Abstract
The imbalance of sex chromosomes between females (XX) and males (XY) necessitates strict regulation of X-linked gene expression. X-Chromosome Inactivation (XCI) selects one X for transcriptional silencing in the early embryo, generating an epigenetically distinct and transcriptionally silent X that is maintained into adulthood. Some genes on the inactive X escape XCI, and human somatic cells have a greater number of escape genes compared to mice. Advances with single-cell technologies have revealed human-specific escape genes in fibroblasts and immune cells, some which exhibit cell and tissue specificity. Here, we review recent discoveries of dynamic XCI in female immune cells, which have changed our understanding of XCI maintenance, and discuss how some X-linked genes might become overexpressed in female-biased autoimmunity.
Keywords: X-chromosome Inactivation, Xist RNA, escape genes, human cells, immune cells
Introduction
Gene dosage is a critical component of proper development and must be tightly regulated. In mammals, regulating gene dosage is especially necessary because of the imbalance of the sex chromosomes between females (XX) and males (XY). If not properly controlled, the additional X chromosome in females has the potential for abnormal increased X-linked gene expression, resulting in developmental dysfunction and disease. Female mammals undergo X-Chromosome Inactivation (XCI), an epigenetic process that transcriptionally silences one of the X chromosomes for dosage compensation [1]. XCI is initiated in the early embryo with up-regulated expression of the long non-coding RNA XIST on one X. Xist RNA spreads across the chromosome to recruit heterochromatic marks and enact large-scale transcriptional and conformational changes to generate an inactive X (Xi) [2][3][4]. These distinct features of the Xi must be maintained to prevent aberrant X-linked gene expression. Previously, it was assumed that all somatic cells maintain XCI with the same mechanisms, where XIST RNA and heterochromatic modifications remain localized on the Xi with each cell cycle throughout the lifespan of the organism [5]. Interestingly, the fidelity of XCI maintenance in somatic cells may be lost with age, where one study followed the activity of X-linked ornithine carbamoyl transferase (OCT) in hepatocytes and found evidence of X-reactivation in aged (14–17 months) animals [6]. Recent work from our lab found that XCI maintenance mechanisms are incredibly diverse in the immune system, which impacts our understanding of the connection between X-linked gene dosage and sex-differences in autoimmunity. Additional research revealed cell and tissue heterogeneity of X-linked gene expression in human immune cells and fibroblasts, also suggestive of diversity with XCI maintenance. Here we review recent advances with XCI maintenance and XCI escape in immune cells and human tissues, and discuss the potential contribution of altered XCI maintenance to disease.
Lose your grip: variable escape from XCI
After XCI is established, organisms can tolerate bi-allelic expression of some X-linked genes. The majority of the genes on the Xi are silenced, yet some genes escape silencing in mice (3%) and humans (15–30%) [7,8]. Tissue-specific XCI escape has been determined with various methods including F1 hybrid mouse models [9][8]. Recent advances with single-cell (sc) profiling have enabled allele-specific X-linked gene expression analyses in human cells. Through a combination of deep scRNAseq with whole genome sequencing, X-linked gene expression from the Xa and Xi was distinguished using human samples containing single nucleotide polymorphisms (SNPs). These experiments used primary fibroblasts and lymphoblastoid cells, and revealed heterogeneity in XCI gene escape. Five novel XCI escape genes (INE2, STK26, UQCRBP1, LINC00630, and TTC3P1) were identified in human fibroblasts. For a patient-derived lymphoblastoid cell line, the authors identified four new escape genes (IDS, SLC9A7, STAG2, STK26) [10]. Remarkably, the likelihood of gene escape is different across individual cells, supporting past observations that human XCI escape is variable between individuals [7][11]. The authors attribute this difference to variable XIST expression during the cell cycle, where fibroblasts in the G0 phase of the cell cycle had higher expression levels of XIST [10]. One interesting interpretation of these results is that fluctuations in XIST expression may impact the fidelity of transcriptional silencing across the X, and that some human cells may tolerate short-lived partial X-reactivation. In support, peripheral blood cells from a female patient with a small ring X chromosome lacked XIST expression and had evidence of bi-allelic expression of ZXDA, despite an intact XIST gene [12]. This observation underscores the necessity of XIST expression for Xi gene silencing, but how bi-allelic expression of X-linked genes is tolerated in certain cell types remains a mystery.
Another recent study profiled the heterogeneity of genes escaping repression from the Xi across different tissues. By comparing male and female expression across 29 tissues, the authors suggest that XCI is incomplete in some tissues, and confirmed that sex-biased expression can be used as a proxy for XCI escape [13]. A more direct assessment using 16 tissue samples from a female with 100% skewed XCI verified that about 23% of X-linked genes escape XCI across these samples, and that the magnitude of escape was similar for most genes [13]. However, a small subset of X-linked genes exhibited tissue-specific bi-allelic expression. ScRNAseq and genotype phasing in lymphoblastoid and dendritic cells identified seven putative escape genes (FHL1, FTX, RBMX2, ATP6AP2, CHM, RPL36A, and PHF8). Interestingly, the amount of gene escape varied across cells and individuals [13], in agreement with recent work using human fibroblasts [10]. Together, these studies demonstrate that XCI maintenance in humans is variable at the single cell level, across cell types, and between individuals. It is still unknown how cells can tolerate short-lived dosage imbalances, or the molecular mechanisms responsible for rapid transcriptional changes. While these studies indicate that escape from XCI is dynamic and cell specific, the functional relevance of this variability remains to be determined.
A delicate sense of balance: disrupted X-dosage and increased escape of X-linked genes in autoimmunity
There are a number of immune-related X-linked genes that become overexpressed in female patients with the autoimmune disease systemic lupus erythematosus (SLE) [14,15], and it is possible that this overexpression arises from the Xi. Using single-molecule RNA FISH in primary human T cells, we recently observed that a small percentage of cells have bi-allelic expression of CXCR3 and CD40L [16]. Healthy human B cell lines also had evidence of bi-allelic expression of CXCR3, CD40L, and TLR7 in a subset of cells [16]. Surprisingly, in SLE patient B cell lines, there were more cells that displayed bi-allelic expression of CXCR3, CD40L, and TLR7 compared to healthy control lines [16]. This was the first indication that altered XCI maintenance in female lymphocytes could have a connection to female-biased SLE disease.
A recent study also found that TLR7 escapes XCI in primary human female immune cells [17]. Using an allele-specific polymerase chain reaction assay, bi-allelic expression of TLR7 was observed in healthy human female B cells, monocytes, and plasmacytoid dendritic cells (p-DCs). The authors also found evidence of bi-allelic expression of TLR7 in these same cell types from men with Klinefelter Syndrome (XXY), who also undergo XCI and contain an Xi. Interestingly, both Klinefelter Syndrome males (XXY) and females (XX) have similar risk ratios for developing the autoimmune diseases SLE and Sjorgrens Syndrome [18–21], which suggests that altered X-linked gene dosage may contribute to autoimmune disease susceptibility. It is possible that increased TLR7 escape from XCI in immune cells could be a contributing factor for elevated risk for SLE. However, it is still unknown how these genes escape XCI in immune cells and whether this could be a cause or consequence of disease.
Special snowflakes: immune cells have unique XCI maintenance mechanisms
XCI initiation generates an Xi enriched with Xist RNA and heterochromatin modifications that can be visualized cytologically. The predominant model was that XCI maintenance is the same across all somatic cells. This model was based on studies that examined XCI features using immortalized or primary fibroblasts, various cancer cell lines, differentiated stem cells, or neural precursor cells [22][23][24][25][26][27][28]. One of the first indications that XCI maintenance might be different in some somatic cells came from studies using mouse lymphocytes. Surprisingly, the authors found that some female progenitor B and T cells lacked Xist RNA ‘clouds’ and most progenitor and mature lymphocytes lacked H3K27me3 foci [29]. Allele-specific expression analyses for Xist and four X-linked genes in pre-B, double positive thymocytes, and mature B and T cells revealed no difference in expression, suggesting that dosage compensation was still maintained despite the absence of H3K27me3 and dispersed Xist RNA signals [29]. These results suggested that cells in the immune system have different features of XCI maintenance without full Xi reactivation, yet details regarding whether any gene escapes XCI was lacking.
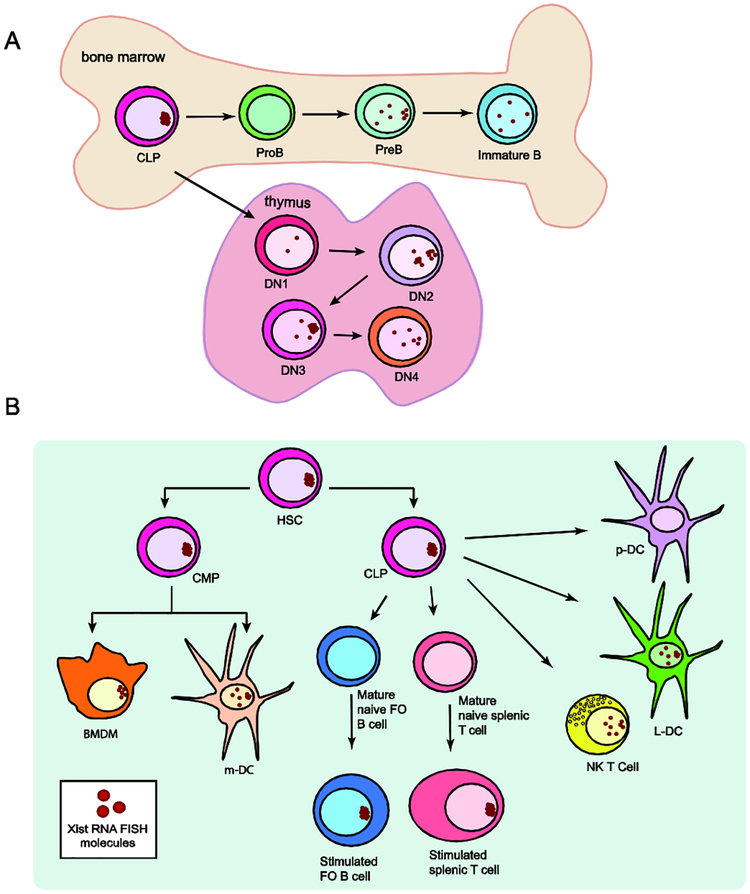
Work from two independent studies examining hematopoietic stem cells and common lymphoid progenitors (CLPs) found canonical localization of Xist RNA transcripts and H3K27me3 similar to fibroblasts [29,30]. We recently reported that Xist RNA was absent from the Xi in pro-B cells, the first B cell progenitor, contradicting earlier reports [29,30]. In pre-B and immature B cell populations, Xist RNA transcripts were not localized at the Xi and were dispersed throughout the nucleus [30] (Fig 1A). Heterochromatin modifications were present at the Xi despite the absence of Xist RNA in pro-B cells, but progressively disappeared from the Xi during subsequent developmental stages. In developing thymocytes, Xist RNA was absent from the Xi in DN1 cells, the first T cell progenitor. However, Xist RNA re-localized to the Xi in DN2 and DN3 thymocytes, then disappeared from the Xi in DN4 thymocytes [31]. While the differences with Xist RNA localization patterns are slight and could be attributed to sorting efficiency and pathogen presence, both studies demonstrate that epigenetic modifications dynamically associate with the Xi during lymphocyte development. The functional relevance for the transient localization of Xist RNA at the Xi and the potential effect on XCI escape for lymphocyte progenitors is currently unknown.
Figure 1: Xist RNA is dynamically localized to the Xi in developing lymphocytes and mature immune cell types.
(A) Schematic cartoon of Xist RNA localization changes in developing B lymphocytes in bone marrow (top) and T cell progenitors in the thymus (bottom). Xist RNA is absent from the Xi in pro-B and DN1 progenitors, and transiently re-appears at the Xi during differentiation. (B) Diversity of Xist RNA localization patterns in immune cell progenitors and seven different mature immune cell types. Xist RNA is not detectable by RNA fluorescence in situ hybridization (FISH) in naïve follicular (FO) B cells, naïve mature CD4+ and CD8+ T cells, and p-DCs. In stimulated B and T lymphocytes, Xist RNA transcripts are tightly clustered to the Xi. Dendritic cells (DCs), bone marrow derived macrophages (BMDMs), and NK T cells have distinct and dispersed patterns of Xist RNA localization. Plasmacytoid dendritic cells (p-DCs), resting and in vitro activated, always lack detectible Xist RNA signals. Red dots represent RNA FISH results for Xist RNA. Abbreviations: HSC (hematopoietic stem cell), CLP (common lymphoid progenitor), CMP (common myeloid progenitor), DN1-DN4 (double negative thymocytes stages 1–4), BMDM (bone marrow derived macrophage), m-DC (myeloid-Dendritic cell), NK T cell (Natural killer T cell), L-DC (lymphoid-Dendritic Cell), p-DC (plasmacytoid dendritic cell).
Interestingly, we recently discovered that mature lymphocytes are the first example of cells that utilize dynamic mechanisms to regulate XCI maintenance. Resting mature T and B cells, from mouse and humans, lacked Xist RNA signals and enrichment of the heterochromatin modifications H3K27me3, H2A-ubiquitin, H4K20me1 and macroH2A on the Xi [16]. In vitro activation of lymphocytes stimulated the return of Xist RNA and some heterochromatin modifications to the Xi, with different kinetics for T or B cells [16]. Xist is continuously transcribed in naïve and activated lymphocytes, therefore Xist RNA transcription and its localization to the Xi are independent processes.
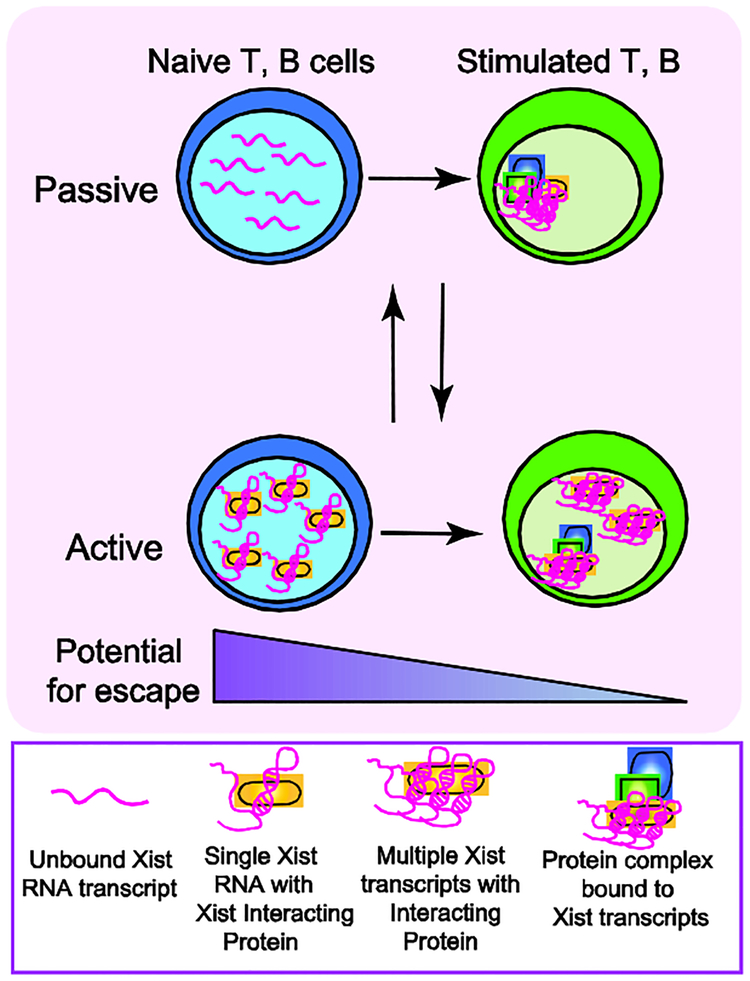
Rapid recruitment of Xist RNA and heterochromatin modifications to the Xi during T or B cell activation is mediated by at least two Xist RNA interacting proteins [32][33]: YY1 and HnRNP-U (SAF-A). Using both mouse and human lymphocytes, we found that these proteins were required to recruit and tether Xist RNA to the Xi during lymphocyte activation, and maintain X-linked gene expression [16,30]. Given the large number of Xist RNA interacting proteins [34–37], it is likely that other factors besides YY1 and HnRNP-U will have similar roles during lymphocyte activation. Incorporating recent discoveries of dynamic XCI maintenance in lymphocytes, we propose a model for the mechanism of Xist RNA localization during lymphocyte activation. This process could be dictated by either passive or active mechanisms (Fig 2). In quiescent mature lymphocytes, newly synthesized Xist RNA transcripts could passively diffuse away from the Xi, and if Xist RNA interacting proteins are limiting, individual and possibly unfolded transcripts could become dispersed across the nucleus. After lymphocyte stimulation, YY1 and HnRNP-U (and likely other XIST RNA Interactome factors) protein levels increase, and these factors recruit Xist RNA transcripts and tether them to the Xi. For the active mechanism in resting lymphocytes, Xist RNA interacting proteins might sequester Xist RNA transcripts away from the Xi, preventing detection by RNA FISH. Upon lymphocyte activation, YY1 and HnRNP-U bring Xist RNA to the Xi and tether the transcripts across this chromosome. It is also possible that YY1 and HnRNP-U interact with other factors for Xist RNA recruitment to the Xi, or that other Xist RNA interacting proteins function independently for recruitment. Why lymphocytes have such a unique regulation of Xist RNA localization is unknown, and the extent to which this could affect X-linked gene dosage is unclear.
Figure 2: Model for active or passive mechanisms that may regulate dynamic Xist RNA localization during lymphocyte activation.
Mature naïve lymphocytes lack Xist RNA on the Xi, and antigen stimulation triggers Xist RNA return to the Xi. We propose that this re-localization process could occur through either a passive (top) or active (bottom) mechanism. Our passive regulation model posits that unfolded/unstructured Xist RNA transcripts are dispersed across the nucleus, preventing cytological detection. Lymphocyte activation increases levels of Xist RNA interacting proteins, including YY1 and HnRNP-U, which could fold and tether Xist RNA transcripts to the Xi. Alternatively, re-localization could occur via an active process, where in naïve cells we propose that Xist RNA interacting proteins bind and sequester Xist RNA transcripts (perhaps in 1:1 stoichiometry), thereby preventing visualization by RNA FISH. Lymphocyte activation increases the concentrations of Xist RNA interacting proteins, including YY1 and HnRNP-U, which could bind to either one or multiple Xist transcripts. Xist RNA interacting factors then recruit Xist RNA to the Xi and tether Xist RNA transcripts across this chromosome. The active and passive mechanisms are not mutually exclusive. The absence of Xist RNA and heterochromatin marks at the Xi in naive cells may “relax” XCI maintenance and increase the number of genes escaping from XCI, such as Tlr7.
We have recently discovered that other immune cells, besides lymphocytes, exhibit diverse patterns of Xist RNA localization, reflecting the presence of alternative mechanisms for XCI maintenance (Fig 1B) [38]. Myeloid lineage derived cells, including myeloid dendritic cells (DCs) and bone-marrow derived macrophages (BMDM), had robust Xist RNA signals at the Xi, similar to fibroblasts (Fig 1B). Lymphoid-derived natural killer (NK) T cells and lymphoid dendritic cells (L-DCs) had a mixture of dispersed and localized Xist RNA patterns. In contrast, plasmacytoid DCs (p-DCs) completely lacked Xist RNA signals, and Xist RNA did not return to the Xi after in vitro stimulation. Surprisingly, the X-linked gene Tlr7 was bi-allelically expressed in a proportion of p-DCs, yet these cells still maintained dosage compensation as evidenced by similar expression levels of X-linked genes between females and males [38]. It is still unknown how dosage compensation is maintained despite the absence of enriched Xist RNA at the Xi in these distinct cell types, nor the amount of XCI escape in each cellular context. It is possible that cells that never localize Xist RNA to the Xi, such as p-DCs, have more XCI escape genes relative to cells that have Xist RNA localized at the Xi, and this escape may be necessary for cellular function. Dosage compensation of X-linked genes is likely occurring in immune cells, yet the molecular details that enable escape at some regions while maintaining transcriptional repression at others remains to be determined.
Conclusions
The recent findings in the field of XCI maintenance is challenging our understanding of the epigenetic mechanisms involved in allele-specific chromosome-wide silencing and XCI escape. New sequencing technologies for profiling transcription at the single-cell level has revealed that gene escape from the Xi is heterogeneous, supporting the idea that XCI maintenance is variable across individual cells and tissues [9,11]. The reason for this heterogeneity in X-linked gene escape remains an open question. Future work investigating how tissue specific escape is regulated and how it impacts the function of these cells is necessary for understanding the origins of increased X-linked expression in autoimmunity [17][14]. It may not be surprising that immune cells maintain XCI with distinct and dynamic mechanisms [16,30,38], as there are many autoimmune diseases with a strong female bias that also correlate with overexpression of some X-linked genes. The evidence of selective XCI escape in immune cells [16,17,38], combined with the variation of Xist RNA localization patterns among immune cells, suggests that dynamic XCI maintenance might enable some genes to become reactivated from the Xi (Fig 2). How immune cells maintain dosage compensation despite the absence of Xist RNA and heterochromatin marks on the Xi in still unknown. DNA methylation could possibly maintain transcriptional repression in the absence of these epigenetic modifications [39]. We speculate that DNA methylation may act as an imprint for Xist RNA and other modifications to return to the Xi during lymphocyte activation. XCI continues to be an exciting model for understanding epigenetic gene regulation and the mechanisms underlying the inheritance of an allele-specific transcriptional state.
Acknowledgments
We would like to thank C. Syrett, Z. Beethem, and A. Dubin for helpful discussions and comments on the models proposed here. This research was supported by a University Research Foundation grant, the National Institute of Health (AI124084, HD085848–03, AI134834, T32 AI-055428) the McCabe Foundation, and the American Chemical Society.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors have declared that there is no conflict of interest.
References & Recommended Reading
• of special interest
•• of outstanding interest
- 1.Lyon MF: Gene action in the X-chromosome of the mouse (mus musculus L.). Nature 1961, 190:372–373. [DOI] [PubMed] [Google Scholar]
- 2.Clemson CM, McNeil JA, Willard HF, Lawrence JB: XIST RNA paints the inactive X chromosome at interphase: Evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol 1996, 132:259–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N: Requirement for Xist in X chromosome inactivation. Nature 1996, 379:131–137. [DOI] [PubMed] [Google Scholar]
- 4.Heard E, Rougeulle C, Arnaud D, Avner P, Allis CD, Spector DL: Methylation of histone H3 at Lys-9 Is an early mark on the X chromosome during X inactivation. Cell 2001, 107:727–738. [DOI] [PubMed] [Google Scholar]
- 5.Jonkers I, Monkhorst K, Rentmeester E, Grootegoed JA, Grosveld F, Gribnau J: Xist RNA Is Confined to the Nuclear Territory of the Silenced X Chromosome throughout the Cell Cycle. Mol Cell Biol 2008, 28:5583–5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wareham KA, Lyont MF, Glenistert PH, Williams ED: Age related reactivation of an X-linked gene. Nature 1987, [DOI] [PubMed] [Google Scholar]
- 7.Carrel L, Willard HF: X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 2005, 434. [DOI] [PubMed] [Google Scholar]
- 8.Yang F, Babak T, Shendure J, Disteche CM: Global survey of escape from X inactivation by RNA-sequencing in mouse. Genome Res 2010, 20:614–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berletch JB, Ma W, Yang F, Shendure J, Noble WS: Escape from X Inactivation Varies in Mouse Tissues. PLoS Genet 2015, doi: 10.1371/journal.pgen.1005079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10•.Garieri M, Stamoulis G, Falconnet E, Ribaux P, Borel C, Santoni FA, Antonarakis S: Extensive cellular heterogeneity of X inactivation revealed by single-cell allele-specific expression in human fibroblasts. PNAS 2018, doi: 10.1101/298984.This work used deep scRNAseq to probe allele-specific escape from XCI. The authors find novel escape genes in both primary fibroblasts and lymphoblastoid cells. Escape of these genes were variable, and the hypothesis proposed suggests differences in Xist expression across the cell cycle acts as a regulatory mechanism of escape.
- 11.Cotton AM, Ge B, Light N, Adoue V, Pastinen T, Brown CJ: Analysis of expressed SNPs identifies variable extents of expression from the human inactive X chromosome. Genome Biol 2013, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomkins DJ, Mcdonald HL, Farrell SA, Brown CJ: Lack of expression of XIST from a small ring X chromosome containing the XIST locus in a girl with short stature, facial dysmorphism and developmental delay. 2002, doi: 10.1038/sj/ejhg/5200757. [DOI] [PubMed] [Google Scholar]
- 13•.Tukiainen T, Villani A-C, Yen A, Rivas MA, Marshall JL, Satija R, Aguirre M, Gauthier L, Fleharty M, Kirby A, et al. : Landscape of X chromosome inactivation across human tissues. Nature 2017, 550:244–248.This study used multiple approaches to probe escape from XCI. Comparison of female and male X-linked gene expression, use of primary tissues from a skewed XCI female, and scRNAseq discovered seven novel escape genes. The authors use these powerful approaches to support other work suggestive of variable and tissue specific escape of X-linked genes and show their efficacy in examining XCI maintenance in human samples.
- 14.Hewagama A, Gorelik G, Patel D, Liyanarachchi P, Joseph McCune W, Somers E, Gonzalez-Rivera T, The Michigan Lupus Cohort, Strickland F, Richardson B: Overexpression of X-Linked genes in T cells from women with lupus. J Autoimmun 2013, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu Q, Wu A, Tesmer L, Ray D, Yousif N, Richardson B: Demethylation of CD40LG on the Inactive X in T Cells from Women with Lupus. J Immunol 2007, 179:6352–6358. [DOI] [PubMed] [Google Scholar]
- 16.Wang J, Syrett CM, Kramer MC, Basu A, Atchison ML, Anguera MC: Unusual maintenance of X chromosome inactivation predisposes female lymphocytes for increased expression from the inactive X. Proc Natl Acad Sci 2016, doi: 10.1073/pnas.1520113113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17••.Souyris M, Cenac C, Azar P, Daviaud D, Canivet A, Grunenwald S, Pienkowski C, Chaumeil J, Mejía JE, Guéry J: TLR7 escapes X chromosome inactivation in immune cells. Sci Immunol 2018, 8855:1–11.This study used a powerful polymerase chain reaction method to determine allele-specific expression of X-linked genes in primary human immune cells. The authors find escape of the X-linked immune gene TLR7 in human female B cells, monocytes, and plasmacytoid dendritic cells. As well, this is the first study to show escape of TLR7 in autoimmune prone Klinefelter Disease (XXY) samples. This study makes the important connection between escape of TLR7 and increased risk of autoimmune diseases.
- 18.Cooney CM, Bruner GR, Aberle T, Namjou-Khales B, Myers LK, Feo L, Li S, D’Souza A, Ramirez A, Harley JB, et al. : 46,X,del(X)(q13) Turner’s syndrome women with systemic lupus erythematosus in a pedigree multiplex for SLE. Genes Immun 2009, 10:478–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harris VM, Sharma R, Cavett J, Kurien BT, Liu K, Koelsch KA, Rasmussen A, Radfar L, Lewis D, Stone DU, et al. : Klinefelter’s syndrome (47,XXY) is in excess among men with Sjögren’s syndrome. Clin Immunol 2016, 168:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dillon S, Aggarwal R, Harding JW, Li LJ, Weissman MH, Li S, Cavett JW, Sevier ST, Ojwang JW, D’Souza A, et al. : Klinefelter’s syndrome (47,XXY) among men with systemic lupus erythematosus. Acta Paediatr Int J Paediatr 2011, 100:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scofield RH, Bruner GR, Namjou B, Kimberly RP, Ramsey-Goldman R, Petri M, Reveille JD, Alarcón GS, Vilá LM, Reid J, et al. : Klinefelter’s syndrome (47,XXY) in male systemic lupus erythematosus patients: Support for the notion of a gene-dose effect from the X chromosome. Arthritis Rheum 2008, 58:2511–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wutz A, Jaenisch R: A Shift from Reversible to Irreversible X Inactivation Is Triggered during ES Cell Differentiation. Mol Cell 2000, 5:695–705. [DOI] [PubMed] [Google Scholar]
- 23.Chan KM, Zhang H, Malureanu L, van Deursen J, Zhang Z: Diverse factors are involved in maintaining X chromosome inactivation. Proc Natl Acad Sci 2011, 108:16699–16704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaenisch R, Csankovszki G, Panning B, Bates B, Pehrson JR: Conditional deletion of Xist disrupts histone macroH2A localization but not maintenance of X inactivation. Nat Genet 1999, 22:323–324. [DOI] [PubMed] [Google Scholar]
- 25.Chow JC, Hall LL, Clemson CM, Lawrence JB, Brown CJ: Characterization of expression at the human XIST locus in somatic, embryonal carcinoma, and transgenic cell lines. Genomics 2003, 82:309–322. [DOI] [PubMed] [Google Scholar]
- 26.Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF: A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 1991, 349:38–44. [DOI] [PubMed] [Google Scholar]
- 27.Paterno GD, McBurney MW: X chromosome inactivation during induced differentiation of a female mouse embryonal carcinoma cell line. J Cell Sci 1985, 75:149–163. [DOI] [PubMed] [Google Scholar]
- 28.Corbel C, Diabangouaya P, Gendrel A-V, Chow JC, Heard E: Unusual chromatin status and organization of the inactive X chromosome in murine trophoblast giant cells. Development 2013, 140:861–872. [DOI] [PubMed] [Google Scholar]
- 29.Savarese F, Flahndorfer K, Jaenisch R, Busslinger M, Wutz A: Hematopoietic Precursor Cells Transiently Reestablish Permissiveness for X Inactivation. Mol Cell Biol 2006, 26:7167–7177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30••.Syrett CM, Sindhava V, Hodawadekar S, Myles A, Liang G, Zhang Y, Nandi S, Cancro M, Atchison M, Anguera MC: Loss of Xist RNA from the inactive X during B cell development is restored in a dynamic YY1-dependent two-step process in activated B cells. PLoS Genet 2017, 13:1–28.Previous work displayed unique mechanisms of XCI maintenace in lymphocytes[14,23]. Work from this paper expands on these findings through examination of the dynamic localization of Xist RNA in B cell progenitors. In addition, this work expandsour mechanistic knowledge by determining the requirement of YY1 for the return of Xist RNA in mature activated B lymphocytes.
- 31••.Syrett CM, Paneru B, Sandoval-Heglund D, Wang J, Banerjee S, Sindhava V, Behrens EM, Atchison M, Anguera MC: Altered X-chromosome inactivation in T cells may promote sex-biased autoimmune diseases. J Clin Investig Insight 2019, 4:e126751.In this work, the authors examine Xist RNA localization in developing T cell progenitor cells. They find that T cell progenitors display unique Xist RNA localization, where Xist RNA is localized at the Xi in some stages of T cell development and absent from the Xi in others. This work also goes on to explore the relationship between XCI maintenance in T cells and development of the female-biased disease Systemic Lupus Erythematosus.
- 32.Jeon Y, Lee JT: YY1 Tethers Xist RNA to the inactive X nucleation center. Cell 2011, 146:119–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hasegawa Y, Brockdorff N, Kawano S, Tsutui K, Tsutui K, Nakagawa S: The matrix protein hnRNP U is required for chromosomal localization of xist RNA. Dev Cell 2010, 19:469–476. [DOI] [PubMed] [Google Scholar]
- 34.Minajigi A, Froberg JE, Wei C, Sunwoo H, Kesner B, Colognori D, Lessing D, Payer B, Boukhali M, Haas W, et al. : A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science (80-) 2015, 349:aab2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McHugh CA, Chen CK, Chow A, Surka CF, Tran C, McDonel P, Pandya-Jones A, Blanco M, Burghard C, Moradian A, et al. : The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 2015, 521:232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chu C, Zhang QC, Da Rocha ST, Flynn RA, Bharadwaj M, Calabrese JM, Magnuson T, Heard E, Chang HY: Systematic discovery of Xist RNA binding proteins. Cell 2015, 161:404–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moindrot B, Cerase A, Coker H, Masui O, Grijzenhout A, Pintacuda G, Schermelleh L, Nesterova TB, Brockdorff N: A Pooled shRNA Screen Identifies Rbm15, Spen, and Wtap as Factors Required for Xist RNA-Mediated Silencing. Cell Rep 2015, 12:562–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38••.Syrett CM, Sindhava V, Sierra I, Dubin AH, Atchison M: Diversity of Epigenetic Features of the Inactive X-Chromosome in NK Cells, Dendritic Cells, and Macrophages. Front Immunol 2019, 9:1–10.This work examines Xist RNA localization and H3K27me3 enrichment across different immune cell lineages. Interestingly, immune cells show strong diversity in Xist RNA localization, with plasmacytoid dendritic cells having no visible Xist RNA in the nucleus. Surprisingly, this work shows bi-allelic expression of Tlr7 in plasmacytoid dendritic cells, but overall these cells are still dosage compensated. This study reveals the extensive diversity of XCI maintenance in immune cells.
- 39.Sharp AJ, Stathaki E, Migliavacca E, Brahmachary M, Montgomery SB, Dupre Y, Antonarakis SE: DNA methylation profiles of human active and inactive X chromosomes. Genome Res 2011, 21:1592–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]