Type VI secretion systems (T6SSs) are highly conserved and complex protein secretion systems that deliver effector proteins into eukaryotic hosts or other bacteria. T6SSs are regulated precisely by a variety of regulatory systems, which enables bacteria to adapt to varied environments.
KEYWORDS: Fur, S. Typhimurium, type VI secretion system, clpV, regulation
ABSTRACT
Type VI secretion systems (T6SSs) are highly conserved and complex protein secretion systems that deliver effector proteins into eukaryotic hosts or other bacteria. T6SSs are regulated precisely by a variety of regulatory systems, which enables bacteria to adapt to varied environments. A T6SS within Salmonella pathogenicity island 6 (SPI-6) is activated during infection, and it contributes to the pathogenesis, as well as interbacterial competition, of Salmonella enterica serovar Typhimurium (S. Typhimurium). However, the regulation of the SPI-6 T6SS in S. Typhimurium is not well understood. In this study, we found that the SPI-6 T6SS core gene clpV was significantly upregulated in response to the iron-depleted condition and during infection. The global ferric uptake regulator (Fur) was shown to repress the clpV expression in the iron-replete medium. Moreover, electrophoretic mobility shift and DNase I footprinting assays revealed that Fur binds directly to the clpV promoter region at multiple sites spanning the transcriptional start site. We also observed that the relieving of Fur-mediated repression on clpV contributed to the interbacterial competition activity and pathogenicity of S. Typhimurium. These findings provide insights into the direct regulation of Fur in the expression and functional activity of SPI-6 T6SS in S. Typhimurium and thus help to elucidate the mechanisms of bacterial adaptability and virulence.
INTRODUCTION
Pathogenic organisms use several common strategies to compete with other species and accomplish systemic infections. The conserved type VI secretion system (T6SS) is present in more than one-fourth of Gram-negative pathogens (1, 2). The structure of the T6SS apparatus is composed of 13 core components, and additional components may be involved in effector translocation. The transenvelope complex of the T6SS consists of stacked Hcp protein hexamers, which are capped by the VgrG protein. Moreover, Hcp and VgrG act not only as secreted effector proteins but also as machine components. The ClpV ATPase is essential for T6SS secretion and the contraction of the surrounding sheath (3–8). Pathogens such as Vibrio cholerae (V. cholerae), Pseudomonas aeruginosa (P. aeruginosa), and pathogenic Escherichia coli employ T6SSs to deliver effector proteins into eukaryotic or prokaryotic cells (9), and these effectors play a role in a broad variety of functions, including interbacterial interaction, stress sensing, biofilm formation, and virulence (10–20). Salmonella enterica serovar Typhimurium (S. Typhimurium) is an important human and animal pathogen that causes human gastroenteritis and a typhoid-like disease in mice. Moreover, it is a leading etiological agent associated with foodborne illness outbreaks worldwide, which commonly result in heavy economic losses globally (21–23). Five different T6SSs encoded within Salmonella pathogenicity islands (SPIs) were identified among Salmonella lineages, whereas S. Typhimurium only harbors a single SPI-6-encoded T6SS (24–28). Recent studies demonstrated that the SPI-6 T6SS is encoded by two divergent operons with opposite orientations (24, 26, 29). It has been reported that SPI-6 T6SS expression is activated during infection, which is associated with the intracellular replication, systemic dissemination, and virulence of S. Typhimurium. Moreover, the bactericidal activity mediated by the SPI-6 T6SS contributes to the survival and interbacterial competition of S. Typhimurium in vitro and in vivo (19, 25, 29–31).
To adapt to various environments or stresses, bacteria can precisely control gene expression through intricate regulatory networks. In addition, the physiological control of virulence gene expression is essential for bacterial pathogenesis. Previous studies have indicated that T6SSs are tightly regulated (4, 32). Various regulatory mechanisms for T6SSs at transcriptional, translational, and posttranslational levels have been identified and characterized (4, 33). P. aeruginosa regulates T6SS transcripts posttranscriptionally via the Gac/Rsm pathway, whereas the threonine phosphorylation pathway activates the T6SS (34, 35). A broad variety of regulators, such as the quorum-sensing system, alternative sigma factors, H-NS, and ferric uptake regulator (Fur), were found to directly or indirectly modify the transcription of T6SSs (29, 36–42). Quorum-sensing regulators were shown to positively or negatively regulate the expression of T6SSs based on cell population density. It has been shown that T6SS gene clusters are positively controlled by the alternative sigma factor RpoN, sigma factor 54, and cognate bacterial enhancer binding proteins (36–38, 43). In Yersinia pseudotuberculosis, the two-component system regulator OmpR positively regulates T6SS-4 to maintain intracellular pH homeostasis (39). The PhoB/PhoR two-component system and Fur act as an activator and a repressor, respectively, for T6SS expression in Edwardsiella tarda (E. tarda) (40). H-NS is a global regulator that controls the expression of multiple secretion systems and virulence genes. S. Typhimurium SPI-6 T6SS gene cluster is repressed by the H-HS through preventing the binding of RNA polymerase (29, 41).
Iron is essential for bacterial survival and pathogenesis. Bacteria modulate gene expression by various regulators that are sensitive to environmental cues, such as iron concentration. Fur is a global regulator that controls genes involved in iron homeostasis and virulence in many bacteria. Fur regulates gene expression through Fe2+-dependent binding to a specific sequence, namely, the Fur box, in the promoter regions of the regulated genes. The Fur box, which contains a 19-bp consensus sequence, is well characterized. The Fur regulon contains genes involved in iron acquisition, flagellar chemotaxis, the tricarboxylic acid cycle, type III secretion systems (T3SSs) and T6SSs, and it is essential for bacterial multiplication and pathogenicity (44–48). In S. Typhimurium, Fur was shown to directly activate SPI-1 expression and repress SPI-2 expression. Moreover, Fur indirectly regulates SPI-1 expression by repressing H-NS (49–51). Previous evidence showed that T6SS expression is repressed directly by Fur in E. tarda, P. aeruginosa, enteroaggregative E. coli (EAEC) and avian pathogenic E. coli (APEC) (20, 38, 40, 52). This prompted us to determine the regulation of the SPI-6 T6SS mediated by Fur in S. Typhimurium.
In the present study, we demonstrated that Fur bound directly to the clpV promoter region and repressed its expression. Furthermore, we showed that the Fur-mediated regulation of SPI-6 T6SS expression was important for the interbacterial competition and virulence of S. Typhimurium. Taken together, these observations reveal that Fur plays a negative regulatory role in clpV expression and T6SS function in S. Typhimurium.
RESULTS
T6SS core gene clpV expression is upregulated during infection and under iron-depleted conditions.
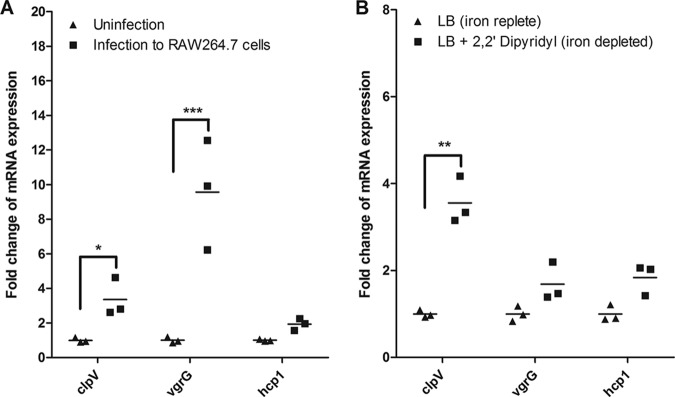
It has been indicated that the expression of SPI-6 T6SS was increased during infection, suggesting that T6SS contributes to the pathogenesis of S. Typhimurium (19, 25, 30, 53). In an initial experiment, we analyzed the transcript levels of T6SS core genes during infection in RAW 264.7 cells by quantitative real-time PCR (qRT-PCR). Similarly, the expressions of SPI-6 T6SS core genes clpV and vgrG were significantly enhanced during infection (P < 0.05 or P < 0.001) (Fig. 1A). During infection, the low-iron environment could induce the expression of T6SS required for survival and virulence. Thus, we next sought to determine whether T6SS gene expression was upregulated in the iron-depleted medium. The expression levels of clpV, vgrG, and hcp1 genes were compared in Luria-Bertani (LB) medium with or without iron chelator 2,2′-dipyridyl. The depletion of free iron leads to significantly increased clpV expression (P < 0.01) (Fig. 1B). These results suggested that T6SS clpV gene expression was increased during infection and in iron-depleted medium.
FIG 1.
Infection and iron-depleted conditions significantly upregulate SPI-6 T6SS core gene clpV expression in S. Typhimurium. (A) The transcript levels of T6SS core genes during infection in RAW 264.7 cells were analyzed by qRT-PCR. The expressions of SPI-6 T6SS core genes clpV and vgrG were significantly enhanced during infection of macrophages. (B) Effect of culture in iron-depleted medium on T6SS core gene expression. The supplementation of iron chelator 2,2′-dipyridyl lead to significantly increased clpV expression. Statistical significance was assessed by using one-way ANOVA (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Fur represses the expression of clpV in S. Typhimurium under iron-replete conditions.
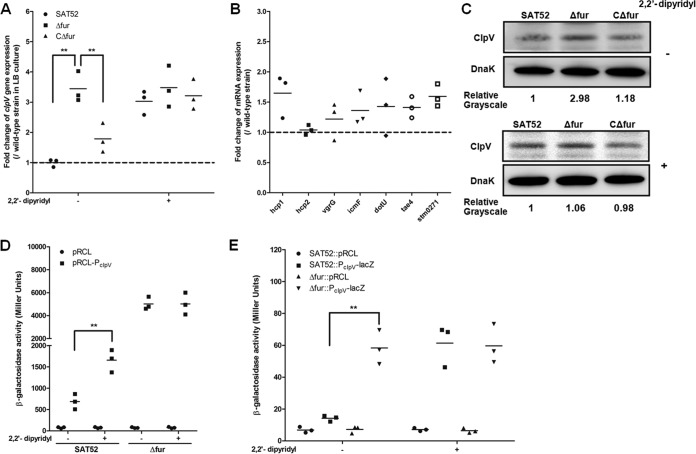
The regulator Fur has been known as the major regulator of iron metabolism. Fur complexes with iron and binds to the Fur box, and it subsequently participates in the regulation of various genes involved in environmental tolerance and virulence (44, 46–48). Hence, we intend to determine whether SPI-6 T6SS genes are potentially regulated by Fur. We constructed the fur gene deletion and complemented strains. Then, the expressions of the T6SS core genes were monitored by qRT-PCR. Inactivation of fur led to significantly higher transcription of clpV (P < 0.01), which is essential and provides energy for the secretion process of the T6SS. Moreover, complementation of the fur gene restored clpV transcription (P < 0.01) (Fig. 2A). However, the transcript levels of other T6SS core genes, namely, dotU, vgrG, icmF, hcp1, hcp2, tae4, and stm0271, were slightly upregulated (<2-fold) in the Δfur mutant strain (Fig. 2B), possibly because these core genes are transcribed by different potential promoters (29). In addition, we performed Western blotting to further validate the production levels of ClpV in these strains. Consistent with the qRT-PCR results, the ClpV production was enhanced in the Δfur mutant strain compared to the wild-type and complemented strains. These results indicate that Fur is a negative regulator of clpV expression. Previous studies have indicated that Fur acts as a regulator for the increased T6SS-1 transcription in response to iron limitation in EAEC (20). Thus, we wanted to test whether the transcription of clpV was enhanced by relieving Fur-mediated repression under iron-depleted conditions. Our results showed that the transcript level of clpV was increased in iron-depleted medium (Fig. 2A). Moreover, Fur was not able to repress clpV transcription and expression under the iron limitation condition (Fig. 2A and C).
FIG 2.
Fur represses the expression of the T6SS clpV gene in S. Typhimurium. (A) The transcript levels of clpV under the indicated conditions were analyzed by qRT-PCR. The results are shown as relative expression ratios compared to that of the wild-type strain SAT52 in LB medium. (B) qRT-PCR analysis for the transcriptional levels of the SPI-6 T6SS core genes hcp1, hcp2, vgrG, icmF, dotU, tae4, and stm0271 in wild-type and fur mutant strains. (C) The production levels of ClpV in Salmonella strains were determined by Western blotting. An anti-DnaK antibody was used as a control. The expression levels of the ClpV protein were determined by quantifying the integrated optical density using ImageJ software. (D) β-Galactosidase activities of bacteria containing plasmid-encoded promoterless pRCL-lacZ fusions and PclpV-lacZ fusions with or without treatment by 2,2′-dipyridyl. (E) The β-galactosidase activities of the chromosomal PclpV-lacZ reporter fusions were measured in wild-type and fur mutant strains in iron-replete and iron-depleted media. Statistical significance was assessed by using one-way ANOVA (**, P < 0.01).
Furthermore, we constructed a lacZ transcriptional reporter fusion plasmid in which lacZ is under the control of the clpV promoter, and it was transformed into wild-type and fur mutant strains. Similarly, deletion of fur significantly activated the transcription of clpV (P < 0.01). Moreover, the activity of the PclpV-lacZ reporter fusion increased in the presence of 2,2′-dipyridyl (Fig. 2D). To avoid artifactual effects of the plasmid on lacZ fusion activity, we constructed a chromosomal PclpV-lacZ transcriptional reporter fusion in the wild-type and fur mutant strains. As expected, the activity of the PclpV-lacZ fusion was significantly higher in the Δfur mutant strain than in the wild-type strain SAT52 in the LB rich medium but not in iron-depleted medium (P < 0.01) (Fig. 2E). Taken together, these findings suggest that Fur represses the expression of clpV in S. Typhimurium under iron-replete conditions.
The Fur protein binds directly to the clpV promoter region.
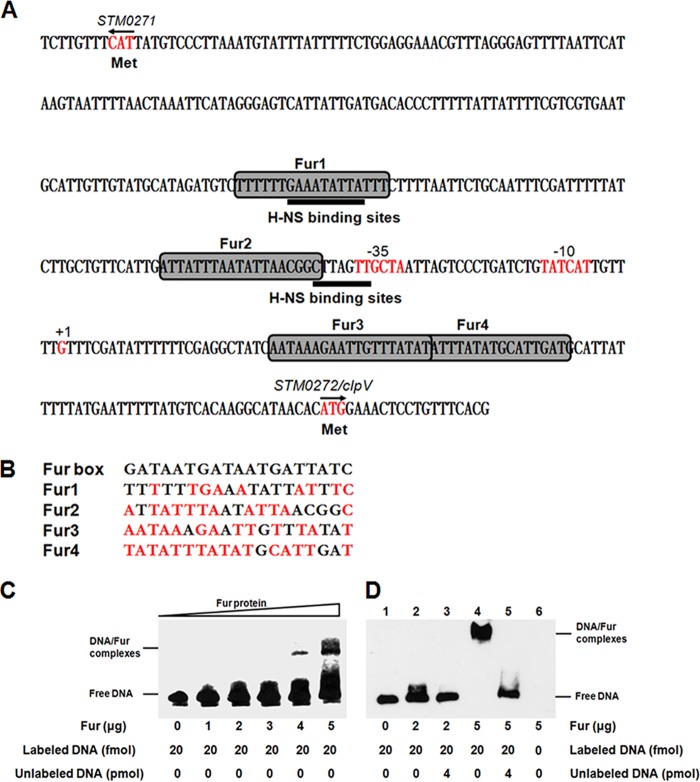
To investigate whether Fur regulation of clpV expression is direct or indirect, we first analyzed the putative Fur binding boxes in the promoter sequence of the SPI-6 T6SS clusters in S. Typhimurium strain SAT52. Our inspection revealed that four putative Fur binding boxes were located upstream of the clpV open reading frame. Moreover, a double Fur binding box was present at positions +25 to +59 downstream of the clpV transcriptional start site (Fig. 3A). However, no Fur binding box was found in other promoters of the SPI-6 T6SS cluster. These putative Fur binding boxes showed 63 to 84% homology to the consensus Fur binding site (Fig. 3B). Thus, the sequence analysis suggests that Fur may bind directly to the clpV promoter and regulate its expression.
FIG 3.
Fur binds directly to the clpV promoter region. (A) In silico analysis for Fur binding box prediction in the clpV promoter region. The putative Fur binding boxes are indicated as gray boxes. H-NS binding sites are indicated by solid lines. The transcriptional initiation site, the corresponding −10 and −35 boxes, and the translational start site are indicated in red letters. (B) Sequence alignment of these putative Fur binding boxes with the Fur box consensus sequence. Bases identical to the consensus are shown in red. (C) EMSA. The clpV promoter DNA fragment (270 bp) was amplified, biotin labeled, and used as a probe, and the biotin-labeled probe was mixed with increasing amounts of purified Fur. (D) Competitive EMSA. The Fur protein was incubated either with the biotin-labeled probe or with both the biotin-labeled and the unlabeled DNA probe. The biotin-labeled DNA was detected using a chemiluminescent substrate. The concentrations of Fur and the probes are shown below the figure.
To verify the direct binding of Fur to the clpV promoter region, a PCR fragment, including the four putative Fur binding boxes, was amplified, biotin labeled, and subjected to an electrophoretic mobility shift assay (EMSA) using purified Fur. As shown in Fig. 3C, the clpV promoter probe was retarded in the presence of increasing amounts of Fur protein. In addition, the binding specificity was confirmed via a competitive EMSA using a specific competitor consisting of unlabeled clpV promoter fragments. The binding of Fur to biotin-labeled probes was abolished by an excess of the specific competitor (Fig. 3D). Collectively, these results demonstrate that Fur could directly bind to the clpV promoter region in S. Typhimurium.
Determination of Fur binding sites required for regulating clpV expression.
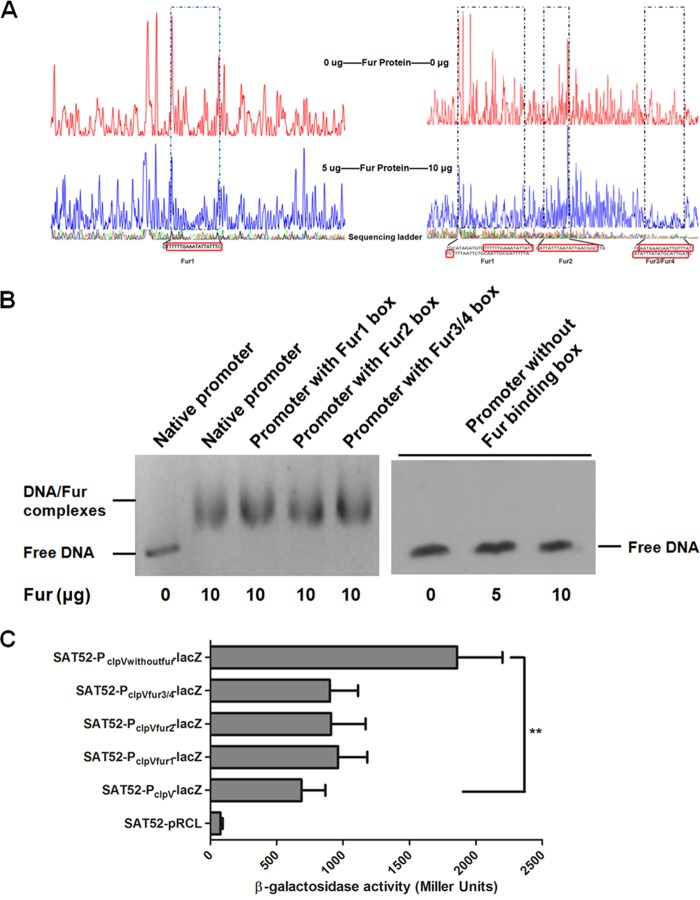
To identify the precise binding sites of Fur in the clpV promoter region, DNase I footprinting assays were performed. The results revealed that Fur bound to a 20-bp region (the Fur1 binding box) of the clpV promoter at a low concentration (5 μg). In contrast, three separate protected regions (positions −129 to −73, positions −58 to −36, and positions +24 to +61, relative to the clpV transcriptional start site), containing the four predicted Fur binding boxes, were identified in the clpV promoter region at a higher concentration (10 μg) of Fur (Fig. 4A). The results indicate that these Fur binding boxes are recognized by Fur; however, the affinity of Fur for the Fur1 binding box was higher than that of the other Fur binding boxes. Interestingly, the upstream and downstream regions of the Fur1 binding box were protected at higher Fur concentrations, suggesting that Fur may bind cooperatively to an initial, high-affinity site and then spread to adjacent sites. Moreover, Fur could bind upstream and downstream of the clpV transcriptional start site (Fur2 and Fur3/4 binding boxes) (Fig. 3A) and regulate the clpV gene transcription. These regulation characteristics of Fur have already been studied in previous works (48).
FIG 4.
Determination of Fur binding sites in the clpV promoter region required for binding and regulation. (A) Mapping Fur binding sites in the clpV promoter region by DNase I footprinting assays. The 5-carboxyfluorescein (FAM)-labeled DNA fragment corresponding to the clpV promoter was incubated with (blue) or without (red) purified Fur. The base sequences of the protected regions are indicated, and the putative Fur binding boxes are shown in the red panes. (B) EMSA demonstrated that the Fur boxes are required for the Fur binding activity. The fragments containing mutated clpV promoter were synthesized and used for the EMSA analysis. Probe with deletion of these Fur binding sites could not be shifted by Fur protein. (C) Effect of Fur binding box mutation on PclpV-lacZ fusion activity. The lacZ transcriptional reporter fusion plasmids containing these mutated clpV promoters were introduced into S. Typhimurium SAT52, and the β-galactosidase activity was analyzed. Statistical significance was assessed by using one-way ANOVA (**, P < 0.01).
To further demonstrate whether these Fur binding boxes are required for binding activity and regulation, we synthesized the clpV promoter fragments and constructed PclpV-lacZ fusion plasmids containing mutations in these Fur binding boxes. The binding of Fur to the mutant clpV promoter probes was determined by EMSA, which demonstrated that Fur protein could bind directly to these putative Fur binding boxes. However, the clpV promoter fragment without the Fur binding box was no longer shifted by the Fur protein (Fig. 4B). Then, the activities of these mutant PclpV-lacZ fusions were compared, which indicated that deletion of Fur binding boxes led to a significantly increased lacZ activity in LB medium (Fig. 4C). Overall, these results demonstrate that Fur could repress the clpV expression by directly binding to these Fur binding sites in the clpV promoter region of S. Typhimurium.
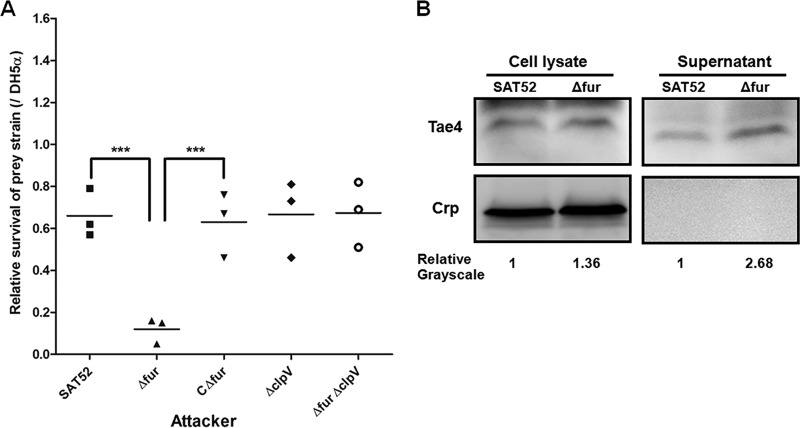
Relieving the Fur-mediated repression of clpV contributes to the interbacterial competition activity of S. Typhimurium.
Recently, it was reported that T6SSs provide a fitness advantage to bacteria, including P. aeruginosa, V. cholerae, EAEC, and S. Typhimurium, through their antibacterial activities (11, 12, 15, 16, 18). Indeed, S. Typhimurium utilizes the SPI-6 T6SS as an antibacterial weapon to compete with other microorganisms in vitro and in colonization of the mouse gut (29, 53). Our finding that the expression of the SPI-6 T6SS core gene clpV was regulated by Fur in S. Typhimurium prompted us to investigate whether antibacterial activity was affected in the Δfur mutant strain. We first determined the interbacterial competition activity of S. Typhimurium using E. coli DH5α strain as prey in LB medium. When cocultured with the wild-type strain SAT52, the prey strain E. coli DH5α did not show significantly decreased survival. This might be due to the repression and weak expression of the SPI-6 T6SS in S. Typhimurium, which is consistent with previous studies (29, 53). In contrast, DH5α survival was significantly reduced when the Δfur mutant was used as an attacker strain compared to the wild-type and complemented strains (P < 0.001). As expected, the mutant ΔclpV and Δfur ΔclpV strains had no impact on DH5α growth, suggesting that the intoxication of prey strain DH5α by S. Typhimurium Δfur mutant strain is due to the SPI-6 T6SS (Fig. 5A). The interbacterial competition activity of S. Typhimurium was further tested in iron-depleted medium. No significant difference in the interbacterial competition activity was observed among the fur mutant, wild-type, and complemented strains; this might be due to the derepression of T6SS mediated by Fur under iron-depleted conditions. Similarly, the ΔclpV and Δfur ΔclpV strains had no impact on the growth and survival of the prey strain DH5α (see Fig. S1 in the supplemental material).
FIG 5.
Derepression of clpV mediated by Fur contributes to the interbacterial competition activity of S. Typhimurium. (A) Interbacterial competition assay. The prey strain E. coli DH5α was mixed with the indicated attacker cells at a ratio of 1:4. After incubation for 4 h at 37°C, the surviving E. coli DH5α cells were recovered and enumerated. The ratio compared to the prey strain E. coli DH5α was calculated as the relative survival. Statistical significance was assessed by using one-way ANOVA (***, P < 0.001). (B) Expression and secretion of SPI-6 T6SS antibacterial effector Tae4 was increased in the mutant Δfur strain. Crp (cyclic AMP receptor protein) is used as the cytoplasmic protein marker.
The T6SS component proteins form a complex membrane tubular structure, which delivers the effectors targeting eukaryotic hosts or bacteria. The effector Tae4 has been demonstrated previously to function via antibacterial amidase activity in S. Typhimurium (53). Moreover, our results demonstrate that Fur directly represses the expression of T6SS clpV in S. Typhimurium. Hence, we sought to determine whether the decreased interbacterial activity of the Δfur mutant strain was due to changes in Tae4 expression and secretion. The tae4 gene was introduced into the plasmid pBAD/Myc-His (Invitrogen) and transformed into wild-type and fur mutant strains. The effect of Fur on the expression and secretion of the Tae4-His fusion protein was then assessed. The expression of Tae4 increased to 1.36-fold in the fur mutant strain, consistent with the qRT-PCR results (Fig. 2B). Moreover, increased Tae4-His production was detected in culture supernatants of the Δfur mutant strain (Fig. 5B). These data demonstrate that relieving the Fur-mediated repression of clpV facilitates the assembly, secretion, and antibacterial activity of SPI-6 T6SS in S. Typhimurium.
Fur and ClpV are required for the infection and pathogenicity of S. Typhimurium.
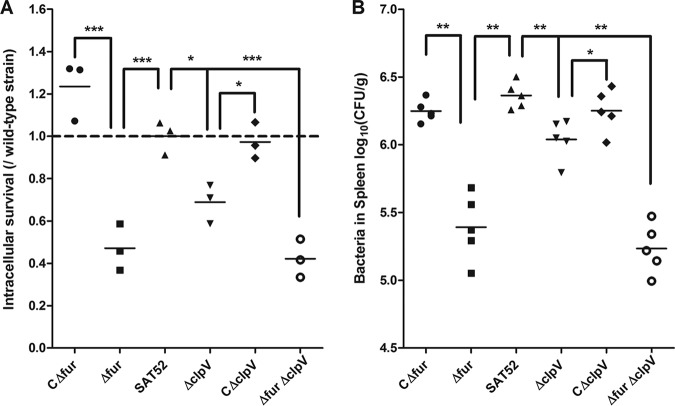
Our results and previous data indicated that the SPI-6 T6SS expression was enhanced and involved in the pathogenesis of S. Typhimurium (19, 25, 30, 53). Thus, we investigated the influence of clpV and fur on bacterial survival in RAW 264.7 macrophages. Consistent with a previous study (19), the clpV and fur mutant strains were defective in intramacrophage survival compared to S. Typhimurium wild-type and complemented strains (Fig. 6A). We also tested whether addition of free iron (FeCl2) affects the intramacrophage survival of S. Typhimurium, which showed similar results (Fig. S2). The intracellular iron concentration is relatively stable; thus, the addition of free iron might not be able to significantly affect intramacrophage survival of S. Typhimurium.
FIG 6.
Bacterial intramacrophage survival and systemic dissemination in mice. (A) Assessment of intramacrophage survival. Intracellular survival of mutant and complemented strains in RAW 264.7 cells was expressed as a fraction of the wild-type strain at 10 h postinvasion. Decreased intracellular survival for the mutant strain was observed, whereas the complemented strain restored the capacity of intracellular survival. Statistical significance was assessed by using one-way ANOVA. (B) Bacterial systemic dissemination in mice. Mice were infected intraperitoneally with S. Typhimurium variants. At 2 days postinfection, the bacterial loads in the spleens were determined. Horizontal bars indicate the means for each sample group. Statistical analyses were performed by using a Mann-Whitney test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
It has been shown that the antibacterial activity of SPI-6 T6SS contributes to early colonization within the host gut for S. Typhimurium (53). Here, we determined the roles of T6SS ClpV in the virulence and colonization capacity of S. Typhimurium during systemic infection. As shown in Table 1, the median lethal dose (LD50) values of the wild-type strain SAT52 and the Δfur and ΔclpV mutant strains were 3.16 × 104 CFU/mouse, 7.50 × 105 CFU/mouse, and 1.33 × 105 CFU/mouse, respectively. Moreover, the mortality rates of the mutant strains were restored by transcomplementation with the fur or clpV genes. These observations further demonstrate that Fur and ClpV are involved in the virulence of S. Typhimurium. Notably, the deletion of both fur and clpV genes led to attenuated virulence compared to single-gene mutant strains (Table 1). Although inactivation of Fur derepressed ClpV function, which subsequently decreased the pathogenicity of the Δfur ΔclpV dual gene mutant strain, the roles of other virulence factors, such as that of the SPI-1 T3SS, might also be affected. Furthermore, the single or dual deletions of the fur and clpV genes decreased bacterial systemic dissemination in vivo. The bacterial loads of the mutant strains in the spleen were significantly lower than those of wild-type and complemented strains (P < 0.05 or P < 0.01) (Fig. 6B).
TABLE 1.
LD50 calculations for each S. Typhimurium strain
| Straina | No. dead/no. tested at challenge dose |
LD50 | ||||
|---|---|---|---|---|---|---|
| 108 CFU | 107 CFU | 106 CFU | 105 CFU | 104 CFU | ||
| SAT52 | 8/8 | 8/8 | 8/8 | 5/8 | 3/8 | 3.16 × 104 |
| Δfur | 8/8 | 7/8 | 6/8 | 0/8 | 0/8 | 7.50 × 105 |
| CΔfur | 8/8 | 8/8 | 8/8 | 6/8 | 0/8 | 5.62 × 104 |
| ΔclpV | 8/8 | 8/8 | 7/8 | 3/8 | 0/8 | 1.33 × 105 |
| CΔclpV | 8/8 | 8/8 | 7/8 | 6/8 | 0/8 | 7.50 × 104 |
| Δfur ΔclpV | 8/8 | 7/8 | 4/8 | 0/8 | 0/8 | 1.33 × 106 |
C, complemented strain.
DISCUSSION
Bacteria precisely modulate gene expression to facilitate their survival in various environments or host cells. T6SSs are present in more than one-fourth of Gram-negative bacteria, and they are involved in a variety of processes, including signal transduction, motility, colonization, secretion of virulence factors, and interbacterial competition (10–20). Previous studies have highlighted that T6SSs are tightly regulated (4, 32). Indeed, it has been shown that multiple regulatory networks, including quorum-sensing systems, two-component systems, alternative sigma factors, H-NS, and Fur, regulate T6SS expression (4, 29, 33, 36–43). Among these regulators, Fur is an extensively investigated global regulator that controls gene expression by binding to the Fur binding box in the promoters of target genes under iron-rich conditions (44, 47). Recently, Fur binding boxes were identified in the promoter regions of the T6SSs in EAEC, APEC, and E. tarda, and their expression was under the control of Fur (20, 38, 40, 52). It has been shown that the SPI-6 T6SS is essential for intramacrophage survival, virulence, and interbacterial competition of S. Typhimurium (19, 25, 30, 53). However, the roles of Fur in the regulation of the SPI-6 T6SS in S. Typhimurium remained unknown. Here, we provided evidence that Fur binds directly to the clpV promoter region and represses the expression and function of the SPI-6 T6SS in S. Typhimurium.
Our and previous studies indicated that T6SS expression was upregulated during infection (19, 25, 30, 53), in which the environments are characterized by low iron. Moreover, we found that the expression of T6SS core gene clpV was enhanced in the iron-depleted medium. Our current knowledge of Fur is as a well-known global regulator facilitating bacterial iron availability and virulence (44–48). Thus, we intended to determine whether Fur regulates the expression of T6SS and clpV. The expression of clpV was significantly higher in the fur gene mutant strain than in the wild-type strain, as determined by qRT-PCR and Western blot analyses. To verify our results, the β-galactosidase activities of different lacZ fusion reporter strains were measured. As expected, the deletion of fur activated PclpV-lacZ expression. In addition, the clpV transcription was significantly enhanced in iron limitation medium; this might be due to the relieving of Fur-mediated repression. These observations demonstrate that Fur plays a negative regulatory role in clpV expression in S. Typhimurium. Similarly, the repression of the T6SS by regulator Fur was also identified in EAEC, APEC, E. tarda, and P. aeruginosa (20, 38, 40, 52).
Furthermore, this work demonstrates by using EMSA and DNase I footprinting analyses that Fur directly binds the clpV promoter. Extensive evidence showed that Fur works as a transcriptional regulator by binding to cognate sites located at the promoter region (44, 54). This study identified four Fur binding boxes in the clpV promoter region of the SPI-6 T6SS in S. Typhimurium. These Fur binding sites were distributed in the clpV promoter region, suggesting that Fur acts cooperatively to repress gene transcription. Among these binding sites, Fur2 and Fur3/4 bind to and spread on the transcriptional start site in the clpV promoter region; this observation is consistent with the mechanism of Fur-mediated repression (44, 54). In contrast, the Fur1 box is located upstream of the promoter but displayed high affinity for the Fur protein. Thus, it might interact initially with Fur and then stimulate further Fur binding at the adjacent downstream binding sites. Previous studies also showed similar observations, indicating that Fur could bind upstream of the promoter and repress gene expression by unknown mechanisms (20, 50). The presence of multiple Fur binding sites in the clpV gene promoter region suggests that several Fur dimers bind to this region. Indeed, several Fur dimers can bind to an extended promoter region, which might efficiently and rapidly alter gene expression to enable bacteria to adapt to the environment (44). Notably, some of these Fur binding boxes overlapped with the H-NS binding sites (29). Moreover, the SPI-6 T6SS gene cluster was shown to be silenced by H-NS via its binding to the clpV promoter region (29). These observations suggest that Fur and H-NS might synergistically control clpV expression in S. Typhimurium. Furthermore, experimental evidence has validated the interplay between Fur and H-NS in controlling SPI-1 gene expression and virulence in S. Typhimurium (49, 50, 55). In addition, Fur could counteract the repression of H-NS by removing H-NS from the repressive site in E. coli. However, H-NS cannot displace Fur from the ftnA promoter, even at high concentrations, indicating that Fur acts as an H-NS antagonist (56). Fur binds to the promoter region of the hns gene, and Fur might be able to indirectly control gene expression by repressing hns gene expression (50). Indeed, expression of the hns gene was upregulated 2.4-fold in the fur mutant strain compared to wild-type strain SAT52. Overall, these data suggest that the expression of SPI-6 T6SS genes might be tightly regulated by a complex regulatory network to optimize gene expression in different environments.
The use of T6SSs is a common strategy for V. cholerae, P. aeruginosa, EAEC, and Salmonella to mediate successful infections in eukaryotic hosts. Moreover, pathogens use this antibacterial weapon to compete efficiently with indigenous microbiota for limited resources (10–20, 53). We therefore sought to determine whether Fur-mediated T6SS regulation provides competition and pathogenesis fitness to S. Typhimurium. Consistent with previous studies (29), the wild-type strain SAT52 did not display sufficient antibacterial activity, which might be repressed by Fur and H-NS. On the other hand, the derepression of the clpV gene in the fur mutant strain caused the lysis of prey E. coli cells. The ClpV ATPase recycles the contracted sheath and was necessary for T6SS assembly and injection. Our results showed that ClpV was required for the interbacterial competition function of SPI-6 T6SS in S. Typhimurium. The relieving of Fur-mediated repression on clpV gene expression might produce and assemble a functionally T6SS structure that is used to deliver the effectors targeting bacteria. Indeed, the Δfur mutant strain showed increased secretion of effector Tae4, which is responsible for its enhanced antibacterial competition activity. In addition, Fur regulates the genes involved in iron acquisition, fimbria, flagella, the SPI secretion system, and metabolism (44–48), which might provide the competition fitness for bacterial growth of the Δfur mutant strain. Our and previous data demonstrated that the low-iron environment inside the host could relieve Fur-mediated transcriptional repression of clpV, strongly suggesting that SPI-6 T6SS contributes to the survival and infection of S. Typhimurium (19, 25). Indeed, increased expression of the SPI-6 T6SS is required for intramacrophage survival and the successful establishment of S. Typhimurium in the gut during infection (25, 31, 53). Similarly, our data showed that the disruption of the SPI-6 T6SS core component ClpV resulted in defective intramacrophage survival, attenuated virulence, and diminished systemic dissemination in mice. Despite the derepression of the clpV gene in the fur mutant strain, the deletion of global regulator Fur extensively dysregulated the expression profile of virulence and metabolic genes (44–48), which might be responsible for the attenuated virulence of the Δfur mutant strain.
Based on our data and previous studies (29, 49–51), we propose a model to illustrate the regulation of the SPI-6 T6SS by Fur in S. Typhimurium (Fig. 7). Under iron-rich conditions, Fur binds to the SPI-6 T6SS clpV promoter region and represses its expression stably. In contrast, iron is scarce inside host cells. Accordingly, Fur is displaced from the promoter region, and the expression of the SPI-6 T6SS increases to promote the interbacterial competition, survival, and infection of S. Typhimurium. In addition, Fur could directly or indirectly control the gene expression of SPIs and other virulence genes, which is also essential for the peak fitness of bacteria in various environments. Taken together, these findings demonstrate that the expression of clpV in the SPI-6 T6SS is strictly controlled by Fur. In response to environmental signals, bacteria could optimize their gene expression through the combinatorial and synergistic actions of multiple regulators. Thus, elucidating the mechanisms of multiple regulatory networks on gene expression will be crucial for understanding bacterial adaptability and virulence.
FIG 7.
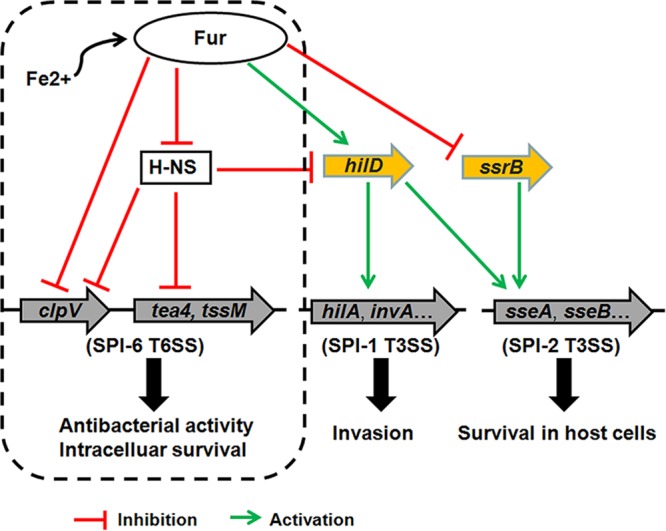
Schematic diagram of the Fur-mediated regulation of SPIs in S. Typhimurium. The regulatory mechanisms by which Fur controls the expression of T3SSs (SPI-1 and SPI-2) and H-NS represses the SPI-6 T6SS have been elucidated previously (29, 49–51). Based on our data, Fur could repress directly the expression of the SPI-6 T6SS clpV gene cluster under iron-rich conditions to keep the energy and source for S. Typhimurium. However, iron-limited environments during infection may relieve the Fur-mediated transcriptional repression of clpV and activate the SPI-6 T6SS as an antibacterial weapon, subsequently facilitating the survival and virulence of S. Typhimurium.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains and plasmids used in this study are listed in Table 2. S. Typhimurium isolate SAT52 was used for the functional analysis (57). E. coli DH5α was used for cloning procedures and competition assays, and E. coli BL21(DE3) was used for the expression procedures. E. coli and Salmonella strains were grown routinely in LB medium at 37°C with aeration. When necessary, ampicillin (100 μg ml−1) or chloramphenicol (30 μg ml−1) was added to the medium. Iron-depleted medium was prepared by supplementing the iron chelator 2,2′-dipyridyl (200 μmol liter−1) in LB medium.
TABLE 2.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristics | Source or reference |
|---|---|---|
| Strains | ||
| SAT52 | Wild-type strain | 57 |
| Δfur | fur deletion mutant of SAT52 | This study |
| CΔfur | Δfur mutant with plasmid pSTV28-fur | This study |
| ΔclpV | clpV deletion mutant of SAT52 | This study |
| CΔclpV | ΔclpV mutant with plasmid pSTV28-clpV | This study |
| Δfur ΔclpV | fur and clpV deletion mutant of SAT52 | This study |
| SAT52-pRCL | SAT52 with plasmid pRCL | This study |
| SAT52-pRCL-PclpV | SAT52 with plasmid pRCL-PclpV | This study |
| Δfur-pRCL | Δfur mutant with plasmid pRCL | This study |
| Δfur-pRCL-PclpV | Δfur mutant with plasmid pRCL-PclpV | This study |
| SAT52:: pRCL-lacZ | pRCL-lacZ transcriptional reporter fusion in SAT52 | This study |
| SAT52::PclpV-lacZ | pRCL-PclpV-lacZ transcriptional reporter fusion in SAT52 | This study |
| Δfur::pRCL-lacZ | pRCL-lacZ transcriptional reporter fusion in Δfur mutant | This study |
| Δfur::PclpV-lacZ | pRCL-PclpV-lacZ transcriptional reporter fusion in Δfur mutant | This study |
| SAT52-pBAD-tae4 | SAT52 with plasmid pBAD-tae4 | This study |
| Δfur-pBAD-tae4 | Δfur mutant with plasmid pBAD-tae4 | This study |
| DH5α | F– Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK– mK+) phoA supE44 λ– | Tiangen |
| BL21(DE3) | F– ompT hsdS(rB– mB–) gal dcm (DE3) | Tiangen |
| Plasmids | ||
| pET28a(+) | Kan, F1 origin, His tag | Novagen |
| pET28a-fur | pET28a(+) carrying fur gene | This study |
| pET28a-clpV | pET28a(+) carrying clpV gene | This study |
| pSTV28 | Cm; lacZ | Takara |
| pSTV28-fur | pSTV28 derivative harboring fur gene | This study |
| pSTV28-clpV | pSTV28 derivative harboring clpV gene | This study |
| pRCL | Promoterless lacZ; Cm gene | 65 |
| pRCL-PclpV | pRCL harboring clpV promoter | This study |
| pBAD/Myc-HisA | Amp; ColE1 derivative cloning vector, pBAD (ara) promoter | Invitrogen |
| pBAD-tae4 | pBAD/Myc-HisA-expressing Tae4-His from the pBAD (ara) promoter | This study |
| pKD46 | Amp; expresses λ red recombinase | 58 |
| pKD3 | Cm gene; template plasmid | 58 |
| pCP20 | Cm Amp; yeast Flp recombinase gene, FLP | 58 |
DNA and protein sequence analyses.
DNA sequences were analyzed using the online Basic Local Alignment Search Tool (BLAST) programs BLASTN and BLASTX from the National Center for Biotechnology Information website (http://blast.ncbi.nlm.nih.gov/). Promoter prediction analyses were conducted with the prediction program tools available at http://www.fruitfly.org/seq_tools/promoter.html. The 500-bp upstream sequence of the clpV gene (from positions −500 to 0 relative to the translation initiation site) was used to search the Fur binding box.
Expression and purification of recombinant proteins.
The open reading frames of the fur and clpV genes of S. Typhimurium SAT52 were amplified and respectively cloned into the pET28a(+) vector. The recombinant proteins were expressed in E. coli BL21(DE3) cells after IPTG (isopropyl-β-d-thiogalactopyranoside) induction at a final concentration of 1 mM. Protein purification was performed using a HisTrap high-performance column (GE Healthcare) according to the manufacturer’s guidelines. The concentrations of the purified proteins were assessed using Pierce BCA protein assay reagent (Thermo Fisher Scientific).
Construction of mutant and complemented strains and plasmids.
The gene deletion mutants were constructed by the Lambda Red recombinase method as described previously (58), with some modification and appropriate primers (Table 3). To generate the chromosomal transcriptional reporter strains, the clpV gene was replaced with a promoterless lacZ gene in wild-type and fur mutant strains using the same procedure. For complementation, the gene coding sequences and their putative promoter regions were amplified and cloned into plasmid pSTV28. The resulting plasmids were transformed into the corresponding mutants to generate complemented strains. The obtained mutant and complemented strains were confirmed using PCR and sequence analysis. To determine the expression and secretion of effector Tae4, the tae4 gene was cloned into plasmid pBAD/Myc-His (Invitrogen, Carlsbad, CA) and transformed into wild-type and fur mutant strains. The growth kinetics of all strains were determined as described previously (17).
TABLE 3.
Primers used in this study
| Analysis and primer | Sequence (5′ to 3′)a | Target gene(s) | Product size (bp) |
|---|---|---|---|
| Gene expression, deletion, and complementation | |||
| clpVEx-F | TCCGAATTCGAAACTCCTGTTTCACGCAG | clpV | 2,634 |
| clpVEx-R | GTGCTCGAGGCAAACGATCTCAAAAACAA | clpV | |
| furEx-F | TCCGAATTCATGACTGACAACAATACCGC | fur | 453 |
| furEx-R | GTGGTCGACTTATTTAGTCGCGTCATCGTGCG | fur | |
| furMu-UF | GCTGTTTCCGCATATCTGCC | Upstream region of fur | 912 |
| furMu-UR | GAAGCAGCTCCAGCCTACACCATGCGGAATCTGTCCTGTT | fur | |
| furMu-CF | AACAGGACAGATTCCGCATGGTGTAGGCTGGAGCTGCTTC | pKD3 | 1,014 |
| furMu-CR | CTTCGAAAGATTTACACTTACATATGAATATCCTCCTTAG | pKD3 | |
| furMu-DF | CTAAGGAGGATATTCATATGTAAGTGTAAATCTTTCGAAG | fur | 870 |
| furMu-DR | CGCGTCGTTATCCAGATGAG | Downstream region of fur | |
| furup-F | AACTCAACGCACAAGCGGTTG | Upstream region of fur | 2,704 |
| furdown-R | CCTATCACCAGTACATCAACC | Downstream region of fur | |
| fur-F | TTTGACGATGCCGGTATCGT | fur | 219 |
| fur-R | AGCGCAGTGGCCGTAAAGATA | fur | |
| furCo-F | TACGAATTCCGCATCAATAGACAAGACC | Upstream region of eivC | 726 |
| furCo-R | GCCAAGCTTGGCTCTTCGAAAGATTTAC | Downstream region of eivC | |
| clpVMu-UF | TTCTGCGGATTGAGCTGTT | Upstream region of clpV | 792 |
| clpVMu-UR | GAAGCAGCTCCAGCCTACACCATGTGTTATGCCTTGTGACA | clpV | |
| clpVMu-CF | TGTCACAAGGCATAACACATGGTGTAGGCTGGAGCTGCTTC | pKD3 | 912 |
| clpVMu-CR | ACACTTCGAACGGCCGGTTTCACATATGAATATCCTCCTTAG | pKD3 | |
| clpVMu-DF | CTAAGGAGGATATTCATATGTGAAACCGGCCGTTCGAAGTGT | clpV | 1,014 |
| clpVMu-DR | CGGTTGCCTGCATATTACTG | Downstream region of clpV | |
| clpVup-F | CATATCCTGTAACGTGTCGGT | Upstream region of clpV | 969 |
| clpVdown-R | TGGGTAGTCTTGGGTCGGAA | Downstream region of clpV | |
| clpVCo-F | TACGAATTCCTGCGGATTGAGCTGTTGTT | Upstream region of clpV | 3,810 |
| clpVCo-R | TACGTCGACAACTGACCTTCACCCGTCAG | Downstream region of clpV | |
| tae4-F | GAGCTCGAGAATGAACAGACCTTCATTCAA | tae4 | 496 |
| tae4-R | GTCAAGCTTCGGCAGTATCCACAGTGTC | tae4 | |
| qRT-PCR | |||
| rpoD RT-F | GTGAAATGGGCACTGTTGAACTG | rpoD | 131 |
| rpoD RT-R | TTCCAGCAGATAGGTAATGGCTTC | rpoD | |
| clpV RT-F | CCAGCGCCATTAGTGATTTTTC | clpV | 219 |
| clpV RT-R | CGATCAACGAGGGCAGTATTTC | clpV | |
| hcp1 RT-F | TAAGGTCTCCGTCACCAACC | hcp1 | 240 |
| hcp1 RT-R | AAGGAGAGCTCCACCGTTTC | hcp1 | |
| hcp2 RT-F | TCTTCCGGTAAGCACATTCC | hcp2 | 195 |
| hcp2 RT-R | CACCACGTATTCCTGCTTC | hcp2 | |
| dotU RT-F | ACCAGCACAGTAGGGTGGTC | dotU | 198 |
| dotU RT-R | CCGACAGTACCAGGTTGGAT | dotU | |
| icmF RT-F | GCTGGCGTAAAATCTTCGAG | icmF | 200 |
| icmF RT-R | GGTAAACCACCAGTCGCAGT | icmF | |
| vgrG RT-F | TGGCGGTAAACGACATATC | vgrG | 183 |
| vgrG RT-R | TATTCCGCCAGAACCTCATC | vgrG | |
| tae4 RT-F | CTCTCCGTATGCAAAGGTTAGT | tae4 | 105 |
| tae4 RT-R | ATAAGATCAGGCTTGCCCATAG | tae4 | |
| STM0271 RT-F | GCGGCTGAATGATGGAGATA | STM0271 | 104 |
| STM0271 RT-R | GAACCAACTGAGGAAACGAAAC | STM0271 | |
| EMSA and DNase I footprinting | |||
| furEMSA-F | CGTCGTGAATGCATTGTTGTA | Upstream region of clpV | 253 |
| furEMSA-R | CATGTGTTATGCCTTGTGACA | clpV | |
| lacZ fusion | |||
| clpVlacZ-UF | CCAGTCCAGATCCAGGTACTCAGC | Upstream region of clpV | 759 |
| clpVlacZ-UR | GAATCCGTAATCATGGTCATGTGTTATGCCTTGTGACATA | clpV | |
| lacZ-F | TATGTCACAAGGCATAACACATGACCATGATTACGGATTC | lacZ | 3,075 |
| lacZ-R | GAAGCAGCTCCAGCCTACACGTCGACATAAAAAAGGGGACCT | lacZ | |
| clpVlacZ-CF | AGGTCCCCTTTTTTATGTCGACGTGTAGGCTGGAGCTGCTTC | pKD3 | 1,014 |
| clpVlacZ-CF | GACACTTCGAACGGCCGGTT CATATGAATATCCTCCTTAGTT | pKD3 | |
| clpVlacZ-DF | AACTAAGGAGGATATTCATATG AACCGGCCGTTCGAAGTGTC | clpV | 812 |
| clpVlacZ-DR | CAGCGTCTGCACCGCGGCTTCCACG | Downstream region of fur | |
| PclpV-F | TTAAAGCTTCGTCGTGAATGCATTGTTGTA | Upstream region of clpV | 275 |
| PclpV-R | CAGGGATCCCACTGCGTGAAACAGGAG | clpV |
Restriction sites are underlined.
To construct the transcriptional lacZ fusion plasmid, the promoter of the clpV gene (from bp −150 to bp +125 relative to the transcriptional start site) was amplified and cloned into the promoterless lacZ plasmid pRCL. The recombinant plasmid, in which the lacZ gene is under the control of clpV promoter, was transformed into wild-type and fur mutant strains. The promoterless plasmid pRCL was used as a control.
β-Galactosidase assays.
β-Galactosidase activity was measured as described previously (59). Briefly, the bacteria were collected and resuspended in Z buffer. A Miller assay for β-galactosidase activity was conducted using ortho-nitrophenyl-β-galactoside as the substrate. This assay was performed three times in triplicate.
EMSA.
To determine the binding of Fur to the clpV promoter, an EMSA was conducted using an EMSA kit (Thermo Fisher Scientific) according to the manufacturer’s protocol. Briefly, the sequence of the clpV promoter region was amplified and labeled with biotin, purified, and quantified. An EMSA was performed in a 20-μl reaction volume that contained biotin-labeled DNA probe (20 fmol) and various amounts of purified Fur in binding buffer [10 mM Tris, 50 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol, 0.1 mM MnCl2, 2.5% glycerol, and 50 ng/μl poly(dI-dC)]. After incubation for 30 min at room temperature, the reaction system was loaded onto a 2% agarose gel, electrophoresed, and transferred to a nylon membrane. The biotin-labeled DNA was detected using a chemiluminescent substrate (Amersham Pharmacia Biotech). A competitive EMSA was performed by simultaneously incubating the biotin-labeled clpV promoter region and the unlabeled clpV promoter region with the Fur protein.
DNase I footprinting assay.
The clpV promoter fragment was amplified and cloned into the pMD18T vector to generate plasmid pMD18T-PclpV. To prepare fluorescent FAM-labeled probes, the promoter region of the clpV gene was PCR amplified with Dpx DNA polymerase from the plasmid pMD18T-PclpV using the primers M13F-47 (FAM labeled) and M13R-48. Then, the FAM-labeled probes were purified and quantified using a NanoDrop 2000C spectrophotometer (Thermo Fisher Scientific). DNase I footprinting assays were performed as described previously (60). The FAM-labeled probe (200 ng) was incubated with different concentrations of the Fur protein for 30 min at 25°C in binding buffer. DNase I digestion was performed by adding 10 μl of DNase I (0.015 U; Promega) and 100 nmol of CaCl2. After incubation for 1 min at 25°C, the reaction was stopped by adding 140 μl of DNase I stop solution (200 mM sodium acetate, 30 mM EDTA, and 0.15% sodium dodecyl sulfate). DNA extraction and electrophoresis were performed as described previously (60).
qRT-PCR.
The expression levels of T6SS genes were investigated using qRT-PCR as described previously (17, 61). Briefly, total RNA was extracted using TRIzol reagent (Invitrogen). RNase-free DNase I (TaKaRa) was added to the RNA samples to remove the contaminating DNA, followed by cDNA synthesis using the PrimeScript RT reagent kit (TaKaRa) according to the manufacturer’s protocol. qRT-PCR was conducted using gene-specific primers (Table 3) as described previously. Data analysis was conducted according to the ΔΔCT method (62) via normalization to the expression of the reference gene rpoD. The relative expression level is shown as the ratio compared to that of the wild-type strain SAT52.
Preparation of secretory proteins.
Secretory proteins were prepared as described previously with modifications (17, 63). Fresh bacteria were diluted 1:100 in LB medium at 37°C with shaking. Bacterial cultures were centrifuged at 12,000 rpm for 15 min at 4°C. The supernatants were then collected and filtered through a 0.22-μm-pore-size membrane to remove bacterial cells. Secretory proteins were precipitated using 10% trichloroacetic acid and washed with acetone. The quality of secretory proteins was verified for the absence of the cytosolic marker cyclic AMP receptor protein (Crp).
Antibody production and Western blotting.
Polyclonal anti-ClpV antiserum was raised in BALB/c mice by subcutaneous immunization with purified ClpV protein mixed with an equal volume of Montanide ISA 206 adjuvant (SEPPIC). The mice were immunized twice, and serum samples were collected 10 days later. For Western blotting, bacterial cultures were concentrated (100 times) by centrifugation at 12,000 rpm and boiled for 10 min. The protein samples were subjected to SDS-PAGE and transferred onto a polyvinylidene fluoride membrane (Amersham Pharmacia Biotech) as described previously (17, 64). Anti-ClpV, anti-DnaK (Enzo Life Sciences), anti-His (Cell Signaling Technology), and anti-Crp (Santa Cruz Biotechnology) were used as primary antibodies. An IRDye 680RD-conjugated donkey anti-mouse polyclonal antibody (LI-COR Biosciences) was used as the secondary antibody. The antigen-antibody complex was visualized with the Odyssey two-color infrared imaging system (LI-COR Biosciences).
Bacterial competition assay.
A bacterial competition assay was performed as described previously (52, 65) with some modifications. In brief, the bacterial strains were grown and adjusted to an optical density at 600 nm of 0.5, and then the fresh attacker (S. Typhimurium) and prey (E. coli DH5α) were mixed at a 4:1 ratio, followed by incubation in LB low-salt plates with nitrocellulose membranes at 37°C for 4 h. Bacteria were collected, diluted, and spotted onto LB plates with or without antibiotics that allowed the selection of attacker or prey bacteria. The survival of prey strain was then calculated. Assays were performed three times in triplicate.
Cell infections.
The intracellular survival assays were carried out as described previously (19) with some modifications. Briefly, murine macrophage RAW 264.7 cells were infected at a multiplicity of infection of 10 for 1 h at 37°C and 5% CO2. Extracellular bacteria were killed with Dulbecco modified Eagle medium (DMEM) containing gentamicin (100 μg/ml) for 1 h, following culture with DMEM containing 10 μg/ml gentamicin for 10 h. The infected cells were then washed and lysed with 0.5% Triton X-100, and surviving bacteria were enumerated by plating on LB agar plates.
Ethical statement.
All animal experiments were conducted in strict accordance with the Guidelines on the Humane Treatment of Laboratory Animals (Ministry of Science and Technology of the People’s Republic of China, policy 2006 398) and were approved by the Institutional Animal Care and Use Committee at the Shanghai Veterinary Research Institute (permit Shvri-Mo-0980). Mice were cared for under specific-pathogen-free conditions at the Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences. To minimize suffering, dying mice were euthanized humanely with an intravenous injection of sodium pentobarbital.
Animal infection experiments.
The LD50 was measured to determine the differences in virulence among the S. Typhimurium strains. Exponential-phase cultures were collected, washed, and adjusted to the appropriate doses in phosphate-buffered saline. Eight-week-old BALB/c mice (n = 8 per group) were infected intraperitoneally with bacterial suspensions containing different numbers of CFU. The numbers of CFU in the injected inoculum were confirmed by plating on LB agar. Negative controls were injected with phosphate-buffered saline. Mortality was monitored daily until 14 days postinfection.
To determine the systemic dissemination of S. Typhimurium, bacterial loads in the spleens of infected mice were measured as described previously, with some modifications (17, 57, 61, 64). Briefly, mice infected with S. Typhimurium were euthanized and dissected at 2 days postinfection. The spleens were collected, weighed, and homogenized in phosphate-buffered saline containing 0.5% Triton X-100. Serial 10-fold dilutions of the homogenates were diluted and plated onto LB agar to count the bacteria.
Statistical analyses.
Statistical analyses were conducted using the GraphPad Software package. One-way analysis of variance (ANOVA) and two-way ANOVA were used to analyze the qRT-PCR, Western blotting, β-galactosidase activity, and bacterial competition assays. Analysis of the animal infection study was performed using a nonparametric Mann-Whitney U test.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (no. 31572523) and the National Key Research and Development Program of China (no. 2016YFD0500800). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
No potential conflicts of interest were disclosed.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00562-19.
REFERENCES
- 1.Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, Heidelberg JF, Mekalanos JJ. 2006. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci U S A 103:1528–1533. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shrivastava S, Mande SS. 2008. Identification and functional characterization of gene components of type VI secretion system in bacterial genomes. PLoS One 3:e2955. doi: 10.1371/journal.pone.0002955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Filloux A. 2009. The type VI secretion system: a tubular story. EMBO J 28:309–310. doi: 10.1038/emboj.2008.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silverman JM, Brunet YR, Cascales E, Mougous JD. 2012. Structure and regulation of the type VI secretion system. Annu Rev Microbiol 66:453–472. doi: 10.1146/annurev-micro-121809-151619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zoued A, Brunet YR, Durand E, Aschtgen MS, Logger L, Douzi B, Journet L, Cambillau C, Cascales E. 2014. Architecture and assembly of the type VI secretion system. Biochim Biophys Acta 1843:1664–1673. doi: 10.1016/j.bbamcr.2014.03.018. [DOI] [PubMed] [Google Scholar]
- 6.Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. 2012. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483:182–186. doi: 10.1038/nature10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durand E, Nguyen VS, Zoued A, Logger L, Pehau-Arnaudet G, Aschtgen MS, Spinelli S, Desmyter A, Bardiaux B, Dujeancourt A, Roussel A, Cambillau C, Cascales E, Fronzes R. 2015. Biogenesis and structure of a type VI secretion membrane core complex. Nature 523:555–560. doi: 10.1038/nature14667. [DOI] [PubMed] [Google Scholar]
- 8.Kapitein N, Bonemann G, Pietrosiuk A, Seyffer F, Hausser I, Locker JK, Mogk A. 2013. ClpV recycles VipA/VipB tubules and prevents non-productive tubule formation to ensure efficient type VI protein secretion. Mol Microbiol 87:1013–1028. doi: 10.1111/mmi.12147. [DOI] [PubMed] [Google Scholar]
- 9.Hachani A, Wood TE, Filloux A. 2016. Type VI secretion and anti-host effectors. Curr Opin Microbiol 29:81–93. doi: 10.1016/j.mib.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 10.Pukatzki S, Ma AT, Revel AT, Sturtevant D, Mekalanos JJ. 2007. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc Natl Acad Sci U S A 104:15508–15513. doi: 10.1073/pnas.0706532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hood RD, Singh P, Hsu F, Guvener T, Carl MA, Trinidad RR, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, Li M, Schwarz S, Wang WY, Merz AJ, Goodlett DR, Mougous JD. 2010. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7:25–37. doi: 10.1016/j.chom.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murdoch SL, Trunk K, English G, Fritsch MJ, Pourkarimi E, Coulthurst SJ. 2011. The opportunistic pathogen Serratia marcescens utilizes type VI secretion to target bacterial competitors. J Bacteriol 193:6057–6069. doi: 10.1128/JB.05671-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang M, Luo Z, Du H, Xu S, Ni B, Zhang H, Sheng X, Xu H, Huang X. 2011. Molecular characterization of a functional type VI secretion system in Salmonella enterica serovar Typhi. Curr Microbiol 63:22–31. doi: 10.1007/s00284-011-9935-z. [DOI] [PubMed] [Google Scholar]
- 14.Ma J, Bao Y, Sun M, Dong W, Pan Z, Zhang W, Lu C, Yao H. 2014. Two functional type VI secretion systems in avian pathogenic Escherichia coli are involved in different pathogenic pathways. Infect Immun 82:3867–3879. doi: 10.1128/IAI.01769-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma LS, Hachani A, Lin JS, Filloux A, Lai EM. 2014. Agrobacterium tumefaciens deploys a superfamily of type VI secretion DNase effectors as weapons for interbacterial competition in planta. Cell Host Microbe 16:94–104. doi: 10.1016/j.chom.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Unterweger D, Miyata ST, Bachmann V, Brooks TM, Mullins T, Kostiuk B, Provenzano D, Pukatzki S. 2014. The Vibrio cholerae type VI secretion system employs diverse effector modules for intraspecific competition. Nat Commun 5:3549. doi: 10.1038/ncomms4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang S, Dai J, Meng Q, Han X, Han Y, Zhao Y, Yang D, Ding C, Yu S. 2014. DotU expression is highly induced during in vivo infection and responsible for virulence and Hcp1 secretion in avian pathogenic Escherichia coli. Front Microbiol 5:588. doi: 10.3389/fmicb.2014.00588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Decoin V, Gallique M, Barbey C, Le Mauff F, Poc CD, Feuilloley MG, Orange N, Merieau A. 2015. A Pseudomonas fluorescens type 6 secretion system is related to mucoidy, motility, and bacterial competition. BMC Microbiol 15:72. doi: 10.1186/s12866-015-0405-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mulder DT, Cooper CA, Coombes BK. 2012. Type VI secretion system-associated gene clusters contribute to pathogenesis of Salmonella enterica serovar Typhimurium. Infect Immun 80:1996–2007. doi: 10.1128/IAI.06205-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brunet YR, Bernard CS, Gavioli M, Lloubes R, Cascales E. 2011. An epigenetic switch involving overlapping fur and DNA methylation optimizes expression of a type VI secretion gene cluster. PLoS Genet 7:e1002205. doi: 10.1371/journal.pgen.1002205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galanis E, Wong DMALF, Patrick ME, Binsztein N, Cieslik A, Chalermchaikit T, Aidara-Kane A, Ellis A, Angulo FJ, Wegener HC, World Health Organization. 2006. Web-based surveillance and global Salmonella distribution, 2000-2002. Emerg Infect Dis 12:381–388. doi: 10.3201/eid1205.050854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pang T, Bhutta ZA, Finlay BB, Altwegg M. 1995. Typhoid fever and other salmonellosis: a continuing challenge. Trends Microbiol 3:253–255. doi: 10.1016/S0966-842X(00)88937-4. [DOI] [PubMed] [Google Scholar]
- 23.Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O’Brien SJ, Jones TF, Fazil A, Hoekstra RM, International Collaboration on Enteric Disease “Burden of Illness Study.” 2010. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis 50:882–889. doi: 10.1086/650733. [DOI] [PubMed] [Google Scholar]
- 24.Blondel CJ, Jimenez JC, Contreras I, Santiviago CA. 2009. Comparative genomic analysis uncovers 3 novel loci encoding type six secretion systems differentially distributed in Salmonella serotypes. BMC Genomics 10:354. doi: 10.1186/1471-2164-10-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu J, Guo JT, Li YG, Johnston RN, Liu GR, Liu SL. 2013. The type VI secretion system gene cluster of Salmonella typhimurium: required for full virulence in mice. J Basic Microbiol 53:600–607. doi: 10.1002/jobm.201200047. [DOI] [PubMed] [Google Scholar]
- 26.Folkesson A, Lofdahl S, Normark S. 2002. The Salmonella enterica subspecies I specific centisome 7 genomic island encodes novel protein families present in bacteria living in close contact with eukaryotic cells. Res Microbiol 153:537–545. doi: 10.1016/S0923-2508(02)01348-7. [DOI] [PubMed] [Google Scholar]
- 27.Pezoa D, Blondel CJ, Silva CA, Yang HJ, Andrews-Polymenis H, Santiviago CA, Contreras I. 2014. Only one of the two type VI secretion systems encoded in the Salmonella enterica serotype Dublin genome is involved in colonization of the avian and murine hosts. Vet Res 45:2. doi: 10.1186/1297-9716-45-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fookes M, Schroeder GN, Langridge GC, Blondel CJ, Mammina C, Connor TR, Seth-Smith H, Vernikos GS, Robinson KS, Sanders M, Petty NK, Kingsley RA, Baumler AJ, Nuccio SP, Contreras I, Santiviago CA, Maskell D, Barrow P, Humphrey T, Nastasi A, Roberts M, Frankel G, Parkhill J, Dougan G, Thomson NR. 2011. Salmonella bongori provides insights into the evolution of the salmonellae. PLoS Pathog 7:e1002191. doi: 10.1371/journal.ppat.1002191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brunet YR, Khodr A, Logger L, Aussel L, Mignot T, Rimsky S, Cascales E. 2015. H-NS silencing of the Salmonella pathogenicity island 6-encoded type VI secretion system limits Salmonella enterica serovar Typhimurium interbacterial killing. Infect Immun 83:2738–2750. doi: 10.1128/IAI.00198-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parsons DA, Heffron F. 2005. sciS, an icmF homolog in Salmonella enterica serovar Typhimurium, limits intracellular replication and decreases virulence. Infect Immun 73:4338–4345. doi: 10.1128/IAI.73.7.4338-4345.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pezoa D, Yang HJ, Blondel CJ, Santiviago CA, Andrews-Polymenis HL, Contreras I. 2013. The type VI secretion system encoded in SPI-6 plays a role in gastrointestinal colonization and systemic spread of Salmonella enterica serovar Typhimurium in the chicken. PLoS One 8:e63917. doi: 10.1371/journal.pone.0063917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyata ST, Bachmann V, Pukatzki S. 2013. Type VI secretion system regulation as a consequence of evolutionary pressure. J Med Microbiol 62:663–676. doi: 10.1099/jmm.0.053983-0. [DOI] [PubMed] [Google Scholar]
- 33.Leung KY, Siame BA, Snowball H, Mok YK. 2011. Type VI secretion regulation: crosstalk and intracellular communication. Curr Opin Microbiol 14:9–15. doi: 10.1016/j.mib.2010.09.017. [DOI] [PubMed] [Google Scholar]
- 34.Brencic A, Lory S. 2009. Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol Microbiol 72:612–632. doi: 10.1111/j.1365-2958.2009.06670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mougous JD, Gifford CA, Ramsdell TL, Mekalanos JJ. 2007. Threonine phosphorylation posttranslationally regulates protein secretion in Pseudomonas aeruginosa. Nat Cell Biol 9:797–803. doi: 10.1038/ncb1605. [DOI] [PubMed] [Google Scholar]
- 36.Sheng L, Gu D, Wang Q, Liu Q, Zhang Y. 2012. Quorum sensing and alternative sigma factor RpoN regulate type VI secretion system I (T6SSVA1) in fish pathogen Vibrio alginolyticus. Arch Microbiol 194:379–390. doi: 10.1007/s00203-011-0780-z. [DOI] [PubMed] [Google Scholar]
- 37.Ishikawa T, Rompikuntal PK, Lindmark B, Milton DL, Wai SN. 2009. Quorum sensing regulation of the two hcp alleles in Vibrio cholerae O1 strains. PLoS One 4:e6734. doi: 10.1371/journal.pone.0006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sana TG, Hachani A, Bucior I, Soscia C, Garvis S, Termine E, Engel J, Filloux A, Bleves S. 2012. The second type VI secretion system of Pseudomonas aeruginosa strain PAO1 is regulated by quorum sensing and Fur and modulates internalization in epithelial cells. J Biol Chem 287:27095–27105. doi: 10.1074/jbc.M112.376368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang W, Wang Y, Song Y, Wang T, Xu S, Peng Z, Lin X, Zhang L, Shen X. 2013. A type VI secretion system regulated by OmpR in Yersinia pseudotuberculosis functions to maintain intracellular pH homeostasis. Environ Microbiol 15:557–569. doi: 10.1111/1462-2920.12005. [DOI] [PubMed] [Google Scholar]
- 40.Chakraborty S, Sivaraman J, Leung KY, Mok YK. 2011. Two-component PhoB-PhoR regulatory system and ferric uptake regulator sense phosphate and iron to control virulence genes in type III and VI secretion systems of Edwardsiella tarda. J Biol Chem 286:39417–39430. doi: 10.1074/jbc.M111.295188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salomon D, Klimko JA, Orth K. 2014. H-NS regulates the Vibrio parahaemolyticus type VI secretion system 1. Microbiology 160:1867–1873. doi: 10.1099/mic.0.080028-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kitaoka M, Miyata ST, Brooks TM, Unterweger D, Pukatzki S. 2011. VasH is a transcriptional regulator of the type VI secretion system functional in endemic and pandemic Vibrio cholerae. J Bacteriol 193:6471–6482. doi: 10.1128/JB.05414-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bernard CS, Brunet YR, Gavioli M, Lloubes R, Cascales E. 2011. Regulation of type VI secretion gene clusters by sigma54 and cognate enhancer binding proteins. J Bacteriol 193:2158–2167. doi: 10.1128/JB.00029-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andrews SC, Robinson AK, Rodriguez QF. 2003. Bacterial iron homeostasis. FEMS Microbiol Rev 27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 45.Schaible UE, Kaufmann SH. 2004. Iron and microbial infection. Nat Rev Microbiol 2:946–953. doi: 10.1038/nrmicro1046. [DOI] [PubMed] [Google Scholar]
- 46.Tsolis RM, Baumler AJ, Stojiljkovic I, Heffron F. 1995. Fur regulon of Salmonella typhimurium: identification of new iron-regulated genes. J Bacteriol 177:4628–4637. doi: 10.1128/jb.177.16.4628-4637.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baichoo N, Helmann JD. 2002. Recognition of DNA by Fur: a reinterpretation of the Fur box consensus sequence. J Bacteriol 184:5826–5832. doi: 10.1128/jb.184.21.5826-5832.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Troxell B, Fink RC, Porwollik S, McClelland M, Hassan HM. 2011. The Fur regulon in anaerobically grown Salmonella enterica sv. Typhimurium: identification of new Fur targets. BMC Microbiol 11:236. doi: 10.1186/1471-2180-11-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Teixido L, Carrasco B, Alonso JC, Barbe J, Campoy S. 2011. Fur activates the expression of Salmonella enterica pathogenicity island 1 by directly interacting with the hilD operator in vivo and in vitro. PLoS One 6:e19711. doi: 10.1371/journal.pone.0019711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Troxell B, Sikes ML, Fink RC, Vazquez-Torres A, Jones-Carson J, Hassan HM. 2011. Fur negatively regulates hns and is required for the expression of HilA and virulence in Salmonella enterica serovar Typhimurium. J Bacteriol 193:497–505. doi: 10.1128/JB.00942-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choi E, Kim H, Lee H, Nam D, Choi J, Shin D. 2014. The iron-sensing Fur regulator controls expression timing and levels of Salmonella pathogenicity island 2 genes in the course of environmental acidification. Infect Immun 82:2203–2210. doi: 10.1128/IAI.01625-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma J, Sun M, Pan Z, Song W, Lu C, Yao H. 2018. Three Hcp homologs with divergent extended loop regions exhibit different functions in avian pathogenic Escherichia coli. Emerg Microbes Infect 7:1. doi: 10.1038/s41426-018-0042-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sana TG, Flaugnatti N, Lugo KA, Lam LH, Jacobson A, Baylot V, Durand E, Journet L, Cascales E, Monack DM. 2016. Salmonella Typhimurium utilizes a T6SS-mediated antibacterial weapon to establish in the host gut. Proc Natl Acad Sci U S A 113:E5044–E5051. doi: 10.1073/pnas.1608858113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Escolar L, Perez-Martin J, de Lorenzo V. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J Bacteriol 181:6223–6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Prajapat MK, Saini S. 2012. Interplay between Fur and HNS in controlling virulence gene expression in Salmonella typhimurium. Comput Biol Med 42:1133–1140. doi: 10.1016/j.compbiomed.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 56.Nandal A, Huggins CC, Woodhall MR, McHugh J, Rodriguez-Quinones F, Quail MA, Guest JR, Andrews SC. 2010. Induction of the ferritin gene (ftnA) of Escherichia coli by Fe2+-Fur is mediated by reversal of H-NS silencing and is RyhB independent. Mol Microbiol 75:637–657. doi: 10.1111/j.1365-2958.2009.06977.x. [DOI] [PubMed] [Google Scholar]
- 57.Wang S, Yang D, Wu X, Wang Y, Wang D, Tian M, Li T, Qi J, Wang X, Ding C, Yu S. 2018. Autotransporter MisL of Salmonella enterica serotype Typhimurium facilitates bacterial aggregation and biofilm formation. FEMS Microbiol Lett 365:fny142. doi: 10.1093/femsle/fny142. [DOI] [PubMed] [Google Scholar]
- 58.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cai W, Wannemuehler Y, Dell’Anna G, Nicholson B, Barbieri NL, Kariyawasam S, Feng Y, Logue CM, Nolan LK, Li G. 2013. A novel two-component signaling system facilitates uropathogenic Escherichia coli’s ability to exploit abundant host metabolites. PLoS Pathog 9:e1003428. doi: 10.1371/journal.ppat.1003428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Y, Cen XF, Zhao GP, Wang J. 2012. Characterization of a new GlnR binding box in the promoter of amtB in Streptomyces coelicolor inferred a PhoP/GlnR competitive binding mechanism for transcriptional regulation of amtB. J Bacteriol 194:5237–5244. doi: 10.1128/JB.00989-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang S, Liu X, Xu X, Yang D, Wang D, Han X, Shi Y, Tian M, Ding C, Peng D, Yu S. 2016. Escherichia coli type III secretion system 2 ATPase EivC is involved in the motility and virulence of avian pathogenic Escherichia coli. Front Microbiol 7:1387. doi: 10.3389/fmicb.2016.01387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 63.Zhou Y, Tao J, Yu H, Ni J, Zeng L, Teng Q, Kim KS, Zhao GP, Guo X, Yao Y. 2012. Hcp family proteins secreted via the type VI secretion system coordinately regulate Escherichia coli K1 interaction with human brain microvascular endothelial cells. Infect Immun 80:1243–1251. doi: 10.1128/IAI.05994-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang S, Niu C, Shi Z, Xia Y, Yaqoob M, Dai J, Lu C. 2011. Effects of ibeA deletion on virulence and biofilm formation of avian pathogenic Escherichia coli. Infect Immun 79:279–287. doi: 10.1128/IAI.00821-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yi Z, Wang D, Xin S, Zhou D, Li T, Tian M, Qi J, Ding C, Wang S, Yu S. 2019. The CpxR regulates type VI secretion system 2 expression and facilitates the interbacterial competition activity and virulence of avian pathogenic Escherichia coli. Vet Res 50:40. doi: 10.1186/s13567-019-0658-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.