Abstract
Although evidence exists that activation of the Rho family GTPase Cdc42 affects axonal development, its specific roles within a growth cone are not well delineated. To evaluate the model that Cdc42 activation regulates growth cone navigation by promoting filopodial activity, we adopted a live analysis strategy that uses transgenicDrosophila lines in which neurons coexpressed constitutively active Cdc42 (Cdc42V12) and membrane-targeted green fluorescent protein. We found that growth cones that displayed pathfinding defects exhibited little change in their filopodial activity, whereas others without pathfinding defects exhibited an ∼50% increase in their filopodial activity. Moreover, effector loop mutations that were added to the constitutively active Cdc42 (Cdc42V12C40 and Cdc42V12A37) exerted little influence over filopodial activity caused by Cdc42 activation but suppressed the pathfinding defects of the growth cones. Together, these data suggest that Cdc42 controls filopodial activity in axonal growth cones independently of its effects on their pathfinding.
Keywords: axon guidance, Cdc42, Drosophila, filopodia, GFP, growth cone, in vivo, pathfinding
Neural network formation relies on the ability of axonal growth cones to navigate in response to localized cues in their microenvironments. During pathfinding, receptors for guidance cues activate signaling cascades in the growth cone that promote cytoskeletal remodeling and consequently alter growth cone morphology and migration. Biochemical and genetic studies have identified receptor-mediated regulation of the monomeric GTPases of the Rho subfamily, such as Rho, Rac, and Cdc42, as a key mechanism for regulating cytoskeletal dynamics (Luo et al., 1996; Jin and Strittmatter, 1997; Van Aelst and D'Souza-Schorey, 1997; Clark et al., 1998; Suter and Forscher, 1998; Ruchhoeft et al., 1999; Vastrik et al., 1999; Wahl et al., 2000). Such guidance receptors as the integrins, plexins, eph, and robo receptors have been shown to associate directly with either these GTPases or their regulators (Jin and Strittmatter, 1997; Clark et al., 1998; Vastrik et al., 1999; Wahl et al., 2000;Driessens et al., 2001; Hu et al., 2001; Wong et al., 2001).
Each Rho-family GTPase contains a membrane-attachment domain, a GTP/GDP binding domain, and an effector loop. These molecules cycle between the GTP-bound (active) and GDP-bound (inactive) states in response to guanine nucleotide exchange factors (GEFs) and GTPase activating proteins (GAPs) that are associated with developmentally and cell type-specific signal inputs. In their active state, these GTPases then bind characteristic sets of effector proteins, primarily via a short loop. These GTPases therefore both integrate multiple upstream signals and regulate multiple downstream pathways.
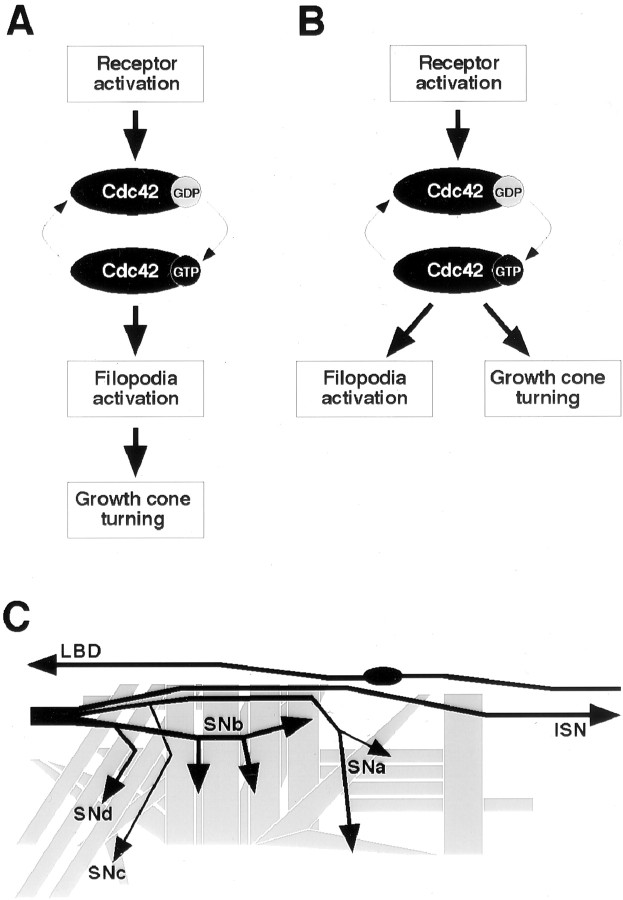
Studies using non-neuronal migratory cells demonstrate that activating Cdc42, Rac, or Rho each induce the assembly of characteristic F-actin-rich structures and that these GTPases may act sequentially to regulate cell motility and adhesion (Hall, 1998). Activation of Cdc42 in particular increases filopodial activity and promotes migration in fibroblasts. In neurons, in which axonal growth cones display filopodia and extend through complex environments, regulation of cytoskeletal dynamics is thought to be critical during axon pathfinding (Bentley and O'Connor, 1994; C. H. Lin et al., 1994). Previous studies on the Drosophila embryonic nervous system, in which Rac1 and Cdc42 were either inactivated or constitutively activated, provide evidence that their well regulated activities are essential for normal axon development (Luo et al., 1994; Sone et al., 1997; Kaufmann et al., 1998). These observations have been interpreted to support the idea that changes in actin polymerization caused by Cdc42 or other monomeric GTPases play an integral part in controlling the directionality of growth cone migration. This view is depicted as the “linear pathway” model (see Fig. 1A).
Fig. 1.
Models of Cdc42 activation. During its activation, Cdc42 switches from a GDP-bound inactive state to a GTP-bound active state (thin arrows). A, In the linear pathway model, the GTP-bound Cdc42 binds to downstream effectors, controlling filopodial activity and, as a direct consequence, leading to directed growth cone navigation. B, In the parallel pathways model, Cdc42 activation initiates several separate downstream events, with one being responsible for filopodial activation and the other regulating cytoskeletal and membrane dynamics during growth cone turning. Arrows do not necessarily indicate direct signaling but rather hierarchical events. C, Stereotyped motor and sensory axon pathways in the Drosophilaembryo. In each abdominal half-segment, the motoneuron axons are grouped into five nerves (ISN, SNa,SNb, SNc, andSNd). The LBD sensory neuron extends an afferent axon (LBD) via its own pathway near the segment border into the CNS. The CNS is to the left, and nine distal (dorsal) muscle cells are not shown.
Although there is little doubt about the importance of Cdc42, at least two unresolved issues surround the roles of Cdc42 during axon development. First, Cdc42 works through a number of potential effectors the subsequent signaling of which can influence events ranging from transcriptional to post-translational regulations (Aspenstrom, 1999;Boettner and Van Aelst, 1999). Among cytoplasmic events likely involved during minute-to-minute pathfinding decisions of a migrating growth cone, actin polymerization may be only one of several means by which effectors of Cdc42 can mediate growth cone behaviors. Second, whether regulation of filopodial activity by Cdc42 is important for growth cone turning has not yet been examined. There is in vitroevidence that growth cones exhibit an asymmetry in filopodial distribution before turning (Zheng et al., 1996). However, this does not seem to hold true in more complex cellular environments during development. The selective invasion and retention of microtubules in filopodia have been proposed as a major correlate to growth cone guidance in situ (Sabry et al., 1991; Myers and Bastiani, 1993; Isbister and O'Connor, 1999). Therefore, it is reasonable to propose an alternative “parallel pathways” model in which Cdc42 activation feeds into parallel downstream pathways where filopodial activity and growth cone pathfinding are not directly linked (see Fig.1B).
In this study, we examined the effects of expressing Cdc42 mutant proteins within live, developing neurons in real time. We analyzed filopodial activity and pathfinding in several types of growth cones expressing mutant Cdc42 proteins and discovered that they are not correlated. These results suggest that Cdc42 controls filopodial activity and pathfinding independently, presumably by activating distinct pathways. The significance of the results will be discussed in the context of cytoplasmic signaling that controls axonal behavior.
MATERIALS AND METHODS
Fly stocks. Neuronal expression of Dcdc42 and membrane-targeted green fluorescent protein (gapGFP) relied on the GAL4/UAS misexpression method (Brand et al., 1994). GAL4 expression was controlled by pan-neuronal GAL4 drivers,GAL4C155 [source: C. Goodman, University of California, Berkeley; D. M. Lin et al. (1994)] orelav'-GAL4 (source: L. Luo, Stanford University).UAS-Dcdc42V12 (a constitutively active Dcdc42) and UAS-Dcdc42N17 (a dominant negative Dcdc42) are as described (Luo et al., 1994).UAS-myc:: Dcdc42V12(myc-tagged Cdc42 with constitutive activation),UAS-myc:: Dcdc42V12A37, andUAS-myc:: Dcdc42V12C40(myc-tagged Cdc42 with both constitutive activation and an additional effector-loop mutation, F37A or Y40C) were newly constructed (see below). UAS-gapGFP was constructed in our lab previously (Ritzenthaler et al., 2000). CantonSstrain was used as a wild-type control.
Site-directed mutagenesis. Mutations in the effector loop coding region were introduced into a cDNA encodingDcdc42V12 tagged with two copies of themyc epitope (source: L. Luo) using QuikChange (Stratagene). After being confirmed by DNA sequencing, mutant Dcdc42 cDNAs were inserted into the pUAST transformation vector (Brand et al., 1994).
Immunocytochemistry. Immunocytochemistry was performed on embryos with monoclonal antibody (mAb) 1D4 [1:40 dilution; source: C. Goodman, University of California; Grenningloh et al. (1991)] and/or anti-myc mAb 1–9E10.2 [1:1000 dilution; source: Developmental Studies Hybridoma Bank, University of Iowa; Evan et al. (1985)] by following the method described (Wolf et al., 1998; Ritzenthaler et al., 2000).
Live filopodia visualization. For live filopodial analysis, we examined the easily visualized LBD sensory and SNb motor growth cones. GFP-expressing embryos were fillet-dissected live in insect saline (140 mm NaCl, 5 mm KCl, 5 mmCaCl2, 4 mmNaHCO3, 1 mmMgCl2, 5 mm TES, 5 mm trehalose, 100 mmsucrose, pH 7.2) and visualized with an EGFP filter set (Chroma Technology, Brattleboro, VT) and a fluorescent microscope (ZeissAxioskop) with an electronic shutter (UniBlitz, Vincent Associates, Rochester, NY) and a cooled CCD camera (C5985, Hamamatsu Photonics, Hamamatsu, Japan). Frames were captured every 15 sec, typically with 1.6–2.2 sec integration time per frame, for a 20–60 min duration and stored on computer using IPLab software (Scanalytics, Fairfax, VA). Individual filopodia were digitally traced and their lengths were measured in each frame using IPLab software. Statistical analysis was aided by Microsoft Excel.
RESULTS
Cdc42 activation affects axon pathfinding
Cdc42 controls the formation of filopodia (Hall, 1998), and filopodia are necessary during growth cone outgrowth (Bentley and Toroian-Raymond, 1986; Chien et al., 1993). However, whether filopodial contacts with substrate play an instructive role with respect to the direction of the growth cone or are merely permissive for migration is not clear (Fig. 1). We therefore examined the axonal defects and changes in filopodial activity resulting from introducing a constitutively active form of Cdc42 into embryonic neurons (see Material and Methods).
Under the control of a pan-neural enhancer,GAL4C155 or elav'-GAL4, constitutively active Cdc42 (Dcdc42V12) was expressed in neurons approximately at hour 8 (stage 11) through at least the completion (stage 17) of embryogenesis. Expression was confirmed by using myc-tagged Dcdc42 constructs. Immunostaining with myc antibody revealed that the proteins were ubiquitously expressed throughout the CNS (data not shown) and within the neurons that were examined (Fig.2A,B). The level of protein expression was higher under the control ofelav'-GAL4 than with GAL4C155(as confirmed by fluorescent intensities of the GFP reporter driven by these two GAL4 drivers; data not shown). The embryos were immunoprocessed with mAb 1D4 (anti-fasciclin II) and examined at hour 16–18 (stage 16), at which time the majority of axons in wild-type embryos should have completed their pathfinding. The antibody labels the axons of a subset of interneurons within the CNS, as well as those of motoneurons and specific sensory neurons in the peripheral nervous system. The wild-type pathways of these axons are stereotypic and well described, in many cases, to the level of individual cells (Jan and Jan, 1994; Keshishian et al., 1996; Landgraf et al., 1997). This allows assessment of the extent and variability of the defects seen among different axons as well as within each individually identified axon.
Fig. 2.
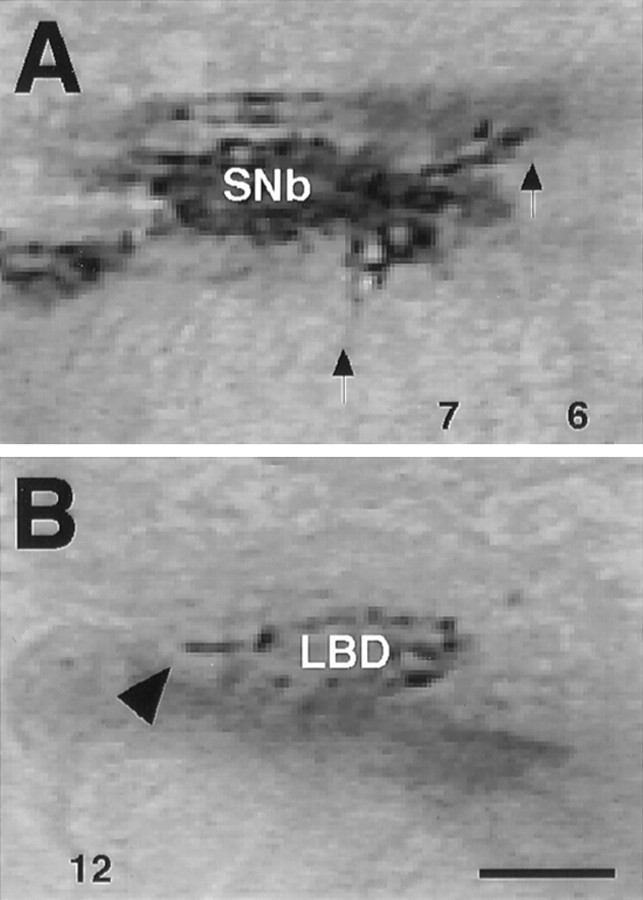
Expression of Cdc42 proteins. Embryos expressing Cdc42 proteins were examined with α-myc immunocytochemistry in hour 12 embryos. A, Localization of the protein can be seen throughout the SNb growth cones and within individual processes (arrows). B, In the LBD neuron, the protein is localized to the growing axon (arrowhead). Apparent discrepancies in expression level can be attributed to the protein localizing to the growth cone, which is much larger in the case of SNb. Scale bar, 5 μm.
The motoneuron axons are classified into groups based on their patterns of fasciculation on leaving the CNS and the relative positions of their target muscles (Fig. 1C). When the constitutively active Cdc42 was present in the neurons, motor axons exhibited navigation patterns ranging from virtually normal to clearly abnormal (Fig.3B–D,I–L). The frequency of abnormalities in each group was higher withelav'-GAL4 than with GAL4C155(Fig. 3I–L). These observations were generally consistent with varied thresholds for Cdc42-dependent signaling among individual neurons.
Fig. 3.
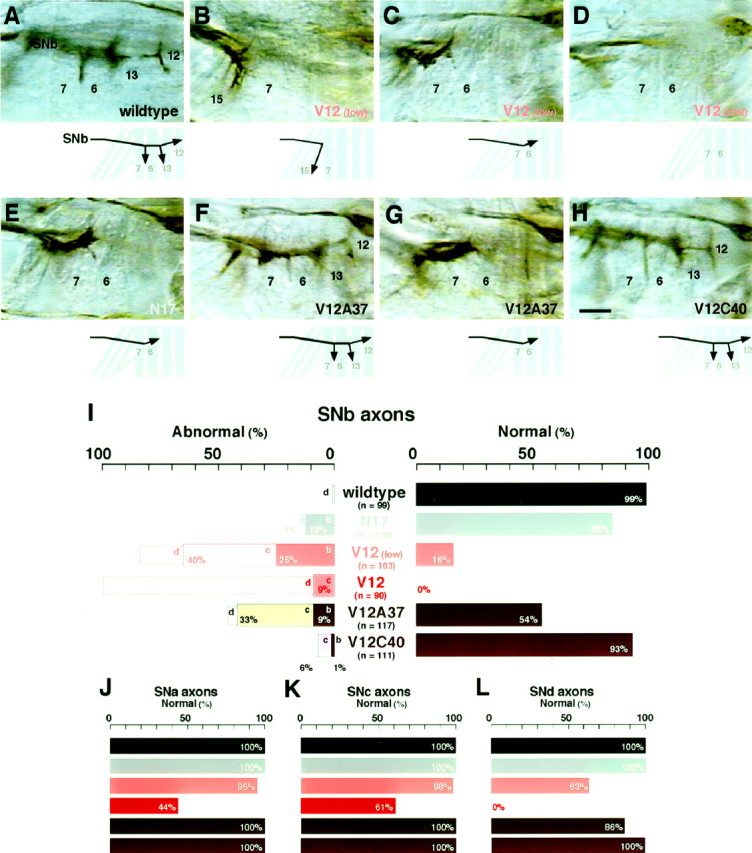
Pathfinding of motoneuron axons.A–H, SNb motoneuron axons were examined with mAb 1D4 immunocytochemistry in hour 16–18 embryos. In wild-type controls (wildtype; CantonS strain), SNb axons, after reaching the ventrolateral muscle cells 6, 7, 12, and 13, exhibit stereotypic growth cone turnings: a posteriorly directed turn between muscles 6 and 7, a bi-directional turn at the proximal edge of muscle 13 (out of focus), and a bi-directional “T-shape” turn at the proximal edge of muscle 12 (A). Although this immunovisualization method does not distinguish individual axons, previous dye-labeling studies have shown the five motoneuron axons that are responsible for innervating these muscles at this stage: RP3 growth cone turns between muscles 6 and 7, both RP1 and RP4 growth cones turn at the edge of muscle 13, and both RP5 and V growth cones terminate at the edge of muscle 12 (Landgraf et al., 1997). In contrast, SNb axons in embryos with pan-neuronal expression of constitutively active Cdc42 [V12 (low)] often displayed a range of pathfinding defects categorized as either turning posteriorly in unison at an incorrect site, e.g., muscle 15 (B), stalling at or before muscle 6 (C), or absence in the proximal (ventral) musculature (D). The absence of normal innervation of SNb occurred when the expression of Cdc42V12 was driven by the high-expression driver (I). Qualitatively similar defects of SNb axons resulted when a dominant negative Cdc42 was expressed in the neurons (N17) (E). With the expression of Dcdc42V12 that carries the additional F37A effector mutation (V12A37), SNb axons either exhibited the normal pathfinding pattern (F) or stalled at or before muscle 6 (G). More dramatic reversion of the SNb pathfinding was observed with the Y40C effector mutation (V12C40). Most of the cases examined showed the normal pathfinding (H). Scale bar, 10 μm. I–L, The frequencies of motor axon pathfinding defects are summarized for SNb, SNa, SNc, and SNd axons. For SNb axons (I), Normal pathfinding was defined as described in A, whereasAbnormal pathfindings were categorized as incorrect turning (b), stalling (c), or absence (d), as defined with more detail in a corresponding manner in B–D. The other groups of axons were scored only for the frequencies ofNormal pathfinding: the distally (ventrolaterally) extending axons forming a fascicle distinct from that of ISN axons, reaching the lateral oblique and transverse muscles, and forking into one distally directed and another posteriorly directed branch (SNa; J), the axons turning posteriorly in the proximal (ventral) region to reach superficial layers of muscles behind all other motor axons (SNc;K), and the axons extending for a short distance along with SNb axons before turning posteriorly to reach proximal (ventral) muscles with oblique orientations (SNd;L). The genotypes examined were wild-type control (wildtype), dominant negative Cdc42 [(N17); GAL4C155/+; UAS-Dcdc42N17/+)], constitutively active Cdc42 [V12(low); GAL4C155/+; UAS-Dcdc42V12 /+], constitutively active Cdc42 with the high-expression GAL4 driver (V12;elav'-GAL4/UAS-Dcdc42V12, elav'-GAL4/UAS-myc:: Dcdc42V12), Cdc42 with the F37A mutation (V12A37;UAS-myc:: Dcdc42V12A37/+; elav'-GAL4/+) and Cdc42 with the Y40C mutation (V12C40; UAS-myc:: Dcdc42V12C40 /+; elav'-GAL4/+). Sample sizes (n) are given for each case as the number of abdominal hemisegments examined. In addition to CantonS, all parental controls (GAL4C155, elav'-GAL4, UAS-Dcdc42V12, UAS-myc:: Dcdc42V12 /TM3, UAS-myc:: Dcdc42V12A37, UAS-myc:: Dcdc42V12C40) were also examined; except for SNb axons in GAL4C155 embryos (87% normal pathfinding; n = 100 half-segments), their scores were indistinguishable from CantonS (97–100% normal pathfinding for each group of axons; n = 84–104 half-segments).
SNb motoneuron axons
In wild type, on approaching the ventrolateral muscle cells, SNb motoneuron axons defasciculate from one another at stereotypic sites and turn either posteriorly or anteriorly before targeting specific muscle cells (Fig. 3A). The sites and directionality of growth cone turning for these axons are invariant (Fig.3I).
In contrast, SNb axons expressingGAL4C155-driven Dcdc42V12 displayed abnormal pathfinding patterns in 84% of the cases (Fig. 3I). Their phenotypes were categorized as follows: turning posteriorly in unison at an incorrect site, i.e., muscle 7, 15, or 16 (25%) (Fig.3B); stalling at or before muscle 6 (40%) (Fig.3C); or absence in the ventral musculature attributable presumably to either stalling within the CNS or bypassing the SNb pathway after leaving the CNS (19%) (Fig. 3D). While examining SNb axons through embryogenesis, we noted abnormal turning as early as hour 12 (stage 14), when these axons in wild type would be navigating around muscles 15 and 16 and beginning to contact the more distal muscles 6 and 7. When a high-expression elav'-GAL4driver was used, the most common phenotype of SNb axons shifted from abnormal turning to a complete absence in the ventral musculature, although there were still cases (9%) in which SNb axons displayed abnormal turns at muscle 15 or 16 (Fig. 3I). The fact that axon pathfinding defects occurred within the CNS (see below) complicated interpretations of the effects in this case. When the neurons expressed a dominant negative form of Cdc42 (Dcdc42N17), SNb axons occasionally (15%) displayed phenotypes similar to those observed with constitutively active Cdc42 (Fig. 3E,I). These observations suggest that out-of-context activation or deactivation of Cdc42 in axons raises their susceptibility to pathfinding errors.
LBD sensory neuron axons
mAb 1D4 also labels the LBD sensory neuron located near the segment border. Before reaching the lateral edge of the CNS by hour 16 (stage 16), its proximally directed axon navigates through the regions adjacent to where SNb axons navigate (Fig.4A).
Fig. 4.
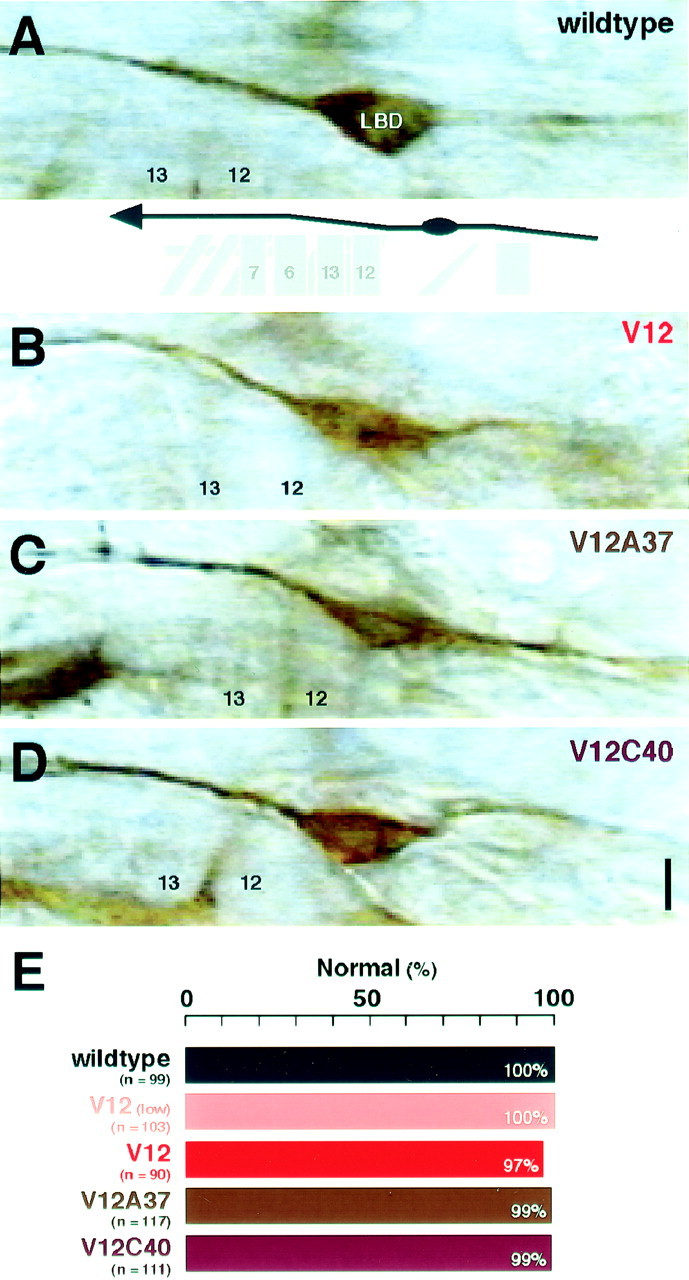
Pathfinding of LBD sensory neuron axons.A–D, LBD sensory neuron axons were examined with mAb 1D4 immunocytochemistry in hour 16–18 embryos. In wild-type controls (wildtype), the LBD axon navigates along a stereotypic pathway toward the CNS (A). The LBD axon exhibited normal pathfinding with the presence of Dcdc42V12 (V12; B), Dcdc42V12A37 (V12A37;C), or Dcdc42 (V12C40; D). Scale bar, 10 μm. E, The frequencies of pathfinding defects are summarized. Normal pathfinding was defined as described for the wild type in A. All parental controls were indistinguishable fromCantonS (100% normal pathfinding; n = 87–108 hemisegments).
With constitutively active Cdc42 expressed in the neurons, the LBD axonal trajectory remained intact (Fig. 4B). This was true with either the elav'-GAL4 or theGAL4C155 driver (Fig.4E). Expression of dominant negative Cdc42 also had little effect on LBD pathfinding (data not shown). Examination of earlier stages offered no evidence that LBD axons navigate at a speed different from wild-type controls (data not shown). The absence of any detectable navigation error cannot be attributed to lack of Dcdc42V12 expression in this neuron (as confirmed by visualizing the myc-tagged version of the transgene), and as we show below, the LBD axons did exhibit a phenotype aside from navigation errors. It should be noted that there are mutations (e.g., PS integrin mutants) that cause LBD axons to make erroneous turns (B. Hoang and A. Chiba, unpublished observations). The LBD axons represent an example of axons that show few pathfinding defects under conditions (constitutive activation of Cdc42) in which other neurons would show such phenotypes.
Cdc42 activation affects filopodial activity
Cdc42 activation induces filopodial activity (Hall, 1998), and filopodial dynamics are proposed to play a role in navigating axons (Bentley and Toroian-Raymond, 1986; Chien et al., 1993). Our primary goal was to evaluate the “linear pathway” model in which constitutive activation of Cdc42 leads to increased filopodial activity in a growth cone leading to erroneous steering behavior of a growth cone (Fig. 1A). We therefore monitored the filopodial behavior of SNb and LBD growth cones in situ by developing a new live analysis strategy (see Materials and Methods). We coexpressed Dcdc42V12 and a membrane-targeted GFP (gapGFP) in neurons and fillet-dissected the transgenic embryos at hour 12–14 (stage 15), when the axons were still engaged in pathfinding. The expression of gapGFP and myc-tagged Dcdc42V12 (visualized with anti-GFP and anti-myc immunocytochemistry) paralleled in relative strengths among individual neurons (data not shown). Phototoxicity of GFP was negligible, particularly when compared with fluorescent cell labeling by DiI application (data not shown), permitting continual monitoring of individual growth cones with a relatively high temporal resolution. This facilitated the live behavioral analysis of growth cones and their filopodia in situ.
LBD sensory neuron axons
GFP labeling revealed morphological dynamics of an individual LBD growth cone (Fig.5A,B). At the time of visualization, LBD neurons exhibited a very critical window of activity before extending its axon forward. The location at which this occurred is typically the area in which LBD axons make incorrect turns in certain mutants. However, Cdc42 activation did not cause inappropriate turning.
Fig. 5.
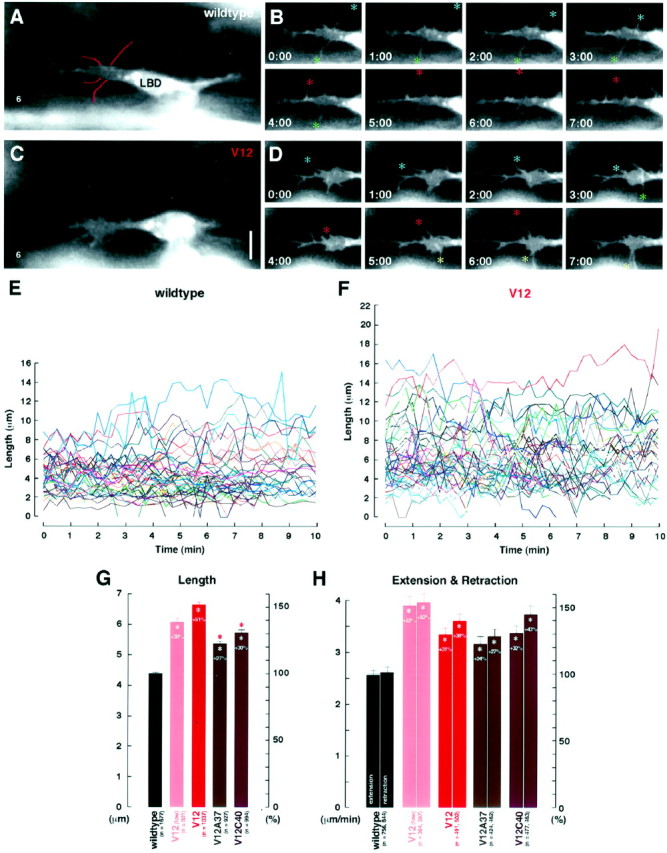
Time-lapse analysis of LBD growth cones.A–D, Time-lapse video images of GFP-labeled neurons show dynamic filopodial activity in LBD growth cones in an hour 12 wild-type embryo (wildtype;A, B) and an hour 12 embryo expressing constitutively active Cdc42 (V12; C,D). B and D each show a 7 min clip from a movie of A and C, respectively. Colored asterisks in B andD follow some individual filopodia through their extensions and retractions. The filopodia were digitally traced (A, red lines) in every frame and measured (see Materials and Methods). Scale bar, 5 μm.E, F, Line graphsdepicting the changes in lengths of individual filopodia in LBD growth cones over the course of 10 min in hour 12 wild-type embryos (wildtype; E) and hour 12 embryos expressing constitutively active Cdc42 (V12;F). G, Filopodial lengths (mean ± SEM) in various genotypes: wildtype control (wildtype; GAL4C155/+; UAS-gapGFP/+), constitutively active Cdc42 [V12(low); GAL4C155/+; UAS-gapGFP/+; UAS-Dcdc42V12/+], constitutively active Cdc42 with the high-expression driver (V12; UAS-gapGFP/+; elav'-GAL4/UAS-Dcdc42V12), Cdc42 with the F37A mutation (V12A37; UAS-myc:: Dcdc42V12A37 /+; UAS-gapGFP/+;elav'-GAL4/+), and Cdc42 with the Y40C mutation (V12C40; UAS-myc:: Dcdc42V12C40 /UAS-gapGFP;elav'-GAL4/+). Sample sizes (n) were those of the filopodia measured in all frames available from the movies (5–10 movies per genotype). H, Filopodial extension and retraction rates in various genotypes.White asterisks in G and Hindicate statistically significant deviation from the wild-type controls, and percentage changes (%) from wild-type levels (gray lines) are indicated; pink asterisksindicate significant deviations from embryos expressing Cdc42V12 (Table 1).
In wild-type controls, LBD growth cones had filopodia that, over the course of their existence (≤600 sec), extended up to 15.1 μm and maintained an average length of 4.40 ± 0.06 μm (Fig.5E,G, Table1). Their rates of extension and retraction, the parameters more informative of actin polymerization controls, were 2.63 ± 0.10 and −2.69 ± 0.10 μm/min, respectively (Fig. 5H, Table 1). It was noteworthy that variants for these parameters were very small among a total of 62 individual filopodia analyzed in 14 embryos.
Table 1.
Filopodial activity in LBD growth cones1-a
| Genotype | Wild type1-b | V12 (low) | V12 | V12A37 | V12C40 |
|---|---|---|---|---|---|
| Length (μm)1-c | |||||
| Mean | 4.40 | 6.09 | 6.66 | 5.38 | 5.74 |
| SEM | 0.06 | 0.13 | 0.09 | 0.07 | 0.09 |
| Maximum | 15.1 | 19.8 | 17.2 | 12.9 | 16.6 |
| n | 1677 | 801 | 1037 | 927 | 994 |
| p versus wild type | <0.01 | <0.01 | <0.01 | <0.01 | |
| pversus V12 | <0.01 | <0.01 | <0.01 | <0.01 | |
| Extension (μm/min)1-d | |||||
| Mean | 2.63 | 4.01 | 3.45 | 3.26 | 3.47 |
| SEM | 0.10 | 0.19 | 0.13 | 0.15 | 0.15 |
| Maximum | 23.0 | 21.3 | 19.4 | 19.9 | 31.8 |
| n | 756 | 364 | 491 | 424 | 477 |
| p versus wild type | <0.01 | <0.01 | <0.01 | <0.01 | |
| pversus V12 | <0.01 | 0.02 | 0.36 | 0.92 | |
| Retraction (μm/min)1-d | |||||
| Mean | −2.69 | −4.08 | −3.71 | −3.41 | −3.84 |
| SEM | 0.10 | 0.18 | 0.14 | 0.14 | 0.17 |
| Maximum | −36.0 | −22.8 | −18.6 | −19.5 | −24.0 |
| n | 844 | 397 | 502 | 462 | 463 |
| p versus wild type | <0.01 | <0.01 | <0.01 | <0.01 | |
| pversus V12 | <0.01 | 0.07 | 0.11 | 0.59 |
Wild type, GAL4C155/+; UAS-gapGFP/+, UAS-gapGFP/+; elav′-GAL4/+; V12 (low), GAL4C155/+; UAS-gapGFP/+; UAS-Dcdc42V12/+; V12, UAS-gapGFP/+; elav′-GAL4/UAS-Dcdc42V12; V12A37, UAS-Dcdc42V12A37/+; UAS-gapGFP/+; elav′-GAL4/+; V12C40, UAS-Dcdc42V12C40/UAS-gapGFP; elav′-GAL4/+.
Hour 12–14 embryos coexpressing the membrane-targeted GFP (gapGFP) and one of the mutant Cdc42 proteins (Dcdc42V12, Dcdc42V12A37, or Dcdc42V12C40) were fillet-dissected, and filopodial activity of LBD growth cones was monitored in situ with time-lapse video microscopy (30–35 filopodia in 5–10 movies per genotype).
Two parental controls produced similar results (p > 0.4 with two-tailedt test) and were combined as wild type.
For each genotype, the mean filopodial length (mean), the SEM values, the maximum length observed (Maximum), the total number of filopodia measurements (n), as well as the results of the two-tailed t tests against the wild-type control (p versus wild type) and the constitutively active Cdc42 (p versus V12) are given. The minimum length for a valid filopodium was 1.0 μm.
The rates were calculated on the basis of the difference between each pair of consecutive frames that had, respectively, a positive (Extension) and negative (Retraction) value.
An enhancement of filopodial activity in LBD growth cones expressing constitutively active Cdc42 was immediately apparent from viewing time-lapse videos (Fig.5C,D,F). When theDcdc42V12 transgene was under the control of GAL4C155, filopodial extension and retraction rates both increased by 52%, and the filopodia length increased by 38% (Fig. 5G,H). Similarly, with elav'-GAL4, a stronger driver, there was a 51% increase in the filopodia length and 31% and 38% increases, respectively, in the extension and retraction rates (Fig.5G,H). In both cases, each parameter measured presented a highly significant increase over wild type (Table1).
SNb motoneuron axons
In wild-type controls at hour 12–14 (stage 15), SNb growth cones spread out their filopodia in the region near muscles 6 and 7 (Fig.6A,B). They extended and retracted filopodia at rates of 3.59 ± 0.10 and −3.54 ± 0.10 μm/min, respectively, while maintaining an average length of 6.44 ± 0.06 μm (Fig.6E,F, Table2). SNb filopodia are therefore, on average, longer and more dynamic than those of LBD growth cones at the same developmental stage.
Fig. 6.
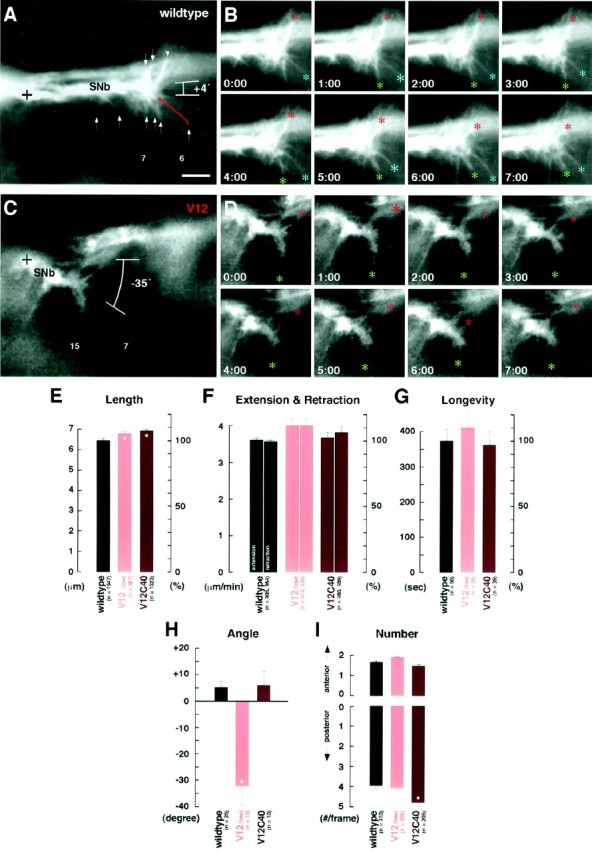
Time-lapse analysis of SNb growth cones.A–D, Time-lapse video images of GFP-labeled neurons show dynamic filopodial activity in SNb growth cones in an hour 12 wild-type embryo (wildtype;A, B) and an hour 12 embryo expressing constitutively active Cdc42 (V12; C,D). Scale bar, 5 μm. E, Filopodial lengths in various genotypes (see Fig. 4 for genotypes; 4–8 movies per genotype). F, Filopodial extension and retraction rates in various genotypes. G, Longevities of individual filopodia were determined by following them throughout the duration of the movie. H, Angles of growth cone extension were determined by taking the baseline at a right angle to the anterior–posterior axis of the ventral longitudinal muscles (muscles 6, 7, 12, and 13) and measuring the angle of the line that connects the ventralmost point of the SNb nerve (A, C,crosses). I, The number of filopodia that extended to either the anterior (A, down arrows) or posterior (A, up arrows) and were longer than 2.5 μm were counted.Asterisks in E, H, andI indicate statistically significant deviation from wild type (see Table 2).
Table 2.
Filopodial activity in SNb growth cones2-a
| Genotype2-b | Wild type2-c | V12 (low) | V12C40 |
|---|---|---|---|
| Length (μm)2-d | |||
| Mean | 6.44 | 6.79 | 6.92 |
| SEM | 0.06 | 0.08 | 0.09 |
| Maximum | 15.6 | 14.3 | 18.5 |
| n | 1947 | 887 | 1023 |
| p versus wild type | <0.01 | <0.01 | |
| Extension (μm/min)2-d | |||
| Mean | 3.59 | 3.98 | 3.64 |
| SEM | 0.10 | 0.17 | 0.15 |
| Maximum | 17.9 | 19.9 | 22.6 |
| n | 905 | 414 | 480 |
| p versus wild type | 0.13 | 0.78 | |
| Retraction (μm/min)2-d | |||
| Mean | −3.54 | −3.97 | −3.79 |
| SEM | 0.10 | 0.16 | 0.14 |
| Maximum | −20.0 | −21.4 | −21.8 |
| n | 964 | 434 | 489 |
| p versus wild type | 0.02 | 0.14 | |
| Longevity (sec)2-e | |||
| Mean | 363 | 395 | 351 |
| SEM | 33 | 42 | 35 |
| n | 50 | 25 | 39 |
| p versus wild type | 0.56 | 0.81 | |
| Angle (°)2-f | |||
| Mean | +4.7 | −32.8 | +5.0 |
| SEM | 1.9 | 6.1 | 4.7 |
| Maximum | +29 | +10 | +20 |
| Minimum | −13 | −57 | −21 |
| n | 25 | 10 | 10 |
| p versus wild type | <0.01 | 0.93 | |
| Number (#)2-g | |||
| Mean (anterior) | 1.67 | 1.93 | 1.47 |
| SEM (anterior) | 0.10 | 0.06 | 0.13 |
| p versus wild type | 0.07 | 0.23 | |
| Mean (posterior) | 3.97 | 4.08 | 4.75 |
| SEM (posterior) | 0.09 | 0.08 | 0.13 |
| p versus wild type | 0.43 | <0.01 | |
| n | 410 | 205 | 205 |
Hour 12–14 embryos coexpressing gapGFP and one of the mutant Cdc42s were dissected, and filopodial activity of SNb growth cones was monitored (32–39 filopodia in four to eight movies per genotype).
See Table 1 for genotypes.
Two parental controls produced similar results (p > 0.5) and were combined together as wild type.
See Table 1 for measurement methods.
Based on monitoring individual filopodia through consecutive frames.
See Figure 6 for measurement methods.
The total number of filopodia (length ≥ 2.5 μm). Anterior and posterior directions are as defined in Figure 6.
In embryos expressing constitutively active Cdc42 under the control ofGAL4C155, SNb growth cones at hour 12–14 were found to consistently deviate from the wild-type pathway, directing their growth cones toward muscles 15 and 16 (Fig.6C,D). In 8 of 10 time-lapse movies (80%), the SNb axons were directed posteriorly at angles greater than −20° [(Fig. 6C) mean = −32.8 ± 6.1°; range = −57° to 10° (Table 2)]. This is in contrast to wild type, in which the SNb axons maintained an angle of 4.7 ± 1.9° (Fig.6A), and no case (n = 25) had angles greater than −20° (Fig. 6H). Despite this, filopodial extension and retraction rates were similar to those of wild-type controls, and the length of filopodia increased by a mere 5% (Fig. 6E,F, Table 2). Furthermore, with or without constitutively active Cdc42, there was no significant difference in either the number or longevity of individual filopodia directed posteriorly versus anteriorly (Fig.6G,I, Table 2).
The lack of a clear correlation between increased filopodial activity and pathfinding defects, both of which resulted from expressing constitutively active Cdc42 in neurons, suggested that these two aspects of growth cone behavior may be governed separately, as in the linear pathway model (Fig. 1B).
Cdc42 effector mutants can suppress axon pathfinding defects associated with Cdc42 activation
In the parallel pathways model, Cdc42 activation would lead to two separable downstream pathways, one being primarily responsible for filopodial activity enhancement and the other for growth cone steering (Fig. 1B). In vertebrate systems, mutations in the effector loop have revealed that different Cdc42 effector proteins interact with Cdc42 at distinct sites within the loop (Lamarche et al., 1996). Replacement of tyrosine with cysteine at amino acid 40 (Y40C) of activated Cdc42 disrupts association with Cdc42/Rac interactive binding (CRIB)-domain effector proteins, which in neurons include the serine-threonine kinase PAK and the tyrosine kinase ACK (Lamarche et al., 1996; Owen et al., 2000). A phenylalanine-to-alanine substitution at amino acid 37 (F37A) inhibits binding of activated Cdc42 to ACK and weakens its association with PAK (Owen et al., 2000). Interaction with the adapter protein N-WASP, a specific effector of activated Cdc42 often proposed to control the actin cytoskeleton in neurons, is independent of the Y40C mutation (Miki et al., 1998). Drosophila Cdc42 shares both F37 and Y40 with human Cdc42 (Luo et al., 1994). Furthermore,Drosophila PAK, ACK, and WASP, which share high degrees of structural homologies with their respective vertebrate counterparts, have been identified, although their in vivo association and cooperation with Dcdc42 are yet to be demonstrated (Harden et al., 1996; Hing et al., 1999; Morrison et al., 2000; Ben-Yaacov et al., 2001).
We set out to evaluate the parallel pathways model by adopting the effector loop mutation strategy (see Materials and Methods). We took the Drosophila Cdc42 gene and added either the Y40C or F37A mutation to the constitutively active G12V mutation. Using the resultant Dcdc42V12C40 andDcdc42V12A37 transgenes, we examined whether these effector loop mutations could suppress the axon pathfinding defects that were associated with Cdc42 activation and whether they could achieve this independent of filopodial activity changes, as predicted by the model (Fig. 1B).
SNb motoneuron axons
SNb axons exhibited abnormal pathfinding patterns in 100% of the cases when the neurons expressed constitutively active Cdc42 (Fig.3I). When the same elav'-GAL4 drove expression of the effector mutant Dcdc42V12C40 instead, navigation patterns of SNb axons became indistinguishable from wild type in 93% of the cases (Fig. 3H,I). Motoneuron axons of the other groups appeared wild type as well (Fig.3J–L). Dcdc42V12C40expression in the neurons was confirmed by detecting the mycepitope that had been added to the construct (Fig.2A). The fact that expression at this level caused very severe SNb pathfinding defects when theDcdc42V12 gene was expressed (Fig.3I) further strengthens the argument that the Y40C mutation is an effective blocker of such axon pathfinding defects. Expressing the other effector mutant Dcdc42V12A37 resulted in a partial reversion of SNb pathfinding to a 54% wild-type level (Fig.3F,G,I). Therefore, the addition of the Y40C mutation, and to a lesser extent the F37A mutation, suppressed the axon pathfinding defects caused by constitutive activation of Cdc42.
CNS interneuron axons
Axon fascicles within the longitudinal connectives of the CNS exhibited abnormal patterns when the neurons expressed constitutively active Cdc42 (Fig. 7B). The severity of the defects, low with GAL4C155(42% abnormal) and high with elav'-GAL4 (85% abnormal) (Fig. 7E), was correlated with the expression level of Dcdc42V12 under each driver. The type of defects observed (e.g., fasciculation errors, incorrect midline crossing) suggest that Cdc42 might be playing a role in adhesion as well as in navigation. Nearly all defects were suppressed when the same driver was used to express Dcdc42V12C40(Fig. 7D,E). In a parallel experiment with Dcdc42V12A37, CNS fascicles appeared wild type in 71% of the cases (Fig.7C,E). These data further supported the notion that the axon pathfinding defects resulting from constitutive activation of Cdc42 could be suppressed by the Y40C mutation in Dcdc42V12, and also that the F37A mutation could achieve similar results to a lesser extent.
Fig. 7.
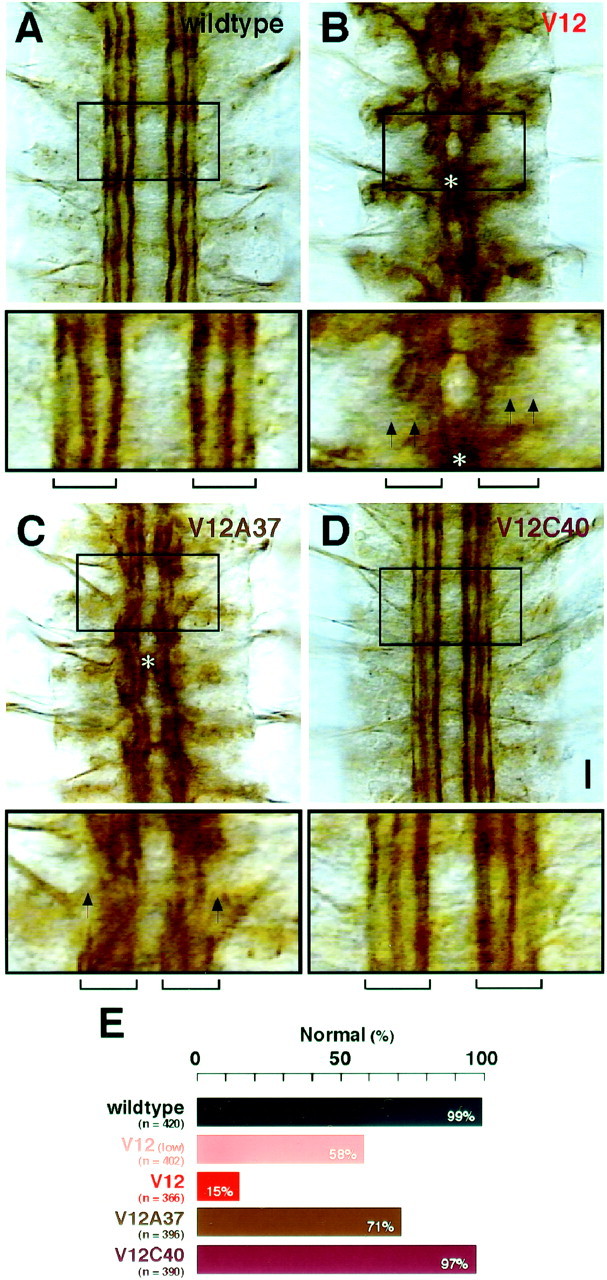
Pathfinding of CNS interneuron axons. A–D, CNS interneuron axon fascicles were examined with mAb 1D4 immunocytochemistry in hour 16–18 embryos. For each panel, the area indicated by a box is shown at the bottom. In wild-type controls (wildtype), three distinct axon fascicles (indicated with the bracket at the bottom) were clearly visible on each side of the CNS midline in each segment (A). However, with Dcdc42V12expression (V12; B), these fascicles were disrupted (arrows), suggesting either fused axon fascicles or stalled axons. In some cases, the two sides of the CNS exhibited fusion of the fascicles at the midline (asterisk). With the Dcdc42V12A37expression (V12A37; C), similar axon defects still occurred (arrows andasterisk) but at lower frequencies (E). Virtually normal CNS axon fascicles formed when the transgene Dcdc42V12C40(V12C40) was expressed (D). Scale bar, 10 μm. E, The frequencies of CNS pathfinding defects. Normal pathfinding in these fascicles was defined as described for wild type in A. See Figure 3for genotypes. All parental controls were indistinguishable fromCanton S (98–100% normal pathfinding;n = 378–420 fascicles in 123–140 hemisegments).
Cdc42 effector mutants do not affect filopodial activity
The parallel pathways model predicts that the Cdc42 effector mutations that suppress pathfinding defects will have little effect on filopodial activity (Fig. 1B). Therefore, we monitored the filopodial activity of LBD and SNb growth cones by coexpressing gapGFP along with either Dcdc42V12C40 or Dcdc42V12A37 in hour 12–14 (stage 15) embryos.
Constitutive activation of Cdc42 enhanced filopodial activity of LBD growth cones (Fig.5C,D,F). The rates of extension and retraction increased 31 and 38%, compared with wild type (Fig. 5H). Expressing the Y40C effector mutant (Dcdc42V12C40) under the control of the same driver (elav'-GAL4) gave rates of 3.47 ± 0.15 and −3.84 ± 0.17 μm/min, respectively, still remaining 32 and 43% above wild-type level (Fig. 5H). With Dcdc42V12A37, these rates stayed at 24 and 27% above wild-type level, respectively (Fig. 5H, Table 1). Neither the differences in the results between the two effector mutants nor those between each effector and the constitutively active Cdc42 were significant (Table 1). In all cases, we noted no axon pathfinding errors by LBD growth cones (Fig. 4C–E).
On the other hand, SNb growth cones expressing Dcdc42V12C40 reverted to the wild-type pathfinding (Fig. 3H,I) but showed no significant change in filopodial activity from wild type (Fig. 6E–H). The only exception was a 20% increase in the number of filopodia directed posteriorly, the significance of which was unknown (Fig. 6I, Table2). Overall, these observations supported the conclusion that this effector mutant of Cdc42 (Dcdc42V12C40) does not affect the downstream pathways that enhance filopodial activity.
DISCUSSION
Cdc42 regulates growth cone pathfinding and filopodial dynamics through separable downstream pathways
In this study, we used an in vivo analysis approach to examine whether the role of the monomeric GTPase Cdc42 in regulating filopodial activity is related to its role in growth cone steering. The activation of Cdc42 is thought to link membrane receptor activity to cytoskeletal dynamics within a navigating axon. The working model that we set out to test was the linear pathway model in which activation of Cdc42 leads to its association with effectors that enhance filopodial activity and that this is directly responsible for coordinating growth cone navigation (Fig. 1A).
Our analysis revealed that activation of Cdc42 can hyperactivate filopodia. However, we also found that this does not necessarily lead to erroneous navigation of a growth cone. The difference in phenotypic effect of constitutively active Cdc42 on these two aspects of growth cone behavior was first noticed during detailed analysis of two sets of growth cones navigating within the embryonic peripheral nervous system. Expression of constitutively active Cdc42 (Dcdc42V12) in LBD growth cones, despite causing a significant enhancement of filopodial activity, did not result in aberrant pathfinding (Fig.8A). Conversely, SNb growth cones expressing the same Dcdc42V12displayed little change in filopodial behavior, but did exhibit severe pathfinding defects (Fig. 8B). SNb growth cones did not start out with any obvious delay in migration but, as they developed, frequently exhibited random stalls or turning at incorrect sites. In the subsequent analysis, we used the effector loop mutation strategy adopted from fibroblasts studies (Joneson et al., 1996;Lamarche et al., 1996). There we demonstrated that upregulated filopodial activity and recurrent navigation errors were separable within an individual growth cone. Although navigation errors of the SNb growth cone were suppressed by the Y40C mutation, the same mutation exerted little effect on the filopodial activity of either the SNb or the LBD growth cone. These data support the alternative parallel pathways model in which Cdc42 activation feeds into separable downstream pathways, one that promotes filopodial activity and another that affects growth cone navigation (Fig. 1B).
Fig. 8.
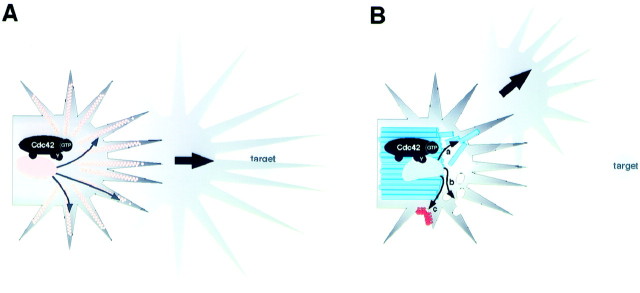
Roles of Cdc42 in a growth cone. A, Cdc42 activation in some growth cones (LBD growth cones) enhances filopodia activity. An effector(s) (oval shape) binds to the GTP-bound Cdc42 and stimulates (arrows inside the growth cone) actin polymerization (peach color) in filopodia. Despite increased length, extension, and retraction rates of filopodia, the growth cone still responds to cues from atarget and navigates normally (Figs. 4, 5).B, In other growth cones (SNb growth cones), activated Cdc42 associates with alternative effector(s) (mushroom-head shape). This association involves a Cdc42 microdomain distinct from that involved in binding to the filopodia-controlling effector(s) (compare with A). The subsequent activation of the alternative effector does not alter the filopodial activity but instead changes microtubule (blue) (a), membrane (b), and cell surface adhesion dynamics in the growth cone (see Discussion for other possibilities). With such an out-of-context alteration of cytoplasmic dynamics, the growth cone turns incorrectly at a random site without an increase in filopodial activity (Figs. 3, 6).
This parallel pathways model does not dismiss the importance of filopodia in growth cone migration. Rather, it separates the Cdc42 signaling cascades that are responsible for the regulation of filopodial behavior from those regulating other subcellular events that contribute to growth cone pathfinding in vivo. Oftentimes, models conceived to explain growth cone guidance end with the Rho subfamily of monomeric GTPases as mediators of local actin reorganization and, ultimately, of growth cone extension and turning. Such models have a tendency to oversimplify the mechanics involved in growth cone pathfinding and underestimate the diverse downstream pathways of Cdc42. The data from this study support the complexity of Cdc42 signaling cascades within a navigating growth cone and, further, implicate cytoplasmic events separate from filopodial activity as key factors in navigational control.
The effector loop mutation strategy
The directionality of axon navigation depends on the spatiotemporal regulation of membrane receptor activation. If Cdc42, a ubiquitous on–off switch, were to play an instructive role in pathfinding, restricting the time and place of its activation would be crucial. Artificially permitting universal out-of-context activation and inactivation, on the other hand, could randomize events that are normally controlled by Cdc42 activities. Consistent with reports elsewhere (Luo et al., 1994; Kaufmann et al., 1998), both the constitutively active Dcdc42V12 and the dominant negative Dcdc42N17 produced pathfinding defects. Although the frequencies of each phenotype differed, growth cones in both cases displayed defects, ranging from stalling to incorrect turning, that were qualitatively similar. This discrepancy of getting similar results from activation and deactivation of the particular signaling molecule can be explained by the fact that both Cdc42 mutants cause growth cones to ignore input signals that would be site and time specific under normal conditions.
Like other GTPases of the Rho subfamily, Cdc42 binds downstream effector proteins via a short evolutionarily conserved loop (amino acids 32–40) that becomes accessible in its active state. Constitutively active Cdc42 that also bears mutations in the effector loop activates a subset of Cdc42-dependent pathways. The Y40C mutation in the effector loop disrupts interactions with effectors that contain the CRIB domain; the F37A mutation primarily affects interactions with other effector types (Lamarche et al., 1996; Owen et al., 2000). Neither mutation affects filopodia formation in fibroblasts (Lamarche et al., 1996).
In Drosophila, within the CNS as well as in the peripheral nervous system, the addition of a Y40C mutation suppressed the growth cone pathfinding defects that were associated with constitutive activation of Dcdc42. The results suggested that the effector proteins whose association with Dcdc42V12 was blocked by the Y40C mutation were responsible for causing the pathfinding defects observed in neurons expressing Dcdc42V12. The wild-type-like phenotype with Dcdc42V12C40 expression presumably reflected the endogenous (wild-type) Dcdc42 working normally with these effectors. Introduction of an F37A mutation also suppressed the pathfinding defects, although to a lesser extent. Our current lack of knowledge concerning the identity of Cdc42 effectors inDrosophila that are susceptible to these mutations prevents us from suggesting a specific explanation of the observed differences in the degrees of phenotypic reversions. Although both effector loop mutants had dramatic effects on growth cone pathfinding, albeit to different degrees, neither Dcdc42V12C40nor Dcdc42V12A37 significantly altered filopodial behavior when compared with situations using Dcdc42V12. The ability of these Cdc42 effector loop mutations to affect pathfinding without altering filopodial control supports the idea that the subcellular events independent of those regulating filopodia are important in controlling axon pathfinding.
In situ behavioral analysis of a growth cone
In vitro studies with cultured neurons indicate that the default navigation pattern for a growth cone is extension in a straight line (Zheng et al., 1996). In a more complex in vivo environment, extracellular cues may activate spatially restricted signaling and cytoskeletal arrangements in the growth cone, resulting in the growth cone turning toward or away from the source of cues. Actin polymerization in filopodia is a dynamic process commonly speculated to be the main target of regulation during growth cone turning (O'Connor and Bentley, 1993; C. H. Lin et al., 1994). A growth cone that has lost its filopodia has difficulty following substrates, supporting the essential requirement of at least some filopodial presence (Gomez and Letourneau, 1994; Zheng et al., 1996). However, the idea that filopodia have roles beyond providing basic motility and signaling capability to the growth cone has not been substantiated in vivo (Bentley and Toroian-Raymond, 1986;Chien et al., 1993). Our study showed an example in which, despite out-of-context upregulation of filopodial activity, growth cones (LBD growth cones) still maintain the normal course of navigation. In another example, growth cones (SNb growth cones) that exhibit a distribution bias, with or without constitutively active Cdc42, make an erroneous turn toward the bias only in the presence of constitutively active Cdc42. These data did not provide support for the instructive role of filopodial activity.
Microtubules have also been shown to be assembled and transported dynamically within a growth cone (Challacombe et al., 1997; Dent et al., 1999; Kabir et al., 2001). Their regulation may dictate the directionality of growth cone navigation (Fig. 8B,a). Also, the plasma membrane may be the subject of active regulation during growth cone turning, with membrane insertion and removal (endocytosis) as opposing means of locally controlling membrane volume (Fig. 8B, b). Furthermore, local control of membrane adhesion can dynamically influence the cytomorphology of a migrating growth cone (Suter and Forscher, 1998) (Fig. 8B, c). Determining which of these or any other subcellular events are primarily engaged when a growth cone makes a turn is a topic of interest.
Live visualization adopted in this study enabled the characterization of dynamic filopodial behavior in growth cones that expressed mutant Cdc42 and membrane-targeted GFP. The practical advantage of GFP was its negligible phototoxicity under the illumination necessary for studies in living cells such as this one. Among the parameters measured were the average longevity of individual filopodia, their correlation with the direction of extension, and the rates of filopodial extension and retraction (Figs. 5, 6). They provided hints for local regulation of actin polymerization that would have been difficult without live analysis. Although not used in the current study, various GFP-based labeling tools are becoming available that will allow live monitoring of different intracellular components within neurons, such as actin filaments (Verkhusha et al., 1999), microtubules (Grieder et al., 2000), endosomes (CLONTECH, 2000), cytosolic calcium concentration (Miyawaki et al., 1997), and GTPases in their activated states (Kraynov et al., 2000; Mochizuki et al., 2001). In future research on the cytoplasmic signaling pathways involved in growth cone pathfinding, experiments that use in situ behavioral analysis on identified growth cones, similar to or perhaps more advanced than the approach taken in this study, are anticipated to prove indispensable.
Footnotes
This work was supported by National Institutes of Health (NIH) Grants GM07283 (M.K.) and NS40954 (P.K.), NIH/National Institute of Neurological Disorders and Stroke Grants NS35049 (A.C.) and NS40420 (A.C.), and National Science Foundation Grant IBN9986067 (A.C.). We thank Xiaomao Zhu (University of Illinois) for help with generating transgenic fly lines, Liqun Luo (Stanford University), Emiko Suzuki (University of Tokyo), and Chihiro Hama (RIKEN Center for Developmental Biology, Japan) for generous gifts of reagents, Linda Van Aelst (Cold Spring Harbor Laboratory) for advice on effector loop mutant strategies, and Huey Hing (University of Illinois) and the current members of the Chiba laboratory for comments on this manuscript.
Correspondence should be addressed to Akira Chiba, B605 CLS Laboratory, 601 South Goodwin Avenue, Urbana, IL 61801. E-mail:a-chiba@uiuc.edu.
REFERENCES
- 1.Aspenstrom P. Effectors for the Rho GTPases. Curr Opin Cell Biol. 1999;11:95–102. doi: 10.1016/s0955-0674(99)80011-8. [DOI] [PubMed] [Google Scholar]
- 2.Ben-Yaacov S, Le Borgne R, Abramson I, Schweisguth F, Schejter ED. Wasp, the Drosophila Wiskott-Aldrich Syndrome gene homologue, is required for cell fate decisions mediated by notch signaling. J Cell Biol. 2001;152:1–13. doi: 10.1083/jcb.152.1.1-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bentley D, O'Connor TP. Cytoskeletal events in growth cone steering. Curr Opin Neurobiol. 1994;4:43–48. doi: 10.1016/0959-4388(94)90030-2. [DOI] [PubMed] [Google Scholar]
- 4.Bentley D, Toroian-Raymond A. Disoriented pathfinding by pioneer neuron growth cones deprived of filopodia by cytochalasin treatment. Nature. 1986;323:712–715. doi: 10.1038/323712a0. [DOI] [PubMed] [Google Scholar]
- 5.Boettner B, Van Aelst L. Rac and Cdc42 effectors. Prog Mol Subcell Biol. 1999;22:135–158. doi: 10.1007/978-3-642-58591-3_7. [DOI] [PubMed] [Google Scholar]
- 6.Brand A, Manoukian AS, Perrimon N. Ectopic expression in Drosophila. In: Goldstein LSB, Fyrberg EA, editors. Methods in cell biology. Academic; San Diego: 1994. pp. 683–696. [DOI] [PubMed] [Google Scholar]
- 7.Challacombe JF, Snow DM, Letourneau PC. Dynamic microtubule ends are required for growth cone turning to avoid an inhibitory guidance cue. J Neurosci. 1997;17:3085–3095. doi: 10.1523/JNEUROSCI.17-09-03085.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chien CB, Rosenthal DE, Harris WA, Holt CE. Navigational errors made by growth cones without filopodia in the embryonic Xenopus brain. Neuron. 1993;11:237–251. doi: 10.1016/0896-6273(93)90181-p. [DOI] [PubMed] [Google Scholar]
- 9.Clark EA, King WG, Brugge JS, Symons M, Hynes RO. Integrin-mediated signals regulated by members of the Rho family of GTPases. J Cell Biol. 1998;142:573–586. doi: 10.1083/jcb.142.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.CLONTECH. Living colors endosome localization vectors. CLONTECHniques. 2000;15:24–25. [Google Scholar]
- 11.Dent EW, Callaway JL, Szebenyi G, Baas PW, Kalil K. Reorganization and movement of microtubules in axonal growth cones and developing interstitial branches. J Neurosci. 1999;19:8894–8908. doi: 10.1523/JNEUROSCI.19-20-08894.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Driessens MHE, Hu H, Nobes CD, Self A, Jordens I, Goodman CS, Hall A. Plexin-B semaphorin receptors interact directly with active Rac and regulate the actin cytoskeleton by activating Rho. Curr Biol. 2001;11:339–344. doi: 10.1016/s0960-9822(01)00092-6. [DOI] [PubMed] [Google Scholar]
- 13.Evan G, Lewis G, Ramsay G, Bishop J. Isolation of monoclonal antibodies specific for c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gomez TM, Letourneau PC. Filopodia initiate choice made by sensory neuron growth cones at laminin/fibronectin borders in vitro. J Neurosci. 1994;14:5959–5972. doi: 10.1523/JNEUROSCI.14-10-05959.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grenningloh G, Rehm EJ, Goodman CS. Genetic analysis of growth cone guidance in Drosophila: fasciclin II functions as a neuronal recognition molecule. Cell. 1991;67:45–57. doi: 10.1016/0092-8674(91)90571-f. [DOI] [PubMed] [Google Scholar]
- 16.Grieder NC, de Cuevas M, Spradling AC. The fusome organizes the microtubule network during oocyte differentiation in Drosophila. Development. 2000;127:4253–4264. doi: 10.1242/dev.127.19.4253. [DOI] [PubMed] [Google Scholar]
- 17.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 18.Harden N, Lee J, Loh HY, Ong YM, Tan I, Leung T, Manser E, Lim L. A Drosophila homolog of the Rac- and Cdc42-activated serine/threonine kinase PAK is a potential focal adhesion and focal complex protein that colocalizes with dynamic actin structures. Mol Cell Biol. 1996;16:1896–1908. doi: 10.1128/mcb.16.5.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hing H, Xiao J, Harden N, Lim L, Zipursky SL. Pak functions downstream of Dock to regulate photoreceptor axon guidance in Drosophila. Cell. 1999;97:853–863. doi: 10.1016/s0092-8674(00)80798-9. [DOI] [PubMed] [Google Scholar]
- 20.Hu H, Marton T, Goodman CS. Plexin B mediates axon guidance in Drosophila by simultaneously inhibiting active Rac and enhancing RhoA signaling. Neuron. 2001;32:39–51. doi: 10.1016/s0896-6273(01)00453-6. [DOI] [PubMed] [Google Scholar]
- 21.Isbister CM, O'Connor TP. Filopodial adhesion does not predict growth cone steering events in vivo. J Neurosci. 1999;19:2589–2600. doi: 10.1523/JNEUROSCI.19-07-02589.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jan YN, Jan LY. Genetic control of cell fate specification in Drosophila peripheral nervous system. Annu Rev Genet. 1994;28:373–393. doi: 10.1146/annurev.ge.28.120194.002105. [DOI] [PubMed] [Google Scholar]
- 23.Jin Z, Strittmatter SM. Rac1 mediates collapsin-1-induced growth cone collapse. J Neurosci. 1997;17:6256–6263. doi: 10.1523/JNEUROSCI.17-16-06256.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joneson T, McDonough M, Bar-Sagi D, Van Aelst L. RAC regulations of actin polymerization and proliferation by a pathway distinct from Jun kinase. Science. 1996;274:1374–1376. doi: 10.1126/science.274.5291.1374. [DOI] [PubMed] [Google Scholar]
- 25.Kabir N, Schaefer AW, Nakhost A, Sossin WS, Forscher P. Protein kinase C activation promotes microtubule advance in neuronal growth cones by increasing average microtubule growth lifetimes. J Cell Biol. 2001;152:1033–1043. doi: 10.1083/jcb.152.5.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaufmann N, Wills ZP, Van Vactor D. Drosophila Rac1 controls motor axon guidance. Development. 1998;125:453–461. doi: 10.1242/dev.125.3.453. [DOI] [PubMed] [Google Scholar]
- 27.Keshishian H, Broadie K, Chiba A, Bate M. The Drosophila neuromuscular junction: a model for studying synaptic development and function. Annu Rev Neurosci. 1996;19:545–575. doi: 10.1146/annurev.ne.19.030196.002553. [DOI] [PubMed] [Google Scholar]
- 28.Kraynov VS, Chamberlain C, Bokoch GM, Schwartz MA, Slabaugh S, Hahn KM. Localized Rac activation dynamics visualized in living cells. Science. 2000;290:333–337. doi: 10.1126/science.290.5490.333. [DOI] [PubMed] [Google Scholar]
- 29.Lamarche N, Tapon N, Stowers L, Burbelo PD, Aspenstrom P, Bridges T, Chant J, Hall A. Rac and Cdc42 induce actin polymerization and G1 cell cycle progression independently of p65PAK and the JNK/SAPK MAP kinase cascade. Cell. 1996;87:519–529. doi: 10.1016/s0092-8674(00)81371-9. [DOI] [PubMed] [Google Scholar]
- 30.Landgraf M, Bossing T, Technau GM, Bate M. The origin, location, and projections of the embryonic abdominal motoneurons of Drosophila. J Neurosci. 1997;17:9642–9655. doi: 10.1523/JNEUROSCI.17-24-09642.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin CH, Thompson CA, Forscher P. Cytoskeletal reorganization underlying growth cone motility. Curr Opin Neurobiol. 1994;4:640–647. doi: 10.1016/0959-4388(94)90004-3. [DOI] [PubMed] [Google Scholar]
- 32.Lin DM, Fetter RD, Kopczynski C, Grenningloh G, Goodman CS. Genetic analysis of fasciclin II in Drosophila: defasciculation, refasciculation and altered fasciculation. Neuron. 1994;13:1055–1069. doi: 10.1016/0896-6273(94)90045-0. [DOI] [PubMed] [Google Scholar]
- 33.Luo L, Liao YJ, Jan LY, Jan YN. Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast function. Genes Dev. 1994;8:1787–1802. doi: 10.1101/gad.8.15.1787. [DOI] [PubMed] [Google Scholar]
- 34.Luo L, Jan LY, Jan YN. Small GTPases in axon outgrowth. Prospect Dev Neurobiol. 1996;4:199–204. [PubMed] [Google Scholar]
- 35.Miki H, Sasaki T, Takai Y, Takenawa T. Induction of filopodium formation by a WASP-related actin-depolymerizing protein N-WASP. Nature. 1998;391:93–96. doi: 10.1038/34208. [DOI] [PubMed] [Google Scholar]
- 36.Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Idura M, Tsien RY. Fluorescent indicators of Ca2+ based on green fluorescent proteins and calmodulin. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 37.Mochizuki N, Yamashita S, Kurokawa K, Ohba Y, Nagai T, Miyawaki A, Matsuda M. Spatio-temporal images of growth-factor-induced activation of Ras and Rap1. Nature. 2001;411:1065–1068. doi: 10.1038/35082594. [DOI] [PubMed] [Google Scholar]
- 38.Morrison DK, Murakami MS, Cleghon V. Protein kinases and phosphatases in the Drosophila genome. J Cell Biol. 2000;150:F57–F62. doi: 10.1083/jcb.150.2.f57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Myers PZ, Bastiani MJ. Growth cone dynamics during the migration of an identified commissural growth cone. J Neurosci. 1993;13:127–143. doi: 10.1523/JNEUROSCI.13-01-00127.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Connor TP, Bentley D. Accumulation of actin in subsets of pioneer growth cone filopodia in response to neural and epithelial guidance cues in situ. J Cell Biol. 1993;123:935–948. doi: 10.1083/jcb.123.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Owen D, Mott HR, Laue ED, Lowe PN. Residues in Cdc42 that specify binding to individual CRIB effector proteins. Biochemistry. 2000;39:1243–1250. doi: 10.1021/bi991567z. [DOI] [PubMed] [Google Scholar]
- 42.Ritzenthaler S, Suzuki E, Chiba A. Postsynaptic filopodia in muscle cells that interact with innervating motoneuron axons. Nat Neurosci. 2000;3:1012–1017. doi: 10.1038/79833. [DOI] [PubMed] [Google Scholar]
- 43.Ruchhoeft ML, Ohnuma S, McNeill L, Holt CE, Harris WA. The neuronal architecture of Xenopus retinal ganglion cells is sculpted by Rho-family GTPases in vivo. J Neurosci. 1999;19:8454–8463. doi: 10.1523/JNEUROSCI.19-19-08454.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sabry JH, O'Connor TP, Evans L, Rotoian-Raymond A, Kirschner M, Bentley D. Microtubule behavior during guidance of pioneer neuron growth cones in situ. J Cell Biol. 1991;115:381–395. doi: 10.1083/jcb.115.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sone M, Hoshino M, Suzuki E, Kuroda S, Kaibuchi K, Nakagoshi H, Saigo K, Nabeshima Y, Hama C. Still life: a protein in synaptic terminals of Drosophila homologous to GDP-GTP exchangers. Science. 1997;275:543–547. doi: 10.1126/science.275.5299.543. [DOI] [PubMed] [Google Scholar]
- 46.Suter DM, Forscher P. An emerging link between cytoskeletal dynamics and cell adhesion molecules in growth cone guidance. Curr Opin Neurobiol. 1998;8:106–116. doi: 10.1016/s0959-4388(98)80014-7. [DOI] [PubMed] [Google Scholar]
- 47.Van Aelst L, D'Souza-Schorey C. Rho GTPases and signaling networks. Genes Dev. 1997;11:2295–9322. doi: 10.1101/gad.11.18.2295. [DOI] [PubMed] [Google Scholar]
- 48.Vastrik I, Eickholt BJ, Walsh FS, Ridley A, Doherty P. Sema3A-induced growth-cone collapse is mediated by Rac1 amino acids 17–32. Curr Biol. 1999;9:991–998. doi: 10.1016/s0960-9822(99)80447-3. [DOI] [PubMed] [Google Scholar]
- 49.Verkhusha VV, Tsukita S, Oda H. Actin dynamics in lamellipodia of migrating border cells in Drosophila ovary revealed by a GFP-actin fusion protein. FEBS Lett. 1999;445:395–401. doi: 10.1016/s0014-5793(99)00124-6. [DOI] [PubMed] [Google Scholar]
- 50.Wahl S, Barth H, Ciossek T, Aktories K, Mueller BK. Ephrin-A5 induces collapse of growth cones by activating Rho and Rho kinase. J Cell Biol. 2000;149:263–270. doi: 10.1083/jcb.149.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wolf B, Seeger MA, Chiba A. Commissureless endocytosis is correlated with initiation of neuromuscular synaptogenesis. Development. 1998;125:3853–3863. doi: 10.1242/dev.125.19.3853. [DOI] [PubMed] [Google Scholar]
- 52.Wong K, Ren XR, Huang YZ, Xie Y, Liu G, Saito H, Tang H, Wen L, Brady-Kalnay SM, Mei L, Wu JY, Xiong WC, Rao Y. Signal transduction in neuronal migration: roles of GTPase activating proteins and the small GTPase Cdc42 in the Slit-Robo pathway. Cell. 2001;107:209–221. doi: 10.1016/s0092-8674(01)00530-x. [DOI] [PubMed] [Google Scholar]
- 53.Zheng JQ, Wan J-J, Poo M-M. Essential role of filopodia in chemotropic turning of nerve growth cone induced by a glutamate gradient. J Neurosci. 1996;16:1140–1149. doi: 10.1523/JNEUROSCI.16-03-01140.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]