Abstract
Genome structural variations, including duplications, deletions, insertions, and inversions, are central in the evolution of eukaryotic genomes. However, structural variations present challenges for high-quality genome assembly, hampering efforts to understand the evolution of gene families and genome architecture. An example is the genome of the pea aphid (Acyrthosiphon pisum) for which the current assembly is composed of thousands of short scaffolds, many of which are known to be misassembled. Here, we present an improved version of the A. pisum genome based on the use of two long-range proximity ligation methods. The new assembly contains four long scaffolds (40–170 Mb), corresponding to the three autosomes and the X chromosome of A. pisum, and encompassing 86% of the new assembly. Assembly accuracy is supported by several quality assessments. Using this assembly, we identify the chromosomal locations and relative ages of duplication events, and the locations of horizontally acquired genes. The improved assembly illuminates the mode of gene family evolution by providing proximity information between paralogs. By estimating nucleotide polymorphism and coverage depth from resequencing data, we determined that many short scaffolds not assembling to chromosomes represent hemizygous regions, which are especially frequent on the highly repetitive X chromosome. Aligning the X-linked aphicarus region, responsible for male wing dimorphism, to the new assembly revealed a 50-kb deletion that cosegregates with the winged male phenotype in some clones. These results show that long-range scaffolding methods can substantially improve assemblies of repetitive genomes and facilitate study of gene family evolution and structural variation.
Keywords: proximity ligation, structural variation, duplication, paralogs, insect genome
Introduction
The feasibility of inexpensive sequencing offers the chance to understand genome evolution and genome architecture across the Tree of Life. Some of the most important events leading to adaptation and diversification involve genomic changes of a larger scale than single nucleotide substitutions (Feuk et al. 2006; Neafsey et al. 2014; Sudmant et al. 2015; Long et al. 2018; Waterhouse et al. 2018). Key changes in the evolution of most lineages include insertions, deletions, inversions, translocations, and duplications of genomic regions (Fawcett et al. 2009; Lien et al. 2016; Riehle et al. 2017; Matthews et al. 2018) as well as acquisition of horizontally transferred genes (Moran and Jarvik 2010; Moran et al. 2012; Crisp et al. 2015; Peccoud et al. 2017). To detect these kinds of changes requires high-quality and complete genome assemblies. Many animal genomes have been sequenced in the last decade using next-generation sequencing technologies that provide cheaper alternatives to the long-dominant Sanger method (Genome 10K Community 2009; i5K Consortium 2013). Although these methods allow deep coverage of genomes, most fall short with respect to assembly of highly repetitive regions or copy number variation resulting from duplication (Treangen and Salzberg 2012; Jiao and Schneeberger 2017). Next-generation sequencing techniques can be combined with other long-read sequencing and long-range scaffolding methods (Lee et al. 2016; Jiao and Schneeberger 2017) to enable chromosome-scale assembly, which in turn can shed light on the genomic changes underlying adaptation and phenotypic variation (Lewin et al. 2009).
A central model for understanding adaptation and diversification is the pea aphid (Acyrthosiphon pisum). This species has been a model for studies on speciation (Hawthorne and Via 2001), sex chromosome evolution (Jaquiéry et al. 2018), horizontal gene transfer (HGT) into animal genomes (Nikoh and Nakabachi 2009; Moran and Jarvik 2010; Nikoh et al. 2010), obligate symbiosis with bacteria (Hansen and Moran 2011; Shigenobu and Stern 2013; Duncan et al. 2016), host plant adaptation (Jaquiéry et al. 2012), developmental polymorphism (Brisson 2010; Shigenobu et al. 2010), and other evolutionary and ecological questions (Brisson and Stern 2006). The sequencing of the A. pisum genome revealed an unusually high level of gene family expansion (IAGC 2010), a result confirmed by other studies (Smadja et al. 2009; Dahan et al. 2015; Duncan et al. 2016). The A. pisum genome also contains genes horizontally transferred from bacteria (Nikoh et al. 2010) and from fungi (Moran and Jarvik 2010), with subsequent duplications of transferred genes in each case. The current version of the genome, Acyr 2.0 (GenBank accession GCA_000142985.2), contains 12,969 scaffolds longer than 1 kb, and an N50 and L50 of 519 kb and 280 scaffolds, respectively, indicating a highly fragmented assembly and reflecting the extensive duplication and expansion of repetitive sequences in this genome.
Even more problematic, many or most of the large scaffolds contain assembly errors and falsely join regions from the same or different chromosomes. For example, to identify regions corresponding to the X chromosome, Jaquiéry et al. (2018) mapped short Illumina reads for males and females of the same aphid clone. Since aphids have XO sex determination, males retain the maternal diploid genotype except for elimination of an X chromosome. Thus, the X chromosome should have half the coverage in males relative to females. Based on this feature, Jaquiéry et al. (2018) identified X-linked regions as those for which male genomes have depth of coverage half that of females. Their results revealed that 56% of the long (≥150 kb) Acyr 2.0 scaffolds combine regions from both X chromosome and autosomes, indicating false assembly. Additionally, other studies examined a set of loci involved both in carotenoid biosynthesis and in male wing dimorphism and uncovered assembly errors in the corresponding scaffolds in each case (Moran and Jarvik 2010; Mandrioli et al. 2016; B. Li et al. 2017). The evident low quality of the current A. pisum genome assembly is an obstacle for many kinds of genetic and evolutionary studies, and it prevents the understanding of larger scale changes in genome architecture.
Here, we applied long-range scaffolding approaches, based on proximity ligation, to improve the genomic assembly of A. pisum. Under this approach, high-quality genomic DNA is self-ligated so that short-reads pair up with a probability dependent on relative proximity within a chromosome. The paired regions are then isolated and sequenced as paired end reads using next-generation sequencing technology (Putnam et al. 2016). Read pair proximity information then is used to scaffold the genome. These methods yielded four large scaffolds corresponding to the X chromosome and the three autosomes of A. pisum, as documented in karyotype studies (Blackman 1980; Mandrioli et al. 2016) and genetic linkage analyses (Jaquiéry et al. 2014). We validated the assembly using several approaches: the content of benchmarking single-copy genes, microsatellite markers that have been mapped genetically (Jaquiéry et al. 2014), and relative sequencing depth in males versus females to verify assignment to the X. Although most of the genome assembled into scaffolds corresponding to the four chromosomes, 14.3% of the assembled sequence was comprised of short scaffolds not assigned to any chromosome. Most of the longest short scaffolds appear to reside on the X chromosome, which contains an elevated proportion of repetitive regions. In addition, our results illuminate the history of gene duplications and the evolution of gene families that function in coloration, male wing dimorphism, and symbiont interactions.
Results
Genome Assembly and Verification
We performed genome assembly of the A. pisum AL4 clone (fig. 1) based on 182 million Chicago read pairs and 212 million HiC read pairs (Putnam et al. 2016) of this clone and on sequence data from Acyr 2.0, which is derived from the parental genotype LSR1.AC.G1 (hereafter called LSR1). The resulting new assembly had a total length of 540 Mb, close to the Acyr 2.0 assembly length and the estimated haploid genome size based on flow cytometry (Bennett et al. 2003; IAGC 2010). The four chromosome-level scaffolds contained 85.7% of the assembled sequence, corresponding to the known chromosome number of A. pisum (three autosomes and the X chromosome) (table 1). As we combined proximity ligation data from the AL4 clone with the LSR1 A. pisum draft genome, both LSR1 and AUS genomes could contribute to the new assembly. Our assembly required 7,735 breaks to be introduced into Acyr 2.0, implying rampant assembly errors in Acyr 2.0. To further investigate the breaks, we mapped the coordinates of Acyr 2.0 genome features to the AL4 assembly. Regions containing mRNA, ncRNA and introns showed higher unmapped rates (4.26–4.55%) than did regions containing exons, CDS and tRNAs (0.19–1.22%) (supplementary table S1, Supplementary Material online), indicating that the misassemblies in Acyr 2.0 are mostly in noncoding regions. Thus, the AL4 assembly is expected to be a vastly improved representation of the A. pisum genome architecture, but to have relatively small effect on the annotated set of protein-coding genes.
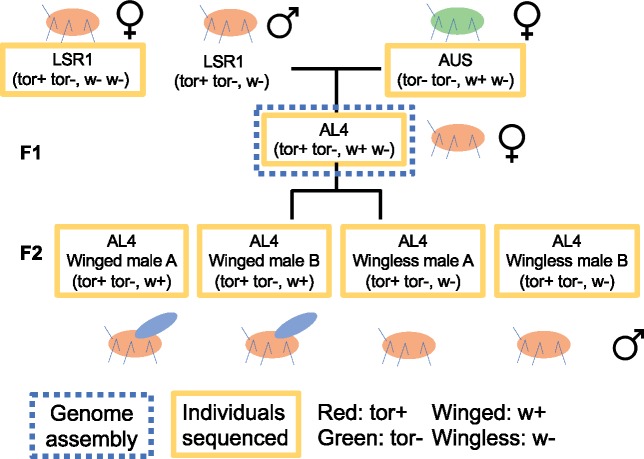
Fig. 1.
Relationships of the aphid samples used in this experiment. AL4 individuals (in the dotted blue box) are the clonal female aphids used for genome assembly. Other individuals (in the solid yellow boxes) were used for resequencing purpose. The w+ w-genotype of AUS is inferred based on the ability of its progeny AL4 to produce winged males, as AUS itself does not produce males.
Table 1.
Statistics Comparing Pea Aphid Genome Assemblies Acyr 2.0 and AL4.
| Acyr 2.0 | AL4 | |
|---|---|---|
| Total size | 541,692,442 | 542,664,571 |
| Total size (without N) | 499,908,163 | 499,891,192 |
| N50 | 518,546 | 132,544,852 |
| N90 | 43,176 | 43,172 |
| Longest scaffold | 3,073,041 | 170,740,645 |
| Shortest scaffold | 200 | 200 |
| L50 | 280 | 2 |
| L90 | 1,511 | 317 |
To evaluate the completeness of the assembly, we performed Benchmarking Universal Single-Copy Orthologs (BUSCOs) assessment (Simão et al. 2015), which showed that the current assembly contains 93.5% of the complete single-copy orthologous genes in Insecta. This BUSCO completeness is similar to that of Acyr 2.0 (table 2). The four chromosome-level scaffolds alone covered a similar number of BUSCOs (93.3% complete BUSCOs), indicating that the short scaffolds contain few BUSCOs.
Table 2.
BUSCO Assessment Based on the Pea Aphid Genome Acyr 2.0, AL4 Assembly, and the Four Chromosome-Level Scaffolds in AL4 Assembly.
| Acyr 2.0 (%) | AL4 (%) | AL4 (4 Chromosome-Level Scaffolds) (%) | ||
|---|---|---|---|---|
| Complete BUSCOs | Single copy | 89.1 | 89.6 | 89.7 |
| Duplicated | 4.8 | 3.9 | 3.6 | |
| Fragmented | 1.4 | 1.8 | 1.9 | |
| Missing | 4.7 | 4.7 | 4.8 | |
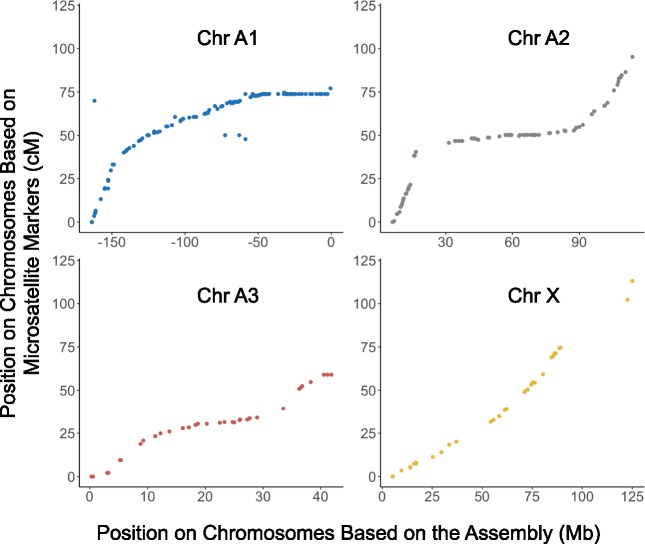
To confirm the overall validity of the assembly, we located previously mapped microsatellite markers (Jaquiéry et al. 2014) on the AL4 assembly. We were able to locate 96.1% (293 out of 305) microsatellite primer pairs to the four chromosome-level scaffolds (supplementary table S2, Supplementary Material online). The overall pattern reveals near-perfect congruence of the new assembly with the linkage map (fig. 2). Based on comparison of genetic distance and physical distance of microsatellite markers, the X chromosome shows high levels of recombination, whereas most autosomal regions experience low recombination rates. Based on the linkage relationship of the microsatellite markers and the length of the chromosome-level scaffolds, we assigned the scaffolds to the corresponding chromosomes (A1, A2, A3, and X) (supplementary table S2, Supplementary Material online).
Fig. 2.
The position of microsatellite markers (Jaquiéry et al. 2014) on the genetic map (centimorgan, cM) and on the AL4 assembly (megabase, Mb).
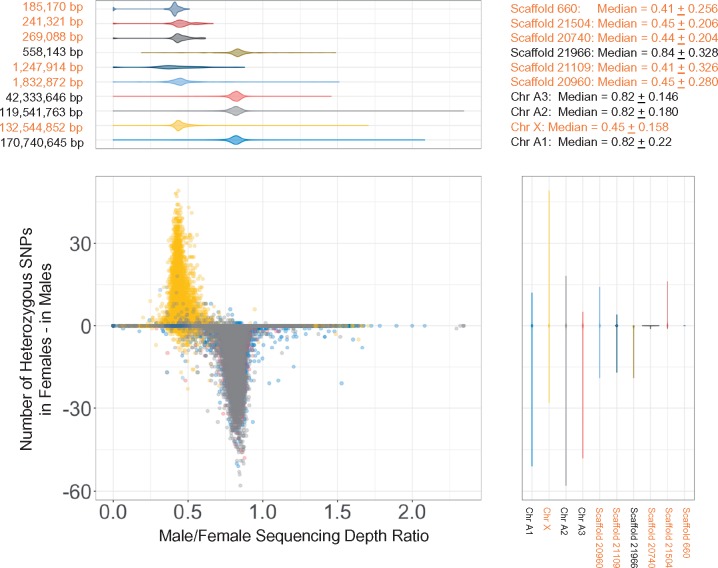
Aphids reproduce clonally during much of their life cycle. Their sexual phase involves a special meiosis in which males retain the maternal diploid genotype except for elimination of an X chromosome. Thus, the X chromosome should have half the coverage in males relative to females. Based on this expectation, a previous study attempted to assign Acyr 2.0 scaffolds to the X, but many longer scaffolds were found to be chimeric, containing sequences from both autosomes and the X chromosome (Jaquiéry et al. 2018). We used the same approach to assess the extent of chimeric scaffolds in our AL4 assembly. We obtained short read Illumina sequences for male and female individuals and estimated the sequencing depth ratio between males and females across the AL4 assembly using 10-kb sliding windows. Because of the XO sex determination, the sequencing depth for the X chromosome in females should be twice the value observed for males. Indeed, we found a bimodal distribution for this ratio with one peak, representing the X (median = 0.45 ± 0.079), centered on a value that is about half the value of the other, larger peak, representing the autosomes (median = 0.82 ± 0.100) (fig. 3 and supplementary fig. S1, Supplementary Material online). For the three other scaffolds, distributions were similar with median close to 0.82, implying that these are the three autosomes (fig. 3). The short scaffolds showed a bimodal distribution, indicating that a large proportion of the short scaffolds came from the X chromosome. For the six longest of the short scaffolds, we determined five to be of X chromosomal origin and one to be of autosomal origin, based on male to female sequencing depth (fig. 3). Furthermore, the X chromosome assignment inferred on this basis was confirmed by the lack of heterozygous single nucleotide polymorphisms (SNPs) in males.
Fig. 3.
The distribution of sequencing depth and heterozygous SNPs on scaffolds >500 kb using 10k-bp sliding windows. The difference of heterozygous SNPs was calculated as the minimum number of heterozygous SNPs in females − the maximum number of heterozygous SNPs in males. The median depth ± two standard deviations of the four chromosomes are next to the plot. The colors of plots were coded based on different scaffolds. Scaffold names in orange indicate the scaffolds are from Chr X based on the distribution of sequencing depth.
We also estimated the overall heterozygosity of the seven AL4 individuals for which we obtained Illumina resequencing data. A previous study (Brisson et al. 2009) found that A. pisum populations have a polymorphism of about 0.13%, based on a 1,608 nucleotide intergenic region. Using the resequencing data, 0.063% of positions in protein-coding gene regions were heterozygous in the paternal LSR1 clone, whereas 0.085% of positions were heterozygous in the maternal AUS clone. The lower value for LSR1 may reflect the fact that this strain underwent one generation of inbreeding prior to sequencing (IAGC 2010). The progeny clone AL4 showed an intermediate heterozygosity of 0.068%. The four males showed nearly identical heterozygosity (0.047%, 0.047%, 0.048%, and 0.048%) with values lower than those observed for females (range 0.063–0.085%). The lower values are expected since males have only one copy of the X chromosome and thus lack heterozygous sites for this portion of the genome.
Genome Annotation and Gene Duplications
We used WQ-MAKER (Hazekamp et al. 2018) to annotate genes in the AL4 assembly, incorporating the protein annotation information derived from the Acyr 2.0 assembly. WQ-MAKER initially identified 91,619 sequences with homology to previously annotated genes. Of these, 40,090 proteins identified in AL4 were found to have ≥98% similarity to 30,991 proteins in the Acyr 2.0 annotation, based on BlastP. This result (a larger number of encoded proteins in AL4) suggests that some of the AL4 genes result from recent duplications that were collapsed into single copies in the Acyr 2.0 assembly. Also, some genes may be misassembled as a single locus in Acyr 2.0, but separated into distinct loci by the improved assembly of AL4. Of the genes from the AL4 annotation that were assignable at 98% protein identity to Acyr 2.0 annotated proteins, many were shorter than 100 amino acids (supplementary fig. S2, Supplementary Material online), which suggests that most are pseudogenes or false positive annotations. For the genes longer than 100 amino acids, we found that 88% (27,496 out of 31,418) of them were ≥98% identical to genes in the AL4 assembly. Thus, the Acyr 2.0 annotation identified the large majority of distinct proteins. The main distinction between the two annotations is that genes that were misassembled or collapsed in Acyr 2.0 are assembled more accurately in AL4.
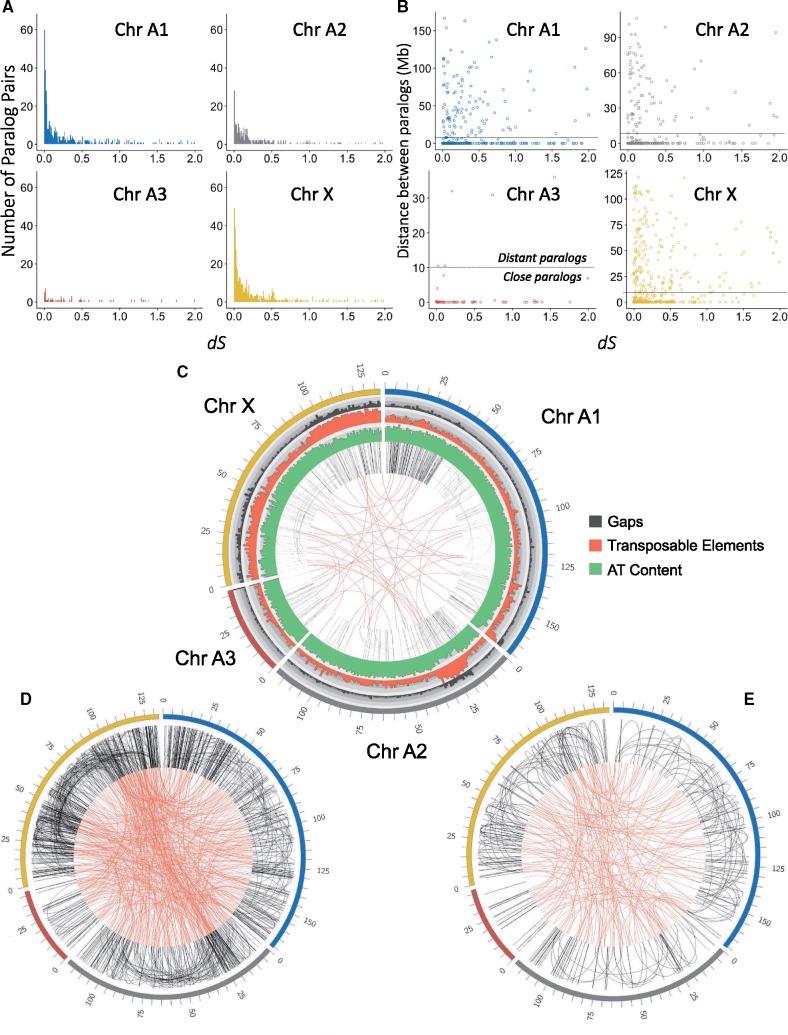
The A. pisum genome was already known to contain extensive gene duplications (IAGC 2010; Duncan et al. 2016), and these are a likely cause of poor assembly in Acyr 2.0. We investigated the distributions of paralogs in AL4 to better understand the history of gene duplication in this genome. In the AL4 genome assembly, we identified 4,502 paralog pairs based on reciprocal best hits from BlastP. We found 1,594 paralog pairs on the same chromosome-level scaffolds, 1,276 paralog pairs were on different chromosome-level scaffolds, and 1,081 paralog pairs were on both short and chromosome-level scaffolds. Among the chromosomes, Chromosome X showed the most within-chromosome paralogs (449 paralogs), and Chromosome A3, the shortest autosome, showed the fewest within-chromosome paralogs (62 paralogs) (fig. 4A). Chromosomes A1 and A2 have 413 and 261 within-chromosome paralogs, respectively. We plotted the frequencies of different paralog pairs with respect to dS values, to determine whether some of the pairs may reflect past duplications of larger chromosome segments spanning multiple genes. We found that paralog pairs on the X chromosome show a peak of ancestral gene duplications corresponding to dS = 0.5, suggesting duplication of a large part of this chromosome in the past. Chromosome A2 contained a high proportion of more recent gene duplications with dS near 0.2. Paralog pairs on the same chromosome-level scaffold with dS ≤ 1.0 ranged from very close (close duplications) to distant from one another (distant duplications), whereas paralog pairs with dS > 1.0 were relatively few with higher proportion to be distant from one another (fig. 4B).
Fig. 4.
(A) The number of paralog pairs and (B) distance between paralog pairs on each chromosome in the AL4 assembly. The x axes show the synonymous divergence (dS) between paralog pairs. The dotted lines split the paralogs into distant paralogs (paralog distance >= 10 Mb) and close paralogs (paralog distance < 10 Mb). (C) Homologous regions within chromosomes (black links) and between chromosomes (red links) based on LAST alignment. (D) The location of younger paralog pairs (0.0 ≤ dS < 0.3) and (E) the location of older paralog pairs (0.3 ≤ dS < 0.6) on chromosomes. Chromosomes in (D) and (E) are color coded in the same way as in (C).
Visualizing the locations of paralog pairs on the chromosomes showed that close duplications were prevalent on the start of Chromosome A1 and some regions of Chromosomes A3 and X (fig. 4C). Distant duplications located on Chromosomes A1, A2, and X. The range of distant duplications were up to ∼50–90 Mb on Chromosome X based on LAST alignment (fig. 4C). Two regions with elevated numbers of duplications, located at the beginning of Chromosome A2 (20–35 Mb) and end of Chromosome X (90–130 Mb), also showed high frequencies of transposable elements, which potentially mediate duplication events. To determine whether recent and ancient paralog pairs have different relative locations, we defined close paralog pairs as separated <10 Mb on a chromosome and distant paralog pairs as those separated by ≥10 Mb on a chromosome. For recent paralog pairs (dS < 0.3), 73% were close and 27% were distant (fig. 4D), whereas for older pairs (0.3 ≤ dS < 0.6), 60% were close and 40% were distant (fig. 4E and supplementary table S3, Supplementary Material online). We found significant correlations between dS and distance on Chromosomes A1 and X, although the correlation coefficients were low (0.19 and 0.12, respectively). We further investigated other potential factors relating to close and distant paralog pairs, such as dN, coding sequence (CDS) length between paralog pairs, numbers of CDS between paralog pairs (supplementary figs. S3–S5, Supplementary Material online). None of these factors correlate strongly with the distance between paralog pairs (supplementary table S4, Supplementary Material online). The elevated numbers of repeats on one end of Chromosome A1 (0–50 Mb) is due to younger duplications, whereas the elevated numbers of repeats on the X is found for both young and old paralog pairs (fig. 4D and E). Because the divergence of these regions was high, possibly obscuring nucleotide-based homology, we used homology searching with protein-coding genes as anchors (MCscan analysis), and the result is consistent with LAST alignment (supplementary fig. S6, Supplementary Material online).
Genome Architecture of the AL4 Assembly
We searched the assembly using four sets of gene families: the carotenoid biosynthetic genes (Moran and Jarvik 2010; Mandrioli et al. 2016), gene regions underlying male wing dimorphism (aphicarus) (Braendle et al. 2005; B. Li et al. 2017), and two sets of genes related to the obligate symbiosis with the bacterium Buchnera aphidicola (Nikoh et al. 2010; Shigenobu and Stern 2013). These include numerous genes previously shown to have been acquired in ancestral aphids through HGT from bacteria or fungi. We were able to align the majority of these gene families to chromosome-level scaffolds.
Aphids are among the rare cases of animals that can make their own carotenoids, due to ancestral HGT of carotenoid biosynthetic genes from fungi, followed by duplications (Moran and Jarvik 2010). In A. pisum, one of the carotenoid desaturase loci underlies a red/green color polymorphism involving the production of the red pigment torulene in individuals possessing the tor+ allele. It was previously shown that green (tor−) individuals lack a region of at least 30 kb that includes this carotenoid desaturase gene copy and that is present as a single copy in heterozygous tor+/tor− genotypes such as AL4 or LSR1 (Moran and Jarvik 2010). In the Acyr 2.0 assembly, the carotenoid biosynthetic genes are located on several scaffolds, and some scaffolds were shown to be chimeric due to misassembly (Moran and Jarvik 2010). In the AL4 assembly, we located three carotenoid synthase/cyclase genes, three carotenoid desaturase genes and a pseudogene of carotenoid desaturase, all clustered near the end of the scaffold designated as Chromosome A1 (supplementary fig. S7, Supplementary Material online), which is consistent with previous in situ polymerase chain reaction (PCR) results (Mandrioli et al. 2016). However, tor itself, though located in the same chromosomal region based on in situ data, was located on a separate short scaffold in the AL4 assembly.
Aphid bacteriocytes are specialized aphid cells containing the obligate bacterial symbiont Buchnera and are characterized by strong upregulation of numerous genes (Hansen and Moran 2011; Shigenobu and Stern 2013) including some genes that were acquired by horizontal transfer from bacterial sources (Nikoh et al. 2010). We aligned these symbiosis-related genes, including bacteriocyte-specific cysteine-rich proteins (BCR) genes and secreted protein (SP) genes to corresponding scaffolds. BCR1, BCR4, and BCR5 clustered on Chromosome A1 (supplementary fig. S8, Supplementary Material online), consistent with suggestions by Shigenobu and Stern (2013). We located BCR2 next to the other three BCR genes, whereas in the Acyr 2.0 genome assembly BCR2 was on a separate scaffold. Shigenobu and Stern (2013) suggested that BCR1, BCR3, BCR4, and BCR5 may have originated from tandem gene duplication events. Their hypothesis is supported by the close location of the four genes in our assembly. We located other BCR genes and SP genes on Chromosomes A1, A2, A3, or X (supplementary fig. S8 and table S5, Supplementary Material online).
We also found all the bacteriocyte-expressed, HGT genes (three ldcA genes, five rlpA genes, amiD, and bLys genes) on Chromosomes A1 or A2 (supplementary table S5, Supplementary Material online). The ldcA2 and ldcA1 genes aligned to the same location (supplementary fig. S9, Supplementary Material online), consistent with the existence of a single locus with ldcA2 reflecting a misassembly within Acyr 2.0. The previous scaffolds for ldcA1 and ldcA2 genes shared 97% similarity (Nikoh and Nakabachi 2009; Nikoh et al. 2010), suggesting these are allelic variants. PCR and Sanger sequencing results confirmed that the AL4 assembly is correct in resolving them as a single locus (supplementary fig. S10, Supplementary Material online).
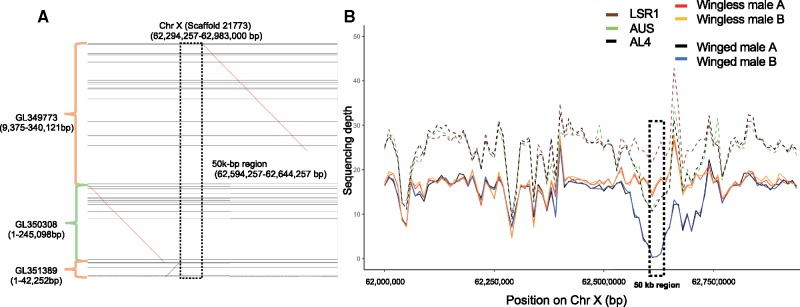
To locate the api regions in our assembly, we broke the three Acyr 2.0 scaffolds containing api sequences (totaling 1,170,040 bp) into 40 contigs based on gaps (“N” characters). Using BlastN searches to locate these contigs in our assembly, we found that Acyr 2.0 scaffolds containing api regions aligned to Chromosome X in the same order as in previous findings (fig. 5A) (B. Li et al. 2017). In addition, portions of the long Acyr 2.0 scaffold containing api regions (scaffold GL349773) aligned elsewhere on the X, or on other autosomes or short scaffolds (mapping results can be retrieved by running the cmd.sh script under step_4 on https://github.com/lyy005/Aphid_AL4_chromosome_assembly; last accessed Feb 9, 2019), implying that regions before 9,375 bp and after 340,121 bp do not belong to the api region. This finding is consistent with a previous finding of misassembly after the 350-kb position in GL349773 (B. Li et al. 2017). In addition, we found a region of ∼50 kb inserted within the api regions (fig. 5B). This 50-kb region was assembled as a separate scaffold in Acyr 2.0 (LSR1 strain) but had not been identified as part of the api region (B. Li et al. 2017).
Fig. 5.
(A) The alignment of the api regions to the sequence of Chromosome X. (B) Sequencing depth and number of genotypes of the api regions for two winged males and two wingless males of clone AL4. Dashed lines represent the diploid female genotypes, including parentals LSR1 (homozygous wingless), AUS (heterozygous), and the sequenced AL4 (heterozygous).
To further investigate the api region, we estimated its sequencing depth using a 10-kb sliding window based on resequencing data. Overall, the api region showed consistent coverage with similar depth for winged and wingless males, except in the ∼50-kb region. Among the seven sequenced individuals, LSR1 females showed the highest sequencing depth in this region; the AUS female, the AL4 female, and the two wingless males showed intermediate depth, and the two winged males showed the lowest depth, near 0 (fig. 5). This indicates that LSR1 is homozygous for the region; AUS and AL4 are heterozygous; wingless males contain a copy of this region on their X, and winged males lack the region entirely. Analysis of sequencing depth using sliding windows along the whole X chromosome also showed that the api region is overrepresented among the top 20 windows with the largest difference in coverage between wingless and winged males (supplementary table S6, Supplementary Material online). PCR validations on the several individual males confirmed that the region was present on the X underlying winglessness and absent from the X underlying wingedness, for the AL4 clone (supplementary figs. S11 and S12, Supplementary Material online). Based on PCR surveys of other A. pisum clones, we found that most sampled clones contain the 50-kb region, regardless of whether they produce winged or wingless males. The region was lacking only in a single clone (ORPG), which produces only winged males (supplementary fig. S13, Supplementary Material online). Both the sequencing depth analysis and PCR results suggest that this 50-kb region is linked to the allele for male winglessness on the LSR1 X chromosome.
Discussion
The publication of the first aphid genome sequence revealed several unusual evolutionary features, including an extraordinary degree of gene family expansion (IAGC 2010) as well as the first verified cases of HGT from microbial sources into animal genomes (Moran and Jarvik 2010; Nikoh et al. 2010). The genome sequence has advanced research on this important and unique group of insects, leading to discoveries involving host–symbiont interactions, HGT, color polymorphism, sex chromosome evolution, and utilization of alternative host plants. However, the extensive duplication within this genome has posed major hurdles for its assembly, limiting the ability to address questions from variant calling to large-scale genome architecture. Largely as a result of the extensive gene family expansion, the existing Acyr 2.0 assembly is fragmented and often incorrect. Our new assembly, based on proximity ligation approaches (HiC and Chicago), has enabled a vast improvement in contiguity and accuracy. In turn, this enables insight into the history of duplication and HGT within this genome.
Repetitive Genome Assembly Using Proximity Ligation Approaches
One of the biggest challenges for assembly of eukaryotic genomes, including the aphid genome, is repetitive sequences (IAGC 2010; Duncan et al. 2016). Repeats lead to fragmentation, misassemblies, and misalignments, especially for assemblies based on short reads of next-generation sequencing techniques (Treangen and Salzberg 2012; Jiao and Schneeberger 2017). Proximity ligation approaches (Putnam et al. 2016) enabled us to achieve chromosome-level assemblies represented by four long scaffolds. Different quality assessments confirmed the quality of the new assembly. The AL4 and Acyr 2.0 assemblies contain similar high proportions of complete BUSCOs. However, AL4 has slightly fewer duplicated BUSCOs, suggesting that allelic variants represented as separate loci in Acyr 2.0 were resolved as single genes by the proximity ligation approach. An example is ldcA, which is represented by two putative loci, ldcA1 and ldcA2, on different scaffolds in Acyr 2.0 (Nikoh and Nakabachi 2009; Nikoh et al. 2010). In the AL4 assembly, these two copies assembled as a single locus, and this result was validated by PCR and Sanger sequencing of amplicons. This result is consistent with previous studies showing a large gap in ldcA2 and lack of ldcA2 expression (Nikoh and Nakabachi 2009; Nikoh et al. 2010).
Improving the Understanding of Gene Family Evolution
Our study demonstrates how chromosome-level scaffolds can facilitate studies of gene family evolution. The A. pisum genome is notable for its unusually high level of gene duplication, reflecting both recent and ancient duplications (IAGC 2010); however, the poor assembly quality has impeded analysis of the sizes or physical distribution of these duplications. We explored gene family evolution through two approaches, including analysis of particular gene families of interest based on their functional roles and global analysis of paralog pairs across the genome.
For the first approach, we were able to identify locations for genes previously shown to have been acquired through HGT followed by gene family expansion through duplication (Moran and Jarvik 2010; Nikoh et al. 2010). Almost all of these genes were located on the four chromosome-level scaffolds, and the multiple copies appear to have arisen through tandem duplications of small genomic regions.
Aphids acquired carotenoid biosynthetic genes by HGT from fungi followed by varying numbers of duplications among aphid lineages, resulting in a variety of carotenoid profiles across species (Nováková and Moran 2012). In the AL4 assembly, these genes are located on Chromosome A1, where they form a tandem array of three pairs, each consisting of a carotenoid synthase/cyclase gene and a carotenoid desaturase gene in divergent orientation. This arrangement matches that of the pair of homologous genes in the donor group of fungi, except that intergenic spacers and introns are greatly expanded in the aphid versions (supplementary fig. S2 in Moran and Jarvik [2010]). The finding that these genes are located together supports the hypothesis that two tandem duplication events subsequent to the HGT resulted in the three copies of the carotenoid gene pairs in the A. pisum genome. An additional duplication of the carotenoid desaturase gene gave rise to tor, which is located on a short scaffold and thus exemplifies a gene that could not be assigned to a chromosome-level scaffold. This region likely failed to assemble because it is hemizygous in AL4 (fig. 1). Many of the other short scaffolds likely also represent hemizygous variants. In these cases, combining long-read data or more resequencing data could help to extend the assembly.
Several other gene families are of particular interest due to their roles in the aphid symbiosis with the obligate endosymbiont, Buchnera aphidicola. In particular, a group of short peptide-encoding genes, the BCR are expressed at high levels only in cells housing Buchnera or in adjacent sheath cells; these have been hypothesized to play roles in controlling proliferation of the intracellular symbiont population (Shigenobu and Stern 2013). Although highly divergent in sequence, the BCR1, BCR2, BCR4, and BCR5 loci were proposed to be paralogs undergoing rapid sequence evolution (Shigenobu and Stern 2013). We found that indeed these loci are located in close proximity on the same AL4 scaffold, supporting their origins through duplication.
Our whole-genome analysis of paralog pairs using the AL4 assembly reveals a long history of ongoing gene duplication on all chromosomes. The older duplications, represented by paralog pairs with dS > 0.4, correspond to events during the early evolution of aphids, based on comparison to dS values for ortholog pairs of divergent aphid species (IAGC 2010). Based on divergence dates estimated from the fossil record for aphids (Żyła et al. 2017), our analysis spans ∼100–200 My of gene duplications.
We found that the abundance of paralogs in the A. pisum genome reflects primarily small-scale duplications involving one or few genes. We found no evidence for whole-genome duplication and almost no evidence for large-scale duplications during the evolution of aphids (fig. 4). The exception is an excess of paralog pairs with dS near 0.5 for the X chromosome (fig. 4A), a result that potentially arises from a large-scale duplication of many genes. However, an excess of duplicates of similar divergence levels can result from other processes. Chromosome-scale assembly can help to resolve whether large, multigene duplication played a role, since a prediction of a large duplication is synteny of genes in the descendant genomic regions (Nakatani and McLysaght 2019). Such synteny is not evident from our assembly (fig. 4E). Thus, the peak observed for dS values of paralogs on the X chromosome likely represents a spurt of small-scale duplications; alternatively, it could reflect an ancient large-scale duplication for which synteny is obscured by later rearrangements.
Based on the distance between paralog pairs on chromosome assemblies, recent duplications could be categorized as either close or distant. Recent duplications, identified by having low dS values, include both close and distant paralog pairs; this observation is consistent with previous evidence from karyotype studies that chromosomal rearrangements are common in aphids (Blackman 1980; Mandrioli et al. 2016). One possible explanation is that aphids have holocentric chromosomes that have diffused microtubule attachments along the chromosomes. DNA fragments could be kept and inherited by cells through attaching to these microtubule attachments along the chromosomes (Mandrioli and Carlo Manicardi 2012). A high rate of chromosome rearrangement has also been found in other holocentric organisms (Coghlan and Wolfe 2002; d’Alencon et al. 2010). These rearrangements may lead to relocation of one member of a paralog pair, resulting in distant paralogs.
Our assembly reveals an elevated incidence of both young and old gene duplications on the X chromosome as compared with autosomes (fig. 4), as has also been found for sex chromosomes in other organisms (Vicoso and Charlesworth 2006; Bellott et al. 2010; Meisel et al. 2010). The end of Chromosome X contains a large number of paralogs and transposable elements. However, the length or number of CDS between paralogs does not correlate strongly with paralog distance. Thus, most paralogs appear to be mediated by DNA duplication events rather than by mRNA and reverse transcriptase (which would lead to the intronless paralogs) (Kaessmann et al. 2009). Previously, the X chromosome of A. pisum has been shown to undergo a distinctive pattern of gene sequence evolution, consistent with population genetic expectations based on its lower effective population size (Jaquiéry et al. 2018). We further found that the X chromosome has a high rate of recombination along its length, based on the physical distribution of microsatellite markers (fig. 2), as also suggested by Jaquiéry et al. (2014). This high rate of recombination may reflect a reduction or redistribution of centromeric proteins that prevent recombination on autosomal regions. In turn, high recombination rates may lead to increased numbers of duplication events.
Discovering Structural Variations Using Chromosome-Level Assembly
Structural variants, including deletions, insertions, duplications, and inversions, can underlie phenotypes, sex determination, and speciation (Rieseberg 2001; Eichler and Sankoff 2003; Murata et al. 2011). Chromosome-level assembly can enable the discovery of structural variations linked to phenotypes. For example, by comparing winged and wingless male individuals in the context of our chromosome-level assembly for the A. pisum genome, we documented potential hemizygous loci (insertions and deletions) (supplementary table S6, Supplementary Material online) on the X chromosome. In particular, we found a ∼50-kb deletion flanked by the api regions within winged males. Although this deletion appears not to underlie male wing dimorphism, this result illustrates how chromosome-level assembly can be used to discover structural variations. Similar approaches might be used for discovering structural genomic variation underlying other ecologically significant phenotypes. For example, genome resequencing, in combination with the assembly, could be used to address the basis for different host plant associations, which underlie the formation of host plant races in A. pisum (Jaquiéry et al. 2012), or to address the genetic basis for differences in ability to produce sexual forms. This AL4 assembly is limited by the failure to include ∼14% of the sequence on the chromosomes; these may represent highly divergent or hemizygous regions that may specify interesting ecological phenotypes. Interestingly, these unassembled regions appear to mostly reside on the X chromosome, based on our analysis of resequencing depth in males versus females and on the absence of SNPs in males (fig. 3). In the future, long-read sequencing data could help to improve the genome assembly by connecting short scaffolds or filling gaps in the genome.
A limitation of our assembly as a reference for the A. pisum genome is that it lacks any regions not present in the sequenced AL4 or LSR1 clones. Since these are both members of the alfalfa host race, the assembly does not include any regions that are specific to other host plant–associated populations. In the future, building a representative (structural variation) map based on this assembly by including population resequencing data could be especially helpful for discovering the genetic basis of ecologically relevant phenotypes (Sudmant et al. 2015).
Materials and Methods
Aphid Culture and Sample Preparation
The AL4 clone chosen for the assembly is an F1 offspring of paternal clone LSR1.AC.G1, which was used for the aphid genome project (IAGC 2010) and maternal clone AUS (Chong and Moran 2016). Both AL4 and LSR1 are clones adapted to Medicago sativa (alfalfa), and thus represent a single host plant race of A. pisum. LSR1 is a red clone, heterozygous for tor, the carotenoid desaturase gene that underlies the production of the red carotenoid torulene and that confers red body color. “AUS” is green and thus homozygous (tor−/tor−). Progeny of this cross were half red and half green individuals as expected by Mendelian ratios (Moran and Jarvik 2010). AL4 is red and thus heterozygous (tor+/tor−) (fig. 1). When induced to produce sexual forms, AL4 yields approximately even numbers of winged and wingless males and thus is dimorphic for the X-linked locus aphicarus (api) (B. Li et al. 2017), which determines wing dimorphism in males (which are XO).
We collected, froze and shipped wingless asexual females from an AL4 lab colony to Dovetail Genomics (Santa Cruz, CA) for DNA extraction, proximity ligation methods (HiC and Chicago library preparation) (Putnam et al. 2016), and Illumina HiSeq X sequencing. All aphid clonal lines were raised separately on fava bean seedlings and maintained at 20 or 15 °C constant temperature with a 16L/8D daily light cycle. Males were generated by placing clones at short day length, as described below.
De Novo Assembly and Verification
We first assembled the AL4 genome using Chicago data based on the Acyr 2.0 assembly using the HiRise assembler (Putnam et al. 2016). The HiRise assembler breaks the original assembly when it conflicts with proximity ligation results. We then used the result from the Chicago assembly as a basis for a further assembly using the HiC data. We filtered bacteria contaminations in the final AL4 assembly using NCBI VecScreen. We then masked potential contaminated sequences with “N” characters. After masking, we removed “N”s at the beginning or at the end of scaffolds.
To assess the genome assembly quality, we used the BUSCOs version 3.0.2 (Simão et al. 2015) on both Acyr 2.0 and the AL4 assembly. We ran the BUSCO analysis using 1,658 Insecta near-universal single-copy orthologs from OrthoDB v9 (Zdobnov et al. 2017) as the benchmark gene set in “genome” mode with default parameters.
To evaluate potential large-scale misassemblies, we mapped the known microsatellite markers of A. pisum and tested if these makers were consistent with linkage groups identified (Jaquiéry et al. 2014). Specifically, we mapped primer sequences for 305 microsatellite markers (Jaquiéry et al. 2014, primer sequences can be found in the https://github.com/lyy005/Aphid_AL4_chromosome_assembly; last accessed Feb 9, 2019) to the AL4 assembly using BLAST+ v2.2.28 (Altschul et al. 1990) with BlastN command, “blastn-short” mode, and e-value = 1. We counted the microsatellite primer pairs as a match if both forward and reverse primers of the same microsatellite locus could be aligned to the same scaffold with 100% similarity, 100% primer sequence length, and in the correct orientation. We scored BLAST hits for whether they were unique matches within the genome and whether allowing for a single base difference between the primer and scaffold sequence resulted in matches. We identified Chromosome X based on linkage group information. Chromosome A3 was identified as the scaffold with the shortest map length and assembly length (Jaquiéry et al. 2014). Chromosomes A1 and A2 were identified based on their relative lengths, with the longer scaffold identified as Chromosome A1. In addition, genes for carotenoid biosynthesis were also found on the longer scaffold (Chromosome A1), which is consistent with findings from in situ localization of these genes on the longer autosome designated as Chromosome A1 in the aphid karyotype (supplementary table S5, Supplementary Material online) (Mandrioli et al. 2016).
Sequencing Depth Evaluation
To further discover potential misassemblies of regions from autosomes and the X chromosome, we resequenced seven A. pisum individuals, including one LSR1 female, one AUS female, one AL4 asexual female, two wingless AL4 males, and two winged AL4 males (fig. 1). We induced AL4 sexual morphs by subjecting asexual female aphids reared at 16 °C to increasingly longer nights, decreasing the daylight hours by 15 min every Monday, Wednesday, and Friday until 12L/12D was reached (spanning two months). When males appeared, we preserved single males, winged or wingless, in separate tubes for later sequencing. We flash froze the individuals and stored at −80 °C. Later, we extracted genomic DNA with the DNeasy Blood & Tissue Kit (Qiagen) insect protocol, using 1.5-ml mortar and pestle tubes and eluting twice with 50 µl of buffer AE. The University of Texas at Austin Genomic Sequencing and Analysis Facility performed library preparation and sequencing using the NEBNext Ultra II DNA library prep kit (New England Biolabs) and Illumina HiSeq 2500 100-bp single-ended sequencing.
We first aligned reads to the AL4 assembly using Bowtie2 version 2.2.6 (Langmead and Salzberg 2012) with default parameters. The resulting SAM files were converted into BAM files, sorted, and indexed using SAMtools version 1.9 (Li et al. 2009). We then estimated the depth of coverage using sliding windows of 10 kb with 2-kb steps as suggested by Jaquiéry et al. (2018). Sequencing depth measures of male and female individuals were normalized based on the median of male or female individuals with the lowest sequencing depth, respectively. For each window, we calculated the sequencing depth ratio as: the ratio between male sequencing depth to female sequencing depth using mosdepth version 0.2.3 (Pedersen and Quinlan 2018). Sequencing depths were normalized among male individuals and female individuals separately. Male individuals were normalized based on the median of the sequencing depth of the individuals with the lowest sequencing depth (wingless male individual A). Female individuals were normalized based on the LSR1 female individual. Given the XO sex determination system, we expected the male to female ratio of median coverage depth to be approximately two times larger for autosomes than for X chromosome in aphids.
As an additional evaluation of assignment to the X versus autosomes, regions were checked for polymorphism in females versus males. We identified SNPs using FreeBayes version 1.2.0 (Garrison and Marth 2012) and filtered as described in the using programs vcffilter, vcfallelicprimitives in FreeBayes (Garrison and Marth 2012) and VCFtools version 0.1.16 (Danecek et al. 2011) (https://github.com/lyy005/Aphid_AL4_chromosome_assembly; last accessed Feb 9, 2019).
Using the resequencing data, we also calculated the heterozygosity of different individuals. Given the large amount of repetitive sequences in the aphid genome, which could interfere with assignment of homologous positions, we only calculated heterozygosity for coding gene regions. The heterozygosity for gene regions of each individual was calculated as (number of heterozygous SNP sites)/(total number of sites in the gene region). We extract the SNPs in gene regions based on the GFF file from genome annotation and the VCF file from FreeBayes using “intersect” command in BEDTools version 2.26.0 (Quinlan and Hall 2010). We counted the number of heterozygous SNP sites in the resulting VCF file. We used the total length of gene regions as the total number of sites.
Genome Annotation
We annotated the assembly using WQ-MAKER version 2.31.9 (Hazekamp et al. 2018) on the Jetstream (Towns et al. 2014; Stewart et al. 2015) in combination with existing coding sequences and protein sequences of the Acyr 2.0 genome annotation 2.1b from AphidBase (IAGC 2010; Legeai et al. 2010). The WQ-MAKER is a modified annotation program, MAKER (Cantarel et al. 2008; Holt and Yandell 2011), on distributed computing resources. We then associated the functions of predicted genes to the existing functional annotation of the Acyr 2.0 genome using BlastP in BLAST+ v2.2.28 (Altschul et al. 1990).
We compared annotations of the two assemblies, in order to estimate the number of genes in the Acyr 2.0 genome that were broken and rearranged in the AL4 assembly and to better understand the distribution of transposable elements in the AL4 assembly. For this purpose, we used the Acyr 2.0 annotations for genes (http://bipaa.genouest.org/sp/acyrthosiphon_pisum/download/annotation/ncbi_2.1/ncbi_annotation_v2.1.gff3; last accessed Feb 9, 2019) and for transposable elements (http://bipaa.genouest.org/sp/acyrthosiphon_pisum/download/tracks/REPET_all.gff3; last accessed Feb 9, 2019) To convert the annotations between the assemblies, we first converted the Dovetail assembly coordinate file to an agp file using a Perl script (https://github.com/Nucleomics-VIB/bionano-tools/blob/master/general-tools/dovetail2agp.pl; last accessed Feb 9, 2019). Then we converted the agp file to a chain file using chain.py from jcvi tool version v0.8.12 (Tang et al. 2015). The chain file was then used to convert annotations from Acyr 2.0 to the AL4 assembly using CrossMap version 0.3.4 (Zhao et al. 2014).
Gene Duplication Analyses
To investigate the history of gene duplication in A. pisum, we reconstructed the paralogous relationship of the annotated genes. First only the longest CDS of each gene was used for paralog prediction. Then, we performed all-to-all BlastP search within the annotated genes with e-value = 1e-10, similarity ≥30%, and alignment length ≥150 aa, following criteria suggested by previous studies (Fawcett et al. 2009; Mathers et al. 2017). Reciprocal best BLAST hit pairs were retained as paralogs. For each paralog pair, we aligned the codon sequences using codon alignment (Y. Li et al. 2017) and removed poorly aligned regions using Gblocks version 0.91b (Castresana 2000). We then calculated pairwise dN and dS values using KaKs Calculator version 2.0 (Zhang et al. 2006) with the “Model Averaging” method. We also calculated the distance between genes by estimating the distance of mRNA annotation between paralog pairs in the gff file. We visualized the data using ggplot2 package version 3.0 (Wickham 2016) in R (R Core Team 2016). We tested the correlation between paralog distance on chromosomes and dS, dN, CDS length, and CDS number using Kendall’s Tau in cor.test function of R (R Core Team 2016).
To look for potential homologous regions in the genome, we located potential duplications on the genome assembly. We used LAST version 956 (Kielbasa et al. 2011) to perform all-to-all search on all the scaffolds. We plotted alignments longer than 5 kb using Circos version 0.69-6 (Krzywinski et al. 2009). We also used MCscan in the JCVI tool kit version 0.8.12 (https://github.com/tanghaibao/jcvi; last accessed Feb 9, 2019) (Wang et al. 2012; Tang et al. 2015) to search for homologous regions using protein-coding genes as anchors.
Location of Functional Genes
To locate genes of interest in the assembly, we annotated the carotenoid biosynthetic genes from the AL4 assembly. We aligned the amino acid sequences of four carotenoid desaturase genes, three carotenoid synthase genes and one carotenoid synthase pseudogene (Mandrioli et al. 2016) to the AL4 assembly using TBlastN with e-value = 1e-10. We used the same parameters for several sets of genes hypothesized to be involved in the symbiosis with Buchnera, based on expression patterns (Nikoh et al. 2010; Shigenobu and Stern 2013). These symbiosis-related genes included BCR genes, SP genes and HGT genes (three ldcA genes, five rlpA genes, amiD, and bLys genes). We visualized the locations of the genes in Sushi package version 1.16 (Phanstiel et al. 2014) in R version 3.4.4 (R Core Team 2016). Details of the genes and sequences can be found at https://github.com/lyy005/Aphid_AL4_chromosome_assembly; last accessed Feb 9, 2019.
Previous genetic mapping studies (Braendle et al. 2005; B. Li et al. 2017) have shown that a region called aphicarus (api), located near one end of the X chromosome, underlies male wing dimorphism. Clone AL4 produces both winged and wingless males, and thus is heterozygous for this region (fig. 1). In Acyr 2.0, the predicted region was assembled into three scaffolds (B. Li et al. 2017), of which one scaffold (NCBI accession number GL349773.1) has been shown to be misassembled after the 350-kb position (B. Li et al. 2017). We determined the locations and confirmed the potential misassemblies of these regions using BlastN with the three scaffolds (NCBI accession numbers GL349773.1, GL350308.1, and GL351389.1) as queries. To determine the potential misassemblies, we broke the scaffolds into contigs at gaps (Ns) and aligned the contigs with the current assembly.
To further investigate genomic differences between winged and wingless males, we plotted the sequencing depth of the aphicarus gene regions in the assemblies among the individuals. We calculated the differences in sequencing depth between winged males and wingless males as (sequencing depth winged male A + winged male B)/(wingless male A + wingless male B). We discovered potential hemizygous regions differing between X chromosomes of winged and wingless males by ranking the difference in this ratio for the 10-kb windows. To achieve better resolution, we also applied a 1-kb sliding window on the aphicarus region to confirm our findings.
To confirm the assembly, we designed two sets of PCR primers for the ldcA2 gene and two sets of primers for the 50-kb insertion within the api region. Amplicons of ldcA2 genes were sequenced using Applied Biosystems 3730 DNA Analyzers and BigDye Terminator v3.1 chemistry at University of Texas at Austin Genomic Sequencing and Analysis Facility. We picked one of the two api region primer pairs to amplify the region in other clones known to produce winged or wingless males.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by National Science Foundation grant DEB 1551092 to N.A.M. This work used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation grant ACI-1548562. We received support from the Texas Advanced Computing Center. We thank Julie Jaquiéry, Fabrice Legeai, Denis Tagu, and Jean-Christophe Simon from INRA, UMR 1349 IGEPP for sharing microsatellite primer data and for helpful discussions. The genome assembly and sequencing data are available on National Center for Biotechnology Information, NIH, under the BioProject ID PRJNA496478. The bioinformatic scripts used in this study are available on GitHub (https://github.com/lyy005/Aphid_AL4_chromosome_assembly, last accessed Feb 9, 2019).
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 2153:403–410. [DOI] [PubMed] [Google Scholar]
- Bellott DW, Skaletsky H, Pyntikova T, Mardis ER, Graves T, Kremitzki C, Brown LG, Rozen S, Warren WC, Wilson RK, et al. 2010. Convergent evolution of chicken Z and human X chromosomes by expansion and gene acquisition. Nature 4667306:612–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MD, Leitch IJ, Price HJ, Johnston JS.. 2003. Comparisons with Caenorhabditis (100 Mb) and Drosophila (175 Mb) using flow cytometry show genome size in Arabidopsis to be 157 Mb and thus 25% larger than the Arabidopsis genome initiative estimate of 125 Mb. Ann Bot. 915:547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman R. 1980. Chromosome numbers in the Aphididae and their taxonomic significance. Syst Entomol. 51:7–25. [Google Scholar]
- Braendle C, Caillaud M, Stern D.. 2005. Genetic mapping of aphicarus—a sex-linked locus controlling a wing polymorphism in the pea aphid (Acyrthosiphon pisum). Heredity 944:435.. [DOI] [PubMed] [Google Scholar]
- Brisson JA. 2010. Aphid wing dimorphisms: linking environmental and genetic control of trait variation. Philos Trans R Soc B 3651540:605–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisson JA, Nuzhdin SV, Stern DL.. 2009. Similar patterns of linkage disequilibrium and nucleotide diversity in native and introduced populations of the pea aphid, Acyrthosiphon pisum. BMC Genet. 10:22.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisson JA, Stern DL.. 2006. The pea aphid, Acyrthosiphon pisum: an emerging genomic model system for ecological, developmental and evolutionary studies. Bioessays 287:747–755. [DOI] [PubMed] [Google Scholar]
- Cantarel BL, Korf I, Robb SMC, Parra G, Ross E, Moore B, Holt C, Sánchez Alvarado A, Yandell M.. 2008. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 181:188–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 174:540–552. [DOI] [PubMed] [Google Scholar]
- Chong RA, Moran NA.. 2016. Intraspecific genetic variation in hosts affects regulation of obligate heritable symbionts. Proc Natl Acad Sci U S A. 11346:13114–13119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coghlan A, Wolfe KH.. 2002. Fourfold faster rate of genome rearrangement in nematodes than in Drosophila. Genome Res. 126:857–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp A, Boschetti C, Perry M, Tunnacliffe A, Micklem G.. 2015. Expression of multiple horizontally acquired genes is a hallmark of both vertebrate and invertebrate genomes. Genome Biol. 161:50.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahan RA, Duncan RP, Wilson AC, Dávalos LM.. 2015. Amino acid transporter expansions associated with the evolution of obligate endosymbiosis in sap-feeding insects (Hemiptera: Sternorrhyncha). BMC Evol Biol. 15:52.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Alencon E, Sezutsu H, Legeai F, Permal E, Bernard-Samain S, Gimenez S, Gagneur C, Cousserans F, Shimomura M, Brun-Barale A, et al. 2010. Extensive synteny conservation of holocentric chromosomes in Lepidoptera despite high rates of local genome rearrangements. Proc Natl Acad Sci U S A. 10717:7680–7685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. 2011. The variant call format and VCFtools. Bioinformatics 2715:2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan RP, Feng H, Nguyen DM, Wilson AC.. 2016. Gene family expansions in aphids maintained by endosymbiotic and nonsymbiotic traits. Genome Biol Evol. 83:753–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler EE, Sankoff D.. 2003. Structural dynamics of eukaryotic chromosome evolution. Science 3015634:793–797. [DOI] [PubMed] [Google Scholar]
- Fawcett JA, Maere S, Van de Peer Y.. 2009. Plants with double genomes might have had a better chance to survive the Cretaceous–Tertiary extinction event. Proc Natl Acad Sci U S A. 10614:5737–5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuk L, Carson AR, Scherer SW.. 2006. Structural variation in the human genome. Nat Rev Genet. 72:85.. [DOI] [PubMed] [Google Scholar]
- Garrison E, Marth G.. 2012. Haplotype-based variant detection from short-read sequencing. unpublished data, https://arxiv.org/abs/1207.3907, last accessed Feb 9, 2019.
- Genome 10K Community. 2009. Genome 10K: a proposal to obtain whole-genome sequence for 10 000 vertebrate species. J Hered. 100:659–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AK, Moran NA.. 2011. Aphid genome expression reveals host–symbiont cooperation in the production of amino acids. Proc Natl Acad Sci U S A. 1087:2849–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawthorne DJ, Via S.. 2001. Genetic linkage of ecological specialization and reproductive isolation in pea aphids. Nature 4126850:904.. [DOI] [PubMed] [Google Scholar]
- Hazekamp NL, Devisetty UK, Merchant N, Thain D.. 2018. MAKER as a service: moving HPC applications to Jetstream Cloud. 2018 IEEE International Conference on Cloud Engineering (IC2E). p. 72–78. Orlando, FL, USA: IEEE. 2018 IEEE International Conference on Cloud Engineering (IC2E).
- Holt C, Yandell M.. 2011. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12:491.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- i5K Consortium. 2013. The i5K initiative: advancing arthropod genomics for knowledge, human health, agriculture, and the environment. J Hered. 104:595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IAGC. 2010. Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 8:e1000313.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaquiéry J, Peccoud J, Ouisse T, Legeai F, Prunier-Leterme N, Gouin A, Nouhaud P, Brisson JA, Bickel R, Purandare S, et al. 2018. Disentangling the causes for faster-X evolution in aphids. Genome Biol Evol. 102:507–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaquiéry J, Stoeckel S, Larose C, Nouhaud P, Rispe C, Mieuzet L, Bonhomme J, Mahéo F, Legeai F, Gauthier J-P, et al. 2014. Genetic control of contagious asexuality in the pea aphid. PLoS Genet. 1012:e1004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaquiéry J, Stoeckel S, Nouhaud P, Mieuzet L, Mahéo F, Legeai F, Bernard N, Bonvoisin A, Vitalis R, Simon J-C, et al. 2012. Genome scans reveal candidate regions involved in the adaptation to host plant in the pea aphid complex. Mol Ecol. 2121:5251–5264. [DOI] [PubMed] [Google Scholar]
- Jiao W-B, Schneeberger K.. 2017. The impact of third generation genomic technologies on plant genome assembly. Curr Opin Plant Biol. 36:64–70. [DOI] [PubMed] [Google Scholar]
- Kaessmann H, Vinckenbosch N, Long M.. 2009. RNA-based gene duplication: mechanistic and evolutionary insights. Nat Rev Genet. 101:19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielbasa SM, Wan R, Sato K, Horton P, Frith M.. 2011. Adaptive seeds tame genomic sequence comparison. Genome Res. 213:487–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA.. 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 199:1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL.. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 94:357.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Gurtowski J, Yoo S, Nattestad M, Marcus S, Goodwin S, McCombie WR, Schatz MC.. 2016. Third-generation sequencing and the future of genomics. Unpublished data, https://www.biorxiv.org/content/10.1101/048603v1, last accessed Feb, 2019.
- Legeai F, Shigenobu S, Gauthier J-P, Colbourne J, Rispe C, Collin O, Richards S, Wilson ACC, Murphy T, Tagu D, et al. 2010. AphidBase: a centralized bioinformatic resource for annotation of the pea aphid genome. Insect Mol Biol. 19:5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin HA, Larkin DM, Pontius J, O’Brien SJ.. 2009. Every genome sequence needs a good map. Genome Res. 1911:1925–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Bickel RD, Parker BJ, Vellichirammal NN, Grantham M, Simon J-C, Stern DL, Brisson JA.. 2017. Unravelling the genomic basis and evolution of the pea aphid male wing dimorphism. Unpublished data, https://www.biorxiv.org/content/10.1101/156133v1, last accessed Feb 9, 2019.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R.. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 2516:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhang R, Liu S, Donath A, Peters RS, Ware J, Misof B, Niehuis O, Pfrender ME, Zhou X, et al. 2017. The molecular evolutionary dynamics of oxidative phosphorylation (OXPHOS) genes in Hymenoptera. BMC Evol Biol. 171:269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien S, Koop BF, Sandve SR, Miller JR, Kent MP, Nome T, Hvidsten TR, Leong JS, Minkley DR, Zimin A, et al. 2016. The Atlantic salmon genome provides insights into rediploidization. Nature 5337602:200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long E, Evans C, Chaston J, Udall JA.. 2018. Genomic structural variations within five continental populations of Drosophila melanogaster. G3 (Bethesda) 810:3247–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandrioli M, Carlo Manicardi G.. 2012. Unlocking holocentric chromosomes: new perspectives from comparative and functional genomics? Curr Genomics. 135:343–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandrioli M, Rivi V, Nardelli A, Manicardi GC.. 2016. Genomic and cytogenetic localization of the carotenoid genes in the aphid genome. Cytogenet Genome Res. 1493:207–217. [DOI] [PubMed] [Google Scholar]
- Mathers TC, Chen Y, Kaithakottil G, Legeai F, Mugford ST, Baa-Puyoulet P, Bretaudeau A, Clavijo B, Colella S, Collin O, et al. 2017. Rapid transcriptional plasticity of duplicated gene clusters enables a clonally reproducing aphid to colonise diverse plant species. Genome Biol. 181:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews BJ, Dudchenko O, Kingan SB, Koren S, Antoshechkin I, Crawford JE, Glassford WJ, Herre M, Redmond SN, Rose NH, et al. 2018. Improved reference genome of Aedes aegypti informs arbovirus vector control. Nature 5637732:501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel RP, Hilldorfer BB, Koch JL, Lockton S, Schaeffer SW.. 2010. Adaptive evolution of genes duplicated from the Drosophila pseudoobscura neo-X chromosome. Mol Biol Evol. 278:1963–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, Jarvik T.. 2010. Lateral transfer of genes from fungi underlies carotenoid production in aphids. Science 3285978:624–627. [DOI] [PubMed] [Google Scholar]
- Moran Y, Fredman D, Szczesny P, Grynberg M, Technau U.. 2012. Recurrent horizontal transfer of bacterial toxin genes to eukaryotes. Mol Biol Evol. 299:2223–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata C, Ogura G, Kuroiwa A.. 2011. A primer set to determine sex in the small Indian mongoose, Herpestes auropunctatus. Mol Ecol Resour. 112:386–388. [DOI] [PubMed] [Google Scholar]
- Nakatani Y, McLysaght A.. 2019. Macrosynteny analysis shows the absence of ancient whole-genome duplication in lepidopteran insects. Proc Natl Acad Sci U S A. 1166:1816–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neafsey DE, Waterhouse RM, Abai MR, Aganezov SS, Alekseyev MA, Allen JE, Amon J, Arcà B, Arensburger P, Artemov G, et al. 2014. Highly evolvable malaria vectors: the genomes of 16 Anopheles mosquitoes. Science 3476217:1258522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoh N, McCutcheon JP, Kudo T, Miyagishima S-y, Moran NA, Nakabachi A.. 2010. Bacterial genes in the aphid genome: absence of functional gene transfer from Buchnera to its host. PLoS Genet. 62:e1000827.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoh N, Nakabachi A.. 2009. Aphids acquired symbiotic genes via lateral gene transfer. BMC Biol. 7:12.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nováková E, Moran NA.. 2012. Diversification of genes for carotenoid biosynthesis in aphids following an ancient transfer from a fungus. Mol Biol Evol. 291:313–323. [DOI] [PubMed] [Google Scholar]
- Peccoud J, Loiseau V, Cordaux R, Gilbert C.. 2017. Massive horizontal transfer of transposable elements in insects. Proc Natl Acad Sci U S A. 11418:4721–4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen BS, Quinlan AR.. 2018. mosdepth: quick coverage calculation for genomes and exomes. Bioinformatics 345:867–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phanstiel DH., Boyle AP, Araya CL, Snyder MP. 2014. Sushi.R: flexible, quantitative and integrative genomic visualizations for publication-quality multi-panel figures. Bioinformatics 30(19):2808–10. [DOI] [PMC free article] [PubMed]
- Putnam NH, O’Connell BL, Stites JC, Rice BJ, Blanchette M, Calef R, Troll CJ, Fields A, Hartley PD, Sugnet CW, et al. 2016. Chromosome-scale shotgun assembly using an in vitro method for long-range linkage. Genome Res. 263:342–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM.. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 266:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2016. R: a language and environment for statistical computing. Vienna (Austria: ): R Foundation for Statistical Computing; Available from: https://www.R-project.org/Last accessed on Feb 2019. [Google Scholar]
- Riehle MM, et al. 2017. The Anopheles gambiae 2La chromosome inversion is associated with susceptibility to Plasmodium falciparum in Africa. eLife 6:e25813.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg LH. 2001. Chromosomal rearrangements and speciation. Trends Ecol Evol. 167:351–358. [DOI] [PubMed] [Google Scholar]
- Shigenobu S, Bickel RD, Brisson JA, Butts T, Chang C-C, Christiaens O, Davis GK, Duncan EJ, Ferrier DEK, Iga M, et al. 2010. Comprehensive survey of developmental genes in the pea aphid, Acyrthosiphon pisum: frequent lineage-specific duplications and losses of developmental genes. Insect Mol Biol. 19:47–62. [DOI] [PubMed] [Google Scholar]
- Shigenobu S, Stern DL.. 2013. Aphids evolved novel secreted proteins for symbiosis with bacterial endosymbiont. Proc Biol Sci. 2801750:20121952.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 3119:3210–3212. [DOI] [PubMed] [Google Scholar]
- Smadja C, Shi P, Butlin RK, Robertson HM.. 2009. Large gene family expansions and adaptive evolution for odorant and gustatory receptors in the pea aphid, Acyrthosiphon pisum. Mol Biol Evol. 269:2073–2086. [DOI] [PubMed] [Google Scholar]
- Stewart CA, Cockerill T, Foster I, Hancock DY, Merchant N, Skidmore E, Stanzione D, Taylor J, Tuecke S, Turner G, et al. 2015. Jetstream: a self-provisioned, scalable science and engineering cloud environment. Proceedings of the 2015 XSEDE Conference: Scientific Advancements Enabled by Enhanced Cyberinfrastructure.XSEDE ’15 ACM, New York, NY. St. Louis: Association for Computing Machinery. p. 29:1–29:8.
- Sudmant PH, Rausch T, Gardner EJ, Handsaker RE, Abyzov A, Huddleston J, Zhang Y, Ye K, Jun G, Fritz MH-Y, et al. 2015. An integrated map of structural variation in 2,504 human genomes. Nature 5267571:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Zhang X, Miao C, Zhang J, Ming R, Schnable JC, Schnable PS, Lyons E, Lu J.. 2015. ALLMAPS: robust scaffold ordering based on multiple maps. Genome Biol. 16:3.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towns J, Cockerill T, Dahan M, Foster I, Gaither K, Grimshaw A, Hazlewood V, Lathrop S, Lifka D, Peterson GD, et al. 2014. XSEDE: accelerating scientific discovery. Comput Sci Eng. 16:62–74. [Google Scholar]
- Treangen TJ, Salzberg SL.. 2012. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat Rev Genet. 131:36.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Charlesworth B.. 2006. Evolution on the X chromosome: unusual patterns and processes. Nat Rev Genet. 78:645.. [DOI] [PubMed] [Google Scholar]
- Wang Y, Tang H, DeBarry JD, Tan X, Li J, Wang X, Lee T-h, Jin H, Marler B, Guo H, et al. 2012. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 407:e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse RM, Aganezov S, Anselmetti Y, Lee J, Ruzzante L, Reijnders MJMF, Bérard S, George P, Hahn MW, Howell PI, et al. 2018. Leveraging evolutionary relationships to improve Anopheles genome assemblies. unpublished data, https://www.biorxiv.org/content/10.1101/434670v3, last accessed Feb 6, 2019.
- Wickham H. 2016. ggplot2: elegant graphics for data analysis. New York: Springer-Verlag; Available from: http://ggplot2.org. [Google Scholar]
- Zdobnov EM, Tegenfeldt F, Kuznetsov D, Waterhouse RM, Simão FA, Ioannidis P, Seppey M, Loetscher A, Kriventseva EV.. 2017. OrthoDB v9. 1: cataloging evolutionary and functional annotations for animal, fungal, plant, archaeal, bacterial and viral orthologs. Nucleic Acids Res. 45(D1):D744–D749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Li J, Zhao X-Q, Wang J, Wong GK-S, Yu J.. 2006. KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinformatics. 44:259–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Sun Z, Wang J, Huang H, Kocher J-P, Wang L.. 2014. CrossMap: a versatile tool for coordinate conversion between genome assemblies. Bioinformatics 307:1006–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Żyła D, Homan A, Wegierek P.. 2017. Polyphyly of the extinct family Oviparosiphidae and its implications for inferring aphid evolution (Hemiptera, Sternorrhyncha). PLoS One 12:1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.