [Google Scholar]
- 21.Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 22.Glossop NR, Lyons LC, Hardin PE. Interlocked feedback loops within the Drosophila circadian oscillator. Science. 1999;286:766–768. doi: 10.1126/science.286.5440.766. [DOI] [PubMed] [Google Scholar]
- 23.Green CB, Besharse JC, Zatz M. Tryptophan hydroxylase mRNA levels are regulated by the circadian clock, temperature, and cAMP in chick pineal cells. Brain Res. 1996;738:1–7. doi: 10.1016/0006-8993(96)00743-3. [DOI] [PubMed] [Google Scholar]
- 24.Green CB, Liang MY, Steenhard BM, Besharse JC. Ontogeny of circadian and light regulation of melatonin release in Xenopus laevis embryos. Brain Res Dev Brain Res. 1999;117:109–116. doi: 10.1016/s0165-3806(99)00109-1. [DOI] [PubMed] [Google Scholar]
- 25.Hida A, Koike N, Hirose M, Hattori M, Sakaki Y, Tei H. The human and mouse Period1 genes: five well-conserved E-boxes additively contribute to the enhancement of mPer1 transcription. Genomics. 2000;65:224–233. doi: 10.1006/geno.2000.6166. [DOI] [PubMed] [Google Scholar]
- 26.Iuvone PM, Besharse JC. Dopamine receptor-mediated inhibition of serotonin N-acetyltransferase activity in retina. Brain Res. 1986;369:168–176. doi: 10.1016/0006-8993(86)90525-1. [DOI] [PubMed] [Google Scholar]
- 27.Jin X, Shearman LP, Weaver DR, Zylka MJ, de Vries GJ, Reppert SM. A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell. 1999;96:57–68. doi: 10.1016/s0092-8674(00)80959-9. [DOI] [PubMed] [Google Scholar]
- 28.King DP, Zhao Y, Sangoram AM, Wilsbacher LD, Tanaka M, Antoch MP, Steeves TD, Vitaterna MH, Kornhauser JM, Lowrey PL, Turek FW, Takahashi JS. Positional cloning of the mouse circadian clock gene. Cell. 1997;89:641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klein DC, Coon SL, Roseboom PH, Weller JL, Bernard M, Gastel JA, Zatz M, Iuvone PM, Rodriguez IR, Begay V, Falcon J, Cahill GM, Cassone VM, Baler R. The melatonin rhythm-generating enzyme: molecular regulation of serotonin N-acetyltransferase in the pineal gland. Recent Prog Horm Res. 1997;52:307–357. [PubMed] [Google Scholar]
- 30.Kroll KL, Amaya E. Transgenic Xenopus embryos from sperm nuclear transplantations reveal FGF signaling requirements during gastrulation. Development. 1996;122:3173–3183. doi: 10.1242/dev.122.10.3173. [DOI] [PubMed] [Google Scholar]
- 31.Lee C, Bae K, Edery I. The Drosophila CLOCK protein undergoes daily rhythms in abundance, phosphorylation, and interactions with the PER-TIM complex. Neuron. 1998;21:857–867. doi: 10.1016/s0896-6273(00)80601-7. [DOI] [PubMed] [Google Scholar]
- 32.Manglapus M, Parshley M, Stewart K, Engbreston G, Knox B, Barlow R. Circadian changes of ERG responses in the Xenopus laevis. Invest Ophthalmol Vis Sci. 1999;40:S610. [Google Scholar]
- 33.Park JH, Helfrich-Forster C, Lee G, Liu L, Rosbash M, Hall JC. Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proc Natl Acad Sci USA. 2000;97:3608–3613. doi: 10.1073/pnas.070036197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plautz JD, Straume M, Stanewsky R, Jamison CF, Brandes C, Dowse HB, Hall JC, Kay SA. Quantitative analysis of Drosophila period gene transcription in living animals. J Biol Rhythms. 1997;12:204–217. doi: 10.1177/074873049701200302. [DOI] [PubMed] [Google Scholar]
- 35.Rollag MD, Niswender GD. Radioimmunoassay of serum concentrations of melatonin in sheep exposed to different lighting regimens. Endocrinology. 1976;98:482–489. doi: 10.1210/endo-98-2-482. [DOI] [PubMed] [Google Scholar]
- 36.Rutila JE, Suri V, Le M, So WV, Rosbash M, Hall JC. CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell. 1998;93:805–814. doi: 10.1016/s0092-8674(00)81441-5. [DOI] [PubMed] [Google Scholar]
- 37.Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M. Entrainment of the circadian clock in the liver by feeding. Science. 2001;291:490–493. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- 38.Vitaterna MH, King DP, Chang AM, Kornhauser JM, Lowrey PL, McDonald JD, Dove WF, Pinto LH, Turek FW, Takahashi JS. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science. 1994;264:719–725. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zatz M. Light and norepinephrine similarly prevent damping of the melatonin rhythm in cultured chick pineal cells: regulation of coupling between the pacemaker and overt rhythms? J Biol Rhythms. 1991;6:137–147. doi: 10.1177/074873049100600204. [DOI] [PubMed] [Google Scholar]
- 40.Zhu H, Green CB. Three cryptochromes are rhythmically expressed in Xenopus laevis retinal photoreceptors. Mol Vis. 2001;7:210–215. [PubMed] [Google Scholar]
- 41.Zhu H, LaRue S, Whiteley A, Steeves TD, Takahashi JS, Green CB. The Xenopus Clock gene is constitutively expressed in retinal photoreceptors. Brain Res Mol Brain Res. 2000;75:303–308. doi: 10.1016/s0169-328x(99)00309-5. [DOI] [PubMed] [Google Scholar]