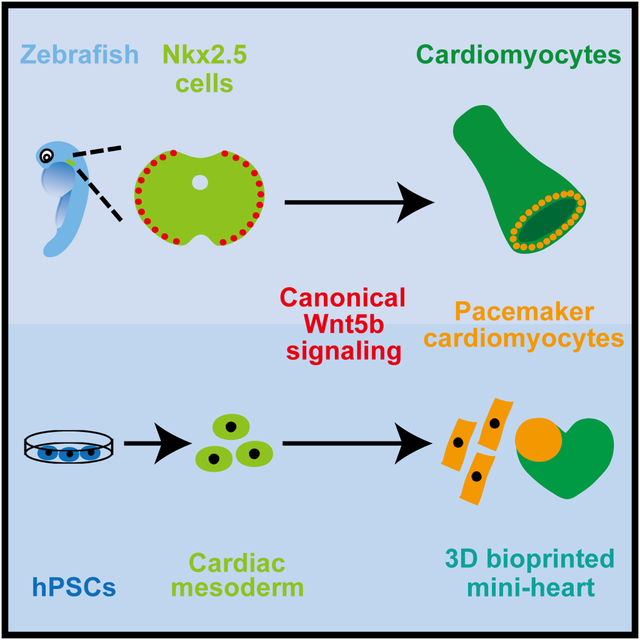
Summary
Pacemaker cardiomyocytes that create the sinoatrial node are essential for the initiation and maintenance of proper heart rhythm. However, illuminating developmental cues that direct their differentiation has remained particularly challenging due to the unclear cellular origins of these specialized cardiomyocytes. By discovering the origins of pacemaker cardiomyocytes, we reveal an evolutionarily conserved Wnt signaling mechanism that coordinates gene regulatory changes directing mesoderm cell fate decisions which lead to the differentiation of pacemaker cardiomyocytes. We show that in zebrafish, pacemaker cardiomyocytes derive from a subset of Nkx2.5+ mesoderm that responds to canonical Wnt5b signaling to initiate the cardiac pacemaker program, including activation of pacemaker cell differentiation transcription factors Isl1 and Tbx18, and silencing of Nkx2.5. Moreover, applying these developmental findings to human pluripotent stem cells (hPSCs) notably results in the creation of hPSC-pacemaker cardiomyocytes, which successfully pace three-dimensional bioprinted hPSC-cardiomyocytes, thus providing potential strategies for biological cardiac pacemaker therapy.
Keywords: pacemaker cardiomyocytes, differentiation, canonical Wnt signaling, Wnt5b, zebrafish, human pluripotent stem cells, 3D bioprinting
Graphical Abstract
eTOC Blurb:
Pacemaker cardiomyocytes are crucial for maintaining heart rate. Ren et al. show that these specialized cardiomyocytes originate from outlying Nkx2.5+ mesoderm that responds to canonical Wnt5b signaling in zebrafish. Applying these findings to human pluripotent stem cells (hPSCs) results in the generation of hPSC-pacemaker cardiomyocytes that can pace bioprinted hPSC-cardiomyocytes.
Introduction
The heart consists of a multitude of diverse cardiomyocyte cell types, including atrial, ventricular and pacemaker cells, which cooperate to ensure proper cardiac function and circulation throughout the body. Because loss or dysfunction of these cell types can lead to severe cardiac arrhythmias or heart failure, increasing efforts have been recently devoted toward understanding how these cardiomyocytes are created in order to develop potential human cardiac regenerative therapies (Laflamme and Murry, 2011). Such endeavors have illuminated not only the origins of many cardiomyocyte lineages (Cai et al., 2003; Stanley et al., 2002) but also key signaling cues and transcriptional regulators which in turn have been employed to efficiently direct various non-cardiac sources including hPSCs and human fibroblasts into specific cardiomyocyte cell types (Cao et al., 2016; Lian et al., 2012; Wada et al., 2013).
Although a large portion of cardiomyocytes derives from two temporally and spatially distinct Nkx2.5+ progenitor populations (Stanley et al., 2002), namely the first and second heart fields, the developmental source of pacemaker cardiomyocytes remains less certain despite the discovery of these specialized pacing cardiomyocytes more than 100 years ago (Keith and Flack, 1907; Trautwein and Uchizono, 1963). Illuminating their developmental origins has been particularly challenging due to their atypical cardiomyocyte characteristics including the lack of Nkx2.5 expression, which distinguishes them from most other cardiomyocytes (Wiese et al., 2009). As a result, recent studies suggest that pacemaker cardiomyocytes may originate from an additional Nkx2.5-negative heart field that is developmentally and molecularly distinct from the aforementioned well-established Nkx2.5+ heart fields (Bressan et al., 2013). However, others propose that they may arise from the outlying regions within these Nkx2.5+ heart fields where the transcription factor Shox2 may reduce Nkx2.5 expression during pacemaker cardiomyocyte differentiation (Lescroart et al., 2012; Liang et al., 2013; Mommersteeg et al., 2010). In line with this notion, Shox2 is specifically expressed in the sinoatrial node (SAN) region and has been shown to be required for pacemaker development through inhibiting Nkx2.5 expression (Blaschke et al., 2007; Espinoza-Lewis et al., 2009). Finally, given the heterogeneity of cardiomyocytes within the SAN (Liang et al., 2015; Sun et al., 2007), pacemaker cardiomyocytes alternatively may derive from multiple progenitor sources.
Along with Shox2, the transcription factors Isl1, Tbx3 and Tbx18 are coordinately expressed to control cardiomyocyte pacemaker differentiation through not only activating genes specific for pacemaker cell differentiation and function but also inhibiting genes involved in the differentiation and function of atrial or ventricular chamber cardiomyocytes (van Weerd and Christoffels, 2016). Although Isl1 regulates the specification of second heart field (SHF) progenitors and is downregulated during the differentiation of these cardiac progenitors into cardiomyocytes (Bu et al., 2009; Cai et al., 2003), Isl1 is also expressed in differentiated pacemaker cardiomyocytes in order to activate genes required for pacemaker development and function including Shox2, Tbx3 and Hcn4 (Liang et al., 2015; Sun et al., 2007; Tessadori et al., 2012; Vedantham et al., 2015). On the other hand, Shox2, Tbx3 and Tbx18 are expressed in pacemaker cardiomyocytes to reinforce the differentiation of these specialized cardiomyocytes through preventing the activation of the cardiac chamber program (Blaschke et al., 2007; Christoffels et al., 2006; Espinoza-Lewis et al., 2009; Kapoor et al., 2013; Mommersteeg et al., 2007; Wiese et al., 2009); however, Tbx3 may also serve to activate genes required for pacemaker cardiomyocyte function as well (Bakker et al., 2012; Hoogaars et al., 2007). Because these factors coordinate a gene regulatory network that ensures proper pacemaker cardiomyocyte formation and function, the combined expression of these factors along with the cardiac conduction ion channel Hcn4 (Stieber et al., 2003) has been utilized in recent pacemaker studies to define pacemaker cardiomyocytes (Birket et al., 2015; Liang et al., 2015; Mommersteeg et al., 2007; Protze et al., 2017; Wiese et al., 2009).
Several signaling pathways have been recently implicated in pacemaker cardiomyocyte development, including BMP and Wnt signaling (Birket et al., 2015; Bressan et al., 2013; Protze et al., 2017). In particular, canonical Wnt signaling has been shown in chick studies to specify mesodermal cells outside the first and second heart fields to the pacemaker lineage during early gastrulation (Bressan et al., 2013), despite its other roles during cardiac development including the induction of cardiac mesoderm formation at pre-gastrulation (Barrow et al., 2007; Liu et al., 1999; Ueno et al., 2007), as well as the specification of SHF progenitors and the inhibition of their differentiation into cardiomyocytes (Ai et al., 2007; Cohen et al., 2007; Kwon et al., 2007; Kwon et al., 2009; Lin et al., 2007; Qyang et al., 2007). Furthermore, previous mouse studies have revealed that canonical Wnt signaling is activated in the SAN during heart development and conditional deletion of b-catenin leads to decreased expression of several cardiac genes including Isl1, Pitx2 and Tbx3 (Lin et al., 2007), which are involved in pacemaker cardiomyocyte development (Bakker et al., 2012; Hoogaars et al., 2007; Liang et al., 2015; Mommersteeg et al., 2007; Tessadori et al., 2012; van Weerd and Christoffels, 2016; Vedantham et al., 2015; Wang et al., 2014; Wiese et al., 2009). Although these findings suggest that canonical Wnt signaling may regulate pacemaker cardiomyocyte differentiation possibly through Isl1, the specific Wnt ligands, the precise mesodermal cells that create these pacemaker cardiomyocyte lineages and the underlying mechanisms by which canonical Wnt signaling along with other signals direct pacemaker cardiomyocyte formation remain to be elucidated.
To clarify the developmental source of pacemaker cardiomyocytes, illuminate the coordinated signaling pathways directing their differentiation and understand how these signaling pathways may activate the pacemaker cardiomyocyte program, we take advantage of the unique strengths of the zebrafish embryo to precisely fate map cardiac mesoderm in vivo using photoconversion lineage tracing strategies and furthermore identify the specific Wnt ligand responsible for directing the differentiation of cardiac progenitor cells to pacemaker cardiomyocytes. Specifically, we reveal that pacemaker cardiomyocytes derive from outlying Nkx2.5+ progenitors that are located at the most lateral regions of the cardiac mesoderm. In response to Wnt5b, these Nkx2.5+ progenitors initiate canonical Wnt signaling to induce pacemaker cardiomyocyte differentiation through not only directly activating pacemaker differentiation transcription factors Isl1 and Tbx18, but also silencing Nkx2.5. Utilizing a combination of inducible genetic and chemical loss and gain of function studies, we further discover that perturbing this canonical Wnt signaling reciprocally alters the allocation of Nkx2.5+ progenitors to prospective Nkx2.5- pacemaker or Nkx2.5+ atrial cardiomyocyte lineages, thus uncovering an important role for Wnt5b in partitioning the outlying regions of Nkx2.5+ mesoderm into a distinct cardiac progenitor pool for the pacemaker cardiomyocyte lineage. Moreover, by applying these developmental findings to hPSCs, we generate hPSC-derived pacemaker-like cardiomyocytes that can pace 3D bioprinted hPSC-cardiomyocytes. Overall, our findings reveal a new role for canonical Wnt signaling during cardiac development, which choreographs gene regulatory changes directing pacemaker cardiomyocyte differentiation.
Results
Pacemaker cardiomyocytes originate from peripherally located Nkx2.5+ progenitors
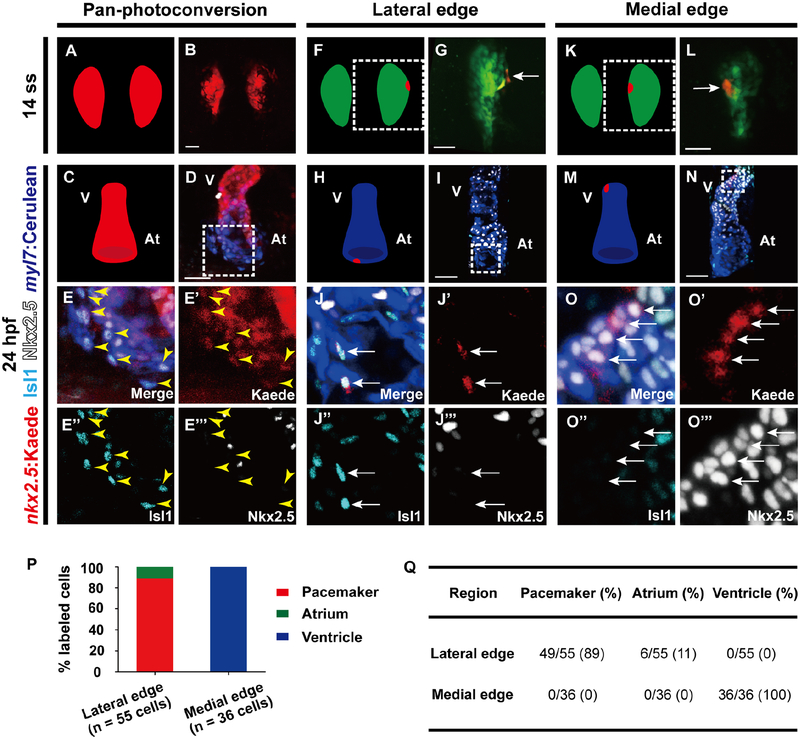
To begin to address the developmental source of pacemaker cardiomyocytes, we used the zebrafish embryo to explore whether pacemaker cardiomyocytes, which typically do not express Nkx2.5 (Wiese et al., 2009), derive from Nkx2.5+ cardiac progenitors prior to their differentiation. To this end, we tracked the fate of specific Nkx2.5+ cells during heart development through spatiotemporally marking distinct Nkx2.5+ cells using the TgBAC(−36nkx2.5:Kaede) [or Tg(nkx2.5:Kaede)] line, which expresses the stable photoconvertible Kaede protein in Nkx2.5+ cells. Photoconversion of all nkx2.5:Kaede+ cells from green to red at the 14 somite stage (ss), the earliest time point in which nkx2.5:Kaede is observable in the anterior lateral plate mesoderm (ALPM) (Guner-Ataman et al., 2013), reveals that Nkx2.5+ cardiac precursors produce not only atrial and ventricular cardiomyocytes but also inflow tract pacemaker cardiomyocytes expressing Isl1, a key transcriptional regulator of pacemaker cardiomyocyte formation (Liang et al., 2015; Tessadori et al., 2012; Vedantham et al., 2015) by 24 hours post fertilization (hpf) (Figures 1A–1E’’’). In addition to not only expressing Isl1 and myl7:Cerulean, a marker for differentiated cardiomyocytes (Yelon et al., 1999), these photoconverted nkx2.5:Kaede+ inflow tract pacemaker cardiomyocytes no longer exhibit the early cardiac developmental marker Nkx2.5, a distinctive feature of pacemaker cardiomyocytes (Figures 1D–1E’’’ yellow arrowheads). To refine the location of these cardiac pacemaker precursors, we next spatially restricted our photoconversion to specific regions of the Nkx2.5+ ALPM domain. Consistent with previous cardiac mesodermal fate mapping studies (Fukui et al., 2018; Mommersteeg et al., 2010), photoconversion at the most lateral edge of this Nkx2.5+ domain marks nkx2.5:Kaede+ cardiac progenitors which become pacemaker cardiomyocytes as well as atrial cardiomyocytes near the inflow region by 24 hpf (Figures 1F–1J’’’, 1P and 1Q). On the other hand, photoconversion along the medial edge labels nkx2.5:Kaede+ cardiac progenitors that primarily develop into ventricular cardiomyocytes that express myl7:Cerulean and Nkx2.5 but not Isl1 (Figures 1K–1O’’’, 1P and 1Q).
Figure 1. Pacemaker cells originate from the most lateral edge of the nkx2.5+ progenitor fields residing within the ALPM.
Confocal imaging of Tg(nkx2.5:Kaede; myl7:Cerulean) embryos shows the location of nkx2.5:Kaede cells immediately after photoconversion at (B, G, L) 14 ss as well as respectively later at (D, E, I, J, N, O) 8 hours post-photoconversion at 24 hpf. Immunostaining detects Isl1 and Nkx2.5 expressing cells. (A, C, F, H, K, M) Schematics illustrate the position of photoconverted (red) and non-photoconverted (green) nkx2.5:Kaede cells as well as myl7:Cerulean cardiomyocytes (blue) for B, D, G, I, L, N images, respectively. (A-E’’’) Pan-photoconversion of Tg(nkx2.5:Kaede; myl7:Cerulean) embryos at 14 ss results in photoconverted nkx2.5:Kaede+ Isl1+ myl7:Cerulean+ pacemaker cardiomyocytes at 24 hpf (n = 201/212 Isl1+ myl7:Cerulean+ pacemaker cardiomyocytes from 8 embryos). (F-O’’’) Localized photoconversion of nkx2.5:Kaede at the (F-J’’’) lateral or (K-O’’’) medial edge at 14 ss reveals that the lateral edge of nkx2.5+ cardiac mesoderm, but not the medial edge, contributes to pacemaker cardiomyocytes at 24 hpf. (E-E’’’, J-J’’’, O-O’’’ and G, L) Insets are magnifications of boxed areas in D, I, N and F, K, respectively. Images E’-E’’’, J’-J’’’, O’-O’’’ are single channels from E, J, O merged images, respectively. Yellow arrowheads point to (E-E’’’) photoconverted nkx2.5:Kaede+ Isl1+ myl7:Cerulean+ pacemaker cardiomyocytes. White arrows point to (G, J-J’’’, L, O-O’’’) photoconverted nkx2.5:Kaede cells. Green – (G, L) non-photoconverted nkx2.5:Kaede; Red – (B, D, E, E’, G, I, J, J’, L, N, O, O’) photoconverted nkx2.5:Kaede; Cyan – (D, E, E’’, I, J, J’’, N, O, O’’) anti-Isl1 immunostaining; White – (D, E, E’’’, I, J, J’’’, N, O, O’’’) anti-Nkx2.5 immunostaining; Blue – (D, E, I, J, N, O) myl7:Cerulean. (P) Quantification of the localized photoconversion studies reveal that 14 ss nkx2.5:Kaede cells photoconverted at the most lateral edge of the nkx2.5+ mesoderm contribute primarily to pacemaker cardiomyocytes (red bar) along with a few atrial cardiomyocytes (green bar), whereas those photoconverted at the medial edge mainly supply ventricular cardiomyocytes (blue bar). (Q) Table summarizes the results of localized nkx2.5:Kaede photoconversion studies at the lateral (n = 55 cells from 24 embryos) and medial edge (n = 36 cells from 8 embryos). At, atrium; V, ventricle. Scale bar, 50 μm. See also Figure S1.
Because these fate mapping studies suggest a dynamic role for Nkx2.5 during pacemaker cardiomyocyte formation, we further examined the developmental window in which Nkx2.5 may influence their development. Using the Tg(hsp70l:nkx2.5-EGFP) [or Tg(hsp70:nkx2.5)] line, we discovered that heat-shock induction of Nkx2.5 at 16 ss, but not at 3, 10 or 23 ss, results in reduced pacemaker cardiomyocyte formation (Figures S1A–S1I’’ and S1L–S1O), suggesting that decreasing Nkx2.5 expression in Nkx2.5+ cardiac progenitors between 16–23 ss is crucial for pacemaker cardiomyocyte development. Supporting these findings, we conversely observed that nkx2.5 −/− mutants display increased Isl1+/myl7:eGFP+ pacemaker cardiomyocytes at the atrial inflow tract region (Figures S1J–S1K’’ and S1P) as recently reported (Colombo et al., 2018). Altogether, these Nkx2.5 fate mapping and genetic studies reveal that pacemaker cardiomyocytes derive from Nkx2.5+ cardiac precursors located at the most lateral regions of the ALPM, and that their differentiation from these precursors involves the coordinated activation of Isl1 and silencing of Nkx2.5 during a critical stage of heart development.
Canonical Wnt5b signaling directs Nkx2.5+ progenitors to pacemaker cardiomyocytes
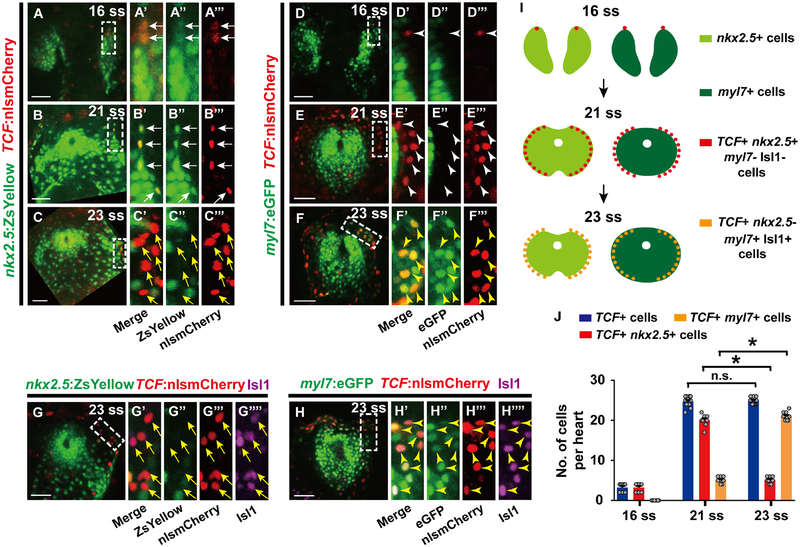
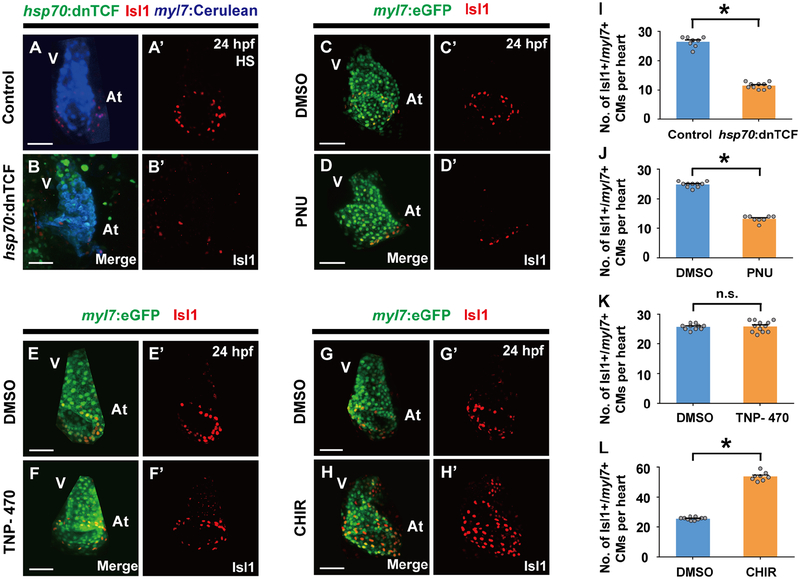
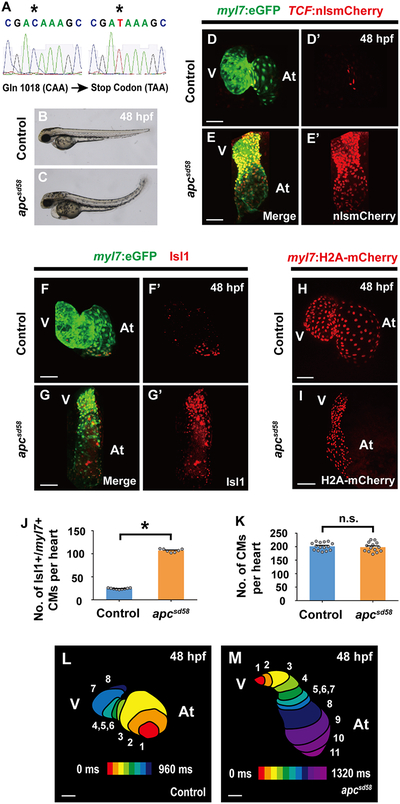
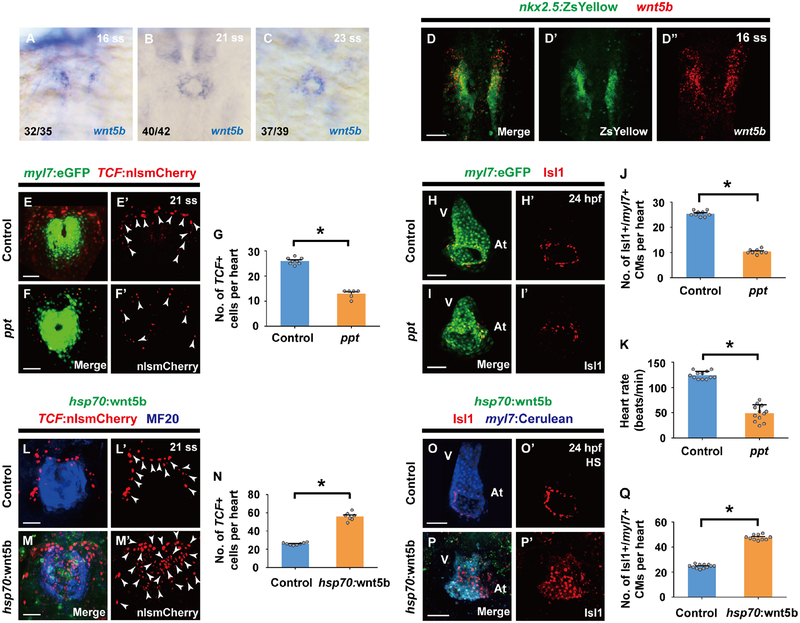
Based on these findings, we next searched for signaling pathways that could assign cardiomyocyte pacemaker fate to the lateral regions of Nkx2.5 expressing mesoderm during this developmental window. Utilizing the Tg(7xTCF-Xla.Siam:nlsmCherry) [or Tg(TCF:nlsmCherry)] line, which reports activated canonical Wnt signaling (Moro et al., 2012), we observed TCF:nlsmCherry Wnt reporter activity in the outlying regions of the cardiac mesoderm (Figures 2A–2F’’’), where cardiac pacemaker precursors reside at a developmental time period (16–23 ss) that is distinct from when canonical Wnt signaling is initiated during the pre-gastrulation stage to form cardiac mesoderm (Figures S2A–S2B’’) (Barrow et al., 2007; Liu et al., 1999; Ueno et al., 2007). At 16 and 21 ss, this TCF:nlsmCherry signal co-localizes with nkx2.5:ZsYellow+ cardiac precursors but not differentiated myl7:eGFP+ cardiomyocytes (Figures 2A–2B’’’ white arrows, 2D–2E’’’ white arrowheads, 2I and 2J). However, by 23 ss, this outlying canonical Wnt activity is primarily activated in myl7:eGFP+ cardiomyocytes that express the cardiac pacemaker marker Isl1 but not in nkx2.5:ZsYellow+ cells (Figures 2C–2C’’’, 2G–2G’’’’ yellow arrows, 2F–2F’’’, 2H–2H’’’’ yellow arrowheads, 2I and 2J), supporting a model in which canonical Wnt signaling is reactivated at this cardiac developmental stage to direct the differentiation of Nkx2.5+ cardiac precursors into pacemaker cardiomyocytes expressing Isl1, myl7:eGFP (a marker for differentiated cardiomyocytes), but not Nkx2.5. Thus, to explore this possibility, we investigated whether altering canonical Wnt activity during this period of heart development (16–23 ss) could affect the formation of pacemaker cardiomyocytes. Using the Tg(hsp70:dnTCF-GFP) [or Tg(hsp70:dnTCF)] line, which can conditionally block canonical Wnt signaling through heat-shock induction of dominant negative TCF (dnTCF), or the canonical Wnt signaling chemical inhibitor PNU-74654 (PNU), we discovered that silencing canonical Wnt signaling at 16 ss leads to reduced pacemaker cardiomyocytes as detected by decreased Isl1+/myl7:eGFP+ cardiomyocytes (Figures 3A–3D’, 3I and 3J) and diminished expression of isl1, tbx18, shox2 and hcn4 genes at 48 hpf, when these genes together mark pacemaker cardiomyocytes in the cardiac inflow tract region (Figures S3A–S3B’’’ and S4A–S4B’’’) (Birket et al., 2015; Liang et al., 2015; Mommersteeg et al., 2007; Protze et al., 2017; van Weerd and Christoffels, 2016; Wiese et al., 2009). However, treating embryos with TNP-470, an inhibitor of β-catenin-independent non-canonical Wnt activation, at 16 ss has no appreciable effect on pacemaker cardiomyocyte formation (Figures 3E–3F’, 3K and S4E–S4F’’’), further suggesting that Wnt signaling acts specifically through canonical pathways to control pacemaker cardiomyocyte development. In support of these findings, constitutively activated canonical Wnt signaling in either zebrafish embryos treated with the glycogen synthase kinase 3 inhibitor CHIR99021 (CHIR) at 16 ss (Figures 3G–3H’, 3L) or apcsd58 mutant embryos (Figures 4A–4E’, 4F–4G’ and 4J) results in increased pacemaker cardiomyocyte formation including in the ventricle, but no effect on overall cardiomyocyte numbers (Figures 4H, 4I and 4K), suggesting that these ectopic pacemaker cardiomyocytes may develop at the expense of chamber cardiomyocytes. Consistent with this notion, hearts with constitutively activated Wnt signaling display an expansion of isl1, tbx18, shox2 and hcn4 (Figures S3C–S3D’’’ and S4C–S4D’’’). Confirming that these ectopic pacemaker cardiomyocytes exhibit cardiac pacing capabilities, we discovered, by analyzing the propagation of cardiac excitation using the Tg(myl7:gCaMP) myocardial-specific calcium indicator line, that apcsd58 mutant hearts harboring pacemaker cardiomyocytes within the ventricle display retrograde cardiac conduction from the ventricular to the atrial chamber at 48 hpf (Figures 4L and 4M) when cardiac conduction typically propagates from atrium to ventricle in wild-type zebrafish embryos (Arrenberg et al., 2010; Chi et al., 2008). Finally, chromatin immunoprecipitation (ChIP) studies reveal that the β-catenin/TCF complex can interact with both Isl1 and Tbx18 enhancers harboring TCF/LEF binding sites (Figure S5), suggesting a possible mechanism by which Wnt signaling may initiate pacemaker differentiation. Overall, these findings support the conclusion that in addition to its previously reported cardiac developmental roles in inducing cardiac mesoderm formation during pre-gastrulation (Barrow et al., 2007; Liu et al., 1999; Ueno et al., 2007) and promoting the specification and expansion of SHF progenitors, which also prevents their differentiation into cardiomyocytes (Ai et al., 2007; Cohen et al., 2007; Kwon et al., 2007; Kwon et al., 2009; Lin et al., 2007; Qyang et al., 2007), canonical Wnt signaling is redeployed in the outlying cardiac mesoderm from 16–23 ss to direct the differentiation of cardiac precursors into pacemaker cardiomyocytes.
Figure 2. Nkx2.5 expression is silenced while canonical Wnt signaling is activated in nkx2.5+ progenitors during pacemaker cardiomyocyte differentiation.
(A-F’’’) Confocal images of (A-C’’’) Tg(nkx2.5:ZsYellow; TCF:nlsmCherry) (n = 8 embryos per stage) or (D-F’’’) Tg(myl7:eGFP; TCF:nlsmCherry) embryos (n = 8, 8, 10 embryos for each respective stage) at (A-A’’’, D-D’’’) 16 ss, (B-B’’’, E-E’’’) 21 ss, or (C-C’’’, F-F’’’) 23 ss reveal that canonical Wnt signaling is activated in outlying Nkx2.5+ mesodermal cells which are decreasing nkx2.5:ZsYellow and increasing myl7:eGFP expression. (G-H’’’’) Anti-Isl1 immunostaining of (G-G’’’’) Tg(nkx2.5:ZsYellow; TCF:nlsmCherry) (n = 9 embryos) and (H-H’’’’) Tg(myl7:eGFP; TCF:nlsmCherry) (n = 10 embryos) embryos at 23 ss shows that canonical Wnt signaling (TCF:nlsmCherry+) is activated in nkx2.5:ZsYellow- Isl1+ cells and myl7:eGFP+ Isl1+ cardiomyocytes. (A’-A’’’, B’-B’’’, C’-C’’’, D’-D’’’, E’-E’’’, F’-F’’’, G’-G’’’’, H’-H’’’’) Insets are magnifications of boxed areas in A, B, C, D, E, F, G, H, respectively. Images A’’-A’’’, B’’-B’’’, C’’-C’’’, D’’-D’’’, E’’-E’’’, F’’-F’’’, G’’-G’’’’, H’’-H’’’’ are single channels from A’, B’, C’, D’, E’, F’, G’, H’ merged images, respectively. (I) Schematics based on A-H’’’’ illustrate the dynamic changes in expression of nkx2.5:ZsYellow, myl7:eGFP, Isl1, and canonical Wnt-activation (TCF:nlsmCherry+) during pacemaker cardiomyocyte differentiation. (J) Further supporting these changes, quantification of TCF:nlsmCherry+ cells, TCF:nlsmCherry+ nkx2.5:ZsYellow+ cells and TCF:nlsmCherry+ myl7:eGFP+ cardiomyocytes (per heart) at 16 ss, 21 ss and 23 ss reveals that TCF:nlsmCherry+ nkx2.5:ZsYellow+ cells are significantly reduced while TCF:nlsmCherry+ myl7:eGFP+ cardiomyocytes are greatly increased from 21–23 ss. However, the total number of TCF:nlsmCherry+ cells is not changed. White arrows point to TCF:nlsmCherry+ nkx2.5:ZsYellow+ cells. Yellow arrows point to TCF:nlsmCherry+ nkx2.5:ZsYellow- cells. White arrowheads point to TCF:nlsmCherry+ myl7:eGFP- cells. Yellow arrowheads point to TCF:nlsmCherry+ myl7:eGFP+ cardiomyocytes. Green – (A-A’’, B-B’’, C-C’’, G-G’’) nkx2.5:ZsYellow, (D-D’’, E-E’’, F-F’’, H-H’’) myl7:eGFP; Red – (A, A’, A’’’, B, B’, B’’’, C, C’, C’’’, D, D’, D’’’, E, E’, E’’’, F, F’, F’’’, G, G’, G’’’, H, H’, H’’’) TCF:nlsmCherry; Magenta – (G, G’, G’’’’, H, H’, H’’’’) anti-Isl1 immunostaining. Scale bar, 50 μm. Mean ± s.e.m. *P< 0.05 by Student’s t-test. n.s. - not significant. See also Figure S2.
Figure 3. Canonical Wnt signaling is required for pacemaker cardiomyocyte formation.
Inhibiting canonical Wnt signaling by (B, B’) heat-shock (HS) induction of hsp70:dnTCF (n = 10 embryos) or (D, D’) PNU treatment (n = 8 embryos) at 16 ss reduces Isl1+/myl7+ pacemaker cardiomyocytes compared to (A, A’) heat-shocked non-transgenic (n = 8 embryos) or (C, C’) DMSO treated controls (n = 10 embryos), respectively. However, inhibiting non-canonical Wnt signaling with (F, F’) TNP-470 treatment (n = 12 embryos) has no discernible effect on pacemaker development, whereas activating canonical Wnt signaling with (H, H’) CHIR (n = 8 embryos) at 16 ss promotes their expansion when compared to (E, E’, G, G’) respective DMSO controls (n = 10, 10 embryos). Images A’-H’ are single channels from A-H merged images, respectively. Green – (B) hsp70:dnTCF, (C-H) myl7:eGFP; Red – (A-H’) anti-Isl1 immunostaining; Blue – (A, B) myl7:Cerulean. (I-L) Bar graphs show the number of Isl1+/myl7+ pacemaker cardiomyocytes (CMs) per heart for A-B’, C-D’, E-F’ and G-H’ conditions, respectively. At, atrium; V, ventricle. Scale bar, 50 μm. Mean ± s.e.m. *P< 0.05 by Student’s t-test. n.s. - not significant. See also Figures S3–S5 and Tables S1 and S2.
Figure 4. A new apc loss-of-function mutant (apcsd58) displays constitutively activated canonical Wnt signaling and ectopic pacemaker formation.
(A) Sequence analysis of sd58 mutants reveals a C to T nonsense mutation at amino acid 1018 in the apc gene, thus resulting in a putative premature truncation. (B, C) Bright-field microscopy of the lateral side of (B) wild-type (n = 12 embryos) and (C) apcsd58 mutant embryos (n = 10 embryos) shows that apcsd58 mutants display an upwards curved tail and edema at 48 hpf. Confocal images of (D, D’, F, F’, H) wild-type and (E, E’, G, G’, I) apcsd58 mutant embryos at 48 hpf reveal that apcsd58 mutant hearts display (E, E’) increased canonical Wnt-activation as detected by TCF:nlsmCherry (n = 8 embryos) and (G, G’) increased number of Isl1+/myl7+ pacemaker cardiomyocytes (n = 7 embryos) but (I) no appreciable change in overall cardiomyocyte numbers (n = 14 embryos) when compared to (D, D’, F, F’, H) respective wild-type control hearts (n = 7, 9, 15 embryos). Images D’-G’ are single channels from D-G merged images, respectively. Green – (D-G) myl7:eGFP; Red – (D-E’) TCF:nlsmCherry, (F-G’) anti-Isl1 immunostaining, (H, I) myl7:H2A-mCherry. (J, K) Bar graphs show the average number of (J) Isl1+/myl7+ pacemaker cardiomyocytes (CMs) from F-G’ or (K) total cardiomyocytes from H and I. (L, M) Optical mapping of the propagation of calcium activation in (L) control (n = 9 embryos) and (M) apcsd58 mutant (n = 7 embryos) hearts at 48 hpf reveals retrograde cardiac conduction from ventricle to atrium in apcsd58 mutants. Numbers at each isochronal line (60 ms intervals) indicate temporal sequence of calcium activation in the heart. At, atrium; V, ventricle. Scale bar, 50 μm. Mean ± s.e.m. *P< 0.05 by Student’s t-test. n.s. - not significant. See also Figure S3.
To identify potential Wnt ligands responsible for controlling pacemaker cardiomyocyte differentiation, we examined the expression of known Wnt ligands by in situ hybridization during heart development and discovered that wnt5b is expressed in a distribution similar to that of Wnt-activated cardiac precursors fated to become pacemaker cardiomyocytes from 16–23 ss (Figures 5A–5C), and furthermore co-localizes with nkx2.5:ZsYellow+ cardiac precursors at 16 ss (Figures 5D–5D’’). Supporting a role for Wnt5b in pacemaker cardiomyocyte development, we discovered that wnt5b loss-of-function (ppt) embryos exhibit fewer TCF:nlsmCherry+ cardiac precursors at the outlying regions of the cardiac mesoderm (Figures 5E–5G), a subsequent reduction in pacemaker cardiomyocytes (Figures 5H–5J) including decreased isl1, tbx18, shox2 and hcn4 pacemaker gene expression in the cardiac inflow tract (Figures S3E–S3F’’’), and significantly reduced heart rate (Figure 5K). Consistent with our findings suggesting that canonical Wnt signaling may affect the decisions of cardiac precursors to choose between an atrial or pacemaker cardiomyocyte fate, wnt5b mutant embryos additionally display an increase in atrial cardiomyocytes but no significant differences in cardiomyocyte cell death or proliferation compared to developmentally stage-matched wild-type embryos (Figure S6). In contrast to these loss-of-function studies, we observed that overexpressing Wnt5b at 16 ss using the heat-shock inducible Tg(hsp70l:wnt5b-GFP) [or Tg(hsp70:wnt5b)] line results in not only increased canonical Wnt signaling activation in the cardiac mesoderm as detected by TCF:nlsmCherry (Figures 5L–5N), but also subsequent ectopic pacemaker cardiomyocyte formation (Figures 5O–5Q) including expanded isl1, tbx18, shox2 and hcn4 pacemaker gene expression throughout the heart (Figures S3G–S3H’’’). Together with our findings that inhibition of the non-canonical Wnt pathway does not affect pacemaker cardiomyocyte formation (Figures 3E–3F’, 3K and S4E–S4F’’’), these results support a role for Wnt5b as a key Wnt ligand that activates canonical Wnt signaling in the outlying regions of Nkx2.5 cardiac mesoderm between 16–23 ss to control gene regulatory programs instructing pacemaker cardiomyocyte fate. Moreover, manipulating canonical Wnt5b signaling can lead to an exchange of fates within these Nkx2.5+ cardiac mesodermal progenitors, suggesting that although these progenitors may be specified to become distinct cardiomyocyte lineages (Yelon et al., 1999), they also maintain the plasticity to switch fates upon exposure to specific signaling cues.
Figure 5. Wnt5b is required for canonical Wnt activation during pacemaker cardiomyocyte development.
(A-C) In situ hybridization analyses reveals wnt5b expression within the ALPM at (A) 16 ss as well as in the cardiac mesoderm at (B) 21 ss and (C) 23 ss. (D-D’’) wnt5b RNAscope in situ hybridization in Tg(nkx2.5:ZsYellow) wild-type embryos (n = 12 embryos) shows that there is an overlap in nkx2.5:ZsYellow and wnt5b expression at 16 ss. (E-F’, H-I’) Confocal images of (E-F’) Tg(myl7:eGFP; TCF:nlsmCherry) (E-E’) wild-type control (n = 8 embryos) or (F-F’) ppt mutant embryos (n = 6 embryos) at 21 ss and (H-I’) anti-Isl1 immunostaining of Tg(myl7:eGFP) (H-H’) wild-type control (n = 9 embryos) or (I-I’) ppt mutant embryos (n = 8 embryos) at 24 hpf show that loss of wnt5b (ppt) leads to less TCF:nlsmCherry+ cells in the outlying regions of the cardiac mesoderm and subsequently fewer Isl1+/myl7+ pacemaker cardiomyocytes at the inflow tract region. Conversely, overexpressing wnt5b by (M-M’, P-P’) heat-shocking Tg(hsp70:wnt5b) embryos (n = 7, 10 embryos) at 16 ss results in increased TCF:nlsmCherry+ cells in the cardiac mesoderm at 21 ss and increased Isl1+/myl7+ pacemaker cardiomyocytes at the inflow tract region at 24 hpf compared to (L-L’, O-O’) non-transgenic heat-shocked sibling controls (n = 8, 12 embryos). Images D’, D’’, E’, F’, H’, I’, L’, M’, O’, P’ are single channels from D, E, F, H, I, L, M, O, P merged images, respectively. Green – (D, D’) nkx2.5:ZsYellow, (E, F, H, I) myl7:eGFP, (M, P) hsp70:wnt5b; Red – (D, D’’) wnt5b RNAscope in situ hybridization probe, (E-F’, L-M’) TCF:nlsmCherry, (H-I’, O-P’) anti-Isl1 immunostaining; Blue – (L, M) anti-MF20 immunostaining, (O, P) myl7:Cerulean. White arrowheads point to TCF:nlsmCherry+ cells. (G, J, N, Q) Bar graphs show the average number of (G, N) TCF:nlsmCherry+ cells per heart for E-F’ and L-M’ conditions, and (J, Q) Isl1+/myl7+ pacemaker cardiomyocytes (CMs) per heart for H-I’ and O-P’ conditions, respectively. (K) Bar graph shows the heart rate for wild-type control (n= 12 embryos) and ppt mutant embryos (n= 12 embryos) at 48 hpf. For in situ hybridization studies in A-C, ratios indicate the number of embryos displaying the respective gene expression pattern versus total number of embryos analyzed. Scale bar, 50 μm. Mean ± s.e.m. *P< 0.05 by Student’s t-test. See also Figure S3 and S6.
Activating canonical Wnt signaling in hPSCs promotes pacemaker-like cardiomyocyte differentiation
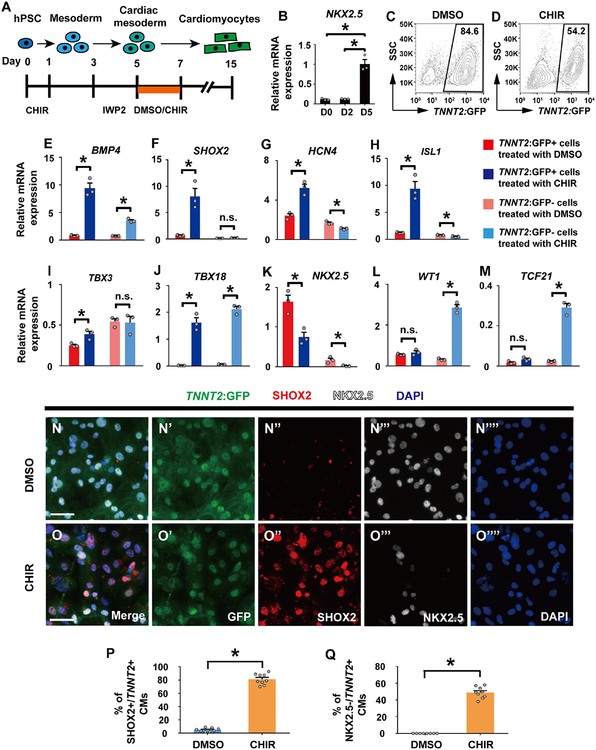
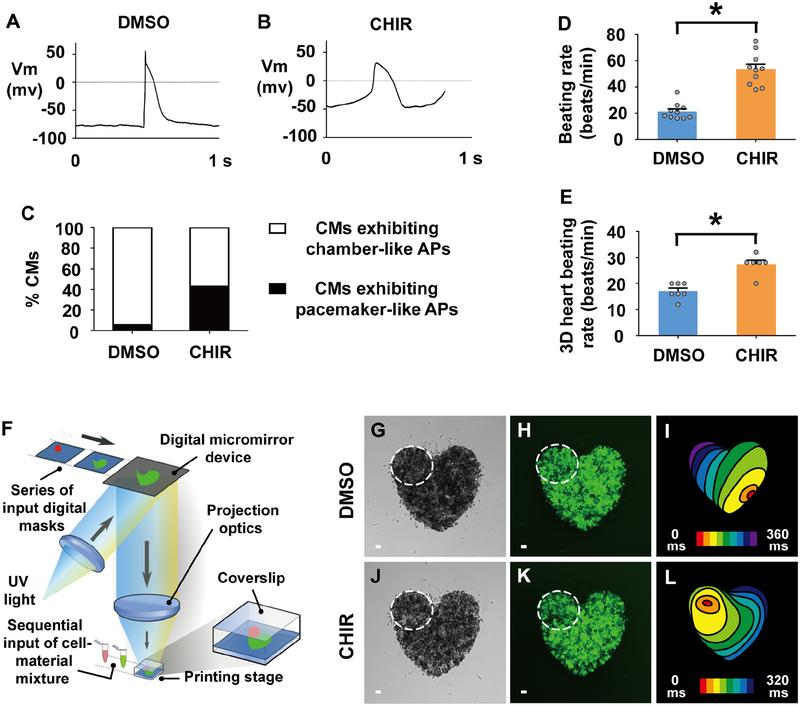
To explore whether these developmental concepts may be applied to hPSCs to create pacemaker cardiomyocytes, we manipulated canonical Wnt signaling at the cardiac precursor stage during the differentiation of hPSCs into cardiomyocytes. Utilizing a highly efficient monolayer hPSC cardiac differentiation protocol (Lian et al., 2012), we initially differentiated H9 hPSCs harboring the TNNT2:GFP cardiac-specific transgene to an early NKX2.5-expressing cardiac precursor stage (Day 5/D5) (Lian et al., 2012) that corresponds to ~14–16 ss in the zebrafish through activating canonical Wnt signaling from D0-D1 and then inhibiting it from D3-D5 (Figures 6A and 6B). Similar to the in vivo zebrafish studies, canonical Wnt signaling was then re-activated in these hPSC-cardiac precursors on D5 by treating them with the small molecule inhibitor CHIR (Figure 6A). Although this canonical Wnt signaling re-activation modestly reduces overall hPSC-cardiomyocyte differentiation efficiency (Figures 6C and 6D), it notably results in TNNT2:GFP+ hPSC-cardiomyocytes displaying pacemaker-like features (Figures 6E–6K, 6N–6Q and 7) and expressing BMP4 (Figure 6E), which has also been recently reported to promote pacemaker cardiomyocyte differentiation (Protze et al., 2017). These Wnt-activated hPSC-cardiomyocytes, which express the cardiomyocyte marker TNNT2, exhibit not only significantly elevated expression of pacemaker-related genes SHOX2, HCN4, ISL1, TBX3 and TBX18, but also a corresponding decrease in NKX2.5 expression (Figures 6F–6K), which were confirmed by SHOX2 and NKX2.5 immunostaining (Figures 6N–6Q). In line with these studies, hPSC-cardiac precursors treated with WNT5B from D5–7 furthermore display significantly increased expression of pacemaker-related genes and decreased expression of NKX2.5 (Figure S7). Functionally supporting these findings, single cell patch clamping studies further revealed that while control (DMSO-treated) cardiomyocytes primarily display action potentials observed in chamber cardiomyocytes including a more consistent and negative resting membrane potential than pacemaker cardiomyocytes (Figures 7A and 7C), Wnt-activated hPSC-cardiomyocytes exhibit pacemaker-like action potentials with less negative resting membrane potentials (Figures 7B and 7C). Corresponding with this change to pacemaker-like characteristics, these Wnt-activated hPSC-cardiomyocytes also display significantly faster beating rates than those detected in control hPSC-cardiomyocytes (Figure 7D), corroborating their increased automaticity. Thus, these molecular and cellular findings support that hPSC-cardiomyocytes, which derive from cardiac precursors where Wnt signaling is re-activated during a critical window of cardiomyocyte differentiation, exhibit key features of pacemaker cardiomyocytes that distinguish them from cardiac progenitor cells, including the presence and absence of a combination of genes specific to pacemaker cardiomyocytes (SHOX2+, HCN4+, ISL1+, TBX3+, TBX18+, but NKX2.5-) (Birket et al., 2015; Liang et al., 2015; Mommersteeg et al., 2007; Protze et al., 2017; van Weerd and Christoffels, 2016; Wiese et al., 2009), their differentiation into cardiomyocytes expressing TNNT2 and their electrophysiologic attributes promoting enhanced cardiac automaticity. Finally, consistent with recent studies showing that temporal modulation of canonical Wnt signaling may induce epicardial cell formation in hPSCs (Bao et al., 2016; Iyer et al., 2015), we notably also found that hPSC TNNT2:GFP-negative non-cardiomyocyte cells derived from these Wnt re-activated hPSC cardiac precursors exhibit significantly increased expression of the epicardial marker genes TBX18, WT1 and TCF21 but decreased expression of NKX2.5 (Figures 6J–6M).
Figure 6. Activating canonical Wnt signaling in human pluripotent stem cell (hPSC)-cardiac progenitors promotes pacemaker-like cardiomyocyte differentiation.
(A) Schematic illustrates the experimental design for inducing human pacemaker-like cardiomyocyte differentiation through activating canonical Wnt signaling in TNNT2:GFP hPSC-cardiac progenitors (i.e., adding 3 μM CHIR from D5–7). (B) NKX2.5 expression was analyzed by qRT-PCR (n = 3 independent cardiomyocyte differentiations) for hPSCs differentiating into cardiomyocytes on D0, D2 and D5. Bar graphs represent average gene expression relative to the housekeeping gene TATA box binding protein (TBP). (C, D) After treating TNNT2:GFP hPSCs with (C) DMSO or (D) 3 μM CHIR during days 5–7 of the monolayer cardiomyocyte differentiation protocol, hPSC-cardiomyocyte differentiation was assessed at day 15 by flow cytometry (n = 3 independent differentiations). (E-M) TNNT2:GFP+ and TNNT2:GFP- cells generated in C, D were sorted and then analyzed for (E) BMP4, (F) SHOX2, (G) HCN4, (H) ISL1, (I) TBX3, (J) TBX18, (K) NKX2.5, (L) WT1 and (M) TCF21 expression by qRT-PCR (n = 3 independent differentiations). Bar graphs represent average gene expression relative to the housekeeping gene TBP. (N-O’’’’) Confocal images of TNNT2:GFP+ cardiomyocytes sorted from D15 cultures and immunostained for SHOX2 and NKX2.5 reveal that treating hPSC-cardiac progenitors with (O-O’’’’) CHIR from D5–7 (n = 3 independent differentiations) results in increased SHOX2+/TNNT2+ and NKX2.5-/TNNT2+ cardiomyocytes compared to (N-N’’’’) DMSO treatment from D5–7 (n = 3 independent differentiations). Images N’-N’’’’, O’-O’’’’ are single channels from N, O merged images, respectively. Green – (N, N’, O, O’) TNNT2:GFP; Red – (N, N’’, O, O’’) anti-SHOX2 immunostaining; White – (N, N’’’, O, O’’’) anti-NKX2.5 immunostaining; Blue – (N, N’’’’, O, O’’’’) DAPI. (P, Q) Bar graphs show the percentage of (P) SHOX2+/TNNT2+ and (Q) NKX2.5-/TNNT2+ cardiomyocytes (CMs) for D5–7 (N-N’’’’) DMSO (n = 8) or (O-O’’’’) CHIR (n = 9) treatment conditions. Scale bar, 50 μm. Mean ± s.e.m. *P< 0.05 by Student’s t-test. n.s. - not significant. See also Figure S7, Tables S1 and S3.
Figure 7. Activating canonical Wnt signaling in human pluripotent stem cell (hPSC)-cardiac progenitors creates functional pacemaker-like cardiomyocytes that can pace hPSC-cardiomyocytes in vitro.
(A, B) Electrophysiology recordings reveal action potentials of individual cardiomyocytes sorted from D15 cultures which were treated with (A) DMSO (n = 18 cardiomyocytes) or (B) CHIR (n = 14 cardiomyocytes) from D5–7. (C) Electrophysiology studies on these TNNT2:GFP+ hPSC-cardiomyocytes (CMs) reveals that a greater percentage of CHIR-treated hPSC-cardiomyocytes (n = 14 cardiomyocytes) exhibit pacemaker-like action potentials (APs) than DMSO-treated hPSC-cardiomyocytes (n = 18 cardiomyocytes). (D) Bar graph shows the average beating rate for cardiomyocytes sorted from D15 cultures which were exposed to either DMSO (n = 10 cardiomyocytes) or CHIR (n = 10 cardiomyocytes) treatment from D5–7. (E-L) hPSC-derived pacemaker-like cardiomyocytes can pace hPSC-cardiomyocyte tissue in vitro. (F) Diagram illustrates 3D bioprinting approach. (G-L) To create multi-cellular in vitro mini-hearts, D15 hPSC-cardiomyocytes treated with (G, H) DMSO or (J, K) CHIR from D5–7 are printed in the upper left region of mini-hearts (dashed lines encircle area), and D15 hPSC-cardiomyocytes are printed outside this encircled region. (G, J) Bright-field and (H, K) GFP microscopy images show printed mini-hearts. (I, L) Optical mapping of the calcium activation in these mini-hearts reveals that cardiac conduction is stably initiated from the site of printed (L) CHIR-treated pacemaker-like cardiomyocytes (n = 6 mini-hearts) but not (I) DMSO-treated control cardiomyocytes (n = 7 mini-hearts). Red/orange – cardiac conduction initiation site; Isochronal lines – 40 milliseconds (ms) intervals. (E) Bar graph shows that the average beating rate is significantly faster in mini-hearts printed with CHIR-treated pacemaker-like cardiomyocytes (n = 6 mini-hearts) compared to those printed with DMSO-treated control cardiomyocytes (n = 7 mini-hearts). Scale bar, 50 μm. Mean ± s.e.m. *P< 0.05 by Student’s t-test. See also Videos S1–S4.
Canonical Wnt-activated hPSC pacemaker-like cardiomyocytes can pace in vitro bioprinted mini-heart models
Employing a rapid 3D bioprinting technology to create in vitro multi-cellular cardiac models (Figure 7F), we next investigated whether these Wnt-activated hPSC-pacemaker-like cardiomyocytes could functionally pace in vitro prefabricated cardiac tissues composed of hPSC-cardiomyocytes. To this end, we bioprinted purified hPSC-cardiomyocytes into a heart-shape configuration (i.e. “mini-hearts”) with a gap in the upper left region (Figures 7G, 7H, 7J and 7K). Within this excluded area, we then immediately bioprinted either purified control (DMSO) or Wnt-activated hPSC-cardiomyocytes (Figures 7G, 7H, 7J and 7K – dashed circle) to examine whether Wnt-activated hPSC-cardiomyocytes specifically display pacemaker activity that can pace the bioprinted hPSC-cardiomyocyte tissues. After confirming recovery and re-synchronization of these bioprinted cardiomyocytes, we then performed optical mapping analyses using calcium imaging on these mini-hearts. In contrast to mini-hearts with bioprinted control hPSC-cardiomyocytes, which display randomly initiated electrical activity (Figure 7I; Video S1), mini-hearts with Wnt-activated hPSC-cardiomyocytes consistently initiate cardiac electrical activity from the upper left region (Figure 7L; Video S2) and exhibit faster paced beating rates (Figure 7E; Videos S3 and S4). Altogether, these results reveal that activating canonical Wnt signaling in hPSC-derived cardiac progenitors can initiate the gene regulatory program that directs their differentiation into human pacemaker-like cardiomyocytes that are capable of pacing human cardiac tissue.
Discussion
Overall, through identifying the origins of pacemaker cardiomyocytes, we discovered that Wnt5b functions as an inductive signaling cue that activates a conserved canonical Wnt signaling pathway which instructs the fate of Nkx2.5+ cardiac progenitors into the cardiac pacemaker lineage. Although previous studies suggest that pacemaker cardiomyocytes may derive from mesodermal progenitor cells not expressing Nkx2.5 (Bressan et al., 2013), our fate mapping studies reveal that they can originate from Nkx2.5+ cardiac precursors, which are specifically located adjacent but also lateral to Nkx2.5+ cardiac mesoderm fated to become atrial cardiomyocytes as suggested in recent mouse studies (Mommersteeg et al., 2010). Consistent with these mouse studies, we further observed Wnt-mediated dynamic silencing of Nkx2.5 during pacemaker cardiomyocyte differentiation, which may explain the recent findings suggesting that pacemaker cardiomyocytes may derive from cardiac mesoderm not expressing Nkx2.5 (Bressan et al., 2013). However, our studies do not preclude the possibility that given the heterogeneity of cells composing the sinoatrial node (Liang et al., 2015; Sun et al., 2007), pacemaker cardiomyocytes may derive from multiple cardiac progenitor cell sources.
Canonical Wnt signaling has been reported to regulate multiple aspects of cardiac development including the induction of cardiac mesoderm formation during pre-gastrulation (Barrow et al., 2007; Liu et al., 1999; Ueno et al., 2007), and the specification and expansion of SHF cardiac progenitor cells at later stages, where canonical Wnt also prevents their differentiation into cardiomyocytes (Ai et al., 2007; Cohen et al., 2007; Kwon et al., 2007; Kwon et al., 2009; Lin et al., 2007; Qyang et al., 2007). In addition to mediating these cardiac developmental events, we have further discovered that canonical Wnt signaling is also redeployed to direct outlying Nkx2.5+ progenitors into pacemaker cardiomyocytes. In contrast to its role for promoting the expansion of SHF cardiac progenitors and inhibiting their differentiation into cardiomyocytes (Ai et al., 2007; Cohen et al., 2007; Kwon et al., 2007; Kwon et al., 2009; Lin et al., 2007; Qyang et al., 2007), we observed that canonical Wnt signaling, which promotes the differentiation of cardiac progenitors into pacemaker cardiomyocytes, functions through not only the activation of a combination of well established markers specific to pacemaker cardiomyocytes (SHOX2, ISL1, TBX18, HCN4, TBX3) (Birket et al., 2015; Liang et al., 2015; Mommersteeg et al., 2007; Protze et al., 2017; van Weerd and Christoffels, 2016; Wiese et al., 2009) but also the expression of cardiomyocyte differentiation markers (myl7 in zebrafish and TNNT2 in hPSCs), which distinguishes these pacemaker cardiomyocytes from cardiac progenitors.
Supporting an additional cardiac developmental role of canonical Wnt signaling in pacemaker cardiomyocyte differentiation that is distinct from its role in specification and expansion of cardiac progenitor cells, we discovered that the ligand responsible for controlling pacemaker cardiomyocyte differentiation is Wnt5b rather than Wnt3 and Wnt8, which have been reported to mediate cardiac progenitor cell formation (Barrow et al., 2007; Jaspard et al., 2000; Liu et al., 1999). Although Wnt5b has been observed to function primarily through non-canonical Wnt signaling pathways (Kilian et al., 2003; Merks et al., 2018), we discovered that similar to lymphatic developmental studies (Nicenboim et al., 2015), it can also induce canonical Wnt signaling in these Nkx2.5+ progenitor cells to both directly activate expression of Isl1 and Tbx18, which are transcriptional regulators involved in pacemaker cardiomyocyte differentiation (Kapoor et al., 2013; Liang et al., 2015; Tessadori et al., 2012; Vedantham et al., 2015; Wiese et al., 2009), as well as inhibit Nkx2.5 expression likely through Isl1-activated expression of Shox2 (Liang et al., 2015; van Weerd and Christoffels, 2016), which has been reported to inhibit Nkx2.5 expression (Espinoza-Lewis et al., 2009). Moreover, our results suggest a role for Wnt5b in partitioning the outlying regions of the Nkx2.5+ cardiac progenitor pool between pacemaker and atrial cardiomyocyte fates, as perturbing Wnt5b activity alters the contributions of these cardiomyocyte cell types to the developing heart. Given that Wnt5b can control Tbx18, which has been reported to regulate not only pacemaker cardiomyocyte (Kapoor et al., 2013; Wiese et al., 2009) but also epicardial cell development (Cai et al., 2008; Christoffels et al., 2009), future investigations into Wnt5b may illuminate whether it also regulates the development of other cardiovascular cell types arising from the outlying domains of the heart fields (Bressan et al., 2013). In line with this possibility, we discovered that temporally activating canonical Wnt signaling during hPSC-cardiomyocyte differentiation not only produced TNNT2+ pacemaker cardiomyocytes but also TNNT2-negative epicardial cells, which is consistent with recent hPSC epicardial cell differentiation studies (Bao et al., 2016; Iyer et al., 2015).
Finally, supporting that this canonical Wnt signaling pathway is evolutionarily conserved during pacemaker cell differentiation, these developmental principles can be applied to hPSC-cardiac precursors to direct their differentiation into pacemaker-like cardiomyocytes that are able to pace other hPSC-cardiomyocytes. Similar to recent hPSC cardiac pacemaker differentiation studies (Birket et al., 2015; Protze et al., 2017), our studies utilize an early inhibition of Wnt signaling to promote cardiac progenitor cell specification. In contrast to these studies that report coordinated manipulation of several signaling pathways (including BMP and RA activation and FGF inhibition) to create pacemaker-like cardiomyocytes (Birket et al., 2015; Protze et al., 2017), we discovered that re-activation of Wnt signaling in these cardiac progenitor cells is sufficient to promote the differentiation of cardiac precursors into pacemaker-like cardiomyocytes that express a similar combination of pacemaker genes (SHOX2+, HCN4+, ISL1+, TBX3+, TBX18+, but NKX2.5-), which have been previously reported in other hPSC pacemaker cardiomyocyte studies (Birket et al., 2015; Protze et al., 2017) or overexpressed in vivo to convert existing working cardiomyocytes into pacemaker-like cells (Bakker et al., 2012; Hoogaars et al., 2007; Kapoor et al., 2013). Supporting that canonical Wnt signaling may promote pacemaker cardiomyocyte differentiation through regulating BMP signaling, we notably observed that this redeployment of canonical Wnt signaling does lead to a substantial increase in the expression of BMP4, which has been reported as a key signal to direct not only cardiac pacemaker cell differentiation in these hPSC pacemaker studies (Protze et al., 2017) but also hPSC epicardial cell differentiation (Witty et al., 2014). Although these complementary findings appear to differ from recent hPSC reports suggesting BMP and FGF inhibition promote pacemaker cardiomyocyte differentiation (Birket et al., 2015), these discrepancies may be due to the timing and coordination of signaling pathways that regulate pacemaker cardiomyocyte differentiation. Overall, our findings support a model in which canonical Wnt signaling is redeployed during cardiac development to mediate the specification of cardiac pacemaker cells through coordinating a hierarchical signaling cascade, which may include BMP signaling and other signaling pathways (FGF, RA), that not only direct cardiac progenitor cells toward cardiac pacemaker fates but also prevent them from differentiating into other cardiomyocyte cell types.
STAR Methods:
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Neil C. Chi (nchi@ucsd.edu). The apcsd58 mutant zebrafish generated in this study is available upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
Zebrafish (Danio rerio) were raised under standard laboratory conditions at 28°C on a 14 hour light /10 hour dark cycle. All the embryos used in this study were collected by natural spawning and incubated in egg water in a 28°C incubator. All animal work was approved by the University of California at San Diego Institutional Animal Care and Use Committee (IACUC). The following established transgenic and mutant lines were used: TgBAC(−36nkx2.5:Kaede)fb9 (Guner-Ataman et al., 2013) abbreviated as Tg(nkx2.5:Kaede); Tg(myl7:Cerulean)co19 (Mathews et al., 2014); Tg(hsp70l:nkx2.5-EGFP)fcu1 (George et al., 2015) abbreviated as Tg(hsp70:nkx2.5); Tg(7xTCF-Xla.Siam:nlsmCherry)ia5 (Moro et al., 2012) abbreviated as Tg(TCF:nlsmCherry); Tg(−36nkx2.5:ZsYellow)fb7 (Zhou et al., 2011) abbreviated as Tg(nkx2.5:ZsYellow); Tg(myl7:eGFP)twu277 (Huang et al., 2003); Tg(hsp70:dnTCF-GFP)w26 (Lewis et al., 2004) abbreviated as Tg(hsp70:dnTCF); Tg(myl7:gCaMP)s878 (Chi et al., 2008); Tg(hsp70l:wnt5b-GFP)w33 (Stoick-Cooper et al., 2007) abbreviated as Tg(hsp70:wnt5b); Tg(myl7:H2A-mCherry)sd12 (Schumacher et al., 2013); Tg(amhc:eGFP)s958 (Zhang et al., 2013); nkx2.5vu179(Targoff et al., 2013) and wnt5b ta98 designated as pipetail (ppt) (Hammerschmidt et al., 1996; Rauch et al., 1997). Homozygous nkx2.5vu179 or ppt mutant embryos were identified using previously characterized defects in cardiac chamber morphology (Targoff et al., 2013) or body axis elongation (Hammerschmidt et al., 1996; Rauch et al., 1997), respectively. The zebrafish embryos used for experiments were less than 48 hpf, a stage at which sex can not be readily determined.
The apcsd58 allele was discovered in a previous forward genetic screen performed at UCSD. Microsatellite markers and bulk segregant analysis were used to map the sd58 allele to chromosome 10. Additional fine genetic mapping using individual homozygous mutant embryos placed the sd58 allele near the microsatellite marker z7316 and the apc gene. Sequencing of the apc gene in sd58 homozygous mutant embryos uncovered a nonsense mutation at amino acid position 1018 (Figure 4A). Furthermore, we found that an independent loss-of-function apc allele, apchu745 (Hurlstone et al., 2003), failed to complement the sd58 allele (data not shown). Increased TCF:nlsmCherry expression confirmed constitutive activation of canonical Wnt signaling in apcsd58 mutants (Figures 4D–4E’).
Human pluripotent stem cells
H9-hTnnTZ-pGZ-D2 human embryonic stem cell line was purchased from WiCell and maintained on Geltrex (Gibco) coated plates in E8 medium. They are female cells and have been authenticated by Short Tandem Repeat (STR) profiling analysis.
METHOD DETAILS
Nkx2.5 photoconversion studies
Tg(nkx2.5:Kaede) embryos were first mounted in 1% low melting point agarose (Lonza) in 35 mm glass bottom petri dishes (MatTek). A selected region of interest was then exposed to UV light for 1 min using a 405 nm blue diode laser on a Leica SP5 confocal laser scanning microscope, with a HCX IRAPO L 25.0X/0.95 water immersion objective. Following photoconversion and imaging, embryos were removed from the agarose, raised in the dark until 24 hpf, confirmed to display wild-type morphology and then fixed for immunofluorescence studies as described below.
Heat shock induction and small molecule inhibitor studies in zebrafish
Heat-shock induction was conducted as previously described (Han et al., 2016). Briefly, embryos containing heat-shock transgenes or wild-type siblings at specified stages were placed into a 37°C incubator for 30 minutes, followed by 3 minutes in a 42°C water bath. Embryos were then returned to a 28°C incubator prior to their subsequent analysis.
For small molecule inhibitor studies, zebrafish embryos were incubated in either 0.1% DMSO (control) or respective small molecule inhibitor between 16 ss and 24 hpf. Specific inhibitors and the concentration used are detailed in Table S1.
in situ hybridization, immunofluorescence, TUNEL assays in zebrafish
Whole mount in situ hybridization and immunofluorescence were performed as previously described (Zhang et al., 2013). The in situ probes and primary antibodies used in this study include: hcn4 (ZDB-GENE-050420–360), isl1 (ZDB-GENE-980526–112), shox2 (ZDB-GENE-040426–1457), tbx18 (ZDB-GENE-020529–2), wnt5b (ZDB-GENE-980526–87), anti-dsRed (rabbit, Clontech, 1:2000), anti-Isl1 (rabbit, GeneTex, 1:1000), anti-Isl1 (mouse, Developmental Studies Hybridoma Bank, 1:100), anti-MF20 (mouse, Developmental Studies Hybridoma Bank, 1:100), anti-Nkx2.5 (rabbit, GeneTex, 1:50), and anti-phospho-histone H3 (rabbit, Upstate, 1:200). in situ probes were detected with Anti-Digoxigenin-AP antibody (Fab fragments, Roche) for in situ hybridization, while primary antibodies were detected with the following appropriate secondary antibodies for immunofluorescence: anti-rabbit IgG-Alexa 568 (goat, Life technologies, 1:250), anti-mouse IgG Cy5 (goat, Life technologies, 1:250), anti-mouse IgG-Alexa 405 (goat, Life technologies, 1:250) and anti-rabbit IgG-Alexa 488 (goat, Life technologies, 1:250).
Cell death was detected using the in situ cell death detection kit (TMR red, Roche). Zebrafish embryos were fixed with 4% paraformaldehyde (PFA) for 2 hours, permeabilized with 0.5% TritonX-100 in PBS and incubated in TUNEL staining solution at 37°C for 2 hours. Fluorescent images were obtained using a Nikon C2 confocal microscope.
RNAscope in situ hybridization
Zebrafish whole mount staining using the RNAscope Multiplex Fluorescent Reagent Kit v2 (ACD Bio) was performed as described previously (Gross-Thebing et al., 2014) with modifications. Briefly, 17 hpf embryos were fixed for 1 hour in 4% paraformaldehyde, washed with PBST (PBS with 0.1%Tween), and dehydrated in MeOH at −20°C overnight. Embryos were dried for 30 minutes at room temperature before Protease III treatment. RNAscope Dr-wnt5b Probe C1 (ACD Bio) was hybridized overnight at 40°C. Embryos were postfixed with 4% paraformaldehyde and washed with 0.2× SSCT before the amplification steps (Amp1–3). To develop the probe signal, embryos were treated with HRP-C1 before Opal 570 dye (Akoya Biosciences, diluted in TSA buffer) incubation, and then treated with HRP-blocker. Fluorescent images were obtained using a Nikon C2 confocal microscope.
Optical mapping for zebrafish embryos
Optical mapping was performed as previously described (Chi et al., 2008). Briefly, individual Tg(myl7:gCaMP) zebrafish embryos at 48 hpf were treated with 10 mM 2,3-butanedione monoxime (Sigma) for 15 minutes to achieve electro-mechanical uncoupling. Embryos were then mounted in 1% low melting point agarose (Lonza) in 35 mm glass bottom petri dishes (MatTek). Epifluorescence images of the heart were obtained with a Nikon Eclipse Ti inverted microscope, using a 20x Plan Apo air objective, a Nikon INTENSILIGHT C-HGFIE Precentered Fiber Illuminator, and standard fluorescein isothiocyanate filter set. Real-time images were captured with an Andor iXon EMCCD camera at a frame rate of 60 ms/frame and processed as described in the Quantification and statistical analysis section.
Chromatin immunoprecipitation (ChIP) studies
Using the GRC Zv9 zebrafish genome assembly, regulatory regions of isl1 or tbx18 genes were interrogated to discover putative TCF/LEF binding sites based on the TCF/LEF consensus sequences CAAAGG (Lin et al., 2007). Identified sites were then matched to previously published regions of H3K27ac and H3K4me1 occupancy (i.e. active enhancers) in the genome of 24 hpf zebrafish embryos (GEO: GSE32483) (Bogdanovic et al., 2012). Primers (see Table S2) were then designed to span these overlapping regions for ChIP quantitative PCR (qPCR) studies. An illustration designating the location of the putative TCF/LEF binding sites (Site 1) and possible active enhancer peaks can be found in Figures S5A–S5D. Negative control primers were located in non-coding regions (Site 2) at least 1.5 kb away from the putative TCF/LEF sites.
ChIP was performed as previously described (Lindeman et al., 2009) with modifications. Briefly, ~800 wild-type embryos at 21–23 ss were collected, homogenized and cross-linked by incubation in 1% formaldehyde. The chromatin was sonicated to generate ~500 base-pair fragments and pre-cleared with protein A agarose beads (Upstate). Equal amounts of soluble chromatin were incubated with anti-β-catenin antibody (mouse, BD Biosciences), which has been previously used for ChIP (Cha et al., 2016), or control IgG antibody (mouse, Invitrogen). After overnight antibody incubations, protein A agarose beads were added to collect the antibody-chromatin complex. Chromatin fragments were then eluted, reverse cross-linked and purified. These DNA fragments were then used for qPCR in combination with primers spanning the putative TCF/LEF (and control) sites as described above. qPCR was performed on a CFX Connect Real-Time PCR Detection System (Bio-Rad) using Power SYBR Green Master Mix (ThermoFisher Scientific). Values were normalized using the percent input method.
Human pluripotent stem cell (hPSC) studies
Differentiation of hPSCs into cardiomyocytes was performed using established protocols as previously described (Lian et al., 2012). Briefly, H9-hTnnTZ-pGZ-D2 (WiCell) human embryonic stem cells (hESCs) were expanded to 80% confluency on Geltrex (Gibco) coated plates in E8 medium. On D0, cells were cultured with RPMI/B27 without insulin (culture media) containing 12 μM CHIR. After 24 hours, CHIR-treated media was removed and cells were cultured for 48 hours in culture media only. On D3, 5 μM IWP2 was then added to culture media. On D5, this culture media was then replaced with culture media containing 3 μM CHIR or 1000 ng/ml WNT5B (R&D systems) to induce pacemaker cardiomyocyte differentiation, or DMSO or HSA (human serum albumin) to induce control cardiomyocyte differentiation. On D7 and onwards, cells were then maintained in culture media + insulin only. Small molecule inhibitors used for the differentiation are detailed in Table S1. Using a BD Influx cell sorter (BD Biosciences), differentiating D15 hPSCs were sorted based on TNNT2:GFP expression and side-scatter to collect TNNT2:GFP+ cardiomyocytes and TNNT2:GFP- non-cardiomyocytes.
For real-time quantitative reverse transcriptase (qRT)-PCR analysis, RNA was extracted from cells (RNeasy mini kit, Qiagen) and used to produce cDNA (Superscript III kit, Invitrogen). qRT-PCR was performed on a CFX Connect Real-Time PCR Detection System (Bio-Rad) using Power SYBR Green Master Mix (ThermoFisher Scientific). RNA expression levels were based on averaging qRT-PCR results measured from three independent cardiomyocyte differentiations. The housekeeping TATA box binding protein (TBP) gene was used to normalize RNA expression levels. Primer sequences are listed in Table S3.
For immunofluorescence staining, sorted D15 TNNT2:GFP+ cells were replated, cultured for another four days and then fixed. Immunofluorescence staining was performed as previously described (Witty et al., 2014) using the following primary antibodies: anti-GFP (chicken, Aves Labs, 1:200), anti-human NKX2–5 (rabbit, cell signaling, 1:800) and anti-human SHOX2 (mouse, Abcam, 1:200). The following secondary antibodies were used: anti-chicken IgG-Alexa 488 (goat, Life technologies, 1:400), anti-rabbit IgG Cy5 (goat, Life technologies, 1:400), and anti-mouse IgG-Alexa 568 (goat, Life technologies, 1:400). DAPI (1 μg/ml, Roche) staining was used to identify nuclei. Stained cells were visualized using a Nikon C2 confocal microscope.
Electrophysiology
Action potentials were recorded as previously described (Veevers et al., 2018). Specifically, TNNT2:GFP+ cells were sorted, replated on plastic bottom dishes and cultured for an additional 4 days as described in the hPSC studies section. Single cell spontaneous action potentials were recorded under whole-cell current-clamp conditions with a patch pipette resistance of 3–5 M. Cells were incubated in an external solution containing: 150 mM NaCl, 5 mM KCl, 1 mM CaCl2, 1 mM MgCl2, 10 mM HEPES and 10 mM glucose (adjusted to pH 7.4, with NaOH). The intracellular pipette solution used for cell electrophysiology studies contained the following: 150 mM KCl, 5 mM NaCl, 1 mM MgCl2, 2 mM EGTA, 1 mM MgATP, 10 mM HEPES (adjusted to pH 7.2 with KOH). Patch clamp recordings were performed using an Axopatch 200B amplifier and pClamp 10.3 software (Molecular devices, LLC). All experiments were performed at room temperature (20–22°C).
3D bioprinting and optical mapping of hPSC-derived cardiomyocyte “mini-hearts”
To bioprint hPSC-derived cardiomyocyte mini-hearts, TNNT2:GFP hPSCs were initially differentiated into D15 cardiomyocytes as described in the hPSC studies section. These hPSC-cardiomyocytes were then FACS sorted based on TNNT2:GFP expression and resuspended at 200 million cardiomyocytes/ml. This hPSC-cardiomyocyte suspension was mixed at a 1:1 ratio with a pre-warmed prepolymer solution consisting of 10% (wt/vol) gelatin methacrylate (GelMA), which was synthesized as previously reported (Ma et al., 2016), and 0.3% (wt/vol) lithium phenyl-2 4 6-trimethylbenzoylphosphinate (LAP) (Ma et al., 2016). The two-step process to bioprint a 3D heart-shaped construct involves printing D15 hPSC-cardiomyocytes that were treated with DMSO from D5–7 into a heart-shape with a circular region missing in the upper left region, followed by printing a complementary circular structure containing D15 hPSC-cardiomyocytes that were either treated with CHIR or DMSO (control) from D5–7 into the empty region. For each printing step, 20 μl of cell-material mixture was pipetted into the space between a methacrylated coverslip fixed on the motion controller stage and a polydimethylsiloxane (PDMS) film attached to a glass slide. UV light (88 mW/cm2) was projected to the stage after loading the first pattern into a digital micromirror device (DMD) chip. This first bioprinted structure was washed three times with warm PBS and aspirated dry. The second cell-material mixture was then pipetted into the space between the same coverslip and PDMS film for a second light exposure. Afterwards, the bioprinted sample was washed in both PBS and media, and then incubated at 37°C with 5% CO2. Culture media was replaced every other day. The height of the construct was set to be 200 μm to allow sufficient oxygen diffusion within the hydrogel encapsulating the cardiomyocytes.
Optical mapping for the 3D bioprinted hearts was performed 7 days after printing. Coverslips with 3D bioprinted mini-hearts were placed into 35 mm glass bottom petri dishes (MatTek). Mini-heart cardiomyocytes were loaded with 10 μM Rhod2-AM (Molecular Probes) for 20 min at 37°C, and then epifluorescence images were obtained with a Nikon Eclipse Ti inverted microscope, using 10x Plan Apo air objective, a Nikon INTENSILIGHT C-HGFIE Precentered Fiber Illuminator, and standard tetramethylrhodamine filter set. Real-time images were acquired with an Andor iXon EMCCD camera at a frame rate of 40 ms/frame and processed as described in the Quantification and statistical analysis section.
Cardiomyocyte beating rate analysis
To examine cardiomyocyte beating rate, TNNT2:GFP+ cells were sorted as described in the hPSC studies section, replated at low density and cultured for another 4 days before the beating rate of individual cardiomyocytes was manually counted. For 3D mini-heart beating rate analysis, the beating rate for each mini-heart was manually counted 7 days after printing when mini-heart beating is synchronized.
QUANTIFICATION AND STATISTICAL ANALYSIS
All fluorescent images were obtained with a Nikon C2 confocal microscope or a Nikon Eclipse Ti inverted fluorescence microscope coupled with an Andor iXon EMCCD camera, and processed using Nikon NIS Elements software, ImageJ and Adobe Photoshop. To count cell numbers, 3D-reconstructions from individual confocal slices were used for manual quantification. To determine the average percentage of Shox2+ or Nkx2.5- hPSC-cardiomyocytes derived from hPSC-cardiac progenitors treated with DMSO or CHIR between D5–7, eight and nine equally-sized microscope images (210 sq microns) were respectively examined from representative regions across three independent cardiomyocyte differentiations for DMSO and CHIR conditions. As a result, a total of 812 and 729 hPSC-cardiomyocytes were analyzed for DMSO or CHIR treatment conditions, respectively. To determine the average percentage of Shox2+ or Nkx2.5- hPSC-cardiomyocytes derived from hPSC-cardiac progenitors treated with HSA or WNT5B between D5–7, eight equally-sized microscope images (210 sq microns) were respectively examined from representative regions across three independent cardiomyocyte differentiations for HSA and WNT5B conditions. As a result, a total of 756 and 612 hPSC-cardiomyocytes were analyzed for HSA or WNT5B treatment conditions, respectively. For optical mapping of calcium activation, images were processed by first identifying the minimum and the maximum values of each pixel across all time points. These values were used to normalize the recorded fluorescence signal intensity at individual time points. Isochronal lines at 60 or 40 ms intervals were drawn as contiguous lines across the maximal fluorescent pixels (Chi et al., 2008). No statistical methods were used to predetermine sample size. Animals were assigned to experimental groups using simple randomization, without investigator blinding. Unpaired two-tailed Student’s t-tests were used to determine statistical significance. P< 0.05 was considered to be statistically significant, as indicated by an asterisk. The standard error of the mean (s.e.m.) was used for error bars.
DATA AND CODE AVAILABILITY
This study did not generate any datasets or code.
Supplementary Material
Video S1. Mini-hearts printed with DMSO-treated control cardiomyocytes in the upper left region randomly initiate cardiac conduction. Related to Figure 7. Representative video of calcium activation in a mini-heart with DMSO-treated control cardiomyocytes in the upper left region shows randomly initiated cardiac conduction. White asterisk labels the site where DMSO-treated control cardiomyocytes are printed. White arrow points to the cardiac conduction initiation site.
Video S2. Mini-hearts printed with CHIR-treated pacemaker-like cardiomyocytes in the upper left region initiate cardiac conduction from the location of CHIR-treated cardiomyocytes. Related to Figure 7. Representative video of calcium activation in a mini-heart printed with CHIR-treated pacemaker-like cardiomyocytes in the upper left region shows that cardiac conduction stably initiates from the site where CHIR-treated cardiomyocytes are printed. White asterisk labels the site where CHIR-treated cardiomyocytes are printed. White arrow points to the cardiac conduction initiation site.
Video S3. Mini-hearts printed with DMSO-treated control cardiomyocytes in the upper left region exhibit slower synchronized beating. Related to Figure 7. Representative bright-field video of a mini-heart printed with DMSO-treated control cardiomyocytes in the upper left region shows slower synchronized beating than those printed with CHIR-treated pacemaker-like cardiomyocytes.
Video S4. Mini-hearts printed with CHIR-treated pacemaker-like cardiomyocytes in the upper left region exhibit faster synchronized beating. Related to Figure 7. Representative bright-field video of a mini-heart printed with CHIR-treated pacemaker-like cardiomyocytes in the upper left region shows faster synchronized beating than those printed with DMSO-treated control cardiomyocytes.
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-dsRed antibody | Clontech | Cat# 632496 |
| Rabbit anti-Isl1 antibody | Genetex | Cat# GTX128201 |
| Mouse anti-Isl1 antibody | DSHB | Cat# 39.4D5 |
| Mouse anti-MF20 antibody | DSHB | Cat# MF20 |
| Rabbit anti-Nkx2.5 anitibody | Genetex | Cat# GTX128357 |
| Rabbit anti-phospho-histone H3 antibody | Upstate | Cat# 06–570 |
| Anti-Digoxigenin-AP, Fab fragments | Sigma Aldrich | Cat# 11093274910 |
| Mouse anti β-catenin antibody | BD Biosciences | Cat# 610153 |
| Mouse IgG antibody | Invitrogen | Cat# 02–6502 |
| Chicken anti-GFP antibody | Aves Labs | Cat# GFP-1020 |
| Rabbit anti-human NKX2–5 antibody | Cell signaling | Cat# 8792 |
| Mouse anti-human SHOX2 antibody | Abcam | Cat# ab55740 |
| Goat anti-rabbit IgG-Alexa 568 | Thermo Fisher Scientific | Cat# A-11011 |
| Goat anti-mouse IgG Cy5 | Thermo Fisher Scientific | Cat# A-10524 |
| Goat anti-mouse IgG-Alexa 405 | Thermo Fisher Scientific | Cat# A-31553 |
| Goat anti-rabbit IgG-Alexa 488 | Thermo Fisher Scientific | Cat# A-11008 |
| Goat anti-chicken IgG-Alexa 488 | Thermo Fisher Scientific | Cat# A-11039 |
| Goat anti-rabbit IgG Cy5 | Thermo Fisher Scientific | Cat# A-10523 |
| Goat anti-mouse IgG-Alexa 568 | Thermo Fisher Scientific | Cat# A-11004 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| PNU-74654 | Sigma Aldrich | Cat# P0052 |
| CHIR99021 | Sigma Aldrich | Cat# SML1046 |
| CHIR99021 | Tocris | Cat# 4423 |
| TNP-470 | Sigma Aldrich | Cat# T1455 |
| IWP2 | Tocris | Cat# 3533 |
| 2,3-butanedione monoxime | Sigma Aldrich | Cat# B0753 |
| Human Wnt5b protein | R&D systems | Cat# 7347-WN |
| Critical Commercial Assays | ||
| in situ cell death detection kit, TMR red | Roche | Cat# 12156792910 |
| RNAscope Multiplex Fluorescent Reagent Kit v2 | ACD Bio | Cat# 323100 |
| RNAscope Dr-wnt5b Probe C1 | ACD Bio | Cat# 579321-C1 |
| RNeasy mini kit | Qiagen | Cat# 74104 |
| Superscript III kit | Invitrogen | Cat# 18080051 |
| Power SYBR Green Master Mix | Thermo Fisher Scientific | Cat# 4367659 |
| Experimental Models: Cell Lines | ||
| Human: H9-hTnnTZ-pGZ-D2 human embryonic stem cells | WiCell | H9-hTnnTZ-pGZ-D2 |
| Experimental Models: Organisms/Strains | ||
| Zebrafish: TgBAC(−36nkx2.5:Kaede) | Burns Lab, Massachusetts General Hospital | fb9Tg |
| Zebrafish: Tg(myl7:Cerulean) | Appel Lab, University of Colorado Denver | co19Tg |
| Zebrafish: Tg(hsp70l:nkx2.5-EGFP) | Targoff Lab, Columbia University | fcu1Tg |
| Zebrafish: Tg(7xTCF-Xla.Siam:nlsmCherry) | Argenton Lab, University of Padova | ia5Tg |
| Zebrafish: Tg(−36nkx2.5:ZsYellow) | Burns Lab, Massachusetts General Hospital | fb7Tg |
| Zebrafish: Tg(myl7:eGFP) | Tsai Lab, National Taiwan University | twu277Tg |
| Zebrafish: Tg(hsp70:dnTCF-GFP) | David Raible Lab, University of Washington | w26Tg |
| Zebrafish: Tg(myl7:gCaMP) | Stainier Lab, Max Planck Institute for Heart and Lung Research | s878Tg |
| Zebrafish: Tg(hsp70l:wnt5b-GFP) | Randall Moon Lab, University of Washington | w33Tg |
| Zebrafish: Tg(myl7:H2A-mCherry) | Yelon Lab, University of California, San Diego | sd12Tg |
| Zebrafish: Tg(amhc:eGFP) | Stainier Lab, Max Planck Institute for Heart and Lung Research | s958Tg |
| Zebrafish: nkx2.5vu179/vu179 | Lila Solnica-Krezel Lab, Vanderbilt University | vu179 |
| Zebrafish: wnt5b ta98/ta98 | Nusslein-Volhard Lab, Planck Institute for Heart and Lung Research | ta98 |
| Zebrafish: apcsd58/sd58 | This manuscript | sd58 |
| Oligonucleotides | ||
| Primers for ChIP-qPCR, see Table S2 | This paper | N/A |
| Primers for qRT-PCR, see Table S3 | This paper | N/A |
| Software and Algorithms | ||
| Nikon NIS Elements software | Nikon | N/A |
| Fiji/ImageJ | NIH | https://imagej.net/Fiji/Downloads |
| Adobe Photoshop CS5 | Adobe | N/A |
| FlowJo version 10 | FlowJo | https://www.flowjo.com/solutions/flowjo/downloads |
| GraphPad Prism 7 | GraphPad | N/A |
| pClamp 10.3 software | Molecular Devices | N/A |
Highlights:
Pacemaker cardiomyocytes derive from a subset of Nkx2.5+ mesoderm
Canonical Wnt5b signaling directs pacemaker cardiomyocyte differentiation in vivo
Wnt5b signaling promotes hPSC-pacemaker cardiomyocyte differentiation in vitro
hPSC-pacemaker cardiomyocytes can pace 3D bioprinted hPSC-cardiomyocytes
Acknowledgements:
We thank N. Tedeschi for fish care; Z. Huang for experimental assistance; S. M. Evans, D. Yelon and N. C. Chi lab members for comments on the manuscript; G. Crump and W. Herzog for plasmids; C. G. Burns for the Nkx2.5 photoconversion line; K. L. Targoff for the heat-shock Nkx2.5 line; F. Argenton for the canonical Wnt signaling reporter line. This work was supported in part by grants from the American Heart Association (15POST23090027) to J.R., the Saving Tiny Hearts Foundation and NIH (R01HL108599) to D.Y., CIRM (RT3–07899) and NIH (R01EB021857) to S.C., and NIH to N.C.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests: The authors declare no competing interests.
References:
- Ai D, Fu X, Wang J, Lu MF, Chen L, Baldini A, Klein WH, and Martin JF (2007). Canonical Wnt signaling functions in second heart field to promote right ventricular growth. Proc. Natl. Acad. Sci. U.S.A 104, 9319–9324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrenberg AB, Stainier DY, Baier H, and Huisken J (2010). Optogenetic control of cardiac function. Science 330, 971–974. [DOI] [PubMed] [Google Scholar]
- Bakker ML, Boink GJ, Boukens BJ, Verkerk AO, van den Boogaard M, den Haan AD, Hoogaars WM, Buermans HP, de Bakker JM, Seppen J, et al. (2012). T-box transcription factor TBX3 reprogrammes mature cardiac myocytes into pacemaker-like cells. Cardiovasc. Res 94, 439–449. [DOI] [PubMed] [Google Scholar]
- Bao X, Lian X, Hacker TA, Schmuck EG, Qian T, Bhute VJ, Han T, Shi M, Drowley L, Plowright A, et al. (2016). Long-term self-renewing human epicardial cells generated from pluripotent stem cells under defined xeno-free conditions. Nat. Biomed. Eng 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrow JR, Howell WD, Rule M, Hayashi S, Thomas KR, Capecchi MR, and McMahon AP (2007). Wnt3 signaling in the epiblast is required for proper orientation of the anteroposterior axis. Dev. Biol 312, 312–320. [DOI] [PubMed] [Google Scholar]
- Birket MJ, Ribeiro MC, Verkerk AO, Ward D, Leitoguinho AR, den Hartogh SC, Orlova VV, Devalla HD, Schwach V, Bellin M, et al. (2015). Expansion and patterning of cardiovascular progenitors derived from human pluripotent stem cells. Nature Biotechnol. 33, 970–979. [DOI] [PubMed] [Google Scholar]
- Blaschke RJ, Hahurij ND, Kuijper S, Just S, Wisse LJ, Deissler K, Maxelon T, Anastassiadis K, Spitzer J, Hardt SE, et al. (2007). Targeted mutation reveals essential functions of the homeodomain transcription factor Shox2 in sinoatrial and pacemaking development. Circulation 115, 1830–1838. [DOI] [PubMed] [Google Scholar]
- Bogdanovic O, Fernandez-Minan A, Tena JJ, de la Calle-Mustienes E, Hidalgo C, van Kruysbergen I, van Heeringen SJ, Veenstra GJ, and Gomez-Skarmeta JL (2012). Dynamics of enhancer chromatin signatures mark the transition from pluripotency to cell specification during embryogenesis. Genome Res. 22, 2043–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressan M, Liu G, and Mikawa T (2013). Early mesodermal cues assign avian cardiac pacemaker fate potential in a tertiary heart field. Science 340, 744–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu L, Jiang X, Martin-Puig S, Caron L, Zhu S, Shao Y, Roberts DJ, Huang PL, Domian IJ, and Chien KR (2009). Human ISL1 heart progenitors generate diverse multipotent cardiovascular cell lineages. Nature 460, 113–117. [DOI] [PubMed] [Google Scholar]
- Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, and Evans S (2003). Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev. Cell 5, 877–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai CL, Martin JC, Sun Y, Cui L, Wang L, Ouyang K, Yang L, Bu L, Liang X, Zhang X, et al. (2008). A myocardial lineage derives from Tbx18 epicardial cells. Nature 454, 104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao N, Huang Y, Zheng J, Spencer CI, Zhang Y, Fu JD, Nie B, Xie M, Zhang M, Wang H, et al. (2016). Conversion of human fibroblasts into functional cardiomyocytes by small molecules. Science 352, 1216–1220. [DOI] [PubMed] [Google Scholar]
- Cha B, Geng X, Mahamud MR, Fu J, Mukherjee A, Kim Y, Jho EH, Kim TH, Kahn ML, Xia L, et al. (2016). Mechanotransduction activates canonical Wnt/beta-catenin signaling to promote lymphatic vascular patterning and the development of lymphatic and lymphovenous valves. Genes Dev. 30, 1454–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY, DeRan MT, Ignatius MS, Grandinetti KB, Clagg R, McCarthy KM, Lobbardi RM, Brockmann J, Keller C, Wu X, et al. (2014). Glycogen synthase kinase 3 inhibitors induce the canonical WNT/beta-catenin pathway to suppress growth and self-renewal in embryonal rhabdomyosarcoma. Proc. Natl. Acad. Sci. U.S.A 111, 5349–5354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi NC, Shaw RM, Jungblut B, Huisken J, Ferrer T, Arnaout R, Scott I, Beis D, Xiao T, Baier H, et al. (2008). Genetic and physiologic dissection of the vertebrate cardiac conduction system. PLoS Biol. 6, e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffels VM, Grieskamp T, Norden J, Mommersteeg MT, Rudat C, and Kispert A (2009). Tbx18 and the fate of epicardial progenitors. Nature 458, E8–9; discussion E9–10. [DOI] [PubMed] [Google Scholar]
- Christoffels VM, Mommersteeg MT, Trowe MO, Prall OW, de Gier-de Vries C, Soufan AT, Bussen M, Schuster-Gossler K, Harvey RP, Moorman AF, et al. (2006). Formation of the venous pole of the heart from an Nkx2–5-negative precursor population requires Tbx18. Circ. Res 98, 1555–1563. [DOI] [PubMed] [Google Scholar]
- Cohen ED, Wang Z, Lepore JJ, Lu MM, Taketo MM, Epstein DJ, and Morrisey EE (2007). Wnt/beta-catenin signaling promotes expansion of Isl-1-positive cardiac progenitor cells through regulation of FGF signaling. J. Clin. Invest 117, 1794–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo S, de Sena-Tomas C, George V, Werdich AA, Kapur S, MacRae CA, and Targoff KL (2018). Nkx genes establish second heart field cardiomyocyte progenitors at the arterial pole and pattern the venous pole through Isl1 repression. Development 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza-Lewis RA, Yu L, He F, Liu H, Tang R, Shi J, Sun X, Martin JF, Wang D, Yang J, et al. (2009). Shox2 is essential for the differentiation of cardiac pacemaker cells by repressing Nkx2–5. Dev. Biol 327, 376–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui H, Miyazaki T, Chow RW, Ishikawa H, Nakajima H, Vermot J, and Mochizuki N (2018). Hippo signaling determines the number of venous pole cells that originate from the anterior lateral plate mesoderm in zebrafish. eLife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George V, Colombo S, and Targoff KL (2015). An early requirement for nkx2.5 ensures the first and second heart field ventricular identity and cardiac function into adulthood. Dev. Biol 400, 10–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross-Thebing T, Paksa A, and Raz E (2014). Simultaneous high-resolution detection of multiple transcripts combined with localization of proteins in whole-mount embryos. BMC Biol 12, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guner-Ataman B, Paffett-Lugassy N, Adams MS, Nevis KR, Jahangiri L, Obregon P, Kikuchi K, Poss KD, Burns CE, and Burns CG (2013). Zebrafish second heart field development relies on progenitor specification in anterior lateral plate mesoderm and nkx2.5 function. Development 140, 1353–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerschmidt M, Pelegri F, Mullins MC, Kane DA, Brand M, van Eeden FJ, Furutani-Seiki M, Granato M, Haffter P, Heisenberg CP, et al. (1996). Mutations affecting morphogenesis during gastrulation and tail formation in the zebrafish, Danio rerio. Development 123, 143–151. [DOI] [PubMed] [Google Scholar]
- Han P, Bloomekatz J, Ren J, Zhang R, Grinstein JD, Zhao L, Burns CG, Burns CE, Anderson RM, and Chi NC (2016). Coordinating cardiomyocyte interactions to direct ventricular chamber morphogenesis. Nature 534, 700–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogaars WM, Engel A, Brons JF, Verkerk AO, de Lange FJ, Wong LY, Bakker ML, Clout DE, Wakker V, Barnett P, et al. (2007). Tbx3 controls the sinoatrial node gene program and imposes pacemaker function on the atria. Genes Dev. 21, 1098–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CJ, Tu CT, Hsiao CD, Hsieh FJ, and Tsai HJ (2003). Germ-line transmission of a myocardium-specific GFP transgene reveals critical regulatory elements in the cardiac myosin light chain 2 promoter of zebrafish. Dev. Dyn 228, 30–40. [DOI] [PubMed] [Google Scholar]
- Hurlstone AF, Haramis AP, Wienholds E, Begthel H, Korving J, Van Eeden F, Cuppen E, Zivkovic D, Plasterk RH, and Clevers H (2003). The Wnt/beta-catenin pathway regulates cardiac valve formation. Nature 425, 633–637. [DOI] [PubMed] [Google Scholar]
- Iyer D, Gambardella L, Bernard WG, Serrano F, Mascetti VL, Pedersen RA, Talasila A, and Sinha S (2015). Robust derivation of epicardium and its differentiated smooth muscle cell progeny from human pluripotent stem cells. Development 142, 1528–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspard B, Couffinhal T, Dufourcq P, Moreau C, and Duplaa C (2000). Expression pattern of mouse sFRP-1 and mWnt-8 gene during heart morphogenesis. Mech. Dev 90, 263–267. [DOI] [PubMed] [Google Scholar]
- Kapoor N, Liang W, Marban E, and Cho HC (2013). Direct conversion of quiescent cardiomyocytes to pacemaker cells by expression of Tbx18. Nature Biotechnol. 31, 54–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith A, and Flack M (1907). The Form and Nature of the Muscular Connections between the Primary Divisions of the Vertebrate Heart. J. Anat. Physiol 41, 172–189. [PMC free article] [PubMed] [Google Scholar]
- Kilian B, Mansukoski H, Barbosa FC, Ulrich F, Tada M, and Heisenberg CP (2003). The role of Ppt/Wnt5 in regulating cell shape and movement during zebrafish gastrulation. Mech. Dev 120, 467–476. [DOI] [PubMed] [Google Scholar]
- Kwon C, Arnold J, Hsiao EC, Taketo MM, Conklin BR, and Srivastava D (2007). Canonical Wnt signaling is a positive regulator of mammalian cardiac progenitors. Proc. Natl. Acad. Sci. U.S.A 104, 10894–10899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon C, Qian L, Cheng P, Nigam V, Arnold J, and Srivastava D (2009). A regulatory pathway involving Notch1/beta-catenin/Isl1 determines cardiac progenitor cell fate. Nat. Cell Biol 11, 951–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laflamme MA, and Murry CE (2011). Heart regeneration. Nature 473, 326–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescroart F, Mohun T, Meilhac SM, Bennett M, and Buckingham M (2012). Lineage tree for the venous pole of the heart: clonal analysis clarifies controversial genealogy based on genetic tracing. Circ. Res 111, 1313–1322. [DOI] [PubMed] [Google Scholar]
- Lewis JL, Bonner J, Modrell M, Ragland JW, Moon RT, Dorsky RI, and Raible DW (2004). Reiterated Wnt signaling during zebrafish neural crest development. Development 131, 1299–1308. [DOI] [PubMed] [Google Scholar]
- Lian X, Hsiao C, Wilson G, Zhu K, Hazeltine LB, Azarin SM, Raval KK, Zhang J, Kamp TJ, and Palecek SP (2012). Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc. Natl. Acad. Sci. U.S.A 109, E1848–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Wang G, Lin L, Lowe J, Zhang Q, Bu L, Chen Y, Chen J, Sun Y, and Evans SM (2013). HCN4 dynamically marks the first heart field and conduction system precursors. Circ. Res 113, 399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Zhang Q, Cattaneo P, Zhuang S, Gong X, Spann NJ, Jiang C, Cao X, Zhao X, Zhang X, et al. (2015). Transcription factor ISL1 is essential for pacemaker development and function. J. Clin. Invest 125, 3256–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Cui L, Zhou W, Dufort D, Zhang X, Cai CL, Bu L, Yang L, Martin J, Kemler R, et al. (2007). Beta-catenin directly regulates Islet1 expression in cardiovascular progenitors and is required for multiple aspects of cardiogenesis. Proc. Natl. Acad. Sci. U.S.A 104, 9313–9318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeman LC, Vogt-Kielland LT, Alestrom P, and Collas P (2009). Fish’n ChIPs: chromatin immunoprecipitation in the zebrafish embryo. Methods Mol. Biol 567, 75–86. [DOI] [PubMed] [Google Scholar]
- Liu P, Wakamiya M, Shea MJ, Albrecht U, Behringer RR, and Bradley A (1999). Requirement for Wnt3 in vertebrate axis formation. Nat. Genet 22, 361–365. [DOI] [PubMed] [Google Scholar]
- Ma X, Qu X, Zhu W, Li YS, Yuan S, Zhang H, Liu J, Wang P, Lai CS, Zanella F, et al. (2016). Deterministically patterned biomimetic human iPSC-derived hepatic model via rapid 3D bioprinting. Proc. Natl. Acad. Sci. U.S.A 113, 2206–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews ES, Mawdsley DJ, Walker M, Hines JH, Pozzoli M, and Appel B (2014). Mutation of 3-hydroxy-3-methylglutaryl CoA synthase I reveals requirements for isoprenoid and cholesterol synthesis in oligodendrocyte migration arrest, axon wrapping, and myelin gene expression. J. Neurosci 34, 3402–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merks AM, Swinarski M, Meyer AM, Muller NV, Ozcan I, Donat S, Burger A, Gilbert S, Mosimann C, Abdelilah-Seyfried S, et al. (2018). Planar cell polarity signalling coordinates heart tube remodelling through tissue-scale polarisation of actomyosin activity. Nat. Commun 9, 2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mommersteeg MT, Dominguez JN, Wiese C, Norden J, de Gier-de Vries C, Burch JB, Kispert A, Brown NA, Moorman AF, and Christoffels VM (2010). The sinus venosus progenitors separate and diversify from the first and second heart fields early in development. Cardiovasc. Res 87, 92–101. [DOI] [PubMed] [Google Scholar]
- Mommersteeg MT, Hoogaars WM, Prall OW, de Gier-de Vries C, Wiese C, Clout DE, Papaioannou VE, Brown NA, Harvey RP, Moorman AF, et al. (2007). Molecular pathway for the localized formation of the sinoatrial node. Circ. Res 100, 354–362. [DOI] [PubMed] [Google Scholar]
- Moro E, Ozhan-Kizil G, Mongera A, Beis D, Wierzbicki C, Young RM, Bournele D, Domenichini A, Valdivia LE, Lum L, et al. (2012). In vivo Wnt signaling tracing through a transgenic biosensor fish reveals novel activity domains. Dev. Biol 366, 327–340. [DOI] [PubMed] [Google Scholar]
- Nicenboim J, Malkinson G, Lupo T, Asaf L, Sela Y, Mayseless O, Gibbs-Bar L, Senderovich N, Hashimshony T, Shin M, et al. (2015). Lymphatic vessels arise from specialized angioblasts within a venous niche. Nature 522, 56–61. [DOI] [PubMed] [Google Scholar]
- Pradhan A, and Olsson PE (2014). Juvenile ovary to testis transition in zebrafish involves inhibition of ptges. Biol. Reprod 91, 33. [DOI] [PubMed] [Google Scholar]
- Protze SI, Liu J, Nussinovitch U, Ohana L, Backx PH, Gepstein L, and Keller GM (2017). Sinoatrial node cardiomyocytes derived from human pluripotent cells function as a biological pacemaker. Nature Biotechnol. 35, 56–68. [DOI] [PubMed] [Google Scholar]
- Qyang Y, Martin-Puig S, Chiravuri M, Chen S, Xu H, Bu L, Jiang X, Lin L, Granger A, Moretti A, et al. (2007). The renewal and differentiation of Isl1+ cardiovascular progenitors are controlled by a Wnt/beta-catenin pathway. Cell Stem Cell 1, 165–179. [DOI] [PubMed] [Google Scholar]
- Rauch GJ, Hammerschmidt M, Blader P, Schauerte HE, Strahle U, Ingham PW, McMahon AP, and Haffter P (1997). Wnt5 is required for tail formation in the zebrafish embryo. Cold Spring Harb. Symp. Quant. Biol 62, 227–234. [PubMed] [Google Scholar]
- Schumacher JA, Bloomekatz J, Garavito-Aguilar ZV, and Yelon D (2013). tal1 Regulates the formation of intercellular junctions and the maintenance of identity in the endocardium. Dev. Biol 383, 214–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley EG, Biben C, Elefanty A, Barnett L, Koentgen F, Robb L, and Harvey RP (2002). Efficient Cre-mediated deletion in cardiac progenitor cells conferred by a 3’UTR-ires-Cre allele of the homeobox gene Nkx2–5. Int. J. Dev. Biol 46, 431–439. [PubMed] [Google Scholar]
- Stieber J, Herrmann S, Feil S, Loster J, Feil R, Biel M, Hofmann F, and Ludwig A (2003). The hyperpolarization-activated channel HCN4 is required for the generation of pacemaker action potentials in the embryonic heart. Proc. Natl. Acad. Sci. U.S.A 100, 15235–15240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoick-Cooper CL, Weidinger G, Riehle KJ, Hubbert C, Major MB, Fausto N, and Moon RT (2007). Distinct Wnt signaling pathways have opposing roles in appendage regeneration. Development 134, 479–489. [DOI] [PubMed] [Google Scholar]
- Sun Y, Liang X, Najafi N, Cass M, Lin L, Cai CL, Chen J, and Evans SM (2007). Islet 1 is expressed in distinct cardiovascular lineages, including pacemaker and coronary vascular cells. Dev. Biol 304, 286–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Targoff KL, Colombo S, George V, Schell T, Kim SH, Solnica-Krezel L, and Yelon D (2013). Nkx genes are essential for maintenance of ventricular identity. Development 140, 4203–4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessadori F, van Weerd JH, Burkhard SB, Verkerk AO, de Pater E, Boukens BJ, Vink A, Christoffels VM, and Bakkers J (2012). Identification and functional characterization of cardiac pacemaker cells in zebrafish. PloS one 7, e47644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautwein W, and Uchizono K (1963). Electron Microscopic and Electrophysiologic Study of the Pacemaker in the Sino-Atrial Node of the Rabbit Heart. Z. Zellforch. Microsk. Anat 61, 96–109. [DOI] [PubMed] [Google Scholar]
- Ueno S, Weidinger G, Osugi T, Kohn AD, Golob JL, Pabon L, Reinecke H, Moon RT, and Murry CE (2007). Biphasic role for Wnt/beta-catenin signaling in cardiac specification in zebrafish and embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A 104, 9685–9690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Weerd JH, and Christoffels VM (2016). The formation and function of the cardiac conduction system. Development 143, 197–210. [DOI] [PubMed] [Google Scholar]
- Vedantham V, Galang G, Evangelista M, Deo RC, and Srivastava D (2015). RNA sequencing of mouse sinoatrial node reveals an upstream regulatory role for Islet-1 in cardiac pacemaker cells. Circ. Res 116, 797–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veevers J, Farah EN, Corselli M, Witty AD, Palomares K, Vidal JG, Emre N, Carson CT, Ouyang K, Liu C, et al. (2018). Cell-Surface Marker Signature for Enrichment of Ventricular Cardiomyocytes Derived from Human Embryonic Stem Cells. Stem Cell Reports 11, 828–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada R, Muraoka N, Inagawa K, Yamakawa H, Miyamoto K, Sadahiro T, Umei T, Kaneda R, Suzuki T, Kamiya K, et al. (2013). Induction of human cardiomyocyte-like cells from fibroblasts by defined factors. Proc. Natl. Acad. Sci. U.S.A 110, 12667–12672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Bai Y, Li N, Ye W, Zhang M, Greene SB, Tao Y, Chen Y, Wehrens XH, and Martin JF (2014). Pitx2-microRNA pathway that delimits sinoatrial node development and inhibits predisposition to atrial fibrillation. Proc. Natl. Acad. Sci. U.S.A 111, 9181–9186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese C, Grieskamp T, Airik R, Mommersteeg MT, Gardiwal A, de Gier-de Vries C, Schuster-Gossler K, Moorman AF, Kispert A, and Christoffels VM (2009). Formation of the sinus node head and differentiation of sinus node myocardium are independently regulated by Tbx18 and Tbx3. Circ. Res 104, 388–397. [DOI] [PubMed] [Google Scholar]
- Witty AD, Mihic A, Tam RY, Fisher SA, Mikryukov A, Shoichet MS, Li RK, Kattman SJ, and Keller G (2014). Generation of the epicardial lineage from human pluripotent stem cells. Nature Biotechnol. 32, 1026–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelon D, Horne SA, and Stainier DY (1999). Restricted expression of cardiac myosin genes reveals regulated aspects of heart tube assembly in zebrafish. Dev. Biol 214, 23–37. [DOI] [PubMed] [Google Scholar]
- Zhang R, Han P, Yang H, Ouyang K, Lee D, Lin YF, Ocorr K, Kang G, Chen J, Stainier DY, et al. (2013). In vivo cardiac reprogramming contributes to zebrafish heart regeneration. Nature 498, 497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Cashman TJ, Nevis KR, Obregon P, Carney SA, Liu Y, Gu A, Mosimann C, Sondalle S, Peterson RE, et al. (2011). Latent TGF-beta binding protein 3 identifies a second heart field in zebrafish. Nature 474, 645–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1. Mini-hearts printed with DMSO-treated control cardiomyocytes in the upper left region randomly initiate cardiac conduction. Related to Figure 7. Representative video of calcium activation in a mini-heart with DMSO-treated control cardiomyocytes in the upper left region shows randomly initiated cardiac conduction. White asterisk labels the site where DMSO-treated control cardiomyocytes are printed. White arrow points to the cardiac conduction initiation site.
Video S2. Mini-hearts printed with CHIR-treated pacemaker-like cardiomyocytes in the upper left region initiate cardiac conduction from the location of CHIR-treated cardiomyocytes. Related to Figure 7. Representative video of calcium activation in a mini-heart printed with CHIR-treated pacemaker-like cardiomyocytes in the upper left region shows that cardiac conduction stably initiates from the site where CHIR-treated cardiomyocytes are printed. White asterisk labels the site where CHIR-treated cardiomyocytes are printed. White arrow points to the cardiac conduction initiation site.
Video S3. Mini-hearts printed with DMSO-treated control cardiomyocytes in the upper left region exhibit slower synchronized beating. Related to Figure 7. Representative bright-field video of a mini-heart printed with DMSO-treated control cardiomyocytes in the upper left region shows slower synchronized beating than those printed with CHIR-treated pacemaker-like cardiomyocytes.
Video S4. Mini-hearts printed with CHIR-treated pacemaker-like cardiomyocytes in the upper left region exhibit faster synchronized beating. Related to Figure 7. Representative bright-field video of a mini-heart printed with CHIR-treated pacemaker-like cardiomyocytes in the upper left region shows faster synchronized beating than those printed with DMSO-treated control cardiomyocytes.
Data Availability Statement
This study did not generate any datasets or code.