Abstract
Current phylogenomic approaches implicitly assume that the predominant phylogenetic signal within a genome reflects the true evolutionary history of organisms, without assessing the confounding effects of postspeciation gene flow that can produce a mosaic of phylogenetic signals that interact with recombinational variation. Here, we tested the validity of this assumption with a phylogenomic analysis of 27 species of the cat family, assessing local effects of recombination rate on species tree inference and divergence time estimation across their genomes. We found that the prevailing phylogenetic signal within the autosomes is not always representative of the most probable speciation history, due to ancient hybridization throughout felid evolution. Instead, phylogenetic signal was concentrated within regions of low recombination, and notably enriched within large X chromosome recombination cold spots that exhibited recurrent patterns of strong genetic differentiation and selective sweeps across mammalian orders. By contrast, regions of high recombination were enriched for signatures of ancient gene flow, and these sequences inflated crown-lineage divergence times by ∼40%. We conclude that existing phylogenomic approaches to infer the Tree of Life may be highly misleading without considering the genomic architecture of phylogenetic signal relative to recombination rate and its interplay with historical hybridization.
Keywords: phylogenomics, recombination, Felidae, X chromosome, hybridization
Introduction
Efforts to resolve the Tree of Life have been empowered by a major shift toward “whole-genome” phylogenetic inference, or phylogenomics (Delsuc et al. 2005; McCormack et al. 2012; Jarvis et al. 2014; Lamichhaney et al. 2015; Prum et al. 2015; Figueiró et al. 2017; Irisarri et al. 2018; Bravo et al. 2019). Phylogenomic studies typically analyze thousands of orthologous loci or whole-genome sequence (wgs) data simultaneously using concatenation or coalescent approaches. Majority rule (or “democratic vote,” sensu Degnan and Rosenberg 2009) approaches, notably those employing concatenation of loci, have long been regarded as the most successful way to mitigate the distorting influences of evolutionary noise at individual loci. They assume that the correct genealogical signal (the “species tree”) will emerge with an ever-increasing amount of data, although it is becoming clear with genomic data matrices that this assumption is often not valid (e.g., Degnan and Rosenberg 2009; Romiguier et al. 2013; Shen et al. 2017; Bravo et al. 2019). Approaches based on the multispecies coalescent model offer an appealing alternative to concatenation approaches by accommodating gene tree heterogeneity across genomes. However, thus far, they do not scale well to fully utilize large whole-genome sequence alignments and tend to possess the caveat of assuming all discordance among genomic loci results from incomplete lineage sorting (or ILS) (Springer and Gatesy 2018; Bravo et al. 2019), an assumption that is debated (Scornavacca and Galtier 2017). Specifically, this latter assumption is untenable in many cases when interspecific gene flow is known to be an important contributor to phylogenomic discordance (e.g., Nadeau et al. 2013; Cahill et al. 2015; Good et al. 2015; Lamichhaney et al. 2015; Nater et al. 2015; Li, Davis, et al. 2016, Martin and Jiggins 2017).
In lineages with an extensive history of hybridization and introgression, signatures of ancient branching events can be depleted from chromosomal segments, notably in regions with high rates of meiotic recombination and more localized effects of linked selection (Brandvain et al. 2014; Schumer et al. 2018). A phylogenomic study of Anopheles mosquitoes provided a vivid illustration of this scenario in which autosomal gene trees were homogenized by rampant postspeciation gene flow, misleading standard phylogenomic inference (Fontaine et al. 2015). The inferred species tree signal was enriched on the smaller X chromosome within a region known to harbor reproductive isolation loci, consistent with the large X-effect, also referred to as the “Second Rule of Speciation” (Coyne and Orr 1989). The large X-effect is the observation that the X chromosome is enriched for genetic elements with large effects on reducing hybrid reproductive fitness (Presgraves 2008; Larson et al. 2017, 2018; Presgraves 2018). Fontaine et al. inferred the most likely species tree by demonstrating that nodes on the X chromosome phylogeny were significantly older than their alternative counterparts in the most frequent autosomal phylogeny. Whether the remarkable genomic depletion of the species tree was exceptional for this clade of mosquitoes or is more broadly generalizable to other taxonomic lineages, is unknown.
Given the demonstrable impact that gene flow has on genome-wide patterns of phylogenetic variation, and the contrasting pattern of genetic differentiation between autosomes and sex chromosomes that is predicted by the large X-effect (Presgraves 2018), we hypothesized that applying standard phylogenomic approaches may be highly misleading when applied to taxa with complex speciation histories. Here, we tested this hypothesis by examining variation in phylogenetic signal across the genomes of 27 species of the cat family (Felidae), a mammalian radiation characterized by notoriously difficult phylogenetic problems (Johnson et al. 2006; Li, Davis, et al. 2016). Previous studies have consistently partitioned the 41 currently recognized felid species into 8 clades that are hypothesized to have radiated rapidly during the mid-late Miocene (Johnson et al. 2006) (fig. 1). However, phylogenies constructed from separate mitogenome, autosomal, and sex chromosome partitions have displayed strongly supported differences within and between the eight clades, which may be driven by complex effects of sex-biased dispersal, gene flow, demography, and/or selection (Johnson et al. 2006; Luo et al. 2014; Li, Davis, et al. 2016). Indeed, studies utilizing nuclear single nucleotide polymorphism (SNP) markers have demonstrated that many discordant phylogeographic patterns within felids inferred from mitochondrial DNA data were likely due to interspecific gene flow (Li, Davis, et al. 2016; Figueiró et al. 2017) or other types of error (Davis et al. 2010; but see Richards et al. [2018]). Likewise, the phylogeny derived from the paternally inherited Y chromosome was similar in many respects to the autosomal tree, but was also strongly influenced by sex-biased dispersal and ancient gene flow (Luo et al. 2014; Li, Davis, et al. 2016), similar to observations in other species (e.g., Nater et al. 2011, 2017). Biogeographic reconstructions of the early felid radiation indicate that the ancestors of all eight extant clades evolved in Eurasia, most likely including periods of sympatry or parapatry, before dispersing across the globe (Li, Davis, et al. 2016). Taken together, previous studies suggest a scenario for the evolution of extant felid clades that is consistent with a history of divergence with gene flow.
Fig. 1.

Previous hypotheses of felid phylogenetic relationships based on 1) maternally inherited mitogenome sequences (Li, Davis, et al. 2016), 2) biparentally inherited SNP markers (Li, Davis, et al. 2016), and 3) paternally inherited Y chromosome sequences (Johnson et al. 2006; Luo et al. 2014).
An important genetic parameter that influences assessments of gene flow, ILS, and genetic diversity is the local rate of recombination (Ortíz-Barrientos et al. 2002; Hobolth et al. 2011; Nachman and Payseur 2012; Pease and Hahn 2013; Mailund et al. 2014; Burri et al. 2015; Payseur and Rieseberg 2016; Kawakami et al. 2017; Haenel et al. 2018). Recombination rate interacts with natural selection to influence how genealogical histories are distributed across chromosomes, as alleles that originate from gene flow are more likely to persist within regions with high-recombination rates because deleterious alleles become effectively unlinked from neutral or positively selected variants (Brandvain et al. 2014; Schumer et al. 2018). However, recombination rate is generally not considered when selecting phylogenetic markers in spite of its predicted influence on genealogical discordance and estimates of divergence times across the genome (Schierup and Hein 2000; Ortíz-Barrientos et al. 2002; Posada and Crandall 2002; Lemey and Posada 2009). Specifically, few empirical studies have examined the direct impact of recombination rate on phylogenetic inference within rapidly diverging, speciose clades with histories of postspeciation gene flow, due to the rarity of recombination maps for the vast majority of organisms (but see Pease and Hahn [2013]).
To more thoroughly understand the interplay between local recombination rates, interspecies gene flow, and phylogenomic resolution in the cat family, we analyzed the fine-scale distribution of phylogenetic signal across the genomes of 27 felid species in the context of recombination rates inferred from domestic cat high-resolution linkage maps (Menotti-Raymond et al. 1999; Menotti-Raymond et al. 2003; Li, Hillier, et al. 2016). We found that phylogenomic signal is strongly partitioned by recombination rate and that the most probable ancient branching events persist within regions of low recombination, with a striking enrichment on the X chromosome as predicted by the large X-effect. Divergence time results were also strongly influenced by interspecies gene flow, leading to distortion of clade ages from regions of high recombination that are enriched for multiple, discordant phylogenies. Our results offer additional evidence that diversification in the presence of gene flow will produce a phylogenomic architecture where the most accurate depiction of ancient branching events persists within only a small fraction of the nuclear genome that possesses historically reduced rates of recombination.
Results
We generated new whole-genome sequences for 14 felid species and aligned these with published genome sequence data for 13 other cats, producing an alignment for 27 species that spanned 1.5 Gb, or 57% of the domestic cat reference assembly (supplementary table S1, Supplementary Material online). To identify regional variation in phylogenetic signal, we partitioned the 18 autosomes and the X chromosome (chrX) into 23,707 nonoverlapping windows of 100 kilobases (kb). For each window of the alignment we calculated maximum likelihood (ML) trees and used their branch lengths to infer divergence times for different clades. We separately calculated levels of support for different topologies from the autosomes and chrX, with each partition divided into regions of low versus high-recombination rate inferred from high-resolution linkage maps (Li, Hillier, et al. 2016) (fig. 2). Analysis of smaller window sizes (50 kb) produced similar results (supplementary fig. S1, Supplementary Material online).
Fig. 2.
(A) Left: the top three most frequently recovered phylogenies for six felid lineages, inferred from window-based ML supermatrix analysis of the 1.5-Gb whole-genome alignment. Right: the frequency of each topology within autosomes and chrX, based on partitioning into low (<0.5 cM/Mb) and high (>2 cM/Mb) recombination rates (Li, Hillier, et al. 2016). Red arrows indicate three lineages in which the most frequent topology in the LRchrX was not the same as the most frequent genome-wide topology. (B) Distribution of different topologies on chrX, and their relationship to recombination rate (bottom).
We present results from analyses performed in two stages. In the first set of analyses, we examined topological frequency and divergence time variation within the six clades for which we had sequence data from at least three species, to identify the most probable branching order among closely related taxa. We reasoned that within-clade analyses of recently diverged species would minimize the impact of homoplasy and putative ancient (interclade) gene flow on observed patterns of variation and estimated divergence times (Posada 2000; Lemey and Posada 2009). Our prediction for these initial analyses was that trees derived from postspeciation introgression would be younger than the actual “species tree” (i.e., the original sequence of branching events) and enriched in regions of high recombination. We then analyzed regional variation in relationships across the whole phylogeny, selecting two species that spanned the oldest crown node of each of the eight clades (with the exception of the Caracal clade, represented only by the serval). For these analyses that considered relationships among the eight lineages of the felid phylogeny, we tested the hypothesis that the impacts of a longer history of gene flow and recombination would produce higher levels of apparent homoplasy, leading to phylogenetic distortion of branch lengths across the tree (Leaché et al. 2014). In this case, one would predict that ages of the crown nodes could be inflated when the ancestors of that lineage had a history of hybridization with more distantly related clades. On the other hand, the deepest nodes in the phylogeny could be shifted toward younger dates when there is a history of reticulation across multiple descendant lineages.
Intraclade Phylogeny
ML estimation of phylogeny for autosomal windows confirmed a highly supported tree for species within each of the six felid clades, with the exception of the Ocelot clade (genus Leopardus) (fig. 2A and supplementary fig. S1, Supplementary Material online). These results corroborated some relationships observed in earlier phylogenetic studies that concatenated autosomal and sex chromosome sequences into a single supermatrix (Johnson et al. 2006). However, in half of the assessed felid clades, the frequency-based rank order of the top three topologies inferred from low-recombining regions of chrX (LRchrX) was strikingly different from the autosomal signal (fig. 2A and supplementary fig. S1, Supplementary Material online). Furthermore, the distribution of the most frequently observed chrX trees was nonrandomly distributed, with an alternative topology enriched within one or more multimegabase spans of extremely reduced recombination (fig. 2B). In such a case, which of the alternative topologies is most likely to be the species tree?
Two observations can aid in this determination. First, low-recombining regions are more likely to retain the species tree because linked selection is more effective at removing larger introgressed segments that harbor even a single deleterious allele through selective sweeps and extended linkage disequilibrium (Nachman and Payseur 2012). Second, alternative trees resulting from hybridization will tend to be enriched within regions displaying high-recombination rates, because introgressed deleterious alleles are more efficiently unlinked from neutral or positively selected variants (Brandvain et al. 2014; Schumer et al. 2018). A third criterion that can be used to determine the most probably ancient branching events is the prediction that alternative topologies derived from postspeciation gene flow between closely related, nonsister taxa will show younger divergence times than the species tree (Joly et al. 2009; Leaché et al. 2014; Fontaine et al. 2015). If postspeciation gene flow is the primary driver of genealogical discordance, then the frequency of these younger, alternative trees could be considerably higher than the species tree (Fontaine et al. 2015). If the topology that predominated within LRchrX had divergence dates that were older than the most common genome-wide topology and enriched throughout the genome in regions of low recombination, then it would likely reflect the original branching event.
Given these predictions, we reasoned that divergence time and recombination rate would be two key criteria in determining the most likely ancient branching events between closely related species (Joly et al. 2009; Fontaine et al. 2015). Therefore, we estimated divergence times for the two main competing hypotheses for each of the three felid lineages in figure 2 that showed a marked genealogical disparity between the autosomes and chrX. We employed a relaxed molecular clock that allows for independent evolutionary rates on different branches of a phylogeny (Drummond et al. 2006) because raw sequence divergence levels may lead to an overestimation of divergence between two taxa in the presence of lineage-specific rate acceleration (due to biological causes or statistical error in gene tree construction).
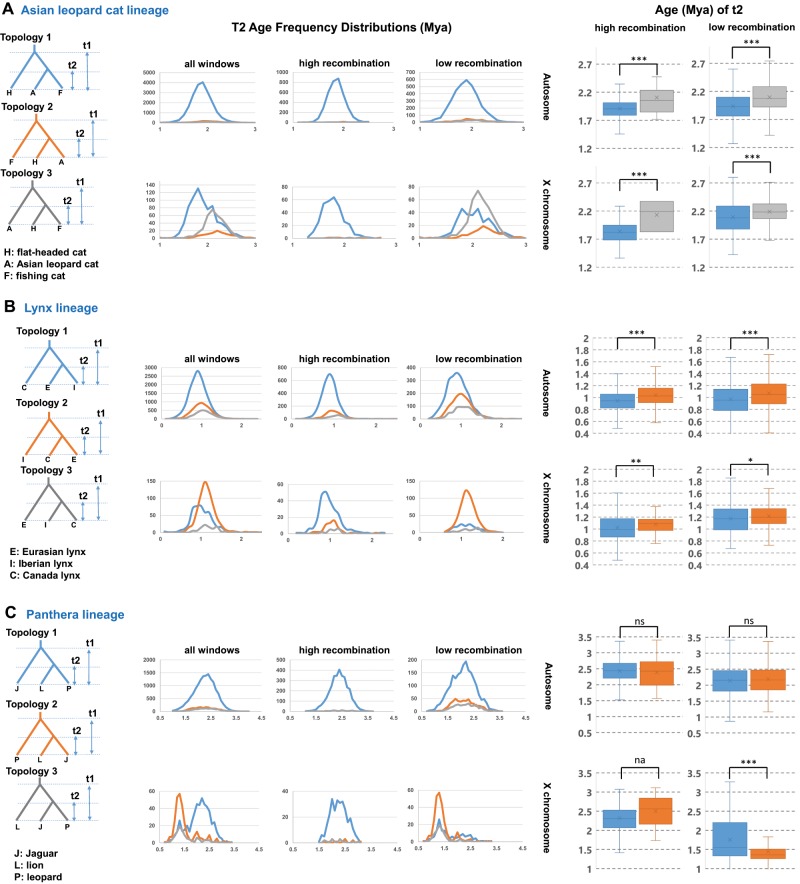
Within the Asian leopard cat and Lynx clades, the mean divergence time for the most common topology in LRchrX (trees 3 and 2, respectively) was significantly older than that of the most common autosomal tree (tree 1) (fig. 3A and B) and was also enriched within regions of low recombination on both the autosomes and chrX (fig. 4). By contrast, the topology that we infer reflects more recent introgression between pairs of nonsister taxa (tree 1) was most frequently found in windows with higher recombination rates (fig. 4 and table 1). In other felid clades that did not show an X-autosome disparity in figure 1, we also found that the inferred species tree was enriched within the LRchrX relative to high-recombination regions (supplementary table S2, Supplementary Material online). Although the windows on the autosomes supporting the inferred speciation history for the Asian leopard cat and Lynx clades were significantly less frequent (fig. 2), they also possessed the oldest divergence times (fig. 3A and B). Taken together, these results support our inference that the oldest tree enriched within low recombination rate regions, and most notably on LRchrX, represents the most probable speciation-driven branching relationships for these two clades.
Fig. 3.
Left: frequency distribution (y axis) of genomic window-based estimates for divergence time (t2) (x axis) for the three felid lineages in which the most common autosomal and chrX phylogenies differed in figure 2. Right: comparison between divergence time estimates for the sister-species pair found in the two most frequent topologies. In the Asian leopard cat (A) and lynx (B) lineages, the topology enriched in the low-recombination cold spot of chrX (gray and orange trees, respectively) had a significantly older divergence time (Mann–Whitney U test, ***P < 0.001, **P < 0.01, *P < 0.05, ns = not significant, na = not analyzed due to too few windows) than topology 1 (blue tree) that was most commonly observed across the genome, and enriched in regions of higher recombination (see fig. 4). Within the Panthera genus (C), trees supporting jaguar + lion are enriched in low-recombining regions of the genome (see fig. 4). In the low recombination regions of chrX, divergence time estimates for the lion + jaguar node from LRchrX are much lower than elsewhere, consistent with published evidence for selective sweeps across this region (Figueiró et al. 2017).
Fig. 4.
Local recombination rate correlates with topology within three felid lineages. Individual plots show the frequency of a particular topology (y axis) within windows of increasing recombination rate (x axis). The tree inferred here to reflect speciation history in each lineage (topology 3 for the Leopard cat clade, topology 2 for the Lynx and Panthera clades) is enriched in regions of low recombination on the autosomes and chrX, whereas the frequency of the most prevalent tree (topology 1 in all three lineages), inferred here to derive from postspeciation admixture, increases with recombination rate.
Table 1.
Recombination Rate Differences between Windows Supporting the Putative Species Tree(ST) and the Most Common Genome-Wide Tree in Figure 3.
| Mean cM/Mb | No. Windows | P Value a | |
|---|---|---|---|
| Asian leopard cat clade | |||
| Topology 1-AUTO | 1.72 | 20,645 | <0.001 |
| Topology 3ST-AUTO | 0.98 | 612 | |
| Topology 1-chrX | 1.78 | 764 | <0.001 |
| Topology 3ST-chrX | 0.12 | 361 | |
| Lynx clade | |||
| Topology 1-AUTO | 1.85 | 14,449 | <0.001 |
| Topology 2ST-AUTO | 1.39 | 5,242 | |
| Topology 1-chrX | 2.28 | 455 | <0.001 |
| Topology 2ST-chrX | 0.56 | 496 | |
| Panthera clade | |||
| Topology 1-AUTO | 1.94 | 14,864 | <0.001 |
| Topology 2ST-AUTO | 1.11 | 1,874 | |
| Topology 1-chrX | 2.25 | 518 | <0.001 |
| Topology 2ST-chrX | 0.23 | 293 | |
Mann–Whitney U test comparing the recombination rate distribution of two topologies.
Within the Panthera clade, trees supporting a sister-group relationship between lion and jaguar are more frequent within low-recombining regions throughout the genome (fig. 4 and table 1) and are also notably enriched across the X chromosome; therefore, under these criteria, it seems most probable that this topology best reflects the species tree. What is notably different about the Panthera clade results is that divergence times across the entire genus were significantly reduced within the largest recombination cold spot, and to a lesser degree in the other cold spots on chromosome X (Figueiró et al. 2017). Using a population genomic scan, Figueiró et al. identified several candidate loci within the large centrally located recombination cold spot that possessed signatures consistent with a selective sweep in jaguars. Figueiró et al. hypothesized that these signatures were consistent with adaptive introgression of specific loci between lion and jaguar, resulting in much lower divergence times across the low-recombining regions of chrX due to linked selection, but not across the vast majority of the genome (fig. 3C). The presence of very compressed trees within this genomic region, but rarely elsewhere, is consistent with patterns expected if selective sweeps occurred in the common ancestor of these two sister species, or even throughout the history of the genus (Nachman and Payseur 2012; O’Fallon 2013). Other complex and nonexclusive processes may also be at play, such as a massive replacement of this LRchrX region with an introgressed segment, subsequently leading to selective rejection of posterior incorporations of genomic fragments originating from the “acceptor” species, while consistently favoring the “donor” species. Another possibility is that the extremely low recombination rates present in these regions may have led to lowered substitution rates in the Panthera lineage that were beyond the reach of the correction provided by our calibrations, further inducing younger ages across this region. Overall, the structured genomic architecture and diversity of signals observed within the Panthera clade illustrate the complexities in accurately inferring evolutionary history in the presence of selection and postspeciation gene flow. Additional genomic sampling of living and extinct pantherine populations will likely aid in better resolving the contribution of these various mechanisms to the complex speciation history of the big cats.
Interclade Phylogeny
Next, we explored the effect of recombination rate and mode of inheritance on the branching relationships of the eight main felid clades. When we divided chrX into low- versus high-recombination regions, concatenated ML trees estimated from the high-recombination regions of chrX were identical to the trees estimated from concatenated autosomal partitions (i.e., subsampled sites from all autosomes, individual chromosomes, and individual 100-kb windows) and possessed very high bootstrap support (fig. 5A). The LRchrX tree relationships were also strongly supported but differed from the chrX high-recombination region trees in both topology (i.e., the position of Asian golden cat and ocelot lineages) and most notably branch lengths (fig. 5A). This result indicated the presence of two highly supported, alternative phylogenetic histories for the cat family that were partitioned by recombination rates. Ninety-five percent of the windows on chrX supported Trees 1–4 (fig. 5B). Interestingly, 99.7% of windows that supported Tree 1 were confined to the two largest recombination coldspots (LRchrX), whereas Trees 3 and 4 were largely restricted to regions characterized by the highest recombination rates, notably the pseudoautosomal region. Tree 2 was restricted to transitional regions between windows with the lowest and highest recombination rates. Therefore, similar to our findings within the different felid clades, the LRchrX possessed a very distinct phylogenetic signal from high-recombining regions on the same chromosome, as well as the autosomes. When we compared the high-resolution recombination and physical maps available for cat to those of human (Nagaraja et al. 1997) and pig (Ma et al. 2010; Fernández et al. 2014), we observed a remarkable conservation of recombination rate profiles across chrX (fig. 6) that span more than 180 My of divergence (Meredith et al. 2011). Although the distribution and pattern of recombination rates have been shown to persist at megabase levels between human and chimpanzees (Paigen and Petkov 2010), there is no precedent for such extreme conservation across deep timescales in mammals. However, similarly recurrent patterns were observed across 50 My of avian evolution (Vijay et al. 2017).
Fig. 5.
The influence of local chrX recombination rate on topology and divergence times. (A) ML trees are shown for the autosomes (concatenation of sites selected every 10 kb across the chromosome alignments) and from all sites on chrX within windows of low (<0.5 cM/Mb) and high (>2.5 cM/Mb) recombination rates. Clade coloring follows figure 1. (B) Top: regional variation in phylogenetic signal density across chrX. The distribution of windows supporting the four most common topologies (color coded according to the key underneath) is shown in relation to recombination rate (in cM/Mb, y axis), plotted underneath the chromosome ideogram. Bottom: timetrees inferred from the four most common trees on chrX, showing the impact of recombination rate on topology and divergence time estimates. Node ages shown for Trees 1–4 are the average of point estimates derived from all 100-kb windows that supported a particular topology: Tree 1 (n = 457), Tree 2 (n = 100), Tree 3 (n = 70), and Tree 4 (n = 64). The timescale below each of the four timetrees indicates millions of years before present (MYBP). Shown above each timetree are two parameters that are distorted by hybridization and recombination, the ratio of external/internal branch length (ebl/ibl), or tree compression, and the shape parameter (α) of the gamma distribution of rate variation among sites. Red arrows indicate the crown node of each felid clade with more than one representative and highlight the differences in the estimated clade ages between regions of low- and high-recombination rate.
Fig. 6.
(A) Remarkable conservation of gene order and recombination rate across eutherian mammal orders. Comparison of colinear recombination maps of chrX in cat (Li, Hillier, et al. 2016) and human (Nagaraja et al. 1997). The human recombination rate image is reproduced (with permission) from the original manuscript figure. In each image, the recombination map (in cM) is plotted along the y axis relative to the physical sequence coordinates on the x axis (in Mb). The red-bounded boxes highlight conserved, syntenic regions of elevated recombination rate, whereas yellow boxes highlight conserved syntenic regions of reduced recombination rate. Lettering in the human chrX image refers to shifts in recombination rate noted in the article. (B) Corresponding boundaries of the conserved chrX low and high-recombination windows in human and pig (data from Fernández et al. [2014]) mapped relative to the domestic cat X chromosome sequence.
Recombination Influences Tree Shape and Divergence Time Estimation
Interspecific gene flow has predictable distorting effects on phylogeny (Posada and Crandall 2002; Lemey and Posada 2009; Leaché et al. 2014), and this information can be used to assist in determining the most likely interclade branching events and divergence times. Frequent hybridization may produce star-like phylogenies when adjacent sites with different histories are combined. Combining these sites into a single analysis has the specific effect of compressing internal branch lengths (ibl) relative to external branches (ebl), with this distortion being proportional to the topological distance between the hybridizing lineages (Schierup and Hein 2000; Lemey and Posada 2009). In addition, simulation studies have demonstrated that phylogenies constructed from these same segments with larger numbers of sites will possess lower values of the alpha (shape) parameter of the gamma distribution of rate variation among sites, because multiple changes will be required at some sites to compensate for apparent homoplasy (Lemey and Posada 2009). Because discordant sites will accumulate more rapidly in regions of high recombination, trees reconstructed from such regions are predicted to be more compressed and show lower alpha values. When we estimated alpha and the ebl/ibl ratio for the four most common topologies on chrX, we found that both parameters were strongly influenced by the local recombination rate (fig. 5B). ML trees from high-recombination windows (Trees 3 and 4) were more star-like with 1.7-fold higher ratios of external to internal branch lengths, and 3-fold lower alpha values than trees produced from recombination cold spots (Trees 1and 2; fig. 5B), confirming the predictions of simulations (Schierup and Hein 2000; Lemey and Posada 2009). Our findings also accord well with previous gene-centric phylogenomic studies which suggested that genes with high GC content (which is positively correlated with meiotic recombination rate) were associated with increased phylogenetic conflict (Romiguier et al. 2013; Shen et al. 2016). In summary, regions with high recombination rates share phylogenetic signatures that are consistent with distorted evolutionary histories, and therefore phylogenies inferred from regions of historically low recombination are more likely to represent the original branching sequence and associated branch lengths.
We then assessed the effects of local recombination rates along chrX on estimation of divergence times for crown felid lineages. As expected, trees from high-recombination rate windows (Trees 3 and 4) produced divergence times for the crown nodes of the terminal felid clades (e.g., Domestic cat clade and Asian leopard cat clade) that were as much as 69% older than estimates derived from the LRchrX (tree 1) (fig. 5B and supplementary table S3, Supplementary Material online). By comparison, divergence times for the same crown nodes that were estimated from the LRchrX are nearly twice as young, and in much stronger agreement with the felid fossil record (supplementary table S3, Supplementary Material online) (Johnson et al. 2006). Near the root of the tree, we observed the opposite effect; divergence estimates are drawn forward in time in regions of high recombination, consistent with the effects of tree compression due to a prolonged period of ancient hybridization between ancestral lineages (i.e., early branches in the tree).
Tests for Interspecies Gene Flow and Genomic Distribution of Admixture Signatures
Although ILS certainly underpins some weakly resolved aspects of discordant branching patterns within and between clades, it is unlikely to be responsible for the striking differences we observed between the autosomes and chrX. We calculated Patterson’s D-statistic for all possible triplets assuming the LRchrX tree as the species phylogeny, as we had concluded that it was the least impacted by the distorting effects of ancient hybridization. We observed that 79% and 46% of ABBA-BABA tests were significant on the autosomes and X chromosome, respectively (fig. 7 and supplementary table S4, Supplementary Material online). These results provide strong evidence for postspeciation gene flow within and between nearly all clades of the Felidae, supporting our contention that introgression, and not ILS, best explains the majority of topological discordance observed across the genome. These results support and extend earlier findings from much sparser nuclear SNP data sets (Walters-Conte et al. 2014; Li, Davis, et al. 2016). For example, the intraclade gene flow evident between the Asian leopard cat and both Fishing cat and Rusty-spotted cat reconciles the discordant phylogeographic patterns of mitochondrial haplotypes that result from mitochondrial capture. Similarly, demographic evidence supported a scenario of historic gene flow between the Iberian lynx and Eurasian lynx (Abascal et al. 2016), consistent with our D-statistics.
Fig. 7.
Heatmap showing statistical support for introgression between pairs of species, based on Patterson’s D-statistic. Each cell in the heatmap indicates the average pairwise Z-score inferred from all possible combinations of species trios including that pair of taxa (see supplementary table S4, Supplementary Material online), assuming the species tree inferred from chrX that is shown above and to the left of the matrix. The tree on the left includes dashed red lines indicating postspeciation admixture identified in the heatmap. Intralineage Z-scores are displayed inside black boxes along the diagonal. Z-scores for the autosomes (upper, red shades) and the X chromosome (lower, blue shades) are shown separately. Using a significance threshold (α = 0.05) that was Bonferroni-corrected for 20 comparisons (the maximum number of trios involving a given pair), Z-scores > 2.81 are considered significant. We conservatively display only chrX Z-scores >5, and autosomal Z-scores >10.
We also quantified and mapped interclade introgressed blocks using five-taxon comparisons performed with the software Dfoil, and observed striking patterns of ancient admixture, indicating that unique signatures of ancient introgression are consistently retained in each descendant species (fig. 8 and supplementary figs. S2–S4, Supplementary Material online). For example, we examined three different inferred episodes of ancient interclade admixture involving the Asian golden cat (AGC) clade, in one case connecting it to the Lynx clade, in another to the Domestic cat clade, and in the third to the Asian leopard cat clade (fig. 8). It is also possible that these introgression episodes occurred even farther back in time, prior to divergence between the assessed clades (e.g., Domestic cat and Lynx clades). As this possibility could not be directly assessed with Dfoil, and would not change the results outlined below (in fact, it would make them stronger), we consider the scenario of three different episodes of admixture. In each case, we focused on the most widespread signatures of introgression, which connected the ancestor of the two extant species of the other clade and each of the sampled species of the AGC clade. Due to a limitation of the Dfoil method, we could not directly assess hybridization between the two ancestral branches (one from each clade), and thus indirectly investigated these episodes via the present-day signatures remaining in each of the two sampled species of the AGC clade. Remarkably, in all three cases, one of the species (marbled cat) consistently retains a larger number of introgressed segments, which are present across all chromosomes but tend to be concentrated in higher recombination, telomeric regions. The other species (Asian golden cat) consistently retains fewer segments from each of the three ancient episodes, and these segments are more evenly distributed across the genome (fig. 8). An alternative explanation would be that hybridization was more recent, postdating the divergence between Asian golden cat and the marbled cat, with each of these species receiving different amounts of introgressed segments from each source (blue and red arrows in fig. 8A). We consider this hypothesis less likely, since it would imply that the same pattern of asymmetric introgression occurred repeatedly in six different episodes of interspecies admixture (i.e., the ancestor of each of the three assessed lineages hybridizing separately with the Asian golden cat and the marbled cat).
Fig. 8.
Quantification and chromosomal distribution of introgressed blocks in the genomes of two felid species (Asian golden cat and marbled cat), resulting from hybridization of their common ancestor with the progenitors of three different lineages of the Felidae. (A) Red and blue arrows show the estimated number of windows which support gene flow between two lineages. (B) Chromosome distribution of windows with signatures of gene flow for different species combinations. (C) Spatial distribution of introgressed blocks derived from the hybridization events shown are shown for chromosome C1 (see supplementary figs. S2–S4, Supplementary Material online, for full results).
Under the differential retention scenario, the consistent differences observed here suggest that the Asian golden cat and the marbled cat have distinct genetic/evolutionary properties (e.g., demographic history and effective population size, Ne) that led them to retain different signatures of ancient introgression episodes preceding their divergence from each other. To investigate this hypothesis, we performed pairwise sequentially Markovian coalescent (PSMC) analyses for these two species (supplementary fig. S5, Supplementary Material online) and observed distinct trajectories, with the Asian golden cat tending to have a larger historical and recent Ne, relative to the marbled cat. Although this past demographic trend may not have had a direct influence on the observed difference in introgression signatures, it raises the hypothesis that Ne differences not only affect the immediate fitness consequences of hybridization (Schumer et al. 2018) but may also shape present-day signatures of ancient hybridization events through their effect on genetic drift and thus retention of alternative haplotypes present in the population. If this hypothesis is affirmed by future assessments, it would imply an additional layer of complexity for phylogenomic inference, since the demographic history of present-day species (and its interplay with selection and recombination) would influence the magnitude and pattern of retention of alternative genealogies, potentially leading to considerably different results depending on the species that is sampled to represent a given clade.
Discussion
Our findings indicate that phylogenetic signal is not randomly distributed across felid genomes but instead is strongly structured by recombination rate and its influence on retention of signatures of gene flow. Specifically, within clades of recently diverged species, trees depicting branching relationships that are consistent with introgression are characterized by younger mean divergence times (fig. 3), significant D-statistics (fig. 7), and are enriched in windows with high rates of recombination rates (fig. 4). In contrast, the branching order reflecting the most likely species tree becomes increasingly common in windows with low recombination rates (fig. 4) and the topologies possess older divergence times. These observations agree with previous studies that predicted that low-recombining regions should retain ancient branching events in the presence of gene flow, as linkage disequilibrium with genes contributing to reproductive isolation that accumulate within such regions would lead to removal of deleterious alleles introduced through hybridization (Nachman and Payseur 2012; Schumer et al. 2018).
However, clades with an extended history of interlineage hybridization will show branch length distortion across the entire phylogeny that is dependent on the age, direction, and breadth of gene flow (Leaché et al. 2014). In the Felidae, we show that a long history of gene flow across the tree leads to compression of the phylogeny (fig. 5B), pulling terminal nodes older and inflating crown-lineage divergence times to be less consistent with fossil estimates, while drawing the basal divergence times forward toward younger dates. This has the effect of creating a more star-like phylogeny that is often used to describe adaptive radiations but may be an artifact driven by recombination-induced distortion of the trees used to infer divergence times. Regions from historically low recombination rates are typically less influenced by this distortion and produce divergence times more consistent with the fossil record. However, these low recombination regions represent a minority of the genome and are enriched in large multimegabase regions on the X chromosome that extend far from the centromere.
If only a small fraction of the genome with historically low recombination rates retains the species tree in taxa that exhibit interspecific gene flow (as in Fontaine et al. [2015]), then this calls into question the validity of most approaches for phylogenomic inference. Both concatenation and coalescent phylogenomic approaches utilize information from all loci, either simultaneously or as sampled gene trees. Coalescent approaches also assume that differences between gene trees are due to ILS (Degnan and Rosenberg 2009). However, when the majority of gene trees from within a genome alignment reflect recent genetic exchange via gene flow, both approaches will incorrectly infer ancestral branching events (Leaché et al. 2014). If hybridization has been rampant throughout the history of the clade, then the tree produced from such an analysis could likely lead to incorrect networks of gene flow between ancient lineages. Given these considerations, it is clear that a priori knowledge about genome architecture, specifically introgression and recombination rates, should be modeled in future phylogenomic analyses (Haenel et al. 2018).
Our results also provide empirical support for the large X-effect (Presgraves 2018) within members of the cat family, where regions of suppressed recombination on chrX are enriched for the putative species tree. Within the domestic cat genome, the multimegabase chrX recombination cold spots are by far the largest, and possess much lower levels of recombination than are observed on any autosomes (Li, Hillier, et al. 2016). These regions were among the most strongly differentiated genomic regions between domestic cats and their wild progenitors (Montague et al. 2014). There are no known inversions that we could attribute to the large chrX recombination cold spots in felids nor in any other eutherian mammal. So what other evolutionary mechanisms might be responsible for such a large region of significantly reduced recombination that retains species branching histories? X chromosome collinearity is unique among eutherian mammal chromosomes in its exceptional conservation, and the gene order across the recombination coldspot is conserved between numerous eutherian mammals (Murphy et al. 1999; Raudsepp et al. 2004; O’Brien et al. 2006; Delgado et al. 2009; Ma et al. 2010). Intriguingly, the largest ∼40-Mb recombination coldspot preserves independent signals of putative selective sweeps with nearly identical, orthologous breakpoints in Panthera and in pigs (Ai et al. 2015; Li, Davis, et al. 2016; Figueiró et al. 2017). The large recombination coldspot also largely overlaps segments in Great Apes marked by selective sweeps, and extended regions of strong genetic differentiation between subspecies and species of rabbits and sheep (Carneiro et al. 2010, 2014; Dutheil et al. 2015; Nam et al. 2015; Chen et al. 2018).
It would appear unlikely that the loci driving these recurrent signatures of genetic differentiation are identical, considering that more than 500 genes reside within this region, which provide a diverse set of targets for linked selection. In the human genome, this region is enriched with ampliconic genes that function in late stages of spermatogenesis (Mueller et al. 2013). In addition, there are notable functional features that are dispersed within and adjacent to the conserved recombination coldspot that merit consideration as drivers of the conserved patterns we observe across divergent mammalian orders. First, the X chromosome inactivation center, which encodes noncoding RNA loci that initiate and maintain female X chromosome silencing, lies within the center of the largest recombination coldspot. Extending outward from the X chromosome inactivation center are multiple, dispersed loci that are known to physically interact to aid in the formation of the 3-dimensional structure of the inactive X chromosome (i.e., the Barr body) in female therian mammals (Deng et al. 2015; Darrow et al. 2016; Giorgetti et al. 2016; Jégu et al. 2017; Wang et al. 2018). It is further noteworthy that these recombination coldspots are also flanked by conserved regions of extremely elevated recombination rate, with highly conserved boundaries in cat, pig and human (fig. 6B). Collectively, these observations suggest that exceptional structural and functional constraints, such as those imparted by X chromosome inactivation, have preserved not only long-range gene order but also megabase-scale recombination patterns over large evolutionary distances (Pál and Hurst 2003), providing a genomic environment that is favorable for the accumulation of reproductive isolation loci and recurrent selective sweeps (Dutheil et al. 2015).
Conclusions
We present empirical evidence of two biases in phylogenomic data sets that may have significant impacts on phylogenetic and divergence time inference at deeper divergences within the Tree of Life. The first bias is in the location and density of sites that support the species tree, which are enriched in historically low recombination regions that may only represent a small minority of a genome. The second is a strong topological bias that results from using phylogenies derived from high-recombination regions that are enriched for signatures of gene flow, supporting observations from previous simulation studies (Posada 2000; Schierup and Hein 2000; Leaché et al. 2014). These effects are dependent on the age, direction and frequency of gene flow throughout the evolutionary history of the group in question. Previous phylogenetic studies of the cat family that concatenated loci from across the nuclear and mitochondrial genomes implied the fossil record underestimated, on average, the first occurrence of crown felid clades by 76% (Johnson et al. 2006; Li, Davis, et al. 2016). The majority of this missing history is removed when regions of low recombination are used to estimate divergence times. Our findings therefore raise the prospect that some described “star-like” phylogenies might be artifacts resulting from combined effects of employing phylogenetic markers located in regions of elevated recombination rate, which may contain mixed histories derived from postspeciation gene flow. Accounting for these two biases may play a central role in resolving conflicts between molecular divergence times and paleontological estimates for taxon originations (Meredith et al. 2011; dos Reis et al. 2012; Liu et al. 2017; Springer et al. 2017), thus contributing to a substantially more accurate reconstruction of the history of life.
Materials and Methods
Sampling and Sequencing
We isolated genomic DNA from cultured fibroblast cells or frozen tissue from 14 wild felid species: Fishing cat (Prionailurus viverrinus), Flat-headed cat (Prionailurus planiceps), Rusty-spotted cat (Prionailurus rubiginosus), Asian golden cat (Catopuma temminckii), Bay cat (Catopuma badia), Marbled cat (Pardofelis marmorata), Canada lynx (Lynx canadensis), Bobcat (Lynx rufus), Jaguarundi (Herpailurus yagouaroundi), Serval (Leptailurus serval), Pampas cat (Leopardus colocola), Ocelot (Leopardus pardalis), Northern Tigrina (Leopardus tigrinus), and Geoffroy’s cat (Leopardus geoffroyi). DNA was extracted with the Qiagen DNeasy kit. Standard Illumina fragment libraries (∼300-bp average insert size) were prepared for each DNA sample and sequenced to ∼30× genome-wide depth of coverage with 2× 125-bp reads on the Illumina HiSeq4000 platform. We combined these new data with published genomes from 13 other felids: domestic cat, Felis catus (Montague et al. 2014); Sand cat, Felis margarita (Li, Davis, et al. 2016); Black-footed cat, Felis nigripes (Li, Davis, et al. 2016); Jungle cat, Felis chaus (Li, Davis, et al. 2016); Leopard cat, Prionailurus bengalensis (Li, Davis, et al. 2016); Eurasian lynx, Lynx lynx (Abascal et al. 2016); Iberian lynx, Lynx pardinus (Abascal et al. 2016); Cheetah, Acinonyx jubatus (Dobrynin et al. 2015); tiger, Panthera tigris (Cho et al. 2013); snow leopard, Panthera uncia (Cho et al. 2013), lion, Panthera leo (Cho et al. 2013); jaguar, Panthera onca (Figueiró et al. 2017); and leopard, Panthera pardus (Figueiró et al. 2017). These 27 species sample the main eight felid clades (Johnson et al. 2006) and comprise 77% of all currently recognized living cat species.
Sequence QC and Read Mapping
All raw reads were trimmed using Trim Galore (http://www.bioinformatics.babraham.ac.uk/ projects/trim_galore; Last accessed June 19, 2019). Trimmed and filtered reads from all wild felids were mapped to a repeatmasked version of the domestic cat genome (v8.0), which was assembled under the guidance of a SNP array-based 58, 055 marker linkage map (Li, Hillier, et al. 2016). Sequence alignment was carried out using BWA with previously described mapping parameters (Li, Davis, et al. 2016). We applied the standard GATK pipeline (https://software.broadinstitute.org/gatk/) to remove polymerase chain reaction duplicates, perform local realignment, and calculate base quality score recalibration.
Genome Alignment and Phylogenomic Analysis
We used ANGSD (Korneliussen et al. 2014) to generate consensus pseudohaploid variant calls for each species, with minimum mapping quality set to 30, and minimum base quality set to 20. Segmental duplications and other highly repetitive regions collapse into single locations in short-read Illumina assemblies and introduce errors into variant calling, which biases branch length estimation. Therefore we applied CNVnator (Abyzov et al. 2011) to identify and exclude all alignment regions with read depth >150% or <50% of the genome-wide average in one or more species.
We performed sliding window-based estimation of phylogeny and divergence times across the 27-species whole-genome alignment. Alignment blocks were created and analyzed in nonoverlapping, 100-kb contiguous windows across the whole-genome alignment. We evaluated smaller (supplementary fig. S1, Supplementary Material online) and larger block sizes, and this had little effect on topology frequencies, therefore the 100-kb block size was used for all subsequent analyses. Although each window was analyzed as a concatenated matrix, by sampling consecutive windows across the genome, we considered the ML tree derived from each window as a reflection of the local density of sites supporting a given topology. We used RAxML (Stamatakis 2014) to perform ML tree searches (GTR + GAMMA model of sequence evolution) for each alignment window. Nodal support for ML trees calculated from each 100-kb window of the aligned sequence matrix was estimated with 200 bootstrap replicates in RAxML. The MCMCTree v4.8a software in the PAML4 package was used to calculate the relative divergence time for the tree imputed from each window. We employed a conservative set of fossil and secondary calibrations (supplementary table S5, Supplementary Material online) used in previous studies to estimate felid divergence times (Johnson et al. 2006; Li, Davis, et al. 2016) that would not constrain nodes impacted by gene flow. MCMCTree analyses were run with the following parameters: 100,000 generations, burn-in set to 10,000 generations, autocorrelated rates.
D Statistics
We used ANGSD (Korneliussen et al. 2014) to calculate D-statistics and Z-scores separately for the autosomes and chrX, for all possible four-taxon combinations across the tree (supplementary table S4, Supplementary Material online). Pseudohaploid sequences were generated from each genome assembly, and we enforced a minimum mapping quality score of 40. The statistical significance of the Z-score for each four-taxon calculation was assessed with weighted block jackknife tests (5-Mb block size for autosomes and 10-Mb block size for chrX). In addition, we used DFOIL (Pease and Hahn 2015) to estimate D-statistics with five-taxon data sets (four ingroup taxa with symmetrical relationships and one outgroup) and 100-kb nonoverlapping windows. We focused on taxon combinations that could test interlineage introgression episodes (hypothesized based on the results from the standard four-taxon ABBA-BABA tests). We used the DFOIL approach to quantify the introgressed blocks (number of 100-kb windows per 10-Mb segment) and to map their chromosomal locations in the domestic cat assembly (supplementary figs. S2–S4, Supplementary Material online).
Demographic History
We applied the PSMC (Li and Durbin 2011) method to infer the historical demography of two cat species (Asian golden cat and marbled cat) that exhibited distinct results in the DFOIL analysis. To call diploid sequences, we generated de novo assemblies using SOAPdenovo2 (Luo et al. 2012) with kmer = 31. Illumina sequences were mapped to their own de novo genome assembly using BWA with default parameter settings. SAMtools was used to estimate mapping coverage, and to call and filter nucleotide variants. Genomic regions with less than half or more than twice the average whole-genome mapping depth were excluded from the final analysis. We applied a mutation rate of 1 × 10−8 and estimated generation times for Asian golden cat (2.2 years) and marbled cat (2 years) (Sunquist M and Sunquist F 2017). The consistency of the PSMC results was assessed by performing 100 bootstrap replicates.
Genome Recombination Rate
We plotted recombination rate variation based on a high-density domestic cat linkage map and the v8 domestic cat genome assembly (Li, Hillier, et al. 2016). We estimated recombination rates in two steps. First, we used the 6,637 framework marker positions from the linkage map to calculate recombination rates (cM/Mb) between markers in the genome assembly. We then assigned those rates 500-kb nonoverlapping windows within each intermarker interval. Second, to mitigate expected changes in fine-scale recombination rates expected over millions of years of species evolution, we calculated and utilized the average recombination rate within larger 2-Mb sliding windows, because previous studies have suggested that broad-scale recombination rate patterns were largely conserved at the megabase scale between species (Auton et al. 2012; Stevison et al. 2016).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We would like to thank Alfred Roca, Nicole Foley, Molly Schumer, and three anonymous reviewers for helpful comments and discussion, and Stephen O’Brien for samples. This work was supported by the US National Science Foundation (DEB-1753760 to W.J.M.), CAPES/Brazil (E.E. and H.V.F.), CNPq/Brazil (E.E. and H.V.F.), and INCT-EECBio/Brazil (E.E. and H.V.F.). The authors declare they have no competing interests.
Whole-genome sequence reads have been deposited under PRJNA407940 in the NCBI short read archive.
References
- Abascal F, Corvelo A, Cruz F, Villanueva-Cañas JL, Vlasova A, Marcet-Houben M, Martínez-Cruz B, Cheng JY, Prieto P, Quesada V.. 2016. Extreme genomic erosion after recurrent demographic bottlenecks in the highly endangered Iberian lynx. Genome Biol. 171:251.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abyzov A, Urban AE, Snyder M, Gerstein M.. 2011. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 216:974–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai H, Fang X, Yang B, Huang Z, Chen H, Mao L, Zhang F, Zhang L, Cui L, He W.. 2015. Adaptation and possible ancient interspecies introgression in pigs identified by whole-genome sequencing. Nat Genet. 473:217–225. [DOI] [PubMed] [Google Scholar]
- Auton A, Fledel-Alon A, Pfeifer S, Venn O, Ségurel L, Street T, Leffler EM, Bowden R, Aneas I, Broxholme J, et al. 2012. A fine-scale chimpanzee genetic map from population resequencing. Science 3366078:193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandvain Y, Kenney AM, Flagel L, Coop G, Sweigart AL.. 2014. Speciation and introgression between Mimulus nasutus and Mimulus guttatus. PLoS Genet. 106:e1004410.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo GA, Antonelli A, Bacon CD, Bartoszek K, Blom MPK, Huynh S, Jones G, Knowles LL, Lamichhaney S, Marcussen T, et al. 2019. Embracing heterogeneity: coalescing the Tree of Life and the future of phylogenomics. PeerJ 7:e6399.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burri R, Nater A, Kawakami T, Mugal CF, Olason PI, Smeds L, Suh A, Dutoit L, Bureš S, Garamszegi LZ, et al. 2015. Linked selection and recombination rate variation drive the evolution of the genomic landscape of differentiation across the speciation continuum of Ficedula flycatchers. Genome Res. 2511:1656–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill JA, Stirling I, Kistler L, Salamzade R, Ersmark E, Fulton TL, Stiller M, Green RE, Shapiro B.. 2015. Genomic evidence of geographically widespread effect of gene flow from polar bears into brown bears. Mol Ecol. 246:1205–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro M, Albert FW, Afonso S, Pereira RJ, Burbano H, Campos R, Melo-Ferreira J, Blanco-Aguiar JA, Villafuerte R, Nachman MW, et al. 2014. The genomic architecture of population divergence between subspecies of the European rabbit. PLoS Genet. 108:e1003519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro M, Blanco-Aguiar JA, Villafuerte R, Ferrand N, Nachman MW.. 2010. Speciation in the European rabbit (Oryctolagus cuniculus): islands of differentiation on the X chromosome and autosomes. Evolution 6412:3443–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z-H, Zhang M, Lv F-H, Ren X, Li W-R, Liu M-J, Nam K, Bruford MW, Li M-H.. 2018. Contrasting patterns of genomic diversity reveal accelerated genetic drift but reduced directional selection on X-chromosome in wild and domestic sheep species. Genome Biol Evol. 105:1282–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YS, Hu L, Hou H, Lee H, Xu J, Kwon S, Oh S, Kim H-M, Jho S, Kim S, et al. 2013. The tiger genome and comparative analysis with lion and snow leopard genomes. Nat Commun. 4:2433.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, Orr HA.. 1989. Two rules of speciation In: Otte D, Endler J, editors. Speciation and its consequences. Sunderland (MA: ): Sinauer; p. 180–207. [Google Scholar]
- Darrow EM, Huntley MH, Dudchenko O, Stamenova EK, Durand NC, Sun Z, Huang S-C, Sanborn AL, Machol I, Shamim M, et al. 2016. Deletion of DXZ4 on the human inactive X chromosome alters higher-order genome architecture. Proc Natl Acad Sci U S A. 11331:E4504–E4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BW, Li G, Murphy WJ.. 2010. Supermatrix and species tree methods resolve phylogenetic relationships within the big cats, Panthera (Carnivora: Felidae). Mol Phylogenet Evol. 561:64–76. [DOI] [PubMed] [Google Scholar]
- Degnan JH, Rosenberg NA.. 2009. Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends Ecol Evol (Amst). 246:332–340. [DOI] [PubMed] [Google Scholar]
- Delgado CLR, Waters PD, Gilbert C, Robinson TJ, Graves J.. 2009. Physical mapping of the elephant X chromosome: conservation of gene order over 105 million years. Chromosome Res. 17:917–926. [DOI] [PubMed] [Google Scholar]
- Delsuc F, Brinkmann H, Philippe H.. 2005. Phylogenomics and the reconstruction of the Tree of Life. Nat Rev Genet. 65:361.. [DOI] [PubMed] [Google Scholar]
- Deng X, Ma W, Ramani V, Hill A, Yang F, Ay F, Berletch JB, Blau CA, Shendure J, Duan Z, et al. 2015. Bipartite structure of the inactive mouse X chromosome. Genome Biol. 16:152.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrynin P, Liu S, Tamazian G, Xiong Z, Yurchenko AA, Krasheninnikova K, Kliver S, Schmidt-Küntzel A, Koepfli K-P, Johnson W, et al. 2015. Genomic legacy of the African cheetah, Acinonyx jubatus. Genome Biol. 16:277.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Reis M, Inoue J, Hasegawa M, Asher RJ, Donoghue PCJ, Yang Z.. 2012. Phylogenomic datasets provide both precision and accuracy in estimating the timescale of placental mammal phylogeny. Proc Biol Sci. 2791742:3491–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Ho SYW, Phillips MJ, Rambaut A.. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol. 45:e88.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutheil JY, Munch K, Nam K, Mailund T, Schierup MH.. 2015. Strong selective sweeps on the X chromosome in the human-chimpanzee ancestor explain its low divergence. PLoS Genet. 118:e1005451.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández AI, Muñoz M, Alves E, Folch JM, Noguera JL, Enciso MP, Rodríguez MDC, Silió L.. 2014. Recombination of the porcine X chromosome: a high density linkage map. BMC Genet. 15:148.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiró HV, Li G, Trindade FJ, Assis J, Pais F, Fernandes G, Santos SHD, Hughes GM, Komissarov A, Antunes A, et al. 2017. Genome-wide signatures of complex introgression and adaptive evolution in the big cats. Sci Adv. 37:e1700299.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine MC, Pease JB, Steele A, Waterhouse RM, Neafsey DE, Sharakhov IV, Jiang X, Hall AB, Catteruccia F, Kakani E, et al. 2015. Mosquito genomics. Extensive introgression in a malaria vector species complex revealed by phylogenomics. Science 3476217:1258524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgetti L, Lajoie BR, Carter AC, Attia M, Zhan Y, Xu J, Chen CJ, Kaplan N, Chang HY, Heard E, et al. 2016. Structural organization of the inactive X chromosome in the mouse. Nature 5357613:575–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good JM, Vanderpool D, Keeble S, Bi K.. 2015. Negligible nuclear introgression despite complete mitochondrial capture between two species of chipmunks. Evolution 698:1961–1972. [DOI] [PubMed] [Google Scholar]
- Haenel Q, Laurentino TG, Roesti M, Berner D.. 2018. Meta-analysis of chromosome-scale crossover rates in eukaryotes and its significance to evolutionary genomics. Mol Ecol. 2711:2477–2497. [DOI] [PubMed] [Google Scholar]
- Hobolth A, Dutheil JY, Hawks J, Schierup MH, Mailund T.. 2011. Incomplete lineage sorting patterns among human, chimpanzee, and orangutan suggest recent orangutan speciation and widespread selection. Genome Res. 213:349–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irisarri I, Singh P, Koblmüller S, Torres-Dowdall J, Henning F, Franchini P, Fischer C, Lemmon AR, Lemmon EM, Thallinger GG, et al. 2018. Phylogenomics uncovers early hybridization and adaptive loci shaping the radiation of Lake Tanganyika cichlid fishes. Nat Commun. 91:3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis ED, Mirarab S, Aberer AJ, Li B, Houde P, Li C, Ho SYW, Faircloth BC, Nabholz B, Howard JT, et al. 2014. Whole-genome analyses resolve early branches in the Tree of Life of modern birds. Science 3466215:1320–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jégu T, Aeby E, Lee JT.. 2017. The X chromosome in space. Nat Rev Genet. 186:377.. [DOI] [PubMed] [Google Scholar]
- Johnson WE, Eizirik E, Pecon SJ, Murphy WJ, Antunes A, Teeling E, O’Brien SJ.. 2006. The late Miocene radiation of modern Felidae: a genetic assessment. Science 3115757:73–77. [DOI] [PubMed] [Google Scholar]
- Joly S, McLenachan PA, Lockhart PJ.. 2009. A statistical approach for distinguishing hybridization and incomplete lineage sorting. Am Nat. 1742:E54–E70. [DOI] [PubMed] [Google Scholar]
- Kawakami T, Mugal CF, Suh A, Nater A, Burri R, Smeds L, Ellegren H.. 2017. Whole-genome patterns of linkage disequilibrium across flycatcher populations clarify the causes and consequences of fine-scale recombination rate variation in birds. Mol Ecol. 2616:4158–4172. [DOI] [PubMed] [Google Scholar]
- Korneliussen TS, Albrechtsen A, Nielsen R.. 2014. ANGSD: Analysis of Next Generation Sequencing Data. BMC Bioinformatics 15:356.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhaney S, Berglund J, Almén MS, Maqbool K, Grabherr M, Martinez-Barrio A, Promerová M, Rubin C-J, Wang C, Zamani N, et al. 2015. Evolution of Darwin’s finches and their beaks revealed by genome sequencing. Nature 5187539:371–375. [DOI] [PubMed] [Google Scholar]
- Larson EL, Keeble S, Vanderpool D, Dean MD, Good JM.. 2017. The composite regulatory basis of the large X-effect in mouse speciation. Mol Biol Evol. 342:282–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson EL, Kopania E, Good JM.. 2018. Spermatogenesis and the evolution of mammalian sex chromosomes. Trends Genet. 349:722–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaché AD, Harris RB, Rannala B, Yang Z.. 2014. The influence of gene flow on species tree estimation: a simulation study. Syst Biol. 631:17–30. [DOI] [PubMed] [Google Scholar]
- Lemey P, Posada D.. 2009. Introduction to recombination detection. In: Lemey P, Salemi M, Vandamme A-M, Lemey P, Salemi M, Vandamme A-M, editors. The phylogenetic handbook. 2nd ed.Cambridge: Cambridge University Press; p. 493–518. [Google Scholar]
- Li G, Davis BW, Eizirik E, Murphy WJ.. 2016. Phylogenomic evidence for ancient hybridization in the genomes of living cats (Felidae). Genome Res. 261:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Hillier LW, Grahn RA, Zimin AV, David VA, Menotti-Raymond M, Middleton R, Hannah S, Hendrickson S, Makunin A, et al. 2016. A high-resolution SNP array-based linkage map anchors a new domestic cat draft genome assembly and provides detailed patterns of recombination. G3 (Bethesda) 6:1607–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2011. Inference of human population history from individual whole-genome sequences. Nature 4757357:493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Zhang J, Rheindt FE, Lei F, Qu Y, Wang Y, Zhang Y, Sullivan C, Nie W, Wang J, et al. 2017. Genomic evidence reveals a radiation of placental mammals uninterrupted by the KPg boundary. Proc Natl Acad Sci U S A. 11435:E7282–E7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 11:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S-J, Zhang Y, Johnson WE, Miao L, Martelli P, Antunes A, Smith JLD, O’Brien SJ.. 2014. Sympatric Asian felid phylogeography reveals a major Indochinese-Sundaic divergence. Mol Ecol. 238:2072–2092. [DOI] [PubMed] [Google Scholar]
- Ma J, Iannuccelli N, Duan Y, Huang W, Guo B, Riquet J, Huang L, Milan D.. 2010. Recombinational landscape of porcine X chromosome and individual variation in female meiotic recombination associated with haplotypes of Chinese pigs. BMC Genomics. 11:159.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailund T, Munch K, Schierup MH.. 2014. Lineage sorting in apes. Annu Rev Genet. 48:519–535. [DOI] [PubMed] [Google Scholar]
- Martin SH, Jiggins CD. 2017. Interpreting the genomic landscape of introgression. Current Opinion in Genetics & Development 47:69–74 [DOI] [PubMed] [Google Scholar]
- McCormack JE, Faircloth BC, Crawford NG, Gowaty PA, Brumfield RT, Glenn TC.. 2012. Ultraconserved elements are novel phylogenomic markers that resolve placental mammal phylogeny when combined with species-tree analysis. Genome Res. 224:746–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menotti-Raymond M, David VA, Lyons LA, Schäffer AA, Tomlin JF, Hutton MK, O’Brien SJ.. 1999. A genetic linkage map of microsatellites in the domestic cat (Felis catus). Genomics. 571:9–23. [DOI] [PubMed] [Google Scholar]
- Menotti-Raymond M, David VA, Roelke ME, Chen ZQ, Menotti KA, Sun S, Schäffer AA, Tomlin JF, Agarwala R, O’Brien SJ., et al. 2003. Second-generation integrated genetic linkage/radiation hybrid maps of the domestic cat (Felis catus) J. Hered. 941:95–106. [DOI] [PubMed] [Google Scholar]
- Meredith RW, Janecka JE, Gatesy J, Ryder OA, Fisher CA, Teeling EC, Goodbla A, Eizirik E, Simao TLL, Stadler T, et al. 2011. Impacts of the Cretaceous Terrestrial Revolution and KPg extinction on mammal diversification. Science 3346055:521–524. [DOI] [PubMed] [Google Scholar]
- Montague MJ, Li G, Gandolfi B, Khan R, Aken BL, Searle SMJ, Minx P, Hillier LW, Koboldt DC, Davis BW, et al. 2014. Comparative analysis of the domestic cat genome reveals genetic signatures underlying feline biology and domestication. Proc Natl Acad Sci U S A. 11148:17230–17235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller JL, Skaletsky H, Brown LG, Zaghlul S, Rock S, Graves T, Auger K, Warren WC, Wilson RK, Page DC.. 2013. Independent specialization of the human and mouse X chromosomes for the male germ line. Nat Genet. 459:1083–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy WJ, Sun S, Chen ZQ, Pecon-Slattery J, O’Brien SJ.. 1999. Extensive conservation of sex chromosome organization between cat and human revealed by parallel radiation hybrid mapping. Genome Res. 912:1223–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachman MW, Payseur BA.. 2012. Recombination rate variation and speciation: theoretical predictions and empirical results from rabbits and mice. Philos Trans R Soc Lond B Biol Sci. 3671587:409–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau NJ, Martin SH, Kozak KM, Salazar C, Dasmahapatra KK, Davey JW, Baxter SW, Blaxter ML, Mallet J, Jiggins CD.. 2013. Genome-wide patterns of divergence and gene flow across a butterfly radiation. Mol Ecol. 223:814–826. [DOI] [PubMed] [Google Scholar]
- Nagaraja R, MacMillan S, Kere J, Jones C, Griffin S, Schmatz M, Terrell J, Shomaker M, Jermak C, Hott C, et al. 1997. X chromosome map at 75-kb STS resolution, revealing extremes of recombination and GC content. Genome Res. 73:210–222. [DOI] [PubMed] [Google Scholar]
- Nam K, Munch K, Hobolth A, Dutheil JY, Veeramah KR, Woerner AE, Hammer MF Great Ape Genome Diversity Project, Mailund T, Schierup MH.. 2015. Extreme selective sweeps independently targeted the X chromosomes of the great apes. Proc Natl Acad Sci U S A. 11220:6413–6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nater A, Burri R, Kawakami T, Smeds L, Ellegren H.. 2015. Resolving evolutionary relationships in closely related species with whole-genome sequencing data. Syst Biol. 646:1000–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nater A, Mattle-Greminger MP, Nurcahyo A, Nowak MG, de Manuel M, Desai T, Groves C, Pybus M, Sonay TB, Roos C, et al. 2017. Morphometric, behavioral, and genomic evidence for a new orangutan species. Curr Biol. 2722:3487–3498. [DOI] [PubMed] [Google Scholar]
- Nater A, Nietlisbach P, Arora N, van Schaik CP, van Noordwijk MA, Willems EP, Singleton I, Wich SA, Goossens B, Warren KS, et al. 2011. Sex-biased dispersal and volcanic activities shaped phylogeographic patterns of extant Orangutans (genus: Pongo). Mol Biol Evol. 288:2275–2288. [DOI] [PubMed] [Google Scholar]
- O’Brien SJ, Menninger JC, Nash WG.. 2006. Atlas of mammalian chromosomes. Hoboken, NJ: John Wiley & Sons. [Google Scholar]
- O’Fallon B. 2013. Purifying selection causes widespread distortions of genealogical structure on the human X chromosome. Genetics 194:485–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortíz-Barrientos D, Reiland J, Hey J, Noor M.. 2002. Recombination and the divergence of hybridizing species. Genetica 116(2-3):167–178. [PubMed] [Google Scholar]
- Paigen K, Petkov P.. 2010. Mammalian recombination hot spots: properties, control and evolution. Nat Rev Genet. 113:221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pál C, Hurst LD.. 2003. Evidence for co-evolution of gene order and recombination rate. Nat Genet. 333:392–395. [DOI] [PubMed] [Google Scholar]
- Payseur BA, Rieseberg LH.. 2016. A genomic perspective on hybridization and speciation. Mol Ecol. 2511:2337–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pease JB, Hahn MW.. 2013. More accurate phylogenies inferred from low-recombination regions in the presence of incomplete lineage sorting. Evolution 678:2376–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pease JB, Hahn MW.. 2015. Detection and polarization of introgression in a five-taxon phylogeny. Syst Biol. 644:651–662. [DOI] [PubMed] [Google Scholar]
- Posada D. 2000. How does recombination affect phylogeny estimation? Trends Ecol. Evol. 1512:489–490. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA.. 2002. The effect of recombination on the accuracy of phylogeny estimation. J Mol Evol. 543:396–402. [DOI] [PubMed] [Google Scholar]
- Presgraves DC. 2008. Sex chromosomes and speciation in Drosophila. Trends Genet. 247:336–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves DC. 2018. Evaluating genomic signatures of “the large X-effect” during complex speciation. Mol Ecol. 2719:3822–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prum RO, Berv JS, Dornburg A, Field DJ, Townsend JP, Lemmon EM, Lemmon AR.. 2015. A comprehensive phylogeny of birds (Aves) using targeted next-generation DNA sequencing. Nature 5267574:569–573. [DOI] [PubMed] [Google Scholar]
- Raudsepp T, Lee E-J, Kata SR, Brinkmeyer C, Mickelson JR, Skow LC, Womack JE, Chowdhary BP.. 2004. Exceptional conservation of horse-human gene order on X chromosome revealed by high-resolution radiation hybrid mapping. Proc Natl Acad Sci U S A. 1018:2386–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards EJ, Brown JM, Barley AJ, Chong RA, Thomson RC.. 2018. Variation across mitochondrial gene trees provides evidence for systematic error: how much gene tree variation is biological? Syst Biol. 675:847–860. [DOI] [PubMed] [Google Scholar]
- Romiguier J, Ranwez V, Delsuc F, Galtier N, Douzery E.. 2013. Less is more in mammalian phylogenomics: AT-rich genes minimize tree conflicts and unravel the root of placental mammals. Mol Biol Evol. 309:2134–2144. [DOI] [PubMed] [Google Scholar]
- Schierup MH, Hein J.. 2000. Consequences of recombination on traditional phylogenetic analysis. Genetics 1562:879–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumer M, Xu C, Powell DL, Durvasula A, Skov L, Holland C, Blazier JC, Sankararaman S, Andolfatto P, Rosenthal GG, et al. 2018. Natural selection interacts with the local recombination rate to shape the evolution of hybrid genomes. Science 3606389:656–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scornavacca C, Galtier N.. 2017. Incomplete lineage sorting in mammalian phylogenomics. Syst Biol. 661:112–120. [DOI] [PubMed] [Google Scholar]
- Shen X-X, Hittinger CT, Rokas A.. 2017. Contentious relationships in phylogenomic studies can be driven by a handful of genes. Nat Ecol Evol. 15:126.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X-X, Salichos L, Rokas A.. 2016. A genome-scale investigation of how sequence, function, and tree-based gene properties influence phylogenetic inference. Genome Biol Evol. 88:2565–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer MS, Emerling CA, Meredith RW, Janečka JE, Eizirik E, Murphy WJ.. 2017. Waking the undead: implications of a soft explosive model for the timing of placental mammal diversification. Mol Phylogenet Evol. 106:86–102. [DOI] [PubMed] [Google Scholar]
- Springer MS, Gatesy J. 2018. Delimiting Coalescence Genes (C-Genes) in Phylogenomic Data Sets. Genes 9(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 309:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevison LS, Woerner AE, Kidd JM, Kelley JL, Veeramah KR, McManus KF Great Ape Genome Project, Bustamante CD, Hammer MF, Wall JD.. 2016. The time scale of recombination rate evolution in great apes. Mol Biol Evol. 334:928–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunquist M, Sunquist F.. 2017. Wild cats of the world. Chicago: University of Chicago Press. [Google Scholar]
- Vijay N, Weissensteiner M, Burri R, Kawakami T, Ellegren H, Wolf J.. 2017. Genomewide patterns of variation in genetic diversity are shared among populations, species and higher-order taxa. Mol Ecol. 2616:4284–4295. [DOI] [PubMed] [Google Scholar]
- Walters-Conte KB, Johnson DLE, Johnson WE, O’Brien SJ, Pecon-Slattery J.. 2014. The dynamic proliferation of CanSINEs mirrors the complex evolution of Feliforms. BMC Evol Biol. 141:137.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C-Y, Jégu T, Chu H-P, Oh HJ, Lee JT.. 2018. SMCHD1 merges chromosome compartments and assists formation of super-structures on the inactive X. Cell 1742:406–421.e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.