Abstract
Subunit vaccines containing one or more target antigens from pathogenic organisms represent safer alternatives to whole pathogen vaccines. However, the antigens by themselves are not sufficiently immunogenic and require additives known as adjuvants to enhance immunogenicity and protective efficacy. Assembly of the antigens into virus-like nanoparticles (VLPs) is a better approach as it allows presentation of the epitopes in a more native context. The repetitive, symmetrical, and high density display of antigens on the VLPs mimic pathogen-associated molecular patterns seen on bacteria and viruses. The antigens, thus, might be better presented to stimulate host’s innate as well as adaptive immune systems thereby eliciting both humoral and cellular immune responses. Bacteriophages such as phage T4 provide excellent platforms to generate the nanoparticle vaccines. The T4 capsid containing two non-essential outer proteins Soc and Hoc allow high density array of antigen epitopes in the form of peptides, domains, full-length proteins, or even multi-subunit complexes. Co-delivery of DNAs, targeting molecules, and/or molecular adjuvants provides additional advantages. Recent studies demonstrate that the phage T4 VLPs are highly immunogenic, do not need an adjuvant, and provide complete protection against bacterial and viral pathogens. Thus, phage T4 could potentially be developed as a “universal” VLP platform to design future multivalent vaccines against complex and emerging pathogens.
Keywords: Vaccines, Virus like particle, Bacteriophage T4, Phage display, Phage assembly, DNA packaging
1. Introduction
Vaccines are one of the most successful and cost-effective medical interventions in the past 100 years. Millions of lives have been saved by mass administration of vaccines [1, 2]. Some of the deadly pathogens such as the smallpox virus that caused millions of deaths have been eradicated [3]. Despite the enormous successes, effective vaccines are still lacking for many complex pathogens that cause serious diseases such as HIV/AIDS, tuberculosis, and malaria [4, 5].
Historically, vaccines were developed using the whole pathogen, either the attenuated (live-attenuated vaccines) or the inactivated (heat- or formalin-killed vaccines) organism [1, 2]. However, there are many technical challenges in continuing these traditional approaches to develop whole pathogen vaccines [5]. Additionally, the whole pathogen vaccines pose significant safety risks including reversion to a pathogenic form, severe reactions in immunocompromised hosts, and adverse effects such as allergic and autoimmune responses [6].
Subunit vaccines containing only the well-defined antigenic parts of the pathogens represent safer alternatives to the whole-pathogen vaccines and also provide unique opportunities to develop vaccines where traditional approaches have failed [7–9]. However, subunit vaccines lack properties associated with whole pathogens and cannot efficiently elicit immune responses needed for protection [8–10]. However, thanks to recent advances, particularly the identification of components capable of stimulating the immune system [11, 12] and efficient vaccine delivery vehicles [13–15], it is possible to rationally design effective subunit vaccines and also ensure long-term protective immunity [16–18].
The immune system has evolved many strategies to efficiently recognize pathogens, such as viruses, and mount strong and protective immune responses to eliminate the infection. The key properties of viruses that are responsible for eliciting such responses may be used as a framework for vaccine design. These include: size, geometry, highly ordered and repeat structure, and multivalent display [18, 19]. Virus-like particles (VLPs) are nanometer scale particles containing similarly assembled viral structural proteins but lack the viral genome and hence the virulence [20, 21]. These nanoparticles by mimicking the structure and organization of the authentic viruses can simulate viral infection and stimulate robust immune responses by the host to antigens associated with the particles [20–22]. Successful VLP vaccines have been developed and commercialized worldwide using this approach. Examples include the hepatitis B vaccine and the human papillomavirus vaccine [20, 21].
A general VLP platform that could incorporate any antigen, or multiple antigens, would greatly streamline the development of the subunit vaccines against many pathogens. It would also dramatically reduce the cost of licensing and manufacture and make the vaccines available on a global scale. Although several viral and synthetic nanoparticles have been under investigation, none has yet emerged as a “universal” platform to assemble foreign antigens [14, 23–27]. Bacteriophages (phages) are highly promising candidates to develop broadly applicable VLP vaccine platforms because of their size, surface structure, safety, stability, biodegradability, and low cost of manufacture [28–33]. In this review, we focus on the bacteriophage T4 which offers unique structural advantages for the assembly of pathogen molecules, both proteins and DNAs, into VLPs and have been shown to elicit potent immune responses.
2. Basic pathways of vaccine-induced immunity
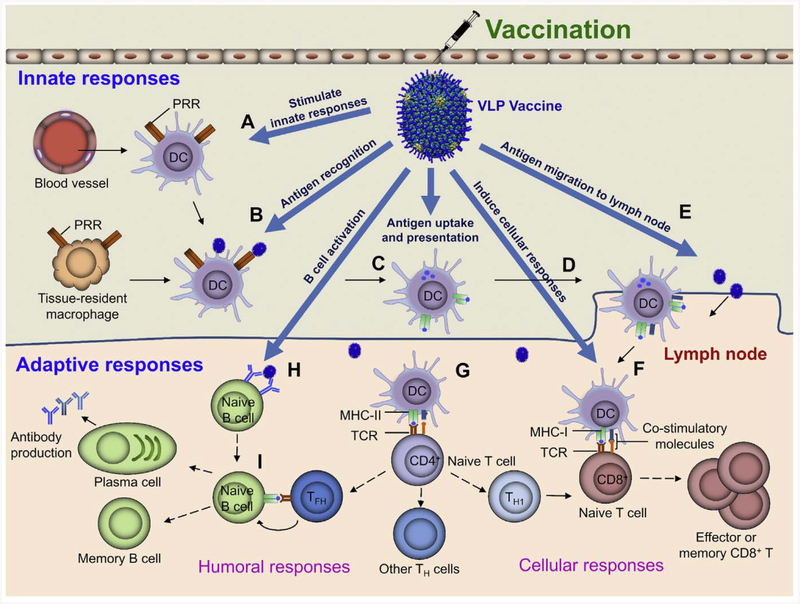
Vaccines stimulate the host immune system to build defenses against a pathogen without a necessary prior exposure to the pathogen, The antigens in the vaccine formulation will be first recognized by the innate immune system which then lead to the induction of adaptive immune responses through the activation of antigen-presenting cells (APCs) such as the dendritic cells (DCs) [10, 19] (Fig. 1). The adaptive immune responses are capable of eliminating the infections more efficiently and specifically than the innate immune responses.
Fig. 1.
Schematic diagram showing potential ways the VLP-based vaccines can stimulate innate and adaptive immune responses. The basic framework of innate and adaptive responses by the host immune system was adapted from Desmet et al. [36]. See text for details of specific advantages provided by VLP vaccines.
2.1. Innate immune responses
The innate immune system has evolved to recognize evolutionarily conserved signatures present in pathogens, called pathogen-associated molecular patterns (PAMPs). Recognition occurs through various pattern-recognition receptors (PRRs) expressed by cells involved in innate immunity such as the macrophages, mast cells, neutrophils, and DCs [34–36]. Engagement of PRRs such as the Toll-like receptors (TLRs) leads to activation of the innate immune cells and creates an inflammatory microenvironment, which then leads to the infiltration of immune cells from circulation to the site of infection, particularly neutrophils and APCs (Fig. 1, A and B) [37]. DCs are professional APCs and widely distributed in all tissues, importantly at the mucosal surfaces and lymphoid organs [38]. Both the tissue resident and migratory DCs efficiently detect pathogens through PRRs and take up antigens from extracellular environment by phagocytosis and micropinocytosis [39]. These lead to maturation of DCs, which involves down-regulation of antigen internalization, upregulation of antigen-processing machinery, and transportation of antigen peptide-loaded major histocompatibility complex (MHC) molecules from intracellular compartments to the surface of the DCs [40], DC maturation is essential for initiation of adaptive immune responses [41]. Only the maturated DCs are able to induce clonal expansion of antigen-specific naive T cells and their concomitant differentiation into effector T cells [41].
Maturation of DCs results in changes in the surface expression of the adhesion molecules and the chemokine receptors such as CCR7, CCR5, CCR1, CXCR1, and CXCR4, which allow DCs to migrate to peripheral lymphoid organs where adaptive immunity will be initiated (Fig. 1C and D) [40]. The whole pathogen vaccines such as the live-attenuated pathogens usually contain multiple PAMPs, hence can be efficiently recognized by PRRs resulting in the elicitation of strong innate immune responses [42]. On the other hand, (non-VLP) subunit vaccines containing well-defined antigens lack the PAMPs, thus requiring the addition of immunostimulators or adjuvants to stimulate strong adaptive immune responses [10].
2.2. Adaptive immune responses
Adaptive immune responses, which mainly depend on T cells and B cells, are initiated in the peripheral lymphoid organs in response to antigens presented by DCs matured during the course of the inmate immune response (Fig. 1C and D) [43]. Activation of T cells is the crucial first step in adaptive immunity. Naive T cells circulating in the bloodstream enter the peripheral lymph organs and sense the pathogen antigens presented by DCs. Naive T cells that do not encounter their specific antigen exit from the lymphoid tissue and return to circulation. Generally, activation of the naive T cells relies on three signals: signal 1 is antigen specific and derived from the interaction of T cell receptor (TCR) with its antigen peptide-MHC complex presented by DCs: signal 2, called the co-stimulatory signal, involves interaction of the co-stimulatory receptor CD28 constitutively expressed by naïve T cells with its ligand, B7, expressed on DCs; and signal 3, a cocktail of cytokines secreted mainly by DCs that direct the differentiation of T cells into subsets of effector T cells [44]. Naive T cells fall into two large groups, CD4+ and CD8+ T cells, based on the presence of the co-receptors of the TCR. The CD8+ T cells recognize antigen peptides presented by MHC class I molecules and differentiate into cytotoxic T lymphocytes (CTLs), which are critical in defense against intracellular pathogens (Fig. 1F) [45], Infection by intracellular pathogens (e.g., viruses) leads to the expression of pathogen antigens inside cells, which will be processed and presented by MHC class I molecules expressed by all nucleated cells and mainly present peptides derived from cytosolic antigens [46]. CTLs can kill these pathogen-infected cells by recognizing the surface-presented antigen peptides in the context of the MHC-1 molecules [45]. Inactivated or subunit vaccines, which cannot infect and express antigens inside cells, are considered as extracellular antigens. These vaccines are normally presented by MHC class II molecules, which are expressed mainly by professional APCs involved in the presentation of extracellular antigens, thus having poor ability to prime CD8+ T cells [46, 47]. However, it is possible that extracellular antigens, particularly the VLP vaccines can be presented by MHC-I through “cross-presentation” to activate CD8+ T cells (Fig. 1C and F) [48]. Many efforts have therefore been devoted to enhance the efficacy of subunit vaccines, particularly against intracellular pathogens by increasing their cross-presentation properties [49].
The CD4+ T cells on the other hand recognize antigen peptides presented by MHC class II molecules (Fig. 1G). Unlike the CD8+ T cells, CD4+ T cells can differentiate into several different kinds of effector T cells (T helper (Th) cells) such as Th1, Th2, Th17, follicular helper T cells (Tfh), and regulatory T cells (Treg) driven by different cytokines [50, 51]. Th1 effector cells are involved in the elimination of intracellular pathogens and triggered mainly by interleukin 12 (IL12) and interferon γ (IFNγ). Th2 cells are mainly against extracellular parasites and triggered mainly by IL4 and IL2. Th17 cells, which are responsible for elimination of extracellular bacteria and fungi, are differentiated mainly by IL6, IL21, IL23, and TGF-β [50, 51]. Tfh cells are located in follicular areas of lymphoid tissue, where they participate in the development of antigen-specific B-cell immunity [50, 51].
B cells, the precursor of antibody-secreting plasma cells, play central roles in humoral immune responses that protect against extracellular pathogens. The activation of B cells usually needs the help of effector Th cells (Fig. 1I). B cells and T cells locate in distinct zones of the lymphoid tissues, B cell zones (primary lymphoid follicles) and T cell zones, respectively [52]. The circulating naive T cells that enter lymphoid tissues get trapped in the T cell zone by recognizing antigen peptide presented by activated DCs and differentiate into Th cells [52]. The circulating naive B cells also enter the T cell zone, where they are activated by Th cells and then migrate to B cell zones [52]. Generally, activation of B cells requires two signals, one from the pathogen antigen and another from the Th cells. B cells can directly bind to pathogen antigen through surface immunoglobulin (also called the B cell receptor or BCR) by recognizing specific epitopes (Fig. 1H), which results in internalization, degradation, and presentation of pathogen peptide on B cell surface through MHC-II molecules (signal 1). Recognition of the peptides bound to MHC-II by Th cells provides the second signal, which includes the interaction of CD40 on B cells with CD40L on Th cells and cytokines secreted by Th cells. Activated B cells then migrate to lymphoid follicles and form germinal centers, where B cells undergo somatic hypermutation and affinity selection [53]. The selected B cells exit germinal centers and differentiate into either the long-lived memory B cells circulating in the periphery or the plasma cells that secrete antibodies [53].
3. Advantages of using bacteriophages as VLP vaccines
Since the report by George Smith in 1985 on the display of foreign peptides on filamentous phage f1 as fusions of the minor capsid gene product (gp) pIII [54], a number of phages have been developed as antigen delivery vehicles. These include in addition to M13 and related filamentous phages [29], phages λ [28], T4 [55, 56], T7 [57], MS2 [33], Qβ [58], and others [59]. Phages are widely distributed and probably constitute the largest proportion of the biomass on Earth [60]. >5000 phages have been studied [61], providing a large pool of phages to choose from for developing vaccine delivery vehicles. The basic principle involves assembly of the targeted pathogen antigen(s) fused to a virus structural protein into a virus-like particle (VLP), which simulates the surface architecture of the mammalian viruses. When injected into a mammalian host, the immune system may recognize the particle as an “invading virus”, efficiently stimulating immune responses against it by the mechanisms described above (Fig. 1). Another advantage of the phage VLPs is that many features of phages could be exploited to enhance immune responses against the antigens, such as surface structure, particle size, and incorporation of multiple targets.
3.1. Surface structure
The molecules found on the surface of most pathogenic viruses and other microbes unlike that on mammalian cells tend to be repetitive in nature [62]. These are recognized by the innate and adaptive immune systems as pathogen-associated geometric patterns similar to PAMPs [62]. The surface patterns found on phage virions are also repetitive in nature and correspond to the subunits of the major capsid protein(s) [63–65], similar to that seen on the surface of the pathogenic mammalian viruses. Antigens fused to the capsid protein will also be exposed on phage in a highly ordered and repetitive format [66]. Thus, phage VLPs are good simulators of pathogenic viruses to stimulate the innate immune system (Fig. 1, A and B). Furthermore, this pattern also allows efficient binding of their epitopes to natural IgM antibodies through high-avidity interactions, leading to the recruitment and activation of complement component 1q (C1q) and the classic pathway of the complement cascade [18]. This was demonstrated in the case of the phage Qβ capsid, which is composed of 90 copies of the capsid protein (CP) dimers. The capsids could bind to natural IgM and fix C1q, leading to their efficient deposition on follicular dendritic cells (FDCs) [67|. In contrast, the soluble CP dimer failed to activate this humoral innate immune system and was not deposited on FDCs [67]. Antigen retention on FDCs is essential for B cell activation and clonal selection within germinal centers [68, 69].
Other properties of the phage surface [15, 18] such as size, shape, charge, and hydrophobicity also mimic the properties of the pathogenic viruses and enhance the efficiency of antigen uptake by APCs. Consequently, phage VLP displayed antigens may be taken up efficiently by the APCs. Barfoot et al. demonstrated that T4 phages can be as efficiently taken up by DCs as the influenza virus [70]. Actually, phage has been used as a model to study the uptake of particulate antigens by macrophages, the professional APCs, since 1960s [71, 72].
3.2. Antigen density
Repetitive and highly localized epitope density of the antigen on the VLP is one important factor for B cell activation [73–75]. High-density promotes cross-linking of the B cell receptors to antigens and facilitates B-cell activation (Fig. 1H and I) [73, 74]. Studies indicate that displaying 60 epitopes per particle at 5–10 nm spacing is an ideal geometry for optimal B cell activation [73]. Indeed most of the licensed viral vaccines contain high density of antigens on the particle surface [76], and low density was suggested as one of the reasons why it has been difficult to develop HIV vaccines [76, 77]. Qβ phage displaying high density of a model peptide (D2) induced higher titers of peptide-specific IgG than phages displaying medium or low density of the same peptide [75]. Since phage capsids usually have hundreds of copies of the capsid protein(s), high density of antigen can be achieved relatively easily. For instance, the phage T4 capsid has 930 copies of major capsid protein gp23, 870 copies of small outer capsid protein (Soc), and 155 copies of highly antigenic outer capsid protein (Hoc) on the surface. Up to 860 copies of the gp41 envelope protein of HIV-1 virus could be displayed on T4 capsid through Soc, which is equivalent to 60–120 times greater density than that present on the HIV virion [78, 79]. Phage λ has 420 copies of the major capsid protein gpE and 405–420 copies of the outer capsid protein gpD. All of the gpD molecules could be used for displaying antigen epitopes.
3.3. Particle size
Adaptive immune responses are initiated in lymph nodes. Therefore, vaccine antigens must be transported to lymph nodes from the site of injection (Fig. 1E). This process is mediated by lymphatic drainage or through immune cell-mediated transport and is dependent on the size of the antigen [15, 18, 80]. Antigen particles that are <200 nm can efficiently enter the lymphatic system by directly crossing lymphatic vessel walls [81]. Upon reaching the lymph nodes, the particulate antigens may be phagocytosed and processed by macrophages resident in subcapsular sinus (SCS) and are further transported to the B cell or T cell zone. The small antigens such as soluble proteins may diffuse directly from SCS to B cell or T cell zones via conduits [18, 80]. Particles with a size of >200–500 nm do not efficiently enter the lymphatic system in free form and require cellular transport by APCs, such as DCs to be delivered to the lymph nodes [18, 80]. The commonly used phage capsids for vaccine delivery are <200 nm, such as Qβ (28 nm) [82], MS2 (26 nm) [33], T7 (56 nm) [83], λ (60 nm) [28], and T4 (120 × 86 nm) [84]. Therefore, phage VLP vaccines can efficiently enter the lymphatic system and are transported to the lymph nodes to stimulate adaptive immune responses.
The uptake and processing of antigens by APCs are also affected by particle size. Soluble antigens are normally presented by MHC class II molecules and have poor ability to be presented by MHC class I molecules [85]. On the other hand, particulate antigens are more likely processed through cross-presentation pathways to activate both CD4+ and CD8+ T cells [85]. Display of soluble antigens on phage, therefore can lead to efficient presentation of the antigen by both MHC class I and class II molecules (Fig. 1C). For instance, p24-specific CD8+ T cells were seen in both spleen and lymph node cells of mice immunized with phage T4 displayed with HIV-1 p24 antigen [86]. On the other hand, significantly lower or no p24-specific CD8+ T cells were identified in mice immunized with soluble p24 [86]. Similarly, immunization of mice with T4 phage displayed with F1mutV antigen of Yersinia pestis (Y. pestis) showed both Th1 and Th2 immune responses whereas immunization with the soluble antigen was biased towards primarily the Th2 responses [55]. This was also directly demonstrated in the case of phage fd where confocal microscopy of B lymphocytes showed that the FITC-labeled fd phage entered both the MHC class I and class II processing pathways [87].
3.4. Co-delivery of antigen and adjuvant
DCs can be activated directly through interaction of pathogen PAMPs with PRRs, or indirectly by exposure to inflammatory mediators. However, while the DCs activated indirectly by inflammatory mediators alone supported CD4+ T cell clonal expansion, it failed to direct T helper cell differentiation [88]. Thus, optimal activation of DCs also seems to require direct stimulation by danger signals imparted by the PAMPs. Linking an antigen to an adjuvant ensures simultaneous delivery of both the components to the same immune cells, such as the APCs and B cells. Therefore, the immune cells will recognize and process the antigens while receiving costimulatory signals from the adjuvant, which could significantly enhance the immune responses. Comes et al. reported that the E7 protein from human papilloma virus induced higher IgG responses when E7 protein was displayed on phage Qβ VLPs packaged with CpG, a ligand for TLR9, compared with a simple mixture of E7 and TLR9 packaged Qβ VLPs [89].
The intact phage itself has the ability to stimulate innate immune response [72], potentially acting as a natural adjuvant. For example, T4 phages induced significant levels of interferon in mice when 2 × 1011 phage particles were injected intravenously. Phage A20/R has a costimulatoiy effect on murine splenocytes activated by suboptimal concentration of concanavalin A [90]. Display of antigens on phages therefore appears to link the antigen to an adjuvant-loaded vaccine delivery system, leading to robust immune responses without any externally added adjuvant. This was supported by a number of studies in which antigens displayed on T4 capsid elicited much stronger immune responses when compared to their soluble counterparts [55].
Furthermore, different parts of the phage could be used to link the antigen and the adjuvant to the same VLP. In phages such as λ [91] both the capsid and the tail have been engineered to deliver foreign molecules. Phage T4 has two outer capsid proteins, Soc and Hoc, both of which can be used to simultaneously display two different foreign molecules, one an antigen and another an adjuvant (e.g. flagellin, the ligand of TLR5). Recently, we have developed a novel vaccine delivery system using only the T4 capsid shell with no tails, which can simultaneously deliver not only two different proteins through Hoc and Soc display, but also DNA molecules packaged inside the capsid [92]. This system therefore has the ability to co-deliver multiple PAMPs and activate multiple innate immune pathways, potentially making it a very robust VLP vaccine delivery system [42, 93]. This was also evident in the live attenuated yellow fever vaccine 17D, one of the most effective vaccines available. This vaccine activates multiple TLRs on DCs to elicit a broad spectrum of innate and adaptive immune responses [93] probably because of the activation of the DC subsets containing different TLRs. The CD103+ DCs mainly expressing TLR3 could only be effectively activated by TLR3 agonist, while CD11b+ DCs mainly expressing TLR7 could only be effectively activated by TLR7 agonist [94]. VLPs containing multiple PAMPs thus might activate all DC subsets containing different PRRs, eliciting a broad spectrum of innate and adaptive immune responses.
3.5. Targeting DCs
DCs play key roles in connecting innate and adaptive immune response (Fig. 1) [38]. Recognition, processing, and presentation of antigens, as well as trafficking of DCs to lymph nodes are key factors that affect vaccine efficacy. Targeting of antigens to DCs, therefore, is one of the promising strategies to enhance vaccine efficacy [95, 96]. An advantage of the phage VLPs is that phages do not have tropism to host cells. Hence, they can be targeted to DCs by displaying a DC-specific targeting molecule along with the antigen molecules. Phage T4 provides one of the best examples because of the flexibility in engineering the capsid using two different capsid proteins with different properties. Our studies show that the T4 nanoparticles could be targeted to DCs by displaying a monoclonal antibody (mAb) attached to Hoc against the DC-specific receptor DEC-205 [92, 97], which is expressed at high levels on lymphoid tissue DCs [98]. Alternatively, phages can be targeted to DCs by displaying a single chain variable fragment of antibody (scFV) against the DEC-205. Sartorius et al. have demonstrated enhanced receptor-mediated uptake of fd phage particles expressing the anti-DEC-205 scFV [99]. Furthermore, many DC targeting peptides were identified by phage display screens, making it possible to target antigen delivery to DCs using these peptides [100–102].
Targeting leads to efficient binding of VLPs to DCs which then leads to enhanced endocytosis [99]. However, endocytosis alone does not induce DC maturation [41]. As discussed above, only matured DCs are able to initiate the adaptive immune response. Triggering of PRRs on DC by danger signals (e.g. PAMPs present on pathogens) is thought to be critical for DC maturation [103]. Since phage structures resemble PAMPs, targeting of phage-based vaccines to DCs not only enhances antigen up-take but also facilitates DC maturation. This was supported by the observation that the phage fd particles displaying the scFV against the DEC-205 induced DC maturation both in vitro and in vivo, while the WT fd particle or fd particles displaying a control scFV (no targeting) could not [99]. Therefore, targeting of phage-based vaccines to DCs can lead to further enhancement of the immune responses through facilitation of DCs maturation. Indeed, a recombinant phage fd displaying an oval-bumin peptide at the NH2-terminus of the pVIII protein and a scFV against the DEC-205 through the pIII protein induced stronger antibody responses when compared to the phage lacking the scFV targeting molecule [99]. Further subsequent studies indicated that DC targeting induced proinflammatory cytokines and type I interferon, which was MyD88 mediated and TLR9 dependent (MyD88 is a universal adapter protein in innate immune signaling used by almost all TLRs) [104]. This might be because the fd phage particles containing a single-strand DNA genome rich in CpG motifs were able to trigger the activation of TLR9, thereby enhancing the immunogenicity of the displayed antigen [104].
3.6. Safety
Live attenuated vaccines are probably the most effective vaccines but have significant safety concerns. For instance, in the case of the attenuated oral polio vaccine (OPV), only a small number of mutations (2 to 10 nucleotides depending on the strain) control the attenuated phenotype [105] and the mutation sites seem to be under strong negative selection [106, 107]. Reversion back to virulent strain was also seen in the attenuated porcine reproductive and respiratory syndrome (PRRS) vaccine [108]. This question does not arise with phage VLP vaccines as they lack the ability to infect mammalian cells [109]. In numerous pre-clinical animal testing including mice [55, 86, 92, 110], rats [55, 92], rabbits [111], and rhesus macaques [112], we found no adverse effects due to immunization with phage T4 particles by intramuscular, intranasal, intravenous, or oral routes. Importantly, no side effects were reported in a human trial where T4 phage was given orally [113]. Similarly, no adverse reactions to phages were reported in human trials where T4-like coliphage cocktail was given orally to healthy Bangladeshi children [114, 115]. Indeed, phages have been used to treat pathogenic bacterial infections by deHerelle in 1919 and although this practice did not continue widely after the discovery of antibiotics, it has been in use in Poland, Georgia, and Russia for nearly a century, indicating the overall safety of phage therapy to humans [116, 117].
4. Bacteriophage T4
The tailed phage T4 has served as a model for >80 years to elucidate the basic mechanisms in molecular biology. At the same time, intensive work on its genetics, biochemistry, and structure has generated deep understanding of the mechanisms involved in head and tail assembly [65, 118], DNA packaging [119], and genome injection [120]. Coupled with this basic knowledge, its unique capsid architecture makes T4 phage a powerful VLP vaccine platform.
4.1. Capsid structure
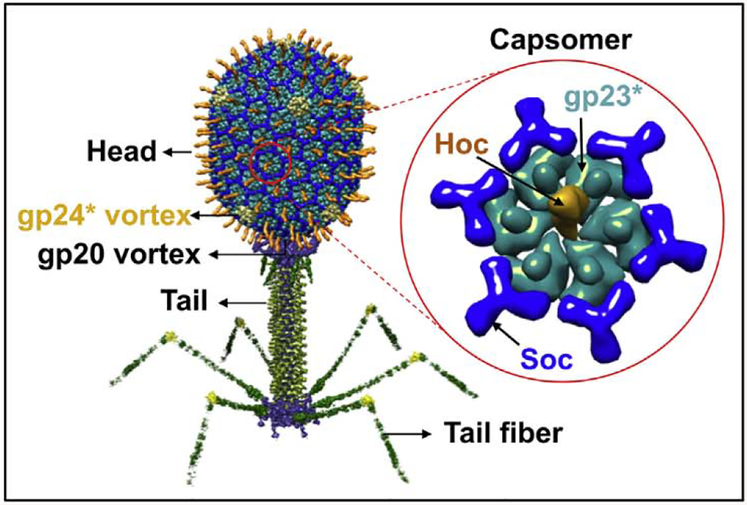
T4 phage consists of three major components; head (or capsid), tail, and tail fibers (Fig. 2). Each of these are assembled by independent pathways and then joined together to form the infectious virion [63]. Application of T4 phage to vaccine development as a VLP platform mainly involves the head. The head is an elongated (prolate) icosahedron, 120 nm long and 86 nm wide, and built with three essential capsid proteins; 930 copies of the major capsid protein gp23* (“*” represents the cleaved form), which form the hexagonal capsid lattice, 55 copies of the vertex protein gp24* which form pentamers at eleven of the twelve vertices, and 12 copies of the portal protein gp20, which form the unique dodecameric portal vertex [63]. The portal vertex is a ring structure with a central channel having a diameter of 3.5–4 nm through which DNA enters the capsid during packaging and exits during infection [121]. Packaged inside the capsid isa 171 kb linear dsDNA genome, the density of which approaches that of the crystalline DNA [122].
Fig. 2.
Structural model of bacteriophage T4. The enlarged capsomer shows the major capsid protein gp23* (cyan; “*” represents the cleaved form) (930 copies), Soc (blue, 870 copies), and Hoc (yellow; 155 copies). Yellow subunits at the five-fold vertices correspond to gp24*. The unique portal vertex (not visible in the picture) connects the head to the tail.
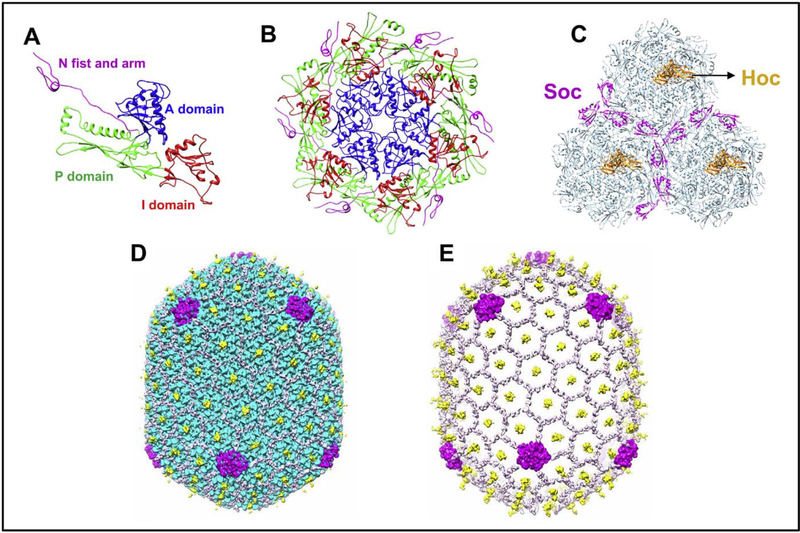
Phage capsids are highly stable structures, a very useful property for designing VLP vaccines. The phage T4 capsid protein gp23* consists of HK97 fold (Fig. 3A), a fold found in all the phage capsid protein structures determined thus far. It has three domains: axial (A), peripheral (P), and insertion (I) domains [84]. The inter-A domain interactions assemble gp23 into hexameric capsomers (Fig. 3B). The subunits are glued together by I domains that cross-link the A domains through a network of noncovalent interactions with the A and P domains of adjacent gp23* subunits. The I-domain is flanked by long linkers that form extensive intra- and inter-capsomer interactions that stabilize the capsid lattice. The inter-capsomer contacts are further reinforced by electrostatic interactions between the P domains of gp23* subunits from adjacent capsomers. Furthermore, the NH2-terminal regions of gp23* interact extensively with four neighboring subunits belonging to two different capsomers (Fig. 3B). These interactions greatly stabilize the capsid structure and allow it to withstand the high internal pressure of the tightly packed genome inside (~25 pN; 2.5 times the pressure in a champagne bottle) [84, 123].
Fig. 3.
Various assembled states of the major capsid protein gp23*; monomer (A), hexameric capsomer (B), and three capsomers (C) of the hexagonal capsid lattice showing Soc subunits (magenta) bound at the interfaces of adjacent capsomers. Hoc monomers (orange) are located at the center of each capsomer. The structures are derived from the cryo-EM structure of the isometric phage T4 capsid [84]. The monomer (A) shows key domains of gp23* that associate through an intricate network of interactions to form the capsomer (B) and the capsid lattice (C). The structure is reinforced by trimeric Soc clamps at the quasi three-fold axes forming a molecular cage around the capsid (D). The capsid subunits are masked in E to depict the Soc molecular cage.
The T4 head also contains two non-essential outer capsid proteins (Figs. 2 and 3), the small outer capsid protein (Soc) and the highly antigenic outer capsid protein (Hoc) [124]. Hoc is a 39 kDa protein containing a string of four Ig-like domains. Shaped as a linear fiber, it binds at the center of each gp23* hexameric capsomer as a monomer (Fig. 3C) [84]. Consequently, 155 copies of symmetrically arranged Hoc fibers emanate from the surface of the T4 head (Figs. 2 and 3D). As the COOH-terminal domain of Hoc contains the capsid binding site, this domain is closest to the capsid wall whereas the NH2-terminus is projected away at ~170 Å distance from the capsid [125]. Soc is a 10 kDa tadpole-shaped molecule and exists as a monomer in solution [126]. However, since it binds to two gp23* molecules between the adjacent capsomers, three molecules of Soc occupy the quasi three-fold axes (Fig. 3C). This arrangement brings the COOH-terminal acidic and basic residues into close proximity. These inter-digitate through electrostatic interactions resulting in trimerization of Soc at the three-fold axes. Consequently, 870 copies of Soc form a molecular cage around the capsid shell and reinforce the capsid by clamping adjacent capsomers (Fig. 3D and E) [126]. This further stabilizes an already stable head and allows it to withstand a harsh external environment in which the phage may be exposed to denaturing agents, high temperature, or extremes of pH (e.g., pH 11) [126]. However, Soc cannot bind between gp23* hexamers and gp24* pentamers at the vertices. Therefore, the vertices do not have Soc reinforcement and hence are structurally the weakest regions of the T4 capsid.
Both Hoc and Soc proteins are non-essential and can be deleted without affecting phage viability or infectivity under laboratory conditions. However, when exposed to pH 11 or higher, the wild-type (WT) phage survives whereas the Soc-phage is unstable and loses viability [126]. Hoc on the other hand provides marginal additional stability to the capsid. Its main function appears to be to sense the external environment and/or adhere to surfaces using its long flexible fiber and Ig-like domains. The latter might increase the efficiency of binding of the phage to bacterial surface thereby facilitating the long tail fibers to capture the receptors on the cell surface [127]. Hoc is also reported to facilitate the binding of phage T4 to mammalian cells, which might be a useful feature for vaccine delivery [128–131]. Additionally, its acidic (pI 4.7) nature creates a highly negatively charged capsid surface that might aid in preventing aggregation and dispersal of phage particles [131].
The above architecture of T4 capsid, decorated with Soc and Hoc, provides several unique features to engineer capsid surface as a VLP vaccine platform. First, it provides a high density of attachment sites, a total of 1025 per capsid, 870 Soc sites and 155 Hoc sites, to display foreign antigens [110, 132]. Second, since the sites are symmetrically distributed on the capsid lattice [84], the surface of T4 phage resembles the PAMPs of the mammalian viruses and probably other pathogenic organisms recognized by the innate immune system. Third, both the NH2-and COOH-termini of Soc and Hoc are exposed to the aqueous medium [125, 126], i.e., not buried in the structure, which allows attachment of antigen molecules without perturbing the structure and folding of the capsid proteins [110, 132]. Fourth, since Soc and Hoc bind to capsid with exquisite specificity and nanomolar affinity, antigens fused to Hoc and Soc can be stably displayed on the capsid surface with high affinity and specificity [55, 110, 126]. Fifth, multiple antigens belonging to one or different pathogens can be displayed on the same capsid to design multivalent vaccines [55, 110, 132]. Finally, the NH2-terminus of the Hoc fiber projected at 170 Å away from the capsid wall provides a platform to display molecules that can target the T4 nanoparticles to specific APCs such as the DCs, and/or co-deliver a molecular adjuvant [92, 125].
4.2. The DNA packaging machine
Assembly of phage head is initiated by the portal assembly formed by twelve molecules of the portal protein gp20 [63]. In phage T4, this occurs on the inner membrane of Escherichia coli (E. coli) with the help of a membrane-bound “chaperone” protein gp40. The major capsid protein gp23 and the major scaffolding protein gp22 along with other scaffolding proteins such as the internal proteins IPI, IPII, and IPIII, gp21 protease, gp68 and gp69 co-assemble to produce the immature prehead [65]. The prehead, thus, is an icosahedral shell formed around a scaffolding core, the shape and size of which is determined by both the shell and the core. The portal remains at the special portal vertex of the assembled head whereas pentamers of the vertex protein gp24 assemble at the other 11 vertices [65]. Most of the inner scaffolding core is degraded by the gp21 protease which is activated once a prehead of precise dimensions is assembled [65]. The protease also cleaves the NH2-terminal regions of gp23 and gp24 subunits at a specific glu-ala peptide bond to generate the mature gp23* and gp24* proteins [65]. The cleaved head with the empty space created inside by scaffold removal is now released into the cytosol.
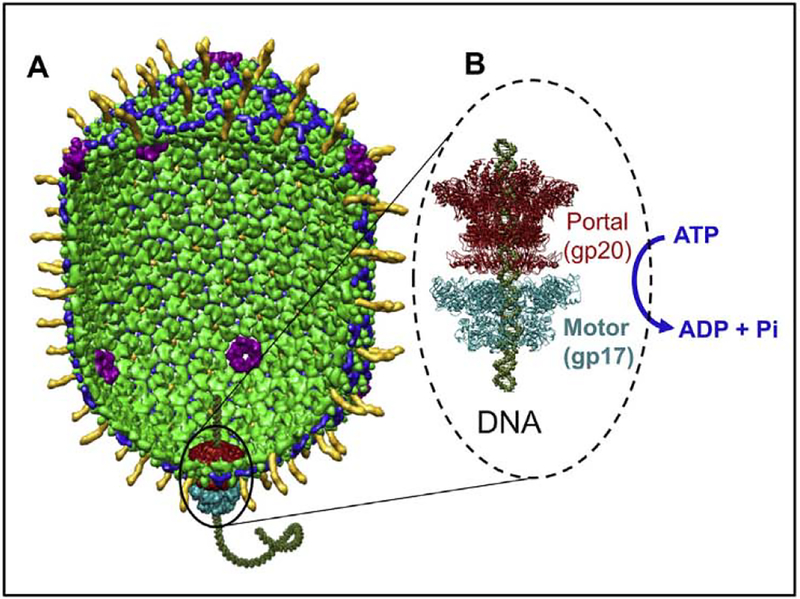
A terminase complex containing two packaging proteins, the motor protein gp17 and the regulator protein gp16, recognizes the concatemeric viral genome and makes a double stranded cut to generate an end (or terminus, hence the name terminase) [119]. The gp17 terminase-DNA complex then assembles on the portal vertex as a pentamer (gp16 is not essential in vitro to form this complex) (Fig. 4) [133]. Two symmetry mismatches are thus created, one between the five-fold capsid vertex and the dodecameric portal and another between the portal and the pentameric packaging motor [119]. Importantly, a DNA packaging machine consisting of the motor, the portal, and the shell is assembled with one end of the DNA inserted into the portal channel (Fig. 4). Packaging is initiated by the motor complex with the DNA getting translocated into the head at the expense of ATP hydrolysis [134]. After about 10–25% of the genome is translocated, the head “expands” in size, by about 15% in both long and short dimensions as a result of a major conformational change in the capsid protein subunits [84, 119]. Expansion is a critical maturation step for several reasons. It increases the inner volume of the capsid by about 50% which is essential i) to accommodate the entire genome, ii) to stabilize the capsid structure, and iii) to create binding sites for Soc and Hoc which, upon binding of Soc and Hoc, further reinforce the capsid structure [65, 119]. Consequently, the capsid can withstand the internal pressure of the tightly packed genome, even under harsh environmental conditions such as pH 11. After about 102–103% of genome is packaged (one “headful”), packaging is terminated when the motor complex makes another cut and dissociates from the packaged head. The motor-DNA complex can then attach to another empty head and continue DNA packaging in a processive fashion whereas the packaged head is sealed off by the attachment of neck and tail proteins at the portal [119].
Fig. 4.
The bacteriophage T4 DNA packaging machine. (A) Structural model of the phage T4 DNA packaging machine. (B) Ribbon model of the pentameric motor (cyan) assembled at the dodecameric portal vertex (dark red). The motor fills the capsid with the phage genome. ~171 kb DN, utilizing the energy from ATP hydrolysis.
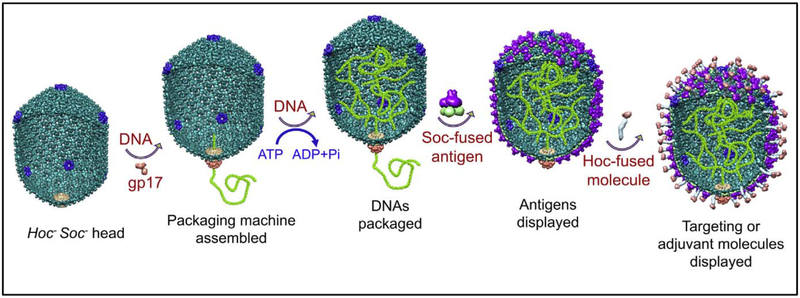
We have been able to reconstitute the DNA packaging machine in vitro using purified components, empty proheads and gp17 (Fig. 4; see below) [134]. Empty proheads are produced from E. coli infected with mutants that are defective in DNA packaging and tail assembly (e.g., 17am18am; gene 18 codes for the tail sheath protein). Alternatively, the DNA packaged heads can be prepared using mutants that are defective in neck and tail assembly (e.g., 10am13am; gene 10 codes for a base-plate protein and gene 13 for a neck protein) and the packaged DNA being unstable can be emptied and digested with DNAse I [135]. Remarkably, these emptied head shells can re-package DNA in vitro as well as the nascent proheads [92, 135]. In fact, these mature phage heads are better for VLP vaccine delivery because these can be produced in much larger quantities and exhibit greater stability than the proheads. A distinct advantage of the T4 packaging system is that its motor protein gp17 alone can function as well as, or better than, when the regulatory protein gp16 is present in vitro and it does not require a specific sequence to initiate or terminate DNA packaging. When mixed with heads, gp17, and ATP, any linear (or circular) DNA can be encapsidated [92, 135]. It could be a single piece of genome length DNA (~171 kb), or a shorter plasmid DNA, or even oligonucleotides as short as 30 bp. In the case of short DNAs, multiple DNA molecules will be packaged inside the same head [92]. This means that, for vaccine delivery, the T4 head-motor complex can be used as a nanoscale vacuum to “stuff’ the head with multiple copies of multiple pathogen genes encoding vaccine antigens.
4.3. In vivo versus in vitro display
The T4 head structure, assembly, and DNA packaging provide extraordinary opportunities for developing a vaccine delivery platform by engineering the surface with antigen proteins and the interior with antigen encoding genes. The first reports of this system used in vivo display wherein short pathogen peptides fused to Hoc or Soc expressed in E. coli cells either from recombinant phage genome or a recombinant plasmid were displayed on phage particles produced in the same cells by hoc−soc− mutant infection. Ren et al. showed that a 43-residue V3 loop peptide of HIV-1 envelope protein gp120 or 312-residue of the poliovirus VP1 capsid protein fused to the COOH-tenninus of Soc bound to T4 phage [136]. Jiang et al. showed that a 36-residue peptide from the PorA protein of Neisseria meningitides fused to the NH2-terminus of either Hoc or Soc was displayed on T4 [137]. These studies demonstrated that both the NH2- and/or COOH-termini of Soc could be used to display pathogen peptides on the capsid surface without compromising its ability to bind to the capsid. Subsequent studies using a variety of antigens fused to Soc further confirmed these results [55, 138, 139]. Indeed, in one study, both the NH2- and COOH-termini of Soc were used at the same time to fuse two different antigens and the dual fusion product was efficiently displayed on the phage capsid, essentially doubling the copy number of the displayed antigens [55, 132]. In the case of Hoc, however, although both the NH2- and COOH-termini can be used to display antigens [86], the COOH-terminus is less desirable because the COOH-terminal domain of Hoc has the capsid binding site [125]. Therefore, the fused antigen creates steric interference for accessing the capsid binding site. Consequently, poor binding was observed for the antigens fused to the NH2-terminus of Hoc [140]. These results were supported by structure determinations of Soc and Hoc [125, 126] which showed that the NH2- and COOH-termini of Soc and the NH2-terminus of Hoc are accessible to solvent and can be effectively used for molecular display.
The in vivo phage display, a hallmark of all phage display systems has certain fundamental limitations, particularly for the development of VLP vaccines. First, the gene for the antigenic peptide fused to Soc or Hoc flanked by several hundred bp of genome must be inserted into a plasmid vector in order to allow for homologous recombination between the T4 genome and the plasmid upon infection with a hue−soc− phage mutant [136, 137]. The recombinant frequency relative to parental phage background is very low, on the order of ~10−4–10−5, even for a highly recombinogenic phage T4. Hence, it requires a complicated selection protocol to identify the recombinant phages, as reported by Ren et al. [136]. The recent demonstration of recombinant generation using the CRISPR-Cas genome editing strategy might alleviate this problem by eliminating the parental phage background [141, 142]. Second, the fusion protein is expressed from a T4 promoter upon infection of E. coli and the resultant protein is displayed on hocsoc− phage in vivo. The phages purified from these infected cells thus contain the displayed antigen and can be used as a VLP vaccine. However, since the fusion protein is produced as part of phage infection, there is no control over the quality of the antigen that is displayed. For instance, truncated or aggregated proteins might assemble on the capsid. Third, the copy number of the displayed antigen cannot be controlled by in vivo phage display. Since different foreign proteins are expressed to different extents, the copy number of the antigen on the VLP could vary greatly and between experiments. Therefore, it would be difficult to control the quality of the manufactured vaccine. Fourth, it would be very difficult to generate multivalent vaccines or to customize the nanoparticle by incorporating different Hoc- and Soc-fused proteins. Finally, since the in vivo display relies entirely on the protein expression in E. coli, it is unable to display proteins that could not fold properly or require certain post-translational modification such as glycosylation for folding and/or antigenicity.
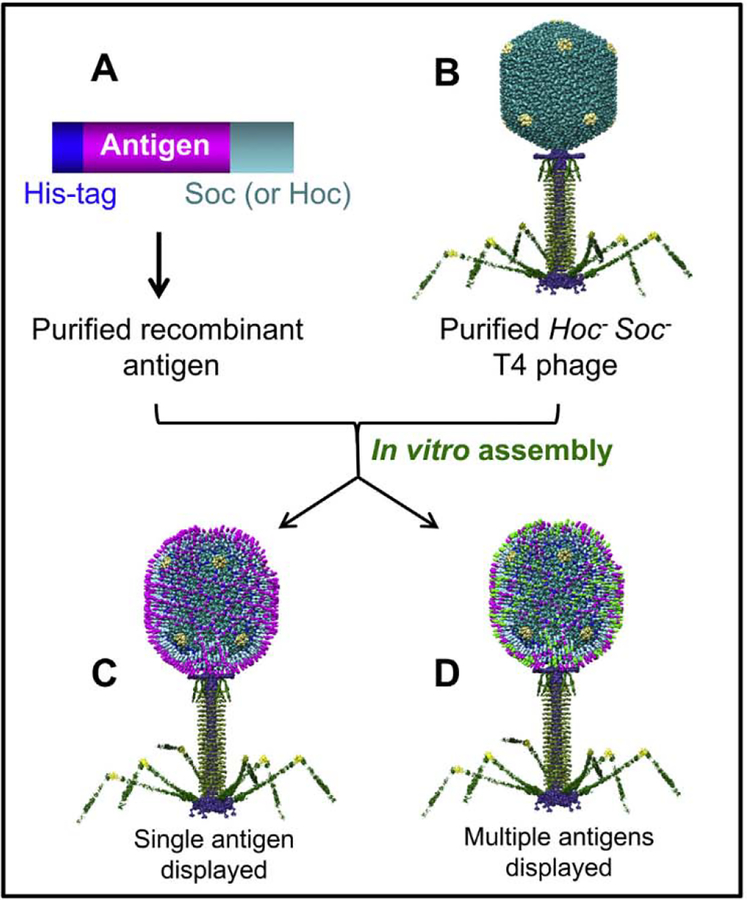
To overcome these problems, we have developed a defined in vitro phage assembly system (Fig. 5) [55, 86, 110, 132]. The Soc- or Hoc-fused antigens with an affinity tag at the NH2- or COOH-termini are expressed in E. coli from a strong promoter such as the phage T7 promoter and purified by a single-step affinity chromatography (Fig. 5A). This could very well be done using a mammalian expression system, especially for antigens requiring specific post-translational modifications. The purified proteins are then functionally and/or antigenically characterized and displayed on phage by mixing the recombinant protein with the purified hocsoc− T4 phage (Fig. 5B). This approach has many advantages. First, it allows a critical step, i.e., biochemical characterization of the antigen, before the display of the antigen on the phage capsid. Consequently, the expression and purification systems can be optimized such that only a pure, functionally well-defined antigen that is most effective in eliciting potent immune responses can be displayed on the phage nanoparticle. For example, we have demonstrated that only the HIV gp41 envelope protein trimers, but not the monomers or nonspecific oligomers, could be selectively displayed using this approach [79]. Second, the composition of the in vitro assembly mixture can be adjusted to generate multivalent vaccines and to control the copy number (Fig. 5C). For example, by adding two different anthrax antigens fused to Soc, LFn-Soc and Soc-PA4, to the reaction mixture, we could generate T4 phage nanoparticles simultaneously displaying both the antigens on the same capsid, and the copy number of each antigen was proportional to the ratio of the proteins to the binding sites added to the binding reaction [132]. Third, large full-length proteins can be displayed on the T4 capsid (Fig. 6). Examples include the 83 kDa protective antigen (PA) [132], 90 kDa lethal factor (LF) [132], 89 kDa edema factor (EF) [140], 66 kDa plague F1mutV [55], and 129 kDa β-galactosidase [92]. Furthermore, the antigens could be assembled sequentially to display macro-molecular complexes on the T4 capsid. For example, the entire tripartite anthrax toxin complex with a molecular size of 600 kDa could be assembled (Fig. 6A to C) [66, 143]. In one sequence, LFn-Soc was first displayed on hoc−soc− phage. Trypsin-nicked PA63 was then assembled as heptamers through specific interaction with the displayed LFn domain. EF was then attached to the unoccupied sites of PA63 heptamers, completing the assembly of the tripartite anthrax toxin. Fourth, in vitro display allows for the preparation of VLPs with targeting capability. As mentioned above, the Hoc fiber with a reach of 170 Å can be used to target the VLP to specific cells such as the antigen presenting DCs by attaching a ligand to the tip of the fiber. This was demonstrated by displaying the DC targeting ligands, DEC205 mAb or CD40, through Hoc (Fig. 7) [92]. Finally, display can be accomplished using empty capsids instead of phage particles [92]. Capsids devoid of packaged DNA and tails might be better alternatives when compared to the phage particles and furthermore, capsids have the additional advantage of filling the empty space with foreign DNA by in vitro DNA packaging. Such particles containing both proteins and DNAs can serve as protein-prime and DNA-boost vaccines.
Fig. 5.
Schematic of phage T4 in vitro display system. The affinity-purified Soc-flised antigen (s) (A) are assembled on purified hoc−soc− T4 phage (B) by mixing the two at 4 °C for 45 min to generate the VIPs [138, 139]. The capsid can be displayed with one antigen (C) or a mixture of different antigens (D; shown in different colors). The same principle is used for the display of Hoc-fused antigens or targeting molecules [86, 92].
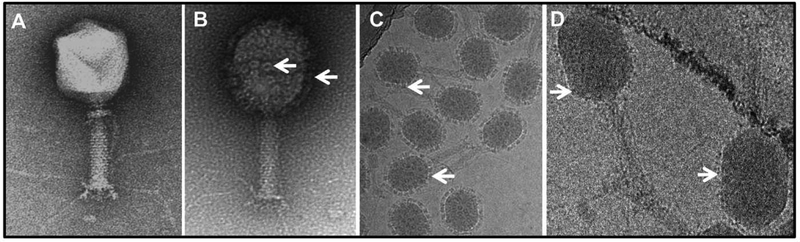
Fig. 6.
Assembly of anthrax toxin complexes and plague antigens on phage T4. Negatively stained images of hoc−soc− T4 phage (A) decorated with anthrax toxin complexes by in vitro display (B). Cryo-electron micrograph of (B) showing rings of anthrax toxin complexes decorating the T4 capsid (C). (D) Cryo-electron micrograph of phage T4 decorated with F1mutV fused to Soc. Due to the small size of Soc-F1mutV relative to anthrax toxin complex, the displayed antigen molecules are seen as a layer of fuzzy projections around the perimeter of the capsid. Some of the displayed anthrax toxin complexes and F1mutV molecules are marked with arrows.
Fig. 7.
Schematic of the assembly of prime-boost T4 nanoparticles. The DNA packaging machine is assembled by binding of gp17 motor at the portal of hoc−soc− phage T4 empty head (the cut-out of the head shows both the exterior and the interior) (A). Using the energy from ATP hydrolysis the motor packages DNA molecules into the head (B). Soc-fused antigens (C) and Hoc-tused targeting molecules (D) are then displayed on the heads.
4.4. T4 VLP vaccines
The above principles and the basic knowledge on T4 structure and assembly have been applied to develop VLP vaccines against a number of pathogenic bacteria and viruses (Table 1), such as Bacillus anthracis [111, 112], Y. pestis [55, 138], HIV-1 [86], foot-and-mouth disease virus (FMDV) [144], classical swine fever virus (CSFV) [145], and bursal disease virus [146].
Table 1.
Pre-clinical studies of T4 nanoparticle vaccine candidates.
| Organism | Antigens (kDa) | Animal model | Capsid proteins | References |
|---|---|---|---|---|
| Bacillus anthracis | PA(83 kDa), LF (89 kDa), Toxin complex (710 kDa) | None | Soc | [132,139,143] |
| Bacillus anthracis | PA(83 kDa), LF (89 kDa), EF (89 kDa) | Mouse | Hoc | [110,140] |
| Bacillus anthracis | PA(83 kDa) | Rabbit | Hoc, Soc | [111] |
| Bacillus anthracis | PA(83 kDa) | Rhesus macaque | Hoc, Soc | [112] |
| Yersinia pestis | LcrV (37 kDa), Caf1 (16 kDa), YscF (11.8 kDa) | Mouse, Rat | Soc | [55, 92, 138] |
| Neisseria meningitidis | PorA peptide (16.9 kDa) | None | Hoc, Soc | [137] |
| HIV-1 | p24 (26 kDa), Nef (31 kDa), gp41 (21 kDa) | Mouse | Soc | [86] |
| HIV-1 | gp41 (39 kDa) | None | Soc | [79] |
| HIV-1 | V3 loop of gp120 (5 kDa) | None | Soc | [136] |
| Foot-and-mouth disease virus | P1 (89 kDa), proteinase 3C (24 kDa) | Mouse, Pig | Soc | [144] |
| Classical swine fever virus | E2 (44 kDa) | Mouse | Hoc, Soc | [145] |
| Bursal disease virus | VP2 (40.0 kDa) | Chicken | Soc | [146] |
| Poliovirus | VP1 (34.5 kDa) | None | Soc | [136] |
4.4.1. Anthrax vaccine
Anthrax, caused by B. anthracis, is a deadly disease and listed as a tier 1 biothreat agent by the United States Center for Disease Control (CDC). The major virulence factors of B. anthracis are the tripartite anthrax toxins [147] consisting of protective antigen (PA; 83 kDa), lethal factor (LF, 90 kDa), and edema factor (EF, 89 kDa). Of these, PA has been the principal target for anthrax vaccine development [148]. It has been well demonstrated that antibodies against PA alone are sufficient to completely protect animals against lethal, aerosolized B. anthracis Ames spore challenge [112, 148]. Although there is a licensed anthrax vaccine (AVA, BioThrax), it is a culture filtrate of PA produced from a non-lethal B. anthracis bacterium adsorbed to Alum adjuvant, which produces significant reactogenicity in vaccinated individuals and is not recommended for general use [149]. Therefore, a safe and effective recombinant subunit vaccine is needed to protect the public against this biothreat.
To construct a T4 VLP vaccine against anthrax, PA was fused to the NH2-terminus of Hoc (PA-Hoc) and assembled on T4 capsid by in vitro display using the hoc−soc− T4 phage nanoparticles [110]. A maximum copy number of ~155 PA-Hoc molecules per capsid was achieved. When intramuscularly injected into mice, the T4 displayed PA without any added adjuvant elicited 6.5-fold higher PA-specific IgG and 4.7-fold higher lethal toxin (LeTx) neutralizing antibody titers compared to those immunized with soluble PA [110]. Similarly, PA was fused to Soc through either the NH2-terminus or the COOH-terminus of Soc and both could be efficiently displayed on T4 phage by in vitro display [132]. The maximum copy number was 350 PA-Soc molecules per capsid. All the 870 binding sites for Soc could not be filled, probably because of the close proximity of Soc to the capsid wall which created steric interference. However, the copy number could be increased by sequential binding of both PA-Hoc and PA-Soc onto the same capsid [111]. This T4-PA VLP vaccine with no adjuvant was highly immunogenic in New Zealand White rabbits inducing high titers of PA-specific antibodies, with endpoint titers of ~106 after two immunizations [111].
The T4-PA VLP vaccine displaying both PA-Hoc and PA-Soc (355 PA molecules per capsid) was tested for its vaccine efficacy in the rhesus macaque model [112]. The animals were intramuscularly injected with the T4 nanoparticles (50 μg T4 displayed PA per dose) three times at 4-week interval. Four weeks after second immunization (i.e., after one boost), the T4 VLPs induced significant amount of PA-specific IgG, which was further increased to about 500 μg/ml after the second boost. The PA-specific and LeTx-neutralizing titers of T4-VLPs without adjuvant were comparable to the soluble PA adjuvanted with Alhydrogel (aluminum hydroxide gel) but lower than that induced by PA mixed with liposomes containing monophosphoryl lipid A. Importantly, the T4-VLPs provided 100% protection when challenged with aerosolized B. anthracis Ames strain spores (up to 85 LD50), and no side effects due to T4 VLP vaccination were observed [112].
4.4.2. Plague vaccine
Plague, caused by Y. pestis, is one of the deadliest infectious diseases known to mankind. It is also classified as a tier 1 biothreat agent by the CDC Currently, there is no FDA approved plague vaccine for public use. Two Y. pestis virulence factors, the capsular protein (Caf1 or F1; 15.6 kDa) and the low calcium response V antigen (LcrV or V; 37.2 kDa) are the main targets for plague vaccines [150]. However, the polymeric nature of F1 with its propensity to aggregate affects vaccine efficacy and generates varied immune responses in humans [151]. We have constructed a mutant F1 antigen that exists as a monomer in solution (F1mut) and was equally effective as an immunogen [55, 138]. F1mut was then fused to the V antigen to generate a bivalent F1mutV which also exists as a monomer [55, 152].
To generate a T4 VLP vaccine against plague, the F1 mutV was fused to the NH2-terminus of Soc and assembled on T4 capsid using our in vitro display system (Fig. 5). About 660 copies of F1mutV decorated the capsid (Fig. 6D) [138]. These T4 VLPs with no adjuvant when immunized in mice elicited strong F1V-specific antibodies and the titers were higher than the soluble counterparts adjuvanted with Alhydrogel [55]. Of particular interest is that the T4 VLP vaccine induced both Th1 and Th2 immune response, whereas the soluble F1mutV vaccine showed a strong bias towards Th2 immune responses and elicited poor Th1 responses, consistent with the potential advantages of VLPs in eliciting innate and adaptive immune responses as described earlier (Fig. 1). This was further supported by the observation that the T4 VLP vaccine induced higher levels of IFN-γ than that of the soluble F1mutV vaccine [55]. Importantly, the T4 VLP vaccine provided 100% protection in mice and brown Norway rats (natural host for Y. pestis) against intranasal challenge with the most lethal Y. pestis CO92 strain up to as high −5000 LD50 dose [55]. Most recently, we have combined the anthrax and plague T4 VLP vaccines to produce a dual vaccine formulation that is efficacious against both inhalation anthrax and pneumonic plague challenges (unpublished).
4.4.3. Viral vaccines
In addition to bacterial vaccines, the T4 phage has also been used to develop viral vaccines. Sathaliyawala et al. have displayed the HIV-1 capsid protein, p24-gag, on T4 capsid through Hoc fusion [86]. The T4 displayed p24 induced robust and long-lasting anti-p24 antibodies (endpoint titers ranging from 64,000 to 72,000) with just 100 ng of p24 whereas the soluble p24 antigen induced poor antibody responses (ranging from 4000 to 9000) even with 10 μg of p24 [86]. Strikingly, the T4-p24 VLPs also induced strong CD4+ and CD84+ T-cell responses as opposed to soluble p24 which elicited poor responses [86]. Together with the results described above, it is clear that the immune responses elicited by T4 VLPs exhibit a distinct pattern when compared to that of the soluble antigens, both in quality and quantity, and includes both humoral and cellular responses. This is beneficial for protective immunity against any pathogen, but particularly so for intracellular pathogens that generally require both antibodies and cellular responses for complete protection. Supporting these findings is another study by Ren et al. using T4 VLPs against the FMDV [144] and CSFV [145]. In the FMDV study, the capsid precursor polyprotein P1 and proteinase 3C were fused to Soc and assembled on T4 capsid by in vivo display. Mice immunized with a mixture of T4 VLPs decorated with P1 or 3C either subcutaneously or orally induced complete protection against FMDV challenge [144]. This study also showed that the T4 VLPs could be used as an oral vaccine, a highly desired property for generating mucosal vaccines [153].
4.5. Prime-boost vaccines
“Prime-boost” immunizations are essential to generate strong immune responses. Two prime-boost strategies have been in use [154], homologous and heterologous. In the homologous approach, the same vaccine formulation is used for both the prime and the boost, whereas in the heterologous prime-boost, the antigen in different forms is used, e.g., prime with the DNA that can stimulate antigen production by host cells, then boost with the protein antigen that is manufactured outside the host. The heterologous approach is powerful as it can generate broader immune responses and has been successfully used in clinical trials including the recent RV144 HIV-1 vaccine trial that showed a modest ~30% protection [154]. The T4 VLP system offers a unique platform wherein both the protein and the DNA components can be incorporated into the same nanoparticle allowing co-delivery in a single immunization (Fig. 7). Here, the delivered protein is expected to act as a “prime” and the DNA as “boost” since the latter expresses the antigen continuously and boosts the immune system for weeks to months. We have developed a simple in vitro assembly system to customize the preparation of prime-boost T4 nanoparticles by sequentially incorporating DNA and protein into VLPs [92]. The DNA corresponding to the antigen gene such as the F1mutV was cloned into a plasmid under the control of the strong CMV promoter and packaged into T4 head using the DNA packaging machine. The F1mutV protein fused to the NH2-terminus of Soc was then displayed by adding the protein to the same reaction mixture. The nanoparticles can then be decorated with a targeting molecule such as the DC-targeting DEC205 mAb attached to Hoc. Such prime-boost nanoparticles by a single immunization elicited strong antibody and cellular responses in mice. A single intramuscular immunization ~5 × 1011 particles per mouse packaged with ~25 μg of F1mut-V DNA and displayed with ~30 μg of F1mut-V protein and ~100 copies per head of DEC205mAb (no adjuvant) induced strong antibody responses. Further, splenocytes isolated from the immunized animals showed high IFN-γ response even without stimulation with the F1-V antigen probably because targeted DNA delivery into DCs induced the expression and presentation of F1-V antigen for a long period leading to continuous stimulation of T cells. These results provide the proof of concept that a single dose prime-boost T4 VLP vaccine can stimulate both arms of the immune system, which can be exploited in future for designing effective multivalent vaccines.
5. Other bacteriophages
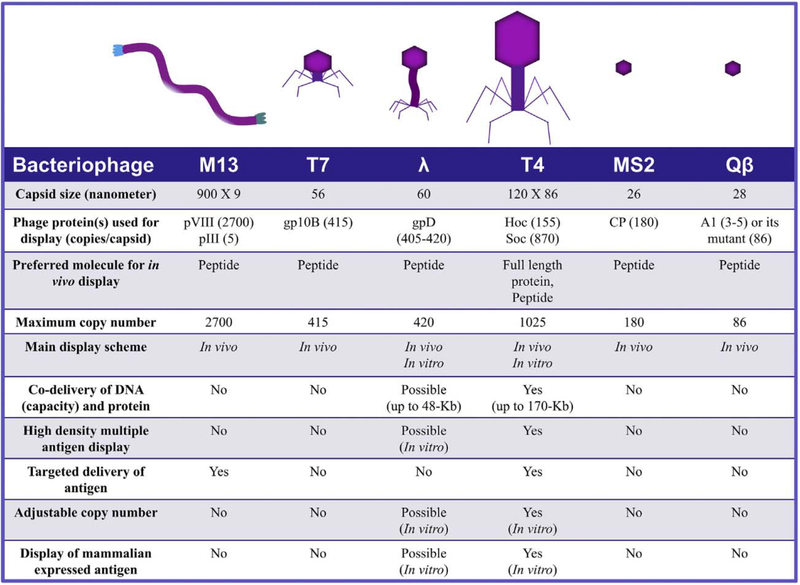
Besides T4, phages such as M13 and other filamentous phages, λ, T7, MS2, and Qβ were also used to develop VLP vaccines (Fig. 8). These phages with different sizes, topology, and/or outer capsid proteins may offer certain advantages for generation of vaccines against particular pathogenic bacteria and viruses.
Fig. 8.
Properties of various bacteriophages used in the development of VLP vaccines.
5.1. Filamentous phages
Filamentous phages M13, fd, and f1 are highly related and have been extensively used for display of short random peptide libraries. These phages have also been used, though to a limited extent, as vaccine platforms to deliver peptide antigens fused to the coat proteins [29]. The ~900 nm long filamentous phage contains approximately 2700 copies of the major coat protein pVIII. One end of the filament contains 5 copies each of the minor capsid proteins pill and pVI, while the other end contains 5 copies of pVII and pIX Although all the 5 phage proteins could be used to display proteins, pVIII is often used to deliver antigen peptides due to its high copy number. Peptides from >20 pathogens have been displayed and tested for their immunogenicity in different animal models, reporting the induction of strong humoral and cellular immune responses [29, 155, 156]. However, since the filamentous phage assembly requires extrusion of the pVIII capsid protein through the E. coli envelope, display is sensitive to the size of the peptide. Generally, short peptides ~8 amino acid residues containing B-cell or T-cell epitopes are optimal for display [30]. However, larger peptides, domains, or even large full-length antigens could be displayed, but the copy number will be quite low as these need to be assembled along with the WT capsid protein that forms the bulk of the capsid structure [157]. Both the pIII and pVIII capsid proteins could be used for display, however the very low copy number of pIII makes it less desirable for the purposes of vaccine delivery.
5.2. Phage λ
The 60 nm icosahedral capsid of λ phage is composed of420 copies of the essential major capsid protein gpE, which forms the hexagonal capsid lattice as well as eleven of the twelve pentameric vertices. In addition, the capsid is decorated with 405–420 copies of gpD, which forms trimers at the quasi three-fold axes [28, 31]. Unlike the phage T4 Soc, gpD is essential to stabilize the capsid against the internal pressure of the packaged 48.5 kb phage genome inside [158]. However, gpD can be dispensable for capsids carrying shorter genomes [158, 159] and has been extensively used for peptide display [160]. Although both the NH2- and COOH-termini could be used for fusion of antigen peptides [28, 31, 161], the apparent interaction of the NH2-terminus with gpE makes it less desirable [162]. Thus, the COOH-terminus is mainly used for peptide/protein display [163, 164]. Gamage et al. showed that λ phage VLPs displaying four immunodominant regions of porcine circovirus 2 capsid protein through fusion to the COOH-terminus of gpD elicited humoral as well as cellular immune responses in pigs [164]. Although display is carried out in vivo, gpD can also be assembled on gpD-minus capsids in vitro [161]. However, folding, proteolysis, and aggregation problems associated with in vivo display and instability of gpD-minus capsids for in vitro display pose limitations for using phage λ as a vaccine delivery platform. For example, Zanghi et al. found that no phage particles could be recovered from infection of E. coli expressing the gpD fusion protein from a plasmid with gpD-deficient phage λ [165]. However, co-expression of both WT gpD and gpD-fusion protein resulted in the recovery of mosaic phages displaying both the WT gpD and the gpD fusion protein [165], suggesting that the fusion protein interfered with phage assembly or prevented the formation of stable phage particles in the absence of the WT gpD protein.
5.3. Phage T7
The 56 nm T7 capsid with 40 kb genome packaged inside contains two forms of the major capsid protein gp10, gp10A (345 amino acids, 36 kDa) and gp10B (398 amino acids, 41 kDa) [83, 166]. The gp10B is a variant of gp10A produced by – 1 frame-shift at the COOH-terminus of the 10A reading frame [83, 166]. However, gp10B is not essential for capsid assembly. Hence, 10B has been used for phage display by fusion of antigen peptides to the COOH-terminus [57, 167, 168]. This system can display antigen peptides up to ~50 amino acids long at a density of up to 415 copies per capsid. Tan et al. have demonstrated that a 46-amino acid immunodominant peptide of the hepatitis B virus small surface antigen displayed on T7 phage is highly immunogenic in rabbits [169]. Xu et al. have shown that T7 phage displaying 40-amino acid GH loop peptide (a major neutralizing epitope) of the FMDV VP1 protein is highly immunogenic and provided 80% protection against virulent homologous virus challenge in pigs [170]. Similarly, the ectodomain (24 amino acids) of the influenza virus channel protein M2 (M2e) displayed on T7 without an adjuvant induced strong humoral and T cell responses that protected mice against both H1N1 (PR8) and H3N2 (X47) virus challenges [171]. The T7 system has also been used to display a library of proteome-wide peptides (56-residue peptide tiles with 28 residue overlaps) from all known human viruses which was then used for serological profiling of human populations exposed to various viruses during their lifetime [168]. Such profiles might hold potential for future vaccine designs [168]. While the above examples illustrate the usefulness of the T7 phage for peptide display, it would be difficult to display large polypeptides and proteins using the T7 system. Although proteins as large as 1200-amino acid protein can be displayed using phage T7, the copy number is greatly reduced, to 0.1–15 copies per capsid [167].
5.4. Phage MS2
With a capsid size of 26 nm and packaging 3.57 kb single stranded RNA genome, the MS2 phage is one of the smallest icosahedral bacteriophages. The capsid is composed of 180 copies of the major coat protein, CP, which alone is sufficient to self-assemble into capsids both in vivo and in vitro [33]. Hence, it provides a simple and convenient system to incorporate antigen peptides into VLPs. Since both the NH2- and COOH-termini are essential for CP assembly, the surface exposed β-hairpin (amino acids 11–17 of CP) of the CP was used for insertion of antigen peptides [172–174], However, the CP assembly is quite sensitive to the position at which the foreign peptide is inserted. Insertion of a 10-amino acids peptide at residue 11 of CP disrupted self-assembly [172, 175]. This can be overcome by covalently joining two CP monomers into a “two-domain” monomer rather than a CP dimer. Then, assembly could tolerate one peptide insertion per CP dimer [172, 175]. Alternatively, insertion of a 25-amino acid peptide between residues 15 and 16 of the CP did not disrupt the CP assembly [173, 176].
Both the above approaches have been used successfully to deliver antigen peptides. Using the “two-domain” strategy, Peabody et al. showed that the MS2 phage displaying a 10-amino acid V3 loop peptide of HIV envelope protein by the two-domain strategy is highly immunogenic [175]. Similarly, 15-amino acid peptides spanning a cross-neutralizing epitope from the minor capsid protein L2 of human papilloma virus (HPV) serotype 16 displayed on MS2 phage induced cross-neutralizing antibodies and protected mice against genital and cutaneous infection by highly diverse HPV pseudovirus types [177]. However, in some cases, the sequence of the inserted peptide affected CP assembly. Basu et al. reported that five of six potential B-cell epitopes (ranging from 11 to 22 amino acids) of the Zika virus envelope protein could not assemble MS2 VLPs because insertion of these peptides disrupted CP assembly [178]. Using the second insertion strategy, Heal et al. showed that the MS2 capsid displaying a protective epitope T1 of the malaria parasite Plasmodium falciparum elicited strong immune responses in mice [176] and Dong et al., showed that MS2 VLPs displaying the FMDV epitope peptide (EP141–160) was highly immunogenic in mice, guinea pigs, and swine, and induced high-titers of neutralizing antibodies [179]. Though the above examples illustrate the usefulness of MS2 VLPs for delivering short antigen peptides, it is apparent that the MS2 system is not suitable for delivering large domains or full-length antigens.
5.5. Phage Qβ
Another small bacteriophage, Qβ, has also been used to deliver antigen peptides [180]. The capsid of Qβ phage is 28 nm in diameter and composed of 180 copies of the major coat protein, CP [82]. It packages 4.2 kb single stranded RNA genome. The infectious Qβ virion in addition contains three to five copies of Al protein, which contains a large 196-amino acid extension at the COOH-terminus of CP as a result of infrequent read-through at the termination codon [82]. Foreign peptides can be displayed on the capsid by fusing the peptide to the read-though domain of A1 [58, 181]. Vasiljeva et al. showed that truncation of the read-though domain to just 6 amino adds at the COOH-terminus increased the copy number of A1 up to 86 copies per capsid [182]. Alternatively, antigens peptides and domains can be chemically coupled to Qβ capsid [75, 183]. Qβ phage displayed antigens were also found to be highly immunogenic [75, 183, 184]. Intranasal immunization of Qβ VLPs displaying the ectodomain of the influenza virus M2 protein elicited strong M2-specific IgG and IgA responses in both serum and bronchus-associated lymphoid tissue in a mouse model and completely protected the mice against a lethal challenge with the influenza virus [184]. In addition, Qβ has been extensively used to generate vaccine candidates targeted to non-infectious diseases such as nicotine addiction, hypertension, cancer, diabetes, allergies, and Alzheimer’s disease [185]. Six of these candidates have completed Phase I or Phase II clinical trials [185].
6. Concluding remarks
Development of vaccines against infectious diseases has been, and still remains, an empirical exercise. However, there is a clear shift from the whole-pathogen vaccines to recombinant subunit vaccines because the latter are equally efficacious but safer and less reactogenic. Recent development of the recombinant shingles and chickenpox vaccines are good examples of this continuing trend. The recombinant vaccines however require the inclusion of a strong adjuvant or an efficient delivery system in order to achieve robust immune responses and superior efficacy. Although adjuvants such as the lipid-based formulations have been enormously successful, design of VLP platforms, particularly those that can be universally employed with any recombinant antigen might provide better, cheaper, and safer alternatives in the future.
Although numerous VLP platforms from viral, bacterial, and synthetic sources have been developed or under investigation, a phage-based platform might be an ideal candidate for “universal” vaccine delivery. Phage VLPs have high stability and can be manufactured relatively easily and cost-effectively. They are safer than their mammalian counterparts and there is no pre-existing immunity in humans. Furthermore, the rich understanding of the genetics, biochemistry, and structure of phages provides enormous advantages for effective vaccine designs.
As described above, many phage VLP platforms have been described and offer different advantages and limitations (Fig. 8). However, most are designed, or primarily suitable, for display of short peptides but not large domains or full-length proteins. Phage T4 by far provides the best option as a universal vaccine delivery vehicle for several reasons. First, the large size of its capsid provides a platform for high density array of antigen molecules. Second, two outer different capsid proteins Soc and Hoc can be used to display antigen molecules of different sizes; peptides, domains, full-length proteins, or multi-subunit complexes. Third, co-delivery of multiple antigens, DNAs, targeting molecules and/or molecular adjuvants provides unique advantages. Finally, research has already demonstrated that phage T4 VLPs provide complete protection against the deadly anthrax and plague infections in animal models including the nonhuman primates.
In conclusion, development of a single universal VLP platform such as phage T4 that can generate vaccines against current and emerging pathogens could greatly accelerate vaccine development in the future. It could reduce time, efforts, and resources as well as streamline the downstream clinical trials, FDA approval, and vaccine manufacture.
Acknowledgments
The authors thank Dr. Victor Padilla-Sanchez for assistance with the illustrations. VBR thanks many current and past post-doctoral fellows and graduate students who contributed to bacteriophage T4 vaccine research, in particular Jennifer Jiang, Laura Abu-Shilbayeh, Zhihong Zhang, Qin Li, Sathish Shivachandra, Taheri Sathaliyawala, Guofen Guo, and Wadad Alsalmi; collaborators Drs. Michael G. Rossmann and Andrei Fokine (Purdue University), Ashok Chopra (University of Texas Medical Branch), Stephen H. Leppla (National Institutes of Health), and Carl Alving, Gary Matyas, and Mangala Rao (Walter Reed Army Institute of Research). The research in VBR’s laboratory has been supported by grants from the National Institute of Allergy and Infectious Diseases (current: AI111538 and AI081726) and National Science Foundation (MCB-1411989).
References
- [1].Plotkin SA, Vaccines: past, present and future, Nat. Med 11 (2005) S5–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Rappuoli R, Pizza M, Del Giudice C, De Gregorio E, Vaccines, new opportunities for a new society, Proc. Natl. Acad. Sci. U. S. A 111 (2014) 12288–12293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Tognotti E, The eradication of smallpox, a success story for modern medicine and public health: what lessons for the future? J. Infect. Dev. Ctries 4 (2010) 264–266. [DOI] [PubMed] [Google Scholar]
- [4].Finco O, Rappuoli R, Designing vaccines for the twenty-first century society, Front Immunol. 5 (2014) 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Delany I, Rappuoli R, De Gregorio E, Vaccines for the 21st century, EMBO Mol. Med 6 (2014) 708–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Yadav DK, Yadav N, Khurana SMP, Vaccines: Present Status and Applications, Academic Press, Waltham, MA, USA, 2014. [Google Scholar]
- [7].Schiller JT, Lowy DR, Raising expectations for subunit vaccine, J. Infect. Dis 211 (2015) 1373–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Moyle PM, Toth I, Modern subunit vaccines: development, components, and research opportunities, ChemMedChem 8 (2013) 360–376. [DOI] [PubMed] [Google Scholar]
- [9].Karch CP, Burkhard P, Vaccine technologies: from whole organisms to rationally designed protein assemblies, Biochem. Pharmacol 120 (2016) 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Pulendran B, Ahmed R, Immunological mechanisms of vaccination, Nat. Immunol 12 (2011) 509–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Levitz SM, Golenbock DT, Beyond empiricism: informing vaccine development through innate immunity research, Cell 148 (2012) 1284–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Coffman RL, Sher A, Seder RA, Vaccine adjuvants: putting innate immunity to work, Immunity 33 (2010) 492–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Yang L, Li W, Kirberger M, Liao W, Ren J, Design of nanomaterial based systems for novel vaccine development, Biomater. Sci 4 (2016) 785–802. [DOI] [PubMed] [Google Scholar]
- [14].Lee KL, Twyman RM, Fiering S, Steinmetz NF, Virus-based nanoparticles as platform technologies for modern vaccines, Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol 8 (2016) 554–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].van Riet E, Ainai A, Suzuki T, Kersten G, Hasegawa H, Combatting infectious diseases; nanotechnology as a platform for rational vaccine design, Adv. Drug Deliv. Rev 74 (2014) 28–34. [DOI] [PubMed] [Google Scholar]
- [16].Mooney M, McWeeney S, Canderan G, Sekaly RP, A systems framework for vaccine design, Curr. Opin. Immunol 25 (2013) 551–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Nabel GJ, Designing tomorrow’s vaccines, N. Engl. J. Med 368 (2013) 551–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bachmann MF, Jennings GT, Vaccine delivery: a matter of size, geometry, kinetics and molecular patterns, Nat Rev. Immunol 10 (2010) 787–796. [DOI] [PubMed] [Google Scholar]
- [19].Zepp F, Principles of vaccine design-lessons from nature, Vaccine 28 (Suppl 3) (2010) C14–C24. [DOI] [PubMed] [Google Scholar]
- [20].Roldao A, Mellado MC, Castilho LR, Carrondo MJ, Alves PM, Virus-like particles in vaccine development, Expert Rev. Vaccines 9 (2010) 1149–1176. [DOI] [PubMed] [Google Scholar]
- [21].Jain NK, Sahni N, Kumru OS, Joshi SB, Volkin DB, Russell Middaugh C, Formulation and stabilization of recombinant protein based virus-like particle vaccines, Adv. Drug Deliv. Rev 93 (2015) 42–55. [DOI] [PubMed] [Google Scholar]
- [22].Orfi E, Szebeni J, The immune system of the gut and potential adverse effects of oral nanocarriers on its function, Adv. Drug Deliv. Rev 106 (2016) 402–409. [DOI] [PubMed] [Google Scholar]
- [23].Frietze KM, Peabody DS, Chackerian B, Engineering virus-like particles as vaccine platforms, Curr. Opin. Virol 18 (2016) 44–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Csaba N, Garcia-Fuentes M, Alonso MJ, Nanoparticles for nasal vaccination, Adv. Drug Deliv. Rev 61 (2009) 140–157. [DOI] [PubMed] [Google Scholar]
- [25].Fan Y, Moon JJ, Particulate delivery systems for vaccination against bioterrorism agents and emerging infectious pathogens, Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol 9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Boraschi D, Italiani P, Palomba R, Decuzzi P, Duschl A, Fadeel B, Moghimi SM, Nanoparticles and innate immunity: new perspectives on host defence, Semin. Immunol 34 (2017) 33–51. [DOI] [PubMed] [Google Scholar]
- [27].Lee EJ, Lee NK, Kim IS, Bioengineered protein-based nanocage for drug delivery, Adv. Drug Deliv. Rev 106 (2016) 157–171. [DOI] [PubMed] [Google Scholar]
- [28].Nicastro J, Sheldon K, Slavcev RA, Bacteriophage lambda display systems: developments and applications, Appl. Microbiol. Biotechnol 98 (2014) 2853–2866. [DOI] [PubMed] [Google Scholar]
- [29].Henry KA, Arbabi-Ghahroudi M, Scott JK, Beyond phage display: non-traditional applications of the filamentous bacteriophage as a vaccine carrier, therapeutic biologic, and bioconjugation scaffold, Front. Microbiol 6 (2015) 755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Prisco A, De Berardinis P, Filamentous bacteriophage fd as an antigen delivery system in vaccination, Int. J. Mol. Sci 13 (2012) 5179–5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Beghetto E, Gargano N, Lambda-display: a powerful tool for antigen discovery, Molecules (Basel, Switzerland) 16 (2011) 3089–3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Jafari N, A. S, Phage particles as vaccine delivery vehicles: concepts, applications and prospects, Asian Pac. J. Cancer Prev 16 (2015) 8019–8029. [DOI] [PubMed] [Google Scholar]
- [33].Fu Y, Li J, A novel delivery platform based on bacteriophage MS2 virus-like particles, Virus Res. 211 (2016) 9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Odendall C, Kagan JC, Activation and pathogenic manipulation of the sensors of the innate immune system. Microbes Infect 19 (2017) 229–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kumar H, Kawai T, Akira S, Pathogen recognition by the innate immune system. Int. Rev. Immunol 30 (2011) 16–34. [DOI] [PubMed] [Google Scholar]
- [36].Desmet CJ, Ishii KJ, Nucleic acid sensing at the interface between innate and adaptive immunity in vaccination, Nat. Rev. Immunol 12 (2012) 479–491. [DOI] [PubMed] [Google Scholar]
- [37].Schenten D, Medzhitov R, The control of adaptive immune responses by the innate immune system. Adv. Immunol 109 (2011) 87–124. [DOI] [PubMed] [Google Scholar]
- [38].Steinman RM, Banchereau J, Taking dendritic cells into medicine, Nature 449 (2007) 419–426. [DOI] [PubMed] [Google Scholar]
- [39].Liu Z, Roche PA, Macropinocytosis in phagocytes: regulation of MHC class-II-restricted antigen presentation in dendritic cells, Front Physiol. 6 (2015) 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Austyn JM, Dendritic cells in the immune system-history, lineages, tissues, tolerance, and immunity, Microbiol. Spectr 4 (2016). [DOI] [PubMed] [Google Scholar]
- [41].Lutz MB, Schuler G, Immature, semi-mature and fully mature dendritic cells: which signals induce tolerance or immunity? Trends Immunol. 23 (2002) 445–449. [DOI] [PubMed] [Google Scholar]
- [42].Delaloye J, Roger T, Steiner-Tardivel QG, Le Roy D, Knaup Reymond M, Akira S, Petrilli V, Gomez CE, Perdiguero B, Tschopp J, Pantaleo G, Esteban M, Calandra T, Innate immune sensing of modified vaccinia virus Ankara (MVA) is mediated by TLR2-TLR6, MDA-5 and the NALP3 inflammasome, PLoS Pathog. 5 (2009), e1000480.. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [43].Banchereau J, Steinman RM, Dendritic cells and the control of immunity, Nature 392 (1998) 245–252. [DOI] [PubMed] [Google Scholar]
- [44].Kenneth M, Casey W, Janeway’s Immunobiology, 9th edition Garland Science, Taylor & Francis Group, New York, 2016. [Google Scholar]
- [45].Zhang N, Bevan MJ, CD8(+) T cells: foot soldiers of the immune system, Immunity 35 (2011)161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Blum JS, Wearsch PA, Cresswell P, Pathways of antigen processing, Annu. Rev. Immunol 31 (2013) 443–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Koup RA, Douek DC, Vaccine design for CD8 T lymphocyte responses, Cold Spring Harb. Perspect. Med 1 (2011) a007252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Joffre OP, Segura E, Savina A, Amigorena S, Cross-presentation by dendritic cells, Nat Rev. Immunol 12 (2012) 557–569. [DOI] [PubMed] [Google Scholar]
- [49].Fehres CM, Unger WW, Garcia-Vallejo JJ, van Kooyk Y, Understanding the biology of antigen cross-presentation for the design of vaccines against cancer, Front. Immunol 5 (2014) 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Zhu J, Yamane H, Paul WE, Differentiation of effector CD4 T cell populations, Annu. Rev. Immunol 28 (2010) 445–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Luckheeram RV, Zhou R, Verma AD, Xia B, CD4(+)T cells: differentiation and functions, Clin. Dev. Immunol 2012 (2012) 925135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Harwood NE, Batista FD, Early events in B cell activation, Annu. Rev. Immunol 28 (2010) 185–210. [DOI] [PubMed] [Google Scholar]
- [53].Nutt SL, Hodgkin PD, Tarlinton DM, Corcoran LM, The generation of antibody-secreting plasma cells, Nat. Rev. Immunol 15 (2015) 160–171. [DOI] [PubMed] [Google Scholar]
- [54].Smith GP, Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface, Science (New York, N.Y.) 228 (1985) 1315–1317. [DOI] [PubMed] [Google Scholar]
- [55].Tao P, Mahalingam M, Kirtley ML, van Lier CJ, Sha J, Yeager LA, Chopra AK, Rao VB, Mutated and bacteriophage T4 nanoparticle arrayed F1-V immunogens from Yersinia pestis as next generation plague vaccines, PLoS Pathog. 9 (2013), e1003495.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Gamkrelidze M, Dabrowska K, T4 bacteriophage as a phage display platform, Arch. Microbiol 196 (2014) 473–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Danner S, Belasco JG, T7 phage display: a novel genetic selection system for cloning RNA-binding proteins from cDNA libraries, Proc. Natl. Acad. Sci. U. S. A 98 (2001) 12954–12959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Kozlovska TM, Cielens I, Vasiljeva I, Strelnikova A, Kazaks A, Dislers A, Dreilina D, Ose V, Gusars I, Pumpens P, RNA phage Q beta coat protein as a carrier for foreign epitopes, Intervirology 39 (1996) 9–15. [DOI] [PubMed] [Google Scholar]
- [59].Tissot AC Renhofa R, Schmitz N, Cielens I, Meijerink E, Ose V, Jennings GT, Saudan P, Pumpens P, Bachmann MF, Versatile virus-like particle carrier for epitope based vaccines, PLoS One 5 (2010), e9809.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Salmond GP, Fineran PC, A century of the phage: past, present and future, Nat. Rev. Microbiol 13 (2015) 777–786. [DOI] [PubMed] [Google Scholar]
- [61].Ackermann HW, Frequency of morphological phage descriptions in the year 2000. Brief review, Arch. Virol 146 (2001) 843–857. [DOI] [PubMed] [Google Scholar]
- [62].Bachmann MF, Zinkernagel RM, Neutralizing antiviral B cell responses, Annu. Rev. Immunol 15 (1997) 235–270. [DOI] [PubMed] [Google Scholar]
- [63].Rao VB, Black LW, Structure and assembly of bacteriophage T4 head, Virol. J 7 (2010) 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Moody MF, Geometry of phage head construction, J. Mol. Biol 293 (1999) 401–433. [DOI] [PubMed] [Google Scholar]
- [65].Black LW, Rao VB, Structure, assembly, and DNA packaging of the bacteriophage T4 head, Adv. Virus Res 82 (2012) 119–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Fokine A, Bowman VD, Battisti AJ, Li Q, Chipman PR, Rao VB, Rossmann MG, Cryo-electron microscopy study of bacteriophage T4 displaying anthrax toxin proteins, Virology 367 (2007) 422–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Link A, Zabel F, Schnetzler Y, Titz A, Brombacher F, Bachmann MF, Innate immunity mediates follicular transport of particulate but not soluble protein antigen, J. Immunol 188 (2012) 3724–3733. [DOI] [PubMed] [Google Scholar]
- [68].Baschong W, Hasler L, Haner M, Kistler J, Aebi U, Repetitive versus monomeric antigen presentation: direct visualization of antibody affinity and specificity, J. Struct. Biol 143 (2003) 258–262. [DOI] [PubMed] [Google Scholar]
- [69].Hinton HJ, Jegerlehner A, Bachmann MF, Pattern recognition by B cells: the role of antigen repetitiveness versus Toll-like receptors, Curr. Top. Microbiol. Immunol 319(2008) 1–15. [DOI] [PubMed] [Google Scholar]
- [70].Barfoot R, Denham S, Gyure LA, Hall J,G, Hobbs SM, Jackson LE, Robertson D, Some properties of dendritic macrophages from peripheral lymph, Immunology 68 (1989) 233–239. [PMC free article] [PubMed] [Google Scholar]
- [71].Aronow R, Danon D, Shahar A, Aronson M, Electron microscopy of in vitro endocytosis of T2 phage by cells from rabbit peritoneal exudate, J. Exp. Med 120 (1964) 943–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Jonczyk-Matysiak E, Weber-Dabrowska B, Owczarek B, Miedzybrodzki R, Lusiak-Szelachowska M, Lodej N, Gorski A, Phage-phagocyte interactions and their implications for phage application as therapeutics, Virus 9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Bachmann MF, Rohrer UH, Kundig TM, Burki K, Hengartner H, Zinkernagel RM, The influence of antigen organization on B cell responsiveness, Science (New York, N.Y.) 262 (1993) 1448–1451. [DOI] [PubMed] [Google Scholar]
- [74].Bachmann MF, Zinkernagel RM, The influence of virus structure on antibody responses and virus serotype formation, Immunol. Today 17 (1996) 553–558. [DOI] [PubMed] [Google Scholar]
- [75].Jegerlehner A, Storni T, Lipowsky G, Schmid M, Pumpens P, Bachmann MF, Regulation of IgG antibody responses by epitope density and CD21-mediated costimulation, Eur. J. Immunol 32 (2002) 3305–3314. [DOI] [PubMed] [Google Scholar]
- [76].Cheng W, The density code for the development of a vaccine? J. Pharm. Sci 105 (2016) 3223–3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Klein JS, Bjorkman PJ, Few and far between: how HIV may be evading antibody avidity. PLoS Pathog. 6 (2010), e1000908.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Bachrach E, Dreja H, L Lin Y, Mettling C, Pinet V, Corbeau P, Piechaczyk M, Effects of virion surface gp120 density on infection by HIV-1 and viral production by infected cells, Virology 332 (2005) 418–429. [DOI] [PubMed] [Google Scholar]
- [79].Gao G, Wieczorek L, Peachman KK, Polonis VR, Alving CR, Rao M, Rao VB, Designing a soluble near full-length HIV-1 gp41 trimer, J. Biol. Chem 288 (2013) 234–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Zabel F.Kundig TM, Bachmann MF, Virus-induced humoral immunity: on how B cell responses are initiated, Curr. Opin. Virol 3 (2013) 357–362. [DOI] [PubMed] [Google Scholar]
- [81].Manolova V, Flace A, Bauer M, Schwarz K, Saudan P, Bachmann MF, Nanoparticles target distinct dendritic cell populations according to their size, Eur. J. Immunol 38 (2008) 1404–1413. [DOI] [PubMed] [Google Scholar]
- [82].Rumnieks J, Tars K, Crystal structure of the maturation protein from bacteriophage Qbeta, J. Mol. Biol, 429 (2017) 688–696. [DOI] [PubMed] [Google Scholar]
- [83].Guo F, Liu Z, Fang PA, Zhang Q, Wright ET Wu W, Zhang C, Vago F, Ren Y, Jakana J, Chiu W, Serwer P, Jiang W, Capsid expansion mechanism of bacteriophage T7 revealed by multistate atomic models derived from cryo-EM reconstructions, Proc. Natl. Acad. Sci. U. S. A 111 (2014) E4606–E4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Chen Z, Sun L, Zhang Z, Fokine A, Padilla-Sanchez V, Hanein D, Jiang W, Rossmann MG, Rao VB, Cryo-EM structure of the bacteriophage T4 isometric head at 3.3-a resolution and its relevance to the assembly of icosahedral viruses, Proc. Natl. Acad. Sci. U. S. A 114 (2017) E8184–E8193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Mantegazza AR, Magalhaes JG, Amigorena S, Marks MS, Presentation of phagocytosed antigens by MHC class I and II, Traffic 14 (2013) 135–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Sathaliyawala T, Rao M, Maclean DM, Birx DL, Alving CR, Rao VB, Assembly of human immunodeficiency virus (HIV) antigens on bacteriophage T4: a novel in vitro approach to construct multicomponent HIV vaccines, J. Virol 80 (2006) 7688–7698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Gaubin M, Fanutti C, Mishal Z, Durrbach A, De Berardinis P, Sartorius R, Del Pozzo G, Guardiola J, Perham RN, Piatier-Tonneau D, Processing of filamentous bacteriophage virions in antigen-presenting cells targets both HLA class I and class II peptide loading compartments, DNA Cell Biol. 22 (2003) 11–18. [DOI] [PubMed] [Google Scholar]
- [88].Sporri R, Reis C Sousa E, Inflammatory mediators are insufficient for full dendritic cell activation and promote expansion of CD4+ T cell populations lacking helper function, Nat. Immunol 6 (2005) 163–170. [DOI] [PubMed] [Google Scholar]
- [89].Gomes AC, Flace A, Saudan P, Zabel F, Cabral-Miranda G, Turabi AE, Manolova V, Bachmann MF, Adjusted particle size eliminates the need of linkage of antigen and adjuvants for appropriated T cell responses in virus-like particle-based vaccines. Front. Immunol 8 (2017) 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Zimecki M, Weber-Dabrowska B, Lusiak-Szelachowska M, Mulczyk M, Boratynski J, Pozniak G, Syper D, Gorski A, Bacteriophages provide regulatory signals in mitogen-induced murine splenocyte proliferation. Cell. Mol. Biol. Lett 8 (2003) 699–711. [PubMed] [Google Scholar]
- [91].Zanghi CN, Sapinoro R, Bradel-Tretheway B, Dewhurst S A tractable method for simultaneous modifications to the head and tail of bacteriophage lambda and its application to enhancing phage-mediated gene delivery, Nucleic Acids Res. 35 (2007) e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Tao P. Mahalingam M, Marasa BS, Zhang Z, Chopra AK, Rao VB, In vitro and in vivo delivery of genes and proteins using the bacteriophage T4 DNA packaging machine, Proc. Natl. Acad. Sci. U. S. A 110 (2013) 5846–5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Querec T, Bennouna S, Alkan S, Laouar Y, Gorden K, Flavell R, Akira S, Ahmed R, Pulendran B, Yellow fever vaccine YF-17D activates multiple dendritic cell subsets via TLR2, 7, 8, and 9 to stimulate polyvalent immunity, J. Exp. Med 203 (2006) 413–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Desch AN, Gibbings SL, Clambey ET, Janssen WJ, Slansky JE, Kedl RM, Henson PM, Jakubzick C, Dendritic cell subsets require cis-activation for cytotoxic CD8 T-cell induction, Nat. Commun 5 (2014) 4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Kastenmuller W, Kastenmuller K, Kurts C, Seder RA, Dendritic cell-targeted vaccines–hope or hype? Nat Rev. Immunol 14 (2014) 705–711. [DOI] [PubMed] [Google Scholar]
- [96].Macri C, Dumont C, Johnston AP, Mintern JD, Targeting dendritic cells: a promising strategy to improve vaccine effectiveness, Transpl. Immunol 5 (2016), e66.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Rao VB, Tao P, Mahalingam M, Marasa BS, Zhang Z, Chopra AK, Delivery of vaccine genes and proteins into dendritic cells using the bacteriophage T4 DNA packaging machine (P3273), J. Immunol 190 (2013) (192.122). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Jiang W, Swiggard WJ, Heufler C, Peng M, Mirza A, Steinman RM, Nussenzweig MC, The receptor DEC-205 expressed by dendritic cells and thymic epithelial cells is involved in antigen processing, Nature 375 (1995) 151–155. [DOI] [PubMed] [Google Scholar]
- [99].Sartorius R, Bettua C, D′Apice L, Caivano A, Trovato M, Russo D, Zanoni I, Granucci F, Mascolo D, Barba P, Del Pozzo G, De Berardinis P, Vaccination with filamentous bacteriophages targeting DEC-205 induces DC maturation and potent anti-tumor T-cell responses in the absence of adjuvants, Eur. J. Immunol 41 (2011) 2573–2584. [DOI] [PubMed] [Google Scholar]
- [100].Sioud M, Skorstad G, Mobergslien A, Saeboe-Larssen S, A novel peptide carrier for efficient targeting of antigens and nucleic acids to dendritic cells, FASEB J. 27 (2013) 3272–3283. [DOI] [PubMed] [Google Scholar]
- [101].Jung SN, Kang SK, Yeo GH, Li HY, Jiang T, Nah JW, Bok JD, Cho CS, Choi YJ, Targeted delivery of vaccine to dendritic cells by chitosan nanoparticles conjugated with a targeting peptide ligand selected by phage display technique, Macromol. Biosci 15 (2015) 395–404. [DOI] [PubMed] [Google Scholar]
- [102].Curiel TJ, Morris C, Brumlik M, Landry SJ, Finstad K, Nelson A, Joshi V, Hawkins C, Alarez X, Lackner A, Mohamadzadeh M, Peptides identified through phage display direct immunogenic antigen to dendritic cells, J. Immunol 172 (2004) 7425–7431. [DOI] [PubMed] [Google Scholar]
- [103].Dalod M, Chelbi R, Malissen B, Lawrence T, Dendritic cell maturation: functional specialization through signaling specificity and transcriptional programming, EMBO J. 33 (2014) 1104–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Sartorius R, D’Apice L, Trovato M, Cuccaro F, Costa V, De Leo MG, Marzullo VM, Biondo C, D’Auria S, De Matteis MA, Ciccodicola A, De Berardinis P, Antigen delivery by filamentous bacteriophage fd displaying an anti-DEC-205 single-chain variable fragment confers adjuvanticity by triggering a TLR9-mediated immune response, EMBO Mol. Med 7 (2015) 973–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Kew OM, Sutter RW, de Gourville EM, Dowdle WR, Pallansch MA, Vaccine-derived polioviruses and the endgame strategy for global polio eradication, Annu. Rev. Microbiol 59 (2005) 587–635. [DOI] [PubMed] [Google Scholar]
- [106].Yoshida H, Horie H, Matsuura K, Kitamura T, Hashizume S, Miyamura T, Prevalence of vaccine-derived polioviruses in the environment, J. Gen. Virol 83 (2002) 1107–1111. [DOI] [PubMed] [Google Scholar]
- [107].Minor PD, Dunn G, The effect of sequences in the 5’ non-coding region on the replication of polioviruses in the human gut, J. Gen. Virol 69 (Pt 5) (1988) 1091–1096. [DOI] [PubMed] [Google Scholar]
- [108].Nielsen HS, Oleksiewicz MB, Forsberg R, Stadejek T, Botner A, Storgaard T, Reversion of a live porcine reproductive and respiratory syndrome virus vaccine investigated by parallel mutations, J. Gen. Virol 82 (2001) 1263–1272. [DOI] [PubMed] [Google Scholar]
- [109].Czapar AE, Steinmetz NF, Plant viruses and bacteriophages for drug delivery in medicine and biotechnology, Curr. Opin. Chem. Biol 38 (2017) 108–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Shivachandra SB, Rao M, Janosi L, Sathaliyawala T, Matyas GR, Alving CR, Leppla SH, Rao VB, In vitro binding of anthrax protective antigen on bacteriophage T4 capsid surface through Hoc-capsid interactions: a strategy for efficient display of large full-length proteins, Virology 345 (2006) 190–198. [DOI] [PubMed] [Google Scholar]
- [111].Peachman KK, Li Q, Matyas GR, Shivachandra SB, Lovchik J, Lyons RC, Alving CR, Rao VB, Rao M, Anthrax vaccine antigen-adjuvant formulations completely protect New Zealand white rabbits against challenge with Bacillus anthracis Ames strain spores, Clin. Vaccine Immunol 19 (2012) 11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Rao M, Peachman KK, Li Q, Matyas GR, Shivachandra SB, Borschel R, Morthole VI, Femandez-Prada C, Alving CR, Rao VB, Highly effective generic adjuvant systems for orphan or poverty-related vaccines, Vaccine 29 (2011) 873–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Bruttin A, Brussow H, Human volunteers receiving Escherichia coli phage T4 orally: a safety test of phage therapy, Antimicrob. Agents Chemother 49 (2005) 2874–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Sarker SA, Berger B, Deng Y, Kieser S, Foata F, Moine D, Descombes P, Sultana S, Huq S, Bardhan PK, Vuillet V, Praplan F, Brussow H, Oral application of Escherichia coli bacteriophage: safety tests in healthy and diarrheal children from Bangladesh, Environ. Microbiol 19 (2017) 237–250. [DOI] [PubMed] [Google Scholar]
- [115].Sarker SA, Sultana S, Reuteler G, Moine D, Descombes P, Charton F, Bourdin G, McCallin S, Ngom-Bru C, Neville T, Akter M, Huq S, Qadri F, Talukdar K, Kassam M, Delley M, Loiseau C, Deng Y, El Aidy S, Berger B, Brussow H, Oral phage therapy of acute bacterial diarrhea with two coliphage preparations: a randomized trial in children from Bangladesh, EBioMedicine 4 (2016) 124–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Sulakvelidze A, Alavidze Z, Morris JG Jr., Bacteriophage therapy, Antimicrob. Agents Chemother 45 (2001) 649–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Cisek AA, Dabrowska I, Gregorczyk KP, Wyzewski Z, Phage therapy in bacterial infections treatment: one hundred years after the discovery of bacteriophages, Curr. Microbiol 74 (2017) 277–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Leiman PG, Arisaka F, van Raaij MJ, Kostyuchenko VA, Aksyuk AA, Kanamaru S, Rossmann MG, Morphogenesis of the T4 tail and tail fibers, Virol. J 7 (2010) 355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Rao VB, Feiss M, Mechanisms of DNA packaging by large double-stranded DNA viruses, Annu. Rev. Virol 2 (2015) 351–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Rossmann MG, Mesyanzhinov VV, Arisaka F, Leiman PG, The bacteriophage T4 DNA injection machine, Curr. Opia Struct. Biol 14 (2004) 171–180. [DOI] [PubMed] [Google Scholar]
- [121].Sun L, Zhang X, Gao S, Rao PA, Padilla-Sanchez V, Chen Z, Sun S, Xiang Y, Subramaniam S, Rao VB, Rossmann MG, Cryo-EM structure of the bacteriophage T4 portal protein assembly at near-atomic resolution, Nat. Commun 6 (2015) 7548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Rao VB, Feiss M, The bacteriophage DNA packaging motor, Annu. Rev. Genet 42 (2008) 647–681. [DOI] [PubMed] [Google Scholar]
- [123].Fuller DN, Raymer DM, Kottadiel VI, Rao VB, Smith DE, Single phage T4 DNA packaging motors exhibit large force generation, high velocity, and dynamic variability, Proc. Natl. Acad. Sri. U. S. A 104 (2007) 16868–16873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Ishii T, Yanagida M, The two dispensable structural proteins (soc and hoc) of the T4 phage capsid; their purification and properties, isolation and characterization of the defective mutants, and their binding with the defective heads in vitro, J. Mol. Biol 109 (1977) 487–514. [DOI] [PubMed] [Google Scholar]
- [125].Fokine A, Islam MZ, Zhang Z, Bowman VD, Rao VB, Rossmann MG, Structure of the three N-terminal immunoglobulin domains of the highly immunogenic outer capsid protein from aT4-like bacteriophage, J. Virol 85 (2011) 8141–8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Qin L, Fokine A, O’Donnell E, Rao VB, Rossmann MG, Structure of the small outer capsid protein, Soc: a clamp for stabilizing capsids of T4-like phages, J. Mol. Biol 395 (2010) 728–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Sathaliyawala T, Islam MZ, Li Q, Fokine A, Rossmann MG, Rao VB, Functional analysis of the highly antigenic outer capsid protein, Hoc, a virus decoration protein from T4-like bacteriophages, Mol. Microbiol 77 (2010) 444–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Dabrowska K, Switala-Jelen K, Opolski A, Gorski A, Possible association between phages, Hoc protein, and the immune system, Arch. Virol 151 (2006) 209–215. [DOI] [PubMed] [Google Scholar]
- [129].Barr JJ, Auro R, Sam-Soon N, Kassegne S, Peters G, Bonilla N, Hatay M, Mourtada S, Bailey B, Youle M, Felts B, Baljon A, Nulton J, Salamon P, Rohwer F, Subdiffusive motion of bacteriophage in mucosal surfaces increases the frequency of bacterial encounters, Proc. Natl. Acad. Sci. U. S. A 112 (2015) 13675–13680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Barr JJ, Auro R, Furlan M, L Whiteson K, L Erb M, Pogliano J, Stotland A, Wolkowicz R, Cutting AS, Doran KS, Salamon P, Youle M, Rohwer F, Bacteriophage adhering to mucus provide a non-host-derived immunity, Proc. Natl. Acad. Sci. U. S. A 110 (2013) 10771–10776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Robertson K, Furukawa Y, Underwood A, Black L, Liu JL, Deletion of the Hoc and Soc capsid proteins affects the surface and cellular uptake properties of bacteriophage T4 derived nanoparticles, Biochem. Biophys. Res. Commun 418 (2012) 537–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Li Q, Shivachandra SB, Zhang Z, Rao VB, Assembly of the small outer capsid protein, Soc, on bacteriophage T4: a novel system for high density display of multiple large anthrax toxins and foreign proteins on phage capsid, J. Mol. Biol 370 (2007) 1006–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Sun S, Kondabagil K, Draper B, Alam T.l., Bowman VD, Zhang Z, Hegde S, Fokine A, Rossmann MG, Rao VB, The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces, Cell 135 (2008) 1251–1262. [DOI] [PubMed] [Google Scholar]
- [134].Kondabagil KR, Zhang Z, Rao VB, The DNA translocating ATPase of bacteriophage T4 packaging motor, J. Mol. Biol 363 (2006) 786–799. [DOI] [PubMed] [Google Scholar]
- [135].Zhang Z, L Kottadiel V, Vafabakhsh R, Dai L, Chemla YR, Ha T, Rao VB, A promiscuous DNA packaging machine from bacteriophage T4, PLoS Biol. 9 (2011), e1000592.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Ren ZJ, Lewis GK, Wingfield PT, Locke EG, Steven AC, Black LW, Phage display of intact domains at high copy number: a system based on SOC, the small outer capsid protein of bacteriophage T4, Protein Sci. 5 (1996) 1833–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Jiang J, Abu-Shilbayeh L, Rao VB, Display of a PorA peptide from Neisseria meningitidis on the bacteriophage T4 capsid surface. Infect. Immun 65 (1997) 4770–4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Tao P, Mahalingam M, Rao VB, Highly effective soluble and bacteriophage T4 nanoparticle plague vaccines against Yersinia pestis. Methods Mol. Biol 1403 (2016)499–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Tao P, Li Q, Shivachandra SB, Rao VB, Bacteriophage T4 as a nanoparticle platform to display and deliver pathogen antigens: construction of an effective anthrax vaccine, Methods Mol. Biol 1581 (2017) 255–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Shivachandra SB, Li Q, Peachman KK, Matyas GR, Leppla SH, Alving CR, Rao M, Rao VB, Multicomponent anthrax toxin display and delivery using bacteriophage T4, Vaccine 25 (2007) 1225–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Tao P, Wu X, Tang WC, Zhu J, Rao VB, Engineering of bacteriophage T4 genome using CRISPR-Cas9, ACS Synth. Biol 6 (2017) 1952–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Tao P, Wu X, Rao VB, Unexpected evolutionary benefit to phages imparted by bacterial CRISPR-Cas9, Sci. Adv 4 (2018), eaar4134.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Li Q, Shivachandra SB, Leppla SH, Rao VB, Bacteriophage T4 capsid: a unique platform for efficient surface assembly of macromolecular complexes, J. Mol. Biol 363 (2006) 577–588. [DOI] [PubMed] [Google Scholar]
- [144].Ren ZJ, Tian CJ, Zhu QS, Zhao MY, Xin AG, Nie WX, Ling SR, Zhu MW, Wu JY, Lan HY, Cao YC, Bi YZ, Orally delivered foot-and-mouth disease virus capsid protomer vaccine displayed on T4 bacteriophage surface: 100% protection from potency challenge in mice, Vaccine 26 (2008) 1471–1481. [DOI] [PubMed] [Google Scholar]
- [145].Wu J, Tu C, Yu X, Zhang M, Zhang N, Zhao M, Nie W, Ren Z, Bacteriophage T4 nanoparticle capsid surface SOC and HOC bipartite display with enhanced classical swine fever virus immunogenicity: a powerful immunological approach, J. Virol. Methods 139 (2007) 50–60. [DOI] [PubMed] [Google Scholar]
- [146].C Cao Y, Shi QC, Ma JY, Xie QM, Bi YZ, Vaccination against very virulent infectious bursal disease virus using recombinant T4 bacteriophage displaying viral protein VP2, Acta Biochim. Biophys. Sin 37 (2005) 657–664. [DOI] [PubMed] [Google Scholar]
- [147].Moayeri M, Leppla SH, Vrentas C, Pomerantsev AP, Liu S, Anthrax pathogenesis, Annu. Rev. Microbiol 69 (2015) 185–208. [DOI] [PubMed] [Google Scholar]
- [148].Williamson ED, Dyson EH, Anthrax prophylaxis: recent advances and future directions, Front. Microbiol 6 (2015) 1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Scorpio A, Blank TE, Day WA, Chabot Anthrax vaccines DJ: Pasteur to the present, Cell. Mol. Life Sci 63 (2006) 2237–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].A Rosenzweig J, jejelowo O, Sha J, Erova TE, Brackman SM, L Kirtley M, van Lier CJ, Chopra AK, Progress on plague vaccine development, Appl. Microbiol. Biotechnol 91 (2011) 265–286. [DOI] [PubMed] [Google Scholar]
- [151].Williamson ED, Flick-Smith HC, Lebutt C, Rowland CA, Jones SM, Waters EL, Gwyther RJ, Miller J, Packer PJ, Irving M, Human immune response to a plague vaccine comprising recombinant F1 and V antigens, Infect Immun. 73 (2005) 3598–3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Tao P, Mahalingam M, Zhu J, Moayeri M, Kirtley ML, Fitts EC, Andersson JA, Lawrence WS, Leppla SH, Chopra AK, Rao VB, A bivalent Anthrax-plague vaccine that can protect against two Tier-1 bioterror pathogens, Bacillus anthracis and Yersinia pestis. Front Immunol. 8 (2017) 687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Vela Ramirez JE, Sharpe LA, Peppas NA, Current state and challenges in developing oral vaccines, Adv. Drug Deliv. Rev 114 (2017) 116–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Lu S, Heterologous prime-boost vaccination, Curr. Opin. Immunol 21 (2009) 346–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Huai Y, Dong S, Zhu Y, Li X, Cao B, Gao X, Yang M, Wang L, Mao C, Genetically engineered virus nanofibers as an efficient vaccine for preventing fungal infection. Adv. Healthc. Mater 5 (2016) 786–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Deng L, Roose K, Job ER, De Rycke R, Van Hamme E, Goncalves A, Parthoens E, Cicchelero L, Sanders N, Fiers W, Saelens X, Oral delivery of Escherichia coli persistently infected with M2e-displaying bacteriophages partially protects against influenza A virus, J. Control. Release 264 (2017) 55–65. [DOI] [PubMed] [Google Scholar]
- [157].Samoylova TI, Braden TD, Spencer JA, Bartol FF, Immunocontraception: filamentous bacteriophage as a platform for vaccine development Curr. Med. Chem 24 (2017) 3907–3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Hemando-Perez M, Lambert S, Nakatani-Webster E, Catalano CE, de Pablo PJ, Cementing proteins provide extra mechanical stabilization to viral cages, Nat. Commun 5 (2014) 4520. [DOI] [PubMed] [Google Scholar]
- [159].Sternberg N, Hoess RH, Display of peptides and proteins on the surface of bacteriophage lambda, Proc. Natl. Acad. Sci. U. S. A 92 (1995) 1609–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Catalano CE, Bacteriophage lambda: the path from biology to theranostic agent, Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol (2018), e1517.. [DOI] [PubMed] [Google Scholar]
- [161].Chang JR, Song EH, Nakatani-Webster E, Monkkonen L, Ratner DM, Catalano CE, Phage lambda capsids as tunable display nanoparticles, Biomacromolecules 15 (2014) 4410–4419. [DOI] [PubMed] [Google Scholar]
- [162].Lander GC, Evilevitch A, Jeembaeva M, Potter CS, Carragher B, Johnson JE, Bacteriophage lambda stabilization by auxiliary protein gpD: timing, location, and mechanism of attachment determined by cryo-EM, Structure 16 (2008) 1399–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Cano PG, Gamage LNA, Marciniuk K, Hayes C, Napper S, Hayes S, Griebel PJ, Lambda display phage as a mucosal vaccine delivery vehicle for peptide antigens, Vaccine 35 (2017) 7256–7263. [DOI] [PubMed] [Google Scholar]
- [164].Gamage LN,Ellis J, Hayes S, Immunogenicity of bacteriophage lambda particles displaying porcine circovirus 2 (PCV2) capsid protein epitopes, Vaccine 27 (2009) 6595–6604. [165] [DOI] [PubMed] [Google Scholar]
- [165].Zanghi CN, Lankes HA, Bradel-Tretheway B, Wegman J, Dewhurst S, A simple method for displaying recalcitrant proteins on the surface of bacteriophage lambda. Nucleic Acids Res. 33 (2005) e160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Condron BG, Atkins JF, Gesteland RF, Frameshifting in gene 10 of bacteriophage T7,J. Bacteriol 173 (1991) 6998–7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Novagen, T7Select® System Manual, Novagen.
- [168].Xu GJ, Kula T, Xu Q, Li MZ, Vernon SD, Ndung’u T, Ruxrungtham K, Sanchez J, Brander C, Chung RT, O’Connor KC, Walker B, Larman HB, Elledge SJ, Viral immunology. Comprehensive serological profiling of human populations using a synthetic human virome, Science (New York, N.Y.) 348 (2015) aaa0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Tan GH, Yusoff K, Seow HF, Tan WS, Antigenicity and immunogenicity of the immunodominant region of hepatitis B surface antigen displayed on bacteriophage T7, J. Med. Virol 77 (2005) 475–480. [DOI] [PubMed] [Google Scholar]
- [170].Xu H, Bao X, Lu Y, Liu Y, Deng B, Wang Y, Xu Y, Hou J, Immunogenicity of T7 bacteriophage nanoparticles displaying G-H loop of foot-and-mouth disease virus (FMDV), Vet Microbiol. 205 (2017) 46–52. [DOI] [PubMed] [Google Scholar]
- [171].Hashemi H, Pouyanfard S, Bandehpour M, Noroozbabaei Z, Kazemi B, Saelens X, Mokhtari-Azad T, Immunization with M2e-displaying T7 bacteriophage nanoparticles protects against influenza A virus challenge, PLoS One 7 (2012), e45765.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Peabody DS, Subunit fusion confers tolerance to peptide insertions in a virus coat protein, Arch. Biochem. Biophys 347 (1997) 85–92. [DOI] [PubMed] [Google Scholar]
- [173].Mastico RA, Talbot SJ Stockley PG, Multiple presentation of foreign peptides on the surface of an RNA-free spherical bacteriophage capsid, J. Gen. Virol 74 (Pt 4) (1993) 541–548. [DOI] [PubMed] [Google Scholar]
- [174].Zhai L, Peabody J, Pang YS, Schiller J, Chackerian B, Tumban E, A novel candidate HPV vaccine: MS2 phage VLP displaying a tandem HPV L2 peptide offers similar protection in mice to Gardasil-9, Antivir. Res 147 (2017) 116–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Peabody DS, Manifold-Wheeler B, Medford A, Jordan SK, do Carmo Caldeira J, Chackerian B, Immunogenic display of diverse peptides on virus-like particles of RNA phage MS2, J. Mol. Biol 380 (2008) 252–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Heal KG, Hill HR, Stockley PG, Hollingdale MR, Taylor-Robinson AW, Expression and immunogenicity of a liver stage malaria epitope presented as a foreign peptide on the surface of RNA-free MS2 bacteriophage capsids, Vaccine 18 (1999) 251–258. [DOI] [PubMed] [Google Scholar]
- [177].Tumban E, Peabody J, Tyler M, Peabody DS, Chackerian B, VLPs displaying a single L2 epitope induce broadly cross-neutralizing antibodies against human papillomavirus, PLoS One 7 (2012), e49751.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Basu R, Zhai L, Contreras A, Tumban E, Immunization with phage virus-like particles displaying Zika virus potential B-cell epitopes neutralizes Zika virus infection of monkey kidney cells, Vaccine 36 (2018) 1256–1264. [DOI] [PubMed] [Google Scholar]
- [179].Dong YM, Zhang GG, Huang XJ, Chen L, Chen HT, Promising MS2 mediated virus-like particle vaccine against foot-and-mouth disease, Antivir. Res 117 (2015) 39–43. [DOI] [PubMed] [Google Scholar]
- [180].Pumpens P, Renhofa R, Dishlers A, Kozlovska T, Ose V, Pushko P, Tars K, Grens E, Bachmann MF, The true story and advantages of RNA phage capsids as nanotools, Intervirology 59 (2016) 74–110. [DOI] [PubMed] [Google Scholar]
- [181].Brown SD, Fiedler JD, Finn MG, Assembly of hybrid bacteriophage Qbeta virus-like particles. Biochemistry 48 (2009) 11155–11157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Vasiljeva I, Kozlovska T, Cielens I, Strelnikova A, A Kazaks V Ose, P. Pumpens, Mosaic Qbeta coats as a new presentation model, FEBS Lett 431 (1998) 7–11. [DOI] [PubMed] [Google Scholar]
- [183].Kundig TM, Senti G, Schnetzler G, Wolf C, Prinz Vavricka BM, Fulurija A, Hennecke F, Sladko K, Jennings GT, Bachmann MF, Der p 1 peptide on virus-like particles is safe and highly immunogenic in healthy adults, J. Allergy Clin. Immunol 117 (2006) 1470–1476. [DOI] [PubMed] [Google Scholar]
- [184].Bessa J, Schmitz N, Hinton HJ, Schwarz K, Jegerlehner A, Bachmann MF, Efficient induction of mucosal and systemic immune responses by virus-like particles administered intranasally: implications for vaccine design, Eur. J. Immunol 38 (2008) 114–126. [DOI] [PubMed] [Google Scholar]
- [185].Huang X, Wang X, Zhang J, Xia N, Zhao Q, Escherichia coli-derived Virus-like Particles in Vaccine Development, 2, npj Vaccines, 2017. [DOI] [PMC free article] [PubMed]