Abstract
Efflux pumps are always at the forefront of bacterial multidrug resistance and account for the failure of antibiotics. The present study explored the potential of 2-(2-Aminophenyl) indole (RP2), an efflux pump inhibitor (EPI) isolated from the soil bacterium, to overcome the efflux-mediated resistance in Staphylococcus aureus. The RP2/antibiotic combination was tested against efflux pump over-expressed S. aureus strains. The compound was further examined for the ethidium bromide (EtBr) uptake and efflux inhibition assay (a hallmark of EPI functionality) and cytoplasmic membrane depolarization. The safety profile of RP2 was investigated using in vitro cytotoxicity assay and Ca2+ channel inhibitory effect. The in vivo efficacy of RP2 was studied in an animal model in combination with ciprofloxacin. RP2 exhibited the synergistic activity with several antibiotics in efflux pump over-expressed strains of S. aureus. In the mechanistic experiments, RP2 increased the accumulation of EtBr, and demonstrated the inhibition of its efflux. The antibiotic-EPI combinations resulted in extended post antibiotic effects as well as a decrease in mutation prevention concentration of antibiotics. Additionally, the in silico docking studies suggested the binding of RP2 to the active site of modeled structure of NorA efflux pump. The compound displayed low mammalian cytotoxicity and had no Ca2+ channel inhibitory effect. In ex vivo experiments, RP2 reduced the intracellular invasion of S. aureus in macrophages. Furthermore, the RP2/ciprofloxacin combination demonstrated remarkable efficacy in a murine thigh infection model. In conclusion, RP2 represents a promising candidate as bacterial EPI, which can be used in the form of a novel therapeutic regimen along with existing and upcoming antibiotics, for the eradication of S. aureus infections.
Keywords: multidrug resistance, efflux pump inhibitor, time-kill kinetics, post-antibiotic effect, membrane potential, combination therapy, natural products
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA), a high priority pathogen (Tacconelli et al., 2018), is responsible for skin and soft tissue infections, endocarditis, and several bloodstream infections (Tong et al., 2015). For the last few decades, there is a substantial decline in the development of new antimicrobial compounds (Boucher et al., 2009). With diminished hope on dwindling discovery pipeline, bridging the gap between widespread multi-drug resistance, and development of new antibiotics requires innovative use of available antibiotic arsenal. Resistance-modifying agents (Abreu et al., 2012) can be used as antibiotic potentiators in combination therapy for the development of novel treatment regimens.
The over-expression of efflux pumps in S. aureus is one of the major cause of antibiotic resistance (Munita and Arias, 2016). Several efflux pumps have been reported in bacteria, of which major facilitator superfamily (MFS) is predominant (Costa et al., 2013), which utilizes a proton gradient for extruding the drugs. In S. aureus, NorA (MFS) is the most studied efflux pump and is reported to be over-expressed in more than 50% of the clinical isolates (DeMarco et al., 2007). The NorA pump exports a broad range of compounds such as fluoroquinolones, detergents, and several dyes like ethidium bromide (EtBr) (Fontaine et al., 2015; Tintino et al., 2016), whereas TetK and MsrA efflux pumps are specific for tetracycline and erythromycin, respectively (Smith et al., 2007).
No efflux pump inhibitor/antibiotic cocktail has been approved hitherto, possibly due to the unnecessary toxicities in human (Stavri et al., 2006). To the best of our knowledge, however, no systematic study has been carried out in the past for EPIs isolated from microbes against S. aureus strains. To compete with the resistance mechanism, antibiotic-producing bacteria have the tendency to counterattack by means of some inhibitors (Wright, 2014). Therefore, the microbes represent a unique bioprospecting opportunity, because there is substantial evidence that bacteria produce inhibitors against efflux pumps (Lee et al., 2001; Whalen et al., 2015).
The current study describes the isolation, identification and assessment of efflux pump inhibitory activity of EPI 2-(2- Aminophenyl) indole from the terrestrial bacterial isolate, Streptomyces Sp. IMTB 2501. The compound was further evaluated for the in vitro toxicity. Additionally, we checked the impact of RP2 on invasive abilities of S. aureus inside macrophages and in a neutropenic murine thigh infection model. The results obtained in this study may aid the development of RP2 as potent EPI against S. aureus efflux pumps.
Materials and Methods
Chemicals, Bacterial Strains, and Growth Conditions
All the antibiotics, chemicals, reagents and dyes used in this study were purchased from Sigma-Aldrich Chemical Co. (St. Louis, MO, United States) unless mentioned otherwise. The strains, SA-1199B (NorA over-expressed) (Kaatz et al., 1987), SA-K1758 (NorA deletion mutant of NCTC 8325-4) (Price et al., 2002), XU212 (clinical isolate, TetK over-expressed) (Gibbons and Udo, 2000), and RN4220-MsrA (transformed with pSK265, containing MsrA efflux gene) (Reynolds et al., 2003) were used in the study. S. aureus ATCC 25923 (Himedia®) was used in mutation studies. Cation-adjusted Mueller-Hinton broth (CA-MHB; Becton-Dickinson, United States) was used to grow the strains.
Purification and Characterization of Bioactive Metabolite
The microbial extract library preparation and screening platform for EPI activity is discussed in Supplementary Material. The 48 h old inoculum of soil isolate IMTB 2501 was grown in R-2A broth (Himedia®, India), supplemented with 1% NaCl, and 0.2% calcium carbonate. After 96 h of incubation, cells were removed by centrifugation and extract was prepared using Diaion® HP-20 resin. The crude extract was fractionated on SephadexTM LH-20 (GE Healthcare) column using a step gradient of methanol: water. The active fractions were pooled and processed further by reverse-phase high-pressure liquid chromatography (RP-HPLC) (Waters, XBridge® BEH C18 OBDTM Prep Column 130 Å, 5 μm, 10 mm × 250 mm). The mobile phase consisted of solvent A as 10 mM ammonium acetate (pH 5.5) and solvent B as 100% acetonitrile. The flow rate was kept constant at 3 mL/min. The following gradient was used; 5% solvent B for 5 min, 5 to 95% B in 45 min, hold at 95% B for 10 min. The peaks were collected and assayed for the bioactivity. The active peak (termed as RP2) was identified and subjected to lyophilization. The purified compound was subjected to gas chromatography (Varian 450-GC) coupled with mass spectrometry (Varian 20-MS). The spectrum was recorded in an electron-ionization mode. NMR spectra (1H, 13C, DEPT, COSY, and HSQC) were recorded on a Bruker 400 MHz spectrometer in DMSO-d6.
Checkerboard Synergy Assay
To examine the synergistic activity of RP2 with the antibiotics, the checkerboard assay was performed. The MIC values of the combinations were used to calculate the fractional inhibitory concentration (FICI) index according to the formula:
The FICI value of ≤0.5 was considered as “Synergy” whereas 0.5 < FICI ≤ 4.0 was observed as an “indifference” effect and the FICI > 4.0 was considered as “antagonistic” effect (Junio et al., 2011).
Ethidium Bromide Accumulation Assay
The fluorometric estimation of EtBr uptake was performed in S. aureus SA-1199B, XU212, and RN4220-MsrA as described previously (Kalia et al., 2012). S. aureus cells (OD550 ∼0.2) were treated with EtBr at 2 μg/mL in the presence of RP2 at its 1/4th MIC (32 μg/mL) or reserpine (20 μg/mL). The accumulation of fluorescence was recorded over a period of 45 min at 3-min intervals at excitation and emission wavelength of 530 and 600 nm, respectively, in a microplate reader (BioTek, United States).
Ethidium Bromide Efflux Inhibition Studies
The effect of the RP2 on EtBr efflux of S. aureus SA-1199B, XU212 and RN4220-MsrA was assessed by fluorometry (Viveiros et al., 2008). A 50 μl of the cell suspension (OD600 ∼0.6) was incubated with 50 μl of phosphate buffer saline (PBS) containing EtBr (2 μg/mL) and RP2 (4–32 μg/mL) or reserpine (20 μg/mL). Post 30 min incubation, the fluorescence was acquired at 530/600 nm. Relative final fluorescence (RFF) index was calculated according to the formula: RFF = (RFtreated – RFuntreated)/RFuntreated where the RFtreated denotes the final fluorescence at the last time point in the presence of RP2 or reserpine; the RFuntreated denotes the final fluorescence at the last time point (45 min) of the experiment in the control well (without EPI) (Machado et al., 2011).
Time-Kill Kinetics
Staphylococcus aureus SA-1199B (106 colony forming units; CFU/mL) was incubated with ciprofloxacin in the presence or absence of RP2 (32 μg/mL). At different time points, the culture was spread plated and the colonies were expressed as CFU/mL. The sample without antibiotic and RP2 was taken as a positive control.
Mutation Prevention Concentration (MPC)
Staphylococcus aureus ATCC 25923 (108 CFU) was plated on MHA plates containing different concentrations of ciprofloxacin, tetracycline or erythromycin. The similar concentrations of antibiotics were also tested in the presence of RP2 (16 and 32 μg/mL). After 48 h, the MPC was determined as the concentration at which no colony appeared. The mutation frequency was calculated as the number of survivors divided by CFU plated (Drugeon et al., 1999).
Post-antibiotic Effect (PAE)
Staphylococcus aureus SA-1199B, XU212, and RN42220 (2 × 108 CFU/mL) were treated with ciprofloxacin, tetracycline, and erythromycin, respectively, at different concentrations, with or without RP2. Post 2 h of incubation, the culture was diluted 1000 times in fresh medium to remove the effect of antibiotics and RP2, followed by determination of viable CFU/mL at different time points. The difference in the time taken for growth between treated culture (T) and the equivalent untreated control (C) to escalate by 1 log10 CFU/mL, was calculated as PAE = T – C (Craig, 1991).
Structure Prediction of NorA and Docking Studies With RP2
The three dimensional structure of NorA protein was predicted using I-TASSER online server (Roy et al., 2010) using PDB IDs 3WDO, 6EXS, 4ZOW, and 4ZPO as templates. This modeled NorA receptor was further used for the docking studies. The ligand and receptor preparation and molecular docking studies were carried out using Schrodinger Suite 2019. The two-dimensional structure of the RP2 was prepared using Maestro. The receptor was prepared for docking using the protein preparation wizard (Sastry et al., 2013) by adding hydrogen atoms to the receptor molecule followed by energy minimization using OLPS3 force field by keeping default 0.3 Å root mean square deviation (RMSD) constraint (Shivakumar et al., 2012). The potential RP2 binding sites in the modeled structure were identified using the SiteMap (Halgren, 2007). The SiteMap analysis identified one potential druggable site with site score of >1 located at the active site of predicted NorA structure. This site was used for grid generation. The grid was generated for docking by keeping box dimension 40 Å encompassing all the protein structure. The molecular docking was performed using Induced Fit Docking module (Farid et al., 2006). The poses were sorted based on the docking score.
Membrane Potential Assay
The membrane depolarization effect of RP2 was quantified using the BacLightTM Bacterial Membrane Potential Kit (Molecular Probes, Life Technologies). S. aureus SA-1199B (107 CFU/mL) was treated with RP2 at concentrations ranging from 4 to 32 μg/mL or carbonyl cyanide 3-chlorophenylhydrazone (CCCP, 2 μg/mL). The dye, 3, 3′-Diethyloxacarbocyanide (DiOC2) was added and the plates were incubated for 30 min. The fluorescence was recorded and change in membrane potential was evaluated.
Determination of Intracellular ATP Levels
The levels of intracellular ATP were evaluated using the ATP Determination Kit (Invitrogen, Life Technologies, United Kingdom) (Machado et al., 2017). S. aureus SA-1199B (105 CFU/mL) was incubated with RP2 (32 μg/mL) in MHB. At different time intervals, the cells were lysed by alternative heat and cold shock method and the supernatant was used for the measurement of total ATP, represented as relative luminescence units.
Mammalian Ca2+ Channel Blocking Assay
Fluo-4 Direct calcium channel assay kit (Life Technologies, Carlsbad, CA, United States) was used, to check the effect of RP2 on human Ca2+ channels (Blanchard et al., 2014). Fluo-4 dye containing probenecid (5 mM) was incubated with HEK 293T cells (5 × 104) for 1 h and fluorescence was measured for 15 s. At 15 s, the cells were treated separately with DMSO (mock), 50 μg/mL of verapamil, or 32 μg/mL of RP2. Then, the Ca2+ channel stimulator carbamylcholine chloride (50 μg/mL) was added at 60 s and the fluorescence was quantified for another 120 s on a microplate reader.
Mammalian Cytotoxicity and Hemolytic Activity
The cytotoxicity of RP2, at a concentration ranging from 62.5 to 1000 μg/mL, was investigated in three cell lines viz; THP-1 (Human Monocytic cell line), HEK-293 (human embryonic kidney cell line) and Hep G2 (human liver cancer cell line). The hemolysis assay was performed with fresh rabbit erythrocytes, as described previously (Jangra et al., 2019).
Macrophage Invasion Assay
The effect of RP2 on the intracellular invasion of S. aureus inside the J774 macrophage cell line was determined (Kalia et al., 2012). S. aureus strains, SA-1199B, SA-1199, and K1758 (106 CFU/well) were used to infect the macrophage (105 cells/well) in the presence or absence of RP2 (32 μg/mL) and incubated for 2 h. The extracellular bacteria were removed with PBS and the cells were further treated with gentamicin (50 μg/mL) for 30 min. Intracellular bacteria were rescued by host cell denaturation using a short exposure of 0.1% saponin and the viability was determined by plating on MHA.
Neutropenic Thigh Infection Model
Female BALB/c mice were made neutropenic by an intraperitoneal administration with cyclophosphamide at 4 (150 mg/kg) and 1 (100 mg/kg) days before infection. An inoculum of 107 CFU/mL S. aureus SA-1199B was resuspended in sterile PBS followed by 50 μL intramuscular injection into the right thighs of mice. At 4 h after infection, one group was euthanized by cervical dislocation. The right thighs were aseptically removed, homogenized in PBS, serially diluted and plated on drug-free MHA for CFU counts. The remaining mice were treated with either RP2 or ciprofloxacin (both at 10 mg/kg) alone or in combination, administered subcutaneously (six mice per group). At 24 h after infection, the mice were sacrificed and CFUs were calculated in the thigh muscle (Ling et al., 2015).
Statistical Analysis
Statistical analysis was carried out using the Student’s t-test. A P value < 0.05 was considered statistically significant and highly significant when ∗∗P < 0.01 and ∗∗∗P < 0.001.
Results
Isolation and Identification of 2-(2-Aminophenyl) Indole
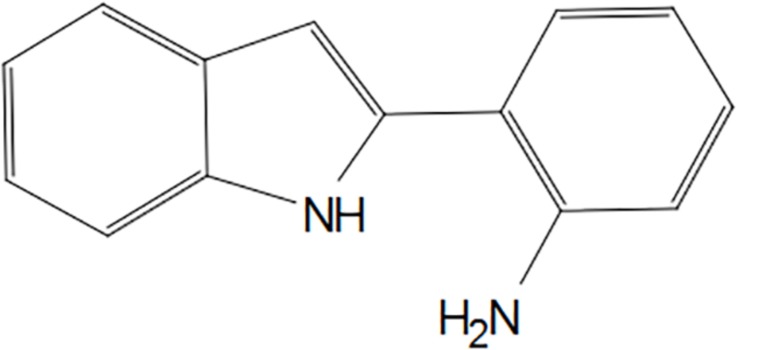
The initial screening identified 50 microbial isolates (Supplementary Table S1), which potentiated the activity of EtBr in S. aureus by at least four folds. Among them, eight isolates (IMTB 2342, IMTB 2501, IMTF 935, IMTF 1118, IMTF 1984, IMTF 2261, IMTF 2413, and IMTF 2454) reduced the MIC of at least one of the antibiotic tested, i.e., norfloxacin, erythromycin and tetracycline by ≥fourfold against S. aureus SA-1199B, XU212 and RN4220-MsrA, respectively (Supplementary Table S2). The isolate IMTB 2501 exhibited the most prominent EPI-like activity and was selected for further study. This strain showed 98.90% similarity with Streptomyces roseochromogenes based on 16S rRNA gene sequence (GenBank accession number MK377074). Bioactivity-guided fractionation of the microbial extract of IMTB 2501 resulted in the isolation of 2-(2- Aminophenyl) indole (C14H12N2) (Figure 1), designated as “RP2.” The structure of RP2 was elucidated on the basis of GC-MS and NMR spectroscopic data (Supplementary Figures S1–S4 and Supplementary Table S3). This article represents the first description of 2-(2-Aminophenyl) indole from a microbial source, with novel EPI functionality.
FIGURE 1.
2D structure of RP2.
Checkerboard Synergy Assay
RP2 exhibited no intrinsic antibacterial activity up to 32 μg/mL, against S. aureus SA-1199B, SA-K1758, XU212 and RN4220-MsrA strains. At this concentration, it reduced the MICs of norfloxacin, ciprofloxacin, moxifloxacin, and chloramphenicol in SA-1199B by 64, 16, 4, and 4 folds, respectively (Table 1), indicating the synergy (FICI ≤ 0.5). On the contrary, SA-K1758 did not show any significant reduction in the MIC. The low MIC value of moxifloxacin in SA-1199B indicates that it is presumably a weaker substrate of NorA efflux pump. RP2 also inhibited TetK and MsrA transporters and reduced the MICs of tetracycline in S. aureus XU212 and erythromycin in RN4220-MsrA by 64 (FICI 0.265) and 32 (FICI 0.281) folds, respectively (Table 1). We also checked synergy of norfloxacin, tetracycline and erythromycin with reserpine, a known efflux pump inhibitor. Reserpine modulated the MICs of norfloxacin and tetracycline by eight- and four-fold, respectively, whereas no synergy was observed with erythromycin (Supplementary Table S4). These results depicted that RP2 also displayed broad efflux pump inhibition spectrum.
TABLE 1.
MICs for numerous antibiotics against S. aureus efflux pump over-expressed/deletion strains in the absence and presence of RP2.
| Compounds |
S. aureus SA1199B (NorA++) |
S. aureus K1758 (NorA–) |
||||
| MICa | MICb | FICIc | MICa | MICb | FICIc | |
| Norfloxacin | 32 | 0.5 (32) | 0.265 | 0.25 | 0.125 (16) | 0.75 |
| 2 (16) | 0.1875 | 0.25 (8) | – | |||
| 4 (8) | 0.1875 | 0.25 (4) | – | |||
| 8 (4) | 0.281 | 0.25 (2) | – | |||
| Ciprofloxacin | 8 | 0.5 (32) | 0.3125 | 0.125 | 0.125 (16) | – |
| 1 (16) | 0.25 | 0.125 (8) | – | |||
| 4 (8) | 0.5625 | 0.125 (4) | – | |||
| Moxifloxacin | 0.25 | 0.0625 (32) | 0.375 | 0.0625 | 0.0625 (16) | – |
| 0.125 (16) | 0.625 | 0.0625 (8) | – | |||
| Chloramphenicol | 8 | 2 (32) | 0.375 | 2 | 2 (16) | – |
| 4 (16) | 0.625 | 2 (8) | – | |||
| RP2 | 128 | n/a | n/a | 64 | n/a | n/a |
| S. aureus XU212 (TetK)∗ | S. aureus RN4220-MsrA∗∗ | |||||
| Tetracycline1/Erythromycin2 | 128 | 2 (32) | 0.265 | 128 | 4 (32) | 0.281 |
| 4 (16) | 0.156 | 16 (16) | 0.25 | |||
| 8 (8) | 0.125 | 32 (8) | 0.3125 | |||
| 32 (4) | 0.281 | 64 (4) | 0.531 | |||
| RP2 | 128 | n/a | n/a | 128 | n/a | n/a |
MICa, minimum inhibitory concentration of compound alone (μg/mL); MICb, MIC of antibiotic in the presence of RP2 (μg/mL in parentheses); FICIc, fractional inhibitory concentration index. The checkerboard synergy assay of RP2 in combination with Tetracycline1 and Erythromycin2 was performed against S. aureus XU212∗ and S. aureus RN4220-MsrA∗∗, respectively.++, NorA over-expressed strain; ––, NorA deletion strain.
Effect of RP2 on Accumulation and Efflux of EtBr
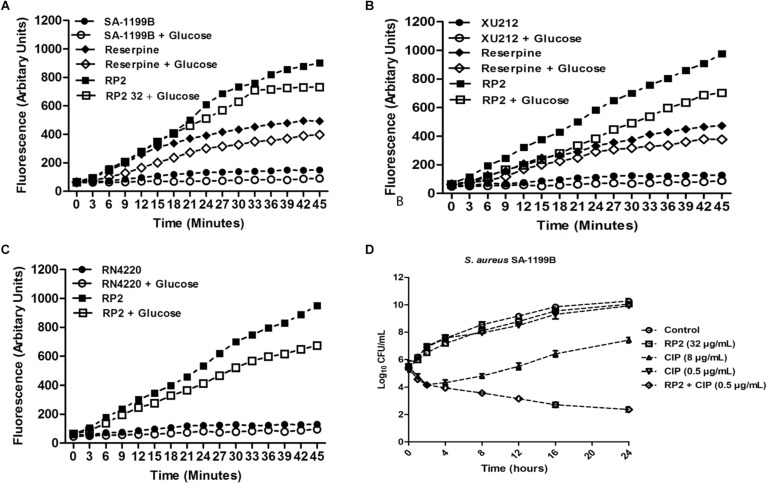
The expression of NorA (SA-1199B), TetK (XU212), and MsrA (RN4220-MsrA) efflux pumps resulted in a slow increase in fluorescence, indicating the negligible accumulation of EtBr (Figures 2A–C). The addition of RP2 enhanced the accumulation significantly, compared to reserpine, a known NorA, and TetK efflux inhibitor (Smith et al., 2007). The presence of glucose reduced the efflux pump inhibitory activity of both RP2 and reserpine, because glucose reenergizes the cells, promoting active efflux. We next evaluated the potential of RP2 in these strains, to inhibit EtBr efflux, and calculated the RFF values (Table 2). The RFF is a measure of efflux inhibition by comparing the final fluorescence of EPI treated vs. untreated cells. The RFF value >1 indicates an enhanced accumulation of EtBr (Machado et al., 2017). At 32 μg/mL, RP2 displayed the RFF values of 11.51, 9.39 and 8.69 against the NorA, TetK, and MsrA efflux pump over-expressed strains, respectively, in comparison to RFF values of reserpine, 6.22 (NorA) and 2.97 (TetK). RP2 demonstrated dose-dependent EtBr accumulation and efflux inhibition.
FIGURE 2.
Effect of RP2 on the accumulation of ethidium bromide by S. aureus: (A) SA-1199B, (B) XU212, and (C) RN4220-MsrA; the concentration of EtBr was 2 μg/mL for all the three tested strains. Assays were performed at 37°C in the presence and absence of glucose. Reserpine was used at 20 μg/mL as the positive control for S. aureus SA-1199B and XU212. (D) Time-kill curve of S. aureus SA-1199B, showing the bactericidal effect of ciprofloxacin in combination with RP2 at 32 μg/mL. Each time point represents the mean log10 CFU ± SD of three independent readings.
TABLE 2.
Relative Final Fluorescence (RFF) values based on the EtBr efflux inhibition for the S. aureus strains in the presence of the efflux inhibitors.
| Compound | Conc. of inhibitor used (μg/mL) |
Relative final fluorescence (RFF ± SD) |
||
| SA1199B | XU212 | RN4220-MsrA | ||
| RP2 | 32 | 11.51 ± 0.16∗∗∗ | 9.39 ± 0.08∗∗∗ | 8.69 ± 0.12∗∗∗ |
| 16 | 8.9 ± 0.14∗∗∗ | 6.8 ± 0.03∗∗∗ | 6.04 ± 0.12∗∗∗ | |
| 8 | 7.27 ± 0.14∗∗∗ | 4.61 ± 0.11∗∗∗ | 4.58 ± 0.06∗∗∗ | |
| 4 | 5.06 ± 0.07∗∗∗ | 2.86 ± 0.08∗∗ | 3.03 ± 0.1∗∗ | |
| Reserpine | 20 | 6.22 ± 0.11∗∗∗ | 2.97 ± 0.05∗∗ | NDa |
The results presented correspond to the average of three independent assays plus standard deviation (±SD). Results were considered highly significant when ∗∗P < 0.01 and ∗∗∗P < 0.001. aND, not determined.
Time-Kill Kinetics and Mutation Frequency Analysis
The time-kill kinetics of S. aureus SA-1199B was examined with the combination of ciprofloxacin (8 μg/mL) and RP2 (32 μg/mL). As shown in Figure 2D, the combination resulted in a reduction of ≥2 log10 CFU in 24 h, in comparison to the antibiotic or RP2 alone. These results prompted us to study the frequency of mutation in S. aureus in the presence of RP2-antibiotic combinations. We observed that RP2 at 16 μg/mL decreased the MPC of ciprofloxacin, tetracycline and erythromycin by twofold each, whereas by 8, 8, and 4 folds, respectively when tested at 32 μg/mL (Table 3). These results highlight the clinical significance of these combinations in limiting the selection of resistant mutants.
TABLE 3.
Mutation frequencies of S. aureus ATCC 25923.
| RP2 (μg/mL) |
Mutation frequency |
|||||||||||
|
Ciprofloxacin |
Tetracycline |
Erythromycin |
||||||||||
| 2 MIC | 4 MIC | 8 MIC | 16 MIC | 2 MIC | 4 MIC | 8 MIC | 16 MIC | 2 MIC | 4 MIC | 8 MIC | 16 MIC | |
| 0 | 5.5 × 10–8 | 3.9 × 10–8 | 1.8 × 10–8 | <10–8 | 5.6 × 10–8 | 4.8 × 10–8 | 2.5 × 10–8 | <10–8 | 6.9 × 10–8 | 5.1 × 10–8 | 3.5 × 10–8 | <10–8 |
| 16 | 3.6 × 10–8 | 1.5 × 10–8 | <10–8 | <10–8 | 2.7 × 10–8 | 1.3 × 10–8 | <10–8 | <10–8 | 3.6 × 10–8 | 1.4 × 10–8 | <10–8 | <10–8 |
| 32 | <10–8 | <10–8 | <10–8 | <10–8 | <10–8 | <10–8 | <10–8 | <10–8 | 1.2 × 10–8 | <10–8 | <10–8 | <10–8 |
The results presented correspond to the average of three independent assays.
Post-antibiotic Effect
The post-antibiotic effect is the process of continued suppression of bacterial growth after a short exposure to antibiotics (Craig, 1991). In the absence of RP2, the PAEs at a 1 × MIC concentration of ciprofloxacin (S. aureus SA-1199B), tetracycline (XU212), and erythromycin (RN4220-MsrA) were 1.43, 2.1, and 1.82 h, respectively (Table 4). The same concentrations of antibiotics in combination with RP2 (32 μg/mL) resulted in significantly prolonged PAEs, ciprofloxacin (3.33 h), tetracycline (3.4 h), and erythromycin (3.32 h). Similar effects were obtained at lower concentrations of antibiotics.
TABLE 4.
PAE of ciprofloxacin, tetracycline, and erythromycin alone and in combination with RP2 against S. aureus SA1199B, XU212 and RN4220-MsrA, respectively.
| Strain | Regimen |
Mean PAE (h) ± SD |
||
| 0.25 MIC | 0.5 MIC | 1 MIC | ||
| S. aureus SA1199B | Ciprofloxacin | 0.43 ± 0.14 | 1.05 ± 0.19 | 1.43 ± 0.16 |
| Ciprofloxacin + RP2 (32 μg/mL) | 1.68 ± 0.13 | 2.15 ± 0.15 | 3.33 ± 0.18 | |
| S. aureus XU212 | Tetracycline | 0.76 ± 0.13 | 1.2 ± 0.17 | 2.1 ± 0.14 |
| Tetracycline + RP2 (32 μg/mL) | 1.93 ± 0.12 | 2.45 ± 0.08 | 3.4 ± 0.25 | |
| S. aureus RN4220-MsrA | Erythromycin | 0.46 ± 0.13 | 1.08 ± 0.17 | 1.82 ± 0.14 |
| Erythromycin + RP2 (32 μg/mL) | 1.77 ± 0.12 | 2.24 ± 0.08 | 3.32 ± 0.25 | |
The results presented correspond to the average of three independent assays plus standard deviation (±SD).
In silico Docking Studies of RP2 With NorA Efflux Pump Model
The predicted structure of NorA consists of 12 transmembrane helices with a wide cavity at the active site located in the center (Figures 3A,B). We used box size of 40 Å encompassing all the protein structure for docking RP2 to identify the potential binding site. 57 out of 65 poses of the docked structure were observed at site 1 and the remaining 8 poses were observed at site 2 (Figure 3 and Supplementary Figures S5A,B). The docking scores for the poses observed in the site 1 were better (<−7.8) compared to the site 2 (>−6.7), therefore we only selected site 1 for further interaction analysis. The docking results suggested that RP2 binding is stabilized by hydrophobic interactions, H-bonding and/or Pi–Pi stacking interactions. The RP2/NorA binding site is lined by two conserved hydrophobic patches at the one end and a hydrophilic patch which covers the other end (Figure 3). These two hydrophobic patches are lined by Ile12, Ile15, Phe16, Leu19, and Ala105, Gly106, Val108, Met109 residues. The hydrophilic patch is composed of Ser333, Thr336, Ser337 and Asn340 residues. Analysis of all 65 different poses suggested that in several poses Gln51 participated in hydrogen bonding with the amino group of RP2. Interestingly, Gln51 has been shown to be involved in stabilizing NorA/inhibitor binding in previous studies as well. In some poses, we also observed Pi-Pi stacking interactions between Phe16/Phe47or Phe140 with benzene ring of RP2. The variations observed in the mode of binding and natures of interactions stabilizing RP2 in selected docked poses are shown in the Supplementary Figures S5C–G. The comparative analysis of RP2 binding with other known NorA inhibitors (analyzed by virtual docking) like ferruginol, totarol, thioxanthene, phenothiazine, levofloxacin, and acridine suggested that they also bind at the same site and share several common interacting residues (Bhaskar et al., 2016). However, unlike other inhibitors, reserpine interacts with only one of the conserved hydrophobic clefts (Ala105, Gly106, Val108, and Met109) (Kalia et al., 2012).
FIGURE 3.
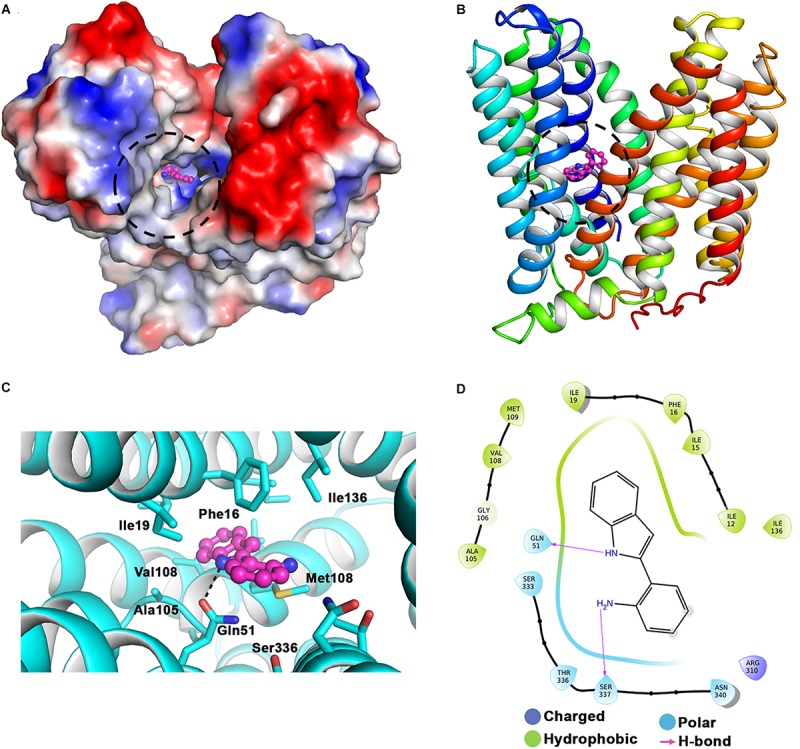
Interaction of RP2 with the active site of NorA. (A) 3D electrostatic potential map and (B) cartoon representation (in rainbow color) of NorA showing RP2 (ball and stick representation in magenta) docked in the active site cleft. (C) The close view of NorA residues, in stick representation, and interacting with RP2. All 12 transmembrane helices of the protein have been colored in rainbow. (D) Ligand interaction diagram in 2D representation showing several interactions involved in NorA/RP2 binding.
Membrane Depolarization
Thirty minutes after the addition of RP2 (4–32 μg/mL), there was no significant change in the percentage of depolarized cells when compared with untreated bacterial cells, indicating no interference of bacterial membrane potential by RP2 (Figure 4A). In contrast, the protonophore CCCP distorted the membrane potential. The percentage of depolarized cells was >90%.
FIGURE 4.
(A) Membrane depolarization. The relative red/green ratio of S. aureus SA-1199B using DIOC2 stained cells after 30 min of exposure to RP2 from 4 to 32 μg/mL. Green and red fluorescence correspond to the depolarized as well as polarized cells, respectively. The changes in the fluorescence were measured at an excitation/emission wavelength of 485/528 nm and 528/590 for green and red fluorescence, respectively. (B) Effect of RP2 on S. aureus ATP levels. S. aureus SA-1199B was exposed to RP2 at 1/4th MIC during 24 h. The ATP levels were quantified using a luciferin-luciferase bioluminescence detection assay. CCCP (2 μg/mL) was included for comparison. The results presented correspond to the mean of three independent readings ± SD. Results were considered highly significant where ∗∗∗P < 0.001.
Effect of RP2 on Intracellular ATP Levels
The effect of RP2 on bacterial ATP synthesis was assessed in S. aureus SA-1199B at a sub-inhibitory concentration (32 μg/mL). The ATP levels remained unaffected during 24 h of exposure with RP2, similar to that of drug-free control (Figure 4B). These results excluded the possibility of EtBr accumulation and efflux inhibition due to sudden ATP depletion. In contrast, the impairment in ATP production was observed when cells were treated with CCCP (positive control) at 2 μg/mL.
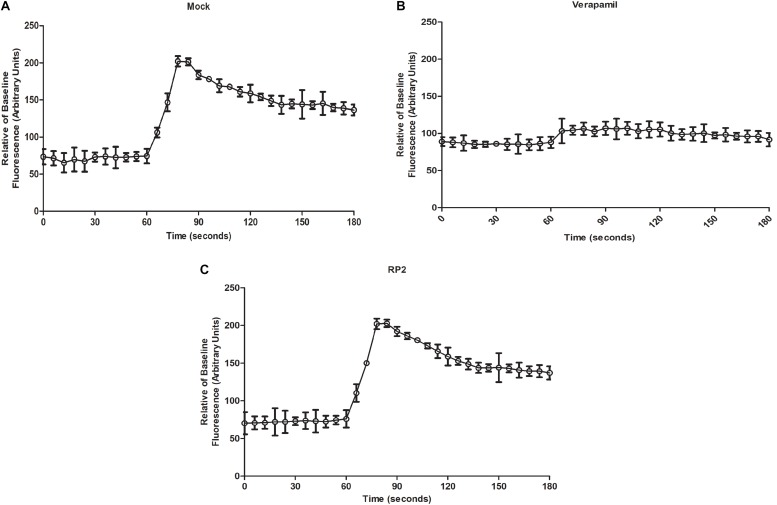
Mammalian Ca2+ Channel Blocking Assays
We evaluated the effect of RP2 on human calcium channel activity by measuring the cytoplasmic Ca2+ levels using Fluo-4 dye. The endoplasmic calcium-channel activity was stimulated by the addition of carbachol which increased the Ca2+ levels by 2.6-folds into the cytoplasm (Figure 5A). On the contrary, the addition of verapamil abolished the Ca2+ ions accumulation inside the cytoplasm (Figure 5B). As shown in Figure 5C, HEK 293T treatment with RP2 at 32 μg/mL did not exhibit any significant effect on mammalian cell Ca2+ channel activity.
FIGURE 5.
Mammalian calcium channel inhibition assays (A) Cytoplasmic Ca2+ measures of mock-treated cells with carbachol added at 60 s (B,C). Same as panel A except that cells were treated with verapamil (50 μg/mL) (C) or RP2 (32 μg/mL), at 45 s pre-carbachol treatment. The results presented correspond to the mean of three independent readings ± SD.
In vitro Toxicity
RP2 exhibited half-maximal inhibition concentration (IC50) at approximately 500 μg/mL for all the three cell lines tested (Figure 6A) and had negligible toxicity at the effective concentration (32 μg/mL). In hemolytic experiments, no significant lysis of red blood cells was observed up to 1000 μg/mL (Figure 6B).
FIGURE 6.
(A) Cytotoxic effect of RP2 on THP-1, HEK-293T, and HEP G2 cell lines. The cells were exposed to RP2 (1000–62.5 μg/mL) during 24 h at 37°C, 5% CO2. The results presented correspond to the mean of three independent readings ± SD. (B) Hemolytic effect of RP2 (1000–62.5 μg/mL) on rabbit erythrocytes. Triton X-100 (0.1%) was included as a positive control. The results presented correspond to the mean of three independent readings ± SD. Results were considered highly significant where ∗∗∗P < 0.001.
Macrophage Invasion Assay
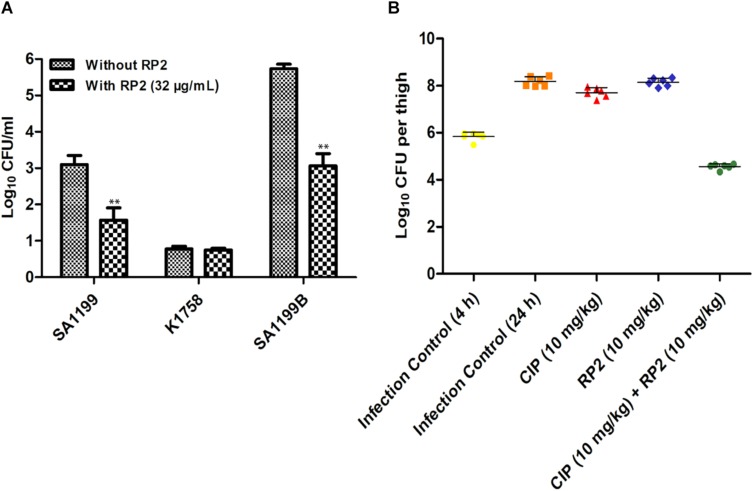
The intracellular invasion of S. aureus SA-1199B (NorA over-expressed) was nearly 2.5 log10 greater as compared to wild type SA-1199. S. aureus KA-1758 (norA gene deletion), attacked macrophages less effectively. In the presence of RP2 (32 μg/mL), the penetration of S. aureus KA-1758 remained unaffected, and whereas the invasiveness of S. aureus SA-1199B was reduced to a larger extent (1.5 log10) compared with the wild-type S. aureus SA-1199 (Figure 7A).
FIGURE 7.
(A) Influence of RP2 (32 μg/mL) on the invasive abilities of S. aureus wild-type SA-1199, NorA over-producing S. aureus SA-1199B and the norA knock-out K1758. The results presented correspond to the mean of three independent readings ± SD. Results were considered significant when ∗P < 0.05 and highly significant when ∗∗P < 0.01 and ∗∗∗P < 0.001. (B) Neutropenic murine thigh infection model. Single dose (subcutaneous; 4 h after infection, 6 mice per group) treatment with ciprofloxacin and RP2 alone and in combination against S. aureus SA-1199B; for drug-treated animals, thigh CFU was determined at 20 h after infection. For controls, CFU in thighs was estimated at 4 and 20 h after infection. The CFU from each thigh are plotted as individual points and error bars represent the SD within an experimental group.
In vivo Efficacy
We investigated the in vivo efficacy of ciprofloxacin in combination with RP2 using a neutropenic murine thigh infection model. We observed that neither ciprofloxacin (10 mg/kg) nor RP2 (10 mg/kg) alone indicated a significant reduction in bacterial counts (Figure 7B). However, the combination of ciprofloxacin and RP2 proved to be very efficient, resulting in a 1.3 and 3.5 log10 reduction in CFU, in comparison to the untreated control at 4 and 24 h, respectively (Figure 7B).
Discussion
For bacterial infection, adherence, secretion of toxins and virulence factors, enzyme inactivation, efflux pumps, etc. are important mechanisms which cause bacterial pathogenicity and resistance to antibiotics. Thus to bowl over effects of pathogenicity in bacteria, identification of inhibitors to target these processes can be an effective strategy (Wang et al., 2014; Song et al., 2017; Imdad et al., 2018; Zhou et al., 2019). The efflux pumps contribute significantly to the development of multidrug resistance in clinical MRSA strains (DeMarco et al., 2007; Costa et al., 2013). The therapeutic failure of current antibiotics encourages the investigation of EPIs as adjunctive therapies. Several phytochemicals have been reported previously as EPIs (Khan et al., 2006; Kalia et al., 2012; Roy et al., 2013; Singh et al., 2017). The present study focused on the exploration of microbial diversity for the discovery of EPIs to combat multi-drug resistance. Our screening campaign identified Streptomyces Sp. IMTB 2501, whose crude fermentation extract displayed the characteristics of S. aureus efflux pump inhibition. Bioassay-directed fractionation yielded an indole metabolite, i.e., 2-(2-Aminophenyl) indole (RP2) previously unreported for the efflux pump inhibitory activity. Though indole derivatives have been reported for NorA efflux pump inhibition (Michalet et al., 2007; Ambrus et al., 2008; Buonerba et al., 2016), their toxicity or in vivo efficacy was not investigated.
RP2 demonstrated synergism with the tested antibiotics against efflux pump over-expressing S. aureus strains, reducing the MICs below their clinical breakpoints. We confirmed the EPI activity of RP2 in real-time fluorescence experiments using EtBr. Additionally, time-kill assay exhibited synergy because we observed >3 log10 reductions in CFU between the combination and most active counterpart (Belley et al., 2008). Moreover, RP2 reduced the frequency of resistant mutants in S. aureus and enhanced the PAE of all the tested antibiotics. Additionally, the in silico docking studies revealed that RP2 binds to active site of NorA efflux pump with a better docking score at the site 1. Comparative interaction analysis of RP2 and other NorA inhibitors like ferruginol, totarol, thioxanthene and phenothiazine docked to NorA suggests that they all target similar binding cleft and hence may adopt similar inhibitory mechanism (Bhaskar et al., 2016).
The ideal EPI should not dissipate the energy source of the pump by disrupting cytoplasmic membrane potential (Lomovskaya et al., 2001). Unlike CCCP, RP2 neither affected the membrane potential nor the total ATP production. Hence the synergistic activity observed was not a consequence of the off-target effect. Furthermore, RP2 had no significant cytotoxicity in mammalian cells, and unlike verapamil, it did not exhibit human Ca2+ channel inhibitory activities. These drawbacks have restricted the development of EPIs in the past (Blanchard et al., 2014). Inhibition of efflux pumps is also associated with other assaults in pathogenic bacteria such as reduced bacterial adherence (Kvist et al., 2008). As reportedly previously (Kalia et al., 2012; Singh et al., 2017), our results also corroborated the role of NorA over-production for the intracellular invasion of S. aureus in J774 macrophages. Involvement of RP2 as an EPI reduced the intracellular invasion. Similar observations have been reported earlier in MexAB-OprM over-producing Pseudomonas aeruginosa (Hirakata et al., 2009). So probably, S. aureus exports some factors required for the invasion using NorA efflux pump and RP2 reduces the invasion by inhibiting the NorA efflux system. The most prominent finding of this study was the efficacy of RP2 and ciprofloxacin combination in the murine thigh infection model.
Conclusion
In conclusion, our docking studies combined with experimental evidences suggest that RP2 is a strong inhibitor of NorA that probably binds and blocks the active site/channel by sterically occluding export of antibiotics/drugs. RP2 represents a remarkable starting point for future medicinal chemistry-based development of lead candidates. The properties of RP2 may help deliver an adjunctive therapy to treat fatal bacterial infections.
Data Availability
The datasets generated for this study, of the analyzed 16S rRNA gene sequence of strain IMTB 2501 can be found in GenBank under the accession number MK377074.
Ethics Statement
This study was carried out in accordance with the recommendations of the Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA). The protocol was approved by the Institutional Animal Ethics Committee (IAEC/18/03) of the Institute of Microbial Technology, Chandigarh, India. Neither randomization nor blinding was considered necessary for the animal infection models.
Author Contributions
RT and HN designed all the experiments, and wrote and edited the manuscript. RT, NM, NC, and MK performed all the microbiological experiments. RT, MJ, and NK performed the animal studies. SC and MR maintained the cell lines and contributed to the cytotoxicity experiments. SJ contributed to the NMR spectroscopy and structure elucidation. DV and KT performed the in silico docking studies. RT, MJ, and HN analyzed the results. RT and HN wrote and edited the manuscript. All the authors approved the manuscript before submission.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge the Director, Institute of Microbial Technology for providing the infrastructure. We thank Dr. Simon Gibbons (UCL College of Pharmacy, London) and Dr. Inshad Ali Khan (CSIR – Indian Institute of Integrative Medicine, Jammu) for sharing the efflux pump over-expressed and deletion strains. We also thank Mr. Deepak Bhatt and Mr. Mohit Paul for their technical assistance in DNA sequencing and mass spectrometry facility.
Footnotes
Funding. This work was supported by the Council of Scientific & Industrial Research (CSIR) Network project BSC-0211G and in-house project of the Institute of Microbial Technology OLP-147.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02153/full#supplementary-material
References
- Abreu A. C., Mcbain A. J., Simoes M. (2012). Plants as sources of new antimicrobials and resistance-modifying agents. Nat. Prod. Rep. 29 1007–1021. 10.1039/c2np20035j [DOI] [PubMed] [Google Scholar]
- Ambrus J. I., Kelso M. J., Bremner J. B., Ball A. R., Casadei G., Lewis K. (2008). Structure–activity relationships of 2-aryl-1H-indole inhibitors of the NorA efflux pump in Staphylococcus aureus. Bioorg. Med. Chem. Lett. 18 4294–4297. 10.1016/j.bmcl.2008.06.093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belley A., Neesham-Grenon E., Arhin F. F., Mckay G. A., Parr T. R., Moeck G. (2008). Assessment by time-kill methodology of the synergistic effects of oritavancin in combination with other antimicrobial agents against Staphylococcus aureus. Antimicrob. Agents Chemother. 52 3820–3822. 10.1128/AAC.00361-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskar B. V., Babu T. M. C., Reddy N. V., Rajendra W. (2016). Homology modeling, molecular dynamics, and virtual screening of NorA efflux pump inhibitors of Staphylococcus aureus. Drug Des. Dev. Ther. 10 3237–3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard C., Barnett P., Perlmutter J., Dunman P. M. (2014). Identification of Acinetobacter baumannii serum-associated antibiotic efflux pump inhibitors. Antimicrob. Agents Chemother. 58 6360–6370. 10.1128/AAC.03535-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher H. W., Talbot G. H., Bradley J. S., Edwards J. E., Gilbert D., Rice L. B., et al. (2009). Bad bugs, no drugs: no ESKAPE! An update from the infectious diseases society of America. Clin. Infect. Dis. 48 1–12. 10.1086/595011 [DOI] [PubMed] [Google Scholar]
- Buonerba F., Lepri S., Goracci L., Schindler B. D., Seo S. M., Kaatz G. W., et al. (2016). Improved potency of indole-based NorA efflux pump inhibitors: from serendipity toward rational design and development. J. Med. Chem. 60 517–523. 10.1021/acs.jmedchem.6b01281 [DOI] [PubMed] [Google Scholar]
- Costa S. S., Viveiros M., Amaral L., Couto I. (2013). Multidrug efflux pumps in Staphylococcus aureus: an update. Open Microbiol. J. 7 59–71. 10.2174/1874285801307010059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig W. A. (1991). The postantibiotic effect. Clin. Microbiol. Newslett. 13 121–124. [Google Scholar]
- DeMarco C. E., Cushing L. A., Frempong-Manso E., Seo S. M., Jaravaza T. A., Kaatz G. W. (2007). Efflux-related resistance to norfloxacin, dyes, and biocides in bloodstream isolates of Staphylococcus aureus. Antimicrob. Agents Chemother. 51 3235–3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drugeon H., Juvin M., Bryskier A. (1999). Relative potential for selection of fluoroquinolone-resistant Streptococcus pneumoniae strains by levofloxacin: comparison with ciprofloxacin, sparfloxacin and ofloxacin. J. Antimicrob. Chemother. 43 55–59. [DOI] [PubMed] [Google Scholar]
- Farid R., Day T., Friesner R. A., Pearlstein R. A. (2006). New insights about HERG blockade obtained from protein modeling, potential energy mapping, and docking studies. Bioorg. Med. Chem. 14 3160–3173. [DOI] [PubMed] [Google Scholar]
- Fontaine F., Héquet A., Voisin-Chiret A.-S., Bouillon A., Lesnard A., Cresteil T., et al. (2015). Boronic species as promising inhibitors of the Staphylococcus aureus NorA efflux pump: study of 6-substituted pyridine-3-boronic acid derivatives. Eur. J. Med. Chem. 95 185–198. 10.1016/j.ejmech.2015.02.056 [DOI] [PubMed] [Google Scholar]
- Gibbons S., Udo E. (2000). The effect of reserpine, a modulator of multidrug efflux pumps, on the in vitro activity of tetracycline against clinical isolates of methicillin resistant Staphylococcus aureus (MRSA) possessing the tet (K) determinant. Phytother. Res. 14 139–140. [DOI] [PubMed] [Google Scholar]
- Halgren T. (2007). New method for fast and accurate binding-site identification and analysis. Chem. Biol. Drug Des. 69 146–148. [DOI] [PubMed] [Google Scholar]
- Hirakata Y., Kondo A., Hoshino K., Yano H., Arai K., Hirotani A., et al. (2009). Efflux pump inhibitors reduce the invasiveness of Pseudomonas aeruginosa. Int. J. Antimicrob. Agents 34 343–346. 10.1016/j.ijantimicag.2009.06.007 [DOI] [PubMed] [Google Scholar]
- Imdad S., Chaurasia A. K., Kim K. K. (2018). Identification and validation of an antivirulence agent targeting HlyU-regulated virulence in Vibrio vulnificus. Front. Cell. Infect. Microbiol. 8:152. 10.3389/fcimb.2018.00152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangra M., Kaur M., Nandanwar H. (2019). In vitro studies on a natural lantibiotic, paenibacillin: a new-generation antibacterial drug candidate to overcome multidrug-resistance. Int. J. Antimicrob. Agents 53 838–843. [DOI] [PubMed] [Google Scholar]
- Junio H. A., Sy-Cordero A. A., Ettefagh K. A., Burns J. T., Micko K. T., Graf T. N., et al. (2011). Synergy-directed fractionation of botanical medicines: a case study with goldenseal (Hydrastis canadensis). J. Natl. Prod. 74 1621–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaatz G., Barriere S., Schaberg D., Fekety R. (1987). The emergence of resistance to ciprofloxacin during therapy of experimental methicillin-susceptible Staphylococcus aureus endocarditis. J. Antimicrob. Chemother. 20 753–758. [DOI] [PubMed] [Google Scholar]
- Kalia N. P., Mahajan P., Mehra R., Nargotra A., Sharma J. P., Koul S., et al. (2012). Capsaicin, a novel inhibitor of the NorA efflux pump, reduces the intracellular invasion of Staphylococcus aureus. J. Antimicrob. Chemother. 67 2401–2408. 10.1093/jac/dks232 [DOI] [PubMed] [Google Scholar]
- Khan I. A., Mirza Z. M., Kumar A., Verma V., Qazi G. N. (2006). Piperine, a phytochemical potentiator of ciprofloxacin against Staphylococcus aureus. Antimicrob. Agents Chemother. 50 810–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvist M., Hancock V., Klemm P. (2008). Inactivation of efflux pumps abolishes bacterial biofilm formation. Appl. Environ. Microbiol. 74 7376–7382. 10.1128/AEM.01310-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. D., Galazzo J. L., Staley A. L., Lee J. C., Warren M. S., Fuernkranz H., et al. (2001). Microbial fermentation-derived inhibitors of efflux-pump-mediated drug resistance. Farmaco 56 81–85. [DOI] [PubMed] [Google Scholar]
- Ling L. L., Schneider T., Peoples A. J., Spoering A. L., Engels I., Conlon B. P., et al. (2015). A new antibiotic kills pathogens without detectable resistance. Nature 517 455–459. 10.1038/nature14098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomovskaya O., Warren M. S., Lee A., Galazzo J., Fronko R., Lee M., et al. (2001). Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob. Agents Chemother. 45 105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado D., Fernandes L., Costa S. S., Cannalire R., Manfroni G., Tabarrini O., et al. (2017). Mode of action of the 2-phenylquinoline efflux inhibitor PQQ4R against Escherichia coli. PeerJ. 5:e3168. 10.7717/peerj.3168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado L., Spengler G., Evaristo M., Handzlik J., Molnár J., Viveiros M., et al. (2011). Biological activity of twenty-three hydantoin derivatives on intrinsic efflux pump system of Salmonella enterica serovar Enteritidis NCTC 13349. In Vivo 25 769–772. [PubMed] [Google Scholar]
- Michalet S., Cartier G., David B., Mariotte A.-M., Dijoux-Franca M.-G., Kaatz G. W., et al. (2007). N-caffeoylphenalkylamide derivatives as bacterial efflux pump inhibitors. Bioorg. Med. Chem. Lett. 17 1755–1758. [DOI] [PubMed] [Google Scholar]
- Munita J. M., Arias C. A. (2016). Mechanisms of antibiotic resistance. Microbiol. Spectr. 4 1–24. 10.1128/microbiolspec.VMBF-0016-2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price C. T., Kaatz G. W., Gustafson J. (2002). The multidrug efflux pump NorA is not required for salicylate-induced reduction in drug accumulation by Staphylococcus aureus. Int. J. Antimicrob. Agents 20 206–213. [DOI] [PubMed] [Google Scholar]
- Reynolds E., Ross J. I., Cove J. H. (2003). Msr (A) and related macrolide/streptogramin resistance determinants: incomplete transporters? Int. J. Antimicrob. Agents 22 228–236. [DOI] [PubMed] [Google Scholar]
- Roy A., Kucukural A., Zhang Y. (2010). I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5 725–738. 10.1038/nprot.2010.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S. K., Kumari N., Pahwa S., Agrahari U. C., Bhutani K. K., Jachak S. M., et al. (2013). NorA efflux pump inhibitory activity of coumarins from Mesua ferrea. Fitoterapia 90 140–150. 10.1016/j.fitote.2013.07.015 [DOI] [PubMed] [Google Scholar]
- Sastry G. M., Adzhigirey M., Day T., Annabhimoju R., Sherman W. (2013). Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J. Comp. Aid. Mol. Des. 27 221–234. 10.1007/s10822-013-9644-8 [DOI] [PubMed] [Google Scholar]
- Shivakumar D., Harder E., Damm W., Friesner R. A., Sherman W. (2012). Improving the prediction of absolute solvation free energies using the next generation OPLS force field. J. Chem. Theory Comput. 8 2553–2558. 10.1021/ct300203w [DOI] [PubMed] [Google Scholar]
- Singh S., Kalia N. P., Joshi P., Kumar A., Sharma P. R., Kumar A., et al. (2017). Boeravinone B, A novel dual inhibitor of NorA bacterial efflux pump of Staphylococcus aureus and human P-glycoprotein, reduces the biofilm formation and intracellular invasion of bacteria. Front. Microbiol. 8:1868. 10.3389/fmicb.2017.01868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E. C., Kaatz G. W., Seo S. M., Wareham N., Williamson E. M., Gibbons S. (2007). The phenolic diterpene totarol inhibits multidrug efflux pump activity in Staphylococcus aureus. Antimicrob. Agents Chemother. 51 4480–4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M., Teng Z., Li M., Niu X., Wang J., Deng X. (2017). Epigallocatechin gallate inhibits Streptococcus pneumoniae virulence by simultaneously targeting pneumolysin and sortase A. J. Cell. Mol. Med. 21 2586–2598. 10.1111/jcmm.13179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavri M., Piddock L. J., Gibbons S. (2006). Bacterial efflux pump inhibitors from natural sources. J. Antimicrob. Chemother. 59 1247–1260. [DOI] [PubMed] [Google Scholar]
- Tacconelli E., Carrara E., Savoldi A., Harbarth S., Mendelson M., Monnet D. L., et al. (2018). Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18 318–327. 10.1016/S1473-3099(17)30753-3 [DOI] [PubMed] [Google Scholar]
- Tintino S. R., Oliveira-Tintino C. D., Campina F. F., Silva R. L., Costa M. D. S., Menezes I. R., et al. (2016). Evaluation of the tannic acid inhibitory effect against the NorA efflux pump of Staphylococcus aureus. Microb. Pathog. 97 9–13. 10.1016/j.micpath.2016.04.003 [DOI] [PubMed] [Google Scholar]
- Tong S. Y., Davis J. S., Eichenberger E., Holland T. L., Fowler V. G. (2015). Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 28 603–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viveiros M., Martins A., Paixão L., Rodrigues L., Martins M., Couto I., et al. (2008). Demonstration of intrinsic efflux activity of Escherichia coli K-12 AG100 by an automated ethidium bromide method. Int. J. Antimicrob. Agents 31 458–462. 10.1016/j.ijantimicag.2007.12.015 [DOI] [PubMed] [Google Scholar]
- Wang J., Qiu J., Tan W., Zhang Y., Wang H., Zhou X., et al. (2014). Fisetin inhibits Listeria monocytogenes virulence by interfering with the oligomerization of listeriolysin O. J. Infect. Dis. 211 1376–1387. 10.1093/infdis/jiu520 [DOI] [PubMed] [Google Scholar]
- Whalen K. E., Poulson-Ellestad K. L., Deering R. W., Rowley D. C., Mincer T. J. (2015). Enhancement of antibiotic activity against multidrug-resistant bacteria by the efflux pump inhibitor 3, 4-dibromopyrrole-2, 5-dione isolated from a Pseudoalteromonas sp. J. Natl. Prod. 78 402–412. 10.1021/np500775e [DOI] [PubMed] [Google Scholar]
- Wright G. D. (2014). Something old, something new: revisiting natural products in antibiotic drug discovery. Can. J. Microbiol. 60 147–154. 10.1139/cjm-2014-0063 [DOI] [PubMed] [Google Scholar]
- Zhou Y., Wang J., Guo Y., Liu X., Liu S., Niu X., et al. (2019). Discovery of a potential MCR-1 inhibitor that reverses polymyxin activity against clinical mcr-1-positive Enterobacteriaceae. J. Infect. 78 364–372. 10.1016/j.jinf.2019.03.004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated for this study, of the analyzed 16S rRNA gene sequence of strain IMTB 2501 can be found in GenBank under the accession number MK377074.