Abstract
We address the challenge of detecting the contribution of noncoding mutations to disease with a deep-learning-based framework that predicts specific regulatory effects and the deleterious impact of genetic variants. Applying this framework to 1,790 Autism Spectrum Disorder (ASD) simplex families reveals disease causality of noncoding mutations: ASD probands harbor both transcriptional and post-transcriptional regulation-disrupting de novo mutations of significantly higher functional impact than unaffected siblings. Further analysis suggests involvement of noncoding mutations in synaptic transmission and neuronal development, and taken together with prior studies reveal a convergent genetic landscape of coding and noncoding mutations in ASD. We demonstrate that sequences carrying prioritized proband mutations possess allele-specific regulatory activity, and highlight a link between noncoding mutations and IQ heterogeneity in ASD probands. Our predictive genomics framework illuminates the role of noncoding mutations in ASD, prioritizes high impact mutations for further study, and is broadly applicable to complex human diseases.
Introduction
Great progress has been made in the past decade in understanding autism spectrum disorder (ASD) genetics, establishing de novo mutations, including copy number variants (CNVs) and point mutations that likely disrupt protein coding genes, as important causes of ASD1,2. Yet all known ASD-associated genes together explain a small fraction of new cases, and it is estimated that overall de novo protein coding mutations, including CNVs, contribute to no more than 30% of simplex ASD cases2,3. The vast majority of identified de novo mutations are located within intronic and intergenic regions, yet little is known regarding their contribution to the genetic architecture of ASD or in disease pathogenesis more generally.
A potential role for noncoding mutations in complex human diseases including ASD has long been speculated. Human regulatory regions show signs of negative selection4, suggesting mutations within these regions lead to deleterious effects, and studies of inherited common variants have shown enriched disease association in noncoding regions5. Furthermore, noncoding mutations affecting gene expression have been discovered to cause Mendelian diseases6 and shown to be enriched in cancer7. Expression dosage effects have also been suggested as underlying the link between CNVs and ASD8. Recently, parentally-inherited structural noncoding variants have been linked to ASD9. Also, on a small cohort of ASD families, some trends with limited sets of mutations have been reported10–12. Likewise, despite the major role RNA-binding proteins (RBPs) play in post-transcriptional regulation, little is known of the pathogenic effect of noncoding mutations affecting RBPs outside of the canonical splice sites. Thus, noncoding mutations could be a cause of ASD, yet no conclusive connection of regulatory de novo noncoding mutations, either transcriptional or post-transcriptional, to ASD etiology has been established.
Recent developments make it possible to perform large-scale studies that reliably identify noncoding de novo mutations at whole genome scale. The Simons Simplex Collection (SSC) whole genome sequencing (WGS) data for 1,790 families differs from many previous large-scale studies in design by including matched unaffected siblings3,13–16. These provide critical background controls for detecting excess of proband mutations, as it is otherwise hard to distinguish disease-relevant excess of mutations from irrelevant biological and technical variation, such as genetic background differences or artificial biases from sequencing, variant calling, and filtering procedures.
However, even with study designs using matched control individuals, detecting the de novo noncoding contribution is still challenging, and establishing the role of the vast noncoding space in the genetic basis of autism remains elusive. Two recent studies17,18 demonstrated that even when considering a wide variety of possible functional annotation categories (e.g. mutations in known regulatory sites, mutations at the location of known histone marks, mutations near ASD- or disease-relevant gene sets), no significant noncoding ASD-proband-specific signal was observed, and that approach would require a very large cohort to detect signal17. This is consistent with the expectation that noncoding mutations, in contrast to loss-of-function coding mutations, can vary highly in functional impact, with potentially only a small fraction of variants having strong effect size. Thus, the challenge is to move beyond simple mutation counts, which are susceptible to both statistical power challenges and confounding factors, such as the rise in mutation counts with parental age. This difficulty is shared in other psychiatric diseases with complex genetic bases, such as intellectual disabilities and schizophrenia. In fact, little is known about the contribution of noncoding rare variants or de novo mutations to human diseases beyond the less common cases with Mendelian inheritance patterns.
To address this challenge, we used a systematic approach (Fig. 1a) that reliably identifies impactful noncoding mutations, analogous to using the genetic codon code to distinguish non-synonymous from synonymous protein coding mutations. This enables comparison of mutational burden between probands and siblings not simply in terms of number of mutations, but in terms of their functional impact. Specifically, we used biochemical data demarcating DNA and RNA binding protein interactions to train and deploy a deep convolutional-neural-network-based framework that predicts the functional and disease impact of de novo mutations in the SSC, with models trained for DNA and RNA. Our framework estimates, with single nucleotide resolution, the quantitative impact of each variant on 2,002 specific transcriptional and 232 specific post-transcriptional regulatory features, including histone marks, transcription factors and RNA-binding protein (RBP) profiles.
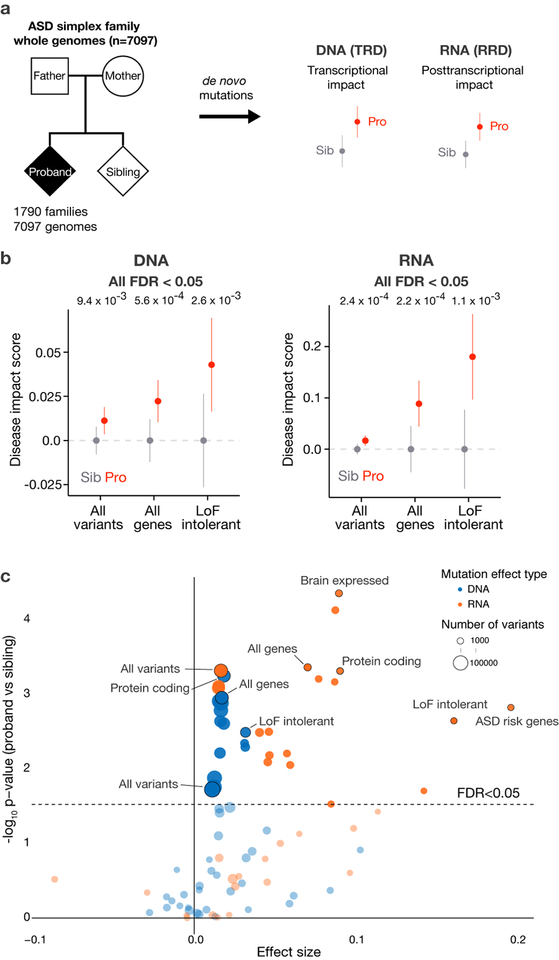
Fig. 1. The elevated noncoding regulatory mutation effect burden in Autism Spectrum Disorder.
a) Overall study design for deciphering the genome-wide de novo noncoding mutation effects contribution to ASD. 1,790 ASD simplex families’ whole genomes were sequenced to identify de novo mutations in the ASD probands and unaffected siblings. SNV de novo mutations were analyzed by their predicted transcriptional (chromatin and TFs) and post-transcriptional (RNA-binding proteins) regulatory effect for comparison between probands and siblings.
b) ASD probands possess mutations with significantly higher predicted disease impact scores compared to their unaffected siblings. We observe significant burden of both transcriptional (DNA - all variants, n = 127,140) and post-transcriptional regulation (RNA - all transcribed variants, n = 77,149) altering mutations in probands. This proband excess is stronger when restricted to mutation near all genes for DNA (n = 69,328) and near alternatively spliced exons for RNA (n = 4,871), and even stronger near ExAC LoF intolerant (DNA n = 14,873, RNA n = 1,355) genes. For analyses that include gene sets, variants were associated with the closest gene within 100kb of the representative TSS for transcriptional regulatory disruption (TRD) analysis. For RNA regulatory disruption (RRD) analysis, variants located in the introns within 400bp of flanking exons in alternative splicing regulatory regions were used. Wilcoxon rank sum test (one-sided) was used for computing the significance levels. All predicted disease impact scores were normalized by subtracting average predicted disease impact scores of sibling mutations for each comparison (mean DIS with the error bars indicate 95% CI). Every result is significant with multiple hypothesis correction (FDR < 0.05) and robust to inclusion or exclusion of protein coding region mutations (Supplementary Fig. 6).
c)Genomic variant set analysis of mutational burden for transcriptional- and posttranscriptional- disruptions. x-axis shows, for each gene set and distance cutoff, the effect size as defined as the difference between average DIS in probands and in siblings. Wilcoxon rank sum test (one-sided) was used for computing the significance levels. Significance level before and after correction for each category is listed in Supplementary Table 2. Categories shown in Fig. 1b are included in the annotation. All gene lists were obtained from Werling et al.17. Distance cutoffs for DNA are 10kb, 50kb, 100kb, 500kb, ∞ to TSS, and distance cutoffs for RNA are 200bp, 400bp, ∞ to all exons or to all alternatively spliced exons. DNA results shown in blue and RNA in orange; dot size corresponds to sample size (number of variants in a category); total sample size n = 127,140. Variant sets with >500 mutations are displayed. Full list of results are available in Supplementary Table 2. Uncorrected p-values are shown in the y-axis and the dashed line indicates categories below FDR 0.05 threshold with the Benjamini-Hochberg method. Results are robust to inclusion or exclusion of protein coding region mutations (Supplementary Fig. 7).
Using this approach, we discovered, at both DNA and RNA regulation levels, a significantly (multiple-hypothesis corrected) elevated burden of disruptive transcriptional-regulatory disrupting (TRD) and RBP-regulatory disrupting (RRD) proband mutations in ASD, providing evidence for causality of noncoding regulatory de novo mutations in autism. Notably, the functional impact difference between proband and sibling mutations is significant when considering de novo mutations genome-wide, with elevated effect sizes observed around loss-of-function intolerant genes (ExAC19). We also identify specific pathways and tissues affected by these mutations, experimentally verify the differential regulatory effect of prioritized variants, and explore a link between the noncoding mutations and IQ in ASD. We provide an interactive interface to explore de novo mutation impact predictions for the biomedical research community at hb.flatironinstitute.org/ASDbrowser.
Results
Contribution of transcriptional and post-transcriptional regulatory mutations to ASD
Analysis of the noncoding mutation contribution to ASD is challenging due to the difficulty of assessing which noncoding mutations are functional, and further, which of those contribute to the disease phenotype. For predicting the regulatory impact of noncoding mutations, we constructed a deep convolutional network-based framework to directly model the functional impact of each mutation and provide a biochemical interpretation including the disruption of transcription factor binding and chromatin mark establishment at the DNA level and of RBP binding at the RNA level (Supplementary Fig. 1, 2). At the DNA level, the framework includes cell-type specific transcriptional regulatory effect models from over 2,000 genome-wide histone marks, transcription factor binding and chromatin accessibility profiles (from ENCODE and Roadmap Epigenomics projects20,21), extending the deep learning-based method that we described previously10 with redesigned architecture (leading to significantly improved performance, p=6.7×10−123, Wilcoxon rank-sum test, Supplementary Fig. 2). At the RNA level, our deep learning-based method was trained on the precise biochemical profiles of over 230 RBP-RNA interactions (derived from CLIP data); such data can identify a wide range of post-transcriptional regulatory binding sites, including those involved in RNA splicing, localization and stability22. At both transcriptional and post-transcriptional levels, our models are accurate and robust in whole chromosome holdout evaluations (Supplementary Fig. 1b). Our models utilize a large sequence context to provide single nucleotide resolution to our predictions, while also capturing dependencies and interactions between various biochemical factors (e.g. histone marks or RBPs). This approach is data-driven, does not rely on known sequence information, such as transcription factor binding motifs, and it can predict impact of any mutation regardless of whether it has been previously observed, which is essential for the analysis of ASD de novo mutations. Finally, to link the biochemical disruption caused by a variant with phenotypic impact, we trained a regularized linear model using a set of curated human disease regulatory noncoding mutations6 (HGMD) and rare variants from healthy individuals in the 1000 Genomes populations23 to generate a predicted disease impact score (DIS) for each autism mutation independently based on its predicted transcriptional and post-transcriptional regulatory effects.
With these approaches, we systematically assessed the functional impact of de novo mutations on regulatory factor binding and chromatin properties, using data derived from 7,097 whole genomes from the SSC cohort with our framework (total 127,140 non-repeat region SNVs, Supplementary Table 1). When considering all de novo mutations, we observed a significantly higher functional impact in probands compared to unaffected siblings, independently at the transcriptional (p=9.4×10−3, one-side Wilcoxon rank-sum test for all; FDR=0.033, corrected for all mutation sets tested) and post-transcriptional (p=2.4×10−4, FDR=0.0049) levels (Fig. 1b, all variants). This analysis is sensitive enough to discover noncoding contribution even if a very small fraction of the noncoding mutations are impactful (see power analysis in Supplementary Fig. 3). Furthermore, our finding is robust and significant directly at the level of biochemical disruptions predicted by DNA and RNA deep learning models as well as with alternative DIS training sets (Supplementary Fig. 4–5) or with inclusion or exclusion of protein coding regions (Supplementary Fig. 6–7).
Werling et al.17 raised the challenge of detecting any significant proband-specific signal even with highly specific subsets of genes or genomic regions, and correspondingly emphasized the need for proper multiple hypothesis correction; this challenge was still not resolved by a larger ASD cohort in a follow-up study18. Notably, our result above does not rely on any selection of variant subsets (e.g. those near predicted ASD-associated genes), is significant even after multiple hypothesis correction, and, unlike the mutation counts, the predicted mutation effects are not correlated with parental age (Supplementary Fig. 8), a confounding factor of mutation count-based analysis.
To gain further insight into the ASD noncoding regulatory landscape, we conducted a comprehensive analysis, with full multiple hypothesis correction for all combinations of 14 gene-sets previously used in Werling et al.17 and 10 genomic regions tested (e.g. TSS or exon proximal). When restricted to genomic regions of higher regulatory potential (i.e. near TSS or alternatively spliced exons), we observed an increased dysregulation effect size (Fig. 1b–c, all genes, TRD p=5.6×10−4, FDR=0.0056; RRD p=2.2×10−4, FDR=0.0048). Among gene sets, we observed an elevated proband burden of high effect mutations close to loss-of-function (LoF) intolerant genes (pLI > 0.9 from ExAC, 3,230 genes, TRD p=2.6×10−3, FDR=0.013; RRD p=1.1×10−3, FDR=0.0078) (Fig. 1b–c, Supplementary Fig. 9), suggesting LoF intolerant genes are highly vulnerable to noncoding disruptive mutations in ASD. This is consistent with the enrichment of coding loss-of-function mutations among the LoF intolerant genes in the SSC cohort24, indicating ASD signal convergence of noncoding and coding de novo mutations. Furthermore, we also find convergent signal at both transcriptional and post-transcriptional levels, thus providing further evidence for the causal role of noncoding effects in ASD (full analysis p-values and FDRs are available in Supplementary Table 2). We observe these signals consistently across SSC cohort subsets that were sequenced in different phases (Supplementary Fig. 10).
Tissue specificity and functional landscape of noncoding ASD-associated de novo mutations
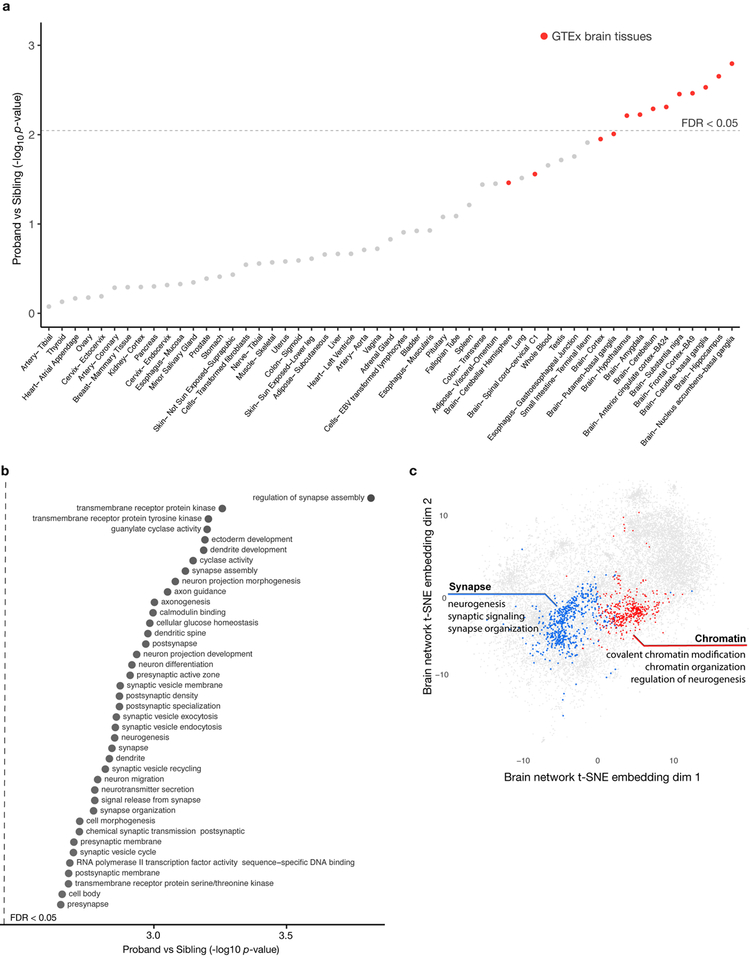
Although one of the hallmarks of autism is altered brain development, a comprehensive tissue association has not been established for de novo noncoding variants. To explore the proband-specific tissue signal, we systematically tested the variant effects for tissue-specific genes derived from all 53 GTEx tissues and cell types25. We observed a consistent significant proband-specific mutation effect associated with brain tissues, with brain regions constituting the top 11 ranked tissues (by difference in proband vs sibling noncoding mutation effect) (Fig. 2a, all with FDR < 0.05). This provides strong evidence that high impact variants from the noncoding genome of ASD probands likely disrupt brain-specific gene regulation, consistent with previous findings for protein coding mutations30.
Fig. 2. Analysis of noncoding mutation effects converges on brain specific signals and neurodevelopmental processes.
a) Brain tissue-specific genes show strongest elevated proband-specific noncoding mutation effect burden. All 53 GTEx tissues are ranked by significance of increased proband mutation burden compared to unaffected siblings in tissue-specific genes (Methods). Uncorrected p-values are shown in the y-axis and the dashed line indicates tissues below the FDR=0.05 threshold corrected with the Benjamini-Hochberg method. Disease impact scores for all mutations within 100kb of representative TSSs (DNA) and intronic mutations within 400bp of exon boundaries (RNA) (n = 71,554) are used for the analysis.
b) Neuronal function and development related processes show significant excess of proband mutation disease impact scores by statistical test NDEA (full list in Supplementary Table 4, see also Methods). Analysis is conducted on the same mutation set as in (a). The top processes (y-axis) and the p-values of proband excess (x-axis) are shown. Uncorrected p-values are shown in the x-axis and all gene sets shown have FDR < 0.05.
c) Genes with significant network neighborhood excess of high-impact proband mutations form two functionally coherent clusters (see annotations for representative enriched gene sets in each cluster, full list is in Supplementary Table 5). Analysis is conducted on the same mutation set as in (a). The brain functional network is visualized by computing two-dimensional embeddings with t-SNE (Methods). Genes, but not network edges, are shown for visualization clarity. Clustering was performed with Louvain community clustering. All genes in the two clusters shown are with FDR < 0.1.
We next investigated the underlying processes and pathways impacted by de novo noncoding mutations in ASD. Such analysis is challenging because in addition to the variability in functional impact of mutations, ASD probands appear highly heterogeneous in underlying causal genetic perturbations26 and single mutations could cause a widespread effect on downstream genes. Thus to detect genes and pathways relevant to the pathogenicity of ASD TRD and RRD mutations, we developed a network-based statistical approach, NDEA (Network-neighborhood Differential Enrichment Analysis) (Supplementary Fig. 11). We used a brain-specific functional network that probabilistically integrates a large compendium of public omics data (e.g. expression, PPI, motifs) to represent how likely two genes are to act together in a biological process27. When applied to ASD de novo mutations, the NDEA approach identifies genes whose functional network neighborhood is significantly enriched for genes with stronger predicted disease impact in proband mutations compared to sibling mutations (Supplementary Table 3).
Globally, NDEA enrichment analysis pointed to a proband-specific role for noncoding mutations in affecting neuronal development, including in synaptic transmission and chromatin regulation (Fig. 2b, Supplementary Table 4), consistent with processes previously associated with ASD based on protein coding variants2, 30. Genes with significant NDEA enrichment were specifically involved in neurogenesis and grouped into two functionally coherent clusters with Louvain community detection algorithm (Fig. 2c, Supplementary Table 5). The synaptic cluster is enriched in ion channels and receptors involved in neurogenesis (p=5.6×10−38), synaptic signaling (p=4.8×10−35) and synapse organization (p=1.5×10−18), including previously known ASD-associated genes such as those involved in synapse organization SHANK2, NLGN2, NRXN2, synaptic signaling NTRK2 and NTRK3, ion channels CACNA1A/C/E/G, KCNQ2, and neurotransmission SYNGAP1, GABRB3, GRIA1, GRIN2A28. The synapse cluster is also significantly enriched for plasma membrane proteins (p=3.9×10−24). In contrast, the chromatin cluster, representing chromatin regulation related processes, displayed an overrepresentation of nucleoplasm (p=2.1×10−9) proteins, with diverse functional roles including covalent chromatin modification (p=2.5×10−9), chromatin organization (5.2×10−8) and regulation of neurogenesis (p=6.4×10−5). The chromatin cluster also includes many known ASD-associated genes such as chromatin remodeling protein CHD8, chromatin modifiers KMT2A, KDM6B, and Parkinson’s disease causal mutation gene PINK129 which is also associated with ASD28 (Supplementary Table 3). Overall, our results demonstrate pathway-level TRD and RRD mutation burden and identify distinct network level hot spots for high impact de novo mutations.
Next, we examined the genetic landscape of ASD-associated de novo noncoding and coding mutations. Specifically, in addition to the network analysis of noncoding mutations at the transcriptional and post-transitional level, we also applied it to the de novo coding mutations2. We compared the gene-specific NDEA statistic of elevated proband-specific noncoding mutation effect burden to that of the coding mutations, finding a significant positive correlation for both TRD and RRD (p=0.004 and Pearson’s r = 0.39 for TRD, p=0.042 and Pearson’s r = 0.30 for RRD; two-sided permutation test). Moreover, by network analysis, TRD and RRD are themselves significantly correlated (p=0.034 two-sided permutation test). This demonstrates that coding and noncoding mutations affect overlapping processes and pathways, indicating a convergent genetic landscape, and highlights the potential of ASD gene discovery by combining coding and noncoding mutations.
Experimental study of ASD noncoding mutation effects on gene regulation
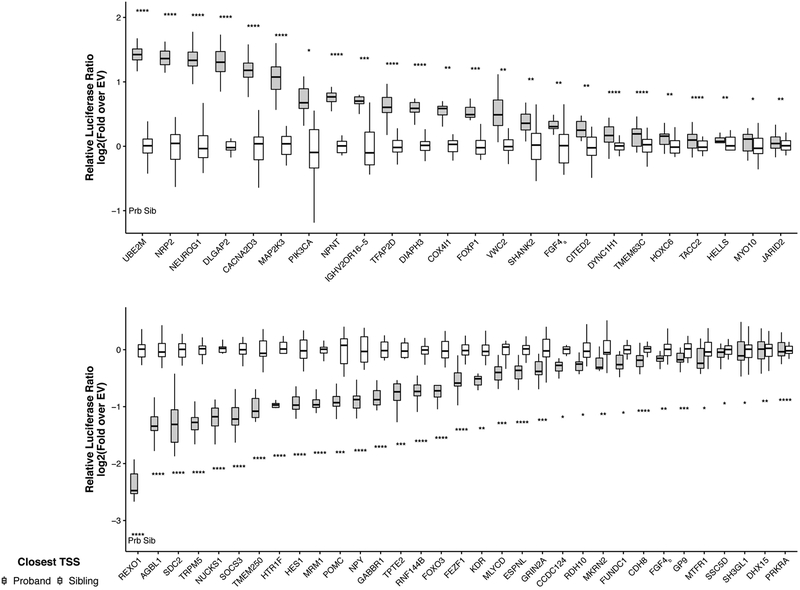
Our analysis identified new candidate noncoding disease mutations with potential impact on ASD through regulation of gene expression. In order to add further evidence to a set of high confidence causal mutations, we experimentally studied allele-specific effects of predicted high-impact mutations in cell-based assays. For TRD mutations, fifty nine genomic regions showed strong transcriptional activity with 96% proband variants (57 variants) showing robust differential activity (Fig. 3, Methods); demonstrating that our prioritized de novo TRD mutations do indeed lie in regions with transcriptional regulatory potential and the predicted effects translate to measurable allele-specific expression effects. Among these genes with the demonstrated strong differential activity mutations, NEUROG1 encodes an important regulator of initiation of neuronal differentiation and in the NDEA analysis had significant network neighborhood proband excess (p=8.5×10−4), and DLGAP2 encodes a guanylate kinase localized to the post-synaptic density in neurons. Mutations near HES1 and FEZF1 also carried significant differential effect on activator activities: neurogenin, HES, and FEZF family transcription factors act in concert during development, both receiving and sending inputs to Wnt and Notch signaling in the developing central nervous system and interestingly, the gut, to control stem cell fate decisions30–34; and Wnt and Notch pathways have been previously associated with autism26,35. SDC2 encodes a synaptic syndecan protein involved in dendritic spine formation and synaptic maturation, and a structural variant near the 3’ end of the gene was reported in an autistic individual (reviewed in Saied-Santiago, 201736). Thus, our method identified alleles of high predicted impact that do indeed show changes in transcriptional regulatory activity in cells. Since many autism genes are under strong evolutionary selection, only effects exerted through (more subtle) gene expression changes may be observable because complete loss of function mutations may be lethal. This implies that further study of the prioritized noncoding regulatory mutations should yield insights into the range of dysregulations associated with autism.
Fig. 3. Allele-specific transcriptional activity of ASD noncoding mutations.
Differential expression by proband or sibling alleles in a dual luciferase assay demonstrated that 57 predicted high TRD disease impact mutations fall in active regulatory elements and the mutations confer substantial changes to the regulatory potential of the sequence. Cells were transfected with pGL4.23-based expression plasmid containing 230nt of genomic region as well as a transfection control, and then luminescence was assayed 42h later (Methods). Y-axis shows the magnitude of transcription activation activity normalized to sibling allele. Significance levels were computed based on t-test and Fisher’s combined probability test (two-sided, stars indicate significance level *: p<0.05, **: p<0.01, ***: p<0.001, ****: p<0.0001; Methods). Sample sizes for all tests are in Supplementary Table 6. Central values of the box plot represent the median; the box extends from the 25th to the 75th percentile; and whiskers extend to the maximum and minimum values no further than 1.5 * IQR from the hinge (where IQR is the inter-quartile range, or distance between the first and third quartiles).
In addition, as a case study for prioritized RRD mutations, we experimentally validated the effect of an ASD proband de novo noncoding mutation laying outside of a canonical splice site that we predicted to disrupt splicing of SMEK1 (ExAC pLI=1.0; Supplementary Fig. 12), Smek1 has previously been shown to regulate cortical neurogenesis through the Wnt signaling pathway37. For this mutation, we observed a >40% reduction in the inclusion of the exon for the ASD proband allele compared to the sibling allele in a minigene assay (Methods), in agreement with the high predicted RRD impact. This demonstrates the highly disruptive biochemical impact a non-splice site de novo mutation can have on RNA splicing.
The individual level clinical relevance of the noncoding de novo mutations
The majority of ASD probands in the SSC do not have a de novo LoF coding mutation1,2 and noncoding mutations outnumber LoF coding mutations by over 500 fold18. While individual impact of noncoding mutations may vary, as a group noncoding mutations could have significant clinical impact. Indeed, we observed a significant increase in ASD risk for individuals with higher burden of impactful de novo mutations (Supplementary Fig. 9, mean DIS per individual, Wilcoxon rank sum test one-sided p=1.4×10−3), with 25% of the SSC ASD probands incurring an aggregate noncoding ASD risk of 1.2 (odds ratio).
Furthermore, the overall contribution of de novo noncoding mutations (explaining 4.3% of the SSC ASD cases) is comparable to that of loss-of-function coding mutations (5.4%) and to that of missense mutations (3.1%) (Supplementary Fig. 13). This analysis leverages the power of the quad simplex design of the SSC cohort, enabling the estimation of causal contribution of each mutation category by correcting for the background occurrence rate among unaffected siblings (see Methods). Thus, our results demonstrate that noncoding de novo mutations have clinical relevance, although not all ASD probands will have impactful noncoding mutations (even in aggregate), and future work will be required to characterize their clinical impact and relationship to phenotypes.
One interesting direction is in association of impacts of noncoding mutations to specific phenotypes, such as IQ heterogeneity among ASD probands. Intellectual disability is estimated to impact 40–60% of autistic children38, and ASD individuals can over-inherit common variants associated with education attainment39. For de novo noncoding mutations analyzed in this study, we observe a significant association between noncoding mutations and IQ in ASD individuals. Specifically, lower IQ ASD individuals have a higher burden of RRDs in intronic regions flanking alternatively spliced exons of ExAC LoF intolerant genes. This provides genetic evidence that aberrant splicing can contribute to the phenotypic heterogeneity observed among ASD probands (Supplementary Fig. 14, p=1.5×10−3), and should be taken into account when projecting clinical outcomes.
Discussion
Even with great strides in understanding the causes of ASD by sequencing and phenotyping of multiple cohorts in the recent years, much of the genetic basis underlying autism remains undiscovered. While a number of coding variants have been associated with ASD, no systematic evidence of de novo noncoding effect has been observed. Here we present a novel deep-learning based approach for quantitatively assessing the impact of noncoding mutations on human disease. Our approach addresses the statistical challenge of detecting the contribution of noncoding mutations by predicting their specific effects on transcriptional and post-transcriptional levels. This approach is general and can be applied to study contributions of noncoding mutations to any complex disease or phenotype.
Here, we apply our strategy to ASD using the 1,790 whole genome sequenced families from the Simons Simplex Collection, and for the first time demonstrate significant proband-specific signal in regulatory de novo noncoding space. Importantly, we independently detect this signal not only at the transcriptional level, but also find significant proband-specific RRD burden. Previously, there’s been limited evidence for disease contribution of mutations disrupting post-transcriptional mechanisms outside of the canonical splice sites. We demonstrate significant ASD disease association at the de novo mutation level for variants impacting a large collection of RBPs regulating post-transcriptional regulation. Overall, our results suggest that both transcriptional and posttranscriptional mechanisms play a significant role in ASD etiology and possibly other complex diseases.
Notably, our study reveals important biological convergences among genetic dysregulations associated with ASD. Our analyses of the disease impact of both DNA and RNA effect mutations point to similar sets of impacted genes and pathways, indicating that the effects of regulatory mutations are convergent. Furthermore, high-impact noncoding regions we find in ASD probands impact the same genes previously found to be impacted by ASD LoF coding mutations. This convergence provides support for the causal contribution of noncoding regulatory mutations to ASD etiology.
Our analyses also demonstrate the potential of predicting disease phenotypes from genetic information, including de novo noncoding mutations. We provide a resource for further research into understanding the mechanism of noncoding impact on ASD, including computationally prioritized TRD and RRD mutations with strong predicted regulatory effects, as well as potentially disease contributing ASD proband mutations with experimentally confirmed effects (Supplementary Table 1,6 and hb.flatironinstitute.org/ASDbrowser). However, there remains much room for further progress in this important area. We expect continuing development of noncoding mutation effect prediction methods will further improve the power of WGS studies in discovering the biological mechanisms of noncoding mutation contribution to autism and other complex human diseases.
Methods
De novo mutation calling and filtering
The Simons Simplex Collection WGS data was made available via Simons Foundation Autism Research Initiative (SFARI), and was processed to generate variant calls via the standard GATK pipeline. The Simons Simplex Collection WGS data can be requested through SFARI Base (https://www.sfari.org/resource/sfari-base/), with the condition that the use of the data is limited to projects related to advancing the field of autism and related neurodevelopmental disorder research (questions on SSC consents should be directed to collections@sfari.org). To call de novo single nucleotide substitutions, inherited mutations were removed, and candidate de novo mutations were selected from the GATK variant calls where the alleles were not present in parents and the parents were homozygous with the same allele. DNMFilter classifier was then used to score each candidate de novo mutation and a threshold of probability > 0.75 was applied for SSC phase1–2 and probability > 0.5 cutoff for phase3 to obtain a comparable number of high-confidence DNM calls across phases.
The DNMFilter40 classifier was trained with an expanded training set combining the original training standards with the verified DNMs from the SSC pilot WGS studies for the initial 40 SSC families. For final analysis, de novo mutation calls within the low complexity repeat regions from UCSC browser table RepeatMasker41 were removed. Also, DNMs appearing in multiple SSC families (i.e. non-singleton DNMs) or individuals with outlier numbers of mutations (> 3 standard deviation more than average) were excluded from the analysis.
Overall genome-wide, we detect 77.7 mutations per individual with Ti/Tv ratio 2.01 with 95% CI [2.00, 2.03] (78.7 for probands with Ti/Tv = 2.02 [1.99, 2.04], 76.7 for siblings with Ti/Tv=2.01 [1.99, 2.03]), with no significant difference in mutation substitution patterns between proband and sibling (Supplementary Fig. 15). The WGS DNM calls were compared against exome sequencing de novo mutations calls and previously validated SSC de novo mutations42: 87.9% of the exome sequencing mutations calls and 90.3% of the validated mutations were rediscovered in our mutation calls.
Training of DNA transcriptional regulatory effects and RNA posttranscriptional effects models
For training the transcriptional regulatory effects model, training labels, such as histone marks, transcription factors, and DNase I profiles, were processed from uniformly processed ENCODE and Roadmap Epigenomics data releases. The training procedure is as described in Zhou and Troyanskaya21 with the following modifications. The model architecture was extended to double the number of convolution layers for increased model depth (see Supplementary Note 1 for details). Similar to our previous model21, all layers except for the last linear layer were shared across all biochemical features. Input features were expanded to include all of the released Roadmap Epigenomics histone marks and DNase I profiles, resulting in 2,002 total features (Supplementary Table 7) compared to 919 original features.
For training the post-transcriptional regulatory effects model, we utilized the DeepSEA network architecture and training procedure with RNA-binding protein (RBP) profiles as training labels (full list of parameters used in model is in Supplementary Note 1). We uniformly processed RNA features composed of 231 CLIP binding profiles for 82 unique RBPs (ENCODE and previously published CLIP datasets) and a branchpoint mapping profile as input features (full list of experimental features listed in Supplementary Table 8). CLIP data processing followed our previously detailed pipeline43, all CLIP peaks with p-value < 0.1 were used for training with an additional filter requirement of two-fold enrichment over input for ENCODE eCLIP data. In contrast to the DeepSEA, only transcribed genic regions were considered as training labels for the post-transcriptional regulatory effects model. Specifically, all gene regions defined by Ensembl (mouse build 80, human build 75) were split into 50nt bins in the transcribed strand sequence. For each sequence bin, RBP profiles that overlapped more than half were assigned a positive label for the corresponding RBP model. Negative labels for a given RBP model were assigned to sequence bins where other RBP’s non-overlapping peaks were observed. Note that our deep learning models, both transcriptional and post-transcriptional, do not use any mutation data for training, thus it can predict impacts for any mutation regardless of whether it has been previously observed.
Disease impact score prediction
We used curated disease regulatory mutations and rare variants from healthy individuals to train a model that prioritizes likely disease-impacting mutations based on the predicted transcriptional or post-transcriptional regulatory impacts of these mutations. As positive examples, we used 4,401 regulatory noncoding mutations curated in the Human Gene Mutation Database (HGMD) with mutation type “regulatory” (DM, DM?, DFP, DP and FP). For negative examples of background mutations, we used 999,668 rare variants that were only observed once within the healthy individuals from the 1000 Genomes project23. We also showed that using common variants with AF>0.01 and location within 100kb to positive as negatives leads to similar conclusions (Supplementary Fig. 5). Absolute predicted probability differences computed by the convolutional network transcriptional regulatory effects model (described above) were used as input features for each of the 2,002 transcriptional regulatory features and for the 232 post-transcriptional regulatory features in the disease impact model. Input features were standardized to unit variance and zero mean before being used for training. We separately trained a L2 regularized logistic regression model for transcriptional effect model (lambda=10) and post-transcriptional effect model (lambda=10, using only genic region variant examples) with the xgboost package (https://github.com/dmlc/xgboost). The positive and negative training samples were separately weighted according to the inverse of the number of samples to address the label imbalance. The predicted probabilities are z-transformed to have mean 0 and standard deviation 1 across all proband and sibling mutations.
Gene sets and resources
All gene sets used are from Werling et al.17. The 14 gene-sets include GENCODE protein coding genes, Antisense, lincRNAs, Pseudogenes, genes with loss-of-function intolerance (pLI) score > 0.9 from ExAC19, predicted ASD risk genes (FDR < 0.3) from Sanders et al.8, FMRP target genes44, Genes associated with developmental delay45,46 andCHD8 target genes47,48. For genes with expression specific to each 53 GTEx tissue, we used expression table from GTEx v7 (gene median TPM per tissue)25, we selected genes for which expression in a given tissue was five times higher than the median expression across all tissues.
We determined the representative TSS for each gene based on FANTOM CAGE transcription initiation counts relative to GENCODE gene models. Specifically, a CAGE peak is associated to a GENCODE gene if it is within 1000bp from a GENCODE v24 annotated transcription start site49,50. Peaks within 1000bp to rRNA, snRNA, snoRNA or tRNA genes were removed to avoid confusion. Next, we selected the most abundant CAGE peak for each gene, and took the TSS position reported for the CAGE peak as the selected representative TSS for the Gene. For genes with no CAGE peaks assigned, we kept the GENCODE annotated gene start position as the representative TSS. FANTOM CAGE peak abundance data were downloaded at http://fantom.gsc.riken.jp/5/datafiles/latest/extra/CAGE_peaks/ and the CAGE read counts were aggregated over all FANTOM 5 tissue or cell types. GENCODE v24 annotation lifted to GRCh37 coordinates were downloaded from http://www.gencodegenes.org/releases/24lift37.html. All chromatin profiles used from ENCODE and Roadmap Epigenomics projects were listed in Supplementary Table 7. The HGMD mutations are from HGMD professional version 2018.1.
Human exons that are alternatively spliced (AS) were obtained from a recent study that has examined publicly available human RNA-seq data to annotate an extensive catalog of AS events51. Internal exon regions (both 5’SS & 3’SS flanking introns), upstream exon (5’SS flanking introns), and downstream terminal exon (3’SS flanking introns) were used for alternative exon definition types of cassette, mutually exclusive, tandem cassette exons. Terminal exon region was used for intron retention, alternative 3’ or 5’ exon AS exon types. All selected exon-flanking intronic regions were collapsed into a final set of genomic intervals used to subset SNVs that are located within alternative splicing exon region (200 or 400nts from exon boundary), illustrated in Supplementary Fig. 16.
Network differential enrichment analysis (NDEA)
Brain-specific functional relationship networks integrate a wide-range of functional genomic data in a tissue-specific manner and predicted the probability of functional association between any pair of genes27. This network was filtered to only include edges with >0.01 probability (above Bayesian prior) to reduce the impact of noisy low-confidence edges.
We use NDEA to test the differential (proband vs sibling) impact of mutations on each gene or gene set. Intuitively, this test generates a p-value that reflects the proband-specific impact of mutations on that gene or gene set, including through its network neighborhood. This also enables statistical assessment of which gene sets (e.g. pathways) are significantly more affected by proband mutations compared to sibling mutations. Technically, NDEA performs a weighted two-sample (proband vs sibling mutations) test, where the weight for each observation is defined based on network connectivity scores (to the gene or gene sets) and two samples are compared based on weighted averages. Each weight is a non-negative constant number that is used to specify the relative contribution of an observation to the test statistic. When all weights are the same, it reduces to regular two-sample t tests; when the weights are different, it adjusted the standard t statistic to use appropriate variance resulting from weighting. Note, unlike some other forms of weighted t-test, the weights are not random variables and do not represent sample sizes. The assumptions of the NDEA test are analogous to those of the standard two-sample t test, including that samples in each set are i.i.d. and the weighted sample means are normally distributed.
For each gene i, the NDEA t statistic is computed by
, in which and are weighted averages of disease impact scores dm of all proband mutations p or all sibling mutations S. Wij(m) is the network edge score (interpreted as functional relationship probability) between gene i and gene j(m) divided by the number of proband (if m is a proband mutation) or sibling (if m is a sibling mutation) mutations gene j(m) associated to, where j(m) indicate the implicated gene of the mutation m. P and S are the set of all proband mutations and the set of all sibling mutations included in the analysis. and are the unbiased estimates of population variance of and and are the effective sample sizes of proband and sibling mutations after network-based weighting for gene i.
Under null hypothesis of the two groups having no difference, the above t statistic approximately follows a t-distribution with the following degree of freedom:
For testing significance difference between proband and sibling mutations, mutations within 100kb of the representative TSS of all genes and all intronic mutations within 400bp to exon boundary were included in this analysis. RNA disease impact scores were used as the mutation score for intronic mutations within 400bp to exon boundary and DNA disease impact scores were used for other mutations.
For gene set level NDEA, we consider the gene set as a meta-node that contains all genes that are annotated to the gene set (e.g. GO term). Then, to any given gene the average of network edge scores for all genes in the meta-node is used as the weights. GO term annotations were pooled from human (EBI 5/9/2017), mouse (MGI 5/26/2017) and rat (RGD 4/8/2017). Query GO terms were obtained from the merged set of curated GO consortium52 slims from Generic, Synapse, ChEMBL, and supplemented by PANTHER53 GO-slim and terms from NIGO54.
For network-based analysis of correlation between coding and noncoding TRD and RRD mutations, we first compute the NDEA t-statistic for every gene for all protein coding mutations from SSC exome sequencing study2,8, all SSC WGS noncoding mutations within 100kb to a gene, and all SSC WGS genic noncoding mutations within 400bp to an exon, respectively. We then compute Pearson correlation across all resulting gene-specific t-statistics between all three pairs of mutation types. For testing statistical significance of the correlation, we permuted proband and sibling labels for all mutations to compute the null distributions of correlations for each pair of mutation types. 1000 permutations were performed.
Network visualization and clustering
For network visualization, we computed a two-dimensional embedding with t-SNE55 by directly taking a distance matrix of all pairs of genes as the input. The distance matrix was computed as -log(probability) from the edge probability score matrix in the brain-specific functional relationship network. The Barnes-Hut t-SNE algorithm implemented in the Rtsne package was used for the computation. Louvain community clustering were performed on the subnetwork containing all protein coding genes with top 10% NDEA FDR.
Selection and cloning of Variant Allele Genomic Regions
All genomic sequences were retrieved from the hg19 human genome assembly. For experimental testing, we selected variants of high predicted disease impact scores larger than 0 and included mutations near genes with evidence for ASD association, including those with coding LoF mutations (e.g. CACNA2D3) and a proximal structural variant (e.g. SDC2). We did not explicitly select mutations based on proximity to TSSs, and the chosen mutations lie from between 7bp and 324kbp away from nearest TSS, with most variants lying farther than 5k from nearest TSS (Supplementary Table 6) For each allele (sibling or proband), we either cloned 230 nucleotides of genomic sequence amplified from proband lymphoblastoid cell lines or used FragmentGenes synthesized by Genewiz (Supplementary Table 6). In both cases, 15 nucleotide flanks on 5’ and 3’ ends matched each flank of the plasmid cloning sites (Supplementary Table 6. Synthesized fragments were cut with KpnI and BglII and cloned into pGL4.23 (Promega) cut with the same enzymes. PCR-amplified genomic DNA was cloned into pGL4.23 blunt-end cut with EcoRV and Eco53kI using GeneArtCloning method from Thermofisher Scientific. All constructs were verified by Sanger sequencing.
Luciferase Reporter Assays
Human neuroblastoma BE(2)-C cells were plated at 2×104 cells/well in 96-well plates and 24 hours later were transfected with Lipofectamine 3000 (L3000–015, Thermofisher Scientific) together with 75ng of Promega pGL4.23 firefly luciferase vector containing the 230nt of human genomic DNA from the loci of interest (Supplementary Table 6), and 4ng of pNL3.1 NanoLuc (shrimp luciferase) plasmid, for normalization of transfection conditions. 42 hours after transfection, luminescence was detected with the Promega NanoGlo Dual Luciferase assay system (N1630) and BioTek Synergy plate reader. Four to six wells per variant were tested in each experiment. Variants were tested in at least two separate experiments. For each sequence tested, the ratio of firefly luminescence (ASD allele) to NanoLuc luminescence (transfection control) was calculated and then normalized to empty vector (pGL4.23 with no insert) on the same plate. Statistics were calculated from fold over empty vector values from each experiment and results from multiple replication experiments are combined with Fisher’s combined probability test. For presentation of the data, we normalized the fold over empty vector value of the proband allele to that of the sibling allele.
SMEK1 minigene assay
To construct the SMEK1 minigene, the genomic region was amplified with primers -- upstream exon + ~1,400nt intron and alternative exon, downstream exon + ~1,400nt intron, then cloned into pSG5 vector (Supplementary Table 6). The mutant minigene was constructed by assembling the PCR amplified vector backbone with synthetic gBlocks (IDT DNA) carrying the desired single base mutation (GRCh37:chr14:g.91932755G>A in RNA). Minigenes (2 μg) were transfected into SH-SY5Y cells and cells were harvested 48 h post-transfection for immunoblotting or RT-qPCR following standard protocols. Three independent experiments were performed for statistical comparison.
Contribution of de novo mutations in ASD SSC
For LoF and missense coding mutations, we use annotations from Supplementary Table 1 of the SSC exome study Iossifov et al2. Out of total 2,508 probands, 331 ASD probands have at least one LoF coding mutation and 1,182 probands have at least one missense mutation. We estimate the expected number of background occurrences in probands using unaffected siblings occurrences adjusted by the overall proband/sibling ratio, resulting in 221.8 for LoF and 1105.0 for missense. The final estimated contribution was determined by the differential between observed and background occurrences (e.g. for LoF, 331 minus 221.8 divided by 2,508 probands, leading to an estimated contribution of 5.4%). For noncoding mutations, we observe 1,086 probands with mean DIS (mean of average DNA DIS and average RNA DIS) > 0, in comparison to a background occurrence of 1,009 per 1,781 individuals (unaffected siblings). The differential of 1,086 minus background 1,009, leads to an estimated contribution of 4.3%.
Statistical analysis
All details of the statistical tests are specified in the associated text or figure legends. The NDEA test is described in detail in the above “Network differential enrichment analysis (NDEA)” section.
Life Sciences Reporting Summary
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Code and data availability
The code is available from https://hb.flatironinstitute.org/asdbrowser/help. ASD WGS data can be obtained from the Simons Foundation Autism Research Initiative (SFARI). All variant predicted scores have been made available as supplementary material and an interactive web interface is available at https://hb.flatironinstitute.org/asdbrowser/.
Supplementary Material
Acknowledgements
We are grateful to the families participating in the Simons Foundation Autism Research Initiative (SFARI) Simplex Collection (SSC). This work is supported by NIH grants R01HG005998, U54HL117798 and R01GM071966, HHS grant HHSN272201000054C and Simons Foundation grant 395506 to O.G.T. and NIH grants 1UM1HG008901, NS034389, NS081706 and NS097404 and Simons Foundation grant SFARI 240432 to R.B.D. STARR Cancer Consortium Award I10–0056 (to C.Y.P and R.B.D.). O.G.T. is a senior fellow of the Genetic Networks program of the Canadian Institute for Advanced Research (CIFAR). R.B.D. is an Investigator of the Howard Hughes Medical Institute. The authors acknowledge all members of the Troyanskaya and Darnell lab for helpful discussions. We also thank the SFARI, Simons Foundation and Flatiron Institute, in particular Natalia Volfovsky and Marta Benedetti. The authors are pleased to acknowledge that a substantial portion of the work in this paper was performed at the TIGRESS high-performance computer center at Princeton University, which is jointly supported by the Princeton Institute for Computational Science and Engineering and the Princeton University Office of Information Technology’s Research Computing department. O.G.T. is a CIFAR fellow.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Sanders SJ et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 485, 237–241 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iossifov I et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature 515, 216–221 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.C Yuen RK et al. Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat. Neurosci 20, 602–611 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernstein BE et al. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Finucane HK et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet (2015). doi: 10.1038/ng.3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stenson PD et al. The Human Gene Mutation Database: 2008 update. Genome Med. 1, 13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feigin ME et al. Recurrent noncoding regulatory mutations in pancreatic ductal adenocarcinoma. Nat. Genet 49, 825–833 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanders SJ et al. Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron 87, 1215–1233 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brandler WM et al. Paternally inherited cis-regulatory structural variants are associated with autism. Science (80-. ). 360, 327LP-331 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turner TN et al. Genome Sequencing of Autism-Affected Families Reveals Disruption of Putative Noncoding Regulatory DNA. Am. J. Hum. Genet 98, 58–74 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turner TN et al. Genomic Patterns of De Novo Mutation in Simplex Autism. Cell 171, 710–722.e12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuen RKC et al. Genome-wide characteristics of de novo mutations in autism. npj Genomic Med. (2016). doi: 10.1038/npjgenmed.2016.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuen RKC et al. Whole-genome sequencing of quartet families with autism spectrum disorder. Nat. Med 21, 185–191 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Michaelson JJ et al. Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell 151, 1431–1442 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang Y et al. Detection of clinically relevant genetic variants in autism spectrum disorder by whole-genome sequencing. Am. J. Hum. Genet 93, 249–63 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kong A et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature 488, 471–475 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Werling DM et al. An analytical framework for whole-genome sequence association studies and its implications for autism spectrum disorder. Nat. Genet 50, 727–736 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.An JY et al. Genome-wide de novo risk score implicates promoter variation in autism spectrum disorder. Science (80-. ). (2018). doi: 10.1126/science.aat6576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lek M et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernstein BE et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat. Biotechnol 28, 1045–8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou J & Troyanskaya OG Predicting effects of noncoding variants with deep learning-based sequence model. Nat. Methods 12, 931–4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ule J, Hwang H-W & Darnell RB The Future of Cross-Linking and Immunoprecipitation (CLIP). Cold Spring Harb. Perspect. Biol 10, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.1000 Genomes Project Consortium et al. A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kosmicki JA et al. Refining the role of de novo protein-truncating variants in neurodevelopmental disorders by using population reference samples. Nat. Genet (2017). doi: 10.1038/ng.3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aguet F et al. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krishnan A et al. Genome-wide prediction and functional characterization of the genetic basis of autism spectrum disorder. Nat. Neurosci 19, 1454–1462 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greene CS et al. Understanding multicellular function and disease with human tissue-specific networks. Nat. Genet 47, 569–576 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iossifov I et al. Low load for disruptive mutations in autism genes and their biased transmission. Proc. Natl. Acad. Sci 112, E5600–E5607 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Valente EM Hereditary Early-Onset Parkinson’s Disease Caused by Mutations in PINK1. Science (80-. ). 304, 1158–1160 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Packer A Neocortical neurogenesis and the etiology of autism spectrum disorder. Neuroscience and Biobehavioral Reviews 64, 185–195 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Kageyama R & Ohtsuka T The Notch-Hes pathway in mammalian neural development. Cell Res 9, 179–188 (1999). [DOI] [PubMed] [Google Scholar]
- 32.Bertrand N, Castro DS & Guillemot F Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci 3, 517–530 (2002). [DOI] [PubMed] [Google Scholar]
- 33.Crosnier C, Stamataki D & Lewis J Organizing cell renewal in the intestine: stem cells, signals and combinatorial control. Nat. Rev. Genet 7, 349–359 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Eckler MJ & Chen B Fez family transcription factors: Controlling neurogenesis and cell fate in the developing mammalian nervous system. BioEssays 36, 788–797 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hormozdiari F, Penn O, Borenstein E & Eichler EE The discovery of integrated gene networks for autism and related disorders. Genome Res. 25, 142–154 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saied-Santiago K & Bülow HE Diverse Roles for Glycosaminoglycans in Neural Patterning. Dev. Dyn (2017). doi: 10.1002/dvdy.24555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang W-H et al. Smek1/2 is a nuclear chaperone and cofactor for cleaved Wnt receptor Ryk, regulating cortical neurogenesis. Proc. Natl. Acad. Sci (2017). doi: 10.1073/pnas.1715772114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walsh CA, Morrow EM & Rubenstein JLR Autism and Brain Development. Cell 135, 396–400 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weiner D, Wigdor E, Ripke S & Robinson E Polygenic transmission disequilibrium confirms that common and rare variation act additively to create risk for autism spectrum disorders. Nat. Genet 49, 978–985 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y, Li B, Tan R, Zhu X & Wang Y A gradient-boosting approach for filtering de novo mutations in parent-offspring trios. Bioinformatics 30, 1830–1836 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smit A, Hubley R & Green P RepeatMasker Open-4.0. 2013–2015. http://www.repeatmasker.org (2013). [Google Scholar]
- 42.de Souza N The ENCODE project. Nat. Methods 9, 1046–1046 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Moore MJ et al. Mapping Argonaute and conventional RNA-binding protein interactions with RNA at single-nucleotide resolution using HITS-CLIP and CIMS analysis. Nat. Protoc 9, 263–293 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Darnell JC et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146, 247–261 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deciphering Developmental Disorders Study. Prevalence and architecture of de novo mutations in developmental disorders. Nature 542, 433–438 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wright CF et al. Genetic diagnosis of developmental disorders in the DDD study: A scalable analysis of genome-wide research data. Lancet (2015). doi: 10.1016/S0140-6736(14)61705-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cotney J et al. The autism-associated chromatin modifier CHD8 regulates other autism risk genes during human neurodevelopment. Nat. Commun (2015). doi: 10.1038/ncomms7404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sugathan A et al. CHD8 regulates neurodevelopmental pathways associated with autism spectrum disorder in neural progenitors. Proc. Natl. Acad. Sci. U. S. A (2014). doi: 10.1073/pnas.1405266111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harrow J et al. GENCODE: The reference human genome annotation for the ENCODE project. Genome Res. 22, 1760–1774 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Forrest ARR et al. A promoter-level mammalian expression atlas. Nature 507, 462–470 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yan Q et al. Systematic discovery of regulated and conserved alternative exons in the mammalian brain reveals NMD modulating chromatin regulators. Proc. Natl. Acad. Sci 112, 3445–3450 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gene Ontology Consortium. Gene Ontology Consortium: going forward. Nucleic Acids Res. 43, D1049–56 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mi H et al. PANTHER version 11: Expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 45, D183–D189 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Geifman N, Monsonego A & Rubin E The Neural/Immune Gene Ontology: Clipping the Gene Ontology for neurological and immunological systems. BMC Bioinformatics 11, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maaten L Van Der & Hinton G. Visualizing Data using t-SNE. J. Mach. Learn. Res 1 620, 267–84 (2008). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The code is available from https://hb.flatironinstitute.org/asdbrowser/help. ASD WGS data can be obtained from the Simons Foundation Autism Research Initiative (SFARI). All variant predicted scores have been made available as supplementary material and an interactive web interface is available at https://hb.flatironinstitute.org/asdbrowser/.