Abstract
Long-label retention has been used by many to prove Cairns’ immortal strand hypothesis and to identify potential stem cells. Here, we describe two strategies using 5-ethynl-2’-deoxyuridine (EdU) to identify and understand the distribution of long-label-retaining mammary epithelial cells during formation of the mouse mammary ductal system. First, EdU was given upon two consecutive days per week during weeks 4 through 10 and analyzed for label retention at 13 weeks of age. Alternatively, EdU was given for 14 consecutive days beginning at 28 days of age and ending at 42 days of age. Analyses were conducted at greater than 91 days of age (13 weeks). Many more LREC were detected following the second labeling method and their distribution among the subsequently developed ducts. This finding indicated that the early-labeled cells that retained their label were distributed into portions of the gland that developed after the ending of EdU treatment (i.e. 42 -> 91 days). These observations may have important meaning with respect to the previously demonstrated retention of regenerative capacity throughout the mouse mammary gland despite age or reproductive history. These results suggest LREC may represent long-lived progenitor cells that are responsible for mammary gland homeostasis. Additionally, these cells may act as multipotent stem cells capable of mammary gland regeneration upon random fragment transplantation into epithelium-denuded mammary fat pads.
Keywords: mammary, stem cell, long-label-retaining, development, EDU-labeling
Introduction
It has been postulated that the stem cells of somatic tissues protect themselves from mutation and cancer risk by selective segregation of their template DNA strands [1]. The mouse mammary ductal system develops primarily after the onset of puberty (~28 days) and is completed approximately 84 days after birth. Therefore, allometric growth of the ducts occurs exclusively within this time period. Previously, we demonstrated that self-renewing mammary epithelial stem cells could be labeled using [3H]- thymidine (3HTdR) during the first two-weeks of allometric growth [2]. After a prolonged chase during which much of the branching duct morphogenesis was completed, 3HTdR-label retaining epithelial cells (LREC) were detected among the epithelium of the maturing glands. Labeling newly synthesized DNA in these glands with a different marker, 5-bromodeoxyuridine (5BrdU), resulted in the appearance of doubly labeled nuclei in a large percentage of the LREC [2, 3]. In contrast, label-retaining cells within the associated lymph nodes did not incorporate 5BrdU during the pulse, indicating that they were not traversing the cell cycle. Upon chase, the second label (5BrdU) was distributed from the double-labeled LREC to the progeny of these doubly-labeled cells, while 3HTdR was retained. These results demonstrate that mammary LREC selectively retain their 3HTdR-labeled template DNA strands and pass newly synthesized 5BrdU-labeled DNA to their progeny during asymmetric divisions. An important aspect of this observation is that the capacity of mouse mammary fragments to regenerate an entire functional mammary gland upon transplantation is never lost despite age or reproductive history [4, 5]. Similar results were obtained in mammary transplants containing self-renewing, LacZ-positive epithelial cells suggesting that cells capable of expansive self-renewal may repopulate new mammary stem cell niches during the allometric growth of new mammary ducts [6].
LRECs have been reported among the epithelium of the murine mammary gland using both 3HTdR and 5BrdU [7-9]. It was reported that as many as 50% of mammary epithelial cells are labeled with 3HTdR after three consecutive injections and much of this label is lost after 2 weeks, consistent with the loss of label by semi-conservative exponential cell divisions [7, 8]. Some cells retained label following this 2-week period and had autoradiographic grain counts similar to cells immediately following 3HTdR injection. A greater number of these cells were obtained when 3HTdR injection was made just at estrus or met-estrus during the estrus cycle [7, 8]. These authors chased the label for just two weeks and used adult females 9-16 weeks of age. In preliminary studies they determined that no heavily labeled cells were present after 5 weeks. In a quite different approach, Welm et al. labeled mice with 5BrdU delivered from an implanted Alzet pump for 14 days beginning at 3 weeks of age [9]. Subsequently the pump was removed and the number and location of labeled mammary cells was analyzed at weekly periods for 9 more weeks. These investigators found that the number of labeled epithelial cells decreased quite rapidly reaching <5% by 9 weeks. Only ~1.5% of the label-retaining cells remaining at 9 weeks expressed progesterone receptor (PR). The remaining label-retaining cells were also stained for keratin 14 (K14) or K18, myoepithelial and luminal epithelial cell markers respectively, revealing a small population of label-retaining cells, which did not express either epithelial marker. In addition, these authors found that the LREC epithelial population at 9 weeks was more prevalent in side population (SP) cells after fluorescence-activated-cell sorting (FACS), suggesting that they may represent mammary epithelial stem cells.
In this manuscript, we employ two different labeling protocols to understand the production and distribution of long-label-retaining epithelial cells (LREC) in the mouse mammary gland. One schedule sought to label LREC during ductal allometric growth in post-pubertal female mice by applying EdU every week over a 48-hour period each week during the development of the branching mammary ductal system. Alternatively, EDU was applied continuously during the first two weeks of puberty and early ductal growth. To our surprise, the second labeling schedule demonstrated that early continuous labeling identified a greater number of LREC than the intermittent labeling throughout ductal elongation, demonstrating that LRECs labeled during the first two weeks of ductal development were distributed among the ductal epithelium several weeks after labeling had ceased. This indicated that the early-labeled LREC were transported and distributed along the newly formed ducts during allometric growth. This is a seminal observation, which suggests that LREC represent reserve long-lived progenitor cells that are responsible for mammary epithelial cell homeostasis. Upon transplantation into epithelium-denuded mammary fat pads, LREC may also act as multipotent and bipotent antecedents for mammary gland regeneration.
Materials and Methods
Animal Housing and Dosing
Balb/C and athymic nude mice were purchased from NCI-Frederick. All mice were housed in in Association for Assessment and Accreditation of Laboratory Animal Care-accredited facilities in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. The National Cancer Institute Animal Care and Use Committee approved all experimental procedures.
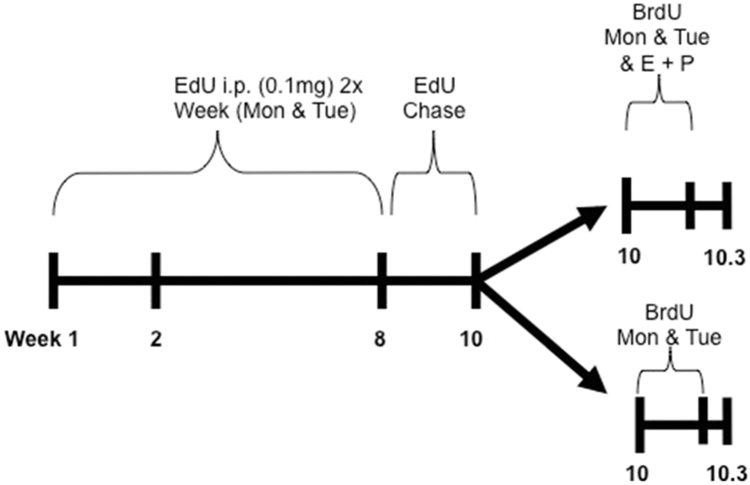
For initial studies, mice were dosed as outlined in Figure 1. Starting at 4 weeks of age, mice were given a 0.1 mg bolus dose of 5-ethynyl-2’-deoxyuridine (EdU) i.p. once daily for two consecutive days (Monday and Tuesday). This dosing schedule was repeated for 8 weeks until the mice were 12 weeks old. This resulted in a total of 16 doses of EdU over the 8-week period. The mice then underwent a 2-week chase period where no treatment was given. After the 2-week chase, mice were then dosed with 0.1 mg of bromodeoxyuridine (BrdU) i.p. on two consecutive days (Monday and Tuesday) with or without concurrent intraperitoneal Estrogen (1 μg) and Progesterone (1 mg) administration to stimulate luminal cell proliferation [10, 11]. Mice were euthanized and mammary glands were collected for analysis, 24 hours after the final dose of BrdU. For each timepoint, all 4 fat pads (thoracic and inguinal) were evaluated from at least 3 individual mice per group.
Figure 1. Mouse dosing method 1.
15 Balb/c mice were dosed with 0.1mg of EdU by intraperitoneal injection on two consecutive days (Monday and Tuesday). After 4 doses (week 2), two mice were euthanized for analysis. The remaining mice received EdU for a total of 8 weeks. Three mice were euthanized at week 8 for analysis. The remaining mice received no treatment for 2 weeks (chase) and then two consecutive doses (Monday and Tuesday evenings) of Bromodeoxyuridine (BrdU) with or without additional treatment with Estrogen and Progesterone (E + P) and euthanized the following morning (Wednesday).
To determine if EdU+ cells were distributed throughout the gland during development, mouse dosing method 2 was employed. Balb/C female mice were dosed with 0.1 mg of EdU for 14 consecutive days starting at 4 weeks of age. This was followed by an 8-week chase period after which the glands were isolated for analysis. This process was repeated in athymic nude mice. Primary cultures were made of the tissue proximal and distal to the nipple (using the lymph node in the fat pad as the demarcation zone for proximal versus distal to the nipple) upon excision of the inguinal mammary glands following cessation of labeling and later after the 8-week chase (Figure 2). To do this, tissue was taken from the inguinal glands, minced, and digested in collagenase type IA (1 mg/mL, Thermo Fisher) overnight at 37°C. Resulting organoids were triturated and plated in petri dishes to grow before undergoing EdU staining and analysis.
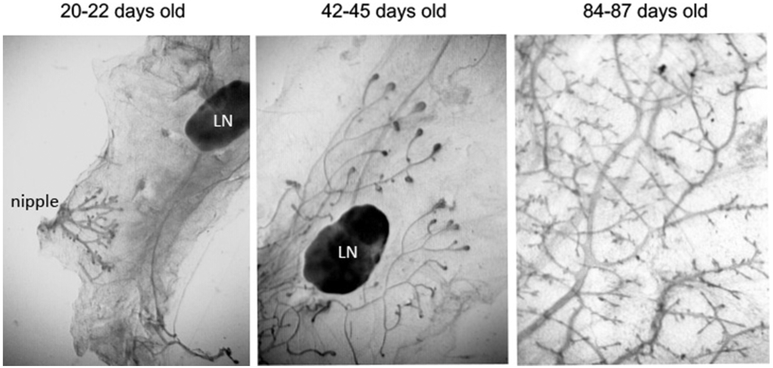
Figure 2. Mammary gland development over the course of twelve weeks.
At twenty to 22 days of age, mice have endogenous epithelium which begins growing from the nipple (left panel) the remaining mammary fat pad is empty, save the lymph node (LN). By 42 to 45 days after birth, the epithelial ductal tree has advanced past the lymph node and into the portion of the gland distal from the nipple (middle panel). The distal portion of the mammary fat pad is filled at approximately 84 days of age (right panel).
Edu Imaging and Immunofluorescence via Confocal Fluorescence Microscopy
Glands were fixed in 4% paraformaldehyde (PFA), paraffin-embedded and sectioned. Prior to staining, sections were subjected to heat-mediated antigen retrieval for 25 mins in boiling pH = 9.0 Tris EDTA containing 0.05% Tween-20. EdU was imaged using the Click-iT EdU kit (Thermo Fisher Scientific) per the manufacturer’s instructions. For BrdU imaging, slides were denatured with 2N HCl for 10 minutes. Immunostaining was performed using primary antibodies to BrdU (Thermo Fisher B35128; 1:100), smooth muscle actin (Thermo Fisher MA1-06110; 1:100), ki67 (Abeam ab16667; 1:100), cytokeratin 8 (K8, Abeam ab53280; 1:200), and cytokeratin 14 (K14, Abeam ab51054; 1:200). Alexa Fluor 568 conjugated goat anti-mouse (A-1104) or rabbit (A-11011) secondary antibodies were used for visualization. The Zenon Alexa Fluor 594 rabbit IgG labeling kit (Thermo Fisher Z2307) was also used for visualization. All sections were counterstained with DAPI. Quantification was done by manually counting a minimum of 10 randomly chosen images across 3-5 samples. Results were statistically compared by a one-way analysis of variance (ANOVA) with a Tukey post-hoc test.
Edu Staining and Immunocytochemistry via Confocal Light Microscopy
Inguinal mammary glands were divided to include only the area between the nipple and the lymph node (LN; proximal) and the area distal from the LN (distal). The mammary tissues were digested in 1 mg/mL collagenase Type IA (Thermo Fisher) overnight at 37°C. Organoids were pelleted, washed, and plated onto 6 cm dishes. Once colonies formed, cells were washed with PBS and fixed in 4% PFA for 15 minutes at room temperature. The fixative was washed away with 3% BSA/PBS and cells were permeabilized with 0.5% Triton X-100 in PBS (as per manufacturer’s instruction) for 20 minutes at room temperature. The cultures were washed with PBS before staining with the Click-iT EdU kit (Thermo Fisher Scientific). Immunocytochemistry was performed using primary antibodies to Alexa Fluor 488 (Thermo Fisher A-11094; 1:200), Ki67 (Abcam AB9260; 1:100), and Smooth Muscle Actin (Sigma A2547; 1:100). Secondary antibodies from R.T.U Vectastain Universal Elite ABC kit (Vector PK7200) were used with DAB Peroxidase Substrate (Vector SK4100). All samples were counterstained with Mayer’s hematoxylin (Sigma MHS1). Colonies with positive EdU staining were quantified.
Flow Cytometry
Mammary glands were removed and subjected to 1 hr digestion collagenase / hyaluronidase (0.1% each, Sigma type 1A #C-9891 and Sigma #H-3506) solution at 37°C under gentle agitation. Mammary organoids were then pelleted, washed, and digested to single cells in pronase (1.25%, CalBioChem #53702). Clumps were removed by a 40 μm filter, and cells were fixed in 70% ethanol before permeabilization in 0.1% Triton X-100. Cells were treated with RNASE and stained for EdU (according to the manufacturer’s protocol; Click-iT EdU kit, Thermo Fisher Scientific) and with 7-aminoactinomycin D (7-AAD). Cell cycle analysis was performed on cells gated for EdU levels to determine cell cycle distribution of EdU+ cells. Doublets were removed by gating forward scatter height vs area. Analysis was performed using Flowjo software. Experiments were performed on pooled mammary cells from 5 individual mice from each group.
Results
Our initial inoculation procedure was to use the label (EdU) to mark cells in the mammary epithelium that were in the DNA synthesis (S) phase of the cell cycle. To accomplish this, we reasoned that labeling for two consecutive days each week during allometric ductal growth would provide the optimal coverage of epithelial cells entering the S phase of the cell cycle. However, this proved to be a poor strategy for capturing LREC in the mammary epithelial population. In the initial studies, we used Balb/C female mice at approximately 3 weeks of age and then switched to FVB/N females when our initial strategy was unsatisfactory. However, in a conversation with Dr. Rebecca Morris, we found that both Balb/C and FVB/N have a deletion of bone morphogenetic protein 5 (BMP5) on Chromosome 9, which results in decreased LREC in epidermal epithelium [12]. Therefore, we continued our studies by labeling the mammary epithelium for 14 consecutive days beginning at day 28 followed by a seven-week chase in both Balb/C and athymic nude females for comparison.
LREC labeling using Method 1 (twice weekly for 7 weeks)
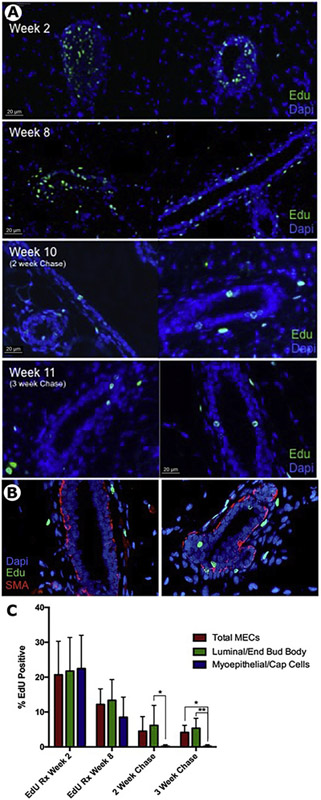
The hypothesis was that label (EdU) administration at intervals during ductal growth and elongation would provide the best coverage for the production of LREC. Figure 1 shows the time course used for this labeling strategy. EdU was given on Monday and Tuesday afternoons beginning at 4 weeks of age and continued for seven additional weeks on Monday and Tuesday. We postulated that in this way EdU would be incorporated in multiple epithelial cells during allometric growth of the branching mammary ductal system. The efficacy of this labeling methodology was evaluated at week 2, week 8, and after a 2-week (week 10) and a 3-week (week 11) chase period (Figure 3A). After two weeks of EdU administration, the terminal end buds (TEB) at the ends of the growing ducts were most heavily labeled (Figure 3A; top panel). Both cap cells and body cells were labeled in the TEB and some luminal and basal cells were labeled in the subtending ducts. After 5 additional weeks of label, both ductal luminal and basal epithelia were positive for EdU (Figure 3B; second panel). Following a 2-week or 3-week chase at the cessation of EdU treatment only luminal epithelial cells remained that were positive for EdU (Figure 3A, B, C). This indicated that the basal cells continued to divide and distributed their EdU semi-conservatively to daughter cells until no EdU could be detected (Figure 3A). This was confirmed by co-staining with smooth muscle actin (SMA), a marker for basal epithelium (Figure 3B). Also shown in Figures 3A and 3B are periductal LRC (not positive for epithelial markers), which we have described earlier [2, 13]. Quantitation of EdU positive LRECs demonstrated no significant change in the amount of LRECs between the 2 week and 3-week chase, and that a 2-week chase was sufficient to eliminate all EdU label from basal/myoepithelial cells (Figure 3C).
Figure 3. EdU incorporates during ductal elongation and is retained in a subset of epithelial cells following chase.
A) Representative images of EdU (green) incorporation at 2 weeks, 8 weeks, 10 weeks and 11 weeks. At week 2, EdU positive cells were seen in both body and cap cells of end buds. After final EdU dosing at week 8, EdU cells were seen throughout the epithelial ducts and periductal cells. Following a 2-week chase (Week 10) or 3-week chase (Week 11) EdU+ LRECs remained in a subset of luminal epithelial cells and periductal cells. B) Representative image of a section from a 10-week sample stained with anti-SMA and EdU demonstrating EdU positive LRECs are within the luminal layer. C) Quantitation of total MECs, luminal MECs, and myoepithelial cells at 2, 8, 10, and 11 weeks. EdU positive LRCs are present almost exclusively in the luminal layer following chase.
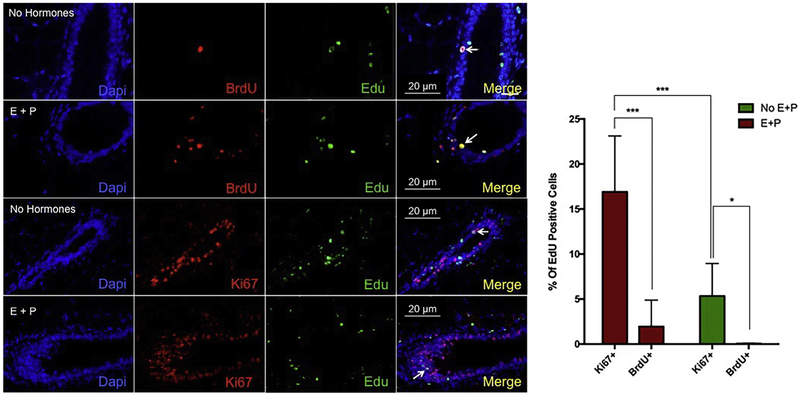
In order to determine how many of these LREC might be in the cell cycle we pulsed with a second thymidine analog (5BrdU) for two consecutive days as with the EdU with or without treatment with estrogen (E) and progesterone (P) to stimulate luminal cell proliferation, as would normally occur during pregnancy. In addition, the tissue was stained for Ki67 to indicate what cells might be in cycle but not necessarily in S phase. Very few EdU-positive cells became double-labeled with 5BrdU however the number was increased significantly following hormone treatment with estrogen and progesterone (Figure 4). Conversely, many LRECs were positive for Ki67. This indicates to us that LREC may have a prolonged cell cycle.
Figure 4. A small subset of EdU positive LRCs incorporate BrdU and/or express ki67.
Representative images of 10-week glands treated with BrdU with or without estrogen (E) and progesterone (P) treatment. EdU (green) and BrdU (red) or ki67 (red) co-positive cells were identified in all cases but E + P treatment increased percentage of both. Arrows indicate double-labeled cells. Right panel shows quantification of the percentage of EdU positive cells that were also positive for Ki67 or BrdU. ***p<0.001; *p<0.05
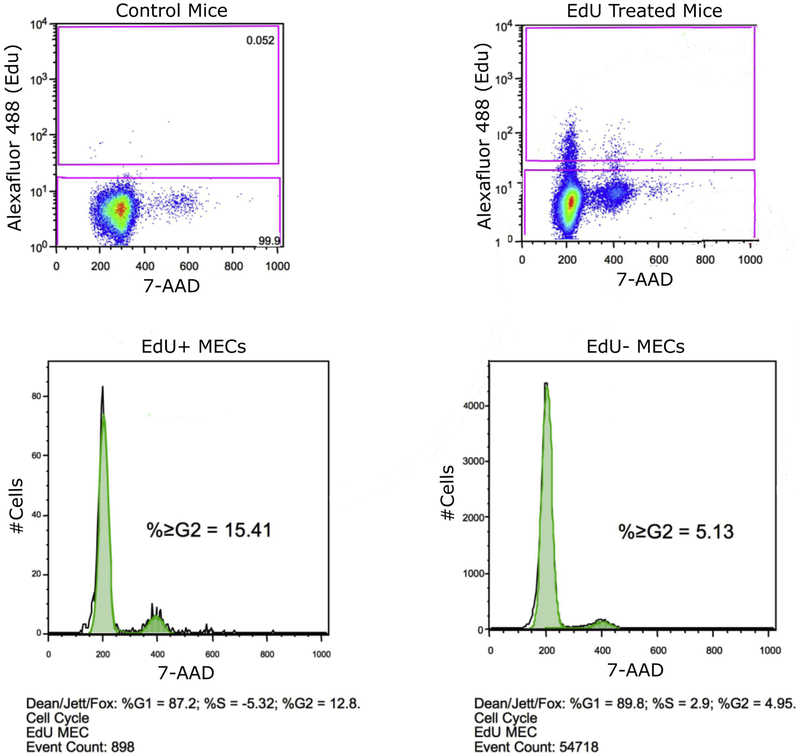
To test this theory further, we isolated single mammary epithelial cells from mice following 2-week chase (week 10) and subjected to flow cytometry to evaluate cell cycle distribution of both EdU+ and EdU− populations (Figure 5). The general EdU− populations showed a normal cell cycle distribution with 89.8% of cells in G1, 2.9% in S, and 4.95% in G2. The EdU+ fraction, however, demonstrated a higher level of cells in the G2 phase (12.8%), with the remaining 87.2% in G1. The differences in distribution were significant as compared by Chi Square Analysis (p<0.001). This suggests many LRECs labeled under this method are locked in the G2 phase of the cell cycle.
Figure 5. A higher percentage of LRECs are in G2 phase of the cell cycle than the general MEC population.
Isolated MECs were taken from mice at week 10 (2-week chase) as described in the text. Samples were gated for EdU fluorescence and cell cycle distribution measured by 7-AAD. Upper left panel shows a negative control sample used to set EdU+ gate. Upper right shows test sample with gates selective for EdU+ and EdU− MECs. Bottom panels show cell cycle distribution of EdU+ fraction (left) and EdU− fraction (right).
LREC Labeling Method 2 (once a day for 14 days)
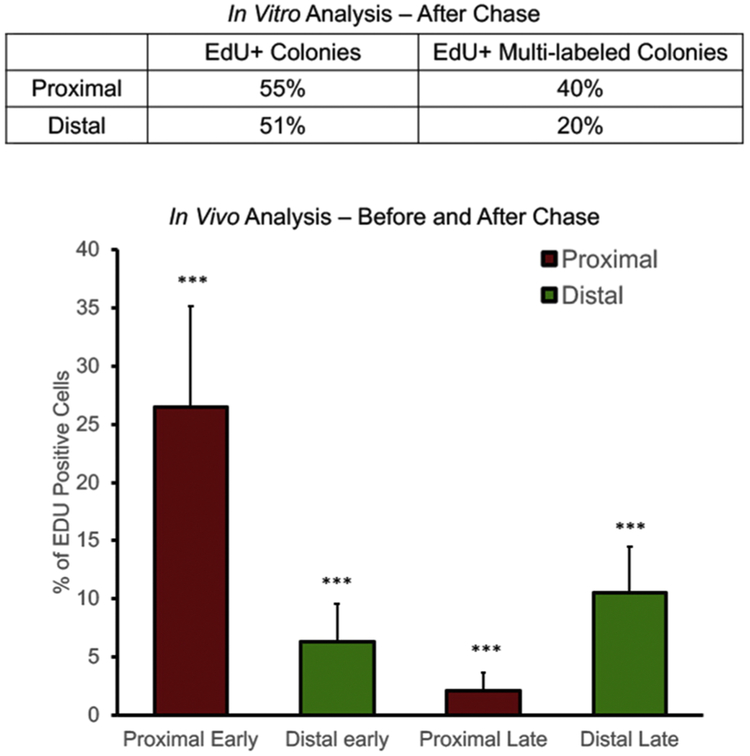
Daily labeling with EdU for 14 consecutive days was to ensure effective labeling of the cells in the growing ducts of four-week-old mice. After an eight-week chase, the mammary glands were collected. This would allow us to track long-label retaining cells through the growth and development of the gland. The distribution of LREC was subsequently determined in two ways: the inguinal glands from some mice were fixed and embedded in paraffin for subsequent sectioning and determination of LREC location (n=8 glands from 4 Balb/C female mice). Alternatively, primary cultures were created from the portion of the inguinal glands proximal to the nipple and that portion distal from the nipple (a region populated by the branching ducts following the cessation of EdU labeling, n=8 glands from 4 Athymic nude females, Figure 2). Subsequently, these cultures were subjected to the Click-iT procedure to visualize cells retaining EdU label in their nuclei. The number of epithelial colonies positively labeled with EdU versus the total number of colonies present were counted after an eight-week chase (Figure 6). Before chase, it was not possible to distinguish long-labeled EdU+ cells from those that would subsequently distribute their label to progeny during symmetric divisions. Therefore, one can only find the LREC (EdU+) following the chase period. In Figure 6 the LREC in the proximal region represent roughly 2.5% of total cells counted in epithelial colonies generated from the proximal portion of the glands, whereas nearly 10% of total cells counted were EdU+ in epithelial colonies generated from the distal portion of the glands.
Figure 6. Comparison of In Vitro and In Vivo analyses for transition of long-label-retaining cells into the distal portion of the gland.
Top panel shows EdU+ colonies in organoids formed by labeling Athymic nude mice. Out of 100 colonies (average of 100 cells per colony) produced from each of two mice after a 15-week chase period, the average percentage of EdU positive colonies is shown (out of 100 colonies counted). Bottom panel shows the analysis of sectioned mammary glands from labeled Balb/C mice. Proximal early and distal early include all EdU+ cells not LREC. Proximal late and distal late only reveal EdU+ cells that retained the label (LREC) over the chase period. ***p<0.001
Upon analysis of sectioned glands, we observe that EdU positive cells appear more frequently in the ducts that are in the area distal from the lymph node (Figure 6). These EdU+ cells distal to the nipple after chase (10.55 ductal cells on average) indicate long-label-retaining epithelial cells which have protected their template while moving into the distal portion of the gland. The large number of EdU+ cells near the nipple (26.5 ductal cells on average) indicate cells which are initially labeled but many of those will not maintain the label through subsequent divisions. The significant increase of EdU+ distal cells represent LREC, which were formed proximally and were re-positioned during the chase period (p = 6.64×10−4; Figure 6). We believe this occurs during the penetration of the growing ducts through the surrounding fat pad.
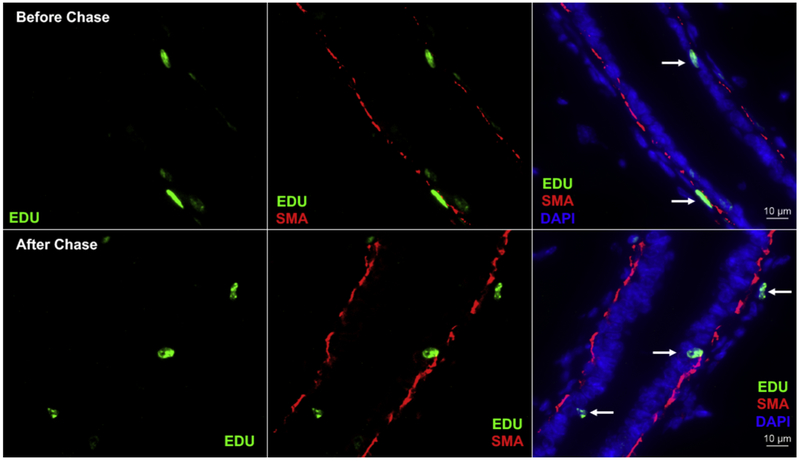
It is of note that a few of the EdU+ cells appear to be periductal when distal from the nipple (Figures 3 and 4). This is confirmed with additional SMA staining before and after the chase period (Figure 7). Long-label retention by periductal cells outside of the basement membrane was reported earlier [2, 11]. These cells are not positive for mammary epithelial cell markers.
Figure 7. SMA (red) is used to indicate where distal EdU+ cells reside within the ductal system.
Before the chase period, all EdU+ cells remain within the duct. However, after the chase period, some EdU+ (sic LREC) become periductal. Additionally, there is an increased number of EdU positive cells (green) distal to the nipple after chase in the inguinal gland, as shown previously. EdU+ cells are indicated with white arrows in the colocalized images. The sections are counterstained with DAPI (blue).
The results obtained by the two distinct labeling methods, i.e. prolonged during ductal epithelial growth over several weeks, compared to intense labeling for 14 days during early ductal expansion, indicate that LREC are produced early and distributed throughout the elongating ducts during subsequent growth. This suggests an important role for LREC in ductal homeostasis and/or alveologenesis subsequent to the onset of pregnancy.
Discussion
In an attempt to understand the production and distribution of long-label-retaining epithelial cells (LREC) in the mouse mammary gland, two strikingly different labeling modes were employed. One schedule sought to label LREC during ductal allometric growth in post-pubertal female mice by applying EdU every week over a 48-hour period each week during the development of the branching mammary ductal system. Alternatively, EdU was applied continuously during the first two weeks of puberty and early ductal growth. To our surprise, the second labeling schedule demonstrated that early continuous labeling identified a greater number of LREC and demonstrated that the LREC labeled during the first two weeks of ductal development were distributed among the ductal epithelium several weeks after labeling had ceased. This indicated that the early-labeled LREC were transported and distributed along the newly formed ducts during allometric growth. This is a seminal observation, which suggests that LREC represent reserve long-lived progenitor cells that are responsible for mammary epithelial cell homeostasis and may function in fragment transplant regeneration as multipotent and bipotent antecedents for mammary gland regeneration in epithelium-denuded mammary fat pads.
In an earlier study of LREC in adult mouse mammary glands labeled by three consecutive 3HTdR injections given at 8-hour intervals upwards of 50% of the cells were labeled immediately after the injections [7]. When contralateral glands were taken from the same mouse two weeks later only 1/50 to 1/1000 cells retained label at the level seen immediately after the 3H pulse. These observations indicate that a considerable amount of cellular turnover occurs in the adult gland. Further study by these authors demonstrated that introducing 3HTdR at estrus and metestrus in cycling virgins provided the greatest number of label-retaining cells 2 weeks later. Despite this, mammary glands labeled in this way contained no detectable LREC when examined after 5 weeks [7]. This result implies that all the cells labeled by this method were exponentially cycling and distributed the labeled DNA in a semi-conservative manner. It is therefore unlikely that this method detects asymmetrically dividing cells because expansive stem cell renewal is not occurring during maintenance of the fully developed mammary ductal system. Conversely, when glands were labeled for 14 consecutive days during the 3rd to the 5th week of life (during active expansion of mammary stem cells in the allometrically growing ducts), LREC were observed even 9 weeks after the cessation of labeling [9]. This method, and the second one employed here, tags stem cells (sic LREC) at their inception and label is retained in these cells through asymmetric divisions even though they may often traverse the cell cycle.
Although cells frequently enter the cell cycle, our initial results suggest the LREC maintain their template strand by spending an elongated time in the G2 phase (Figure 5). Another group obtained similar results, noticing a protection of the template strand via an elongated cell cycle for 3HTdR-labeled intestinal stem cells [14]. In trying to identify stem cells in the transition between the anal canal and rectum in mice, Runck and colleagues described label-retaining cells that were also slow cycling [15]. These results, taken together, suggest prolonged residence in the cell cycle allows for fewer cellular turnovers which perhaps provides a mechanism for stem cells to protect their template strands, as Cairns predicted [1].
We propose that the LREC represent the long-lived stem cells that are maintained at the growing tips of mammary ducts. Additionally, these LREC are redistributed along the subtending ducts where they become reserve multipotent cells that can function as bipotent alveolar progenitors in the intact glands during pregnancy and as multipotent stem cells when random fragments of duct are transplanted to epithelium-cleared mammary fat pads [16]. A recent study of ductal growth and development in situ indicates that the ductal tips also termed terminal end buds (TEB) possess the bulk of cycling epithelial cells, as we also showed in Figure 3 [17]. These authors developed a physical model of ductal growth and branching that demonstrates that stem cells at the termini of growing ducts determine when and where ductal bifurcations occur and when ductal growth ceases. As we show in Figure 6, initially-labeled cells are more numerous than after chase. This is because many cells divide semi-conservatively and initially labeled DNA is distributed evenly between mitotic daughters. After chase, the label remains detectable either if the cells do not divide or if they divide and selectively retain their labeled DNA strands. Fernandez-Gonzalez and colleagues recently made a similar observation, that as many as 80% of luminal cells were labeled during their 2010 study but many lost the label after chase, indicating symmetric division during ductal development [18]. The remaining cells that retained their DNA label, are considered LREC.
Fernandez-Gonzalez and colleagues used a computational microscopy platform to study mammary epithelial cell populations in situ [18, 19]. Multiple cellular features could be mapped to glandular locations at various scales with this new technique. In order to accomplish this, the study made use of an implanted miniosmotic pump to incorporate bromodeoxyuridine (BrdU) in vivo while the mammary gland was undergoing DNA synthesis during puberty. Epithelial cells which maintained the BrdU label in the adult gland were studied as long label-retaining cells (LREC). This labeling approach coupled with the novel microscopic platform, allowed the group to study the cellular composition and architecture of the mouse mammary gland. Luminal label-retaining cells were enriched 3.4-fold in large ducts. Therefore, they postulated that these LRC represent epithelial progenitor cells. These LREC were not only in the large ducts proximal to the nipple, but also found in the distal portion of the mammary gland after chase (as we found in Figure 6). They further demonstrated that epithelial cells isolated from the gland proximal to the nipple compared to the distal portion were enriched for the putative stem cell markers CD24 and CD49f as measured by fluorescence activated cell sorting. These authors did not recognize that the LREC they observed during labeling in early puberty had been transported to distal regions of the gland following cessation of labeling, as we show in Figure 6.
In addition to the LREC, stroma cells were also detected as long label-retaining cells (LRC) by EdU retention, following the chase period (Figure 7). Among these were periductal cells that were negative for all epithelial markers by immune-histochemistry. Nevertheless, as we have previously reported, these periductal LRC (not demonstrably epithelial) cells were replicating during pregnancy, as evidenced by incorporation of a second thymidine analog, and were found proximal to developing secretory acini during early pregnancy suggesting that they could represent LREC which underwent epithelial-mesenchymal transition (EMT) to escape from the mammary ducts during branching [11, 13].
Our proposal is that the LREC represent a specific epithelial cell subpopulation whose function is to divide asymmetrically to produce committed transiently amplifying daughters to replace naturally occurring cell loss among the mammary epithelium. A second proposal is that both multipotent stem cells and lineage-limited progenitors exist within the long-label-retaining population found within the epithelium of intact mammary glands [23]. Accordingly, asymmetric cell division may be a property of stem cells and particularly of stem cells functioning within a tissue-specific stem cell niche, and also lineage-limited epithelial progenitors reviewed by H. Lin [24]. But are LREC multipotent stem cells or simply giving rise to epithelial cells committed to a single epithelial cell lineage? In the current study, it was not possible to determine whether LREC daughters represented epithelial cells committed to one epithelial lineage or to several. In either case, mammary LREC have been shown to be self-renewing by retention of the 3HTdR–labeled DNA [2]. This is apparently accomplished by asymmetric distribution of the old and new DNA strands. Therefore, mammary LREC possess at least one property commonly ascribed to somatic stem cells.
In fact, earlier studies using H2B-GFP to identify LREC have shown that the LREC identified in this manner in mouse mammary and subsequently, isolated and analyzed, express stem cells related markers and possess self-renewal capacity [20, 21]. Both of these groups showed self-renewal capability by transplanting FACS-sorted LREC (sic stem cells) back into cleared mouse mammary fat pads. Collection of LREC labeled with 5BrdU in bovine mammary tissue also indicates that these cells are often characterized by putative mammary stem cell markers [22]. These studies, combined with our findings, suggest LREC may represent mammary somatic stem cells.
Highlights:
DNA labeling shows long-label-retaining epithelial cells (LREC) are made early
After chase, LREC are distributed throughout developing mammary gland
LREC represent long-lived progenitor cells important for mammary homeostasis
Upon transplantation, LREC may act as multipotent stem cells
Acknowledgments
Funding: This work was supported by the National Institutes of Health Intramural Research Program.
Footnotes
Declaration of Interest: The authors have no declarations of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- 1.Cairns J, Mutation selection and the natural history of cancer. Nature, 1975. 255(5505): p. 197–200. [DOI] [PubMed] [Google Scholar]
- 2.Smith GH, Label-retaining epithelial cells in mouse mammary gland divide asymmetrically and retain their template DNA strands. Development, 2005. 132(4): p. 681–7. [DOI] [PubMed] [Google Scholar]
- 3.Smith GH and Boulanger CA, Mammary stem cell repertoire: new insights in aging epithelial populations. Mech Ageing Dev, 2002. 123(11): p. 1505–19. [DOI] [PubMed] [Google Scholar]
- 4.Daniel CW, et al. , The influence of mammogenic hormones on serially transplanted mouse mammary gland. Exp Gerontol, 1971. 6(1): p. 95–101. [DOI] [PubMed] [Google Scholar]
- 5.Raafat A, et al. , Effects of age and parity on mammary gland lesions and progenitor cells in the FVB/N-RC mice, in PLoS One. 2012: United States. p. e43624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boulanger CA, et al. , Interaction with the mammary microenvironment redirects spermatogenic cell fate in vivo. Proc Natl Acad Sci U S A, 2007. 104(10): p. 3871–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeps N, et al. , Detection of a population of long-lived cells in mammary epithelium of the mouse. Cell Tissue Res, 1996. 286(3): p. 525–36. [DOI] [PubMed] [Google Scholar]
- 8.Zeps N, et al. , Estrogen receptor-negative epithelial cells in mouse mammary gland development and growth. Differentiation, 1998. 62(5): p. 221–6. [DOI] [PubMed] [Google Scholar]
- 9.Welm BE, et al. , Sca-1(pos) cells in the mouse mammary gland represent an enriched progenitor cell population. Dev Biol, 2002. 245(1): p. 42–56. [DOI] [PubMed] [Google Scholar]
- 10.Anderson E and Clarke RB, Steroid receptors and cell cycle in normal mammary epithelium. J Mammary Gland Biol Neoplasia, 2004. 9(1): p. 3–13. [DOI] [PubMed] [Google Scholar]
- 11.Booth BW and Smith GH, Estrogen receptor-alpha and progesterone receptor are expressed in label-retaining mammary epithelial cells that divide asymmetrically and retain their template DNA strands. Breast Cancer Res, 2006. 8(4): p. R49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kangsamaksin T and Morris RJ, Bone morphogenetic protein 5 regulates the number of keratinocyte stem cells from the skin of mice. J Invest Dermatol, 2011. 131(3): p. 580–5. [DOI] [PubMed] [Google Scholar]
- 13.Booth BW, Boulanger CA, and Smith GH, Selective segregation of DNA strands persists in long-label-retaining mammary cells during pregnancy. Breast Cancer Res, 2008. 10(5): p. R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Potten CS, Owen G, and Booth D, Intestinal stem cells protect their genome by selective segregation of template DNA strands. J Cell Sci, 2002. 115(Pt 11): p. 2381–8. [DOI] [PubMed] [Google Scholar]
- 15.Runck LA, et al. , Identification of epithelial label-retaining cells at the transition between the anal canal and the rectum in mice. Cell Cycle, 2010. 9(15): p. 3039–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith GH and Medina D, Does the Mouse Mammary Gland Arise from Unipotent or Multipotent Mammary Stem/Progenitor Cells? J Mammary Gland Biol Neoplasia, 2018. 23(1–2): p. 1–3. [DOI] [PubMed] [Google Scholar]
- 17.Scheele CL, et al. , Identity and dynamics of mammary stem cells during branching morphogenesis. Nature, 2017. 542(7641): p. 313–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fernandez-Gonzalez R, et al. , In situ analysis of cell populations: long-term label-retaining cells. Methods Mol Biol, 2010. 621: p. 1–28. [DOI] [PubMed] [Google Scholar]
- 19.Fernandez-Gonzalez R, et al. , Mapping mammary gland architecture using multi-scale in situ analysis. Integr Biol (Camb), 2009. 1(1): p. 80–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.dos Santos CO, et al. , Molecular hierarchy of mammary differentiation yields refined markers of mammary stem cells. Proc Natl Acad Sci U S A, 2013. 110(18): p. 7123–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaanta AS, et al. , Evidence for a multipotent mammary progenitor with pregnancy-specific activity. Breast Cancer Res, 2013. 15(4): p. R65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choudhary V, et al. , Comparison of homogeneous and heterogeneous catalysts for glucose-to-fructose isomerization in aqueous media. ChemSusChem, 2013. 6(12): p. 2369–76. [DOI] [PubMed] [Google Scholar]
- 23.Wuidart A, et al. , Quantitative lineage tracing strategies to resolve multipotency in tissue-specific stem cells. Genes & Development, 2016. 30(11): p. 1261–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin H, The stem-cell niche theory: Lessons from flies. Nature Rev. Genet, 2002. 3: p. 931–939. [DOI] [PubMed] [Google Scholar]