Reproductive tract pathology caused by Chlamydia trachomatis infection is an important global cause of human infertility. To better understand the mechanisms associated with Chlamydia-induced genital tract pathogenesis in humans, we used CRISPR genome editing to disrupt Toll-like receptor 3 (TLR3) function in the human oviduct epithelial (hOE) cell line OE-E6/E7 in order to investigate the possible role(s) of TLR3 signaling in the immune response to Chlamydia.
KEYWORDS: Chlamydia trachomatis, TLR3, epithelial cells, inflammation, oviducts
ABSTRACT
Reproductive tract pathology caused by Chlamydia trachomatis infection is an important global cause of human infertility. To better understand the mechanisms associated with Chlamydia-induced genital tract pathogenesis in humans, we used CRISPR genome editing to disrupt Toll-like receptor 3 (TLR3) function in the human oviduct epithelial (hOE) cell line OE-E6/E7 in order to investigate the possible role(s) of TLR3 signaling in the immune response to Chlamydia. Disruption of TLR3 function in these cells significantly diminished the Chlamydia-induced synthesis of several inflammation biomarkers, including interferon beta (IFN-β), interleukin-6 (IL-6), interleukin-6 receptor alpha (IL-6Rα), soluble interleukin-6 receptor beta (sIL-6Rβ, or gp130), IL-8, IL-20, IL-26, IL-34, soluble tumor necrosis factor receptor 1 (sTNF-R1), tumor necrosis factor ligand superfamily member 13B (TNFSF13B), matrix metalloproteinase 1 (MMP-1), MMP-2, and MMP-3. In contrast, the Chlamydia-induced synthesis of CCL5, IL-29 (IFN-λ1), and IL-28A (IFN-λ2) was significantly increased in TLR3-deficient hOE cells compared to their wild-type counterparts. Our results indicate a role for TLR3 signaling in limiting the genital tract fibrosis, scarring, and chronic inflammation often associated with human chlamydial disease. Interestingly, we saw that Chlamydia infection induced the production of biomarkers associated with persistence, tumor metastasis, and autoimmunity, such as soluble CD163 (sCD163), chitinase-3-like protein 1, osteopontin, and pentraxin-3, in hOE cells; however, their expression levels were significantly dysregulated in TLR3-deficient hOE cells. Finally, we demonstrate using hOE cells that TLR3 deficiency resulted in an increased amount of chlamydial lipopolysaccharide (LPS) within Chlamydia inclusions, which is suggestive that TLR3 deficiency leads to enhanced chlamydial replication and possibly increased genital tract pathogenesis during human infection.
INTRODUCTION
The bacterial pathogen Chlamydia trachomatis has caused 1,526,658 infections in the United States in 2015 (an increase of 6% since 2014), and it is the most commonly reported bacterial sexually transmitted disease (STD) in the United States (1). Genital tract C. trachomatis infections are easily cured with antibiotics if properly diagnosed at early stages of infection. However, because 75 to 90% of women infected with Chlamydia are asymptomatic for clinical disease, opportunities for therapeutic interventions are usually missed. The asymptomatic nature of the clinical symptoms is the major factor contributing to the continuing spread of the disease to uninfected partners and the more severe pathogenesis and sequelae that often lead to infertility in women. Further contributing to the growing rates of infectivity among previously uninfected populations are statistics showing that up to 90% of men infected with Chlamydia exhibit no symptoms (2, 3) and that an effective vaccine remains elusive (4).
Chlamydia infections are also leading causes of pelvic inflammatory disease (PID) (5), tubal occlusion (6), and ectopic pregnancy (7, 8) in women. Interactions between host immunity and Chlamydia infection are thought to be largely responsible for the pathology associated with human chlamydial disease, although the precise pathogenic mechanisms remain unclear (9, 10). As an obligate intracellular pathogen, Chlamydia species are known to interact with host cell pattern recognition receptors (PRRs), including a variety of intracellular cytosolic receptors and Toll-like receptors (TLRs), to trigger the innate immune inflammatory response (11–18). Stimulation of genital tract epithelial cell TLRs (and other PRRs) by chlamydial pathogen-associated molecular patterns (PAMPs) triggers cytokine responses that are critical to the establishment of innate and adaptive immunity. These Chlamydia-induced cytokine responses also include the synthesis of inflammatory mediators that have been implicated as the major culprits in the pathology associated with Chlamydia disease (12, 14, 19–24). The overall goal of these investigations into the interactions between host cell PRRs and Chlamydia infection is to identify the PRRs that trigger specific inflammatory mediators that cause scarring and fibrosis and then define therapeutic measures to prevent this process.
Human genital tract epithelial cells express most of the known TLRs; however, the TLRs are known to vary in their expression levels within the female reproductive tract (depending upon the concentration of specific sex hormones) and their tissue distribution (25). The human fallopian tube-derived epithelial cell line OE-E6/E7 (26) was shown to express functional proteins for TLR1 through -6, of which TLR2 was shown to have a role in the innate immune response to C. trachomatis infection (27, 28). TLR2 has also been shown to have a role in the immune responses to Chlamydia muridarum infection in mice, and it had a significant role in the Chlamydia-induced genital tract pathology observed in infected animals (12, 29–31). In our early investigations into the role of TLR3 in the immune response to genital tract Chlamydia infection, we showed that C. muridarum-infected murine oviduct epithelial (OE) cells secrete interferon beta (IFN-β) in a mostly TLR3-dependent manner and that mouse OE cells deficient in TLR3 show dramatic reductions in the synthesis of other inflammatory immune mediators in addition to IFN-β (13, 15). Our most recent report shows that TLR3-deficient mice have increased chlamydial loads, aberrant genital tract secretion levels of several different inflammatory mediators, altered CD4+ T-cell recruitment, and severe oviduct and uterine horn pathology compared to wild-type (WT) controls (32). Our data indicate a protective role for TLR3 signaling in the immune response to Chlamydia infection in mice. However, the role of TLR3 in the immune response to Chlamydia infection in human oviduct tissue has not yet been investigated and remains unclear. In this study, we used the immortalized human oviduct epithelial (hOE) cell line OE-E6/E7 to assess the role of TLR3 in the immune responses to Chlamydia trachomatis L2 infection.
RESULTS
IFN-β is induced in human OE-E6/7 cells in response to Chlamydia infection.
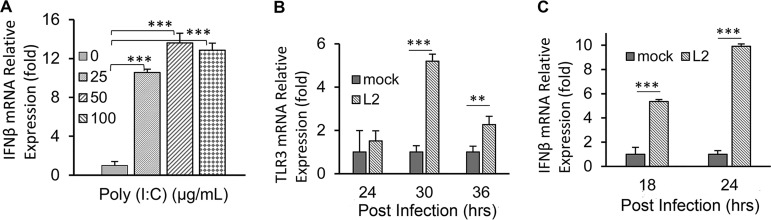
IFN-β is known to be expressed during activation of the TLR3 signaling pathway during certain viral infections and by stimulation via the synthetic double-stranded RNA analog poly(I·C) (33, 34). To confirm the presence of TLR3 and ascertain its function in human OE-E6/E7 cells, hOE cells were incubated in cell culture medium supplemented with increasing concentrations of poly(I·C) for 24 h. Figure 1A shows that the relative IFN-β mRNA expression level was increased at concentrations of 25, 50, and 100 μg/ml compared to untreated controls. These results confirm that TLR3 is functional in hOE cells by demonstrating a dose-dependent increase in IFN-β gene expression in response to poly(I·C) stimulation. To ascertain the impact of Chlamydia infection on IFN-β synthesis in hOE cells, we next infected hOE cells with Chlamydia trachomatis L2 at a multiplicity of infection (MOI) of 10 inclusion-forming units (IFU)/cell for 24 h and measured the mRNA expression levels of both IFN-β and TLR3. As shown in Fig. 1B and C, mRNA expression levels of IFN-β and TLR3 were increased during Chlamydia infection. These data are suggestive that Chlamydia infection of hOE cells induces IFN-β synthesis and upregulates TLR3 gene expression in human oviduct tissue in a manner similar to what we have observed in murine oviduct epithelial cells (14, 24). These findings provide the impetus for us to extrapolate that IFN-β induced during Chlamydia infection in hOE cells will also occur via TLR3-dependent mechanisms, similar to what we have reported for murine OE cells (13).
FIG 1.
TLR3 is present and functional in human OE-E6/7 cells. Human OE-E6/7 cells were seeded into 12-well plates and cultured in DMEM only or DMEM supplemented with poly(I·C) for 24 h or infected with 10 IFU/cell C. trachomatis L2 for 36 h. (A) Poly(I·C) induces the expression of IFN-β mRNA in a dose-dependent manner. (B and C) Relative expression levels of TLR3 mRNA (B) and IFN-β mRNA (C) in response to C. trachomatis L2 infection at the time points indicated. Data are representative of results from at least three independent experiments.
Disruption of TLR3 function in human genital tract epithelial cells and clone identification.
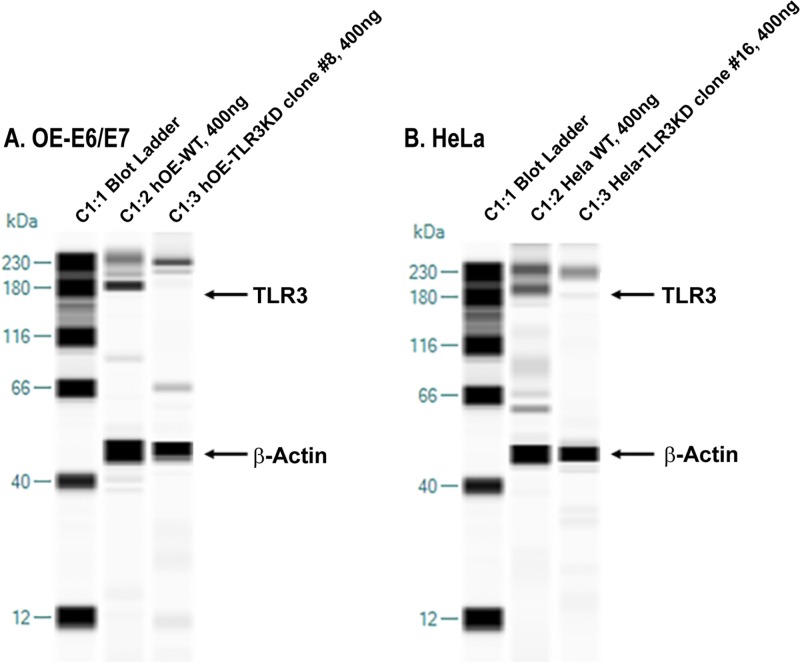
To ascertain the role of TLR3 in the innate immune response to human genital tract Chlamydia infections in vitro, we disrupted TLR3 function in both human oviduct (hOE cells) and cervical (HeLa cells) tissue using the Sigma CRISPR lentivirus system (see Materials and Methods). The CRISPR system consisted of 3 guide RNA (gRNA) sequences that targeted the human TLR3 gene at exon 2, exon 2, and exon 4, respectively (see Fig. S1 in the supplemental material). To ascertain the efficacy of using this approach to disrupt TLR3 function in these cells, we first examined TLR3 protein expression levels in the putative clones by using capillary electrophoresis in the Wes system to identify and quantitate TLR3 protein. In these experiments, we loaded a total of 400 ng of the cell lysate isolated from each of the selected clones and WT control cells. Figure 2 shows the results of capillary electrophoresis in which we simultaneously immunoassayed for the expression of both TLR3 and the β-actin loading control in hOE cells, HeLa cells, and the representative CRISPR-generated TLR3 “knockdown” (TLR3KD) clones generated in each parental cell line. As shown, a major band indicative of TLR3 protein expression was identified in hOE and HeLa cells, with peak molecular weights of 174 kDa and 187 kDa, respectively. Our data are suggestive that the TLR3 protein is posttranslationally modified in different ways in the different cell types, which affects their electrophoretic migration and apparent molecular weight (35).
FIG 2.
Disruption of TLR3 protein expression in human oviduct and cervical cells. The Wes simple Western system was used to confirm the disruption of TLR3 protein expression. (A) Human OE-E6/7 cells. Lane 1, ladder; lane 2, 400 ng WT human OE-E6/7 cell protein; lane 3, 400 ng hOE-TLR3KD cell protein. (B) HeLa cells. Lane 1, ladder; lane 2, 400 ng WT HeLa cell protein; lane 3, 400 ng HeLa TLR3KD cell protein. Data presented are representative data for selected TLR3-deficient clones from both cell types.
A qualitative examination of the data in Fig. 2 shows that TLR3 protein expression levels were substantially lower in both clone 8 of hOE-TLR3KD cells and clone 16 of HeLa TLR3KD cells than in their wild-type counterparts. These clones were selected and used as TLR3-deficient versions of human OE-E6/7 cells and HeLa cells, respectively, and were quantitatively analyzed for actual TLR3 expression levels with Wes Compass software. To ascertain the actual reduction in TLR3 protein expression levels, the protein band peaks were identified and quantified using the chemiluminescence peak area of the Wes-generated data. The ratio of TLR3 protein expression to that of the β-actin loading control was used as the index for calculating the relative TLR3 protein expression level. In hOE cells, the ratios of TLR3/β-actin for the WT and TLR3KD groups, expressed in units of pixel density (PD), were 5,732 PD/41,538 PD (0.138) and 347 PD/16,533 PD (0.021), respectively. The calculated TLR3 protein expression level was decreased about 85% in clone 8 of hOE-TLR3KD cells. In HeLa cells, the ratios of TLR3/β-actin for the WT and TLR3KD groups were 9,254 PD/43,601 PD (0.212) and 310 PD/16,482 PD (0.018), respectively. The calculated expression level of the TLR3 protein was decreased about 94% in clone 16 of HeLa TLR3KD cells. Collectively, our findings demonstrate that the CRISPR lentivirus system was very effective in disrupting TLR3 protein expression in both of these cell types.
TLR3 deficiency in human genital tract epithelial cells results in decreased IFN-β synthesis in response to poly(I·C) stimulation and chlamydial infection.
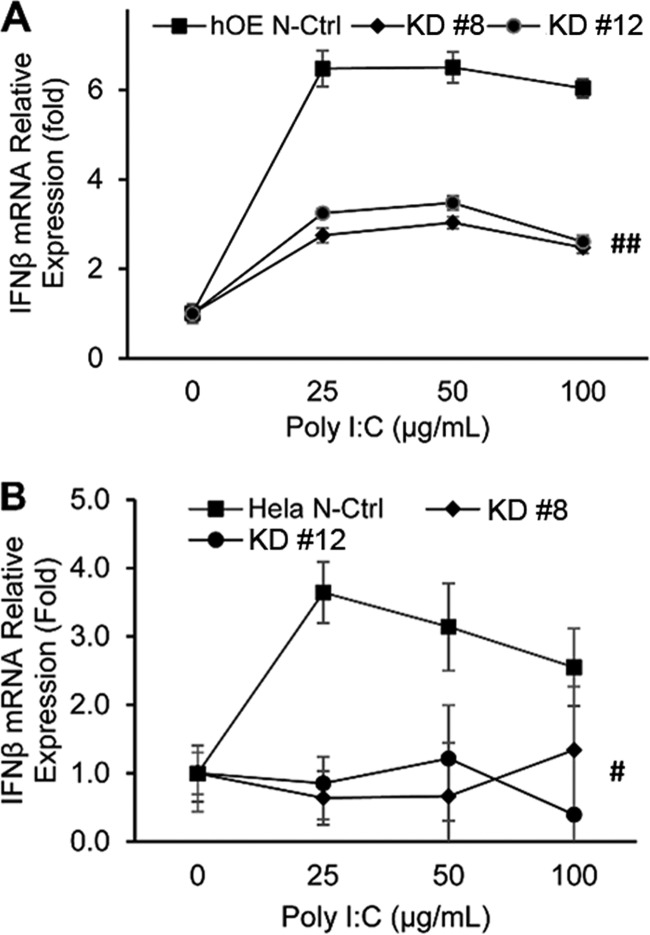
To determine whether the TLR3-deficient clones representing human oviduct and cervical tissue were also deficient in their ability to elicit appropriate TLR3-dependent innate immune responses, we first treated the clone representative of each cell type with 0, 25, 50, and 100 μg/ml of the TLR3 agonist poly(I·C) for 24 h and assessed TLR3’s functionality by measuring the induction of IFN-β gene expression (Fig. 3). As shown in Fig. 3A, hOE-TLR3KD cells exhibited significantly lower levels of IFN-β gene expression in response to poly(I·C) induction at all concentrations tested than in nontarget CRISPR control cells. We observed similar reductions in the induction of IFN-β transcription in response to poly(I·C) in the representative HeLa TLR3KD clones (Fig. 3B). However, the fold difference in IFN-β gene induction between the TLR3-deficient clones and the nontarget CRISPR control for HeLa cells seemed to decrease at higher poly(I·C) concentrations. We observed no noticeable differences in poly(I·C) induction of IFN-β transcription between the WT cells and their nontarget CRISPR control counterparts for either cell type (data not shown). These findings show a successful disruption of TLR3 function in TLR3-deficient epithelial cell clones representing human oviduct and cervical tissues.
FIG 3.
Disruption of TLR3 function by CRISPR dramatically decreases IFN-β mRNA expression in response to poly(I·C) stimulation. Selected clones representing TLR3-deficient oviduct (hOE) (A) and cervical (HeLa) (B) cells were treated with 0, 25, 50, and 100 μg/ml poly(I·C) for 24 h. The cells were harvested for total cell RNA isolation. The response to poly(I·C) was determined by measuring the induction of IFN-β mRNA synthesis via quantitative PCR. The relative gene expression levels of each clone compared to their respective nontarget CRISPR controls are shown. Data are representative of results from at least three independent experiments.
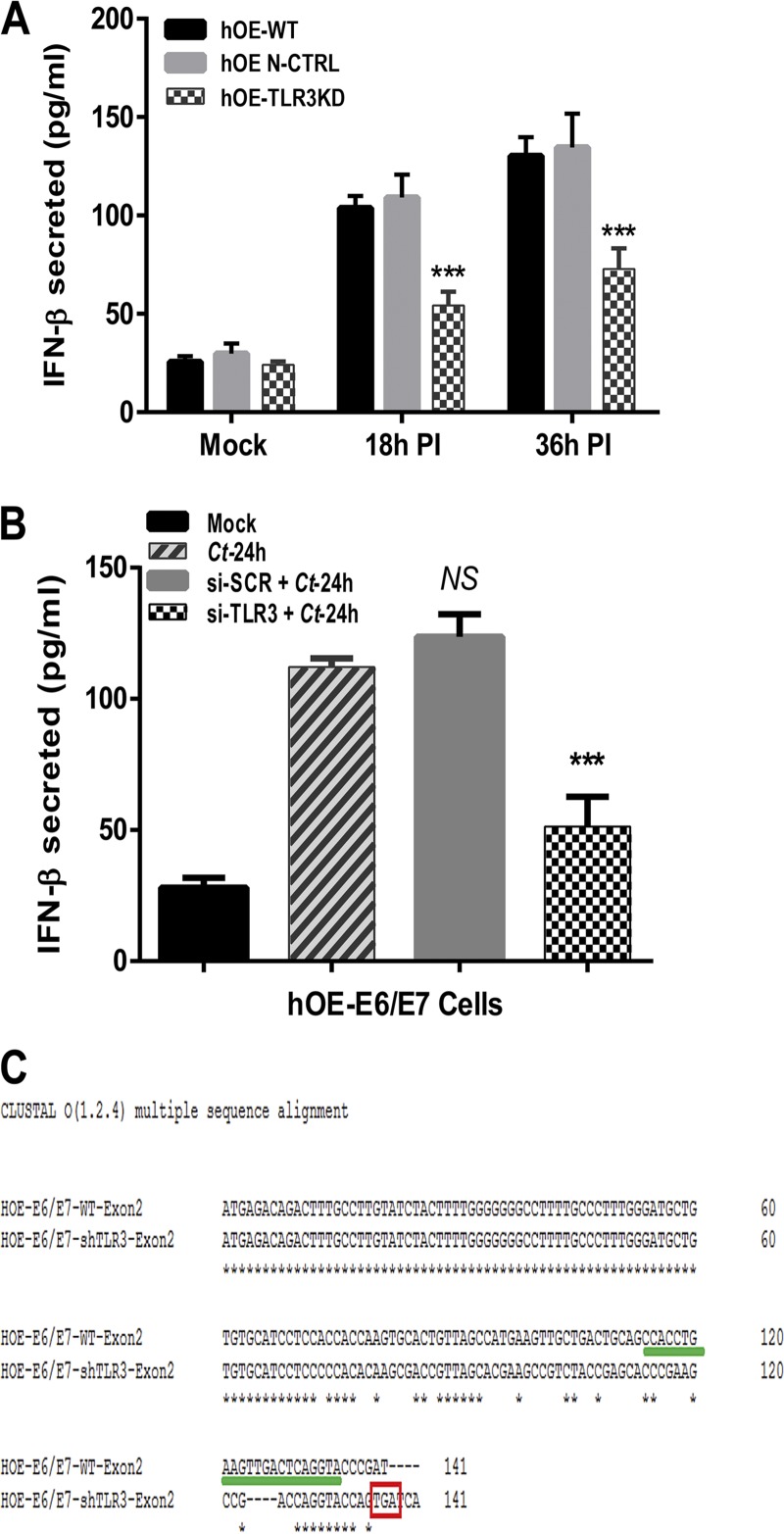
In order to make comparisons to what we have previously reported for the role of TLR3 in the innate immune response to Chlamydia infection in murine oviduct epithelial cells, we put more emphasis on hOE cells in this study and selected hOE-TLR3KD clone 8 for use in the remainder of this report. We first sought to ascertain the impact of TLR3 deficiency on the C. trachomatis-induced synthesis of IFN-β in human oviduct epithelial cells. We infected wild-type hOE (hOE-WT), hOE nontemplate CRISPR control (hOE-N-Ctrl), and hOE-TLR3KD cells with 10 IFU C. trachomatis L2 before harvesting cell supernatants at 18 and 36 h postinfection to measure the amount of IFN-β secreted. Figure 4A shows a significant reduction in the Chlamydia-induced synthesis of IFN-β in TLR3-deficient hOE cells compared to both the WT and nontemplate control cells. Our data show a 40 to 50% reduction in the amount of IFN-β synthesized at both time points and are suggestive that TLR3 plays a significant role in the optimal synthesis of IFN-β in C. trachomatis-infected hOE cells.
FIG 4.
TLR3 deficiency results in decreased synthesis of IFN-β during C. trachomatis infection of the oviduct epithelium. (A) WT, hOE-N-Ctrl, and TLR3-deficient hOE cells were either mock infected or infected with C. trachomatis L2 at an MOI of 10 IFU/cell for up to 36 h. Supernatants were collected from cells representing mock infection and 18 h and 36 h postinfection (PI) before measurement of chlamydia-induced synthesis of IFN-β by a standard ELISA. (B) WT hOE cells were infected with 10 IFU/cell C. trachomatis L2 (Ct) 24 h after treatment with 2.5 μg/ml of either scrambled siRNA (si-SCR) or TLR3-specific siRNA (si-TLR3). Supernatants were harvested at 24 h postinfection to measure the chlamydia-induced synthesis of IFN-β by a standard ELISA. (C) CLUSTAL-O multiple-sequence alignment of a sequenced PCR product containing exon 2 of human TLR3 reveals an early stop codon (TGA) at position 141. The TGA stop codon is inside the red box, and gRNA sequences are highlighted with a green line. Data are representative of results from at least 3 independent experiments. Statistically significant differences are shown by asterisks (***, P < 0.005; NS, not significant).
We next assessed whether our limiting-dilution clonal expansion procedure introduced “founder effects,” such as causing a global dysregulation in TLR signaling that leads to diminished synthesis of inflammatory cytokines during Chlamydia infection in hOE-TLR3KD cells. To test whether there were founder effects introduced that negatively impacted TLR signaling in a global manner, we treated WT and TLR3-deficient hOE cells with ultrapure preparations of ligands for TLR2 (peptidoglycan), TLR5 (flagellin), and TLR9 CpG oligodeoxynucleotides (ODN-CpG) for 24 h before testing supernatants for interleukin-6 (IL-6) synthesis by an enzyme-linked immunosorbent assay (ELISA) (14). As shown in Fig. S2, we saw no significant differences between WT and TLR3-deficient hOE cells in the synthesis of IL-6 when the cells were treated with agonists for either TLR2, TLR5, or TLR9. To address the possibility that the reduced IFN-β synthesis observed in hOE-TLR3KD cells was due to pathways unrelated to TLR3 that may have been disrupted by CRISPR, we used small interfering RNA (siRNA) as an alternative method to transiently disrupt TLR3 gene expression and protein function in WT hOE cells. Figure 4B demonstrates significant reductions in the amounts of IFN-β secreted into the supernatants of C. trachomatis L2-infected hOE cells that were pretreated with TLR3-specific siRNA 24 h prior to infection.
To discern the exact nature of the TLR3 gene defect introduced by CRISPR-Cas9, clone 8 of hOE-TLR3KD cells was selected for gene sequencing and gene sequence alignment with CLUSTAL-O. Analysis of the TLR3 gene sequencing data revealed that clone 8 of hOE-TLR3KD cells was heterozygous for TLR3 gene disruption at exon 2 and that the resultant knockout allele incorporated a premature stop codon (TGA) at nucleotide position 141 (Fig. 4C). The CRISPR-Cas9 method of gene disruption resulted in the translation of a truncated TLR3 protein that is likely nonfunctional, and our collective data examining TLR3 protein expression and function (Fig. 2 and 4 and Fig. S2) are suggestive that there is a minimal contribution from the TLR3 allele that was not disrupted by CRISPR-Cas9 at exon 2. Collectively, these findings show that the CRISPR-Cas9 methodology was effective in disrupting TLR3 gene function in hOE cells and corroborate our previous reports that implicate TLR3 as a significant contributor to the Chlamydia-induced synthesis of IFN-β during Chlamydia infection of oviduct epithelial cells (13, 36).
TLR3 deficiency alters Chlamydia-induced synthesis of acute inflammatory biomarkers in human oviduct epithelial cells.
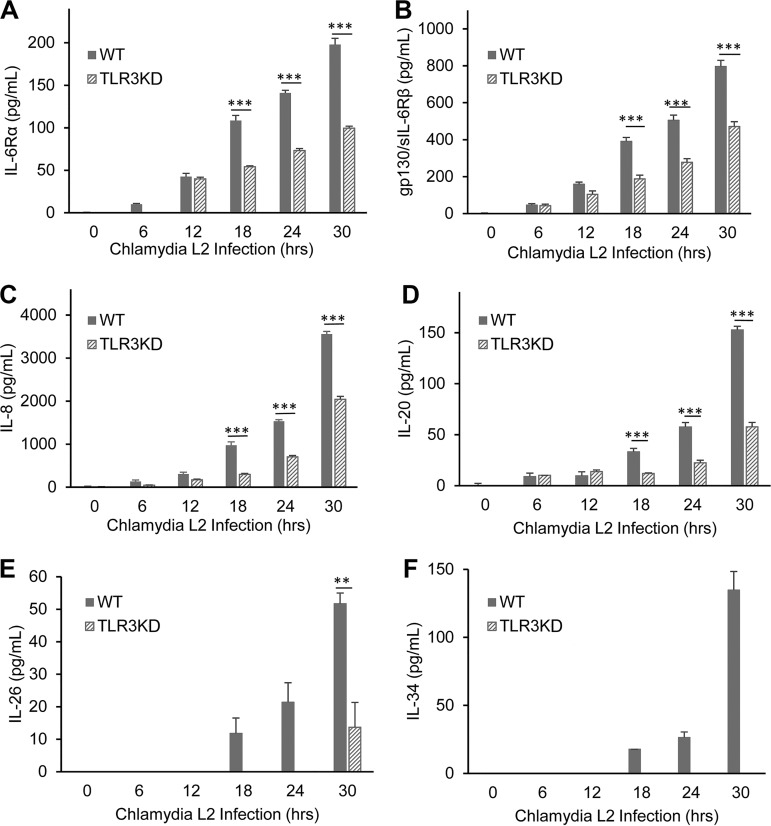
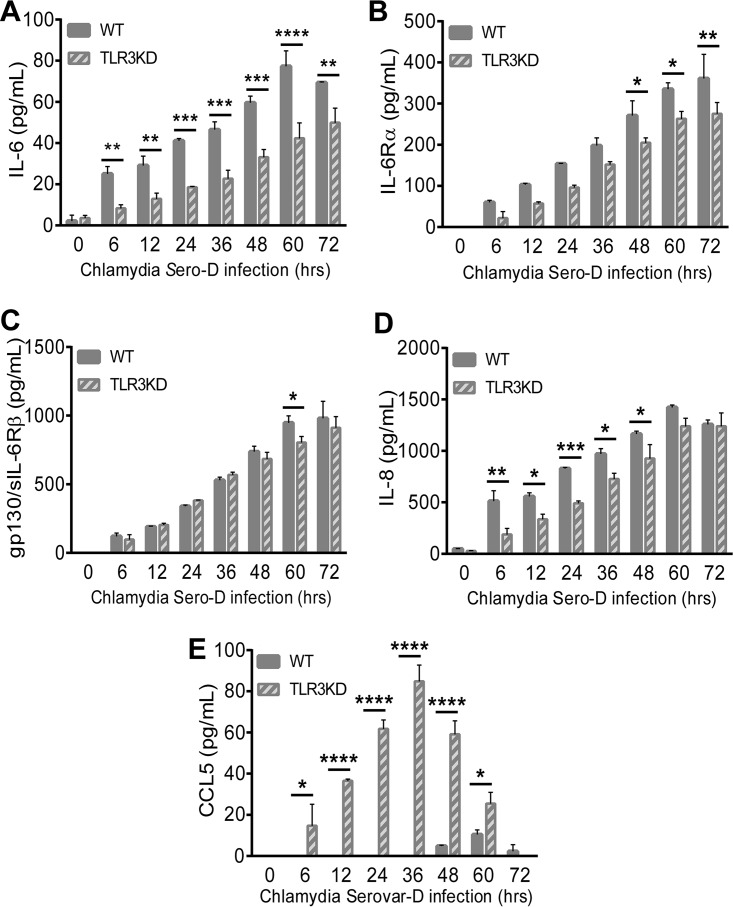
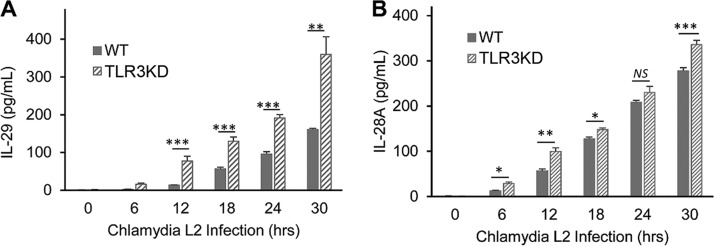
We previously reported that murine OE cells showed dysregulation in the C. muridarum-induced synthesis of a multitude of acute inflammatory mediators and subsequently showed that TLR3-deficient mice exhibited increased chlamydial shedding, aberrant T-cell recruitment, and severe genital tract pathology compared to WT mice (15, 32). To assess whether the absence of TLR3 is associated with the chlamydia-induced synthesis of key biomarkers of inflammation in human oviduct epithelial cells, we performed a multiplex ELISA for the detection and quantification of key human innate immune inflammatory biomarkers. hOE-WT and hOE-TLR3KD cells were either mock infected, infected with C. trachomatis L2 for up to 30 h, or infected with C. trachomatis serovar D for up to 72 h before multiplex analyses of the cell supernatants were performed. As shown in Fig. 5 and 6, TLR3-deficient hOE cells secreted significantly reduced levels of the acute inflammatory markers IL-6, IL-6Rα, sIL-6Rβ (gp130), IL-8, IL-20, IL-26, and IL-34 into the supernatants of C. trachomatis-infected cells throughout infection. Conversely, TLR3 deficiency in hOE cells resulted in the increased synthesis of CCL5 and the type III interferon IL-29 (IFN-λ1) throughout infection and IL-28A (IFN-λ2) late in infection (Fig. 6 and 7). Collectively, these data demonstrate a putative role for TLR3 in the acute phase of the innate immune response to Chlamydia that is known to occur early during infection in vivo (17, 37–40) and that TLR3 has some role in modulating mediators of the adaptive immune response (41).
FIG 5.
TLR3 deficiency results in attenuation of many of the acute-phase inflammatory mediators. WT and TLR3-deficient hOE cells were infected with C. trachomatis L2 at an MOI of 10 IFU/cell for up to 30 h. Supernatants were collected from individual wells every 6 h for multiplex ELISAs to measure the expression of IL-6Rα (A), sIL-6Rβ (gp130) (B), IL-8 (C), IL-20 (D), IL-26 (E), and IL-34 (F). Statistically significant differences are shown by asterisks (**, P < 0.01; ***, P < 0.001). Data are representative of results from three independent experiments.
FIG 6.
TLR3 deficiency alters acute-phase inflammatory mediator synthesis during infection with C. trachomatis serovar D. WT and TLR3-deficient hOE cells were infected with C. trachomatis serovar D at an MOI of 10 IFU/cell for up to 72 h. Supernatants were collected from individual wells at the times indicated for multiplex ELISAs to measure the expression of IL-6 (A), IL-6Rα (B), sIL-6Rβ (gp130) (C), IL-8 (D), and CCL5 (E). Statistically significant differences are shown by asterisks (*, P < 0.05; **, P < 0.01; ***, P < 0.005; ****, P < 0.001). Data are representative of results from three independent experiments.
FIG 7.
TLR3 deficiency causes increased expression of type III IFNs. WT and TLR3-deficient hOE cells were infected with C. trachomatis L2 at an MOI of 10 IFU/cell for up to 30 h. Supernatants were collected from individual wells every 6 h for multiplex ELISAs to measure the expression of IL-29 (A) and IL-28A (B). Statistically significant differences are shown by asterisks (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Data are representative of results from three independent experiments.
TLR3 has a functional role in C. trachomatis-induced synthesis of factors associated with genital tract fibrosis, scarring, and chronic inflammation.
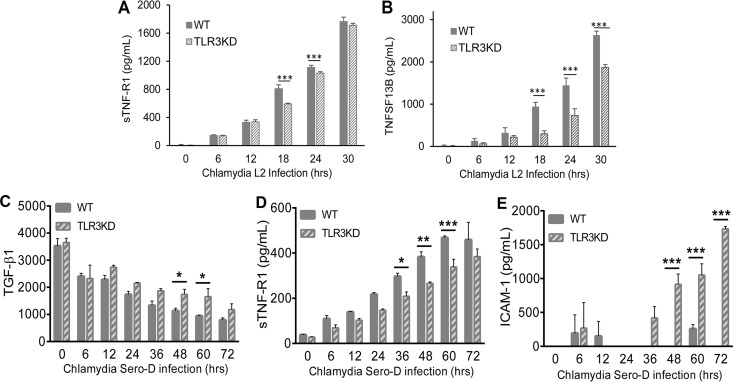
In our previous investigations into the role of TLR3 in the pathogenesis of genital infections in mice, one key aspect of TLR3 deficiency that we observed was that mice deficient in TLR3 appeared to exhibit indicators of more pronounced chronic sequelae, such as lymphocytic endometritis and hydrosalpinx (32). To examine whether TLR3 has a similar role in the pathogenesis of genital tract Chlamydia infections in humans, we next measured the Chlamydia-induced synthesis of biomarkers associated with chronic inflammatory disease and tissue necrosis in WT and TLR3-deficient hOE cells. As shown in Fig. 8A and D, C. trachomatis infection induces the synthesis of soluble tumor necrosis factor receptor 1 (sTNF-R1) in both cell types; however, its level of synthesis was significantly lower in TLR3-deficient hOE cells at middle to late times during infection. The exact role of sTNF-R1 in Chlamydia infection is not clear, but it is a known negative regulator of tumor necrosis factor alpha (TNF-α), which is a cytokine associated with severe genital tract sequelae in mice (42). Figure 8B shows that TLR3 deficiency leads to a similar impact on the chlamydia-induced synthesis of tumor necrosis factor ligand superfamily member 13B (TNFSF13B), a cytokine that belongs to the tumor necrosis factor family that acts as a potent B-cell activator (43). Interestingly, Fig. 8C and E show that the synthesis of transforming growth factor β1 (TGF-β1) and intercellular adhesion molecule 1 (ICAM-1) is increased during C. trachomatis infection of hOE-TLR3KD cells, implicating TLR3 in the negative regulation of key components of the pathophysiological process of fibrogenesis (44).
FIG 8.
TLR3 deficiency alters the synthesis of cytokines associated with cellular adhesion and tissue integrity during genital tract infection with C. trachomatis. WT and TLR3-deficient hOE cells were infected with either C. trachomatis L2 (A and B) or C. trachomatis serovar D (C to E) at an MOI of 10 IFU/cell for up to 72 h. Supernatants were collected from individual wells at the times indicated for multiplex ELISAs to measure the expression of sTNF-R1 (A and D), TNFSF13B (B), TGF-β1 (C), and ICAM-1 (E). Statistically significant differences are shown by asterisks (*, P < 0.05; **, P < 0.01; ***, P < 0.005). Data are representative of results from three independent experiments.
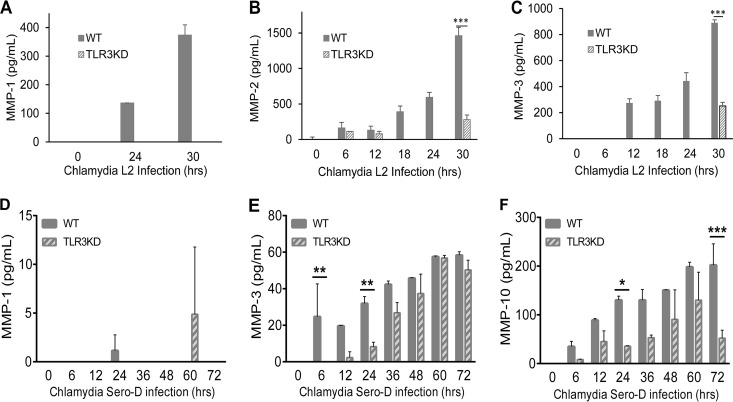
The matrix metalloproteinases (MMPs) are a tightly regulated family of proteins that are involved in the breakdown of extracellular matrix in normal physiological processes and are known to have a regulatory role in the inflammatory immune response, wound healing, cell migration, and embryonic development (45). Dysregulation of these proteins during Chlamydia infection has been demonstrated to play a role in the pathogenesis of fallopian tube damage during genital tract infections and in corneal scarring in patients with trachoma (46, 47). To ascertain whether MMPs are synthesized in response to C. trachomatis infection in hOE cells, and to determine whether TLR3 deficiency impacts their protein expression levels, we next measured the secretion of candidate MMPs into the supernatants of C. trachomatis-infected cells in our multiplex ELISA. As shown in Fig. 9, Chlamydia infection induced the production of MMP-1, MMP-2, MMP-3, and MMP-10 throughout infection, supporting the observations of others regarding the induction of MMPs in genital tract infections (46, 48–50). The Chlamydia-induced synthesis of these MMPs was either completely absent or severely diminished in TLR3-deficient hOE cells relative to hOE-WT cells infected with the L2 serovar (Fig. 8A to C). However, the synthesis levels were more similar when these cells were infected with serovar D albeit in significantly smaller amounts in hOE-TLR3KD cells at various points postinfection (Fig. 8D to F). Taken together, data from Fig. 8 and 9 are suggestive that TLR3 plays some role in regulating the synthesis of immune factors involved in modulating the genital tract pathologies associated with Chlamydia infection in humans.
FIG 9.
TLR3 regulates Chlamydia-induced expression of matrix metalloproteinases during genital tract infections with C. trachomatis. WT and TLR3-deficient hOE cells were infected with 10 IFU/cell of either C. trachomatis L2 (A to C) or C. trachomatis serovar D (D to F) for up to 72 h. Supernatants were collected from individual wells at the times indicated for multiplex ELISAs to measure the expression of MMP-1 (A and D), MMP-2 (B), MMP-3 (C and E), and MMP-10 (F). Statistically significant differences are shown by asterisks (*, P < 0.05; **, P < 0.01; ***, P < 0.005). Data are representative of results from three independent experiments.
TLR3 signaling regulates Chlamydia-induced synthesis of biomarkers associated with persistence, metastasis, and autoimmunity.
A major factor in the protective immune response to C. trachomatis infection is the synthesis of IFN-γ, which inhibits the growth of chlamydial reticulate bodies (RBs) via mechanisms that lead to tryptophan starvation, chlamydial death, and eventual clearance of the bacteria (51). However, recent evidence of chlamydial reticulate bodies being able to substantially alter their gene transcription, decrease metabolism, and enter into what is known as a “persistent” state suggests a survival mechanism that Chlamydia has evolved to evade immune surveillance (52–54). Persistence of microbial infections is often implicated in the triggering of autoimmune reactions, and this was demonstrated in studies investigating the role that Chlamydia persistence plays in triggering self-immune reactions in infected male rodents (55).
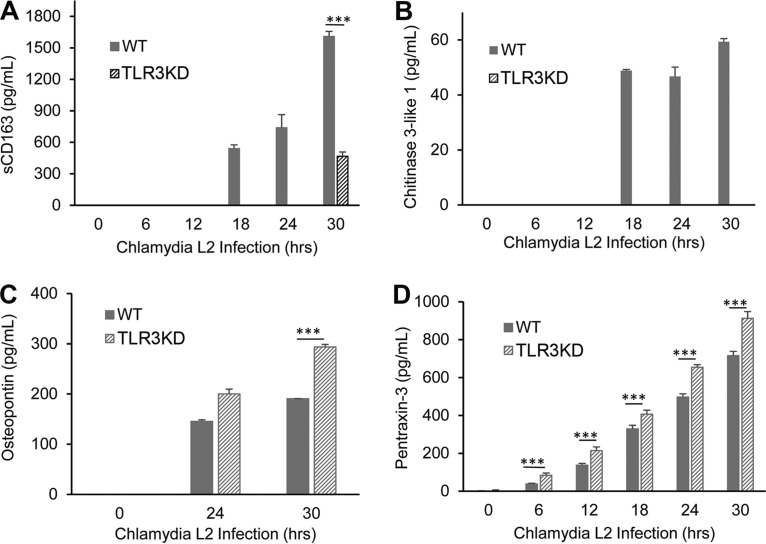
To determine whether Chlamydia infection induces a cellular response that signals a state of persistence in infected hOE cells, our multiplex analyses included several biomarkers for persistence and autoimmunity that are known to be secreted by cells in various clinical syndromes. Figures 10 and 11 show the results of chlamydial induction of soluble CD163 (sCD163), chitinase-3-like protein 1, lipocalin-2, osteopontin (OPN), and pentraxin-3 throughout infection in WT and TLR3-deficient hOE cells. As shown, Chlamydia induces the synthesis of sCD163 (a factor associated with long-term chronic inflammatory diseases such as rheumatoid arthritis) and antiapoptotic chitinase-3-like protein 1 in hOE cells. However, our data show that the protein secretion levels of these biomarkers were significantly reduced in the absence of TLR3 when these cells were infected with the L2 serovar. Although secretion levels of sCD163 were much higher during infection with serovar D, there was no significant reduction during TLR3 deficiency as was observed for L2 infection.
FIG 10.
TLR3 plays a role in regulating Chlamydia-induced synthesis of biomarkers associated with persistence and autoimmunity. Secreted protein levels of sCD163 (A), chitinase-3-like protein 1 (B), osteopontin (C), and pentraxin-3 (D) were measured in the supernatants of WT and TLR3-deficient hOE cells that were infected with C. trachomatis L2 at an MOI of 10 IFU/cell for up to 30 h. The supernatants were collected from individual wells for multiplex ELISAs at the indicated time points postinfection. Statistically significant differences are denoted by asterisks (***, P < 0.001). Data are representative of results from three independent experiments.
FIG 11.
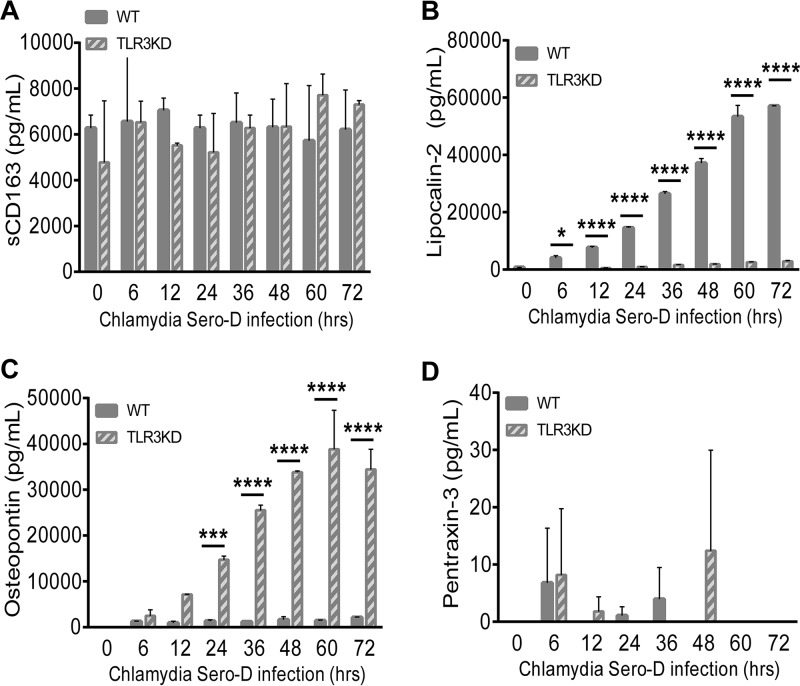
TLR3 plays a role in regulating Chlamydia-induced synthesis of biomarkers associated with iron sequestration, persistence, and autoimmunity during infection with C. trachomatis serovar D. WT and TLR3-deficient hOE cells were infected with C. trachomatis serovar D at an MOI of 10 IFU/cell for up to 72 h. Supernatants were collected from individual wells at the times indicated for multiplex ELISAs to measure the expression of sCD163 (A), lipocalin-2 (B), osteopontin (C), and pentraxin-3 (D). Statistically significant differences are shown by asterisks (*, P < 0.05; ***, P < 0.005; ****, P < 0.001). Data are representative of results from three independent experiments.
Figures 10 and 11 show that TLR3 deficiency leads to significantly increased levels of osteopontin and pentraxin-3. The role of these proteins in Chlamydia infection is not clear; however, osteopontin is an inflammatory mediator often associated with autoimmune disease, chronic inflammatory disorders, and progression of tumor cells (56), while pentraxin-3 is a known biomarker for PID and is also associated with autoimmune diseases (57, 58). Collectively, our findings indicate that TLR3 may have a functional role in regulating the expression of biomarkers symbolic of long-term or persistent disease states in Chlamydia-infected hOE cells, which is a precondition that often correlates with the initiation of autoimmune responses and chronic sequelae (59–62).
TLR3 deficiency alters the size of chlamydial inclusions.
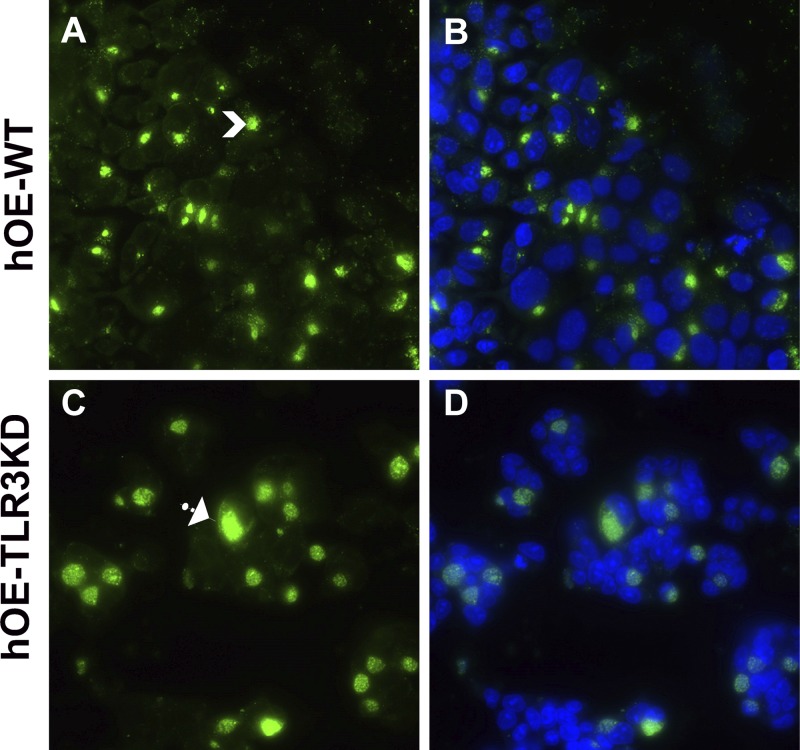
Our previous investigations into mechanisms that impact the chlamydia-induced synthesis of IFN-β showed that disruption of IFN-β had a significant impact on chlamydial inclusion size and chlamydial replication. In that regard, we showed that C. muridarum replication in murine OE cells deficient in either TLR3 or STAT-1 was more robust and that the inclusions were larger and aberrantly shaped (15, 63) (Fig. S3). To determine the physiological consequences of TLR3 deficiency for Chlamydia inclusion size in human oviduct epithelial tissue, we infected WT and TLR3-deficient hOE cells with C. trachomatis serovar D at an MOI of 10 IFU/cell for 36 h. We next stained the cells for chlamydial lipopolysaccharide (LPS) using the EVI-HI anti-Chlamydia LPS antibody and examined the infected cells for chlamydial inclusion size by fluorescence microscopy and multispectral flow cytometry. We first examined C. trachomatis serovar D-infected hOE-WT and hOE-TLR3KD cells by immunofluorescence microscopy to get a qualitative comparison of the chlamydial inclusion size in order to ascertain whether TLR3 has any impact on Chlamydia development. As demonstrated in the representative capture shown in Fig. 12, we routinely saw that the chlamydial inclusions in TLR3-deficient hOE cells were much larger, amorphously shaped, and more diffusely stained with punctate patterns throughout the inclusion. In contrast, the inclusions in hOE-WT cells were generally more compact and more uniformly stained and had a higher staining intensity per pixel than TLR3-deficient hOE cells. We saw very similar trends in C. muridarum-infected WT and TLR3-deficient OE cells derived from mice (Fig. S3).
FIG 12.
TLR3 deficiency in murine OE cells leads to large and aberrantly shaped chlamydial inclusions. hOE-WT cells (A and B) and hOE-TLR3KD cells (C and D) were either mock treated or infected with C. trachomatis serovar D at an MOI of 10 IFU/cell for 36 h. Chlamydial inclusions were stained using anti-chlamydial LPS monoclonal antibody and detected via an Alexa Fluor 488-conjugated secondary antibody. Nuclei were visualized via DAPI staining (B and D). Data shown are representative. Arrows show smaller versus larger inclusions. Magnification, ×60.
We next examined C. trachomatis L2-infected hOE-WT and hOE-TLR3KD cells to quantitatively determine the impact of TLR3 deficiency on chlamydial inclusion development and size via multispectral imaging flow cytometry using the Amnis Image StreamX MKII instrument. Figure S4 shows a representative image from multispectral imaging flow cytometry, in which we scanned (in triplicate) 10,000 cells each of the hOE-WT and hOE-TLR3KD cell lines that were either mock infected or infected with C. trachomatis L2 for 24 h. Analyses of the multispectral imaging flow cytometry data with IDEAS software revealed that the average Chlamydia inclusion diameters were 15.3 μm and 17.2 μm in hOE-WT and hOE-TLR3KD cells, respectively. Collectively, the immunofluorescence analyses and imaging flow cytometry results indicate that TLR3 deficiency in the human oviduct epithelium leads to increased chlamydial inclusion size.
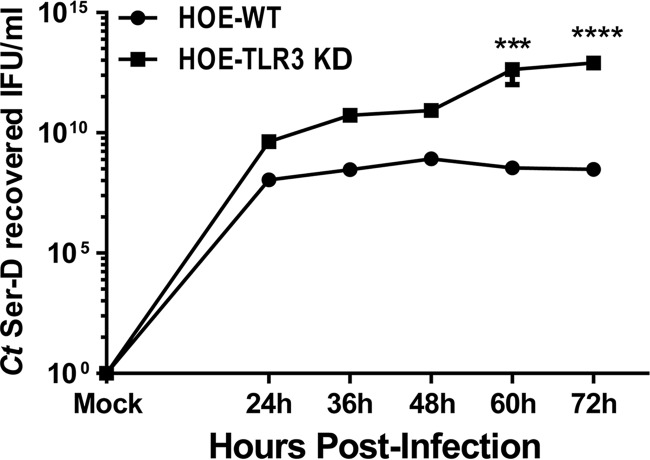
To examine whether Chlamydia replication during TLR3 deficiency correlates with the increased inclusion size, we next measured chlamydial replication in wild-type and TLR3-deficient hOE cells that were infected with 5 IFU/cell C. trachomatis serovar D. As shown in Fig. 13, C. trachomatis replication was greater at all time points in hOE-TLR3KD cells than in wild-type cells, suggesting that stimulation of TLR3 by C. trachomatis results in an immune response that negatively affects Chlamydia growth. Interestingly, our data in Fig. 13 show that chlamydial replication in hOE-WT cells peaks at around 48 h postinfection before the recovery of infectious progeny begins to drop. In contrast, the recovery of infectious elementary bodies (EBs) in Chlamydia-infected hOE-TLR3KD cells continued to rise past the 48-h time point (Fig. S5). Collectively, our data indicate that TLR3 plays a role in limiting Chlamydia replication in human oviduct tissue, corroborate our previous studies in mice, and thus implicate TLR3 in a protective role against genital tract Chlamydia infections in both species.
FIG 13.
Measuring chlamydial replication in infected WT and TLR3-deficient hOE cells. hOE-WT and hOE-TLR3KD cells were either mock treated or infected with C. trachomatis serovar D (in triplicate) at an MOI of 5 IFU/cell for 72 h. The cells were disrupted in sucrose-phosphate-glutamate (SPG) buffer at the indicated time points as described in Materials and Methods, lysates were collected and sonicated, and the titer was determined on fresh HeLa cell monolayers. The data presented are representative of results from three different experiments. Statistically significant differences are shown by asterisks (***, P < 0.005; ****, P < 0.001).
DISCUSSION
Our research focuses on the impact of TLR3 signaling on the immune response to chlamydial infection in the oviduct epithelium, and we were the first to demonstrate more severe genital tract pathogenesis in mice deficient in TLR3, confirming our hypothesis that TLR3 has a protective role in the immune responses to murine genital tract infections (32). In this investigation, our goal was to ascertain whether TLR3 had a similar protective role in the immune response to genital tract Chlamydia infection in humans and to more precisely delineate the immune responses in oviduct epithelial cells that contribute to the fibrosis and scarring that lead to reproductive sequelae in clinical disease. The OE-E6/E7 cells used in this report were derived from human fallopian tubes and were immortalized by the human papillomavirus 16 (HPV16) E6/E7 open reading frame (ORF) by retroviral expression (26). Although these are not primary oviduct epithelial cells, the cells are immortalized and offer the advantage of being able to be passaged in the laboratory, and they are close enough to primary oviduct epithelial cells that they can serve as an adequate representation of what we believe occurs during in vivo Chlamydia-OE cell interactions during natural genital tract infection.
Because OE-E6/E7 cells express a functional TLR3, we first disrupted its gene expression and subsequent protein function using CRISPR to generate a TLR3-deficient version of the OE-E6/E7 cells, which could then be used to help delineate the immune responses to Chlamydia infection that are directly related to TLR3 function. We were able to generate several clones of TLR3-deficient hOE cells and demonstrated the loss of both TLR3 protein expression and its functional response to the TLR3 agonist poly(I·C). We also demonstrated a significant reduction in the chlamydia-induced synthesis of IFN-β in hOE cells deficient in TLR3 function; however, the reduction was a bit more modest in hOE-TLR3KD cells than in TLR3-deficient OE cells derived from TLR3KO mice (13). The differences that we observed in the reduction in Chlamydia-induced IFN-β synthesis between TLR3-deficient hOE and murine OE cells are likely related to the fact that we were not able to completely disable TLR3 gene expression using CRISPR, whereas functional TLR3 gene expression in the TLR3KO mouse is completely absent. Other possibilities to explain the more modest reduction in the Chlamydia-induced synthesis of IFN-β in hOE cells could be more significant contributions of other pathways identified to enhance to the Chlamydia-induced type I IFN response, such as cyclic GMP-AMP (cGAMP) synthase (cGAS) and STING, in hOE cells (16, 64). The significance of the relatively higher level of IFN-β synthesis observed during Chlamydia infection in TLR3-deficient hOE cells than in TLR3-deficient murine OE cells is not yet known; however, we have shown that IFN-β can regulate the chlamydia-induced synthesis of a multitude of other inflammatory mediators (15).
As sentinels for invasion by microbial pathogens, epithelial cells lining the reproductive tract secrete cytokines and chemokines upon chlamydial infection that function in various facets of innate immunity, such as inflammation, lymphocyte recruitment, polarization, and genital tract scarring (24, 65). We reported previously that TLR3 regulates the syntheses of a multitude of these factors both in vitro and in vivo in murine OE cells and mice and concluded that TLR3 has a regulatory function affecting multiple aspects of both the innate and adaptive immune responses (13, 15, 32). We demonstrate in this report that TLR3 may have a similar role in human oviduct epithelial tissue and extrapolate our findings to speculate that TLR3 deficiency may have a significant role in outcomes of infections in humans. As an example, we show in Fig. 5 to 7 that TLR3 deficiency in hOE cells leads to significant reductions in several factors associated with the acute inflammatory response, such as IL-6, IL-6Rα, sIL-6Rβ (gp130), and IL-8. IL-6Rα and gp130 are the two chains that comprise the IL-6 receptor, which is a type I receptor for the pleiotropic cytokine IL-6. IL-6 represents a keystone cytokine in infection, cancer, and inflammation (66) and has been shown to have a significant role in both inhibiting C. muridarum infection in mice and exacerbating its pathogenicity in the mouse genital tract (67). IL-8 is a neutrophil chemotactic factor known to be induced early during Chlamydia infection and was thought to be associated with preterm delivery complications in pregnant women infected with C. trachomatis (68).
The loss of TLR3 function in hOE cells did not result in a global downregulation of the Chlamydia-induced synthesis of all mediators that shape the immune response, nor was its impact limited to that of the acute inflammatory response. Our data also demonstrate that TLR3 deficiency leads to the downregulation and upregulation of the chlamydia-induced synthesis of several cytokines and chemokines that have an impact at various phases of the adaptive immune response, which can potentially affect long-term outcomes of infection in humans and impact genital tract pathology. Figures 6 and 7 show that TLR3-deficient hOE cells produced significantly higher levels of the leukocyte-recruiting factor CCL5 (69) and the type III interferons IL-28A and IL-29 (70) than did WT hOE cells. The type III IFNs are hypothesized to have a role in the polarization of the immune response to Chlamydia infection by inhibiting the production of IL-13, IL-4, and IL-5, thereby promoting the development of protective Th1 immunity to infection (41). Our investigations into the role of TLR3 in the pathogenesis of C. muridarum infection in mice show that TLR3 deficiency leads to significantly altered CD4+/CD8+ T-cell ratios and increased lymphocytic infiltration into the uterine horns and oviducts by day 21 postinfection (32). The observation of significantly higher levels of the type III interferons IL-28A and IL-29 in TLR3-deficient hOE cells supports the hypothesis that TLR3 may have a role in the polarization of the immune response in humans as well, and it would manifest itself by having an effect on the recruitment of lymphocytes into the female reproductive tract in women infected with C. trachomatis.
We recently reported that TLR3 deficiency resulted in an increased frequency and severity of pronounced chronic sequelae (such as lymphocytic endometritis and hydrosalpinx) during late stages of C. muridarum genital tract infections in mice (32). In this report, we show that TLR3-deficient hOE cells were dysregulated in the Chlamydia-induced synthesis of several biomarkers associated with chronic inflammation, which suggests that chronic clinical outcomes would occur at a higher frequency in humans lacking functional TLR3. Collectively, our data implicate TLR3 in having some impact on regulating the incidence and severity of outcomes of chronic inflammation caused during genital tract Chlamydia infections in humans. TNF-α is involved in systemic chronic inflammation that causes many of the clinical problems associated with autoimmune disorders such as rheumatoid arthritis, ankylosing spondylitis, inflammatory bowel disease, psoriasis, and refractory asthma (71–73). These disorders are sometimes treated by using TNF inhibitors that either mimic the TNF-α receptor (TNF-R1) to bind to and block its activity or are actual monoclonal antibodies that bind TNF-α and block its active site (74). TLR3 deficiency in hOE cells showed defective synthesis of soluble tumor necrosis factor receptor 1 (sTNF-R1) during C. trachomatis infection. sTNF-R1 binds to and inactivates TNF-α, a major cytokine associated with scarring of oviduct tissue and severe genital tract sequelae in C. muridarum-infected mice (42). This finding suggests that the diminished presence of sTNF-R1 in TLR3-deficient cells would result in reduced effectiveness at inactivating Chlamydia-induced TNF-α and a subsequent increased incidence of TNF-α-mediated scarring. In this regard, dysregulation of the expression of matrix metalloproteinases (MMPs) is known to directly impact the severity of genital tract fibrosis and scarring in mice infected with Chlamydia (48–50). The Chlamydia-induced synthesis of MMP-1, MMP-2, MMP3, and MMP-10 was shown to be diminished in TLR3-deficient hOE cells (Fig. 9), suggesting that the normal physiological process of breaking down extracellular matrix proteins by the MMPs would be attenuated and will likely result in the increased fibrosis and scarring observed when the expression levels of certain MMPs are not sufficient (75, 76).
Our data showed that TLR3 deficiency in hOE cells significantly altered the Chlamydia-induced expression levels of biomarkers for chronic inflammation, persistence, and autoimmunity, such as soluble CD163 (sCD163), chitinase-3-like protein 1, osteopontin (OPN), and pentraxin-3 (56–58, 77–80). OPN was first identified in osteoclasts as an extracellular structural protein of bone and is known as an essential factor in bone remodeling (81). However, subsequent to the initial identification of OPN as a structural component of bone tissue, OPN was shown to be expressed in a wide range of immune cells, including macrophages, neutrophils, dendritic cells, and various lymphocyte types, and is now known to have functions in several aspects of host immunity (82). The exact role that OPN plays in the immune response to Chlamydia infection is poorly understood; however, recent studies have linked OPN to persistent inflammation and have hypothesized a role for OPN in cell transformation during persistent Chlamydia infection (83, 84). Our data showing significant increases in OPN production in TLR3-deficient hOE cells during C. trachomatis infection suggest that TLR3 deficiency may lead to increased incidences of persistent infections and indicate a role for TLR3 in the prevention of long-term chronic inflammation and possible cellular transformation. A link between sCD163, chitinase-3-like protein 1, pentraxin-3, and Chlamydia infection has not yet been established; however, the dysregulation of these known biomarkers for chronic inflammation, persistent infection, and autoimmunity supports a role for TLR3 in limiting the clinical symptoms of chronic inflammation during infection.
Finally, we examined the possible impact that TLR3 deficiency in hOE cells may have on chlamydial replication and inclusion structures. We previously showed that C. muridarum replication was more robust, the inclusions were larger and more aberrantly shaped in TLR3-deficient murine OE cells, and TLR3-deficient mice sustained significantly higher bacterial burdens than WT mice during early infection and midinfection (15, 32) (see Fig. S3 in the supplemental material). We hypothesized from those studies that the overall negative effect on C. muridarum biology was largely due to the host-beneficial impact of TLR3-dependent IFN-β synthesis, and we showed that chlamydial replication in TLR3-deficient OE cells pretreated with exogenous recombinant IFN-β was significantly diminished (15). Here, we corroborate the mouse studies by showing that the inclusions were substantially larger and that they were stained in a more punctate and diffuse pattern in TLR3-deficient hOE cells 36 h after infection with C. trachomatis serovar D. Imaging flow cytometry revealed that TLR3-deficient hOE cells had higher levels of Chlamydia LPS within chlamydial inclusions at 24 h postinfection when infected with C. trachomatis L2 (not shown), which was highly suggestive that the larger inclusions contain more bacterial particles. We confirmed the multispectral imaging flow cytometry data by demonstrating that there were consistently higher levels of Chlamydia replication in TLR3-deficient hOE cells at all time points between 24 and 72 h postinfection (Fig. 13 and Fig. S5) and that the differences were statistically significant after 48 h postinfection. Our data showing that TLR3 deficiency in hOE cells leads to significant decreases in the chlamydia-induced synthesis of IFN-β implicate this cytokine in the control of Chlamydia infection in human genital tract epithelial cells, as was observed in mice. However, our data in Fig. 11 show that TLR3 deficiency in hOE cells also resulted in drastic reductions in the chlamydia-induced secretion of lipocalin-2, which is an innate immune protein that limits bacterial growth by sequestering iron-containing siderophores (85). Although it is unclear exactly which mechanism exerts the greatest impact on regulating chlamydial growth in hOE cells, our data are suggestive that there are likely redundant mechanisms that control Chlamydia replication that are disrupted during TLR3 deficiency.
Collectively, all of our previous investigations into the impact of TLR3 on the immune response to Chlamydia infection in mice have demonstrated a protective role for TLR3 in regard to genital tract pathology, and our data represent the first of such in regard to Chlamydia infection. Although the exact mechanism(s) that TLR3 invokes to elicit this protective immunity is unknown and needs further study, our preliminary investigations implicate TLR3 function in upregulating the gene expression of other TLRs known to have an impact on genital tract pathology (such as TLR2 [12]), through pathways involving IFN-β synthesis (15). In this report, we expanded our investigations into examining the impact of TLR3 on the immune response to Chlamydia infection in human oviduct epithelial cells, and our initial results thus far show a similar role for TLR3 in the protective immune response in humans. Studies are under way to further investigate the role of this enigmatic Toll-like receptor in human genital tract Chlamydia infection.
MATERIALS AND METHODS
Cell culture.
Human OE-E6/E7 (hOE) cells and HeLa cells were maintained in high-glucose Dulbecco’s modified Eagle’s medium (DMEM; Life Technologies, Inc.) supplemented with 10% bovine calf serum (HyClone) in an incubator at 37°C with 5% CO2. The hOE cells were originally derived from fallopian tube tissue and were immortalized by expressing the HPV16 E6/E7 open reading frame in a retroviral expression system (26).
Reagents.
The following TLR agonists were purchased from InvivoGen (San Diego, CA): (i) peptidoglycan from Escherichia coli serotype O111:B4 (125 endotoxin units [EU]/mg); (ii) ultrapure flagellin purified from Bacillus subtilis (>95% purity); and (iii) ODN 2216, a synthetic oligonucleotide (ODN) that preferentially binds human TLR9, and ODN 2243, an ODN 2216 control without CpGs. Wild-type (WT) and TLR3-deficient hOE cells were grown to confluence in 24-well plates before being treated with the appropriate TLR ligand at the concentrations specified in the text. Supernatants were harvested at the 24-h posttreatment time point and analyzed for cytokine content using an ELISA for IL-6 (R&D Systems, Minneapolis, MN) according to the manufacturer’s protocol.
Generation of TLR3-deficient human epithelial cell lines.
Human OE-E6/E7 cells and HeLa cells were grown to 60 to 70% confluence before being transduced with either human TLR3 CRISPR knockout lentivirus or CRISPR lentivirus nontargeting control transduction particles and 4 μg/μl Polybrene. The CRISPR lentivirus transduction particles (pLV-U6g-EPCG-TLR3), all-in-one ready-to-use Cas9 and guide RNA (gRNA), and CRISPR negative controls were purchased from Sigma-Aldrich (St. Louis, MO). The CRISPR system consisted of 3 gRNA sequences (CCACCTGAAGTTGACTCAGGTA, CCAACTTCACAAGGTATAGCCA, and CAGGGTGTTTTCACGCAATTGG), which targeted the human TLR3 gene (GenBank accession number NM_003265) at exon 2, exon 2, and exon 4, respectively. The CRISPR universal negative control targets no known human, mouse, or rat gene. Transduced human OE-E6/E7 cells and HeLa cells were subjected to 5-μg/ml puromycin selection, and the puromycin-resistant cells were sorted by using a BD FACSAria cell sorter. The brightest green fluorescent protein (GFP)-positive cells were collected and cultured for further single-cell cloning with glass cloning cylinders. Individual cell colonies were isolated and expanded. Higher-GFP-expressing cell clones were further selected by using a BD Accuri C6 flow cytometer. Confirmation of TLR3 gene deletion, protein expression, and receptor function in CRISPR Cas9 TLR3 “knockdown” (TLR3KD) cells was assessed using PCR, Western blotting, and an ELISA, respectively.
Genomic DNA purification and PCR amplification.
Genomic DNA (gDNA) from TLR3-deficient human OE cells (hOE-TLR3KD) was purified by using the PureLink genomic DNA minikit from Invitrogen (catalog number K1820-01). Gene-specific primer pairs (forward primer 5′-ACA AGG AAT ATA CCA ATG CAT TTG-3′ and reverse primer 5′-GAT ATT TAG ATA GTA AGT CTA AGG-3′) were designed approximately 300 bp up- and downstream of the gRNA sequence (CCACCTGAAGTTGACTCAGGTA) spanning exon 2 of the TLR3 gene. The purified gDNA as a template and Platinum Super-Fi PCR master mix (catalog number 12358-010; Invitrogen) were used to amplify the PCR product (599 bp). The cycling conditions were an initial denaturation step at 95°C for 2 min followed by 32 cycles of denaturation at 95°C for 45 s, annealing at 50.5°C for 45 s, elongation at 72°C for 45 s, and a final elongation step at 72°C for 2 min. The amplified PCR products were gel purified using the QIAquick gel extraction kit (Qiagen, Germantown, MD). The PCR product from hOE-WT cells was used as the control sample throughout this experiment. A portion of the amplified PCR products was sent for sequencing at the IUSM Bioinformatics Core Sequencing Facility, while the rest of the PCR products were used in the cloning experiment.
Cloning and plasmid purification.
An additional adenine overhang was added to the portion of the PCR product to be cloned by using high-fidelity Taq polymerase and dATP at 70°C for 20 min. The reaction mixtures were then ligated into the pGEM-T Easy vector (catalog number A1360; Promega) overnight at 4°C and then transformed into TOP10 competent E. coli cells. The transformed E. coli cells were plated onto LB agar plates containing ampicillin (60 μg/ml) and supplemented with isopropyl-β-d-thiogalactopyranoside (IPTG). The blue and white colonies were screened and inoculated into 5 ml LB medium containing ampicillin (final concentration, 60 μg/ml). The putative positive cultures were used for plasmid purification by using a QiaSpin miniprep kit (catalog number 27106), and inserts were confirmed by PCR before plasmid sequencing.
Chlamydia trachomatis preparation.
The C. trachomatis L2-434/Bu (L2) and C. trachomatis UW‐3CX serovar D strains were generously provided by David E. Nelson. The C. trachomatis serovar D and L2 mother pools used in these experiments were subsequently grown and titrated as described previously (86), whereby antibody specific for chlamydial LPS was used to identify chlamydial inclusions in infected HeLa cells. Alexa Fluor 488 and Alexa Fluor 594 anti-mouse IgG antibodies used in these experiments were purchased from Invitrogen/Life Technologies (Carlsbad, CA), and immunostaining results for titration of infectious chlamydial elementary bodies (EBs) were scanned and recorded by using the Evos FL auto cell imaging system (Thermo Fisher, Pittsburgh, PA). The corresponding isotype controls were used as negative controls.
hOE and HeLa cell infections.
OE-E6/E7 and HeLa cells were plated into either 12- or 6-well tissue culture plates and grown to 80 to 90% confluence. For all experiments (unless stated otherwise), the cells were infected with 10 inclusion-forming units (IFU) of C. trachomatis/cell in cell culture medium. Immediately after the addition of Chlamydia, the tissue culture plates were gently agitated, centrifuged at 1,200 rpm (200 × g) in a tabletop centrifuge for 1 h, and then incubated at 37°C in a 5% CO2 humidified incubator without a subsequent change of the medium until the time of cell harvest. Mock-infected control cells received an equivalent volume of epithelial cell culture medium but lacked any viable C. trachomatis cells.
Protein extraction and evaluation of protein expression levels.
Epithelial cell lysates were prepared via incubation of phosphate-buffered saline (PBS)-washed cell monolayers in radioimmunoprecipitation assay (RIPA) buffer (EMD Millipore, Burlington, MA) with the addition of 1 mM phenylmethylsulfonyl fluoride (PMSF) and 1× protease inhibitor cocktail (Sigma). The total protein concentration of each sample was determined by using the Pierce bicinchoninic acid (BCA) protein assay (Thermo Scientific). Analyses of protein expression were performed by using the Wes simple Western system (ProteinSimple, San Jose, CA). Endogenous TLR3 protein expression in WT and CRISPR-modified epithelial cells was detected using a 1:50 dilution of TLR3 monoclonal antibody (Abcam, Cambridge, MA). Relative TLR3 protein expression levels between WT and CRISPR-modified cells were obtained by measuring TLR3’s expression against that of the intracellular β-actin control protein bound to an anti-β-actin monoclonal antibody (1:300; Sigma). Protein detection with the Wes system was accomplished according to the manufacturer’s protocol using a streptavidin-horseradish peroxidase (HRP)-based methodology (ProteinSimple). The positive control for testing antibody specificity for TLR3 expression included loading (in separate reaction mixtures) of either 50 ng or 100 ng of the HEK293 cell lysate overexpressing human TLR3 (Novus).
RNA purification and real-time quantitative PCR.
Total cell RNA was isolated from mock- and C. trachomatis-infected WT and TLR3-deficient epithelial cells using the RNeasy kit plus (Qiagen, Valencia, CA). The DNA-free RNA was quantified using the NanoDrop spectrophotometer (Thermo Scientific), and cDNA was obtained with the Applied Biosystems high-capacity cDNA reverse transcription kit (Thermo Fisher). Target mRNA was quantified using an Applied Biosystems TaqMan gene expression master kit with reaction mixtures containing human TLR3 primers, human IFN-β primers, and/or β-actin control primers. Quantitative measurements were performed via the ABI7500 real-time PCR detection system (Thermo Fisher).
Standard and multiplex ELISAs.
The human IFN-β ELISA kit (catalog number 41410-1) and IL-6 ELISA kit (catalog number D6050), purchased from R&D Systems, were used to measure Chlamydia-induced IFN-β and IL-6, respectively, according to the manufacturer’s protocol. The standard ELISA kits were used to measure single cytokines secreted into the supernatants of hOE-WT, hOE-N-Ctrl, and hOE-TLR3KD cells that were either mock treated, infected with C. trachomatis, or treated with various TLR agonists.
To measure the chlamydia-induced synthesis of several immune factors simultaneously, WT and TLR3-deficient hOE (hOE-TLR3KD) cells were either mock treated or infected at an MOI of 10 IFU/cell with either serovar D or the L2 strain of C. trachomatis. For C. trachomatis L2 infections, the supernatants were harvested at 0 h (mock), 6 h, 12 h, 18 h, 24 h, and 30 h postinfection. The supernatants from the L2 infections were subjected to multiplex ELISAs in triplicate using Bio-Plex Pro 37-plex human inflammation panel 1 (catalog number 171AL001M; Bio-Rad, Hercules, CA) according to the manufacturer’s instructions. For infections with C. trachomatis serovar D, the supernatants were harvested at 0 h (mock), 6 h, 12 h, 24 h, 36 h, 48 h, 60 h, and 72 h postinfection. The supernatants from the C. trachomatis serovar D infections were subjected to multiplex ELISAs in triplicate using a custom-designed 27-plex human magnetic Luminex assay (catalog number LXSAHM; R&D Systems) according to the manufacturer’s instructions. Analyses of the data were performed in concert with the Indiana University Multiplex Analysis Core located in the Melvin and Bren Simon Cancer Center.
RNA interference.
Transfections of the TLR3-specific siRNA (catalog number AM16708; Ambion/Thermo Fisher) and the scrambled control RNA (Silencer negative control; Thermo Fisher) were done using Lipofectamine RNAiMAX (Thermo Fisher) as described previously (87). Briefly, 75 to 80% confluent hOE-WT cells were transfected with 2.5 μg of each siRNA for 24 h at 37°C with 5% CO2. After the 24-h period, cell supernatants were replaced with fresh medium prior to being infected with 10 IFU/cell C. trachomatis L2. The level of IFN-β expression was determined at the specified times postinfection by an ELISA as described above.
Flow cytometric analysis and multispectral imaging flow cytometry.
WT and TLR3-deficient epithelial cells were either mock treated or infected at an MOI of 10 IFU/cell with Chlamydia trachomatis L2 for 24 h. At the 24-h time point, monolayers were washed once with PBS–2 mM EDTA before being gently removed from the plate with trypsin-Versene and washed 3 times in ice-cold PBS-EDTA, and cell pellets were resuspended in 4% formalin for 30 min at room temperature. The cells were washed 3 times with PBS-EDTA and permeabilized in 0.3% Triton X-100–PBS–EDTA for 5 min at room temperature. The cells were blocked in blocking buffer (5% fetal bovine serum [FBS]–0.1% bovine serum albumin [BSA]–0.1%Triton X-100–PBS–EDTA) at room temperature for 60 min. The cells were stained with anti-chlamydia LPS antibody (from David E. Nelson) in blocking buffer (1:5) for 60 min at room temperature. The cells were washed 3 times with PBS-EDTA. The cells were further stained with allophycocyanin (APC) anti-mouse IgG(H+L) secondary antibody (1:1,000; Thermo-Fisher) in blocking buffer for 30 min before being washed 3 times with PBS-EDTA. Finally, the cells were suspended in 0.6 ml 2% FBS–PBS–EDTA for flow cytometry analysis. Cellular responses to Chlamydia infection were analyzed via a BD LSRFortessa cell analyzer (Becton, Dickinson, Franklin Lakes, NJ) or by an Amnis Image StreamX MKII multispectral imaging flow cytometer (EMD Millipore). Data were interpreted by using FlowJo v10 (FlowJo, LLC) and IDEAS (EMD Millipore) software.
Immunofluorescence staining.
WT and TLR3-deficient hOE cells were seeded into a 96-well μ-plate (Ibidi, Fitchburg, WI) and allowed to grow to 90% confluence before being either mock treated or infected at an MOI of 5 IFU/cell with C. trachomatis serovar D. At 36 h postinfection, the infected cells were then fixed with 200 μl of methanol and incubated at room temperature for 10 min. The fixed cells were stained with a 1:100 dilution of anti-chlamydial LPS antibody (EVI-HI; provided to us by David E. Nelson) and incubated for 1 h at room temperature. The stained cells were washed 3 times with PBS. Detection was accomplished with a secondary stain of Alexa Fluor 488 and Alexa Fluor 594 anti-mouse IgG antibodies (Invitrogen). Nuclei were counterstained with 4,6-diamidino-2-phenylindole (DAPI; Life Technologies) according to the manufacturer’s protocol and imaged at a ×60 magnification under oil immersion using a Nikon Eclipse Ti microscope.
Statistical analysis.
Numerical data are presented as means ± standard errors of the means (SEM). All experiments were repeated at least three times, and statistical significance was determined using either 2-way analysis of variance (ANOVA) in GraphPad Prism or Student’s two-tailed t test. Statistically significant differences in the figures are shown by asterisks, with the minimum criterion being a P value of <0.05.
Supplementary Material
ACKNOWLEDGMENTS
We thank David Nelson and his lab members for providing the C. trachomatis L2 strain, serovar D, and Chlamydia LPS antibody; for the use of the Evos system; and for their thoughtful discussion and support. We also thank Cheikh Seye for access to the Wes simple Western system. We thank the Multiplex Analysis Core at Indiana University’s Melvin and Bren Simon Cancer Center for providing support in analysis of samples and interpretation of data.
This work was supported by NIH grant AI104944 to W.A.D.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00483-19.
REFERENCES
- 1.CDC. 2017. 2015 sexually transmitted diseases survey. CDC, Atlanta, GA. [Google Scholar]
- 2.Farley TA, Cohen DA, Elkins W. 2003. Asymptomatic sexually transmitted diseases: the case for screening. Prev Med 36:502–509. doi: 10.1016/S0091-7435(02)00058-0. [DOI] [PubMed] [Google Scholar]
- 3.Korenromp EL, Sudaryo MK, de Vlas SJ, Gray RH, Sewankambo NK, Serwadda D, Wawer MJ, Habbema JD. 2002. What proportion of episodes of gonorrhoea and chlamydia becomes symptomatic? Int J STD AIDS 13:91–101. doi: 10.1258/0956462021924712. [DOI] [PubMed] [Google Scholar]
- 4.Elwell C, Mirrashidi K, Engel J. 2016. Chlamydia cell biology and pathogenesis. Nat Rev Microbiol 14:385–400. doi: 10.1038/nrmicro.2016.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nanni C, Marangoni A, Quarta C, Di Pierro D, Rizzello A, Trespidi S, D’Ambrosio D, Ambrosini V, Donati M, Aldini R, Zanotti-Fregonara P, Grassetto G, Rubello D, Fanti S, Cevenini R. 2009. Small animal PET for the evaluation of an animal model of genital infection. Clin Physiol Funct Imaging 29:187–192. doi: 10.1111/j.1475-097X.2008.00854.x. [DOI] [PubMed] [Google Scholar]
- 6.Jerchel S, Knebel G, Konig P, Bohlmann MK, Rupp J. 2012. A human fallopian tube model for investigation of C. trachomatis infections. J Vis Exp 2012:4036. doi: 10.3791/4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chow JM, Yonekura ML, Richwald GA, Greenland S, Sweet RL, Schachter J. 1990. The association between Chlamydia trachomatis and ectopic pregnancy. A matched-pair, case-control study. JAMA 263:3164–3167. doi: 10.1001/jama.1990.03440230060033. [DOI] [PubMed] [Google Scholar]
- 8.Naderi T, Kazerani F, Bahraminpoor A. 2012. Comparison of chlamydia infection prevalence between patients with and without ectopic pregnancy using the PCR method. Ginekol Pol 83:819–821. [PubMed] [Google Scholar]
- 9.Chen L, Lei L, Chang X, Li Z, Lu C, Zhang X, Wu Y, Yeh IT, Zhong G. 2010. Mice deficient in MyD88 develop a Th2-dominant response and severe pathology in the upper genital tract following Chlamydia muridarum infection. J Immunol 184:2602–2610. doi: 10.4049/jimmunol.0901593. [DOI] [PubMed] [Google Scholar]
- 10.Stephens RS. 2003. The cellular paradigm of chlamydial pathogenesis. Trends Microbiol 11:44–51. doi: 10.1016/S0966-842X(02)00011-2. [DOI] [PubMed] [Google Scholar]
- 11.Bakken TE, Roddey JC, Djurovic S, Akshoomoff N, Amaral DG, Bloss CS, Casey BJ, Chang L, Ernst TM, Gruen JR, Jernigan TL, Kaufmann WE, Kenet T, Kennedy DN, Kuperman JM, Murray SS, Sowell ER, Rimol LM, Mattingsdal M, Melle I, Agartz I, Andreassen OA, Schork NJ, Dale AM. 2012. Association of common genetic variants in GPCPD1 with scaling of visual cortical surface area in humans. Proc Natl Acad Sci U S A 109:3985–3990. doi: 10.1073/pnas.1105829109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darville T, O’Neill JM, Andrews CW Jr, Nagarajan UM, Stahl L, Ojcius DM. 2003. Toll-like receptor-2, but not Toll-like receptor-4, is essential for development of oviduct pathology in chlamydial genital tract infection. J Immunol 171:6187–6197. doi: 10.4049/jimmunol.171.11.6187. [DOI] [PubMed] [Google Scholar]
- 13.Derbigny WA, Johnson RM, Toomey KS, Ofner S, Jayarapu K. 2010. The Chlamydia muridarum-induced IFN-beta response is TLR3-dependent in murine oviduct epithelial cells. J Immunol 185:6689–6697. doi: 10.4049/jimmunol.1001548. [DOI] [PubMed] [Google Scholar]
- 14.Derbigny WA, Kerr MS, Johnson RM. 2005. Pattern recognition molecules activated by Chlamydia muridarum infection of cloned murine oviduct epithelial cell lines. J Immunol 175:6065–6075. doi: 10.4049/jimmunol.175.9.6065. [DOI] [PubMed] [Google Scholar]
- 15.Derbigny WA, Shobe LR, Kamran JC, Toomey KS, Ofner S. 2012. Identifying a role for Toll-like receptor 3 in the innate immune response to Chlamydia muridarum infection in murine oviduct epithelial cells. Infect Immun 80:254–265. doi: 10.1128/IAI.05549-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prantner D, Darville T, Nagarajan UM. 2010. Stimulator of IFN gene is critical for induction of IFN-beta during Chlamydia muridarum infection. J Immunol 184:2551–2560. doi: 10.4049/jimmunol.0903704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rank RG, Lacy HM, Goodwin A, Sikes J, Whittimore J, Wyrick PB, Nagarajan UM. 2010. Host chemokine and cytokine response in the endocervix within the first developmental cycle of Chlamydia muridarum. Infect Immun 78:536–544. doi: 10.1128/IAI.00772-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Welter-Stahl L, Ojcius DM, Viala J, Girardin S, Liu W, Delarbre C, Philpott D, Kelly KA, Darville T. 2006. Stimulation of the cytosolic receptor for peptidoglycan, Nod1, by infection with Chlamydia trachomatis or Chlamydia muridarum. Cell Microbiol 8:1047–1057. doi: 10.1111/j.1462-5822.2006.00686.x. [DOI] [PubMed] [Google Scholar]
- 19.Da Costa CU, Wantia N, Kirschning CJ, Busch DH, Rodriguez N, Wagner H, Miethke T. 2004. Heat shock protein 60 from Chlamydia pneumoniae elicits an unusual set of inflammatory responses via Toll-like receptor 2 and 4 in vivo. Eur J Immunol 34:2874–2884. doi: 10.1002/eji.200425101. [DOI] [PubMed] [Google Scholar]
- 20.Erridge C, Pridmore A, Eley A, Stewart J, Poxton IR. 2004. Lipopolysaccharides of Bacteroides fragilis, Chlamydia trachomatis and Pseudomonas aeruginosa signal via Toll-like receptor 2. J Med Microbiol 53:735–740. doi: 10.1099/jmm.0.45598-0. [DOI] [PubMed] [Google Scholar]
- 21.Netea MG, Kullberg BJ, Galama JM, Stalenhoef AF, Dinarello CA, Van der Meer JW. 2002. Non-LPS components of Chlamydia pneumoniae stimulate cytokine production through Toll-like receptor 2-dependent pathways. Eur J Immunol 32:1188–1195. doi:. [DOI] [PubMed] [Google Scholar]
- 22.Prebeck S, Kirschning C, Durr S, da Costa C, Donath B, Brand K, Redecke V, Wagner H, Miethke T. 2001. Predominant role of Toll-like receptor 2 versus 4 in Chlamydia pneumoniae-induced activation of dendritic cells. J Immunol 167:3316–3323. doi: 10.4049/jimmunol.167.6.3316. [DOI] [PubMed] [Google Scholar]
- 23.Aziz A. 2006. A study on immunopathogenetic mechanisms of atherosclerotic process caused by chronic infection of Chlamydia pneumoniae in rats (Ratus novergicus). Acta Med Indones 38:206–212. [PubMed] [Google Scholar]
- 24.Johnson RM. 2004. Murine oviduct epithelial cell cytokine responses to Chlamydia muridarum infection include interleukin-12-p70 secretion. Infect Immun 72:3951–3960. doi: 10.1128/IAI.72.7.3951-3960.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li HW, Liao SB, Chiu PC, Tam WW, Ho JC, Ng EH, Ho PC, Yeung WS, Tang F, O WS. 2010. Expression of adrenomedullin in human oviduct, its regulation by the hormonal cycle and contact with spermatozoa, and its effect on ciliary beat frequency of the oviductal epithelium. J Clin Endocrinol Metab 95:E18–E25. doi: 10.1210/jc.2010-0273. [DOI] [PubMed] [Google Scholar]
- 26.Lee YL, Lee KF, Xu JS, Wang YL, Tsao SW, Yeung WS. 2001. Establishment and characterization of an immortalized human oviductal cell line. Mol Reprod Dev 59:400–409. doi: 10.1002/mrd.1046. [DOI] [PubMed] [Google Scholar]
- 27.Zandieh Z, Ashrafi M, Jameie B, Amanpour S, Mosaffa N, Salman Yazdi R, Pacey A, Aflatoonian R. 2015. Evaluation of immunological interaction between spermatozoa and fallopian tube epithelial cells. Andrologia 47:1120–1130. doi: 10.1111/and.12391. [DOI] [PubMed] [Google Scholar]
- 28.Shaw JL, Wills GS, Lee KF, Horner PJ, McClure MO, Abrahams VM, Wheelhouse N, Jabbour HN, Critchley HO, Entrican G, Horne AW. 2011. Chlamydia trachomatis infection increases fallopian tube PROKR2 via TLR2 and NFkappaB activation resulting in a microenvironment predisposed to ectopic pregnancy. Am J Pathol 178:253–260. doi: 10.1016/j.ajpath.2010.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagarajan UM, Ojcius DM, Stahl L, Rank RG, Darville T. 2005. Chlamydia trachomatis induces expression of IFN-gamma-inducible protein 10 and IFN-beta independent of TLR2 and TLR4, but largely dependent on MyD88. J Immunol 175:450–460. doi: 10.4049/jimmunol.175.1.450. [DOI] [PubMed] [Google Scholar]
- 30.O’Connell CM, Ionova IA, Quayle AJ, Visintin A, Ingalls RR. 2006. Localization of TLR2 and MyD88 to Chlamydia trachomatis inclusions. Evidence for signaling by intracellular TLR2 during infection with an obligate intracellular pathogen. J Biol Chem 281:1652–1659. doi: 10.1074/jbc.M510182200. [DOI] [PubMed] [Google Scholar]
- 31.Yang X, Coriolan D, Schultz K, Golenbock DT, Beasley D. 2005. Toll-like receptor 2 mediates persistent chemokine release by Chlamydia pneumoniae-infected vascular smooth muscle cells. Arterioscler Thromb Vasc Biol 25:2308–2314. doi: 10.1161/01.ATV.0000187468.00675.a3. [DOI] [PubMed] [Google Scholar]
- 32.Carrasco SE, Hu S, Imai DM, Kumar R, Sandusky GE, Yang XF, Derbigny WA. 2018. Toll-like receptor 3 (TLR3) promotes the resolution of Chlamydia muridarum genital tract infection in congenic C57BL/6N mice. PLoS One 13:e0195165. doi: 10.1371/journal.pone.0195165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsumoto M, Funami K, Tanabe M, Oshiumi H, Shingai M, Seto Y, Yamamoto A, Seya T. 2003. Subcellular localization of Toll-like receptor 3 in human dendritic cells. J Immunol 171:3154–3162. doi: 10.4049/jimmunol.171.6.3154. [DOI] [PubMed] [Google Scholar]
- 34.Matsumoto M, Kikkawa S, Kohase M, Miyake K, Seya T. 2002. Establishment of a monoclonal antibody against human Toll-like receptor 3 that blocks double-stranded RNA-mediated signaling. Biochem Biophys Res Commun 293:1364–1369. doi: 10.1016/S0006-291X(02)00380-7. [DOI] [PubMed] [Google Scholar]
- 35.Carpenter S, O’Neill LAJ. 2009. Recent insights into the structure of Toll-like receptors and post-translational modifications of their associated signalling proteins. Biochem J 422:1–10. doi: 10.1042/BJ20090616. [DOI] [PubMed] [Google Scholar]
- 36.Derbigny WA, Hong SC, Kerr MS, Temkit M, Johnson RM. 2007. Chlamydia muridarum infection elicits a beta interferon response in murine oviduct epithelial cells dependent on interferon regulatory factor 3 and TRIF. Infect Immun 75:1280–1290. doi: 10.1128/IAI.01525-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng W, Shivshankar P, Li Z, Chen L, Yeh IT, Zhong G. 2008. Caspase-1 contributes to Chlamydia trachomatis-induced upper urogenital tract inflammatory pathologies without affecting the course of infection. Infect Immun 76:515–522. doi: 10.1128/IAI.01064-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Darville T, Andrews CW Jr, Sikes JD, Fraley PL, Rank RG. 2001. Early local cytokine profiles in strains of mice with different outcomes from chlamydial genital tract infection. Infect Immun 69:3556–3561. doi: 10.1128/IAI.69.6.3556-3561.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maxion HK, Kelly KA. 2002. Chemokine expression patterns differ within anatomically distinct regions of the genital tract during Chlamydia trachomatis infection. Infect Immun 70:1538–1546. doi: 10.1128/IAI.70.3.1538-1546.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tseng CT, Rank RG. 1998. Role of NK cells in early host response to chlamydial genital infection. Infect Immun 66:5867–5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wan C, Latter JL, Amirshahi A, Symonds I, Finnie J, Bowden N, Scott RJ, Cunningham KA, Timms P, Beagley KW. 2014. Progesterone activates multiple innate immune pathways in Chlamydia trachomatis-infected endocervical cells. Am J Reprod Immunol 71:165–177. doi: 10.1111/aji.12168. [DOI] [PubMed] [Google Scholar]
- 42.Murthy AK, Li W, Chaganty BK, Kamalakaran S, Guentzel MN, Seshu J, Forsthuber TG, Zhong G, Arulanandam BP. 2011. Tumor necrosis factor alpha production from CD8+ T cells mediates oviduct pathological sequelae following primary genital Chlamydia muridarum infection. Infect Immun 79:2928–2935. doi: 10.1128/IAI.05022-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schneider P, MacKay F, Steiner V, Hofmann K, Bodmer JL, Holler N, Ambrose C, Lawton P, Bixler S, Acha-Orbea H, Valmori D, Romero P, Werner-Favre C, Zubler RH, Browning JL, Tschopp J. 1999. BAFF, a novel ligand of the tumor necrosis factor family, stimulates B cell growth. J Exp Med 189:1747–1756. doi: 10.1084/jem.189.11.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Igietseme JU, Omosun Y, Nagy T, Stuchlik O, Reed MS, He Q, Partin J, Joseph K, Ellerson D, George Z, Goldstein J, Eko FO, Bandea C, Pohl J, Black CM. 2018. Molecular pathogenesis of Chlamydia disease complications: epithelial-mesenchymal transition and fibrosis. Infect Immun 86:e00585-17. doi: 10.1128/IAI.00585-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagase H, Woessner JF Jr.. 1999. Matrix metalloproteinases. J Biol Chem 274:21491–21494. doi: 10.1074/jbc.274.31.21491. [DOI] [PubMed] [Google Scholar]
- 46.Ault KA, Kelly KA, Ruther PE, Izzo AA, Izzo LS, Sigar IM, Ramsey KH. 2002. Chlamydia trachomatis enhances the expression of matrix metalloproteinases in an in vitro model of the human fallopian tube infection. Am J Obstet Gynecol 187:1377–1383. doi: 10.1067/mob.2002.126850. [DOI] [PubMed] [Google Scholar]
- 47.El-Asrar AM, Geboes K, Al-Kharashi SA, Al-Mosallam AA, Missotten L, Paemen L, Opdenakker G. 2000. Expression of gelatinase B in trachomatous conjunctivitis. Br J Ophthalmol 84:85–91. doi: 10.1136/bjo.84.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Imtiaz MT, Distelhorst JT, Schripsema JH, Sigar IM, Kasimos JN, Lacy SR, Ramsey KH. 2007. A role for matrix metalloproteinase-9 in pathogenesis of urogenital Chlamydia muridarum infection in mice. Microbes Infect 9:1561–1566. doi: 10.1016/j.micinf.2007.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Imtiaz MT, Schripsema JH, Sigar IM, Kasimos JN, Ramsey KH. 2006. Inhibition of matrix metalloproteinases protects mice from ascending infection and chronic disease manifestations resulting from urogenital Chlamydia muridarum infection. Infect Immun 74:5513–5521. doi: 10.1128/IAI.00730-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pal S, Schmidt AP, Peterson EM, Wilson CL, de la Maza LM. 2006. Role of matrix metalloproteinase-7 in the modulation of a Chlamydia trachomatis infection. Immunology 117:213–219. doi: 10.1111/j.1365-2567.2005.02281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brunham RC, Rey-Ladino J. 2005. Immunology of Chlamydia infection: implications for a Chlamydia trachomatis vaccine. Nat Rev Immunol 5:149–161. doi: 10.1038/nri1551. [DOI] [PubMed] [Google Scholar]
- 52.Witkin SS, Minis E, Athanasiou A, Leizer J, Linhares IM. 2017. Chlamydia trachomatis: the persistent pathogen. Clin Vaccine Immunol 24:e00203-17. doi: 10.1128/CVI.00203-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beatty WL, Belanger TA, Desai AA, Morrison RP, Byrne GI. 1994. Tryptophan depletion as a mechanism of gamma interferon-mediated chlamydial persistence. Infect Immun 62:3705–3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Beatty WL, Byrne GI, Morrison RP. 1993. Morphologic and antigenic characterization of interferon gamma-mediated persistent Chlamydia trachomatis infection in vitro. Proc Natl Acad Sci U S A 90:3998–4002. doi: 10.1073/pnas.90.9.3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mackern-Oberti JP, Motrich RD, Breser ML, Cejas H, Cuffini C, Maccioni M, Rivero VE. 2011. Male rodent genital tract infection with Chlamydia muridarum: persistence in the prostate gland that triggers self-immune reactions in genetically susceptible hosts. J Urol 186:1100–1106. doi: 10.1016/j.juro.2011.04.086. [DOI] [PubMed] [Google Scholar]
- 56.Rittling SR, Singh R. 2015. Osteopontin in immune-mediated diseases. J Dent Res 94:1638–1645. doi: 10.1177/0022034515605270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang XL, Zhang L, Duan Y, Wang YJ, Wang J. 2016. Association of pentraxin 3 with autoimmune diseases: a systematic review and meta-analysis. Arch Med Res 47:223–231. doi: 10.1016/j.arcmed.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 58.Yang SF, Wu TF, Tsai HT, Lin LY, Wang PH. 2014. New markers in pelvic inflammatory disease. Clin Chim Acta 431:118–124. doi: 10.1016/j.cca.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 59.Baio P, Brucato A, Buskila D, Gershwin ME, Giacomazzi D, Lopez LR, Luzzati R, Matsuura E, Selmi C, Sarzi-Puttini P, Atzeni F. 2008. Autoimmune diseases and infections: controversial issues. Clin Exp Rheumatol 26:S74–S80. [PubMed] [Google Scholar]
- 60.Teixeira AR, Hecht MM, Guimaro MC, Sousa AO, Nitz N. 2011. Pathogenesis of Chagas’ disease: parasite persistence and autoimmunity. Clin Microbiol Rev 24:592–630. doi: 10.1128/CMR.00063-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dolcino M, Puccetti A, Barbieri A, Bason C, Tinazzi E, Ottria A, Patuzzo G, Martinelli N, Lunardi C. 2015. Infections and autoimmunity: role of human cytomegalovirus in autoimmune endothelial cell damage. Lupus 24:419–432. doi: 10.1177/0961203314558677. [DOI] [PubMed] [Google Scholar]
- 62.Sebastiani GD, Galeazzi M. 2009. Infection-genetics relationship in systemic lupus erythematosus. Lupus 18:1169–1175. doi: 10.1177/0961203309345737. [DOI] [PubMed] [Google Scholar]
- 63.Hosey KL, Hu S, Derbigny WA. 2015. Role of STAT1 in Chlamydia-induced type-1 interferon production in oviduct epithelial cells. J Interferon Cytokine Res 35:901–916. doi: 10.1089/jir.2015.0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Y, Yeruva L, Marinov A, Prantner D, Wyrick PB, Lupashin V, Nagarajan UM. 2014. The DNA sensor, cyclic GMP-AMP synthase, is essential for induction of IFN-beta during Chlamydia trachomatis infection. J Immunol 193:2394–2404. doi: 10.4049/jimmunol.1302718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kagnoff MF, Eckmann L. 1997. Epithelial cells as sensors for microbial infection. J Clin Invest 100:6–10. doi: 10.1172/JCI119522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hunter CA, Jones SA. 2015. IL-6 as a keystone cytokine in health and disease. Nat Immunol 16:448–457. doi: 10.1038/ni.3153. [DOI] [PubMed] [Google Scholar]
- 67.Sun X, Tian Q, Wang L, Xue M, Zhong G. 2017. IL-6-mediated signaling pathways limit Chlamydia muridarum infection and exacerbate its pathogenicity in the mouse genital tract. Microbes Infect 19:536–545. doi: 10.1016/j.micinf.2017.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vogel I, Thorsen P, Curry A, Sandager P, Uldbjerg N. 2005. Biomarkers for the prediction of preterm delivery. Acta Obstet Gynecol Scand 84:516–525. doi: 10.1111/j.0001-6349.2005.00771.x. [DOI] [PubMed] [Google Scholar]
- 69.Maghazachi AA, Al-Aoukaty A, Schall TJ. 1996. CC chemokines induce the generation of killer cells from CD56+ cells. Eur J Immunol 26:315–319. doi: 10.1002/eji.1830260207. [DOI] [PubMed] [Google Scholar]
- 70.Li M, Liu X, Zhou Y, Su SB. 2009. Interferon-lambdas: the modulators of antivirus, antitumor, and immune responses. J Leukoc Biol 86:23–32. doi: 10.1189/jlb.1208761. [DOI] [PubMed] [Google Scholar]
- 71.Billmeier U, Dieterich W, Neurath MF, Atreya R. 2016. Molecular mechanism of action of anti-tumor necrosis factor antibodies in inflammatory bowel diseases. World J Gastroenterol 22:9300–9313. doi: 10.3748/wjg.v22.i42.9300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Deodhar A, Yu D. 2017. Switching tumor necrosis factor inhibitors in the treatment of axial spondyloarthritis. Semin Arthritis Rheum 47:343–350. doi: 10.1016/j.semarthrit.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 73.Kalden JR, Schulze-Koops H. 2017. Immunogenicity and loss of response to TNF inhibitors: implications for rheumatoid arthritis treatment. Nat Rev Rheumatol 13:707–718. doi: 10.1038/nrrheum.2017.187. [DOI] [PubMed] [Google Scholar]
- 74.Haraoui B, Bykerk V. 2007. Etanercept in the treatment of rheumatoid arthritis. Ther Clin Risk Manag 3:99–105. doi: 10.2147/tcrm.2007.3.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee DE, Trowbridge RM, Ayoub NT, Agrawal DK. 2015. High-mobility group box protein-1, matrix metalloproteinases, and vitamin D in keloids and hypertrophic scars. Plast Reconstr Surg Glob Open 3:e425. doi: 10.1097/GOX.0000000000000391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rohani MG, Parks WC. 2015. Matrix remodeling by MMPs during wound repair. Matrix Biol 44–46:113–121. doi: 10.1016/j.matbio.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 77.Kirkegaard-Klitbo DM, Mejer N, Knudsen TB, Moller HJ, Moestrup SK, Poulsen SD, Kronborg G, Benfield T. 2017. Soluble CD163 predicts incident chronic lung, kidney and liver disease in HIV infection. AIDS 31:981–988. doi: 10.1097/QAD.0000000000001432. [DOI] [PubMed] [Google Scholar]
- 78.Zhi Y, Gao P, Xin X, Li W, Ji L, Zhang L, Zhang X, Zhang J. 2017. Clinical significance of sCD163 and its possible role in asthma (review). Mol Med Rep 15:2931–2939. doi: 10.3892/mmr.2017.6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Di Rosa M, Distefano G, Zorena K, Malaguarnera L. 2016. Chitinases and immunity: ancestral molecules with new functions. Immunobiology 221:399–411. doi: 10.1016/j.imbio.2015.11.014. [DOI] [PubMed] [Google Scholar]
- 80.Nathan N, Corvol H, Amselem S, Clement A. 2015. Biomarkers in interstitial lung diseases. Paediatr Respir Rev 16:219–224. doi: 10.1016/j.prrv.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 81.Choi ST, Kim JH, Kang EJ, Lee SW, Park MC, Park YB, Lee SK. 2008. Osteopontin might be involved in bone remodelling rather than in inflammation in ankylosing spondylitis. Rheumatology (Oxford) 47:1775–1779. doi: 10.1093/rheumatology/ken385. [DOI] [PubMed] [Google Scholar]
- 82.Wang KX, Denhardt DT. 2008. Osteopontin: role in immune regulation and stress responses. Cytokine Growth Factor Rev 19:333–345. doi: 10.1016/j.cytogfr.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 83.De Filippis A, Buommino E, Domenico MD, Feola A, Brunetti-Pierri R, Rizzo A. 2017. Chlamydia trachomatis induces an upregulation of molecular biomarkers podoplanin, Wilms’ tumour gene 1, osteopontin and inflammatory cytokines in human mesothelial cells. Microbiology 163:654–663. doi: 10.1099/mic.0.000465. [DOI] [PubMed] [Google Scholar]
- 84.Rizzo A, Carratelli CR, De Filippis A, Bevilacqua N, Tufano MA, Buommino E. 2014. Transforming activities of Chlamydia pneumoniae in human mesothelial cells. Int Microbiol 17:185–193. doi: 10.2436/20.1501.01.221. [DOI] [PubMed] [Google Scholar]
- 85.Flo TH, Smith KD, Sato S, Rodriguez DJ, Holmes MA, Strong RK, Akira S, Aderem A. 2004. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature 432:917–921. doi: 10.1038/nature03104. [DOI] [PubMed] [Google Scholar]
- 86.Brothwell JA, Muramatsu MK, Toh E, Rockey DD, Putman TE, Barta ML, Hefty PS, Suchland RJ, Nelson DE. 2016. Interrogating genes that mediate Chlamydia trachomatis survival in cell culture using conditional mutants and recombination. J Bacteriol 198:2131–2139. doi: 10.1128/JB.00161-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Skovdahl HK, van Beelen Granlund A, Østvik AE, Bruland T, Bakke I, Torp SH, Damås JK, Sandvik AK. 2015. Expression of CCL20 and its corresponding receptor CCR6 is enhanced in active inflammatory bowel disease, and TLR3 mediates CCL20 expression in colonic epithelial cells. PLoS One 10:e0141710. doi: 10.1371/journal.pone.0141710. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.