Abstract
Identifying the genetic architecture of complex phenotypes is a central goal of modern biology, particularly for disease-related traits. Genome-wide association methods are a classical approach for identifying the genomic basis of variation in disease phenotypes, but such analyses are particularly challenging in natural populations due to sample size difficulties. Extensive mark-recapture data, strong linkage disequilibrium, and a lethal transmissible cancer make the Tasmanian devil (Sarcophilus harrisii) an ideal model for such an association study. We used a RAD-capture approach to genotype 624 devils at ~16,000 loci and then used association analyses to assess the heritability of three cancer-related phenotypes: infection case-control (where cases were infected devils and controls were devils that were never infected), age of first infection, and survival following infection. The SNP array explained much of the phenotypic variance for female survival (>80%) and female case-control (>61%). We found that a few large-effect SNPs explained much of the variance for female survival (~5 SNPs explained >61% of the total variance) whereas more SNPs (~56) of smaller effect explained less of the variance for female case-control (~23% of the total variance). By contrast, these same SNPs did not account for a significant proportion of phenotypic variance in males, suggesting that the genetic bases of these traits and/or selection differ across sexes. Loci involved with cell adhesion and cell-cycle regulation underlay trait variation, suggesting that the devil immune system is rapidly evolving to recognize and potentially suppress cancer growth through these pathways. Overall, our study provided necessary data for genomics-based conservation and management in Tasmanian devils.
Keywords: genotype-phenotype, effect size, cancer, adaptation, GWAS
Introduction
A longstanding and significant problem in biology is understanding the genotype-phenotype relationship, and the recent development of genomic techniques has allowed researchers to address this problem with increasing sophistication. Because the genetic basis of polygenic traits, however, has been difficult to characterize, the genetics underlying many ecologically-important and disease-related traits is often unknown (Savolainen, Lascoux, & Merila, 2013; Schork et al., 2013; Shao et al., 2008; Wellenreuther & Hansson, 2016). To date, genome-wide association studies (GWASes) have largely been used to identify variants associated with complex phenotypes (Gibson, 2012; Manolio et al., 2009), particularly those related to disease. Although GWASes have successfully identified candidate loci, discovered variants often explain only a small proportion of the phenotypic variance (Eichler et al., 2010; Marjoram, Zubair & Nuzhdin, 2014; Park et al., 2010; Schork et al., 2013) and, therefore, possess low predictive power (Chatterjee et al., 2010). Indeed, most variants in humans exhibit small effect sizes and explain only a small proportion of heritability (Hindorff et al., 2009; Yang et al., 2010, 2012) despite a bias toward detecting and publishing larger effect sizes (i.e., “Winner’s Curse”; Dembeck et al., 2015; Gibson, 2012; Park et al., 2010).
A handful of studies have found evidence of large-effect SNPs for certain traits in humans (e.g., ~12 variants or fewer explained ~50% of the phenotypic variance), such as pigmentation (Sulem et al., 2007), age-related macular degeneration (Jakobsdottir et al. 2009), hypoxia adaptation (Simonson et al., 2010), lung cancer risk (Wang et al., 2014), and lipid levels associated with coronary heart disease (Helgadottir et al., 2016). Other studies, however, have argued that the polygenic nature of complex traits often require thousands of SNPs to explain a large proportion of the heritability; for example, ~9,500 variants explained ~50% of the variation in human height (Wood et al., 2014). Even in these case studies, much of the phenotypic variance is still unaccounted for (Yang et al., 2010, 2015). This missing heritability could be the result of many variants of small effect being missed due to significance thresholds, unsampled rare variants of large effect, and/or causal variants of any effect size not being captured by the SNP array (e.g., causal variants not being in linkage disequilibrium (LD) with the genotyped variants; Gibson, 2012; Hindorff et al., 2009; Manolio et al., 2009). Therefore, even in humans for which we have extensive genomic resources and genotypic information for millions of individuals, it remains unclear whether most quantitative traits are determined by many loci of small effect or a few loci of large effect.
Identifying the genetic basis of ecologically-important traits in threatened and endangered wildlife populations is becoming increasingly important for management and conservation, such as to guide captive breeding programs. Emerging infectious diseases (EIDs) are now considered a major cause of species’ declines and endangerment, and GWAS methods are a classical approach for identifying the genomic basis of variation in disease phenotypes (e.g., Hindorff et al., 2009). GWASes in natural populations, however, are particularly challenging owing to difficulties in achieving sufficient sample sizes to attain appropriate statistical power (Kardos et al. 2016). For example, identifying the genetic basis of a complex trait such as survival following infection requires extensive mark-recapture data that is often difficult to obtain. As a result, the rare examples of large-effect loci in wildlife species [e.g., color variation in mice (Linnen et al., 2013) and armor plating in sticklebacks (Colosimo et al., 2005)] have not typically been associated with disease and have been discovered using methods other than GWAS approaches. Although detecting variants of an appreciable effect in relatively large natural populations typically requires several thousand samples and extensive sampling of the genome (Yang et al., 2015), simulations have demonstrated that these variants can be reliably detected with far less sampling in relatively small populations with strong LD (e.g., LD >50 kb; Kardos et al. 2016). The Tasmanian devil (Sarcophilus harrisii) matches these criteria; with extensive mark-recapture field data, strong LD (~200 kb; Epstein et al., 2016), and a species-specific, nearly 100% lethal infectious cancer (Hamede et al., 2015), the devil is an ideal model for a GWAS in a natural population.
The Tasmanian devil is the largest extant marsupial carnivore, and facial tumors were first discovered in the northeastern part of the island in 1996. The disease is caused by an infectious cell line and is, therefore, a transmissible cancer (Pearse & Swift, 2006). Such cancers are extremely rare, with the only other natural cases found in dogs (Murgia, Pritchard, Kim, Fassati & Weiss, 2006) and bivalves (Metzger et al., 2016). Since 1996, devil facial tumor disease (DFTD) has spread approximately 80% of the way across Tasmania, caused upwards of 95% declines in populations affected the longest, and reduced the total population size by 80% (McCallum, 2008; McCallum et al., 2009). The cancer is spread via biting, which is common during social interactions (Hamede, McCallum & Jones, 2013). Low genetic diversity in devils due to historic population bottlenecks (3–5 k years ago; Brüniche-Olsen et al., 2014; Hendricks et al., 2017; Miller et al., 2011) and silencing of cell surface MHC molecules by DFTD have led to what appears to be universal susceptibility (Siddle et al., 2013). Simple epidemiological models have predicted devil extinction 25–30 years following disease arrival (McCallum et al., 2009), but the longest-diseased populations persist, suggesting devils may be responding to the strong selection imposed by DFTD (Jones et al., 2008). Indeed, recent work has discovered that some devils exhibited an immune response to DFTD (Pye et al., 2016) and, in rare cases, even tumor regression (Wright et al., 2017). Further, time-series genome scan analyses across three populations pre- and post-DFTD emergence found evidence for rapid evolution in genes related to immune function and cancer risk (Epstein et al., 2016). Taken together, these results suggest the evolution of resistance and/or tolerance to DFTD.
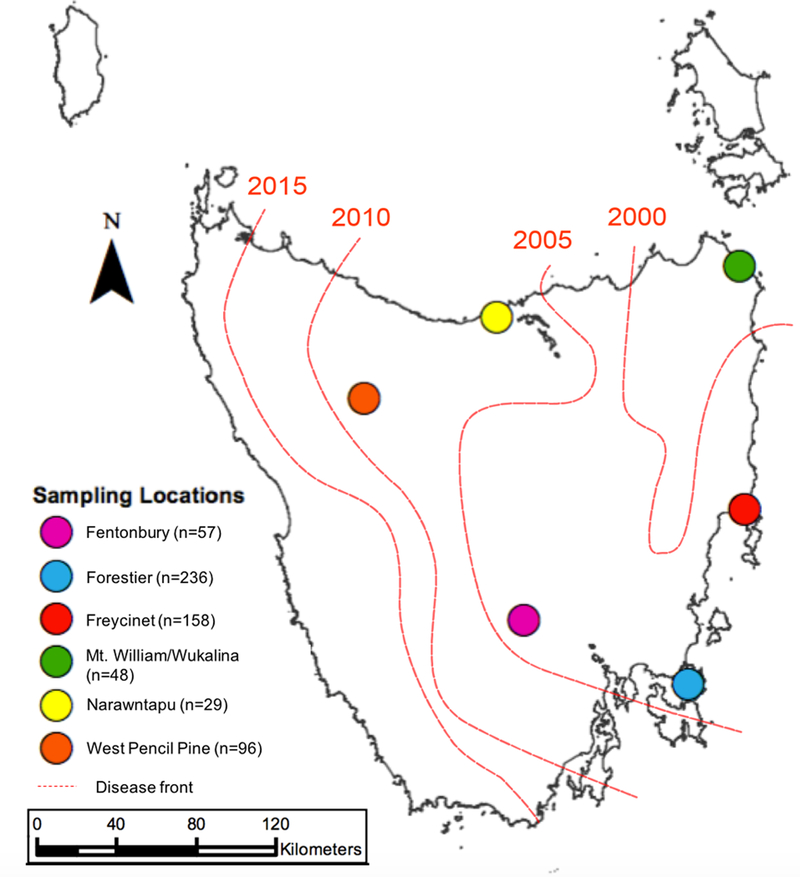
Although genomic regions showing a signature of selection have been identified (Epstein et al., 2016), the relationship of these markers to specific DFTD-related phenotypes as well as effect sizes of particular variants is unknown. We used a restriction-site associated DNA (RAD)-capture (“Rapture”) approach (Ali et al., 2016) to genotype 624 individuals from six localities (Figure 1) at approximately 16,000 RAD loci; loci were selected for homology with mammalian immune function, cancer recognition, and to provide broad coverage of the genome. We then used association analyses to assess heritability and identify loci underlying three devil phenotypes/phenotype proxies: infection case-control (where cases were infected devils and controls were devils that were never infected), age of first infection, and survival following infection.
Figure 1.
Sampling localities in Tasmania. We genotyped and phenotyped 624 Tasmanian devils from six collection sites. Red lines indicate the approximate location of the disease front from 2000 to 2015.
Materials and Methods
Trapping and phenotypic data
Tasmanian devils were trapped from 2000–2016 using custom-built traps constructed of 300 mm polypropylene pipe. All traps were baited with meat. Trapping sessions were carried out with 40–120 traps over 7–10 consecutive nights in a capture–mark–recapture framework. Traps were checked daily beginning at dawn; details of field methods were previously described (Hamede et al., 2015). Following initial capture, devils were individually tagged with microchip transponders (Allflex NZ Ltd, Palmerstone North, New Zealand). Devils were aged using a combination of head width (a linear measure of body size), molar eruption, molar tooth wear, and canine over-eruption. Most individuals were trapped as juveniles and, therefore, the age was known. DFTD status was categorized from histopathological confirmation of tumor biopsies. All devils were released following data collection (see below) except for nine devils from Forestier that were euthanized for health reasons; these devils were not included in survival analyses (see below), although their inclusion did not affect results (data not shown).
We performed association analyses (described below) for three phenotypes: 1) case-control where “cases” were infected individuals, and “controls” were individuals that were never infected and were captured (uninfected) ≥ 800 days from the estimated date of birth for the GEMMA analyses and ≥ 1000 days for the ANGSD analysis (see below), 2) the estimated age of an individual (in days) when it was first observed with DFTD, and; 3) length of known time to be alive (in days) after being observed with DFTD, our proxy for survival. Because observing the endpoint of death in a mark-recapture trapping framework is impossible (i.e., cannot trap a dead individual), we estimated survival as the difference in days between the first time an individual was observed with DFTD and the last time it was observed at all; we required at least two capture events following infection and that the individual must have survived ≥ 40 days to allow for re-capture to be possible. We recognize that our survival estimate was a simplified proxy for true survival, but we did not possess the necessary longitudinal data across all sampled sites to more robustly model true survival as previously described (Wells et al., 2017). Mark-recapture frameworks estimate survival for classes of individuals (e.g., McDonald 2018), but individual phenotype estimates are required for GWASes. We therefore chose to maximize sample size and statistical power by using the simplified survival proxy described above. To complement this simplified survival metric, we calculated an additional estimate for a single sampling locality (West Pencil Pine) for which we possessed the necessary longitudinal data to do so. West Pencil Pine is the most intensely and consistently sampled locality and has been sampled at three-month intervals since the outbreak of the disease in 2006. Additionally, tumor growth models and robust survival estimates (Hamede et al., 2017, Wells et al., 2017) have been calculated only for this locality. We used our individual tumor measurements and the logistic tumor growth curves from Wells et al. (2017) to back-calculate from the first observation of a tumor on an individual to the time when the tumor was at a volume of 3 mm3 (representing the size at which tumors are first observable) for 60 individuals from West Pencil Pine. We then followed the approach of Kéry and Schuab (2012) to test for differences in recapture probabilities while controlling for infection status and seasonality. We did not detect any significant differences in recapture probabilities between seasons or disease status (data not shown). Therefore, any adjustments made to estimate survival beyond the last capture would be made equally to diseased (i.e., cases) and non-diseased (i.e., controls) individuals and would have no effect on the association analyses. The new West Pencil Pine-specific survival proxy was the time in days from the back-calculated date of infection to the date of final capture. We use “survival” to refer to the simplified proxy (i.e., the difference in days between the first time an individual was observed with DFTD and the last time it was observed at all) throughout the manuscript and “West Pencil Pine-specific survival” to refer to the back-calculated survival estimate for the 60 West Pencil Pine individuals. The simplified survival proxy and the West Pencil Pine-specific estimates showed a significant, positive correlation (P<0.0001, R2=0.7117, R=0.8050; Figure S1), indicating that our simplified proxy provided a fair estimate of survival following infection. For age of first infection and case/control, we only included individuals that were born during or after the first year of DFTD in their respective population. Because the West Pencil Pine site was not strongly impacted by disease from 2006 – 2011 due to the presence of a tetraploid tumor associated with low prevalence rates (Hamede et al., 2015), we used 2011 as the year of disease arrival for this population. All phenotype data are provided in Table S1.
RAD-capture array development
We used the data (i.e., 360 individuals sequenced for 90,000 loci) from Epstein et al. (2016) to develop a RAD-capture array (Ali et al., 2016); the details of data processing and genotyping of the original RAD loci have been previously described (Epstein et al., 2016). RAD-capture extends traditional RADseq, which amplifies all loci adjacent to restriction enzyme cut sites, by adding a sequence capture step to the end of the RADseq protocol (Ali et al., 2016). We targeted 7,108 RAD loci that were genotyped in more than half the individuals, had ≤ 3 non-singleton SNPs, and had a SNP with a minor allele frequency (MAF) ≥ 0.05. To improve coverage of the genome, each locus was ≥ 20 kb away from other targeted loci. Additionally, we targeted 6,315 loci that had a non-singleton SNP within 50 kb of an immune related gene, had ≤ 4 non-singleton SNPs, and were genotyped in ≥ 1/3 of the individuals. Finally, we targeted 3,316 loci that showed some preliminary evidence of association with DFTD susceptibility and had ≤ 5 non-singleton SNPs (Epstein et al., 2016). In total, we targeted 15,898 RAD loci (there was some overlap among criteria). All targeted restriction cut sites are provided in Table S2.
Sequencing and data processing
RAD-capture libraries were constructed using the pstI restriction enzyme for 624 S. harrisii from six localities (Figure 1). Libraries were sequenced on an Illumina NextSeq at the University of Oregon Genomics and Cell Characterization Core Facility. Reads were de-multiplexed, and low-quality reads were removed using process_radtags from Stacks (v1.21; Catchen et al. 2013); this step also removed reads without recognizable barcodes or cut sites. The clone_filter program was used to remove potential PCR duplicates. Reads were then aligned to the reference genome (downloaded from Ensembl June 2014; Murchison et al., 2012) using bowtie2 (Langmead & Salzberg, 2012) with the --sensitive, --end-to-end, and -X 900 settings. Reads were retained if they aligned to an expected locus or were the mate of a read that aligned to an expected locus. Regions on the X chromosome were excluded from all analyses due to reduced genotyping accuracy and power (Wise, Gyi, & Manolio, 2013); only 350 of the 15,898 targets occurred on the X chromosome. Plots of number of loci covered per individual, number of individuals with coverage per locus, and mean depth of coverage per individual are provided in Figure S2.
Because GEMMA association analyses (see below) required individual genotype calls, genotype likelihoods for each potential segregating position were calculated with ANGSD (described in detail below; Korneliussen, Albrechtsen, & Nielsen, 2014); missing genotypes were imputed, and genotype probabilities were calculated in BEAGLE (Howie, Donnelly & Marchini, 2009). Imputation was conducted using a larger data set containing 3,568 individuals (data not shown), and each locality and chromosome were imputed separately; imputed genotypes are available upon request. The parameters and settings used in ANGSD (v0.910) are provided in Table S3.
GEMMA association tests
We fit a Bayesian Sparse Linear Mixed Model (BSLMM; Zhou, Carbonetto & Stephens, 2013) implemented in GEMMA (Zhou & Stephens, 2012) for case-control, age of first infection, survival following infection, and the West Pencil Pine-specific survival estimate to characterize the genetic basis of each trait. Because preliminary models indicated a greater predictive power for some phenotypes when males and females were examined separately (data not shown), sexes were analyzed collectively as well as independently. BSLMMs are a hybrid between linear mixed and sparse regression models and work under the assumption that most SNPs have a very small effect on the phenotype and a few SNPs have a larger effect. The model estimates an effect-size term for every SNP, the number of large-effect SNPs, the proportion of phenotypic variance explained by all SNPs (and other similar hyperparameters), and the proportion of phenotypic variance explained by only large-effect SNPs. The effect sizes are estimated simultaneously for all SNPs after accounting for relatedness via a K-matrix as well as background effects of all loci.
We used the imputed genotypes and genotype probabilities from BEAGLE (described above) as input for GEMMA. We ran GEMMA on SNPs with MAF > 0.05 and ≤ 5% missing data following imputation. We ran ≥ five million iterations following a 500,000-iteration burn-in, and we chose the linear BSLMM option except for the case-control phenotype, for which we chose the probit model. Similar to previous work (Lind et al., 2017), we used the posterior probability of a SNP having a large effect on a phenotype (after accounting for the effects of other SNPs in the genome) to identify candidate genes (see below) because we felt that this metric aptly captured a variant’s contribution to that particular trait. SNP counts and sample sizes are given in Table 1.
Table 1:
Number of SNPs and individuals included for each association test. Because different numbers of capture events were required for each phenotype, sample sizes vary by trait. Because sample sizes vary by trait, SNP filtering, particularly filtering for minor allele frequency, varied by trait, resulting in different numbers of SNPs for each analysis within a phenotype.
| Analysis | SNPs | Samples |
|---|---|---|
| GEMMA: age at infection | 10,777 (males) | 213 (males) |
| 11,503 (females) | 205 (females) | |
| 10,569 (both) | 418 (both) | |
| GEMMA: case/control | 10,777 (males) | 275 (males) |
| 11,503 (females) | 289 (females) | |
| 10,461 (both) | 564 (both) | |
| GEMMA: survival after infection | 10,777 (males) | 41 (males) |
| 11,503 (females) | 69 (females) | |
| 11,875 (both) | 110 (both) | |
| ANGSD: age at infection | 11,417 | 418 |
| ANGSD: case/control | 11,964 | 468 |
| ANGSD: survival after infection | 5,428 | 110 |
Association tests with ANGSD
To complement our GEMMA analyses, we used ANGSD to run generalized linear model association tests (Skotte, Korneliussen & Albrechtsen, 2012) on the same phenotypes described above except for the West Pencil Pine-specific survival estimate (not analyzed in ANGSD). Although ANGSD does not calculate chip heritability and other genetic architecture statistics (e.g., effect size) as the BSLMM in GEMMA does, ANGSD directly incorporates genotyping uncertainty into the analysis by estimating the posterior probability of each possible genotype (using estimates of the population allele frequency); the generalized linear models used to test for an association between genotype and phenotype are summed over the possible genotypes and weighted by the posterior probabilities. ANGSD also directly calculates p-values for each per-SNP association whereas GEMMA does not. The same settings used for the allele frequency estimation were used here (Table S3). We used an additive association model and estimated the genotype probability using the allele frequency as a prior. Only sites with MAF ≥ 5% (as estimated by ANGSD) were included.
Sex was included as a covariate for all analyses. For the survival proxy, we also used age at first infection as a covariate. Because population structure can lead to inflated p-values in association testing (Xu & Shete, 2005), we conducted a PCA on genotypes and included principal component (PC) axes as covariates in the analyses. First, we obtained a genetic covariance matrix from the genotype likelihoods using ngsCovar (Fumagalli, Vieira, Linderoth & Nielsen, 2014) and extracted the PCs. We next calculated Tracy-Widom significance values for the PCs (Patterson, Price & Reich, 2006). Except for the survival phenotype (Pearson’s correlation between first PC and phenotype; p=0.04), PCs were not significantly correlated with phenotypes. For completeness, we ran the association analysis for each phenotype with no PCs and with a number of PCs chosen based on the appearance of scree plots (1–7, depending on the phenotype). We found that, without including any PCs, age at first infection exhibited a low inflation factor (1.02), and the QQ plot indicated a nearly flat distribution of p-values (Figure S3). For the case-control analysis, there was some inflation (inflation factor = 1.21), but including PCs resulted in little improvement in the inflation factor or the shape of the curve (inflation factor with PCs ranged from 1.18 – 2.18; Figure S3). For survival after infection, however, there was a clear improvement by including five PCs as covariates (Figure S3). Visualization of QQ plots was achieved using the qqman R package (Turner, 2014).
Following the recommendations of François, Martins, Cave & Schoville (2016), we adjusted the p-values of ANGSD results based on a genomic inflation factor correction. The genomic inflation factor is the ratio between the median Z-scores and the expected median Z-scores for a χ2 distribution with one degree of freedom (Devlin & Roeder, 1999), and works under the assumption that most SNPs are not strongly associated with the phenotype of interest. To perform the adjustment, we divided the raw Z-scores by the inflation factor. All p-values were adjusted before using them to identify candidate genes (Figure S4).
Identification of candidate genes
Candidate SNPs were identified as the top 0.1% of SNPs for each phenotype. Tasmanian devil genomes have extensive LD (~200 kb; Epstein et al., 2016), and we used bedtools (Quinlan & Hall, 2010) to conservatively identify candidate genes within 100 kb of the top SNPs. Putative gene functions were identified using GeneCards (www.genecards.org) and/or NCBI.
Results
The genetic basis of cancer-resistant phenotypes
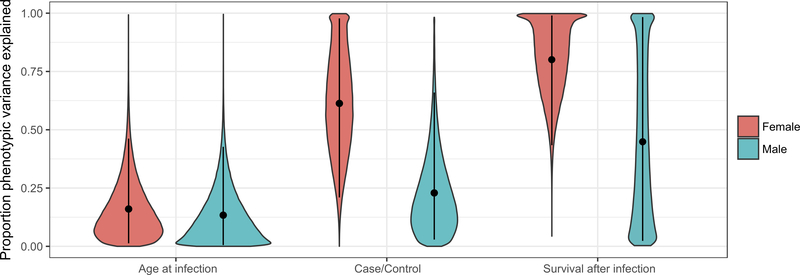
We first conducted joint-sex association tests using a BSLMM in GEMMA and found that the mean of the posterior distribution of the proportion of phenotypic variance explained (PVE; a measure of the additive effect of all interrogated SNPs, or “chip heritability”; Zhou, Carbonetto & Stephens, 2013) accounted for a substantial proportion of the variance for survival following infection (0.709; 95% CI=0.276–0.999; Table 2). The survival proxy (the length of time known to be alive after being observed with DFTD) was associated with a few SNPs of large-effect (~7), and these SNPs accounted for 60.6% of the PVE accounted for by all SNPs (or ~43% of the total PVE; Table 2). Mean PVE was less in the case-control phenotype (0.263; 95% CI=0.088–0.502; Table 2), and this trait was associated with more SNPs of smaller effect (~63; Table 2). PVE credible intervals were large and approximately overlapped zero for age at first infection (Table 2). Although mean PVE was large for the West Pencil Pine-specific survival estimate (0.537), PVE credible intervals were also large (95% CI = 0.020–0.998; Figure S5; Table S4).
Table 2:
GEMMA association results and genetic architecture statistics. The mean variance explained by all genotypes represents the mean of the posterior distribution of the proportion of phenotypic variance explained. The mean variance explained by large-effect SNPs represents the percentage of the total variance explained by only large-effect SNPs (e.g., large-effect SNPs associated with female age at first infection explained 36.5% of the 16.1% total variance explained by all SNPs, or approximately 5.9% of the total variance). Parentheses present 95% posterior credible intervals from the posterior distributions.
| Sex | Phenotype | Mean Variance Explained By All Genotypes (%) | Median Variance Explained By All Genotypes (%) | Number large-effect SNPs | Mean Variance Explained By Large-effect SNPs (%) |
|---|---|---|---|---|---|
| Both | Age at infection | 7.1 (0.3–21.6) | 5.7 | 50.0 (0–270) | 39.1 (0–96.2) |
| Both | Case/control | 26.3 (8.8–50.2) | 25.1 | 63.3 (0–269) | 37.2 (0–96.1) |
| Both | Survival after infection | 70.9 (27.6–99.9) | 73.3 | 7.0 (1–34) | 60.6 (26.2–97.3) |
| Female | Age at infection | 16.1 (0.9 – 47.1) | 13.1 | 38.1 (0 – 203) | 36.5 (0 – 95.5) |
| Female | Case /control | 61.4 (21.4 – 98.4) | 61.3 | 56.1 (0 – 261) | 38.3 (0 – 95.6) |
| Female | Survival after infection | 80.1 (46.7 – 99.9) | 82.4 | 4.8 (1 – 14) | 76.5 (43.3 – 98.6) |
| Male | Age at infection | 13.3 (0.4 – 42.5) | 10.2 | 44.1 (0 – 246) | 42.0 (0 – 96.4) |
| Male | Case /control | 23.0 (1.4 – 64.9) | 19.4 | 52.7 (0 – 261) | 39.6 (0 – 96.3) |
| Male | Survival after infection | 44.8 (1.1 – 99.6) | 39.2 | 37.0 (0 – 196) | 43.4 (0 – 96.6) |
Because preliminary models indicated a greater predictive power for some phenotypes when males and females were examined separately, we next conducted sex-specific association tests using a BSLMM in GEMMA and found that the mean PVE was substantial for female case-control (0.614; 95% CI = 0.214–0.984) and female survival following infection (0.801; 95% CI = 0.467–0.999; Figure 2; Table 2). Female survival was found to be associated with a few SNPs of large-effect (~ 5; Table 2); these SNPs accounted for 76.5% of the PVE accounted for by all SNPs (e.g., five SNPs associated with female survival explained 76.5% of the 80.1% total PVE explained by all SNPs, or ~ 61% of the total PVE). Female case-control was associated with more SNPs of smaller effect (~56 SNPs accounted for ~ 23% of the total PVE; Table 2). PVE credible intervals were large and overlapped zero (or nearly so) for all traits in males (including survival following infection) as well as age of first infection in females (Figure 2; Table 2), potentially owing to small sample sizes and other confounding factors (Table 1; see Discussion for detail). The large PVE credible intervals for male survival and male case-control, however, indicated that the substantial amount of variance explained in the initial joint-sex association tests for survival and, to a lesser extent, case-control, were driven by females. Consistent with the joint-sex analyses above, mean PVE as well as PVE credible intervals were large for the West Pencil Pine-specific survival estimate in females (0.682, 95% CI= 0.052–0.999) and males (0.494, 95% CI = 0.015–0.998; Figure S5; Table S4), suggesting that our simplified survival proxy was necessary to achieve greater sample sizes and, therefore, power for our association analyses.
Figure 2.
The proportion of the phenotypic variance explained by all sequenced genotypes for three disease-related traits. We plotted the gemma posterior distribution of the proportion of phenotypic variance explained by all SNPs (y-axis) for each phenotype (x-axis) for females (pink) and males (blue) independently. Points indicate the mean of the distribution, and bars represent 95% posterior credible intervals.
Cancer-resistant candidate genes
To determine which specific loci were associated with female survival and female case-control, we identified genes within 100 kb of the top 0.1% of SNPs from the GEMMA analyses, given that LD is strong at this scale within the genome (i.e., ~200 kb; Epstein et al., 2016); this approach identified candidate genes linked to variants with the largest effect for each trait. For female case-control, nine genes were identified within 100 kb of a top SNP, and five of these genes had putative functions; one gene, secretory carrier membrane protein 1, is implicated in immune function (Table 3). The largest posterior probability of any top SNP being of large-effect was 0.193 and shared across four genes, again indicating that female case-control was associated with more SNPs of smaller effect (Table S5). Eight genes were within 100 kb of a top SNP related to female survival, and two of these genes had putative functions; one gene, ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 (ST8SIA2), is implicated in immune function. ST8SIA2 is related to chronic inflammation following infection and possessed the largest posterior probability of being a large-effect SNP for female survival (0.995; Table S5).
Table 3:
Candidate genes within 100 kb of the top 0.1% SNPs for female case/control and female survival following infection.
| Ensembl ID | Gene name | Putative function |
|---|---|---|
| Female case/control | ||
| ENSSHAG00000011304 | Forkhead box G1 | Repressor |
| ENSSHAG00000001038 | A kinase (PRKA) anchor protein 10 | Binds to regulatory subunits of proteinase K |
| ENSSHAG00000003186 | Unc-51 like autophagy activating kinase 2 | Autophagy |
| ENSSHAG00000016943* | Secretory carrier membrane protein 1* | Recycling carrier to cell surface; implicated in immune function* |
| ENSSHAG00000017224 | Adaptor-related protein complex 3, beta 1 subunit | Organelle biogenesis |
| Female survival after infection | ||
| ENSSHAG00000010546* | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2* | Catalyzes sialic acid transfer; related to chronic inflammation due to infection* |
| ENSSHAG00000013531 | Solute carrier family 12, member 8 | Cation/chloride cotransporter |
Only genes with a putative function were included. Genes with functions related to immune response and/or cancer risk were indicated with an asterisk.
For the four phenotypes for which the PVE credible intervals approximately overlapped zero (all three traits in males and female age at first infection), all SNPs exhibited posterior probabilities of being a large-effect SNP ≤ 0.034, except for male age at first infection (≤ 0.069). Similarly, all SNPs associated with West Pencil Pine-specific survival exhibited posterior probabilities of being a large-effect SNP ≤ 0.018 for females and ≤0.056 for males. All posteriors for SNPs associated with these traits were substantially lower than those found for the two female phenotypes discussed above, indicating that the posterior probability of being a large-effect SNP reflected a variant’s “significance” to a particular phenotype.
To complement the GEMMA results described above, we identified candidate genes within 100 kb of the top 0.1% of the SNPs identified in the ANGSD analyses for survival, age at first infection, and case-control (Table S5). For case-control, 17 genes were identified within 100 kb of a top SNP, and 15 of these genes had putative functions. Nine of these genes are implicated in immune/tumor function and are listed in Table S5; candidates included genes involved with apoptosis [e.g., ADAMTS-like 4 (p<0.0001), myeloid cell leukemia 1 (p<0.0001)], cell adhesion and signaling [integrin alpha 10 (p=0.0021)], and non-self DNA recognition [RNA Polymerase III Subunit C (p=0.0021)]. For survival following infection, five genes were identified within 100 kb of a top SNP (Table S5). Four of these genes had putative functions, but none were implicated in immune and/or tumor function. For age at first infection, we identified nine genes within 100 kb of a top SNP (Table S5), and seven of these genes had putative functions. Four of these genes were implicated in immune/tumor function.
The top 1% of SNPs (and corresponding genes and statistics) from the nine GEMMA analyses and three ANGSD analyses are provided in Table S5; the top SNPs for the West Pencil Pine-specific survival estimate are not included.
Discussion
We identified the genetic basis underlying key cancer-resistant/tolerant phenotypes in Tasmanian devils and found that few loci of large effect explained a large proportion of the phenotypic variance for female survival. Female case-control was associated with more SNPs of smaller effect, although relatively few loci (56 rather than thousands of variants) still accounted for a substantial amount of the total variance (23%; all genotypes accounted for 61.4%). Given the recent discovery of DFTD and subsequent evidence for a rapid evolutionary response in devils (approximately 4–6 generations; Epstein et al., 2016), we might expect that selection acted on standing genetic variation in the form of a few, large-effect loci. First-step (i.e., initial) mutations are often of large-effect and confer a large fitness advantage because these mutations outcompete other, less beneficial mutations in the population (Rokyta et al. 2005), especially when the population is far from the phenotypic optimum as would be expected when selection is imposed by a novel disease. Novel beneficial mutations are unlikely to arise over short timescales, but large-effect variants can be segregating in the population neutrally prior to the onset of novel selective pressures (i.e., prior to DFTD arrival). Our results, at least for female survival, were consistent with these expectations. In contrast, PVE confidence intervals approximately overlapped zero for all three traits in males and age of first infection in females.
A smaller male sample size for survival (41 males versus 69 females; Table 1) suggested that a lack of power for the male survival association test may explain this sex-specific difference. However, similar sample sizes among sexes for case-control (275 males at 10,777 SNPs versus 289 females at 11,503 SNPs) suggested that the difference in case-control PVE across males and females was biological rather than an artifact of our sampling. Whether this difference in case-control PVE represents a difference in genomic architecture and/or DFTD-imposed selection strength (e.g., due to differences in fecundity or other life history traits) across males and females remains uncertain. Sex chromosomes offer one possible molecular mechanism underlying this case-control difference. Sex chromosomes can enable the rapid evolution of sexual dimorphism, and both the Y-chromosome (Kutch & Fedorka, 2015) and the inactivated X-chromosome in females (Wang et al., 2016) have been shown to influence genome-wide expression, particularly for loci with an immune-related function. Kutch and Fedorka (2017) recently detected significant Y-chromosome-by-genetic-background epistatic effects following infection, including evidence of sign epistasis (i.e., reversal of fitness values). If a beneficial female allele is deleterious in males for a shared trait such as case-control, the trait heritability in males could be significantly reduced, consistent with our results. Additionally, males may experience stronger selection because of higher variance in reproductive success, reducing the segregating genetic variation for male-expressed, DFTD-related traits. Further work is needed to test these hypotheses and identify the mechanism underlying the sex-based differences in PVE we identified in this study.
Prior work showed a rapid evolutionary response in two small genomic regions on chromosomes 3 and 4 in genes associated with cell-cycle regulation, cell adhesion, and immune response (Epstein et al., 2016), but the relationship of these genomic regions to specific phenotypes was unknown. Consistent with this previous work, we found that loci involved with immunity, cell-cycle regulation, and cell adhesion underlay variation in female survival and may thereby drive cancer resistance (or tolerance; Wells et al., 2017). For example, Epstein et al. (2016) identified CD146 as a candidate gene, and we identified ST8SIA2 as a top candidate gene associated with female survival. Both of these genes are involved with cell adhesion and often regulate inflammatory response. Collectively, the functions of these candidate genes, along with the phenotypes they are associated with, indicate that the devil immune system is evolving to recognize, and potentially suppress, the growth of tumor cells, providing a potential mechanism for the recently identified tumor regression in specific devil populations (Wright et al., 2017).
As global biodiversity is increasingly threatened by anthropogenically-driven changes such as EIDs, effective conservation management will likely benefit from an understanding of the genomic signatures of adaptation in natural populations. Here, we showed that genomic studies can be applied to natural populations to guide conservation and management. We found that few loci of large effect explain variation in survival in the face of a lethal EID. In such cases, clear management recommendations emerge, such as basing the selection of individuals for captive breeding programs on this genetic information to ensure the survival of the Tasmanian devil. Discovery of EIDs continues to increase, and together with predictions of rapid and dramatic global change, will necessitate rapid responses in terms of conservation and management.
Supplementary Material
Acknowledgments
This work was funded under NSF grant DEB 1316549 as part of the joint NSF-NIH-USDA Ecology and Evolution of Infectious Diseases program to AS, PAH, MJ and HM, the Australian Research Council (ARC) Future Fellowship (FT100100250) to MJ, ARC Large Grants (A00000162) to MJ, Linkage (LP0561120) to MJ and HM, and Discovery (DP110102656) to MJ and HM. Field work was additionally supported by Eric Guiler grants from the Save the Tasmanian Devil Appeal – University of Tasmania Foundation, the Ian Potter Foundation, the Australian Academy of Science, Estate of W.V. Scott, the National Geographic Society, the Mohammed bin Zayed Conservation Fund, the Holsworth Wildlife Trust, the Tasmanian Government, and the Commonwealth Government of Australia. Animal use was approved by the IACUC at Washington State University (ASAF#04392), the University of Tasmania Animal Ethics Committee (A0008588, A0010296, A0011696, A0013326, A0015835), and the Department of Primary Industries, Parks, Water and Environment Animal Ethics Committee. Additional support was provided by NIH P30 GM103324.
Footnotes
Data accessibility
The sequence data has been deposited at NCBI under BioProject PRJNA306495 (http://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA306495); BioSample accessions are provided in Table S6. All phenotypic data are provided in Table S1.
References
- 1.Ali OA, O’Rourke SM, Amish SJ, Meek MH, Luikart G, Jeffres C, & Miller MR (2016). RAD capture (Rapture): flexible and efficient sequence-based genotyping. Genetics, 202(2), 389–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brüniche-Olsen A, Jones ME, Austin JJ, Burridge CP, & Holland BR (2014). Extensive population decline in the Tasmanian devil predates European settlement and devil facial tumour disease. Biology Letters, 10(11), 20140619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Catchen J, Hohenlohe PA, Bassham S, Amores A, & Cresko WA (2013). Stacks: an analysis tool set for population genomics. Molecular Ecology, 22(11), 3124–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chatterjee N, Wheeler B, Sampson J, Hartge P, Chanock SJ, & Park JH (2013). Projecting the performance of risk prediction based on polygenic analyses of genome-wide association studies. Nature Genetics, 45(4), 400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colosimo PF, Hosemann KE, Balabhadra S, Villarreal G, Dickson M, Grimwood J, Schmutz J, Myers RM, Schluter D & Kingsley DM (2005). Widespread parallel evolution in sticklebacks by repeated fixation of ectodysplasin alleles. Science, 307(5717), 1928–1933. [DOI] [PubMed] [Google Scholar]
- 6.Dembeck LM, Huang W, Magwire MM, Lawrence F, Lyman RF, & Mackay TF (2015). Genetic architecture of abdominal pigmentation in Drosophila melanogaster. PLoS Genetics, 11(5), e1005163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devlin B, & Roeder K (1999). Genomic control for association studies. Biometrics, 55(4), 997–1004. [DOI] [PubMed] [Google Scholar]
- 8.Eichler EE, Flint J, Gibson G, Kong A, Leal SM, Moore JH, & Nadeau JH (2010). Missing heritability and strategies for finding the underlying causes of complex disease. Nature Reviews Genetics, 11(6), 446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Epstein B, Jones M, Hamede R, Hendricks S, McCallum H, Murchison EP, Schönfeld B, Wiench C, Hohenlohe P & Storfer A (2016). Rapid evolutionary response to a transmissible cancer in Tasmanian devils. Nature Communications, 7, 12684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.François O, Martins H, Caye K, & Schoville SD (2016). Controlling false discoveries in genome scans for selection. Molecular Ecology, 25(2), 454–469. [DOI] [PubMed] [Google Scholar]
- 11.Fumagalli M, Vieira FG, Linderoth T, & Nielsen R (2014). ngsTools: methods for population genetics analyses from next-generation sequencing data. Bioinformatics, 30(10), 1486–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibson G (2012). Rare and common variants: twenty arguments. Nature Reviews Genetics, 13(2), 135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamede RK, McCallum H, & Jones M (2013). Biting injuries and transmission of Tasmanian devil facial tumour disease. Journal of Animal Ecology, 82(1), 182–190. [DOI] [PubMed] [Google Scholar]
- 14.Hamede RK, Pearse AM, Swift K, Barmuta LA, Murchison EP, & Jones ME (2015). Transmissible cancer in Tasmanian devils: localized lineage replacement and host population response. Proceedings of the Royal Society B: Biological Sciences, 282, 20151468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamede RK, Beeton NJ, Carver S & Jones ME (2017). Untangling the model muddle: empirical tumour growth in Tasmanian devil facial tumour disease. Scientific Reports, 7(1), 6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helgadottir A, Gretarsdottir S, Thorleifsson G, Hjartarson E, Sigurdsson A, Magnusdottir A, Jonasdottir A, Kristjansson H, Sulem P, Oddsson A & Sveinbjornsson G (2016). Variants with large effects on blood lipids and the role of cholesterol and triglycerides in coronary disease. Nature Genetics, 48(6), 634–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hendricks S, Epstein B, Schönfeld B, Wiench C, Hamede R, Jones M, Storfer A & Hohenlohe P (2017). Conservation implications of limited genetic diversity and population structure in Tasmanian devils (Sarcophilus harrisii). Conservation Genetics, 18(4), 977–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, & Manolio TA (2009). Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proceedings of the National Academy of Sciences, 106(23), 9362–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howie BN, Donnelly P, & Marchini J (2009). A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genetics, 5(6), e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jakobsdottir J, Gorin MB, Conley YP, Ferrell RE, & Weeks DE (2009). Interpretation of genetic association studies: markers with replicated highly significant odds ratios may be poor classifiers. PLoS Genetics, 5(2), e1000337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones ME, Cockburn A, Hamede R, Hawkins C, Hesterman H, Lachish S, Mann D, McCallum H & Pemberton D (2008). Life-history change in disease-ravaged Tasmanian devil populations. Proceedings of the National Academy of Sciences, 105(29), 10023–10027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kardos M, Husby A, McFarlane SE, Qvarnström A, & Ellegren H (2016). Whole-genome resequencing of extreme phenotypes in collared flycatchers highlights the difficulty of detecting quantitative trait loci in natural populations. Molecular Ecology Resources, 16(3), 727–741. [DOI] [PubMed] [Google Scholar]
- 23.Kéry M & Schaub M (2011). Bayesian population analysis using WinBUGS: a hierarchical perspective. Academic Press. [Google Scholar]
- 24.Korneliussen TS, Albrechtsen A, & Nielsen R (2014). ANGSD: analysis of next generation sequencing data. BMC bioinformatics, 15(1), 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kutch IC, & Fedorka KM (2015). Y-linked variation for autosomal immune gene regulation has the potential to shape sexually dimorphic immunity. Proceedings of the Royal Society B: Biological Sciences. 282, 20151301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kutch IC, & Fedorka KM (2017). A test for Y-linked additive and epistatic effects on surviving bacterial infections in Drosophila melanogaster. Journal of evolutionary biology, 30, 1400–1408. [DOI] [PubMed] [Google Scholar]
- 27.Langmead B, & Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nature Methods, 9(4), 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lind BM, Friedline CJ, Wegrzyn JL, Maloney PE, Vogler DR, Neale DB, & Eckert AJ (2017). Water availability drives signatures of local adaptation in whitebark pine (Pinus albicaulis Engelm.) across fine spatial scales of the Lake Taho Basin, USA. Molecular Ecology, 26(12), 3168–3185. [DOI] [PubMed] [Google Scholar]
- 29.Linnen CR, Poh YP, Peterson BK, Barrett RD, Larson JG, Jensen JD, & Hoekstra HE (2013). Adaptive evolution of multiple traits through multiple mutations at a single gene. Science, 339(6125), 1312–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A & Cho JH (2009). Finding the missing heritability of complex diseases. Nature, 461(7265), 747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marjoram P, Zubair A, & Nuzhdin SV (2014). Post-GWAS: where next? More samples, more SNPs or more biology? Heredity, 112(1), 79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCallum H (2008). Tasmanian devil facial tumour disease: lessons for conservation biology. Trends in Ecology & Evolution, 23(11), 631–637. [DOI] [PubMed] [Google Scholar]
- 33.McCallum H, Jones M, Hawkins C, Hamede R, Lachish S, Sinn DL, Beeton N & Lazenby B (2009). Transmission dynamics of Tasmanian devil facial tumor disease may lead to disease-induced extinction. Ecology, 90(12), 3379–3392. [DOI] [PubMed] [Google Scholar]
- 34.McDonald T (2018). mra: Mark-Recapture Analysis. R Package version 2.16.11. https://CRAN.R-project.org/package=mra
- 35.Metzger MJ, Villalba A, Carballal MJ, Iglesias D, Sherry J, Reinisch C, Muttray AF, Baldwin SA & Goff SP (2016). Widespread transmission of independent cancer lineages within multiple bivalve species. Nature, 534(7609), 705–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller W, Hayes VM, Ratan A, Petersen DC, Wittekindt NE, Miller J, Walenz B, Knight J, Qi J, Zhao F & Wang Q (2011). Genetic diversity and population structure of the endangered marsupial Sarcophilus harrisii (Tasmanian devil). Proceedings of the National Academy of Sciences, 108(30), 12348–12353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murchison EP, Schulz-Trieglaff OB, Ning Z, Alexandrov LB, Bauer MJ, Fu B, Hims M, Ding Z, Ivakhno S, Stewart C & Ng BL (2012). Genome sequencing and analysis of the Tasmanian devil and its transmissible cancer. Cell, 148(4), 780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murgia C, Pritchard JK, Kim SY, Fassati A, & Weiss RA (2006). Clonal origin and evolution of a transmissible cancer. Cell, 126(3), 477–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park JH, Wacholder S, Gail MH, Peters U, Jacobs KB, Chanock SJ, & Chatterjee N (2010). Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nature Genetics, 42(7), 570–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patterson N, Price AL, & Reich D (2006). Population structure and eigenanalysis. PLoS Genetics, 2(12), e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pearse AM, & Swift K (2006). Allograft theory: transmission of devil facial-tumour disease. Nature, 439(7076), 549. [DOI] [PubMed] [Google Scholar]
- 42.Pye RJ, Pemberton D, Tovar C, Tubio JM, Dun KA, Fox S, Darby J, Hayes D, Knowles GW, Kreiss A & Siddle HV (2016). A second transmissible cancer in Tasmanian devils. Proceedings of the National Academy of Sciences, 113(2), 374–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quinlan AR, & Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics, 26(6), 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rokyta DR, Joyce P, Caudle SB, & Wichman HA (2005). An empirical test of the mutational landscape model of adaptation using a single-stranded DNA virus. Nature Genetics, 37(4), 441. [DOI] [PubMed] [Google Scholar]
- 45.Savolainen O, Lascoux M, & Merilä J (2013). Ecological genomics of local adaptation. Nature Reviews Genetics, 14(11), 807–820. [DOI] [PubMed] [Google Scholar]
- 46.Schork AJ, Thompson WK, Pham P, Torkamani A, Roddey JC, Sullivan PF, Kelsoe JR, O’Donovan MC, Furberg H, Schork NJ & Andreassen OA (2013). All SNPs are not created equal: genome-wide association studies reveal a consistent pattern of enrichment among functionally annotated SNPs. PLoS Genetics, 9(4), e1003449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shao H, Burrage LC, Sinasac DS, Hill AE, Ernest SR, O’Brien W, Courtland HW, Jepsen KJ, Kirby A, Kulbokas EJ & Daly MJ (2008). Genetic architecture of complex traits: large phenotypic effects and pervasive epistasis. Proceedings of the National Academy of Sciences, 105(50), 19910–19914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Siddle HV, Kreiss A, Tovar C, Yuen CK, Cheng Y, Belov K, Swift K, Pearse AM, Hamede R, Jones ME & Skjødt K (2013). Reversible epigenetic down-regulation of MHC molecules by devil facial tumour disease illustrates immune escape by a contagious cancer. Proceedings of the National Academy of Sciences, 110(13), 5103–5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simonson TS, Yang Y, Huff CD, Yun H, Qin G, Witherspoon DJ, Bai Z, Lorenzo FR, Xing J, Jorde LB & Prchal JT (2010). Genetic evidence for high-altitude adaptation in Tibet. Science, 329(5987), 72–75. [DOI] [PubMed] [Google Scholar]
- 50.Skotte L, Korneliussen TS, & Albrechtsen A (2012). Association Testing for Next-Generation Sequencing Data Using Score Statistics. Genetic Epidemiology, 36(5), 430–437. [DOI] [PubMed] [Google Scholar]
- 51.Sulem P, Gudbjartsson DF, Stacey SN, Helgason A, Rafnar T, Magnusson KP, Manolescu A, Karason A, Palsson A, Thorleifsson G & Jakobsdottir M (2007). Genetic determinants of hair, eye and skin pigmentation in Europeans. Nature Genetics, 39(12), 1443–1452. [DOI] [PubMed] [Google Scholar]
- 52.Turner SD (2014). qqman: an R package for visualizing GWAS results using QQ and manhattan plots. BioRxiv, 005165. [Google Scholar]
- 53.Wang J, Syrett CM, Kramer MC, Basu A, Atchison ML, & Anguera MC (2016). Unusual maintenance of X chromosome inactivation predisposes female lymphocytes for increased expression from the inactive X. Proceedings of the National Academy of Sciences, 113(14), E2029–E2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Y, McKay JD, Rafnar T, Wang Z, Timofeeva MN, Broderick P, Zong X, Laplana M, Wei Y, Han Y & Lloyd A (2014). Rare variants of large effect in BRCA2 and CHEK2 affect risk of lung cancer. Nature Genetics, 46(7), 736–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wellenreuther M, & Hansson B (2016). Detecting polygenic evolution: problems, pitfalls, and promises. Trends in Genetics, 32(3), 155–164. [DOI] [PubMed] [Google Scholar]
- 56.Wells K, Hamede RK, Kerlin DH, Storfer A, Hohenlohe PA, Jones ME, & McCallum HI (2017). Infection of the fittest: devil facial tumour disease has greatest effect on individuals with highest reproductive output. Ecology Letters, 20(6), 770–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wise AL, Gyi L, & Manolio TA (2013) eXclusion: toward integrating the X chromosome in genome-wide association analyses. The American Journal of Human Genetics, 92(5), 643–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wood AR, Esko T, Yang J, Vedantam S, Pers TH, Gustafsson S, Chu AY, Estrada K, Luan JA, Kutalik Z & Amin N (2014). Defining the role of common variation in the genomic and biological architecture of adult human height. Nature Genetics, 46(11), 1173–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wright B, Willet CE, Hamede R, Jones M, Belov K, & Wade CM (2017). Variants in the host genome may inhibit tumour growth in devil facial tumours: evidence from genome-wide association. Scientific Reports, 7(1), 423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu H, & Shete S (2005). Effects of population structure on genetic association studies. BMC Genetics, 7(1), S109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Madden PA, Heath AC, Martin NG, Montgomery GW & Goddard ME (2010). Common SNPs explain a large proportion of the heritability for human height. Nature Genetics, 42(7), 565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang J, Ferreira T, Morris AP, Medland SE, Madden PA, Heath AC, Martin NG, Montgomery GW, Weedon MN, Loos RJ & Frayling TM (2012). Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nature Genetics, 44(4), 369–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang J, Bakshi A, Zhu Z, Hemani G, Vinkhuyzen AA, Lee SH, Robinson MR, Perry JR, Nolte IM, van Vliet-Ostaptchouk JV & Snieder H (2015). Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nature Genetics, 47(10), 1114–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou X, Carbonetto P, & Stephens M (2013). Polygenic modeling with Bayesian sparse linear mixed models. PLoS Genetics, 9(2), e1003264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou X, & Stephens M (2012). Genome-wide efficient mixed-model analysis for association studies. Nature Genetics, 44(7), 821–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.