Nipah virus is a zoonotic biosafety level 4 agent with high mortality rates in humans. The genus to which Nipah virus belongs, Henipavirus, includes five officially recognized pathogens; however, over 20 species have been identified in multiple continents within the last several years. As there are still no vaccines or treatments for NiV infection, elucidating its process of viral particle production is imperative both for targeted drug design as well as for particle-based vaccine development. Developments in high-throughput technologies make proteomic analysis of isolated viral particles a highly insightful approach to understanding the life cycle of pathogens such as Nipah virus.
Keywords: Nipah virus, paramyxovirus, proteomics, vesicular trafficking, endocytosis, cytoskeleton, host-pathogen interaction
ABSTRACT
Classified as a biosafety level 4 (BSL4) select agent, Nipah virus (NiV) is a deadly henipavirus in the Paramyxoviridae family, with a nearly 75% mortality rate in humans, underscoring its global and animal health importance. Elucidating the process of viral particle production in host cells is imperative both for targeted drug design and viral particle-based vaccine development. However, little is understood concerning the functions of cellular machinery in paramyxoviral and henipaviral assembly and budding. Recent studies showed evidence for the involvement of multiple NiV proteins in viral particle formation, in contrast to the mechanisms understood for several paramyxoviruses as being reliant on the matrix (M) protein alone. Further, the levels and purposes of cellular factor incorporation into viral particles are largely unexplored for the paramyxoviruses. To better understand the involvement of cellular machinery and the major structural viral fusion (F), attachment (G), and matrix (M) proteins, we performed proteomics analyses on virus-like particles (VLPs) produced from several combinations of these NiV proteins. Our findings indicate that NiV VLPs incorporate vesicular trafficking and actin cytoskeletal factors. The involvement of these biological processes was validated by experiments indicating that the perturbation of key factors in these cellular processes substantially modulated viral particle formation. These effects were most impacted for NiV-F-modulated viral particle formation either autonomously or in combination with other NiV proteins, indicating that NiV-F budding relies heavily on these cellular processes. These findings indicate a significant involvement of the NiV fusion protein, vesicular trafficking, and actin cytoskeletal processes in efficient viral particle formation.
IMPORTANCE Nipah virus is a zoonotic biosafety level 4 agent with high mortality rates in humans. The genus to which Nipah virus belongs, Henipavirus, includes five officially recognized pathogens; however, over 20 species have been identified in multiple continents within the last several years. As there are still no vaccines or treatments for NiV infection, elucidating its process of viral particle production is imperative both for targeted drug design as well as for particle-based vaccine development. Developments in high-throughput technologies make proteomic analysis of isolated viral particles a highly insightful approach to understanding the life cycle of pathogens such as Nipah virus.
INTRODUCTION
Nipah virus (NiV) is a pathogen in the family Paramyxoviridae, which includes measles and mumps viruses. It is highly virulent and capable of infecting numerous species of mammals. NiV particles are enveloped and transmissible to humans from fruit bat reservoirs as well as from livestock and other humans. Infection in humans leads to respiratory disease, severe encephalitis, and a case mortality rate between 40% and 100% (http://www.searo.who.int/entity/emerging_diseases/links/nipah_virus_outbreaks_sear/en/). Based on the absence of approved vaccines or treatments with the emerging threat that these viruses pose to human health, NiV and the closely related Hendra virus (HeV) are classified as biosafety level 4 (BSL4) agents and are included within the genus Henipavirus. Moreover, the World Health Organization recently listed NiV among the pathogens most likely to cause a major pandemic (1). Importantly, at least 20 new henipavirus-like viruses have been discovered in the last decade, underscoring the potential threat of NiV and related pathogens (2–4). The identification of pathogen-host interactions and elucidation of how these pathogens produce viral particles may yield novel antiviral therapeutics or support the development of particle-based vaccines such as those approved by the FDA for hepatitis B virus and human papillomavirus (5–7).
The formation of infectious paramyxovirus particles must include the incorporation of two kinds of transmembrane proteins, the fusion (F) and attachment (termed G for NiV and other henipaviruses) glycoproteins, which cooperate for viral entry in addition to the viral ribonucleoprotein complex, which includes the single-stranded, negative-sense viral genome and several viral proteins needed for transcription and replication (8). All paramyxoviruses additionally produce a matrix (M) protein which is generally thought to be central to the assembly and budding of viral components into new particles (9). Specifically, several paramyxovirus M proteins drive budding largely through oligomerization into a scaffolding array capable of inducing membrane curvature (10). While for some paramyxoviruses, the matrix protein is solely required to produce particles (11, 12), for Sendai virus, mumps virus, and simian virus 5, one or both glycoproteins are supportive of or essential for particle formation (13–16). Interestingly, live NiV containing a deletion of the M gene yielded a dramatic reduction in, but not complete loss of, infectivity (17). Further, potential roles for NiV proteins other than M in viral production have begun to be identified (18–21).
Unlike paramyxovirus matrix proteins, which appear to primarily use electrostatic interactions between M monomers to drive budding (10, 22, 23), other paramyxovirus proteins appear to instead reappropriate cellular machinery to accomplish budding (15, 19–21, 24). For example, proteins of the endosomal sorting complexes required for transport (ESCRTs) and associated factors are specifically targeted for recruitment to budding sites by numerous enveloped viruses which encode conserved motifs, termed late domains, within their viral proteins (19, 25–32). Generally, the removal or mutation of these late domains drastically reduces particle formation efficiency. Viral utilization of cellular factors other than ESCRTs for viral budding have also been reported, including Rab11 GTPase, involved in recycling endosome function, and known Rab11-interacting factors (33–35). Another cellular machinery known to be involved in several mechanisms of assembly and/or egress is the actin cytoskeleton; however, its roles vary greatly and, for many viruses, are poorly understood (24, 36–39), highlighting the many strategies viruses have adopted to ensure their replication and spread.
An approach to the identification and elucidation of roles for cellular machinery in enveloped particle formation is the use of proteomics to identify the host proteins enriched in viral particles and the cellular processes involved (40–42). A recent study by Vera-Velasco et al. focused on the identification of host cellular proteins in NiV virus-like particles (VLPs) produced from cotransfection of the NiV F, G, and M proteins (43). Among the insights gained in this study, 67 human proteins were identified that were primarily associated with vesicular transport and sorting. However, the importance of these cellular factors during viral particle formation and the involvement of each F, G, and M viral protein in the recruitment or incorporation of such cellular factors into VLPs remain elusive.
Based on two studies indicating roles of the NiV and HeV F proteins in particle formation (18, 44), we expanded this VLP proteomics approach to include several combinations of particles produced from the coexpression of F, G, and M. Our subsequent finding that F- but not M-derived VLPs incorporate numerous cellular factors adds support to the notion that henipaviral F proteins are involved in viral assembly and budding with a heavy reliance on cellular factors. Our results validate the importance of the most enriched processes, vesicular trafficking and the cytoskeleton, as modulatory of NiV particle release primarily through F-driven budding.
RESULTS
Overview of workflow and results from VLP and cellular proteomics.
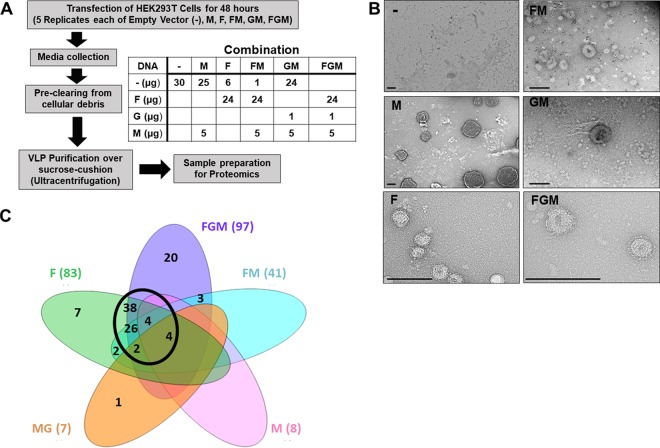
To help identify specific cellular factors and machinery incorporated during NiV budding, we transfected human embryonic kidney (HEK293T) cells with plasmid constructs coding for individual structural NiV proteins, F, G, and M. To best elucidate the relative importance of the F and M proteins for incorporation of cellular factors, five combinations of transfections (F alone, M alone, F and M [FM], G and M [GM], and F, G, and M [FGM]) were completed in addition to an empty vector control, a background for naturally expressed extracellular vesicles (e.g., exosomes and microvesicles) (Fig. 1A). Forty-eight hours after transfection, VLPs and their corresponding transfected cells were separately isolated and prepared for proteomic analyses (Fig. 1A). As described in Materials and Methods, the observed peptide counts were used to remove replicates indicating contamination from overflow, resulting in 3 to 5 biological replicates per transfection type being used for further analyses. Average peptide counts were used to identify which cellular factors were incorporated into VLPs using two thresholds, a minimum average peptide count of 4.33 for the replicates of a given transfection type (e.g., F only), based on the average observed peptide counts of the viral proteins themselves, as well as a maximum average count of 2 in empty vector controls to remove proteins highly expressed in background extracellular vesicles. To verify that this approach produces vesicles and to visualize vesicles for each combination, negative staining and transmission electron microscopy were used (Fig. 1B). Importantly, spikes were most apparent in combinations containing F and/or G, supporting the incorporation of these proteins, whereas M-only VLPs lacked spikes and tended to include high-contrast structures internally (Fig. 1B). From the VLP proteomics, the following numbers of cellular proteins were identified for each VLP sample type: 8 (M), 83 (F), 41 (FM), 7 (MG), and 97 (FGM) (Fig. 1C). A Venn diagram was constructed to help identify cellular proteins that were common to all, some, or only one VLP combination.
FIG 1.
Overview of workflow and results from VLP and cellular omics. (A) Human embryonic kidney (HEK293T) cells were transfected with combinations of constructs coding for the Nipah virus fusion (F), attachment (G), and matrix (M) proteins. Media and cells were collected after 48 h. The medium supernatant was used to isolate virus-like particles (VLPs), and then both VLPs and cells were prepared for proteomics analysis. (B) VLPs produced from the listed combinations of viral protein transfection were isolated and negatively stained with 1.5% uranyl acetate. Scale bars = 250 nm. (C) Venn diagram summarizing the number of proteins identified as enriched in each VLP combination and their distribution of overlap. The black circle highlights the significant overlap between combinations including F.
VLP proteomics highlighted by enrichment of cytoskeletal and endosomal trafficking machinery.
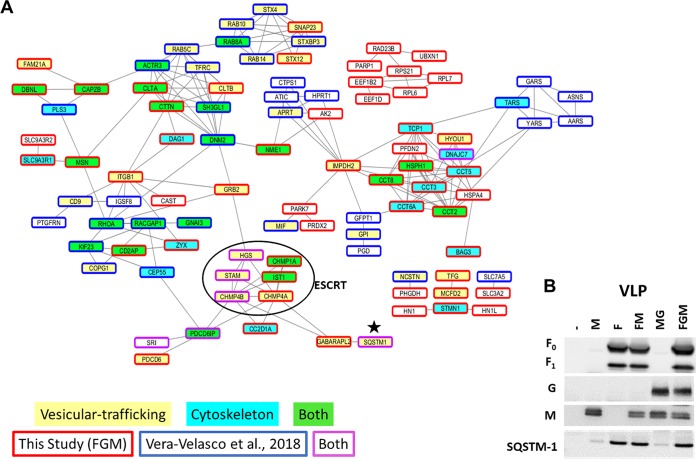
We next created a protein interaction map by combining our FGM VLP proteomics data with the data from a recent proteomics composition study on NiV FGM VLPs (43) to help us identify protein clusters consistently incorporated into VLPs. Overall, the enriched processes and protein-protein interaction sets have a high degree of overlap; however, specific proteins identified in common are relatively few. Small differences in specific protein compositions for VLPs are likely influenced by differences in methodologies; however, the consistent enrichment of several processes suggests their underlying involvement. Namely, two of the most enriched functional groups of proteins were vesicle-mediated transport and the actin cytoskeleton (Fig. 2A). Sequestosome-1 (SQSTM1), an adapter protein involved in vesicle-mediated transport, namely, macroautophagy (45), was found to be among the most enriched proteins, particularly in VLPs produced from cells expressing F. To support the validity of our VLP proteomics platform, the VLP samples used in proteomics were also analyzed by Western blotting, and the pattern of SQSTM1 incorporation primarily into VLPs containing NiV-F was confirmed (Fig. 2B). A full list of VLP-incorporated proteins is shown in Table 1, comparing the proteins identified in different types of VLPs (F, M, etc.) with those in FGM VLPs in a study by Vera-Velasco et al. (43) and further identifying whether each protein is associated with vesicle-mediated transport or the cytoskeleton. Comparisons between the lists of cellular proteins incorporated into each of the VLP types (Table 1) was primarily done to elucidate how much the budding mechanisms of F and M rely on cellular machinery.
FIG 2.
VLP proteomics highlighted by enrichment of cytoskeletal and endosomal trafficking machinery. (A) STRING protein interaction map comparing proteins identified in the FGM VLPs described by Vera-Velasco et al. (43) (blue box) and this study (red boxes). This map identifies which protein clusters have associated factors included in one or both (purple boxes) studies. Among the most enriched groups based on gene ontological analyses of the combined data set are vesicular trafficking (yellow) and the cytoskeleton (light blue), with proteins that belong to both also designated (green). ESCRTs which somewhat overlap both processes were observed in both studies and are in a black circle. Note that the clusters associated with these processes tend to have included proteins identified in both studies analyzed. Importantly, only proteins with at least one interaction were shown on this map; a full list can be found in Table 1. One of the most clearly enriched proteins in VLP proteomics for this study and that by Vera-Velasco et al. (43) was the autophagy adapter protein, sequestosome-1 (SQSTM1, designated by small black star). (B) Using Western blot analysis of VLPs for each combination, sequestosome-1 was used to validate the accuracy of our VLP proteomics, since this protein exhibited an expression pattern similar to the mass spectrometry results. The Western blot is representative of at least three replicates.
TABLE 1.
Genes and number of VLP incorporated proteins
| Gene | Full name | Accession no. | Avg. no. of observed peptides fora: |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| VMT (65) | Cytoskeleton (54) | Negative control | M (8) | F (83) | FM (41) | MG (7) | FGM (97) | Vera-Velasco et al. (67) | |||
| AARS | Alanyl-tRNA synthetase | P49588 | |||||||||
| ABCE1 | ATP binding cassette subfamily E member 1 | P61221 | |||||||||
| ACTR3 | ARP3 actin-related protein 3 homolog | F8WEW2 | |||||||||
| AK2 | Adenylate kinase 2 | F8VZG5 | 0.8 | 0.7 | 6.0 | 4.3 | 1.4 | 5.5 | |||
| ALCAM | Activated leukocyte cell adhesion molecule | F5GXJ9 | 0.3 | 1.0 | 7.3 | 3.0 | 1.8 | 4.3 | |||
| AMOT | Angiomotin | A2BDD9 | 0.3 | 0.0 | 6.0 | 1.3 | 0.4 | 9.0 | |||
| ANXA6 | Annexin A6 | E5RFF0 | |||||||||
| APRT | Adenine phosphoribosyltransferase | H3BQB1 | |||||||||
| ASNS | Asparagine synthetase | C9J605 | |||||||||
| ATIC | 5-Aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase | V9HWH7 | |||||||||
| BAG3 | BCL2-associated athanogene 3 | O95817 | 0.0 | 0.0 | 19.3 | 9.7 | 0.2 | 23.3 | |||
| CA2 | Carbonic anhydrase 2 | V9HW21 | 0.3 | 2.7 | 2.5 | 3.3 | 3.8 | 5.5 | |||
| CALCOCO2 | Calcium binding and coiled-coil domain 2 | H0YBP4 | 0.0 | 0.0 | 10.0 | 4.7 | 0.8 | 10.3 | |||
| CALM1 | Calmodulin 1 | Q96HY3 | 2.0 | 4.0 | 5.0 | 4.3 | 4.0 | 4.3 | |||
| CALU | Calumenin | A0A024R755 | 0.0 | 0.0 | 8.0 | 3.3 | 2.0 | 8.3 | |||
| CAPZA1 | Capping actin protein of muscle Z-line alpha subunit 1 | P52907 | 0.5 | 0.7 | 4.5 | 2.3 | 1.2 | 4.3 | |||
| CAPZB | Capping actin protein of muscle Z-line beta subunit | B1AK87 | 2.0 | 1.7 | 6.3 | 3.3 | 2.4 | 5.0 | |||
| CAST | Calpastatin | E7ES10 | 0.0 | 0.0 | 3.5 | 1.0 | 0.4 | 5.0 | |||
| CBR1 | Carbonyl reductase 1 | E9PQ63 | |||||||||
| cc2d1a | Coiled-coil and C2 domain containing 1A | K7EJY5 | 0.0 | 0.0 | 5.5 | 3.3 | 0.2 | 7.5 | |||
| CCT2 | Chaperonin containing TCP1 subunit 2 | F8VQ14 | 0.5 | 1.3 | 4.8 | 4.3 | 2.2 | 6.3 | |||
| CCT3 | Chaperonin containing TCP1 subunit 3 | P49368 | 1.3 | 2.0 | 5.8 | 4.3 | 2.6 | 5.5 | |||
| CCT5 | Chaperonin containing TCP1 subunit 5 | E7ENZ3 | 1.5 | 3.3 | 6.3 | 5.7 | 3.2 | 7.3 | |||
| CCT6A | Chaperonin containing TCP1 subunit 6A | P40227 | 0.8 | 0.3 | 4.3 | 4.7 | 1.8 | 5.5 | |||
| CCT8 | Chaperonin containing TCP1 subunit 8 | H7C2U0 | 1.5 | 0.7 | 5.0 | 4.3 | 1.8 | 7.5 | |||
| CD276 | CD276 molecule | H0YN85 | |||||||||
| CD2AP | CD2-associated protein | Q9Y5K6 | 0.0 | 1.3 | 8.0 | 3.3 | 1.0 | 10.5 | |||
| CD9 | CD9 molecule | B4DK09 | |||||||||
| CDC37 | Cell division cycle 37 | Q16543 | 0.0 | 1.0 | 2.3 | 2.7 | 0.2 | 5.0 | |||
| CDV3 | CDV3 homolog | D6R973 | 0.0 | 1.0 | 6.3 | 4.3 | 1.8 | 7.5 | |||
| CEP55 | Centrosomal protein 55 | Q53EZ4 | |||||||||
| CHMP1A | Charged multivesicular body protein 1A | F8VUA2 | 0.0 | 0.0 | 4.0 | 2.7 | 0.6 | 4.5 | |||
| CHMP4A | Charged multivesicular body protein 4A | Q9BY43 | 0.3 | 1.0 | 5.3 | 3.0 | 1.2 | 6.3 | |||
| CHMP4B | Charged multivesicular body protein 4B | Q9H444 | 1.3 | 2.3 | 10.0 | 7.3 | 3.2 | 10.5 | |||
| CLIC4 | Chloride intracellular channel 4 | Q9Y696 | |||||||||
| CLTA | Clathrin light chain A | C9J8P9 | 0.8 | 2.7 | 5.8 | 3.7 | 4.2 | 6.3 | |||
| CLTB | Clathrin light chain B | D6RJD1 | 0.0 | 0.7 | 3.0 | 1.7 | 0.4 | 5.0 | |||
| COPG1 | Coatomer protein complex subunit gamma 1 | D6RG17 | |||||||||
| CPNE1 | Copine-1 | Q99829 | |||||||||
| CPNE3 | Copine-3 | E5RFT7 | |||||||||
| CTPS1 | CTP synthase 1 | P17812 | |||||||||
| CTTN | Cortactin | H0YCD9 | 0.3 | 3.7 | 13.3 | 6.7 | 4.2 | 14.5 | |||
| CXADR | Coxsackievirus and adenovirus receptor | P78310 | 0.3 | 2.0 | 5.5 | 3.3 | 1.2 | 5.5 | |||
| DAG1 | Dystroglycan 1 | C9JQL4 | 0.0 | 1.7 | 5.5 | 2.7 | 0.6 | 8.3 | |||
| DBI | Diazepam binding inhibitor, acyl-CoA binding proteinb | B8ZWD8 | 1.8 | 6.0 | 7.8 | 4.3 | 2.6 | 5.0 | |||
| DBN1 | Drebrin 1 | D6RCR4 | 0.3 | 1.3 | 3.8 | 1.0 | 2.0 | 4.5 | |||
| DBNL | Drebrin-like | H7C111 | 0.0 | 0.7 | 4.8 | 1.7 | 1.0 | 4.5 | |||
| DNAJC7 | DnaJ heat shock protein family | K7EQ73 | 0.0 | 0.0 | 5.0 | 2.7 | 0.0 | 6.0 | |||
| DNM2 | Dynamin 2 | K7EMR9 | |||||||||
| DSG2 | Desmoglein 2 | J3KSI6 | 1.3 | 5.3 | 16.5 | 10.0 | 4.8 | 18.5 | |||
| DYNC1LI1 | Dynein cytoplasmic 1 light-intermediate-chain 1 | Q9Y6G9 | |||||||||
| EEF1B2 | Eukaryotic translation elongation factor 1 beta 2 | F8WFC9 | 1.5 | 5.3 | 8.3 | 6.3 | 5.6 | 7.8 | |||
| EEF1D | Eukaryotic translation elongation factor 1 delta | Q9BW34 | 1.8 | 4.0 | 10.3 | 5.7 | 3.4 | 9.8 | |||
| EHD4 | EH domain containing 4 | Q9H223 | |||||||||
| EPB41 | Erythrocyte membrane protein band 4.1 | Q4VB87 | |||||||||
| EPB41L2 | Erythrocyte membrane protein band 4.1-like 2 | E9PMG5 | 0.8 | 1.3 | 3.3 | 1.7 | 3.0 | 5.5 | |||
| ERP29 | Endoplasmic reticulum protein 29 | F8W1G0 | 0.5 | 1.7 | 7.8 | 4.3 | 0.6 | 7.5 | |||
| fam21a | Family with sequence similarity 21 member A | Q641Q2 | 0.0 | 0.0 | 6.8 | 2.0 | 1.8 | 8.3 | |||
| FLOT2 | Flotillin 2 | Q14254 | |||||||||
| GABARAPL2 | GABA type A receptor-associated protein-like 2 | H3BU36 | 0.0 | 0.0 | 6.0 | 3.0 | 0.2 | 5.3 | |||
| GARS | Glycyl-tRNA synthetase | A0A090N8G0 | |||||||||
| GFPT1 | Glutamine–fructose-6-phosphate transaminase 1 | Q06210 | |||||||||
| GIPC1 | GIPC PDZ domain containing family member 1 | K7EJ33 | 0.0 | 1.0 | 3.5 | 1.7 | 1.2 | 5.5 | |||
| gnai3 | G protein subunit alpha i3 | P08754 | |||||||||
| GNAS | GNAS complex locus | Q5JWF2 | |||||||||
| GOLM1 | Golgi membrane protein 1 | C9J941 | 0.5 | 1.3 | 5.3 | 3.0 | 2.0 | 5.3 | |||
| GPI | Glucose-6-phosphate isomerase | A0A0J9YXP8 | |||||||||
| GRB2 | Growth factor receptor-bound protein 2 | J3QRL5 | 0.5 | 0.0 | 5.5 | 2.0 | 0.8 | 5.8 | |||
| HGS | Hepatocyte growth factor-regulated tyrosine kinase substrate | I3L1E3 | 0.0 | 0.3 | 4.3 | 2.0 | 2.8 | 5.8 | |||
| HINT1 | Histidine triad nucleotide binding protein 1 | H0YC49 | 0.0 | 2.3 | 4.5 | 2.7 | 2.0 | 4.0 | |||
| HN1 | hematological and neurological expressed 1 | J3KT51 | 0.8 | 2.3 | 4.8 | 3.0 | 2.4 | 4.5 | |||
| HN1L | Hematological and neurological expressed 1-like | Q9H910 | 0.3 | 1.7 | 4.5 | 4.0 | 1.6 | 5.3 | |||
| HPRT1 | Hypoxanthine phosphoribosyltransferase 1 | P00492 | |||||||||
| HSP90AB3P | Heat shock protein 90 alpha family class B member 3, pseudogene | Q58FF7 | 1.0 | 1.0 | 4.8 | 3.3 | 2.2 | 5.3 | |||
| HSPA4 | Heat shock protein family A | P34932 | 1.0 | 3.0 | 4.5 | 2.3 | 1.6 | 4.8 | |||
| HSPH1 | heat shock protein family H | A0A024RDS1 | 0.8 | 1.3 | 7.5 | 4.3 | 1.6 | 8.3 | |||
| HYOU1 | Hypoxia upregulated 1 | A0A087WW13 | 1.3 | 1.3 | 4.3 | 3.0 | 1.0 | 5.8 | |||
| IGSF8 | Immunoglobulin superfamily member 8 | Q969P0 | |||||||||
| IMPDH2 | Inosine monophosphate dehydrogenase 2 | Q6QE17 | 0.3 | 2.3 | 8.8 | 5.0 | 2.8 | 6.3 | |||
| IPO5 | Importin 5 | C9J875 | |||||||||
| IST1 | IST1, ESCRT-III-associated factor | J3QQP8 | 0.3 | 2.3 | 8.5 | 3.3 | 1.4 | 8.0 | |||
| ITGB1 | Integrin subunit beta 1 | P05556 | 1.3 | 2.3 | 4.3 | 4.3 | 2.4 | 6.8 | |||
| KHSRP | KH-type splicing regulatory protein | M0QYH3 | 1.3 | 2.7 | 4.3 | 4.3 | 1.4 | 7.8 | |||
| KIF23 | Kinesin family member 23 | H7BYN4 | |||||||||
| LMNB1 | Lamin B1 | B4DZT3 | 0.5 | 2.0 | 2.3 | 4.0 | 0.8 | 5.8 | |||
| MAPRE1 | Microtubule-associated protein RP/EB family member 1 | A2VCR0 | 0.0 | 0.3 | 6.0 | 3.0 | 1.6 | 4.0 | |||
| MARCKSL1 | MARCKS-like 1 | P49006 | |||||||||
| MCFD2 | Multiple coagulation factor deficiency 2 | H7BZ18 | 0.3 | 3.3 | 7.8 | 5.0 | 2.6 | 5.8 | |||
| MIF | Macrophage migration inhibitory factor | I4AY87 | |||||||||
| MSN | Moesin | V9HWC0 | 0.5 | 4.0 | 5.3 | 5.3 | 3.6 | 6.3 | |||
| NASP | Nuclear autoantigenic sperm protein | E9PKR5 | 0.0 | 0.0 | 2.8 | 1.0 | 1.8 | 5.0 | |||
| NCSTN | Nicastrin | Q5T205 | 0.3 | 1.3 | 2.8 | 2.3 | 1.0 | 2.8 | |||
| NDRG1 | N-myc downstream-regulated 1 | Q8N959 | 0.0 | 0.3 | 6.3 | 3.3 | 1.2 | 7.5 | |||
| NENF | Neudesin neurotrophic factor | Q9UMX5 | 0.3 | 1.0 | 4.5 | 2.0 | 0.4 | 6.0 | |||
| NME1 | NME/NM23 nucleoside diphosphate kinase 1 | E7ERL0 | 0.5 | 2.3 | 7.0 | 3.3 | 2.2 | 5.5 | |||
| NSFL1C | NSFL1 cofactor | R4GNE6 | 0.0 | 0.7 | 6.5 | 1.3 | 0.4 | 6.8 | |||
| NUCB2 | Nucleobindin 2 | E9PLR0 | 1.5 | 3.7 | 9.8 | 7.0 | 3.0 | 10.3 | |||
| OLA1 | Obg-like ATPase 1 | J3KQ32 | |||||||||
| PARK7 | Parkinsonism-associated deglycase | K7EN27 | 0.0 | 2.7 | 8.0 | 5.0 | 2.6 | 7.5 | |||
| PARP1 | Poly (ADP-ribose) polymerase 1 | Q5VX85 | 0.5 | 4.7 | 5.8 | 7.3 | 2.8 | 11.0 | |||
| PDCD6 | Programmed cell death 6 | A0A024QZ42 | 0.8 | 1.7 | 5.5 | 5.3 | 1.4 | 4.5 | |||
| PDCD6IP | Programmed cell death 6-interacting protein | F8WBR8 | 1.5 | 2.3 | 10.3 | 11.7 | 4.4 | 11.8 | |||
| PFDN2 | Prefoldin subunit 2 | Q9UHV9 | 0.0 | 1.0 | 4.5 | 2.7 | 1.8 | 5.0 | |||
| PGD | Phosphogluconate dehydrogenase | K7EMN2 | |||||||||
| PHGDH | Phosphoglycerate dehydrogenase | Q5SZU1 | 0.8 | 1.0 | 4.3 | 3.7 | 1.4 | 5.0 | |||
| PLD3 | Phospholipase D family member 3 | Q8IV08 | |||||||||
| PLS3 | Plastin 3 | H7C4N2 | |||||||||
| PPA1 | Pyrophosphatase | Q5SQT6 | 0.0 | 0.0 | 4.0 | 1.7 | 1.4 | 4.8 | |||
| PPIB | Peptidylprolyl isomerase B | P23284 | 1.0 | 2.3 | 6.3 | 2.7 | 1.0 | 5.0 | |||
| PRDX2 | Peroxiredoxin 2 | A0A024R7F2 | 0.0 | 1.0 | 9.5 | 3.3 | 1.6 | 6.0 | |||
| PRDX4 | Peroxiredoxin 4 | A6NG45 | 1.8 | 5.7 | 10.5 | 7.0 | 1.6 | 7.8 | |||
| PRKCSH | Protein kinase C substrate 80K-H | A0A0S2Z4D8 | 0.0 | 0.7 | 6.8 | 2.3 | 0.6 | 5.5 | |||
| PSMB6 | Proteasome subunit beta 6 | Q6IAT9 | |||||||||
| PTGFRN | Prostaglandin F2 receptor inhibitor | A4QPA1 | |||||||||
| RAB10 | RAB10, member of RAS oncogene family | Q9UL28 | |||||||||
| RAB14 | RAB14, member of RAS oncogene family | P61106 | |||||||||
| RAB5C | RAB5C, member of RAS oncogene family | F8VVK3 | |||||||||
| RAB8A | RAB8A, member of RAS oncogene family | P61006 | |||||||||
| RACGAP1 | Rac GTPase-activating protein 1 | F8VZ66 | |||||||||
| RAD23B | RAD23 homolog B, nucleotide excision repair protein | Q5W0S5 | 0.8 | 5.0 | 12.5 | 6.0 | 3.2 | 10.3 | |||
| RCN1 | Reticulocalbin 1 | E9PLM2 | 0.0 | 0.0 | 3.8 | 2.0 | 1.6 | 5.3 | |||
| RCN2 | Reticulocalbin 2 | L8E7F0 | 0.0 | 0.0 | 6.0 | 2.3 | 2.0 | 4.5 | |||
| RHOA | ras homolog family member A | C9JX21 | |||||||||
| RPL15 | Ribosomal protein L15 | E7ERA2 | 2.0 | 3.7 | 6.0 | 3.3 | 3.2 | 3.3 | |||
| RPL6 | Ribosomal protein L6 | F8VU16 | 1.8 | 3.3 | 8.0 | 7.7 | 5.6 | 9.5 | |||
| RPL7 | Ribosomal protein L7 | P18124 | 1.5 | 2.0 | 5.5 | 5.3 | 3.0 | 6.3 | |||
| RPS18 | Ribosomal protein S18 | J3JS69 | 1.3 | 3.7 | 5.0 | 4.3 | 3.6 | 3.3 | |||
| RPS21 | Ribosomal protein S21 | Q6FGH5 | 0.5 | 2.0 | 4.3 | 2.7 | 1.4 | 5.0 | |||
| SDCBP | Syndecan binding protein | E9PBU7 | |||||||||
| SDF4 | Stromal cell-derived factor 4 | Q9BRK5 | 0.0 | 1.3 | 6.5 | 4.0 | 2.2 | 7.8 | |||
| SH3GL1 | SH3 domain containing GRB2-like 1, endophilin A2 | Q9BVL7 | |||||||||
| SLC12A2 | Solute carrier family 12 member 2 | B7ZM24 | 1.5 | 2.3 | 4.8 | 1.0 | 1.2 | 5.0 | |||
| SLC16A1 | Solute carrier family 16 member 1 | A0A024R0H1 | |||||||||
| SLC39A14 | Solute carrier family 39 member 14 | A0A0S2Z5C8 | |||||||||
| SLC3A2 | Solute carrier family 3 member 2 | F5GZS6 | 1.0 | 3.3 | 6.5 | 4.0 | 3.8 | 8.8 | |||
| SLC7A5 | Solute carrier family 7 member 5 | Q01650 | |||||||||
| SLC9A3R1 | SLC9A3 regulator 1 | O14745 | 0.8 | 5.0 | 15.0 | 5.0 | 5.2 | 12.0 | |||
| slc9a3r2 | SLC9A3 regulator 2 | Q15599 | 0.0 | 0.0 | 1.8 | 0.3 | 0.8 | 6.0 | |||
| SNAP23 | Synaptosome-associated protein 23 | O00161 | 0.3 | 2.0 | 13.0 | 7.0 | 3.2 | 12.5 | |||
| SQSTM1 | Sequestosome 1 | E7EMC7 | 0.0 | 1.7 | 12.8 | 5.0 | 1.2 | 10.5 | |||
| SRI | Sorcin | C9J0K6 | 0.3 | 1.3 | 8.0 | 5.0 | 2.4 | 7.8 | |||
| STAM | Signal-transducing adapter molecule | B4DZT2 | 0.0 | 1.3 | 3.8 | 2.3 | 2.0 | 5.0 | |||
| STMN1 | Stathmin 1 | A2A2D0 | 1.3 | 8.0 | 8.5 | 8.0 | 4.4 | 12.0 | |||
| STX12 | Syntaxin 12 | Q86Y82 | 0.0 | 0.7 | 6.8 | 4.0 | 0.6 | 7.3 | |||
| STX4 | Syntaxin 4 | Q12846 | |||||||||
| STXBP3 | Syntaxin binding protein 3 | O00186 | |||||||||
| TARS | Threonyl-tRNA synthetase | D6RCS6 | |||||||||
| TAX1BP1 | Tax1 binding protein 1 | C9JBZ7 | 0.0 | 0.0 | 8.5 | 3.0 | 0.8 | 7.0 | |||
| TBCA | Tubulin folding cofactor A | O75347 | 0.5 | 2.0 | 3.8 | 2.7 | 3.6 | 6.5 | |||
| TCP1 | T-complex 1 | F5H7Y1 | 0.8 | 0.3 | 4.8 | 4.3 | 2.4 | 6.8 | |||
| TF | Transferrin | C9JB55 | 1.5 | 1.3 | 1.8 | 2.7 | 4.8 | 2.0 | |||
| TFG | TRK-fused gene | Q92734 | 0.0 | 4.0 | 12.8 | 7.7 | 4.2 | 12.5 | |||
| TFRC | Transferrin receptor | A8K6Q8 | |||||||||
| TOLLIP | Toll-interacting protein | Q6FIE9 | 0.3 | 1.0 | 5.3 | 3.7 | 1.2 | 3.5 | |||
| TOM1 | Target of Myb1 membrane trafficking protein | H7BYN7 | 0.0 | 0.3 | 6.0 | 4.7 | 1.6 | 6.3 | |||
| TPD52L2 | Tumor protein D52-like 2 | O43399 | 0.0 | 1.7 | 5.3 | 2.3 | 1.8 | 4.5 | |||
| TSPAN3 | Tetraspanin 3 | L8E893 | |||||||||
| TSPAN6 | Tetraspanin 6 | A0A024RCI0 | |||||||||
| TXNDC12 | Thioredoxin domain containing 12 | V9GY50 | 0.5 | 0.7 | 5.5 | 3.0 | 0.6 | 4.5 | |||
| UBXN1 | UBX domain protein 1 | Q04323 | 0.0 | 0.7 | 5.0 | 1.3 | 0.8 | 5.0 | |||
| UCHL1 | Ubiquitin C-terminal hydrolase L1 | P09936 | 0.0 | 4.0 | 8.5 | 3.7 | 3.2 | 6.8 | |||
| USP5 | Ubiquitin-specific peptidase 5 | A0A140VJZ1 | |||||||||
| VPS35 | VPS35, retromer complex component | Q96QK1 | |||||||||
| VTA1 | Vesicle trafficking 1 | Q9NP79 | 0.0 | 0.0 | 5.8 | 1.7 | 1.0 | 2.5 | |||
| YARS | Tyrosyl-tRNA synthetase | A0A0S2Z4R1 | |||||||||
| ZYX | Zyxin | H7C3R3 | 0.0 | 1.0 | 3.8 | 3.7 | 2.4 | 8.5 | |||
Proteins identified in VLPs from at least one type of transfection and/or Vera-Velasco et al. (43) are listed along with their UniProt identifier, totaling 166 proteins. The inclusion of any given protein in any of these sample sets or in the Gene Ontology (GO) groups of vesicle-mediated transport (VMT) or cytoskeleton is indicated by gray shading. White shading indicates that the protein of interest was not identified in that group. The number of proteins identified in each transfection or GO set from the full list of 166 proteins is indicated in the header for the transfection or GO group.
CoA, coenzyme A; EB, elementary body; RP, replicative phase; GABA, gamma-aminobutyric acid.
Nipah virus F-driven budding is reduced upon inhibition of endocytic machinery.
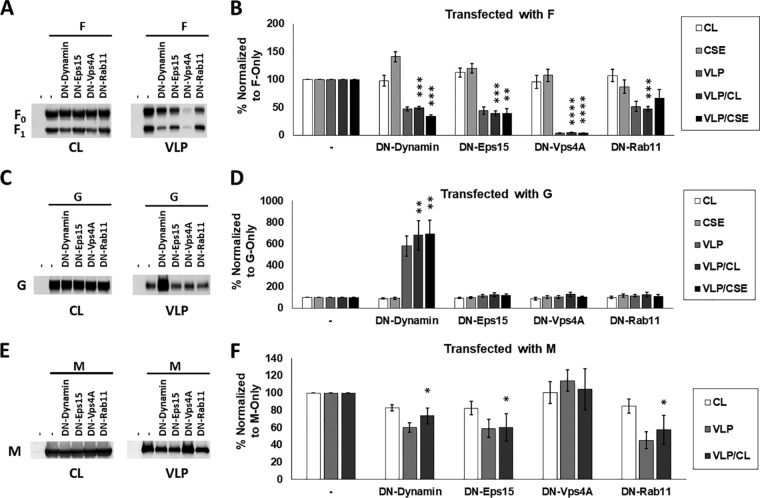
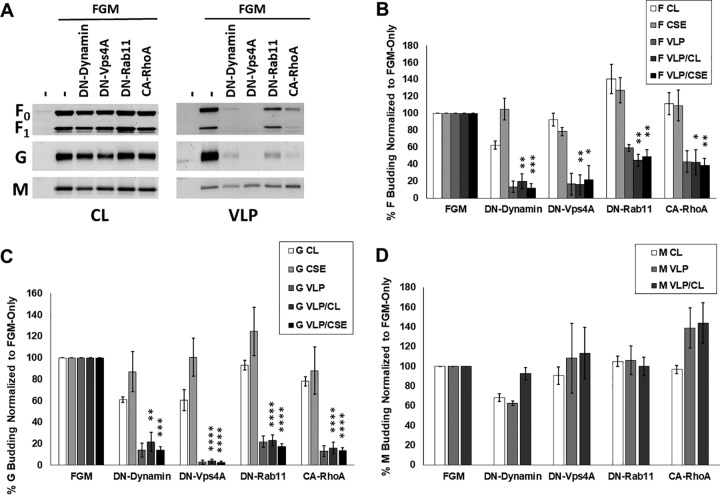
Based on our proteomics analyses, F seemed more likely than M to utilize cellular machinery for its budding function. To further validate proteomics and determine whether NiV F-driven budding is dependent on machinery involved in vesicular trafficking, we measured F expression in cell lysates (CL), at the cell surface (CSE), and in viral particles (VLP) after cotransfection with an empty vector (positive control, set to 100%) or the following dominant-negative (DN) constructs to inhibit several pathways of vesicular trafficking: DN-dynamin (various endocytic pathways), DN-Eps15 (clathrin-mediated endocytosis), DN-Vps4A (function of ESCRT complexes involved in multivesicular body formation and several mechanisms of viral budding), and DN-Rab11 (primarily recycling from endosomal system to the plasma membrane) (Fig. 3A). After quantification of these parameters with flow cytometry or densitometry, two budding indices, one based on each measurement of cellular expression, were calculated to assess the actual budding efficiencies under each transfection condition, VLP/CL and VLP/CSE, with the positive control set to 100% for each budding index.
FIG 3.
Nipah virus budding is significantly modulated upon the inhibition of endocytosis, recycling, and ESCRT function. (A) HEK293T cells were transfected with NiV F alone or with dominant-negative (DN) mutants of the following key vesicular trafficking factors: dynamin (endocytosis), Eps15 (clathrin-mediated endocytosis), VPS4A (ESCRT function), or Rab11 (recycling). Total cell lysates (CL) and virus-like particle (VLP) fractions were prepared 24 h after transfection and used for SDS-PAGE and Western blot analysis, whereas F cell surface expression (CSE) was measured using flow cytometry. (B) Using densitometry to assess the expression of the F protein in VLPs along with corresponding total cell lysate and CSE values, two indices were determined, VLP/CL and VLP/CSE. (C and D) As with panels A and B, a construct encoding G was coexpressed with each DN factor and the following CL, CSE, VLP, and index values elucidated. Similarly, coexpression with these constructs were tested for effects on M expression in cell lysates and VLPs but not at the cell surface due to it being a cytoplasmic protein. (E and F) Representative Western blots (E) and quantification (F) are shown. The results are representative of at least three experiments, with error bars indicating the standard error of the mean. One-way Student t tests were used to assess significant differences in budding efficiency indices compared to when they are cotransfected with pcDNA3.1, an empty vector (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
We report that F budding VLP/CL and/or VLP/CSE efficiencies were significantly reduced to a range of 4 to 49% compared to the positive controls (assessed with one-sample t tests) after coexpression with each of these constructs (Fig. 3B). The only index not meeting significance was VLP/CSE for DN-Rab11, which still appeared to trend toward a reduction (reduced to 66%, P = 0.13), with the VLP/CL reduction for DN-Rab11 being to 52.67%, P = 0.0009). F budding was most affected by DN-Vps4A, indicating the importance of ESCRT function in its budding mechanism, whether directly or indirectly. Since this study additionally focused on how these factors might affect G and M budding efficiencies, they were similarly tested except that CSE cannot be measured for NiV M, which is not a transmembrane protein. Of significant interest, NiV G budding efficiency increased almost 7-fold based on both indices in the case of cotransfection with DN-dynamin but did not change under any other condition (Fig. 3C and D). This result was entirely unexpected and suggests a novel function of dynamin, direct or indirect, in the unelucidated and normally relatively inefficient process of G-driven particle formation (Fig. 3C) (18, 46). Compared to F and G, the effects of these constructs on M budding were less dramatic, though DN-dynamin, DN-Eps15, and DN-Rab11 coexpression led to significant (P < 0.05) M budding efficiency reductions by 27%, 40%, and 42%, respectively (Fig. 3E and F). Importantly, the finding that DN-Vps4a does not affect NiV M budding efficiency is consistent with prior literature (47).
Nipah virus budding is significantly affected by actin cytoskeletal manipulation through mutant RhoGTPases.
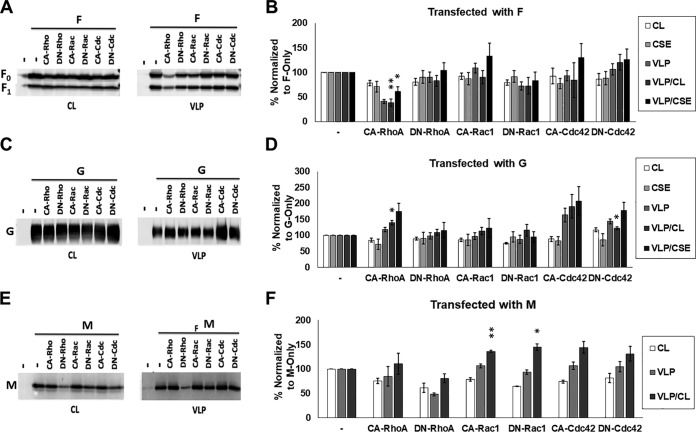
Actin cytoskeletal factor enrichment was largely specific to F alone and FGM VLPs in our proteomics analysis, which suggests that these factors may be recruited specifically by the presence of F to sites of assembly and budding. The actin cytoskeleton is involved in many cellular processes (48, 49), as well as assembly and/or egress for some viruses; however, these roles vary greatly and, for many viruses, are poorly understood (24, 36–39). The actin cytoskeleton is composed of monomeric globular (G-actin) and polymeric filamentous actin (F-actin), which are interconverted and organized by remodeling proteins, including RhoGTPases, to form specific microdomains within a cell (50–53). Major reorganization of the actin cytoskeleton into structures such as stress fibers, lamellipodia, and filopodia is regulated by the RhoGTPases RhoA, Rac1, and Cdc42, respectively (40). For an initial test of whether NiV F budding is modulated by the activity of RhoGTPases, we assessed whether F budding efficiency indices (VLP/CL and VLP/CSE) are altered during the coexpression of constitutively active (CA) and dominant-negative (DN) mutants of RhoA, Rac1, or Cdc42. We found that CA-RhoA reduced the levels of F in VLPs (VLP/CL to 54% [P < 0.01] and VLP/CSE to 61% [P < 0.05]) (Fig. 4A and B). We also tested the effects of these constructs on G- and M-driven budding. The G VLP/CL budding indexes following CA-RhoA and DN-Cdc42 coexpression were 140% and 122%, respectively (Fig. 4C and D). While the significance of inefficient G-driven budding is not understood, these findings may yield some insight into the processes involved. Interestingly, M budding was found to be increased by both CA-Rac1 and DN-Rac1, suggesting that perturbation of the function of this GTPase may support particle formation (Fig. 4E and F).
FIG 4.
Nipah virus budding is significantly affected by actin cytoskeletal manipulation through mutant RhoGTPases. (A and B) HEK293T cells were transfected with NiV F alone or with dominant-negative (DN) or constitutively active (CA) constructs for RhoA, Rac1, and Cdc42. As with Fig. 3, CL, CSE, and VLPs were quantified and both budding indices assessed. SDS-PAGE and Western blot analysis. (C to F) The effects of coexpressing these mutants were assessed for G-driven (C and D) and M-driven (E and F) budding. The results are representative of at least three experiments, with error bars indicating the standard error of the mean. One-way Student t tests were used to assess significant differences in budding efficiency indices compared to when they are cotransfected with pcDNA3.1, an empty vector (*, P < 0.05; **, P < 0.01).
Inhibition of Nipah virus fusion protein budding reduces attachment protein incorporation regardless of matrix protein expression.
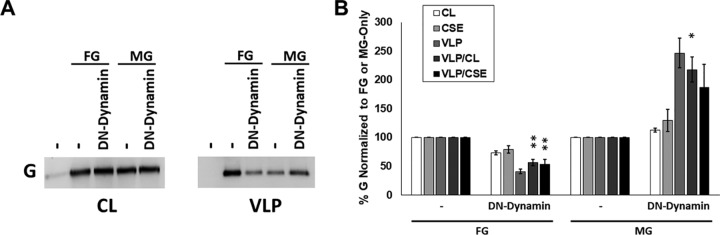
To better elucidate the importance of the mutant factors tested so far, several of the most interesting factors were also assessed in the context of F, G, and M coexpression, which should better emulate particles produced in live NiV infections (Fig. 5A). While the FGM particles produced from this combination lack the internal viral genome ribonucleoprotein (vRNP) complex responsible for infection, they would contain all viral machinery needed for both entry (both F and G) and incorporation (M) of the vRNP. Despite the presence of the M protein, generally considered to be the most important viral protein in the process of paramyxovirus budding, we found that DN-dynamin, DN-Vps4a, DN-Rab11, and CA-RhoA all significantly and consistently reduced both indices of F budding efficiency by 51% to 88% (Fig. 5A and B). More strikingly still, we report that these reductions were also seen for NiV G in the context of F and M expression (FGM samples), which exhibited no less than a 77% reduction in budding efficiency in any tested combination (Fig. 5A and C). This was particularly interesting since only DN-dynamin and CA-RhoA appeared to previously affect G-driven budding, and both led to increased budding efficiency. M budding efficiencies, on the other hand, were not significantly affected by any coexpressed cellular factor while in this combination (Fig. 5A and D). Importantly, these findings support the model depicted by our prior studies that G incorporation into VLPs may be more tied to the budding activity of NiV F than that of M (18). To further investigate this model and elucidate whether the change from increased (G-only) to decreased (FGM) G incorporation during coexpression of DN-dynamin was driven by the presence of F or M, the budding efficiency of G in both the FG and MG combinations was next assessed with and without cotransfection of DN-dynamin (Fig. 6). Supporting our model, these experiments demonstrated that the addition of DN-dynamin substantially reduces G incorporation in the FG combination and either does not change or supports increased G incorporation in the MG combination (Fig. 6A and B).
FIG 5.
Inhibition of Nipah virus fusion protein budding reduces attachment protein incorporation despite the presence of normally budding matrix protein. (A) To assess the effects that manipulation of vesicular trafficking (Fig. 3) and the actin cytoskeleton (Fig. 4) have on more complete budding particles, NiV F, G, and M were cotransfected with or without DN-dynamin, DN-Vps4a, DN-Rab11, or CA-RhoA. The expression of each viral protein in cell lysates, on the cell surface for F and G, and in VLPs was quantified and used to produce budding indices as done previously. (B to D) These values are summarized for F (B), G (C), and M (D). The results are representative of at least three experiments, with error bars indicating the standard error of the mean. One-way Student t tests were used to assess significant differences in budding efficiencies for each viral protein compared to FGM coexpression alone (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
FIG 6.
Loss of increased G incorporation from DN-dynamin expression occurs with expression of F but not G. HEK293T cells were transfected with constructs for NiV G and either F or M with or without the additional expression of DN-dynamin. As with Fig. 3 and 5, VLPs were collected compared with total and surface expression to assess the G budding efficiency. (A and B) Western blots (A) along with densitometric and surface expression (B) were quantified. The results are representative of at least three experiments, with error bars indicating the standard error of the mean. One-way Student t tests were used to assess significant differences in budding efficiencies for each viral protein compared to FGM coexpression alone (*, P < 0.05; **, P < 0.01).
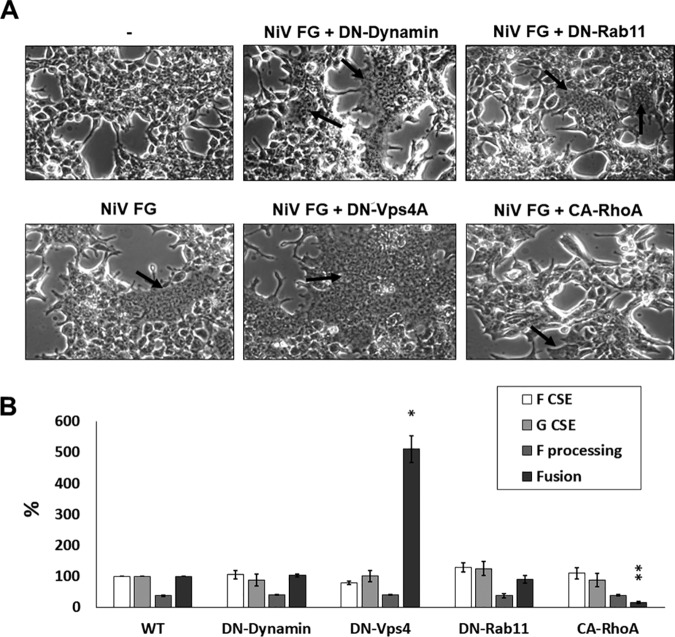
Select mutant cellular factors involved in vesicular trafficking and cytoskeletal organization affect F budding and cell-cell fusion without affecting F processing.
While we have so far shown that mutants of several cellular factors involved in vesicular trafficking and in cytoskeletal organization impact NiV budding in the hopes of validating and expanding upon VLP proteomics, the full importance of these major cellular processes in the life cycle of NiV remains unclear. Since proteasomal cleavage of NiV F by endolysosomal cathepsin B or L has been shown to be essential for its fusogenic activity (54–56), we tested whether some of the constructs affected F fusogenicity using a cell-cell fusion assay. While we report that neither DN-dynamin nor DN-Rab11 significantly affected fusion, DN-Vps4a dramatically increased fusion (by 410%, P < 0.05), while CA-RhoA significantly decreased fusion (to 16%, P < 0.01) (Fig. 6A). Notably, data from FGM cotransfection experiments (Fig. 5) showed that none of the tested constructs significantly affected F or G CSE nor F processing levels; thus, these parameters were not responsible for the fusogenicity phenotypes observed (Fig. 7B). These findings not only suggest that NiV-F can be internalized or trafficked through the endolysosomal system through dynamin-independent means but also indicate that ESCRT and cytoskeleton machinery are able to modulate both sites of budding and fusion, either directly or indirectly.
FIG 7.
Expression of select mutant factors that inhibit F budding does not affect processing but can modulate fusion. (A) HEK293T cells were transfected with an empty vector or with NiV F and G with or without the listed cellular factors. Light microscope images were taken at ×200 magnification. (B) By counting nuclei in fused syncytia, levels of fusion were quantified, with F and G cotransfection alone set to 100%. Levels of F processing as well as F and G CSE levels were quantified from FGM cotransfections. The results are representative of at least three experiments, with error bars indicating standard error of the mean. One-way Student t tests were used to assess significant differences in budding efficiencies for each viral protein compared to FGM coexpression alone (*, P < 0.05; **, P < 0.01).
DISCUSSION
While recent studies have helped to clarify how Nipah virus assembly and budding occur, many gaps in our understanding remain. While paramyxovirus matrix proteins are generally accepted as crucial for driving particle formation, incorporating all required viral components as well as supporting the stability and infectivity of produced virions, significantly less is understood concerning how other viral proteins may support particle formation (9, 17). Unlike most paramyxoviruses, NiV F, G, and M have all been reported to autonomously produce VLPs to various degrees upon expression alone, suggesting potential roles as supportive in budding (46). From this finding, it is likely that particles lacking one or more viral components are produced during live infection. While no biological roles have so far been shown for NiV particles lacking F, G, or the internal viral machinery, some such particles may serve a role during infection. One possibility could be the use of FG particles lacking M and/or the vRNP as decoys to attract neutralizing antibodies. Our recent study further suggested a novel role for NiV F as capable of increasing the VLP incorporation of G (18). Moreover, our study implicated the cytoplasmic tail of NiV F as containing several specific motifs important for this budding function (37, 57). The exact function(s) of these motifs remain unclear; however, the presence of a YxxL motif may suggest interaction with the ESCRT-associated protein ALIX and/or other vesicular trafficking machinery (21, 58–60).
In a recent study published by Vera-Velasco et al. (43), VLPs produced from the coexpression of NiV F, G, and M in HEK293 cells were analyzed using liquid chromatography-tandem mass spectrometry (LC-MS/MS)-based proteomics, identifying proteins involved in vesicular trafficking as significantly incorporated (43). While this study gave an exciting first look into cellular proteins and machinery potentially involved in NiV particle formation, the importance of F, G, and M for cellular protein incorporation remained unknown. Additionally, the importance of the incorporated proteins and machinery identified in this study remained unelucidated. Here, we analyzed by proteomics the VLPs produced from either individual expression or various combinations of F, G, and M coexpression, and we validated the most significant processes. Corroborating the findings from the study by Vera-Velasco et al. (43), we report that proteins incorporated into FGM VLPs were most significantly associated with vesicular trafficking (Fig. 2B). We also found that proteins involved in the cytoskeleton showed significant incorporation as a group. Further, we reported that while M-only VLPs incorporated few cellular proteins, all VLP combinations containing F (F alone and FM) largely recapitulated the levels and patterns of cellular factor incorporation observed during FGM expression (Fig. 2). Together, these findings support a model where the matrix protein induces budding based on its ability to form higher-order scaffolds, whereas the fusion protein likely relies on the recruitment of cellular machinery to sites of particle formation and, in some cases, to virions (23, 29, 59). Future proteomics studies focused on posttranslational modifications of viral and cellular factors incorporated into VLPs may further clarify the processes of assembly and budding.
To attempt to validate those VLP proteomic analyses conducted previously (43) and those described here, we targeted several aspects of vesicular trafficking and actin cytoskeletal organization. Supporting our findings that VLPs made with combinations that include F or M tend to incorporate cellular factors associated with vesicle transport (Fig. 2), we report that dominant-negative constructs of dynamin, Eps15, and Rab11 each inhibit F and M budding when coexpressed (Fig. 3). Confirming further reports of M independence from ESCRT machinery (47), we also report that DN-Vps4A significantly reduced F but not M budding. The potential importance of ESCRTs in NiV budding is greatly supported by the identification of multiple ESCRT proteins both in this study and in that by Vera-Velasco et al. (43). Very recently, live NiV infection was shown to be significantly reduced by knockdown of ESCRT machinery, with a new role for the small NiV C protein as a potential ESCRT recruitment factor. As with several retroviruses, numerous viral proteins and motifs (61–64) can be important for ESCRT recruitment, and since neither our study nor that published previously included C, there remains a significant potential for F-driven budding to rely on the direct action of ESCRTs.
Reduced F and M budding during DN-Rab11 cotransfection is further corroborated by previous findings with the closely related Hendra virus (HeV) for which both the F and M proteins have been shown to interact with Rab11-positive recycling endosomes, and their budding is at least partially dependent on Rab11 activity (44). The full function of Rab11 during henipavirus budding is unknown; however, the importance of Rab11 and Rab11-interacting proteins has also been recently highlighted for several more distantly related enveloped viruses, including influenza A virus and respiratory syncytial virus (33, 35).
Our findings from the coexpression of constitutively active and dominant-negative RhoGTPases suggest that the actin cytoskeleton modulates NiV budding and validate the significance of cytoskeletal factor incorporation into NiV VLPs. Further studies are needed to enrich our understanding of potential roles that actin remodeling has in henipavirus particle formation. The potential involvement of the actin cytoskeleton may be in trafficking, assembly, or in the budding process itself (36, 38). Specific RhoGTPases have also been implicated as having roles in different pathways of endocytosis (65–68), which may explain the variation of effects on F, G, and M budding efficiencies (Fig. 4).
To better understand the importance of our budding phenotypes upon vesicular trafficking or cytoskeletal perturbation, FGM VLPs were characterized. We found a very clear phenotype suggesting the dependence of G incorporation on the budding of F regardless of M budding (Fig. 5). Interestingly, these results were supported by our previous findings using a budding-defective F mutant (18). Most striking was the clear shift from DN-dynamin expression increasing G-only budding by almost 7-fold (Fig. 3) to a reduction by nearly the same magnitude when F was expressed (Fig. 5), suggesting that the budding activity of F overrides that of G when they are coexpressed. A potential explanation for this is the observed shift in trafficking profiles of these proteins in a comparison of individual expression and coexpression; however, these changes require further study (58, 69).
Since vesicular trafficking has been previously associated with henipavirus F protein maturation, we also sought to understand how our constructs that most clearly altered budding would affect F processing and fusogenic capacities (56). We found that neither processing nor levels of F or G CSE were altered substantially after coexpression of any of the following constructs tested: DN-dynamin, DN-Vps4A, DN-Rab11, or CA-RhoA. Despite no clear change to processing or CSE, DN-Vps4A and CA-RhoA coexpression led to clear phenotypes of increased and reduced fusion, respectively (Fig. 7).
Somewhat similar reductions in fusion seen with CA-RhoA have been observed with HeV but were not consistent when assessed for parainfluenza 5, another paramyxovirus (70). Thus, the roles of actin cytoskeletal dynamics are not well understood for any aspect of the NiV life cycle. Future study is needed to understand the functions of the cytoskeleton during the life cycles of paramyxoviruses, though these mechanisms are likely to be obscured by differences between cell types and will vary by viral species.
Our observation of increased fusion during DN-Vps4A was completely unexpected and does not seem to be due to increased F processing nor to F or G CSE increases. The cause of this phenotype is unknown but we speculate could be explained by a change in F and G localization within the cell that is more optimal for cellular fusion. Superresolution microscopy may shed further light into the effects of Vps4A in cell-cell fusion.
Here, we have described a VLP proteomics approach of identifying cellular factors and processes potentially involved in NiV assembly and budding. Further, this was accomplished for VLPs produced from several combinations of NiV proteins to clarify the function of several viral proteins in cellular factor incorporation. We validated our VLP proteomics results by demonstrating that perturbation of the most significantly enriched functional groups, vesicular trafficking and the actin cytoskeleton, led to substantial modulation of VLP budding, with the clearest phenotypes being reductions in F-driven budding. The importance of functional F budding during the incorporation of the G protein into VLPs was further demonstrated using these mutant constructs. Together, these findings support a significant involvement of the NiV fusion protein and uncover the involvement of vesicular trafficking and actin cytoskeletal processes in efficient Nipah virus assembly and budding.
MATERIALS AND METHODS
Cells, DNA plasmids, and antibodies.
HEK293T cells were obtained from the ATCC and used for all experiments. These cells were grown in Dulbecco’s modified Eagle medium (DMEM; Corning) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (Gibco) at 37°C with 5% CO2. Previously published DNA constructs for NiV proteins, pcDNA3.1-F-AU1, pcDNA3.1-G-HA, and pCMV-3XFLAG-M, were used (18). These constructs were also used to make pCAGGS-NiV-F-AU1 and pCAGGS-NiV-M-3XFLAG. A dominant-negative (DN; S25N) mutant of Rab11 (71) inserted into a pGL vector was generously provided by Ruth Collins (Cornell University). pEGFP constructs containing previously described dominant-negative mutants of dynamin-2 (DN-dynamin; K44A) and eps15 (DN-Eps15; Δ95/295) were used (72–74). pcDNA3.1 constructs containing constitutively active (CA) and dominant-negative RhoA, Rac1, and Cdc42 were used (75). The dominant-negative Vps4A (DN-Vps4A) mutant (76) was synthesized and inserted into a pcDNA3.1 vector (Biomatik). Antibodies used against NiV F were rabbit polyclonal 835term for flow cytometry (77) and mouse monoclonal anti-AU1 for Western blotting (BioLegend). Also used were primary antibodies against G, mouse monoclonal anti-hemagglutinin (HA) for flow cytometry (BioLegend) and rabbit polyclonal anti-HA for Western blotting (Bethyl). Mouse monoclonal M2 anti-FLAG was used to detect NiV M for Western blotting (Sigma-Aldrich). Mouse monoclonal anti-sequestosome-1 antibodies were used to validate VLP proteomics (Abcam, no. ab56416). Goat anti-mouse and anti-rabbit 647 and 488 secondary antibodies (Thermo Fisher Scientific) were used for Western blotting and flow cytometry, as described previously (18).
NiV VLP production and sample preparation for proteomic analyses.
Five 15-cm2 dishes confluent with HEK293T cells were transfected using polyethylenimine (PEI; Polysciences, Inc.) at 1 mg/ml for each of the following combinations of plasmid constructs: pcDNA3.1, F, M, MG, FM, and FGM at an FGM DNA ratio of 24:1:5 F:G:M) (the table in Fig. 1A shows this as well). For each dish, 30 μg of total DNA was transfected at a 4:1 ratio of PEI to DNA, as described previously (78). For these experiments, pCAGGS versions of NiV-F and NiV-M constructs were used for optimal expression. After 48 h, the medium was collected and cleared of cellular debris by centrifugation at 376 × g for 10 min, as previously described (18). The resulting supernatant was collected and layered over 20% sucrose in NaCl-Tris-EDTA (NTE) buffer and spun at 110,000 × g for 90 min. After this VLP purification spin, VLPs were resuspended with 5% sucrose in NTE buffer and shipped to Pacific Northwest National Laboratories for VLP proteomics analyses, which are described below.
Sample preparation for VLP proteomics.
Purified viral-like particle samples were lysed and extracted by adding 1.0 ml of 2:1 (vol/vol) chloroform-methanol and vortexing before storing on ice. The samples were then placed in a centrifugal concentrator and dried completely overnight. Proteins were solubilized with the addition of 10 μl of homogenization buffer (8 M urea, 10 mM dithiothreitol, 50 mM Tris [pH 8]), followed by 30 s in a bath sonicator. The lysate was then incubated at 60°C for 30 min to denature and reduce the proteins. The protein solution was quantitatively transferred to a low-retention LC vial (Waters) using 25 μl of 50 mM Tris (pH 8). SNP and AFLP Package for Phylogenetic (SNaPP) analysis was carried out as described previously (79, 80). Briefly, 25 μl of sample was injected onto a 150-μm by 2-cm immobilized enzyme reactor for digestion. The digestion column was syringe packed using Poroszyme immobilized trypsin (Applied Biosystems). Following digestion, peptides were separated using a 100-min gradient on an in-house-packed 50-μm by 75-cm C18 analytical column (Phenomenex). The SNaPP system was coupled to a Q Exactive Plus mass spectrometer (Thermo Scientific) operated at a mass resolution of 70 K for MS1 and 17.5 for MS2 collection. Data were collected in data-dependent acquisition (DDA) mode with a top 12 method and a dynamic exclusion window of 30 s. The maximum ion time was increased to 200 ms for MS2 to maximize identifications from low sample loadings.
Proteomics.
Peptide samples (5 μl) were analyzed by LC-MS/MS using a Waters nano-Acquity M-class dual-pumping ultraperformance liquid chromatography (UPLC) system (Milford, MA) configured for on-line trapping at 5 μl/min for 8 min, followed by gradient elution through a reversed-phase analytical column at 300 nl/min. Columns were packed in-house using 360-μm outside diameter (o.d.) fused silica (Polymicro Technologies, Inc.) and contained Jupiter C18 medium (Phenomenex) in 5-μm particle size for the trapping column (100 μm inside diameter [i.d.] by 4 cm long) and 3-μm particle size for the analytical column (75 μm i.d. by 70 cm long). Mobile phases consisted of (MP-A) 0.1% formic acid in water and (MP-B) 0.1% formic acid in acetonitrile with the following gradient profile: 0 min, 1% MP-B; 2 min, 8% MP-B; 20 min, 12% MP-B; 75 min, 30% MP-B; 97 min, 45% MP-B; 100 min, 95% MP-B; and 110 min, 95% MP-B.
MS analysis was performed using a Q-Exactive high-fidelity (HF) mass spectrometer (Thermo Scientific, San Jose, CA). Electrospray emitters were prepared in-house using 150-μm o.d. by 20-μm i.d. chemically etched fused silica (89), subsequently attached to the column using a metal union, and coupled to the mass spectrometer via a custom-built nanospray source. Electrospray voltage (2.2 kV) was applied at the metal union providing ions to the heated (325°C) ion transfer tube entrance of the MS. Data collection was started 20 min after the gradient began and continued for a total acquisition time of 100 min. Precursor mass spectra (MS) were acquired from m/z 400 to 2,000 at a resolution of 60 K (automatic gain control [AGC] target, 3e6; max IT, 20 ms), followed by data-dependent MS/MS spectra of the top 12 most abundant ions from the precursor spectrum with an isolation window of m/z 2.0 and at a resolution of 15 K (AGC target 1e5; max IT, 200 ms) using a normalized collision energy of 30 and a 45-s exclusion time.
VLP proteomics analyses.
Modern mass spectrometers can observe very low-abundance contaminants in low-complexity samples, such as VLPs. Further, the experimental enrichment of very small particles with subsequent sample preparation by SNaPP analysis results in an approximation of loading protein quantity. Hence, samples are batched as sets and then randomized within sets to allow for the interpretation of potential carryover from abundant proteins from one VLP composition measurement to another. Relative quantification was performed via a spectral counting approach where the number of tandem mass spectra identified as matching a protein is used as a proxy for abundance that can be compared across data sets.
Further, the identification of peptides corresponding to the three NiV proteins, F, G, and M, was used to evaluate the quality of measurements. LC-MS/MS analyses considered for further analysis were included or excluded based on expected viral protein compositions. Specifically, we set the requirement that a replicate would only be included in the next stages of analysis if it contained no more than 2 observed peptides corresponding to unexpected viral protein carryover (e.g., M peptides in an F sample replicate). Similarly, we also set the requirement that each included replicate would require at least two observed peptides for each expected viral protein. The potential limitations of this were that the viral proteins did have different observation rates due to different lengths and expression levels; however, levels of peptides for viral proteins within samples they were expected were almost always much greater than in samples in which they were unexpected, regardless of the viral protein.
To identify host-derived proteins considered to be incorporated into VLPs of each combination, an approach based on averaged observed peptides was used. All proteins considered incorporated in any type of VLP were not allowed to have any more than an average observed peptide count of 2 in the control samples (VLPs isolated from cells transfected with an empty vector) and were required to have at least an average of 4.33 observed peptides in the VLP type of interest. By setting a low maximum threshold for observed peptides in the empty vector control, we sought to remove cellular factors highly enriched in background exosomes and microvesicles. An average observed peptide count of 4.33 in VLPs was set as the threshold, as this corresponded to the lowest level of average peptide incorporation for any expected viral protein in any of the VLP combination types.
After the incorporated host-protein protein lists were determined for each VLP combination, comparisons were done using InteractiVenn (81). Further, protein-protein interaction mapping was completed using the STRING and Cytoscape softwares (82, 83). Gene ontological designations used for Table 1 and the protein interaction map were conducted using GOTermFinder (84).
VLP budding assay.
HEK293T cells were transfected at between 65% and 85% confluence using 1 mg/ml PEI. After 24 h, medium was collected and precleared by centrifugation as described above, while the transfected cells were harvested with 10 mM EDTA (from 0.5 M stock; VWR) in Dulbecco’s phosphate-buffered saline (DPBS; Corning) and split into two groups, the cell lysate and cell surface expression. For both groups, cells were pelleted by centrifugation at 376 × g for 10 min and the supernatant removed. For the cell lysate fraction, 1× radioimmunoprecipitation assay (RIPA) buffer (Sigma-Aldrich) was prepared in tissue culture-grade water (Corning) with the cOmplete Ultra protease inhibitor (Roche) and added to the pellet prior to 30 min of vortexing and rotating at 4°C. The lysate supernatant was collected and prepared for 10% SDS-PAGE (Protogel), transferred to an activated polyvinylidene difluoride (PVDF) membrane (Thermo Fisher Scientific), and blotted with antibodies against AU1, HA, and FLAG tags, as described previously (18). For the group to be assessed for cell surface expression, the pellet was resuspended with FACS buffer (1% FBS in DPBS) and stained with rabbit polyclonal 835term (for F) and mouse monoclonal anti-HA (for G) for 1 h prior to three washes with FACS buffer, secondary antibody staining for 30 min, two final FACS washes, and analysis with a Millipore Guava easyCyte flow cytometer 8HT; 10,000 or more cells were analyzed per sample, as described previously (18).
Transmission electron microscopy sample preparation and imaging.
One 15-cm dish of HEK293T cells was transfected using PEI, as discussed above, for each combination of F, G, and M. VLPs were harvested and negatively stained with 1% uranyl acetate and visualized using a Tecnai T12 Spirit microscope (FEI) at 120 kV located in the Cornell University Center for Materials Research.
Cell-cell fusion assay.
HEK293T cells were transfected at 80 to 95% confluence with NiV F, G, and either pcDNA3.1 or one of the mutant cellular factors as described at a DNA:PEI ratio of 1:3 with a total transfection of 2.1 μg of DNA (F:G:pcDNA or cell factor ratio of 3:1:3). After 12 h, the cells were fixed in 1% paraformaldehyde at 37°C for 1 h and imaged on a Leica bright-field microscope. Five fields of view at ×200 magnification were used to quantify levels of cell-cell fusion, where each nucleus associated with a syncytium of four or more nuclei was counted. For each transfected sample, the fields of view were averaged, reduced by background syncytial levels (empty vector control), and normalized to the positive control (set at 100%) (85–88).
ACKNOWLEDGMENTS
This work was supported by the Department of Homeland Security (DHS) Science and Technology Directorate (S&T) under contract HSHQPM-14-X-00238 to J.N.A. and Katrina Waters, with a subcontract to H.A.C., and by NIH grant R01 IA109022 to H.C.A. Work was partially performed in the Environmental Molecular Sciences Laboratory (EMSL), a DOE-BER national scientific user facility at Pacific Northwest National Laboratory (PNNL). PNNL is a multi-program national laboratory operated by Battelle Memorial Institute for the DOE under contract DE-AC05-76RLO 1830.
The funders had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
G.P.J., H.C.A., B.B.-T., J.R.T., J.N.A., and E.M.C. designed the experiments; G.P.J., B.B.-T., B.N.R.L., K.M.M., N.T.U., J.L.R.Z., V.O., H.C.A., P.D.P., and H.M.B. performed the experiments; G.P.J., J.N.A., and J.P.W. conducted data and statistical analyses; and G.P.J., H.C.A., and J.N.A. wrote the manuscript.
REFERENCES
- 1.World Health Organization. 2015. Blueprint for R&D preparedness and response to public health emergencies due to highly infectious pathogens. World Health Organization, Geneva, Switzerland: https://www.who.int/docs/default-source/blue-print/blueprint-for-r-d-preparedness-and-response-meeting-report.pdf?sfvrsn=156d23be_2. [Google Scholar]
- 2.Drexler JF, Corman VM, Müller MA, Maganga GD, Vallo P, Binger T, Gloza-Rausch F, Cottontail VM, Rasche A, Yordanov S, Seebens A, Knörnschild M, Oppong S, Sarkodie YA, Pongombo C, Lukashev AN, Schmidt-Chanasit J, Stöcker A, Carneiro AJB, Erbar S, Maisner A, Fronhoffs F, Buettner R, Kalko EKV, Kruppa T, Franke CR, Kallies R, Yandoko ERN, Herrler G, Reusken C, Hassanin A, Krüger DH, Matthee S, Ulrich RG, Leroy EM, Drosten C. 2012. Bats host major mammalian paramyxoviruses. Nat Commun 3:796. doi: 10.1038/ncomms1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu Z, Yang L, Yang F, Ren X, Jiang J, Dong J, Sun L, Zhu Y, Zhou H, Jin Q. 2014. Novel henipa-like virus, Mojiang paramyxovirus, in rats, China, 2012. Emerg Infect Dis 20:1064–1066. doi: 10.3201/eid2006.131022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marsh GA, de Jong C, Barr JA, Tachedjian M, Smith C, Middleton D, Yu M, Todd S, Foord AJ, Haring V, Payne J, Robinson R, Broz I, Crameri G, Field HE, Wang L-F. 2012. Cedar virus: a novel henipavirus isolated from Australian bats. PLoS Pathog 8:e1002836. doi: 10.1371/journal.ppat.1002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chackerian B. 2007. Virus-like particles: flexible platforms for vaccine development. Expert Rev Vaccines 6:381–390. doi: 10.1586/14760584.6.3.381. [DOI] [PubMed] [Google Scholar]
- 6.Noad R, Roy P. 2003. Virus-like particles as immunogens. Trends Microbiol 11:438–444. doi: 10.1016/S0966-842X(03)00208-7. [DOI] [PubMed] [Google Scholar]
- 7.Grgacic EVL, Anderson DA. 2006. Virus-like particles: passport to immune recognition. Methods 40:60–65. doi: 10.1016/j.ymeth.2006.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Samal SK. (ed). 2011. The biology of paramyxoviruses. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 9.Harrison MS, Sakaguchi T, Schmitt AP. 2010. Paramyxovirus assembly and budding: building particles that transmit infections. Int J Biochem Cell Biol 42:1416–1429. doi: 10.1016/j.biocel.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Battisti AJ, Meng G, Winkler DC, McGinnes LW, Plevka P, Steven AC, Morrison TG, Rossmann MG. 2012. Structure and assembly of a paramyxovirus matrix protein. Proc Natl Acad Sci U S A 109:13996–14000. doi: 10.1073/pnas.1210275109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coronel EC, Murti KG, Takimoto T, Portner A. 1999. Human parainfluenza virus type 1 matrix and nucleoprotein genes transiently expressed in mammalian cells induce the release of virus-like particles containing nucleocapsid-like structures. J Virol 73:7035–7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pantua HD, McGinnes LW, Peeples ME, Morrison TG. 2006. Requirements for the assembly and release of Newcastle disease virus-like particles. J Virol 80:11062–11073. doi: 10.1128/JVI.00726-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takimoto T, Murti KG, Bousse T, Scroggs RA, Portner A. 2001. Role of matrix and fusion proteins in budding of Sendai virus. J Virol 75:11384–11391. doi: 10.1128/JVI.75.23.11384-11391.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sugahara F, Uchiyama T, Watanabe H, Shimazu Y, Kuwayama M, Fujii Y, Kiyotani K, Adachi A, Kohno N, Yoshida T, Sakaguchi T. 2004. Paramyxovirus Sendai virus-like particle formation by expression of multiple viral proteins and acceleration of its release by C protein. Virology 325:1–10. doi: 10.1016/j.virol.2004.04.019. [DOI] [PubMed] [Google Scholar]
- 15.Li M, Schmitt PT, Li Z, McCrory TS, He B, Schmitt AP. 2009. Mumps virus matrix, fusion, and nucleocapsid proteins cooperate for efficient production of virus-like particles. J Virol 83:7261–7272. doi: 10.1128/JVI.00421-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmitt AP, Leser GP, Waning DL, Lamb RA. 2002. Requirements for budding of paramyxovirus simian virus 5 virus-like particles. J Virol 76:3952–3964. doi: 10.1128/jvi.76.8.3952-3964.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dietzel E, Kolesnikova L, Sawatsky B, Heiner A, Weis M, Kobinger GP, Becker S, von Messling V, Maisner A. 2016. Nipah virus matrix protein influences fusogenicity and is essential for particle infectivity and stability. J Virol 90:2514–2522. doi: 10.1128/JVI.02920-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnston GP, Contreras EM, Dabundo J, Henderson BA, Matz KM, Ortega V, Ramirez A, Park A, Aguilar HC. 2017. Cytoplasmic motifs in the Nipah virus fusion protein modulate virus particle assembly and egress. J Virol 91:e02150-16. doi: 10.1128/JVI.02150-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park A, Yun T, Vigant F, Pernet O, Won ST, Dawes BE, Bartkowski W, Freiberg AN, Lee B. 2016. Nipah virus C protein recruits Tsg101 to promote the efficient release of virus in an ESCRT-dependent pathway. PLoS Pathog 12:e1005659. doi: 10.1371/journal.ppat.1005659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sakaguchi T, Kato A, Sugahara F, Shimazu Y, Inoue M, Kiyotani K, Nagai Y, Yoshida T. 2005. AIP1/ALIX is a binding partner of Sendai virus C protein and facilitates virus budding. J Virol 79:8933–8941. doi: 10.1128/JVI.79.14.8933-8941.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Irie T, Nagata N, Yoshida T, Sakaguchi T. 2008. Recruitment of ALIX/AIP1 to the plasma membrane by Sendai virus C protein facilitates budding of virus-like particles. Virology 371:108–120. doi: 10.1016/j.virol.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 22.Ke Z, Strauss JD, Hampton CM, Brindley MA, Dillard RS, Leon F, Lamb KM, Plemper RK, Wright ER. 2018. Promotion of virus assembly and organization by the measles virus matrix protein. Nat Commun 9:1736. doi: 10.1038/s41467-018-04058-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu YC, Grusovin J, Adams TE. 2018. Electrostatic interactions between hendra virus matrix proteins are required for efficient virus-like-particle assembly. J Virol 92:e00143-18. doi: 10.1128/JVI.00143-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dietzel E, Kolesnikova L, Maisner A. 2013. Actin filaments disruption and stabilization affect measles virus maturation by different mechanisms. Virol J 10:249. doi: 10.1186/1743-422X-10-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bieniasz PD. 2006. Late budding domains and host proteins in enveloped virus release. Virology 344:55–63. doi: 10.1016/j.virol.2005.09.044. [DOI] [PubMed] [Google Scholar]
- 26.Dilley KA, Gregory D, Johnson MC, Vogt VM. 2010. An LYPSL late domain in the Gag protein contributes to the efficient release and replication of Rous sarcoma virus. J Virol 84:6276–6287. doi: 10.1128/JVI.00238-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dolnik O, Kolesnikova L, Stevermann L, Becker S. 2010. Tsg101 is recruited by a late domain of the nucleocapsid protein to support budding of Marburg virus-like particles. J Virol 84:7847–7856. doi: 10.1128/JVI.00476-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Freed EO. 2002. Viral late domains. J Virol 76:4679–4687. doi: 10.1128/jvi.76.10.4679-4687.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heidecker G, Lloyd PA, Fox K, Nagashima K, Derse D. 2004. Late assembly motifs of human T-cell leukemia virus type 1 and their relative roles in particle release. J Virol 78:6636–6648. doi: 10.1128/JVI.78.12.6636-6648.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Licata JM, Simpson-Holley M, Wright NT, Han Z, Paragas J, Harty RN. 2003. Overlapping motifs (PTAP and PPEY) within the Ebola virus VP40 protein function independently as late budding domains: involvement of host proteins TSG101 and VPS-4. J Virol 77:1812–1819. doi: 10.1128/jvi.77.3.1812-1819.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmitt AP, Leser GP, Morita E, Sundquist WI, Lamb RA. 2005. Evidence for a new viral late-domain core sequence, FPIV, necessary for budding of a paramyxovirus. J Virol 79:2988–2997. doi: 10.1128/JVI.79.5.2988-2997.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Votteler J, Sundquist WI. 2013. Virus budding and the ESCRT pathway. Cell Host Microbe 14:232–241. doi: 10.1016/j.chom.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rossman JS, Jing X, Leser GP, Lamb RA. 2010. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell 142:902–913. doi: 10.1016/j.cell.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen BJ, Lamb RA. 2008. Mechanisms for enveloped virus budding: can some viruses do without an ESCRT? Virology 372:221–232. doi: 10.1016/j.virol.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Utley TJ, Ducharme NA, Varthakavi V, Shepherd BE, Santangelo PJ, Lindquist ME, Goldenring JR, Crowe JE. 2008. Respiratory syncytial virus uses a Vps4-independent budding mechanism controlled by Rab11-FIP2. Proc Natl Acad Sci U S A 105:10209–10214. doi: 10.1073/pnas.0712144105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taylor MP, Koyuncu OO, Enquist LW. 2011. Subversion of the actin cytoskeleton during viral infection. Nat Rev Microbiol 9:427–439. doi: 10.1038/nrmicro2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miazza V, Mottet-Osman G, Startchick S, Chaponnier C, Roux L. 2011. Sendai virus induced cytoplasmic actin remodeling correlates with efficient virus particle production. Virology 410:7–16. doi: 10.1016/j.virol.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 38.Gladnikoff M, Shimoni E, Gov NS, Rousso I. 2009. Retroviral assembly and budding occur through an actin-driven mechanism. Biophys J 97:2419–2428. doi: 10.1016/j.bpj.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fackler OT, Kräusslich H-G. 2006. Interactions of human retroviruses with the host cell cytoskeleton. Curr Opin Microbiol 9:409–415. doi: 10.1016/j.mib.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 40.Chertova E, Chertov O, Coren LV, Roser JD, Trubey CM, Bess JW Jr, Sowder RC Jr, Barsov E, Hood BL, Fisher RJ, Nagashima K, Conrads TP, Veenstra TD, Lifson JD, Ott DE. 2006. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J Virol 80:9039–9052. doi: 10.1128/JVI.01013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Radhakrishnan A, Yeo D, Brown G, Myaing MZ, Iyer LR, Fleck R, Tan B-H, Aitken J, Sanmun D, Tang K, Yarwood A, Brink J, Sugrue RJ. 2010. Protein analysis of purified respiratory syncytial virus particles reveals an important role for heat shock protein 90 in virus particle assembly. Mol Cell Proteomics 9:1829–1848. doi: 10.1074/mcp.M110.001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ren X, Xue C, Kong Q, Zhang C, Bi Y, Cao Y. 2012. Proteomic analysis of purified Newcastle disease virus particles. Proteome Sci 10:32. doi: 10.1186/1477-5956-10-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vera-Velasco NM, García-Murria MJ, Sánchez del Pino MM, Mingarro I, Martinez-Gil L. 2018. Proteomic composition of Nipah virus-like particles. J Proteomics 172:190–200. doi: 10.1016/j.jprot.2017.10.012. [DOI] [PubMed] [Google Scholar]
- 44.Cifuentes-Muñoz N, Sun W, Ray G, Schmitt PT, Webb S, Gibson K, Dutch RE, Schmitt AP. 2017. Mutations in the transmembrane domain and cytoplasmic tail of hendra virus fusion protein disrupt virus-like-particle assembly. J Virol 91:e00152-17. doi: 10.1128/JVI.00152-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cha-Molstad H, Yu JE, Feng Z, Lee SH, Kim JG, Yang P, Han B, Sung KW, Yoo YD, Hwang J, McGuire T, Shim SM, Song HD, Ganipisetti S, Wang N, Jang JM, Lee MJ, Kim SJ, Lee KH, Hong JT, Ciechanover A, Mook-Jung I, Kim KP, Xie XQ, Kwon YT, Kim BY. 2017. p62/SQSTM1/equestosome-1 is an N-recognin of the N-end rule pathway which modulates autophagosome biogenesis. Nat Commun 8:102. doi: 10.1038/s41467-017-00085-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patch JR, Crameri G, Wang L-F, Eaton BT, Broder CC. 2007. Quantitative analysis of Nipah virus proteins released as virus-like particles reveals central role for the matrix protein. Virol J 4:1. doi: 10.1186/1743-422X-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patch JR, Han Z, McCarthy SE, Yan L, Wang L-F, Harty RN, Broder CC. 2008. The YPLGVG sequence of the Nipah virus matrix protein is required for budding. Virol J 5:137. doi: 10.1186/1743-422X-5-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blanchoin L, Boujemaa-Paterski R, Sykes C, Plastino J. 2014. Actin dynamics, architecture, and mechanics in cell motility. Physiol Rev 94:235–263. doi: 10.1152/physrev.00018.2013. [DOI] [PubMed] [Google Scholar]
- 49.Engl W, Arasi B, Yap LL, Thiery JP, Viasnoff V. 2014. Actin dynamics modulate mechanosensitive immobilization of E-cadherin at adherens junctions. Nat Cell Biol 16:587–594. doi: 10.1038/ncb2973. [DOI] [PubMed] [Google Scholar]
- 50.Sit S-T, Manser E. 2011. Rho GTPases and their role in organizing the actin cytoskeleton. J Cell Sci 124:679–683. doi: 10.1242/jcs.064964. [DOI] [PubMed] [Google Scholar]
- 51.Spiering D, Hodgson L. 2011. Dynamics of the Rho-family small GTPases in actin regulation and motility. Cell Adh Migr 5:170–180. doi: 10.4161/cam.5.2.14403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Etienne-Manneville S, Hall A. 2002. Rho GTPases in cell biology. Nature 420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 53.Hodge RG, Ridley AJ. 2016. Regulating Rho GTPases and their regulators. Nat Rev Mol Cell Biol 17:496–510. doi: 10.1038/nrm.2016.67. [DOI] [PubMed] [Google Scholar]
- 54.Diederich S, Sauerhering L, Weis M, Altmeppen H, Schaschke N, Reinheckel T, Erbar S, Maisner A. 2012. Activation of the Nipah virus fusion protein in MDCK cells is mediated by cathepsin B within the endosome-recycling compartment. J Virol 86:3736–3745. doi: 10.1128/JVI.06628-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weis M, Maisner A. 2015. Nipah virus fusion protein: importance of the cytoplasmic tail for endosomal trafficking and bioactivity. Eur J Cell Biol 94:316–322. doi: 10.1016/j.ejcb.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vogt C, Eickmann M, Diederich S, Moll M, Maisner A. 2005. Endocytosis of the Nipah virus glycoproteins. J Virol 79:3865–3872. doi: 10.1128/JVI.79.6.3865-3872.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rohrer J, Schweizer A, Russell D, Kornfeld S. 1996. The targeting of Lamp1 to lysosomes is dependent on the spacing of its cytoplasmic tail tyrosine sorting motif relative to the membrane. J Cell Biol 132:565–576. doi: 10.1083/jcb.132.4.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mattera R, Farías GG, Mardones GA, Bonifacino JS. 2014. Co-assembly of viral envelope glycoproteins regulates their polarized sorting in neurons. PLoS Pathog 10:e1004107. doi: 10.1371/journal.ppat.1004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Deschambeault J, Lalonde JP, Cervantes-Acosta G, Lodge R, Cohen EA, Lemay G. 1999. Polarized human immunodeficiency virus budding in lymphocytes involves a tyrosine-based signal and favors cell-to-cell viral transmission. J Virol 73:5010–5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lodge R, Lalonde JP, Lemay G, Cohen EA. 1997. The membrane-proximal intracytoplasmic tyrosine residue of HIV-1 envelope glycoprotein is critical for basolateral targeting of viral budding in MDCK cells. EMBO J 16:695–705. doi: 10.1093/emboj/16.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, Wettstein DA, Stray KM, Côté M, Rich RL, Myszka DG, Sundquist WI. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- 62.Mercenne G, Alam SL, Arii J, Lalonde MS, Sundquist WI. 2015. Angiomotin functions in HIV-1 assembly and budding. Elife 4:e03778. doi: 10.7554/eLife.03778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sharma S, Aralaguppe SG, Abrahams M-R, Williamson C, Gray C, Balakrishnan P, Saravanan S, Murugavel KG, Solomon S, Ranga U. 2017. The PTAP sequence duplication in HIV-1 subtype C Gag p6 in drug-naive subjects of India and South Africa. BMC Infect Dis 17:95. doi: 10.1186/s12879-017-2184-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Strack B, Calistri A, Craig S, Popova E, Göttlinger HG. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114:689–699. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- 65.Qualmann B, Mellor H. 2003. Regulation of endocytic traffic by Rho GTPases. Biochem J 371:233–241. doi: 10.1042/BJ20030139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.de Curtis I, Meldolesi J. 2012. Cell surface dynamics–how Rho GTPases orchestrate the interplay between the plasma membrane and the cortical cytoskeleton. J Cell Sci 125:4435–4444. doi: 10.1242/jcs.108266. [DOI] [PubMed] [Google Scholar]
- 67.Symons M, Rusk N. 2003. Control of vesicular trafficking by Rho GTPases. Curr Biol 13:R409–R418. doi: 10.1016/s0960-9822(03)00324-5. [DOI] [PubMed] [Google Scholar]
- 68.Kippert A, Trajkovic K, Rajendran L, Ries J, Simons M. 2007. Rho regulates membrane transport in the endocytic pathway to control plasma membrane specialization in oligodendroglial cells. J Neurosci 27:3560–3570. doi: 10.1523/JNEUROSCI.4926-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lamp B, Dietzel E, Kolesnikova L, Sauerhering L, Erbar S, Weingartl H, Maisner A. 2013. Nipah virus entry and egress from polarized epithelial cells. J Virol 87:3143–3154. doi: 10.1128/JVI.02696-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schowalter RM, Wurth MA, Aguilar HC, Lee B, Moncman CL, McCann RO, Dutch RE. 2006. Rho GTPase activity modulates paramyxovirus fusion protein-mediated cell-cell fusion. Virology 350:323–334. doi: 10.1016/j.virol.2006.01.033. [DOI] [PubMed] [Google Scholar]
- 71.Ren M, Xu G, Zeng J, De Lemos-Chiarandini C, Adesnik M, Sabatini DD. 1998. Hydrolysis of GTP on rab11 is required for the direct delivery of transferrin from the pericentriolar recycling compartment to the cell surface but not from sorting endosomes. Proc Natl Acad Sci U S A 95:6187–6192. doi: 10.1073/pnas.95.11.6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Daecke J, Fackler OT, Dittmar MT, Kräusslich H-G. 2005. Involvement of clathrin-mediated endocytosis in human immunodeficiency virus type 1 entry. J Virol 79:1581–1594. doi: 10.1128/JVI.79.3.1581-1594.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Benmerah A, Bayrou M, Cerf-Bensussan N, Dautry-Varsat A. 1999. Inhibition of clathrin-coated pit assembly by an Eps15 mutant. J Cell Sci 112:1303–1311. [DOI] [PubMed] [Google Scholar]
- 74.van der Bliek AM, Redelmeier TE, Damke H, Tisdale EJ, Meyerowitz EM, Schmid SL. 1993. Mutations in human dynamin block an intermediate stage in coated vesicle formation. J Cell Biol 122:553–563. doi: 10.1083/jcb.122.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sato T, Nakashima A, Guo L, Tamanoi F. 2009. Specific activation of mTORC1 by Rheb G-protein in vitro involves enhanced recruitment of its substrate protein. J Biol Chem 284:12783–12791. doi: 10.1074/jbc.M809207200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fujita H. 2003. A dominant negative form of the AAA ATPase SKD1/VPS4 impairs membrane trafficking out of endosomal/lysosomal compartments: class E vps phenotype in mammalian cells. J Cell Sci 116:401–414. doi: 10.1242/jcs.00213. [DOI] [PubMed] [Google Scholar]
- 77.Aguilar HC, Matreyek KA, Filone CM, Hashimi ST, Levroney EL, Negrete OA, Bertolotti-Ciarlet A, Choi DY, McHardy I, Fulcher JA, Su SV, Wolf MC, Kohatsu L, Baum LG, Lee B. 2006. N-Glycans on Nipah virus fusion protein protect against neutralization but reduce membrane fusion and viral entry. J Virol 80:4878–4889. doi: 10.1128/JVI.80.10.4878-4889.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Landowski M, Dabundo J, Liu Q, Nicola AV, Aguilar HC. 2014. Nipah virion entry kinetics, composition, and conformational changes determined by enzymatic virus-like particles and new flow virometry tools. J Virol 88:14197–14206. doi: 10.1128/JVI.01632-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Clair G, Piehowski PD, Nicola T, Kitzmiller JA, Huang EL, Zink EM, Sontag RL, Orton DJ, Moore RJ, Carson JP, Smith RD, Whitsett JA, Corley RA, Ambalavanan N, Ansong C. 2016. Spatially-resolved proteomics: rapid quantitative analysis of laser capture microdissected alveolar tissue samples. Sci Rep 6:39223. doi: 10.1038/srep39223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huang EL, Piehowski PD, Orton DJ, Moore RJ, Qian W-J, Casey CP, Sun X, Dey SK, Burnum-Johnson KE, Smith RD. 2016. SNaPP: simplified nanoproteomics platform for reproducible global proteomic analysis of nanogram protein quantities. Endocrinology 157:1307–1314. doi: 10.1210/en.2015-1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Heberle H, Meirelles GV, da Silva FR, Telles GP, Minghim R. 2015. InteractiVenn: a Web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics 16:169. doi: 10.1186/s12859-015-0611-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. 2015. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G. 2004. GO::TermFinder–open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Aguilar HC, Matreyek KA, Choi DY, Filone CM, Young S, Lee B. 2007. Polybasic KKR motif in the cytoplasmic tail of Nipah virus fusion protein modulates membrane fusion by inside-out signaling. J Virol 81:4520–4532. doi: 10.1128/JVI.02205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Biering SB, Huang A, Vu AT, Robinson LR, Bradel-Tretheway B, Choi E, Lee B, Aguilar HC. 2012. N-Glycans on the Nipah virus attachment glycoprotein modulate fusion and viral entry as they protect against antibody neutralization. J Virol 86:11991–12002. doi: 10.1128/JVI.01304-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bradel-Tretheway BG, Liu Q, Stone JA, McInally S, Aguilar HC. 2015. Novel functions of hendra virus G N-glycans and comparisons to Nipah virus. J Virol 89:7235–7247. doi: 10.1128/JVI.00773-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu Q, Stone JA, Bradel-Tretheway B, Dabundo J, Benavides Montano JA, Santos-Montanez J, Biering SB, Nicola AV, Iorio RM, Lu X, Aguilar HC. 2013. Unraveling a three-step spatiotemporal mechanism of triggering of receptor-induced Nipah virus fusion and cell entry. PLoS Pathog 9:e1003770. doi: 10.1371/journal.ppat.1003770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kelly RT, Page JS, Luo Q, Moore RJ, Orton DJ, Tang K, Smith RD. 2006. Chemically etched open tubular and monolithic emitters for nanoelectrospray ionization mass spectrometry. Anal Chem 78:7796–7801. doi: 10.1021/ac061133r. [DOI] [PMC free article] [PubMed] [Google Scholar]