Abstract
New genes are of recent origin and only present in a subset of species in a phylogeny. Accumulated evidence suggests that new genes, like old genes that are conserved across species, can also take on important functions and be essential for the survival and reproductive success of organisms. Although there are detailed analyses of the mechanisms underlying new genes’ gaining fertility functions, how new genes rapidly become essential for viability remains unclear. We focused on a young retro-duplicated gene (CG7804, which we named Cocoon) in Drosophila that originated between 4 and 10 Ma. We found that, unlike its evolutionarily conserved parental gene, Cocoon has evolved under positive selection and accumulated many amino acid differences at functional sites from the parental gene. Despite its young age, Cocoon is essential for the survival of Drosophila melanogaster at multiple developmental stages, including the critical embryonic stage, and its expression is essential in different tissues from those of its parental gene. Functional genomic analyses found that Cocoon acquired unique DNA-binding sites and has a contrasting effect on gene expression to that of its parental gene. Importantly, Cocoon binding predominantly locates at genes that have other essential functions and/or have multiple gene–gene interactions, suggesting that Cocoon acquired novel essential function to survival through forming interactions that have large impacts on the gene interaction network. Our study is an important step toward deciphering the evolutionary trajectory by which new genes functionally diverge from parental genes and become essential.
Keywords: retrogene evolution, gene network, development, Drosophila, lethality
Introduction
The genetic basis underlying the diversity of life remains a central question in evolutionary biology. Nucleotide substitutions or indels that change protein coding or regulatory sequences are often observed to contribute to functional, phenotypic, and behavioral polymorphism and divergence within and between species (e.g., Rost et al. 2004; Wittkopp et al. 2009; Linnen et al. 2013; Ding et al. 2016, reviewed in Wray [2007] and Barrett and Hoekstra [2011]). However, in addition to gradual changes at preexisting genes, gene composition turns over rapidly even between closely related species (e.g., Demuth et al. 2006; Zhang et al. 2010, 2011 and reviewed in Kaessmann [2010]) Indeed, although humans and chimpanzees have only diverged 1.5% in their orthologous coding sequences (Chimpanzee Sequencing and Analysis Consortium 2005), they differ by at least 6% of their gene content (Demuth et al. 2006).
The origination of new genes is an important evolutionary process contributing to the dynamic turnover of genes in genomes over the phylogeny. This dynamic gene turnover has been widely documented in Drosophila (Zhang et al. 2010), primates (Demuth et al. 2006; Zhang et al. 2011), and plants (Moore and Purugganan 2005) (reviewed in Kaessmann [2010]). Because of their recent origin, new genes are only present in a subset of species in a phylogeny and the prevailing view was that they have dispensable functions and are not essential to an organism’s fitness (e.g., Jacob 1977; Ashburner et al. 1999). However, recent evidence in a variety of eukaryotic species shows that new genes can quickly become essential for an organism’s viability and fertility (Chen et al. 2010; Cooper and Kehrer-Sawatzki 2011; Charrier et al. 2012; Dennis et al. 2012; Ding et al. 2012; Ranz and Parsch 2012; Reinhardt et al. 2013; Ross et al. 2013; VanKuren and Long 2018), suggesting that new genes unique to few species can also have essential functions similar to those of highly conserved genes.
One of the mechanisms by which new genes arise is through duplication, in which a copy of a gene is created through either DNA or RNA intermediates. Many evolutionary fates have been predicted for the duplicated (new) and original (parental) genes, grossly pseudofunctionalization, neofunctionalization, or subfunctionalization (Ohno 1970; Lynch and Conery 2000; Innan and Kondrashov 2010). Despite the convenient conceptual distinction, it is often challenging to distinguish between these alternative models due to the fact that the past evolutionary trajectories for duplicated and original genes are usually unknown or hard to decipher. Several in-depth analyses of the evolutionary steps leading to novel fertility functions of duplicated genes (Loppin et al. 2005; Heinen et al. 2009; Ding et al. 2010; Chen et al. 2012; Yeh et al. 2012) have shed light on the initial evolutionary processes leading to gained essential function of new genes. In contrast, few studies have focused on viability (e.g., Ross et al. 2013). Many genes responsible for essential viability functions (e.g., development of body plan in Drosophila embryos [Stauber et al. 1999]) are identified as ancient gene duplicates (reviewed in Chen et al. [2013]), suggesting new genes indeed can gain critical roles in the most essential and core functions of organisms. Yet, the past evolutionary trajectories leading to gained essential viability function of new genes and whether those are similar to those of essential fertility function still need further investigation.
A potential mechanism by which duplicated genes become essential is through forming multiple protein–protein or protein–nucleic acids interactions with preexisting genes and thus being integrated into the cellular genetic network. Indeed, new genes with essential fertility functions can locally or globally reshaped the regulatory network (Matsuno et al. 2009; Ding et al. 2010; Chen et al. 2012). Similarly, a new gene could quickly become essential for survival by gaining multiple interaction partners in a gene network. This hypothesis is consistent with the observations that genes with many interaction partners (hub genes) are more likely to have essential functions (Jeong et al. 2001; Yu et al. 2004; Batada et al. 2007; Blomen et al. 2015 and reviewed in Barabási and Oltvai [2004] and Barabási et al. [2011]). However, comparisons of ancient orthologous genes reported that the accumulation of gene–gene interactions is a slow evolutionary process (Kim et al. 2012). Whether and how, in a short evolutionary time, new genes can gain widespread impacts on gene interaction network is still an open question.
In this study, we characterized the evolutionary history and function of a young duplicated gene that quickly become essential for the survival of Drosophila melanogaster. This young gene (CG7804) duplicated from another essential gene (TBPH, also known as TDP-43 human homolog or CG10327) through retrotransposition between 4 and 10 Ma (Zhang et al. 2010) and is present in few Drosophila species. The especially young age of CG7804 offers a rare opportunity to investigate the initial evolutionary steps underlying new genes’ gaining essentiality. The parental gene, TBPH, is highly conserved among animals (Ayala et al. 2005; Li et al. 2010), and its null mutant was found lethal in Drosophila (Feiguin et al. 2009; Lin et al. 2011; Hazelett et al. 2012). Furthermore, a mutant allele in human has been associated with neuronal diseases (Sreedharan et al. 2008). TBPH is shown to bind to nucleic acids (Kuo et al. 2009), influencing the splicing (Buratti and Baralle 2001; Ayala et al. 2006; Bose et al. 2008) and transcriptional regulation (Ayala et al. 2008) of many genes. On the other hand, little is known about the duplicated gene, CG7804. We found that CG7804 evolved under much faster rates of amino acid substitutions than its parental gene. Despite its young age, functional analyses showed that CG7804 is essential for the survival of D. melanogaster at multiple developmental stages, including the critical embryonic stage. RNA-seq and Chromatin-Immunoprecipitation-sequencing (ChIP-seq) analyses suggest that CG7804 acquired essential function to survival through gaining DNA-binding targets at genes whose expression has other essential functions (i.e., mutant lethal) and/or at genes that engage in a large number of protein–protein/gene–gene interactions. In particular, CG7804 expression is essential in different tissues from those of TBPH and its influence on gene expression (gene activation) is opposite to that of the parental gene. Our study is an important step toward deciphering the evolutionary trajectories by which duplicated genes functionally diverge from its parental gene and become essential.
Results
Choice of Studying CG7804
Our study began with a question: Whether a young gene can quickly become essential for survival in a short evolutionary time through influencing gene interaction network? We specifically aimed to address one of the many plausible scenarios: A young gene acquired multiple nucleic acid–binding sites and thus has a global influence on gene regulatory network in nonreproductive tissues. Accordingly, we used several criteria to narrow down our candidate young duplicated genes whose evolutionary trajectories will help address the question. A duplicated gene needs to have a good annotated gene model (according to Flybase), have expression in nonreproductive tissues (according to modENCODE tissue expression study [Graveley et al. 2011; Brown et al. 2014]), and have a parental gene that binds nucleic acid. We would like to study a duplicated gene that is young, but present in D. melanogaster and other species, which allows estimation of its initial (right after origination) and continuous rates of molecular evolution. Accordingly, we focused on duplicated genes that are present in D. melanogaster, D. simulans, and D. sechellia, but absent in D. yakuba (between 4 and 10 My old) according to Zhang et al. (2010). Ten young duplicated genes met these criteria. We further narrowed down our list to CG7804 by choosing young genes that have lethal and/or semilethal phenotype in previous in vivo RNAi screens (Mummery-Widmer et al. 2009; Neely et al. 2010; Schnorrer et al. 2010). It is worth noting that we did not consider RNAi screen phenotypes in Chen et al. (2010) due to the potential artificial dominant phenotypic effects associated with the RNAi strains used in that particular study (Green et al. 2014).
CG7804 Evolved with Accelerated Rates of Amino Acid Substitution
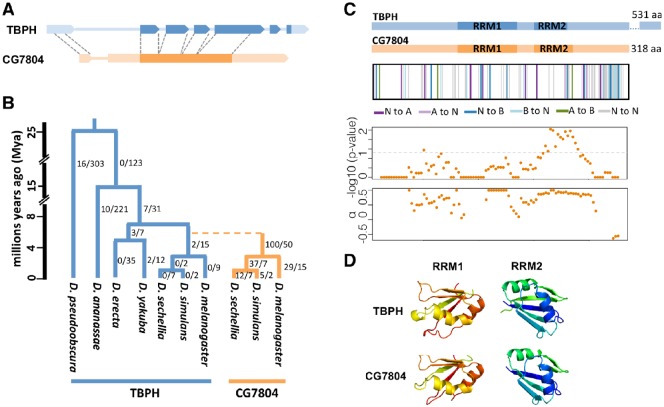
CG7804 (on chr3L) originated 4–10 Ma via an RNA intermediate (retrotransposition) from TBPH (on chr2R). Among the sequenced 12 Drosophila species (Clark et al. 2007), CG7804 is only present in D. melanogaster, D. simulans, and D. sechellia (Zhang et al. 2010) (fig. 1A). Despite its short evolutionary history, we detected a burst of 100 amino acid substitutions during the initial 2 My after the origination of CG7804 by using maximum-likelihood method (Yang 2007) (fig. 1B). Estimated dN/dS ratios on branches of the CG7804 clade (orange in fig. 1B) ranges from 0.38 (D. sechellia terminal branch) to 1.39 (branch leading to the D. simulans and D. sechellia CG7804 ancestor), which are much larger than those of corresponding species in the TBPH clade (blue in fig. 1B, ranges from 0.0001 [majority] to 0.11 [branch leading to D. yakuba and D. erecta TBPH ancestor]). Indeed, a likelihood ratio test found that a model with two dN/dS ratios fit the data better than the model with a single dN/dS ratio (likelihood ratio test, P value < 0.001), suggesting that CG7804 and TBPH evolve with different rates. The dN/dS ratio is also higher for CG7804 clade than for TBPH clade (dN/dS ratio = 0.54 [CG7804 clade] and 0.015 [TBPH clade]). In addition, unpolarized McDonald–Kreitman test (MK test) found a significant excess of amino acid substitutions between D. melanogaster and D. simulans in CG7804 (McDonald and Kreitman 1991; Fisher’s exact test, P value = 0.0011), suggesting that positive selection is acting on either or both of the branches leading to D. melanogaster and D. simulans CG7804. 75.8% of these CG7804 amino acid substitutions are inferred to have been fixed by positive selection (α, Smith and Eyre-Walker 2002). It is worth noting that due to selection on synonymous sites, the estimated proportion of adaptive amino acid substitutions should be considered as an overestimation (Matsumoto et al. 2016). On the contrary, we found no evidence suggesting that TBPH is under positive selection (unpolarized MK test, P value = 0.27). Overall, compared with its parental gene, CG7804 has had faster rates of amino acid substitution than its parental gene and is under positive selection.
Fig. 1.
Structural and evolutionary history of CG7804 and TBPH. (A) Exon–intron structure of TBPH (blue) and CG7804 (orange). Filled boxes represent exons (darker color, coding sequence; lighter color, UTRs), whereas lines represent introns. Because CG7804 originated through a retrotransposition event, it lacks most of the introns of TBPH and some of its noncoding sequences do not share homology with TBPH. (B) The duplication event of CG7804 from TBPH is denoted as a dashed line in the phylogeny. The clades of CG7804 and TBPH are in orange and blue, respectively. The number of amino acid substitutions to number of synonymous substitutions inferred by PAML (see text) is denoted at right to branches. The dating of the species phylogeny is from Obbard et al. [2012]. (C) The structures of the amino acid sequences of TBPH and CG7804 are shown at the upper panel. In the second panel, the divergent amino acids of CG7804 from TBPH are denoted as vertical lines. Different colors indicate different changes in amino acid chemical properties (N, noncharged; A, acidic; B, basic). The third and fourth panels show the results of sliding window MK test of CG7804 (window size 99 bp and step 9 bp), including −log 10 P value of the MK test and the estimated proportion of amino acid fixations between Drosophila melanogaster and D. simulans that were driven by positive selection (α). Note that the coordinates of four panels are aligned. (D) Predicted structures of the RRMs of TBPH and CG7804 are shown. The 3D structures of the first (RRM1) and the second (RRM2) RNA-recognition motifs were predicted using Phyre (Kelley et al. 2015). The rainbow color is from N (red) to C (blue) termini.
TBPH has two RRM (RNA-recognition motif) domains, which has been demonstrated to bind to both RNA and DNA (Kuo et al. 2009). CG7804 is predicted to possess the same nucleic acid–binding domains (amino acid 109–174, 194–239) with similar overall structures (fig. 1C and D). Between 4 and 10 My, large number of amino acid differences has accumulated between CG7804 and TBPH (28.6%, 91 out of 318 amino acids), including high divergence in the two RRM domains (18.8%, 21 out of 112 amino acids of the RRM domains). Many of these amino acid substitutions lead to changes in protein charges (fig. 1C). As a comparison, TBPH paralogs from D. melanogaster and D. yakuba, a species in which CG7804 is absent, diverge only 5.0% in amino acid sequences (27 amino acids). Furthermore, TBPH in D. melanogaster accumulated only two amino acid substitutions since the duplication event of CG7804 (fig. 1B).
Interestingly, sliding window MK test analysis found that regions with significant MK results and/or a large proportion of amino acid substitutions fixed by positive selection overlap with either of the two predicted RRM domains of CG7804 (fig. 1C). Furthermore, at least two amino acid differences between CG7804 and TBPH are likely to have substantial functional effects. A previous study (Lukavsky et al. 2013) experimentally identified that, in human TBPH, Met132 and the salt bridge between residues Arg151 and Asp247 are important for nucleic acid–binding affinity. Both of these residues are highly conserved among TBPH orthologs in animals (supplementary fig. S1, Supplementary Material online). However, in CG7804, these key residues were replaced by amino acids with different charges (hydrophobic Met132 was substituted with positively charged lysine and negatively charged Asp247 was substituted with the positively charged lysine, supplementary fig. S1, Supplementary Material online), both likely led to diverged nucleic acid–binding targets and/or functional role of CG7804 from those of TBPH. These results suggest that, while the parental gene remained highly constrained at the amino acid sequences, CG7804 quickly accumulated many amino acid substitutions, even at functionally important domains and sites.
CG7804 Is Essential for the Survival of D. melanogaster
Because our evolutionary genetic analysis supports positive selection acting on CG7804, we predicted that it gained function since its origination, instead of being pseudogenized. We employed GAL4/UAS system, and first used ubiquitous GAL4 drivers (Act5C-GAL4 and Tub-GAL4) to knockdown the expression of CG7804 and TBPH individually (see Materials and Methods). Consistent with previous studies (Feiguin et al. 2009; Lin et al. 2011; Hazelett et al. 2012), TBPH knockdown analysis found that the gene is essential for D. melanogaster survival (lethality rate: 97.5% with Act5C-GAL4 and 86.6% with Tub-GAL4, see Materials and Methods). Surprisingly, despite originating recently on an evolutionary timescale, expression knockdown of CG7804 also led to very low survival rate (lethality rate: 94.7% with Act5C-GAL4 and 95.5% with Tub-GAL4). For CG7804 knockdown, most of the lethality happens at the stages between pupae and adults (fig. 2A). Indeed, we found that flies could not develop pass the pharate adult stage and identified many eclosion lethal incidences (i.e., flies could not emerge and were stuck and dead half way in pupal cases, fig. 2B). Many other flies that eclosed were dead in Drosophila culture media. A detailed tracking of pupa identified that, while 91.9% of the wildtype pupa successfully eclosed and survived, only 17.7% of the CG7804 knockdown individuals who reached the pupal stage did so (fig. 2C). To test if the high lethality associated with CG7804 knockdown is caused by flies unable to first open the pupal cases, we manually removed the pupal cap for both CG7804-knockdown and wildtype flies. Manual removal of pupal cap led to a slight increase in pupal lethality for wildtype flies (increased from 5% to 12.5%; fig. 2C). On the other hand, pupal cap removal decreased the lethality rate of CG7804-knockdown pupa from 76.6% to 60% (fig. 2C). Yet, even after considering the increased lethality due to pupal cap removal (∼7.5% in wildtype), 52.5% CG7804 knockdown pupa still did not reach adulthood (fig. 2C).
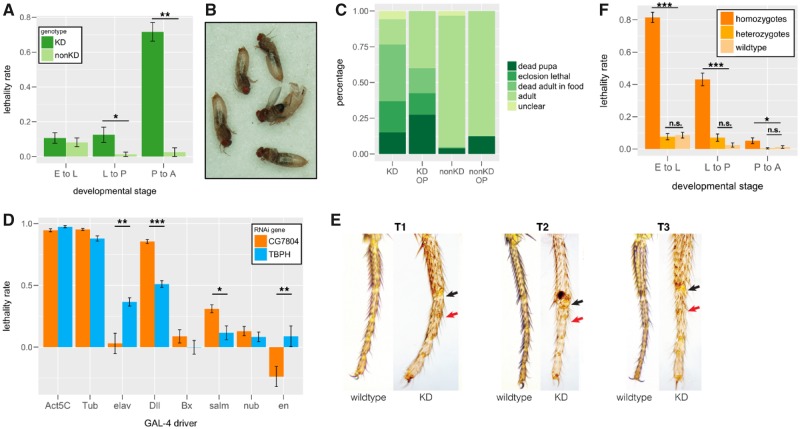
Fig. 2.
Stage-specific lethality associated with CG7804 knockdown and knockout. (A) Expression knockdown of CG7804 using Tub-GAL4 driver results in different lethality rates at different developmental stages. (B) Expression knockdown of CG7804 leads to eclosion lethal. (C) The outcome of pupae with CG7804 expression knockdown. (D) CG7804 expression is essential in different tissues from those of its parental gene, TBPH. Lethality rate is significantly different between CG7804 and TBPH knockdown when using elav, Dll, and en GAL4 drivers. Because the lethality rate is estimated relative to wildtype genotype (see Materials and Methods), negative lethality rate (e.g., en driver knocking down CG7804) means higher survival rate of that particular genotype than the wildtype. (E) Expression knockdown of CG7804 using Dll-GAL4 driver results in completely fused leg joint (red arrow) or semifused leg joint (black arrow). Legs of wildtype (Dll-GAL4 driver strain) individuals are shown side by side with those of knockdown individuals. T1–T3 are first, second, and third legs, respectively. (F) CG7804 knockout homozygotes have significantly higher lethality rate from embryo to larva and from larva to pupa than wildtype individuals. CG7804 knockout heterozygotes have similar lethality rates to those of wildtype individuals. E, embryo; L, third instar larvae (L3); P, pupae. KD, individuals with CG7804 knockdown genotype; nonKD, wildtype individuals; OP, pupae with pupa cased removed (open pupae). Mann–Whitney U test: *P < 0.05, **P < 0.01, and ***P < 0.001.
In addition to different developmental stages, we also investigated in which tissues the expression of CG7804 is essential. According to the modENCODE tissue expression study (Graveley et al. 2011; Brown et al. 2014), TBPH is ubiquitously expressed, whereas CG7804 has high expression mainly in imaginal discs and male-specific tissues. We used tissue-specific GAL4-drivers to knockdown the expression of CG7804 and TBPH in tissues that are not sexually dimorphic. Expression knockdown of TBPH using neuronal-specific elav GAL4-driver leads to much lower survival rate than expression knockdown of CG7804 using the same driver (fig. 2E), which is consistent with previously identified role of TBPH in neuronal functions (Feiguin et al. 2009; Hazelett et al. 2012). On the other hand, expression knockdown of CG7804 using Dll (leg imaginal disc) and salm (imaginal discs) led to lower survival rate than that of TBPH knockdown with the same GAL4-drivers (fig. 2D). Interestingly, expression knockdown of CG7804 at leg imaginal disc (using Dll GAL4 driver) leads to fused leg joints (fig. 2E), which is not observed in TBPH knockdown flies with the same GAL4-driver (supplementary fig. S3, Supplementary Material online). These disparities in tissue-specific knockdown effects suggest that the expression of CG7804 is essential for viability at different tissues from those of its parental gene, TBPH.
We used CRISPR/CAS9 system (Cong et al. 2013; Gratz et al. 2013; Kondo and Ueda 2013) to generate null mutant of CG7804 (see Materials and Methods). Consistent with results using GAL4/RNAi expression knockdown, CG7804 knockout homozygotes have extremely high lethality rate when compared with CG7804 knockout heterozygotes from the same cross (99.4%, see Materials and Methods). In addition, another mutant of CG7804 that was generated by a different approach (insertion of a MIMIC construct in exon [Venken et al. 2011]) also shows extremely high lethality (99.21%), and the two mutants cannot complement each other (supplementary table S1, Supplementary Material online). It is worth noting that CG7804 knockout heterozygotes have similar survival rates as those of wildtype individuals (fig. 2F), suggesting that the effect of CG7804 knockout on viability is largely recessive. Interestingly, the lethality associated with CG7804 knockout happened at earlier stages: mainly at embryo to larva, and larva to pupa stages (fig. 2F), and we did not observe eclosion lethal phenotype with CG7804 knockout pupa (see Discussion for potential causes). Despite low, CG7804 expression at embryonic stage is detected by our RNA-seq experiment (see below), modENCODE developmental time course RNA-seq (Graveley et al. 2011), and Reverse transcription polymerase chain reaction (RT-PCR) assay (supplementary fig. S4, Supplementary Material online). Overall, both our expression knockdown and null mutant analyses support the conclusion that CG7804 is highly essential for the survival of D. melanogaster, despite being young and only present in few species.
Given the high expression of CG7804 in male reproductive tissues (Graveley et al. 2011; Brown et al. 2014), it is natural to wonder whether CG7804 also gained essential functions for male fertility and, like other new genes with gained essential functions in male fertility (e.g., Ding et al. 2010; Chen et al. 2012; VanKuren and Long 2018), whether that could have been the main driving force for CG7804’s fast molecular evolution. We used germline-specific GAL4-driver (Bam-GAL4) to knockdown the expression of CG7804 and TBPH in male testis and tested whether that influence male fertility. While males with CG7804 knockdown have significantly fewer offspring than males without, similar effect was observed for TBPH knockdown (supplementary fig. S2, Supplementary Material online). In fact, there is no difference between males with CG7804 or TBPH knockdown (Mann–Whitney U test, P = 0.970), arguing against that the role of CG7804 in male fertility is a gained function and that drives the positive selection on its amino acid sequences.
CG7804 Knockout Perturbs Expression of Genes with Important Developmental Functions
To investigate the mechanisms by which CG7804 is essential for the survival of D. melanogaster at multiple developmental stages, we sequenced and compared the transcriptomes of CG7804 knockout and wildtype mixed-sex individuals at embryonic, larval, and pupal stages (see Materials and Methods). Comparing between CG7804 knockout and wildtype individuals, 20.0% (embryo), 6.3% (larva), and 8.5% (pupa) of the genes analyzed have significantly differential expression (false discovery rate [FDR] < 0.05) (fig. 3A–C). The observed large number of differentially expressed genes suggests that CG7804 has a global influence on the transcriptome. It is worth noting that the rate of development from eggs to adults is not significantly different between CG7804 knockout and wildtype individuals (supplementary fig. S5, Supplementary Material online), suggesting that the observed global transcriptome differences between these two genotypes are not merely driven by shifts in the developmental rates.
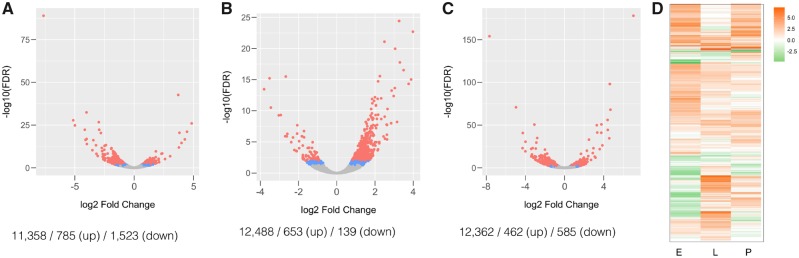
Fig. 3.
Differential expression upon CG7804 knockout. Volcano plots for the log 2 fold change in expression level (x axis) and −log 10 FDR (y axis) for the embryonic stage (A), the larval stage (B), and the pupal stage (C). Red dots represent genes that are differentially expressed with FDR < 0.01 and blue dots for FDR < 0.05. Gray dots represent genes that are not differentially expressed. Numbers under each panel are the number of genes analyzed/number of upregulated genes/number of downregulated genes (D) heatmap for the log 2 fold change in expression level at three developmental stages (E, embryo; L, larva; P, pupa). Each horizontal row represents one gene. Oranges are for positive log 2 fold change (i.e., upregulated with CG7804 knockout), whereas greens are the opposite.
The especially large number of genes influenced at the embryonic stage is consistent with the observed strong embryonic lethality associated with CG7804 knockout. At the embryonic stage, the number of downregulated genes (1,523) in CG7804 knockout is much higher than that of upregulated genes (785, binomial test, P < 10−16, fig. 3A). These downregulated genes are enriched for Gene Ontology (GO) of chitin-related processes (e.g., chitin metabolism and catabolism, chitin-based cuticle development [supplementary table S2, Supplementary Material online]). On the other hand, upregulated genes are enriched for mitosis-related processes (e.g., chromosome condensation and separation, DNA and centrosome replication, and DNA repair, supplementary table S2, Supplementary Material online). In contrast to the observations at the embryonic stage, CG7804 knockout at larval stage leads to more upregulated genes, which are surprisingly also enriched with chitin-related processes (653 [up] vs. 139 [down], binomial test, P < 10−16, fig. 3B, supplementary table S2, Supplementary Material online). Finally, there are slightly fewer up- than down-regulated genes at the pupal stage with CG7804 knockout (462 [up] vs. 585 [down], binomial test, P = 0.00016, fig. 3C). Genes with most significant downregulation again have functions in cuticle development (Cpr72Eb, Lcp65Ad). Downregulated genes are enriched with function in imaginal disc-derived morphogenesis, which is consistent with our findings from tissue-specific expression knockdown analysis (see above, supplementary table S2, Supplementary Material online). Chitin is the basis of critical structures of insects (e.g., the exoskeleton and trachea), and insect growth and morphogenesis heavily depend on the synthesis and remodeling of chitin-based structures (Merzendorfer and Zimoch 2003). It is expected that these process will be especially important for the development of embryos (develop into larvae) and pupa (develop into adults), which is consistent with our GO enrichment analyses at these two developmental stages. On the other hand, the upregulation of genes with chitin-related functions in CG7804 knockout larva is intriguing, which could result from the compensatory regulation in response to the reduced expression of chitin-related genes at embryonic stages. Interestingly, among all genes analyzed, there seems to be little consistency in the directionality of expressional changes across developmental stages (fig. 3D), suggesting that the global influence of CG7804 on the transcriptome is contingent on the gene expression network at specific developmental stages.
CG7804 Gained Novel Functions in Gene Regulatory Network
The two predicted RRM domains, which were shown to bind to both DNAs and RNAs in protein TBPH (Kuo et al. 2009), harbor substitutions that are unique to CG7804 and might have significant functional consequences (see above). Accordingly, we hypothesized that CG7804 quickly become essential through acquiring new nucleic acid–binding targets and/or new functional roles in gene regulation (upregulation/downregulation). Here, we focused on the evolution of potential DNA-binding targets of protein CG7804 and generated transgenic D. melanogaster strains that express GFP-tagged CG7804 and GFP-tagged TBPH under endogenous cis-regulatory sequences to test our hypothesis (see Materials and Methods). It is worth noting that GFP-tagged CG7804 is able to rescue CG7804 null mutants (supplementary table S3, Supplementary Material online), suggesting that GFP-tagging does not perturb the native functions of CG7804 proteins and that the lethality of CG7804 null strains is indeed the result of CG7804 disruption, instead of other unidentified mutations.
The GFP-tagged CG7804 has a nuclear localization, which is similar to that of TBPH and consistent with the predicted function of CG7804 in nucleic acid binding (fig. 4A). We performed ChIP-seq targeting GFP-tagged CG7804 and GFP-tagged TBPH to identify their genomic binding sites at the pupal stage, at which we found high lethality associated with expressional knockdown of CG7804. There are 553 genomic regions enriched with CG7804 binding (Irreproducible rate [IDR] < 0.001, see Materials and Methods), and the majority of them overlap with at least one gene (470, 85.0%). This is significant when compared with randomly selected genomic windows with matching chromosomal distributions and sizes (Permutation test, P = 0.016). Over a quarter of the remaining enriched region (28, 32.5%) are within 2-kb upstream of transcription start sites, and accordingly also have potential roles in regulating gene expression. In total, we identified 649 genes (4.66% of all annotated protein-coding genes) that either are in CG7804 enriched region (628 genes) or have CG7804-binding enrichment in their upstream 2-kb region (358 genes), with 337 genes having CG7804-binding enrichment at both their upstream and gene sequences. On the other hand, 1,864 genes have binding enrichment of the parental gene, TBPH, either over gene body and/or in their upstream 2-kb region.
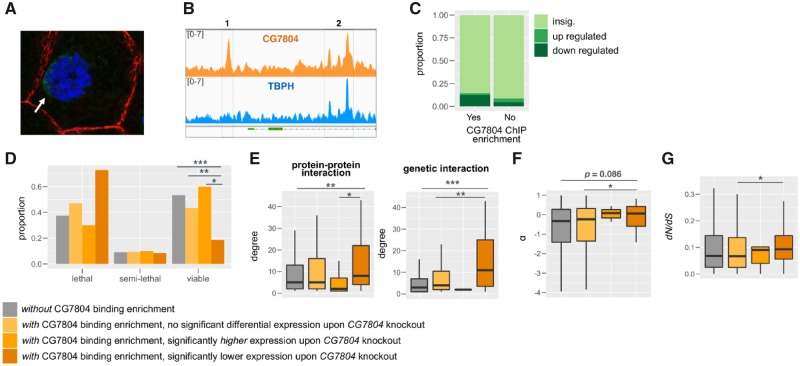
Fig. 4.
Genes with CG7804-binding enrichment are different from other genes in the genome. (A) CG7804 has nuclear localization in the salivary gland of third instar larva. Green, CG7804; blue, DNA; red, cytoskeleton. (B) Examples of a binding region that is unique to CG7804 (1) or is shared between CG7804 and TBPH (2). This example shows that even for some genes bound by both paralogs, CG7804 may still have gained unique binding sites (here, upstream of an essential gene, MyC). (C) Bar plots for the proportion of genes that are differentially expressed (either downregulated or upregulated) for genes with or without CG7804-binding enrichment. Different shade of green colors is whether a gene is differentially expressed upon CG7804 knockout. (D–G) Comparing (D) known mutant phenotype, (E) degree (number of protein–protein interaction/genetic interaction a gene is involved in), (F) α (the proportion of adaptive amino acid substitution), and (G) dN/dS ratio (Drosophila melanogaster linage-specific substitution rates) between genes with/without CG7804-binding enrichment and differential expression upon CG7804 knockout. Three comparisons were performed: 1) between genes with and without CG7804-binding enrichment, 2) among genes with CG7804-binding enrichment, genes with and without differential expression upon CG7804 knockout, and 3) among genes with CG7804-binding enrichment and differential expression upon CG7804 knockout, genes with increased and decreased expression. Mann–Whitney U test: *P < 0.05, **P < 0.01, and ***P < 0.001.
A potential evolutionary scenario is that CG7804 inherited TBPH-binding sites and thus is functionally redundant to TBPH. Despite the fact that a large fraction of genes with CG7804-binding enrichment also has TBPH-binding enrichment (529, 81.5%), 120 genes only have CG7804-binding enrichment. On the other hand, genes with both CG7804- and TBPH-binding represent a smaller proportion of TBPH-binding genes (28.4%), and there are a large number of genes with only TBPH-binding (1,336). The much larger number of genes with TBPH-unique binding than CG7804-unique binding and the highly conserved function of TBPH across the phylogeny suggest a scenario that, after duplication, CG7804 lost many of the ancestral binding sites and gained new binding sites for 120 genes. However, without knowledge of TBPH-binding profiles in species without CG7804, we could not exclude the possibility that TBPH lost the ancestral binding sites of these 120 genes, resulting in them being CG7804-binding specific. Interestingly, even for genes that are enriched for the binding of both CG7804 and TBPH, the location of the enrichment region may diverge, which suggests that the paralogs may have differential regulation of even the same target genes (fig. 4B).
To investigate if CG7804-binding enrichment over the genome has functional consequences, in particular influencing the transcript levels of genes with binding enrichment, we compared our ChIP-seq results with transcriptomes of CG7804 knockout experiment at the pupal stage (see above). Genes that have CG7804-binding enrichment are also more likely to have a significant differential expression (FDR < 0.05, see above) between CG7804 knockouts and wildtype pupa (Fisher’s exact test, P < 10−5, odds ratio = 1.80, fig. 4C), suggesting a direct effect of the CG7804 binding on expression. In particular, among genes that have CG7804-binding enrichment and are differentially expressed between CG7804 knockout and wildtype pupa, there is an excess of genes that have lowered expression (Fisher’s exact test, P < 10−8, odds ratio = 5.5, fig. 4C). For other genes with CG7804-binding enrichment but not identified as with significant differential expression with our stringent threshold upon CG7804 knockout (i.e., genes with FDR ≥ 0.05), they still have lower q-value (i.e., adjusted P value for controlling for multiple tests, Mann–Whitney U test, P < 10−6) and smaller log 2 fold change in expression (i.e., more negative, suggesting lowered expression in knockouts than in wildtype; Mann–Whitney U test, P = 0.0080). These results indicate that the role of CG7804 in gene regulation may be predominantly activation of gene expression, which is opposite to the role of TBPH in gene regulation (mainly downregulation, Hazelett et al. 2012).
A potential mechanism by which CG7804 becomes essential is through influencing the regulation of other essential genes in Drosophila. Consistently, we observed that genes with CG7804-binding enrichment are more likely to have known lethal or semilethal phenotypes (shown by either knockout mutant or expression knockdown, see Materials and Methods) than other genes (Fisher’s exact test, P < 10−8, odds ratio = 1.66, fig. 4D). Furthermore, among genes with CG7804-binding enrichment, genes that are differentially expressed upon CG7804 knockout are even more likely to have known lethal or semilethal phenotype (Fisher’s exact test, P = 0.0034, odds ratio = 2.35, fig. 4D), and this is mainly driven by genes that have lowered expression upon CG7804 knockout (comparing between genes with significant increased or decreased transcript levels, Fisher’s exact test, P = 0.011, odds ratio = 6.31, fig. 4D). It has been repeatedly observed that hub genes, which have many interaction partners in gene–gene interaction networks, tend to have essential functions (Jeong et al. 2001; Yu et al. 2004; Batada et al. 2007; Blomen et al. 2015). Consistent with this view and our hypothesis, genes that have CG7804-binding enrichment have more experimentally validated protein–protein interactions or reported genetic interactions than other genes in the genome (degree in PPI network, Mann–Whitney U test, P < 0.0016; degree in genetic interaction network, Mann–Whitney U test, P < 10−6, fig. 4E). In particular, genes that have CG7804-binding enrichment and lowered expression upon CG7804 knockout have even more genetic interaction partners than other genes with CG7804-binding enrichment but no differential expression (degree in genetic interaction network, Mann–Whitney U test, P = 0.0029, fig. 4E). These observations invite the conclusion that CG7804 becomes essential mainly through regulating the expression of other essential and/or hub genes.
Interestingly, genes with CG7804-binding enrichment are more likely to be under adaptive evolution (shown by rejection of MK test on the divergence between D. melanogaster and D. simulans and an excess of amino acid substitutions compared with the null expectation) than other genes (Fisher’s exact test, P = 0.006, odds ratio = 1.54). They also have marginally significantly larger α (proportion of amino acid substitutions that are under adaptive evolution, Mann–Whitney U test, P = 0.086, fig. 4G), but no faster rates of nonsynonymous substitutions on the D. melanogaster branch (dN/dS ratio, Mann–Whitney U test, P = 0.90, fig. 4G). In particular, among genes with CG7804-binding enrichment, those that have differential expression upon CG7804 knockout have both more adaptive substitutions (α, Mann–Whitney U test, P = 0.041, fig. 4F) and faster rates of protein evolution (dN/dS ratio, Mann–Whitney U test, P = 0.027, fig. 4G) than those that show no differential expression. Our observation revealed that genes with CG7804-binding enrichment, especially those whose expression is expected to be upregulated by CG7804 binding (i.e., have lowered expression upon CG7804 knockout), are more likely to have known lethal phenotype, have multiple interaction partners in protein–protein and/or genetic interaction network, and are more often under adaptive evolution.
GO enrichment analysis shows that genes with CG7804-binding enrichment are significantly enriched with functions related to development (P value < 0.05 after multiple test correction, supplementary table S4, Supplementary Material online). This includes imaginal disc-derived morphogenesis, which is consistent with the observed lethality of tissue-specific knockdown of CG7804 in imaginal discs (fold enrichment = 4.47), as well as eye, trachea, and neuronal development. Interestingly, genes with CG7804-binding enrichment are also enriched with protein binding (fold enrichment = 1.69) and transcription factor activity (fold enrichment = 4.34), both of which are expected to influence a large number of genes and have extensive functional impacts. Even more, genes with CG7804-binding enrichment and lowered expression upon CG7804 knockout show even stronger enrichment for imaginal disc-related morphogenesis (fold enrichment = 6.56) and transcription factor activity (fold enrichment = 4.65, supplementary table S5, Supplementary Material online).
There are 120 genes that only have CG7804-binding enrichment but no TBPH-binding enrichment (18.49% of genes with CG7804 binding). These genes are not different in terms of their expression, protein/genetic interaction network, and evolutionary rates if compared with those that have binding enrichment for both CG7804 and TBPH. Nevertheless, a large fraction of these genes also show differential expression upon CG7804 knockout (19%) and a majority of them have downregulated gene expression (87.5%), which support the regulatory importance of these CG7804-unique binding enrichment.
Discussion
Every gene in an organism’s genome must have arisen at some time point in the past. The origination of new genes is a major contributor to the dynamic turnover of genes over evolutionary time. Despite being young and restricted to few species on a phylogeny, new genes have diverse essential functions and they underlie important adaptive evolution (reviewed in Taylor and Raes [2004], Kaessmann [2010], Ding et al. [2012], Chen et al. [2013], Long et al. [2013], and Ventura and Long [2017]). Yet, how, in a short evolutionary time, new genes become essential is still an open question, and detailed functional dissection of new genes is a natural and important step to address this overarching question.
Despite its young age, our focused duplicated gene, CG7804, is found essential for D. melanogaster viability in both expression knockdown and gene knockout experiment. Interestingly, we identified eclosion lethal (i.e., flies got stuck halfway in pupal cases) as well as fused leg joints phenotype associated with CG7804 knockdown, and the former led to our naming this gene “Cocoon.” Part of the essentiality of Cocoon is likely the result of the accelerated accumulation of amino acid substitutions (dN/dS) compared with its parental gene and the large proportion of adaptive amino acid substitutions (α) between closely related Drosophila species. RNA-seq analysis revealed that Cocoon has a widespread effect on transcriptome at multiple developmental stages, consistent with the observed essentiality of the gene through development. In particular, Cocoon knockout leads to significant changes in expression of a fifth of genes in embryo, in which the functional consequence is expected to impact both somatic and germline cells and has a long-lasting effect through development. Importantly, our ChIP-seq analysis at pupal stage suggests that Cocoon rapidly becomes essential by forming interactions with and regulating the expression of multiple preexisting genes, in particular those that have known lethal phenotypes, have many protein–protein/genetic interaction partners (hub genes), and/or are under adaptive evolution. To the best of our knowledge, this is the first study that detailed the evolutionary steps for how a young (between 4 and 10 My old) new gene becomes essential for viability.
Interestingly, although we identified that Cocoon binding predominantly results in upregulation of targeted genes, the role of TBPH in expression regulation was more often negative (i.e., downregulation) in both flies (Hazelett et al. 2012) and mice (Polymenidou et al. 2011). It is intriguing that such drastic changes could have evolved in a short evolutionary time. This contrasting effect on gene expression could have resulted from different protein interaction partners of CG7804 and TBPH. TDP-43 (human version of TBPH) is known to interact with a large suite of proteins (Freibaum et al. 2010) through its C-terminal region (Buratti et al. 2005), which was lost during the duplication event of CG7804. Furthermore, CG7804’s accumulation of amino acid substitutions is not restricted to the two nucleic acid–binding domains, which could also lead to CG7804’s interactions with a different suite of proteins from that of TBPH. These observations suggest a scenario that while TBPH interacts with repressive proteins that together downregulate their binding targets, CG7804 has evolved to interact with other proteins that instead promote upregulation of gene expression. Alternatively, although the downregulation of gene expression through TBPH binding may be functionally important for the majority of its target genes, this effect at some genes could be deleterious. The accelerated evolution of CG7804 and its opposite effect on gene expression when compared with TBPH could have been driven by selective pressure to resolve this “conflict” (Pavlicev and Wagner 2012). It is worth noting that the resolution of such conflict could be a continuous process. For instance, amino acid changes that were fixed by positive selection right after the origination of CG7804 could have other negative pleiotropic effects. Accordingly, through time, other amino acid substitutions accumulated. This could explain why, instead of the branch leading to the common ancestor of all CG7804, estimated dN/dS ratio is the highest and above one on the branch leading to D. simulans and D. sechellia CG7804 ancestor and why CG7804 amino acid divergence between D. melanogaster and D. simulans is also under positive selection.
According to network theory, “hubs” are central nodes with a larger number of links, and the perturbation of hubs is expected to have more widespread influence on the overall network than the perturbation of peripheral nodes with few links (for biological networks, reviewed in Barabási and Oltvai [2004] and Barabási et al. [2011]). Indeed, in a gene–gene interaction network, hub genes are often essential (Jeong et al. 2001; Yu et al. 2004; Batada et al. 2007; Blomen et al. 2015). Comparisons of ancient orthologous genes found that the essentiality of genes can evolve via gradual increases in the number of interactions (Kim et al. 2012). Similarly, genome-wide analyses in yeast, mouse, and human concluded that, on average, the integration of new genes into preexisting gene–gene interaction network is a gradual process (Capra et al. 2010; Abrusán 2013; Zhang et al. 2015). However, a young transcription factor with novel fertility functions was found to massively reshape the gene interaction network by preferentially binding to and influencing the expression of genes with sex-biased expression (Chen et al. 2012). Similarly, we found that Cocoon, a duplicate from another nucleic acid–binding protein TBPH (Kuo et al. 2009), becomes essential through acquiring many interaction partners in a short evolutionary time. In particular, the binding of Cocoon to other essential and/or hub genes may further expedite the evolution of gained essentiality. These observations suggest a notable exception to the common view that the accumulation of genetic interaction is a slow process, and that new genes play a minimum role in essential cellular functions. This may be especially true for certain classes of genes, such as transcription factors. New genes with these functions may be more likely to acquire multiple new interactions quickly and influence the expression of hundreds of genes.
Although analyses using gene knockout and expression knockdown both supported a strong role of Cocoon in viability, the developmental stages at which Cocoon has the strongest influence and the phenotypic effect vary between perturbation methods. Specifically, Cocoon knockout results in strongest lethality at the embryonic stage, whereas Cocoon knockdown did not show a viability effect until pupal stage. This could be due to the fact that RNAi expression knockdown is rarely complete, as suggested by our RT-PCR assay (supplementary table S6, Supplementary Material online). Reduced, but nevertheless nonzero, Cocoon expression in an RNAi experiment could have been sufficient for its vital function at embryonic and larval stages. On the other hand, incongruence between knockout and knockdown phenotypes has been widely observed in other systems (De Souza et al. 2006; Daude et al. 2012; Kok et al. 2015). A recent zebrafish study observed that genetic compensation in knockout mutants, but not in knockdown individuals, is one of the explanations (Rossi et al. 2015). Such finding suggests that knockout and knockdown experiment should be viewed as complementing approaches. Still, in our experiment, the majority of CG7804 knockout flies died before pupa. The survived knockout individuals may have genetic compensation that influence their phenotypes at later developmental stages, and this could potentially explain why we did not observe high lethality and eclosion lethal with knockout flies that survived past the larval stage. Such scenario would suggest that our transcriptome comparisons between knockout and wildtype flies might have provided a genetic compensation-biased view of CG7804 functions.
The large overlap of binding targets between Cocoon and TBPH invites the conclusion that, since its origination, Cocoon “inherited” the binding targets of TBPH and from its parental gene. However, our evolutionary and functional analyses suggest that Cocoon might have evolved through a more complex process. Tissue-specific expression knockdown analysis found that Cocoon and TBPH support essential functions in different tissues, suggesting that regulatory changes of CG7804 are critical for its gained essentiality. On the other hand, the accumulated amino acid substitutions of Cocoon since its originations, especially at functionally important sites and domains, may also play an important role in the evolution of its function. We found an appreciable number of genes (18.49% of genes with CG7804 binding) with only Cocoon binding. Moreover, Cocoon binding predominantly leads to upregulation of targeted gene, a contrasting effect to the downregulation of gene expression associated with TBPH binding (Polymenidou et al. 2011; Hazelett et al. 2012). These are in contrast to the highly conserved protein sequence and molecular function of TBPH, not only within Drosophila but also across animals. In fact, the human version of TBPH (TDP-43) is able to complement the loss of function TBPH mutant in D. melanogaster (Li et al. 2010), suggesting that the function of TBPH likely has not changed since the origination of Cocoon. Overall, our observations reveal a scenario that Cocoon evolved new function (neofunctionalized) since its duplication from TBPH both through regulatory changes (gained essential expression at different tissues from that of TBPH) and coding sequence changes (gained DNA-binding targets and different functional role in gene expression regulation). Our study provides a novel view for how duplicated new genes can quickly become essential for viability.
Materials and Methods
Evolutionary Genetic Analysis of CG7804 and TBPH
We used coding sequence of CG7804 and TBPH of 12 Drosophila species from Clark et al. (2007) and aligned using Clustal (Sievers et al. 2011), followed by manual curation (see supplementary fig. S6, Supplementary Material online, for alignment). We used CODEML program in PAML (v 4.9, Yang 2007) to estimate dN/dS ratio on each branches and ran a likelihood ratio test (with one degree of freedom) to investigate whether a branch model with two dN/dS ratios (i.e., the two genes evolved with different rates; model = 2) fits better than a branch model with single dN/dS ratio (i.e., the two genes evolved with the same rates; model = 0) assuming F3X4 model of codon frequencies. Tree was specified as (((((D. mel CG7804, (D. sim CG7804, D. sec CG7804)) $1, (D. mel TBPH, (D. sim TBPH, D. sec TBPH))), (D. yak TBPH, D. ere TBPH)), D. ana TBPH), D. pseudo TBPH). The log likelihoods for the two models are −3,803.08 (one dN/dS, model = 0) and −3,663.63 (two dN/dS, model = 2). For MK tests, we used D. melanogaster polymorphism data from Lack et al. (2015) and used D. simulans allele from Hu et al. (2013) as outgroup to perform unpolarized tests. The number of observed nonsynonymous polymorphic sites, nonsynonymous fixed sites, synonymous polymorphic sites, and synonymous fixed sites are respectively 16, 66, 19, and 19 (CG7804) and 15, 4, 43, and 25 (TBPH).
Domains of CG7804 were predicted by Pfam (Finn et al. 2016) and the tertiary structures of predicted domains were computed using Phyre (v 2, Kelley et al. 2015). To have a broad view of the evolutionary conservation among TBPH orthologs, protein sequences from 49 species were retrieved from NCBI, aligned using Clustal (Sievers et al. 2011), followed by manual curation. We then compared the residues found at specific functional sites that were tested experimentally by (Lukavsky et al. 2013) with the diverged residues only found in CG7804.
Generation of Transgenic Strains and Mutants
Design of guide RNA, injection of guide RNA, and screen for CRISPR mutants were done by Genetic Service Inc. (Sudbury, MA). CG7804 mutant has 2-bp deletion in the coding sequence, which is confirmed by Sanger sequencing. Detailed CRISPR design and sequencing confirmation are in supplementary text S1, Supplementary Material online. To further confirm that our CRISPR mutant is a true null mutant, we used another mutant of CG7804 to perform complementation tests. We used a strain (BDSC 36014, supplementary table S7, Supplementary Material online) that has a MIMIC construct (Venken et al. 2011) inserted in the coding sequence of CG7804 and is likely a null allele of CG7804. The presence of the MIMIC insertion was confirmed by PCR (see, supplementary table S8, Supplementary Material online, for primer information). We found CRISPR and the MIMIC strain do not complement each other, which suggests CRISPR CG7804 is a null mutant of CG7804 (supplementary table S1, Supplementary Material online). We balanced CG7804 mutants over balancer chromosomes with ubiquitously expressed GFP for developmental stage-specific lethality analysis (see supplementary table S7, Supplementary Material online, for strains used). It is worth mentioning that, for those few CG7804 knockout individuals that survived to adult, we were able to detect expression of CG7804 either through RT-PCR or through RNA-seq. However, these detected transcripts of CG7804 all have the same frameshift deletion.
Constructs of GFP-tagged CG7804 and GFP-tagged TBPH were generated using BAC-recombineering and P(acman) BACs CH322-116J04 (CG7804, 22,283 bp) and CH321-59A22 (TBPH, 79,765 bp) (Venken et al. 2009) and cloning vector pAV007 (GeneBank # KF411445) by Genome Engineering Core of the University of Chicago (Chicago, IL). The pAV007 was inserted directly after CGAACCAGAGCAGCGGATCTCAAAACGCCGCGGAGAAGTCAAACTTTCTT in CH321-59A22 (TBPH) and after GCATGCATTCATTTAATCCACATGGTTACCAAATGAATCGCGTCATGAAC in CH322-116J04 (CG7804) to generate C-termini GFP-tagged proteins. These constructs were introduced into the genomes of strains with attP docking sites (Bateman et al. 2006; Bischof et al. 2007) (see supplementary table S7, Supplementary Material online, for strains used). Embryo injection of constructs, screening, and balancing were done by Genetivision (Houston, TX). Insertions of BAC constructs were confirmed by PCR following (Venken et al. 2009) (see supplementary table S8, Supplementary Material online, for primer sequences). The expression of GFP-tagged transgenes was confirmed by RT-PCR (supplementary fig. S7, Supplementary Material online). Total RNA was extracted from third instar larvae, adult heads, and testes from TBPH- and CG7804-GFP strains, treated with DNAse (Qiagen), and reverse-transcribed (Invitrogen SuperScript III First-Strand Synthesis Mix). cDNA was used to carry out PCR using forward primers specific to each focused gene, and a reverse primer annealing to the GFP tag (see supplementary table S8, Supplementary Material online, for primer sequences). Additional controls for the PCR included the RNA sample from the larvae without reverse-transcription (to ensure no genomic DNA contamination), cDNA from another genotype (w1118), and water.
Essentiality Analysis
Virgin females of RNAi strain (homozygous) were crossed to males of GAL4 strain. The GAL4 strain is heterozygous for GAL4 construct, which is balanced with visible markers and/or construct of ubiquitously expressed GFP. Expression knockdown and wildtype offspring were recognized by visible markers (adult) or presence/absence of GFP (embryo, larva, and pupa). All comparisons are within crosses (RNAi/+; GAL4/+ vs. RNAi/+; +/balancer). For each cross, the expected number of knockdown individuals was estimated using the number of individuals with other genotypes and with the assumption that alleles were inherited following Mendelian rules. The survival rate of knockdown individuals was estimated as observed number of knockdown individuals divided by the expectation, and the lethality rate is one minus the survival rate. At least 10 independent crosses that have at least 20 adults in each cross were counted. For tracking stage-specific lethality, 20 embryos/larvae/pupa of each genotype were collected and placed on fresh medium and the numbers of next-stage individuals were counted after 5 and 10 days. We collected embryos of mixed stages through standard apple juice plate, larvae at L3 stage, and white prepupa. This experiment was repeated for at least four times for a specific genotype or specific developmental stage. GAL4 and RNAi strains used in the study can be found in supplementary table S7, Supplementary Material online. It is worth noting that RNAi strains used this study was generated by TRiP (Perkins et al. 2015), which does not suffer from similar issues of artificial dominant phenotypic effects as VDRC RNAi strains (Green et al. 2014). RNAi for either CG7804 or TBPH are both predicted to have zero off-target (s19 = 1). Estimation of survival rate of knockout individuals was done by crossing CG7804 knockout heterozygotes and compared the number of CG7804 knockout homozygous and heterozygous F1 offspring in the same cross. Tracking of stage-specific lethality rate for knockout individuals used the same methods as the experiment for knockdown individuals. Genotypes compared were CG7804 knockout homozygotes, CG7804 knockout heterozygotes, and Cas9 strain from which the knockout mutant was generated. All flies were reared with standard Drosophila medium at 25 °C with 12/12 light and dark cycle.
Male Fertility Assay
Expression of CG7804 or TBPH was knocked down using a germline-specific GAL4-driver (Bam-GAL4, from G. Findlay Lab, see supplementary table S7, Supplementary Material online, for strain details), and their progeny was counted. In details, sets of ten virgin females from the GAL4-driver were crossed to ten males of RNAi strain (homozygous) to obtain males with CG7804 or TBPH knockdown. These 3–5 days old virgin males were allowed to cross to two females from strain BDSC36304 (the background strain from which RNAi strains were generated) for 2 days, and then crossed again for 2 days with two additional virgin females. Females were allowed to lay eggs for 7 days, and total progeny was counted after 20 days. Ten to fifteen crosses were used for each genotype tested. In addition to males with CG7804 or TBPH knockdown (CG7804-RNAi/Bam-GAL4 or TBPH-RNAi/Bam-GAL4), males with genotypes Bam-GAL4/+ and RNAi/+ were tested as controls.
Real-Time RT-PCR Analysis
Expression knockdown of CG7804 and TBPH by RNAi with Tub-GAL4 driver was confirmed by real-time RT-PCR analysis (supplementary table S6, Supplementary Material online). RNA samples were extracted in triplicate from 30 third instar larvae using Qiagen RNeasy mini kit, digested with DNAase I (Invitrogen) to remove genomic DNA, and reverse-transcribed to cDNA with SuperScript III Reverse Transcriptase (Invitrogen) using oligo(dT) primers. Real-time RT-PCR was performed using iTaq Universal SYBR Green Supermix (Biorad), with the primers described at supplementary table S8, Supplementary Material online, and with three technical replicates for each biological replicate. Quantitative PCR values were normalized using the ΔΔCT method to two independent control products, Rp49 and Actin.
RNA-Seq Experiment and Analysis
Knockout individuals were collected by crossing individuals that are heterozygous for the null allele (null/GFP-balancer) and collect F1 without GFP (homozygous for the null allele). For the wildtype counterparts, we used the Cas9 strain from which the knockout mutant was generated (see supplementary table S7, Supplementary Material online, for strains used and see Text S2 for discussing of the choice of wildtype counterpart). We collected 0–24-h embryos using standard apple juice plate, wandering L3, and white prepupa. For all three stages, we used mixed-sex individuals and have two biological replicates for each genotype at each developmental stage. Total RNAs were extracted from collected materials using RNeasy Plus kit (Qiagen). RNA-Seq sequencing library was prepared using Illumina TruSeq and sequenced on Illumina Hi-Seq with 100 bp, paired-end reads (IGSB Sequencing core, the University of Chicago).
Raw reads were processed with trim-galore (Anon forthcoming) to remove adaptors and low-quality bases galore (-q 30 –stringency 3 –paired). Processed reads were mapped to D. melanogaster release 6 genome (Hoskins et al. 2015) using splicing-aware aligner, TopHat (v 2.1.1, Trapnell et al. 2009), with default parameters. Htseq-count (v 0.7.0, Anders et al. 2015) with default parameter was used to count the number of reads mapping to exons. DESeq2 (Love et al. 2014, implemented in R) was used to normalize and estimate expressional fold enrichment between two D. melanogaster genotypes with following parameter specification. Only genes with at least ten mean read counts were included in the DESeq2 analysis. We also used the default independent filtering implemented by DESeq2, which uses normalized counts as filter statistics to filter out genes that have little probability of being significant.
Developmental Rate Analysis
To investigate whether the CG7804 knockout affects the fly development time (which could potentially compound the developmental stage-specific RNA-seq analysis), the egg-adult development time was compared between the knockout and wildtype background individuals. Eighty to one hundred inseminated females were allowed to lay eggs in an agar plate for 1 h, and then 20 eggs were transferred to food vials, where they developed at 25 °C. Adult eclosion was scored twice a day until all adults had eclosed.
ChIP-Seq Experiment and Analysis
ChIP was performed using modENCODE protocol (http://www.modencode.org/) using anti-GFP antibody (from Kevin White’s Laboratory) with two biological IP replicates for each genotype (CG7804-GFP and TBPH-GFP). ChIP-Seq sequencing library was prepared using NuGen Ovation Ultralow Library Systems V2 (San Carlos, CA) and sequenced on Illumina Hi-Seq with 100 bp, paired-end reads (IGSB Sequencing core, the University of Chicago).
Adaptor sequence and low-quality bases (below 30) were removed from raw reads using trim-galore (-q 30 –stringency 3 –paired). Processed reads were mapped to D. melanogaster reference genome release 6 (Hoskins et al. 2015) using bwa mem (v 0.7.5, Li and Durbin 2009) with default parameters. Reads with mapping quality lower than 30 were removed from the analysis using Samtools (v 1.3.1, Li 2011). Enrichment (IP with respect to Input) was called using MACS2 with liberal P value threshold (narrow peak mode, –extsize 100 –p 0.5 [Zhang et al. 2008]). The liberal P value threshold is necessary to include both peaks with high and low reproducibility, which provides the information necessary for IDR analysis (Li et al. 2011). We used IDR analysis (v 2.0.2, –input-file-type narrowPeak –rank p.value) to identify enrichment peaks with lower than 1% irreproducibility rate between replicates. Genes overlapping with enrichment peaks were identified using Bedtools (“intersect” function) (Quinlan and Hall 2010).
Analysis of Gene Properties
Degree of each gene in protein–protein network was estimated as the number of experimentally validated, nonredundant protein–protein interaction using data from BioGrid 3.4 (Stark et al. 2006). The degree of each gene in genetic interaction network was estimated as the number of reported genetic interaction on Flybase (release February 2017). MK test (McDonald and Kreitman 1991) and the estimation of α (Smith and Eyre-Walker 2002), the proportion of adaptive substitutions, were done using sequences of Zambia D. melanogaster population (Lack et al. 2015) and the divergence between D. melanogaster and D. simulans (release 2, Hu et al. 2013). Drosophila melanogaster lineage-specific dN/dS ratio was estimated using CODEML program in PAML (v 4.9, branch model, Yang 2007), with D. melanogaster, D. simulans, and D. yakuba alleles. Phenotypic data for all genes annotated were downloaded from Flybase, which were based on either knockout mutants or expressional knockdown analysis. Genes were classified into three categories: lethal (with at least one lethal phenotype observed), semilethal (with no lethal phenotype observed and with at least one semilethal phenotype observed), and viable (with no known lethal or semilethal phenotype). Genes without phenotypic data were excluded from this analysis. GO enrichment analysis was performed using DAVID (v 6.8) with Benjamini–Hochberg correction (Huang et al. 2009). Because not all genes are expressed at all developmental stages, GO enrichment analysis for differentially expressed genes used the list of genes with high enough expression to be included in our RNA-seq analysis as the background list.
Imaging Analysis
Images were acquired with a Zeiss LSM700 confocal using either Plan-Apochromat 20×/0.8 M27 or Plan-Apochromat 40×/NA 1.3 oil-immersion lens. Salivary glands were dissected in PBS, fixed with 4% formaldehyde in PBS for 15 min and stained with rabbit polyclonal anti-GFP (Torrey Pines, 1:1,000) and AlexaFluor-488 conjugated secondary antibodies (1:400, Molecular Probes). Dissected tissues were mounted in SlowFade antifade solution (Invitrogen) after TRITC-phalloidin (Sigma) and DAPI (Molecular Probes) stains.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Jennifer Moran of Genome Engineering core of the University of Chicago, and Alec Victor, Matt Kirkey, Jeffrey Gersch from Kevin White’s lab for hosting Y.C.G.L. for ChIP experiment. We thank Dr Findlay for generously Bam-GAL4 strain and Dr White for sharing GFP antibody. Mia Levine, Claus Kemkemer, Benjamin Krinsky, and Nicholas VanKuren provided helpful discussions of the project. We are also grateful to Josie Reinhardt, Maria Vibranovski, and Li Zhao for critically reading the manuscript. Y.C.G.L. was supported by NIH NRSA F32 GM109676 and Chicago Biomedical Consortium Postdoctoral Research Award PDR-043. I.M.V. was supported by the Science without Borders scholarship (BEX18816/12-6). G.R.R. was supported by NSF Graduate Research Fellowship. M.L. was supported by NSF1051826 and NIH R01GM116113. This provides access to the raw data “ChIP-seq and RNA-seq data have been deposited to GEO with accession number GSE100420”.
References
- Abrusán G. 2013. Integration of new genes into cellular networks, and their structural maturation. Genetics 1954:1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W.. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 312:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anon. Forthcoming. Babraham bioinformatics—trim galore! Available from: http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/; Accessed March 2015.
- Ashburner M, Misra S, Roote J, Lewis SE, Blazej R, Davis T, Doyle C, Galle R, George R, Harris N.. 1999. An exploration of the sequence of a 2.9-Mb region of the genome of Drosophila melanogaster: the Adh region. Genetics 1531:179–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala YM, Misteli T, Baralle FE.. 2008. TDP-43 regulates retinoblastoma protein phosphorylation through the repression of cyclin-dependent kinase 6 expression. Proc Natl Acad Sci U S A. 10510:3785–3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala YM, Pagani F, Baralle FE.. 2006. TDP43 depletion rescues aberrant CFTR exon 9 skipping. FEBS Lett. 5805:1339–1344. [DOI] [PubMed] [Google Scholar]
- Ayala YM, Pantano S, D’Ambrogio A, Buratti E, Brindisi A, Marchetti C, Romano M, Baralle FE.. 2005. Human, Drosophila, and C. elegans TDP43: nucleic acid binding properties and splicing regulatory function. J Mol Biol. 3483:575–588. [DOI] [PubMed] [Google Scholar]
- Barabási A-L, Gulbahce N, Loscalzo J.. 2011. Network medicine: a network-based approach to human disease. Nat Rev Genet. 121:56–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabási A-L, Oltvai ZN.. 2004. Network biology: understanding the cell’s functional organization. Nat Rev Genet. 52:101–113. [DOI] [PubMed] [Google Scholar]
- Barrett RDH, Hoekstra HE.. 2011. Molecular spandrels: tests of adaptation at the genetic level. Nat Rev Genet. 1211:767–780. [DOI] [PubMed] [Google Scholar]
- Batada NN, Urrutia AO, Hurst LD.. 2007. Chromatin remodelling is a major source of coexpression of linked genes in yeast. Trends Genet. 2310:480–484. [DOI] [PubMed] [Google Scholar]
- Bateman JR, Lee AM, Wu C.-T.. 2006. Site-specific transformation of Drosophila via ϕC31 integrase-mediated cassette exchange. Genetics 1732:769–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K.. 2007. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci U S A. 1049:3312–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomen VA, Májek P, Jae LT, Bigenzahn JW, Nieuwenhuis J, Staring J, Sacco R, van Diemen FR, Olk N, Stukalov A, et al. 2015. Gene essentiality and synthetic lethality in haploid human cells. Science 3506264:1092–1096. [DOI] [PubMed] [Google Scholar]
- Bose JK, Wang I-F, Hung L, Tarn W-Y, Shen C-K.. 2008. TDP-43 overexpression enhances exon 7 inclusion during the survival of motor neuron pre-mRNA splicing. J Biol Chem. 28343:28852–28859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JB, Boley N, Eisman R, May GE, Stoiber MH, Duff MO, Booth BW, Wen J, Park S, Suzuki AM.. 2014. Diversity and dynamics of the Drosophila transcriptome. Nature 5127515:393–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti E, Baralle FE.. 2001. Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J Biol Chem. 27639:36337–36343. [DOI] [PubMed] [Google Scholar]
- Buratti E, Brindisi A, Giombi M, Tisminetzky S, Ayala YM, Baralle FE.. 2005. TDP-43 binds heterogeneous nuclear ribonucleoprotein A/B through its C-terminal tail an important region for the inhibition of cystic fibrosis transmembrane conductance regulator exon 9 splicing. J Biol Chem. 28045:37572–37584. [DOI] [PubMed] [Google Scholar]
- Capra JA, Pollard KS, Singh M.. 2010. Novel genes exhibit distinct patterns of function acquisition and network integration. Genome Biol. 1112:R127.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrier C, Joshi K, Coutinho-Budd J, Kim J-E, Lambert N, de Marchena J, Jin W-L, Vanderhaeghen P, Ghosh A, Sassa T, et al. 2012. Inhibition of SRGAP2 function by its human-specific paralogs induces neoteny during spine maturation. Cell 1494:923–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Krinsky BH, Long M.. 2013. New genes as drivers of phenotypic evolution. Nat Rev Genet. 149:645–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Ni X, Krinsky BH, Zhang YE, Vibranovski MD, White KP, Long M.. 2012. Reshaping of global gene expression networks and sex-biased gene expression by integration of a young gene. EMBO J. 31:2798–2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhang YE, Long M.. 2010. New genes in Drosophila quickly become essential. Science 330:1682–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimpanzee Sequencing and Analysis Consortium. 2005. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437:69–87. [DOI] [PubMed] [Google Scholar]
- Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, Kaufman TC, Kellis M, Gelbart W, Iyer VN, et al. 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450:203–218. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 3396121:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DN, Kehrer-Sawatzki H.. 2011. Exploring the potential relevance of human-specific genes to complex disease. Hum Genomics. 52:99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daude N, Wohlgemuth S, Brown R, Pitstick R, Gapeshina H, Yang J, Carlson GA, Westaway D.. 2012. Knockout of the prion protein (PrP)-like Sprn gene does not produce embryonic lethality in combination with PrPC-deficiency. Proc Natl Acad Sci U S A. 10923:9035–9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza AT, Dai X, Spencer AG, Reppen T, Menzie A, Roesch PL, He Y, Caguyong MJ, Bloomer S, Herweijer H, et al. 2006. Transcriptional and phenotypic comparisons of Ppara knockout and siRNA knockdown mice. Nucleic Acids Res. 3416:4486–4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demuth JP, Bie TD, Stajich JE, Cristianini N, Hahn MW.. 2006. The evolution of mammalian gene families. PLoS One 11:e85.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis MY, Nuttle X, Sudmant PH, Antonacci F, Graves TA, Nefedov M, Rosenfeld JA, Sajjadian S, Malig M, Kotkiewicz H, et al. 2012. Evolution of human-specific neural SRGAP2 genes by incomplete segmental duplication. Cell 1494:912–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Berrocal A, Morita T, Longden KD, Stern DL.. 2016. Natural courtship song variation caused by an intronic retroelement in an ion channel gene. Nature 536:nature19093. [DOI] [PubMed] [Google Scholar]
- Ding Y, Zhao L, Yang S, Jiang Y, Chen Y, Zhao R, Zhang Y, Zhang G, Dong Y, Yu H, et al. 2010. A young Drosophila duplicate gene plays essential roles in spermatogenesis by regulating several Y-linked male fertility genes. PLoS Genet. 612:e1001255.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Zhou Q, Wang W.. 2012. Origins of new genes and evolution of their novel functions. Annu Rev Ecol Evol Syst. 431:345–363. [Google Scholar]
- Feiguin F, Godena VK, Romano G, D’Ambrogio A, Klima R, Baralle FE.. 2009. Depletion of TDP-43 affects Drosophila motoneurons terminal synapsis and locomotive behavior. FEBS Lett. 583:1586–1592. [DOI] [PubMed] [Google Scholar]
- Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, et al. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44(D1):D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freibaum BD, Chitta R, High AA, Taylor JP.. 2010. Global analysis of TDP-43 interacting proteins reveals strong association with RNA splicing and translation machinery. J Proteome Res. 92:1104–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz SJ, Cummings AM, Nguyen JN, Hamm DC, Donohue LK, Harrison MM, Wildonger J, O’Connor-Giles KM.. 2013. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 1944:1029–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, et al. 2011. The developmental transcriptome of Drosophila melanogaster. Nature 4717339:473–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green EW, Fedele G, Giorgini F, Kyriacou CP.. 2014. A Drosophila RNAi collection is subject to dominant phenotypic effects. Nat Methods. 113:222–223. [DOI] [PubMed] [Google Scholar]
- Hazelett DJ, Chang J-C, Lakeland DL, Morton DB.. 2012. Comparison of parallel high-throughput RNA sequencing between knockout of TDP-43 and its overexpression reveals primarily nonreciprocal and nonoverlapping gene expression changes in the central nervous system of Drosophila. G3 (Bethesda) 27:789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinen T, Staubach F, Häming D, Tautz D.. 2009. Emergence of a new gene from an intergenic region. Curr Biol. 1918:1527–1531. [DOI] [PubMed] [Google Scholar]
- Hoskins RA, Carlson JW, Wan KH, Park S, Mendez I, Galle SE, Booth BW, Pfeiffer BD, George RA, Svirskas R, et al. 2015. The Release 6 reference sequence of the Drosophila melanogaster genome. Genome Res. 253:445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu TT, Eisen MB, Thornton KR, Andolfatto P.. 2013. A second-generation assembly of the Drosophila simulans genome provides new insights into patterns of lineage-specific divergence. Genome Res. 231:89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA.. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 41:44–57. [DOI] [PubMed] [Google Scholar]
- Innan H, Kondrashov F.. 2010. The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet. 112:97–108. [DOI] [PubMed] [Google Scholar]
- Jacob F. 1977. Evolution and tinkering. Science 1964295:1161–1166. [DOI] [PubMed] [Google Scholar]
- Jeong H, Mason SP, Barabási A-L, Oltvai ZN.. 2001. Lethality and centrality in protein networks. Nature 4116833:41–42. [DOI] [PubMed] [Google Scholar]
- Kaessmann H. 2010. Origins, evolution, and phenotypic impact of new genes. Genome Res. 2010:1313–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg M.. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 106:845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kim I, Han SK, Bowie JU, Kim S.. 2012. Network rewiring is an important mechanism of gene essentiality change. Sci Rep. 2:900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok FO, Shin M, Ni C-W, Gupta A, Grosse AS, van Impel A, Kirchmaier BC, Peterson-Maduro J, Kourkoulis G, Male I, et al. 2015. Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev Cell 321:97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S, Ueda R.. 2013. Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics 1953:715–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo P-H, Doudeva LG, Wang Y-T, Shen C-K, Yuan HS.. 2009. Structural insights into TDP-43 in nucleic-acid binding and domain interactions. Nucleic Acids Res. 376:1799–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lack JB, Cardeno CM, Crepeau MW, Taylor W, Corbett-Detig RB, Stevens KA, Langley CH, Pool JE.. 2015. The Drosophila genome nexus: a population genomic resource of 623 Drosophila melanogaster genomes, including 197 from a single ancestral range population. Genetics 1994:1229–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27:2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2514:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Brown JB, Huang H, Bickel PJ.. 2011. Measuring reproducibility of high-throughput experiments. Ann Appl Stat. 53:1752–1779. [Google Scholar]
- Li Y, Ray P, Rao EJ, Shi C, Guo W, Chen X, Woodruff EA, Fushimi K, Wu JY.. 2010. A Drosophila model for TDP-43 proteinopathy. Proc Natl Acad Sci U S A. 1077:3169–3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M-J, Cheng C-W, Shen C-K.. 2011. Neuronal function and dysfunction of Drosophila dTDP. PLoS One 66:e20371.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnen CR, Poh Y-P, Peterson BK, Barrett RDH, Larson JG, Jensen JD, Hoekstra HE.. 2013. Adaptive evolution of multiple traits through multiple mutations at a single gene. Science 3396125:1312–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M, VanKuren NW, Chen S, Vibranovski MD.. 2013. New gene evolution: little did we know. Annu Rev Genet. 47:307–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loppin B, Lepetit D, Dorus S, Couble P, Karr TL.. 2005. Origin and neofunctionalization of a Drosophila paternal effect gene essential for zygote viability. Curr Biol. 152:87–93. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 1512:550.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukavsky PJ, Daujotyte D, Tollervey JR, Ule J, Stuani C, Buratti E, Baralle FE, Damberger FF, Allain F-T.. 2013. Molecular basis of UG-rich RNA recognition by the human splicing factor TDP-43. Nat Struct Mol Biol. 2012:1443–1449. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS.. 2000. The evolutionary fate and consequences of duplicate genes. Science 2905494:1151–1155. [DOI] [PubMed] [Google Scholar]
- Matsumoto T, John A, Baeza-Centurion P, Li B, Akashi H.. 2016. Codon usage selection can bias estimation of the fraction of adaptive amino acid fixations. Mol Biol Evol. 336:1580–1589. [DOI] [PubMed] [Google Scholar]
- Matsuno M, Compagnon V, Schoch GA, Schmitt M, Debayle D, Bassard J-E, Pollet B, Hehn A, Heintz D, Ullmann P, et al. 2009. Evolution of a novel phenolic pathway for pollen development. Science 3255948:1688–1692. [DOI] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M.. 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 3516328:652–654. [DOI] [PubMed] [Google Scholar]
- Merzendorfer H, Zimoch L.. 2003. Chitin metabolism in insects: structure, function and regulation of chitin synthases and chitinases. J Exp Biol. 206(Pt 24):4393–4412. [DOI] [PubMed] [Google Scholar]
- Moore RC, Purugganan MD.. 2005. The evolutionary dynamics of plant duplicate genes. Curr Opin Plant Biol. 82:122–128. [DOI] [PubMed] [Google Scholar]
- Mummery-Widmer JL, Yamazaki M, Stoeger T, Novatchkova M, Bhalerao S, Chen D, Dietzl G, Dickson BJ, Knoblich JA.. 2009. Genome-wide analysis of Notch signalling in Drosophila by transgenic RNAi. Nature 4587241:987–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neely GG, Hess A, Costigan M, Keene AC, Goulas S, Langeslag M, Griffin RS, Belfer I, Dai F, Smith SB, et al. 2010. A genome-wide Drosophila screen for heat nociception identifies α2δ3 as an evolutionarily conserved pain gene. Cell 1434:628–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obbard DJ, Maclennan J, Kim K-W, Rambaut A, O’Grady PM, Jiggins FM.. 2012. Estimating divergence dates and substitution rates in the Drosophila phylogeny. Mol Biol Evol. 2911:3459–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. 1970. Evolution by gene duplication. Berlin Heidelberg: Springer Science & Business Media. [Google Scholar]
- Pavlicev M, Wagner GP.. 2012. A model of developmental evolution: selection, pleiotropy and compensation. Trends Ecol Evol (Amst). 276:316–322. [DOI] [PubMed] [Google Scholar]
- Perkins LA, Holderbaum L, Tao R, Hu Y, Sopko R, McCall K, Yang-Zhou D, Flockhart I, Binari R, Shim H-S, et al. 2015. The transgenic RNAi project at Harvard Medical School: resources and validation. Genetics 2013:843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymenidou M, Lagier-Tourenne C, Hutt KR, Huelga SC, Moran J, Liang TY, Ling S-C, Sun E, Wancewicz E, Mazur C, et al. 2011. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci. 144:459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM.. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 266:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranz JM, Parsch J.. 2012. Newly evolved genes: moving from comparative genomics to functional studies in model systems. Bioessays 346:477–483. [DOI] [PubMed] [Google Scholar]
- Reinhardt JA, Wanjiru BM, Brant AT, Saelao P, Begun DJ, Jones CD.. 2013. De novo ORFs in Drosophila are important to organismal fitness and evolved rapidly from previously non-coding sequences. PLoS Genet. 910:e1003860.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross BD, Rosin L, Thomae AW, Hiatt MA, Vermaak D, de la Cruz AFA, Imhof A, Mellone BG, Malik HS.. 2013. Stepwise evolution of essential centromere function in a Drosophila neogene. Science 3406137:1211–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi A, Kontarakis Z, Gerri C, Nolte H, Hölper S, Krüger M, Stainier D.. 2015. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature 5247564:230–233. [DOI] [PubMed] [Google Scholar]
- Rost S, Fregin A, Ivaskevicius V, Conzelmann E, Hörtnagel K, Pelz H-J, Lappegard K, Seifried E, Scharrer I, Tuddenham EGD, et al. 2004. Mutations in VKORC1 cause warfarin resistance and multiple coagulation factor deficiency type 2. Nature 4276974:537–541. [DOI] [PubMed] [Google Scholar]
- Schnorrer F, Schönbauer C, Langer CCH, Dietzl G, Novatchkova M, Schernhuber K, Fellner M, Azaryan A, Radolf M, Stark A, et al. 2010. Systematic genetic analysis of muscle morphogenesis and function in Drosophila. Nature 4647286:287–291. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 7:539.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith NGC, Eyre-Walker A.. 2002. Adaptive protein evolution in Drosophila. Nature 4156875:1022–1024. [DOI] [PubMed] [Google Scholar]
- Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, Ackerley S, Durnall JC, Williams KL, Buratti E, et al. 2008. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science 3195870:1668–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C, Breitkreutz B-J, Reguly T, Boucher L, Breitkreutz A, Tyers M.. 2006. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 3490001:D535–D539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauber M, Jäckle H, Schmidt-Ott U.. 1999. The anterior determinant bicoid of Drosophila is a derived Hox class 3 gene. Proc Natl Acad Sci U S A. 967:3786–3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JS, Raes J.. 2004. Duplication and divergence: the evolution of new genes and old ideas. Annu Rev Genet. 38:615–643. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL.. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 259:1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanKuren NW, Long M.. 2018. Gene duplicates resolving sexual conflict rapidly evolved essential gametogenesis functions. Nat Ecol Evol. 24:705–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken KJT, Carlson JW, Schulze KL, Pan H, He Y, Spokony R, Wan KH, Koriabine M, de Jong PJ, White KP, et al. 2009. Versatile P(acman) BAC Libraries for transgenesis studies in Drosophila melanogaster. Nat Methods. 66:431–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken KJT, Schulze KL, Haelterman NA, Pan H, He Y, Evans-Holm M, Carlson JW, Levis RW, Spradling AC, Hoskins RA, et al. 2011. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat Methods. 89:737–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura I, Long M.. 2017. Connecting evolutionary genomics to cell biology In: Encyclopedia of cell biology. Vol. 4 Amsterdam, Netherlands: Elsevier; p. 153–159. [Google Scholar]
- Wittkopp PJ, Stewart EE, Arnold LL, Neidert AH, Haerum BK, Thompson EM, Akhras S, Smith-Winberry G, Shefner L.. 2009. Intraspecific polymorphism to interspecific divergence: genetics of pigmentation in Drosophila. Science 3265952:540–544. [DOI] [PubMed] [Google Scholar]
- Wray GA. 2007. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 83:206.. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol Biol Evol. 248:1586–1591. [DOI] [PubMed] [Google Scholar]
- Yeh S-D, Do T, Chan C, Cordova A, Carranza F, Yamamoto EA, Abbassi M, Gandasetiawan KA, Librado P, Damia E, et al. 2012. Functional evidence that a recently evolved Drosophila sperm-specific gene boosts sperm competition. Proc Natl Acad Sci U S A. 1096:2043–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Greenbaum D, Xin Lu H, Zhu X, Gerstein M.. 2004. Genomic analysis of essentiality within protein networks. Trends Genet. 206:227–231. [DOI] [PubMed] [Google Scholar]
- Zhang W, Landback P, Gschwend AR, Shen B, Long M.. 2015. New genes drive the evolution of gene interaction networks in the human and mouse genomes. Genome Biol. 16:202.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based Analysis of ChIP-Seq (MACS). Genome Biol. 99:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YE, Landback P, Vibranovski MD, Long M.. 2011. Accelerated recruitment of new brain development genes into the human genome. PLoS Biol. 910:e1001179.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YE, Vibranovski MD, Krinsky BH, Long M.. 2010. Age-dependent chromosomal distribution of male-biased genes in Drosophila. Genome Res. 2011:1526–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.