Abstract
To enable the widespread application of genomic medicine, the extraction of genomic DNA from thin sections of archived formalin-fixed and paraffin-embedded (FFPE) tissue blocks for next-generation sequencing (NGS) is often necessary. However, there are currently no guidelines available on which specific regions of the microtome sections to use for macrodissection with respect to the histopathological factors observed under microscopic examination. The aim of this study was to clarify the relationship between histopathological factors and DNA quality, and to standardize the macrodissection method for more efficient implementation of NGS. FFPE tissue specimens of 218 patients from the Biomarker Research for Anti-EGFR Monoclonal Antibodies by Comprehensive Cancer Genomics study were used to investigate the relationship between 15 histopathological factors and the quantitative ratio of double-stranded DNA (dsDNA) to total nucleic acids, as well as the ∆ crossing point value of each tissue specimen. Multivariate logistic regression analysis revealed that specimen storage of ≥3 years was negatively associated with dsDNA quality (P=0.0007, OR: 4.30, 95% CI: 1.85-10.04). In contrast, the presence of a mucus pool was positively associated with dsDNA quality (P=0.0308, OR: 0.23, 95% CI: 0.06-0.87). Metastatic tumors and longer specimen storage periods were significantly associated with lower ∆Cp values (P=0.0007, OR: 4.43, 95% CI: 1.87-10.49; and P=0.0003, OR: 5.51, 95% CI: 2.18-13.95, respectively). Therefore, macrodissection should not be performed on specimens exhibiting histopathological factors associated with poor DNA quality. In particular, the use of tissue blocks with a storage period of <3 years allows the extraction of genomic DNA suitable for NGS.
Keywords: histopathological factors, formalin-fixed and paraffin-embedded tissue specimen, next-generation sequencing, dsDNA, ∆Cp
Introduction
Thin sections from formalin-fixed and paraffin-embedded (FFPE) human cancer tissues are obtained by surgery or biopsy and are routinely used, not only for pathological diagnosis but also for next-generation sequencing (NGS) analysis in clinical genetic laboratories. Thin-sliced sections are typically stained with hematoxylin and eosin (H&E) for observation by light microscopy, which enables pathological diagnosis based on World Health Organization (WHO) classification and TNM staging. Accordingly, FFPE tissue blocks are archived from all cancer patients. The present study hypothesized that certain histopathological characteristics of thin-sliced sections may be more or less suitable for downstream NGS, thereby affecting their application in precision medicine.
The methods for nucleic acid extraction from FFPE tissue specimens have dramatically improved. Consequently, genomic DNA is increasingly being used for NGS instead of Sanger sequencing to detect genetic variants, as well as for quantitative PCR (qPCR) to detect fusion genes. Specific qualitative factors such as the ∆ crossing point (Cp) value are reported to be indicative of DNA quality and various quantitative variables have been shown to affect the integrity of genomic DNA (1). ∆Cp is defined as the cycle number at detection threshold (crossing point). In brief, the measured Cp is the cycle at which PCR amplification begins its exponential phase and is considered, the point that is most reliably proportional to the initial concentration. Several studies have demonstrated that the quality of the genomic DNA extracted from FFPE tissue specimens is critical for performing optimal NGS in a clinical laboratory setting and these studies cite various important factors affecting the yield and quality of the DNA, including fixation conditions and specimen storage time; however, histopathological factors are not discussed (2-5). In addition, previous studies have shown that the low quality of genomic DNA extracted from FFPE tissue specimens poses the risk of introducing critical errors in downstream clinical analyses (6-9). However, it remains unclear whether histopathological factors observed under a light microscope using H&E-stained sections cut from FFPE tissue blocks have any impact on the success of NGS.
The authors previously conducted the collaborative Biomarker Research for Anti-EGFR Monoclonal Antibodies by Comprehensive Cancer Genomics (BREAC) study, which involved several institutes and used NGS to identify a predictive biomarker for the efficacy of cetuximab treatment (10). This study used FFPE tissue samples and genomic DNA was successfully extracted from all specimens and was suitable for NGS analysis. However, to the best of our knowledge there exists no published study that clarifies the relationship between the quality of DNA extracted from FFPE tissue blocks and the histopathological factors identified by microscopic observation using thin-sliced sections stained with H&E during routine pathological diagnosis.
The aim of the present study was to characterize the histopathological factors affecting the level of DNA quality required for NGS and then to standardize the macrodissection method of FFPE tissue blocks based on these histopathological factors to enable the selection of appropriate tissue blocks for NGS during routine pathological diagnosis.
Materials and methods
Patient characteristics
FFPE tissue specimens from 218 patients, including neoplastic tissues, were submitted for the BREAC study (10). A total of 298 patients (age range, 28-85 years) were recruited from seven institutions, including Cancer Institute Hospital of Japanese Foundation for Cancer Research (Tokyo, Japan), Aichi Cancer Center Hospital (Nagoya, Japan), National Cancer Center Hospital East (Kashiwa, Japan), Saitama Cancer Center (Saitama, Japan), Shikoku Cancer Center (Matsuyama, Japan), Shizuoka Cancer Center (Shizuoka, Japan) and Hokkaido University Hospital (Sapporo, Japan), between April 2012 and May 2013 and were initially registered in the present study. Patients with insufficient FFPE tissue specimens (11 patients), target re-sequencing failure (36 patients) and ineligible patients (9 patients) were excluded from the BREAC study according to the eligibility criteria (10). Biopsy specimens from 24 patients were also excluded from this study. Baseline characteristics of the patients were registered with the Office of Translational Research of the Exploratory Oncology Research and Clinical Trial Center (EPOC), National Cancer Center, (Chiba, Japan), by the investigators of each of the participating facilities. Genomic DNA from all specimens was successfully extracted and subjected to whole-genome sequencing. The study was approved by the Ethics Committee of each of the seven institutes [approval numbers: 2011-137 (National Cancer Center Hospital East), 13-009 (Hokkaido University Hospital), 24-15-24-1-5 (Shizuoka Cancer Center), H23-no2 (Shikoku Cancer Center), 2011-1084 (Cancer Institute Hospital of Japanese Foundation for Cancer Research), 20110903 (Saitama Cancer Center), 2016-1-122 (Aichi Cancer Center Hospital)] and the study conformed with the guidelines of the Declaration of Helsinki. Written informed consent was obtained from patients who were alive when initiating this study. For deceased patients and their relatives at that time, the study design on the website of each center was disclosed and the relatives were allowed to approve or deny inclusion in the study. This study was conducted in accordance with the Ethical Guidelines for the human genome and genetic analysis research of the Ministry of Education, Culture, Sports, Science and Technology, Ministry of Health, Labor and Welfare and Ministry of Economy, Trade and Industry.
DNA extraction from the FFPE tissue specimens
Archived FFPE tissue specimens collected during the BREAC study before anti-EGFR antibody treatment were used for DNA extraction. Specimens from either biopsied or surgically resected tissues were submitted to the Division of Translational Research, EPOC. Central pathological diagnosis of thin sections from all FFPE tissue specimens submitted from all seven institutions was conducted by the authorized pathologist (S.F.) under microscopic observation, using the slides stained with H&E (hematoxylin, 7 min and eosin, 2 min at both at room temperature).
As part of the BREAC study, the H&E stained sections were observed with a light microscope to determine whether areas of the tissue exhibiting unfavorable histopathology for high DNA quality could be selectively removed to some extent. These histopathological factors included necrosis, mucous pools, tumor budding, desmoplasia, excessive inflammatory cells, microabscess, poor fixation and burning effect. The region to be used for DNA extraction was determined and marked on the glass slide with a color pen, and then macrodissection of the microscopically specified regions was performed. Genomic DNA was then extracted from both the carcinoma cells and normal cells from the thin-sliced sections (10 µm) with the Absolutely RNA FFPE kit using a modified version of the manufacturer's protocol for DNA extraction (Agilent Technologies, Inc.). For the present study, DNA quality was then determined by estimating the dsDNA/total nucleic acid ratio and ∆Cp value. A NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Inc.) was used to quantify the total nucleic acids and the Quant-iT PicoGreen dsDNA Reagent and kit (Thermo Fisher Scientific, Inc.) was used to quantify the dsDNA. The Infinium HD FFPE QC kit (cat. no. WG-321-1001; Illumina, Inc.) was used for qPCR to determine the ∆Cp value [Cp(intact DNA) – Cp(FFPE tissue DNA)] between the test samples extracted from the FFPE tissue specimens and control intact genomic DNA so as to evaluate the quality of the extracted DNA according to the manufacturer's instructions. The relationships between the ∆Cp value and the dsDNA/total nucleic acid ratio and the histopathological factors of each cancer specimen were then investigated. The thermocycling conditions were as follows: 95˚C for 5 min, followed by 40 cycles of 95˚C for 30 sec, 57˚C for 30 sec and 72˚C for 30 sec.
Histopathological factors presumed to affect DNA quality
The present study focused on the histopathological factors suspected to affect the quality of DNA extracted from the cancer cells and, in turn, the quality of the downstream target sequencing data. The studied histopathological factors were as follows: Intra-tubular necrosis, extra-tubular necrosis, mucus pools, tumor budding, histological differentiation, histological grading according to the WHO classification, desmoplasia, excessive infiltration of inflammatory cells, microabscesses, burning effect, the ratio of tumor cells to the total number of cells, metastatic tumors, the area (mm2) of the tumor region taken from the FFPE section for DNA extraction, the use of laser microdissection and the length of storage of the FFPE tissue specimens. These exploratory variables were then observed under a light microscope by the same authorized pathologist (S.F.) who undertook the central pathological diagnosis in the BREAC study (10). Sections were then selected for high tumor cell ratio, a low number of denatured tumor cells and the exclusion of the histopathological factors that may have a detrimental effect on NGS.
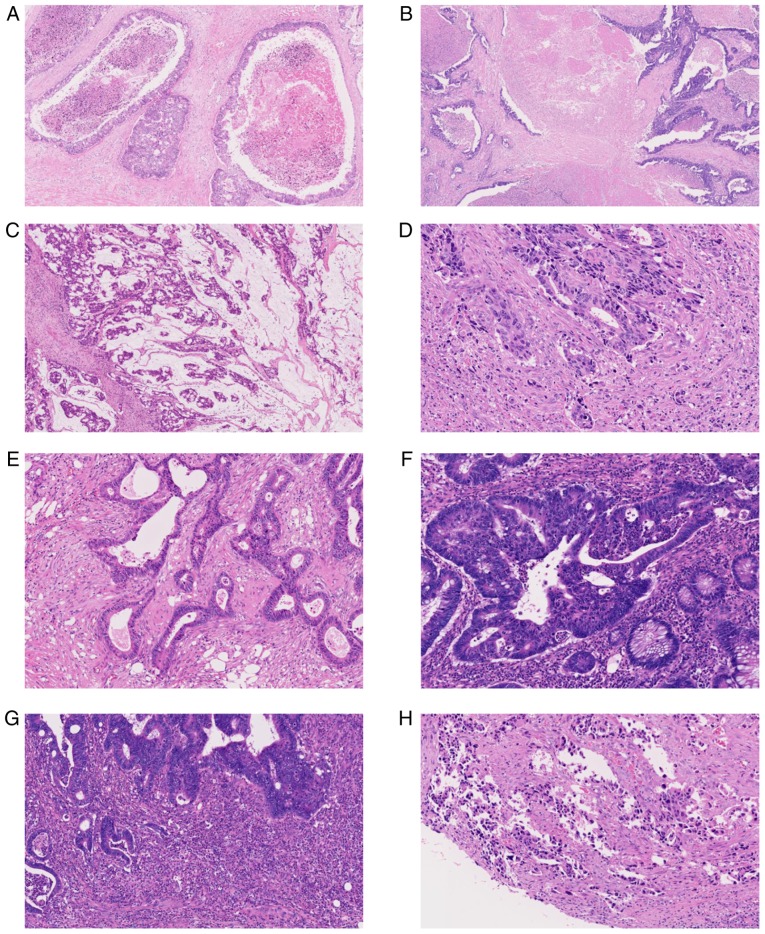
Micrographs of representative histopathological factors presumed to affect DNA quality are shown in Fig. 1. Intra-tubular necrosis refers to localized necrosis in the neoplastic duct formed by tumor cells (Fig. 1A). Extra-tubular necrosis occurs when necrosis does not remain only in the neoplastic ducts formed by the tumor cells and necrotic materials leak out of these ducts and are observed extensively in tumor tissue (Fig. 1B). Mucous pools refer to tumor cells floating within the neoplastic mucus (Fig. 1C). Tumor budding is a process during which individual or small clusters of up to five tumor cells detach from the main tumor mass and invade the surrounding stroma (11) (Fig. 1D). Desmoplasia refers to stromal myofibroblastic proliferation attributable to infiltrating tumor cells (Fig. 1E). Excessive infiltration of inflammatory cells refers to a high degree of inflammatory cell infiltration into the tumor cells; these inflammatory cells are difficult to separate out even by microdissection (Fig. 1F). Microabscesses are a collection of neutrophils (Fig. 1G), whereas the burning effect occurs when tumor cells are degenerated by artificial heat (Fig. 1H).
Figure 1.
Representative micrographs of histological factors possibly related to poor quality of extracted DNA. (A) Intra-tubular necrosis, (B) extra-tubular necrosis in metastatic tumors involving the liver, (C) a mucus pool, (D) tumor budding, (E) desmoplasia, (F) excessive infiltration of inflammatory cells, (G) a microabscess and (H) burning effect.
The region to be used for DNA extraction was demarcated by excluding areas containing these histopathological factors as far as possible and then macrodissection was performed.
Statistical analysis
The present study first investigated whether there was a difference between the seven participating institutes in terms of the quality of the DNA they extracted. Summary statistics and graphical methods (box plots and histograms) were used to compare the ∆Cp values and dsDNA quality.
Continuous variables, including ∆Cp value and dsDNA quality, were dichotomized according to their median since the aim of this study was to explore the histopathological factors related to poor DNA quality. The dichotomized outcome indicators were modeled using univariate and multivariate logistic regression analyses with forward stepwise selection (entry and removal criterion of P<0.05) for the ∆Cp value and dsDNA quality, respectively. Patients with missing outcome variables were excluded. To determine the favorable length of storage of FFPE specimens, the best cut-off value was investigated to discriminate the dichotomized ∆Cp values and dsDNA quality using Fisher's exact test. For the continuous collection area, logarithmic transformation was applied and modeled as itself because no clinically acceptable cut-off value was determined.
All statistical analyses including Fisher's exact test were performed with SAS 9.4 Software (SAS Institute, Inc.). All P-values were obtained from two-tailed statistical tests. P<0.05 was considered to indicate at statically significant difference.
Results
Patient characteristics
The patient characteristics are shown in Table I. All studied specimens were surgically resected from the 218 patients. Among these specimens, 180 (82.6%) were primary tumors and 38 (17.4%) were metastatic. Sites of metastasis were the liver, lungs or lymph nodes and the number of each of these metastatic sites was 32 (84.2%), 4 (10.5%), and 2 (5.3%), respectively.
Table I.
Patient characteristics.
| Characteristic | Patients (n) | Patients (%) |
|---|---|---|
| Total number | 218 | 100.0 |
| Median age (range), years | 61 (28-85) | 100.0 |
| Gender | ||
| Male | 129 | 59.2 |
| Female | 89 | 40.8 |
| Specimen | ||
| Surgical | 218 | 100.0 |
| Biopsy | 0 | 0.0 |
| Tumor | ||
| Primary | 180 | 82.6 |
| Metastatic | 38 | 17.4 |
| Primary tumor site (n=180) | ||
| Colon (n=133) | ||
| Ascending colon | 21 | 11.7 |
| Transverse colon | 12 | 6.7 |
| Descending colon | 7 | 3.9 |
| Sigmoid colon | 73 | 40.5 |
| Rectosigmoid colon | 20 | 11.1 |
| Rectum | 42 | 23.3 |
| Unknown | 5 | 2.8 |
| Metastatic tumor site (n=38) | ||
| Liver | 32 | 84.2 |
| Lung | 4 | 10.5 |
| Lymph nodes | 2 | 5.3 |
| Tumor differentiation | ||
| Good | 93 | 42.7 |
| Moderate | 110 | 50.4 |
| Poor | 15 | 6.9 |
Data are presented as n (%) or median (range).
No difference is observed in the mean dsDNA quality and ∆Cp value among the different institutes participating in the BREAC study
It was necessary to first verify whether there were any differences in the mean dsDNA/total nucleic acid ratio and ∆Cp value among the institutes participating in the BREAC study. Statistical analysis revealed that there was no significant difference in these values among the seven institutes, as shown in Table II, implying that the processes adopted for preparing FFPE tissue blocks at each facility did not affect dsDNA quality or the ∆Cp value and that none of the facilities used irregular fixation or storage methods. Therefore, it was concluded that a total of 218 FFPE tissue specimens were available for the analyses in this study.
Table II.
Facility-dependent dsDNA quantity, ∆Cp value and fixation method.
| dsDNAa | ∆Cpb | Formalin fixation | |||||
|---|---|---|---|---|---|---|---|
| Facility | Mean ± SD | Median (range) | Mean ± SD | Median (range) | Concentration (%) | pH | Fixation time (h) |
| 1 | 9.853±2.752 | 10.223 (3.916-19.006) | -1.641±1.390 | -1.726 (-4.605-1.135) | 20 | 7.4 | 48-72 |
| 2 | 13.083±3.710 | 12.266 (5.829-23.350) | -1.886±1.411 | -1.997 (-6.760-0.670) | 10c | 7.0 | 24-36 |
| 3 | 12.387±5.904 | 11.605 (3.592-28.807) | -1.832±1.221 | -1.430 [-5.570-(-0.077)] | 10 | 7.0 | 24-36 |
| 4 | 15.287±2.723 | 14.424 (11.237-21.515) | -1.389±0.950 | -1.048 [-4.160-(-0.013)] | 20c | 7.4 | 36-48 |
| 5 | 13.929±4.363 | 13.409 (4.288-23.984) | -1.880±0.934 | -1.653 [-4.630-(-0.673)] | 15 | 7.4 | 24 |
| 6 | 12.485±3.435 | 12.007 (7.141-22.570) | -1.150±0.867 | -1.108 (-3.203-0.733) | 10 | 7.0 | 24-102 |
| 7 | 13.074±3.187 | 13.487 (5.545-17.032) | -1.566±0.762 | -1.600 [-3.283-(-0.180)] | 10 | 7.4 | 24-48 |
Data are presented at the mean ± SD or median (range).aQuantitative ratio of dsDNA to total nucleic acids (%);
bratio of ∆Cp to control;
cneutral buffered formalin. Facility 1, Cancer Institute Hospital of Japanese Foundation for Cancer Research (Tokyo, Japan); 2, Aichi Cancer Center Hospital (Nagoya, Japan); 3, National Cancer Center Hospital East (Kashiwa, Japan); 4, Saitama Cancer Center (Saitama, Japan); 5, Shikoku Cancer Center (Matsuyama, Japan); 6, Shizuoka Cancer Center (Shizuoka, Japan); 7, Hokkaido University Hospital (Sapporo, Japan).
Determination of cut-off values for the dsDNA/total nucleic acid ratio and ∆Cp value
The median and average of the dsDNA/total nucleic acid ratio were 12.17 and 12.61, respectively. The samples with dsDNA quality less than the median value were defined as samples with a lower quality of genomic DNA. The median and average of the ∆Cp value were -1.48 and -1.63, respectively. Samples with a ∆Cp lower than the median were defined as samples with a lower quality of genomic DNA. The four patients with missing ∆Cp values were excluded. Therefore, in subsequent analyses, the present study analyzed the relationship between ∆Cp and the histopathological factors in specimens from the remaining 214 patients.
Relationships between the storage period of the FFPE tissue specimens and variables such as dsDNA quality and ∆Cp value
The relationships between the storage period of the FFPE tissue specimens and dsDNA quality and ∆Cp value were assessed using Fisher's exact test, as shown in Table III. The results showed that both dichotomized values became significantly smaller when the storage period was ≥3 years (P<0.001). The cut-off for the storage period was, therefore, set to 3 years for this study.
Table III.
Storage time cut-offs for FFPE tissue blocks based on dsDNA quantity (n=218) and ∆Cp value (n=214).
| A. Depending on dsDNA quantity (n=218). | |||||
|---|---|---|---|---|---|
| N (%) | dsDNA ≥ median (%) | ||||
| Cut-off (years) | <Cut-off | ≥Cut-off | <Cut-off | ≥Cut-off | P-value |
| 2 | 6 (2.75) | 212 (97.25) | 83.3 | 49.1 | 0.1061 |
| 3 | 35 (16.06) | 183 (83.94) | 77.1 | 44.8 | 0.0004 |
| 4 | 73 (33.49) | 145 (66.51) | 69.9 | 40.0 | <0.0001 |
| 5 | 123 (56.42) | 95 (43.58) | 59.4 | 40.7 | 0.0013 |
| 6 | 172 (78.90) | 46 (21.10) | 51.7 | 43.5 | 0.2034 |
| 7 | 192 (88.07) | 26 (11.93) | 50.5 | 46.2 | 0.4174 |
| 8 | 205 (94.00) | 13 (5.96) | 51.2 | 30.8 | 0.1260 |
| B. Depending on ∆Cp value (n=214). | |||||
| N (%) | ΔCp ≥ median (%) | ||||
| Cut-off (years) | <Cut-off | ≥Cut-off | <Cut-off | ≥Cut-off | P-value |
| 2 | 6 (2.80) | 208 (97.20) | 100.0 | 48.6 | 0.0145 |
| 3 | 34 (15.89) | 180 (84.11) | 79.4 | 44.4 | 0.0001 |
| 4 | 71 (33.18) | 143 (66.82) | 64.8 | 42.7 | 0.0018 |
| 5 | 120 (56.07) | 94 (43.93) | 60.0 | 37.2 | 0.0007 |
| 6 | 168 (78.50) | 46 (21.50) | 53.6 | 37.0 | 0.3322 |
| 7 | 188 (87.85) | 26 (12.15) | 53.2 | 26.9 | 0.0100 |
| 8 | 201 (93.93) | 13 (6.07) | 51.2 | 30.8 | 0.1259 |
Histopathological factors influencing dsDNA quality
The regions of the FFPE sections used for DNA extraction were selected based on the presence of as few negative histological factors as possible. Nevertheless, a certain amount of these factors were inevitably included in the regions used for macrodissection. The subsequent analysis was, therefore, still able to elucidate the relationship between the histological factors included in the DNA extraction region and DNA quality.
Univariate and multivariate logistic regression analyses were performed to identify variables influencing dsDNA quality (Table IV). Multivariate regression analysis revealed that the storage of FFPE specimens for ≥3 years was significantly associated with a lower dsDNA/total nucleic acid ratio (P=0.0007, OR: 4.30, 95% CI: 1.85-10.04). Contrary to the hypothesis that some of the histopathological factors analyzed in this study are responsible for poor DNA quality, the presence of mucus pools appeared to be a positive factor in a favorable dsDNA/total nucleic acid ratio (P=0.0308, OR: 0.23, 95% CI: 0.06-0.87). In addition, there was a non-negligible difference in the proportion of samples with a higher dsDNA/total nucleic acid ratio between samples with and without a mucus pool (Fisher's exact test, P=0.0496; Table V). However, there was no difference in the proportion of samples with a higher ∆Cp value between samples with and without a mucus pool (Table SI). These results confirmed that mucus pools are significantly correlated only with dsDNA quality.
Table IV.
Variables influencing dsDNA quality determined using univariate and multivariate analyses (Cox proportional hazard model).
| Univariate analysis | Multivariate analysis | ||||||
|---|---|---|---|---|---|---|---|
| Variable | Cases (n=218)a | OR | 95% CI | P-value | OR | 95% CI | P-value |
| Intra-tubular necrosis | 1.71 | 0.68-4.31 | 0.2553 | ||||
| Absence | 21 | ||||||
| Presence | 197 | ||||||
| Extra-tubular necrosis | 1.20 | 0.71-2.05 | 0.4975 | ||||
| Absence | 117 | ||||||
| Presence | 101 | ||||||
| Mucus pool | 0.25 | 0.07-0.93 | 0.0386 | 0.23 | 0.06-0.87 | 0.0308 | |
| Absence | 204 | ||||||
| Presence | 14 | ||||||
| Tumor budding | 0.57 | 0.23-1.38 | 0.2006 | ||||
| Grade 1 | 148 | ||||||
| Grade 2 | 46 | ||||||
| Grade 3 | 24 | ||||||
| Histological differentiation | 1.15 | 0.40-3.30 | 0.7895 | ||||
| Well/Moderate | 203 | ||||||
| Poor/Undifferentiated | 15 | ||||||
| Grading (WHO classification) | 1.15 | 0.40-3.30 | 0.7895 | ||||
| Grade 1/2 | 203 | ||||||
| Grade 3/4 | 15 | ||||||
| Desmoplasia | 0.89 | 0.51-1.55 | 0.6709 | ||||
| Absence | 141 | ||||||
| Presence | 77 | ||||||
| Excessive infiltration of inflammatory cells | 0.70 | 0.36-1.38 | 0.3042 | ||||
| Absence | 176 | ||||||
| Presence | 42 | ||||||
| Microabscess | 0.65 | 0.33-1.30 | 0.2269 | ||||
| Absence | 177 | ||||||
| Presence | 41 | ||||||
| Burning effect | 0.61 | 0.23-1.64 | 0.3288 | ||||
| Absence | 200 | ||||||
| Presence | 18 | ||||||
| RTCb | 2.15 | 0.83-5.55 | 0.1147 | ||||
| >50% | 21 | ||||||
| ≤50% | 197 | ||||||
| Metastatic tumor | 2.20 | 1.06-4.57 | 0.0349 | ||||
| No | 180 | ||||||
| Yes | 38 | ||||||
| Tumor area using log-scale (mm2) | 0.84 | 0.63-1.12 | 0.2429 | ||||
| Laser microdissection | 0.74 | 0.16-3.40 | 0.7018 | ||||
| No | 211 | ||||||
| Yes | 7 | ||||||
| FFPE storage time (years) | 4.19 | 1.79-9.64 | 0.0009 | 4.30 | 1.85-10.04 | 0.0007 | |
| >3 | 35 | ||||||
| ≤3 | 183 | ||||||
aPatients registered for BREAC trial;
bratio tumor cells to total number of cells.
Table V.
Association of the presence of mucus pools with the dsDNA quantity.
| Quantitative ratio of dsDNA to total nucleic acids (n) | ||||
|---|---|---|---|---|
| Mucus pool | ≤Median | ≥Median | Total (n) | P-value |
| Presence | 11 | 3 | 14 | 0.0496 |
| Absence | 98 | 106 | 204 | |
| Total (n) | 109 | 109 | 218 | |
Histopathological factors influencing the ∆Cp value
Univariate and multivariate logistic regression analyses were performed on samples from 214 patients to identify variables influencing DNA quality based on the ∆Cp value (Table VI). Multivariate analysis revealed that metastatic tumors and the storage of FFPE specimen for ≥3 years were significantly associated with a lower ∆Cp value (Metastatic tumor: P=0.0007, OR: 4.43, 95% CI: 1.87-10.49, storage period of FFPE specimen: P=0.0003, OR: 5.51, 95% CI: 2.18-13.95).
Table VI.
Variables influencing ∆Cp values determined using univariate and multivariate analyses (Cox proportional hazard model).
| Univariate analysis | Multivariate analysis | ||||||
|---|---|---|---|---|---|---|---|
| Variable | Cases (n=214)a | OR | 95% CI | P-value | OR | 95% CI | P-value |
| Intra-tubular necrosis | 2.15 | 0.83-5.56 | 0.1143 | ||||
| Absence | 21 | ||||||
| Presence | 193 | ||||||
| Extra-tubular necrosis | 1.51 | 0.88-2.60 | 0.1322 | ||||
| Absence | 115 | ||||||
| Presence | 99 | ||||||
| Mucus pool | 1.00 | 0.34-2.96 | 1.0000 | ||||
| Absence | 200 | ||||||
| Presence | 14 | ||||||
| Tumor Budding | 0.77 | 0.32-1.83 | 0.8152 | ||||
| Grade 1 | 145 | ||||||
| Grade 2 | 45 | ||||||
| Grade 3 | 24 | ||||||
| Histological differentiation | 1.55 | 0.53-4.51 | 0.4248 | ||||
| Well/Moderate | 199 | ||||||
| Poor/Undifferentiated | 15 | ||||||
| Grading (WHO classification) | 1.55 | 0.53-4.51 | 0.4248 | ||||
| Grade 1/2 | 199 | ||||||
| Grade 3/4 | 15 | ||||||
| Desmoplasia | 0.75 | 0.43-1.32 | 0.3164 | ||||
| Absence | 139 | ||||||
| Presence | 75 | ||||||
| Excessive infiltration of inflammatory cells | 0.83 | 0.42-1.65 | 0.6031 | ||||
| Absence | 173 | ||||||
| Presence | 41 | ||||||
| Microabscess | 0.83 | 0.41-1.66 | 0.5962 | ||||
| Absence | 175 | ||||||
| Presence | 39 | ||||||
| Burning effect | 1.28 | 0.48-3.37 | 0.6230 | ||||
| Absence | 196 | ||||||
| Presence | 18 | ||||||
| RTCb | 0.73 | 0.29-1.80 | 0.4920 | ||||
| >50% | 21 | ||||||
| ≤50% | 193 | ||||||
| Metastatic tumor | 3.86 | 1.72-8.65 | 0.0010 | 4.43 | 1.87-10.49 | 0.0007 | |
| No | 177 | ||||||
| Yes | 37 | ||||||
| Tumor area using log-scale (mm2) | 0.87 | 0.65-1.16 | 0.3307 | ||||
| >58.1 | |||||||
| ≤58.1 | |||||||
| Laser microdissection | 0.49 | 0.09-2.74 | 0.4167 | ||||
| No | 208 | ||||||
| Yes | 6 | ||||||
| Storage time (years) | 4.82 | 2.00-11.64 | 0.0005 | 5.51 | 2.18-13.95 | 0.0003 | |
| >3 | 34 | ||||||
| ≤3 | 180 | ||||||
aFour patients with missing ∆Cp values were excluded;
bratio of tumor cells to the total number of cells.
Discussion
In the BREAC study, DNA extraction from each FFPE tissue specimen was carried out such that the histopathological factors determined through microscopic observation, including necrosis, mucus pools, tumor budding, desmoplasia, excessive inflammatory cells, microabscesses, poor fixation, and burning effect, were excluded as much as possible from the tumor area from which the DNA was extracted (10). However, it was difficult to eliminate these histopathological factors from the tumor area for DNA extraction, which allowed the evaluation of their effect in the present study. The results of the present study on these archived specimens showed that metastatic tumors and FFPE specimen storage of ≥3 years were strongly associated with poor DNA quality. Thus, it would be preferable to extract DNA for NGS purposes from tumor regions where these histopathological factors have been excluded as far as possible by macrodissection. These findings should be incorporated into future standard operating procedures; current guidelines published in several countries, including the USA (http://jmd.amjpathol.org/article/S1525-1578(17)30025-9/fulltext#sec1.2.1), Australia (http://jmd.amjpathol.org/article/S1525-1578(17)30025-9/fulltext#sec1.2.1) and Japan (http://jccls.org/active/MM6-A1_agenda.pdf and http://pathology.or.jp/genome_med/elearning.html) do not include standards for the proper removal of histopathological factors affecting the quality of DNA for NGS from FFPE tissue specimens.
A total of three factors significantly affecting the quality of DNA were identified in this study. Interestingly, mucus pools comprised a positive factor correlated with the extraction of better-quality DNA for NGS. However, it is not yet known how mucus pools exert this effect. To the best of our knowledge, there are no studies on the relationship between mucus pools and DNA quality. Further investigation is, therefore, needed.
In contrast, metastatic tumors comprised an independent negative factor affecting the ∆Cp value. Although the presence of metastatic tumors was not an independent negative factor affecting the dsDNA/total nucleic acid ratio based on multivariate analysis, it was a negative factor for this ratio based on univariate analysis. Advanced colorectal cancer generally shows metastasis to the liver and lung. It is also well known that extensive necrosis is generally observed in metastatic lesions of colorectal cancer. It was difficult to determine the area for DNA extraction in metastatic tumor specimens from which necrosis had been completely excluded, which explains one of the reasons for this contradictory outcome in the current study. Based on this result, it is recommended to select a region with as little necrosis as possible when FFPE specimens of metastatic tumors are processed for DNA extraction.
Storage of the FFPE tissue block for ≥3 years was a negative factor related to DNA quality, as indicated by both the dsDNA/total nucleic acid ratio and the ∆Cp value. A previous study suggested that FFPE specimens stored for several years may have a poorer quality of DNA (5). FFPE tissues are one of the most widely available clinical specimens and have potential utility as a source of DNA for NGS. Archived FFPE tissue specimens are required not only for clinical trials but also for population-based studies. Since older FFPE tissue specimens may be required, especially from patients suffering from cancer for a long time, it is necessary to establish an improved method for storing these specimens that can be incorporated into routine pathological practice. However, methods of targeted NGS using low-quality DNA are constantly improving, which will likely lead to further improvement in the application of NGS to damaged DNA extracted from old FFPE tissue specimens in the future.
The reasons for dividing the samples into two groups based on the median were the following; i) There is no definitive, published cutoff value for dsDNA or ΔCp and the significance associated with fluctuations in these values is unknown. ii) Relatively high-quality specimens were collected in the BREAC trial. Rather than subgrouping a heterogeneous population with the median, in the present study a homogeneous population has been subgrouped with the median. This method was considered to be sensitive enough to search for factors affecting the dsDNA and ΔCp values. iii) The ability to extract factors with statistically higher power when the outcome variables are segregated based on the median and the sample sizes are equal. From the results of this study, it cannot be concluded that the samples judged to be of low quality in this study are completely unsuitable for NGS. However, this study indicated useful guidelines for the optimal extraction of DNA for NGS.
There are several limitations to this study. First, this was a retrospective study. Detailed information of all the specimens from the time the specimens were submitted until they became FFPE tissue specimens used in the present study could not be obtained at each institute, although there was no disparity among these facilities in terms of the quality of extracted DNA. DNA extraction was carried out by first removing the studied histological factors as much as possible. If any of the histopathological factors were quantitatively included, there is a possibility that the quality of DNA was influenced by these factors. In addition, the sequencing parameters were optimized for individual DNA samples, such as the pre- and post-capture PCR cycles, to maximize the gathered genomic data. Therefore, it is impossible to compare the parameters related to the success of DNA sequencing in the BREAC study and the histopathological factors investigated in this study. Finally, only one central pathologist investigated all the tissue specimens in this study. In the near future, the findings of this study should be validated by other pathologists.
The proposal of the present study is that macrodissection of microtomed sections should be performed after the removal of as many negative histological factors as possible and that the use of old FFPE tissue blocks stored for ≥3 years should be avoided. This is all possible during routine observation of H&E-stained sections under a light microscope and it would enable the extraction of high-quality DNA suitable for downstream NGS.
Supplementary Material
Acknowledgements
The authors would like to thank Dr. Wataru Okamoto and Ms. Izumi Miki (Office of Translational Research of the Exploratory Oncology Research and Clinical Trial Center, National Cancer Center, Kashiwa, Chiba, Japan) for organizing the registered patients' information in the BREAC trial.
Funding
The BREAC study was supported by the Adaptable and Seamless Technology Transfer Program through target-driven R&D, Practical Application Development by SME Start-up, the Japan Science and Technology Agency, the National Cancer Center Research and Development Fund (23-A-2), G & G Science Co., Ltd., and Medical & Biological Laboratories Co., Ltd.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
SF, TYo, KT, KMa, YS, KA, HE, MS, NN, MM, YK, YA and AO conceived and designed the study. ES, KYamaz, TYa, KMu, TN, KYamag, SY, KS and HB provided study materials or patients. SN and TYo collected and analyzed the data. SM, CN and KT performed genome sequencing. SF, TYa, KT and SN undertook data analysis and interpretation. SF, KT, SN and TYo wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of each of the seven institutes [approval numbers: 2011-137 (National Cancer Center Hospital East), 13-009 (Hokkaido University Hospital), 24-15-24-1-5 (Shizuoka Cancer Center), H23-no2 (Shikoku Cancer Center), 2011-1084 (Cancer Institute Hospital of Japanese Foundation for Cancer Research), 20110903 (Saitama Cancer Center), 2016-1-122 (Aichi Cancer Center Hospital)] and the study conformed with the guidelines of the Declaration of Helsinki. Written informed consent was obtained from patients who were alive when initiating this study. For deceased patients and their relatives at that time, the study design on the website of each center was disclosed and the relatives were allowed to approve or deny inclusion in the study. This study was conducted in accordance with the Ethical Guidelines for the human genome and genetic analysis research of the Ministry of Education, Culture, Sports, Science and Technology, Ministry of Health, Labour and Welfare and Ministry of Economy, Trade and Industry.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Treece AL, Montgomery ND, Patel NM, Civalier CJ, Dodd LG, Gulley ML, Booker JK, Weck KE. FNA smears as a potential source of DNA for targeted next-generation sequencing of lung adenocarcinomas. Cancer Cytopathol. 2016;124:406–414. doi: 10.1002/cncy.21699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nagahashi M, Shimada Y, Ichikawa H, Nakagawa S, Sato N, Kaneko K, Homma K, Kawasaki T, Kodama K, Lyle S, et al. Formalin-fixed paraffin-embedded sample conditions for deep next generation sequencing. J Surg Res. 2017;220:125–132. doi: 10.1016/j.jss.2017.06.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Einaga N, Yoshida A, Noda H, Suemitsu M, Nakayama Y, Sakurada A, Kawaji Y, Yamaguchi H, Sasaki Y, Tokino T, Esumi M. Assessment of the quality of DNA from various formalin-fixed paraffin-embedded (FFPE) tissues and the use of this DNA for next-generation sequencing (NGS) with no artifactual mutation. PLoS One. 2017;12(e0176280) doi: 10.1371/journal.pone.0176280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho M, Ahn S, Hong M, Bang H, Van Vrancken M, Kim S, Lee J, Park SH, Park JO, Park YS, et al. Tissue recommendations for precision cancer therapy using next generation sequencing: A comprehensive single cancer center's experiences. Oncotarget. 2017;8:42478–42486. doi: 10.18632/oncotarget.17199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carrick DM, Mehaffey MG, Sachs MC, Altekruse S, Camalier C, Chuaqui R, Cozen W, Das B, Hernandez BY, Lih CJ, et al. Robustness of next generation sequencing on older formalin-fixed paraffin-embedded tissue. PLoS One. 2015;10(e0127353) doi: 10.1371/journal.pone.0127353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bettoni F, Koyama FC, de Avelar Carpinetti P, Galante PAF, Camargo AA, Asprino PF. A straightforward assay to evaluate DNA integrity and optimize next-generation sequencing for clinical diagnosis in oncology. Exp Mol Pathol. 2017;103:294–299. doi: 10.1016/j.yexmp.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 7.Bonfiglio S, Vanni I, Rossella V, Truini A, Lazarevic D, Dal Bello MG, Alama A, Mora M, Rijavec E, Genova C, et al. Performance comparison of two commercial human whole-exome capture systems on formalin-fixed paraffin-embedded lung adenocarcinoma samples. BMC Cancer. 2016;16(692) doi: 10.1186/s12885-016-2720-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dumur CI, Almenara JA, Powers CN, Ferreira-Gonzalez A. Quality control material for the detection of somatic mutations in fixed clinical specimens by next-generation sequencing. Diagn Pathol. 2015;10(169) doi: 10.1186/s13000-015-0403-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim S, Park C, Ji Y, Kim DG, Bae H, van Vrancken M, Kim DH, Kim KM. Deamination effects in formalin-fixed, paraffin-embedded tissue samples in the era of precision medicine. J Mol Diagn. 2017;19:137–146. doi: 10.1016/j.jmoldx.2016.09.006. [DOI] [PubMed] [Google Scholar]
- 10.Shinozaki E, Yoshino T, Yamazaki K, Muro K, Yamaguchi K, Nishina T, Yuki S, Shitara K, Bando H, Mimaki S, et al. Clinical significance of BRAF non-V600E mutations on the therapeutic effects of anti-EGFR monoclonal antibody treatment in patients with pretreated metastatic colorectal cancer: The biomarker research for anti-EGFR monoclonal antibodies by comprehensive cancer genomics (BREAC) study. Br J Cancer. 2017;117:1450–1458. doi: 10.1038/bjc.2017.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ueno H, Murphy J, Jass JR, Mochizuki H, Talbot IC. Tumour ‘budding’ as an index to estimate the potential of aggressiveness in rectal cancer. Histopathology. 2002;40:127–132. doi: 10.1046/j.1365-2559.2002.01324.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.