Abstract
Background
Long noncoding RNAs (lncRNAs) play an important role in gastric cancer. In this study, we aimed to uncover the epigenetic regulatory mechanism of lncRNA KCNK15-AS1 in gastric cancer progression.
Patients and methods
Forty patients were included in the study. The expression of KCNK15-AS1 was detected by real-time PCR (RT-PCR), the promoter of KCNK15-AS1 was detected by methylation-specific PCR, and the luciferase assay was performed to detect the relationship between KCNK15-AS1 and miR-21. The relationship of the proteins was explored by an RNA pull-down assay and RNA immunoprecipitation. Chromatin immunoprecipitation was performed to detect the relationship between the promoter and the protein.
Results
The expression of KCNK15-AS1 was lower in the tumor tissue compared to the normal tissue. KCNK15-AS1 interacted with miR-21. Both the overexpression of KCNK15-AS1 and the knockdown of the expression of miR-21 inhibited proliferation and promoted apoptosis and decreased the level of MMP-9, bcl-2, and MMP-2 but increased the level of Bax. In addition, the methylation of KCNK15-AS1 was detected in the tumor tissue but was not detected in the normal tissue. Treatment with 5-azacytidine and chidamide decreased the level of DNMT1 and HDAC1 and increased the level of KCNK15-AS1. The RNA pull-down and RNA immunoprecipitation results showed that KCNK15-AS1 interacted with DNMT1 and HDAC1. The ChIP-seq result showed that the promoter of MAPK interacted with DNMT1, and the promoter of AKT and STAT5 interacted with HDAC1.
Conclusion
In this study, we identified two regulatory axes, namely KCNK15-AS1-DNMT1-MAPK and KCNK15-AS1-HDAC1-AKT, which were associated with gastric cancer progression. Chidamide and 5-azacytidine might provide new modes for treating gastric cancer.
Keywords: lncRNA, KCNK15-AS1, DNMT1, HDAC1, gastric cancer, miR-21
Introduction
Gastric cancer is the third leading cause of cancer-related deaths all over the world, and it is also one of the most common types of cancer. The prognosis of gastric cancer is always poor, which is partly due to the lack of typical early symptoms.1 Although great efforts have been made in exploring the potential mechanism of gastric cancer, finding a novel treatment target is still a challenge.2–5
Many molecules play important roles in the development of gastric cancer, including long noncoding RNAs (lncRNAs).5 For example, lncRNA UFC1 promotes gastric cancer progression, and a high level of lncRNA UFC1 predicts a poor prognosis.6 The knockdown of the expression of lncRNA NNT-AS1 inhibits proliferation and promotes apoptosis both in vivo and in vitro.7 LncRNA KCNK15-AS1 is downregulated in pancreatic cancer, and the overexpression of KCNK15-AS1 inhibits migration and invasion.8 However, the epigenetic regulation of lncRNA KCNK15-AS1 in gastric cancer is still unknown.
Epigenetics is the study of heritable changes in gene expression without any alteration in the DNA sequence. The abnormal methylation of promoter regions plays an important role in tumorigenesis, including human choriocarcinoma and squamous cell lung cancer. In this regard, lncRNA-associated methylation has been receiving increased attention, especially with regard to the role of lncRNA Esrp2 and MEG3 in human breast and lung cancers, respectively.
The methylation of genes at CpG sites as well as methylation-related genes is reported to take part in the occurrence and development of cancer.9 DNA methyltransferase (cytosine-5) 1 (DNMT1), an initiating factor that regulates DNA methylation, is reportedly involved in cancer, such as chronic myeloid leukemia and hepatocellular carcinoma.10,11 Histone modification and HDAC1 also participate in cancer progression, including breast cancer, pancreatic ductal cancer, and renal cell carcinoma.12–14 HDAC inhibitors (HDACis) have gained positive results in treating many types of cancer.15–17 Chidamide is a novel HDACi that is used to treat cutaneous T cell lymphoma.18 However, whether the expression of KCNK15-AS1 and miR-21 is influenced by chidamide in gastric cancer cells remains unknown. Therefore, in this study, we aimed to detect the biological role of KCNK15-AS1 in gastric cancer and to further explore the pathology of gastric cancer and the role of chidamide in treating gastric cancer.
Patients and methods
Patients and tissue samples
Thirty paired gastric cancer patients were included in this study, and the tumor tissues and the adjacent noncancerous tissues (5 cm away from the tumor edge) were obtained from these patients, who were admitted to the Department of General Surgery, China-Japan Union Hospital of Jilin University, between May 2017 and March 2018. All the patients read the consent and signed it, and this study was approved by the institutional ethics committee of China-Japan Union Hospital of Jilin University. This study was conducted in accordance with the Declaration of Helsinki. All the tissues were frozen in liquid nitrogen and were then stored at –80°C for further use.
Cell culture
Two human gastric cancer cell lines (BGC-823 and SGC- 7901) were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The human normal gastric mucosa epithelial cell line GES-1 was purchased from the Shanghai Hong Shun Biotechnology Co., Ltd. (Shanghai, China). The GES-1, BGC-823, and SGC-7901 cells were cultured in RPMI 1640 medium (Thermo Fisher Scientific, Waltham, MA, USA), containing 10% FBS and penicillin/streptomycin. All the cells were cultured in a 37°C incubator with a 5% CO2 atmosphere. The BGC-823 and SGC-7901 cells were seeded into six-well plates (5x105/well) and were treated with different concentrations of 5-azacytidine and chidamide based on its EC50(median effective concentration).
Construction of the lentiviral vector and siRNA vector
The sequence of KCNK15-AS1 was synthesized by Thermo Fisher Scientific and cloned into the overexpression vector (Thermo Fisher Scientific). miR-21 mimics, miR-21 control, miR-21 inhibitors, si-MAPK, si-HDAC1, and siRNA-control were also purchased from Thermo Fisher Scientific.
Cell transfection
The BGC-823 and SGC-7901 cells were transfected with LV-KCNK15-AS1, or LV-control and miR-21 controls or miR-21 inhibitors and transfected with si-MAPK, si-HDAC1 or siRNA-control using Lipofectamine 3000 (Thermo Fisher Scientific).
Cell proliferation assay
After transfection with LV-KCNK15-AS1 or LV-control or transfection with the miR-21 inhibitors or miR-21 controls, we seeded the BGC-823 and SGC-7901 cells into 96-well plates (1x104 cells/well) and added 10 μL of MTT (Sigma- Aldrich Co., St Louis, MO, USA) to each well. The 96-well plates were incubated at 37°C in a humidified 5% CO2 atmosphere for 4 hours. The absorbance was then read at 490 nm in a microplate reader (Thermo Fisher Scientific).
Apoptosis assay
After transfection with LV-KCNK15-AS1 or LV-control or transfection with the miR-21 inhibitors or miR-21 control, the Annexin V–fluorescein isothiocyanate (FITC) and propidium iodide (PI) kit (BD Biosciences, San Jose, CA, USA) was used for analyzing apoptosis.
Real-time PCR (RT-PCR)
The total RNA was isolated using Trizol. The RNA was reverse transcribed into cDNA using the First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). The reverse transcription reactions were conducted under the following conditions: 42°C for 60 minutes, 25°C for 5 minutes, and 70°C for 5 minutes. The quantitative PCR (qPCR) mixtures had a final volume of 20 μL and contained 1 μL of template cDNA, 0.5 μL of each sense and antisense primer, 10 μL of SYBR Green Mix (Thermo Fisher Scientific), and 8 μL of diethyl pyrocarbonate-treated water. The RT-qPCR conditions were as follows: denaturation at 95°C for 5 minutes followed by 45 cycles of 95°C for 15 seconds, 60°C for 35 seconds, and 72°C for 20 seconds in an ABI 7500 Real-Time Rotary Analyzer (Thermo Fisher Scientific). The 2−∇∇CT method was used to calculate the relative gene expression. Each reaction was repeated at least three times independently. The primers were synthesized by Thermo Fisher Scientific and are summarized in Table 1.
Table 1.
Primer sequences for RT-PCR
| Gene | Primer | Product (bp) |
|---|---|---|
| KCNK15-AS1 | Forward: 5′-CTCCCCTTCTAGCGCTCACG-3′ | 154 |
| Reverse: 5′-CTAGCCGCCGTCTATACTACCGGCT-3′ | ||
| DNMT1 | Forward: 5′-AGACACTCGCTCAGCTTCTTG-3′ | 106 |
| Reverse: 5′-CAATTGCTGCTGGGATTCATC-3′ | ||
| HDAC1 | Forward: 5′-CTGCTTTGTATTCCCTTTTGCA-3′ | 131 |
| Reverse: 5′-TTGATTTCTCCTGGCTGTCTC-3′ | ||
| MMP-2 | Forward: 5′-CCTCCCGAGTGAAGTCATCGTGG-3′ | 142 |
| Reverse: 5′-GGACAGGTGCTTCATCAGCTCG-3′ | ||
| MMP-9 | Forward: 5′-ACCACACGTTCCTAAGCTGG-3′ | 119 |
| Reverse: 5′-TCCCTGCACGCAGAGATTTT-3′ | ||
| Bax | Forward: 5′-AACTACAACTTCTTCCCTCGCAA-3′ | 122 |
| Reverse: 5′-CAAAGTTATGTCCACTGTCTCT-3′ | ||
| Bcl-2 | Forward: 5-CATCACCGTAGTCCCATTG-3′ | 171 |
| Reverse: 5-AAATTCCCCACTTCGCTTGG-3′ | ||
| ACTB | Forward: 5′-GAGCTACGAGCTGCCTGAC-3′ | 121 |
| Reverse: 5′-GGTAGTTTCGTGGATGCCACAG-3′ |
Abbreviation: RT-PCR, real-time PCR.
Detection of the miR-21 levels
The RNA from the tissues and the BGC-823, SGC-7901, and GES-1 cell lines was extracted using the RNeasy Mini Kit (Qiagen NV, Venlo, the Netherlands) according to the manufacturer’s instructions. Reverse transcription reactions and qPCR were performed using the miScript Reverse Transcription kit (Qiagen NV) and the miScript SYBR® Green PCR kit (Qiagen NV). The programs for the PCR were as follows: 95°C for 1 minute; and then, 40 cycles of 95°C for 10 seconds, 55°C for 30 seconds, and 70°C for 30 seconds. After normalization to U6, the relative expression level of mature miR-21 was calculated using the 2−∇∇CT method.
Methylation-specific PCR
The genomic DNA was isolated using the DNA isolate kit (Zymo, Beijing, China). The DNA was sulfated using the EZ DNA methylation-Gold kit (Zymo USA Inc, Fleming Island, FL, USA). The bisulfite-modified DNA (2 μL), Zymo Taq PreMix (12.5 μL), water (8.5 μL), upstream primer (1 μL), and downstream primer (1 μL) were included in this reaction. The conditions for the PCR reaction were as follows: 95°C for 10 minutes followed by 35 cycles of 30 seconds at 95°C, 45 seconds at 54°C for annealing, 45 seconds at 72°C, and a final extension step of 7 minutes at 72°C. The PCR products were separated by 2% agarose gel (GoldView; Saibaisheng Bioengineering Co., Ltd., Beijing, China) electrophoresis as follows: KCNK15-AS1 methylated (M) and KCNK15-AS1 unmethylated (U) positive and negative methylation, KCNK15-AS1 M and KCNK15-AS1 U partial methylation, and KCNK15-AS1 M and KCNK15-AS1 U positive and negative unmethylated. The primers were synthesized by Saibaisheng Bioengineering Co., Ltd and are summarized in Table 2.
Table 2.
Primer sequences of the methylated KCNK15-AS1 gene
| Gene | Primer | Product (bp) |
|---|---|---|
| KCNK15-AS1,M-MSP | Forward: 5′-CGTAGCTACTGCTATTATCGTTC-3′ | 162 |
| Reverse: 5′-TTATGCCTTGGGCTACGTACT-3′ | ||
| KCNK15-AS1,U-MSP | Forward:5′-GGTACGTATGCTTGGGCTTATG-3′ | 162 |
| Reverse: 5′-ATTGGCGCTTAGCTAGGCTA-3′ |
Abbreviations: M, methylated; MSP, methylation-specific PCR; U, unmethylated.
Western blotting
We lysed the cells using RIPA buffer. The protein concentration was determined using a BCA protein assay kit (Thermo Fisher Scientific). The protein was separated on 8%–15% SDS-PAGE gels, and the separated bands were electro-transferred to a polyvinylidene difluoride (PVDF) membrane (Amersham Pharmacia, Piscataway, NJ, USA). The PVDF membrane was blocked with 5% nonfat skim milk at room temperature and was probed with antibodies targeting DNMT1 (1:1,000), HDAC1 (1:1,000), MMP-2 (1:1,000), MMP-9 (1:1,000), Bax (1:1,000), bcl-2 (1:1,000), and ATCB (1:5,000; all from Abcam, Cambridge, MA, USA). The diluted corresponding secondary antibody was incubated with the membrane at room temperature for 1 hour, and the membrane was then rinsed three times with PBS(phosphate buffer saline) for 5 minutes each time. The protein expression was examined using the BioSpectrum Imaging System (UVP, LLC, Upland, CA, USA).
Luciferase assay
LncRNA-KCNK15-AS1 (lncRNA-KCNK15-AS1-wt) and its mutant of 3′-UTR, devoid of specific miRNA-binding sites (lncRNA-KCNK15-AS1-mu), were cloned into the Renilla luciferase gene in the vector pRL-TK (Promega Corporation, Fitchburg, WI, USA). Each plasmid was transfected into the cells together with specific miRNA mimics or with a negative control mimic (RiboBio, Guangzhou, China). The firefly luciferase gene in the vector pGL3- control (Promega Corporation) was used as a control for the transfection efficiency. The luciferase assays were performed using the dual-luciferase reporter assay system kit (Promega Corporation) according to the manufacturer’s instructions. The luciferase expression was analyzed by Modulus single-tube multimode reader (Promega Corporation). The relative luciferase expression was equal to the expression of the Renilla luciferase (pRL-TK) divided by the expression of the firefly luciferase. All the assays were repeated at least three times.
RNA pull-down assay
The Pierce Magnetic RNA-Protein Pull-Down Kit (Thermo Fisher Scientific) was used to detect the KCNK15-AS1- binding proteins. The KCNK15-AS1 sense and KCNK15-AS1 antisense lncRNAs were generated and incubated with the protein from the BGC-823 cells. The protein was then collected for the mass spectrometry analysis.
RNA immunoprecipitation
The RNA immunoprecipitation was done using the Magna RIP kit (EMD Millipore, Billerica, MA, USA). We evaluated the connection and configuration of the antibody beads. The antibody beads were suspended and mixed with the sample after thawing and were then incubated at 4°C overnight. The immunoprecipitation was completed after the suspension was placed on a magnetic frame and was washed with buffer at least six times. Finally, the immune coprecipitation products were collected, and the RNA was extracted and purified to determine the abundance of the target RNA.
Chromatin immunoprecipitation
The cells were added into a sufficient amount of formaldehyde for cross-linking, and the DNA–protein complex was immobilized. The cell lysis solution was added to the cell lysis. Ultrasound broke the large DNA fragment into 100-500 bp. The HDAC1 and DNMT1 antibodies and beads were added to precipitate the DNA–protein complex. NaCl, with the cross-linking solution, was used at 65°C to obtain free DNA and protein. A QIAGEN PCR kit was used to purify the DNA, and the chromatin immunoprecipitation and sequencing were performed. The output of the original read filtering, including the contamination, the sequencing of the joint, and the low-quality base ratio of high reads, resulted in clean data. Then, the reads were compared to the reference genome using the alignment software Short Oligonucleotide Analysis Package (SOAP) aligner/soap2 and to the duplication treatment, according to the needs of each library reads. Then, the quality control was assessed to determine whether the sequencing data quality standards passed the quality control standard, and we obtained the sequence alignment as the analysis object, an analysis of the distribution of the reference genome coverage, the depth of the statistical information of the genomic locus, the distribution of the gene function element analysis, the gene function significant enrichment analysis, and the pathway enrichment analysis.
Statistical analyses
Data are expressed as mean±SD and were analyzed using SPSS 19.0 software (IBM Corporation, Armonk, NY, USA). The significant differences between the groups were analyzed using Student’s t-test or one-way ANOVA for more than two subgroups. The chi-squared test was applied to compare the rates, and P<0.05 was considered as statistically significant difference.
Results
The KCNK I5-ASI expression decreases in gastric cancer tissue and overexpression of KCNK I5-ASI inhibits proliferation and promotes apoptosis
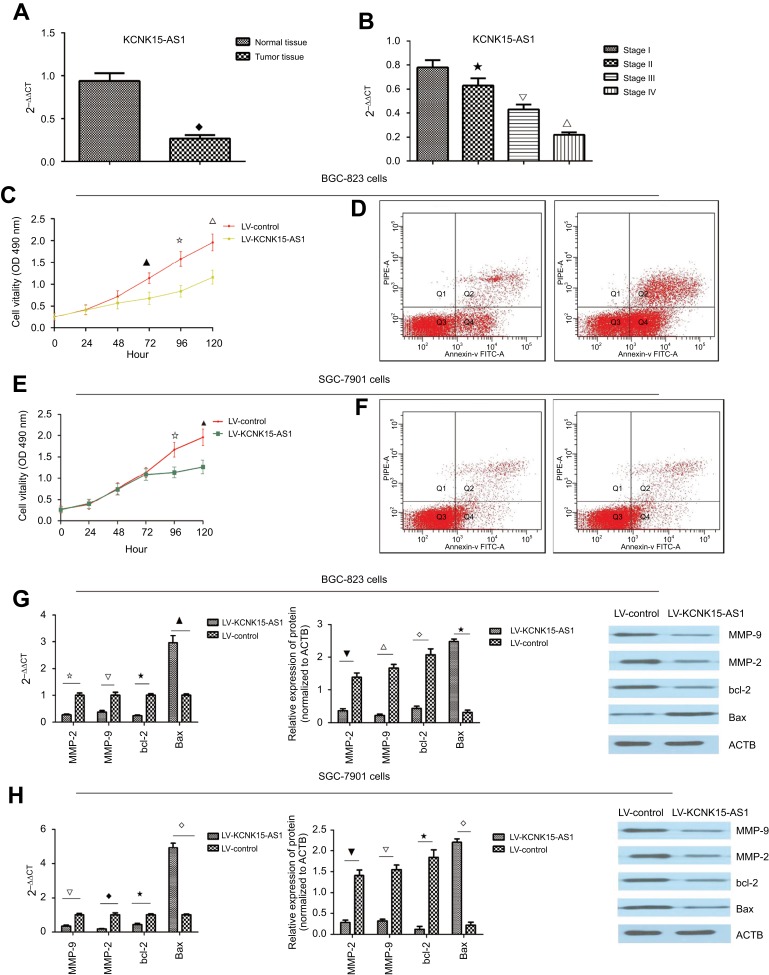
The mRNA level of KCNK15-AS1 was detected by RT-PCR, and the level of KCNK15-AS1 was downregulated in the gastric cancer tissue compared to the adjacent noncancerous tissues (Figure 1A). Moreover, the lowest level of KCNK15-AS1 was found in the clinical stage III and IV samples (Figure 1B).
Figure 1.
(A) The expression level of KCNK15-AS1 was lower in the tumor tissue (♦P<0.05). (B) The expression level of KCNK15-AS1 was lower in the stage III and stage IV cases (⋆△∇P<0.05). (C) KCNK15-AS1 overexpression inhibited proliferation in the BGC-823 cells (▁⢶∇P<0.05). (D) KCNK15-AS1 overexpression promoted apoptosis in the BGC-823 cells. (E) KCNK15-AS1 overexpression inhibited proliferation in the SCG7901 cells (⢶▁P<0.05). (F) KCNK15-AS1 overexpression promoted apoptosis in the SCG7901 cells. (G) KCNK15-AS1 overexpression decreased the mRNA and protein levels of MMP-2, MMP-9, and bcl-2 and increased the level of Bax in the BGC-823 cells (⢶△⋆▁P<0.05; ⎂△⋄⋆P<0.05). (H) KCNK15-AS1 overexpression decreased the mRNA and protein levels of MMP-2, MMP-9, and bcl-2 and increased the level of Bax in the SGC-7901 cells (△♦⋆⋄P<0.05; ⎂△⋆⋄P<0.05).
Abbreviation: FITC, fluorescein isothiocyanate.
To further explore the role of KCNK15-AS1 in gastric cancer, we also detected the level of KCNK15-AS1 in cancer cell lines and found that the mRNA level of KCNK15-AS1 was lower in the BGC-823 and SGC-7901 cells compared to the normal gastric mucosa epithelial cell line GES-1. Thus, we transfected the BGC-823 and SGC-7901 cells with LV- KCNK15 and found that the expression level of KCNK15-AS1 was increased by 7.3-fold and 8.9-fold in BGC-823 and SGC-7901 cells, respectively. Then, an MTT assay was performed to determine the influence of KCNK15-AS1 on proliferation in the BGC-823 and SGC-7901 cells. The overexpression of KNCK15-AS1 inhibited the proliferation of the BGC-823 and SGC-7901 cells at 72, 96, and 120 hours and at 96 and 120 hours, respectively, after transfection (Figure 1C and E). Flow cytometry was performed to explore the influence of KCNK15-AS1 on apoptosis in the BGC-823 and SGC-7901 cells. The overexpression of KNCK15-AS1 promoted apoptosis in BGC-823 and SGC-7901 cells (Figure 1D and F). Western blot and quantitative RT-PCR (qRT-PCR) were performed to examine the effect of the overexpression of KNCK15-AS1 on the levels of bcl-2, Bax, MMP-2, and MMP-9. The overexpression of KNCK15-AS1 decreased the levels of bcl-2, MMP-2, and MMP-9 but increased the level of Bax (Figure 1G and H). These results indicated that the expression level of KCNK15-AS1 was downregulated in gastric cancer and the overexpression of KCNK15-AS1 inhibited proliferation and promoted apoptosis.
Interaction of KCNK I5-ASI with miR-2I
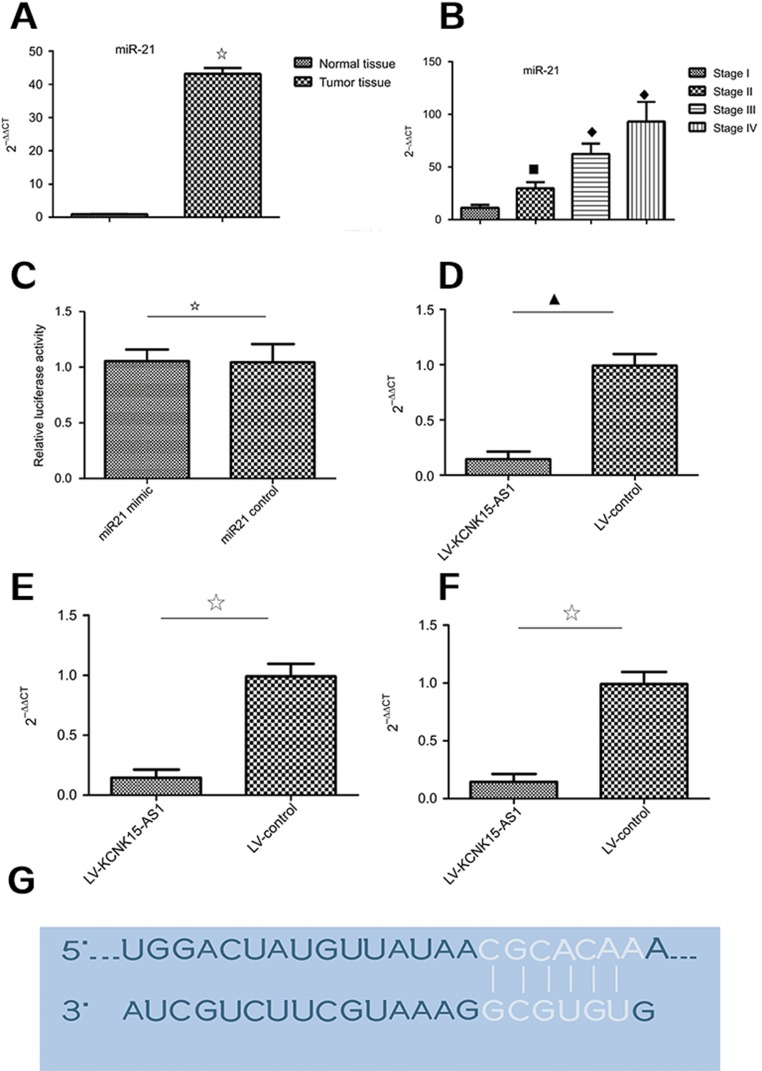
KCNK15-AS1 regulated proliferation and apoptosis of gastric cancer cells, and we speculated that KCNK15-AS1 might interact with miRNA to exert effects on gastric cancer cells. Competing endogenous RNAs are lncRNAs that act as sponges to bind miRNAs and prevent them from performing normal regulatory activities. According to the bioinformatics analysis, the miR-21 was the putative miRNA. The miR-21 expression level was detected in gastric cancer. The miR-21 was increased in gastric cancer compared to normal tissue and was higher in advanced stage (Figure 2A and B). Then, the luciferase reporter assay was performed to determine whether KCNK15-AS1 binds to miR-21. The results indicated that the transfection with the miR-21 mimic decreased the luciferase activity of HOTAIR wild-type plasmids but had no effect on the mutant-type plasmids (Figure 2C and D). Furthermore, KNCK15-AS1 overexpression decreased the expression of miR-21 (Figure 2E and F). The putative binding sequences are shown in Figure 2G. These results revealed that KCNK15-AS1 could act as sponges of miR-21.
Figure 2.
(A) The expression level of miR-21 was higher in the tumor tissue compared to the normal tissue (⢶P<0.05). (B) The expression level of miR-21 was higher in the stage III and stage IV cases compared to the stage I and stage II cases (□♦♦P<0.05). (C) The relative luciferase activity was not different between the miR-21 mimic group and the miR-21 control group (⢶P>0.05). (D) The relative luciferase activity was lower in the miR-21 mimic group compared to the miR-21 control group (⋆P<0.05). (E) In the BGC823 cells, KCNK15-AS1 overexpression decreased the expression level of miR-21 (▁P<0.05). (F) In the SGC7901 cells, KCNK15-AS1 overexpression decreased the expression level of miR-21 (⢶P<0.05). (G) The RNA up algorithm predicted the potential binding of miR-21 to KCNK15-AS1, with a considerable sequence complementary in the indicated regions.
miR-2I knockdown inhibits proliferation and promotes apoptosis
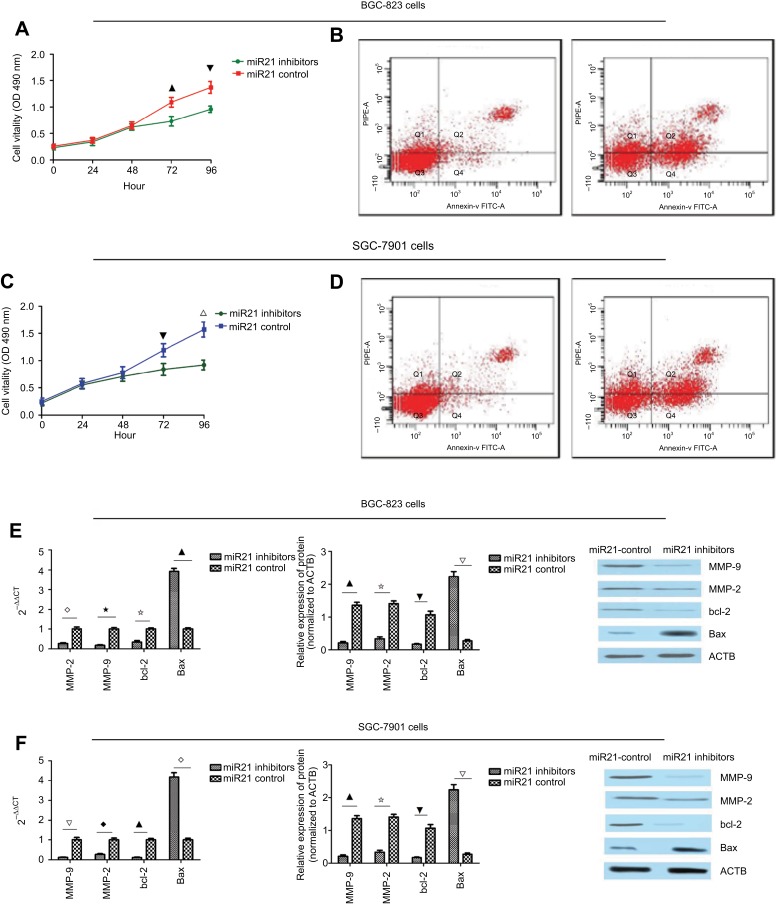
To explore the role of miR-21 in gastric cancer, we first detected the level of miR-21, which was higher in the BGC- 823 and SGC-7901 cells compared to the normal gastric mucosa epithelial cell line GES-1. Thus, we transfected the BGC-823 and SGC-7901 cells with miR-21 inhibitors and found that the expression level of miR-21 was decreased by 20% and 30% in the BGC-823 and SGC-7901 cells, respectively. Then, an MTT was performed to assay the influence of miR-21 on proliferation in the BGC-823 and SGC-7901 cells. The knockdown of miR-21 inhibited the proliferation of the BGC-823 and SGC-7901 cells at 72 hours after transfection (Figure 3A and C). Flow cytometry was performed to detect the influence of miR-21 on apoptosis in the BGC-823 and SGC-7901 cells. The knockdown of miR-21 promoted apoptosis in the BGC-823 and SGC-7901 cells (Figure 3B and D). Western blot and qRT-PCR analyses were performed to determine whether the knockdown of miR-21 altered on the levels of bcl-2, Bax, MMP-2, and MMP-9. miR-21 knockdown decreased the levels of bcl-2, MMP-2, and MMP-9 but increased the level of Bax (Figure 3E and F).
Figure 3.
(A) miR-21 knockdown inhibited proliferation in the BGC-823 cells (▁⎂P<0.05). (B) miR-21 knockdown promoted apoptosis in the BGC-823 cells. (C) miR-21 knockdown inhibited proliferation in the SCG7901 cells (⎂∇P<0.05). (D) miR-21 knockdown promoted apoptosis in the SCG7901 cells. (E) miR-21 knockdown decreased the mRNA and protein levels of MMP-2, MMP-9, and bcl-2 and increased the level of Bax in the BGC-823 cells (⋄⋆⢶▁P<0.05; ▁⢶⎂△P<0.05). (F) miR-21 knockdown decreased the mRNA and protein levels of MMP-2, MMP-9, and bcl-2 and increased the level of Bax in the SGC-7901 cells (△♦▁⋄P<0.05; ▁⢶⎂△P<0.05).
Abbreviation: FITC, fluorescein isothiocyanate.
KCNKI5-ASI promoter methylation status in gastric cancer
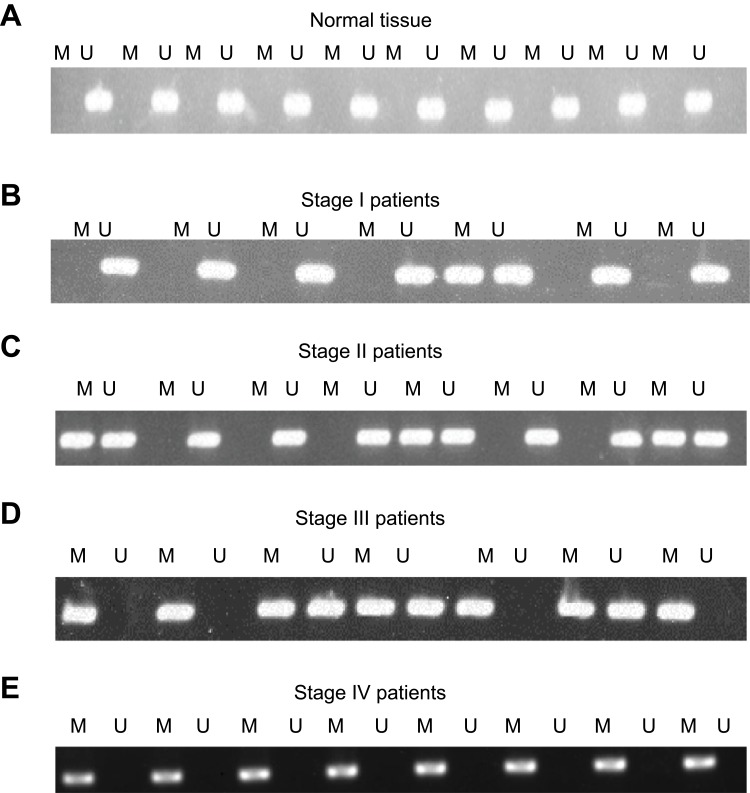
Gene methylation exerts an important function in cancer development. To further investigate the regulation mechanism of KCNK15-AS1 on gastric cancer, the methylation status of KCNK15-AS1 promoter was determined in gastric cancer tissue and cell lines using methylation-specific PCR. The results showed that KCNK15-AS1 promoter methylation was not found in the adjacent normal tissues, whereas it was found in gastric cancer patients from stage I to stage IV (Figure 4A–E). The frequency rate of methylation was higher in stage III and IV than stage I and II. Based on these results, we concluded that the KCNK15-AS1 promoter methylation was existed in gastric cancer tissue.
Figure 4.
(A) The methylation of the KCNK15-AS1 promoter was not detected in the normal controls. (B) The methylation of the KCNK15-AS1 promoter was detected in one of seven stage I patients. (C) The methylation of the KCNK15-AS1 promoter was detected in three of eight stage II patients. (D) The methylation of the KCNK15-AS1 promoter was detected in all the stage III patients. (E) The methylation of the KCNK15-AS1 promoter was detected in all the stage IV patients.
Abbreviations: M, methylated; U, unmethylated.
HDAC1 and DNMT1 participate in KCNK15-AS1 methylation in gastric cancer
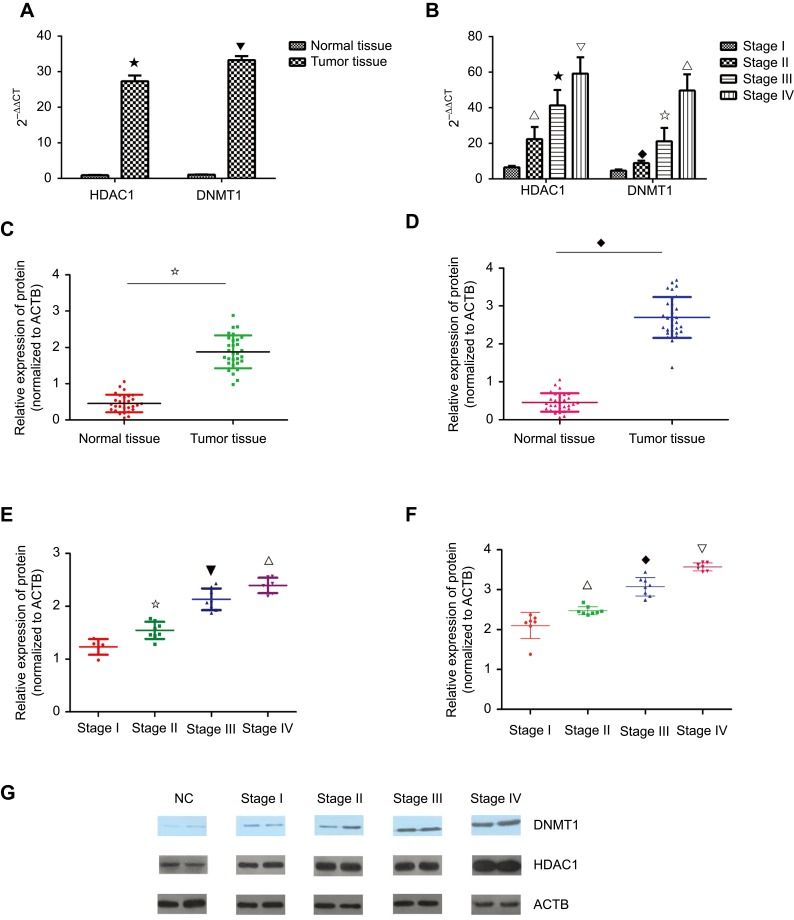
In the abovementioned results, KCNK15-AS1 promoter methylation was found. Thus, the expression changes of methylation and histone modification-related protein were detected such as HDAC1 and DNMT1. The mRNA levels of HDAC1 and DNMT1 were detected by RT-PCR, and the level of HDAC1 and DNMT1 was significantly increased in gastric cancer compared to normal tissue (Figure 5A). Moreover, the highest mRNA level of HDAC1 and DNMT1 was found in the clinical stage III and IV samples (Figure 5B). Consistent with the results of the RT-PCR, the protein level of HDAC1 and DNMT1 was higher in the gastric cancer tissue (Figure 5C and D). Moreover, the highest level of HDAC1 and DNMT1 was found in the clinical stage III and IV samples (Figure 5E–G).
Figure 5.
(A) The expression level of HDAC1 and DNMT1 was higher in the tumor tissue compared to the normal tissue (⋆ ⎂P<0.05). (B) The expression level of HDAC1 and DNMT1 was higher in the stage III and stage IV cases compared to the stage I and stage II cases (∇ ⢶ △P<0.05; ⎂ ♦ ⢶ ∇P<0.05). (C) The protein level of DNMT1 was higher in the tumor tissue compared to the normal tissue (⢶P<0.05). (D) The protein level of HDAC1 was higher in the tumor tissue compared to the normal tissue (♦P<0.05). (E) The protein level of DNMT1 was higher in the high stage than in the low stage (⢶ ⎂ ∇P<0.05). (F) The protein level of DNMT1 was higher in the high stage than the low stage(∇ ♦ △P<0.05). (G) Western blot results showed the protein level of DNMT1 and HDAC1 in different stages.
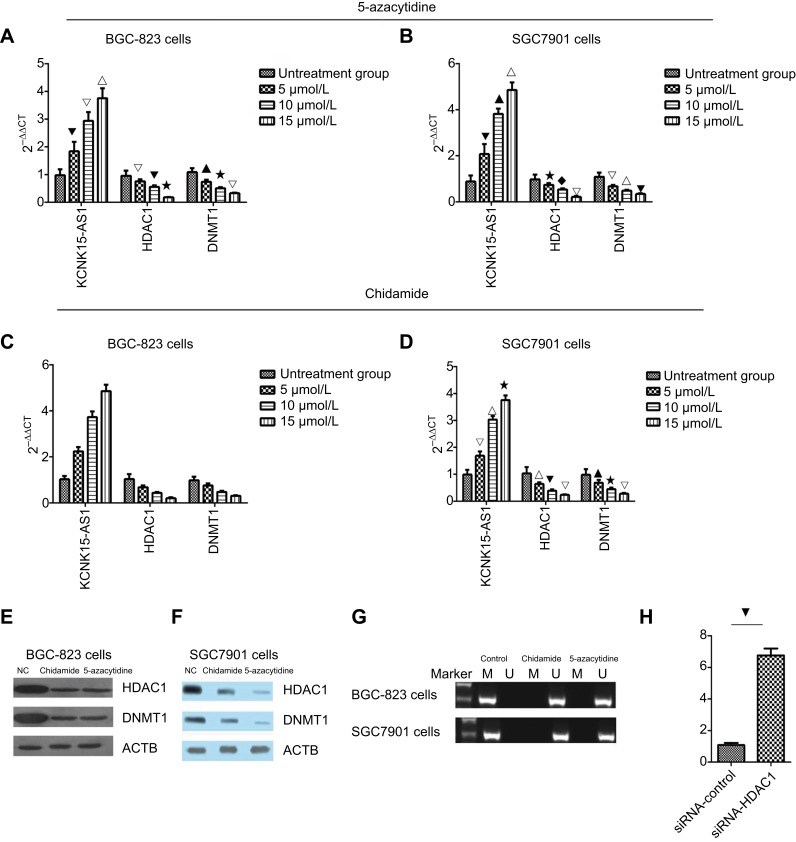
To further explore the role of HDAC1 and DNMT1 in KCNK15-AS1 methylation, the BGC-823 and SGC-7901 cells lines were treated with different concentrations (5, 10, and 15 μmol/L) of chidamide (HDAC inhibitors) and different concentrations (5, 10, and 15 μmol/L) of 5-azacytidine (DNMT inhibitor). Results showed that the mRNA levels of HDAC1 and DNMT1 decreased in a dose-dependent manner in the treated groups compared to the control group, whereas the KCNK15-AS1 mRNA level was increased in the treated groups compared to the control group in BGC-823 cells and SGC7901 cells after treatment with 5-azacytidine and chidamide (Figure 6A–D). The methylation statues of KCNK15-AS1 in BGC-823 cells and SGC7901 cells after treatment with 5-azacytidine and chidamide were also detected.
Figure 6.
(A) The BGC-823 cells were treated with 5-azacytidine, and the expression level of KCNK15-AS1 increased in a dose-dependent manner, and the expression of HDAC1 and DNMT1 decreased in a dose-dependent manner (⎂△∇P<0.05; ▁⋆△P<0.05; △⎂ ⋆P<0.05; ▁△⋆P<0.05). (B) The SCG7901 cells was treated with 5-azacytidine. The expression level of KCNK15-AS1 increased in a dose-dependent manner, and the expression of HDAC1 and DNMT1 decreased in a dose-dependent manner (⎂▁∇P<0.05; ⋆♦△P<0.05; △∇⎂P<0.05; ▁ △ ⋆P<0.05). (C) The BGC-823 cells were treated with chidamide. The expression level of KCNK15-AS1 increased in a dose-dependent manner, and the expression of HDAC1 and DNMT1 decreased in a dose-dependent manner (♦△∇△P<0.05; ⢶⎂∇△P<0.05; △ ⢶▁△P<0.05; ⢶⋆♦△P<0.05). (D) The SCG7901 cells were treated with chidamide. The expression level of KCNK15-AS1 increased in a dose-dependent manner, and the expression of HDAC1 and DNMT1 decreased in a dose-dependent manner (△∇⋆P<0.05; ▁⋆△P<0.05; ∇▁△P<0.05; ▁△⋆P<0.05). (E) Western blot showed the expression levels of HDAC1 and DNMT1 in BGC-823 cells treated with chidamide and 5-azacytidine. (F) Western blot showed the expression levels of HDAC1 and DNMT1 in SCG7901 cells treated with chidamide and 5-azacytidine. (G) After treatment with chidamide and 5-azacytidine, KCNK15-AS1 was unmethylated in both the BGC-823 and SCG7901 cells. (H) The expression of KCNK15-AS1 was decreased after the knockdown of HDAC1 (⎂P<0.05).
Based on the RT-PCR results, we chose the highest concentration of chidamide and 5-azacytidine for the next experiments. The protein levels of HDAC1 and DNMT1 decreased following chidamide treatment compared to the control group (Figure 6E and F). The methylation-specific PCR results showed that the methylation status of KCNK 15- AS1 was detected in the BGC-823 and SGC-7901 cell lines but was not detected after chidamide and 5-azacytidine treatment (Figure 6G). Finally, we knocked down HDAC1 and revealed that the expression of KCKN15-AS1 was significantly increased (Figure 6H). These results indicated that HDAC1 and DNMT1 participated in KCNK15-AS1 methylation in gastric cancer.
ChIP-seq for DNMTI and HDACI
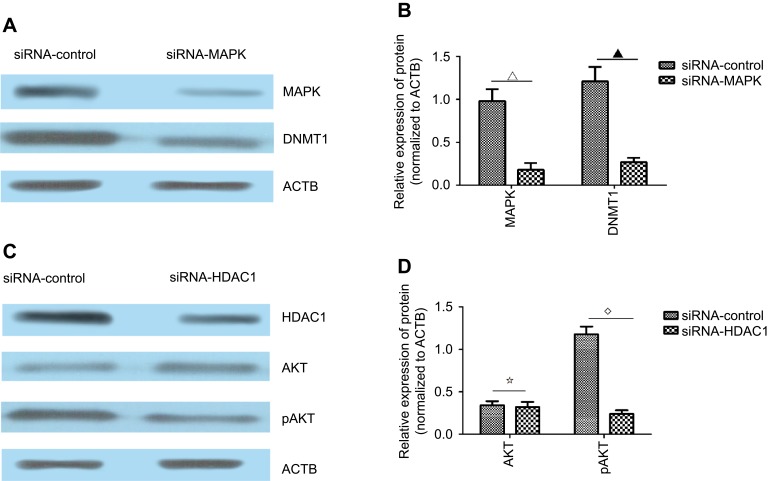
After sequencing the samples, we obtained 17,126,115 original reads. After data filtration, the results showed that the promoter regions of MAPK interacted with DNMT1. In addition, the promoter regions of AKT and STAT5 interacted with HDAC1.
To further assess these findings, we designed and synthesized two siRNAs to knock down the expression of MAPK and HDAC1. We found that the knockdown of MAPK decreased the protein expression of DNMT1 (Figure 7A and B). Furthermore, the knockdown of HDAC1 had no influence on the total protein level of AKT, but decreased the phosphorylation level of AKT (Figure 7C and D). Overall, we showed that KCNK15-AS1 interacted with DNMT1 and HDAC1 to regulate MAPK and AKT.
Figure 7.
(A) Western blot results showed the expression level of MAPK and DNMT1 after the knockdown of MAPK. (B) The expression levels of MAPK and DNMT1 were decreased after the knockdown of MAPK compared to the siRNA-control group (∇▁P<0.05). (C) Western blot results showed the expression level of HDAC1, AKT, and pAKT after the knockdown of HDAC1. (D) The expression levels of AKT and pAKT were decreased after the knockdown of HDAC1 compared to the siRNA-control group (⢶⋄P<0.05).
Discussion
Gastric cancer accounts for ~15.8% and 17.7% of new cancer cases and cancer deaths, respectively. Surgery and chemotherapy are the two main treatments used for gastric cancer patients.19 However, these two treatments are not adequate, which is partly due to the high risk of relapse and chemoresistance. Thus, the prognosis of gastric cancer still remains poor. Therefore, identifying treatment targets remains a challenge in the clinic.20
During the occurrence and development of gastric cancer, the expression of lncRNAs is upregulated or downregulated. MEG3 is downregulated, whereas HOTAIR is upregulated in gastric cancer.21,22 In our study, the expression of KCNK15-AS1 was downregulated in the gastric cancer tissue. Moreover, the lowest level of KCNK15-AS1 was found in the advanced stage of gastric cancer. The overexpression of KCNK15-AS1 inhibited proliferation and promoted apoptosis. Thus, we revealed that a low level of KCNK15-AS1 was associated with the progression of gastric cancer.
miRNAs are a conserved family of small noncoding RNA molecules that posttranscriptionally regulate gene expression.23 miR-21 is a recently identified cancer-related miRNA. Studies showed that miR-21 acts as oncogene or in different types of human cancers. Previous studies have focused on ceRNAs, such as lncRNAs, which act as sponge RNAs that upregulate or downregulate their target miRNAs.24 In our study, miR-21 acted as the target miRNA for KCNK15-AS1. Both the overexpression of KCNK15-AS1 and the knockdown of miR-21 inhibited proliferation and promoted apoptosis. At the same time, the level of bcl-2 and Bax was also determined, and both the overexpression of KCNK15-AS1 and the knockdown of miR-21 decreased the level of bcl-2 and increased the level of Bax. Thus, we speculated that both the overexpression of KCNK15-AS1 and the knockdown of miR-21 inhibited proliferation and promoted apoptosis by decreasing the level of bcl-2. MMP-2 and MMP-9 are two important members of MMP family.25 A high expression level of MMP-2 and MMP-9 is found in many types of cancer, such as ovarian, cervical, hepatocellular, head and neck and thyroid, and squamous cell carcinomas.26,27 In our study, the overexpression of KCNK15-AS1 decreased the levels of MMP-2 and MMP-9.
In this study, we also found that there was KCNK15-AS1 methylation in gastric cancer and the advanced stage showed a high frequency rate of methylation. DNA methylation and histone deacetylation are two key mechanisms that affect gene expression and gene promoters.28 The DNMT family of proteins, which includes DNMT1, is involved in the mediation of CpG island methylation, whereas the HDAC family regulates histone deacetylation.15 Therefore, we first assessed the expression of DNMT1 and HDAC1 in the gastric cancer tissue and found that the highest level of DNMT1 and HDAC1 was found in the advanced stage of gastric cancer. Thus, we speculated that a high level of DNMT1 and HDAC1 was related to the progression of gastric cancer.
Next, we chose two gastric cancer cell lines as a research model and treated the cells with 5-azacytidine and chidamide. After the treatment, the expression levels of HDAC1 and DNMT1 decreased and the level of KCNK15-AS1 increased. Thus, we concluded that the low level of KCNK15-AS1 was regulated not only by DNA methylation but also by histone acetylation.
Many studies focus on RNA-binding proteins in cancer progression. In our study, the RNA pull down and RIP confirmed that DNMT1 and HDAC1 interacted with KCNK15-AS1. Thus, we concluded that DNMT1 and HDAC1 may regulate KCNK15-AS1. To detect DNMT1 and HADC1 in gastric cancer, we performed ChIP-seq. The promoter of MAPK interacted with DNMT1, and the promoter of AKT and STAT5 interacted with HDAC1. We speculated that MAPK regulated DNMT1 and that AKT and STAT5 regulated HDAC1, and these genes may regulate KCNK15-AS1. Thus, targeting DNA methyla- tion and histone acetylation is a promising avenue for the treatment of gastric cancer. Indeed, procaine, which is a local anesthetic, shows potential as DNA methylation inhibitor gastric carcinoma.29 Furthermore, one promising cancer therapy involves epigenetic control. For example, the histone deacetylase inhibitor trichostatin A was used to treat gastric cell lines, and different patterns of histone deacetylase and acetyltransferase mRNA expression were observed. Importantly, there was a downregulation of the oncogene MYC and an upregulation of the tumor suppressor CDKN1A.30 Our findings contribute to the utilization of epigenetic control mechanisms to combat gastric cancer.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Zhao L, Liu Y, Tong D, et al. MeCP2 promotes gastric cancer progression through regulating FOXF1/Wnt5a/p-catenin and MYOD1/Caspase-3 signaling pathways. EBioMedicine. 2017;16:87–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li H, Li W, Liu S, et al. DNMT1, DNMT3A and DNMT3B polymorphisms associated with gastric cancer risk: a systematic review and meta-analysis. EBioMedicine. 2016;13:125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Argentieri MA, Nagarajan S, Seddighzadeh B, Baccarelli AA, Shields AE. Epigenetic pathways in human disease: the impact of DNA methyla- tion on stress-related pathogenesis and current challenges in biomarker development. EBioMedicine. 2017;18:327–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qiu D, Wang Q, Wang Z, et al. RNF185 modulates JWA ubiquitination and promotes gastric cancer metastasis. Biochim Biophys Acta Mol Basis Dis 2018;1864(5 Pt A):1552–1561. [DOI] [PubMed] [Google Scholar]
- 5.Chen B, Zhao Q, Guan L, et al. Long non-coding RNA NNT-AS1 sponges miR-424/E2F1 to promote the tumorigenesis and cell cycle progression of gastric cancer. J Cell Mol Med 2018;22(10):4751–4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang X, Liang W, Liu J, et al. Long non-coding RNA UFC1 promotes gastric cancer progression by regulating miR-498/Lin28b. J Exp Clin Cancer Res. 2018;37(1):134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu YB, Jiang Q, Yang MY, Zhou JX, Zhang Q. Long noncoding RNA NNT-AS1 promotes hepatocellular carcinoma progression and metastasis through miR-363/CDK6 axis. Oncotarget. 2017;8(51):88804–88814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He Y, Hu H, Wang Y, et al. ALKBH5 inhibits pancreatic cancer motility by decreasing long non-coding RNA KCNK15-AS1 methylation. Cell Physiol Biochem. 2018;48(2):838–846. [DOI] [PubMed] [Google Scholar]
- 9.Ma MZ, Lin R, Carrillo J, et al. A DNMT3B4-del contributes to aberrant DNA methylation patterns in lung tumorigenesis. EBioMedicine. 2015;2(10):1340–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zy L, Yang L, Liu XJ, Wang XZ, Pan YX, Luo JM. The long noncoding RNA MEG3 and its target miR-147 regulate JAK/STAT pathway in advanced chronic myeloid leukemia. EBioMedicine. 2018;34:61–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bárcena-Varela M, Caruso S, Llerena S, et al. Dual targeting of histone methyltransferase G9a and DNA-methyltransferase 1 for the treatment of experimental hepatocellular carcinoma Hepatology. Epub 2018. Jul 16. [DOI] [PubMed] [Google Scholar]
- 12.Zhang N, Zhang H, Liu Y, et al. SREBP1, targeted by miR-18a-5p, modulates epithelial-mesenchymal transition in breast cancer via forming a co-repressor complex with Snail and HDAC1/2 Cell Death Differ. 2018;66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shinke G, Yamada D, Eguchi H, et al. Role of histone deacetylase 1 in distant metastasis of pancreatic ductal cancer. Cancer Sci. 2018;109(8):2520–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H, Wang X, Zhang C, et al. HDAC1-induced epigenetic silencing of ASPP2 promotes cell motility, tumour growth and drug resistance in renal cell carcinoma. Cancer Lett. 2018;432:121–131. [DOI] [PubMed] [Google Scholar]
- 15.Zhang T, Li J, Ma X, et al. Inhibition of HDACs-EphA2 signaling axis with WW437 demonstrates promising preclinical antitumor activity in breast cancer. EBioMedicine. 2018;31:276–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vigushin DM, Coombes RC. Histone deacetylase inhibitors in cancer treatment. Anticancer Drugs. 2002;13(1):1–13. [DOI] [PubMed] [Google Scholar]
- 17.Dokmanovic M, Marks PA. Prospects: histone deacetylase inhibitors. J Cell Biochem. 2005;96(2):293–304. [DOI] [PubMed] [Google Scholar]
- 18.Shi Y, Jia B, Xu W, et al. Chidamide in relapsed or refractory peripheral T cell lymphoma: a multicenter real-world study in China. J Hematol Oncol. 2017;10(1):69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurata T, Fushida S, Kinoshita J, et al. Low-dose eribulin mesylate exerts antitumor effects in gastric cancer by inhibiting fibrosis via the suppression of epithelial-mesenchymal transition and acts synergisti- cally with 5-fluorouracil. Cancer Manag Res. 2018;10:2729–2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu X, Zhang J, Liu L, et al. Protein arginine methyltransferase 5-mediated epigenetic silencing of IRX1 contributes to tumorigenicity and metastasis of gastric cancer. Biochim Biophys Acta Mol Basis Dis 2018;1864(9 Pt B):2835–2844. [DOI] [PubMed] [Google Scholar]
- 21.Yan J, Dang Y, Liu S, Zhang Y, Zhang G. LncRNA HOTAIR promotes cisplatin resistance in gastric cancer by targeting miR-126 to activate the PI3K/AKT/MRP1 genes Tumour Biol. Epub 2016. November 13. [DOI] [PubMed] [Google Scholar]
- 22.Dan J, Wang J, Wang Y, et al. LncRNA-MEG3 inhibits proliferation and metastasis by regulating miRNA-21 in gastric cancer. Biomed Pharmacother. 2018;99:931–938. [DOI] [PubMed] [Google Scholar]
- 23.Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107(7):823–826. [DOI] [PubMed] [Google Scholar]
- 24.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3): 353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Osawa Y, Oboki K, Imamura J, et al. Inhibition of cyclic adenosine monophosphate (cAMP)-response element-binding protein (CREB)- binding protein (CBP)/p-Catenin reduces liver fibrosis in mice. EBio- Medicine. 2015;2(11):1751–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y, Huang L, Wang S, et al. WAP four-disulfide core domain protein 2 promotes metastasis of human ovarian cancer by regulation of metastasis-associated genes. J Ovarian Res. 2017;10(1):40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang LL, Sun KX, Wu DD, et al. DLEU1 contributes to ovarian carcinoma tumourigenesis and development by interacting with miR- 490-3p and altering CDK1 expression. J Cell Mol Med. 2017;21(11): 3055–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349(21):2042–2054. [DOI] [PubMed] [Google Scholar]
- 29.Li YC, Wang Y, Li DD, Zhang Y, Zhao TC, Li CF. Procaine is a specific DNA methylation inhibitor with anti-tumor effect for human gastric cancer. J Cell Biochem. 2018;119(2):2440–2449. [DOI] [PubMed] [Google Scholar]
- 30.Wisnieski F, Calcagno DQ, Leal MF, et al. Differential expression of histone deacetylase and acetyltransferase genes in gastric cancer and their modulation by trichostatin A. Tumour Biol. 2014;35(7):6373–6381. [DOI] [PubMed] [Google Scholar]