Abstract
Somatic APC (adenomatous polyposis coli), TP53, KRAS mutations are present in roughly 80%, 60%, and 40%, respectively, of human colorectal cancers (CRCs). Most TP53 mutant alleles in CRCs encode missense mutant proteins with loss-of-function (LOF) of p53’s transcriptional activity and dominant negative (DN) effects on wild type p53 function. Missense mutant p53 proteins have been reported to exert gain-of-function (GOF) effects in cancer. We compared the phenotypic effects of the common human cancer-associated TP53 R273H missense mutation to p53 null status in a genetically engineered mouse CRC model. Inactivation of one allele of Apc together with activation of a Kras mutant allele in mouse colon epithelium instigated development of serrated and hyperplastic epithelium and adenomas (AK mice). Addition of a Trp53R270H or Trp53null mutant allele to the model (AKP mice) led to markedly shortened survival and increased tumor burden relative to that of AK mice, including adenocarcinomas in AKP mice. Comparable life span and tumor burden were seen in AKP mice carrying Trp53R270H or Trp53null alleles, along with similar frequencies of spontaneous metastasis to lymph nodes, lung and liver. The fraction of adenocarcinomas with submucosa or deeper invasion was higher in AKP270/fl mice than in AKPfl/fl mice, but the incidence of adenocarcinomas per mouse did not differ significantly between AKPfl/fl and AKP270/fl mice. In line with their comparable biological behaviors, mouse primary tumors and tumor-derived organoids with the Trp53R270H or Trp53null alleles had highly similar gene expression profiles. Human CRCs with TP53 R273 missense mutant or null alleles also had essentially homogeneous gene expression patterns. Our findings indicate the R270H/R273H p53 mutant protein does not manifest definite GOF biological effects in mouse and human CRCs, suggesting possible GOF effects of mutant p53 in cancer phenotypes are likely allele-specific and/or context-dependent.
Keywords: colon cancer, mouse model, p53, tumor suppressor gene, loss of function, gain of function
Introduction
Accumulated genetic and epigenetic alterations underlie development of colorectal cancer (CRC), which in some patients can metastasize to lymph nodes and distant organs, such as liver and lung [1–4]. Significant molecular and biological heterogeneity is seen in CRCs overall, and comprehensive studies have helped to define molecular subtypes of CRCs and associated clinico-pathological features [4–7]. About 85% of CRCs manifest the microsatellite stable (MSS) or non-hypermutated phenotype, whereas the remainder show the microsatellite instability-high (MSI-H) or hypermutated phenotype. In non-hypermutated CRCs, the most frequently somatically mutated genes are APC, TP53, KRAS, PIK3CA, SMAD4, and FBXW7, with many other recurrently mutated genes found in less than 10% of cases [4–7]. About 80% of non-hypermutated CRCs harbor truncating nonsense or frameshift mutations or splice mutations in the APC (adenomatous polyposis coli) gene that disable its tumor suppressor function [4–7]. Activating KRAS mutations are found in roughly 40% of CRCs. In the absence of other cooperating alterations, KRAS activation promotes only colon epithelial hyperplasia [8]. But, when combined with certain mutations, such as APC mutations, mutant KRAS enhances tumorigenic features [2], and it likely also contributes to advanced cancer phenotypes, as a recent study showed mutant Kras had roles in invasion and metastatic growth in a mouse CRC model [9].
TP53, the most frequently mutated gene across human cancer types [10], is mutated in about 60% of CRCs [5]. The p53 protein is a transcription factor that, in response to cell stress, regulates expression of genes functioning in cell cycle arrest, senescence, apoptosis, and other processes [11, 12]. Most cancer-associated TP53 mutations (>60%) are missense mutations, leading to expression of a stabilized full-length protein that is highly abundant in tumor cells relative to wild type p53 levels in normal cells[13, 14]. Most TP53 missense mutations reside in the DNA binding domain (DBD) and result in loss-of-function (LOF) of p53’s binding to key regulatory elements in target genes [15, 16]. The cancer-associated missense mutant p53 proteins also often exert dominant-negative (DN) effects on wild-type p53 protein in cells, via hetero-oligomerization of mutant p53 with wild type p53 [15].
Many studies have reported missense mutant p53 proteins may exert unique, pro-tumorigenic effects in cancer cells and these effects have often been referred to as p53 “gain-of-function” (GOF) properties [17, 18]. Many studies to assess mutant p53 GOF have been undertaken in cell culture and/or with cell lines implanted as tumor xenografts. Collectively, the work indicates that missense mutant p53 proteins, when expressed endogenously or ectopically from transduced cDNAs can enhance cell proliferation, survival, invasion and migration, cancer stem cell phenotypes, and/or drug resistance [13, 17, 18]. GOF effects of mutant p53 have also been reported in some murine cancer models for skin cancer [19], pancreatic cancer [20], rhabdomyosarcoma [21], and sporadic and chemically induced colitis-associated intestinal tumors [22–25]. In these mouse model studies, compared to Trp53-null tumors, tumors harboring missense mutation in Trp53 (at codon 172, 248 or 270) were described as more invasive [19–25], and some lesions gained metastatic potential [19–21]. In addition, mice carrying Trp53 missense mutation in the germline had differences in their tumor spectrum from Trp53-null mice, including more carcinomas and endothelial tumors [26]. Moreover, in another similar study, mice carrying a heterozygous Trp53 R172H mutation in the germline showed more metastatic osteosarcomas and carcinomas compared to mice heterozygous for a Trp53 null allele, although the overall tumor spectrum was not distinct [27]. A potential GOF effect for missense mutant p53 has also been reported in Li-Fraumeni patients, with earlier tumor onset in patients with germline TP53 missense mutations than in patients with TP53 germline alterations leading to complete loss of p53 expression [28, 29].
In spite of the reported findings on mutant p53 GOF in modulating cancer phenotypes, there is no unifying hypothesis for how missense mutant p53 proteins exert GOF effects, as published studies have presented many seemingly unlinked molecular mechanisms to explain the presumptive GOF effects observed [11, 18]. In addition, a number of the studies undertaken could not distinguish between p53 GOF versus p53 DN effects, as missense mutant p53 proteins were not infrequently studied in cell expressing wild type p53 in tissue culture or in vivo. Moreover, some mouse and human cancer studies have failed to find evidence for mutant p53 GOF effects. Two mouse cancer model studies found the introduction of Trp53 missense mutations into lung or ovarian genetically engineered cancer models did not generate any demonstrable GOF biological activity compared to animals with Trp53 null alleles [30, 31]. Moreover, it was observed that distant metastases were nearly 8-fold more common in patients with ovarian cancers that carried TP53 null mutations compared with those whose tumors had TP53 missense mutations [32]. The lack of a unifying mechanism for the apparent p53 GOF effects in the different studies where effects have been seen, taken together with the absence of demonstrable GOF in some human and mouse cancer studies, suggest that any GOF effects by mutant p53 are likely to be tissue-specific and/or context-dependent. Some recent studies have reported that both Trp53 null and missense alleles can contribute to invasive and metastatic phenotypes in intestinal cells in vivo or in organoid models systems[9, 24, 33–35]. However, the relative biological effects of Trp53 missense and null mutations in collaborating with other accumulated gene defects often contributing to CRCs, such as Apc and Kras alterations, in the in vivo development of mouse colon cancer have not been reported. We present here in-depth and comprehensive studies of the effects of R270H and null Trp53 alleles in a mouse model of CRC reflecting the molecular pathogenesis of a significant fraction of human CRCs.
Material and Methods
Mice
Apcflox/flox (580S) mice[36] and KrasLSL-G12D/+ mice [37] have been described previously. Trp53R270H/+ (01XM1) and Trp53flox/flox (01XC2) mice were purchased from the National Cancer Institute (Bethesda, MD) mouse repository. To target the floxed alleles specifically in mouse colon and cecum, CDX2P-CreERT2 transgenic mice [38] were first intercrossed with Apcflox/flox , and KrasLSL-G12D/+ mice to generate CDX2P-CreERT2 Apcflox/+ KrasLSL-G12D/+ mice. For clarity, Apcflox/+ and KrasLSL-G12D/+ were abbreviated as A and K, respectively. Then CDX2P-CreERT2 AK mice were crossed with Trp53R270H/+ mice (P270/+) and Trp53flox/flox mice (Pfl/fl) to generate the following Cre-positive triple transgenic strains: CDX2P-CreERT2AKP270/+, CDX2P-CreERT2AKP270/fl and CDX2P-CreERT2AKPfl/fl. Animal husbandry and experimental procedures were carried out under approval from the University of Michigan’s Institutional Animal Care and Use Committee (PRO00007181) and according to Michigan state and US federal regulations. All the mice were housed in specific-pathogen free (SPF) conditions. After weaning, rodent 5LOD chow and automatically supplied water were provided ad libitum to mice.
Tamoxifen (TAM) treatment
Adult mice (2–4 months of age) with CDX2P-CreERT2 transgene were injected intraperitoneally with TAM (Sigma-Aldrich, St. Louis, MO) dissolved in corn oil (Sigma-Aldrich). For two daily TAM doses, we administered TAM at 70 mg/kg per dose. Animals were euthanized when the mice reached humane endpoints.
Establishment of colonic organoids
Organoids were derived from proximal colon tumors generated in CDX2P-CreERT2 AKP270/fl mice (n=3) or CDX2P-CreERT2 AKPfl/fl mice (n=2) at 3 months post TAM injection. The organoids derived from the adenomas in the Apcfl/fl mice (Cont 1, after 15 days post TAM injection, n=3) and wild-type colon epithelium (Cont 2, n=4) were used as controls. Mice were first treated with TAM to induce gene targeting and/or tumor formation, and then colonic organoids were generated and propagated using previously described “TMDU protocol” [39] with minor modifications[40]. TMDU medium lacking Wnt3a and R-spondin2 conditional media were used to select for Apc deletion mutation in organoids that carry Apcflox alleles. All the organoids were cultured for 4 days before they were harvested for RNA.
Immunohistochemistry (IHC)
Mouse tissues were fixed in 10% (v/v) buffered formalin and embedded in paraffin. Sections of the paraffin-embedded tissues were subjected to immunohistochemical (IHC) analysis as described[8]. The following primary antibodies were used for IHC analysis: mouse anti–β-catenin (1:800; BD Biosciences, Lexington, KY), rabbit anti-p53 (1:500; Vector Laboratories Inc., Burlingame, CA), goat anti–E-cadherin (1:500; R&D Systems, Minneapolis, MN), Rabbit anti-vimentin (1:500; Epitomics, Burlingame, CA), and mouse anti-CDX2 (1:100; BioGenex, Fremont, CA). The IHC staining with anti-p53 antibody was classified according to the percentage of stained cells. Expression of p53 was considered to be weak if <10% of neoplastic cells were observed to be stained, and strong if >10% of neoplastic cells were stained.
Gene Expression
CDX2P-CreERT2 AKP270/fl mice (n=6) and CDX2P-CreERT2 AKPfl/fl (n=6) mice were treated with TAM to induce tumor formation. The colon adenocarcinomas from these mice were laser capture-microdissected (LCM) and total RNAs were extracted using Arcturus PicoPure RNA Isolation Kit (Applied Biosystems, Foster City, CA). Microdissected wild-type normal colon epithelial cells were included as controls (n=3). We also collected total RNAs from organoids using Trizol (Invitrogen, San Diego, CA), followed by RNeasy columns (Qiagen, Germantown, MD). Tumor organoids were generated from the mouse colon tumors arising in CDX2P-CreERT2 AKP270/fl mice (n=3) or CDX2P-CreERT2 AKPfl/fl mice (n=2) at 3 months post TAM injection. The organoids derived from adenomas in CDX2P-CreERT2 Apcfl/fl mice (Cont 1, 15 days post TAM injection, n=3) and wild-type colon epithelium (Cont 2, n=4) were used as controls. Micro-dissected tumor and normal tissues from mice had transcript abundances measured with Affymetrix Mouse Gene ST 2.1 strips using WT-Pico kits, while organoid samples were run on ST 2.1 plates, using Affymetrix Affy Plus kits. The 41345 probe-sets on the two platforms are identical. Transcript abundances were estimated using the RMA algorithm[41], separately for the two experiments. We analyzed only 24562 probe-sets that had associated Entrez gene IDs. Our raw and processed data as well as our statistical analysis are available in GEO series GSE125262. For enrichment test, we mapped mouse genes to human homologs using only 1-to-1 best homologs from NCBI Homologene build 68, giving 15850 distinct genes. We tested our up and down gene lists for enrichment in 50 Hallmark gene sets from the Molecular Signatures Database (MSigDB v6.1, http://www.broadinstitute.org/gsea/msigdb/index.jsp) [42]. For gene expression analysis of human CRCs, we downloaded normalized counts for mRNA-seq data from Illumina Genome Analyzer assays from Broad Institute TCGA Genome Data Analysis Center (GDAC) Firehose for colorectal cancer samples (COADREAD project) at http://gdac.broadinstitute.org/, and log2-transformed after adding 1 to each count. We also downloaded the Mutation Annotation File, and considered only the 216 tumors that had mutation data. Our analysis of 9 tumors with TP53 codon 270 mutations and 36 with null TP53 mutations is also available in GSE125262.
Statistical Analysis
Kaplan-Meier survival curves were compared by log-rank test. We compared the rates of tumors between different genotypes of mice with exact Poisson tests (using the poisson.test function in R) and used the two-sided Fisher’s exact test to test the association between different p53 mutations and tumor invasion depth. The two-sided Fisher’s exact test was used to compare p53 staining between adenomas and carcinomas generated in CDX2P-CreERT2 AKP270/+ mice.
Results
Missense and null Trp53 mutations promote mouse colon tumor progression in CDX2P-CreERT2 Apcfl/+, KrasLSL-G12D (AK) mice
To develop a mouse colon tumor model instigated by gene defects like those often present in human colon tumors, we generated compound mutant mice carrying one floxed Apc allele (Apcflox, 580S, abbreviated as A) that could be inactivated by Cre recombinase function and a floxed, latently oncogenic allele of Kras (KrasLSL-G12D, abbreviated as K) that could be activated by Cre recombination. Using the CDX2P-CreERT2 transgene and TAM treatment to activate Cre function in colon epithelial cells [38], we targeted the Apc and Kras alleles in epithelium of the distal ileum, cecum and colon. Whereas CDX2P-CreERT2 K mice show serrated and hyperplasic colon epithelium [8], the CDX2P-CreERT2AK (AK) mice had serrated and hyperplastic colon epithelium and also developed an average of 13 adenomas ranging in size from 0.5mm to 2 mm by the time that humane endpoints were reached for the mice at 10–15 months after TAM treatment (Fig. 1a). In six of 10 AK mice, we found one or two adenocarcinomas arising in the cecum. Of the eight total adenocarcinomas found in the 10 AK mice studied, seven tumors invaded into the muscularis propria (MP) or deeper and five reached the sub-serosa or serosa (Fig. 1b, Table 1 and Supplementary Table 1). However, because only 8 of 131 observed tumors in the AK mice were invasive, the findings imply that additional somatic defects besides Apc and Kras mutations were required for development of invasive colon tumors in the AK mice.
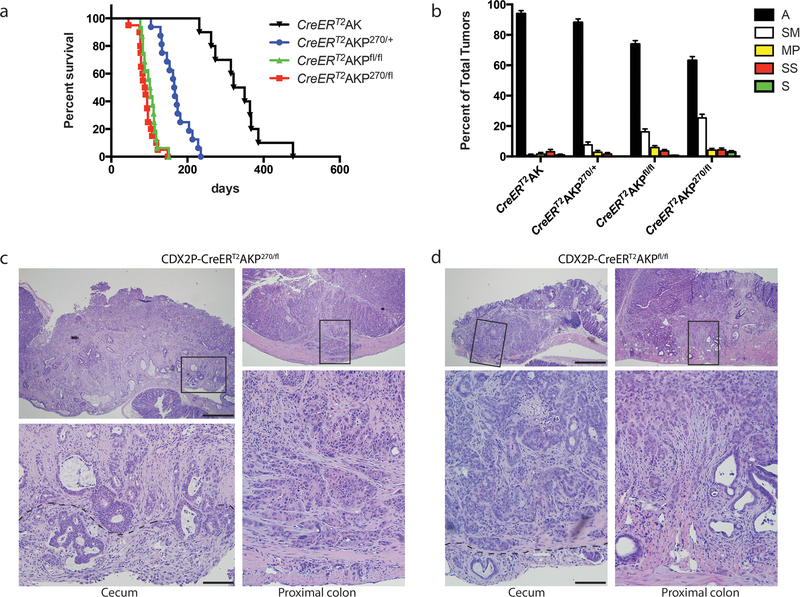
Fig. 1. Missense and null Trp53 mutations promote mouse colon tumor progression in CDX2P-CreERT2 Apc fl/+, Kras LSL-G12D (AK) mice.
a Kaplan-Meier survival curves of CDX2P-CreERT2AK (AK) mice (n=10, median survival = 336 days), CDX2P-CreERT2AKP270/+ (AKP270/+) mice (n=16, median survival = 166 days), CDX2P-CreERT2AKPfl/fl (AKPfl/fl) mice (n=16, median survival = 97 days) and CDX2P-CreERT2AKP270/fl (AKP270/fl) mice (n=20, median survival = 88 days). TAM was administered daily for two days and time zero represents the second of these days. P-values were obtained by log-rank test in the following comparisons: P = 1.4E-6 for AK vs. AKP270/+; P = 1.4E-7 for AKPfl/fl vs. AKP270/+; P = 1.9E-8 for AKP270/fl vs. AKP270/+. b Percent of colon tumors with different depths of invasion from AK mice, AKP270/+ mice, AKPfl/fl mice and AKP270/fl mice. Data are from multiple lesions per mouse (n=10–14 mice per group). P-values based on Fisher Exact tests and Exact Poisson tests are shown in Table 1 and Supplementary Table 1, respectively. Error bars are standard error of the mean. A: adenoma; SM: submucosa; MP: muscularis propria; SS: subserosa; S: serosa. c, d Hematoxylin and eosin (H&E) staining of representative invasive tumors from proximal colon (right) and cecum (left) tissues are shown for AKP270/fl mice (c) and AKPfl/fl mice (d). Tumors in (c) and (d) were collected 3 months after TAM injection. Images with high magnification are shown in the bottom panels and are boxed in the lower magnified areas in the corresponding top panels. The dashed lines indicate the boundary between muscular layer and subserosa. Scale bars: 500 μm for low magnification image (top panels); 100 μm for high magnification images (bottom panels).
Table 1.
Effects of missense and null Trp53 mutations on invasion depth of colon tumors
| Mouse | A | SM | MP | SS | S | Total |
|---|---|---|---|---|---|---|
| AK | 123 (93.9%) | 1 (0.8%) | 2 (1.5%) | 4 (3.1%) | 1 (0.8%) | 131 |
| AKP270/+ | 163 (88.1%) | 14 (7.6%) | 5 (2.7%) | 3 (1.6%) | 0 (0.0%) | 185 |
| AKPfl/fl | 269 (73.9%) | 59 (16.2%) | 21 (5.8%) | 13 (3.6%) | 2 (0.5%) | 364 |
| AKP270/fl | 214 (63.1%) | 86 (25.4%) | 14 (4.1%) | 15 (4.4%) | 10 (2.9%) | 339 |
| Total | 483 | 145 | 35 | 28 | 12 | 703 |
| P-values for Fisher Exact Tests of the proportion of adenomas vs. adenocarcinomas (SM or worse) | |||
|---|---|---|---|
| AKP270/+ | AKPfl/fl | AKP270/fl | |
| AK | 0.12 | 2.90E-07 | 7.20E-13 |
| AKP270/+ | 1.00E-04 | 3.10E-10 | |
| AKPfl/fl | 0.0026 | ||
A: adenoma; SM: submucosa; MP: muscularis propria; SS: subserosa; S: serosa
As noted above, TP53 mutations are found in about 60% of human CRCs and appear to be selected for during adenoma-carcinoma progression [4–7]. We sought to assess cooperation of Trp53 inactivation with Apc and Kras mutations in colon tumor progression in vivo, as well as to determine if potential GOF effects of a Trp53 missense mutant allele could be seen in the model. We introduced a constitutive Trp53R270H mutant allele (the murine equivalent of human R273H, referred to as P270) and/or a conditional null allele Trp53flox (referred to as Pfl) into the AK model. Mice with compound genotypes were generated and their abbreviations are as follows: CDX2P-CreERT2AKP270/+ (AKP270/+ mice), CDX2P-CreERT2AKP270/fl (AKP270/fl mice) and CDX2P-CreERT2AKPfl/fl(AKPfl/fl mice). Cohorts of adult AKP270/+, AKP270/fl, and AKPfl/fl mice were induced for colon tumors via TAM treatment and their health status was monitored. Compared to AK mice (median survival = 336 days), the AKP270/+, AKP270/fl, AKPfl/fl mice all had significantly shortened life spans. The AKP270/fl and AKPfl/fl mice had median survivals of 88 and 97 days, respectively, and AKP270/+ mice had a median survival of 166 days (Fig. 1a). The AKP270/+ mice had significantly longer survival than AKP270/fl and AKPfl/fl mice (P < 0.0001 for each compared to AKP270/+ mice), but no statistically significant survival difference was seen when comparing AKP270/fl and AKPfl/fl mice (Fig. 1a). The addition of the Trp53 mutation(s) (R270H or null) significantly increased the overall tumor incidence relative to that seen in AK mice (P<0.0001 for both AKPfl/fl and AKP270/fl, when compared to AK mice, Supplementary Table 1 bottom left). AKP mice also had multiple colon and cecum tumors that manifested invasive features (7.3 and 8.9 invasive tumors per mouse for AKPfl/fl mice and AKP270/fl mice, respectively) compared to AK mice (0.8 invasive tumors per mouse; Fig. 1b and Supplementary Table 1 top right, P<0.0001 for both AKPfl/fl vs. AK and AKP270/fl vs. AK comparisons).
In-depth analysis of mouse cecum and colon tissues obtained at necropsy defined the extensive tumor burden in the AKP270/+, AKPfl/fl and AKP270/fl mice, with lesions ranging from adenomas to late stage adenocarcinomas (Fig. 1b–d and Table 1). All the AKP270/+, AKPfl/fl and AKP270/fl mice studied carried at least one adenocarcinomas (tumors with submucosa or deeper invasion). Evidence of invasion into smooth muscle and, in some cases, to the serosa was found in both cecum and proximal colon regions of the AKP270/fl and AKPfl/fl mice (Fig. 1c and d, respectively). The AKP270/+ mice, where Cre-targeted epithelial cells initially retain one functional wild-type Trp53 allele, had a longer latency period for tumor development and the overall tumor burden (18.5 per mouse) and the percentages of tumors with deeper invasion (11.9%) were significantly lower when compared to AKP270/fl or AKPfl/fl mice (Supplementary Table 1 and Table 1, P < 0.01 compared to AKP270/fl or AKPfl/fl mice for tumor burden per mouse and the percentages of adenocarcinomas). These findings support the notion wild type p53 has a tumor suppressor function in colon tumorigenesis even in the context of a Trp53 missense mutant allele. When all the observed tumors were compared between AKP270/fl and AKPfl/fl mice, we found the proportion of adenocarcinomas with submucosa or deeper invasion was higher in AKP270/fl mice than in AKPfl/fl mice (36.9% and 26.1% for AKP270/fl and AKPfl/fl mice, respectively; Table 1, P=0.0026), suggesting a possible modest gain-of-function effect for the R270H Trp53 allele in enhancing tissue invasion. However, the modest increase in invasive potential in AKP270/fl mice may be offset, because the average tumor burden per mouse trended higher in AKPfl/fl mice than AKP270/fl mice (28.0 vs 24.2 per mouse, P = 0.054; Supplementary Table 1), and the AKPfl/fl and AKP270/fl mice did not display significant differences in the incidence of adenocarcinomas (7.3 vs 8.9 adenocarcinomas per mouse, Supplementary Table 1 top right, P = 0.16). Furthermore, invasion of some tumors into muscularis propria, subserosa, or serosa was seen in both AKP270/fl and AKPfl/fl mice (Fig. 1b and Table 1). Because only about 30% of the tumors from AKP270/fl or AKPfl/fl mice progressed to adenocarcinomas during the study period, additional somatic alterations besides Apc, Kras and Trp53 defects are likely critical in generating carcinomas in the AKP mice.
Tumor histology and metastasis in AKP270/fl or AKPfl/fl mice
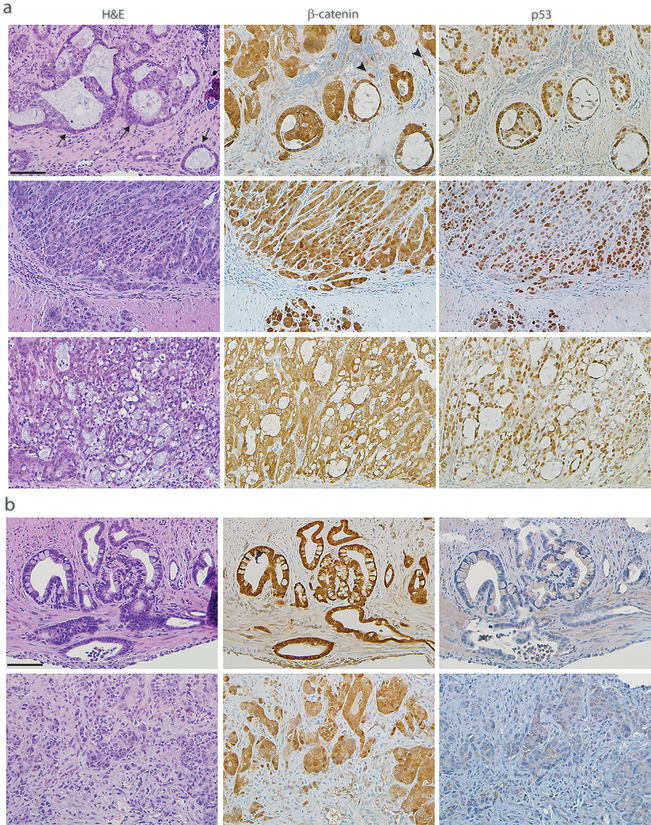
Each of the AKP270/fl and AKPfl/fl mice developed one or more adenocarcinomas by the time humane endpoints were reached. In AKP270/fl mice, more than 80% of the adenocarcinomas showed well to moderately differentiated features, with tumor budding/sprouting at the invasion front and occasional mucin secretion (Fig. 2a top). About 15–20% of the AKP270/fl adenocarcinomas were poorly differentiated, with trabecular type solid morphology (Fig. 2a middle). Rare signet-ring cell carcinomas were seen in AKP270/fl mice (Fig. 2a bottom). In AKPfl/fl mice, about 50% of the adenocarcinomas were well to moderately differentiated with mucin secretion (Fig. 2b top); the remainder were poorly differentiated (Fig. 2b bottom). Strong nuclear and cytoplasmic staining of β-catenin was seen in invasive AKP270/fl and AKPfl/fl tumors (Fig. 2 middle), consistent with defective Wnt signaling arising from Apc inactivation. Strong nuclear staining for p53 was observed in AKP270/fl tumor cells. As expected, no p53 staining was seen in AKPfl/fl tumor cells (Fig. 2 right).
Fig. 2. Histological types of colon adenocarcinomas from p53 mutant mice.
H&E stains (left panels) and immunohistochemical stains for β-catenin (middle panels) and p53 (right panels) are shown for independent colon tumors of representative histological types from the AKP270/fl mice (a) and AKPfl/fl mice (b). Tumors were collected 3 months after TAM induction. Scale bars: 50 μm.
At necropsy, lymph node metastases were found in 3 of 13 AKPfl/fl mice and 5 of 14 AKP270/fl mice (Table 2). Among the three AKPfl/fl mice that each had a metastasis to an abdominal lymph node, one mouse had also multiple metastases to the lung and another mouse had multiple metastases to the liver (Table 2). Among the five AKP270/fl mice each with a metastasis to an abdominal lymph node, one mouse had multiple metastases to the lung and another had metastatic lesions in both lung and liver (Table 2). No metastatic lesions were found in AK or AKP270/+ mice (Table 2). Our data indicate mouse CRCs with missense or null mutations in Trp53 can give rise to spontaneous lymph node and distant metastases at comparable frequencies, arguing against a GOF effect for missense Trp53 mutations in enhancing metastasis in our mouse CRC model.
Table 2.
Effects of missense and null Trp53 mutations on mouse colon tumor progression in AKP mice
| Mouse | No. of mice | Lymph node metastasis(n/N) | Lung metastasis(n/N) | Liver metastasis(n/N) |
|---|---|---|---|---|
| AK | 10 | 1/10 | 0/10 | 0/10 |
| AKP270/+ | 10 | 0/10 | 0/10 | 0/10 |
| AKPfl/fl | 13 | 3/13 | 1/13 | 1/13 |
| AKP270/fl | 14 | 5/14 | 2/14 | 1/14 |
Epithelial-mesenchymal transition in metastatic CRCs with missense or null Trp53
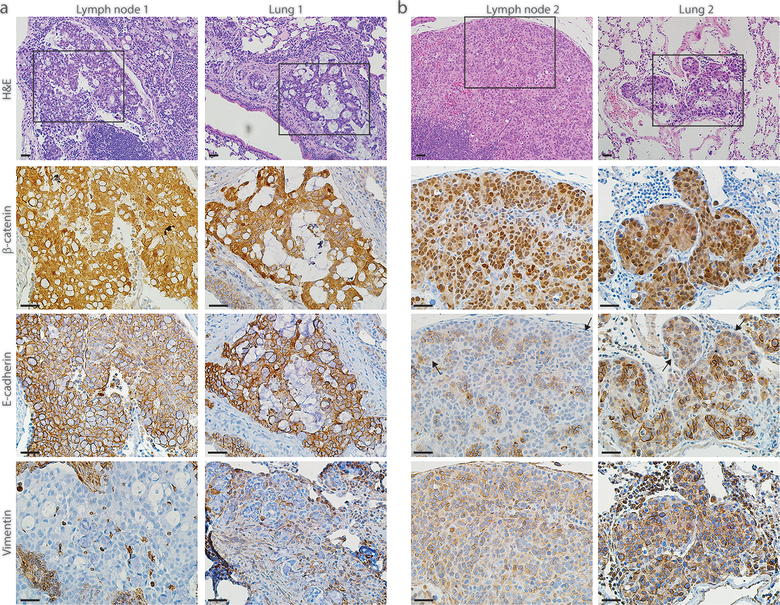
In the individual lymph node metastatic deposits found in each of five different AKP270/fl mice, four lesions showed moderately differentiated adenocarcinoma and one lesion was poorly differentiated (Fig. 3a, b top panels). All of the moderately differentiated lymph node metastatic lesions had strong nuclear β-catenin staining as well as strong membrane staining for E-cadherin and absent vimentin staining (Fig. 3a), suggesting retention of epithelial properties in AKP270/fl metastatic cells that retained moderately differentiated features. In the one AKP270/fl mouse where the lymph node metastasis had poorly differentiated features, adenocarcinoma lesions with poorly differentiated morphology were also found in the lung, liver, and cecum (Fig. 3b and Supplementary Fig. 1). The poorly differentiated cecum lesion and the metastatic lesions showed strong nuclear β-catenin staining and loss of E-cadherin expression and strong vimentin expression, consistent with epithelial-mesenchymal transition (EMT) features (Fig. 3b and Supplementary Fig. 1). All metastatic lesions in AKP270/fl mice stained positively for CDX2 (Supplementary Fig. 2), consistent with their colonic epithelial origin.
Fig. 3. Colon-derived metastatic tumors with missense Trp53 mutations can undergo epithelial-mesenchymal transition (EMT).
Photomicrographs of H&E-stained sections showing the lymph node and lung metastases found in two representative AKP270/fl mice (a, b top panels). Immunohistochemical stains showing strong nuclear expression of β-catenin, strong membrane expression of E-cadherin and absence of vimentin expression for the moderately differentiated lesions found in one representative mouse (a, bottom three panels). In contrast, the poorly differentiated lesions found in another mouse (b) show strong nuclear expression of β-catenin, loss of E-cadherin expression and strong expression of vimentin, indicating that the metastatic lesions found in this mouse have undergone EMT (b bottom three panels). The representative photomicrographs of H&E-stained sections were shown with low power magnification for orientation; the serial sections were subjected to immunohistochemical staining and the boxed area with stains for β-catenin, E-cadherin and vimentin were shown as high power magnification. Scale bars: 20 μm for all the images.
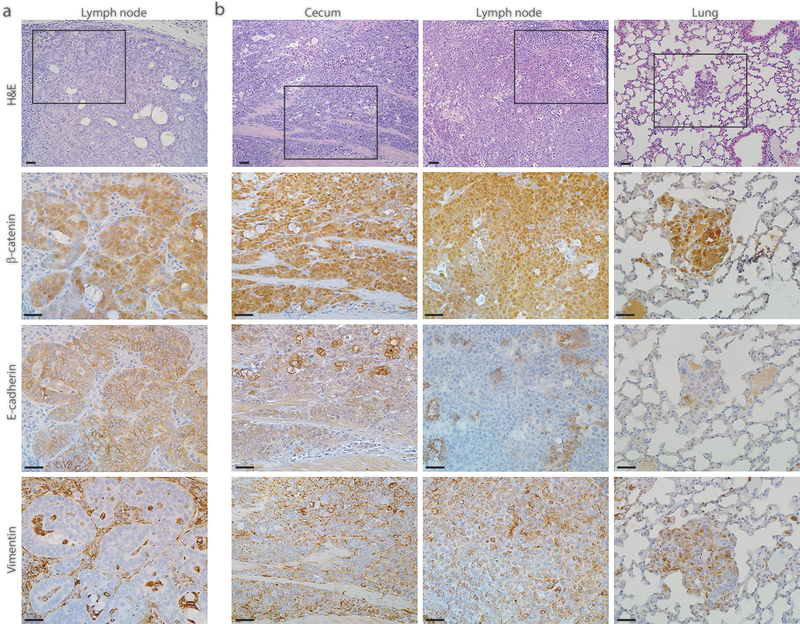
Two of the three lymph node metastases from tumor-bearing AKPfl/fl mice were moderately differentiated and one was poorly differentiated (Fig. 4a, b, top panels). The AKPfl/fl mouse with liver metastasis had a moderately differentiated lymph node metastasis. In this mouse with liver metastasis, the presumptive index colon primary adenocarcinoma as well as the lymph node and liver metastatic lesions all showed strong nuclear expression of β-catenin and strong membrane expression of E-cadherin (Supplementary Fig. 3). In the single AKPfl/fl mouse with a poorly differentiated lymph node metastasis, multiple lesions with similar morphology were also found in its lung (Fig. 4b). Immunohistochemical studies showed that the lung metastatic lesions and the presumptive index primary adenocarcinoma in the cecum all had strong nuclear expression of β-catenin and reduced E-cadherin expression and strong vimentin expression, consistent with EMT in the primary cancer and metastatic cells (Fig. 4b). The metastatic lesions in AKPfl/fl mice were all CDX2-positive (Supplementary Fig. 4). Taken together, our data indicate there is no significant difference in the histological features of metastatic lesions arising in AKP270/fl mice or AKPfl/fl mice, and the poorly differentiated subset of the primary tumors and metastatic lesions with missense or null Trp53 mutations can manifest EMT features.
Fig. 4. Colon-derived metastatic tumors with null Trp53 mutations can undergo epithelial-mesenchymal transition (EMT).
a Photomicrograph of an H&E-stained section showing a moderately differentiated tumor lesion found in the lymph node of a representative AKPfl/fl mouse 2.5 months post TAM injection (top panel). Immunohistochemical stains for this lesion show strong nuclear expression of β-catenin, strong membrane expression of E-cadherin and absence of vimentin expression (bottom three panels). b Photomicrographs of H&E-stained sections showing the poorly differentiated lesions found in the cecum, lymph node, and lung of another AKPfl/fl mouse 4 months post TAM injection (top panel). All these lesions show strong nuclear expression of β-catenin, loss of E-cadherin expression and strong expression of vimentin, indicating that the primary and metastatic lesions found in this mouse have undergone EMT (bottom three panels). The representative photomicrographs of H&E-stained sections were shown with low power magnification for orientation; the serial sections were subjected to immunohistochemical staining and the boxed area with stains for β-catenin, E-cadherin and vimentin were shown as high power magnification. Scale bars: 20 μm for all the images.
Stabilization of missense mutant p53 protein is associated with loss of wild-type p53 and disease progression
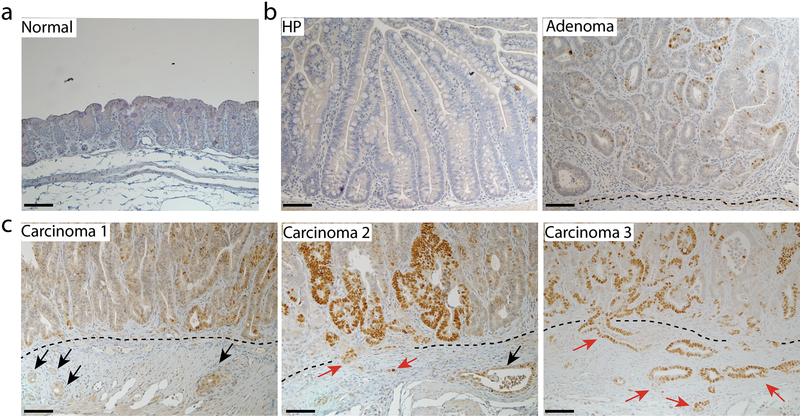
The majority of human tumors with missense TP53 mutations have been reported to show TP53 loss of heterozygosity (LOH)[43–46], and one study indicated that TP53 LOH is a prerequisite for missense mutant p53 stabilization in both mouse sarcomas and breast carcinomas [43]. We evaluated p53 staining patterns at various stages of tumor development in AKP270/+ mice after induction of tumorigenesis by TAM injection, because the Cre-targeted colon epithelium in AKP270/+ mice initially has one functional Trp53 allele. In hyperplastic polyps in AKP270/+ mice, where Kras is activated and Apc LOH had not yet occurred, p53 staining is non-detectable (Fig. 5b, panel HP), akin to the negative staining for p53 seen in the epithelium of wild-type mice (Fig. 5a), even though the p53 R270H missense mutant protein is constitutionally expressed in colon tissues of the AKP270/+ mice. Some p53 immunoreactivity was clearly seen in adenomas (Fig. 5b, panel adenoma and Table 3, n=185) and carcinomas (Fig. 5c and Table 3, n=28) arising in AKP270/+ mice. Among these tumors, 94% of the adenomas and 43% of the carcinomas showed weak p53 staining (Fig. 5 and Table 3, 5–10% positivity), whereas 6% of adenomas and 57% of carcinomas had strong p53 staining (Fig. 5 and Table 3, >10% positivity, P < 0.0001). The majority of tumors with strong p53 staining showed p53 LOH, while the tumors with weak p53 staining still retained the Trp53 wild-type allele (Fig. 6). Our results are similar to those seen in recent work on p53 missense protein expression in mouse sarcomas and breast carcinoma [43]. Our data demonstrate that akin to the situation in human colon tumorigenesis [1], expression of a stabilized missense mutant p53 protein is closely correlated with loss of wild type p53 function and progression to invasion.
Fig. 5. The stabilization of p53R270H mutant protein during tumor progression from hyperplasia to carcinoma in AKP270/+ mice.
Immunohistochemical staining of p53 in a representative hyperplastic polyp (HP, b left panel), an adenoma (b right panel) and three carcinomas (c) arising in the cecum of AKP270/+ mice. A section from the cecum of a wild-type mouse was used as a control (a). The p53 staining ranges from non-detectable staining in the normal (a) and HP (b) sections, to weak staining (<10% positivity) in the adenoma (b) and carcinoma 1(c left panel), and to strong staining in carcinomas 2 and 3 (>10% positivity). The dashed lines indicate the muscularis mucosae. Black arrows indicate the invasive glands with weak p53 staining; red arrows indicate the invasive gland with strong p53 staining. Scale bars: 50 μm.
Table 3.
Tumor progression from adenoma to carcinoma is associated with stabilization of p53R270H mutant in AKP270/+ mice
| p53 staining | |||
|---|---|---|---|
| Weak | Strong | Total | |
| Adenomas | 174 | 11 | 185 |
| Carcinomas | 12 | 16 | 28 |
| Total | 186 | 27 | 213 |
*P =4E-10 by two-sided Fisher’s Exact Test
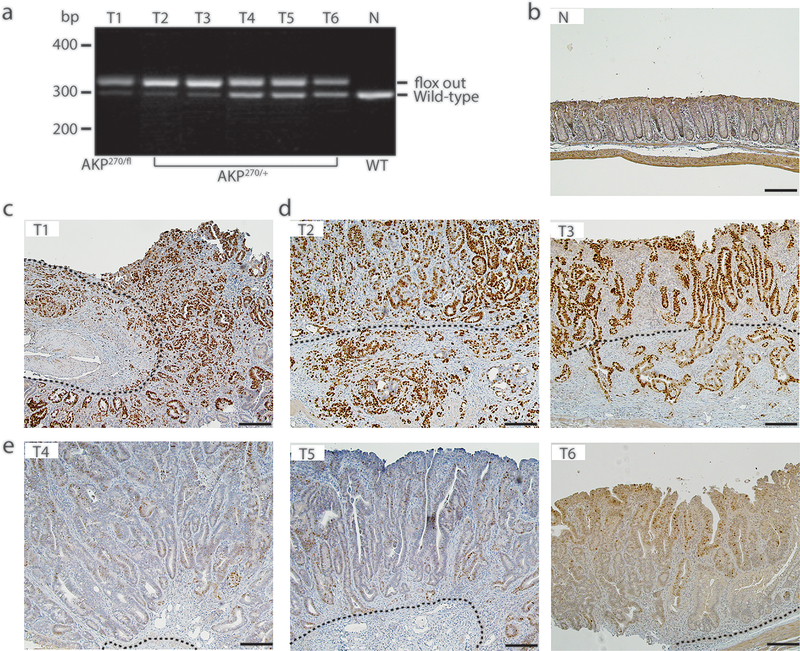
Fig. 6. The stabilization of p53R270H missense mutant is correlated with p53 loss-of-heterozygosity (LOH).
a Genotyping of colon tumors (T2-T6) arising in AKP270/+ mice 5 months after TAM induction. A tumor arising in an AKP270/fl mouse (T1) and colon tissue from a wild-type mouse (N) are used as a positive and negative control, respectively. The presence of the Trp53 wild-type allele and recombined Trp53 mutant allele (flox out) were detected by PCR. b-e Immunohistochemical staining of p53 in colon tissues described in (a) show a strong correlation between mutant p53 stabilization (d, T2 and T3) and p53 loss-of-heterozygosity, compared to the weak p53 staining and the presence of Trp53 wild-type allele in tumors 4–6 (e). The dashed lines indicate the muscularis mucosae. Scale bars: 100 μm.
Global gene expression profiling reveals few differences between mouse or human colon tumor cells harboring R270H/R273H missense mutant and null Trp53/TP53 alleles
We profiled gene expression in colon adenocarcinoma tissues from AKP270/fl mice (n=6) or AKPfl/fl (n=6) mice, using Affymetrix Mouse ST 2.1 Arrays with 24562 probe-sets. We isolated RNA following laser capture microdissection (LCM) of cancer cells from one invasive primary tumor from each mouse. As controls, microdissected normal colon epithelial cells from wild type mice (n = 3) were included. We fit ANOVA models to these 15 samples with terms for 3 means. Comparisons of the tumor tissues to normal tissues gave over 2400 probe-sets with P < 0.001 for both AKP270/fl and AKPfl/fl tumors, but comparison of the two tumor types to each other gave only 30 probe-sets, where we expect 24.6 probe-sets (24562*0.001) merely by chance. This latter result indicates most of the 30 probe-sets found are false positives (Table 4, top portion), and there were few if any significant observable differences in gene expression between colon tumors with Trp53 missense mutations and those with Trp53 null mutations. Consistent with this notion, we found the AKP270/fl and AKPfl/fl adenocarcinomas were indistinguishable in principal component (PC) analyses of gene expression, although both groups of tumors have gene expression patterns that are clearly distinct from wild-type colon epithelium (Supplementary Figure 5).
Table 4.
Number of small P-values in three array experiments
| Fairly weak | Very weak | |||||||
|---|---|---|---|---|---|---|---|---|
| P <.01, FC >1.3 | P <.05, FC >1.3 | |||||||
| Experiment | Comparison | P <.01 | P <.001 | up | down | up | down | |
| tissues | AKPfl/fl vs. AKP270/fl | 259 | 30 | 110 | 94 | 439 | 395 | |
| tissues | AKPfl/fl vs. WT control | 4416 | 2455 | 2065 | 2253 | 3139 | 3213 | |
| tissues | AKP270/fl vs. WT control | 4306 | 2431 | 1902 | 2311 | 3053 | 3209 | |
| organoids | AKPfl/fl vs. AKP270/fl | 232 | 35 | 90 | 87 | 370 | 363 | |
| organoids | AKPfl/fl vs. Cont 1 | 1642 | 435 | 740 | 780 | 1592 | 1734 | |
| organoids | AKP270/fl vs. Cont 1 | 2091 | 609 | 874 | 982 | 1677 | 1928 | |
| organoids | AKPfl/fl vs. Cont 2 | 2794 | 960 | 1358 | 1213 | 2457 | 2180 | |
| organoids | AKP270/fl vs. Cont 2 | 3398 | 1332 | 1595 | 1464 | 2646 | 2390 | |
| organoids | Cont 1 vs. Cont 2 | 1456 | 404 | 651 | 612 | 1484 | 1266 | |
| Expected by chance | 245.6 | 24.6 | (24562 probe-sets) | |||||
| P <.01, FC >1.3 | P <.05, FC >1.3 | |||||||
| up | down | up | down | |||||
| TCGA | TP53 null vs 273 mutation | 244 | 30 | 63 | 157 | 249 | 573 | |
| Expected by chance | 205 | 20.5 | (20531 human genes) | |||||
AKPfl/fl: CDX2P-CreERT2 Apcfl/+ Kras LSL-G12D/+Trp53fl/fl, 3 months post TAM injections
AKP270/fl: CDX2P-CreERT2 Apcfl/+ KrasLSL-G12D/+Trp53R270H/fl, 3 months post TAM injections
WT: Wild-type
Cont 1: CDX2P-CreERT2 Apcfl/fl, 15 days post TAM injections
Cont 2: Wild-type control
We also conducted Affymetrix gene expression analysis on organoids derived from colon tumors arising in AKP270/fl mice (n=3) or AKPfl/fl mice (n=2). Organoids derived from adenomas in Apcfl/fl mice (Cont 1, n=3) or from wild-type colon epithelium (Cont 2, n=4) were used as controls. We fit an ANOVA model to the four groups of organoids. We found 435 probe-sets with P < 0.001 for AKPfl/fl organoids and 609 probe-sets for AKP270/fl organoids, when compared to Apc-null organoids (Cont 1, Table 4). In addition, more differentially expressed probe-sets were identified (960 for AKPfl/fl and 1332 for AKP270/fl, P < 0.001) when AKPfl/fl or AKP270/fl organoids were compared to normal colon organoids (Cont2, Table 4). We performed pathway enrichment test on genes that were differentially expressed in both AKPfl/fl and AKP270/fl tumors and organoids compared to normal colon epithelium. We found that a variety of pathways were significantly altered in both p53 null and missense mutant tumor-derived cells, with genes involved in G2M-checkpoint, cell growth, stress response and cell metabolism as the most significantly changed gene sets (Supplementary Table 2). However, we obtained only 35 probe-sets with P < 0.001 for AKPfl/fl vs. AKP270/fl comparison (Table 4), where again 24.6 were expected by chance, showing that it was difficult to find gene expression differences between the organoids with null and missense Trp53 mutations. PC plots showed AKPfl/fl and AKP270/fl samples always clustered together, and were separated from organoids derived from Apc-mutant adenomas (Cont1, Supplementary Figure 6) or normal colon epithelium (Cont2, Supplementary Figure 6).
To test whether there were demonstrable differences in gene expression patterns between human CRCs with missense and null TP53 mutations, we analyzed log2-transformed normalized counts from RNA-seq data for CRCs in The Cancer Genome Atlas (TCGA) [5], comparing 9 tumors with TP53 codon 273 missense mutations to 36 tumors with null mutations (caused by frame shift, splice site, and nonsense mutations, see Supplementary Table 3 for details) by two-sample T-test for 20531 genes. We found just 244 genes with P < 0.01 and 30 genes with P < 0.001, which were only slightly more than the number expected by chance (205 and 20.5, respectively, Table 4). TP53 itself gave the most significant difference in gene expression, being almost 5-fold lower on average in tumors with null mutations vs. TP53 codon 273 mutations (Supplementary Table 4, P = 2×10−10). Again, we found little separation of the two sets of tumors in PC analysis (Supplementary Figure 7). Consistent with the similar transcriptional signature, we fit Cox proportional hazards models to 43 patients with survival data, using age, stage and TP53 status in the model, and did not find any significant association between patient outcome and TP53 mutation status (R273 missense mutation vs. null mutation, Supplementary Table 5, P = 0.67 by Wald test). It is worth mentioning that with only 7 deaths among these patients, the power to detect differences was quite low. Finally, we looked at the intersection of genes giving P < 0.05 in any of the three data sets described above (i.e. human and mouse colon tumors, mouse colon organoids) that changed in the same direction for Trp53/TP53 missense vs. null comparison. In Supplementary Table 4 we show genes that gave P < 0.05 and fold-changes of at least 1.3 in any two of the 3 data sets. We found only Trp53/TP53 that changed in the same direction in all three studies (Supplementary Table 4; the detailed statistics for each gene in all three data sets is available in our public GEO series). Taken together, our gene expression analyses for mouse and human CRCs indicate that colon tumors with R270/R273 missense or null Trp53/TP53 mutations have very similar gene expression profiles, which is consistent with the absence of any major differences in biological features in the colon tumors and phenotypes arising in vivo in AKP270/fl and AKPfl/fl mice.
Discussion
TP53 mutations are found in about 60% of human CRCs, and the vast majority of the TP53 mutations in CRCs result in missense substitutions in the DNA-binding domain of the p53 protein [4–7]. These missense mutant p53 proteins in cancer cells have LOF of p53’s transcriptional regulatory role and DN effects on wild type p53 protein in cells [15, 16]. Prior studies have suggested GOF roles in the cancer process for various p53 missense mutant proteins, but definitive insights into a unifying molecular mechanism by which missense mutant p53 proteins, when endogenously expressed, have GOF effects in cancer cells have been elusive [11, 17, 18]. Some prior studies have also not yielded clear evidence in support of GOF contributions of p53 missense mutant proteins in cancer, including some mouse genetic studies in lung and ovarian cancer models as well as studies of spontaneous tumors developing in mice carrying a constitutional (germline) hot spot Trp53 mutation (R246S) [30, 31, 47, 48]. These mouse genetic model findings argue against the notion that missense mutant forms of p53 invariably exert GOF effects. Moreover, a recent comprehensive analysis of TP53 mutations in human cancer cells yielded evidence indicating that missense mutations in TP53 exert LOF and DN effects in human cancer cells, but no evidence of GOF for the missense mutant alleles in the human cancer cells was found in the assays and endpoints studied [15]. As such, arguments to generalize broadly the possible p53 GOF effects in the cancer process in vivo need to be considered carefully, including recognizing caveats in published studies about exogenous versus endogenous expression of any TP53 or Trp53 mutant allele, the in vivo relevance of the tumor phenotypes being studied for potential GOF effects, the robustness of the assays and data pursued with respect to those phenotypes, as well as whether the studies undertaken could definitively distinguish p53 GOF effects from possible DN effects on wild type p53.
To address the role of Trp53 mutations in colon tumor progression in vivo and whether evidence of a GOF effect for a common missense variant of Trp53 could be obtained, we directly compared the effects of a Trp53 missense (R270H) and a Trp53 null mutation in a mouse model of CRC based on combined somatic mutations of the murine homologues of genes frequently mutated in human CRCs, namely the APC, KRAS, and TP53 genes. We found that in concert with Apc and Kras mutations, tumors with the R270H missense Trp53 allele and a null Trp53 allele or tumors with only Trp53 null alleles were associated with comparable mouse survival, tumor burden, likelihood of lymph node and distant metastasis, and EMT features. The subtly higher fraction of adenocarcinomas with submucosa or deeper invasion in AKP270/fl mice than in AKPfl/fl mice suggested a possible modest GOF effect for the R270H Trp53 allele in enhancing tissue invasion, but the number of adenocarcinomas per mouse was not significantly different between the AKPfl/fl and AKP270/fl mice. In line with the similar biological features of AKP270/fl and AKPfl/fl tumors, primary tumor tissues and tumor-derived organoids harboring the Trp53R270H or Trp53null alleles had highly similar gene expression profiles. We also found that human CRCs with codon 273 missense or null TP53 alleles had homogeneous gene expression patterns and no apparent effects of codon 273 missense status on the likelihood of death from CRC were seen for the 43 CRC patients studied with codon 273 missense versus null p53 status. Our studies indicate that definite GOF properties for the common missense variant p53 protein R270H/R273H were not evident in the mouse and human CRCs we studied, and our findings suggest GOF effects of mutant TP53 alleles in cancer are likely to be allele-specific and context-dependent.
The lack of definite mutant p53 GOF effects in our mouse CRC model contrasts with findings reported in some previous mouse genetic work where apparent mutant p53 GOF activity in tumorigenesis models was pursued [20–25]. Two different groups working with mouse intestinal tumorigenesis models reported that a Trp53 missense mutation, at Trp53 codon 172 in one paper and at Trp53 codon 270 in the other, but not a Trp53-null mutation, contributed to enhanced invasiveness of intestinal tumors instigated solely by Apc deletion [23, 24]. The study by Muller et al. reported that all mice with one R172H mutant Trp53 allele and one deletion mutant Apc allele developed invasive adenocarcinomas, whereas only 25% of mice with deletion of one allele of Apc and one Trp53 null allele developed adenocarcinomas. The authors did not find significant differences between the two genotypes in overall tumor incidence and latency. In the study by Nakayama et al. the authors reported that, in the context of a mutant Apc allele, mice carrying heterozygous R270H mutation in Trp53 have a marked increase in the proportion of invasive adenocarcinomas and a significantly shorter life span compared to mice with heterozygous deletion of Trp53. Consistent with the study by Muller et al., we found both AKP270/fl and AKPfl/fl mice developed colon tumors with similar incidence and latency. However, in contrast to the findings in the two prior studies, we found that AKP270/fl and AKPfl/fl mice had similar overall survival and all mice of either Trp53 genotype developed at least 2 adenocarcinomas. In addition, compared to the mutant p53 GOF activity in tissue invasion reported in the prior Muller and Nakayama studies, evidence for any mutant p53 GOF effect in tissue invasion in our studies was at best modest and mixed. We found the fraction of adenocarcinomas with submucosa or deeper invasion was higher in AKP270/fl mice than in AKPfl/fl mice, but the incidence of adenocarcinomas per mouse did not differ significantly between AKPfl/fl and AKP270/fl mice, and AKPfl/fl mice had an increase in the average tumor number per mouse relative to AKP270/fl mice.
Among the factors that might account for the differences in phenotypes reported in the prior studies of Muller and colleagues and Nakayama and colleagues compared to our findings include the specific gene mutation combinations used, as neither of the prior manuscripts examined the effects of a mutant Trp53 allele in concert with Apc and Kras mutations, focusing instead only on studying how Apc and Trp53 mutations might collaborate. Moreover, as both prior studies examined the effects of the mutant Trp53 allele in mice and tissues that carried a wild type Trp53 allele, the prior studies may be emphasizing DN effects of mutant p53 on the remaining wild-type p53, with respect to the enhanced tissue invasion phenotypes reported. In contrast, in our studies, we sought to evaluate possible p53 GOF effects in the context of no wild-type p53 by comparing the mice with one R270H mutant allele and one null allele of Trp53 to mice with two Trp53-null alleles. In addition, compared to the CDX2P-CreERT2 targeting approach that we used to instigate tumor formation chiefly in the cecum and proximal colon of the mice, the Cre-targeting approaches in the prior work (i.e., AhCre or Villin-CreERT2 transgenic alleles) generate a significant burden of small intestinal tumors. The location(s) in the intestinal tract where the invasive tumors arose in the mice was not fully specified in the two prior manuscripts. In addition, the Muller et al. and Nakayama et al. manuscripts do not fully address details of tumor phenotyping, as the per mouse incidence of invasive tumors was not reported in each paper, and detailed data on the histological features, presence or absence of adenocarcinoma, and depth of invasion in each of the multiple tumors that would be expected to arise in the intestinal tract of each mouse following AhCre or Villin-CreERT2 targeting of Apc was not presented. As we found in our work, only about 30% of the mouse colon tumors arising in vivo with inactivation of Apc, activation of Kras, and bi-allelic defects in Trp53 were invasive. As such, it seems not unlikely that a similarly small or even smaller fraction of the intestinal tumors arising in the mice described in the publications of Muller et al. and Nakayama et al. were likely invasive following targeting of only Apc and Trp53.
In our studies, we also assessed the ability of wild type Trp53 function to inhibit tumorigenesis. Specifically, though the R270H Trp53 mutant allele was constitutively expressed in all tissues of AKP270/+ mice, including colon epithelium, only 12.7% of the tumors arising in these mice showed strong p53 staining (Table 3). In contrast, 97.6 % of colon tumors arising in the AKP270/fl mice showed strong staining for p53. The apparent repressive effects of the wild-type Trp53 allele are not only revealed in its ability to affect the levels of the stabilized missense mutant p53 protein in AKP270/+ tumors, but also by the effects of wild type Trp53 in longer survival, as a result of reduced tumor burden and the delay in disease progression in AKP270/+ mice relative to AKP270/fl mice. Consistent with the apparently significant tumor-suppressive effect of wild-type p53 in AKP270/+ mouse tumors, strong staining for the stabilized missense mutant p53 protein was associated with loss of the remaining wild-type p53 allele via LOH and progression from adenoma to carcinoma. The potent inhibitory effects that we observed for the wild-type p53 protein on the expression of a missense mutant p53 protein and tumor progression phenotypes has also been observed in other mouse tumor models [24, 47, 48]. The observed effects reflect the ability of the wild type p53 protein to function in key transcriptional responses, including p53’s function in activating expression of Mdm2, a known negative feedback regulator that plays a role in promoting ubiquitin-mediated degradation for both wild type and mutant p53 proteins. Our data, suggest that a strong DN effect of mutant p53 is not readily apparent in normal colon epithelium or benign colon tumor cells harboring a heterozygous missense mutation in p53. The results offer further support for the notion that a certain threshold level of mutant p53 is required for DN effect of mutant p53 in vivo, as has also been reported from other mouse models and in some cell line studies [47, 48].
Interestingly, our global gene expression analyses of p53 mutant and null tumors and tumor-derived organoids reveal very similar transcriptional signatures. Our pathway enrichment test shows that a variety of pathways are altered in both p53 null and missense mutant tumor cells compared to normal colon epithelium, which includes genes involved in G2M-checkpoint, cell growth, cell metabolism, hypoxia, stress response, immune response, and epithelial mesenchymal transition (EMT). Our findings contrast to data from a previous lung cancer model that reported that even though the p53 mutant lung tumors did not show GOF phenotypic effects (e.g., increased invasiveness or metastatic potential), the p53 mutants exhibited evidence of transcriptional GOF [48]. The difference between our findings and those in the Turrell et al. manuscript may reflect differences between lung and colon epithelium and also the models from which the total RNAs are collected. In our study, we compare the RNAs directly from tumors and tumor-derived organoids, which more closely reflect the in vivo setting. However in the lung cancer study mentioned above[48], the RNAs were collected from tumor-derived cell lines, which have been selected to grow under in vitro conditions, and may thus go through more selective pressure. Of note, in their list of molecular functions significantly altered in p53 mutant tumors relative to p53-null tumors, genes involved in cell death and survival, cellular movement, and cell cycle are among the top. However, the GOF transcriptional signatures of p53 mutants did not yield any measurable GOF cellular phenotypes in their study [48]. These results, together with ours, reinforce the notion that both functional and transcriptional GOFs of mutant p53 should be viewed with respect to tissue- and context-dependence.
The most prevalent mutation at TP53 codon 273 in human CRCs is an Arg to His substitution (R273H), but other TP53 codon 273 mutations are found in CRCs at lower frequency, such as R273C mutation[5, 15]. Different TP53 missense substitutions at a particular codon may have differing prevalence among cancer types, with the R273H mutation enriched in intestine cancers and R273L potentially enriched in lung cancers[15]. These apparent tissue-preferential TP53 mutation patterns may reflect in part mutational signatures in the relevant tissues of origin, biological differences among different mutant p53 proteins, and possibly both factors[15]. A prior study showed TP53 R273H and R273C mutations, but not an R273G mutation, exhibited some potential GOF activities (e.g., evidence for enhanced cell proliferation, invasion, and drug resistance) when the mutant TP53 alleles were overexpressed in TP53 null cancer cell lines, though in other assays all three TP53 R273 mutants displayed LOF[49]. Given the potential for differential biological effects of certain TP53 mutations on cancer phenotypes, the mutation-type specificity of TP53 mutations (even at the same codon) needs to be considered when DN, LOF and GOF properties of p53 are studied in the future.
Our work is the first to demonstrate that in a mouse colon cancer model, compared to p53-null tumors, p53 R270 missense mutant-expressing tumors do not display definitive GOF phenotypic effects, such as enhanced metastatic potential or an altered transcriptional signature or decreased overall mouse survival. Small molecule and peptide “reactivators” of mutant p53, such as PRIMA-1 and pCAPs, have been shown to be able to restore wild-type-like tumor suppressive functions to mutant p53 proteins through their binding and induction of a wild-type-like conformation[50]. The potent tumor suppressive effects of wild-type p53 when co-expressed in colon tumors with mutant p53 suggest that restoration of wild-type-like p53 activity by such p53 “reactivators” might have relevance for treating the significant fraction of CRCs with missense p53 alleles, because our findings indicate that R270H/R273H mutant p53 proteins do not show GOF functional effects when endogenously expressed in CRCs and there is a large pool of stabilized mutant p53 protein available to the reactivator approach and even restoration of a subset of the available p53 protein pool might have significant tumor suppressor effects.
Supplementary Material
Acknowledgements
This paper was supported by University of Michigan Cancer Center Support Grant - P30CA46592. Jinyu Tang wants to thank the China Scholarship Council for supporting her study in the Fearon Lab at the University of Michigan.
Footnotes
Disclosure/Conflict of Interest
The authors have no duality of interest to declare.
Supplementary information is available at Laboratory Investigation’s website
References
- 1.Fearon ER. Molecular genetics of colorectal cancer. Annu Rev Pathol 2011;6:479–507. [DOI] [PubMed] [Google Scholar]
- 2.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell 1990;61:759–767. [DOI] [PubMed] [Google Scholar]
- 3.Jones S, Chen WD, Parmigiani G, Diehl F, Beerenwinkel N, Antal T, et al. Comparative lesion sequencing provides insights into tumor evolution. Proc Natl Acad Sci U S A 2008;105:4283–4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yaeger R, Chatila WK, Lipsyc MD, Hechtman JF, Cercek A, Sanchez-Vega F, et al. Clinical sequencing defines the genomic landscape of metastatic colorectal cancer. Cancer Cell 2018;33:125–136 e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas N Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giannakis M, Mu XJ, Shukla SA, Qian ZR, Cohen O, Nishihara R, et al. Genomic correlates of immune-cell infiltrates in colorectal carcinoma. Cell Rep 2016;15:857–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr., Kinzler KW. Cancer genome landscapes. Science 2013;339:1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng Y, Bommer GT, Zhao J, Green M, Sands E, Zhai Y, et al. Mutant KRAS promotes hyperplasia and alters differentiation in the colon epithelium but does not expand the presumptive stem cell pool. Gastroenterology 2011;141:1003–1013 e1001–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boutin AT, Liao WT, Wang M, Hwang SS, Karpinets TV, Cheung H, et al. Oncogenic Kras drives invasion and maintains metastases in colorectal cancer. Genes Dev 2017;31:370–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, et al. Mutational landscape and significance across 12 major cancer types. Nature 2013;502:333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kastenhuber ER, Lowe SW. Putting p53 in context. Cell 2017;170:1062–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mello SS, Attardi LD. Deciphering p53 signaling in tumor suppression. Curr Opin Cell Biol 2018;51:65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer 2009;9:701–713. [DOI] [PubMed] [Google Scholar]
- 14.Shirole NH, Pal D, Kastenhuber ER, Senturk S, Boroda J, Pisterzi P, et al. TP53 exon-6 truncating mutations produce separation of function isoforms with pro-tumorigenic functions. Elife 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giacomelli AO, Yang X, Lintner RE, McFarland JM, Duby M, Kim J, et al. Mutational processes shape the landscape of TP53 mutations in human cancer. Nat Genet 2018;50:1381–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baugh EH, Ke H, Levine AJ, Bonneau RA, Chan CS. Why are there hotspot mutations in the TP53 gene in human cancers? Cell Death Differ 2018;25:154–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muller PA, Vousden KH. Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell 2014;25:304–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aschauer L, Muller PA. Novel targets and interaction partners of mutant p53 Gain-Of-Function. Biochem Soc Trans 2016;44:460–466. [DOI] [PubMed] [Google Scholar]
- 19.Caulin C, Nguyen T, Lang GA, Goepfert TM, Brinkley BR, Cai WW, et al. An inducible mouse model for skin cancer reveals distinct roles for gain- and loss-of-function p53 mutations. J Clin Invest 2007;117:1893–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morton JP, Timpson P, Karim SA, Ridgway RA, Athineos D, Doyle B, et al. Mutant p53 drives metastasis and overcomes growth arrest/senescence in pancreatic cancer. Proc Natl Acad Sci U S A 2010;107:246–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doyle B, Morton JP, Delaney DW, Ridgway RA, Wilkins JA, Sansom OJ. p53 mutation and loss have different effects on tumourigenesis in a novel mouse model of pleomorphic rhabdomyosarcoma. J Pathol 2010;222:129–137. [DOI] [PubMed] [Google Scholar]
- 22.Cooks T, Pateras IS, Tarcic O, Solomon H, Schetter AJ, Wilder S, et al. Mutant p53 prolongs NF-kappaB activation and promotes chronic inflammation and inflammation-associated colorectal cancer. Cancer Cell 2013;23:634–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muller PA, Caswell PT, Doyle B, Iwanicki MP, Tan EH, Karim S, et al. Mutant p53 drives invasion by promoting integrin recycling. Cell 2009;139:1327–1341. [DOI] [PubMed] [Google Scholar]
- 24.Nakayama M, Sakai E, Echizen K, Yamada Y, Oshima H, Han TS, et al. Intestinal cancer progression by mutant p53 through the acquisition of invasiveness associated with complex glandular formation. Oncogene 2017;36:5885–5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schulz-Heddergott R, Stark N, Edmunds SJ, Li J, Conradi LC, Bohnenberger H, et al. Therapeutic ablation of gain-of-function mutant p53 in colorectal cancer inhibits Stat3-mediated tumor growth and invasion. Cancer Cell 2018;34:298–314 e297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell 2004;119:847–860. [DOI] [PubMed] [Google Scholar]
- 27.Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell 2004;119:861–872. [DOI] [PubMed] [Google Scholar]
- 28.Bougeard G, Sesboue R, Baert-Desurmont S, Vasseur S, Martin C, Tinat J, et al. Molecular basis of the Li-Fraumeni syndrome: an update from the French LFS families. J Med Genet 2008;45:535–538. [DOI] [PubMed] [Google Scholar]
- 29.Zerdoumi Y, Aury-Landas J, Bonaiti-Pellie C, Derambure C, Sesboue R, Renaux-Petel M, et al. Drastic effect of germline TP53 missense mutations in Li-Fraumeni patients. Hum Mutat 2013;34:453–461. [DOI] [PubMed] [Google Scholar]
- 30.Jackson EL, Olive KP, Tuveson DA, Bronson R, Crowley D, Brown M, et al. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res 2005;65:10280–10288. [DOI] [PubMed] [Google Scholar]
- 31.Wu R, Baker SJ, Hu TC, Norman KM, Fearon ER, Cho KR. Type I to type II ovarian carcinoma progression: mutant Trp53 or Pik3ca confers a more aggressive tumor phenotype in a mouse model of ovarian cancer. Am J Pathol 2013;182:1391–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sood AK, Sorosky JI, Dolan M, Anderson B, Buller RE. Distant metastases in ovarian cancer: association with p53 mutations. Clin Cancer Res 1999;5:2485–2490. [PubMed] [Google Scholar]
- 33.Dow LE, O’Rourke KP, Simon J, Tschaharganeh DF, van Es JH, Clevers H, et al. Apc restoration promotes cellular differentiation and reestablishes crypt homeostasis in colorectal cancer. Cell 2015;161:1539–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Rourke KP, Loizou E, Livshits G, Schatoff EM, Baslan T, Manchado E, et al. Transplantation of engineered organoids enables rapid generation of metastatic mouse models of colorectal cancer. Nat Biotechnol 2017;35:577–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roper J, Tammela T, Cetinbas NM, Akkad A, Roghanian A, Rickelt S, et al. In vivo genome editing and organoid transplantation models of colorectal cancer and metastasis. Nat Biotechnol 2017;35:569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shibata H, Toyama K, Shioya H, Ito M, Hirota M, Hasegawa S, et al. Rapid colorectal adenoma formation initiated by conditional targeting of the Apc gene. Science 1997;278:120–123. [DOI] [PubMed] [Google Scholar]
- 37.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev 2001;15:3243–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feng Y, Sentani K, Wiese A, Sands E, Green M, Bommer GT, et al. Sox9 induction, ectopic Paneth cells, and mitotic spindle axis defects in mouse colon adenomatous epithelium arising from conditional biallelic Apc inactivation. Am J Pathol 2013;183:493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yui S, Nakamura T, Sato T, Nemoto Y, Mizutani T, Zheng X, et al. Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5(+) stem cell. Nat Med 2012;18:618–623. [DOI] [PubMed] [Google Scholar]
- 40.Sakamoto N, Feng Y, Stolfi C, Kurosu Y, Green M, Lin J, et al. BRAF(V600E) cooperates with CDX2 inactivation to promote serrated colorectal tumorigenesis. Elife 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003;4:249–264. [DOI] [PubMed] [Google Scholar]
- 42.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011;27:1739–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alexandrova EM, Mirza SA, Xu S, Schulz-Heddergott R, Marchenko ND, Moll UM. p53 loss-of-heterozygosity is a necessary prerequisite for mutant p53 stabilization and gain-of-function in vivo. Cell Death Dis 2017;8:e2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parikh N, Hilsenbeck S, Creighton CJ, Dayaram T, Shuck R, Shinbrot E, et al. Effects of TP53 mutational status on gene expression patterns across 10 human cancer types. J Pathol 2014;232:522–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Silwal-Pandit L, Vollan HK, Chin SF, Rueda OM, McKinney S, Osako T, et al. TP53 mutation spectrum in breast cancer is subtype specific and has distinct prognostic relevance. Clin Cancer Res. 2014;20:3569–3580. [DOI] [PubMed] [Google Scholar]
- 46.Varley JM, Evans DG, Birch JM. Li-Fraumeni syndrome--a molecular and clinical review. Br J Cancer 1997;76:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee MK, Teoh WW, Phang BH, Tong WM, Wang ZQ, Sabapathy K. Cell-type, dose, and mutation-type specificity dictate mutant p53 functions in vivo. Cancer Cell 2012;22:751–764. [DOI] [PubMed] [Google Scholar]
- 48.Turrell FK, Kerr EM, Gao M, Thorpe H, Doherty GJ, Cridge J, et al. Lung tumors with distinct p53 mutations respond similarly to p53 targeted therapy but exhibit genotype-specific statin sensitivity. Genes De. 2017;31:1339–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li J, Yang L, Gaur S, Zhang K, Wu X, Yuan YC, et al. Mutants TP53 p.R273H and p.R273C but not p.R273G enhance cancer cell malignancy. Hum Mutat 2014;35:575–584. [DOI] [PubMed] [Google Scholar]
- 50.Schulz-Heddergott R, Moll UM. Gain-of-function (GOF) mutant p53 as actionable therapeutic target. Cancers (Basel). 2018;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.