Abstract
A mammalian cell houses two genomes located separately in the nucleus and mitochondria. During evolution, communications and adaptations between these two genomes occur extensively to achieve and sustain homeostasis for cellular functions and regeneration. Mitochondria provide the major cellular energy and contribute to gene regulation in the nucleus, whereas more than 98% of mitochondrial proteins are encoded by the nuclear genome. Such two-way signaling traffic presents an orchestrated dynamic between energy metabolism and consumption in cells. Recent reports have elucidated the way how mitochondrial bioenergetics synchronizes with the energy consumption for cell cycle progression mediated by cyclin B1/CDK1 as the communicator. This review is to recapitulate cyclin B1/CDK1 mediated mitochondrial activities in cell cycle progression and stress response as well as its potential link to reprogram energy metabolism in tumor adaptive resistance. Cyclin B1/CDK1-mediated mitochondrial bioenergetics is applied as an example to show how mitochondria could timely sense the cellular fuel demand and then coordinate ATP output. Such nucleus-mitochondria oscillation may play key roles in the flexible bioenergetics required for tumor cell survival and compromising the efficacy of anti-cancer therapy. Further deciphering the cyclin B1/CDK1-controlled mitochondrial metabolism may invent effect targets to treat resistant cancers.
Keywords: metabolism, mitochondria, CDK, cell cycle, tumor resistance
1. Introduction
In addition to the functions in signaling transduction, mitochondria in all organisms including singular or multiple cell forms provide the major biofuel in the form of adenosine triphosphate (ATP), the energy currency mainly generated through oxidative phosphorylation (OXPHOS) by coupling of electron transport with proton pumping, for the energy consumption required for cell proliferation and organ development. Instead of its own genome, more than 98% of mitochondrial proteins are transcribed by the genes located in the nuclear genome [1], and only 13 out of ~1500 mitochondrial proteins/factors remain to be encoded by mitochondrial DNA [2, 3]. Such coordinative pattern of two genomes in the same cell illustrates a potential evolution trend in which an organelle is adapted to a host in order to keep the homeostatic cellular functions under the control of the leading genome. It can therefore be assumed that the nuclear genome gradually rules over the mitochondrial functions so as to provide timely and economically energy supply required for different cellular functions and organism regeneration.
This two-way signaling traffic between mitochondria and the nucleus is further illustrated by accumulating evidence including that nucleus-coded proteins control the mitochondrial DNA segregation [4], dynamics, function, and autophagy [5]; whereas mitochondrial dysfunction leads to nuclear genomic instability [6], tumorigenesis [7–9], tumor growth [10, 11], therapeutic resistance [12], and tumor metastasis [13, 14]. Over functional mitochondria are also implied in different stress conditions including the adaptive response to radiation in cancer cells [15–18]. In addition, mitochondria-assisted cell cycle progression is confirmed by blocking mitochondrial fission that damages cell cycle progression and causes apoptosis [19]. Recent results suggest that mitochondria are the key cellular organelle targeted by CDKs (cylcin-dependent kinases) in compensating cell cycle regulation. In such studies, CDK4 is shown to upregulate mitochondrial antioxidant MnSOD [20], cyclin D1 inhibits mitochondrial activity in B cells [21], cyclin B1/CDK1 not only coordinates mitochondrial biogenetics for G2/M progression [22], but also mediates SIRT3 activation to enhance mitochondrial function and tumor radioresistance [23], and phosphorylates mitochondrial antioxidant MnSOD in cell adaptive response to radiation stress [24]. These results further confirm the concept that healthy mitochondria are indeed required for normal cell functions, deficiency or over function will cause different pathological conditions in cells such as cell transformation and tumor aggressiveness.
In this review, we aim to illustrate the cyclin B1/CDK1-modulated mitochondrial activities in cell cycle progression and proliferation. Taking a backward approach, we want to reveal a potential mechanism on how mitochondrial energy metabolism coordinates with cell cycle such as G2/M transition and tumor aggressive phenotype. Further elucidation of the mechanisms underlying mitochondria-regulated cell behaviors will help to understand the network on energy generation and consumption within a cell and define unknown mechanisms in balancing energy consumption in normal and tumor cells.
2. CDK1-DRP1 pathway in regulation of mitochondrial dynamics
Mitochondrial proliferation origins from existing mitochondria via complementary fission and fusion events [29], these two opposing processes dynamically and harmoniously coordinated to maintain the average size of mitochondria, plays critical roles in maintaining mitochondria function and cell division, and closely links with human diseases [25–28]. An optimal balance between fission and fusion could be critical in maintaining mitochondrial membrane dynamics and different cellular functions [30, 31]. The fusion events are carried out by a mitochondrial transmembrane GTPase known as Mitofusin (Mfn) [32] whereas dynamin related protein 1 (DRP1) is responsible for mitochondrial fission events [33], which is dependent on the communication between DRP1 and GTPase. It has been shown that DRP1 determines GTPase activity during mitochondrial fission [34, 35]. DRP1 contains at least 5 phosphorylation sites at serine residues including Ser585 (in rat cells), Ser616, Ser637, Ser656 and Ser693 (in human cells), etc. These sites were suggested to be modified by different kinases, in which only Ser585 and Ser616 can be modified by cyclin B1/CDK1.Taguchi et al. has demonstrated that in addition to chromatid segregation, cyclin B1/CDK1 regulates the mitotic mitochondrial fragmentation, a cell cycle-regulated mitochondrial fission [36]. Interestingly, phosphorylation of DRP1 by cyclin B1/CDK1 at Ser-585 residue during mitosis is found to be required to translocate DRP1 from cytosol to the mitochondrial outer membrane which is required for mitochondrial fission [36, 37]. The activated DRP1 then punctuates spots on the mitochondrial membrane to proceed with membrane constriction and fission directed by Fis1 [38, 39]. The energy-sensing adenosine monophosphate (AMP)-activated protein kinase (AMPK) is also involved in mitochondrial fragmentation when mitochondrial respiration via electron transfer chain is inactivated. Recently, a specific substrate of AMPK is identified as mitochondrial fission factor (MFF) that is identified to be the outer-membrane receptor for DRP1 required for DRP1-mediated fission process [40]. DRP1 mediated mitochondrial fragmentation is believed to allow equal distribution of the mother mitochondria into two daughter cells as the fission events occur during the cell cycle. Deficiency of DRP1 causes elongated mitochondrial filaments with reduced mitochondrial fragmentation [36, 41], and leads to mitochondrial dysfunction, loss of mtDNA and decrease of cellular ATP generation [42, 43]. As such the dynamic equilibrium between fission and fusion in mitochondrial dynamic plays critical roles in maintaining mitochondria function and cell division [44]. The cyclin B1/CDK1-mediated activation DRP-1 and mitochondrial fission contribute to the balance between fission and fusion may coordinate with cyclin B1/CDK1-mediated mitochondrial bioenergetics only in the fused mitochondria which is to be further elucidated.
Another proof in cyclin B1/CDK1-mediated mitochondrial dynamics is supported by Ras-related protein Ral-A (RALA) and its effector RalA-binding protein 1 (RALBP1). The mitotic kinase, Aurora A, phosphorylates RALA at Ser-194 to enhance the translocation of RALA into mitochondria, in which RAPLA concentrates RALBP1 and promotes mitochondrial localization of DRP1 [45]. RALBP1 binds to cyclin B1/CDK1 to enhance DRP1 phosphorylation and activation at Ser-616, which controls the fission and proper segregation of mitochondria during cell division and maintains appropriate mitochondrial and cellular function [45, 46]. However, in addition to DRP1-guided mitochondrial fragmentation linked with reduced mitochondrial bioenergetics, DRP1 is also required for mitochondrial metabolic activation, supported by the report that ATP production is severely impaired in DRP1−/− deficient cells [43]. Phosphorylation at Ser-637 of DRP1 by cAMP-dependent protein kinase (PKA) blocks its GTPase activity by reducing the intra-molecular interaction that drive GTP hydrolysis [46, 47], which can be lifted by the phosphatase Calcineurin [48]. Data discussed in the following sections on CDK1-mediated mitochondrial bioenergetics in cell cycle progression and tumor cell behavior will highlight the concept that an enhanced mitochondrial bioenergetics is required for G2/M transition and cell proliferation. Further elucidating the precise mitochondrial dynamics linked with metabolic activity may reveal more insights on CDK1/DRP1-mediated mitochondrial functions that drive different cell behaviors.
3. CDK1 regulates mitochondrial functions
A specific subset of CDKs with their corresponding partner cyclins orchestrate the precise cell cycle progression [49]. CDK1, among the well-defined cell cycle check point proteins, is involved in regulation of an array of fundamental cellular functions for cell proliferative growth [50, 51]. Mouse embryos lacking CDK2, CDK3, CDK4, and CDK6 are still able to undergo organogenesis whereas fail without CDK1. CDK1 is able to bind to different cyclins and is sufficient to regulate all the steps required for cell division [52]. This report together with many other results indicates that CDK1 is the most critical cell cycle element for cell proliferation and organ development. CDK1 is involved not only in mitochondrial dynamics as mentioned above [36, 45], but also in mitochondrial protein influx and bioenergetics [22, 54, 55], targeting CDK1 initiates mitochondria-initiated apoptosis [53],. Another cell cycle-related complex, cyclin D1/CDK4 is also found relocating to the mitochondria and phosphorylate the major antioxidant enzyme MnSOD at Serine-106, whereby enhancing MnSOD enzymatic activity to promote mitochondrial homeostasis and prevent cellular genotoxic stress [20]. Further elucidating how mitochondrial bioenergetics is differently regulated by varied complexes of cyclins/CDKs during progression of different cycle phases may reveal more unknown mechanisms underlying cell cycle-related mitochondrial adjustment.
Cyclin B1 and its catalytic partner CDK1 belong to the fundamental kinase machinery regulating the progression from G2 to mitosis. Also known as the mitotic promoting factor (MPF), cyclin B1/CDK1 phosphorylation governs key steps for mitotic entrance featured by the nuclear envelope breakdown, spindle formation, and chromatin condensation [56]. However, the mitochondrial transition of cyclin B1/CDK1 seems to be dependent on the total levels of cellular cyclin B1 and CDK1 [57]. Under normal growth conditions, mitochondrial localization of cyclin B1/CDK1 is attenuated during G1 phase due to a lack of cyclin B1 accumulation.
3.1. Mitochondrial translocation of CDK1
The mitochondrial matrix localization of cyclin B1/CDK1 complex has recently been identified in multiple human cells, especially shows increase in those experiencing cell cycle transition or exposed to DNA-damaging stress conditions [22, 57]. Under genomic toxic conditions such as anti-cancer chemotherapy and radiation, cyclin B1/CDK1 expression is induced to cause the cell cycle G2/M arrest, which is believed either to extend the time period for cells figuring and fixing DNA damages to survive or to warrant a time required to initiate and process apoptosis [58, 59]. Following genotoxic stress such as ionizing radiation (IR), mitochondrial translocation of cyclin B1/CDK1 is enhanced [57]. As shown in Figure 1, relocation of cyclin B1/CDK1 to mitochondria could results from an increased overall level of cyclin B1/CDK1 in cytoplasm at the peak time of cell cycle progression during G2/M transition, or leaded by a specific mechanism to allow mitochondria to sense the increased demand of energy consumption needs. Since present and being activated at the prophase of mitosis [60], mitochondrial CDK1 phosphorylates substrates to promote mitosis. The activation of CDK1 in mitochondria is supported by the report that Cdc25c, the phosphatase activator of CDK1 [61], is identified by mapping of the mitochondrial intermembrane space [62]; both reports support that cyclin B1/CDK1 can be fully activated to boost ATP generation whereas the CDK1 proteins in cytoplasm and nucleus remain inactive for progression into prophase although the mechanism causing such segregations is to be revealed.
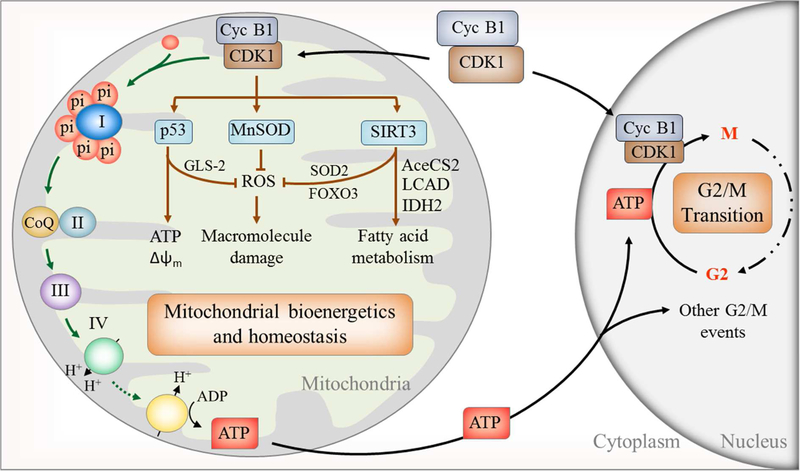
Figure 1. A model of nuclear and mitochondrial cooperation in regulating mitochondrial bioenergetics for G2/M transition.
Cyclin B1/CDK1 relocates to mitochondria during G2/M phase to phosphorylate and activate an array of substrates including multiple subunits of mitochondrial CI to enhance mitochondrial function and energy production which drives energy-sensitive G2/M transition. In addition, cyclin B1/CDK1 activates p53, MnSOD, and SIRT3 to eliminate ROS and protect and maintain mitochondrial homeostasis. CDK1-SIRT3-mediated fatty acid metabolism and mitochondrial homeostasis remain to be further elucidated. Cyc B1, cyclin B1. Also see other mitochondrial substrates of cyclin B1/CDK1 in Table 1.
Proteins and polypeptides with mitochondrial leading sequences are transported into mitochondria by the translocase of the outer mitochondrial membrane (TOM) complex and translocase of mitochondrial inner membrane 23 (TIM23) complex [63]. However, like many other mitochondrial proteins, CDK1 does not hold the leading sequence for mitochondrial translocation. Many other proteins without mitochondrial leading sequences are translocated to mitochondria by chaperons [64]. The translocation of cyclin B1/CDK1 is possibly conducted with chaperons such as HSP70, HSP-90, and Cdc37 [65–68]. Harbauer et al show that clb3-activated CDK1 is able to phosphorylate the cytosolic Tom6 precursor to enhance the import of Tom6 into mitochondria, which accelerates Tom40 assembly and mitochondrial protein influx [54, 55]. Additionally, 14–3-3 family proteins, well-defined as mitochondrial chaperons with mitochondrial targeting sequences in their ligands [69–71], can dynamically balance cyclin B1/CDK1 inhibition and activation to control G2/M checkpoint maintenance and release [72]. 14–3-3ζ binds and transports Cdc25c to mitochondria to activate CDK1. Both 14–3-3ζ and cyclin B1 are upregulated in radiation-resistant breast cancer cells [16] and knockdown of 14–3-3ζ enhances radiosensitivity and radio-induced apoptosis in CD133 (+) liver cancer stem cells [73]. Further elucidation is appreciated to reveal the mechanistic insight underlying cyclin B1/CDK1 mitochondrial translocation in normal and tumor cells.
3.2. CDK1 substrates in mitochondria
Reversible protein phosphorylation is a fundamental post-translational modification required to guide different mitochondrial functions [74, 75]. CDK1 belongs to the serine/threonine (S/T) kinase family catalyzing the transfer of phosphate from ATP to proline (P)-oriented serine (S) or threonine (T) residues. Its substrates contain either an optimal (S/T*-P-x-K/R; x, any residue) or a minimal (S/T*-P) consensus motif. It is well-defined that in mammals the oscillations in activation of different CDKs govern cell reproduction by catalyzing the transfer of phosphate from ATP to specific protein substrates. Ubersax et al has identified 200 substrates in whole-cell extracts of budding yeast that are directly phosphorylated by CDK1 [76]. However, CDK1 is shown to phosphorylate mitochondrial proteins isolated from G0/G1 cells that possessed low endogenous cyclin B1/CDK1 activity and dozens of mitochondrial proteins contain at least one consensus site or motif that can be directly phosphorylated by CDK1. These substrates involve in multiple bio-functions including electron transport (CI-CV), tricarboxylic acid cycle, amino acid, and lipid metabolism (Table 1) [22]. A cluster of CI subunits is identified to be the CDK1 substrates in OXPHOS (NDUFV1 at T-383, NDUFV2 at T-164, NDUFV3 at S-530, NDUFS2 at S-364, NDUFAF1 at S-450, NDUFA12 at T-142, NDUFB5 at S-128 and NDUFB6 at S-550), and CIII subunit (UQCRC1 at T-266). Expression of mitochondria-targeted cyclin B1/CDK1 increases CI activity and ATP generation and G2/M-associated mitochondrial enhancement is deficient by expression of mitochondria-targeted mutant CDK1 [22]. MnSOD, the primary mitochondrial antioxidant in detoxifying superoxide for mitochondrial homeostasis is found to contain CDK1 phosphorylation consensus (Serine/Threonine-proline [Ser/ThrPro]) at Ser-106 [24, 77] and cyclin B1/CDK1-mediated MnSOD phosphorylation is required for MnSOD tetrameric conformation, enzymatic activity, and protein stability [24]. Interestingly, another critical substrate of mitochondrial cyclin B1/CDK1 is the tumor suppressor p53 that is identified at Serine-315 [57] which is to be discussed below. Cyclin B1/CDK1 also phosphorylate SIRT3, an essential NAD+-dependent deacetylase for mitochondrial functions and homeostasis [78], at Thr-150 and-Ser159 [23]. An array of SIRT3-modified proteins including AceCS2 (K642) [79], LCAD (K42) [80], IDH2 (K413) [81], SOD2 (K53/89) [82], FOXO3 (K271/290) [83], and HMGCS2 (K310/447/473) [84] have been identified and suggested to govern mitochondrial metabolism via SIRT3 regulation (Figure 1). The cyclin B1/CDK1-mediated SIRT3 activation is another example how cyclin B1/CDK1 is applied to adjust mitochondrial activity and homeostatic status. Since SIRT3 activity is well-defined to play a key role in for preventing mitochondria-associated carcinogenesis and aging [85], this link between cylcin B1/CDK1 and SIRT further supports the close relation of cell cycle events causing ging and cell transformation.
Table 1.
Substrates of CDK1 in mitochondria detected by in vitro kinase assay
| Function | Gene symbol | Protein | Nucleus/Mitochondria-encoded | Optimal phsphorylation site and position | Minimal phosphorylation site number |
|---|---|---|---|---|---|
| OXPHOS (complex I) | NDUFV1, UQOR1 | NADH dehydrogenase [ubiquinone] flavoprotein 1 | Nucleus | T383 P C R | 0 |
| NDUFV2 | NADH dehydrogenase [ubiquinone] flavoprotein 2 | Nucleus | T164 P D K | 4 | |
| NDUFV3 | NADH dehydrogenase [ubiquinone] flavoprotein 3 | Nucleus | S530 P P K | 1 | |
| NDUFS2 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 2 | Nucleus | S364 P P K | 1 | |
| NDUFB5 | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 5 | Nucleus | S550 P W R | 2 | |
| NDUFB6 | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 6 | Nucleus | S550 P W R | 2 | |
| NDUFA12, DAP13 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 12 | Nucleus | T142 P Y K | 1 | |
| NDUFAF1, CIA30, CGI-65 | Complex I intermediate-associated protein 30 | Nucleus | S450 P G K | 1 | |
| OXPHOS (complex III subunit 1) | UQCRC1 | Cytochrome b-c1 complex subunit 1 | Nucleus | T266 P C R | 3 |
| OXPHOS (complex III subunit 5) | UQCRFS1 | Cytochrome b-c1 complex subunit Rieske | Nucleus | 0 | 6 |
| OXPHOS (complex V) | ATP5A1, ATP5F1A ATP5A, ATP5AL2, ATPM | ATP synthase subunit alpha | Nucleus | 0 | 1 |
| ATP5B, ATP5F1B, ATPMB, ATPSB | ATP synthase subunit beta | Nucleus | 0 | 1 | |
| Amino acid metabolism | GLUD1, GLUD | Glutamate dehydrogenase 1 | Nucleus | T144 P C K | 2 |
| GATM, AGAT | Glycine amidinotransferase | Nucleus | 0 | 3 | |
| GOT2 | Aspartate aminotransferase | Nucleus | 0 | 2 | |
| ABAT, GABAT | 4-aminobutyrate aminotransferase | Nucleus | 0 | 1 | |
| BCKDHB | 2-oxoisovalerate dehydrogenase subunit beta | Nucleus | 0 | 5 | |
| AGMAT | Agmatinase | Nucleus | 0 | 4 | |
| OTC | Ornithine carbamoyltransferase | Nucleus | 0 | 3 | |
| SHMT2 Serine | Serine hydroxymethyltransferase | Nucleus | S266 P F K | 3 | |
| Carbohydrate metabolism | ALDH1B1, ALDH5, ALDHX | Aldehyde dehydrogenase X | Nucleus | S91 P W R | 4 |
| ALDH4A1, ALDH4, P5CDH | Delta-1-pyrroline-5-carboxylate dehydrogenase | Nucleus | S44 P E R | 3 | |
| ALDH6A1, MMSDH | Methylmalonate-semialdehyde dehydrogenase [acylating] | Nucleus | 0 | 2 | |
| ALDH2, ALDM | Aldehyde dehydrogenase | Nucleus | S91 P W R | 3 | |
| Lipid metabolism | HMGCS2 | Hydroxymethylglutaryl-CoA synthase | Nucleus | T6 P V K | 4 |
| SUCLG2 Succinyl-CoA | Succinate--CoA ligase [GDP-forming] subunit beta | Nucleus | 0 | 4 | |
| ACADSB Short/branched | Short/branched chain specific acyl-CoA dehydrogenase | Nucleus | 0 | 1 | |
| ACADM medium-chain | Medium-chain specific acyl-CoA dehydrogenase | Nucleus | 0 | 1 | |
| ACOT1, CTE1 | Acyl-coenzyme A thioesterase 1 | Nucleus | 0 | 2 | |
| Lipid metabolism | ACAT1, ACAT, MAT | Acetyl-CoA acetyltransferase | Nucleus | 0 | 5 |
| SUCLG2 | Succinate--CoA ligase [GDP-forming] subunit beta | Nucleus | 0 | 4 | |
| ACADS | Short-chain specific acyl-CoA dehydrogenase | Nucleus | 0 | 2 | |
| HSD17B10, ERAB, HADH2, MRPP2, SCHAD, SDR5C1, XH98G2 | 3-hydroxyacyl-CoA dehydrogenase type-2 | Nucleus | 0 | 1 | |
| HSD17B8, FABGL, HKE6, RING2, SDR30C1 | Estradiol 17-beta-dehydrogenase 8 | Nucleus | 0 | 1 | |
| ECH1 | Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase | Nucleus | 0 | 1 | |
| ACSL1, FACL1, FACL2, LACS, LACS1, LACS2 | Long-chain-fatty-acid --CoA ligase 1 | Nucleus | 0 | 1 | |
| Redox | SOD2 | Superoxide dismutase [Mn] | Nucleus | 0 | 1 |
| PRDX3, AOP1 | Thioredoxin-dependent peroxide reductase | Nucleus | T128 P R K | 3 | |
| MPST, TST2 | 3-mercaptopyruvate sulfurtransferase | Nucleus | 0 | 2 | |
| TST Thiosulfate | Thiosulfate sulfurtransferase | Nucleus | 0 | 2 | |
| NNT | NAD(P) transhydrogenase | Nucleus | S400 P D K | 4 | |
| T467 P F R | |||||
| TCA cycle | PCK2, PEPCK2 | Phosphoenolpyruvate carboxykinase [GTP] | Nucleus | 0 | 7 |
| MDH2 | Malate dehydrogenase | Nucleus | 0 | 4 | |
| FH, FH1 | Fumarate hydratase | Nucleus | 0 | 1 | |
| Protein targeting | HSPD1, HSP60 | 60 kDa heat shock protein | Nucleus | 4 | |
| 14–3-3 epsilon | 14–3-3 protein epsilon | Nucleus | S187 P D R | 0 | |
| Protein synthesis | TUFM | Elongation factor Tu | Nucleus | 0 | 2 |
3.3. CDK1 enhances OXPHOS in G2/M progression
Metabolic activity and energy supply are believed to be a crucial determinant for cell division and proliferation. However, the exact mechanism how mitochondria sense the increased cellular energy demands by different cycling phases was unknown [86]. ATP is generated in mitochondria by the electron transport chain (ETC) carried by the respiratory chain complexes I–IV transfering electrons i n a stepwise pattern until they finally reduce oxygen to form water. Mitochondria are in charge of processing both the tricarboxylic acid cycle (TCA) and OXPHOS whereby providing the major cellular energy source. OXPHOS supplies more than 90% of cellular ATP required for eukaryotic cells [87], and it is irrefutably critical for the progression of the cell cycle as the level of ATP determines whether cells are ready for division [88]. It is assumed that G1/S and G2/M phases are energy-sensitive processes in which pronounced energy supply is demanded for increasing biomass for cell cycle transition [89]. For this function, all of five large protein complexes (CI-CV) must be functional in the respiration chain [90, 91] and many mitochondrial function-related diseases are linked to the status of OXPHOS [92–95]. CI is the largest complex containing 46 subunits, and serves as the major entry point of electrons into OXPHOS. A functionally efficient CI is required for OXPHOS function [96] and for successful cell-cycle progression [97]. As reported. the D-type cyclins and CDKs are associated with the metabolic crosstalk [86]. By searching mitochondrial protein databases for the CDK1 consensus phosphorylation motif (S/T*-P-x-K/R) [98] among all the subunits of mitochondrial respiration chain (Complex I – V), interestingly, 12 subunits, inclu ding 8 components from CI (NADH ubiquinone oxidoreductase) [99], can be potentially phosphorylated by cyclin B1/CDK1 (Table 1) [22], indicating that cyclin B1/CDK1 functions as a switcher to control the surge of ATP output. A cluster of other mitochondrial factors are also targeted by cyclin B1/CDK1-phosphorylation that includes proteins in carbohydrate and lipid metabolism of mitochondria including TCA cycle. The dual functions of cyclin B1/CDK1 in both the nucleus and the mitochondria mark a tight connection between cell cycle progression and mitochondrial cooperation, and raise the question of whether this coordination is linked in the different metabolisms in normal and cancer cells.
4. Cyclin B1/CDK1 in mitochondria-associated apoptosis
Mitochondria-regulated apoptosis plays a key role in tumor response to anti-cancer therapies. Although several studies report that cyclin B1/CDK1 is responsible for initiating mitochondria-mediated apoptosis under cell damage conditions by its phosphorylation of several pro- (Bcl-2, BAD and Bcl-xL cytosol [100, 101]) and anti-apoptotic proteins (Mcl1 [102]) in normal and cancer cells, more emerging evidence demonstrate that cyclin B1/CDK1 is pivotal in inhibition of apoptosis in tumors. Cyclin B1/CDK1-mediated phosphorylation of pro-caspase-9 and survivin (BIRC5) lead to inhibition of apoptosis [103, 104] and Intact cyclin B1 with mutant CDK1 is shown to compromise CDK1-mediated substrates phosphorylation, mitochondrial function as well as cell viability [57] [22].. Cyclin B1, as the most important catalytic partner of CDK1, has been unearthed to be abnormally activated and aberrantly expressed in a number of human cancers including esophageal squamous cell carcinoma, laryngeal squamous cell carcinoma, and colorectal carcinomas [105–107]. Besides, cyclin B1 is linked with anti-apoptosis and tumor resistance in head and neck cancer [108, 109], one of the most aggressive cancers [110], triggers tumor proliferation and links with poor prognosis in esophageal squamous cell carcinoma [111], non-small cell lung cancer [112], and colorectal carcinoma [113]. Consistently, deficiency of cyclin B1 causes inhibition of cell proliferation and activation of apoptosis [114] and activated cyclin B1 leads to a pro-survival machinery mediated by the NF- κB signaling network [115]. These different results suggest that mitochondrial energy is required for both cell death and cell cycle. Cyclin B1/CDK1 not only regulates mitochondrial fuel output for normal cell cycle progression but also can initiate mitochondria-mediated apoptosis by modifying several pro-apoptosis proteins and anti-apoptotic proteins when cells exposed to excessive damage stresses.
4.1. CDK1-mediated mitochondrial bioenergetics for DNA repair
Radiation-generated ROS induces mitochondrial and nuclear DNA strand break [116] and potentially leads to cell apoptosis [117]. In this process, cellular energy supply, DNA repair capacity, and apoptosis play important roles in determining cell fate [118]. Timely and efficient DNA repair, ,an energy-consuming process [119–121], is necessary for cell survival [122]. However, it remains unclear how cellular energy supplement is coordinated in DNA repair. A study in Jurkat cells indicates that both glycolysis and OXPHOS provide the energy supply of apoptosis [123]. A group of stress responsive proteins including MnSOD, cyclin B1/CDK1, cyclin D1/CDK4, and survivin [20, 124, 125] (Figure 2) inflow into mitochondria to induce cellular adaptive protection. The CI in the respiration chain is identified to be a key cyclin B1/CDK1 target in mitochondria [22]. Mitochondrial relocation of cyclin B1/CDK1 and nuclear DNA repair is correlated with oxygen consumption and ATP production in normal human cells after radiation [126]. The basal and radiation-induced mitochondrial ATP generation is reduced significantly in cells harboring CDK1 phosphorylation-deficient mutant CI subunits. Similarly, mitochondrial ATP generation and nuclear DNA repair are also compromised severely in cells harboring mitochondria-targeted, kinase-deficient CDK1 [126]. Together, these results implicate that cyclin B1/CDK1 functions to deliver signals to mitochondria to boost ATP generation for DNA repair and cell survival under genotoxic stress conditions.
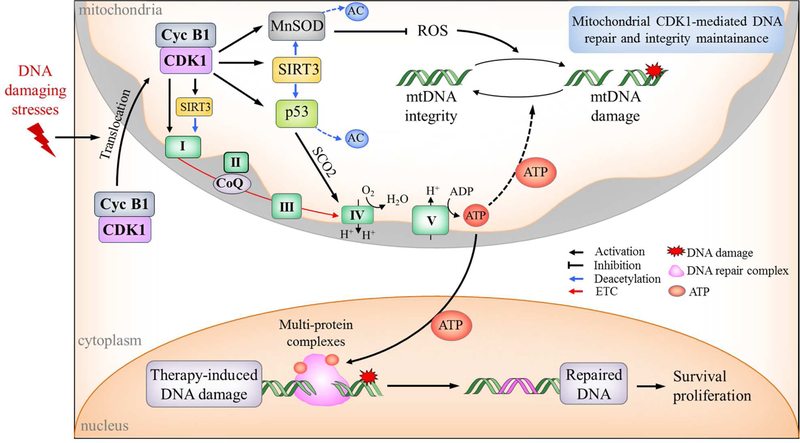
Figure 2. Cyclin B1/CDK1-mediated mitochondria bioenergetics in DNA repair and tumor resistance.
DNA-damaging agents including chemotherapy and radiotherapy enhance the translocation of cyclin B1/CDK1 to mitochondria causing phosphorylation and activation of OXPHOS leading to enhanced ATP output required to repair DNA damages for cell survival. CDK1 also directly phosphorylates MnSOD to eliminate ROS, activates p53 to promote energy generation and maintain mitochondrial DNA integrity, or indirectly enhances the activity of MnSOD and p53 via SIRT3-mediated deacetylation, which as a whole improve mitochondrial homeostasis and function. NDUFA9, one of the subunits of CI, can also be deacetylated by SIRT3 to promote OXPHOS. Thus, our model applies to the situation in which under non-stressful conditions tumor cells may relay on the energy generated by predominant oxidative glycolysis, whereas under genotoxic crisis such as chemotherapy and radiation, cyclin B1/CDK1-mediated OXHPOS is activated to meet the extra energy demands for repairing damages and enhancing cell survival.
4.2. CDK1- p53 pathway in anti-apoptosis
The p53 protein is well-characterized to regulate mitochondria-mediated apoptosis at protein and mRNA levels [127, 128]. p53 initiates the apoptotic cascade by inducing the expression or interacting directly with cytoplasmic Bcl-2 family proteins [129]. Mitochondrial function of p53 has been associated with mtDNA transcription, DNA repair, mitochondrial bioenergetics [130–132], as well as ATP production since p53 also regulates mitochondrial respiratory gene, SCO2 and GLS-2 [133]. It is generally believed that localization of p53 to mitochondria resembles a major starting signal for mitochondria-mediated apoptosis [134]. However, it has been proposed that mitochondrial p53 may not necessarily induce apoptosis [135]. Activation of CDK via Spy1 is indicated for the anti-apoptotic response via p53 induced by intrinsic DNA damage response such as in cell division [136]. Mice that lack of critical effectors of p53-induced apoptosis do not develop tumors spontaneously [137]. Therefore, the role of mitochondria-localized p53 should be considered broadly, depending on its cooperative and differential phosphorylation in addition to its apoptotic signal [138]. p53 can also be phosphorylated by mitochondrial cyclin B1/CDK1 [57]. Mitochondrial p53, cyclin B1 and CDK1 are elevated after radiation treatment, enhancing the phosphorylation of mitochondrial p53 at Ser-315, the only putative site for cyclin B1/CDK1 phosphorylation [139]. In contrast with p53-mediated pro-apoptotic signaling, this phosphorylation suppresses the mitochondrial apoptosis by sequestering p53 from binding to Bcl-2 and Bcl-xL [57]. This finding reinstates the role of cyclin B1/CDK1 in non-cell cycle functions. The pattern of cyclin B1/CDK1-mediated p53-apoptotic inhibition demonstrates a feedback signaling pathway for mitochondria-initiated apoptosis. The decision of pro- or anti-apoptotic response may be made based on the degree of DNA damage in the nucleus and the length of period cell arrested.
5. Mitochondrial bioenergetics in reprogramming energy metabolism in tumor cells.
Increasing interests have been attracted in cancer energy metabolism [140]. The high acidity in tumor tissues due to enhanced lactate generation even under oxygenated condition suggests that tumor cells generate ATP via glycolysis (aerobic glycolysis, Warburg Effect). Although ATP generation via glycolysis is less efficacious compared to OXPHOS, glycolysis generates fundamental elements for cellular function and proliferation such as pyruvate and NADPH that are required for ribose production and protein glycosylation [141–143]. The Warburg assumption is that mitochondria are damaged, and glycolysis is primarily used as the major energy-supply for cellular proliferation and survival even under oxygenated conditions. However, this theory was challenged by a series of evidence since 1950s [144–148], which indicate that mitochondria in some cancer cells remain intact structure and function [149], and mitochondria are actively involved in tumor response to anti-cancer therapy. This apparent contradiction between glycolysis-dominated energy reprogramming and the OXPHOS-activated bioenergetics in tumor cells may reflect unknown dynamics of the flexible re-/deprogramming energy metabolism in cancer cells which is required to adapt and survive under different genotoxic conditions.
5.1. Mitochondria are still active and have potential role in tumor cells.
Contrasted to the prevailing thoughts of aerobic glycolysis, increasing results demonstrated functional mitochondria exist in tumors. Heat-shock-protein-90 (HSP90)-involved mitochondrial protein folding is required for cellular bioenergetics in tumor cells [150]. By using the isotopically labeled glucose, lactic acid, and palmitic acid, Weinhouse et al show that oxidation of glucose and fatty acid in tumor is similarly to normal tissues [147]. Similarly, Wenner et al observe that enzyme activities of the TCA cycle in tumor cells are at a similar active level in normal tissues [148]. Upon vivo experiment, Whitaker-Menezes report that epithelial cancer cells enhance OXPHOS for generation of high amounts of ATP. The transcriptional upregulation of genes in mitochondrial OXPHOS is identified in a report with over 2,000 breast cancer patients [151]. Others also report that mitochondrial bioenergetics may be reactivated to provide additional energy supply in the maintenance and function of cancer stem cells [18, 152–155], hematopoietic cells, as well as lymphoma and leukemia cells [155, 156]. However, the exact mechanism guiding such mitochondrial activation is yet to be elucidated. Other studies also reveal that cancer cells remain OXPHOS capacity to provide the vital amount of cellular fuels for energy consumption required for the fast proliferation of cancer cells [157, 158] and adaption of metabolic stress such as nutrient deficiency and hypoxia [159, 160]. On the other hand, pharmacological glycolysis inhibitors such as 2-DG, fail to provide significant therapeutic benefits in vivo test [161, 162]. These results together suggest that mitochondria in cancer cells can dynamically activate or inactivate its functions with different cellular energy consumption conditions and in different cancer types. Such plasticity in adjusting mitochondrial bioenergetics of tumor cells challenges the anticancer modalities that aim to inhibit tumor growth by blocking glycolysis.
5.2. Mitochondrial bioenergetics in tumor resistance to therapy
Mitochondrial metabolism has been linked with tumor therapeutic resistance [18, 150, 163–169]. Mitochondrial bioenergetics is enhanced when cellular energy demand is increased for repairing DNA damages to allow cancer cells to survive from therapeutic genotoxicity [23, 167]. Overexpressing MnSOD, a key enzyme for maintaining mitochondrial homeostasis, induce radioresistance in breast cancer MCF7 cells [16, 170]. The mammalian target of rapamycin complex 1 (mTORC1) that serves as an sensor of extracellular nutrient levels and intracellular bioenergetics [171], maintains cellular bioenergetics under glucose deficiency [172]. mTOR, as the core element in mTORC1 enhances OXPHOS with reduced glycolysis for tumor cells to survive IR [167]. The apparent “quiet” mitochondria in tumor cells can function as a backup line to boost up cellular fuel supply under genotoxic anticancer modalities such as ionizing radiation and chemotherapy (Figure 2 legend). This “backing-up” mechanism proposed here is to demonstr ate a flexible feature in the energy metabolism of cancer cells under genotoxic and “normal” growth conditions. In addition, mitochondrial metabolism is identified in tumor cells and actively involved in tumor metastasis [173–175]. These observations are further supported by findings that reprogramming the mitochondrial trafficking can help to fuel the tumor cell invasion [14, 176]. Most importantly, mitochondrial energy metabolism is recently linked with the aggressive phenotype of triple-negative breast cancer (TNBC) [174]. Therefore, further elucidation on the dynamic alterations in mitochondrial energy metabolism in tumor cells especially IR-associated energy dynamics will provide critical information on invention of effective metabolic targets to treat cancer cells.
5.3. CDK1-mediated mitochondrial bioenergetics in cancer cells plays a pivotal role in tumor radiation response.
As discussed above, in addition to cyclin B1/CDK-mediated morphological alterations via DRP1 regulation and mitochondrial fission [36,177], CDK1 is able enhance mitochondrial metabolism in cell cycle progression of normal cells and in radiation-induced stress. Cyclin B1/CDK1 promotes the activity of OXPHOS to induce anti-apoptotic response in human colon cancer HCT116 cells via cyclin B1/CDK1-mediated SIRT3 activation together with phosphorylation of p53 at Ser-315 leading to an increased mitochondrial ATP production and reduced mitochondrial apoptosis [23, 57]. Besides, cyclin B1/CDK1 are found to specifically modify MnSOD that can enhance resistance of breast cancer cells in genotoxic conditions such as IR [15, 16] and increase mitochondrial homeostasis, biosynthesis, and signaling aggressive phenotype of cancer cells [10, 178–180]. MnSOD, located in mitochondria, is important in balancing the ROS (reactive oxygen species) and protecting mitochondrial function [116]. Down-regular of MnSOD expression or inhibit of its enzymatic activity in normal cells have been proven leading to a high risk of cell transformation [181,182]. Increased expression of MnSOD may be beneficial to cancer cells to survive from radiation therapy [183], as well stimulate metastatic behavior [184–187]. When exposed under ionizing radiation, mitochondria recruit cyclin B1/CDK1 in cancer cells enhancing MnSOD activity and stability along with improved mitochondrial function and cellular adaptive response [24]. Similarly, CDK4, the key kinase for G1/S progression, activates MnSOD to improve mitochondrial function and serve as an important pathway in cellular adaptive protection [20]. Therefore, a cluster of mitochondrial proteins post-transcriptionally modified by cyclin B1/CDK1 can instantly upregulates mitochondrial energy output for tumor cell survival under genotoxic cancer therapy (Figure 2). However, it is currently unknown how such cyclin B1/CDK1-boosted mitochondrial energy supply could be mechanistically associated with specific tumor cells such as cancer stem cells under anti-cancer therapy. Future exploring in cyclin B1/CDK1-mediated mitochondrial bioenergetics in different therapeutic modalities may invent effect targets to timely block the cellular energy reprogramming to eliminate resistant tumor cells.
6. Concluding remarks
We illustrate here that mitochondrial energy metabolism is critically regulated in cell proliferation and tumor growth [188–190]. Beside their direct functions in regulation of cell cycle progression, the complex of cyclin B1/CDK1 is able to coordinate the cell cycle events with enhanced mitochondrial bioenergetics. Such cyclin B1/CDK1 regulated mitochondrial metabolism is highlighted in this review to demonstrate how the two genomes are functionally cooperated to control the balance of energy supply with the timely demanded cellular fuel consumption. However, it remains unclear how cyclin B1/CDK1-mediated mitochondrial homeostasis orchestrates different cell cycle phases. It is also to be further elucidated whether the mitochondrial bioenergetics is required for tumor cell survival under anti-cancer treatments such as chemotherapy and radiation. In addition to mitochondrial targets, cyclin B1/CDK1-controlled cytoplasmic components for cell cycle progression must coordinate timely with mitochondrial functions, and way around, the oscillation for mitochondrial bioenergetics should meet the specific requirements of cell division and proliferation. To define specific metabolic targets in tumor cells, outstanding questions still need to be addressed. First, it remains unclear if a specific cluster of cyclin B1/CDK1-regulated metabolic enzymes is required for the different metabolic status in normal and cancer cells [86, 191, 192]. Second, cross-talking among the cyclin B1/CDK1-targeted proteins may guide the mitochondrial bioenergetics based on the different stages of cell cycle progression and tumor growth status such as tumor metastasis. Since different CDKs are required in transition of each cell cycle stage, it would be highly appreciated if mitochondrial energy metabolism can be precisely oscillated with each cycle phase. In addition, the mechanistic insights guiding the signaling traffics between the nucleus and mitochondria under physiological and pathological conditions needs to be further deciphered. These studies will help to understand the flexible mitochondrial bioenergetics in cancer cells under different stress conditions and thus to invent new metabolic targets to treat cancer.
HIGHLIGHTS.
Two genomes located in the nucleus and mitochondria coordinate to achieve homeostasis.
Such two-way signaling traffics present an orchestrated dynamics between energy metabolism and consumption.
Cyclin B1/CDK1 regulates mitochondrial bioenergetics for cell cycle progression and tumor response to therapy.
Deciphering the cyclin B1/CDK1-controlled mitochondrial metabolism may invent effect targets to treat resistant cancers.
ACKNOWLEDGEMENTS
We apologize to the authors whose publications could not be included in this review due to limited space. The authors acknowledge Dr. Aris Alexandrou for reviewing the article. The research in the JJ Li’s laboratory at University of California Davis is supported by National Institutes of Health Grant R01 CA213830.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST STATEMENT
The authors declare no financial conflict.
REFERENCES
- 1.Ryan MT, Hoogenraad NJ, Mitochondrial-nuclear communications, Annu. Rev. Biochem 76 (2007) 701–722. [DOI] [PubMed] [Google Scholar]
- 2.Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG, Sequence and organization of the human mitochondrial genome, Nature 290 (1981) 457–465. [DOI] [PubMed] [Google Scholar]
- 3.Boore JL, Animal mitochondrial genomes, Nucleic Acids Res 27 (1999) 1767–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Battersby BJ, Loredo-Osti JC, Shoubridge EA, Nuclear genetic control of mitochondrial DNA segregation, Nat. Genet 33 (2003) 183–186. [DOI] [PubMed] [Google Scholar]
- 5.Thomas KJ, McCoy MK, Blackinton J, Beilina A, van der Brug M, Sandebring A, Miller D, Maric D, Cedazo-Minguez A, Cookson MR, DJ-1 acts in parallel to the PINK1/parkin pathway to control mitochondrial function and autophagy, Hum. Mol. Genet 20 (2011) 40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Veatch JR, McMurray MA, Nelson ZW, Gottschling DE, Mitochondrial dysfunction leads to nuclear genome instability via an iron-sulfur cluster defect, Cell 137 (2009) 1247–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chourasia AH, Boland ML, Macleod KF, Mitophagy and cancer, Cancer Metab 3 (2015) 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K, Mangal D, Yu KH, Yeo CJ, Calhoun ES, Scrimieri F, Winter JM, Hruban RH, Iacobuzio-Donahue C, Kern SE, Blair IA, Tuveson DA, Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis, Nature 475 (2011) 106–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li XJ, Jiang YH, Meisenhelder J, Yang WW, Hawke DH, Zheng YH, Xia Y, Aldape K, He J, Hunter T, Wang LW, Lu ZM, Mitochondria-Translocated PGK1 Functions as a Protein Kinase to Coordinate Glycolysis and the TCA Cycle in Tumorigenesis, Mol. Cell 61 (2016) 705–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wallace DC, Mitochondria and cancer, Nat. Rev. Cancer 12 (2012) 685–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kashatus JA, Nascimento A, Myers LJ, Sher A, Byrne FL, Hoehn KL, Counter CM, Kashatus DF, Erk2 phosphorylation of Drp1 promotes mitochondrial fission and MAPK-driven tumor growth, Mol. Cell 57 (2015) 537–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghosh JC, Siegelin MD, Vaira V, Faversani A, Tavecchio M, Chae YC, Lisanti S, Rampini P, Giroda M, Caino MC, Seo JH, Kossenkov AV, Michalek RD, Schultz DC, Bosari S, Languino LR, Altieri DC, Adaptive Mitochondrial Reprogramming and Resistance to PI3K Therapy, J. Natl. Cancer I 1 107 (2015): dju502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Porporato PE, Payen VL, Perez-Escuredo J, De Saedeleer CJ, Danhier P, Copetti T, Dhup S, Tardy M, Vazeille T, Bouzin C, Feron O, Michiels C, Gallez B, Sonveaux P, A Mitochondrial Switch Promotes Tumor Metastasis, Cell Rep 8 (2014) 754–766. [DOI] [PubMed] [Google Scholar]
- 14.LeBleu VS, O’Connell JT, Gonzalez Herrera KN, Wikman H, Pantel K, Haigis MC, de Carvalho FM, Damascena A, Domingos Chinen LT, Rocha RM, Asara JM, Kalluri R, PGC-1alpha mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis, Nat. Cell Biol 16 (2014) 992–1003, 1001–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li JJ, Oberley LW, Clair D St., Ridnour LA, Oberley TD, Phenotypic changes induced in human breast cancer cells by overexpression of manganese-containing superoxide dismutase cDNA., Oncogene 10 (1995) 1989–2000. [PubMed] [Google Scholar]
- 16.Guo G, Yan-Sanders Y, Lyn-Cook BD, Wang T, Tamae D, Ogi J, Khaletskiy A, Li Z, Weydert C, Longmate JA, Huang TT, Spitz DR, Oberley LW, Li JJ, Manganese superoxide dismutase-mediated gene expression in radiation-induced adaptive responses, Mol. Cell Biol 23 (2003) 2362–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sotgia F, Whitaker-Menezes D, Martinez-Outschoorn UE, Salem AF, Tsirigos A, Lamb R, Sneddon S, Hulit J, Howell A, Lisanti MP, Mitochondria “fuel” breast cancer metabolism: Fifteen markers of mitochondrial biogenesis label epithelial cancer cells, but are excluded from adjacent stromal cells, Cell Cycle 11 (2012) 4390–4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Candas D, Lu CL, Fan M, Chuang FY, Sweeney C, Borowsky AD, Li JJ, Mitochondrial MKP1 is a target for therapy-resistant HER2-positive breast cancer cells, Cancer Res 74 (2014) 7498–7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rehman J, Zhang HJ, Toth PT, Zhang Y, Marsboom G, Hong Z, Salgia R, Husain AN, Wietholt C, Archer SL, Inhibition of mitochondrial fission prevents cell cycle progression in lung cancer, FASEB J 26 (2012) 2175–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin C, Qin L, Shi Y, Candas D, Fan M, Lu CL, Vaughan AT, Shen R, Wu LS, Liu R, Li RF, Murley JS, Woloschak G, Grdina DJ, Li JJ, CDK4-mediated MnSOD activation and mitochondrial homeostasis in radioadaptive protection, Free Radic. Biol Med 81 (2015) 77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tchakarska G, Roussel M, Troussard X, Sola B, Cyclin D1 inhibits mitochondrial activity in B cells, Cancer Res 71 (2011) 1690–1699. [DOI] [PubMed] [Google Scholar]
- 22.Wang Z, Fan M, Candas D, Zhang TQ, Qin L, Eldridge A, Wachsmann-Hogiu S, Ahmed KM, Chromy BA, Nantajit D, Duru N, He F, Chen M, Finkel T, Weinstein LS, Li JJ, Cyclin B1/Cdk1 coordinates mitochondrial respiration for cell-cycle G2/M progression, Dev. Cell 29 (2014) 217–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu R, Fan M, Candas D, Qin L, Zhang X, Eldridge A, Zou JX, Zhang T, Juma S, Jin C, Li RF, Perks J, Sun LQ, Vaughan AT, Hai CX, Gius DR, Li JJ, CDK1-Mediated SIRT3 Activation Enhances Mitochondrial Function and Tumor Radioresistance, Mol. Cancer Ther 14 (2015) 2090–2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Candas D, Fan M, Nantajit D, Vaughan AT, Murley JS, Woloschak GE, Grdina DJ, Li JJ, CyclinB1/Cdk1 phosphorylates mitochondrial antioxidant MnSOD in cell adaptive response to radiation stress, J. Mol. Cell Biol 5 (2013) 166–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen H, Chan DC, Emerging functions of mammalian mitochondrial fusion and fission, Hum. Mol. Genet 14 Spec No. 2 (2005) R283–289. [DOI] [PubMed] [Google Scholar]
- 26.Chan DC, Mitochondria: dynamic organelles in disease, aging, and development, Cell 125 (2006) 1241–1252. [DOI] [PubMed] [Google Scholar]
- 27.Loson OC, Liu R, Rome ME, Meng S, Kaiser JT, Shan SO, Chan DC, The mitochondrial fission receptor MiD51 requires ADP as a cofactor, Structure 22 (2014) 367–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen KH, Dasgupta A, Lin J, Potus F, Bonnet S, Iremonger J, Fu J, Mewburn J, Wu D, Dunham-Snary K, Theilmann AL, Jing ZC, Hindmarch C, Ormiston ML, Lawrie A, Archer SL, Epigenetic Dysregulation of the Drp1 Binding Partners MiD49 and MiD51 Increases Mitotic Mitochondrial Fission and Promotes Pulmonary Arterial Hypertension: Mechanistic and Therapeutic Implications, Circulation (2018) [DOI] [PMC free article] [PubMed]
- 29.Posakony JW, England JM, Attardi G, Mitochondrial growth and division during the cell cycle in HeLa cells, J. Cell Biol 74 (1977) 468–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nunnari J, Marshall WF, Straight A, Murray A, Sedat JW, Walter P, Mitochondrial transmission during mating in Saccharomyces cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mitochondrial DNA, Mol. Biol. Cell 8 (1997) 1233–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Westermann B, Merging mitochondria matters: cellular role and molecular machinery of mitochondrial fusion, EMBO Rep 3 (2002) 527–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eura Y, Ishihara N, Yokota S, Mihara K, Two mitofusin proteins, mammalian homologues of FZO, with distinct functions are both required for mitochondrial fusion, J. Biochem 134 (2003) 333–344. [DOI] [PubMed] [Google Scholar]
- 33.Smirnova E, Shurland DL, Ryazantsev SN, van der Bliek AM, A human dynamin-related protein controls the distribution of mitochondria, J. Cell Biol 143 (1998) 351–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santel A, Frank S, Shaping mitochondria: The complex posttranslational regulation of the mitochondrial fission protein DRP1, IUBMB Life 60 (2008) 448–455. [DOI] [PubMed] [Google Scholar]
- 35.Chang CR, Blackstone C, Dynamic regulation of mitochondrial fission through modification of the dynamin-related protein Drp1, Ann. Ny. Acad. Sci 1201 (2010) 34–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taguchi N, Ishihara N, Jofuku A, Oka T, Mihara K, Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission, J. Biol. Chem 282 (2007) 11521–11529. [DOI] [PubMed] [Google Scholar]
- 37.Smirnova E, Griparic L, Shurland DL, van der Bliek AM, Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells, Mol Biol Cell 12 (2001) 2245–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chan DC, Mitochondrial fusion and fission in mammals, Annu. Rev. Cell. Dev. Biol 22 (2006) 79–99. [DOI] [PubMed] [Google Scholar]
- 39.Lee YJ, Jeong SY, Karbowski M, Smith CL, Youle RJ, Roles of the mammalian mitochondrial fission and fusion mediators Fis1, Drp1, and Opa1 in apoptosis, Mol. Biol. Cell 15 (2004) 5001–5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toyama EQ, Herzig S, Courchet J, Lewis TL Jr., Loson OC, Hellberg K, Young NP, Chen H, Polleux F, Chan DC, Shaw RJ, Metabolism. AMP-activated protein kinase mediates mitochondrial fission in response to energy stress, Science 351 (2016) 275–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bleazard W, McCaffery JM, King EJ, Bale S, Mozdy A, Tieu Q, Nunnari J, Shaw JM, The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast, Nat. Cell Biol 1 (1999) 298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parone PA, Da Cruz S, Tondera D, Mattenberger Y, James DI, Maechler P, Barja F, Martinou JC, Preventing mitochondrial fission impairs mitochondrial function and leads to loss of mitochondrial DNA, PloS one 3 (2008) e3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benard G, Bellance N, James D, Parrone P, Fernandez H, Letellier T, Rossignol R, Mitochondrial bioenergetics and structural network organization, J. Cell Sci 120 (2007) 838–848. [DOI] [PubMed] [Google Scholar]
- 44.Youle RJ, van der Bliek AM, Mitochondrial fission, fusion, and stress, Science 337 (2012) 1062–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kashatus DF, Lim KH, Brady DC, Pershing NLK, Cox AD, Counter CM, RALA and RALBP1 regulate mitochondrial fission at mitosis, Nat. Cell Biol 13 (2011) 1108–U1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang CR, Blackstone C, Cyclic AMP-dependent protein kinase phosphorylation of Drp1 regulates its GTPase activity and mitochondrial morphology, J. Biol. Chem 282 (2007) 21583–21587. [DOI] [PubMed] [Google Scholar]
- 47.Knott AB, Perkins G, Schwarzenbacher R, Bossy-Wetzel E, Mitochondrial fragmentation in neurodegeneration, Nat. Rev. Neurosci 9 (2008) 505–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cribbs JT, Strack S, Reversible phosphorylation of Drp1 by cyclic AMP-dependent protein kinase and calcineurin regulates mitochondrial fission and cell death, EMBO Rep 8 (2007) 939–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hochegger H, Takeda S, Hunt T, Cyclin-dependent kinases and cell-cycle transitions: does one fit all?, Nat. Rev. Mol. Cell Biol 9 (2008) 910–916. [DOI] [PubMed] [Google Scholar]
- 50.Truman AW, Kristjansdottir K, Wolfgeher D, Hasin N, Polier S, Zhang H, Perrett S, Prodromou C, Jones GW, Kron SJ, CDK-dependent Hsp70 Phosphorylation controls G1 cyclin abundance and cell-cycle progression, Cell 151 (2012) 1308 −1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mohamed TMA, Ang YS, Radzinsky E, Zhou P, Huang Y, Elfenbein A, Foley A, Magnitsky S, Srivastava D, Regulation of Cell Cycle to Stimulate Adult Cardiomyocyte Proliferation and Cardiac Regeneration, Cell 173 (2018) 104–116 e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Santamaria D, Barriere C, Cerqueira A, Hunt S, Tardy C, Newton K, Caceres JF, Dubus P, Malumbres M, Barbacid M, Cdk1 is sufficient to drive the mammalian cell cycle, Nature 448 (2007) 811–815. [DOI] [PubMed] [Google Scholar]
- 53.Chou LC, Yang JS, Huang LJ, Wu HC, Lu CC, Chiang JH, Chen KT, Kuo SC, Chung JG, The synthesized 2-(2-fluorophenyl)-6,7-methylenedioxyquinolin-4-one (CHM-1) promoted G2/M arrest through inhibition of CDK1 and induced apoptosis through the mitochondrial-dependent pathway in CT −26 murine colorectal adenocarcinoma cells, J. Gastroenterol 44 (2009) 1055–1063. [DOI] [PubMed] [Google Scholar]
- 54.Harbauer AB, Opalinska M, Gerbeth C, Herman JS, Rao S, Schonfisch B, Guiard B, Schmidt O, Pfanner N, Meisinger C, Mitochondria. Cell cycle-dependent regulation of mitochondrial preprotein translocase, Science 346 (2014) 1109–1113. [DOI] [PubMed] [Google Scholar]
- 55.Harbauer AB, Zahedi RP, Sickmann A, Pfanner N, Meisinger C, The protein import machinery of mitochondria-a regulatory hub in metabolism, stress, and disease, Cell Met 19 (2014) 357–372. [DOI] [PubMed] [Google Scholar]
- 56.Gautier J, Minshull J, Lohka M, Glotzer M, Hunt T, Maller JL, Cyclin is a component of maturation-promoting factor from Xenopus, Cell 60 (1990) 487–494. [DOI] [PubMed] [Google Scholar]
- 57.Nantajit D, Fan M, Duru N, Wen Y, Reed JC, Li JJ, Cyclin B1/Cdk1 phosphorylation of mitochondrial p53 induces anti-apoptotic response, PloS one 5 (2010) e12341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sherwood SW, Rush DF, Kung AL, Schimke RT, Cyclin B1 expression in HeLa S3 cells studied by flow cytometry, Exp. Cell Res 211 (1994) 275–281. [DOI] [PubMed] [Google Scholar]
- 59.Golsteyn RM, Cdk1 and Cdk2 complexes (cyclin dependent kinases) in apoptosis: a role beyond the cell cycle, Cancer Lett 217 (2005) 129–138. [DOI] [PubMed] [Google Scholar]
- 60.Jackman M, Lindon C, Nigg EA, Pines J, Active cyclin B1-Cdk1 first appears on centrosomes in prophase, Nat. Cell Biol 5 (2003) 143–148. [DOI] [PubMed] [Google Scholar]
- 61.Takizawa CG, Morgan DO, Control of mitosis by changes in the subcellular location of cyclin-B1-Cdk1 and Cdc25C, Curr. Opin. Cell Biol 12 (2000) 658–665. [DOI] [PubMed] [Google Scholar]
- 62.Hung V, Zou P, Rhee HW, Udeshi ND, Cracan V, Svinkina T, Carr SA, Mootha VK, Ting AY, Proteomic mapping of the human mitochondrial intermembrane space in live cells via ratiometric APEX tagging, Mol. Cell 55 (2014) 332–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van der Laan M, Hutu DP, Rehling P, On the mechanism of preprotein import by the mitochondrial presequence translocase, Biochim. Biophys. Acta 1803 (2010) 732–739. [DOI] [PubMed] [Google Scholar]
- 64.Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N, Importing mitochondrial proteins: machineries and mechanisms, Cell 138 (2009) 628–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smith BJ, Yaffe MP, A Mutation in the Yeast Heat-Shock Factor Gene Causes Temperature-Sensitive Defects in Both Mitochondrial Protein Import and the Cell-Cycle, Mol. Cell Biol 11 (1991) 2647–2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhu DH, Dix DJ, Eddy EM, HSP70–2 is required for CDC2 kinase activity in meiosis I of mouse spermatocytes, Development 124 (1997) 3007–3014. [DOI] [PubMed] [Google Scholar]
- 67.Munoz MJ, Jimenez J, Genetic interactions between Hsp90 and the Cdc2 mitotic machinery in the fission yeast Schizosaccharomyces pombe, Mol. Gen. Genet 261 (1999) 242–250. [DOI] [PubMed] [Google Scholar]
- 68.Turnbull EL, Martin IV, Fantes PA, Activity of Cdc2 and its interaction with the cyclin Cdc13 depend on the molecular chaperone Cdc37 in Schizosaccharomyces pombe, J. Cell Sci 119 (2006) 292–302. [DOI] [PubMed] [Google Scholar]
- 69.Macha MA, Matta A, Chauhan S, Siu KM, Ralhan R, 14–3-3 zeta is a molecular target in guggulsterone induced apoptosis in head and neck cancer cells, BMC Cancer 10 (2010) 655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tang Z, Baykal AT, Gao H, Quezada HC, Zhang H, Bereczki E, Serhatli M, Baykal B, Acioglu C, Wang S, Ioja E, Ji X, Zhang Y, Guan Z, Winblad B, Pei JJ, mTor is a signaling hub in cell survival: a mass-spectrometry-based proteomics investigation, J. Proteome Res 13 (2014) 2433–2444. [DOI] [PubMed] [Google Scholar]
- 71.Fu HA, Subramanian RR, Masters SC, 14–3-3 proteins: Structure, function, and regulation, Annu. Rev. Pharmacol 40 (2000) 617–647. [DOI] [PubMed] [Google Scholar]
- 72.Gardino AK, Yaffe MB, 14–3-3 proteins as signaling integration points for cell cycle control and apoptosis, Semin. Cell Dev. Biol 22 (2011) 688–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee YK, Hur W, Lee SW, Hong SW, Kim SW, Choi JE, Yoon SK, Knockdown of 14–3-3zeta enhances radiosensitivity and radio-induced apoptosis in CD133(+) liver cancer stem cells, Exp. Mol. Med 46 (2014) e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Danial NN, Gramm CF, Scorrano L, Zhang CY, Krauss S, Ranger AM, Datta SR, Greenberg ME, Licklider LJ, Lowell BB, Gygi SP, Korsmeyer SJ, BAD and glucokinase reside in a mitochondrial complex that integrates glycolysis and apoptosis, Nature 424 (2003) 952–956. [DOI] [PubMed] [Google Scholar]
- 75.Pagliarini DJ, Wiley SE, Kimple ME, Dixon JR, Kelly P, Worby CA, Casey PJ, Dixon JE, Involvement of a mitochondrial phosphatase in the regulation of ATP production and insulin secretion in pancreatic beta cells, Mol. Cell 19 (2005) 197–207. [DOI] [PubMed] [Google Scholar]
- 76.Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, Shokat KM, Morgan DO, Targets of the cyclin-dependent kinase Cdk1, Nature 425 (2003) 859–864. [DOI] [PubMed] [Google Scholar]
- 77.Candas D, Li JJ, MnSOD in oxidative stress response-potential regulation via mitochondrial protein influx, Antioxid. Redox Signal 20 (2014) 1599–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, Yang Y, Chen Y, Hirschey MD, Bronson RT, Haigis M, Guarente LP, Farese RV Jr., Weissman S, Verdin E, Schwer B, Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation, Mol. Cell Biol 27 (2007) 8807–8814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hallows WC, Lee S, Denu JM, Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases, Proc. Natl. Acad. Sci. U. S. A 103 (2006) 10230–10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, Stevens RD, Li Y, Saha AK, Ruderman NB, Bain JR, Newgard CB, Farese RV Jr., Alt FW, Kahn CR, Verdin E, SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation, Nature 464 (2010) 121–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Someya S, Yu W, Hallows WC, Xu J, Vann JM, Leeuwenburgh C, Tanokura M, Denu JM, Prolla TA, Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction, Cell 143 (2010) 802–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Qiu X, Brown K, Hirschey MD, Verdin E, Chen D, Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation, Cell Met 12 (2010) 662–667. [DOI] [PubMed] [Google Scholar]
- 83.Tseng AH, Shieh SS, Wang DL, SIRT3 deacetylates FOXO3 to protect mitochondria against oxidative damage, Free Radic. Biol. Med 63C (2013) 222–234. [DOI] [PubMed] [Google Scholar]
- 84.Shimazu T, Hirschey MD, Hua L, Dittenhafer-Reed KE, Schwer B, Lombard DB, Li Y, Bunkenborg J, Alt FW, Denu JM, Jacobson MP, Verdin E, SIRT3 deacetylates mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase 2 and regulates ketone body production, Cell Met 12 (2010) 654–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tao R, Vassilopoulos A, Parisiadou L, Yan Y, Gius D, Regulation of MnSOD Enzymatic Activity by Sirt3 Connects the Mitochondrial Acetylome Signaling Networks to Aging and Carcinogenesis, Antioxid. Redox Signal (2013) [DOI] [PMC free article] [PubMed]
- 86.Buchakjian MR, Kornbluth S, The engine driving the ship: metabolic steering of cell proliferation and death, Nat. Rev. Mol. Cell Biol 11 (2010) 715–727. [DOI] [PubMed] [Google Scholar]
- 87.Hatefi Y, The mitochondrial electron transport and oxidative phosphorylation system, Annu. Rev. Biochem 54 (1985) 1015–1069. [DOI] [PubMed] [Google Scholar]
- 88.McBride HM, Neuspiel M, Wasiak S, Mitochondria: more than just a powerhouse, Curr. Biol 16 (2006) R551–560. [DOI] [PubMed] [Google Scholar]
- 89.Blanchet E, Annicotte JS, Lagarrigue S, Aguilar V, Clape C, Chavey C, Fritz V, Casas F, Apparailly F, Auwerx J, Fajas L, E2F transcription factor-1 regulates oxidative metabolism, Nat. Cell Biol 13 (2011) 1146–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Guo R, Zong S, Wu M, Gu J, Yang M, Architecture of Human Mitochondrial Respiratory Megacomplex I2III2IV2, Cell 170 (2017) 1247–1257 e1212. [DOI] [PubMed] [Google Scholar]
- 91.Letts JA, Sazanov LA, Clarifying the supercomplex: the higher-order organization of the mitochondrial electron transport chain, Nat. Struct. Mol. Biol 24 (2017) 800–808. [DOI] [PubMed] [Google Scholar]
- 92.Kawamata H, Manfredi G, Proteinopathies and OXPHOS dysfunction in neurodegenerative diseases, J. Cell Biol 216 (2017) 3917–3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mailloux RJ, Lemire J, Appanna VD, Hepatic response to aluminum toxicity: dyslipidemia and liver diseases, Exp. Cell Res 317 (2011) 2231–2238. [DOI] [PubMed] [Google Scholar]
- 94.Suzuki T, Nagao A, Suzuki T, Human mitochondrial tRNAs: biogenesis, function, structural aspects, and diseases, Annu. Rev. Genet 45 (2011) 299–329. [DOI] [PubMed] [Google Scholar]
- 95.Torraco A, Peralta S, Iommarini L, Diaz F, Mitochondrial Diseases Part I: mouse models of OXPHOS deficiencies caused by defects in respiratory complex subunits or assembly factors, Mitochondrion 21 (2015) 76–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Roessler MM, King MS, Robinson AJ, Armstrong FA, Harmer J, Hirst J, Direct assignment of EPR spectra to structurally defined iron-sulfur clusters in complex I by double electron-electron resonance, Proc. Natl. Acad. Sci U. S. A 107 1930–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Salazar-Roa M, Malumbres M, Fueling the Cell Division Cycle, Trends in cell biology 27 (2017) 69–81. [DOI] [PubMed] [Google Scholar]
- 98.Songyang Z, Blechner S, Hoagland N, Hoekstra MF, Piwnica-Worms H, Cantley LC, Use of an oriented peptide library to determine the optimal substrates of protein kinases, Curr. Biol 4 (1994) 973–982. [DOI] [PubMed] [Google Scholar]
- 99.Ugalde C, Vogel R, Huijbens R, Van Den Heuvel B, Smeitink J, Nijtmans L, Human mitochondrial complex I assembles through the combination of evolutionary conserved modules: a framework to interpret complex I deficiencies, Hum. Mol. Genet 13 (2004) 2461–2472. [DOI] [PubMed] [Google Scholar]
- 100.Konishi Y, Lehtinen M, Donovan N, Bonni A, Cdc2 phosphorylation of BAD links the cell cycle to the cell death machinery, Mol. Cell 9 (2002) 1005–1016. [DOI] [PubMed] [Google Scholar]
- 101.Terrano DT, Upreti M, Chambers TC, Cyclin-dependent kinase 1-mediated Bcl-xL/Bcl-2 phosphorylation acts as a functional link coupling mitotic arrest and apoptosis, Mol. Cell Biol 30 (2010) 640–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Harley ME, Allan LA, Sanderson HS, Clarke PR, Phosphorylation of Mcl-1 by CDK1-cyclin B1 initiates its Cdc20-dependent destruction during mitotic arrest, EMBO J 29 (2010) 2407–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Allan LA, Clarke PR, Phosphorylation of caspase-9 by CDK1/cyclin B1 protects mitotic cells against apoptosis, Mol. Cell 26 (2007) 301–310. [DOI] [PubMed] [Google Scholar]
- 104.O’Connor DS, Grossman D, Plescia J, Li FZ, Zhang H, Villa A, Tognin S, Marchisio PC, Altieri DC, Regulation of apoptosis at cell division by p34(cdc2) phosphorylation of survivin, Proc. Natl. Acad. Sci. U. S. A 97 (2000) 13103–13107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Takeno S, Noguchi T, Kikuchi R, Uchida Y, Yokoyama S, Muller W, Prognostic value of cyclin B1 in patients with esophageal squamous cell carcinoma, Cancer 94 (2002) 2874–2881. [DOI] [PubMed] [Google Scholar]
- 106.Li JQ, Kubo A, Wu F, Usuki H, Fujita J, Bandoh S, Masaki T, Saoo K, Takeuchi H, Kobayashi S, Imaida K, Maeta H, Ishida T, Kuriyama S, Cyclin B1, unlike cyclin G1, increases significantly during colorectal carcinogenesis and during later metastasis to lymph nodes, Int. J. Oncol 22 (2003) 1101–1110. [PubMed] [Google Scholar]
- 107.Dong Y, Sui L, Watanabe Y, Sugimoto K, Tokuda M, Clinical relevance of cyclin B1 overexpression in laryngeal squamous cell carcinoma, Cancer Lett 177 (2002) 13–19. [DOI] [PubMed] [Google Scholar]
- 108.Hassan KA, El-Naggar AK, Soria JC, Liu D, Hong WK, Mao L, Clinical significance of cyclin B1 protein expression in squamous cell carcinoma of the tongue, Clin. Cancer Res 7 (2001) 2458–2462. [PubMed] [Google Scholar]
- 109.Hassan KA, Ang KK, El-Naggar AK, Story MD, Lee JI, Liu D, Hong WK, Mao L, Cyclin B1 overexpression and resistance to radiotherapy in head and neck squamous cell carcinoma, Cancer Res 62 (2002) 6414–6417. [PubMed] [Google Scholar]
- 110.Rew DA, Wilson GD, Cell production rates in human tissues and tumours and their significance. Part II: clinical data, Eur. J. Surg. Oncol 26 (2000) 405–417. [DOI] [PubMed] [Google Scholar]
- 111.Murakami H, Furihata M, Ohtsuki Y, Ogoshi S, Determination of the prognostic significance of cyclin B1 overexpression in patients with esophageal squamous cell carcinoma, Virchows Arch 434 (1999) 153–158. [DOI] [PubMed] [Google Scholar]
- 112.Soria JC, Jang SJ, Khuri FR, Hassan K, Liu D, Hong WK, Mao L, Overexpression of cyclin B1 in early-stage non-small cell lung cancer and its clinical implication, Cancer Res 60 (2000) 4000–4004. [PubMed] [Google Scholar]
- 113.Wang A, Yoshimi N, Ino N, Tanaka T, Mori H, Overexpression of cyclin B1 in human colorectal cancers, J. Cancer Res. Clin. Oncol 123 (1997) 124–127. [DOI] [PubMed] [Google Scholar]
- 114.Yuan J, Yan R, Kramer A, Eckerdt F, Roller M, Kaufmann M, Strebhardt K, Cyclin B1 depletion inhibits proliferation and induces apoptosis in human tumor cells, Oncogene 23 (2004) 5843–5852. [DOI] [PubMed] [Google Scholar]
- 115.Ozeki M, Tamae D, Hou DX, Wang T, Lebon T, Spitz DR, Li JJ, Response of cyclin B1 to ionizing radiation: regulation by NF-kappaB and mitochondrial antioxidant enzyme MnSOD, Anticancer Res 24 (2004) 2657–2663. [PMC free article] [PubMed] [Google Scholar]
- 116.Spitz DR, Azzam EI, Li JJ, Gius D, Metabolic oxidation/reduction reactions and cellular responses to ionizing radiation: a unifying concept in stress response biology, Cancer Metastasis Rev 23 (2004) 311–322. [DOI] [PubMed] [Google Scholar]
- 117.Shao L, Luo Y, Zhou D, Hematopoietic stem cell injury induced by ionizing radiation, Antioxid. Redox Signal 20 (2014) 1447–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bernstein C, Bernstein H, Payne CM, Garewal H, DNA repair/pro-apoptotic dual-role proteins in five major DNA repair pathways: fail-safe protection against carcinogenesis, Mutat. Res 511 (2002) 145–178. [DOI] [PubMed] [Google Scholar]
- 119.Bakkenist CJ, Kastan MB, Initiating cellular stress responses, Cell 118 (2004) 9–17. [DOI] [PubMed] [Google Scholar]
- 120.Kulkarni R, Thomas RA, Tucker JD, Expression of DNA repair and apoptosis genes in mitochondrial mutant and normal cells following exposure to ionizing radiation, Environ. Mol. Mutagen 52 (2011) 229–237. [DOI] [PubMed] [Google Scholar]
- 121.Ward I, Chen J, Early events in the DNA damage response, Curr. Top. Dev. Biol 63 (2004) 1–35. [DOI] [PubMed] [Google Scholar]
- 122.Hoeijmakers JH, DNA damage, aging, and cancer, New Engl. J. Med 361 (2009) 1475–1485. [DOI] [PubMed] [Google Scholar]
- 123.Eguchi Y, Shimizu S, Tsujimoto Y, Intracellular ATP levels determine cell death fate by apoptosis or necrosis, Cancer Res 57 (1997) 1835–1840. [PubMed] [Google Scholar]
- 124.Horbinski C, Chu CT, Kinase signaling cascades in the mitochondrion: a matter of life or death, Free Radic. Biol. Med 38 (2005) 2–11. [DOI] [PubMed] [Google Scholar]
- 125.Pagliarini DJ, Dixon JE, Mitochondrial modulation: reversible phosphorylation takes center stage?, Trends Biochem. Sci 31 (2006) 26–34. [DOI] [PubMed] [Google Scholar]
- 126.Qin L, Fan M, Candas D, Jiang G, Papadopoulos S, Tian L, Woloschak G, Grdina DJ, Li JJ, CDK1 Enhances Mitochondrial Bioenergetics for Radiation-Induced DNA Repair, Cell Reports 13 (2015) 2056–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, Pancoska P, Moll UM, p53 has a direct apoptogenic role at the mitochondria, Mol. Cell 11 (2003) 577–590. [DOI] [PubMed] [Google Scholar]
- 128.Chipuk JE, Kuwana T, Bouchier-Hayes L, Droin NM, Newmeyer DD, Schuler M, Green DR, Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis, Science 303 (2004) 1010–1014. [DOI] [PubMed] [Google Scholar]
- 129.Chipuk JE, Green DR, Dissecting p53-dependent apoptosis, Cell Death Differ 13 (2006) 994–1002. [DOI] [PubMed] [Google Scholar]
- 130.Saleem A, Adhihetty PJ, Hood DA, Role of p53 in mitochondrial biogenesis and apoptosis in skeletal muscle, Physiol. Genomics 37 (2009) 58–66. [DOI] [PubMed] [Google Scholar]
- 131.Bakhanashvili M, Grinberg S, Bonda E, Simon AJ, Moshitch-Moshkovitz S, Rahav G, p53 in mitochondria enhances the accuracy of DNA synthesis, Cell Death Differ 15 (2008) 1865–1874. [DOI] [PubMed] [Google Scholar]
- 132.Yoshida Y, Izumi H, Torigoe T, Ishiguchi H, Itoh H, Kang D, Kohno K, P53 physically interacts with mitochondrial transcription factor A and differentially regulates binding to damaged DNA, Cancer Res 63 (2003) 3729–3734. [PubMed] [Google Scholar]
- 133.Suzuki S, Tanaka T, Poyurovsky MV, Nagano H, Mayama T, Ohkubo S, Lokshin M, Hosokawa H, Nakayama T, Suzuki Y, Sugano S, Sato E, Nagao T, Yokote K, Tatsuno I, Prives C, Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species, Proc. Natl. Acad. Sci. U. S. A 107 (2010) 7461–7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhao Y, Chaiswing L, Velez JM, Batinic-Haberle I, Colburn NH, Oberley TD, St Clair DK, p53 translocation to mitochondria precedes its nuclear translocation and targets mitochondrial oxidative defense protein-manganese superoxide dismutase, Cancer Res 65 (2005) 3745–3750. [DOI] [PubMed] [Google Scholar]
- 135.Ferecatu I, Bergeaud M, Rodriguez-Enfedaque A, Le Floch N, Oliver L, Rincheval V, Renaud F, Vallette FM, Mignotte B, Vayssiere JL, Mitochondrial localization of the low level p53 protein in proliferative cells, Biochem. Biophys. Res. Commun 387 (2009) 772–777. [DOI] [PubMed] [Google Scholar]
- 136.McAndrew CW, Gastwirt RF, Donoghue DJ, The atypical CDK activator Spy1 regulates the intrinsic DNA damage response and is dependent upon p53 to inhibit apoptosis, Cell cycle 8 (2009) 66–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Aubrey BJ, Kelly GL, Janic A, Herold MJ, Strasser A, How does p53 induce apoptosis and how does this relate to p53-mediated tumour suppression?, Cell Death Differ 25 (2018) 104–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kapoor M, Hamm R, Yan W, Taya Y, Lozano G, Cooperative phosphorylation at multiple sites is required to activate p53 in response to UV radiation, Oncogene 19 (2000) 358–364. [DOI] [PubMed] [Google Scholar]
- 139.Bischoff JR, Friedman PN, Marshak DR, Prives C, Beach D, Human p53 is phosphorylated by p60-cdc2 and cyclin B-cdc2, Proc. Natl. Acad. Sci. U. S. A 87 (1990) 4766–4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Dang CV, Links between metabolism and cancer, Genes Dev 26 (2012) 877–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC, The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth, Nature 452 (2008) 230–233. [DOI] [PubMed] [Google Scholar]
- 142.Hitosugi T, Zhou L, Elf S, Fan J, Kang HB, Seo JH, Shan C, Dai Q, Zhang L, Xie J, Gu TL, Jin P, Aleckovic M, LeRoy G, Kang Y, Sudderth JA, DeBerardinis RJ, Luan CH, Chen GZ, Muller S, Shin DM, Owonikoko TK, Lonial S, Arellano ML, Khoury HJ, Khuri FR, Lee BH, Ye K, Boggon TJ, Kang S, He C, Chen J, Phosphoglycerate mutase 1 coordinates glycolysis and biosynthesis to promote tumor growth, Cancer Cell 22 (2012) 585–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Vander Heiden MG, Locasale JW, Swanson KD, Sharfi H, Heffron GJ, Amador-Noguez D, Christofk HR, Wagner G, Rabinowitz JD, Asara JM, Cantley LC, Evidence for an alternative glycolytic pathway in rapidly proliferating cells, Science 329 (2010) 1492–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Pasto A, Bellio C, Pilotto G, Ciminale V, Silic-Benussi M, Guzzo G, Rasola A, Frasson C, Nardo G, Zulato E, Nicoletto MO, Manicone M, Indraccolo S, Amadori A, Cancer stem cells from epithelial ovarian cancer patients privilege oxidative phosphorylation, and resist glucose deprivation, Oncotarget 5 (2014) 4305–4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Porporato PE, Filigheddu N, Pedro JMB, Kroemer G, Galluzzi L, Mitochondrial metabolism and cancer, Cell Res 28 (2018) 265–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wang T, Fahrmann JF, Lee H, Li YJ, Tripathi SC, Yue C, Zhang C, Lifshitz V, Song J, Yuan Y, Somlo G, Jandial R, Ann D, Hanash S, Jove R, Yu H, JAK/STAT3-Regulated Fatty Acid beta-Oxidation Is Critical for Breast Cancer Stem Cell Self-Renewal and Chemoresistance, Cell Metab 27 (2018) 1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Weinhouse S, Millington RH, Wenner CE, Metabolism of neoplastic tissue. I. The oxidation of carbohydrate and fatty acids in transplanted tumors, Cancer Res 11 (1951) 845–850. [PubMed] [Google Scholar]
- 148.Wenner CE, Spirtes MA, Weinhouse S, Metabolism of neoplastic tissue. II. A survey of enzymes of the citric acid cycle in transplanted tumors, Cancer Res 12 (1952) 44–49. [PubMed] [Google Scholar]
- 149.Villa AM, Doglia SM, Mitochondria in tumor cells studied by laser scanning confocal microscopy, J. Biomed. Opt 9 (2004) 385–394. [DOI] [PubMed] [Google Scholar]
- 150.Chae YC, Caino MC, Lisanti S, Ghosh JC, Dohi T, Danial NN, Villanueva J, Ferrero S,Vaira V, Santambrogio L, Bosari S, Languino LR, Herlyn M, Altieri DC, Control of tumor bioenergetics and survival stress signaling by mitochondrial HSP90s, Cancer Cell 22 (2012) 331–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Whitaker-Menezes D, Martinez-Outschoorn UE, Flomenberg N, Birbe RC, Witkiewicz AK,Howell A, Pavlides S, Tsirigos A, Ertel A, Pestell RG, Broda P, Minetti C, Lisanti MP, Sotgia F, Hyperactivation of oxidative mitochondrial metabolism in epithelial cancer cells in situ: visualizing the therapeutic effects of metformin in tumor tissue, Cell Cycle 10 (2011) 4047–4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gan B, Hu J, Jiang S, Liu Y, Sahin E, Zhuang L, Fletcher -Sananikone E, Colla S, Wang YA, Chin L, Depinho RA, Lkb1 regulates quiescence and metabolic homeostasis of haematopoietic stem cells, Nature 468 (2010) 701–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ito K, Carracedo A, Weiss D, Arai F, Ala U, Avigan DE, Schafer ZT, Evans RM, Suda T, Lee CH, Pandolfi PP, A PML-PPAR-delta pathway for fatty acid oxidation regulates hematopoietic stem cell maintenance, Nat. Med 18 (2012) 1350–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Knobloch M, Braun SM, Zurkirchen L, von Schoultz C, Zamboni N, Arauzo-Bravo MJ, Kovacs WJ, Karalay O, Suter U, Machado RA, Roccio M, Lutolf MP, Semenkovich CF, Jessberger S, Metabolic control of adult neural stem cell activity by Fasn-dependent lipogenesis, Nature 493 (2013) 226–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Samudio I, Harmancey R, Fiegl M, Kantarjian H, Konopleva M, Korchin B, Kaluarachchi K, Bornmann W, Duvvuri S, Taegtmeyer H, Andreeff M, Pharmacologic inhibition of fatty acid oxidation sensitizes human leukemia cells to apoptosis induction, J. Clin. Invest 120 (2010) 142–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Monti S, Savage KJ, Kutok JL, Feuerhake F, Kurtin P, Mihm M, Wu B, Pasqualucci L, Neuberg D, Aguiar RC, Dal Cin P, Ladd C, Pinkus GS, Salles G, Harris NL, Dalla-Favera R, Habermann TM, Aster JC, Golub TR, Shipp MA, Molecular profiling of diffuse large B-cell lymphoma identifies robust subtypes including one characterized by host inflammatory response, Blood 105 (2005) 1851–1861. [DOI] [PubMed] [Google Scholar]
- 157.Metallo CM, Vander Heiden MG, Understanding metabolic regulation and its influence on cell physiology, Mol. Cell 49 (2013) 388–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ward PS, Thompson CB, Metabolic reprogramming: a cancer hallmark even warburg did not anticipate, Cancer Cell 21 (2012) 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Vyas S, Zaganjor E, Haigis MC, Mitochondria and Cancer, Cell 166 (2016) 555–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wallace DC, Mitochondria and cancer, Nature reviews. Cancer 12 (2012) 685–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Pusapati RV, Daemen A, Wilson C, Sandoval W, Gao M, Haley B, Baudy AR, Hatzivassiliou G, Evangelista M, Settleman J, mTORC1-Dependent Metabolic Reprogramming Underlies Escape from Glycolysis Addiction in Cancer Cells, Cancer Cell 29 (2016) 548–562. [DOI] [PubMed] [Google Scholar]
- 162.Rodriguez-Enriquez S, Marin-Hernandez A, Gallardo-Perez JC, Carreno-Fuentes L, Moreno-Sanchez R, Targeting of cancer energy metabolism, Mol. Nutr. Food Res 53 (2009) 29–48. [DOI] [PubMed] [Google Scholar]
- 163.Dreier A, Barth S, Goswami A, Weis J, Cetuximab induces mitochondrial translocalization of EGFRvIII, but not EGFR: involvement of mitochondria in tumor drug resistance?, Tumour Biol 33 (2012) 85–94. [DOI] [PubMed] [Google Scholar]
- 164.Kulikov AV, Vdovin AS, Zhivotovsky B, Gogvadze V, Targeting mitochondria by alpha-tocopheryl succinate overcomes hypoxia-mediated tumor cell resistance to treatment, Cell. Mol. Life Sci 71 (2014) 2325–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Duru N, Candas D, Jiang G, Li JJ, Breast cancer adaptive resistance: HER2 and cancer stem cell repopulation in a heterogeneous tumor society, J. Cancer Res. Clin. Oncol 140 (2014) 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Duru N, Fan M, Candas D, Menaa C, Liu HC, Nantajit D, Wen Y, Xiao K, Eldridge A, Chromy BA, Li S, Spitz DR, Lam KS, Wicha MS, Li JJ, HER2-associated radioresistance of breast cancer stem cells isolated from HER2-negative breast cancer cells, Clin. Cancer Res 18 (2012) 6634–6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Lu CL, Qin L, Liu HC, Candas D, Fan M, Li JJ, Tumor cells switch to mitochondrial oxidative phosphorylation under radiation via mTOR-mediated hexokinase II inhibition--a Warburg-reversing effect, PloS one 10 (2015) e0121046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Obre E, Rossignol R, Emerging concepts in bioenergetics and cancer research: metabolic flexibility, coupling, symbiosis, switch, oxidative tumors, metabolic remodeling, signaling and bioenergetic therapy, Int. J. Biochem. Cell Biol 59 (2015) 167–181. [DOI] [PubMed] [Google Scholar]
- 169.Kang R, Tang D, Schapiro NE, Loux T, Livesey KM, Billiar TR, Wang H, Van Houten B, Lotze MT, Zeh HJ, The HMGB1/RAGE inflammatory pathway promotes pancreatic tumor growth by regulating mitochondrial bioenergetics, Oncogene 33 (2014) 567–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Li JJ, Oberley LW, St Clair DK, Ridnour LA, Oberley TD, Phenotypic changes induced in human breast cancer cells by overexpression of manganese-containing superoxide dismutase, Oncogene 10 (1995) 1989–2000. [PubMed] [Google Scholar]
- 171.Wullschleger S, Loewith R, Hall MN, TOR signaling in growth and metabolism, Cell 124 (2006) 471–484. [DOI] [PubMed] [Google Scholar]
- 172.Young RM, Ackerman D, Quinn ZL, Mancuso A, Gruber M, Liu L, Giannoukos DN, Bobrovnikova-Marjon E, Diehl JA, Keith B, Simon MC, Dysregulated mTORC1 renders cells critically dependent on desaturated lipids for survival under tumor-like stress, Genes Dev 27 (2013) 1115–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zhang Q, Raje V, Yakovlev VA, Yacoub A, Szczepanek K, Meier J, Derecka M, Chen Q, Hu Y, Sisler J, Hamed H, Lesnefsky EJ, Valerie K, Dent P, Larner AC, Mitochondrial localized Stat3 promotes breast cancer growth via phosphorylation of serine 727, J. Biol. Chem 288 (2013) 31280–31288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Park JH, Vithayathil S, Kumar S, Sung PL, Dobrolecki LE, Putluri V, Bhat VB, Bhowmik SK, Gupta V, Arora K, Wu D, Tsouko E, Zhang Y, Maity S, Donti TR, Graham BH, Frigo DE, Coarfa C, Yotnda P, Putluri N, Sreekumar A, Lewis MT, Creighton CJ, Wong LJ, Kaipparettu BA, Fatty Acid Oxidation-Driven Src Links Mitochondrial Energy Reprogramming and Oncogenic Properties in Triple-Negative Breast Cancer, Cell Rep 14 (2016) 2154–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Pascual G, Avgustinova A, Mejetta S, Martin M, Castellanos A, Attolini CS, Berenguer A, Prats N, Toll A, Hueto JA, Bescos C, Di Croce L, Benitah SA, Targeting metastasis-initiating cells through the fatty acid receptor CD36, Nature 541 (2017) 41–45. [DOI] [PubMed] [Google Scholar]
- 176.Caino MC, Ghosh JC, Chae YC, Vaira V, Rivadeneira DB, Faversani A, Rampini P, Kossenkov AV, Aird KM, Zhang R, Webster MR, Weeraratna AT, Bosari S, Languino LR, Altieri DC, PI3K therapy reprograms mitochondrial trafficking to fuel tumor cell invasion, Proc. Natl. Acad. Sci. U. S. A 112 (2015) 8638–8643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Strack S, Wilson TJ, Cribbs JT, Cyclin-dependent kinases regulate splice-specific targeting of dynamin-related protein 1 to microtubules, The Journal of cell biology 201 (2013) 1037–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Weinberg SE, Chandel NS, Targeting mitochondria metabolism for cancer therapy, Nat. Chem. Biol 11 (2015) 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Weinberg F, Hamanaka R, Wheaton WW, Weinberg S, Joseph J, Lopez M, Kalyanaraman B, Mutlu GM, Budinger GR, Chandel NS, Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity, Proc. Natl. Acad. Sci. USA 107 (2010) 8788–8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, Kamphorst JJ, Chen G, Lemons JM, Karantza V, Coller HA, Dipaola RS, Gelinas C, Rabinowitz JD, White E, Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis, Genes Dev 25 (2011) 460–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Oberley LW, McCormick ML, Sierra-Rivera E, Kasemset-St Clair D, Manganese superoxide dismutase in normal and transformed human embryonic lung fibroblasts, Free Radic. Biol. Med 6 (1989) 379–384. [DOI] [PubMed] [Google Scholar]
- 182.St Clair DK, Holland JC, Complementary DNA encoding human colon cancer manganese superoxide dismutase and the expression of its gene in human cells, Cancer Res 51 (1991) 939–943. [PubMed] [Google Scholar]
- 183.Murley JS, Nantajit D, Baker KL, Kataoka Y, Li JJ, Grdina DJ, Maintenance of manganese superoxide dismutase (SOD2)-mediated delayed radioprotection induced by repeated administration of the free thiol form of amifostine, Radiat. Res 169 (2008) 495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Connor KM, Hempel N, Nelson KK, Dabiri G, Gamarra A, Belarmino J, Van De Water L, Mian BM, Melendez JA, Manganese superoxide dismutase enhances the invasive and migratory activity of tumor cells, Cancer Res 67 (2007) 10260–10267. [DOI] [PubMed] [Google Scholar]
- 185.Hempel N, Carrico PM, Melendez JA, Manganese superoxide dismutase (Sod2) and redox-control of signaling events that drive metastasis, Anticancer Agents Med. Chem 11 (2011) 191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Holley AK, Kiningham KK, Spitz DR, Edwards DP, Jenkins JT, Moore MR, Progestin stimulation of manganese superoxide dismutase and invasive properties in T47D human breast cancer cells, J. Steroid Biochem. Mol. Biol 117 (2009) 23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Nelson KK, Ranganathan AC, Mansouri J, Rodriguez AM, Providence KM, Rutter JL, Pumiglia K, Bennett JA, Melendez JA, Elevated sod2 activity augments matrix metalloproteinase expression: evidence for the involvement of endogenous hydrogen peroxide in regulating metastasis, Clin. Cancer Res 9 (2003) 424–432. [PubMed] [Google Scholar]
- 188.Nakada D, Saunders TL, Morrison SJ, Lkb1 regulates cell cycle and energy metabolism in haematopoietic stem cells, Nature 468 (2010) 653–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Amoedo ND, El-Bacha T, Rodrigues MF, Rumjanek FD, Cell cycle and energy metabolism in tumor cells: strategies for drug therapy, Recent Pat. Anticancer Drug Discov 6 (2011) 15–25. [DOI] [PubMed] [Google Scholar]
- 190.Alexandrou AT, Li JJ, Cell cycle regulators guide mitochondrial activity in radiation-induced adaptive response, Antioxid. Redox Signal 20 (2014) 1463–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Fiske BP, Vander Heiden MG, Seeing the Warburg effect in the developing retina, Nat. Cell Biol 14 (2012) 790–791. [DOI] [PubMed] [Google Scholar]
- 192.Lunt SY, Vander Heiden MG, Aerobic glycolysis: meeting the metabolic requirements of cell proliferation, Annu. Rev. Cell Dev. Biol 27 (2011) 441–464. [DOI] [PubMed] [Google Scholar]