Abstract
Background
Imbalance of intestinal microbiota was closely related to colitis. Under these circumstances, regulation of enteric flora may be beneficial to the repair of inflammation. We aimed to investigate the effects of probiotics (Bifidobacterium and Lactobacillus), prebiotics and their combination on inflammation, and microflora in mice of acute colitis.
Methods
C57BL/6J mice were divided into six groups randomly (blank control group, model control group, probiotics group, synbiotics group, lactitol group and probiotics + lactitol group). Each group was given 2.5% dextran sulfate sodium drinking water for 5 days other than the blank control group. Except for the model control group, the other four groups were intervened with probiotics, synbiotics (probiotics and inulin), lactitol, and probiotics + lactitol. Mice were sacrificed after 1 week of gavage, and pathologic scores were calculated. The feces of different periods and intestinal mucosa samples were collected to analyze the differences of intestinal microbiota by 16S rRNA sequencing. Differences of two groups or multiple groups were statistically examined through unpaired Student t test and analysis of variance (ANOVA), respectively. ANOVA, Tukey, Anosim, and metastats analysis were used to compare differences of microbiota among different groups.
Results
After gavage for 1 week, the pathologic scores of groups with the intervention were significantly lower than those in the model control group, and the difference was statistically significant (P < 0.05). The model control group was higher in the genus of Bacteroides (relative abundance: 0.3679 vs. 0.0099, P = 0.0016) and lower in Lactobacillus (relative abundance: 0.0020 vs. 0.0122, P = 0.0188), Roseburia (relative abundance: 0.0004 vs. 0.0109, P = 0.0157), compared with the blank control group. However, the same phenomenon was not found in groups gavaged with probiotics and lactitol. Compared with model control group, mice with intervention were increased with Bifidobacterium (relative abundance: 0.0172 vs. 0.0039, P = 0.0139), Lachnospiraceae_NK4A136_group (relative abundance: 0.1139 vs. 0.0320, P = 0.0344), Lachnospiraceae_UCG-006 (relative abundance: 0.0432 vs. 0.0054, P = 0.0454), and decreased with Alistipes (relative abundance: 0.0036 vs. 0.0105, P = 0.0207) in varying degrees. The mucosal flora was more abundant than the fecal flora, and genus of Mucispirillum (relative abundance: 0.0207 vs. 0.0001, P = 0.0034) was more common in the mucosa. Lactitol group showed higher level of Akkermansia than model control group (relative abundance: 0.0138 vs. 0.0055, P = 0.0415), probiotics group (relative abundance: 0.0138 vs. 0.0022, P = 0.0041), and synbiotics group (relative abundance: 0.0138 vs. 0.0011, P = 0.0034), while probiotics + lactitol group had more abundant Akkermansia than synbiotics group (relative abundance: 0.0215 vs. 0.0013, P = 0.0315).
Conclusions
Probiotics and prebiotics reduce the degree of inflammation in acute colitis mice obviously. Mice with acute colitis show reduced beneficial genera and increased harmful genera. Supplementation of probiotics and prebiotics display the advantage of increasing the proportion of helpful bacteria and regulating the balance of intestinal microbiota. Lactitol might promote the proliferation of Akkermansia.
Keywords: Probiotics, Lactitol, Intestinal microbiota, Akkermansia
Introduction
Gut micro-ecosystem was the largest micro-ecosystem in the human body.[1] Under normal conditions, a large number of bacteria formed a microbial barrier to provide energy and nutrition, protect the intestinal structure, maintain the intestinal immune homeostasis, and resist the invasion of pathogenic bacteria.[2] Generally speaking, host and intestinal flora were in dynamic balance. Once the balance was destroyed, it could lead to various diseases. For these patients, supplementation of probiotics was beneficial to the recovery and reconstruction of intestinal microbiota.[3]
Probiotics, prebiotics, and synbiotics could supplement probiotics directly or indirectly.[4,5] It was worth mentioning that the effectiveness of probiotics may be influenced by colonization ability and survival rate of viable bacteria. Taking these into account, prebiotics was also a good choice. Prebiotics could not be absorbed by the host, but could promote the proliferation of one or more beneficial bacteria selectively. What commonly used were oligosaccharides, lactulose, inulin, and so forth.[4] Besides, lactitol was applied widely in the treatment of hepatic encephalopathy and chronic constipation. Meanwhile, it conformed to the definition of prebiotics and had an impact on the regulation of bacterial flora.
In recent years, more and more studies have shown that except for heredity, immunity, and environment, the occurrence of ulcerative colitis (UC) was closely related to an imbalance of enteric flora.[6,7] Normally, more than 90% of the intestinal bacteria belonged to Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria. However, in patients with UC, there was an obvious trend that Firmicutes decreased while Bacteroidetes and Proteobacteria increased.[8] The classical treatment for UC included 5-amino alicylic acid, glucocorticoid, immunosuppressant. However, these drugs would inevitably bring side effects while exerting their efficacy. Since the pathogenesis of UC involved imbalance of bacteria, the treatment of probiotics had been put on the agenda.[9]
The rapid development of high throughput sequencing technology provided new ideas for the study of intestinal flora. Through sequencing the DNA sequences which encoding ribosome 16S rRNA in the bacterial genome, we could analyze the abundance and classification of bacteria and explore meaningful changes of microbiota.[10] In addition, acute colitis mice model induced by dextran sulfate sodium (DSS) was similar to UC in symptoms and pathologic manifestations.[11] Therefore, it was a simple and scientific choice to use this model to study intestinal flora.
In addition to traditional probiotics, we also focused on the effects of prebiotics. The purpose of our study was to explore the effects of probiotics, prebiotics and their combination on inflammation, and intestinal microflora of acute colitis mice, to have a deeper understanding of microbiota and colitis.
Methods
Ethical approval
This study was approved by the Animal Care Ethics and Use Committee of Peking Union Medical College Hospital (PUMCH, No. XHDW-2015-0032).
Experimental animals
Sixty male, 6 to 8 weeks old, C57BL/6J mice weighing 18 to 20 g (purchased from Beijing Vital River Laboratory Animal Technology Company, No. SCXK2014-0004[11401300066549]) were housed in specific pathogen-free conditions. They were divided into six groups randomly. Except for blank control group, other five groups were given 2.5% DSS drinking water for 5 days. Blank control and model control group were not given intragastric administration, while other groups were intervened with probiotics, synbiotics (probiotics and inulin), lactitol, and probiotics + lactitol, respectively.
Mice were sacrificed after 1 week of gavage. Feces together with distal intestinal mucosa samples, before the intervention, during the intervention, at the end of gavage, were collected to analyze the differences of intestinal microbiota by 16S rRNA sequencing.
Probiotics, prebiotics, and synbiotics
Probiotics was composed of Lactobacillus acidophilus, L. Rhamnosus, and Bifidobacterium lactis, and was given 1.0 × 109 colony-forming units (CFU) per day per mice.[12] Synbiotics was consist of the above-mentioned probiotics and inulin, and was administered 5 × 108 CFU/day. As for lactitol, each mouse was given 6.6 g/kg per day. To observe synergistic effects, the dose of single component was reduced to half, namely, 5 × 108 CFU/day probiotics and 3.3 g/kg per day lactitol. All the regents were administrated through gavage. Lactitol was provided by Zhengda Tianqing Pharmaceutical Limited Company (Nanjing, China), while other reagents were provided by Beijing Macro-Union Pharmaceutical Limited Corporation (Beijing, China).
Evaluation of inflammation
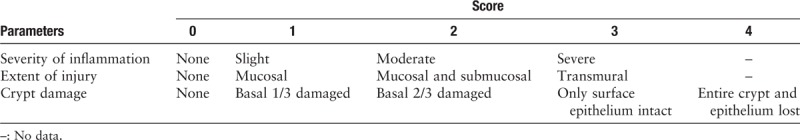
Mucosal specimens were dehydrated, paraffin embedded, sectioned, and stained with hematoxylin-eosin. Histologic evaluation was performed by a pathologist according to the scoring criteria in Table 1.[13] Finally, each score was multiplied by the coefficient on the basis of percentage of tissue involved (0–25%: ×1, 26–50%: ×2, 51–75%: ×3, 76–100%: ×4).
Table 1.
Scoring criteria of inflammation in acute colitis mice.
Sequencing and analysis of intestinal microbiota
Feces and intestinal mucosa adjacent to the rectum were sent to Allwegene Science and Technology Limited Company (Beijing, China) to detect microbiota by 16S rRNA amplification through MiSeq PE300 sequencing platform.
With sequencing results, the specific analysis included the following three aspects. Firstly, alpha diversity analysis. Operational taxonomic unit (OTU) was the basic unit of taxonomy and relative abundance analysis, the most important part of which was alpha diversity analysis. Alpha diversity index could reflect the richness and uniformity of the microbial community, among which observed species was widely used because it referred to the actual number of OTUs in the sample. Secondly, principal component analysis (PCA). The difference of specimen was reflected in the two-dimensional coordinate diagram; the more similar sample composition was, the closer the distance in PCA diagram. Through Anosim test, in case the difference between groups was greater than difference within the group, and P < 0.05, implying the existence of statistically significant genus. Thirdly, taxonomic analysis. On the basis of PCA, the metastats analysis was applied to discover specific and meaningful genus. We defined that abundance >1% and P < 0.05 to be significantly different.
Statistical analysis
Continuous variables were described as mean ± standard deviation, while categorical variables were presented as numbers and proportions. Differences between two groups or multiple groups were statistically examined through unpaired Student t test and analysis of variance (ANOVA), respectively. ANOVA, Tukey, Anosim, and metastats analysis were used to compare differences of microbiota among different groups. P values were two-tailed, and P < 0.05 was considered statistically significant. All analyses were performed with Statistical Package for Social Sciences (SPSS; version 19, SPSS Inc., Chicago, IL, USA).
Results
Inflammation of mice in each group
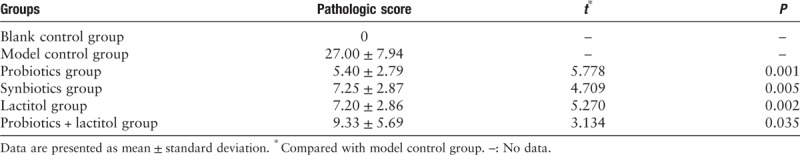
Mice were killed after 1 week of gavage, pathologic scores in intervention groups were statistically decreased compared with model control group, and there was no statistical difference among different intervention groups [Figure 1 and Table 2].
Figure 1.
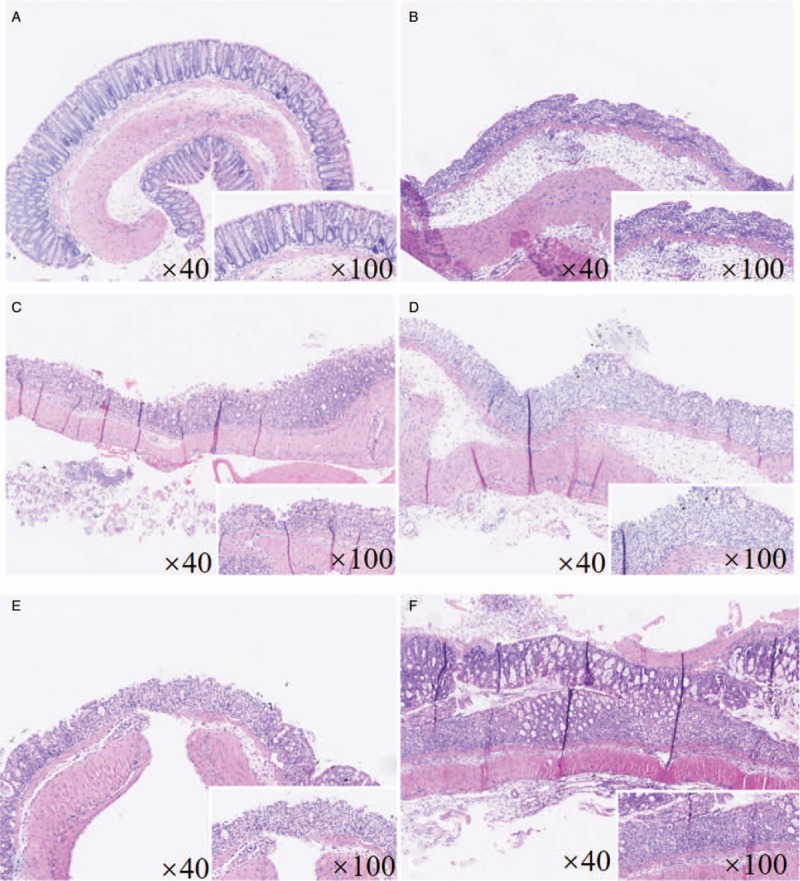
Hematoxylin-eosin dyeing of colonic mucosa (original magnification: ×40, ×100, respectively). (A) Blank control group, with normal gland shape and regular structure. (B) Model control group, with obvious crypt destruction and heavy degree of inflammation, damage scope involved submucosa or even deeper. (C) Probiotics group. (D) Synbiotics group. (E) Lactitol group. (F) Probiotics + lactitol group, with different degrees of inflammation, the extent of injury and crypt damage was alleviated than model control group.
Table 2.
Pathologic scores of different groups.
General condition of mice
On the third day of giving 2.5% DSS, mice showed manifestations of loose and bloody stool, accompanied by weight loss, gloomy hair loss, and fatigue. As for model control group, body weight decreased progressively with aggravated symptoms, these indicators began to improve when exchanged for drinking water on the sixth day. Other intervention group got the lowest body weight on the fourth day, and since then weight gradually increased.
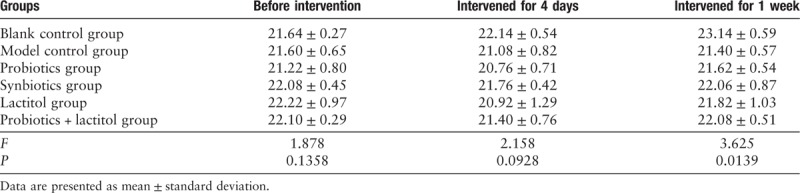
After 1 week of gavage, the body weight of each DSS group was lower than blank control group, which difference was statistically significant (P = 0.0139). However, except for blank control group, there was no significant difference among the remaining groups [Table 3].
Table 3.
Body weight of different groups (g).
Analysis of intestinal microbiota
Comparison of fecal flora in each group before the experiment
Before the intervention, collect feces (W0 feces). Alpha diversity analysis showed no significant difference in fecal microbiota among the six groups (P = 0.1343) [Figure 2A]. PCA of six groups displayed no distinct difference [Figure 2B]. These suggested that the baseline of fecal flora was consistent with each other, thus provided a basis for further analysis.
Figure 2.
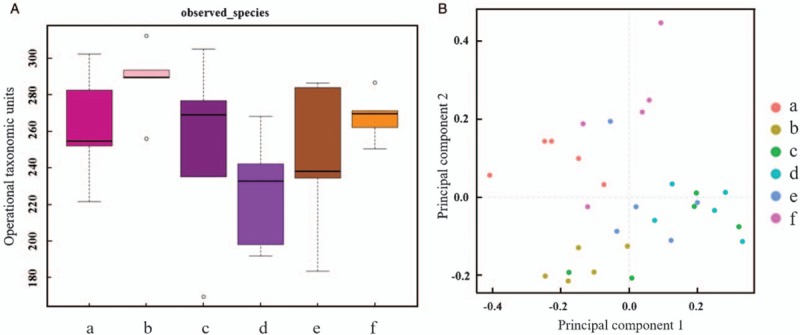
Comparison of fecal flora in each group before the experiment. (A) Alpha diversity analysis. (B) Principal component analysis. The baseline of fecal flora was consistent with each other. (a) Blank control group. (b) Model control group. (c) Probiotics group. (d) Synbiotics group. (e) Lactitol group. (f) Probiotics + lactitol group.
Comparison of fecal flora in each group in the middle of the experiment
On the fourth day of the intervention, mice of intragastric groups exhibited the lowest weight and heaviest symptom, we defined specimen at this time point as D4 feces. Compared with blank control group, alpha diversity of the other five groups decreased significantly (P = 0.0050, 0.0002, 0.0106, 0.0002, and 0.0003, respectively) [Figure 3A], indicating the decline in alpha diversity under inflammation. With regard to PCA, there was no significantly different genus between synbiotics group and lactitol group (P = 0.5615), while other groups revealed distinctly different genus [Figure 3B]. The next step was taxonomic analysis. Lactobacillus was decreased in model control group compared with blank control group (relative abundance: 0.0020 vs. 0.0122, P = 0.0188), while other intragastric groups did not show this reduction. Five DSS model groups showed higher Bacteroides than blank control group (relative abundance: 0.3519 vs. 0.0208, P = 0.0002; 0.3366 vs. 0.0208, P = 0.0001; 0.2381 vs. 0.0208, P = 0.0011; 0.2308 vs. 0.0208, P = 0.0001; 0.2442 vs. 0.0208, P = 0.0027). Besides, probiotics + lactitol group displayed higher Akkermansia than blank control group (relative abundance: 0.0404 vs. 0.0087, P = 0.0178), and more Faecalibacterium than model control group (relative abundance: 0.2854 vs. 0.0589, P = 0.0215) [Figure 3C].
Figure 3.
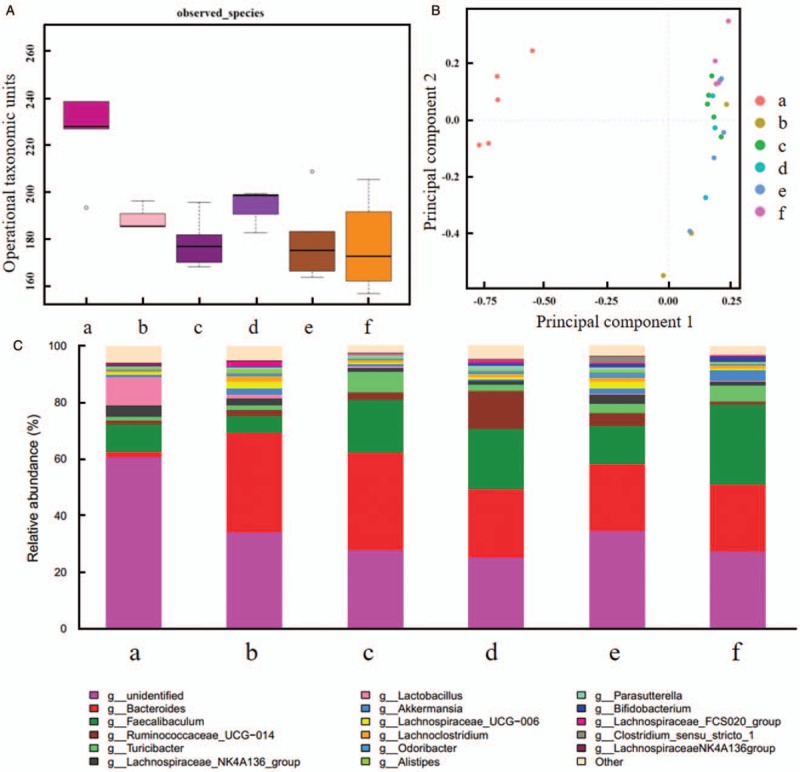
Comparison of intestinal flora for the fourth day of intervention (D4 feces). (A) Alpha diversity analysis. (B) Principal component analysis. (C) Taxonomic analysis. (a) Blank control group. (b) Model control group. (c) Probiotics group. (d) Synbiotics group. (e) Lactitol group. (f) Probiotics + lactitol group. Compared with blank control group, alpha diversity of the other five groups decreased significantly, there was no significantly different genus between synbiotics group and lactitol group, while other groups revealed distinctly different genus.
Comparison of fecal flora in each group at the end of the experiment
After intervention for 1 week, specimens were collected (W1 feces). Similar to D4 feces, alpha diversity of five inflammatory groups decreased distinctly (P = 0.0177, 0.0232, 0.0006, 0.0008, 0.0008, respectively) [Figure 4A]. Compared probiotics group with lactitol group and probiotics + lactitol group, lactitol group with synbiotics group, and probiotics + lactitol group, PCA demonstrated no statistically significant genus (P = 0.3201, 0.1944, 0.2036, 0.0761, respectively) [Figure 4B]. Other groups showed obvious changes when compared with each other. Bacteroides increased obviously in inflammatory groups compared with blank control group (relative abundance: 0.3679 vs. 0.0099, P = 0.0016; 0.2008 vs. 0.0099, P = 0.0006; 0.2871 vs. 0.0099, P = 0.0005; 0.2775 vs. 0.0099, P = 0.0003; 0.2101 vs. 0.0099, P = 0.0025), probiotics + lactitol group was more abundant in Akkermansia than synbiotics group (relative abundance: 0.0215 vs. 0.0013, P = 0.0315). Compared with model control group, Lachnospiraceae_NK4A136_group was richer in probiotics group (relative abundance: 0.2010 vs. 0.0320, P = 0.0352), synbiotics group (relative abundance: 0.1170 vs. 0.0320, P = 0.0401), and lactitol group (relative abundance: 0.1139 vs. 0.0320, P = 0.0344) [Figure 4C].
Figure 4.
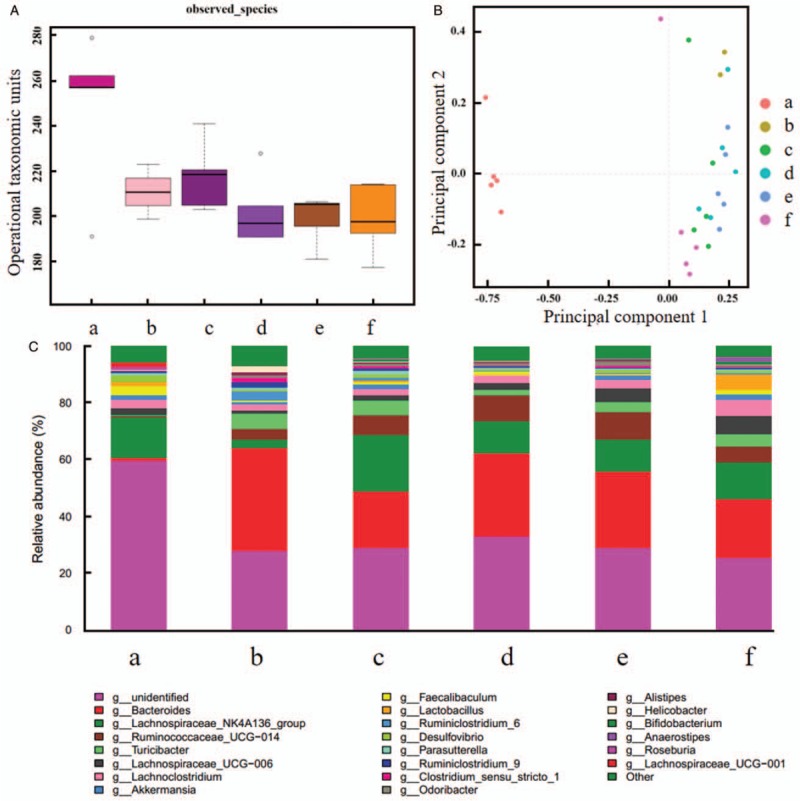
Comparison of intestinal flora after intervention for 1 week (W1 feces). (A) alpha diversity analysis. (B) Principal component analysis. (C) Taxonomic analysis. (a) Blank control group. (b) Model control group. (c) Probiotics group. (d) Synbiotics group. (e) Lactitol group. (f) Probiotics + lactitol group. Alpha diversity of five inflammatory groups decreased distinctly. Compared probiotics group with lactitol group and probiotics + lactitol group, lactitol group with synbiotics group and probiotics + lactitol group, PCA demonstrated no statistically significant genus.
Comparison of mucosal flora in each group at the end of the experiment
Alpha diversity of mucosa in five inflammatory groups was reduced than blank control group (P < 0.001). In addition, alpha diversity was more abundant in lactitol group than model control group, probiotics group, and synbiotics group (P = 0.0038, 0.0177, and 0.0183, respectively) [Figure 5A]. There was no statistically significant genus between probiotics group and model control group, between synbiotics group and probiotics group (P = 0.8621 and 0.3936, respectively). However, PCA indicated different genus among other groups [Figure 5B]. Compared with blank control group, Faecalibacterium was decreased in model control group (relative abundance: 0.0009 vs. 0.0265, P = 0.0131), probiotics group (relative abundance: 0.0039 vs. 0.0265, P = 0.0152), and synbiotics group (relative abundance: 0.0075 vs. 0.0265, P = 0.0305). Lactitol group was higher in Akkermansia than model control group (relative abundance: 0.0138 vs. 0.0055, P = 0.0415), probiotics group (relative abundance: 0.0138 vs. 0.0022, P = 0.0041), and synbiotics group (relative abundance: 0.0138 vs. 0.0011, P = 0.0034) [Figure 5C].
Figure 5.
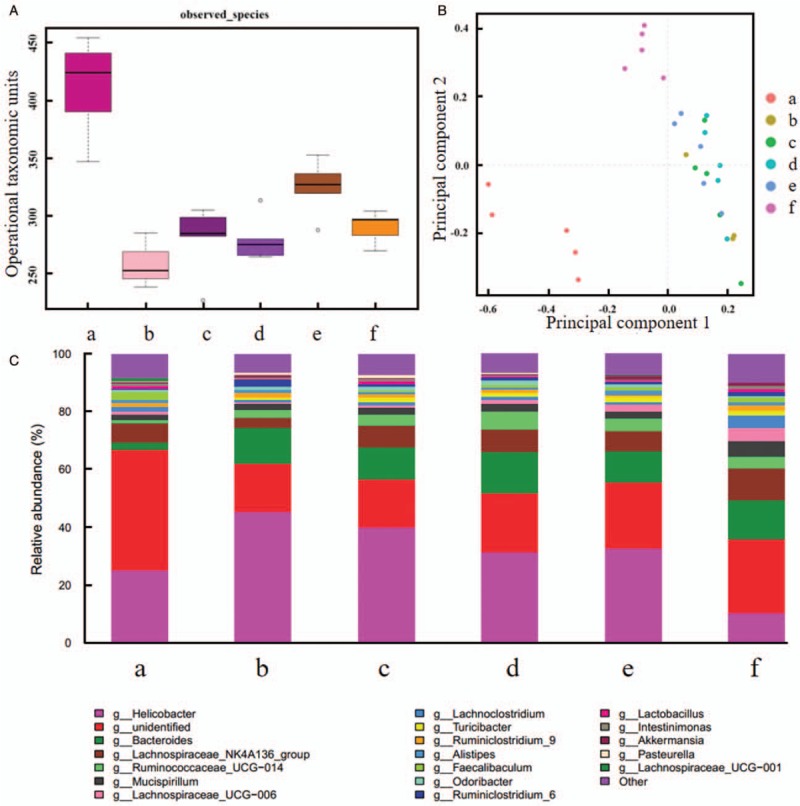
Comparison of intestinal flora for mucosa. (A) Alpha diversity analysis. (B) Principal component analysis. (C) Taxonomic analysis. (a) Blank control group. (b) Model control group. (c) Probiotics group. (d) Synbiotics group. (e) Lactitol group. (f) Probiotics + lactitol group. Alpha diversity of mucosa in five inflammatory groups was reduced than blank control group. There was no statistically significant genus between probiotics group and model control group, between synbiotics group and probiotics group. However, PCA indicated different genus among other groups.
Comparison of intestinal microbiota for different periods in each group
Alpha diversity of mucosa was more abundant than feces in blank control group, synbiotics group, lactitol group, probiotics + lactitol group, and Mucispirillum was increased in mucosa. Even if no intervention was accepted, genera in blank control group were changed over time. As for D4 feces, Lachnospiraceae_NK4A136_group and Ruminiclostridium decreased obviously. In model control group, Lactobacillus showed a decreasing trend of various periods. D4 feces displayed reduced Lactobacillus than W0 feces in other four groups (relative abundance: 0.0050 vs. 0.0946, P = 0.0027; 0.0007 vs. 0.0392, P = 0.0044; 0.0012 vs. 0.1363, P = 0.0005; 0.0013 vs. 0.0391, P = 0.0002; respectively); however, the decreased trend was not appeared when compared D4 feces with W1 feces, suggesting the intervention was effective.
Discussion
Balance of intestinal microbiota played an important role in constructing mucosal barrier and maintaining normal immune function, which mechanisms included producing antibacterial substances, competing with harmful bacteria, and regulating host immunity. Immunodeficiency animal model was constructed with the application of gene knockout, mice showed no intestinal inflammation in a sterile environment. Nevertheless, colitis appeared after recovery of enteric flora, which brought about the theory of “no microbiota, no inflammation.”[14] In recent years, the incidence of UC increased gradually, and more attention was paid to the close relationship between the disease and intestinal flora.[15] Damage of intestinal ecology not only participated in the launch and continuous of UC, but also promoted the progression and caused serious complications such as colorectal cancer. The mechanism may be related to stimulation of abnormal immune response, production of inflammatory factors, and activation of inflammatory pathways. According to the recent literature, patients with intestinal inflammation had a lower level of beneficial bacteria such as Lactobacillus and Bifidobacterium compared with healthy control group, while Enterococcus and Escherichia increased distinctly, thus triggered the occurrence and development of inflammation.[16] Correspondingly, supplement of probiotics was beneficial for remission of UC.[17] Many randomized double-blind trials showed that Bifidobacteria, Lactobacillus, and VSL#3 could help induce remission and prolong maintenance time. Similarly, there were some results in colitis model, suggesting that administration of probiotics could inhibit inflammation and prevent the occurrence of dysplasia. Prebiotics could promote the growth of probiotics thus provided probiotics indirectly. There was a large amount of research on probiotics previously, while the impact of prebiotics on intestinal microbiota was rarely studied.
Analyzing the results of our experiment, we could see the increase and decrease of many genera. Among them, changes of Akkermansia and Faecalibacterium were most significant. Lactitol group showed a higher level of Akkermansia than model control group, probiotics group, and synbiotics group, while probiotics + lactitol group had more abundant Akkermansia than blank control group and synbiotics group. The above results showed promotion of lactitol on the proliferation of Akkermansia. As kind of strict anaerobic enteric bacteria, Akkermansia consisted approximately 1% to 4% of intestinal microbiota, and could degrade mucin, produce short-chain fatty acids, and propionic acid, and provide energy for the host.[18] In recent years, Akkermansia has received extensive attention from scholars. Studies found that its abundance was negatively correlated with levels of free fatty acids and IL-6 in serum.[19]Akkermansia could also ameliorate inflammatory response and insulin resistance in obese and diabetic patients,[20] protect intestinal epithelial cells and enhance mucosal barrier function.[21] Besides, recent research presented it could restore the response of epithelial tumor mouse model to the inhibitor of programmed death-1.[22] The genome of Akkermansia had been proved to be able to encode a variety of secretory proteins such as sulfates, proteases, and glycohydrolyzases.[23] Therefore, we speculated that it might decompose lactitol and promote its own proliferation.
In addition to Akkermansia, the change of Faecalibacterium was also been concerned. Faecalibacterium in model control group, probiotics group, synbiotics group, and lactitol group was lower when compared with blank control group. Named in 2002, Faecalibacterium belonged to Firmicutes, and was strictly anaerobic.[24] It was one of the main bacteria producing butyrate, could up-regulate the function of regulatory T cells, and secrete anti-inflammatory factors to alleviate intestinal inflammation. Faecalibacterium could mitigate colitis induced by TNBS in mice,[25] and decreased evidently in patients with IBD. Besides, it was reported that the abundance of Faecalibacterium was negatively correlated with the incidence of IBD and colorectal cancer.[26]
In addition to the above results, there were other discoveries worth mentioning. Mucosal flora was different from the fecal flora of same period in both abundance and species. Many groups suggested that alpha diversity of mucosal microbiota was higher than that of feces, and the proportion of Mucispirillum was greater in mucosa than feces. Mucispirillum was been found and isolated in recent years, and mainly colonized in mucous layer of intestinal membrane, which was consistent with the results of our study.[27,28] Compared with blank control group, Bacteroides in five DSS model groups was significantly increased when compared with blank control group. Model control group had less Lactobacillus than blank control group, while other four intragastric intervention groups showed no similar trend. As for blank control group, no gastric intervention was given, and environment and diet remained unchanged during the whole process. Nevertheless, microbial diversity and composition changed greatly over time. This phenomenon might mainly due to variation of age in mice.
Analysis was carried out at genus level because of inadequacy in species level. We did not dynamically observe the trend of mucosal microbiota. With regard to duration after stopping lavage, we did not penetrate into discussion in depth.
The role of prebiotics in clinical patients has also been reported. For example, fructooligosaccharide could regulate microbiota in patients with metabolic syndrome,[29] lactitol could increase the number of Lactobacillus and Bifidobacterium, and reduce the level of endotoxin in chronic hepatitis patients. However, there were few studies on lactitol in patients with UC, thus further validation was still needed. Synbiotics was compound of probiotics and prebiotics, in which prebiotics could selectively promote the growth of probiotics, thus played a better role than single component. The most commonly used combination was Lactobacillus, Bifidobacterium, and fructooligosaccharide.[30,31] Taking synergistic action into consideration, we chose half dose of single component in probiotics + lactitol group, but result was not shown as expected. The mixture was self-combined by us, and lactitol might not specifically promote the proliferation of Lactobacillus and Bifidobacterium, and also had effects on other intestinal flora. In addition, osmotic pressure of combinations could be harmful to mice. This suggests that animal model could provide direction for clinical research to some extent, but could not completely simulate physiologic and pathologic state of human. In the study, the effects of probiotics and prebiotics on intestinal microbiota in acute colitis mice are preliminarily explored. Influence of different preparations on clinical patients and its mechanism still need to be further studied to be better applied to patients.
Funding
This work was supported by grants from the National Natural Science Foundation of China (No. 81370500 and No. 81770559).
Conflicts of interest
None.
Footnotes
How to cite this article: Wang YN, Meng XC, Dong YF, Zhao XH, Qian JM, Wang HY, Li JN. Effects of probiotics and prebiotics on intestinal microbiota in mice with acute colitis based on 16S rRNA gene sequencing. Chin Med J 2019;132:1833–1842. doi: 10.1097/CM9.0000000000000308
References
- 1.Leclercq S, Matamoros S, Cani PD, Neyrinck AM, Jamar F, Stärkel P, et al. Intestinal permeability, gut-bacterial dysbiosis, and behavioral markers of alcohol-dependence severity. Proc Natl Acad Sci U S A 2014; 111:E4485–E4493. doi: 10.1073/pnas.1415174111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marchesi JR, Adams DH, Fava F, Hermes GD, Hirschfield GM, Hold G, et al. The gut microbiota and host health: a new clinical frontier. Gut 2016; 65:330–339. doi: 10.1136/gutjnl-2015-309990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kahrstrom CT, Pariente N, Weiss U. Intestinal microbiota in health and disease. Nature 2016; 535:47.doi: 10.1038/535047a. [DOI] [PubMed] [Google Scholar]
- 4.Markowiak P, Slizewska K. Effects of probiotics, prebiotics, and synbiotics on human health. Nutrients 2017; 9: doi: 10.3390/nu9091021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Louis P, Flint HJ, Michel C. How to manipulate the microbiota: prebiotics. Adv Exp Med Biol 2016; 902:119–142. doi: 10.1007/978-3-319-31248-4_9. [DOI] [PubMed] [Google Scholar]
- 6.McLean MH, Dieguez DJ, Miller LM, Young HA. Does the microbiota play a role in the pathogenesis of autoimmune diseases? Gut 2015; 64:332–341. doi: 10.1136/gutjnl-2014-308514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arrieta MC, Stiemsma LT, Amenyogbe N, Brown EM, Finlay B. The intestinal microbiome in early life: health and disease. Front Immunol 2014; 5:427.doi: 10.3389/fimmu.2014.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fiocchi C. Inflammatory bowel disease pathogenesis: where are we? J Gastroenterol Hepatol 2015; 30 Suppl 1:12–18. doi: 10.1111/jgh.12751. [DOI] [PubMed] [Google Scholar]
- 9.Derikx LA, Dieleman LA, Hoentjen F. Probiotics and prebiotics in ulcerative colitis. Best Pract Res Clin Gastroenterol 2016; 30:55–71. doi: 10.1016/j.bpg.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 10.Xu Y, Xie Z, Wang H, Shen Z, Guo Y, Gao Y, et al. Bacterial diversity of intestinal microbiota in patients with substance use disorders revealed by 16S rRNA gene deep sequencing. Sci Rep 2017; 7:3628.doi: 10.1038/s41598-017-03706-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chassaing B, Aitken JD, Malleshappa M, Vijay-Kumar M. Dextran sulfate sodium (DSS)-induced colitis in mice. Curr Protoc Immunol 2014; 104:15–25. doi: 10.1002/0471142735.im1525s104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toumi R, Soufli I, Rafa H, Belkhelfa M, Biad A, Touil-Boukoffa C. Probiotic bacteria Lactobacillus and Bifidobacterium attenuate inflammation in dextran sulfate sodium-induced experimental colitis in mice. Int J Immunopathol Pharmacol 2014; 27:615–627. doi: 10.1177/039463201402700418. [DOI] [PubMed] [Google Scholar]
- 13.Zhao H, Zhang H, Wu H, Li H, Liu L, Guo J, et al. Protective role of 1,25(OH)2 vitamin D3 in the mucosal injury and epithelial barrier disruption in DSS-induced acute colitis in mice. BMC Gastroenterol 2012; 12:57.doi: 10.1186/1471-230X-12-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sellon RK, Tonkonogy S, Schultz M, Dieleman LA, Grenther W, Balish E, et al. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect Immun 1998; 66:5224–5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen L, Wang W, Zhou R, Ng SC, Li J, Huang M, et al. Characteristics of fecal and mucosa-associated microbiota in Chinese patients with inflammatory bowel disease. Medicine (Baltimore) 2014; 93:e51.doi: 10.1097/MD.0000000000000051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, et al. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut 2006; 55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen WX, Ren LH, Shi RH. Enteric microbiota leads to new therapeutic strategies for ulcerative colitis. World J Gastroenterol 2014; 20:15657–15663. doi: 10.3748/wjg.v20.i42.15657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collado MC, Derrien M, Isolauri E, de Vos WM, Salminen S. Intestinal integrity and Akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly. Appl Environ Microbiol 2007; 73:7767–7770. doi: 10.1128/AEM.01477-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodriguez-Carrio J, Salazar N, Margolles A, González S, Gueimonde M, de Los Reyes-Gavilán CG, et al. Free fatty acids profiles are related to gut microbiota signatures and short-chain fatty acids. Front Immunol 2017; 8:823.doi: 10.3389/fimmu.2017.00823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansen CH, Krych L, Nielsen DS, Vogensen FK, Hansen LH, Sørensen SJ, et al. Early life treatment with vancomycin propagates Akkermansia muciniphila and reduces diabetes incidence in the NOD mouse. Diabetologia 2012; 55:2285–2294. doi: 10.1007/s00125-012-2564-7. [DOI] [PubMed] [Google Scholar]
- 21.Reunanen J, Kainulainen V, Huuskonen L, Ottman N, Belzer C, Huhtinen H, et al. Akkermansia muciniphila adheres to enterocytes and strengthens the integrity of the epithelial cell layer. Appl Environ Microbiol 2015; 81:3655–3662. doi: 10.1128/AEM.04050-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018; 359:91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 23.Singla V, Chakkaravarthi S. Applications of prebiotics in food industry: a review. Food Sci Technol Int 2017; 23:649–667. doi: 10.1177/1082013217721769. [DOI] [PubMed] [Google Scholar]
- 24.Duncan SH, Hold GL, Harmsen HJ, Stewart CS, Flint HJ. Growth requirements and fermentation products of Fusobacterium prausnitzii, and a proposal to reclassify it as Faecalibacterium prausnitzii gen. nov., comb. nov. Int J Syst Evol Microbiol 2002; 52 (Pt 6):2141–2146. doi: 10.1099/00207713-52-6-2141. [DOI] [PubMed] [Google Scholar]
- 25.Miquel S, Martin R, Rossi O, Bermúdez-Humarán LG, Chatel JM, Sokol H, et al. Faecalibacterium prausnitzii and human intestinal health. Curr Opin Microbiol 2013; 16:255–261. doi: 10.1016/j.mib.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 26.Ferreira-Halder CV, Faria A, Andrade SS. Action and function of Faecalibacterium prausnitzii in health and disease. Best Pract Res Clin Gastroenterol 2017; 31:643–648. doi: 10.1016/j.bpg.2017.09.011. [DOI] [PubMed] [Google Scholar]
- 27.Robertson BR, O’Rourke JL, Neilan BA, Vandamme P, On SL, Fox JG, et al. Mucispirillum schaedleri gen. nov., sp. nov., a spiral-shaped bacterium colonizing the mucus layer of the gastrointestinal tract of laboratory rodents. Int J Syst Evol Microbiol 2005; 55 (Pt 3):1199–1204. doi: 10.1099/ijs.0.63472-0. [DOI] [PubMed] [Google Scholar]
- 28.Loy A, Pfann C, Steinberger M, Hanson B, Herp S, Brugiroux S, et al. Lifestyle and horizontal gene transfer-mediated evolution of Mucispirillum schaedleri, a core member of the murine gut microbiota. mSystems 2017; 2: doi: 10.1128/mSystems.00171-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Cossio LF, Fourrier C, Sauvant J, Everard A, Capuron L, Cani PD, et al. Impact of prebiotics on metabolic and behavioral alterations in a mouse model of metabolic syndrome. Brain Behav Immun 2017; 64:33–49. doi: 10.1016/j.bbi.2016.12.022. [DOI] [PubMed] [Google Scholar]
- 30.Krumbeck JA, Walter J, Hutkins RW. Synbiotics for improved human health: recent developments, challenges, and opportunities. Annu Rev Food Sci Technol 2018; 9:451–479. doi: 10.1146/annurev-food-030117-012757. [DOI] [PubMed] [Google Scholar]
- 31.Huang LS, Kong C, Gao RY, Yan X, Yu HJ, Wen B, et al. Analysis of fecal microbiota in patients with functional constipation undergoing treatment with synbiotics. Eur J Clin Microbiol Infect Dis 2018; 37:555–563. doi: 10.1007/s10096-017-3149-7. [DOI] [PubMed] [Google Scholar]


