Abstract
Circular RNAs (circRNAs) are expressed at high levels in the brain and are involved in various CNS diseases. However, the potential role of circRNAs in ischemic stroke-associated neuronal injury remains largely unknown. Here, we investigated the important functions of circRNA TLK1 (circTLK1) in this process. The levels of circTLK1 were significantly increased in brain tissues in a mouse model of focal cerebral ischemia and reperfusion. Knockdown of circTLK1 significantly decreased infarct volumes, attenuated neuronal injury, and improved neurological deficits. Furthermore, circTLK1 functioned as an endogenous miR-335-3p sponge to inhibit miR-335-3p activity, resulting in the increase of 2,3,7,8-tetrachlorodibenzo-p-dioxin-inducible poly (ADP-ribose) polymerase expression and a subsequent exacerbation of neuronal injury. Clinical studies confirmed increased levels of circTLK1 in the plasma of patients with acute ischemic stroke (59 males and 12 females). Our findings reveal a detrimental role of circTLK1 in ischemic brain injury.
SIGNIFICANCE STATEMENT The extent of neuronal injury after brain ischemia is a primary factor determining stroke outcomes. However, the molecular switches that control the death of ischemic neurons are poorly understood. While our previous studies indicated the involvement of circRNAs in ischemic stroke, the potential role of circRNAs in neuronal injury remains largely unknown. The levels of circTLK1 were significantly increased in the brain tissue and plasma isolated from animal models of ischemic stroke and patients. Knockdown of circTLK1 significantly decreased infarct volumes, attenuated neuronal injury, and improved subsequent long-term neurological deficits. To our knowledge, these results provide the first definitive evidence that circTLK1 is detrimental in ischemic stroke.
Keywords: circular RNA TLK1, miR-335, neuronal injury, stroke, TIPARP
Introduction
Ischemic stroke is a common neurological disorder worldwide and a major cause of permanent disability (Murphy and Corbett, 2009; Dimyan and Cohen, 2011). Clinical treatments, such as tissue plasminogen activator-mediated thrombolysis, are often restricted by a narrow therapeutic time window (Goldstein, 2007) and insufficient long-term effects (George and Steinberg, 2015). The extent of neuronal injury after brain ischemia is a primary factor determining outcomes. Our current understanding of neuronal injury is based on experimental models and clinical studies. Multiple destructive mechanisms, including oxidative stress, cell apoptosis, excitotoxicity, and the inflammatory response, are responsible for neuronal damage (George and Steinberg, 2015). During the hours after ischemic stroke, neurons will become permanently damaged and undergo cell death (Lai et al., 2014). In addition to the loss of neurons located within the core area immediately affected by the loss of blood supply, neuronal loss is also observed in the surrounding area of the brain, resulting in a sudden loss of function (Rumpel, 2018). Therefore, therapeutic options that rescue damaged neurons are important; however, to date, only a few therapeutic agents have been reported to relieve neurological deficits after stroke (Buchli and Schwab, 2005). Novel therapeutic approaches that decrease neuronal injury are urgently needed (Cramer, 2018).
Circular RNAs (circRNAs), which are newly identified endogenous noncoding RNAs (ncRNAs), are characterized by back-splicing resulting from covalently closed continuous loops. Many circRNAs are abundantly expressed in the brain, as evidenced by the enrichment of circRim2, circEl2, and circDym in the cerebellum and high expression of circPlxnd1 in the cortex (Rybak-Wolf et al., 2015). Although their physiological functions are not yet completely defined, dynamic changes in the abundance of several circRNAs, such as circHomer1a, have been observed in the brain during the process of synapse formation in development (You et al., 2015). As shown in our previous studies, circHECTD1 aggravates ischemic damage by regulating astrocyte activation (Han et al., 2018b), and circDLGAP4 overexpression improves the blood–brain barrier integrity in subjects with stroke (Bai et al., 2018). In the present study, circRNA-mm9_circ_009932, also known as circTLK1, was upregulated during the acute period after focal ischemia (GEO accession number GSE115697; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE115697). circTLK1 is expressed in neurons, but its function remains unclear. A genome-wide bioinformatics analysis revealed that circTLK1 acts as a sponge for miR-335-3p. Moreover, 2,3,7,8-tetrachlorodibenzo-p-dioxin-inducible poly (ADP-ribose) polymerase (TIPARP), a predicted target of miR-335-3p, is upregulated in ischemic brain tissues. However, researchers have not determined whether the circTLK1/miR-335-3p/TIPARP axis is involved in cerebral ischemic injury, particularly neuronal injury.
Therefore, in this context, we aimed to investigate the role of circTLK1 in neuronal injury after transient middle cerebral artery occlusion (tMCAO) in adult mice. We hypothesized that upregulated circTLK1 binds to miR-335-3p and acts as an endogenous miR-335-3p sponge to increase the expression of miR-335-3p target genes, which results in neuronal injury and thus contributes to neurological deficits. Downregulation of circTLK1 expression significantly improved neurological deficits in tMCAO mice (Fig. 1). Our results reveal a detrimental role of circTLK1 in neuronal injury after brain ischemia.
Figure 1.
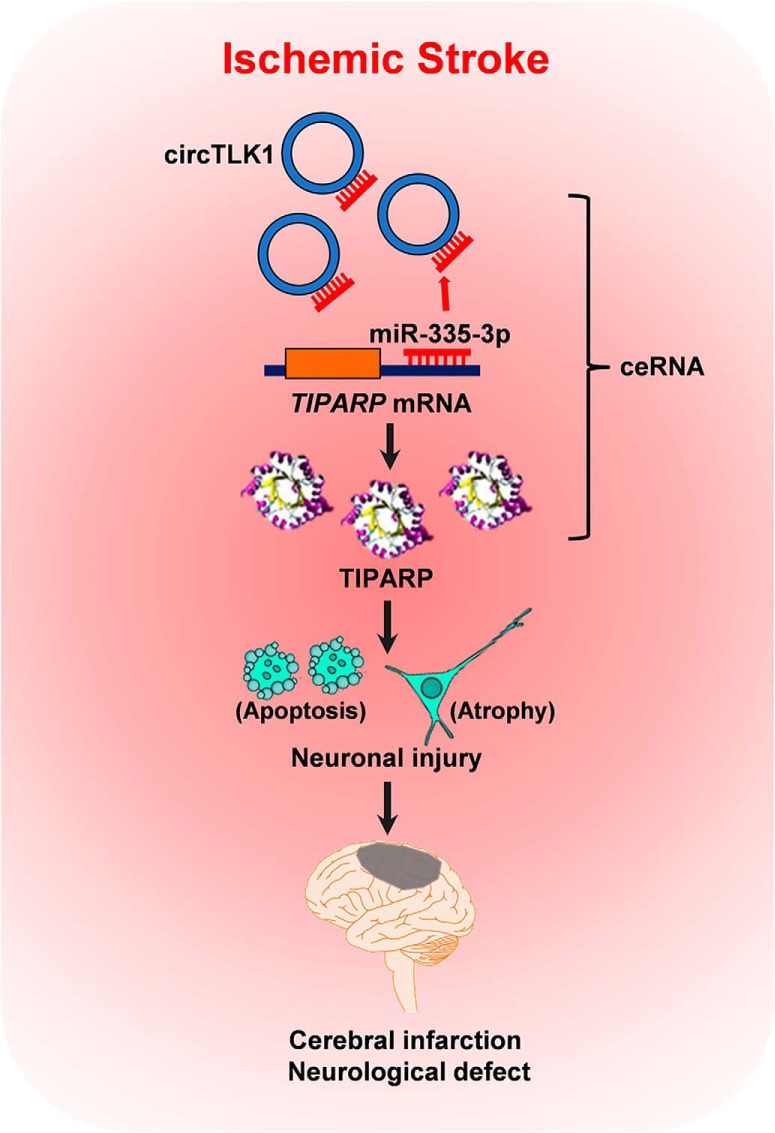
The proposed pathway by which circTLK1 regulates stroke progression. Upregulated circTLK1 directly binds to miR-335-3p and acts as an endogenous miR-335-3p sponge to inhibit miR-335-3p activity and concomitantly increase the expression of the downstream intermediate TIPARP, which results in neuronal injury and contributes to cerebral infarction and neurological deficit. ceRNA, competing endogenous RNA.
Materials and Methods
Study approval and patient recruitment.
The ethics committees at Jiangsu Province Hospital and Drum Tower Hospital approved this research protocol (approval IDs 2016-SR-235 and 2016-123-02, respectively). Participants or their legally authorized representatives provided written informed consent to participate in the study. Patients with acute ischemic stroke (AIS) were recruited from two academic stroke centers: the Emergency Department at the Affiliated Jiangsu Province Hospital from April 2016 to April 2017 and the Neurology Department at the Affiliated Drum Tower Hospital from March 2016 to August 2017.
Human plasma collection.
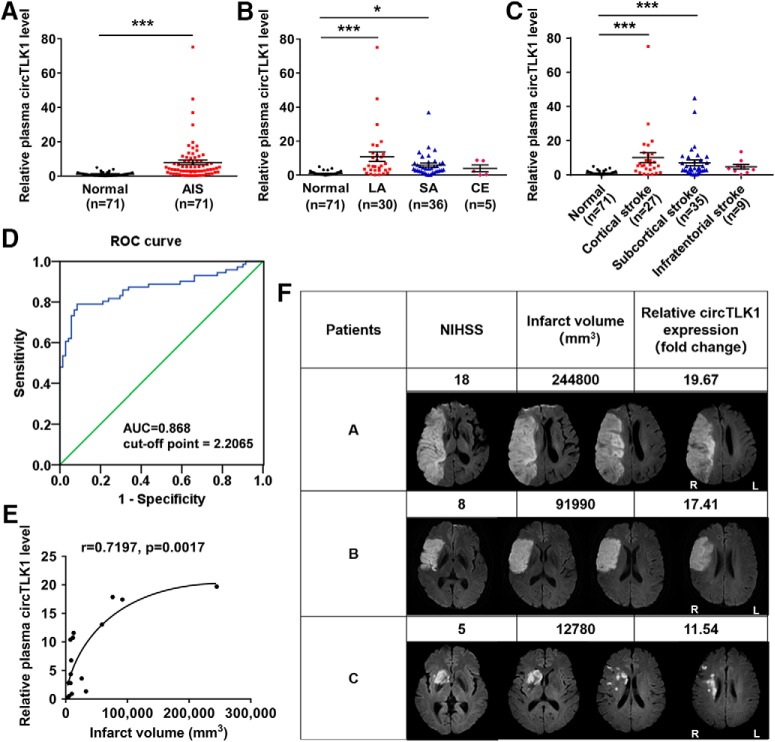
Seventy-one patients with ischemic stroke were recruited. The diagnosis of ischemic stroke was based on the clinical presentation in combination with a diffusion-weighted imaging (DWI) lesion on MRI or a new lesion on a delayed CT scan. Brain imaging (either CT or MRI) was performed routinely within 24 h of admission. Experienced neuroradiologists reviewed the imaging studies to confirm the diagnosis and used the National Institutes of Health Stroke Scale (NIHSS) to determine the severity of the patients' neurological deficits. Patients with recurrent stroke or a stroke onset longer than 48 h, intracerebral hemorrhaging, active malignant disease, hematological disease, inflammatory or infectious diseases, renal or liver failure, tumors, surgery within the past 3 months, prior treatment with low molecular weight or unfractionated heparin within the past month, or unknown diseases were excluded from the study.
Patients were classified into three groups by experienced neuroradiologists according to the Trial of Org 10172 in Acute Stroke Treatment classification: large-artery arteriosclerosis (LA), cardioembolism, and small-artery occlusion (Adams et al., 1993). In addition, the CT or MRI data were analyzed by experienced neuroradiologists who categorized the infarcts into three groups: cortical infarcts, subcortical infarcts, and infratentorial infarcts (Wardlaw et al., 2013; Tan et al., 2016). Patients with LA were recruited for infarct volume measurements. MRI with DWI data were available for 16 patients with LA (53.3%). DWI lesion volumes were determined by one experienced neurologist who was unaware of the patients' clinical and laboratory results. The infarct volume was calculated using the ABC/2 method (A and B represent the largest diameter of the infarct and its largest perpendicular diameter, respectively, whereas C represents the thickness of the slices with a visible infarct lesion) (Sims et al., 2009).
Nonstroke controls were recruited from patients who underwent an annual medical examination at the hospital. The demographic and clinical characteristics of the 71 patients and 71 nonstroke controls are provided in Table 1. The subjects were matched for age and gender. Stroke risk factors were defined as follows: (1) hypertension: blood pressure ≥ 140/90 mmHg; and (2) diabetes mellitus: fasting glucose level ≥ 7.0 mm or 2 h postprandial blood glucose level ≥ 11.1 mm.
Table 1.
Baseline characteristics of the study participantsa
| Characteristic | Control (n = 71) | AIS (n = 71) | p |
|---|---|---|---|
| Age (yr) | 68.56 ± 1.07 | 67.86 ± 1.30 | 0.676 |
| Male sex, n (%) | 59 (83.10) | 59 (83.10) | 1.000 |
| BMI (kg/m2) | 24.01 ± 0.26 | 24.48 ± 0.21 | 0.152 |
| Smoking, n (%) | 22 (30.99) | 33 (46.48) | 0.085 |
| Drinking, n (%) | 16 (22.53) | 24 (33.80) | 0.191 |
| Hypertension, n (%) | 35 (49.30) | 45 (63.38) | 0.128 |
| Diabetes mellitus, n (%) | 14 (19.72) | 25 (35.21) | 0.059 |
| Total cholesterol (mm) | 4.287 ± 0.12 | 4.15 ± 0.12 | 0.434 |
| Triglycerides (mm) | 1.36 ± 0.10 | 1.55 ± 0.08 | 0.136 |
| LDL (mm) | 2.54 ± 0.10 | 2.57 ± 0.10 | 0.872 |
| HDL (mm) | 1.12 ± 0.04 | 1.06 ± 0.04 | 0.234 |
| LPA (g/L) | 0.26 ± 0.03 | 0.36 ± 0.04 | 0.023 |
| NIHSS score | — | 29 (40.85%) | — |
| 1–4 | — | — | — |
| 5–15 | — | 34 (47.89%) | — |
| 16–20 | — | 7 (9.86%) | — |
| 21–42 | — | 1 (1.41%) | — |
aBMI, Body mass index; LDL, low-density lipoprotein; HDL, high-density lipoprotein; LPA: lipoprotein (a). Data are mean ± SEM. p values were calculated using Fisher's exact test or Student's t test, as appropriate.
Plasma samples were collected from patients who did not receive drug therapy. The mean time of the blood draw at enrollment was 19.6 ± 1.7 h after stroke onset. Peripheral blood samples (5 ml) were collected in EDTA-K2 anticoagulant tubes (BD Biosciences) immediately after patient admission and centrifuged at 2500 rpm for 25 min at 4°C. The prepared supernatant was then transferred to RNase/DNase-free tubes and stored at −80°C until further processing.
Animals.
Adult male C57BL/6J mice (24.0–26.0 g, 8–10 weeks old) were purchased from the Model Animal Research Center of Nanjing University (Nanjing, China) and randomly assigned to experimental groups. All animals were housed at a constant temperature and humidity under a 12 h light/12 h dark cycle, with the lights turned on at 7:00 A.M. Food and water were available ad libitum. All animal experiments were approved by the Institutional Animal Care and Use Committee at the Medical School of Southeast University (approval ID SYXK-2010.4987) and performed in accordance with the Animal Research: Reporting of In Vivo Experiments guidelines.
Lentiviral vector construction.
The plasmids pHBLV-U6-ZsGreen-Puro with circTLK1 shRNA (shRNA-circTLK1), miR-335-3p mimic, antago miR-335-3p (anti-miR-335-3p), TIPARP shRNA (shRNA-TIPARP), and their control (Con) sequences were purchased from Hanbio and inserted into lentiviruses. The shRNA sense sequences were shRNA-circTLK1, 5′-GAATCAGAGGTGCAATGGA-3′ and shRNA-TIPARP, 5′-GCCTCCTTCTCCTTCTGAT-3′.
Adeno-associated virus (AAV) shRNA vector construction.
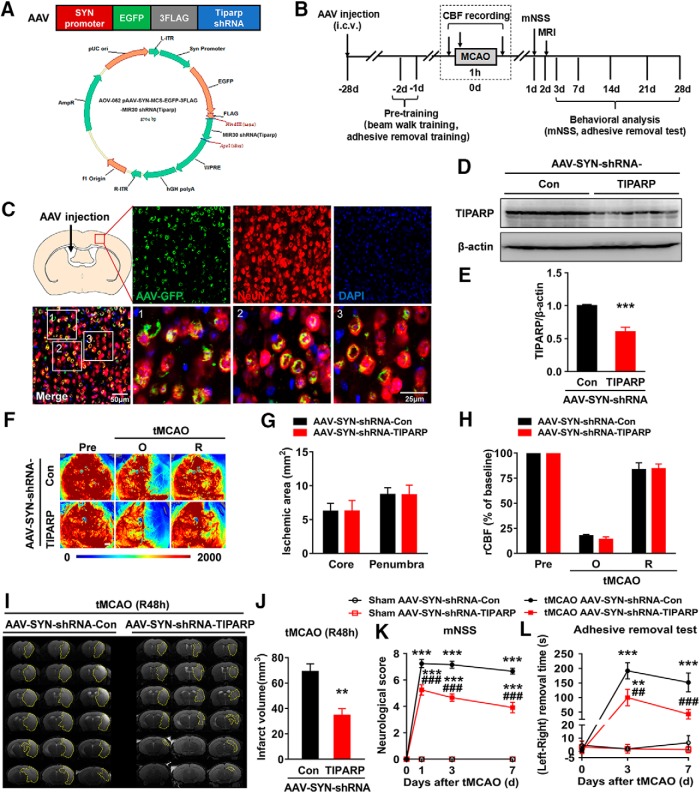
Three AAV vectors (AAV2/8 serotype), AAV-SYN-EGFP-shRNA-Con (AAV-SYN-shRNA-Con), AAV-SYN-EGFP-shRNA-circTLK1 (AAV-SYN-shRNA-circTLK1), and AAV-SYN-EGFP-shRNA-TIPARP (AAV-SYN-shRNA-TIPARP), were constructed and packaged by OBio. All viral particles express EGFP under the synapsin I (SYN) promoter (a neuron-specific promoter) flanked by the woodchuck hepatitis post-transcriptional regulatory element. In the AAV vector, the shRNA sequences were cloned into human miR-30-based stem loops via polymerase-mediated extension of overlapping DNA oligonucleotides. The packaging cassettes were flanked by inverted terminal repeats for the AAV cassettes. These sequences were also cloned into human miR-30-based stem loops through the polymerase extension of overlapping DNA oligonucleotides. For the cloning of shRNA vectors for AAV production, the following target sequences were used for RNAi: shRNA-circTLK1, 5′-GAATCAGAGGTGCAATGGA-3′ and shRNA-TIPARP, 5′-GCCTCCTTCTCCTTCTGAT-3′.
Microinjection of circTLK1 shRNA lentivirus, neuron-specific circTLK1 shRNA AAV, and neuron-specific TIPARP shRNA AAV.
The left lateral ventricle of 5-week-old C57BL/6J mice was microinjected with either the circControl shRNA-GFP (shRNA-Con) lentivirus or circTLK1 shRNA-GFP (shRNA-circTLK1) lentivirus (2 μl of 1 × 109 TU/ml, Hanbio) at the following microinjection coordinates: AP, −0.3 mm; lateral, 1.0 mm; and ventral, 2.2 mm. At 1 or 3 weeks after the microinjection, we divided the mice into the following groups to evaluate the effect of circTLK1 shRNA lentiviruses on the tMCAO model: sham + shRNA-Con, sham + shRNA-circTLK1, tMCAO + shRNA-Con, and tMCAO + shRNA-circTLK1.
For intracerebroventricular microinjections of AAV, the neuron-specific control shRNA-GFP AAV (AAV-SYN-shRNA-Con), circTLK1 shRNA-GFP AAV (AAV-SYN-shRNA-circTLK1), or TIPARP shRNA-GFP AAV (AAV-SYN-shRNA-TIPARP) (viral preparations for 4 × 1010 viral genomes/mouse in a volume of 1 μl, Obio) was infused into the left lateral ventricle at a rate of 0.1 μl/min, followed by 10 min of rest to allow diffusion. At 4 weeks after the microinjection, we divided the mice into the following 4 groups to evaluate the effect of neuron-specific circTLK1 shRNA AAV on the tMCAO model: sham + AAV-SYN-shRNA-Con, sham + AAV-SYN-shRNA-circTLK1, tMCAO + AAV-SYN-shRNA-Con, and tMCAO + AAV-SYN-shRNA-circTLK1. The same experiment was also designed to evaluate the effect of neuron-specific TIPARP shRNA AAV on the neurological outcomes in the tMCAO model.
tMCAO.
tMCAO was performed using methods described in a previous study (Stetler et al., 2008), with some modifications. Briefly, anesthesia was induced with 3% isoflurane and maintained with 1.5% isoflurane in 30% oxygen and 70% nitrous oxide using a face mask. The right external carotid artery was exposed, and a silicone rubber-coated 6–0 nylon filament (Doccol) was inserted and advanced 9–10 mm to the carotid bifurcation along the internal carotid artery and to the origin of the middle cerebral artery. One hour after the occlusion, the filament was removed to restore blood flow to the middle cerebral artery territory. Systolic blood pressure was monitored during surgery using a noninvasive tail-cuff (BP-2010, Softron Beijing Biotechnology). Arterial blood-gas was measured 15 min after the onset of reperfusion using a calibrated blood-gas analyzer, according to the manufacturer's instructions (Edan i15 VET, Edan Instrument). The rectal temperature was measured using a rectal probe and maintained at 37.0 ± 0.5°C during the surgery and recovery period with a temperature-controlled heating pad. Physiological variables were monitored in companion cohorts for all groups. In sham-operated mice, the external carotid artery was surgically prepared for insertion of the filament, but the filament was not inserted. Sham-operated mice served as controls.
Cerebral blood flow (CBF) measurements.
Cortical CBF was monitored using a moorFLPI-2 Full-Field Laser Perfusion Imager, according to the manufacturer's instructions (Moor Instruments). Briefly, a charge-coupled device camera was placed above the head of the anesthetized mouse, and the intact skull surface was illuminated with a laser diode (785 nm) to allow laser penetration through the brain in a diffuse manner. CBF was measured in both cerebral hemispheres and recorded 15 min before MCAO, throughout the ischemic period and up to 15 min after the onset of reperfusion. The animal was not exposed to direct sunlight or infrared radiation, and the room temperature was maintained at 26°C. We established an ROI, including the right cortical infarct region, which is posterior to the coronal suture and medial to the linear temporalis, to evaluate changes in CBF. Animals that did not show a reduction in CBF of at least 75% of the baseline level and animals that suddenly died immediately after ischemic induction (<10%) were excluded from further experimentation (Shi et al., 2016) since the probability of infarction is >95% if the early CBF is <25% of the control (Heiss et al., 2001). The areas of the ischemic core (0%–20% residual CBF) and the penumbra region (20%–30% residual CBF) were defined as described in previous studies (Leak et al., 2013; Shi et al., 2016) and were measured from laser speckle images.
In vivo MRI scanning.
In vivo MRI was performed using a 7.0 tesla small animal MRI scanner (Bruker PharmaScan) (Bai et al., 2014; Chang et al., 2015). The mice were anesthetized with 2% isoflurane delivered through a nose cone, and the respiratory rate and body temperature were monitored using a physiology monitor. T2-weighted imaging was conducted 24 h, 48 h, or 28 d after tMCAO using a 2D fast-spin echo sequence (2500/33 ms of repetition time/echo time, 1 average). Twelve axial slices with a slice thickness of 1 mm, a matrix of 256 × 256, and an FOV of 20 × 20 mm were positioned over the brain, excluding the olfactory bulb. MRIs were then scanned and quantified with National Institutes of Health ImageJ software (http://rsbweb.nih.gov/ij/), and the hemisphere volume was calculated as follows: ∑hemisphere area × slice thickness. The lesion volume was acquired based on the high signal area of the T2-weighted images, from which the artifact of brain edema was subtracted. The lesion volume was calculated as the volume of contralateral hemisphere minus the ipsilateral noninfarct hemisphere. Volume loss (brain atrophy) at 28 d after tMCAO was computed as the volume of the contralateral hemisphere minus the ipsilateral hemisphere.
Neurological deficits and behavioral tests.
Neurological deficits were evaluated 24 h after tMCAO. Neurological function was determined using the modified Neurological Severity Score test (Li et al., 2000) and performed by a researcher who was blinded to the experimental groups. The score was graded on a scale of 0–14 points (normal score, 0 points; maximum score, 14 points). One point was awarded for the inability to perform a test or for the lack of a tested reflex; thus, a higher score indicates a more severe injury.
Behavioral tests were performed before MCAO and 3, 7, 14, 21, and 28 d after tMCAO by an independent investigator who was blinded to the experimental groups. In addition, mice that died during the experiments were excluded from further data and statistical analyses. Mortality was assessed 28 d after the induction of tMCAO.
For the adhesive removal somatosensory test, all mice were familiarized with the testing environment. In the initial test, 2 small pieces of adhesive-backed paper dots (of equal size, 25 mm2) were used as bilateral tactile stimuli occupying the distal-radial region on the wrist of each forelimb. The time it took for the mice to remove each stimulus from the forelimb was recorded in three trials per day. Individual trials were separated by at least 5 min. Before surgery, the animals were trained for 3 d. Once the mice were able to remove the dots within 10 s, they were subjected to tMCAO.
The cylinder test encourages the use of the forelimbs for vertical wall exploration/press in a cylinder. Mice were placed in a plastic cylinder (15 cm tall with a diameter of 10 cm) and videotaped for 5 min.
Triphenyltetrazolium chloride staining and cerebral infarction measurement.
The infarct volume was evaluated 24 h after tMCAO. Each brain was coronally sectioned into five 1 mm slices with a brain matrix. The brain slices were incubated with 2% triphenyltetrazolium chloride (Sigma-Aldrich, T8877) at 37°C for 10 min to determine the size and extent of the infarction. Images were analyzed using ImageJ software. We determined the infarct volume by subtracting the volume of noninfarcted tissue in the ipsilateral hemisphere from the volume of noninfarcted tissue in the intact contralateral hemisphere to correct for brain edema. The infarct volume was calculated by summing the infarct volumes for all slices of each brain (Arumugam et al., 2006).
Immunostaining and analysis of images of sections.
Tissues were cut into 30 μm coronal slices with a cryostat. Sections were subsequently incubated with 0.3% Triton X-100 in PBS for 15 min and blocked with 10% normal goat serum (ZSGB-BIO, ZLI-0956) in 0.3% Triton X-100 for 1 h at room temperature. Next, sections were incubated with a rabbit anti-NeuN antibody (1:250, Proteintech Group, 26975-1-AP), rabbit anti-GFAP antibody (1:100, Proteintech Group, 16825-1-AP), or rabbit anti-Iba-1 antibody (1:500, Wako Pure Chemicals, 019-19741) to examine the colocalization with GFP-positive cells after microinjection, as well as a mouse anti-NeuN antibody (1:500, Abcam, ab104224) and a rabbit anti-TIPARP antibody (1:100, Abcam, ab170817) overnight at 4°C for double immunofluorescence staining. On the next day, sections were incubated with an AlexaFluor-594-conjugated goat anti-rabbit IgG (1:400, Invitrogen, A-11037), AlexaFluor-488-conjugated goat anti-rabbit IgG (1:400, Invitrogen, A-11034), or AlexaFluor-594-conjugated goat anti-mouse IgG (1:400, Invitrogen, A-11005) for 1 h. After a final washing step with PBS, sections were mounted onto glass slides. Images were captured using a microscope (Carl Zeiss, LSM700).
Immunohistochemistry and image analysis.
The 30-μm-thick coronal cryostat sections were incubated with a mouse anti-MAP2 antibody (1:250, Abcam, ab11267) overnight at 4°C and then incubated with HRP-conjugated goat anti-mouse IgG secondary antibody (1:2000, Cell Signaling Technology, 7076P2). Thereafter, the sections were incubated with the DAB chromogen (Vector Laboratories, SK-4100). Images were captured using a microscope (Olympus, DP73). The areas of brain atrophy, the contralateral hemisphere and ipsilateral hemisphere, as well as the total cross-sectional brain area were measured in seven equally spaced MAP2-stained sections encompassing the MCA territory by an observer who was blinded to the groups using the National Institutes of Health Image program, and areas were multiplied by the distance between sections to obtain the respective volumes. Volume loss (brain atrophy) at 28 d after tMCAO was computed as the volume of the contralateral hemisphere minus the ipsilateral hemisphere (Shi et al., 2016).
Golgi-Cox staining, imaging, and analysis.
Freshly dissected mouse brains were used for Golgi-Cox staining with the FD Rapid GolgiStain kit (FD Neuro Technologies) according to the manufacturer's protocol. Briefly, mouse brains were incubated in Golgi solution A + B for 7 d followed by 1 d incubation in solution C. Coronal sections (150 μm) were cut with a cryostat (CM1950, Leica Microsystems). Sections were then stained, dehydrated, and cleared with xylene according to the manufacturer's protocol.
Images of Golgi-stained neurons were acquired with a microscope in z-stack images (Carl Zeiss, ImagerM2). Only pyramidal neurons in the peri-infarct cortex (within 200 μm of the infarct) were included in our analyses (Tang et al., 2017). For a neuron to be selected, the following three criteria must be met: (1) the neuron was located in the peri-infarct cortex; (2) the neuron was distinct from neighboring neurons to allow dendrite identification; and (3) the neuron had consistent and dark staining throughout its structure (Gould et al., 1990). The computer-based cell tracing software Neurolucida 360 (MBF Bioscience) was used for 3D reconstruction of neurons. NeuroExplorer (MBF Bioscience) was used to measure the dendrite length and for Sholl and spine density analyses. The total length of dendrites was calculated as the sum of the dendritic length from one neuron. Sholl analysis was used to assess the complexity of neural dendrites by placing 3D concentric circles in 20 μm increments starting at 10 μm from the soma. The number of dendritic intersections with each ring was counted, and the results are reported as the number of intersections per radial distance from the soma and the total number of intersections. Twenty neurons from 4 animals per group were used for the Sholl analysis, which was performed in a blinded manner. We traced and measured the length of dendrites and counted the number of spines along the dendrites to calculate the spine density, according to the manufacturer's protocol. Spine density was calculated from the number of spines divided by the length of the dendrite. Fifty neurons from 6 animals per group were analyzed in a blinded manner.
Quantitative RNA sequencing.
Mice subjected to tMCAO were killed at 24 h and 28 d after tMCAO. Total RNA was extracted from the ischemic cortex of each mouse and used in RNA-Seq experiments. RNA-Seq experiments were performed according to the manufacturer's protocol, and data were analyzed by LC Biotech (http://www.lc-bio.com/). Briefly, total RNA was extracted using TRIzol reagent (Invitrogen, 15596026) according to the manufacturer's procedure. The total RNA quantity and purity were analysis of Bioanalyzer 2100 and RNA 1000 Nano LabChip Kit (Agilent Technologies) with RIN number > 7.0. Poly(A) RNA was purified from total RNA (5 μg) using poly-T oligo-conjugated magnetic beads using two rounds of purification. Following purification, the mRNA was fragmented into small pieces using divalent cations at a high temperature. Then, the cleaved RNA fragments were reverse-transcribed to generate cDNAs, which were subsequently used to synthesize U-labeled second-stranded DNAs with Escherichia coli DNA polymerase I, RNase H, and dUTP. An A-base was then added to the blunt ends of each strand and ligated to modified Illumina multiplex barcode adapters, including custom Unique Molecular Identifiers for minimizing sequence-dependent bias and amplification noise as described in a previous study (Shiroguchi et al., 2012). Size selection was performed with AMPureXP beads. After the heat-labile UDG enzyme treatment of the U-labeled second-stranded DNAs, the ligated products were amplified with PCR using the following conditions: initial denaturation at 95°C for 3 min; 8 cycles of denaturation at 98°C for 15 s, annealing at 60°C for 15 s, and extension at 72°C for 30 s; and then a final extension step at 72°C for 5 min. The average insert size for the final cDNA library was 300 bp (±50 bp). Finally, we performed paired-end sequencing on an Illumina Novaseq 6000 instrument (LC Biotech) according to the manufacturer's recommended protocol. Then, the expression levels of all transcripts were evaluated by calculating the fragments per kilobase per million reads. The threshold of significantly differential expression was set to p < 0.05 and |log2(fold change)| ≥ 1. The Kyoto Encyclopedia of Genes and Genomes (KEGG) database was used to explore the biological pathways in which the differentially expressed genes (DEGs) were involved. After KEGG annotation, the KEGG pathway enrichment analysis was performed.
Cell cultures.
Cultures of primary neurons from the cerebral cortex were obtained from C57BL/6J mouse embryos (E15-E16). Briefly, the cortices were dissected from mouse embryos and incubated with trypsin-EDTA (Invitrogen, 25200056) for 10 min; next, neurons were dissociated and plated at a density of 0.3 million cells per well in Neurobasal medium (Invitrogen, 21103049) supplemented with 0.5% GlutaMAX supplement (Invitrogen, 35050061) and 2% B27 supplement (Invitrogen, 17504044). Glial growth was suppressed by the addition of cytosine-β-d-arabinofuranoside (10 μm, Sigma-Aldrich, C1768) 1 d after plating, yielding cultured cells containing >90% neurons, as confirmed by staining for NeuN (neuron marker) and GFAP (astrocyte marker). The cells were maintained at 37°C in humidified air containing 5% CO2. The medium was changed every 3 d.
Primary mouse astrocytes and microglial cells were obtained from postnatal (P)1 to P2 C57BL/6J mice. The dissected brain cortices were placed in medium supplemented with PBS. After digestion with trypsin-EDTA, cells were plated on poly-L-lysine-precoated cell culture flasks containing DMEM supplemented with fetal FBS (10% v/v) and penicillin/streptomycin (1% v/v). Cultures were maintained in a humidified incubator (37°C, 5% CO2). For primary cortical astrocyte cultures, the astrocytes were harvested by trypsinization after 7–10 d of culture. For primary cortical microglia cultures, CSF2/GM-CSF (colony-stimulating factor 2 [granulocyte-macrophage]; 0.25 ng/ml) was added to the medium 7 d later to promote microglial proliferation. The microglia were detached from the flasks by shaking and collected from the cell medium by centrifugation at 1500 × g for 5 min.
HEK293T cells were grown in a 5% CO2 atmosphere in DMEM (Corning, 10-013-CVR) supplemented with FCS (10% v/v, Invitrogen, 10099-141) and penicillin/streptomycin (1% v/v, Invitrogen, 10378-016).
Lentiviral transduction of cortical neurons.
Neurons were transduced with the circControl shRNA, circTLK1 shRNA, miR-control, miR-335-3p, anti-miR-control, anti-miR-335-3p, control shRNA, or TIPARP shRNA lentiviruses (Hanbio) at 4 DIV. Cultured neurons were infected with lentiviruses at a multiplicity of infection of 5. Cotransductions were performed using two lentiviruses, each at a multiplicity of infection of 5. Infections were performed by directly adding lentiviruses to the growth medium. Infections proceeded for 7 d before oxygen-glucose deprivation treatment at 11 DIV.
Oxygen glucose deprivation/reperfusion (OGD/R) treatment.
In all in vitro experiments, neurons were used after 11 DIV. The OGD/R treatment was performed as previously described (Harms et al., 2004). Briefly, the cells were cultured with glucose-free deoxygenated DMEM (Invitrogen, 11966–025) in an incubator (Thermo Fisher Scientific) with a premixed gas (95% N2 and 5% CO2) for 2 h and then returned to 95% air, 5% CO2, and normal Neurobasal (B27) medium. Cultures in the control group were cultured with normal Neurobasal (B27) medium for the same incubation time. Neurons were subsequently collected for cell viability assays.
Cell viability assay.
The Cell Counting Kit 8 (CCK8, Dojindo, CK04) is widely used to assess cell viability. Neuronal injury was measured 6 h after OGD/R using the CCK8 assay according to the manufacturer's protocol. Cells were divided into three groups: (a) the blank control group, (b) the normal control group, and (c) the experimental group. The blank control group contained the Neurobasal medium, the normal control group contained neurons and Neurobasal medium, and the experimental group contained the lentivirus-treated neurons and Neurobasal medium. The experimental group was treated as described above. Cell viability is calculated according to the following formula: [(c − a)/(b − a)]. Values represent the fold changes in cell viability induced by OGD/R. The microplate reader was used to measure the OD450. Neuronal viability was estimated based on the optical density values.
Immunostaining and analysis of images of neuron cultures.
In the neuronal morphology experiments, neurons were fixed with 4% (w/v) PFA in PBS 1 week after the lentiviral infection and OGD/R treatment. The following primary antibodies were used: a mouse anti-MAP-2 antibody (1:250, Abcam, ab11267) and a mouse anti-SMI312 antibody (1:500, BioLegend, 837904). The secondary antibody was AlexaFluor-594-conjugated goat anti-mouse IgG (1:250, Invitrogen, A-11005). Prolong Gold anti-fade reagent containing DAPI (Southern Biotechnology, 0100–20) was applied to visualize the nuclei. Immunofluorescence images were captured using a microscope (Olympus, DP73). The NeuronJ and advanced Sholl analysis plug-ins in ImageJ were used to calculate the total dendritic length and dendritic complexity. Synapse images were captured using a microscope in z-stack images (Carl Zeiss, LSM700). Spine density was calculated as the number of spines divided by the dendritic length using the reconstructed images of LV-GFP-infected neuronal dendritic spines from ImageJ.
Real-time PCR.
Real-time PCR was performed according to our previous studies in an Applied Biosystems Real-Time PCR System (Yao et al., 2014). First, total RNA was extracted using TRIzol reagent (Invitrogen, 15596026). For mature miRNAs, the RNA was reverse-transcribed with a stem-loop RT primer (RiboBio) using a HiScript Q Select RT SuperMix for qPCR Kit (Vazyme, R133–01) and quantified using AceQ qPCR SYBR Green Master Mix (Vazyme, R141–02). U6 was used as an internal control. Bulge-loop miRNA qRT-PCR primer sets (one RT primer and a pair of qPCR primers for each set) specific for miR-335-3p, miR-142–3p, miR-129-5p, and miR-320-3p were designed by RiboBio. The circRNAs and mRNAs were reverse-transcribed using a HiScript Q RT SuperMix for qPCR Kit (Vazyme, R123–01) and quantified using SYBR Green Real-time PCR Master Mix (Vazyme, R141–02). GAPDH was used as an internal control. The primers used to amplify the circRNA, and mRNA transcripts were synthesized by Invitrogen. The sequences of the primers are listed in Table 2.
Table 2.
The primer sequences used for RT-PCR analyses
| List of oligonucleotide sequences | 5′-3′ |
|---|---|
| GAPDH (human)-forward | ACCATCTTCCAGGAGCGAGAT |
| GAPDH (human)-reverse | GGGCAGAGATGATGACCCTTT |
| GAPDH (mouse)-forward | AGGTCGGTGTGAACGGATTTG |
| GAPDH (mouse)-reverse | TGTAGACCATGTAGTTGAGGTCA |
| circKLHL2 (human)-forward | AGGTGAGATGAGTGAGAGCC |
| circKLHL2 (human)-reverse | CTGGGCAGTGTTTTTCGGTA |
| circTLK1 (human)-forward | ACAGTTTTGGAAGCTTGGGATCT |
| circTLK1 (human)-reverse | TGCTCCCACTTGCAACTCCA |
| circTLK1 (mouse)-forward | GACAAAGAATCAGAGGTGCAATGG |
| circTLK1 (mouse)-reverse | ACTCCCAGTACTCCCAGTAGC |
| TLK1 (mouse)-forward | CAGGGAAGGTGCAATGGATG |
| TLK1 (mouse)-reverse | AGCTTTTGCTCCAACACTGC |
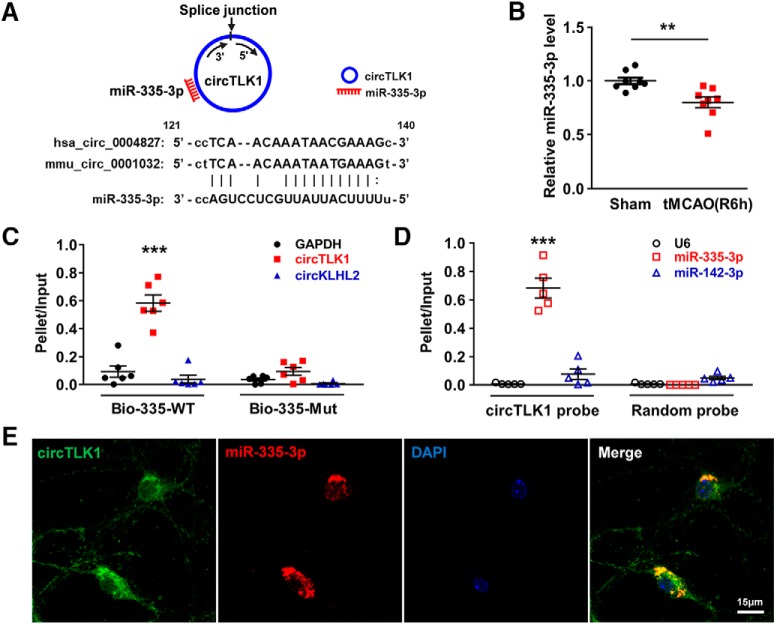
FISH.
As described in our previous study (Yao et al., 2014), neurons cultured on coverslips were fixed with 4% PFA and permeabilized with 0.25% Triton X-100 in PBS for 15 min. The samples were prehybridized in hybridization buffer (50% formamide, 10 mm Tris-HCl, pH 8.0, 200 μg/ml yeast tRNA, 1× Denhardt's solution, 600 mm NaCl, 0.25% SDS, 1 mm EDTA, and 10% dextran sulfate) for 1 h before an overnight incubation with hybridization buffer containing 50 nm biotin-labeled circTLK1 probes (Invitrogen) or 25 nm digoxigenin-labeled miR-335-3p probes (Invitrogen) at 37°C. On the second day, after several washing steps, the samples were blocked with a solution of 1% BSA and 3% NGS in PBS for 1 h at room temperature and then incubated with FITC-streptavidin (1:200, Invitrogen, 434311) and an anti-digoxigenin-poly-POD antibody (1:200, Roche Diagnostics, 11207733910) overnight at 4°C. After the coverslips were washed three times with TBS, signals were amplified by incubating the sections with a TSA Cy5 kit (PerkinElmer, NEL745001KT) for 10 min at room temperature. Then, the coverslips were mounted with Prolong Gold anti-fade reagent containing DAPI. Images were captured using a microscope (Carl Zeiss, LSM700).
FISH in combination with immunostaining was performed as described in our previous study (Zhang et al., 2016). After permeabilization and prehybridization in hybridization buffer, the brain sections or coverslips were incubated with hybridization buffer containing 50 nm biotin-labeled circTLK1 probes (Invitrogen) at 37°C overnight. After washes and blocking, samples were incubated with FITC-streptavidin (1:200, Invitrogen, 434311), a mouse anti-NeuN antibody (1:500, Abcam, ab104224), or a mouse anti-MAP2 antibody (1:250, Abcam, ab11267) overnight at 4°C. Then, the samples were incubated with AlexaFluor-594-conjugated goat anti-mouse IgG (1:250, Invitrogen, A-11005) and mounted with Prolong Gold anti-fade reagent containing DAPI. Immunofluorescence images were captured using a confocal microscope (Carl Zeiss, LSM700). The mouse circTLK1 probe sequence, which was also biotinylated at the 5′-end, was 5′-AGCTCATCCATTGCACCTCTGATTCTTTGTCA-3′. The digoxigenin-labeled miR-335-3p probe sequence was 5′-GGTCAGGAGCAATAATGAAAAA-3′.
Affinity isolation assay with biotinylated miRNA.
As described in our previous study (Han et al., 2018b), 2 × 106 HEK293T cells were seeded into plates 1 d before transfection. On the next day, HEK293T cells were transfected with 3′-biotinylated WT miR-335-3p (Bio-335-WT) or mutant miR-335-3p (Bio-335-Mut) at a final concentration of 50 nm for 36 h. Cells were incubated with lysis buffer [20 mm Tris, pH 7.5, 200 mm NaCl, 2.5 mm MgCl2, 0.05% Igepal (Sigma-Aldrich, 18896), 60 U/ml Superase-In (Invitrogen, AM2694), 1 mm DTT (Sigma-Aldrich, 43816), and protease inhibitors] on ice for 10 min. Lysates were precleared by centrifugation, and 50 μl aliquots of the samples were prepared as input. The remaining lysates were incubated with M-280 streptavidin magnetic beads (Invitrogen, 11205D). The beads were precoated with yeast tRNA (Invitrogen, 54016) to prevent the nonspecific binding of RNA and protein complexes. The beads were incubated at 4°C for 1.5 h and washed twice with ice-cold lysis buffer, twice with low-salt buffer (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl, pH 8.0, and 150 mm NaCl), and once with high-salt buffer (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl, pH 8.0, and 500 mm NaCl). The bound RNAs were purified with TRIzol to measure circTLK1, circKLHL2, and GAPDH levels. For the total input of each sample, cells were incubated with 500 μl of lysis buffer, and 50 μl of the supernatant of cell lysates was collected after centrifugation for RNA extraction. Then, 100 ng of RNA was used as the template for reverse transcription in 10 μl reaction system. Next, 2 μl of RT products was used for qPCR in a 20 μl reaction system. The 3′-biotinylated WT miR-335-3p sequence was 5′-UUUUUCAUUAUUGCUCCUGACC-3′. The 3′-biotinylated mutant miR-335-3p sequence was 5′-UGGCCAAGUAUUGCUCCUGACC-3′. The 3′-biotinylated miRNAs were all synthesized by GenePharma.
Affinity isolation assay with biotinylated DNA probes.
Due to the low background expression level of miR-335-3p in HEK293T cells, stably transfected cell lines were used in this experiment. Briefly, HEK293T cells were seeded and transfected with the miR-335-3p mimic lentivirus. After a 24 h incubation, the virus solution was removed and replaced with complete medium. At 72 h after transduction, we added puromycin to the medium at a concentration of 5 μg/ml to establish stably transfected cells. After antibiotic selection for 3 weeks, the blank control cells had all died, and the corresponding stably transfected cell lines were obtained.
As described in our previous study (Han et al., 2018b), a biotinylated DNA probe complementary to circTLK1 was synthesized and dissolved in 500 μl of wash/binding buffer (0.5 m NaCl, 20 mm Tris-HCl, pH 7.5, and 1 mm EDTA). The probe was incubated with M-280 streptavidin magnetic beads at 4°C for 3 h to generate probe-coated magnetic beads. HEK293T cell lysates were incubated with probe-coated beads at 25°C for 1.5 h; and after washing with the wash/binding buffer, the RNA complexes bound to the beads were eluted and extracted for real-time PCR analysis. Relative miR-335-3p, miR-142–3p, and U6 levels were measured. For the total input of each sample, cells were incubated with 500 μl of lysis buffer, and 50 μl of the supernatant of cell lysates was collected after centrifugation for RNA extraction. Then, 100 ng of RNA was used as the template for reverse transcription in a 10 μl reaction system. Next, 2 μl of RT products was used for qPCR in a 20 μl reaction system. The 5′-biotinylated sequence of the circTLK1 pull-down probe was 5′-AGCTCATCCATTGCACCTCTGATTCTTTGTCA-3′, and the 5′-biotinylated sequence of the random pull-down probe was 5′-AAAGCTGCTGCTAGTGCAGTGTCCGTACCAGTCGG-3′. These probes were all synthesized by Invitrogen.
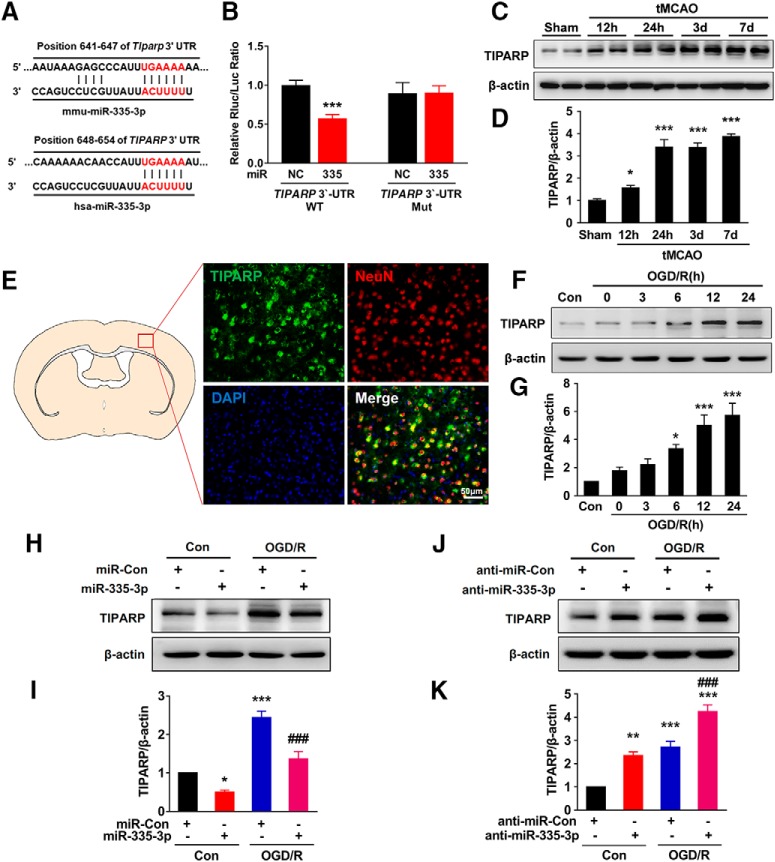
Luciferase activity assays.
The 3′-UTR of the 708 bp human TIPARP gene containing the putative miR-335-3p target site was PCR amplified from human genomic DNA using forward (5′-GCGGCTCGAGATGTATCAGCCAACCAGAG-3′) and reverse (5′-AATGCGGCCGCCCACAAACATTACCAGGAG-3′) primers, and the DNA fragment was cloned into the XhoI and NotI sites at the 3′-end of the luc2 gene in the pmiR-RB-REPORT vector (RiboBio). For the pmiR-RB-TIPARP-3′-UTR-miR-335-3p-target-mutant vector, the miR-335-3p target site (TGAAAA) within the TIPARP 3′-UTR was changed to ACTTTT via PCR-based mutagenesis with the primers TIPARP-miR-335-3p-F (5′-CAACCATTACTTTTATTTTTGTCACCAGCAAA-3′) and TIPARP-miR-335-3p-R (5′-ACAAAAATAAAAGTAATGGTTGTTTTTTGTTG-3′). Briefly, HEK293T cells were transfected with a miR-335-3p mimic (RiboBio) and a target plasmid, pmiR-RB-TIPARP-3′-UTR or pmiR-RB-TIPARP-3′-UTR-miR-335-3p-target-mutant, at a molar ratio of 50:1. An miRNA control was used as a negative control. Luciferase activity was determined at 24 h after transfection, and reporter assays were performed according to the manufacturer's protocol (Promega, E2920). Renilla luciferase activity was normalized to firefly luciferase activity and reported as a percentage of the control.
Western blot analysis.
As described in our previous studies (Yao et al., 2011; Bai et al., 2016), proteins were extracted with RIPA lysis buffer (Beyotime, P0013B), separated on SDS polyacrylamide gels (10% and 12%), and electrophoretically transferred onto PVDF membranes. Membranes were probed with antibodies recognizing TLK1 (1:1000, Proteintech Group, 13564–1-AP), Bax (1:1000, Cell Signaling Technology, 2772S), Bcl-xl (1:1000, Cell Signaling Technology, 2764S), caspase-3 (1:1000, Cell Signaling Technology, 9662S), PSD95 (1:1000, Abcam, ab76115), SYN (1:250, Santa Cruz Biotechnology, sc-8295), TIPARP (1:1,00, Abcam, ab170817), and β-actin (1:1000, CMCTAG, AT0001) overnight at 4°C. Membranes were then incubated with an HRP-conjugated goat anti-mouse/rabbit IgG secondary antibody (1:2000, Cell Signaling Technology, 7076P2/7074P2) and a donkey anti-goat IgG secondary antibody (1:5000, Abcam, ab6885). Signals were detected using chemiluminescence and imaged with a Microchemi 4.2 (DNR) digital image scanner. The individual protein bands were quantified using densitometry with ImageJ software.
Experimental design and statistical analysis.
All mice were male, and the experimental groups were age-matched. Power analysis software (J. Hintze, NCSS; www.ncss.com) was used to calculate the sample power required for the clinical study. The sample size required for the animal study was empirically determined based on the results of previous experiments and similar to the size generally used in the field.
Data are presented as mean ± SEM. p < 0.05 was considered to indicate a significant difference. Statistical analyses were performed using GraphPad Prism 6 Software. Shapiro–Wilk tests were used to assess the normality of the distribution for each group. Correlations were determined by calculating Pearson's correlation coefficients. Interaction effects were analyzed using nonlinear regression models. The mortality rate was determined using a χ2 analysis.
Significance was assessed with Student's t test (two-tailed) for comparisons of 2 groups. The differences in neurological deficits caused by tMCAO (R24 h), infarct volumes after tMCAO (R24 or 48 h), and brain atrophy after tMCAO (R28 d) between groups were examined using Student's t test (shRNA-Con-microinjected groups vs shRNA-circTLK1-microinjected groups, AAV-shRNA-Con-microinjected groups vs AAV-shRNA-circTLK1-microinjected groups, and AAV-shRNA-Con-microinjected groups vs AAV-shRNA-TIPARP-microinjected groups, respectively). The same analysis was also performed to examine differences between the sham and tMCAO groups, and between normal controls and patients with AIS. The expression of circTLK1 in shRNA-Con and shRNA-circTLK-microinjected groups in vivo, as well as shRNA-Con and shRNA-circTLK lentivirus-transduced groups in vitro, was examined using Student's t test, and the expression of TIPARP in cells transduced with shRNA-Con and shRNA-TIPARP lentivirus or AAV was also analyzed.
One- or two-way ANOVA followed by Holm–Sidak tests were used to compare 3 or more groups. Analyses of differences in circTLK1 or TIPARP expression between control and OGD/R-treated groups, and TIPARP expression between Sham and tMCAO mice were conducted with a one-way ANOVA followed by the Holm–Sidak test. Behavioral data collected at multiple repeating time points were analyzed using two-way repeated-measures ANOVA, followed by Holm–Sidak post hoc tests. The results were statistically significant if p < 0.05 according to the ANOVA. p values, the statistical analyses used, and sample sizes are described in the figure legends.
Results
Knockdown of circTLK1 expression reduces brain infarction in tMCAO mice
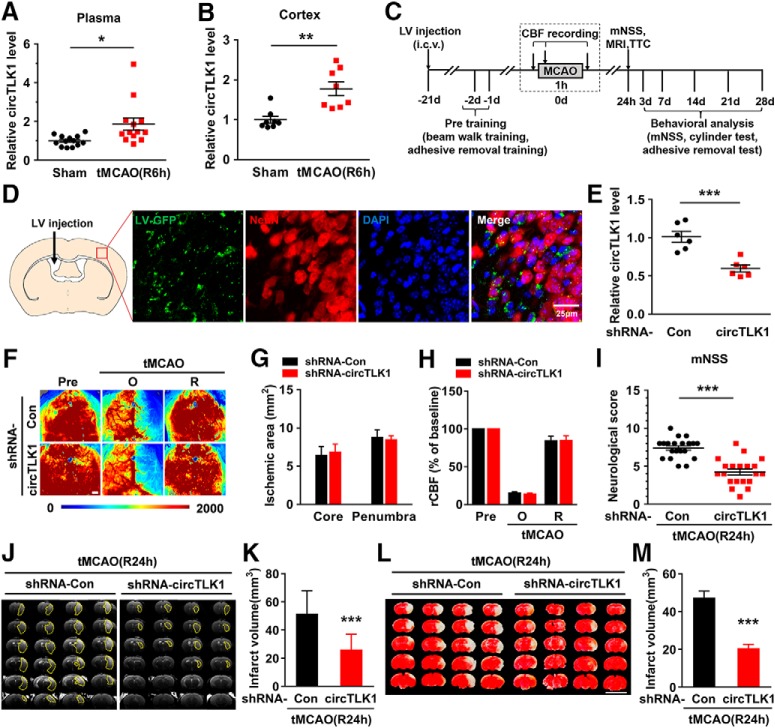
Our previous circRNA microarray study in an established tMCAO stroke model revealed significantly increased levels of 5 of 1178 circRNAs (by >1.5-fold) in ischemic tissues compared with the sham group (GEO accession number GSE115697; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE115697). circTLK1, which is derived from exons 2 and 3 of the TLK1 (tousled like kinase 1) gene, is one of these upregulated circRNAs (Fig. 2-1A). Using real-time PCR (Fig. 2-1B,C), we verified the expression of circTLK1 in different mouse organs, including the brain, heart, liver, spleen, lung, and kidney. As shown in Fig. 2-1D, circTLK1 was expressed at high levels in the brain, spleen, and lung.
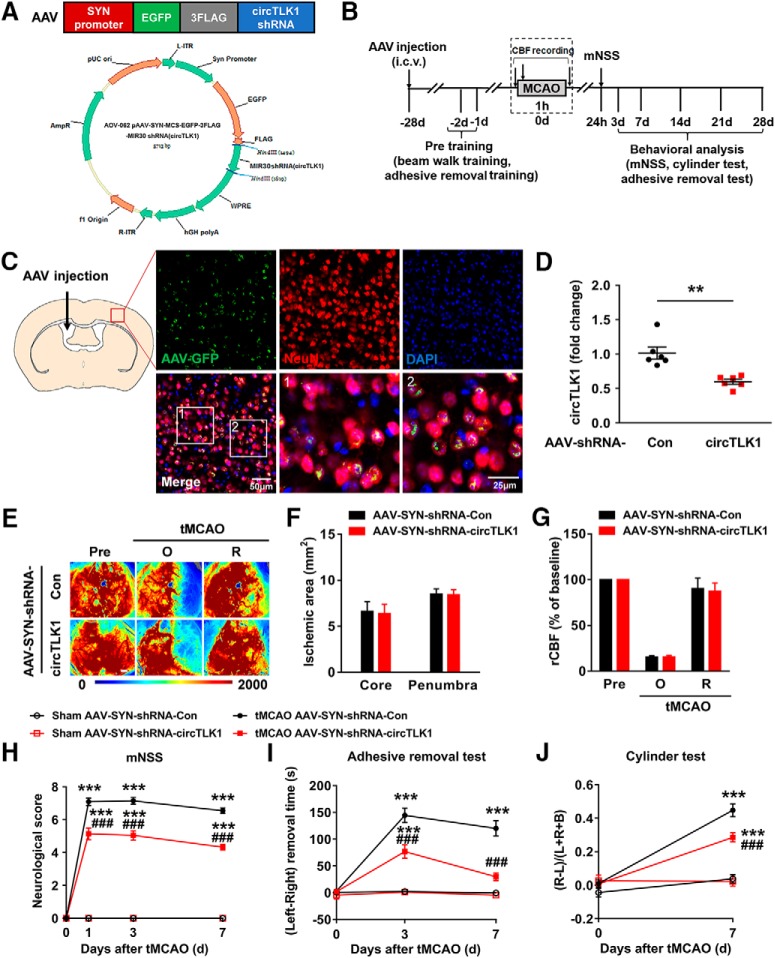
Consistent with the microarray data, we observed elevated levels of circTLK1 in mouse plasma (Fig. 2A) and the ipsilateral cortex (Fig. 2B) 6 h after tMCAO reperfusion. However, the levels of the TLK1 mRNA and protein in the ipsilateral cortex were significantly decreased 6 h after tMCAO (Fig. 2-2). Next, we sought to validate the role of circTLK1 in the pathogenesis of stroke in vivo. The lateral ventricles of mice were microinjected with either the circControl shRNA lentivirus (shRNA-Con) or circTLK1 shRNA lentivirus (shRNA-circTLK1) (Fig. 2C; Fig. 2-3). We first examined the efficacy of shRNA-circTLK1-GFP lentivirus transduction in vivo. GFP was widely expressed in the cortex 21 d after injection and colocalized with NeuN, a neuronal marker, in certain cells (Fig. 2D; Fig. 2-4). As expected, decreased circTLK1 expression was observed in circTLK1 shRNA-injected mice compared with circControl shRNA-injected mice (Fig. 2E). Three weeks after the lentivirus microinjections, the mice were subjected to tMCAO. Treatment with the circTLK1 shRNA lentivirus did not alter the CBF (Fig. 2F–H) or physiological parameters (Table 3) of these mice. The neurological deficits were significantly alleviated in the circTLK1 shRNA-injected mice compared with the circControl shRNA-injected mice after 24 h of reperfusion (Fig. 2I). The infarct size, as evaluated using MRI, was significantly decreased at 24 h after reperfusion in the circTLK1 shRNA-injected mice compared with the circControl shRNA-injected mice (Fig. 2J,K). We determined the brain infarct volume at 24 h after surgery using 2,3,5-triphenyltetrazolium chloride staining to verify this observation. The infarct volume was significantly decreased in the circTLK1 shRNA-injected mice than in the circControl shRNA-injected mice (Fig. 2L,M).
Figure 2.
Knockdown of circTLK1 expression reduces brain infarction in tMCAO mice. A, Relative plasma circTLK1 levels in tMCAO and sham mice at 6 h after reperfusion, as measured by real-time PCR. n = 13 animals per group. t(24) = 2.664. *p = 0.0136 (two-tailed t test). B, Relative circTLK1 levels in the ischemic cortex of tMCAO and sham mice at 6 h after reperfusion, as measured by real-time PCR. n = 8 animals per group. t(14) = 4.013. **p= 0.0013 (two-tailed t test). C, Illustration of the experimental procedure. D, Distribution of circTLK1 shRNA-GFP lentiviruses in the cortex after lentivirus microinjection. Representative images obtained 3 weeks after the microinjection of circTLK1 shRNA lentiviruses into the lateral ventricle. Green represents circTLK1 shRNA lentivirus-infected GFP-positive cells. Red represents the neuronal marker NeuN. Blue represents nuclei stained with DAPI. Scale bar, 25 μm. E, Relative expression of circTLK1 in the ipsilateral cortex 3 weeks after circTLK1 shRNA lentivirus microinjection, as determined by real-time PCR. n = 6 animals per group. t(10) = 4.910. ***p = 0.0006 (two-tailed t test). F–H, Regional CBF was monitored using 2D laser speckle imaging techniques 15 min before tMCAO (Pre), 15 min after the onset of tMCAO (O), and after 15 min of reperfusion (R). F, Representative images from the shRNA-Con and shRNA-circTLK1 groups. G, Ischemic areas measured from laser speckle images of either the ischemic core (0%–20% residual CBF) or the penumbra (20%–30% residual CBF) were not affected by the transduction of circTLK1 shRNA lentiviruses. H, Quantification of CBF. Data are presented as percentages of the baseline levels before ischemia (Pre). n = 6 animals per group. Scale bar, 1 mm. I, Neurological deficits were measured at 24 h after tMCAO by the modified Neurological Severity Score. n = 20 animals per group. t(38) = 6.453. ***p < 0.0001 (two-tailed t test). J, K, Representative T2-weighted MRI of shRNA-Con-microinjected and shRNA-circTLK1-microinjected tMCAO mice at 24 h after reperfusion. Dashed line indicates the infarct area. n = 13 or 14 animals per group. t(25) = 4.834. ***p < 0.0001 (two-tailed t test). L, M, Brain infarct volume at 24 h after tMCAO, as measured in triphenyltetrazolium chloride-stained coronal sections. n = 11 animals per group. t(20) = 6.694. ***p < 0.0001 (two-tailed t test). Scale bar, 1 cm. mNSS, Modified Neurological Severity Score. For genetic information and primer sequence of circTLK1, see Figure 2-1. For the expression of TLK1 in the ischemic cortex after tMCAO in mice, see Figure 2-2. For the structures and sequence of circTLK1 shRNA, see Figure 2-3. For the distribution of shRNA circTLK1-GFP lentiviruses in the cortex after lentivirus microinjection, see Figure 2-4. TTC, 2,3,5-triphenyltetrazolium chloride.
Table 3.
Physiological parameters and blood-gas analysis of micea
| LV-shRNA-Con |
LV-shRNA-circTLK1 |
AAV-SYN-shRNA-Con |
AAV-SYN-shRNA-circTLK1 |
AAV-SYN-shRNA-TIPARP |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sham (7) | tMCAO (7) | Sham (5) | tMCAO (7) | Sham (7) | tMCAO (7) | Sham (7) | tMCAO (7) | Sham (6) | tMCAO (7) | |
| Weight (g) | 23.73 ± 0.36 | 23.27 ± 0.61 | 24.16 ± 0.91 | 23.39 ± 0.55 | 23.66 ± 0.71 | 23.46 ± 0.35 | 23.77 ± 0.46 | 23.97 ± 0.77 | 23.25 ± 0.62 | 23.30 ± 0.97 |
| Body temperature (°C) | 36.73 ± 0.21 | 36.81 ± 0.17 | 36.80 ± 0.18 | 36.80 ± 0.21 | 36.76 ± 0.19 | 37.01 ± 0.09 | 36.87 ± 0.09 | 36.83 ± 0.10 | 36.68 ± 0.13 | 36.81 ± 0.16 |
| SBP (B) (mmHg) | 100.3 ± 5.2 | 94.3 ± 6.3 | 100.8 ± 4.7 | 98.6 ± 5.9 | 102.3 ± 3.6 | 107.7 ± 4.9 | 102.0 ± 2.1 | 98.2 ± 4.4 | 102.4 ± 4.1 | 97.2 ± 2.4 |
| SBP (A) (mmHg) | 95.9 ± 7.6 | 93.9 ± 6.5 | 108.8 ± 4.0 | 97.9 ± 3.5 | 99.7 ± 8.1 | 99.4 ± 5.3 | 104.0 ± 7.5 | 95.8 ± 4.1 | 99.2 ± 5.5 | 95.1 ± 4.5 |
| pH (mmHg) | 7.34 ± 0.03 | 7.34 ± 0.02 | 7.35 ± 0.02 | 7.35 ± 0.02 | 7.36 ± 0.01 | 7.35 ± 0.02 | 7.35 ± 0.03 | 7.35 ± 0.02 | 7.35 ± 0.02 | 7.35 ± 0.02 |
| pO2 (mmHg) | 177.71 ± 7.44 | 174.43 ± 8.44 | 165.80 ± 12.24 | 174.00 ± 15.10 | 167.14 ± 6.22 | 171.14 ± 14.37 | 170.30 ± 2.40 | 175.10 ± 7.08 | 169.33 ± 10.45 | 170.86 ± 4.57 |
| pCO2 (mmHg) | 37.71 ± 1.13 | 36.03 ± 2.40 | 38.76 ± 2.62 | 41.16 ± 1.44 | 40.46 ± 1.84 | 40.57 ± 2.72 | 39.89 ± 2.61 | 37.56 ± 3.34 | 37.67 ± 1.87 | 41.66 ± 2.48 |
aThe mice were subjected to 60 min of MCAO, followed by reperfusion. Body weight was measured 15 min after the onset of reperfusion or 15 min after the sham operation. Body temperature was controlled and maintained using a feedback temperature-control unit. Systolic blood pressure (SBP) was measured before ischemia (B) and 15 min after the onset of reperfusion (A). A total of 200 μl of arterial blood was collected 15 min after the onset of reperfusion for the blood-gas determination (pH, pO2, and pCO2). Data are mean ± SEM. The number of mice in each group is shown in parentheses. No significant differences in any of the parameters were observed between treatment groups.
Genetic information and primer sequence of circTLK1. (A) The exon composition of circTLK1. (B) The divergent primer of circTLK1 contains the back-splicing site. (C) Divergent primers amplified circTLK1 from cDNA, but not genomic DNA (gDNA). GAPDH, linear control. cDNA used for convergent primer was derived from RNA pretreated with RNase R before reverse transcription. (D) Relative expression of circTLK1 in different mouse organs. n = 4 animals per group. Download Figure 2-1, TIF file (229.5KB, tif)
Expression of TLK1 in the ischemic cortex after tMCAO in mice. (A) Relative expression of Tlk1 mRNA in the ipsilateral cortex 6 h after reperfusion in mice as determined by real-time PCR. n = 8 animals per group. t(14) = 2.574, *p = 0.0221, two-tailed t-test. (B and C) Western blot analysis of TLK1 expression in the ipsilateral cortex after tMCAO. (C) Bar graph showing a significant decrease in TLK1 in the ipsilateral cortex 6 h after reperfusion. n = 6 animals per group. t(10) = 3.371, **p = 0.0071, two-tailed t-test. Images of whole blots presented in the Fig. 2-2B were shown in Fig. 2-5. Download Figure 2-2, TIF file (765.6KB, tif)
Structures and sequence of circTLK1 shRNA. The siRNA sequences of circTLK1 contains the back-splicing site. Download Figure 2-3, TIF file (157.5KB, tif)
Distribution of shRNA circTLK1-GFP lentiviruses in the cortex after lentivirus microinjection. Representative images obtained three weeks after the microinjection of circTLK1 shRNA lentiviruses into the lateral ventricle. Green: GFP positive cells infected by circTLK1 shRNA lentiviruses. Red: the astrocyte marker GFAP (A), microglia marker Iba-1 (B), oligodendrocyte marker galactocerebroside (GalC) (C) and vascular cell marker caveolin-1 (D). Blue: nuclei stained with DAPI. Scale bar: 25 μm. (E) Quantification of the proportion of GFP positive cells co-localizing with NeuN+, GFAP+, Iba-1+, GalC+ or Caveolin-1+ profiles. At least 1500 cells were counted for the quantification from 3 different animals. Download Figure 2-4, TIF file (304.5KB, tif)
Images of whole blots presented in the Figure 2-2B. Download Figure 2-5, TIF file (148.3KB, tif)
circTLK1 knockdown ameliorates neurological deficits after focal ischemia in mice
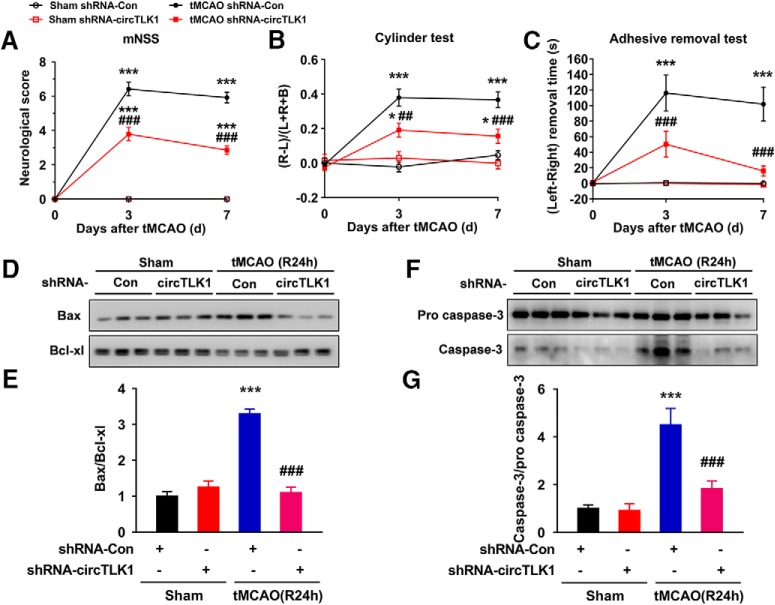
We next evaluated the role of circTLK1 in neuronal deficits after stroke. Neurological deficit scoring, an adhesive removal test, and a cylinder test were conducted before and 3 and 7 d after tMCAO. Microinjection of the circTLK1 shRNA lentivirus significantly ameliorated neurological deficits (Fig. 3A) and improved somatosensory functions (cylinder test and adhesive removal test) (Fig. 3B,C) after tMCAO. Consistent with these behavioral improvements, the circTLK1 shRNA lentivirus treatment also ameliorated the substantially decreased Bax/Bcl-xl ratio and caspase-3/pro caspase-3 ratio induced by tMCAO (Fig. 3D–G; Fig. 3-1).
Figure 3.
circTLK1 knockdown ameliorates neurological deficits after focal ischemia in mice. A, Neurological deficits were measured by calculating the modified Neurological Severity Scores at 3 and 7 d after tMCAO. n = 9, 9, 12, and 14 animals in the sham + shRNA-Con, sham + shRNA-circTLK1, tMCAO + shRNA-Con, and tMCAO + shRNA-circTLK1 groups, respectively. shRNA-circTLK1: F(3,40) = 133.8, p < 0.0001; time: F(2,80) = 176.9, p < 0.0001; interaction: F(6,80) = 69.42, p < 0.0001; subjects (matching): F(40,80) = 2.037, p = 0.0035. ***p < 0.001 versus the sham + shRNA-Con group. ###p < 0.001 versus the tMCAO + shRNA-Con group (two-way repeated-measures ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). B, Cylinder test of forelimb symmetry after stroke. L, Left; R, right; B, both forepaws. n = 9, 9, 12, and 14 animals in the sham + shRNA-Con, sham + shRNA-circTLK1, tMCAO + shRNA-Con, and tMCAO + shRNA-circTLK1 groups, respectively. shRNA-circTLK1: F(3,40) = 30.08, p < 0.0001; time: F(2,80) = 20.1, p < 0.0001; interaction: F(6,80) = 7.897, p < 0.0001; subjects (matching): F(40,80) = 0.8624, p = 0.6925. *p < 0.05 versus the sham + shRNA-Con group. ***p < 0.001 versus the sham + shRNA-Con group. ##p < 0.01 versus the tMCAO + shRNA-Con group (two-way repeated-measures ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). ###p < 0.001 versus the tMCAO + shRNA-Con group (two-way repeated-measures ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). C, Adhesive removal test of forelimb function at 3 and 7 d after tMCAO. n = 9, 9, 12, and 14 animals in the sham + shRNA-Con, sham + shRNA-circTLK1, tMCAO + shRNA-Con, and tMCAO + shRNA-circTLK1 groups, respectively. shRNA-circTLK1: F(3,40) = 21, p < 0.0001; time: F(2,80) = 12.62, p < 0.0001; interaction: F(6,80) = 6.75, p < 0.0001; subjects (matching): F(40,80) = 1.222, p = 0.2218. ***p < 0.001 versus the sham + shRNA-Con group. ###p < 0.001 versus the tMCAO + shRNA-Con group (two-way repeated-measures ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). D, E, Western blot analysis of Bax and Bcl-xl levels in the ischemic cortex at 24 h after tMCAO. Representative images are shown in D. E, Transduction with circTLK1 shRNA lentiviruses attenuated the tMCAO-induced increase in the Bax/Bcl-xl ratio. n = 6 animals per group. Bax/Bcl-xl (shRNA-circTLK1: F(1,20) = 41.81, p < 0.0001; tMCAO: F(1,20) = 51.37, p < 0.0001; interaction: F(1,20) = 66.52, p < 0.0001). ***p < 0.001 versus the sham + shRNA-Con group. ###p < 0.001 versus the tMCAO + shRNA-Con group (two-way ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). F, G, Western blot analysis of caspase-3 levels in the ischemic cortex at 24 h after tMCAO. Representative images are shown in F. G, Transduction of the circTLK1 shRNA lentiviruses attenuated the tMCAO-induced increase in the caspase-3/pro caspase-3 ratio. n = 6 animals per group. Caspase-3/pro caspase-3 (shRNA-circTLK1: F(1,20) = 10.98, p = 0.0035; tMCAO: F(1,20) = 28.35, p < 0.0001; interaction: F(1,20) = 9.811, p = 0.0052). ***p < 0.001 versus the sham + shRNA-Con group. ###p < 0.001 versus the tMCAO + shRNA-Con group (two-way ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). Images of whole blots presented in D, F are shown in Figure 3-1.
Images of whole blots presented in the Figure 3D and F. Download Figure 3-1, TIF file (97.7KB, tif)
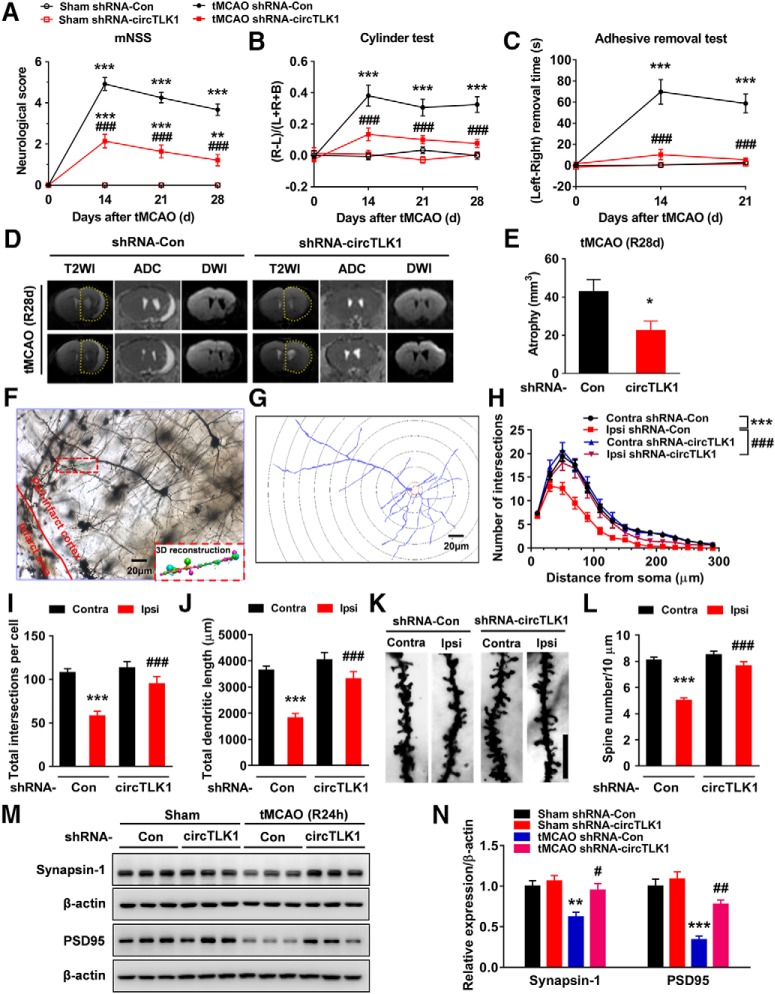
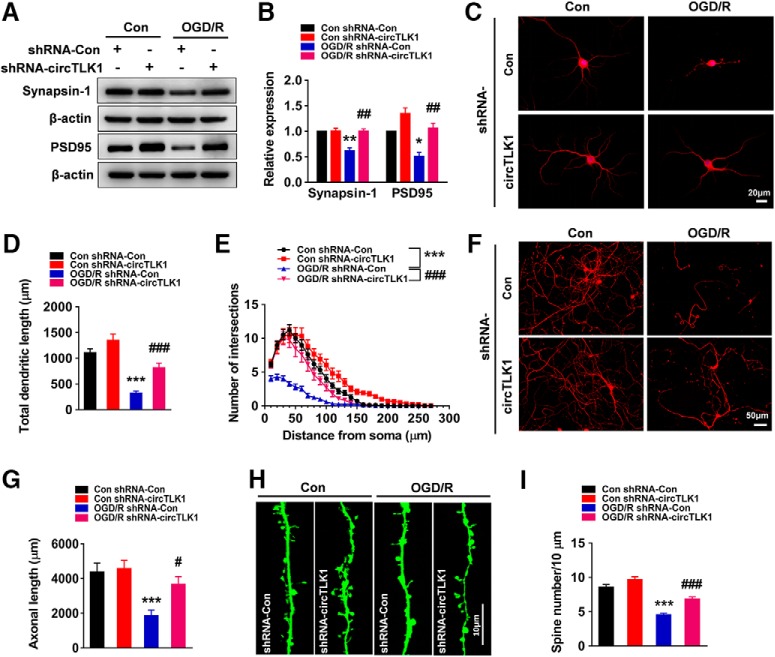
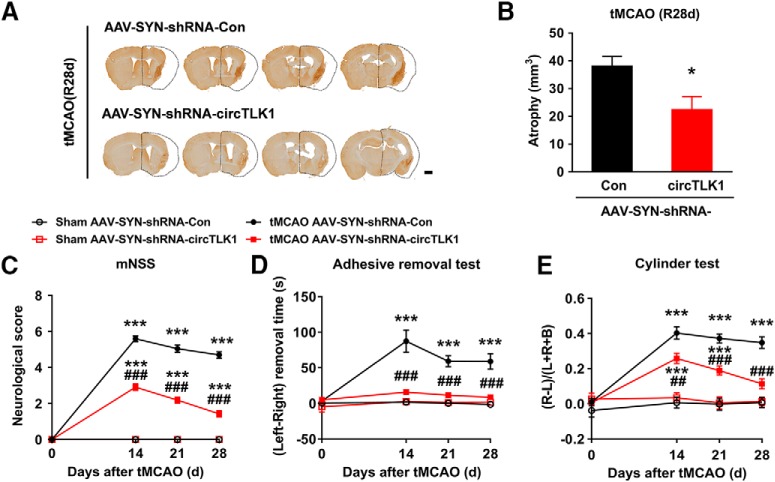
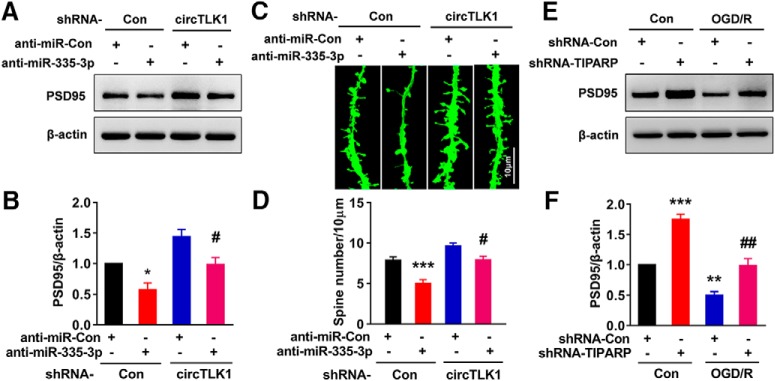
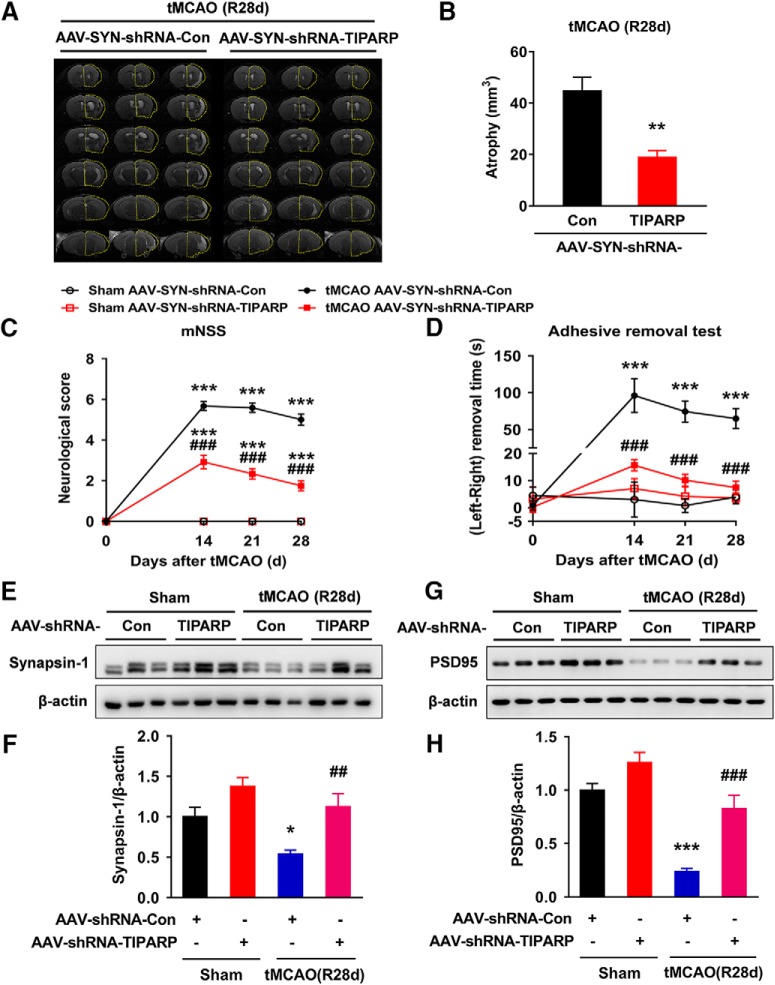
Furthermore, we continued to perform neurological deficit scoring, the adhesive removal test, and the cylinder test before and 14, 21, and 28 d after tMCAO. As shown in Figure 4A–C, microinjection of the circTLK1 shRNA lentivirus significantly ameliorated neurological deficits (Fig. 4A) and improved somatosensory functions (cylinder test and adhesive removal test) (Fig. 4B,C) after tMCAO. A significant difference in the mortality rate was not observed among the groups at 28 d after tMCAO (Fig. 4-1). Brain atrophy evaluated by MRI at 28 d after tMCAO was significantly reduced in the shRNA-circTLK1 lentivirus-injected mice with stroke compared with the circControl shRNA lentivirus-injected mice (Fig. 4D,E). Compared with the circControl shRNA lentivirus treatment, the circTLK1 shRNA lentivirus treatment significantly increased the dendritic complexity in the peri-infarct cortex at 28 d after tMCAO (Fig. 4F–J). Consistent with the behavioral changes, tMCAO significantly decreased the number of spines in the peri-infarct cortex of mice at 28 d after tMCAO, which was ameliorated by the shRNA circTKL1 treatment (Fig. 4K,L). Moreover, the levels of the synapse-associated proteins SYN and PSD95 were decreased in the tMCAO group, changes that were significantly alleviated by the circTLK1 shRNA lentivirus microinjection (Fig. 4M,N; Fig. 4-3).
Figure 4.
circTLK1 knockdown ameliorates long-term neuronal deficits and injury after focal ischemia in mice. A, Long-term neurological deficits were measured by calculating the modified Neurological Severity Scores at 14, 21, and 28 d after tMCAO. n = 9, 9, 12, and 14 animals in the sham + shRNA-Con, sham + shRNA-circTLK1, tMCAO + shRNA-Con, and tMCAO + shRNA-circTLK1 groups, respectively. shRNA-circTLK1: F(3,40) = 91.05, p < 0.0001; time: F(3,120) = 77.18, p < 0.0001; interaction: F(9,120) = 35.38, p < 0.0001; subjects (matching): F(40,120) = 3.281, p < 0.0001. **p < 0.01 versus the sham + shRNA-Con group. ***p < 0.001 versus the sham + shRNA-Con group. ###p < 0.001 versus the tMCAO + shRNA-Con group (two-way repeated-measures ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). B, Cylinder test of forelimb symmetry at 14, 21, and 28 d after tMCAO. L, Left; R, right; B, both forepaws. n = 9, 9, 12, and 14 animals in the sham + shRNA-Con, sham + shRNA-circTLK1, tMCAO + shRNA-Con, and tMCAO + shRNA-circTLK1 groups, respectively. shRNA-circTLK1: F(3,40) = 39.93, p < 0.0001; time: F(3,120) = 10.56, p < 0.0001; interaction: F(9,120) = 5.682, p < 0.0001; subjects (matching): F(40,120) = 1.073, p = 0.3755. ***p < 0.001 versus the sham + shRNA-Con group. ###p < 0.001 versus the tMCAO + shRNA-Con group (two-way repeated-measures ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). C, Adhesive removal test of forelimb function at 14 and 21 d after tMCAO. n = 9, 9, 12, and 14 animals in the sham + shRNA-Con, sham + shRNA-circTLK1, tMCAO + shRNA-Con, and tMCAO + shRNA-circTLK1 groups, respectively. shRNA-circTLK1: F(3,40) = 34.17, p < 0.0001; time: F(2,80) = 21.66, p < 0.0001; interaction: F(6,80) = 16.29, p < 0.0001; subjects (matching): F(40,80) = 1.89, p = 0.0080. ***p < 0.001 versus the sham + shRNA-Con group. ###p < 0.001 versus the tMCAO + shRNA-Con group (two-way repeated-measures ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). D, The T2WI, ADC, and DWI MRI data from the tMCAO mice at 28 d after reperfusion. Dashed lines indicate the relative area of the uninjured contralateral hemisphere to illustrate the area of ipsilateral atrophy for comparison. T2WI, T2-weighted imaging. ADC, apparent diffusion coefficient. E, Brain atrophy was measured in shRNA-Con lentivirus-microinjected and shRNA-circTLK1 lentivirus-microinjected tMCAO mice at 28 d after stroke from T2WI images. n = 5 animals per group. t(8) = 2.515. *p = 0.0361 (two-tailed t test). F–J, Representative images of z-stack projections of Golgi-Cox-stained neurons in the peri-infarct cortex at 28 d after tMCAO, followed by 3D reconstruction of spines (F, bottom right) and dendrites (G) as well as the Sholl analysis. H, Sholl analysis of the dendritic complexity of Golgi-stained neurons in the peri-infarct cortex at 28 d after tMCAO. Significant differences were observed at 50–130 μm from the soma. The average total numbers of intersections per cell are shown in I. Quantification of the total dendritic length per cell is shown in J. Twenty neurons from 4 animals per condition were analyzed. Total intersections (shRNA-circTLK1: F(1,76) = 10.18, p = 0.0021; Ipsi: F(1,76) = 25.56, p < 0.0001; interaction: F(1,76) = 5.613, p = 0.0204). Total dendritic length (shRNA-circTLK1: F(1,76) = 16.77, p = 0.0001; Ipsi: F(1,76) = 30.76, p < 0.0001; interaction: F(1,76) = 5.715, p = 0.0193). ***p < 0.001 versus the Contra group. ###p < 0.001 versus the shRNA-Con Ipsi group (two-way ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). Scale bar, 20 μm. Contra, Contralateral cortex; Ipsi, ipsilateral cortex. K, L, Images and bar graph represent the dendritic spine density in the peri-infarct cortex at 28 d after tMCAO. Fifty neurons from 6 animals per condition were analyzed. shRNA-circTLK1: F(1,196) = 36.49, p < 0.0001; Ipsi: F(1,196) = 61.24, p < 0.0001; interaction: F(1,196) = 20.10, p < 0.0001. ***p < 0.001 versus the Contra group. ###p < 0.001 versus the shRNA-Con Ipsi group (two-way ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). Scale bar, 10 μm. M, N, Western blot analysis of SYN and PSD95 levels in the ischemic cortex at 24 h after tMCAO. Representative images are shown in M. N, Transduction with circTLK1 shRNA lentiviruses attenuated the tMCAO-induced decrease in SYN and PSD95 levels. n = 6 animals per group. SYN (shRNA-circTLK1: F(1,20) = 7.835, p = 0.0111; tMCAO: F(1,20) = 12.47, p = 0.0021; interaction: F(1,20) = 3.659, p = 0.0702). PSD95 (shRNA-circTLK1: F(1,20) = 13.99, p = 0.0013; tMCAO: F(1,20) = 47.22, p < 0.0001; interaction: F(1,20) = 6.279, p = 0.0210). **p < 0.01 versus the sham + shRNA-Con group. ***p < 0.001 versus the sham + shRNA-Con group. #p < 0.05 versus the tMCAO + shRNA-Con group (two-way ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). ##p < 0.01 versus the tMCAO + shRNA-Con group (two-way ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). For the effect of shRNA-circTLK1 lentivirus microinjection on mortality after focal cerebral ischemia, see Figure 4-1. For the effect of poststroke knockdown of circTLK1 on neurological deficits after focal ischemia in mice, see Figure 4-2. Images of whole blots presented in M are shown in Figure 4-3.
Effect of shRNA-circTLK1 lentivirus microinjection on mortality after focal cerebral ischemia. (A) Animal survival rate from the same experiments was recorded daily for 28 d after tMCAO. n=9 animals for both sham groups, and n=17 animals for both tMCAO-treated groups. p = 0.1238 by log-rank test. (B) The mortality rate was calculated as the number of dead mice at 28 d after tMCAO in the shRNA-Con lentivirus and shRNA-circTLK1 lentivirus treated mice. Mice that died during the experiments were excluded from further data analyses and statistics. Download Figure 4-1, TIF file (1.1MB, tif)
Post-stroke knockdown of circTLK1 ameliorates neurological deficits after focal ischemia in mice. (A) Relative expression level of circTLK1 in ipsilateral cortex before (Pre) and 3, 7, 14 d after the shRNA-circTLK1 lentivirus injected into lateral ventricle. Microinjection of shRNA circTLK1 lentivirus significantly decreased the expression of circTLK1 at 14 d after microinjection. n = 6 animals per group. F (3, 20) = 2.556, P = 0.0841. *p < 0.05 versus the pre- microinjection group. (B) Illustration of the experimental procedure designed for C-G. Mice were microinjected with the shRNA circTLK1 lentivirus at 7 d before tMCAO, which will decrease the expression of circTLK1 at 7 d after tMCAO. (C) Animal survival rate from the same experiments was recorded daily for 28 d after tMCAO. n=11, 10, 30 and 30 animals for the sham + shRNA-Con, sham + shRNA-circTLK1, tMCAO + shRNA-Con and tMCAO + shRNA-circTLK1 groups, respectively. p = 0.0657 by log-rank test. (D) The mortality rate was calculated as the number of dead mice at 28 d after tMCAO in the tMCAO + shRNA-Con and tMCAO + shRNA-circTLK1 group. (E) Long-term neurological deficit was measured by mNSS at different time points. Neurological deficit scores were measured at 1, 3, 7, 14, 21 and 28 d after tMCAO. n = 11, 10, 21 and 24 animals for the sham + shRNA-Con, sham + shRNA-circTLK1, tMCAO + shRNA-Con and tMCAO + shRNA-circTLK1 groups, respectively. shRNA-circTLK1: F (3, 62) = 888.6, P < 0.0001; time: F (6, 372) = 286.3, P < 0.0001; interaction: F (18, 372) = 107.9, P < 0.0001; subjects (matching): F (62, 372) = 3.009, P < 0.0001. ***p < 0.001 versus the sham + shRNA-Con group; ###p < 0.001 versus the tMCAO + shRNA-Con group (two-way repeated measures ANOVA followed by Holm-Sidak post hoc multiple comparison tests). (F) Cylinder test of forelimb symmetry at 3, 7, 14, 21 and 28 d after tMCAO. L: left; R: right; B: both forepaws in F. n = 11, 10, 21 and 24 animals for the sham + shRNA-Con, sham + shRNA-circTLK1, tMCAO + shRNA-Con and tMCAO + shRNA-circTLK1 groups, respectively. shRNA-circTLK1: F (3, 62) = 66.44, P < 0.0001; time: F (5, 310) = 19.26, P < 0.0001; interaction: F (15, 310) = 8.377, P < 0.0001; subjects (matching): F (62, 310) = 1.226, P = 0.1357. ***p < 0.001 versus the sham + shRNA-Con group; ##p < 0.01 and ###p < 0.001 versus the tMCAO + shRNA-Con group (two-way repeated measures ANOVA followed by Holm-Sidak post hoc multiple comparison tests). (G) Adhesive removal test of forelimb function at different time points after stroke. Adhesive removal tests were performed at 3, 7, 14, 21 and 28 d after tMCAO. n = 11, 10, 21 and 24 animals for the sham + shRNA-Con, sham + shRNA-circTLK1, tMCAO + shRNA-Con and tMCAO + shRNA-circTLK1 groups, respectively. shRNA-circTLK1: F (3, 62) = 23.32, P < 0.0001; time: F (5, 310) = 9.857, P < 0.0001; interaction: F (15, 310) = 4.844, P < 0.0001; subjects (matching): F (62, 310) = 1.721, P = 0.0015. *p < 0.05, **p < 0.01 and ***p < 0.001 versus the sham + shRNA-Con group; ##p < 0.01 versus the tMCAO + shRNA-Con group (two-way repeated measures ANOVA followed by Holm-Sidak post hoc multiple comparison tests). Download Figure 4-2, TIF file (159.6KB, tif)
Images of whole blots presented in the Figure 4M. Download Figure 4-3, TIF file (230KB, tif)
Next, we further examined the efficacy of shRNA-circTLK1 lentivirus at different time points. Microinjection of the shRNA-circTLK1 lentivirus significantly decreased the expression of circTLK1 at 14 d after microinjection (Fig. 4-2A). In mice receiving microinjection of shRNA-circTLK1 lentivirus at 7 d before tMCAO, the expression of circTLK1 was reduced at 7 d after tMCAO (Fig. 4-2B). A significant difference in the mortality rate was not observed between the groups at 28 d after tMCAO (Fig. 4-2C,D). As shown in Fig. 4-2E–G, knockdown of circTLK1 expression after tMCAO significantly ameliorated the neurological deficits and improved somatosensory functions (cylinder test and adhesive removal test) at 14, 21, and 28 d after tMCAO.
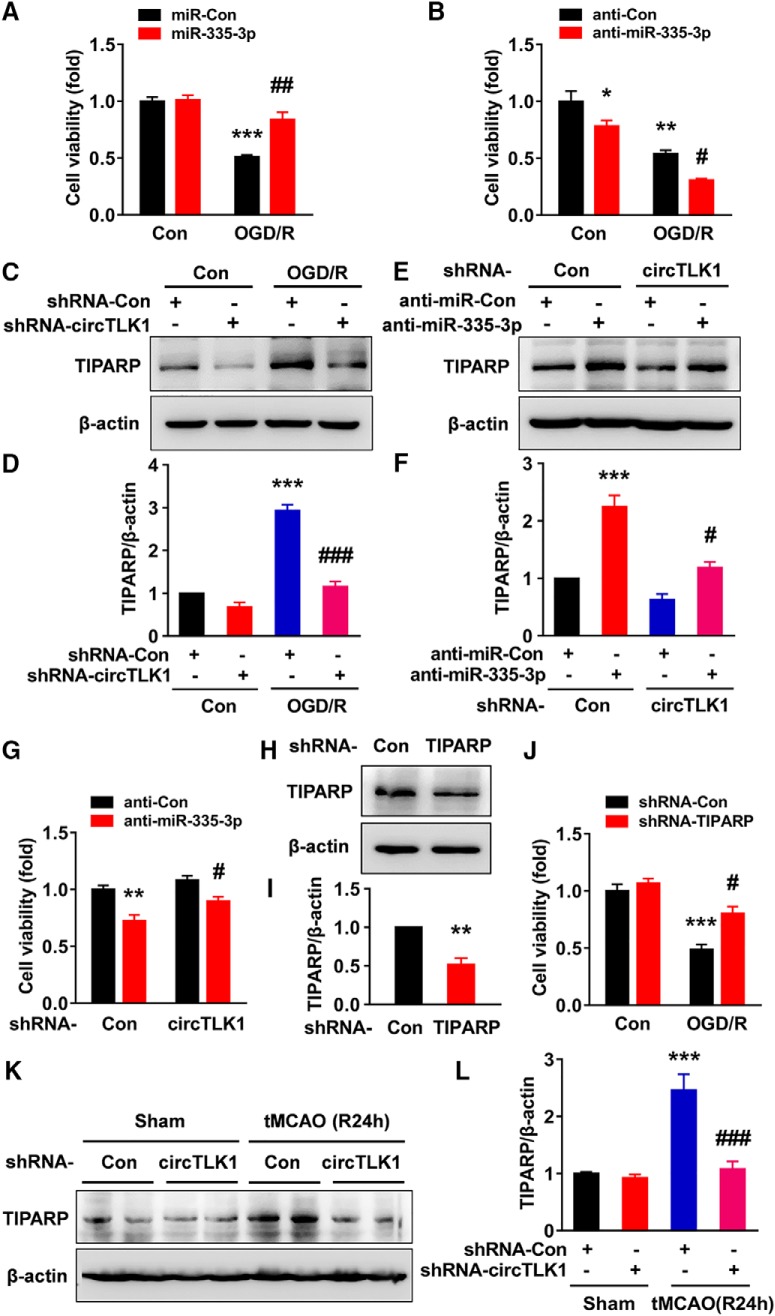
circTLK1 knockdown ameliorates neuronal injury in vitro
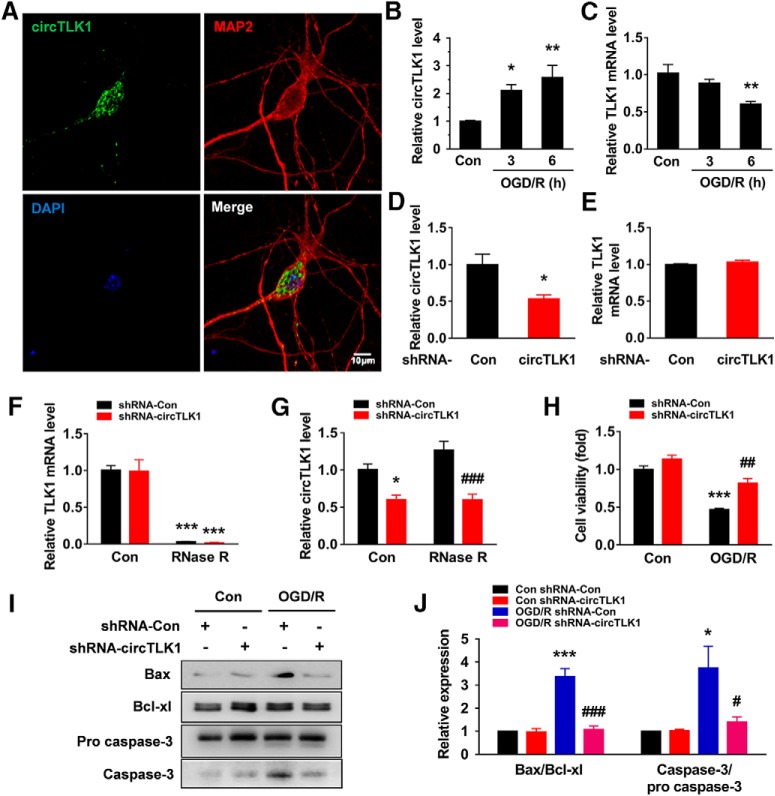
Having determined that circTLK1 plays a critical role in stroke, further experiments were needed to dissect the detailed mechanisms. First, we examined the localization of circTLK1 by in situ hybridization (Fig. 5-1A); circTLK1 colocalized with NeuN (Fig. 5-1B). This finding was further confirmed by examining the expression of circTLK1 in different types of primary cultured cells from the brain, including neurons, astrocytes, and microglia, using real-time PCR. As shown in Fig. 5-1C, circTLK1 was expressed at high levels in neurons. As shown in Figure 5A, circTLK1 was expressed in primary cultured cortical neurons. The expression of circTLK1 was significantly increased in primary cortical neurons treated with OGD/R (Fig. 5B). However, the expression of TLK1 mRNA in cortical neurons was significantly decreased after OGD/R (Fig. 5C). Transduction of primary cortical neurons with the circTLK1 shRNA lentivirus decreased the expression of circTLK1 (Fig. 5D), while no change in TLK1 mRNA expression was observed (Fig. 5E). We treated RNA samples with RNase R, which cleaves linear RNAs, to confirm that the transduction of the circTLK1 shRNA lentivirus only affected circTLK1. As shown in Figure 5F, the RNase R treatment resulted in a decrease in the level of linear TLK1 mRNA. However, transduction with circTLK1 shRNA decreased circTLK1 levels in cells treated with RNase R, confirming that the circTLK1 shRNA specifically targeted circTLK1 (Fig. 5G). We examined the effects of circTLK1 on OGD/R-induced neuronal survival. As shown in Figure 5H, the circTLK1 shRNA lentivirus treatment significantly inhibited the decrease in neuronal survival induced by the OGD/R treatment. Western blots revealed increases in the Bax/Bcl-xl ratio and the caspase-3/pro caspase-3 ratio at 3–24 h after OGD (Fig. 5-2A–C), and these effects were abolished by the circTLK1 shRNA at 6 h after OGD/R (Fig. 5I,J; Fig. 5-3).
Figure 5.
circTLK1 knockdown ameliorates neuronal injury in vitro. A, circTLK1 expression in primary cortical neurons. The outline of the neuronal soma was identified using an antibody against MAP-2. Green represents circTLK1. Red represents MAP-2. Blue represents DAPI. Scale bar, 10 μm. B, Relative expression of circTLK1 in primary neurons after OGD/R. The cells were treated with OGD for 2 h and subjected to reperfusion for 3 or 6 h. n = 5. F(2,12) = 8.299, p = 0.0055. *p < 0.05 versus the control group (one-way ANOVA followed by the Holm–Sidak test). **p < 0.01 versus the control group (one-way ANOVA followed by the Holm–Sidak test). C, Relative expression of the TLK1 mRNA in primary neurons after OGD/R. Cells were treated with OGD for 2 h and subjected to reperfusion for 3 or 6 h. n = 4. F(2,9) = 7.848, p = 0.0106. **p < 0.01 versus the control group (one-way ANOVA followed by the Holm–Sidak test). D, Relative expression of circTLK1 in primary cortical neurons after circTLK1 shRNA lentivirus transduction, as determined by real-time PCR. Cells were transduced with the circTLK1 shRNA lentivirus for 7 d, and then the expression of circTLK1 was measured. n = 6. t(10) = 3.034. *p = 0.0126 (two-tailed t test). E, Relative expression of the TLK1 mRNA in primary mouse cortical neurons after circTLK1 shRNA lentivirus transduction, as measured using real-time PCR. Cells were transduced with the circTLK1 shRNA lentivirus for 7 d, and the expression of the TLK1 mRNA was then measured. Transduction of the circTLK1 shRNA lentivirus did not alter levels of the TLK1 mRNA. n = 3. F, G, Total RNA was extracted from circControl and circTLK1 shRNA lentivirus-transduced neurons and incubated with or without RNase R, followed by real-time PCR. The relative expression levels of the TLK1 mRNA (F) and circTLK1 (G) are shown. n = 4. TLK1 mRNA (shRNA-circTLK1: F(1,12) = 0.02594, p = 0.8747; RNase R: F(1,12) = 137.8, p < 0.0001; interaction: F(1,12) = 8.15e-005; p = 0.9929). circTLK1 (shRNA-circTLK1: F(1,12) = 41.38, p < 0.0001; RNase R: F(1,12) = 2.387, p = 0.1483; interaction: F(1,12) = 2.495, p = 0.1402). *p < 0.05 versus the no RNase R-treated (control) shRNA-Con group. ***p < 0.001 versus the no RNase R-treated (control) shRNA-Con group. ###p < 0.001 versus the RNase R-treated shRNA-Con group (two-way ANOVA followed by the Holm–Sidak test). H, The circTLK1 shRNA lentivirus attenuated the OGD/R-induced increase in cell injury, as measured by the CCK8 assay. Cells were transduced with circTLK1 shRNA lentiviruses for 7 d, treated with OGD for 2 h, and subjected to reperfusion for 6 h. n = 3. shRNA-circTLK1: F(1,8) = 25.88, p = 0.0009; OGD/R: F(1,8) = 79.74, p < 0.0001; interaction: F(1,8) = 5.086, p = 0.0541. ***p < 0.001 versus the control shRNA-Con group. ##p < 0.01 versus the OGD/R-treated shRNA-Con group (two-way ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). I, J, The circTLK1 shRNA lentivirus attenuated the OGD/R-induced increase in the Bax/Bcl-xl ratio and caspase-3/pro caspase-3 ratio. Cells were transduced with circTLK1 shRNA lentiviruses for 7 d, treated with OGD for 2 h, and subjected to reperfusion for 6 h. n = 3. Bax/Bcl-xl (shRNA-circTLK1: F(1,8) = 31.85, p = 0.0005; OGD/R: F(1,8) = 35.80, p = 0.0003; interaction: F(1,8) = 30.48, p = 0.0006). Caspase-3/pro caspase-3 (shRNA-circTLK1: F(1,8) = 5.588, p = 0.0457; OGD/R: F(1,8) = 10.04, p = 0.0132; interaction: F(1,8) = 5.836, p = 0.0421). *p < 0.05 versus the control shRNA-Con group. ***p < 0.001 versus the control shRNA-Con group. #p < 0.05 versus the OGD/R-treated shRNA-Con group (two-way ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). ###p < 0.001 versus the OGD/R-treated shRNA-Con group (two-way ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). For the expression of circTK1 in the ischemic cortex and primary cell cultures in mice, see Figure 5-1. For the effect of OGD/R on neuronal injury in primary cortical neurons, see Figure 5-2. Images of whole blots presented in I are shown in Figure 5-3.
Expression of circTK1 in the ischemic cortex and primary cell cultures in mice. (A) Structures and probe sequence of circTLK1 used for FISH, the probe was circRNA specific (back splicing site). B: biotinylated. (B) Colocalization of NeuN and circTLK1 in the cortex of mice. The areas labeled with the different numbers in the rectangles are enlarged on the bottom or right side of the image. Green: circTLK1. Red: NeuN. Blue: DAPI. Scale bar: 25 μm. (C) Relative expression of circTLK1 in different cell types of primary cultures from the mouse cortex by qPCR. n = 3 samples per group. Download Figure 5-1, TIF file (127.8KB, tif)
Effect of OGD/R on neuronal injury in primary cortical neurons. (A) Western blot analysis of Bcl-xl, Bax, and caspase-3 expression after OGD/R. (B and C) Bar graph showing a significant increase in the Bax/Bcl-xl ratio (B) and the caspase-3/pro caspase-3 ratio (C) in neurons after OGD/R. n = 3. Bax/Bcl-xl: F (5, 12) = 9.409, P = 0.0008. Caspase-3/pro caspase-3: F (5, 12) = 6.014, P = 0.0052. *p < 0.05, **p < 0.01, and ***p < 0.001 versus the control group (one-way ANOVA followed by the Holm-Sidak test). (D) Western blot analysis of synapsin-1 and PSD95 expression after OGD/R. (E and F) Bar graph showing a significant decrease in synapsin-1 (E) and PSD95 (F) in neurons after OGD/R. n = 3. Synapsin-1: F (5, 12) = 130.3, P < 0.0001; PSD95: F (5, 12) = 62.03, P < 0.0001. *p < 0.05, **p < 0.01, and ***p < 0.001 versus the control group (one-way ANOVA followed by the Holm-Sidak test). Images of whole blots presented in the Fig. 5-2A and D were shown in Fig. 5-4. Download Figure 5-2, TIF file (422.7KB, tif)
Images of whole blots presented in the Figure 5I. Download Figure 5-3, TIF file (188.4KB, tif)
Images of whole blots presented in the Figure 5-2A and D. Download Figure 5-4, TIF file (359.5KB, tif)
In addition, we examined the effect of circTLK1 on the levels of SYN and PSD95. The levels of these proteins were decreased at 0–24 h after OGD (Fig. 5-2D–F). As shown in Figure 6A, B (Fig. 6-1) cells transduced with the circTLK1 shRNA lentivirus exhibited significant increases in SYN and PSD95 levels at 6 h after OGD/R. The neuroprotective effects of circTLK1 shRNA were also confirmed by immunostaining for MAP-2. As shown in Figure 6C–E, a marked loss of MAP-2 immunoreactivity and decreased length of neuronal processes were observed in OGD/R-treated neurons, and these effects were ameliorated by transduction with the circTLK1 shRNA lentivirus. Similarly, axonal staining with SMI-312 (a panaxonal neurofilament marker) revealed decreased axonal outgrowth at 6 h after OGD/R, which was rescued by circTLK1 knockdown (Fig. 6F,G). Cortical neurons were then exposed to OGD/R and subjected to confocal microscopy. Similar to the in vivo findings, OGD/R exposure resulted in a significant decrease in the density of dendritic spines, which was ameliorated by transduction of the circTLK1 shRNA lentivirus (Fig. 6H,I).
Figure 6.
Effect of circTLK1 on neuronal plasticity in primary cortical neurons after OGD/R. A, B, The circTLK1 shRNA lentivirus attenuated the OGD/R-induced decrease in SYN and PSD95 levels. Cells were transduced with the circTLK1 shRNA lentivirus for 7 d and subjected to OGD for 2 h and reperfusion for 6 h. n = 3. SYN (shRNA-circTLK1: F(1,8) = 20.70, p = 0.0019; OGD/R: F(1,8) = 19.67, p = 0.0022; interaction: F(1,8) = 18.67, p = 0.0025). PSD95 (shRNA-circTLK1: F(1,8) = 30.54, p = 0.0006; OGD/R: F(1,8) = 22.48, p = 0.0015; interaction: F(1,8) = 1.582, p = 0.2439). *p < 0.05 versus the control shRNA-Con group. **p < 0.01 versus the control shRNA-Con group. ##p < 0.01 versus the OGD/R-treated shRNA-Con group (two-way ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). C, Representative images of cultured cortical neurons. D, Total dendritic length per neuron. E, Morphological characteristics of dendrites from neurons subjected to OGD for 2 h and reperfusion for 6 h were measured by Sholl analysis. The circTLK1 shRNA lentivirus attenuated OGD/R-induced decreases in dendritic complexity. Significant differences were observed at 10–100 μm from the soma. Fifteen neurons from three independent experiments were quantified. Total dendritic length (shRNA-circTLK1: F(1,56) = 18.86, p < 0.0001; OGD/R: F(1,56) = 60.25, p < 0.0001; interaction: F(1,56) = 2.297, p = 0.1352). ***p < 0.001 versus the control shRNA-Con group. ###p < 0.001 versus the OGD/R-treated shRNA-Con group (two-way ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). Scale bar, 20 μm. F, G, Images (F) and bar graph (G) represent axonal outgrowth in primary cortical neurons after OGD/R. Axon length was measured 6 h after OGD. Fifteen images from three independent experiments were quantified. shRNA-circTLK1: F(1,56) = 5.168, p = 0.0269; OGD/R: F(1,56) = 15.12, p = 0.0003; interaction: F(1,56) = 3.308, p = 0.0743. ***p < 0.001 versus the control shRNA-Con group. #p < 0.05 versus the OGD/R-treated shRNA-Con group (two-way ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). Scale bar, 50 μm. H, I, Images (H) and bar graph (I) represent the dendritic spine density of representative neurons. Twenty-six to 34 neurons from five independent experiments were quantified. shRNA-circTLK1: F(1,120) = 23.11, p < 0.0001; OGD/R: F(1,120) = 93.68, p < 0.0001; interaction: F(1,120) = 2.780, p = 0.0980. ***p < 0.001 versus the control shRNA-Con group. ###p < 0.001 versus the OGD/R-treated shRNA-Con group (two-way ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). Scale bar, 10 μm. Images of whole blots presented in A are shown in Figure 6-1.
Images of whole blots presented in the Figure 6A. Download Figure 6-1, TIF file (325.3KB, tif)
Neuron-specific knockdown of circTLK1 improves neurological deficits after stroke in mice
We used neuron-specific AAV (AAV2/8 serotype) with an SYN promoter carrying the shRNA to inhibit the expression of circTLK1 in neurons and confirm the effect of circTLK1 on neuronal injury induced by ischemic stroke in vivo. AAV-SYN-shRNA-circTLK1 was microinjected into the lateral ventricle to selectively inhibit the expression of circTLK1 in neurons. The circControl-GFP AAV (AAV-SYN-shRNA-Con) was used as the control (Fig. 7A). The mice were monitored to determine the role of circTLK1 in stroke pathogenesis (Fig. 7B). GFP was widely expressed and colocalized with NeuN-positive cells, but not other cell types, such as astrocytes, oligodendrocytes, microglial cells, and endothelial cells (Fig. 7C; Fig. 7-1), indicating successful AAV-SYN-shRNA-circTLK1 transduction in neurons. Decreased expression of circTLK1 was observed in shRNA-circTLK1-injected mice compared with AAV-SYN-shRNA-Con-injected mice (Fig. 7D). Treatment with the neuron-specific circTLK1 shRNA AAV did not alter the CBF (Fig. 7E–G) or physiological parameters (Table 3) of these mice. Next, we investigated whether neuronal-specific knockdown of circTLK1 resulted in functional improvements in mice with stroke. Microinjection of the circTLK1 shRNA AAV resulted in a significant recovery of neurological deficits (Fig. 7H) and somatosensory and sensorimotor functions (Fig. 7I,J) after tMCAO.
Figure 7.
Neuron-specific knockdown of circTLK1 improves neurological deficits after stroke in mice. A, Structure of the AAV virus (AAV2/8 serotype) used in the experiments presented in B–J. B, Experimental procedure shown in C–J. C, The distribution of shRNA-circTLK1-GFP-AAV in the cortex after microinjection. Representative images obtained 4 weeks after the microinjection of the circTLK1 shRNA into the lateral ventricle. Green represents GFP-circTLK1-shRNA-AAV. Red represents the neuronal marker NeuN. Blue represents nuclei stained with DAPI. Scale bar, 25 or 50 μm. D, The expression of circTLK1 in the ipsilateral cortex at 28 d after the circTLK1-shRNA-AAV microinjection was determined by real-time PCR. n = 6 animals per group. t(10) = 4.438. **p = 0.0013 (two-tailed t test). E–G, Regional CBF was monitored using 2D laser speckle imaging techniques 15 min before tMCAO (Pre), 15 min after the onset of tMCAO (O), and after 15 min of reperfusion (R). E, Representative images of the AAV-SYN-shRNA-Con and AAV-SYN-shRNA-circTLK1 groups. F, Ischemic areas measured from laser speckle images of either the ischemic core (0%–20% residual CBF) or penumbra (20%–30% residual CBF) were not affected by AAV-SYN-shRNA-circTLK1. G, Quantification of CBF. Data are presented as percentages of the baseline levels before tMCAO (Pre). n = 6 animals per group. Scale bar, 1 mm. H, Neurological deficits were measured by the modified Neurological Severity Scores at different time points. Neurological deficit scores measured at 1, 3, and 7 d after tMCAO. n = 10, 10, 20, and 21 animals in the sham + AAV-SYN-shRNA-Con, sham + AAV-SYN-shRNA-circTLK1, tMCAO + AAV-SYN-shRNA-Con, and tMCAO + AAV-SYN-shRNA-circTLK1 groups, respectively. AAV-shRNA-circTLK1: F(3,57) = 286.5, p < 0.0001; time: F(3,171) = 288.2, p < 0.0001; interaction: F(9,171) = 104.5, p < 0.0001; subjects (matching): F(57,171) = 3.246, p < 0.0001. ***p < 0.001 versus the sham + AAV-SYN-shRNA-Con group. ###p < 0.001 versus the tMCAO + AAV-SYN-shRNA-Con group (two-way repeated-measures ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). I, Adhesive removal test of forelimb function at different time points after stroke. The adhesive removal test was performed at 3 and 7 d after tMCAO. n = 10, 10, 20, and 21 animals in the sham + AAV-SYN-shRNA-Con, sham + AAV-SYN-shRNA-circTLK1, tMCAO + AAV-SYN-shRNA-Con, and tMCAO + AAV-SYN-shRNA-circTLK1 groups, respectively. AAV-shRNA-circTLK1: F(3,57) = 52.77, p < 0.0001; time: F(2,114) = 30.23, p < 0.0001; interaction: F(6,114) = 14.02, p < 0.0001; subjects (matching): F(57,114) = 1.092, p = 0.3417. ***p < 0.001 versus the sham + AAV-SYN-shRNA-Con group. ###p < 0.001 versus the tMCAO + AAV-SYN-shRNA-Con group (two-way repeated-measures ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). J, Cylinder test of forelimb symmetry at 7 d after stroke. L, Left; R, right; B, both forepaws in J. n = 10, 10, 20, and 21 animals in the sham + AAV-SYN-shRNA-Con, sham + AAV-SYN-shRNA-circTLK1, tMCAO + AAV-SYN-shRNA-Con, and tMCAO + AAV-SYN-shRNA-circTLK1 groups, respectively. AAV-shRNA-circTLK1: F(3,57) = 24.03, p < 0.0001; time: F(1,57) = 73.88, p < 0.0001; interaction: F(3,57) = 19.52, p < 0.0001; subjects (matching): F(57,57) = 0.9259, p = 0.6139. ***p < 0.001 versus the sham + AAV-SYN-shRNA-Con group. ###p < 0.001 versus the tMCAO + AAV-SYN-shRNA-Con group (two-way repeated-measures ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). For the distribution of shRNA-GFP in the cortex after neuron-specific AAV microinjection, see Figure 7-1.
Distribution of shRNA-GFP in the cortex after neuron-specific AAV microinjection. Representative images obtained four weeks after the microinjection of circTLK1 shRNA neuron-specific AAV into the lateral ventricle. Green: GFP positive cells infected with circTLK1 shRNA AAV. Red: the astrocyte marker GFAP (A), microglia marker Iba-1 (B), oligodendrocyte marker galactocerebroside (GalC) (C) and vascular cell marker caveolin-1 (D). Blue: nuclei stained with DAPI. Scale bar: 25 μm. Download Figure 7-1, TIF file (214.7KB, tif)
Moreover, MAP-2 staining revealed less brain atrophy in shRNA-circTLK1-AAV-treated mice with stroke than in shRNA-Con AAV-treated mice with stroke (Fig. 8A,B). Next, we investigated whether neuron-specific knockdown of circTLK1 led to functional improvements at 14, 21, and 28 d after tMCAO. As shown in Figure 8C–E, microinjection of the circTLK1 shRNA AAV resulted in a significant recovery of neurological deficits (Fig. 8C) and somatosensory and sensorimotor functions after tMCAO (Fig. 8D,E). A significant difference in the mortality rate was not observed among the groups at 28 d after tMCAO (Fig. 8-1).
Figure 8.
Neuron-specific knockdown of circTLK1 improves long-term neurological functions after stroke in mice. A, B, Images (A) and bar graph (B) represent brain atrophy quantified in MAP2-stained coronal sections at 28 d after tMCAO. Dashed lines indicate the relative area of the uninjured contralateral hemisphere to illustrate the area of ipsilateral atrophy for comparison. n = 6 animals per group. t(10) = 2.666. *p = 0.0237 (two-tailed t test). Scale bar, 1 mm. C, Long-term neurological deficits were measured using the modified Neurological Severity Scores at different time points. Neurological deficit scores were measured at 14, 21, and 28 d after tMCAO. n = 10, 10, 20, and 21 animals in the sham + AAV-SYN-shRNA-Con, sham + AAV-SYN-shRNA-circTLK1, tMCAO + AAV-SYN-shRNA-Con, and tMCAO + AAV-SYN-shRNA-circTLK1 groups, respectively. AAV-shRNA-circTLK1: F(3,57) = 289.6, p < 0.0001; time: F(3,171) = 232.4, p < 0.0001; interaction: F(9,171) = 113, p < 0.0001; subjects (matching): F(57,171) = 3.32, p < 0.0001. ***p < 0.001 versus the sham + AAV-SYN-shRNA-Con group. ###p < 0.001 versus the tMCAO + AAV-SYN-shRNA-Con group (two-way repeated-measures ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). D, Adhesive removal test of forelimb function at different time points after stroke. Adhesive removal tests were performed at 14, 21, and 28 d after tMCAO. n = 10, 10, 20, and 21 animals in the sham + AAV-SYN-shRNA-Con, sham + AAV-SYN-shRNA-circTLK1, tMCAO + AAV-SYN-shRNA-Con, and tMCAO + AAV-SYN-shRNA-circTLK1 groups, respectively. AAV-shRNA-circTLK1: F(3,57) = 37.70, p < 0.0001; time: F(3,171) = 8.885, p < 0.0001; interaction: F(9,171) = 6.747, p < 0.0001; subjects (matching): F(57,171) = 1.551, p = 0.0165. ***p < 0.001 versus the sham + AAV-SYN-shRNA-Con group. ###p < 0.001 versus the tMCAO + AAV-SYN-shRNA-Con group (two-way repeated-measures ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). E, Cylinder test of forelimb symmetry at 14, 21, and 28 d after tMCAO. L, Left; R, right; B, both forepaws in E. n = 10, 10, 20, and 21 animals in the sham + AAV-SYN-shRNA-Con, sham + AAV-SYN-shRNA-circTLK1, tMCAO + AAV-SYN-shRNA-Con, and tMCAO + AAV-SYN-shRNA-circTLK1 groups, respectively. AAV-shRNA-circTLK1: F(3,57) = 66.61, p < 0.0001; time: F(3,171) = 23.1, p < 0.0001; interaction: F(9,171) = 8.519, p < 0.0001; subjects (matching): F(57,171) = 1.164, p = 0.2283. ***p < 0.001 versus the sham + AAV-SYN-shRNA-Con group. ##p < 0.01 versus the tMCAO + AAV-SYN-shRNA-Con group (two-way repeated-measures ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). ###p < 0.001 versus the tMCAO + AAV-SYN-shRNA-Con group (two-way repeated-measures ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). For the effect of neuron-specific shRNA-circTLK1 AAV microinjection on mortality after focal cerebral ischemia, see Figure 8-1.
Effect of neuron-specific shRNA-circTLK1 AAV microinjection on mortality after focal cerebral ischemia. (A) Animal survival rate from the same experiments was recorded daily for 28 d after tMCAO. n=10 animals for both sham groups, n=25 and 26 animals for tMCAO AAV-SYN-shRNA-Con and tMCAO AAV-SYN-shRNA-circTLK1 groups, respectively. p = 0.2239 by log-rank test. (B) The mortality rate was calculated as the number of dead mice at 28 d after tMCAO in the AAV-SYN-shRNA-Con and AAV-SYN-shRNA-circTLK1 groups. Mice that died during the experiments were excluded from further data analyses and statistics. Download Figure 8-1, TIF file (353.2KB, tif)
circTLK1 regulated TIPARP by binding to miR-335-3p
Since circRNAs function as ncRNA sponges to interact with miRNAs and increase the expression of a target protein, we sought to determine the downstream target protein displaying increased levels in the ipsilateral side of tMCAO mice that served as an inducer of ischemic brain injury. We first searched our microarray data from tMCAO mouse brains and found that TIPARP mRNA levels were significantly increased in the brain infarct and surrounding areas (GEO accession number GSE98319; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE98319). Importantly, TIPARP has been shown to be involved in ischemic brain injury (Han et al., 2018b). The next logical step is to search for miRNAs that target both circTLK1 and TIPARP. Using the bioinformatics program RNAhybrid, the miRNA target prediction software from Arraystar, TargetScan (Enright et al., 2003) and miRanda (Pasquinelli, 2012), only three miRNAs, miR-335-3p, miR-320-3p, and miR-129-5p, bound to circTLK1 and the 3′-UTR of the TIPARP mRNA (Fig. 9A; Table 4). Among these 3 miRNAs, only the levels of miR-335-3p, but not miR-320-3p or miR-129-5p (Fig. 9-1), were significantly decreased in the ischemic cortex at 6 h after tMCAO (Fig. 9B). We next sought to determine whether circTLK1 physically bound to miR-335-3p. Therefore, a biotin-coupled miR-335-3p mimic was then applied to test whether miR-335-3p pulled down circTLK1. We observed an enrichment of circTLK1 in the miR-335-3p-captured fraction, but not in the fractions captured by mutant miR-335-3p mimics with disrupted base paring between circTLK1 and miR-335-3p (Fig. 9C). Another circRNA that was not predicted to bind to miR-335-3p, circKLHL2, was not captured by miR-335-3p (Fig. 9C). These findings were confirmed in an inverse affinity isolation assay for miR-335-3p using a biotin-labeled specific circTLK1 probe, but not with a random probe, or an unpredicted miRNA, such as miR-142–3p (Fig. 9D). We further examined the localization of circTLK1 and miR-335-3p in primary cortical neurons using a FISH analysis with specific probes. FISH revealed the colocalization of circTLK1 and miR-335-3p in the cytoplasm of primary cortical neurons (Fig. 9E).
Figure 9.
circTLK1 binds to miR-335-3p. A, Notably, circTLK1 contains one site complementary to miR-335-3p, as analyzed by RNAhybrid. B, Effect of tMCAO on miR-335-3p expression in the ischemic cortex. The expression of miR-335-3p was detected in the ischemic cortex 6 h after tMCAO. n = 8 animals per group. t(14) = 3.436. **p = 0.0040 (two-tailed t test). C, RNA was pulled down by biotinylated WT miR-335-3p (Bio-335-WT) or biotinylated mutant miR-335-3p (Bio-335-mut), and levels of circTLK1, circKLHL2, and the GAPDH mRNA were quantified by real-time PCR. n = 6. ***p < 0.001 versus the bio-335-Mut circTLK1 group. t(10) = 7.529. ***p < 0.0001 (two-tailed t test). D, The miRNAs were pulled down by the circTLK1 probe or a random probe, and miR-335-3p, miR-142–3p, and U6 levels were analyzed by real-time PCR. n = 5. ***p < 0.001 versus the random probe miR-335-3p group. t(8) = 9.780. ***p < 0.0001 (two-tailed t test). E, FISH assay showing the location of circTLK1 and miR-335-3p in primary neurons. Green represents circTLK1. Red represents miR-335-3p. Blue represents DAPI. Scale bar, 15 μm. For relative expression of miRNAs in the ischemic cortex after tMCAO in mice, see Figure 9-1. WT, wild type.
Table 4.
The target miRNAs of mmu_circRNA_009932 (circTLK1) predicted using the miRNA target prediction software from Arraystar based on TargetScan and miRanda
| Seq1 | Seq2 | Sites | TargetScan |
miRanda |
||
|---|---|---|---|---|---|---|
| Context+ | Context | Structure | Energy | |||
| mmu-miR-129-5p | mmu_circRNA_009932 | 1 | — | — | 143 | −15.34 |
| mmu-miR-320-3p | mmu_circRNA_009932 | 1 | — | — | 141 | −13.31 |
| mmu-miR-335-3p | mmu_circRNA_009932 | 1 | — | −0.067 | 145 | −10.42 |
Relative expression of miRNAs in the ischemic cortex after tMCAO in mice. Relative expression levels of miR-129-5p (A) and miR-320-3p (B) in the ischemic cortex 6 h after reperfusion. n = 8 animals per group. miR-129-5p: t(14) =1.091, p = 0.2935; miR-320-3p: t(14) =1.467, p = 0.1644; two-tailed t test. Download Figure 9-1, TIF file (275.2KB, tif)
As described above, the 3′-UTR of the human and mouse TIPARP mRNA contains a conserved miR-335-3p binding site (Fig. 10A). Cotransfection of an miR-335-3p-overexpressing vector and a pmiR-GLO plasmid with the WT TIPARP 3′-UTR resulted in the downregulation of luciferase activity, and this effect was reversed in HEK293T cells transfected with a mutated TIPARP 3′-UTR (Fig. 10B). The level of TIPARP was significantly increased in the ischemic tissue 12 h to 7 d after tMCAO (Fig. 10C,D). As shown in Figure 10E, TIPARP was highly colocalized with NeuN in the cortex. Additionally, the in vitro study confirmed that TIPARP expression was significantly increased in primary cortical neurons treated with OGD/R (Fig. 10F,G). Transduction of cells with the mimic miR-335-3p lentivirus decreased TIPARP expression (Fig. 10H,I), whereas transduction with the anti-miR-335-3p lentivirus increased the level of the TIPARP protein in primary cortical neurons treated with or without OGD/R (Fig. 10J,K; Fig. 10-1).
Figure 10.
TIPARP is the downstream target of miR-335-3p. A, Putative miR-335-3p binding sites in the TIPARP mRNA. Red represents the potential complementary residues. B, Relative luciferase activity of WT and 3′-UTR mutant constructs of the TIPARP mRNA cotransfected with the miR-335-3p mimic and the miRNA negative control. n = 5. ***p < 0.001 versus the miRNA negative control (NC) group. t(8) = 5.251. ***p = 0.0008 (two-tailed t test). C, D, Images (C) and bar graph (D) represent Western blot analyses of TIPARP levels in the ischemic cortex of tMCAO mice. n = 6 animals per group. F(4,25) = 42.75, p < 0.0001. *p < 0.05 versus the sham group (one-way ANOVA followed by the Holm–Sidak test). ***p < 0.001 versus the sham group (one-way ANOVA followed by the Holm–Sidak test). E, Colocalization of NeuN and TIPARP in the mouse cortex. Green represents TIPARP. Red represents NeuN. Blue represents DAPI. Scale bar, 50 μm. F, G, Images (F) and bar graph (G) represent Western blot analyses of TIPARP levels in primary cortical neurons after OGD/R. n = 3. F(5,12) = 12.84, p = 0.0002. *p < 0.05 versus the control group (one-way ANOVA followed by the Holm–Sidak test). ***p < 0.001 versus the control group (one-way ANOVA followed by the Holm–Sidak test). H, I, Images (H) and bar graph (I) represent Western blot analyses of TIPARP levels in primary cortical neurons transduced with the mimic miR-335-3p lentivirus. Cells were transduced with the mimic miR-335-3p lentiviruses for 7 d, treated with OGD for 2 h, and subjected to reperfusion for 6 h. n = 4. Mimic miR-335-3p: F(1,12) = 35.37, p < 0.0001; OGD/R: F(1,12) = 76.23, p < 0.0001; interaction: F(1,12) = 4.735, p = 0.0502. *p < 0.05 versus the control miR-Con group. ***p < 0.001 versus the control miR-Con group. ###p < 0.001 versus the OGD/R-treated miR-Con group (two-way ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). J, K, Images (J) and bar graph (K) represent Western blot analyses of TIPARP levels in primary cortical neurons transduced with the anti-miR-335-3p lentivirus. Cells were transduced with anti-miR-335-3p lentiviruses for 7 d, treated with OGD for 2 h, and subjected to reperfusion for 6 h. n = 4. Anti-miR-335-3p: F(1,12) = 47.18, p < 0.0001; OGD/R: F(1,12) = 74.94, p < 0.0001; interaction: F(1,12) = 0.1660, p = 0.6909. **p < 0.01 versus the control anti-miR-Con group. ***p < 0.001 versus the control anti-miR-Con group. ###p < 0.001 versus the OGD/R-treated anti-miR-Con group (two-way ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). Images of whole blots presented in C, F, H, J are shown in Figure 10-1. UTR, untranslated region.
Images of whole blots presented in the Figure 10C, F, H and J. Download Figure 10-1, TIF file (732.7KB, tif)
The circTLK1/miR-335-3p axis aggravates neuronal injury via the downstream protein TIPARP
Having determined the relationship between circTLK1/miR-335-3p with TIPARP, we next examined the functional role of circTLK1/miR-335-3p in neuronal injury. Transduction with the mimic miR-335-3p lentivirus significantly inhibited the decrease in neuronal survival observed in the OGD/R-treated group (Fig. 11A; Fig. 11-1A–C). In contrast, transducing cells with the anti-miR-335-3p lentivirus reduced neuronal survival in OGD/R-challenged neurons (Fig. 11B; Fig. 11-1D,E). Furthermore, transducing neurons with the mimic miR-335-3p lentivirus significantly increased the levels of SYN and PSD95 in neurons and ameliorated the OGD/R-induced decrease in SYN and PSD95 levels (Fig. 11-2A–C). In contrast, transducing cells with the anti-miR-335-3p lentivirus reduced the levels of SYN and PSD95 in neurons and aggravated the loss of SYN and PSD95 in OGD/R-challenged neurons (Fig. 11-2D–F).
Figure 11.
The circTLK1/miR-335-3p axis aggravates neuronal injury via the downstream protein TIPARP. A, Neurons transduced with the mimic miR-335-3p lentivirus showed an attenuated OGD/R-induced increase in cell injury, as measured by the CCK8 assay. Cells were transduced with mimic miR-335-3p lentiviruses for 7 d, treated with OGD for 2 h, and subjected to reperfusion for 6 h. n = 3. Mimic miR-335-3p: F(1,8) = 15.54, p = 0.0043; OGD/R: F(1,8) = 59.18, p < 0.0001; interaction: F(1,8) = 13.53, p = 0.0062. ***p < 0.001 versus the control miR-Con group. ##p < 0.01 versus the OGD/R-treated miR-Con group (two-way ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). B, Neurons transduced with the anti-miR-335-3p lentivirus exhibited an exacerbation of the OGD/R-induced increase in cell injury, as measured by the CCK8 assay. Cells were transduced with anti-miR-335-3p lentiviruses for 7 d, treated with OGD for 2 h, and subjected to reperfusion for 6 h. n = 3. Anti-miR-335-3p: F(1,8) = 17.44, p = 0.0031; OGD/R: F(1,8) = 75.21, p < 0.0001; interaction: F(1,8) = 0.02178, p = 0.8863. *p < 0.05 versus the control anti-miR-Con group. **p < 0.01 versus the control anti-miR-Con group. #p < 0.05 versus the OGD/R-treated anti-miR-Con group (two-way ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). C, D, Images (C) and bar graph (D) represent that the circTLK1 shRNA lentivirus attenuated the OGD/R-induced increase in TIPARP levels in primary cortical neurons, as determined by Western blot analyses. Cells were transduced with circTLK1 shRNA lentiviruses for 7 d, treated with OGD for 2 h, and subjected to reperfusion for 6 h. n = 3. shRNA-circTLK1: F(1,8) = 97.11, p < 0.0001; OGD/R: F(1,8) = 127.6, p < 0.0001; interaction: F(1,8) = 46.77, p = 0.0001. ***p < 0.001 versus the control shRNA-Con group. ###p < 0.001 versus the OGD/R-treated shRNA-Con group (two-way ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). E, F, Images (E) and bar graph (F) represent that the transduction of cortical neurons with the anti-miR-335-3p lentivirus significantly attenuated the circTLK1 shRNA-induced decrease in TIPARP levels, as determined by Western blot analyses. n = 4. shRNA-circTLK1: F(1,12) = 35.09, p < 0.0001; anti-miR-335-3p: F(1,12) = 55.75, p < 0.0001; interaction: F(1,12) = 8.062, p = 0.0149. ***p < 0.001 versus the group cotransduced with the circControl shRNA and anti-miR-Con. #p < 0.05 versus the group cotransduced with the circTLK1 shRNA and anti-miR-Con (two-way ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). G, Transduction of cortical neurons with the anti-miR-335-3p lentivirus significantly reduced the circTLK1 shRNA-induced increase in cell viability, as determined by the CCK8 assay. n = 3. shRNA-circTLK1: F(1,8) = 10.20, p = 0.0127; anti-miR-335-3p: F(1,8) = 32.64, p = 0.0004; interaction: F(1,8) = 1.213, p = 0.3028. **p < 0.01 versus the group cotransduced with the circControl shRNA and anti-miR-Con. #p < 0.05 versus the group cotransduced with the circTLK1 shRNA and anti-miR-Con (two-way ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). H, I, Images (H) and bar graph (I) represent TIPARP levels in primary mouse cortical neurons after TIPARP shRNA lentivirus transduction, as measured by Western blotting. n = 3. t(4) = 5.745. **p = 0.0045 (two-tailed t test). J, The TIPARP shRNA lentivirus attenuated the OGD/R-induced decrease in cell viability, as determined by the CCK8 assay. Cells were transduced with TIPARP shRNA lentiviruses for 7 d, treated with OGD for 2 h, and subjected to reperfusion for 6 h. n = 3. shRNA-TIPARP: F(1,8) = 13.70, p = 0.0060; OGD/R: F(1,8) = 56.40, p < 0.0001; interaction: F(1,8) = 5.632, p = 0.0450. ***p < 0.001 versus the control shRNA-Con group. #p < 0.05 versus the OGD/R-treated shRNA-Con group (two-way ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). K, L, Images (K) and bar graph (L) represent Western blot analyses of TIPARP levels in the ischemic cortex after the circTLK1 shRNA lentivirus injection at 24 h after tMCAO. n = 4 animals per group. shRNA-circTLK1: F(1,12) = 21.18, p = 0.0006; tMCAO: F(1,12) = 25.81, p = 0.0003; interaction: F(1,12) = 16.92, p = 0.0014. ***p < 0.001 versus the sham + shRNA-Con group. ###p < 0.001 versus the tMCAO + shRNA-Con group (two-way ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). For the effect of miR-335-3p on neuronal injury after OGD/R in primary cortical neurons, see Figure 11-1. For the effect of miR-335-3p on neuronal plasticity after OGD/R in primary cortical neurons, see Figure 11-2. Images of whole blots presented in C, E, H, K are shown in Figure 11-3.
Effect of miR-335-3p on neuronal injury after OGD/R in primary cortical neurons. (A-C) Neurons transduced with the mimic miR-335-3p lentivirus attenuated the OGD/R-induced increase in the Bax/Bcl-xl ratio and caspase-3/pro caspase-3 ratio. The cells were transduced with mimic miR-335-3p lentiviruses for 7 d, treated with OGD for 2 h and subjected to reperfusion for 6 h. n = 3. Bax/Bcl-xl (mimic miR-335-3p: F (1, 8) = 17.27, P = 0.0032; OGD/R: F (1, 8) = 32.9, P = 0.0004; interaction: F (1, 8) = 14.63, P = 0.0051). Caspase-3/pro caspase-3 (mimic miR-335-3p: F (1, 8) = 6.088, P = 0.0389; OGD/R: F (1, 8) = 19.93, P = 0.0021; interaction: F (1, 8) = 7.008, P = 0.0294). **p < 0.01 and ***p < 0.001 versus the control miR-Con group; #p < 0.05 and ##p < 0.01 versus the OGD/R-treated miR-Con group (two-way ANOVA followed by Holm-Sidak post hoc multiple comparison tests). (D-F) Neurons transduced with the anti-miR-335-3p lentivirus aggravated the OGD/R-induced increase in the Bax/Bcl-xl ratio and caspase-3/pro caspase-3 ratio. The cells were transduced with anti-miR-335-3p lentiviruses for 7 d, treated with OGD for 2 h and subjected to reperfusion for 6 h. n = 3. Bax/Bcl-xl (anti-miR-335-3p: F (1, 8) = 31.33, P = 0.0005; OGD/R: F (1, 8) = 94.28, P < 0.0001; interaction: F (1, 8) = 15.51, P = 0.0043). Caspase-3/pro caspase-3 (anti-miR-335-3p: F (1, 8) = 8.411, P = 0.0199; OGD/R: F (1, 8) = 44.8, P = 0.0002; interaction: F (1, 8) = 4.57, P = 0.0650). *p < 0.05 and ***p < 0.001 versus the control anti-miR-Con group; #p < 0.05 and ###p < 0.001 versus the OGD/R-treated anti-miR-Con group (two-way ANOVA followed by Holm-Sidak post hoc multiple comparison tests). Images of whole blots presented in the Fig. 11-1A and D were shown in Fig. 11-4. Download Figure 11-1, TIF file (136.4KB, tif)
Effect of miR-335-3p on neuronal plasticity after OGD/R in primary cortical neurons. (A-C) Neurons transduced with the mimic miR-335-3p lentivirus attenuated the OGD/R-induced decrease in synapsin-1 and PSD95. The cells were transduced with mimic miR-335-3p lentiviruses for 7 d, treated with OGD for 2 h and subjected to reperfusion for 6 h. n = 3. Synapsin-1 (mimic miR-335-3p: F (1, 8) = 25.33, P = 0.0010; OGD/R: F (1, 8) = 38.63, P = 0.0003; interaction: F (1, 8) = 0.6650, P = 0.4384). PSD95 (mimic miR-335-3p: F (1, 8) = 27.28, P = 0.0008; OGD/R: F (1, 8) = 52.57, P < 0.0001; interaction: F (1, 8) = 0.1417, P = 0.7164). *p < 0.05 and **p < 0.01 versus the control miR-Con group; #p < 0.05 versus the OGD/R-treated miR-Con group (two-way ANOVA followed by Holm-Sidak post hoc multiple comparison tests). (D-F) Neurons transduced with the anti-miR-335-3p lentivirus aggravated the OGD/R-induced decrease in synapsin-1 and PSD95. The cells were transduced with anti-miR-335-3p lentiviruses for 7 d, treated with OGD for 2 h and subjected to reperfusion for 6 h. n = 3. Synapsin-1 (anti-miR-335-3p: F (1, 8) = 61.78, P < 0.0001; OGD/R: F (1, 8) = 216.4, P < 0.0001; interaction: F (1, 8) = 0.2554, P = 0.6269). PSD95 (anti-miR-335-3p: F (1, 8) = 48.60, P = 0.0001; OGD/R: F (1, 8) = 224.5, P < 0.0001; interaction: F (1, 8) = 0.4629, P = 0.5155). **p < 0.01 and ***p < 0.001 versus the control anti-miR-Con group; ##p < 0.01 versus the OGD/R-treated anti-miR-Con group (two-way ANOVA followed by Holm-Sidak post hoc multiple comparison tests). Images of whole blots presented in the Fig. 11-2A and D were shown in Fig. 11-5. Download Figure 11-2, TIF file (77.8KB, tif)
Images of whole blots presented in the Figure 11C, E, H and K. Download Figure 11-3, TIF file (580.3KB, tif)
Images of whole blots presented in the Figure 11-1A and D. Download Figure 11-4, TIF file (317.8KB, tif)
Images of whole blots presented in the Figure 11-2A and D. Download Figure 11-5, TIF file (308.9KB, tif)
Moreover, transduction with the circTLK1 shRNA lentivirus significantly inhibited the OGD/R-induced increase in TIPARP levels in mouse cortical neurons (Fig. 11C,D). We cotransduced primary mouse cortical neurons with the circTLK1 shRNA lentivirus and anti-miR-335-3p lentivirus to further verify that miR-335-3p acts as a mediator of circTLK1 to control TIPARP expression. Transduction with the circTLK1 shRNA lentivirus attenuated the increase in TIPARP expression induced by the anti-miR-335-3p lentivirus (Fig. 11E,F). Next, we cotransduced primary mouse cortical neurons with the circTLK1 shRNA lentivirus and anti-miR-335-3p lentivirus to determine whether circTLK1 ameliorated neuronal injury via miR-335-3p. As shown in Figure 11G, transduction with the circTLK1 shRNA lentivirus attenuated the increase in neuronal injury induced by the anti-miR-335-3p lentivirus. We next sought to examine the role of TIPARP in neuronal injury using a genetic approach. Transduction of primary mouse neurons with the TIPARP shRNA lentivirus efficiently decreased TIPARP expression (Fig. 11H,I). Knockdown of TIPARP expression significantly rescued the OGD/R-induced decrease in neuronal survival (Fig. 11J). This finding was further confirmed in vivo, as that microinjection of the circTLK1 shRNA lentivirus significantly inhibited the increase in TIPARP expression observed in tMCAO mice at 24 h after reperfusion (Fig. 11K,L), suggesting that TIPARP expression was regulated by circTLK1 (Fig. 11-3).
Next, we further examined the effect of circTLK1 on neuronal function. As shown in Figure 12A, B, transduction with the anti-miR-335-3p lentivirus significantly inhibited the increases in PSD95 expression induced by the circTLK1 shRNA lentivirus. Moreover, transduction of neurons with the circTLK1 shRNA lentivirus increased the spine density, and this effect was inhibited in neurons cotransduced with the circTLK1 shRNA lentivirus and anti-miR-335-3p lentivirus (Fig. 12C,D). Knockdown of TIPARP expression significantly rescued the OGD/R-induced decrease in PSD95 levels (Fig. 12E,F; Fig. 12-1).
Figure 12.
The circTLK1/miR-335-3p axis impairs neuronal plasticity via the downstream protein TIPARP. A, B, Images (A) and bar graph (B) represent that the transduction of cortex neurons with the anti-miR-335-3p lentivirus significantly reduced the circTLK1 shRNA-induced increase in PSD95 levels, as determined by Western blot analyses. n = 4. shRNA-circTLK1: F(1,12) = 18.33, p = 0.0011; anti-miR-335-3p: F(1,12) = 19.82, p = 0.0008; interaction: F(1,12) = 0.02298, p = 0.8820. *p < 0.05 versus the group cotransduced with the circControl shRNA and anti-miR-Con. #p < 0.05 versus the group cotransduced with the circTLK1 shRNA and anti-miR-Con (two-way ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). C, D, Images (C) and bar graph (D) represent that the transduction of cortical neurons with the anti-miR-335-3p lentivirus significantly reduced the circTLK1 shRNA-induced increase in spine density. n = 8. shRNA-circTLK1: F(1,28) = 29.84, p < 0.0001; anti-miR-335-3p: F(1,28) = 29.20, p < 0.0001; interaction: F(1,28) = 1.825, p = 0.1875. ***p < 0.001 versus the group cotransduced with circControl shRNA and anti-miR-Con. #p < 0.05 versus the group cotransduced with the circTLK1 shRNA and anti-miR-Con (two-way ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). Scale bar, 10 μm. E, F, Images (E) and bar graph (F) represent that the TIPARP shRNA lentivirus attenuated the OGD/R-induced decrease in PSD95 levels. Cells were transduced with TIPARP shRNA lentiviruses for 7 d, treated with OGD for 2 h, and subjected to reperfusion for 6 h. n = 3. shRNA-TIPARP: F(1,8) = 62.21, p < 0.0001; OGD/R: F(1,8) = 65.63, p < 0.0001; interaction: F(1,8) = 2.909, p = 0.1265. **p < 0.01 versus the control shRNA-Con group. ***p < 0.001 versus the control shRNA-Con group. ##p < 0.01 versus the OGD/R-treated shRNA-Con group (two-way ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). Images of whole blots presented in A, E are shown in Figure 12-1.
Images of whole blots presented in the Figure 12A and E. Download Figure 12-1, TIF file (563.5KB, tif)
Neuron-specific knockdown of TIPARP improves neurological deficits after stroke in mice
We used neuron-specific AAV (AAV2/8 serotype) with an SYN promoter carrying a TIPARP shRNA to inhibit the expression of TIPARP in neurons and confirm the effect of TIPARP on neuronal injury induced by ischemic stroke in vivo (Fig. 13A). The mice were monitored to determine the role of neuronal TIPARP in stroke pathogenesis (Fig. 13B). GFP was widely expressed and colocalized with NeuN-positive cells (Fig. 13C), indicating successful neuron-specific TIPARP shRNA AAV (AAV-SYN-shRNA-TIPARP) transduction. As shown in Figure 13D, E (Fig. 13-2) the protein level of TIPARP was significantly decreased in the cortex at 28 d after the AAV microinjection. When tMCAO was induced, the neuron-specific TIPARP shRNA AAV treatment did not change the CBF or physiological parameters of these mice (Fig. 13F–H; Table 3). MRI scanning performed at 48 h after tMCAO showed that, compared with the control shRNA AAV (AAV-SYN-shRNA-Con) treatment, the neuron-specific TIPARP shRNA AAV treatment significantly reduced the brain infarction volume (Fig. 13I,J). Microinjection of the neuron-specific TIPARP shRNA AAV resulted in significant improvements in neurological scores (Fig. 13K) and forepaw function, as revealed by the adhesive removal test (Fig. 13L). Furthermore, TIPARP knockdown significantly decreased the levels of the apoptotic proteins Bax and caspase-3 at 24 h after tMCAO (Fig. 13-1A–D). We performed RNA-Seq of the ipsilateral cortex at 24 h after tMCAO to investigate the underlying mechanisms of AAV shRNA-mediated knockdown of TIPARP in neuronal injury (GEO accession number GSE131712; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE131712). At an expression fold change cutoff of 2.0 and p < 0.05, we identified 516 DEGs between the tMCAO + AAV-SYN-shRNA-TIPARP and tMCAO + AAV-SYN-shRNA-Con groups at 24 h after tMCAO, among which 77 genes showed higher expression and 439 genes showed lower expression (Fig. 13-1E). We identified several DEGs involved in the regulation of cell processes and signaling pathways that clustered together in the comparison of AAV-SYN-shRNA-Con and AAV-SYN-shRNA-TIPARP (Fig. 13-1F). Upon the KEGG pathway analysis, the significantly differential genes were enriched in those involved in biological processes that are critical for cell processes, signaling pathways, and neurology, including apoptosis, p53 signaling pathway, calcium signaling pathways, and neuroactive ligand–receptor interaction (Fig. 13-1G).
Figure 13.
Neuron-specific knockdown of TIPARP improves neurological deficits after stroke in mice. A, Schematic representation of AAV constructs (AAV2/8 serotype). B, Schematic illustrating the experimental procedure used in C–L. C, The distribution of the neuron-specific TIPARP shRNA-GFP AAV in the cortex after microinjection. Representative images obtained 4 weeks after microinjection of AAV-SYN-shRNA-TIPARP into the lateral ventricle. Green represents neuron-specific TIPARP shRNA AAV-infected GFP-positive cells. Red represents the neuronal marker NeuN. Blue represents nuclei stained with DAPI. Scale bar, 25 or 50 μm. D, E, Western blot analysis (D) and bar graph (E) represent a significant decrease in the level of the TIPARP protein in the cortex at 28 d after microinjection. n = 6. t(10) = 5.715. ***p = 0.0002 (two-tailed t test). F–H, Regional CBF was monitored using 2D laser speckle imaging techniques 15 min before tMCAO (Pre), 15 min after the onset of tMCAO (O), and after 15 min of reperfusion (R). F, Representative images of the AAV-SYN-shRNA-Con and AAV-SYN-shRNA-TIPARP groups. G, Ischemic areas measured from laser speckle images of either the ischemic core (0%–20% residual CBF) or the penumbra (20%–30% residual CBF) were not affected by AAV-SYN-shRNA-TIPARP. H, Quantification of CBF. Data are presented as percentages of the baseline levels before tMCAO (Pre). n = 6 animals per group. Scale bar, 1 mm. I, J, Representative T2-weighted MRI (I) and infarct volumes (J) of AAV-SYN-shRNA-Con-microinjected and AAV-SYN-shRNA-TIPARP-microinjected tMCAO mice at 48 h after reperfusion. n = 5 animals per group. t(8) = 4.302. **p = 0.0026 (two-tailed t test). K, Neurological deficits were measured by the modified Neurological Severity Scores 1, 3, and 7 d after tMCAO. n = 8, 8, 12, and 12 animals in the sham + AAV-SYN-shRNA-Con, sham + AAV-SYN-shRNA-TIPARP, tMCAO + AAV-SYN-shRNA-Con, and tMCAO + AAV-SYN-shRNA-TIPARP groups, respectively. AAV-shRNA-TIPARP: F(3,36) = 293.4, p < 0.0001; time: F(3,108) = 189.2, p < 0.0001; interaction: F(9,108) = 70.17, p < 0.0001; subjects (matching): F(36,108) = 2.104, p = 0.0018. ***p < 0.001 versus the sham + AAV-SYN-shRNA-Con group. ###p < 0.001 versus the tMCAO + AAV-SYN-shRNA-Con group (two-way repeated-measures ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). L, Adhesive removal test of forelimb function performed 3 and 7 d after tMCAO. n = 8, 8, 12, and 12 animals in the sham + AAV-SYN-shRNA-Con, sham + AAV-SYN-shRNA-TIPARP, tMCAO + AAV-SYN-shRNA-Con, and tMCAO + AAV-SYN-shRNA-TIPARP groups, respectively. AAV-shRNA-TIPARP: F(3,36) = 18.85, p < 0.0001; time: F(2,72) = 16.42, p < 0.0001; interaction: F(6,72) = 7.916, p < 0.0001; subjects (matching): F(36,72) = 1.453, p = 0.0893. **p < 0.01 versus the sham + AAV-SYN-shRNA-Con group. ***p < 0.001 versus the sham + AAV-SYN-shRNA-Con group. ##p < 0.01 versus the tMCAO + AAV-SYN-shRNA-Con group (two-way repeated-measures ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). ###p < 0.001 versus the tMCAO + AAV-SYN-shRNA-Con group (two-way repeated-measures ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). For potential mechanisms underlying neuroprotective effect of neuron-specific knockdown of TIPARP in ischemic cortex after stroke, see Figure 13-1. Images of whole blots presented in D are shown in Figure 13-2.
The potential mechanisms underlying neuroprotective effect of neuron-specific knockdown of TIPARP in ischemic cortex after stroke. (A and B) Western blot analysis of Bax and Bcl-xl expression in the ischemic cortex at 24 h after tMCAO. Representative images are shown in A. B shows that neuron-specific knockdown of TIPARP attenuated the tMCAO-induced increase in the ratio of Bax/Bcl-xl. n = 6 animals per group. AAV-shRNA-TIPARP: F (1, 20) = 31.68, P < 0.0001; tMCAO: F (1, 20) = 40.9, P < 0.0001; interaction: F (1, 20) = 24.31, P < 0.0001. ***p < 0.001 versus the sham + AAV-shRNA-Con group; ###p < 0.001 versus the tMCAO + AAV-SYN-shRNA-Con group (two-way ANOVA followed by Holm-Sidak post hoc multiple comparison tests). (C and D) Western blot analysis of caspase-3 expression in the ischemic cortex at 24 h after tMCAO. Representative images are shown in C. D shows that neuron-specific knockdown of TIPARP attenuated the tMCAO-induced increase in activated caspase-3. n = 6 animals per group. AAV-shRNA-TIPARP: F (1, 20) = 16.1, P = 0.0007; tMCAO: F (1, 20) = 23.63, P < 0.0001; interaction: F (1, 20) = 21.57, P = 0.0002. ***p < 0.001 versus the sham + AAV-SYN-shRNA-Con group; ###p < 0.001 versus the tMCAO + AAV-SYN-shRNA-Con group (two-way ANOVA followed by Holm-Sidak post hoc multiple comparison tests). (E-G) RNA-seq analysis of differentially expressed genes (DEGs) in ipsilateral cortex at 24 h after stroke. Differences in gene expression among groups are shown in E. Red represents genes upregulated ≥ 2.0-fold and p < 0.05. Blue represents genes downregulated ≥ 2.0-fold and p < 0.05. F, Heatmap and unsupervised clustering of transcriptomes depicting DEGs, involved in the regulation of cell processes and signaling pathways, determined by RNA sequencing in the ischemic cortex among groups. Each row indicates a DEG. Each line represents the gene expression per animal. Red represents upregulated gene. Green represents downregulated gene. G, Kyoto Encyclopedia of Genes and Genomes (KEGG) annotation of DEGs as a result of neuron-specific knockdown of TIPARP 24 h after stroke. Enriched KEGG pathways of DEGs regulated in the tMCAO + AAV-SYN-shRNA-TIPARP group versus the tMCAO + AAV-SYN-shRNA-Con group. The KEGG pathways were summarized in three main categories: group A, cellular processes; group B, signaling pathways; group C, neurology. Rich factor represents the ratio of the number of enriched DEGs in the KEGG pathway and the number of annotated background genes in this pathway. The x-axis shows rich factor of each pathway. The y-axis refers to KEGG pathways. Color of the dots represents P-value, size of the dots represents the number of genes. n = 3 animals per group. Sham_C: sham + AAV-SYN-shRNA-Con; Sham_T: sham + AAV-SYN-shRNA-TIPARP; R24h_C: tMCAO (R24h) + AAV-SYN-shRNA-Con; R24h_T: tMCAO (R24h) + AAV-SYN-shRNA-TIPARP. Numbers: the sample number. Images of whole blots presented in the Fig. 13-1A and C were shown in Fig. 13-3. Download Figure 13-1, TIF file (398.6KB, tif)
Images of whole blots presented in the Figure 13D. Download Figure 13-2, TIF file (406.6KB, tif)
Images of whole blots presented in the Figure 13-1A and C. Download Figure 13-3, TIF file (288.5KB, tif)
Regarding the long-term effect of this treatment, microinjection of the neuron-specific TIPARP shRNA AAV significantly decreased the brain atrophy volume at 28 d after tMCAO (Fig. 14A,B) and resulted in significant improvements in neurological scores (Fig. 14C) and forepaw function, as revealed by the adhesive removal test (Fig. 14D). A significant difference in mortality rate was not observed among the groups at 28 d after tMCAO (Fig. 14-1). Moreover, the levels of the synapse-associated proteins SYN and PSD95 were decreased in the tMCAO group, changes that were significantly alleviated by the neuron-specific TIPARP shRNA AAV microinjection (Fig. 14E–H; Fig. 14-3). We performed RNA-Seq of the ipsilateral cortex at 28 d after tMCAO to investigate the underlying mechanisms and long-term effects of AAV shRNA-mediated knockdown of TIPARP (GEO accession number GSE131712; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE131712). At an expression fold change cutoff of 2.0 and p < 0.05, we identified 192 DEGs between the tMCAO + AAV-SYN-shRNA-TIPARP and tMCAO + AAV-SYN-shRNA-Con groups at 28 d after tMCAO, among which 159 genes showed higher expression and 33 genes showed lower expression (Fig. 14-2A). We identified several DEGs involved in the regulation of neurology, immunology, and signaling pathways that clustered together in the comparison of AAV-SYN-shRNA-Con and AAV-SYN-shRNA-TIPARP (Fig. 14-2B). Upon the KEGG pathway analysis, the significantly differential genes were enriched in those with functions in biological processes involved in the regulation of neurology, signaling pathways, and immunology, such as neuroactive ligand–receptor interaction, Toll-like receptor signaling pathway, and cytokine-cytokine receptor interaction (Fig. 14-2C).
Figure 14.
Neuron-specific knockdown of TIPARP improved long-term outcomes after stroke in mice. A, B, Representative T2-weighted MRI (A) and brain atrophy volume (B) of AAV-SYN-shRNA-Con-microinjected and AAV-SYN-shRNA-TIPARP-microinjected tMCAO mice at 28 d after reperfusion. Dashed lines indicate the relative area of the uninjured contralateral hemisphere to illustrate the area of ipsilateral atrophy for comparison. n = 5 animals per group. t(8) = 4.265. **p = 0.0027 (two-tailed t test). C, Long-term neurological deficits were measured by the modified Neurological Severity Scores at 14, 21, and 28 d after tMCAO. n = 8, 8, 12, and 12 animals in the sham + AAV-SYN-shRNA-Con, sham + AAV-SYN-shRNA-TIPARP, tMCAO + AAV-SYN-shRNA-Con, and tMCAO + AAV-SYN-shRNA-TIPARP groups, respectively. AAV-shRNA-TIPARP: F(3,36) = 257.9, p < 0.0001; time: F(3,108) = 117.6, p < 0.0001; interaction: F(9,108) = 54.5, p < 0.0001; subjects (matching): F(36,108) = 1.839, p = 0.0087. ***p < 0.001 versus the sham + AAV-SYN-shRNA-Con group. ###p < 0.001 versus the tMCAO + AAV-SYN-shRNA-Con group (two-way repeated-measures ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). D, Adhesive removal test of forelimb function performed 14, 21, and 28 d after tMCAO. n = 8, 8, 12, and 12 animals in the sham + AAV-SYN-shRNA-Con, sham + AAV-SYN-shRNA-TIPARP, tMCAO + AAV-SYN-shRNA-Con, and tMCAO + AAV-SYN-shRNA-TIPARP groups, respectively. AAV-shRNA-TIPARP: F(3,36) = 25.72, p < 0.0001; time: F(3,108) = 6.846, p = 0.0003; interaction: F(9,108) = 5.243, p < 0.0001; subjects (matching): F(36,108) = 1.563, p = 0.0412. ***p < 0.001 versus the sham + AAV-SYN-shRNA-Con group. ###p < 0.001 versus the tMCAO + AAV-SYN-shRNA-Con group (two-way repeated-measures ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). E, F, Western blot analysis of SYN levels in the ischemic cortex 28 d after tMCAO. Representative images are shown in E. F, Neuron-specific knockdown of TIPARP attenuated the tMCAO-induced decrease in SYN levels. n = 6 animals per group. AAV-shRNA-TIPARP: F(1,20) = 17.13, p = 0.0005; tMCAO: F(1,20) = 9.36, p = 0.0062; interaction: F(1,20) = 0.8395, p = 0.3705. *p < 0.05 versus the sham + AAV-SYN-shRNA-Con group. ##p < 0.01 versus the tMCAO + AAV-SYN-shRNA-Con group (two-way ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). G, H, Western blot analysis of PSD95 levels in the ischemic cortex 28 d after tMCAO. Representative images are shown in G. H, Neuron-specific knockdown of TIPARP attenuated the tMCAO-induced decrease in PSD95 levels. n = 6 animals per group. AAV-shRNA-TIPARP: F(1,20) = 24.6, p < 0.0001; tMCAO: F(1,20) = 48.79, p < 0.0001; interaction: F(1,20) = 3.812, p = 0.0650. ***p < 0.001 versus the sham + AAV-SYN-shRNA-Con group. ###p < 0.001 versus the tMCAO + AAV-SYN-shRNA-Con group (two-way ANOVA followed by Holm–Sidak post hoc multiple-comparison tests). For the effect of neuron-specific shRNA-TIPARP AAV microinjection on mortality after focal cerebral ischemia, see Figure 14-1. For potential mechanisms underlying long-term effects of neuron-specific knockdown of TIPARP in ischemic cortex after stroke, see Figure 14-2. Images of whole blots presented in E, G are shown in Figure 14-3.
Effect of neuron-specific shRNA-TIPARP AAV microinjection on mortality after focal cerebral ischemia. (A) Animal survival rate from the same experiments was recorded daily for 28 d after tMCAO. n=8 animals for both sham groups, n=18 and 17 animals for the tMCAO + AAV-SYN-shRNA-Con and tMCAO + AAV-SYN-shRNA-TIPARP groups, respectively. p = 0.1030 by log-rank test. (B) The mortality rate was calculated as the number of dead mice at 28 d after tMCAO in the AAV-SYN-shRNA-Con and AAV-SYN-shRNA-TIPARP groups. Mice that died during the experiments were excluded from further data analyses and statistics. Download Figure 14-1, TIF file (1MB, tif)
The potential mechanisms underlying long-term effect of neuron-specific knockdown of TIPARP in ischemic cortex after stroke. RNA-seq analysis of DEGs was performed in ipsilateral cortex at 28 d after stroke. (A) Differences in gene expression among groups. Red represents genes upregulated ≥ 2.0-fold and p < 0.05. Blue represents genes downregulated ≥ 2.0-fold and p < 0.05. (B) Heatmap and unsupervised clustering of transcriptomes depicting DEGs, involved in the regulation of signaling pathways, nervous system, and immunology, determined by RNA sequencing in the ischemic cortex among groups. Each row indicates a DEG. Each line represents the gene expression per animal. Red represents upregulated gene. Green represents downregulated gene. (C) KEGG annotation of DEGs as a result of neuron-specific knockdown of TIPARP at 28 d after stroke. Enriched KEGG pathways of DEGs regulated in the tMCAO + AAV-SYN-shRNA-TIPARP versus tMCAO + AAV-SYN-shRNA-Con groups. The KEGG pathways were summarized in three main categories: A, neurology; B, signaling pathways; C, immunology. Rich factor represents the ratio of the number of enriched DEGs in the KEGG pathway and the number of annotated background genes in this pathway. The x-axis shows rich factor of each pathway. The y-axis refers to KEGG pathways. Color of the dots represents P-value, size of the dots represents the number of genes. n = 3 animals per group. Sham_C: sham + AAV-SYN-shRNA-Con; Sham_T: sham + AAV-SYN-shRNA-TIPARP; R28d_C: tMCAO (R28d) + AAV-SYN-shRNA-Con; R28d_T: tMCAO (R28d) + AAV-SYN-shRNA-TIPARP. Numbers: the sample number. Download Figure 14-2, TIF file (136KB, tif)
Images of whole blots presented in the Figure 14E and G. Download Figure 14-3, TIF file (240.6KB, tif)
circTLK1 is upregulated in the plasma of patients with AIS
We further examined the level of circTLK1 in patients with AIS (age 67.86 ± 1.30 years). The sociodemographic and clinical characteristics of these patients are listed in Table 1. The levels of circTLK1 in these subjects were significantly increased (p < 0.001, n = 71) compared with age- and gender-matched cognitively healthy controls (n = 71) (Fig. 15A). AIS includes different subtypes, such as LA, small-artery occlusion, and cardioembolism, and a subsequent analysis indicated that plasma circTLK1 levels were significantly increased in patients with LA and small-artery occlusion (Fig. 15B). We analyze the differences among lesion localizations, such as cortical, subcortical, or infratentorial strokes, and found that plasma circTLK1 levels were significantly increased in both patients with cortical-infarcted and subcortical-infarcted AIS, but the levels were not significantly changed in infratentorial-infarcted patients (Fig. 15C). An analysis of the receiver operating characteristic curve revealed an area under the curve of circTLK1 of 0.868, with a sensitivity of 0.789 and specificity of 0.915 (Fig. 15D). Further regression analysis revealed larger infarct volumes in patients with AIS presenting higher circTLK1 expression (r = 0.7197, p = 0.0017; Fig. 15E). Data from three sample patients with different lesion patterns, together with their NIHSS scores, lesion volumes, and circTLK1 fold changes, are shown in Figure 15F.
Figure 15.
circTLK1 is upregulated in the plasma of patients with AIS. A, Relative plasma circTLK1 levels in patients with AIS (n = 71) and healthy controls (n = 71), as measured by real-time PCR. t(140) = 5.067. ***p < 0.0001 (two-tailed t test). B, Relative plasma circTLK1 levels in patients with stroke classified according to the Trial of Org 10172 in Acute Stroke Treatment subtypes and healthy controls. F(3,138) = 11.45, p < 0.0001. *p < 0.05 versus the normal control group (one-way ANOVA followed by the Holm–Sidak test). ***p < 0.001 versus the normal control group (one-way ANOVA followed by the Holm–Sidak test). C, Relative plasma circTLK1 levels in patients with stroke categorized by lesion locations: Group A, cortical infarcts; Group B, subcortical infarcts; and Group C, infratentorial infarcts; and healthy controls. F(3,138) = 9.936, p < 0.0001. ***p < 0.001 versus the normal control group (one-way ANOVA followed by the Holm–Sidak test). D, Receiver operating characteristic curve of plasma circTLK1 levels for the diagnosis of AIS. E, The correlation between plasma circTLK1 levels and infarct volume. Patients with LA were recruited for infarct volume measurements. Correlations were estimated by calculating Pearson's correlation coefficients (r = 0.7197, p = 0.0017). Solid line was generated with a nonlinear regression analysis using Prism 6.0 (GraphPad). F, Representative data from 3 patients with different lesion patterns showing their NIHSS scores, lesion volumes, and relative plasma circTLK1 levels (fold change). R, Right hemisphere; L, left hemisphere. CE, cardioembolism; SA, small-artery occlusion; ROC, receiver operating characteristic; AUC, area under the curve.
Discussion
circRNAs are abundant in the mammalian brain, and their roles in many nervous system diseases, such as Alzheimer's disease (Lukiw, 2013), multiple system atrophy (Chen et al., 2016), neuropathic pain (Zhou et al., 2017), major depressive disorder (Cui et al., 2016), drug abuse (Huang et al., 2017), and ischemic stroke (Bai et al., 2018), have been explored. Notably, circRNAs regulate gene expression through different regulatory mechanisms, such as functioning as microRNA sponges (Hansen et al., 2013), competing with linear splicing (Ashwal-Fluss et al., 2014), and binding to RNA-associated proteins (Yang et al., 2017). Based on the available evidence, circRNAs exert regulatory effects on different diseases via different mechanisms (Han et al., 2018a). The expression of circRNAs is altered after stroke, but the functional importance of circRNAs in poststroke outcomes is currently unknown. Our study elucidated the role of the circTLK1/miR-335-3p/TIPARP axis in stroke-associated neuronal injury and neurological deficits. Our findings regarding the expression and function of circTLK1 indicate that it represents a promising biomarker for evaluating the damage caused by ischemic insults.
Current stroke research is centered on the development of acute neuroprotective agents. However, the failure of these agents in clinical trials suggests that future stroke research must expand its horizons to include both acute and long-term neuroprotection (Rumpel, 2018). In patients with stroke, neuronal damage correlates with the infarct size, and treatments that are capable of decreasing the infarct size often also attenuate neuronal injury (Bi et al., 2017; Chassagnon et al., 2017). As shown in the present study, strategies targeting early brain damage prevent neuronal injury and inhibit the cascades of damage induced by ischemia/reperfusion, thereby eliciting long-term improvements in function. One potential limitation of the study is that the expression of circTLK1 was knocked down before tMCAO, and our findings were not restricted to the effect of this molecule on the late stage of stroke. Therefore, we are unable to exclude the potential effects exerted during the early stage of stroke. However, based on our findings, strategies targeting the very early pathological events occurring after stroke halt the progression of the injury, thereby conferring prolonged protection against neuronal injury.
We further examined the efficacy of shRNA circTLK1 lentivirus at different time points. Microinjection of shRNA circTLK1 lentivirus significantly decreased the expression of circTLK1 at 14 d after microinjection (Fig. 4-2). Microinjection shRNA-circTLK1 lentivirus at 7 d before tMCAO led to reduced circTLK1 expression at day 7 after ischemia, while the brain infarct is mature after the acute stages of ischemic injury. The mechanism underlying this improvement of outcomes from stroke may be due to enhanced cell survival and neuroplasticity as reported in previous studies (Murphy and Corbett, 2009; Li et al., 2015; Lin et al., 2017). Multiple pathophysiological events occur in the peri-infarct area over days to weeks in the delayed phase after stroke (George and Steinberg, 2015), our findings reveal a previously unrecognized role of circTLK1 in neural injury after stroke.
The mechanism underlying the action of circTLK1 is currently unknown; however, circTLK1 bound to miR-335-3p. As shown in previous studies, circRNAs may act as endogenous sponges to interact with miRNAs and regulate the expression of miRNA target genes. The circRNA HRCR functions as an miR-223 sponge to regulate cardiac hypertrophy and heart failure (Wang et al., 2016). Consistent with these findings, circTLK1 functioned as an miR-335-3p sponge to regulate neuronal injury in the present study. The interaction between miR-335-3p and circTLK1 was confirmed using a miRNA affinity isolation assay.
According to numerous studies, miR-335-5p and miR-335-3p synergize to inhibit estrogen receptor α expression and promote tamoxifen resistance (Martin et al., 2017). Recent evidence also indicated that miR-335-3p lies downstream of β-catenin to regulate gastric cancer (Dong et al., 2015). In addition to cell proliferation, miR-335-3p plays a critical role in cell differentiation (Meyer et al., 2015). In our study, the level of miR-335-3p was decreased in the ischemic cortex of tMCAO mice. To determine whether the circTLK1-mediated functional effects specifically depend on miR-335-3p, primary cortical neurons were cotransduced with miR-335-3p and circTLK1. AntagomiR-335-3p significantly weakened the improvement in neuronal injury after circTLK1 knockdown. Although miR-335-3p was sponged by circTLK1 in the present study, our results do not exclude direct or indirect involvement of other circRNAs or molecules in the regulation of miR-335-3p activity. Further investigations are warranted to determine whether miRNAs other than miR-335-3p bind to circTLK1 and modulate neuronal damage and plasticity.
TIPARP, a critical target induced by constitutively active aryl hydrocarbon receptor signaling (Yamada et al., 2016), is expressed in many different tissues, including the brain, heart, liver, spleen, and reproductive organs (Ma et al., 2001). TIPARP contains a CCCH-type zinc finger, a tryptophan-tryptophan-glutamate domain involved in protein–protein interactions, and a C-terminal catalytic domain (Hottiger et al., 2010). As a member of the poly (ADP)-ribose polymerase family, TIPARP transfers mono-ADP-ribose, but not poly-ADP-ribose, to its substrates (Vyas et al., 2014). ADP-ribosylation is a post-translational modification that is involved in several biological processes, such as immune cell function, transcription regulation, and DNA repair (Kraus and Hottiger, 2013). TIPARP expression is induced by nuclear hormone receptors (Sasse et al., 2013), PDGF (Chen et al., 2004), and viral infection (Atasheva et al., 2012). Therefore, TIPARP likely exerts many effects that are independent of the aryl hydrocarbon receptor (Ahmed et al., 2015). In the present study, the miR-335-3p/TIPARP axis played a critical role in neuronal injury in stroke. The detailed mechanism by which TIPARP regulates neuronal injury, particularly which target transcription factors are involved in this process, requires further examination in future studies.
Consistent with the findings from the animal model, the plasma circTLK1 levels were significantly increased in patients with AIS. When the patients were stratified into AIS subtypes, the level of circTLK1 was significantly increased in the LA and small-artery occlusion subtypes, but not in the cardioembolism subtype. The plasma circTLK1 level positively correlated with the lesion volume. Given the small sample size examined in these studies, the correlation of circTLK1 levels with the lesion volume requires further study in larger samples.
Collectively, our findings identified the miR-335-3p/TIPARP regulatory mechanism of circTLK1 that is involved in ischemic stroke-associated neuronal injury. As our results provide the first evidence that links a pure circRNA to ischemic neuronal injury, this study supplies new insights of molecular events that occur after stroke.
Footnotes
This work was supported by National Natural Science Foundation of China Grants 81473190, 81402910, 81673410, and 81603090, and Jiangsu Specially Appointed Professor and Major State Basic Research Development Program of China (973 Program: 2013CB733800 and 2013CB733803).
The authors declare no competing financial interests.
References
- Adams HP Jr, Bendixen BH, Kappelle LJ, Biller J, Love BB, Gordon DL, Marsh EE 3rd (1993) Classification of subtype of acute ischemic stroke: definitions for use in a multicenter clinical trial. TOAST. Trial of Org 10172 in Acute Stroke Treatment. Stroke 24:35–41. 10.1161/01.STR.24.1.35 [DOI] [PubMed] [Google Scholar]
- Ahmed S, Bott D, Gomez A, Tamblyn L, Rasheed A, Cho T, MacPherson L, Sugamori KS, Yang Y, Grant DM, Cummins CL, Matthews J (2015) Loss of the mono-ADP-ribosyltransferase, Tiparp, increases sensitivity to dioxin-induced steatohepatitis and lethality. J Biol Chem 290:16824–16840. 10.1074/jbc.M115.660100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam TV, Chan SL, Jo DG, Yilmaz G, Tang SC, Cheng A, Gleichmann M, Okun E, Dixit VD, Chigurupati S, Mughal MR, Ouyang X, Miele L, Magnus T, Poosala S, Granger DN, Mattson MP (2006) Gamma secretase-mediated notch signaling worsens brain damage and functional outcome in ischemic stroke. Nat Med 12:621–623. 10.1038/nm1403 [DOI] [PubMed] [Google Scholar]
- Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S (2014) circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 56:55–66. 10.1016/j.molcel.2014.08.019 [DOI] [PubMed] [Google Scholar]
- Atasheva S, Akhrymuk M, Frolova EI, Frolov I (2012) New PARP gene with an anti-alphavirus function. J Virol 86:8147–8160. 10.1128/JVI.00733-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai YY, Gao X, Wang YC, Peng XG, Chang D, Zheng S, Li C, Ju S (2014) Image-guided pro-angiogenic therapy in diabetic stroke mouse models using a multi-modal nanoprobe. Theranostics 4:787–797. 10.7150/thno.9525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Zhang Y, Hua J, Yang X, Zhang X, Duan M, Zhu X, Huang W, Chao J, Zhou R, Hu G, Yao H (2016) Silencing microRNA-143 protects the integrity of the blood–brain barrier: implications for methamphetamine abuse. Sci Rep 6:35642. 10.1038/srep35642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Zhang Y, Han B, Yang L, Chen X, Huang R, Wu F, Chao J, Liu P, Hu G, Zhang JH, Yao H (2018) Circular RNA DLGAP4 ameliorates ischemic stroke outcomes by targeting miR-143 to regulate endothelial-mesenchymal transition associated with blood–brain barrier integrity. J Neurosci 38:32–50. 10.1523/JNEUROSCI.1348-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi M, Gladbach A, van Eersel J, Ittner A, Przybyla M, van Hummel A, Chua SW, van der Hoven J, Lee WS, Müller J, Parmar J, Jonquieres GV, Stefen H, Guccione E, Fath T, Housley GD, Klugmann M, Ke YD, Ittner LM (2017) Tau exacerbates excitotoxic brain damage in an animal model of stroke. Nat Commun 8:473. 10.1038/s41467-017-00618-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchli AD, Schwab ME (2005) Inhibition of nogo: a key strategy to increase regeneration, plasticity and functional recovery of the lesioned central nervous system. Ann Med 37:556–567. 10.1080/07853890500407520 [DOI] [PubMed] [Google Scholar]
- Chang D, Wang YC, Bai YY, Lu CQ, Xu TT, Zhu L, Ju S (2015) Role of P38 MAPK on MMP activity in photothrombotic stroke mice as measured using an ultrafast MMP activatable probe. Sci Rep 5:16951. 10.1038/srep16951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassagnon IR, McCarthy CA, Chin YK, Pineda SS, Keramidas A, Mobli M, Pham V, De Silva TM, Lynch JW, Widdop RE, Rash LD, King GF (2017) Potent neuroprotection after stroke afforded by a double-knot spider-venom peptide that inhibits acid-sensing ion channel 1a. Proc Natl Acad Sci U S A 114:3750–3755. 10.1073/pnas.1614728114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BJ, Mills JD, Takenaka K, Bliim N, Halliday GM, Janitz M (2016) Characterization of circular RNAs landscape in multiple system atrophy brain. J Neurochem 139:485–496. 10.1111/jnc.13752 [DOI] [PubMed] [Google Scholar]
- Chen WV, Delrow J, Corrin PD, Frazier JP, Soriano P (2004) Identification and validation of PDGF transcriptional targets by microarray-coupled gene-trap mutagenesis. Nat Genet 36:304–312. 10.1038/ng1306 [DOI] [PubMed] [Google Scholar]
- Cramer SC. (2018) Treatments to promote neural repair after stroke. J Stroke 20:57–70. 10.5853/jos.2017.02796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Niu W, Kong L, He M, Jiang K, Chen S, Zhong A, Li W, Lu J, Zhang L (2016) hsa_circRNA_103636: potential novel diagnostic and therapeutic biomarker in major depressive disorder. Biomark Med 10:943–952. 10.2217/bmm-2016-0130 [DOI] [PubMed] [Google Scholar]
- Dimyan MA, Cohen LG (2011) Neuroplasticity in the context of motor rehabilitation after stroke. Nat Rev Neurol 7:76–85. 10.1038/nrneurol.2010.200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L, Deng J, Sun ZM, Pan AP, Xiang XJ, Zhang L, Yu F, Chen J, Sun Z, Feng M, Xiong JP (2015) Interference with the beta-catenin gene in gastric cancer induces changes to the miRNA expression profile. Tumour Biol 36:6973–6983. 10.1007/s13277-015-3415-1 [DOI] [PubMed] [Google Scholar]
- Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS (2003) MicroRNA targets in Drosophila. Genome Biol 5:R1. 10.1186/gb-2003-5-1-r1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George PM, Steinberg GK (2015) Novel stroke therapeutics: unraveling stroke pathophysiology and its impact on clinical treatments. Neuron 87:297–309. 10.1016/j.neuron.2015.05.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein LB. (2007) Acute ischemic stroke treatment in 2007. Circulation 116:1504–1514. 10.1161/CIRCULATIONAHA.106.670885 [DOI] [PubMed] [Google Scholar]
- Gould E, Woolley CS, Frankfurt M, McEwen BS (1990) Gonadal steroids regulate dendritic spine density in hippocampal pyramidal cells in adulthood. J Neurosci 10:1286–1291. 10.1523/JNEUROSCI.10-04-01286.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han B, Chao J, Yao H (2018a) Circular RNA and its mechanisms in disease: from the bench to the clinic. Pharmacol Ther 187:31–44. 10.1016/j.pharmthera.2018.01.010 [DOI] [PubMed] [Google Scholar]
- Han B, Zhang Y, Zhang Y, Bai Y, Chen X, Huang R, Wu F, Leng S, Chao J, Zhang JH, Hu G, Yao H (2018b) Novel insight into circular RNA HECTD1 in astrocyte activation via autophagy by targeting MIR142-TIPARP: implications for cerebral ischemic stroke. Autophagy 14:1164–1184. 10.1080/15548627.2018.1458173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495:384–388. 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- Harms C, Bösel J, Lautenschlager M, Harms U, Braun JS, Hörtnagl H, Dirnagl U, Kwiatkowski DJ, Fink K, Endres M (2004) Neuronal gelsolin prevents apoptosis by enhancing actin depolymerization. Mol Cell Neurosci 25:69–82. 10.1016/j.mcn.2003.09.012 [DOI] [PubMed] [Google Scholar]
- Heiss WD, Kracht LW, Thiel A, Grond M, Pawlik G (2001) Penumbral probability thresholds of cortical flumazenil binding and blood flow predicting tissue outcome in patients with cerebral ischaemia. Brain 124:20–29. 10.1093/brain/124.1.20 [DOI] [PubMed] [Google Scholar]
- Hottiger MO, Hassa PO, Lüscher B, Schuler H, Koch-Nolte F (2010) Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem Sci 35:208–219. 10.1016/j.tibs.2009.12.003 [DOI] [PubMed] [Google Scholar]
- Huang R, Zhang Y, Han B, Bai Y, Zhou R, Gan G, Chao J, Hu G, Yao H (2017) Circular RNA HIPK2 regulates astrocyte activation via cooperation of autophagy and ER stress by targeting MIR124–2HG. Autophagy 13:1722–1741. 10.1080/15548627.2017.1356975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus WL, Hottiger MO (2013) PARP-1 and gene regulation: progress and puzzles. Mol Aspects Med 34:1109–1123. 10.1016/j.mam.2013.01.005 [DOI] [PubMed] [Google Scholar]
- Lai TW, Zhang S, Wang YT (2014) Excitotoxicity and stroke: identifying novel targets for neuroprotection. Prog Neurobiol 115:157–188. 10.1016/j.pneurobio.2013.11.006 [DOI] [PubMed] [Google Scholar]
- Leak RK, Zhang L, Stetler RA, Weng Z, Li P, Atkins GB, Gao Y, Chen J (2013) HSP27 protects the blood–brain barrier against ischemia-induced loss of integrity. CNS Neurol Disord Drug Targets 12:325–337. 10.2174/1871527311312030006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Nie EH, Yin Y, Benowitz LI, Tung S, Vinters HV, Bahjat FR, Stenzel-Poore MP, Kawaguchi R, Coppola G, Carmichael ST (2015) GDF10 is a signal for axonal sprouting and functional recovery after stroke. Nat Neurosci 18:1737–1745. 10.1038/nn.4146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Chopp M, Chen J, Wang L, Gautam SC, Xu YX, Zhang Z (2000) Intrastriatal transplantation of bone marrow nonhematopoietic cells improves functional recovery after stroke in adult mice. J Cereb Blood Flow Metab 20:1311–1319. 10.1097/00004647-200009000-00006 [DOI] [PubMed] [Google Scholar]
- Lin YH, Dong J, Tang Y, Ni HY, Zhang Y, Su P, Liang HY, Yao MC, Yuan HJ, Wang DL, Chang L, Wu HY, Luo CX, Zhu DY (2017) Opening a new time window for treatment of stroke by targeting HDAC2. J Neurosci 37:6712–6728. 10.1523/JNEUROSCI.0341-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukiw WJ. (2013) Circular RNA (circRNA) in Alzheimer's disease (AD). Front Genet 4:307. 10.3389/fgene.2013.00307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Baldwin KT, Renzelli AJ, McDaniel A, Dong L (2001) TCDD-inducible poly(ADP-ribose) polymerase: a novel response to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Biochem Biophys Res Commun 289:499–506. 10.1006/bbrc.2001.5987 [DOI] [PubMed] [Google Scholar]
- Martin EC, Conger AK, Yan TJ, Hoang VT, Miller DF, Buechlein A, Rusch DB, Nephew KP, Collins-Burow BM, Burow ME (2017) MicroRNA-335–5p and -3p synergize to inhibit estrogen receptor alpha expression and promote tamoxifen resistance. FEBS Lett 591:382–392. 10.1002/1873-3468.12538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer SU, Sass S, Mueller NS, Krebs S, Bauersachs S, Kaiser S, Blum H, Thirion C, Krause S, Theis FJ, Pfaffl MW (2015) Integrative analysis of microRNA and mRNA data reveals an orchestrated function of microRNAs in skeletal myocyte differentiation in response to TNF-alpha or IGF1. PLoS One 10:e0135284. 10.1371/journal.pone.0135284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy TH, Corbett D (2009) Plasticity during stroke recovery: from synapse to behaviour. Nat Rev Neurosci 10:861–872. 10.1038/nrn2735 [DOI] [PubMed] [Google Scholar]
- Pasquinelli AE. (2012) MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet 13:271–282. 10.1038/nrg3162 [DOI] [PubMed] [Google Scholar]
- Rumpel S. (2018) Supporting recovery from brain injury. Science 360:30–31. 10.1126/science.aat2450 [DOI] [PubMed] [Google Scholar]
- Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, Herzog M, Schreyer L, Papavasileiou P, Ivanov A, Öhman M, Refojo D, Kadener S, Rajewsky N (2015) Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell 58:870–885. 10.1016/j.molcel.2015.03.027 [DOI] [PubMed] [Google Scholar]
- Sasse SK, Mailloux CM, Barczak AJ, Wang Q, Altonsy MO, Jain MK, Haldar SM, Gerber AN (2013) The glucocorticoid receptor and KLF15 regulate gene expression dynamics and integrate signals through feed-forward circuitry. Mol Cell Biol 33:2104–2115. 10.1128/MCB.01474-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Zhang L, Pu H, Mao L, Hu X, Jiang X, Xu N, Stetler RA, Zhang F, Liu X, Leak RK, Keep RF, Ji X, Chen J (2016) Rapid endothelial cytoskeletal reorganization enables early blood–brain barrier disruption and long-term ischaemic reperfusion brain injury. Nat Commun 7:10523. 10.1038/ncomms10523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiroguchi K, Jia TZ, Sims PA, Xie XS (2012) Digital RNA sequencing minimizes sequence-dependent bias and amplification noise with optimized single-molecule barcodes. Proc Natl Acad Sci U S A 109:1347–1352. 10.1073/pnas.1118018109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims JR, Gharai LR, Schaefer PW, Vangel M, Rosenthal ES, Lev MH, Schwamm LH (2009) ABC/2 for rapid clinical estimate of infarct, perfusion, and mismatch volumes. Neurology 72:2104–2110. 10.1212/WNL.0b013e3181aa5329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetler RA, Cao G, Gao Y, Zhang F, Wang S, Weng Z, Vosler P, Zhang L, Signore A, Graham SH, Chen J (2008) Hsp27 protects against ischemic brain injury via attenuation of a novel stress-response cascade upstream of mitochondrial cell death signaling. J Neurosci 28:13038–13055. 10.1523/JNEUROSCI.4407-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan MY, Singhal S, Ma H, Chandra RV, Cheong J, Clissold BB, Ly J, Srikanth V, Phan TG (2016) Examining subcortical infarcts in the era of acute multimodality CT imaging. Front Neurol 7:220. 10.3389/fneur.2016.00220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Lin YH, Ni HY, Dong J, Yuan HJ, Zhang Y, Liang HY, Yao MC, Zhou QG, Wu HY, Chang L, Luo CX, Zhu DY (2017) Inhibiting histone deacetylase 2 (HDAC2) promotes functional recovery from stroke. J Am Heart Assoc 6:e007236. 10.1161/JAHA.117.007236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas S, Matic I, Uchima L, Rood J, Zaja R, Hay RT, Ahel I, Chang P (2014) Family-wide analysis of poly(ADP-ribose) polymerase activity. Nat Commun 5:4426. 10.1038/ncomms5426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B, Zhou LY, Sun T, Wang M, Yu T, Gong Y, Liu J, Dong YH, Li N, Li PF (2016) A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J 37:2602–2611. 10.1093/eurheartj/ehv713 [DOI] [PubMed] [Google Scholar]
- Wardlaw JM, Smith EE, Biessels GJ, Cordonnier C, Fazekas F, Frayne R, Lindley RI, O'Brien JT, Barkhof F, Benavente OR, Black SE, Brayne C, Breteler M, Chabriat H, Decarli C, de Leeuw FE, Doubal F, Duering M, Fox NC, Greenberg S, et al. (2013) Neuroimaging standards for research into small vessel disease and its contribution to ageing and neurodegeneration. Lancet Neurol 12:822–838. 10.1016/S1474-4422(13)70124-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Horimoto H, Kameyama T, Hayakawa S, Yamato H, Dazai M, Takada A, Kida H, Bott D, Zhou AC, Hutin D, Watts TH, Asaka M, Matthews J, Takaoka A (2016) Constitutive aryl hydrocarbon receptor signaling constrains type I interferon-mediated antiviral innate defense. Nat Immunol 17:687–694. 10.1038/ni.3422 [DOI] [PubMed] [Google Scholar]
- Yang Q, Du WW, Wu N, Yang W, Awan FM, Fang L, Ma J, Li X, Zeng Y, Yang Z, Dong J, Khorshidi A, Yang BB (2017) A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ 24:1609–1620. 10.1038/cdd.2017.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H, Duan M, Buch S (2011) Cocaine-mediated induction of platelet-derived growth factor: implication for increased vascular permeability. Blood 117:2538–2547. 10.1182/blood-2010-10-313593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H, Ma R, Yang L, Hu G, Chen X, Duan M, Kook Y, Niu F, Liao K, Fu M, Hu G, Kolattukudy P, Buch S (2014) MiR-9 promotes microglial activation by targeting MCPIP1. Nat Commun 5:4386. 10.1038/ncomms5386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, Wang X, Hou J, Liu H, Sun W, Sambandan S, Chen T, Schuman EM, Chen W (2015) Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci 18:603–610. 10.1038/nn.3975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Shen K, Bai Y, Lv X, Huang R, Zhang W, Chao J, Nguyen LK, Hua J, Gan G, Hu G, Yao H (2016) Mir143-BBC3 cascade reduces microglial survival via interplay between apoptosis and autophagy: implications for methamphetamine-mediated neurotoxicity. Autophagy 12:1538–1559. 10.1080/15548627.2016.1191723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Xiong Q, Chen H, Yang C, Fan Y (2017) Identification of the spinal expression profile of noncoding RNAs involved in neuropathic pain following spared nerve injury by sequence analysis. Front Mol Neurosci 10:91. 10.3389/fnmol.2017.00091 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genetic information and primer sequence of circTLK1. (A) The exon composition of circTLK1. (B) The divergent primer of circTLK1 contains the back-splicing site. (C) Divergent primers amplified circTLK1 from cDNA, but not genomic DNA (gDNA). GAPDH, linear control. cDNA used for convergent primer was derived from RNA pretreated with RNase R before reverse transcription. (D) Relative expression of circTLK1 in different mouse organs. n = 4 animals per group. Download Figure 2-1, TIF file (229.5KB, tif)
Expression of TLK1 in the ischemic cortex after tMCAO in mice. (A) Relative expression of Tlk1 mRNA in the ipsilateral cortex 6 h after reperfusion in mice as determined by real-time PCR. n = 8 animals per group. t(14) = 2.574, *p = 0.0221, two-tailed t-test. (B and C) Western blot analysis of TLK1 expression in the ipsilateral cortex after tMCAO. (C) Bar graph showing a significant decrease in TLK1 in the ipsilateral cortex 6 h after reperfusion. n = 6 animals per group. t(10) = 3.371, **p = 0.0071, two-tailed t-test. Images of whole blots presented in the Fig. 2-2B were shown in Fig. 2-5. Download Figure 2-2, TIF file (765.6KB, tif)
Structures and sequence of circTLK1 shRNA. The siRNA sequences of circTLK1 contains the back-splicing site. Download Figure 2-3, TIF file (157.5KB, tif)
Distribution of shRNA circTLK1-GFP lentiviruses in the cortex after lentivirus microinjection. Representative images obtained three weeks after the microinjection of circTLK1 shRNA lentiviruses into the lateral ventricle. Green: GFP positive cells infected by circTLK1 shRNA lentiviruses. Red: the astrocyte marker GFAP (A), microglia marker Iba-1 (B), oligodendrocyte marker galactocerebroside (GalC) (C) and vascular cell marker caveolin-1 (D). Blue: nuclei stained with DAPI. Scale bar: 25 μm. (E) Quantification of the proportion of GFP positive cells co-localizing with NeuN+, GFAP+, Iba-1+, GalC+ or Caveolin-1+ profiles. At least 1500 cells were counted for the quantification from 3 different animals. Download Figure 2-4, TIF file (304.5KB, tif)
Images of whole blots presented in the Figure 2-2B. Download Figure 2-5, TIF file (148.3KB, tif)
Images of whole blots presented in the Figure 3D and F. Download Figure 3-1, TIF file (97.7KB, tif)
Effect of shRNA-circTLK1 lentivirus microinjection on mortality after focal cerebral ischemia. (A) Animal survival rate from the same experiments was recorded daily for 28 d after tMCAO. n=9 animals for both sham groups, and n=17 animals for both tMCAO-treated groups. p = 0.1238 by log-rank test. (B) The mortality rate was calculated as the number of dead mice at 28 d after tMCAO in the shRNA-Con lentivirus and shRNA-circTLK1 lentivirus treated mice. Mice that died during the experiments were excluded from further data analyses and statistics. Download Figure 4-1, TIF file (1.1MB, tif)
Post-stroke knockdown of circTLK1 ameliorates neurological deficits after focal ischemia in mice. (A) Relative expression level of circTLK1 in ipsilateral cortex before (Pre) and 3, 7, 14 d after the shRNA-circTLK1 lentivirus injected into lateral ventricle. Microinjection of shRNA circTLK1 lentivirus significantly decreased the expression of circTLK1 at 14 d after microinjection. n = 6 animals per group. F (3, 20) = 2.556, P = 0.0841. *p < 0.05 versus the pre- microinjection group. (B) Illustration of the experimental procedure designed for C-G. Mice were microinjected with the shRNA circTLK1 lentivirus at 7 d before tMCAO, which will decrease the expression of circTLK1 at 7 d after tMCAO. (C) Animal survival rate from the same experiments was recorded daily for 28 d after tMCAO. n=11, 10, 30 and 30 animals for the sham + shRNA-Con, sham + shRNA-circTLK1, tMCAO + shRNA-Con and tMCAO + shRNA-circTLK1 groups, respectively. p = 0.0657 by log-rank test. (D) The mortality rate was calculated as the number of dead mice at 28 d after tMCAO in the tMCAO + shRNA-Con and tMCAO + shRNA-circTLK1 group. (E) Long-term neurological deficit was measured by mNSS at different time points. Neurological deficit scores were measured at 1, 3, 7, 14, 21 and 28 d after tMCAO. n = 11, 10, 21 and 24 animals for the sham + shRNA-Con, sham + shRNA-circTLK1, tMCAO + shRNA-Con and tMCAO + shRNA-circTLK1 groups, respectively. shRNA-circTLK1: F (3, 62) = 888.6, P < 0.0001; time: F (6, 372) = 286.3, P < 0.0001; interaction: F (18, 372) = 107.9, P < 0.0001; subjects (matching): F (62, 372) = 3.009, P < 0.0001. ***p < 0.001 versus the sham + shRNA-Con group; ###p < 0.001 versus the tMCAO + shRNA-Con group (two-way repeated measures ANOVA followed by Holm-Sidak post hoc multiple comparison tests). (F) Cylinder test of forelimb symmetry at 3, 7, 14, 21 and 28 d after tMCAO. L: left; R: right; B: both forepaws in F. n = 11, 10, 21 and 24 animals for the sham + shRNA-Con, sham + shRNA-circTLK1, tMCAO + shRNA-Con and tMCAO + shRNA-circTLK1 groups, respectively. shRNA-circTLK1: F (3, 62) = 66.44, P < 0.0001; time: F (5, 310) = 19.26, P < 0.0001; interaction: F (15, 310) = 8.377, P < 0.0001; subjects (matching): F (62, 310) = 1.226, P = 0.1357. ***p < 0.001 versus the sham + shRNA-Con group; ##p < 0.01 and ###p < 0.001 versus the tMCAO + shRNA-Con group (two-way repeated measures ANOVA followed by Holm-Sidak post hoc multiple comparison tests). (G) Adhesive removal test of forelimb function at different time points after stroke. Adhesive removal tests were performed at 3, 7, 14, 21 and 28 d after tMCAO. n = 11, 10, 21 and 24 animals for the sham + shRNA-Con, sham + shRNA-circTLK1, tMCAO + shRNA-Con and tMCAO + shRNA-circTLK1 groups, respectively. shRNA-circTLK1: F (3, 62) = 23.32, P < 0.0001; time: F (5, 310) = 9.857, P < 0.0001; interaction: F (15, 310) = 4.844, P < 0.0001; subjects (matching): F (62, 310) = 1.721, P = 0.0015. *p < 0.05, **p < 0.01 and ***p < 0.001 versus the sham + shRNA-Con group; ##p < 0.01 versus the tMCAO + shRNA-Con group (two-way repeated measures ANOVA followed by Holm-Sidak post hoc multiple comparison tests). Download Figure 4-2, TIF file (159.6KB, tif)
Images of whole blots presented in the Figure 4M. Download Figure 4-3, TIF file (230KB, tif)
Expression of circTK1 in the ischemic cortex and primary cell cultures in mice. (A) Structures and probe sequence of circTLK1 used for FISH, the probe was circRNA specific (back splicing site). B: biotinylated. (B) Colocalization of NeuN and circTLK1 in the cortex of mice. The areas labeled with the different numbers in the rectangles are enlarged on the bottom or right side of the image. Green: circTLK1. Red: NeuN. Blue: DAPI. Scale bar: 25 μm. (C) Relative expression of circTLK1 in different cell types of primary cultures from the mouse cortex by qPCR. n = 3 samples per group. Download Figure 5-1, TIF file (127.8KB, tif)
Effect of OGD/R on neuronal injury in primary cortical neurons. (A) Western blot analysis of Bcl-xl, Bax, and caspase-3 expression after OGD/R. (B and C) Bar graph showing a significant increase in the Bax/Bcl-xl ratio (B) and the caspase-3/pro caspase-3 ratio (C) in neurons after OGD/R. n = 3. Bax/Bcl-xl: F (5, 12) = 9.409, P = 0.0008. Caspase-3/pro caspase-3: F (5, 12) = 6.014, P = 0.0052. *p < 0.05, **p < 0.01, and ***p < 0.001 versus the control group (one-way ANOVA followed by the Holm-Sidak test). (D) Western blot analysis of synapsin-1 and PSD95 expression after OGD/R. (E and F) Bar graph showing a significant decrease in synapsin-1 (E) and PSD95 (F) in neurons after OGD/R. n = 3. Synapsin-1: F (5, 12) = 130.3, P < 0.0001; PSD95: F (5, 12) = 62.03, P < 0.0001. *p < 0.05, **p < 0.01, and ***p < 0.001 versus the control group (one-way ANOVA followed by the Holm-Sidak test). Images of whole blots presented in the Fig. 5-2A and D were shown in Fig. 5-4. Download Figure 5-2, TIF file (422.7KB, tif)
Images of whole blots presented in the Figure 5I. Download Figure 5-3, TIF file (188.4KB, tif)
Images of whole blots presented in the Figure 5-2A and D. Download Figure 5-4, TIF file (359.5KB, tif)
Images of whole blots presented in the Figure 6A. Download Figure 6-1, TIF file (325.3KB, tif)
Distribution of shRNA-GFP in the cortex after neuron-specific AAV microinjection. Representative images obtained four weeks after the microinjection of circTLK1 shRNA neuron-specific AAV into the lateral ventricle. Green: GFP positive cells infected with circTLK1 shRNA AAV. Red: the astrocyte marker GFAP (A), microglia marker Iba-1 (B), oligodendrocyte marker galactocerebroside (GalC) (C) and vascular cell marker caveolin-1 (D). Blue: nuclei stained with DAPI. Scale bar: 25 μm. Download Figure 7-1, TIF file (214.7KB, tif)
Effect of neuron-specific shRNA-circTLK1 AAV microinjection on mortality after focal cerebral ischemia. (A) Animal survival rate from the same experiments was recorded daily for 28 d after tMCAO. n=10 animals for both sham groups, n=25 and 26 animals for tMCAO AAV-SYN-shRNA-Con and tMCAO AAV-SYN-shRNA-circTLK1 groups, respectively. p = 0.2239 by log-rank test. (B) The mortality rate was calculated as the number of dead mice at 28 d after tMCAO in the AAV-SYN-shRNA-Con and AAV-SYN-shRNA-circTLK1 groups. Mice that died during the experiments were excluded from further data analyses and statistics. Download Figure 8-1, TIF file (353.2KB, tif)
Relative expression of miRNAs in the ischemic cortex after tMCAO in mice. Relative expression levels of miR-129-5p (A) and miR-320-3p (B) in the ischemic cortex 6 h after reperfusion. n = 8 animals per group. miR-129-5p: t(14) =1.091, p = 0.2935; miR-320-3p: t(14) =1.467, p = 0.1644; two-tailed t test. Download Figure 9-1, TIF file (275.2KB, tif)
Images of whole blots presented in the Figure 10C, F, H and J. Download Figure 10-1, TIF file (732.7KB, tif)
Effect of miR-335-3p on neuronal injury after OGD/R in primary cortical neurons. (A-C) Neurons transduced with the mimic miR-335-3p lentivirus attenuated the OGD/R-induced increase in the Bax/Bcl-xl ratio and caspase-3/pro caspase-3 ratio. The cells were transduced with mimic miR-335-3p lentiviruses for 7 d, treated with OGD for 2 h and subjected to reperfusion for 6 h. n = 3. Bax/Bcl-xl (mimic miR-335-3p: F (1, 8) = 17.27, P = 0.0032; OGD/R: F (1, 8) = 32.9, P = 0.0004; interaction: F (1, 8) = 14.63, P = 0.0051). Caspase-3/pro caspase-3 (mimic miR-335-3p: F (1, 8) = 6.088, P = 0.0389; OGD/R: F (1, 8) = 19.93, P = 0.0021; interaction: F (1, 8) = 7.008, P = 0.0294). **p < 0.01 and ***p < 0.001 versus the control miR-Con group; #p < 0.05 and ##p < 0.01 versus the OGD/R-treated miR-Con group (two-way ANOVA followed by Holm-Sidak post hoc multiple comparison tests). (D-F) Neurons transduced with the anti-miR-335-3p lentivirus aggravated the OGD/R-induced increase in the Bax/Bcl-xl ratio and caspase-3/pro caspase-3 ratio. The cells were transduced with anti-miR-335-3p lentiviruses for 7 d, treated with OGD for 2 h and subjected to reperfusion for 6 h. n = 3. Bax/Bcl-xl (anti-miR-335-3p: F (1, 8) = 31.33, P = 0.0005; OGD/R: F (1, 8) = 94.28, P < 0.0001; interaction: F (1, 8) = 15.51, P = 0.0043). Caspase-3/pro caspase-3 (anti-miR-335-3p: F (1, 8) = 8.411, P = 0.0199; OGD/R: F (1, 8) = 44.8, P = 0.0002; interaction: F (1, 8) = 4.57, P = 0.0650). *p < 0.05 and ***p < 0.001 versus the control anti-miR-Con group; #p < 0.05 and ###p < 0.001 versus the OGD/R-treated anti-miR-Con group (two-way ANOVA followed by Holm-Sidak post hoc multiple comparison tests). Images of whole blots presented in the Fig. 11-1A and D were shown in Fig. 11-4. Download Figure 11-1, TIF file (136.4KB, tif)
Effect of miR-335-3p on neuronal plasticity after OGD/R in primary cortical neurons. (A-C) Neurons transduced with the mimic miR-335-3p lentivirus attenuated the OGD/R-induced decrease in synapsin-1 and PSD95. The cells were transduced with mimic miR-335-3p lentiviruses for 7 d, treated with OGD for 2 h and subjected to reperfusion for 6 h. n = 3. Synapsin-1 (mimic miR-335-3p: F (1, 8) = 25.33, P = 0.0010; OGD/R: F (1, 8) = 38.63, P = 0.0003; interaction: F (1, 8) = 0.6650, P = 0.4384). PSD95 (mimic miR-335-3p: F (1, 8) = 27.28, P = 0.0008; OGD/R: F (1, 8) = 52.57, P < 0.0001; interaction: F (1, 8) = 0.1417, P = 0.7164). *p < 0.05 and **p < 0.01 versus the control miR-Con group; #p < 0.05 versus the OGD/R-treated miR-Con group (two-way ANOVA followed by Holm-Sidak post hoc multiple comparison tests). (D-F) Neurons transduced with the anti-miR-335-3p lentivirus aggravated the OGD/R-induced decrease in synapsin-1 and PSD95. The cells were transduced with anti-miR-335-3p lentiviruses for 7 d, treated with OGD for 2 h and subjected to reperfusion for 6 h. n = 3. Synapsin-1 (anti-miR-335-3p: F (1, 8) = 61.78, P < 0.0001; OGD/R: F (1, 8) = 216.4, P < 0.0001; interaction: F (1, 8) = 0.2554, P = 0.6269). PSD95 (anti-miR-335-3p: F (1, 8) = 48.60, P = 0.0001; OGD/R: F (1, 8) = 224.5, P < 0.0001; interaction: F (1, 8) = 0.4629, P = 0.5155). **p < 0.01 and ***p < 0.001 versus the control anti-miR-Con group; ##p < 0.01 versus the OGD/R-treated anti-miR-Con group (two-way ANOVA followed by Holm-Sidak post hoc multiple comparison tests). Images of whole blots presented in the Fig. 11-2A and D were shown in Fig. 11-5. Download Figure 11-2, TIF file (77.8KB, tif)
Images of whole blots presented in the Figure 11C, E, H and K. Download Figure 11-3, TIF file (580.3KB, tif)
Images of whole blots presented in the Figure 11-1A and D. Download Figure 11-4, TIF file (317.8KB, tif)
Images of whole blots presented in the Figure 11-2A and D. Download Figure 11-5, TIF file (308.9KB, tif)
Images of whole blots presented in the Figure 12A and E. Download Figure 12-1, TIF file (563.5KB, tif)
The potential mechanisms underlying neuroprotective effect of neuron-specific knockdown of TIPARP in ischemic cortex after stroke. (A and B) Western blot analysis of Bax and Bcl-xl expression in the ischemic cortex at 24 h after tMCAO. Representative images are shown in A. B shows that neuron-specific knockdown of TIPARP attenuated the tMCAO-induced increase in the ratio of Bax/Bcl-xl. n = 6 animals per group. AAV-shRNA-TIPARP: F (1, 20) = 31.68, P < 0.0001; tMCAO: F (1, 20) = 40.9, P < 0.0001; interaction: F (1, 20) = 24.31, P < 0.0001. ***p < 0.001 versus the sham + AAV-shRNA-Con group; ###p < 0.001 versus the tMCAO + AAV-SYN-shRNA-Con group (two-way ANOVA followed by Holm-Sidak post hoc multiple comparison tests). (C and D) Western blot analysis of caspase-3 expression in the ischemic cortex at 24 h after tMCAO. Representative images are shown in C. D shows that neuron-specific knockdown of TIPARP attenuated the tMCAO-induced increase in activated caspase-3. n = 6 animals per group. AAV-shRNA-TIPARP: F (1, 20) = 16.1, P = 0.0007; tMCAO: F (1, 20) = 23.63, P < 0.0001; interaction: F (1, 20) = 21.57, P = 0.0002. ***p < 0.001 versus the sham + AAV-SYN-shRNA-Con group; ###p < 0.001 versus the tMCAO + AAV-SYN-shRNA-Con group (two-way ANOVA followed by Holm-Sidak post hoc multiple comparison tests). (E-G) RNA-seq analysis of differentially expressed genes (DEGs) in ipsilateral cortex at 24 h after stroke. Differences in gene expression among groups are shown in E. Red represents genes upregulated ≥ 2.0-fold and p < 0.05. Blue represents genes downregulated ≥ 2.0-fold and p < 0.05. F, Heatmap and unsupervised clustering of transcriptomes depicting DEGs, involved in the regulation of cell processes and signaling pathways, determined by RNA sequencing in the ischemic cortex among groups. Each row indicates a DEG. Each line represents the gene expression per animal. Red represents upregulated gene. Green represents downregulated gene. G, Kyoto Encyclopedia of Genes and Genomes (KEGG) annotation of DEGs as a result of neuron-specific knockdown of TIPARP 24 h after stroke. Enriched KEGG pathways of DEGs regulated in the tMCAO + AAV-SYN-shRNA-TIPARP group versus the tMCAO + AAV-SYN-shRNA-Con group. The KEGG pathways were summarized in three main categories: group A, cellular processes; group B, signaling pathways; group C, neurology. Rich factor represents the ratio of the number of enriched DEGs in the KEGG pathway and the number of annotated background genes in this pathway. The x-axis shows rich factor of each pathway. The y-axis refers to KEGG pathways. Color of the dots represents P-value, size of the dots represents the number of genes. n = 3 animals per group. Sham_C: sham + AAV-SYN-shRNA-Con; Sham_T: sham + AAV-SYN-shRNA-TIPARP; R24h_C: tMCAO (R24h) + AAV-SYN-shRNA-Con; R24h_T: tMCAO (R24h) + AAV-SYN-shRNA-TIPARP. Numbers: the sample number. Images of whole blots presented in the Fig. 13-1A and C were shown in Fig. 13-3. Download Figure 13-1, TIF file (398.6KB, tif)
Images of whole blots presented in the Figure 13D. Download Figure 13-2, TIF file (406.6KB, tif)
Images of whole blots presented in the Figure 13-1A and C. Download Figure 13-3, TIF file (288.5KB, tif)
Effect of neuron-specific shRNA-TIPARP AAV microinjection on mortality after focal cerebral ischemia. (A) Animal survival rate from the same experiments was recorded daily for 28 d after tMCAO. n=8 animals for both sham groups, n=18 and 17 animals for the tMCAO + AAV-SYN-shRNA-Con and tMCAO + AAV-SYN-shRNA-TIPARP groups, respectively. p = 0.1030 by log-rank test. (B) The mortality rate was calculated as the number of dead mice at 28 d after tMCAO in the AAV-SYN-shRNA-Con and AAV-SYN-shRNA-TIPARP groups. Mice that died during the experiments were excluded from further data analyses and statistics. Download Figure 14-1, TIF file (1MB, tif)
The potential mechanisms underlying long-term effect of neuron-specific knockdown of TIPARP in ischemic cortex after stroke. RNA-seq analysis of DEGs was performed in ipsilateral cortex at 28 d after stroke. (A) Differences in gene expression among groups. Red represents genes upregulated ≥ 2.0-fold and p < 0.05. Blue represents genes downregulated ≥ 2.0-fold and p < 0.05. (B) Heatmap and unsupervised clustering of transcriptomes depicting DEGs, involved in the regulation of signaling pathways, nervous system, and immunology, determined by RNA sequencing in the ischemic cortex among groups. Each row indicates a DEG. Each line represents the gene expression per animal. Red represents upregulated gene. Green represents downregulated gene. (C) KEGG annotation of DEGs as a result of neuron-specific knockdown of TIPARP at 28 d after stroke. Enriched KEGG pathways of DEGs regulated in the tMCAO + AAV-SYN-shRNA-TIPARP versus tMCAO + AAV-SYN-shRNA-Con groups. The KEGG pathways were summarized in three main categories: A, neurology; B, signaling pathways; C, immunology. Rich factor represents the ratio of the number of enriched DEGs in the KEGG pathway and the number of annotated background genes in this pathway. The x-axis shows rich factor of each pathway. The y-axis refers to KEGG pathways. Color of the dots represents P-value, size of the dots represents the number of genes. n = 3 animals per group. Sham_C: sham + AAV-SYN-shRNA-Con; Sham_T: sham + AAV-SYN-shRNA-TIPARP; R28d_C: tMCAO (R28d) + AAV-SYN-shRNA-Con; R28d_T: tMCAO (R28d) + AAV-SYN-shRNA-TIPARP. Numbers: the sample number. Download Figure 14-2, TIF file (136KB, tif)
Images of whole blots presented in the Figure 14E and G. Download Figure 14-3, TIF file (240.6KB, tif)