Abstract
Stable cell lines can continuously produce a recombinant protein without the need to repeatedly engineer the genome. In a previous study HIPK1, Homeodomain-interacting Protein Kinase 1, was found to be a target of the microRNA miR-22 that, when repressed, improved expression of both an intracellular and a secreted protein. In this report, HEK293 cells stably over-expressing miR-22 were compared with HEK293 with knockout of HIPK1, executed by CRISPR/Cas9, for their ability to improve recombinant protein expression. In this model case of luciferase, over-expression of miR-22 improved overall activity 2.4-fold while the HIPK1 knockout improved overall activity 4.7-fold.
Keywords: miR-22, CRISPR/Cas9, HIPK1, stable, protein expression
Introduction
Improved recombinant protein expression from mammalian cells is a long sought-after objective that is being accomplished in various ways [1–3]. Recently, this aim is also being explored utilizing small non-coding regulatory RNAs, especially microRNAs [4–6]. MicroRNAs, are short non-coding regulatory RNAs about 22 nucleotides long [7] that affect the expression of genes by binding to their mRNA [8, 9]. A single microRNA can affect the expression of several genes and currently over 2000 different human microRNAs have been identified [10]. Compared with transcription factors, regulatory proteins or kinases, microRNAs, as a result of their lower burden on the translational machinery, exert reduced metabolic load in the host cell, allowing cell resources to be used more efficiently towards recombinant protein production [11, 12]. In recent studies, microRNAs have been implemented to improve recombinant protein expression in mammalian cells by creating stable, high producing cell lines [4–6, 11, 13]. For example, over-expression of miR-17 was shown to increase specific and overall recombinant human erythropoietin fusion protein (EpoFc) titer in Chinese hamster ovary (CHO) cells [14] and overexpression of miR-557 and miR-1287 increased productivity in CHO-IgG cell lines while miR-557 helped with some difficult-to-express proteins [15, 16]. Another microRNA, miR-183, related to cell cycle and proliferation, was shown to improve specific productivity of CHO cells [17]. The converse approach of stably depleting a specific microRNA through sponges and decoys has also been successfully used to improve productivity [11, 18, 19]. While CHO cells are currently the most widely used for recombinant protein expression, human cell lines, such as human embryonic kidney cells (HEK293) have the advantage of human post-translational modifications, which makes them useful production tools for certain human proteins [20].
Some options for creating stable cell lines based on results obtained from a microRNA effect include over-expressing the identified microRNA or the deletion of a specific target gene or genes [15–17, 21, 22]. However, since a single microRNA can affect the expression of several genes, either approach has its drawbacks: it is possible that over-expression of a single microRNA will affect unrelated cell functions, and unless a specific gene has been identified, it is not practical to delete or inactivate several target genes at the same time. In a previous high-throughput screen of 875 human microRNAs, miR-22 was identified as a top candidate for improving the expression of several proteins, including two membrane proteins, a secreted protein and a reporter protein [23]. Following this information by implementing a high-throughput siRNA screen [24] and a microarray analysis [25] it was possible to identify the Homeodomain-interacting Protein Kinase 1 (HIPK1) as a target of miR-22 that, when repressed, improved expression of both a reporter protein, firefly luciferase, and a secreted protein, glypican-3 human (h) Fc fusion protein. The identification of a microRNA and one of its specific targets provided the opportunity to compare the protein expression capability of stable HEK293 cell lines over-expressing miR-22 with stable HEK293 cell lines with HIPK1 knocked out using CRISPR/Cas9. In this model case of firefly luciferase, over-expression of miR-22 improved overall activity by 2.4-fold while the HIPK1 knockout improved overall activity 4.7-fold.
Materials and Methods
Cell lines and cultures
A CMV-LUC2-Hygro HEK293 cell line (Luc-HEK cells) constitutively expressing Photinus pyralis firefly luciferase was purchased from Promega (Madison, WI, USA). Anchorage-dependent cells were maintained in 10% Fetal Bovine Serum (FBS, Atlanta Biologicals, Flowery Branch, GA, USA). Dulbecco’s Modified Eagle Medium (DMEM10, Gibco, Gaithersburg, MD, USA) and cells adapted to suspension were maintained in Freestyle medium (Gibco, Gaithersburg, MD, USA) on a shaker at 130 rpm. Experiments were completed with cells between passage number 3 and 50. Cells were kept in a humidified incubator set at 5% CO2 and 37°C.
Stable microRNA-22 transfection
Luc-HEK cells were transfected with the pCMV-miR-22 vector or pCMV-miR-negative control vector (Origene Technologies, Rockville, MD, USA) (see Supplementary Figure 1) in a 24-well plate with Lipofectamine3000 (Life Technologies, Carlsbad, CA, USA), according to the manufacturer’s protocol. Cells were selected with G418 Genticin (Life Technologies, Carlsbad, CA, USA) and clones were sorted with green fluorescent protein (GFP)-based fluorescence-activated cell sorting (FACS) single cell sorting (FACSAria 2, Becton Dickinson, San Jose, CA; a 488nm laser operating at 100 mW was used for excitation of the GFP and the fluorescence of the GFP was detected in two channels using a 515/20 bandpass filter in one channel and a 576/25 bandpass filter in the second) then selected with luciferase and cell viability assays (see below).
Luciferase activity, western blot, and cell viability assays
Luciferase expressing cells were harvested and transferred to 96-well plate assays, where luciferase was assayed with ONE-Glo™ Reagent (Promega, Madison, WI, USA) and viability measured with CellTiter-Glo Reagent (Promega, Madison, WI, USA), using a SpectraMax i3 plate reader (Molecular Devices, San Jose, CA, USA) according to the manufacturer’s protocol. The ‘per cell luciferase production’ was calculated from overall luciferase activity and viable cell number. P-values were calculated with a two-sample unpaired t-test assuming unequal variances with the data analysis package in Excel. For luciferase activity and western blot, harvest was performed 72 h after seeding for 3 consecutive passages after clones reached confluency in a T25 plate following single cell cloning. For growth studies, harvest was performed daily as described below. For the western blot, luciferase expressing cells were transfected as above in duplicates and lysed using radioimmunoprecipitation buffer (RIPA) buffer with protease and phosphatase inhibitor cocktail (halt™ Protease and phosphatase inhibitor cocktail (100x) # 78440 Thermo Fisher Scientific, Waltham, MA, USA). Proteins were separated with aNuPAGE 4–12% bis-tris gel (Thermo Fisher Scientific, Waltham, MA, USA) and transferred to a nitrocellulose membrane using the iBlot Gel Transfer System (Invitrogen, Carlsbad, CA, USA). This was then used for immunodetection with mouse anti-luciferase at a 1:1,500 dilution (Thermo Fisher, # PA1–179, Rockford, IL, USA) and mouse anti-β-actin at a 1:1,000 dilution (Sigma-Aldrich, # A2228 St. Louis, MO, USA) as primary antibodies and a horseradish peroxidase (HRP) conjugated goat anti-mouse secondary antibody at a 1:5,000 (#474–1806, KPL, Sera Care Milford, MA, USA). The membrane was stripped between detection of luciferase and β-actin using Restore Plus stripping buffer (Thermo Fisher Scientific, Waltham, MA, USA). Signals were detected with an ECL Plus chemiluminescence reagent (Thermo Fisher Scientific, Waltham, MA, USA). Densitometry fold change calculations were performed using the gel analysis feature of ImageJ software (National Institutes of Health, Bethesda, MD, USA). Background was subtracted and the luciferase value was normalized by the B-actin for loading control. For molecular weight markers the MagicmarkTM XP western protein standard (Invitrogen) was used.
RNA and DNA extraction
Total RNA was extracted from the cell pellets with the miRNEasy kit (Qiagen, Hilden, Germany) with DNase Digestion following the manufacturer’s protocol with an extra RPE buffer (Qiagen) wash. Genomic DNA was extracted from the cell pellets using the DNEasy Blood and Tissue Kit (Qiagen) following the manufacturer’s protocol. RNA and DNA concentrations and quality were determined with the NanoDrop 2000 or NanoDropOne Spectrophotometer (Thermo Fisher Scientific).
qRT PCR
For microRNA expression analysis, the miScript PCR starter kit (Qiagen) was used following the manufacturer’s instructions with 100 ng RNA and measured on the 7500 Fast Real Time PCR System (Applied Biosystems, Foster City, CA, USA). Relative gene expression was calculated using the 2−ΔΔCT method with human RNU6B as the reference gene [26]. For Luciferase expression analysis, the Maxima First strand cDNA synthesis (Thermo Fisher Scientific) and Sybr Green (Applied Biosystems) were used according to the manufacturer’s instructions with 500 ng of RNA and measured on the CFX96 Touch (Bio-Rad Laboratories, Hercules, CA, USA). Relative gene expression was calculated using the 2−ΔΔCT method with human GAPDH as the reference gene. Primer sequences can be found in Supplementary Table 1.
Growth Studies
Suspension cells were seeded at 15,000 cells per mL in a 125-mL shake flask. A 1.5 mL sample was taken daily and measured for glucose and lactate measurement using the YSI (Yellow Springs Instrument Co., Yellow Springs, OH, USA), cell count using the Cedex HiRes (Roche, Basel, Switzerland) and luciferase and cell viability as described above.
Stable HIPK1 knockout
Luc-HEK cells were transduced using a lentiviral CRISPR/Cas9 system (Sigma-Aldrich, St. Louis, MO, USA) to knock out HIPK1 (see Supplementary Figure 2) or a non-targeting control according to the manufacturer’s protocol. Cells were maintained with puromycin (Life Technologies, Carlsbad, CA, USA) pressure and clones were sorted with GFP based -FACS single cell sorting then selected with luciferase and cell viability assays.
Sequencing Analysis
The section of HIPK1 targeted by the gRNA of the CRISPR lentivirus was amplified from the genomic DNA using PCR with Phusion High Fidelity PCR master mix (New England Biolabs, Ipswich, MA) following the manufacturer’s protocol. PCR primers are listed in Supplemental Table 1. For each sample, 2-50 μL PCR reactions were purified using the QIAquick gel extraction kit (Qiagen, Hilden, Germany) after a gel electrophoresis in a 0.8% agarose gel. The Center for Biologies Evaluation and Research at the Food and Drug Administration (Silver Spring, MD, USA) then sequenced these samples.
TOPO cloning
Using the same PCR amplification and gel extraction described in the sequencing analysis, PCR products with blunt ends were produced, cloned into the pCR-Blunt-TOPO vector (Invitrogen, Carlsbad, CA) and transformed into One shot competent E. coli following the manufacturer recommendations. For each sample, 10 colonies were selected, cultured overnight in 2 mL of LB media with 50 μg/ml kanamycin and then the plasmids were extracted with the QIAprep Spin Miniprep kit (Qiagen) and sequenced by the Center for Biologics Evaluation and Research at the Food and Drug Administration.
Adaption to suspension
Anchorage dependent cells were gradually adapted to suspension in a stepwise manner, decreasing the concentration of the medium from DMEM10 and adapting the cells to a chemically defined Freestyle (Gibco, Gaithersburg, MD, USA) medium, 20% each passage, keeping the concentration the same for multiple passages if needed for the adaption. Once the cells were adapted to chemically defined media, they were added to non-tissue culture treated T flasks, placed on a shaker at 125 rpm and finally transferred to a 125 mL shake-flask.
Results
Effect of stable over-expression of miR-22 on luciferase expression
To evaluate the effect of stable over-expression of miR-22 on protein expression, cells constitutively expressing firefly luciferase were transfected with a miR-22 plasmid or a negative control vector. Cells over-expressing the miR-22 plasmid were selected with antibiotic and then sorted with GFP-based FACS for single cell cloning. The clone selected was based on luciferase and cell viability assays and was compared to the parental luciferase cells (Luc-HEK), to the miR-22 expressing pool and the negative control (Luc-HEK-miR-NC) (Figure 1A). Overall luciferase activity in the selected cell line was 2.4-fold higher and luciferase per cell was 2.7- fold higher than the parental Luc-HEK and 2.0-fold higher than the negative control. The overall luciferase activity of the unsorted pool over-expressing miR-22 was also higher than both the negative control and the parental cells. The western blot (Figure 1B) demonstrated that the luciferase protein expression was increased by a fold change of 2.7 in the mir-22 clone and 2.9-fold higher in the mir-22 pool compared with Luc-HEK (as determined by densitometry analysis), and quantitative PCR supported the over-expression of miR-22 (Figure 2); both precursor miR-22 and mature miR-22–3p, which target HIPK1, were over-expressed compared with the negative control and the parental Luc-HEK cells. A growth and viability study (Supplementary Figure 3 A, B) showed a slightly lower rate in the cells with over-expressed miR-22. Glucose consumption and lactate production were in accordance with the growth data (Supplementary Figure 3 C, D).
Figure 1:
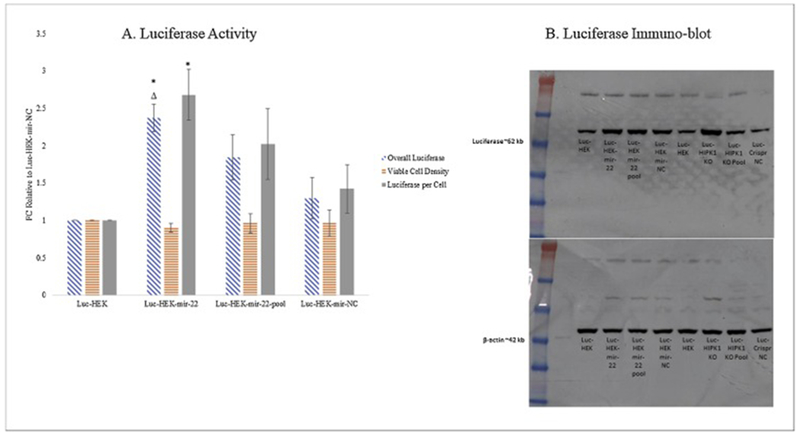
Effect of over-expression of miR-22 in luciferase-expressing HEK cells.
A) Fold change (FC) of overall luciferase (blue), cell viability (orange), and luciferase per cell (grey) of Luc-HEK, Luc-HEK-miR-22, Luc-HEK-miR-22 pool and Luc-HEK-miR-NC cells relative to Luc-HEK demonstrates improved luciferase activity with microRNA 22 over-expression. Error bars represent Standard Error of the Mean (SEM) from triplicate measurements. * indicates P ≤ 0.05 relative to Luc-HEK and Δ indicates P ≤ 0.05 relative to Luc-HEK-miR-NC calculated using two-sample unpaired t-test assuming unequal variances. B) Western blot analysis confirms that over-expression of miR-22 improves luciferase expression in Luc-HEK-miR-22 (2.7 FC) and the Luc-HEK-miR-22 pool (2.9 FC) Luc-HEK-HIPK1 KO (2.4 FC), and the Luc-HEK-HIPK1 KO pool (1.5 FC) compared to Luc-HEK. A monoclonal antibody against firefly luciferase was used to detect the protein, with antibodies against β-actin as an endogenous control.
Figure 2:
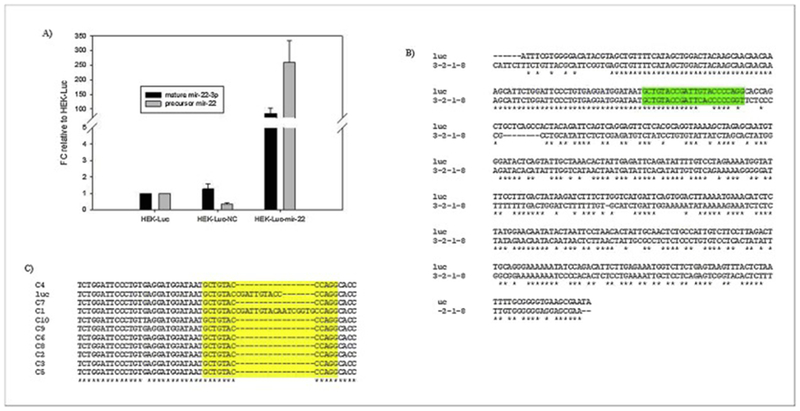
Verification of miR-22 overexpression and HIPK1 knockout
A) Real time qPCR of reverse-transcribed total cellular mRNA, using primers targeting precursor miR-22 and mature miR-22-3p confirm increased transcription levels of miR-22 in Luc-HEK-miR-22. RNU6B was used as an endogenous control. B) Clustal Omega DNA sequence alignment of CRISPR HIPK1 guide RNA target and surrounding regions, showing mutations in the CRISPR/CAS9 treated cell line Luc-HEK-HIPK1-KO (3-1-2-8) compared to the parental Luc-HEK cell line. Highlighted region is the gRNA target sequence. C) Clustal Omega DNA sequence alignment from 10 colonies of TOPO cloned Luc-HEK-HIPK1 cells demonstrate that the mutation is biallelic.
Effect of stable HIPK1 knockout on luciferase expression
To determine the effect of stable HIPK1 knockout on protein expression, the CRISPR/Cas9 lentiviral system was used to transduce luciferase expressing HEK cells. The cells were then put under antibiotic pressure and sorted with GFP-based FACS single cell cloning. The clone selected was based on overall luciferase and cell viability assays (Figure 3) and showed a 4.7-fold higher luciferase activity, and 4.4-fold higher specific luciferase activity per cell than the parental Luc-HEK cells. The HIPK1 KO clone also had a 3.6-fold higher specific activity per cell than the negative control. The overall luciferase activity of the unsorted pool was 1.5-fold higher than the Luc-HEK cells and 1.3-fold higher than the negative control. The western blot (Figure 1B) confirmed the improved luciferase expression in the HIPK1 KO of 2.4-fold and the pool was 1.5-fold higher than the Luc-HEK (as determined by densitometry analysis). To confirm HIPK1 gene mutation, the section targeted by the gRNA was amplified with PCR, gel purified and sequenced (Figure 2B). Then TOPO cloning, and sequencing was performed to confirm a double stranded break (Figure 2C). Transcriptional and translational analyses determined that the CRISPR-mediated mutation in the Luc-HEK HIPK1 KO clone introduces a premature stop codon into the HIPK1 gene, leading to a predicted truncated protein of 736 amino acids; the full-length protein is 1230 amino acids long (Supplementary Figure 4). This truncated protein would be deficient in some important functions such as nuclear localization and kinase activity, likely making the protein functionally inactive. A growth and viability study showed that the parental cells grew faster than the HIPK1 KO cells (Supplementary Figure 5A and B) and glucose consumption and lactate production were measured and were in accordance with the growth data (Supplementary Figure 5C and D).
Figure 3:
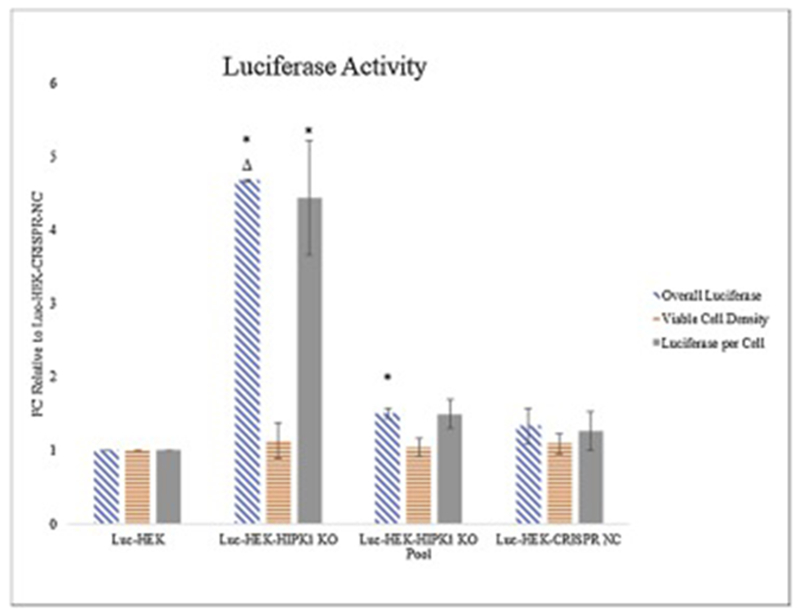
Effect of knock-out of HIPK1 in luciferase-expressing HEK cells
Fold change (FC) of the overall luciferase (blue), cell viability (orange), and luciferase per cell (grey) of Luc-HEK, Luc-HEK-HIPK1 KO, Luc-HEK-HIPK1 KO pool, and Luc-HEK-CRISPR-NC cells relative to Luc-HEK demonstrates improved luciferase activity with knockout of HIPK1. Error bars represent Standard Error of the Mean (SEM) from triplicate measurements. * indicates P ≤ 0.05 relative to Luc-HEK and Δ indicates P ≤ 0.05 relative to Luc-HEK-CRISPR-NC calculated using two-sample unpaired t-Test assuming unequal variances.
Adaption to anchorage independent culture conditions
Since suspension cells are easily scalable, and can be grown in a bioreactor, the anchorage-dependent over-expressing miR-22 and HIPK1 knockout clones along with the parental Luc-HEK cell line were adapted to suspension culture in a stepwise process. Supplementary Figure 6 shows the comparison of viable cell density and cell viability, glucose consumption, and lactate production between the different cell lines. The growth rates were similar but a little slower in the modified than the parental cells. Glucose consumption and lactate production were in accordance with the growth data.
Discussion
Large amounts of recombinant protein are needed for a variety of industrial and research purposes and improving protein expression by stable over-expression of microRNAs has been successfully implemented in the Chinese hamster ovary (CHO) cells [14–17]. MicroRNAs are non-coding RNAs and thus the translational burden of over-expression is reduced which makes them advantageous for cell engineering [11]. However, since microRNAs regulate multiple genes [27, 28] there is the potential for some advantageously regulated genes to be outweighed by some that inhibit recombinant protein expression. Most microRNAs including miR-22 are not completely elucidated and while there are many predicted targets, most have not been validated [29, 30]. For this reason, it could be argued that knocking down or knocking out a single gene that has been shown to improve recombinant protein expression is a better method. In this study the effects were compared of a stable over-expressing miR-22 clone and a stable knockout of HIPK1, which is the identified target of miR 22 (a protein kinase with co-repressive effects on transcription), on HEK cells expressing luciferase [31, 32]. While both stable cell lines improved luciferase expression compared to the parental cell lines and the negative contol, the HIPK1 KO showed higher luciferase expression in the HEK cells. The growth rates of both the microRNA over-expressing cells and the HIPK1 knockout cells were lower than those of the parental cell lines which may suggest that some of their growth machinery has been redirected towards production, but could additionally be due to the antibiotics used for selection.
The comparison shows that both stable over-expression of microRNA 22 and stable knockout of HIPK1 improve recombinant protein expression in HEK293 cells. In the example provided here, knockout of HIPK1 improves the expression of recombinant protein to a greater extent than that achieved by overexpressing microRNA-22, but the process of generating and confirming a CRISPR knockout is more time consuming. In addition, CRISPR/Cas9 editing has often been associated with off-target effects which could affect the phenotype, hence the guide and protospacer adjacent motif (PAM) sequences as well as delivery methods, need to be carefully selected [33]. With advancements in CRISPR technology this may be streamlined and more efficient in the future. A knockout of a single gene also allows more understanding of the improved process in the cell than overexpressing the microRNA.
Supplementary Material
Highlights.
Mir-22 improved luciferase expression 2.4-fold, while improvement by HIPK1 knockout was 4.7-fold.
Both HEK293 cell lines were able to be adapted to suspension growth conditions
Although CRISPR/Cas9 knockout is more effective, it is time consuming and labor intensive.
Acknowledgments
The authors thank Mr. B. Inwood for critical reading of the manuscript and editorial assistance. The research was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK/NIH).
Abbreviations:
- CHO
Chinese hamster ovary
- CRISPR
Clustered Regularly Interspaced Short Palendromic Repeats
- DMEM
Dulbecco’s Modified Eagle Medium
- EpoFC
recombinant human erythropoietin fusion protein
- FBS
fetal bovine serum
- FC
fold change
- gRNA
guide RNA
- GFP
green fluorescent protein
- FACS
fluorescence-activated cell sorting
- HEK293
Human Embryonic Kidney cells
- HIPK1
Homeodomain-interacting Protein Kinase 1
- HIPK1 KO
HIPK1 knockout
- HRP
horseradish peroxidase
- miR
microRNA
- Luc-HEK
Luciferase expressing Human Embryonic Kidney cells
- NC
negative control
- PAM
protospacer adjacent motif
- RIPA
radioimmunoprecipitation
- siRNA
small interfering RNA
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The authors declare no financial or commercial conflict of interest.
References
- [1].Hacker DL, Balasubramanian S. Recombinant protein production from stable mammalian cell lines and pools. Curr Opin Struct Biol 2016;38:129–36. doi: 10.1016/j.sbi.2016.06.005. [DOI] [PubMed] [Google Scholar]
- [2].Collins JH, Young EM. Genetic engineering of host organisms for pharmaceutical synthesis. Curr Opin Biotechnol 2018;53:191–200. doi: 10.1016/j.copbio.2018.02.001. [DOI] [PubMed] [Google Scholar]
- [3].Palomares LA, Estrada-Mondaca S, Ramirez O. Production of Recombinant Proteins: Challenges and Solutions. Methods Mol Biol 2004;276:15–52. doi: 10.1385/1-59259-774-2:015. [DOI] [PubMed] [Google Scholar]
- [4].Ambros V microRNAs: Tiny regulators with great potential. Cell 2001;107:823–6. doi: 10.1016/S0092-8674(01)00616-X. [DOI] [PubMed] [Google Scholar]
- [5].Barron N, Sanchez N, Kelly P, Clynes M. MicroRNAs: Tiny targets for engineering CHO cell phenotypes? Biotechnol Lett 2011. doi: 10.1007/s10529-010-0415-5. [DOI] [PubMed] [Google Scholar]
- [6].Druz A, Betenbaugh M, Shiloach J. MicroRNAs as engineering targets: Pathway manipulation to impact bioprocess phenotypes. MicroRNAs as Tools Biopharm. Prod, vol. 9789400751, 2012, p. 65–85. doi: 10.1007/978-94-007-5128-6_5. [DOI] [Google Scholar]
- [7].Moran Y, Agron M, Praher D, Technau U. The evolutionary origin of plant and animal microRNAs. Nat Ecol Evol 2017;1:27. doi: 10.1038/s41559-016-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Steinkraus BR, Toegel M, Fulga TA. Tiny giants of gene regulation: Experimental strategies for microRNA functional studies. Wiley Interdiscip Rev Dev Biol 2016;5:311–62. doi: 10.1002/wdev.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Nelson P, Kiriakidou M, Sharma A, Maniataki E, Mourelatos Z. The microRNA world: small is mighty. Trends Biochem Sci 2003;28:534–40. [DOI] [PubMed] [Google Scholar]
- [10].Wong N, Wang X. miRDB: An online resource for microRNA target prediction and functional annotations. Nucleic Acids Res 2015;43:D146–52. doi: 10.1093/nar/gku1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kelly PS, Gallagher C, Clynes M, Barron N. Conserved microRNA function as a basis for Chinese hamster ovary cell engineering. Biotechnol Lett 2015;37:787–98. doi: 10.1007/s10529-014-1751-7. [DOI] [PubMed] [Google Scholar]
- [12].Jadhav V, Hackl M, Druz A, Shridhar S, Chung CY, Heffner KM, et al. CHO microRNA engineering is growing up: Recent successes and future challenges. Biotechnol Adv 2013;31:1501–13. doi: 10.1016/j.biotechadv.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Inwood S, Betenbaugh MJ, Shiloach J. Methods for using small non-coding mas to improve recombinant protein expression in mammalian cells. Genes (Basel) 2018;9:209–19. doi: 10.3390/genes9010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Jadhav V, Hackl M, Klanert G, Hernandez Bort JA, Kunert R, Grillari J, et al. Stable overexpression of miR-17 enhances recombinant protein production of CHO cells. J Biotechnol 2014;175:38–44. doi: 10.1016/j.jbiotec.2014.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Strotbek M, Florin L, Koenitzer J, Tolstrup A, Kaufmann H, Hausser A, et al. Stable microRNA expression enhances therapeutic antibody productivity of Chinese hamster ovary cells. Metab Eng 2013;20:157–66. doi: 10.1016/j.ymben.2013.10.005. [DOI] [PubMed] [Google Scholar]
- [16].Fischer S, Marquart KF, Pieper LA, Fieder J, Gamer M, Gorr I, et al. miRNA engineering of CHO cells facilitates production of difficult-to-express proteins and increases success in cell line development. Biotechnol Bioeng 2017;114:1495–510. doi: 10.1002/bit.26280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Pfizenmaier J, Junghans L, Teleki A, Takors R. Hyperosmotic stimulus study discloses benefits in ATP supply and reveals miRNA/mRNA targets to improve recombinant protein production of CHO cells. Biotechnol J 2016;11:1037–47. doi: 10.1002/biot.201500606. [DOI] [PubMed] [Google Scholar]
- [18].Sanchez N, Kelly P, Gallagher C, Lao NT, Clarke C, Clynes M, et al. CHO cell culture longevity and recombinant protein yield are enhanced by depletion of miR-7 activity via sponge decoy vectors. Biotechnol J 2014;9:396–404. doi: 10.1002/biot.201300325. [DOI] [PubMed] [Google Scholar]
- [19].Druz A, Son YJ, Betenbaugh M, Shiloach J. Stable inhibition of mmu-miR-466h-5p improves apoptosis resistance and protein production in CHO cells. Metab Eng 2013;16:87–94. doi: 10.1016/j.ymben.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Dumont J, Euwart D, Mei B, Estes S, Kshirsagar R. Human cell lines for biopharmaceutical manufacturing: history, status, and future perspectives. Crit Rev Biotechnol 2016;36:1110–22. doi: 10.3109/07388551.2015.1084266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Schoellhom M, Fischer S, Wagner A, Handrick R, Otte K. miR-143 targets MAPK7 in CHO cells and induces a hyperproductive phenotype to enhance production of difficult-to-express proteins. Biotechnol Prog 2017;33:1046–58. doi: 10.1002/btpr.2475. [DOI] [PubMed] [Google Scholar]
- [22].Fischer S, Paul AJ, Wagner A, Mathias S, Geiss M, Schandock F, et al. miR-2861 as novel HDAC5 inhibitor in CHO cells enhances productivity while maintaining product quality. Biotechnol Bioeng 2015;112:2142–53. doi: 10.1002/bit.25626. [DOI] [PubMed] [Google Scholar]
- [23].Xiao S, Chen YC, Betenbaugh MJ, Martin SE, Shiloach J. MiRNA mimic screen for improved expression of functional neurotensin receptor from HEK 293 cells. Biotechnol Bioeng 2015;112:1632–43. doi: 10.1002/bit.25567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Xiao S, Chen YC, Buehler E, Mandal S, Mandal A, Betenbaugh M, et al. Genome-scale RNA interference screen identifies antizyme 1 (OAZ1) as a target for improvement of recombinant protein production in mammalian cells. Biotechnol Bioeng 2016;113:2403–15. doi: 10.1002/bit.26017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Inwood S, Buehler E, Betenbaugh M, Lai M, Shiloach J. Identifying HIPK1 as Target of miR-22– 3p Enhancing Recombinant Protein Production From HEK 293 Cell by Using Microarray and HTP siRNA Screen. Biotechnol J 2018;13. doi: 10.1002/biot.201700342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-AACT method. Methods 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- [27].Bhat SS, Jarmolowski A, Szweykowska-Kulihska Z. MicroRNA biogenesis: Epigenetic modifications as another layer of complexity in the microRNA expression regulation. Acta Biochim Pol 2016;63:717–23. doi: 10.18388/abp.2016_1370. [DOI] [PubMed] [Google Scholar]
- [28].Catalanotto C, Cogoni C, Zardo G. MicroRNA in control of gene expression: An overview of nuclear functions. Int J Mol Sci 2016; 17:1712. doi: 10.3390/ijms17101712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Shukla V, Varghese VK, Kabekkodu SP, Mallya S, Satyamoorthy K. A compilation of Web-based research tools for miRNA analysis. Brief Funct Genomics 2017;16:249–73. doi: 10.1093/bfgp/elw042. [DOI] [PubMed] [Google Scholar]
- [30].Hunt EA, Broyles D, Head T, Deo SK. MicroRNA Detection: Current Technology and Research Strategies. Annu Rev Anal Chem 2015;8:217–37. doi: 10.1146/annurev-anchem-071114-040343. [DOI] [PubMed] [Google Scholar]
- [31].Matre V, Nordgard O, Alm-Kristiansen AH, Ledsaak M, Gabrielsen OS. HIPK1 interacts with c-Myb and modulates its activity through phosphorylation. Biochem Biophys Res Commun 2009;388:150–4. doi: 10.1016/j.bbrc.2009.07.139. [DOI] [PubMed] [Google Scholar]
- [32].Isono K -i., Nemoto K, Li Y, Takada Y, Suzuki R, Katsuki M, et al. Overlapping Roles for Homeodomain-Interacting Protein Kinases Hipk1 and Hipk2 in the Mediation of Cell Growth in Response to Morphogenetic and Genotoxic Signals. Mol Cell Biol 2014;26:2758–71.doi: 10.1128/mcb.00609-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhang XH, Tee LY, Wang XG, Huang QS, Yang SH. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol Ther - Nucleic Acids 2015;4:e214. doi: 10.1038/mtna.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


